Abstract
Purpose of review:
The field of neurotrauma research faces a reproducibility crisis. In response, research leaders in traumatic brain injury (TBI) and spinal cord injury (SCI) are leveraging data curation and analytics methods to encourage transparency, and improve the rigor and reproducibility. Here we review the current challenges and opportunities that come from efforts to transform neurotrauma’s big data to knowledge.
Recent Findings:
Three parallel movements are driving data-driven-discovery in neurotrauma. First, large multicenter consortia are collecting large quantities of neurotrauma data, refining common data elements (CDEs) that can be used across studies. Investigators are now testing the validity of CDEs in diverse research settings. Second, data sharing initiatives are working to make neurotrauma data findable, accessible, interoperable and reusable (FAIR). These efforts are reflected by recent open data repository projects for preclinical and clinical neurotrauma. Third, machine learning analytics are allowing researchers to uncover novel data-driven-hypotheses and test new therapeutics in multidimensional outcome space.
Summary:
We are on the threshold of a new era in data collection, curation, and analysis. The next phase of big data in neurotrauma research will require responsible data stewardship, a culture of data-sharing, and the illumination of ‘dark data’.
Keywords: DATA SCIENCE, PRECISION MEDICINE, TRANSLATION, BASIC SCIENCE, LONG-TAIL DATA
Introduction
As the push for more transparency, rigor, and reproducibility in neuroscience research accelerates with mandates from funders and journals, it is worth considering how to best achieve these goals in complex neurological disorders such as neurotrauma (TBI and SCI). We argue that one solution is for researchers and clinicians to recognize neurotrauma as a “Big Data” problem. The term “Big Data” was first characterized in the internet technology field in the early 21st century as data that is difficult to work with because it is ‘big’ according to at least one of three attributes (the 3V’s): Volume, Velocity, and Variety [1]. In the context of neurotrauma informatics, each of the 3Vs are represented to varying degrees. The mapping of complete genomes presents an issue of volume, where a single high-throughput sequencing experiment can produce several terabytes (1012 bytes) of data [2]. Real-time critical care multi-monitoring allows data collection at a higher velocity than ever before (e.g., 60 data-points/sec) [3,4]. However, we argue that for the world of neurotrauma research, the key to transforming big-data-to-knowledge lies in the way we handle data variety [5,6]. The complexity and heterogeneity of neurotrauma, both between individuals and within individuals as injury processes evolve over time, require analytical methods that can leverage this complexity to inform decision making. Here we will discuss the current strategies for harmonizing large clinical and preclinical data collection efforts, and recent trends in multivariate analyses and machine learning aimed at integrating multimodal neurotrauma datasets.
Exploiting Data Sharing to Improve Reproducibility
Over the past 25 years, the burgeoning field of neuroinformatics has made great strides by bringing the large volume and variety of data together from across research institutions and disciplines. Shared public data repositories, including the Allen Brain Atlas, the US BRAIN Initiative, the European Human Brain Project, and the Big-Data-To-Knowledge (BD2K) initiative have increased in size and scope in recent years [7–10]. Further, in the past 10 years public neuroscience databases have been federated through the Neuroscience Information Framework (NIF), a vast public resource developed through NIH Blueprint Consortium which now houses the world’s largest searchable collection of neuroscience data (http://neuinfo.org). NIF developed its infrastructure into SciCrunch, a scalable technology that allows scientific communities to create custom portals for curating, searching, accessing, and sharing their data with one another [11]. NIF/SciCrunch incorporate the data stewardship principles of findability, accessibility, interoperability, and reusability (FAIR) that are the guiding elements of data sharing efforts throughout the broader scientific community [12]. In the realm of neurotrauma, one successful example of community-driven FAIR data stewardship is coming from the preclinical spinal cord injury research community. In a series of NIH- and NGO-sponsored workshops and symposia at international neuroscience conferences over the last 3 years, leaders in basic and clinical neurotrauma research, along with bioinformaticians and industry representatives, have developed a data sharing community. As a result of those meetings, the Open Data Commons for Spinal Cord Injury (ODC-SCI) is being developed with multiple non-profit and federal stakeholders, and though still nascent, it is growing in size and breadth of datasets, demonstrating grass-roots community engagement [6].
Within the TBI research community, data sharing has been driven at the federal level in the form of the Federal Interagency TBI Research (FITBIR) informatics system and repository, a joint venture of the US Department of Defense and NIH. FITBIR contains clinical TBI research data with data deposition mandated as a condition of funding of clinical studies and data collection guided by the expert-consensus-driven NINDS TBI common data elements (CDEs) [13,14]. This effort has now been extended to preclinical TBI research, providing common data collection standards that may enable easier data sharing in the future [15,16].
The development of ODC-SCI, FITBIR and other data sharing communities allows researchers to not only share published data, but also unpublished data (‘dark-data’) which may include pilot studies, parametric or metadata from published studies, or results from failed studies [5]. Bringing dark data to light is an important concept for the future of data sharing and knowledge discovery. Given that most published findings are highly distilled and compressed into a narrative in order to present the most impressive of positive results, we are left with a ‘long tail of dark data’ that, while not deemed essential for a high-impact publication, represent the vast silent majority of data collected (Fig.1). By sharing this siloed data across the scientific community, we reduce experimental redundancy and waste, as well as create exciting new opportunities for data-driven discovery through multidimensional analysis of pooled data resources. In the following section we will highlight some of the challenges and recent successes in this approach.
Figure 1.
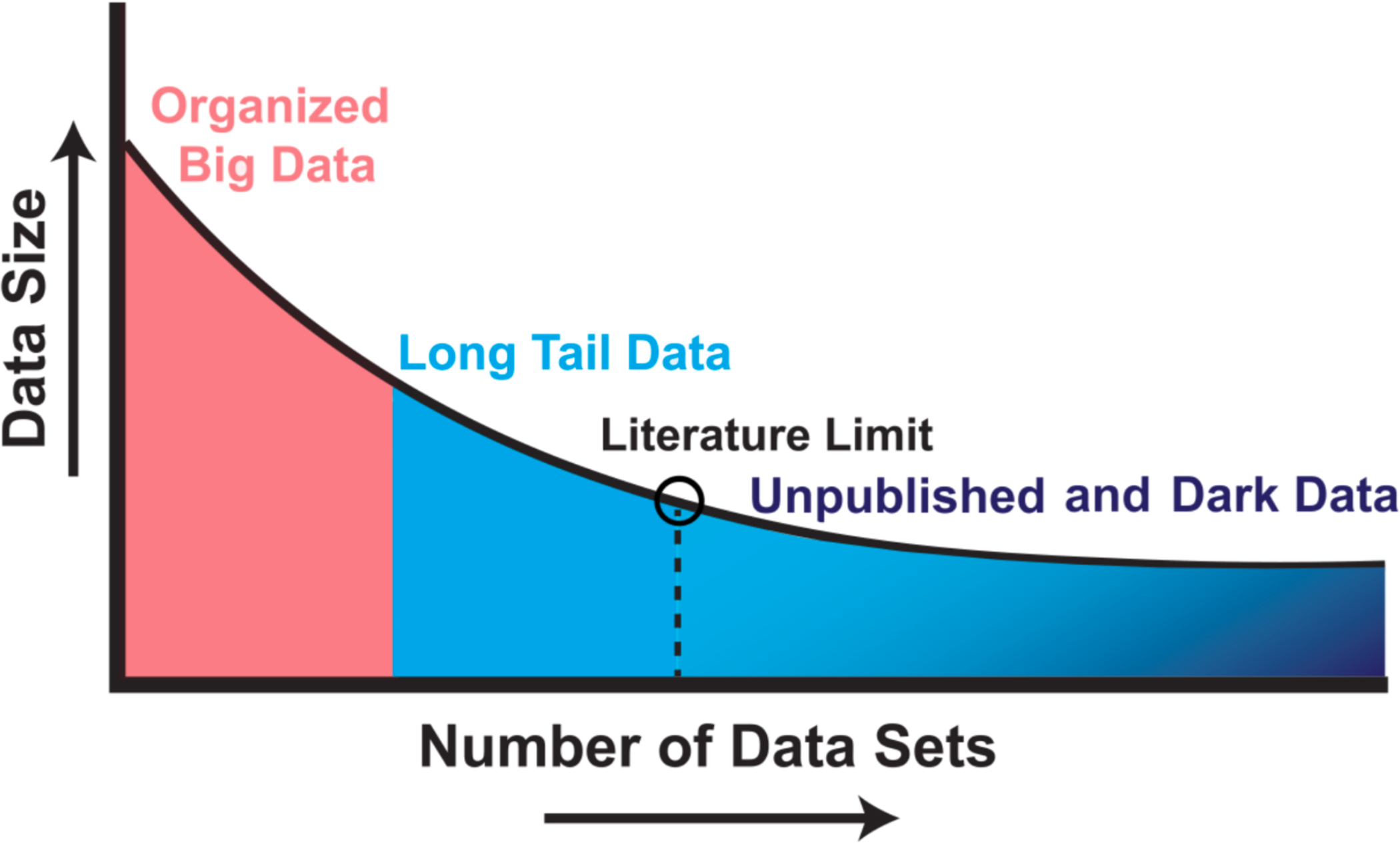
Illustration of long-tail and ‘dark’ data. Studies that have plotted data set size against the number of data sources reliably uncover a skewed distribution. Well-organized big data efforts featuring homogenous, well-organized data represent only a small proportion of the total data collected by scientists. A very large proportion of scientific data falls in the long-tail of the distribution, with numerous small independent research efforts yielding a rich variety of specialty research data sets. The extreme right portion of the long tail includes data that are unpublished; such as siloed databases, null findings, laboratory notes, animal care records, etc. These dark data hold a potential wealth of knowledge but are often inaccessible to the outside world. Modified from Ferguson et al., 2014.
Refining the TBI Common Data Elements
Given the wide variety of TBI data that is now being collected, an important issue for handling this data is ensuring that there are consistent measurement standards across studies. In response to the lack of success in a number of previous TBI clinical trials, the International Mission for Prognosis and Analysis of Clinical Trials in TBI (IMPACT) aggregated long-tail data from thousands of TBI patients from previous trials [17–19]. Among the successful outcomes from this effort has been the development of draft common data elements, which were built upon, sanctioned and codified by the National Institute of Health as the TBI-CDEs [20,21]. The development and implementation of TBI-CDEs for TBI research has been crucial for the harmonization of datasets from different trials and studies. The TBI-CDEs have undergone revisions over the course of the past 10 years, guided in part by prospective clinical observational studies Transforming Research and Clinical Knowledge for TBI (TRACK-TBI) [22] and the TBI Endpoints Development (TED) Initiative [23]. This led to TBI-CDE v2.0, and currently consists of variables across demographic, neuroimaging, fluid biomarkers, genomics, neurocritical care monitoring, and measures of behavioral and cognitive recovery. In the past year, the NINDS also partnered with the Department of Defense to develop a new set of CDEs specifically for sports-related concussion (SRC), one of the leading causes of TBI in the US [24,25].
While it is clear that data standardization has been necessary to adequately compare results within and between studies, recent work has been aimed at comparing and validating measures in order to further refine the CDEs, and eliminate redundancies that may be present in the over 900 measures included in CDE v2.0. For example, Harburg et al., recently investigated the rating variables for computed tomography (CT) imaging that are included in the CDEs [26]. Given that CT findings are among the most important factors for clinical prognosis and TBI trial inclusion, researchers sought to determine which of the 22 CT CDE characteristics were most reliable. They found the inter-rater reliability of some CT measures (Marshall Grade, Sub- and Extradural Hemmorrhage, Midline Shift) to be high, even when comparing novice raters to experienced neuroradiologists. In contrast, other TBI-CDE measures such as the Fisher Grade, Depressed Fracture, and Intracranial Air were much less reliable. Similarly, Ngwenya et al. investigated the extent to which CDEs for subjective cognitive complaints after mild TBI (mTBI) were in concordance with one another. They compared the acute concussion evaluation (ACE) and the Rivermead Post Concussion Symptoms Questionnaire (RPQ) of the 228 mTBI patients, and found only moderate agreement between these two measures [27]. Although these instruments are meant to measure the same construct, the authors determined the RPQ was a better surrogate for cognitive impairment. These results demonstrate the need to be vigilant when testing the convergent and discriminant validity of the many CDEs that overlap, and that more parametric studies like these are needed in order to continue refining the TBI-CDEs [28].
Shedding Light on Dark-Data through Analytics
The recent multicenter TBI data collection and curation efforts are now presenting an opportunity to thoughtfully apply analytical approaches that will be both sensitive and robust. Given the attention to statistical rigor brought by the current reproducibility crisis in neurotrauma research [29,30], it is crucial that the great wealth of data that comes from these large TBI clinical studies be analyzed and interpreted in ways that yield stable, generalizable findings. A typical neurotrauma research article is likely to include a handful of statistically significant univariate measures among many tests that were run. Despite a presumably large amount of data collected from a number of domains, often only those analyses that best fit the a priori hypothesis are shown, making replication that much more improbable. We have gained valuable insight for how best to analyze these types of multimodal datasets from recent preclinical work. Haefeli et al., analyzed multiple preclinical TBI drug therapy experiments, and succinctly demonstrated the lurking problem inherent in running a multitude of univariate tests [31]. With a total of 202 rat subjects and 30 outcome variables collected per subject, analysis would require 6,000 pair-wise t-tests, or 300 main effects and interactions by ANOVA to completely capture every possible point of significance (Fig.2) [31]. This illustrates that if one had reported any small handful of the significant effects found (as would be expected in a typical scientific report), one would run the high risk of capitalizing on chance and reporting a false-positive finding (statistically known as ‘family-wise Type I error’) [32]. To address this issue, Haefeli et al. used a machine learning approach of non-linear principal component analysis (NL-PCA) to integrate raw source data from multiple outcome measures across multiple studies. This approach provides the opportunity to observe significant relationships between multiple diverse outcome measures that might otherwise be missed by conventional univariate analysis of each outcome analyzed separately. In this case, Haefeli et al. were able to reveal a synergistic effect between a neurotrophic factor and an inflammatory agent, and a further interaction with physical training after TBI.
Figure 2.
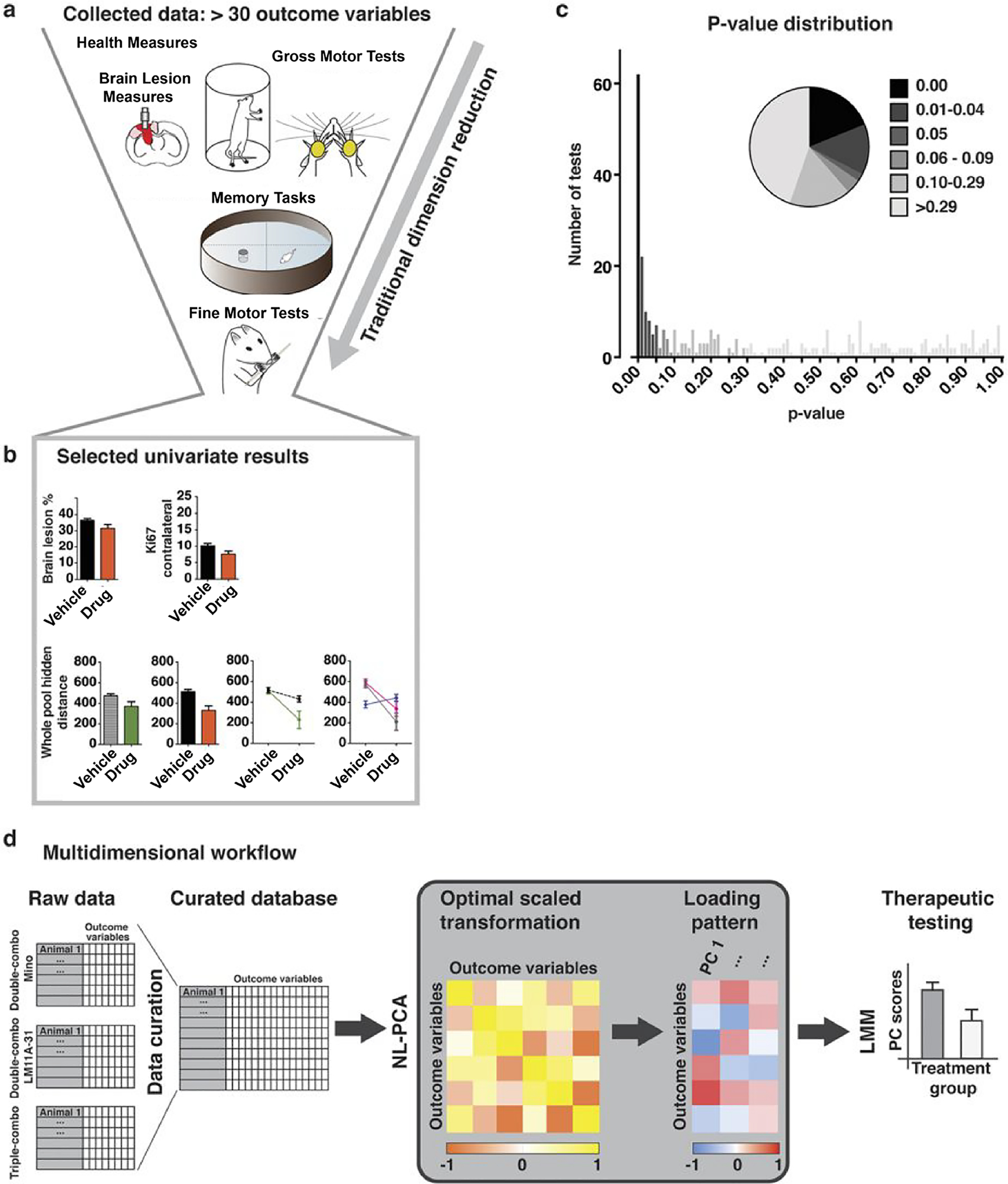
Univariate versus Multidimensional Analytical Workflows. (a) Collected outcomes measures in a preclinical model of TBI. Measures span information about lesion characteristics, motor, cognitive and general health domains. (b) Variables selected in an arbitrary fashion. Bar graphs reflect estimated marginal means of significant main effects and line graphs reflect significant interactions. (c) Frequency distribution and piechart of univariate p-values, illustrating the hundreds univariate analyses that could come this diverse dataset. (d) A model workflow to analyze all outcome measures from a multivariate approach. Outcome variables of all 202 rats are fed into a non-linear principal component analysis (NL-PCA). NL-PCA handles different analysis levels (e.g., ordinal and numeric) in the dataset by optimal-scaling transformations. The NL-PCA loading pattern shows the weight of every outcome variable on the obtained PCs. Individual subject-level PC scores are calculated by summing the optimally-transformed data variable values weighted by loadings. A linear mixed model (LMM) can then be used to run a single hypothesis test on the multidimensional outcome measure (i.e., PC score). Modified from Haefeli et al., 2017.
This multivariate approach is now proving useful in clinical TBI studies as well. TRACK-TBI investigators recently demonstrated the utility of analyzing a large panel of blood-based biomarkers or imaging biomarkers collected early after TBI as a multivariate ensemble rather than testing the significance of each one individually [33–36]. This approach provides a descriptive insight into which candidate markers were most closely cross-correlated, as well as the predictive value of these clusters as an ensemble on diagnostic and prognostic measures. Data-driven approaches are also being used to validate groups of CDEs as multivariate clusters that may better capture the underlying construct better than individual measures. For instance, the Glasgow Outcome Scale- Extended (GOS-E) is the most used outcome CDE in TBI, in part due to its simplicity and ease of use. But there is a tradeoff between simplicity and specificity, as many have noted that the GOS-E may be too crude and non-specific to accurately represent the complex and heterogenous nature of recovery from TBI [37–39]. Nelson et al., tested whether a multidimensional assessment battery may be more useful and informative. By integrating other domains, such as emotional and cognitive tests into a multivariate cluster, they uncovered that a significant number of TBI patients who had reached the ceiling of the GOS-E measure (score of 8, Full Recovery) could be still be classified as impaired by the multivariate measure [28].
Another powerful feature of a multivariate approach is that by starting from a data-driven perspective, new hypotheses can be generated. Nielson et al. recently used a machine learning tool called topological data analysis (TDA) to investigate novel predictors of outcome after mild TBI [40]. Rather than use a typical regression model that tests predictors of a single outcome measure (such as GOS-E), the TDA approach places each patient in the multidimensional ‘syndromic space’ of all outcome measures, and then by mapping this topology to different predictors, one can rapidly determine which predictors most clearly reflect the patient distribution in this space (Fig.3). Using this approach, they observed that specific genetic polymorphisms were predictive of unfavorable outcome after mild TBI. In this way, they were able to discover novel stratification of patients based on underlying genetic predisposition, and provided an avenue for future hypothesis testing of these polymorphisms as potential mild TBI biomarkers.
Figure 3. Integrating Diverse Clinical TBI Data.
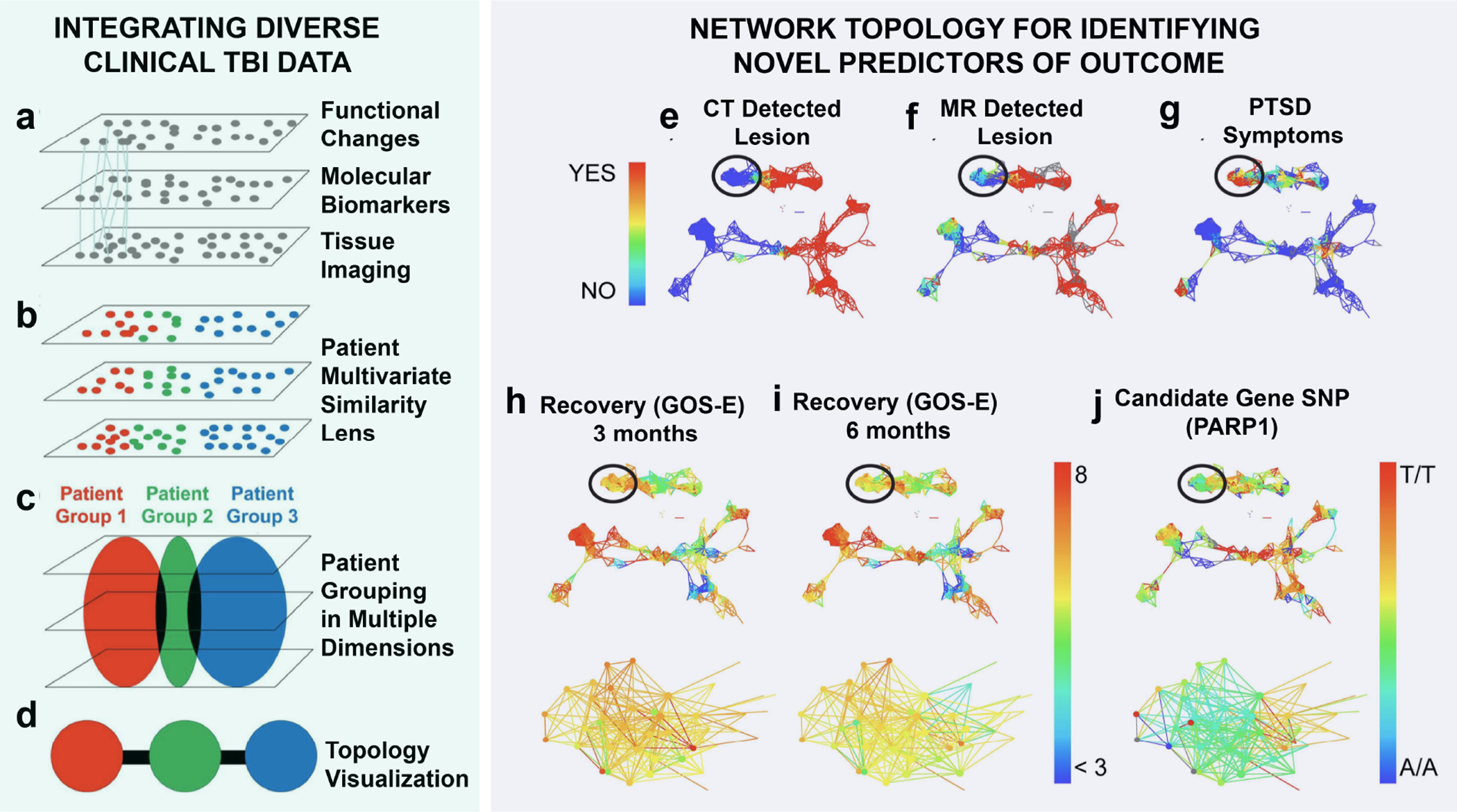
Model of multivariate dimension reduction. (a) Patients are tracked across multiple domains (function, biomarkers, imaging, providing connections (lines) across domains to improve patient classification using the full syndromic space. (b) Multivariate pattern detection lens can be used to categorize (colors) patients across all domains. (c) Patient grouping by multivariate lens. (d) Topological visualization renders patient groups into individual nodes, colored by the multivariate lens. Edges (black lines) indicate individuals appearing in both groups producing a syndromic map of patient clusters. Network Topology For Identifying Novel Predictors of Outcome. Patients with traumatic brain injury were mapped onto a topological data network, highlighting color schemes for CT (e) and MRI (f) brain lesion pathology and whether they had a confirmed diagnosis of PTSD (DSM IV) at 6 months post-TBI (g). Patients in the circled regions of the network were identified due to substantial dysfunction measured by the GOS-E both at 3 months (h) and 6 months (i) post-TBI, compared with other patients in the network with no CT pathology and no diagnosis of PTSD. Data-driven exploration of these patients in the network revealed a significant categorical enrichment for the PARP1 SNP (g), Panels e-g yes (1 = red) vs, no (0 = blue); Panels h-i GOS-E range from less than 3 (blue) to 8 (red); Panel j PARP1 allele A/A = 1 = blue, A/T = 2 = yellow/green, T/T = 3 = red. Modified from Nielson et al., 2017.
Limitations and Analytical Challenges
As the volume, velocity and variety of neurotrauma data continues to increase, so too will the probability that data fields are missing values. This data-feature, known in statistical literature as ‘missingness’, can arise for multiple reasons, from equipment failures to subject-related issues such as loss-to-follow-up. Data-management decisions made (either consciously or unconsciously) to deal with missing data can have a major effect on the power and validity of results from neurotrauma big data. Missing values analysis (MVA) is a sub-discipline within data-science that has its roots in the 1970s [41,42], but the principles of MVA are well-suited to be applied to the heterogenous and often incomplete data collected in our current neurotrauma research. However, MVA has yet to be systematically incorporated into neurotrauma research. One of the most fundamental questions addressed by MVA is whether data is missing at random, or if there is perhaps a systematic, latent factor (e.g. cognitive decline causing loss-to-follow-up) that is responsible for the pattern of missingness. Understanding this issue informs the subsequent decision-making process for how the missing data is handled.
Conclusions
As the field of neurotrauma continues to scale up in data collection efforts over the next decade, taking a big-data framework will ensure that our curation and analytic approaches are designed to embrace the heterogeneity of neurotrauma data, not as an obstacle to overcome, but an advantage that can be leveraged to facilitate precision in knowledge-discovery. Continued refinement and validation of common data elements will be needed to streamline data harmonization across centers. Likewise, analyses that are sensitive to the multidimensionality of neurotrauma data will be necessary to facilitate reproducibility across studies. Current open data initiatives are bringing the disparate knowledge domains together and democratizing research, and we are beginning to see the fruits of these labors. Our efforts must be agile and adaptable as technology advances, and our minds must remain open to ideas that are increasingly driven by data.
Key Points.
The Big Data problem in neurotrauma research is characterized by the wide variety of data collected in preclinical and clinical studies.
Data sharing between research centers and the continued refinement of common data elements are beginning to have a positive impact on research findings.
Acknowledgements
Financial Support and sponsorship. This work was supported by grants from the NIH (NS088475, NS106899 to ARF), the US Department of Veterans Affairs (1I01RX002245-01 to ARF), Wings for Life (WFL-US-006/14 to ARF) and the Craig H. Neilsen Foundation.
Footnotes
Conflicts of interest: None.
References
- 1.Laney D. 3D data management: Controlling data volume, velocity and variety. META Group Research Note 2001; 6. [Google Scholar]
- 2.Ekblom R, Wolf JB. A field guide to whole‐genome sequencing, assembly and annotation. Evolutionary Applications 2014; 7:1026–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Celi LA, Mark RG, Stone DJ, Montgomery RA. “Big Data” in the Intensive Care Unit. Closing the Data Loop American Journal of Respiratory and Critical Care Medicine. American Thoracic Society; 2013; 187:1157–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.DonnellyJoseph AZ, SmielewskiPeter KM, CzosnykaMarek AHJ. A Description of a New Continuous Physiological Index in Traumatic Brain Injury Using the Correlation between Pulse Amplitude of Intracranial Pressure and Cerebral Perfusion Pressure. Journal of Neurotrauma 2018; 35:963–74. [DOI] [PubMed] [Google Scholar]
- 5.Ferguson AR, Nielson JL, Cragin MH, et al. Big data from small data: data-sharing in the ‘long tail’of neuroscience. Nature Neuroscience 2014;17:1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *6.Callahan A, Anderson KD, Beattie MS, et al. Developing a data sharing community for spinal cord injury research. Exp Neurol Academic Press 2017; 295:135–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jones AR, Overly CC, Sunkin SM. The Allen Brain Atlas: 5 years and beyond. Nat Rev Neurosci 2009; 10:821–8. [DOI] [PubMed] [Google Scholar]
- 8.Jorgenson LA, Newsome WT, Anderson DJ, et al. The BRAIN Initiative: developing technology to catalyse neuroscience discovery. Phil Trans R Soc B 2015; 370:20140164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Amunts K, Ebell C, Muller J, et al. The Human Brain Project: Creating a European Research Infrastructure to Decode the Human Brain. Neuron 2016; 92:574–81. [DOI] [PubMed] [Google Scholar]
- 10.Bui AAT, Van Horn JD. Envisioning the future of “big data” biomedicine. Journal of Biomedical Informatics 2017; 69:115–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grethe JS, Bandrowski A, Banks DE, et al. SciCrunch: A cooperative and collaborative data and resource discovery platform for scientific communities. InFront. Neuroinform Conference Abstract: Neuroinformatics 2014. [Google Scholar]
- 12.Wilkinson MD, Dumontier M, Aalbersberg IJ, et al. The FAIR Guiding Principles for scientific data management and stewardship. Scientific Data 2016; 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saatman KE, Duhaime AC, Bullock R, et al. Classification of traumatic brain injury for targeted therapies. Journal of Neurotrauma 2008; 25:719–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Manley GT, Maas AIR. Traumatic brain injury: an international knowledge-based approach. JAMA 2013; 310:473–4. [DOI] [PubMed] [Google Scholar]
- 15.Smith DH, Hicks RR, Johnson VE, et al. Pre-clinical traumatic brain injury common data elements: toward a common language across laboratories. Journal of Neurotrauma 2015; 32:1725–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *16.DeWitt D, Hawkins BE, Dixon CE, et al. Preclinical Testing of Therapies for Traumatic Brain Injury. Journal of Neurotrauma 2018;(ja). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Steyerberg EW, Mushkudiani N, Perel P, et al. Predicting outcome after traumatic brain injury: development and international validation of prognostic scores based on admission characteristics. PLoS Medicine 2008; 5:e165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maas AI, Harrison-Felix CL, Menon D, et al. Common data elements for traumatic brain injury: recommendations from the interagency working group on demographics and clinical assessment. Archives of Physical Medicine and Rehabilitation 2010; 91:1641–9. [DOI] [PubMed] [Google Scholar]
- 19.Maas AI, Murray GD, Roozenbeek B, et al. International Mission on Prognosis Analysis of Clinical Trials in Traumatic Brain Injury (IMPACT) Study Group. Advancing care for traumatic brain injury: findings from the IMPACT studies and perspectives on future research. The Lancet Neurology 2013; 12:1200–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saatman KE, Duhaime AC, Bullock R, et al. Classification of traumatic brain injury for targeted therapies. Journal of Neurotrauma 2008; 25:719–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Whyte J, Vasterling J, Manley GT. Common data elements for research on traumatic brain injury and psychological health: current status and future development. Archives of Physical Medicine and Rehabilitation 2010; 91:1692–6. [DOI] [PubMed] [Google Scholar]
- 22.Yue JK, Vassar MJ, Lingsma HF, et al. Transforming research and clinical knowledge in traumatic brain injury pilot: multicenter implementation of the common data elements for traumatic brain injury. Journal of Neurotrauma 2013; 30:1831–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **23.Manley GT, Mac Donald CL, Markowitz AJ, et al. The Traumatic Brain Injury Endpoints Development (TED) initiative: progress on a public-private regulatory collaboration to accelerate diagnosis and treatment of traumatic brain injury. Journal of Neurotrauma 2017; 34:2721–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Joseph K, Alai S, Esterlitz J, et al. Streamlining Clinical Research in Sports-Related Concussion: The National Institute of Neurological Disorders and Stroke (NINDS)/National Institutes of Health (NIH), and Department of Defense (DOD) Sport-Related Concussion Common Data Elements (CDEs)(P2. 014). Neurology 2018; 90:P2.014. [Google Scholar]
- 25.Broglio SP, Kontos AP, Levin H, et al. The National Institute of Neurological Disorders and Stroke and Department of Defense Sport-Related Concussion Common Data Elements Version 1.0 Recommendations. Journal of Neurotrauma 2018;(ja). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harburg McCormick, Kenney, et al. Reliability of the NINDS common data elements cranial tomography (CT) rating variables for traumatic brain injury (TBI). Brain InjuryTaylor & Francis; 2017. January 5;31(2):174–84. [DOI] [PubMed] [Google Scholar]
- *27.Ngwenya LB, Gardner RC, Yue JK, et al. Concordance of common data elements for assessment of subjective cognitive complaints after mild-traumatic brain injury: a TRACK-TBI Pilot Study. Brain Injury 2018:1–8. [DOI] [PubMed] [Google Scholar]
- *28.Nelson LD, Ranson J, Ferguson AR, et al. Validating Multi-Dimensional Outcome Assessment Using the Traumatic Brain Injury Common Data Elements: An Analysis of the TRACK-TBI Pilot Study Sample. Journal of Neurotrauma 2017; 34:3158–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Steward O, Balice-Gordon R. Rigor or mortis: best practices for preclinical research in neuroscience. Neuron 2014; 84:572–81. [DOI] [PubMed] [Google Scholar]
- 30.Steward O. A Rhumba of “R’s”: Replication, Reproducibility, Rigor, Robustness: What Does a Failure to Replicate Mean?. eNeuro 2016; 3:ENEURO-0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **31.Haefeli J, Ferguson AR, Bingham D, et al. A data-driven approach for evaluating multi-modal therapy in traumatic brain injury. Scientific Reports 2017; 7:42474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krzywinski M, Altman N. Points of significance: Comparing samples—part II. Nature Methods 2014; 11:355–6. [DOI] [PubMed] [Google Scholar]
- **33.Huie JR, Diaz-Arrastia R, Yue J, et al. Validation of multivariate proteomic panel for TBI biomarker discovery: A TRACK-TBI Pilot study. Journal of Neurotrauma 2018. (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- *34.Korley FK, Yue JK, Wilson D, et al. Performance Evaluation of a Multiplex Assay for Simultaneous Detection of Four Clinically Relevant TBI Biomarkers. Journal of Neurotrauma 2018:(ja). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yuh EL, Mukherjee P, Lingsma HF, et al. Magnetic resonance imaging improves 3‐month outcome prediction in mild traumatic brain injury. Annals of Neurology 2013; 73:224–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yuh EL, Cooper SR, Ferguson AR, et al. Quantitative CT improves outcome prediction in acute traumatic brain injury. Journal of Neurotrauma 2012; 29:735–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lingsma HF, Roozenbeek B, Steyerberg EW, et al. Early prognosis in traumatic brain injury: from prophecies to predictions. The Lancet Neurology 2010; 9:543–54. [DOI] [PubMed] [Google Scholar]
- 38.Beers SR, Wisniewski SR, Garcia-Filion P, et al. Validity of a pediatric version of the Glasgow Outcome Scale–Extended. Journal of Neurotrauma 2012; 29:1126–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *39.Maas AI, Menon DK, Adelson PD, et al. Traumatic brain injury: integrated approaches to improve prevention, clinical care, and research. The Lancet Neurology 2017; 16:987–1048. [DOI] [PubMed] [Google Scholar]
- **40.Nielson JL, Cooper SR, Yue JK, et al. Uncovering precision phenotype-biomarker associations in traumatic brain injury using topological data analysis. PloS one 2017; 12:e0169490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rubin DB. Inference and missing data. Biometrika 1976; 63:81–92. [Google Scholar]
- 42.Little RJ & Rubin DB. The analysis of social science data with missing values. Sociological Methods & Research, 1989; 18, 292–326. [Google Scholar]


