Abstract
Lung cancer is the most common malignant tumor, accounting for 25% of cancer-related deaths and 14% of new cancers worldwide. Lung adenocarcinoma is the most common type of pulmonary cancer. Although there have been some improvements in the traditional therapy of lung cancer, the outcome and prognosis of patients remain poor. Lung cancer is the leading cause of cancer-related deaths worldwide, with 1.8 million new cases being diagnosed each year. Precision medicine based on genetic alterations is considered a new strategy of lung cancer treatment that requires highly specific biomarkers for precision diagnosis and treatment. Fibrinogen-like protein 2 (FGL2) plays important roles in both innate and adaptive immunity. However, the diagnostic value of FGL2 in lung cancer is largely unknown. In this study, we systematically investigated the expression profile and potential functions of FGL2 in lung adenocarcinoma. We used the TCGA and Oncomine datasets to compare the FGL2 expression levels between lung adenocarcinoma and adjacent normal tissues. We utilized the GEPIA, PrognoScan and Kaplan-Meier plotter databases to analyze the relationship between FGL2 expression and the survival of lung adenocarcinoma patients. Then, we investigated the potential roles of FGL2 in lung adenocarcinoma with the TIMER database and functional enrichment analyses. We found that FGL2 expression was significantly lower in lung adenocarcinoma tissue compared with adjacent normal tissue. A high expression level of FGL2 was correlated with better prognostic outcomes of lung adenocarcinoma patients, including overall survival and progression-free survival. FGL2 was positively correlated with the infiltration of immune cells, including dendritic cells, CD8+ T cells, macrophages, B cells, and CD4+ T cells, in lung adenocarcinoma. Functional enrichment analyses also showed that a high expression level of FGL2 was positively correlated with enhanced T cell activities, especially CD8+ T cell activation. Thus, we propose that high FGL2 expression, which is positively associated with enhanced antitumor activities mediated by T cells, is a beneficial marker for lung adenocarcinoma treatment outcomes.
Keywords: FGL2, Lung cancer, Bioinformatics, Prognosis, Tumor infiltrating lymphocyte
Introduction
Lung cancer is a malignant lung tumor with unrestrained cell proliferation in the lung (Schwartz & Cote, 2016). As a highly prevalent and invasive disorder in women and men, more than 1.8 million people are diagnosed with lung cancer every year (Torre, Siegel & Jemal, 2016). Lung cancer accounts for almost 25% of cancer-related deaths and 14% of new cancers worldwide. Although there have been some improvements in the therapy and diagnosis of lung cancer, the outcome and prognosis of patients remain poor. The 5-year survival rate varies from 4–17% depending on regional and stage differences. Almost 1.6 million people die from lung cancer yearly. The lack of effective first-line drugs, the nonoptimal administration route and the formation of resistant tumors might be correlated with the poor prognostic survival of lung cancer patients (Pilkington et al., 2015).
Lung cancer is generally classified into two kinds of histopathology groups: non-small cell lung cancer and small cell lung cancer (Latimer, 2018). As the most common type of lung cancer, lung adenocarcinoma belongs to the non-small cell lung cancer group. Fifty percent of non-small cell lung cancer patients have a lung adenocarcinoma. It is also the most common form of lung cancer in Asian countries. The symptoms and signs of lung adenocarcinoma are similar to those of other types of lung cancer (Zappa & Mousa, 2016). Shortness of breath and persistent cough are the main symptoms of lung adenocarcinoma patients. The category of lung adenocarcinoma includes a variety of subtypes, such as acinar predominant adenocarcinoma, lepidic predominant adenocarcinoma, and micropapillary predominant adenocarcinoma. Radiotherapy, chemotherapy, surgical resection, and immunotherapy are common therapies employed to treat lung adenocarcinoma. However, because of the difficulties in diagnosing early lung adenocarcinoma, the average five-year survival rate is only approximately 18% (Hirsch et al., 2017).
Fibrinogen-like protein 2 (FGL2) plays important roles in both innate and adaptive immunity (Liu, Liu & Chen, 2017). The FGL2 protein is located on the surface of endothelial cells and macrophages (Yang & Hooper, 2013). CD8+ and CD4+ T cells constitutively secrete FGL2 protein to induce an inflammatory response. Several disorders, including severe acute respiratory syndrome (SARS), abortion and allograft rejection, are correlated with FGL2 (Hsieh et al., 2010; Zhao et al., 2013). In the area of cancer research, previous studies have found that altered FGL2 gene expression contributes to immune surveillance evasion in murine renal carcinoma (Birkhäuser et al., 2013). Moreover, FGL2 contributes to glioblastoma multiforme (GBM) progression by stimulating immunosuppression mechanisms (Yan et al., 2015). However, the diagnostic value of FGL2 in lung cancer is largely unknown.
In this study, we systematically explored the potential roles of FGL2 in lung adenocarcinoma. Data downloaded from the TCGA dataset and PNAS were used to compare the FGL2 expression levels between lung adenocarcinoma and adjacent normal tissues. Three bioinformatics databases, including GEPIA, PrognoScan and Kaplan–Meier plotter, were adopted to analyze the relationship of FGL2 expression and the survival of lung adenocarcinoma patients. The TIMER database was used to discover the association between the immune status and FGL2 expression in lung adenocarcinoma. Functional enrichment analyses, including Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway and GSEA, were used to explore the potential functions of FGL2 in lung adenocarcinoma development.
Methods
Bioinformatic evaluation of FGL2 gene expression data
The normalized FPKM (fragments per kilobase per million mapped reads) values were downloaded from The Cancer Genome Atlas (TCGA) Data Portal (https://portal.gdc.cancer.gov). Normalized RNA-Seq datasets were used as input. Microarray mRNA data of lung adenocarcinoma were downloaded from Proc. Natl. Acad. Sci. USA (PNAS) (https://www.pnas.org/) (Bhattacharjee et al., 2001) and the GEO database (GSE32863). The microarray data were log2 transformed. FGL2 expression was compared between lung cancer and normal adjacent tissues. Statistical significance was calculated with SPSS 20.0. Detailed information of included patients are listed in Table S1.
Analysis of prognostic potential
The GEPIA, PrognoScan and Kaplan–Meier plotter databases were used to evaluate the prognostic potential of FGL2 in lung adenocarcinoma. The GEPIA (Gene Expression Profiling Interactive Analysis) database is a new web server (http://gepia.cancer-pku.cn/) for cancer and normal gene expression profiling and interactive analyses. GSEA was first introduced at 2003. Some concerns appeared immediately after GSEA was proposed (Tamayo et al., 2016). The concerns or limitations were list as follows: the null distribution of GSEA is superfluous and very hard to be worth calculating. The Kolmogorov–Smirnov-like statistic is not as sensitive as original. The results of GSEA are dependent on the algorithm clusters the genes, and the number of clusters being analyzed. The PrognoScan database is a new database (http://dna00.bio.kyutech.ac.jp/PrognoScan/) used to explore the relation between patient prognosis and gene expression with large collections of tumor microarray datasets. It is a useful platform to evaluate potential tumor markers in cancer research. The Kaplan–Meier plotter database (http://kmplot.com/analysis/) is a useful online tool used to assess the effects of specific genes on cancer prognosis and can estimate survival from lifetime data. Detailed information of included patients are listed in Table S1.
TIMER database analysis
The TIMER database is a useful web tool that can be used to conduct a comprehensive analysis of tumor-infiltrating immune cells. This tool can evaluate the relationship between the immune status and FGL2 mRNA expression in the lung adenocarcinoma microenvironment. The immune status includes inflammatory cells and the immune gene marker sets of immune cells. The TIMER database was used to measure the correlation between FGL2 mRNA expression and the infiltration of immune cells, including B cells, CDT cells, CD4+ cells, macrophages and dendritic cells. Furthermore, the TIMER database was used to measure the correlation between FGL2 mRNA expression and the immune gene marker sets of immune cells.
Functional enrichment analysis
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses were used to analyze the potential function of FGL2 with the Database for Annotation, Visualization and Integrated Discovery (DAVID). GO analysis is a powerful bioinformatics tool used to determine biological processes (describing the physiological or cellular role carried out by the FGL2), cellular components (CC) and molecular functions (MF).
Results
The mRNA expression level of FGL2 in lung adenocarcinoma
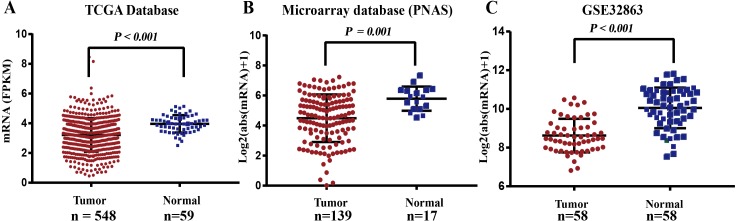
Related data were downloaded from the bioinformatics database TCGA Data Portal and used to compare the mRNA expression level of FGL2 in lung adenocarcinoma and normal adjacent tissues. The results showed that the mean FPKM value of FGL2 in lung adenocarcinoma was 3.266, which was significantly lower than that in normal adjacent tissue (3.958, P < 0.001) (Fig. 1A). Microarray data from PNAS and the GEO also showed that the FGL2 mRNA expression level was lower in lung adenocarcinoma tissue than in normal adjacent tissue (Figs. 1B–1C).
Figure 1. The mRNA expression level of FGL2 in lung adenocarcinoma.
(A) There was a significant difference between the FPKM value of FGL2 mRNA expression in lung adenocarcinoma and normal adjacent tissue (P < 0.001). The left bar indicates the FPKM value of FGL2 mRNA expression in lung adenocarcinoma tissue (mean FPKM value =3.958). The right bar indicates the FPKM value of FGL2 expression level in normal adjacent tissue (mean FPKM value = 3.266). (B) The microarray database from PNAS showed a significant difference in the FGL2 mRNA levels between lung adenocarcinoma and normal adjacent tissues (P = 0.001). (C) The GSE32863 database showed that the FGL2 expression level in lung adenocarcinoma tissue was significantly lower than that in normal adjacent tissue (P < 0.001).
The prognostic value of FGL2 in lung adenocarcinoma
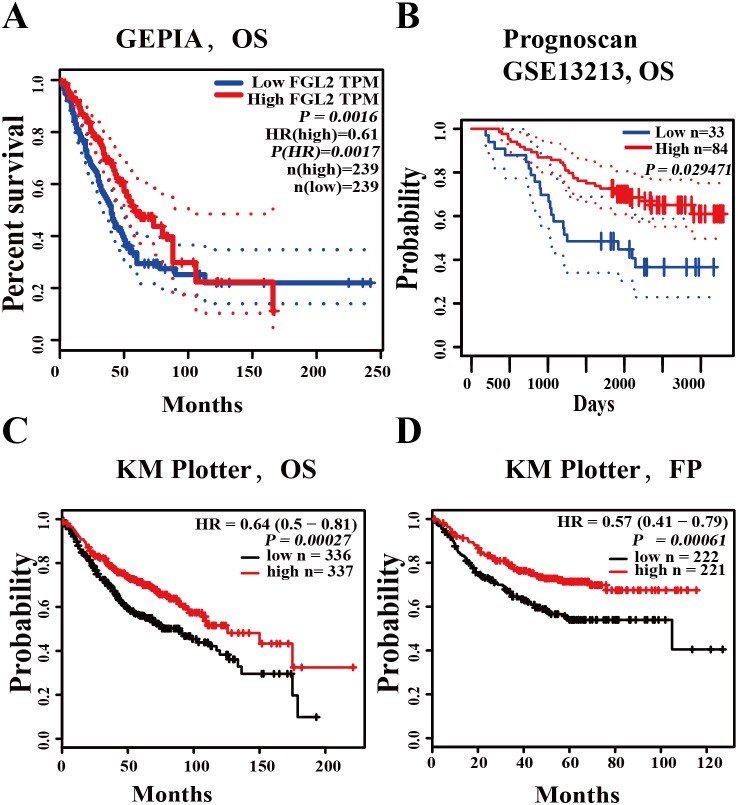
We used three databases (PrognoScan, GEPIA, Kaplan–Meier plotter) to analyze the prognostic value of FGL2 in lung adenocarcinoma. In the GEPIA database, high FGL2 expression was correlated with better overall survival (OS) in lung cancer (OS HR = 0.61, log-rank P = 0.0016, cutoff-high = 50%) (Fig. 2A). In the PrognoScan database, an analysis of the cohort GSE13213 showed that a high FGL2 mRNA level was related to better overall survival in lung adenocarcinoma (OS HR = 0.68, 95% CI = 0.48 to 0.96, Cox P = 0.029471) (Fig. 2B). In the Kaplan–Meier plotter database, high FGL2 mRNA expression was correlated with better overall survival and progression-free survival in lung adenocarcinoma patients (OS HR = 0.64, 95% CI = 0.50 to 0.81, log-rank P = 0.00027; FP HR = 0.57, 95% CI = 0.41 to 0.79, log-rank P = 0.00061) (Figs. 2C–2D). These results suggest that a high FGL2 expression level is correlated better outcomes of lung adenocarcinoma.
Figure 2. The prognostic value of FGL2 in lung adenocarcinoma.
(A) The GEPIA database showed that a high FGL2 mRNA level was correlated with better overall survival (OS) in lung adenocarcinoma. (B) In the PrognoScan database, the cohort GSE13213 was used to evaluate the relationship between the FGL2 expression level and overall survival in lung adenocarcinoma patients. (C–D) The Kaplan–Meier plotter database was used to analyze the relationship between the FGL2 expression level and OS and progression-free survival in lung adenocarcinoma patients.
The correlation between immune cell infiltration and FGL2 expression in lung adenocarcinoma
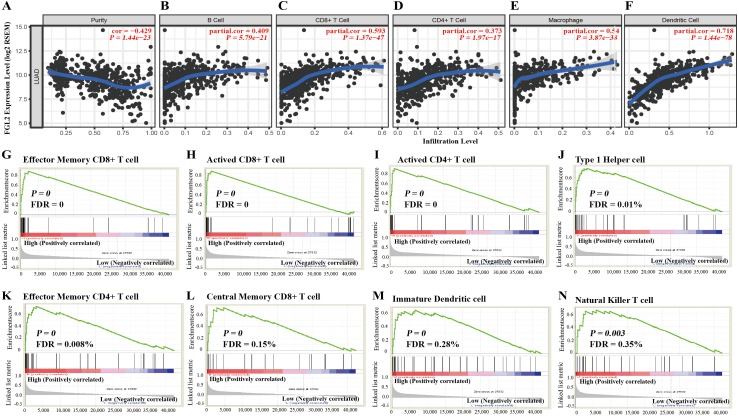
We used the TIMER database to explore the correlation between immune cell infiltration and FGL2 expression in lung adenocarcinoma. As shown in Fig. 3A, the FGL2 expression level was positively correlated with B cell infiltration (r = 0.409, P = 5.79e−21), CD8+ T cell infiltration (r = 0.539, P = 1.37e−47), CD4+ T cell infiltration (r = 0.379, P = 1.97e−17), macrophage infiltration (r = 0.540, P = 3.87e−38) and dendritic cell infiltration (r = 0.718, P = 1.44e−78) in lung adenocarcinoma.
Figure 3. The correlation between immune cell infiltration and FGL2 expression in lungadenocarcinoma.
(A–F) The TIMER database showed that FGL2 expression was positively correlated with the infiltration of B cells, CD8+ T cells, CD4+ T cells, macrophages, neutrophils and dendritic cells in lung adenocarcinoma. (G–N) GSEA showed that FGL2 expression was positively correlated with effector memory CD8+ T cells, activated CD8+ T cells, activated CD4+ T cells, type 1 T helper cells, effector memory CD4+ T cells, central memory CD8+ T cells, immature dendritic cells and natural killer T cells.
Then, we further investigated the correlation between the FGL2 mRNA level and the subtypes of immune cell infiltration with GSEA (Fig. 3B). FGL2 expression was positively correlated with the infiltration of effector memory CD8+T cells, activated CD8+ T cells, activated CD4+ T cells, type 1 T helper cells, effector memory CD4+T cells, central memory CD8+ T cells, immature dendritic cells and natural killer T cells.
The correlation between the immune marker sets of immune cells and FGL2 expression in lung adenocarcinoma
To validate the relationship between FGL2 expression and immune cell infiltration, we also investigated the correlation between the immune marker sets of immune cells and FGL2 expression in lung adenocarcinoma with the TIMER and GEPIA databases. As shown in Table 1, data from the TIMER database indicated that FGL2 expression was positively correlated with most of the immune marker sets. For example, FGL2 was positively corelated with the T cell gene markers CD3D (Cor = 0.532, P = 1.97e−37), CD3E (Cor = 0.583, P = 3.25e−46), and CD2 (Cor = 0.646, P = 1.59e−59); CD8+ T cell (cytotoxic T lymphocyte, CTL) gene markers CD8A (Cor = 0.561, P = 3.93E−42) and CD8B (Cor = 0.456, P = 1.03E−26), dendritic cell gene markers HLA-DPB1 (Cor = 0.635, P = 6.07E−57), HLA-DRA (Cor = 0.683, P = 6.79E−69), and HLA-DPA1 (Cor = 0.681, P = 1.45E−68); and Th17 gene markers ICOS (Cor = 0.667, P = 1.00E−64) and IL1B (Cor = 0.428, P = 2.26E−23).
Table 1. Correlation between immune marker sets of immune cells and FGL2 expression in lung adenocarcinoma based data from TIMER database.
| Description | Gene markers | Cor | P | Description | Gene markers | Cor | P |
|---|---|---|---|---|---|---|---|
| T cell (general) | CD3D | 0.532 | 1.97E−37 | CTL (Cytotoxic T Lymphocytes ) | CD8A | 0.561 | 3.93E−42 |
| CD3E | 0.583 | 3.25E−46 | CD8B | 0.456 | 1.03E−26 | ||
| CD2 | 0.646 | 1.59E−59 | GZMB | 0.325 | 1.45E−13 | ||
| B cell | CD19 | 0.232 | 2.01E−07 | Dendritic cell | HLA-DPB1 | 0.635 | 6.07E−57 |
| CD79A | 0.21 | 2.54E−06 | HLA-DQB1 | 0.386 | 5.34E−19 | ||
| CD79B | 0.316 | 7.03E−13 | HLA-DRA | 0.683 | 6.79E−69 | ||
| CD22 | 0.314 | 7.18E−15 | HLA-DPA1 | 0.681 | 1.45E−68 | ||
| M1 Macrophage | INOS | 0.039 | 3.85E−01 | DEC-205 | 0.471 | 1.19E−28 | |
| CIITA | 0.544 | 2.73E−39 | BDCA-1 | 0.424 | 6.01E−23 | ||
| IRF5 | 0.307 | 3.39E−12 | BDCA-4 | 0.257 | 6.66E−09 | ||
| COX2 | −0.127 | 4.72E−03 | BDCA-2 | 0.48 | 8.93E−30 | ||
| M2 Macrophage | CD163 | 0.618 | 2.61E−53 | CD11c | 0.46 | 3.07E−27 | |
| IRF4 | 0.375 | 6.49E−18 | Th1 | CD38 | 0.211 | 2.18E−06 | |
| VSIG4 | 0.604 | 3.00E−50 | T-bet | 0.48 | 8.15E−30 | ||
| MS4A4A | 0.685 | 1.40E−69 | STAT4 | 0.409 | 2.78E−21 | ||
| TAM | CCL2 | 0.35 | 1.16E−15 | STAT1 | 0.461 | 2.63E−27 | |
| CCL5 | 0.506 | 2.07E−33 | IFN-γ | 0.432 | 7.89E−24 | ||
| CD68 | 0.554 | 4.95E−41 | TNF-α | 0.295 | 2.51E−11 | ||
| IL10 | 0.603 | 3.42E−50 | Th2 | GATA3 | 0.313 | 1.22E−12 | |
| Neutrophils | CD66b (CEACAM8) | 0.237 | 1.06E−07 | IL13 | 0.142 | 1.63E−03 | |
| CD15 | 0.135 | 2.74E−03 | STAT6 | 0.077 | 8.69E−02 | ||
| CD11b (ITGAM) | 0.587 | 5.45E−47 | Tfh | BCL6 | −0.03 | 5.09E−01 | |
| CCR7 | 0.434 | 4.99E−24 | CD200 | 0.275 | 5.64E−10 | ||
| Natural killer cell | NKp46 | 0.405 | 7.42E−21 | IL21 | 0.319 | 4.27E−13 | |
| NKp44 | 0.109 | 1.52E−02 | ICOS | 0.667 | 1.00E−64 | ||
| NKp30 | 0.481 | 7.29E−30 | Th17 | STAT3 | 0.035 | 4.39E−01 | |
| FCGR3A | 0.682 | 1.36E−68 | IL17A | 0.183 | 4.19E−05 | ||
| FCGR3B | 0.326 | 1.13E−13 | IL1A | 0.227 | 3.36E−07 | ||
| NKG2A (KLRC1) | 0.351 | 1.01E−15 | IL1B | 0.428 | 2.26E−23 | ||
| KIR2DL1 | 0.151 | 7.73E−04 | CCL20 | −0.052 | 2.53E−01 | ||
| KIR2DL3 | 0.22 | 7.87E−07 | Treg | FOXP3 | 0.456 | 1.04E−26 | |
| KIR3DL1 | 0.169 | 1.64E−04 | CCR8 | 0.572 | 3.47E−44 | ||
| TGFβ | 0.365 | 5.36E−17 |
Data from the GEPIA database also showed similar results to that of the TIMER database. Detailed information is listed in Table 2.
Table 2. Correlation between immune marker sets of immune cells and FGL2 expression in lung adenocarcinoma based data from GEPIA database.
| Description | Gene markers | Cor | P | Description | Gene markers | Cor | P |
|---|---|---|---|---|---|---|---|
| T cell (general) | CD3D | 0.500 | 0 | CTL (Cytotoxic T Lymphocytes ) | CD8A | 0.530 | 0 |
| CD3E | 0.580 | 0 | CD8B | 0.390 | 0 | ||
| CD2 | 0.640 | 0 | GZMB | 0.280 | 3.50E−10 | ||
| B cell | CD19 | 0.240 | 1.30E−07 | Dendritic cell | HLA-DPB1 | 0.510 | 0 |
| CD79A | 0.220 | 1.70E−06 | HLA-DQB1 | 0.230 | 3.00E−07 | ||
| CD79B | 0.290 | 1.70E−10 | HLA-DRA | 0.610 | 0 | ||
| CD22 | 0.290 | 1.50E−10 | HLA-DPA1 | 0.610 | 0 | ||
| M1 Macrophage | INOS | 0.040 | 3.80E−01 | DEC-205 | 0.450 | 0 | |
| CIITA | 0.570 | 0 | BDCA-1 | 0.290 | 1.10E−10 | ||
| IRF5 | 0.300 | 3.70E−11 | BDCA-4 | 0.250 | 2.90E−08 | ||
| COX2 | −0.110 | 2.00E−02 | BDCA-2 | 0.420 | 0 | ||
| M2 Macrophage | CD163 | 0.460 | 0 | CD11c | 0.410 | 0 | |
| IRF4 | 0.300 | 1.70E−11 | Th1 | CD38 | 0.160 | 3.30E−04 | |
| VSIG4 | 0.500 | 0 | T-bet | 0.130 | 3.00E−03 | ||
| MS4A4A | 0.610 | 0 | STAT4 | 0.320 | 3.30E−13 | ||
| TAM | CCL2 | 0.300 | 2.40E−11 | STAT1 | 0.510 | 0 | |
| CCL5 | 0.430 | 0 | IFN-γ | 0.470 | 0 | ||
| CD68 | 0.550 | 0 | TNF-α | 0.320 | 6.90E−13 | ||
| IL10 | 0.580 | 0 | Th2 | GATA3 | 0.017 | 7.10E−01 | |
| Neutrophils | CD66b (CEACAM8) | 0.057 | 2.10E−01 | IL13 | 0.240 | 8.30E−08 | |
| CD15 | 0.110 | 1.50E−02 | STAT6 | 0.100 | 2.50E−02 | ||
| CD11b (ITGAM) | 0.560 | 0 | Tfh | BCL6 | 0.085 | 6.10E−02 | |
| CCR7 | 0.410 | 0 | CD200 | 0.370 | 0 | ||
| Natural killer cell | NKp46 | 0.390 | 0 | IL21 | 0.560 | 0 | |
| NKp44 | 0.068 | 1.40E−01 | ICOS | 0.650 | 0 | ||
| NKp30 | 0.440 | 0 | Th17 | STAT3 | 0.210 | 2.90E−06 | |
| FCGR3A | 0.660 | 0 | IL17A | 0.220 | 8.70E−07 | ||
| FCGR3B | 0.430 | 0 | IL1A | −0.003 | 9.50E−01 | ||
| NKG2A (KLRC1) | 0.230 | 5.00E−07 | IL1B | 0.380 | 0 | ||
| KIR2DL1 | −0.054 | 2.30E−01 | CCL20 | −0.046 | 3.10E−01 | ||
| KIR2DL3 | 0.130 | 5.70E−03 | Treg | FOXP3 | 0.500 | 0 | |
| KIR3DL1 | 0.150 | 1.30E−03 | CCR8 | 0.580 | 0 | ||
| TGFβ | 0.330 | 1.80E−13 |
Functional enrichment analyses of FGL2-correlated genes
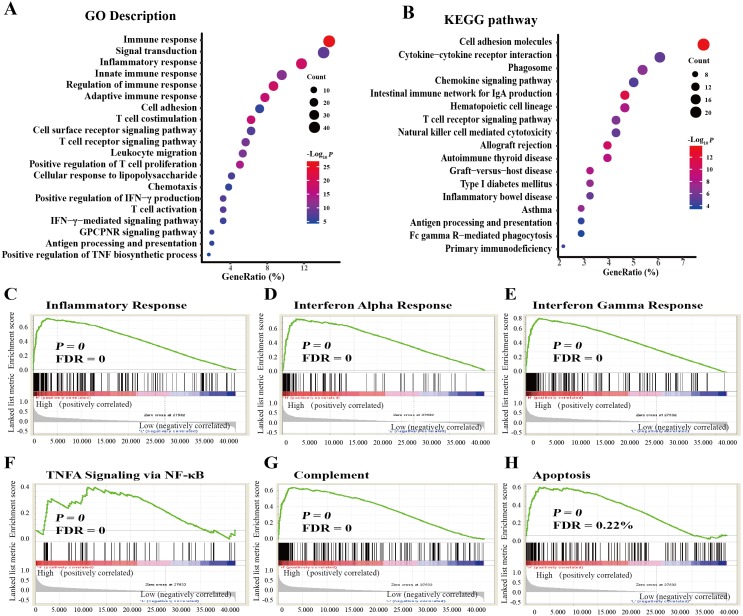
We performed GO and KEGG pathway analyses with data obtained from the TCGA dataset. GO analysis indicated that FGL2-correlated genes were enriched in the immune response, the adaptive immune response, the positive regulation of T cell proliferation, the positive regulation of interferon-gamma production, the positive regulation of tumor necrosis factor production, T cell activation, the interferon-gamma-mediated signaling pathway, T cell costimulation, T cell differentiation, the T cell receptor signaling pathway, antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-dependent, etc (Fig. 4A). KEGG pathway analysis showed that FGL2 was correlated with genes involved in cell adhesion, natural killer cell-mediated cytotoxicity, the TNF signaling pathway, the T cell receptor signaling pathway, antigen processing and presentation, etc. (Fig. 4B).
Figure 4. Functional enrichment analysis of FGL2-correlated genes in lung adenocarcinoma.
(A) GO analysis of FGL2-correlated genes in lung adenocarcinoma with the TCGA dataset. (B) KEGG analysis of FGL2-correlated genes in lung adenocarcinoma with the TCGA dataset. (C–H) GSEA was performed to explore the biological functions of FGL2 in lung adenocarcinoma.
We also conducted GSEA to explore the biological functions of FGL2 in lung adenocarcinoma. FGL2 expression was positively related to the inflammatory response, the interferon alpha response, the interferon gamma response, TNF alpha signaling via NF-κB, complement, and apoptosis (Fig. 4C).
Discussion
Lung adenocarcinoma is a type of non-small cell lung cancer and the most common histologic type of lung cancer. In lung adenocarcinoma, there is a variety of divergent molecular, pathologic, and clinical spectra (Testa, Castelli & Pelosi, 2018). The common signs of lung adenocarcinoma include weight loss, dyspnea, chest pain, and cough. The extrapulmonary manifestations include hypercalcemia of the malignancy and hypertrophic pulmonary osteoarthropathy. Tobacco smoking is a risk factor for lung adenocarcinoma (Song et al., 2017). In addition to smoking, gene mutations are also important mutagenic factors of lung adenocarcinoma (Ding et al., 2008). Somatic mutations might influence tumor suppressor genes and oncogenes in lung adenocarcinoma. Recent opinions on the treatment of lung adenocarcinoma have changed from traditional chemotherapy to precision medicine based on the genetic alterations of cancer (Herbst, Morgensztern & Boshoff, 2018). Previous studies have found that many gene mutations contribute to lung adenocarcinoma. The mutated genes include EGFR, KRAS, TP53, STK11, NF1, and KEAP1. The EGFR-activating mutation frequency varies depending on ethnicity and region. EGFR is correlated with the cell proliferation, invasion, survival, and angiogenesis of tumors (Sharma et al., 2007). Oral EGFR tyrosine kinase inhibitors such as gefitinib and erlotinib prolonged progression-free survival and the objective response rate compared with traditional chemotherapy (Mok et al., 2009; Shepherd et al., 2005). The inactivation of KEAP1 in KRAS mutations is related to the inhibition of glutaminase in lung cancer (Romero et al., 2017). TP53 mutations are commonly found in advanced-grade lung adenocarcinoma (Ahrendt et al., 2003). Precision medicine also provides genes that are beneficial for the progression and survival of cancers. FGL2 is an important factor in regulating the immune system. FGL2 is upregulated in GBM, promoting GBM development by suppressing dendritic cell activities (Yan et al., 2015). However, the diagnostic value of FGL2 in lung cancer is largely unknown. In this study, we systematically investigated the expression profile and potential functions of FGL2 in lung adenocarcinoma.
First, we evaluated the expression of FGL2 in lung adenocarcinoma tissue and adjacent normal tissue. Data obtained from the TCGA dataset and the Oncomine database indicated that FGL2 expression was significantly lower in lung adenocarcinoma tissue than in adjacent normal tissue. This implied that FGL2 might be a beneficial biomarker of lung adenocarcinoma.
Second, we analyzed the relationship of the FGL2 level and the prognostic survival of lung adenocarcinoma patients with three bioinformatics datasets. A high FGL2 mRNA level was correlated with better prognostic outcomes of lung adenocarcinoma, including overall survival and progression-free survival. These results indicate that FGL2 might negatively regulate the progression of lung adenocarcinoma.
Then, we explored the correlation between FGL2 expression and the immune status in the tumor microenvironment of lung adenocarcinoma. FGL2 expression was positively correlated with immune cell infiltration and immune marker sets in lung adenocarcinoma. The immune cells included CD8+ T cells, CD4+ T cells, macrophages, B cells and dendritic cells. CD8+ T cells (often called cytotoxic T lymphocytes or CTLs) are very important for tumor surveillance. CD8+ T cells use three major mechanisms to kill tumor cells. The first is the secretion of cytokines (primarily TNF-α and IFN-γ). Our results showed that high FGL2 expression was positively correlated with IFN-γ production and signaling and TNF-α production and signaling. The second major function is the production and release of cytotoxic granules, which mainly contain perforin 1 (PRF1) and granzymes. Our results showed that the expression level of FGL2 was positively correlated with the expression levels of PRF1, granzyme K (GZMK), GZMA, GZMH, GZMB, and GZMM. The third antitumor function of CD8+ T cells is to induce the apoptosis of tumor cells via Fas/Fas ligand (FasLG) interactions. Our results showed that the expression level of FGL2 was positively correlated with the expression levels of Fas and FasLG. These results show that high FGL2 expression is closely related to enhanced CD8+ T cell-mediated antitumor activities. Dendritic cells (DCs) are considered important factors that provide protective immunity against lung adenocarcinoma (Wang, Huang & Li, 2019). Inactive DCs are correlated with the poor prognosis of lung cancer patients. DCs present antigens to activate antitumor T cells. Mature DCs in lung cancer express high levels of cytokines and costimulatory molecules (CD40/80/86) to activate T cells (Macri et al., 2018). MHC type II molecules on DCs promote the activation of CD4+ T cells. MHC type 1 molecules on DCs promote the activation of CD8+ T cells (Liu & Cao, 2015). Except for activating T cells, DCs can also recruit and activate NK cells by secreting C-C chemokine receptor type 5 (CCR5) at the tumor site (Liu et al., 2008). In our study, we found that FGL2 expression was positively corelated to the infiltration of CD4+ T cells, CD8+ T cells, macrophages, B cells and DCs in lung adenocarcinoma. In addition, the FGL2 level was positively correlated with several subtypes of immune cells, including effector memory CD8+ T cells, activated CD8+ T cells, activated CD4+ T cells, type 1 T helper cells, effector memory CD4+ T cells, central memory CD8+ T cells, immature dendritic cells and natural killer T cells. These results indicate that FGL2 plays an important role in antitumor immunity by enhancing antitumor activities in lung adenocarcinoma.
There were some low correlation values for certain gene markers assayed to ascertain correlation between immune markers and FGL2 expression. As for B cells, Table 1 indicated the correlation of B cells gene markers varied from 0.21–0.316. In Table 2, the correlation of B cells gene markers varied from 0.24–0.29. In addition, Fig. 3A indicated the correlation between FGL2 and B cells was 0.409. The results showed that FGL2 has not possessed strong correlation with B cells in the tumor microenvironment of lung adenocarcinoma. Tumor infiltrating B cells appear in every stage of lung cancer and play critical roles in shaping tumor progression. However, the B cells functions in antitumor immunity of lung cancer are controversial (Wang et al., 2019). Some studies demonstrated that tumor infiltrating B cells have protective effects on anti-tumor immunity in lung cancer, while other studies revealed that tumor infiltrating B cells have inhibitory effects on antitumor immunity in lung cancer. Owning to the B cells functions in antitumor immunity is dualistic, FGL2 might possessed poor or moderate correlation with B cells in the tumor microenvironment of lung adenocarcinoma. Apart from B cells, Fig. 3A showed correlation between FGL2 and CD4+ T cells was 0.373. This result indicated that the correlation between FGL2 and CD4+ T cells was moderate at best. Previous studies found that the role of CD4+ T cells in antitumor activity of lung cancer is dualistic (Zheng, Hu & Yao, 2017). Some types of CD4+ T cells impair the functions of cytotoxic T lymphocytes to promote the tumor development, while some types of CD4+ T cells induce the activation of cytotoxic T lymphocytes to exert antitumor immune response. This might explain the reason that correlation between FGL2 and CD4+ T cells was poor and moderate in lung adenocarcinoma.
Finally, we performed functional enrichment analysis to explore the biological functions of FGL2 in lung adenocarcinoma. GO and KEGG analyses indicated that FGL2-correlated genes are mainly enriched in pathways involved in T cell proliferation/differentiation/activation and antigen processing and presentation. GSEA showed that FGL2 expression was positively correlated with enhanced tumor killing. These results further indicate that FGL2 enhances antitumor activities in lung adenocarcinoma.
In the research conducted by Zhu et al. titled “Stroma-derived fibrinogen-like protein 2 activates cancer-associated fibroblasts to promote tumor growth in lung cancer”, they found that FGL2 could promote tumor growth of lung cancer by activating cancer-associated fibroblasts using a mouse model of Lewis lung carcinoma (Zhu et al., 2017). In the research conducted by Yan et al. (2019) titled “FGL2 promotes tumor progression in the CNS by suppressing CD103+ dendritic cell differentiation”, they found FGL2 accelerated tumor progression of GBM by suppressing CD103+ dendritic cell differentiation. In our study, we extracted data of lung adenocarcinoma patients from different clinical databases such as TCGA, PrognoScan, and TIMER. We found that FGL2 expression was significantly lower in lung adenocarcinoma tissue compared with adjacent normal tissue. A high expression level of FGL2 was correlated with better prognostic outcomes of lung adenocarcinoma patients. We speculated that FGL2 might play differential roles in distinct models of cancer.
In the mouse model of Lewis lung carcinoma, FGL2 induced an activated and pro-tumorigenic phenotype of cancer-associated fibroblasts in the tumor microenvironment (TME). Cancer-associated fibroblasts, also known as tumor-associated fibroblast, promote tumorigenic features by producing cytokines, or initiating extracellular matrix remodeling in the tumor microenvironment (Erdogan & Webb, 2017). The cytokines could disrupt normal cell functions, such as normal cell cycle regulation to active their pro-tumor actions (Öhlund, Elyada & Tuveson, 2014). In addition, cancer-associated fibroblasts produce and secret angiogenic factors such as fibroblast growth factor (FGF), and vascular endothelial growth factor (VEGF) to stimulate angiogenesis supporting the formation of tumors and the proliferation of cancer cells and metastasis (Shiga et al., 2015).
In the mouse model of brain tumor, Yan et al. demonstrated that FGL2 promotes GBM tumor progression by suppressing CD103+ dendritic cell differentiation. The function of dendritic cells and their subtypes in GBM has not been elucidated clearly (Srivastava et al., 2019). Dendritic cells might interplay with other types of immune cells including macrophages, T cells, and microglia in the tumor microenvironment (TME). Some researchers considered that certain subsets of dendritic cells recognize and present tumor antigens to induce the T cells immune responses (D’Agostino et al., 2012). If the CD103+ dendritic cell differentiation was suppressed, the antigen processing functions of dendritic cells might be impaired in GBM. So, FGL2 might promote GBM tumor progression by suppressing CD103+ dendritic cell differentiation.
In our study, we demonstrated FGL2 was positively correlated with T cells, especially CD8+T cells activation in the tumor microenvironment of lung adenocarcinoma patients. CD8+T cells, also known as cytotoxic T lymphocytes (CTLs), play important role of antitumor immune response in the tumor microenvironment (Aerts & Hegmans, 2013). CTLs could recognize and kill tumor cells by the complex formation of T-cell receptor (TCR) and human leukocyte antigen class (HLA). TCR and related signaling molecules could activate the transduction cascade to induce immune synapse and stimulate antitumor responses of CTLs (Farhood, Najafi & Mortezaee, 2019). Then, the activated CTLs produce and secret cytotoxic granules such as granzymes and perforin into the targeted tumor cells. Adhesive and co-stimulatory molecules including CD80/CD86, CD11a/CD18, and lymphocyte function-associated antigen 1 (LFA-1) are important in the process of TCR-mediated CTLs antitumor effects (Durgeau et al., 2018). CTL-associated antigen 4 (CTLA-4) and programmed death-1 receptor (PD-1)-ligand (PD-L1) are checkpoint receptors could be targeted to relieve CTLs exhaustion. Antigen presenting cells (APCs) such as dendritic cells (DCs) and macrophages could activate naïve CD8+ T cells by binding the TCRs with CD3 and other costimulatory molecules. FGL2 is mainly expressed in APCs such as DCs and macrophages. So, we infer that FGL2 might activate CTLs functions to exert anti-tumor effects by stimulating APCs to bind naïve CD8+ T cells.
In brief, FGL2 might play different roles in different types of cancer models. In lung cancer animal model conducted by Zhu et al., FGL2 might promote tumor progress by activating cancer-associated fibroblasts in tumor microenvironment. In GBM animal model, FGL2 promotes GBM progress by suppressing CD103+ dendritic cell differentiation. In clinical databases of lung cancer patients, FGL2 exhibited antitumor activities by activating CTLs in the tumor microenvironment of lung cancer.
There are no significant differences in the expression levels of FGL2 between patients with or without EGFR or KRAS mutations in the TCGA database. There is no significant correlation between FGL2 and other genes implicated in lung adenocarcinoma such as EGFR, KRAS, TP53, STK11, NF1, and KEAP1. We speculated that FGL2 might indirectly affect those genes in lung adenocarcinoma by changing the immune status in the tumor environment. EGFR regulates several signaling transduction cascades such as MAPK, JNK, and Akt signaling pathways, leading to tumor cell proliferation, cell cycle progression, angiogenesis, and metastasis (Bethune et al., 2019). EGFR-targeted therapy such as EGFR tyrosine kinase inhibitors (EGFR-TKIs) alters the tumor microenvironment in lung cancer (Matsumoto et al., 2019). EGFR-TKIs could increase cytotoxic CD8+ T cells and dendritic cells in the tumor environment of lung cancer (Jia et al., 2019). FGL2 also increase cytotoxic CD8+ T cells and dendritic cells in the tumor environment of lung cancer. So, FGL2 might affect EGFR by influencing immune status in tumor environment of lung cancer. KRAS is important in promoting cell survival and growth in tumor cells. Almost 30% patients with lung adenocarcinoma are positive for KRAS gene mutation (Tomasini et al., 2016). KRAS is a strong initiator of tumorigenesis in lung adenocarcinoma. It is also a predictive response to targeted therapy of lung adenocarcinoma (Dias Carvalho et al., 2018). KRAS is related with immune-suppressed state by regulating components of adaptive and innate immune response (Cullis, Das & Bar-Sagi, 2018). Mutant KRAS could up-regulate immunosuppressive cells in tumor such as myeloid-derived suppressor cells (MDSCs), CD4+FoxP3+ T regulatory cells, and CD19+IL10+ B regulatory cells. These immunosuppressive cells could suppress the activities of tumoricidal cells such as CD8+ T cytotoxic cells, natural killer (NK) cells in the tumor microenvironment. On the contrary, FGL2 could increase the levels of tumoricidal cells such as CD8+ T cytotoxic cells, natural killer (NK) cells in the tumor microenvironment. So, FGL2 affect KRAS by influencing immune status in tumor environment of lung cancer. TP53 is regarded as tumor suppressor gene that it could prevent genome mutation. TP53 could activate DNA repair process and arrest cell proliferation by holding cell cycle (Uehara & Tanaka, 2018). Mutation of TP53 leads to tumor escape from senescence and apoptosis. Activation of TP53 could increase the levels of tumor-infiltrating leukocytes such as CD8+ T cells in tumor microenvironment (Guo et al., 2017). FGL2 also increases the levels of tumor-infiltrating leukocytes such as CD8+ T cells in tumor microenvironment. So, FGL2 possesses synergic effects with TP53 to enhance antitumor immunity in tumor microenvironment. STK11, known as liver kinase B1 (LKB1), regulates cell polarity and regarded as tumor suppressor. Loss of STK11 would lead to cell polarity disorganization and induce tumor growth. Koyama et al. found that loss of STK11/LKB1 induced neutrophil recruitment and inflammatory mediator production to suppress the T cells in tumor environment of lung cancer (Koyama et al., 2016). In addition, STK11/LKB1 inactivated mutations were related with reduced expression of PD-1 ligand PD-L1 in tumor cells. So, we infer that FGL2 exert synergic effects with STK11 to enhance the cytotoxic T lymphocytes activities in tumor environment of lung adenocarcinoma. NF1 is regarded as a tumor suppressor in lung cancer negatively regulates RAS signaling pathway. NF1 mutations present in similar patients populations with KRAS mutation (Redig et al., 2016). We infer that FGL2 affect KRAS by influencing immune status in tumor environment of lung adenocarcinoma. So, FGL2 might affect NF1 function by influencing immune status in tumor environment of lung adenocarcinoma. KEAP1 has been shown to interact with nuclear factor erythroid 2-related factor 2 (NRF2). KEAP1-NRF2 pathway play important role in oxidative response by inducing anti-inflammatory and antioxidant effects. KEAP1 mutation is correlated with poor prognosis of lung cancer (Frank et al., 2018). Aberrant KEAP1-NRF2 pathway activity alters the immune microenvironment of lung adenocarcinoma. KEAP1 mutation is associated reduced leukocyte infiltration of tumor microenvironment in lung adenocarcinoma (Thorsson et al., 2018). We infer that FGL2 affects the KEAP1 effect by influencing immune status in tumor environment of lung adenocarcinoma. So, we infer some indirect correlations might explain the mechanism of FGL2 affecting other genes. FGL2 might affect other genes functions by influencing immune status in tumor environment of lung adenocarcinoma.
Conclusion
In this study, we explored the expression profile and potential effects of FGL2 in lung adenocarcinoma. We found that FGL2 expression was significantly lower in lung adenocarcinoma tissue than in adjacent normal tissue. High FGL2 mRNA expression was correlated with better prognostic outcomes of lung adenocarcinoma patients, including overall survival and progression-free survival. These results indicate that FGL2 might function as a negative regulator of lung adenocarcinoma. Then, we investigated the potential mechanism of FGL2 in lung adenocarcinoma. FGL2 was positively correlated with the infiltration of immune cells, including CD8+ T cells, CD4+ T cells, macrophages, B cells and dendritic cells, in lung adenocarcinoma. These results imply that FGL2 exerts its antitumor effects by enhancing immune cell infiltration in lung adenocarcinoma. GO and KEGG functional enrichment analyses and GSEA also showed that FGL2 expression was positively correlated with enhanced tumor killing. Thus, we propose that FGL2, which is positively associated with enhanced antitumor activities mediated by T cells, is a beneficial marker for lung adenocarcinoma treatment outcomes. In this study, we used bioinformatic analysis to discover the potential roles of FGL2 in lung adenocarcinoma. In the future studies, in vivo and in vitro experiments will carry out to demonstrate the role of FGL2 in modulating the T cell-mediated immune response in lung adenocarcinoma.
Supplemental Information
Funding Statement
This work was supported by the National Natural Science Foundation of China [grant numbers 81430099, 31500704, and 81904142], Projects of International Cooperation and Exchanges [grant number 2014DFA32950] and research program from Beijing University of Chinese Medicine [grant numbers 1000041510049, BUCM-2019-JCRC006 and 2019-JYB-TD013]. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Contributor Information
Guangrui Huang, Email: hgr@bucm.edu.cn.
Anlong Xu, Email: xuanlong@bucm.edu.cn.
Additional Information and Declarations
Competing Interests
The authors declare there are no competing interests.
Author Contributions
Kai Yuan and Yanyan Feng performed the experiments, analyzed the data, authored or reviewed drafts of the paper, and approved the final draft.
Hesong Wang, Lu Zhao, Wei Wang, Ting Wang and Yuyin Feng analyzed the data, prepared figures and/or tables, and approved the final draft.
Guangrui Huang and Anlong Xu conceived and designed the experiments, prepared figures and/or tables, and approved the final draft.
Data Availability
The following information was supplied regarding data availability:
The code is available in the Supplemental Files.
References
- Aerts & Hegmans (2013).Aerts JG, Hegmans JP. Tumor-specific cytotoxic T cells are crucial for efficacy of immunomodulatory antibodies in patients with lung cancer. Cancer Research. 2013;73:2381–2388. doi: 10.1158/0008-5472.CAN-12-3932. [DOI] [PubMed] [Google Scholar]
- Ahrendt et al. (2003).Ahrendt SA, Hu Y, Buta M, McDermott MP, Benoit N, Yang SC, Wu L, Sidransky D. p53 mutations and survival in stage I non-small-cell lung cancer: results of a prospective study. Journal of the National Cancer Institute. 2003;95:961–970. doi: 10.1093/jnci/95.13.961. [DOI] [PubMed] [Google Scholar]
- Bethune et al. (2019).Bethune G, Bethune D, Ridgway N, Xu Z. Epidermal growth factor receptor (EGFR) in lung cancer: an overview and update. International Journal of Cancer. 2019;145:1432–1444. [PMC free article] [PubMed] [Google Scholar]
- Bhattacharjee et al. (2001).Bhattacharjee A, Richards WG, Staunton J, Li C, Monti S, Vasa P, Ladd C, Beheshti J, Bueno R, Gillette M, Loda M, Weber G, Mark EJ, Lander ES, Wong W, Johnson BE, Golub TR, Sugarbaker DJ, Meyerson M. Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:13790–13795. doi: 10.1073/pnas.191502998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkhäuser et al. (2013).Birkhäuser FD, Koya RC, Neufeld C, Rampersaud EN, Lu X, Micewicz ED, Chodon T, Atefi M, Kroeger N, Chandramouli GV, Li G, Said JW, McBride WH, Kabbinavar FF, Ribas A, Pantuck AJ, Belldegrun AS, Riss J. Dendritic cell-based immunotherapy in prevention and treatment of renal cell carcinoma: efficacy, safety, and activity of Ad-GM⋅CAIX in immunocompetent mouse models. Journal of Immunotherapy. 2013;36:102–111. doi: 10.1097/CJI.0b013e31827bec97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullis, Das & Bar-Sagi (2018).Cullis J, Das S, Bar-Sagi D. Kras and tumor immunity: friend or foe? Cold Spring Harbor Perspectives in Medicine. 2018;8:a031849. doi: 10.1101/cshperspect.a031849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Agostino et al. (2012).D’Agostino PM, Gottfried-Blackmore A, Anandasabapathy N, Bulloch K. Brain dendritic cells: biology and pathology. Acta Neuropathologica. 2012;124:599–614. doi: 10.1007/s00401-012-1018-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias Carvalho et al. (2018).Dias Carvalho P, Guimarães CF, Cardoso AP, Mendonça S, Costa ÂM, Oliveira MJ, Velho S. KRAS oncogenic signaling extends beyond cancer cells to orchestrate the microenvironment. Cancer Research. 2018;78:7–14. doi: 10.1158/0008-5472.CAN-17-2084. [DOI] [PubMed] [Google Scholar]
- Ding et al. (2008).Ding L, Getz G, Wheeler DA, Mardis ER, McLellan MD, Cibulskis K, Sougnez C, Greulich H, Muzny DM, Morgan MB, Fulton L, Fulton RS, Zhang Q, Wendl MC, Lawrence MS, Larson DE, Chen K, Dooling DJ, Sabo A, Hawes AC, Shen H, Jhangiani SN, Lewis LR, Hall O, Zhu Y, Mathew T, Ren Y, Yao J, Scherer SE, Clerc K, Metcalf GA, Ng B, Milosavljevic A, Gonzalez-Garay ML, Osborne JR, Meyer R, Shi X, Tang Y, Koboldt DC, Lin L, Abbott R, Miner TL, Pohl C, Fewell G, Haipek C, Schmidt H, Dunford-Shore BH, Kraja A, Crosby SD, Sawyer CS, Vickery T, Sander S, Robinson J, Winckler W, Baldwin J, Chirieac LR, Dutt A, Fennell T, Hanna M, Johnson BE, Onofrio RC, Thomas RK, Tonon G, Weir BA, Zhao X, Ziaugra L, Zody MC, Giordano T, Orringer MB, Roth JA, Spitz MR, Wistuba II, Ozenberger B, Good PJ, Chang AC, Beer DG, Watson MA, Ladanyi M, Broderick S, Yoshizawa A, Travis WD, Pao W, Province MA, Weinstock GM, Varmus HE, Gabriel SB, Lander ES, Gibbs RA, Meyerson M, Wilson RK. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durgeau et al. (2018).Durgeau A, Virk Y, Corgnac S, Mami-Chouaib F. Recent advances in targeting CD8 T-cell immunity for more effective cancer immunotherapy. Frontiers in Immunology. 2018;9:1–14. doi: 10.3389/fimmu.2018.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdogan & Webb (2017).Erdogan B, Webb DJ. Cancer-associated fibroblasts modulate growth factor signaling and extracellular matrix remodeling to regulate tumor metastasis. Biochemical Society Transactions. 2017;45:229–236. doi: 10.1042/BST20160387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farhood, Najafi & Mortezaee (2019).Farhood B, Najafi M, Mortezaee K. CD8+ cytotoxic T lymphocytes in cancer immunotherapy: a review. Journal of Cellular Physiology. 2019;234:8509–8521. doi: 10.1002/jcp.27782. [DOI] [PubMed] [Google Scholar]
- Frank et al. (2018).Frank R, Scheffler M, Merkelbach-Bruse S, Ihle MA, Kron A, Rauer M, Ueckeroth F, König K, Michels S, Fischer R, Eisert A, Fassunke J, Heydt C, Serke M, Ko YD, Gerigk U, Geist T, Kaminsky B, Heukamp LC, Clement-Ziza M, Büttner R, Wolf J. Clinical and pathological characteristics of KEAP1- and NFE2L2-mutated non-small cell lung carcinoma (NSCLC) Clinical Cancer Research. 2018;24:3087–3096. doi: 10.1158/1078-0432.CCR-17-3416. [DOI] [PubMed] [Google Scholar]
- Guo et al. (2017).Guo G, Yu M, Xiao W, Celis E, Cui Y. Local activation of p53 in the tumor microenvironment overcomes immune suppression and enhances antitumor immunity. Cancer Research. 2017;77:2292–2305. doi: 10.1158/0008-5472.CAN-16-2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbst, Morgensztern & Boshoff (2018).Herbst RS, Morgensztern D, Boshoff C. The biology and management of non-small cell lung cancer. Nature. 2018;553:446–454. doi: 10.1038/nature25183. [DOI] [PubMed] [Google Scholar]
- Hirsch et al. (2017).Hirsch FR, Scagliotti GV, Mulshine JL, Kwon R, Curran Jr WJ, Wu YL, Paz-Ares L. Lung cancer: current therapies and new targeted treatments. Lancet. 2017;389:299–311. doi: 10.1016/S0140-6736(16)30958-8. [DOI] [PubMed] [Google Scholar]
- Hsieh et al. (2010).Hsieh YH, Chen CW, Schmitz SF, King CC, Chen WJ, Wu YC, Ho MS. Candidate genes associated with susceptibility for SARS-coronavirus. Bulletin of Mathematical Biology. 2010;72:122–132. doi: 10.1007/s11538-009-9440-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia et al. (2019).Jia Y, Li X, Jiang T, Zhao S, Zhao C, Zhang L, Liu X, Shi J, Qiao M, Luo J, Liu S, Han R, Su C, Ren S, Zhou C. EGFR-targeted therapy alters the tumor microenvironment in EGFR-driven lung tumors: implications for combination therapies. International Journal of Cancer. 2019;145:1432–1444. doi: 10.1002/ijc.32191. [DOI] [PubMed] [Google Scholar]
- Koyama et al. (2016).Koyama S, Akbay EA, Li YY, Aref AR, Skoulidis F, Herter-Sprie GS, Buczkowski KA, Liu Y, Awad MM, Denning WL, Diao L, Wang J, Parra-Cuentas ER, Wistuba II, Soucheray M, Thai T, Asahina H, Kitajima S, Altabef A, Cavanaugh JD, Rhee K, Gao P, Zhang H, Fecci PE, Shimamura T, Hellmann MD, Heymach JV, Hodi FS, Freeman GJ, Barbie DA, Dranoff G, Hammerman PS, Wong KK. STK11/LKB1 deficiency promotes neutrophil recruitment and proinflammatory cytokine production to suppress T-cell activity in the lung tumor microenvironment. Cancer Research. 2016;76:999–1008. doi: 10.1158/0008-5472.CAN-15-1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latimer (2018).Latimer KM. Lung cancer: clinical presentation and diagnosis. FP Essent. 2018;464:23–26. [PubMed] [Google Scholar]
- Liu et al. (2008).Liu C, Lou Y, Lizée G, Qin H, Liu S, Rabinovich B, Kim GJ, Wang YH, Ye Y, Sikora AG, Overwijk WW, Liu YJ, Wang G, Hwu P. Plasmacytoid dendritic cells induce NK cell-dependent, tumor antigen-specific T cell cross-priming and tumor regression in mice. Journal of Clinical Investigation. 2008;118:1165–1175. doi: 10.1172/JCI33583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Liu & Chen (2017).Liu XG, Liu Y, Chen F. Soluble fibrinogen like protein 2 (sFGL2), the novel effector molecule for immunoregulation. Oncotarget. 2017;8:3711–3723. doi: 10.18632/oncotarget.12533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu & Cao (2015).Liu Y, Cao X. Intratumoral dendritic cells in the anti-tumor immune response. Cellular & Molecular Immunology. 2015;12:387–390. doi: 10.1038/cmi.2014.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macri et al. (2018).Macri C, Pang ES, Patton T, O’Keeffe M. Dendritic cell subsets. Seminars in Cell and Developmental Biology. 2018;84:11–21. doi: 10.1016/j.semcdb.2017.12.009. [DOI] [PubMed] [Google Scholar]
- Matsumoto et al. (2019).Matsumoto Y, Sawa K, Fukui M, Oyanagi J, Izumi M, Ogawa K, Suzumura T, Watanabe T, Kaneda H, Mitsuoka S, Asai K, Kimura T, Yamamoto N, Koh Y, Kawaguchi T. Impact of tumor microenvironment on the efficacy of epidermal growth factor receptor-tyrosine kinase inhibitors in patients with EGFR-mutant non-small cell lung cancer. Cancer Science. 2019;110:3244–3254. doi: 10.1111/cas.14156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok et al. (2009).Mok TS, Wu YL, Thongprasert S, Yang CH, Chu DT, Saijo N, Sunpaweravong P, Han B, Margono B, Ichinose Y, Nishiwaki Y, Ohe Y, Yang JJ, Chewaskulyong B, Jiang H, Duffield EL, Watkins CL, Armour AA, Fukuoka M. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. The New England Journal of Medicine. 2009;361:947–957. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- Öhlund, Elyada & Tuveson (2014).Öhlund D, Elyada E, Tuveson D. Fibroblast heterogeneity in the cancer wound. Journal of Experimental Medicine. 2014;211:1503–1523. doi: 10.1084/jem.20140692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilkington et al. (2015).Pilkington G, Boland A, Brown T, Oyee J, Bagust A, Dickson R. A systematic review of the clinical effectiveness of first-line chemotherapy for adult patients with locally advanced or metastatic non-small cell lung cancer. Thorax. 2015;70:359–367. doi: 10.1136/thoraxjnl-2014-205914. [DOI] [PubMed] [Google Scholar]
- Redig et al. (2016).Redig AJ, Capelletti M, Dahlberg SE, Sholl LM, Mach S, Fontes C, Shi Y, Chalasani P, Jänne PA. Clinical and molecular characteristics of nf1-mutant lung cancer. Clinical Cancer Research. 2016;22:3148–3156. doi: 10.1158/1078-0432.CCR-15-2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero et al. (2017).Romero R, Sayin VI, Davidson SM, Bauer MR, Singh SX, LeBoeuf SE, Karakousi TR, Ellis DC, Bhutkar A, Sánchez-Rivera FJ, Subbaraj L, Martinez B, Bronson RT, Prigge JR, Schmidt EE, Thomas CJ, Goparaju C, Davies A, Dolgalev I, Heguy A, Allaj V, Poirier JT, Moreira AL, Rudin CM, Pass HI, Van der Heiden MG, Jacks T, Papagiannakopoulos T. Keap1 loss promotes Kras-driven lung cancer and results in dependence on glutaminolysis. Nature Medicine. 2017;23:1362–1368. doi: 10.1038/nm.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz & Cote (2016).Schwartz AG, Cote ML. Epidemiology of lung cancer. Advances in Experimental Medicine and Biology. 2016;893:21–41. doi: 10.1007/978-3-319-24223-1_2. [DOI] [PubMed] [Google Scholar]
- Sharma et al. (2007).Sharma SV, Bell DW, Settleman J, Haber DA. Epidermal growth factor receptor mutations in lung cancer. Nature Reviews Cancer. 2007;7(3):169–181. doi: 10.1038/nrc2088. [DOI] [PubMed] [Google Scholar]
- Shepherd et al. (2005).Shepherd FA, Rodrigues Pereira J, Ciuleanu T, Tan EH, Hirsh V, Thongprasert S, Campos D, Maoleekoonpiroj S, Smylie M, Martins R, Van Kooten M, Dediu M, Findlay B, Tu D, Johnston D, Bezjak A, Clark G, Santabárbara P. Erlotinib in previously treated non-small-cell lung cancer. The New England Journal of Medicine. 2005;353(2):123–132. doi: 10.1056/NEJMoa050753. [DOI] [PubMed] [Google Scholar]
- Shiga et al. (2015).Shiga K, Hara M, Nagasaki T, Sato T, Takahashi H, Takeyama H. Cancer-associated fibroblasts: their characteristics and their roles in tumor growth. Cancer. 2015;7:2443–2458. doi: 10.3390/cancers7040902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song et al. (2017).Song MA, Benowitz NL, Berman M, Brasky TM, Cummings KM, Hatsukami DK, Marian C, O’Connor R, Rees VW, Woroszylo C, Shields PG. Cigarette filter ventilation and its relationship to increasing rates of lung adenocarcinoma. Journal of the National Cancer Institute. 2017;109(12):1–18. doi: 10.1093/jnci/djx075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava et al. (2019).Srivastava S, Jackson C, Kim T, Choi J, Lim M. A characterization of dendritic cells and their role in immunotherapy in glioblastoma: from preclinical studies to clinical trials. Cancer. 2019;11:E537. doi: 10.3390/cancers11040537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamayo et al. (2016).Tamayo P, Steinhardt G, Liberzon A, Mesirov JP, Mesirov JP. The limitations of simple gene set enrichment analysis assuming gene independence. Statistical Methods in Medical Research. 2016;25:472–487. doi: 10.1177/0962280212460441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Testa, Castelli & Pelosi (2018).Testa U, Castelli G, Pelosi E. Lung cancers: molecular characterization, clonal heterogeneity and evolution, and cancer stem cells. Cancer. 2018;10:E248. doi: 10.3390/cancers10080248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorsson et al. (2018).Thorsson V, Gibbs DL, Brown SD, Wolf D, Bortone DS, Ou Yang TH, Porta-Pardo E, Gao GF, Plaisier CL, Eddy JA, Ziv E, Culhane AC, Paull EO, Sivakumar IKA, Gentles AJ, Malhotra R, Farshidfar F, Colaprico A, Parker JS, Mose LE, Vo NS, Liu J, Liu Y, Rader J, Dhankani V, Reynolds SM, Bowlby R, Califano A, Cherniack AD, Anastassiou D, Bedognetti D, Mokrab Y, Newman AM, Rao A, Chen K, Krasnitz A, Hu H, Malta TM, Noushmehr H, Pedamallu CS, Bullman S, Ojesina AI, Lamb A, Zhou W, Shen H, Choueiri TK, Weinstein JN, Guinney J, Saltz J, Holt RA. The immune landscape of cancer. Immunity. 2018;48:812–830. doi: 10.1016/j.immuni.2018.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasini et al. (2016).Tomasini P, Walia P, Labbe C, Jao K, Leighl NB. Targeting the KRAS pathway in non-small cell lung cancer. Oncologist. 2016;21:1450–1460. doi: 10.1634/theoncologist.2015-0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torre, Siegel & Jemal (2016).Torre LA, Siegel RL, Jemal A. Lung cancer statistics. Advances in Experimental Medicine and Biology. 2016;893:1–19. doi: 10.1007/978-3-319-24223-1_1. [DOI] [PubMed] [Google Scholar]
- Uehara & Tanaka (2018).Uehara I, Tanaka N. Role of p53 in the regulation of the inflammatory tumor microenvironment and tumor suppression. Cancer. 2018;10:E219. doi: 10.3390/cancers10070219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Huang & Li (2019).Wang JB, Huang X, Li FR. Impaired dendritic cell functions in lung cancer: a review of recent advances and future perspectives. Cancer Communications. 2019;39(1):43–53. doi: 10.1186/s40880-019-0387-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang et al. (2019).Wang SS, Liu W, Ly D, Xu H, Qu L, Zhang L. Tumor-infiltrating B cells: their role and application in anti-tumor immunity in lung cancer. Cellular & Molecular Immunology. 2019;16:6–18. doi: 10.1038/s41423-018-0027-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan et al. (2015).Yan J, Kong LY, Hu J, Gabrusiewicz K, Dibra D, Xia X, Heimberger AB, Li S. FGL2 as a multimodality regulator of tumor-mediated immune suppression and therapeutic target in gliomas. Journal of the National Cancer Institute. 2015;107(8):djv137. doi: 10.1093/jnci/djv137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan et al. (2019).Yan J, Zhao Q, Gabrusiewicz K, Kong LY, Xia X, Wang J, Ott M, Xu J, Davis RE, Huo L, Rao G, Sun SC, Watowich SS, Heimberger AB, Li S. FGL2 promotes tumor progression in the CNS by suppressing CD103+ dendritic cell differentiation. Nature Communications. 2019;10:448–462. doi: 10.1038/s41467-018-08271-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang & Hooper (2013).Yang G, Hooper WC. Physiological functions and clinical implications of fibrinogen-like 2: a review. World Journal of Clinical Infectious Diseases. 2013;3:37–46. doi: 10.5495/wjcid.v3.i3.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zappa & Mousa (2016).Zappa C, Mousa SA. Non-small cell lung cancer: current treatment and future advances. Translational Lung Cancer Research. 2016;5:288–300. doi: 10.21037/tlcr.2016.06.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao et al. (2013).Zhao Z, Wang L, Yang C, Zhao T, Li L, Hu L, Wu D, Rong R, Xu M, Zhu T. Soluble FGL2 induced by tumor necrosis factor-α and interferon-γ in CD4+ T cells through MAPK pathway in human renal allograft acute rejection. Journal of Surgical Research. 2013;184:1114–1122. doi: 10.1016/j.jss.2013.04.011. [DOI] [PubMed] [Google Scholar]
- Zheng, Hu & Yao (2017).Zheng X, Hu Y, Yao C. The paradoxical role of tumor-infiltrating immune cells in lung cancer. Intractable and Rare Diseases Research. 2017;6:234–241. doi: 10.5582/irdr.2017.01059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu et al. (2017).Zhu Y, Zhang L, Zha H, Yang F, Hu C, Chen L, Guo B, Zhu B. Stroma-derived fibrinogen-like protein 2 activates cancer-associated fibroblasts to promote tumor growth in lung cancer. International Journal of Biological Sciences. 2017;13:804–814. doi: 10.7150/ijbs.19398. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The following information was supplied regarding data availability:
The code is available in the Supplemental Files.