Abstract
Background
VP4 [P] genotype binding specificities of rotaviruses and differential expression of histo-blood group antigens (HBGAs) between populations may contribute to reduced efficacy against severe rotavirus disease. P[6]-based rotavirus vaccines could broaden protection in such settings, particularly in Africa, where the Lewis-negative phenotype and P[6] rotavirus strains are common.
Methods
The association between HBGA status and G3P[6] rotavirus vaccine (RV3-BB) take was investigated in a phase 2A study of RV3-BB vaccine involving 46 individuals in Dunedin, New Zealand, during 2012–2014. FUT2 and FUT3 genotypes were determined from DNA extracted from stool specimens, and frequencies of positive cumulative vaccine take, defined as an RV3-BB serum immune response (either immunoglobulin A or serum neutralizing antibody) and/or stool excretion of the vaccine strain, stratified by HBGA status were determined.
Results
RV3-BB produced positive cumulative vaccine take in 29 of 32 individuals (91%) who expressed a functional FUT2 enzyme (the secretor group), 13 of 13 (100%) who were FUT2 null (the nonsecretor group), and 1 of 1 with reduced FUT2 activity (i.e., a weak secretor); in 37 of 40 individuals (93%) who expressed a functional FUT3 enzyme (the Lewis-positive group) and 3 of 3 who were FUT3 null (the Lewis-negative group); and in 25 of 28 Lewis-positive secretors (89%), 12 of 12 Lewis-positive nonsecretors (100%), 2 of 2 Lewis-negative secretors, and 1 of 1 Lewis-negative weak secretor.
Conclusions
RV3-BB produced positive cumulative vaccine take irrespective of HBGA status. RV3-BB has the potential to provide an improved level of protection in settings where P[6] rotavirus disease is endemic, irrespective of the HBGA profile of the population.
Keywords: Rotavirus, RV3-BB, histo-blood group antigens, neonatal vaccination, vaccine take, Lewis antibodies, secretor status
Rotavirus vaccines have made a major impact on rotavirus hospitalizations and death due to rotavirus gastroenteritis. More than 98 countries have now introduced rotavirus vaccines nationally or subnationally within their national immunization programs. Despite this success, both Rotarix and RotaTeq vaccines have reduced efficacy against severe rotavirus disease in low-income countries (49.2%–72.2% efficacy for Rotarix and 64.2% efficacy for RotaTeq), compared with high-income countries (>88% efficacy for each vaccine) [1–4]. The genotypic diversity of rotavirus is greater in Africa, compared with other continents. In sub-Saharan Africa, there is an increased proportion of P[6] rotavirus strains causing severe disease; P[6] strains accounted for 22.6% of rotaviruses causing disease between 2006 and 2016, whereas they have been detected only sporadically in higher-income countries globally [5–7]. Population differences in histo-blood group antigen (HBGA) status could contribute to these differences in genotypes circulating within a population and the level of protection provided by vaccines based on P[8] strains.
HBGAs are neutrally charged carbohydrates that are expressed in humans on red blood cells; on the mucosal epithelia of the digestive, respiratory, and genitourinary tracts; and, in soluble form, in secretions such as saliva and breast milk. Enteric pathogens frequently use HBGAs as the first step in the cell attachment and entry process and, consequently, can be key determinants of a pathogen’s host range and tissue tropism [8–11]. For rotavirus, infectivity is mediated by trypsin cleavage of the VP4 outer capsid protein to produce the VP8* and VP5* subunits; initial attachment to host cells is mediated by VP8*, whereas VP5* is required for cellular entry [12, 13]. The binding of the VP8* of some human rotavirus strains to HBGAs has been demonstrated, strongly suggesting that HBGAs are important host factors or cellular receptors [14–18].
The biosynthesis of type 1 HBGAs occurs by the stepwise addition of monosaccharide units to precursor disaccharide molecules. This process is catalyzed by the enzymes fucosyltransferase 2 (FUT2) and fucosyltransferase 3 (FUT3), encoded by FUT2 and FUT3, respectively. Both genes have dominant alleles (Se for FUT2 and Le for FUT3) that encode functional enzymes, and recessive alleles (se and le, respectively), caused by specific single-nucleotide polymorphisms (SNPs), that do not code functional enzymes. Individuals who express a functional FUT2 enzyme are referred to as “secretors” and have the genotype Se/Se or Se/se, whereas those who are FUT2 null are known as “nonsecretors” and have the genotype se/se. Similarly, individuals who express a functional FUT3 enzyme are categorized as “Lewis positive” and have the genotype Le/Le or Le/le, whereas FUT3 null individuals are classified as “Lewis negative” and have the genotype le/le [19].
The HBGA phenotype of an individual represents the different combinations of the presence or absence of functional FUT2 and FUT3 enzymes and can be determined by assaying for the resulting antigens that are detectable in secretions. Lewis-positive secretors have functional FUT2 and FUT3 and produce Leb antigen, Lewis-positive nonsecretors have functional FUT3 but not FUT2 and produce Lea antigen, and Lewis-negative secretors have functional FUT2 but not FUT3 and produce H type 1 antigen; in Lewis-negative nonsecretors, neither FUT2 nor FUT3 is functional, and thus Leb, Lea, and H type 1 antigens are not produced. The prevalence of the HBGA phenotypes varies between populations; approximately 75% of Europeans, 50%–60% of Africans, and 42% of Asians are Lewis-positive secretors, whereas only 20% of Europeans are Lewis-positive nonsecretors. The Lewis-negative phenotype is less common in Europeans and Asians (8% and 7%, respectively), whereas it was detected at a higher rate (32%) in Burkina Faso in West Africa [20–22]. The phenotype for individuals categorized as “weak secretors,” in which the enzyme activity of FUT2 is decreased because of a specific missense mutation at nucleotide position 385 (A > T), occurs in 10%–20% of Southeast and East Asian populations [23, 24].
The Lewis and secretor status of an individual may mediate susceptibility to rotavirus infection, including vaccination with a live viral vaccine, as the binding specificity of rotavirus to HBGAs may be VP4 [P] genotype dependent. For P[8] rotavirus strains, including the P[8]-based Rotarix vaccine, secretors have been observed to be more susceptible to infection and vaccine take than nonsecretors. vaccine take than nonsecretors [25–33]. The role of Lewis status is less clear, but the Lewis-negative phenotype was more common in infants who developed P[6] rotavirus gastroenteritis following a full 2-dose course of Rotarix, compared with community controls (odds ratio, 3.2; 95% confidence interval, 1.4–7.2) [28]. For P[6] rotaviruses, limited epidemiological studies and in vitro binding assays have demonstrated differential HBGA receptor specificity when compared to P[8] and P[4] strains [34]. The VP4 [P] genotype–dependent binding specificity of rotaviruses and the differential expression of HBGAs between populations could contribute to the reduced efficacy against severe rotavirus disease for Rotarix and RotaTeq observed in low-income settings with a high burden of rotavirus disease. It is plausible that a P[6]-based rotavirus vaccine could play an important role in broadening protection in Africa, where the Lewis-negative phenotype is more prevalent and where P[6] rotavirus strains are endemic.
The RV3-BB human neonatal rotavirus vaccine is based on an isolate of a G3P[6] human neonatal rotavirus strain that circulated among healthy newborns in obstetric hospitals in Melbourne, Australia [35]. A phase 2A double-blinded, randomized, placebo-controlled, single-center, 3-arm parallel group study of oral RV3-BB rotavirus vaccine was conducted at a single center in Dunedin, New Zealand, between 13 January 2012 and 17 April 2014, which has been previously described (Australian New Zealand Clinical Trials Registry identifier ACTRN12611001212943) [36]. Vaccine take was demonstrated in >90% of participants in this trial after administration of 3 doses of RV3-BB vaccine, when the first dose was administered 0–5 days after birth (neonatal schedule) or at approximately 8 weeks of age (infant schedule). The aim of the current study was to determine whether Lewis and secretor status influenced vaccine take after vaccination with the G3P[6] human neonatal rotavirus vaccine RV3-BB.
METHODS
Subjects and Samples
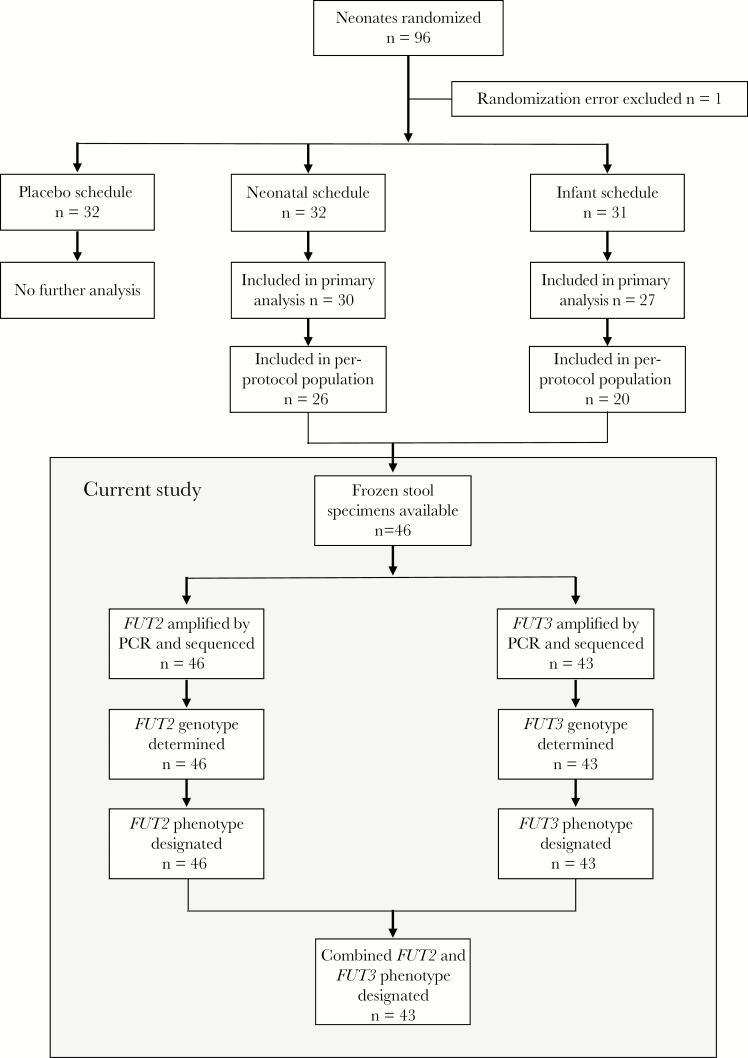
This study was performed on frozen stool samples collected from all participants in the per protocol population in the phase 2A trial in New Zealand for whom written informed consent was provided by the parent(s)/guardian for future evaluation of studies of rotavirus (n = 46; Figure 1). The protocol was approved by the Lower South Region Ethics Committee, New Zealand; the Human Research Ethics Committee, Royal Children’s Hospital, Australia; and the New Zealand Medicines and Medical Devices Safety Authority.
Figure 1.
Flow of participants through the study selection process and summary of stool specimen analyses. PCR, polymerase chain reaction. FUT2, gene encoding fucosyltransferase 2; FUT3, gene encoding fucosyltransferase 3.
DNA Extraction From Frozen Stool Specimens
DNA was extracted from a 200-mg stool specimen from each participant, using the NucleoSpin DNA Stool kit (Macherey-Nagel, Düren, Germany) according to the manufacturer’s instructions, with the following modifications. For the disruption and homogenization step, samples were agitated for 30 seconds and rested for 1 minute, for 4 rounds, at room temperature, using the Mini-Beadbeater (Biospec Products, Bartlesville, OK). An intermediate volume of elution buffer (100 µL) was used to elute DNA, to dilute residual polymerase chain reaction (PCR) inhibitors such as bile salts and complex polysaccharides that are inherent to stool. Eluted DNA was stored at −30°C.
FUT2 and FUT3 Amplification
A 1184-bp region and a 1491-bp region spanning the enzyme-coding regions of FUT2 and FUT3, respectively, were amplified by PCR. The FUT2 primers were 5′-CTAACGTGTCCCGTTTTCCTC-3′ (forward) and 5′-CCCAACGCATCTTCACAGA-3′ (reverse). The FUT3 primers were 5′-GGAGCTTTGGTAAGCAGGAG-3′ (forward) and 5′-TCAGTGTGGCAAGGTCTCTG-3′ (reverse). The forward primer sequences have been described previously [33, 37], and reverse primers were designed for this project. Lyophilized high-performance liquid chromatography–purified primers (Merck, Darmstadt, Germany) were resuspended to 10-µM working stocks, using UltraPure DNase/RNase-free distilled water (catalog no. 10977-015; Invitrogen).
PCR Amplification and Complementary DNA Purification
PCR amplification was performed for each amplicon, using the PrimeSTAR GXL DNA polymerase kit (Takara, Kusatsu, Japan), according to manufacturer’s instructions. Briefly, 50 µL PCR reactions were prepared, with each containing nuclease-free water, 1× PrimeSTAR GXL buffer with 5 mM MgCl2, 200 µM dNTPs, 0.2 µM each of the forward and reverse primers, 1.25 U of PrimeSTAR GXL DNA polymerase, and 5 µL of genomic DNA. PCR was performed as described previously [33]: 1 cycle at 94°C for 2 minutes; 25 cycles at 94°C for 30 seconds, at 65°C for 30 seconds, and at 72°C for 90 seconds; and 20 cycles at 94°C for 30 seconds, at 55°C for 30 seconds, and at 72°C for 90 seconds). Amplicons were purified using the QIAquick Gel Extraction Kit (Qiagen, Hilden, Germany).
FUT2 and FUT3 Genotyping
The nucleotide sequence of purified FUT2 and FUT3 amplicons was determined by next-generation sequencing (NGS; Centre for Genomics Medicine—Sequencing Service and Development Platform, Victorian Clinical Genetic Services/Murdoch Children’s Research Institute). In brief, complementary DNA was amplified and purified with AMpure magnetic beads (Beckman Coulter), and libraries for NGS were then prepared using the Nextera XT library prep kit (Illumina). Libraries were pooled in equimolar ratios and sequenced on the MiSeq system (Illumina), using 2 × 150-bp sequencing.
Before alignment, paired-end fastq files were initially screened for potential quality problems with FastQC, version 11.8, and the human reference (genome) assembly GRCh38 was indexed with SAMtools, version 1.5. Alignment of reads to the reference genome was performed using the Burrows-Wheeler Alignment tool, version 7.15, with the mem algorithm. Aligned reads were then prepared for SNP calling by converting SAM files to BAM files and were sorted with SAMtools. SNPs were then called with the SAMtools mpileup function and the BCFtools, version 1.5, call function. SNPs were then filtered and annotated with BCFtools.
Inferring Lewis and Secretor Status
The Lewis and secretor phenotype for each participant was designated by analysis of the SNPs identified in the coding regions of each gene. For FUT2, participants who were homozygous mutants for the G428A (rs601338) nonsense and G739A (rs602662) missense SNPs were designated as nonsecretors, and those who were homozygous mutants at the A385T (rs1047781) missense SNP were designated as weak secretors. Conversely, those who were wild type or heterozygous mutants for these SNPs were designated as secretors. For FUT3, participants who were homozygous mutants for at least one of the T202C (rs812936), G508A (rs3745635) or T1067A (rs3894326) missense SNPs were designated as Lewis negative. Conversely, those that were wild-type or heterozygous mutants for these SNPs were designated as Lewis positive. Participants who were homozygous mutants for the T59G (rs28362459) reducing or the C314T (rs778986) missense SNPs were designated Lewis negative only if these mutations were seen in addition to the T202C, G508A or T1067A missense SNPs.
Statistical Analyses
Frequencies of positive cumulative vaccine take by Lewis, secretor and combined Lewis and secretor status were expressed as proportions (and percentages). Cumulative vaccine take was defined as a serum immune response (ie, a ≥3-fold increase in titer from baseline) of anti-rotavirus immunoglobulin A (IgA) or serum neutralizing antibodies 28 days following dose administration or as detection of RV3-BB virus excretion by reverse transcription PCR analysis of stool specimens at least once during days 3–7 following dose administration, as previously described [36]. χ 2 analysis and relative risks (RRs) were used to compare frequencies of vaccine take between variables, using Stata, version 15.1 (StataCorp, College Station, TX). Differences were considered statistically significant at a P value of < .05.
RESULTS
The demographic characteristics of the study subset were similar to those previously described for the intention-to-treat cohort: age at first dose of investigational product, sex, race, gestational age, and birth weight (Supplementary Table 1) [36]. DNA was extracted from stool specimens from all 46 participants, and FUT2 and FUT3 were amplified by PCR for 46 of 46 and 43 of 46, respectively. Amplicons for both genes were obtained for 43 of 46 participants. The nucleotide sequence was determined by NGS, and SNPs were called on the basis of alignment with a human reference genome.
FUT2 and FUT3 Genotypes and Inferring Phenotypes
For FUT2, 13 participants (28%) were homozygous mutants for both the nonsense variant G428A (rs601338) and the missense variant G739A (rs602662) and were designated as nonsecretors (Table 1). One participant (2%) was a homozygous mutant for the missense variant A385T (rs1047781) and was designated as a weak secretor. The remaining 32 participants (70%) were either wild type or heterozygous mutants for these SNPs and were designated as secretors. Supplementary Table 2 shows the distribution of FUT2 SNPs and the allele frequencies detected in the cohort, and Supplementary Table 3 shows FUT2 genotypes and phenotype designations for each participant.
Table 1.
Distribution of Lewis and Secretor Phenotypes
| Phenotype | Participants, No. (%) |
|---|---|
| Secretor phenotype (n = 46) | |
| Secretor | 32 (70) |
| Nonsecretor | 13 (28) |
| Weak secretor | 1 (2) |
| Not determined | 0 |
| Lewis phenotype (n = 46) | |
| Positive | 40 (86) |
| Negative | 3 (7) |
| Not determined | 3 (7) |
| Combined phenotype (n = 46) | |
| Le, Se (Lewis positive, secretor) | 28 (61) |
| Le, se (Lewis positive, nonsecretor) | 12 (26) |
| le, Se (Lewis negative, secretor) | 2 (4) |
| le, sew (Lewis negative, weak secretor) | 1 (2) |
| le, se (Lewis negative, nonsecretor) | 0 |
| Not determined | 3 (7) |
Phenotypes were inferred from genotype type data. See “Methods” for descriptions of secretor and Lewis phenotypes.
Abbreviations: le, nonfunctional FUT3; Le, at least 1 functional gene encoding FUT3; RR, relative risk; se, nonfunctional FUT2; Se, at least 1 functional gene encoding FUT2.
For FUT3, 3 participants (7%) were designated as Lewis negative (Table 1). Of these, 1 was a homozygous mutant for the T202C (rs812936) and C314T (rs778986) missense variants, 1 was a homozygous mutant for the T59G (rs28362459) reducing and G508A (rs3745635) missense variants, and 1 was homozygous mutant for the T59G (rs28362459) reducing and T1067A (rs3894326) missense variants. Forty participants (86%) were either wild type or heterozygous mutants for these SNPs and were designated as Lewis positive. Lewis phenotype was not designated for 3 participants, owing to failure of FUT3 amplification by PCR. Supplementary Table 4 shows the distribution of FUT3 SNPs and the allele frequencies detected in the cohort, and Supplementary Table 5 shows FUT3 genotypes and phenotype designations for each participant.
Overall, 28 participants (61%) were designated as Lewis-positive secretors, 12 (26%) as Lewis-positive nonsecretors, 2 (4%) as Lewis-negative secretors, and 1 (2%) as a Lewis-negative weak secretor (Table 1). There were no Lewis-negative nonsecretors in the cohort.
Vaccine Take, by Lewis and Secretor Status
RV3-BB produced positive cumulative vaccine take, irrespective of the secretor, Lewis, and combined Lewis and secretor status of participants in the cohort (Table 2). Vaccine take was detected in 29 of 32 secretors (91%), 13 of 13 nonsecretors (100%), and 1 of 1 weak secretor (100%); 37 of 40 Lewis-positive participants (93%) and 3 of 3 Lewis-negative participants (100%); and 25 of 28 Lewis-positive secretors (89%), 12 of 12 Lewis-positive nonsecretors (100%), 2 of 2 Lewis-negative secretors (100%), and 1 of 1 Lewis-negative weak secretor (100%). There were no Lewis-negative nonsecretors in the cohort, so vaccine take could not be assessed for this phenotype.
Table 2.
Proportion (%) Positive Vaccine Take by Lewis and Secretor Status
| Positive Vaccine Take | Positive for Serum IgA and/or SNA Response | Positive for Serum IgA Response | Positive for SNA Response | Positive for RV3-BB Virus Excretion | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Phenotypea | Participants, Proportion (%) | RR (95% CI) | P | Participants, Proportion (%) | RR (95% CI) | P | Participants, Proportion (%) | RR (95% CI) | P | Participants, Proportion (%) | RR (95% CI) | P | Participants, Proportion (%) | RR (95% CI) | P |
| Secretor phenotype (n = 46) | |||||||||||||||
| Secretor | 29/32 (91) | Reference | 21/32 (66) | Reference | 21/32 (66) | Reference | 4/31 (13) | Reference | 23/32 (72) | Reference | |||||
| Nonsecretor | 13/13 (100) | 0.91 (.81–1.01) | .25 | 11/13 (85) | 0.76 (.55–1.09) | .20 | 11/13 (85) | 0.78 (.55–1.09) | .20 | 3/11 (27) | 0.47 (.13–1.79) | .27 | 10/13 (77) | 0.93 (.65–1.35) | .73 |
| Weak secretor | 1/1 (100) | … | 1/1 (100) | … | 1/1 (100) | … | 0/1 (0) | … | 1/1 (100) | … | |||||
| Lewis phenotype (n = 43) | |||||||||||||||
| Positive | 37/40 (93) | Reference | 29/40 (73) | … | 29/40 (73) | … | 7/38 (18) | … | 28/40 (70) | … | |||||
| Negative | 3/3 (100) | 0.93 (.85–1.01) | .62 | 1/3 (33) | … | 1/3 (33) | … | 0/3 (0) | … | 3/3 (100) | … | ||||
| Combined phenotype (n = 43) | |||||||||||||||
| Le, Se (Lewis positive, secretor) | 25/28 (89) | Reference | 19/28 (68) | Reference | 19/28 (68) | Reference | 4/27 (15)j | Reference | 19/28 (68) | Reference | |||||
| Le, se (Lewis positive, nonsecretor) | 12/12 (100) | 0.89 (.79–1.01) | .24 | 10/12 (83) | 0.81 (.57–1.17) | .32 | 10/12 (83) | 0.81 (.57–1.17) | .32 | 3/11 (27)j | 0.54 (.15–2.04) | .37 | 9/12 (75) | 0.91 (.60–1.37) | .65 |
| le, Se (Lewis negative, secretor) | 2/2 (100) | … | 0/2 (0) | … | 0/2 (0) | … | 0/2 (0) | … | 2/2 (100) | … | |||||
| le, sew (Lewis negative, weak secretor) | 1/1 (100) | … | 1/1 (100) | … | 1/1 (100) | … | 0/1 (0) | … | 1/1 (100) | … | |||||
| le, se (Lewis negative, nonsecretor) | 0 | … | 0 | … | 0 | … | 0 | … | 0 | … | |||||
Phenotypes were inferred from genotype type data. See Methods for descriptions of secretor and Lewis phenotypes.
Abbreviations: CI, confidence interval; IgA, immunoglobulin A; le, nonfunctional FUT3; Le, at least 1 functional gene encoding FUT3; RR, relative risk; se, nonfunctional FUT2; Se, at least 1 functional gene encoding FUT2; SNA, serum neutralizing antibody.
When vaccine take was broken down into its components of serum response and RV3-BB virus excretion, no difference was observed by secretor, Lewis, or combined Lewis and secretor status. Among secretors and nonsecretors, a serum response was detected in 21 of 32 (66%) and 11 of 13 (85%), respectively, and excretion was observed in 23 of 32 (72%) and 10 of 13 (77%), respectively. With respect to Lewis status, a serum response was detected in 29 of 40 Lewis-positive participants (73%) and 1 of 3 Lewis-negative participants (33%), whereas excretion was observed in 28 of 40 (70%) and 3 of 3 (100%), respectively. Stratification by both phenotypes combined revealed that a serum response was present in 19 of 28 Lewis-positive secretors (68%) and 10 of 12 Lewis-positive nonsecretors (83%), and excretion was found in 19 of 28 (68%) and 9 of 12 (75%), respectively. Both Lewis-negative secretors were positive for excretion but not a serum response, and the Lewis-negative weak secretor was positive for both a serum response and excretion. No difference in HBGA status was observed when serum response was separated by serum IgA and Serum Neutralising Antibody (SNA) responses. χ 2 analyses and RRs calculated to compare frequencies of vaccine take between HBGA groups showed no significant differences (P > .05 for all comparisons).
DISCUSSION
This study demonstrated that the G3P[6] human neonatal vaccine RV3-BB produced positive cumulative vaccine take, irrespective of HBGA status. We observed no difference in positive vaccine take by secretor status, by Lewis status, or by combined Lewis and secretor status. The sample size was small, and there were only 3 Lewis-negative individuals in the study cohort, warranting cautious interpretation of these results. This is the first study to assess whether HBGA status influences take of a rotavirus vaccine based on a P[6] strain.
Importantly, Lewis positivity was not a restriction factor for the RV3-BB vaccine, with 37 of 40 Lewis-positive individuals (92.5%) and 3 of 3 Lewis-negative individuals (100%) positive vaccine take. This contrasts with observations from studies with disease-causing wild-type P[6] strains and may be a result of the intrinsic functional and structural characteristics of asymptomatic neonatal P[6] strains: RV3 has been shown to have a unique neonatal P[6] VP8*, which may be adapted to the neonatal gut to cause infection independent of HBGA status [38, 39]. In Burkina Faso, P[6] strains were observed to preferentially (but not exclusively) infect Lewis-negative children (of 27 infected children, 18 were Lewis negative, compared with 9 who were Lewis positive; odds ratio, 5.5; P < .0001), irrespective of secretor status [33]. Consistent with our study, secretor status has not been consistently associated with susceptibility to P[6] rotavirus infection (odds ratio, 0.4; 95% confidence interval, 0–4.1) [40]. In one study, in Swedish children, the geometric mean SNA titers to the G4P[6] ST3 strain were similar in secretors and nonsecretors [41].
Studies investigating P[8] rotavirus vaccine take and HBGA status have produced varying results, which seem to be population dependent. Epidemiological studies in the United States and France have shown nonsecretor status to be a restriction factor for P[8] rotavirus infection in infants [29, 30]. In Pakistan and Ghana, the higher rates of seroconversion to Rotarix was observed in secretors, with no difference in seroconversion based on Lewis status [25, 26]. In Nicaragua, the Lea phenotype (present in Lewis-positive nonsecretors) was found to be a restriction factor to seroconversion in children after 1 dose of either Rotarix or RotaTeq [27]. In in vitro binding assays, the expressed and purified VP8* of P[8] rotaviruses bound synthetically expressed H-type 1 antigen (present in Lewis-negative secretors) and Leb antigen (present in Lewis-positive secretors) but not Lea antigen (present in Lewis-positive nonsecretors) [34]. In contrast, no difference in Rotarix take was observed by secretor or Lewis status in a Malawian cohort, and in Bangladesh, secretors and nonsecretors in the vaccine group of a Rotarix efficacy trial were protected similarly [28, 32]. In Tunisian infants, P[8] rotaviruses were able to infect secretors and nonsecretors, although numbers of nonsecretors in the study were low [31].
A strength of this study was that genotyping analysis was conducted using NGS, where the entire coding regions of interest for FUT2 and FUT3 were examined for known SNPs. Compared with methods such as PCR/restriction fragment–length polymorphism analysis, in which a limited number of specific SNPs are targeted, the “unbiased” approach used in this study gave the best possible chance of accurately designating the genotype of the individual, even without phenotypic confirmation. In this study, confirmation of Lewis and secretor genotypes by phenotypic analysis of saliva or red blood cells was not possible, as these samples were not collected as part of the vaccine trial. However, the genotypic analysis of samples collected from neonates and infants is more reliable than phenotypic analysis alone [42]. Furthermore, HBGA phenotype can vary with developmental stage, and while there is concordance of genotype and salivary phenotype, phenotyping of red blood cells may be problematic [43, 44].
This study provides evidence that the P[6]-based RV3-BB vaccine induces take, irrespective of HBGA status. This vaccine could address suboptimal efficacy of the P[8]-based vaccines in regions where the burden of P[6] rotavirus disease is high, regardless of the HGBA phenotype profile of the population. Extrapolation of results of this study to different settings requires further study. Such studies are underway in Indonesia and Malawi.
This study has some limitations. It was conducted in a relatively small sample size in a homogenous population with a small number of Lewis-negative and secretor-negative participants. The high rate of vaccine take (>90%) in this population limited the ability to explore the HBGA status of participants without vaccine take. Further studies are underway in Indonesia and Malawi to broaden the populations and improve sample sizes and power for statistical analyses.
In summary, we found that the human neonatal vaccine RV3-BB (G3P[6]) produced a positive cumulative vaccine take, irrespective of HBGA status. We observed no difference in positive vaccine take by secretor status, by Lewis status, or by combined Lewis and secretor status. This is the first study to assess whether HBGA status influences vaccine take following receipt of a rotavirus vaccine based on a P[6] strain. The RV3-BB vaccine has the potential to provide an improved level of protection, particularly in Africa, where the Lewis-negative phenotype is more prevalent and where P[6] rotavirus strains causing disease are endemic.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. We thank the participants and their families, for taking part in this study; the midwives and antenatal staff at the Dunedin Hospital, for facilitating invitation into the study; Barry Taylor and Pam Jackson, as well as the study staff from both the Department of Women’s and Children’s Health, Dunedin School of Medicine, University of Otago, New Zealand, and the RV3 Rotavirus Vaccine Program, Murdoch Children’s Research Institute (MCRI), Victoria, Australia; Amanda Handley and Fran Justice from MCRI, for help with data management; and the Centre for Genomics Medicine—Sequencing Service and Development Platform, Victorian Clinical Genetic Services/MCRI, for their assistance.
Financial support. This work was supported by the Australian National Health and Medical Research Council (project grant ID491239), the New Zealand Health Research Council International Investment Opportunities Fund Trans-Tasman Clinical Trials Collaborative Initiative (08_T02), and the Victorian Government’s Operational Infrastructure Support Program.
Potential conflicts of interest. J. E. B is director of the Australian Rotavirus Surveillance Program, which is supported by research grants from GlaxoSmithKline, as well as the Commonwealth Department of Health and Aging. C. D. K. is currently employed as Senior Program Officer, Enteric and Diarrheal Disease, Bill and Melinda Gates Foundation. D. C. is currently employed by ViiV Healthcare: all work on this project was completed while at the Murdoch Children’s Research Institute. C. D. K. and Murdoch Children’s Research Institute hold a provisional patent for the RV3-BB vaccine. All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Armah GE, Breiman RF, Tapia MD, et al. Immunogenicity of the pentavalent rotavirus vaccine in African infants. Vaccine 2012; 30(Suppl 1):A86–93. [DOI] [PubMed] [Google Scholar]
- 2. Madhi SA, Cunliffe NA, Steele D, et al. Effect of human rotavirus vaccine on severe diarrhea in African infants. N Engl J Med 2010; 362:289–98. [DOI] [PubMed] [Google Scholar]
- 3. Vesikari T, Karvonen A, Prymula R, et al. Efficacy of human rotavirus vaccine against rotavirus gastroenteritis during the first 2 years of life in European infants: randomised, double-blind controlled study. Lancet 2007; 370:1757–63. [DOI] [PubMed] [Google Scholar]
- 4. Vesikari T, Matson DO, Dennehy P, et al. ; Rotavirus Efficacy and Safety Trial (REST) Study Team Safety and efficacy of a pentavalent human-bovine (WC3) reassortant rotavirus vaccine. N Engl J Med 2006; 354:23–33. [DOI] [PubMed] [Google Scholar]
- 5. Ouermi D, Soubeiga D, Nadembega WMC, et al. Molecular epidemiology of rotavirus in children under five in Africa (2006–2016): a systematic review. Pak J Biol Sci 2017; 20:59–69. [DOI] [PubMed] [Google Scholar]
- 6. Roczo-Farkas S, Kirkwood CD, Cowley D, et al. The impact of rotavirus vaccines on genotype diversity: a comprehensive analysis of 2 decades of Australian surveillance data. J Infect Dis 2018; 218:546–54. [DOI] [PubMed] [Google Scholar]
- 7. Iturriza-Gomara M, Dallman T, Banyai K, et al. Rotavirus genotypes co-circulating in Europe between 2006 and 2009 as determined by EuroRotaNet, a pan-European collaborative strain surveillance network. Epidemiol Infect 2011; 139:895–909. [DOI] [PubMed] [Google Scholar]
- 8. Arifuzzaman M, Ahmed T, Rahman MA, et al. Individuals with Le(a+b-) blood group have increased susceptibility to symptomatic vibrio cholerae O1 infection. PLoS Negl Trop Dis 2011; 5:e1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hutson AM, Atmar RL, Graham DY, Estes MK. Norwalk virus infection and disease is associated with ABO histo-blood group type. J Infect Dis 2002; 185:1335–7. [DOI] [PubMed] [Google Scholar]
- 10. Borén T, Falk P, Roth KA, Larson G, Normark S. Attachment of Helicobacter pylori to human gastric epithelium mediated by blood group antigens. Science 1993; 262:1892–5. [DOI] [PubMed] [Google Scholar]
- 11. Chaudhuri A, DasAdhikary CR. Possible role of blood-group secretory substances in the aetiology of cholera. Trans R Soc Trop Med Hyg 1978; 72:664–5. [DOI] [PubMed] [Google Scholar]
- 12. Fiore L, Greenberg HB, Mackow ER. The VP8 fragment of VP4 is the rhesus rotavirus hemagglutinin. Virology 1991; 181:553–63. [DOI] [PubMed] [Google Scholar]
- 13. Dowling W, Denisova E, LaMonica R, Mackow ER. Selective membrane permeabilization by the rotavirus VP5* protein is abrogated by mutations in an internal hydrophobic domain. J Virol 2000; 74:6368–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Barbé L, Le Moullac-Vaidye B, Echasserieau K, et al. Histo-blood group antigen-binding specificities of human rotaviruses are associated with gastroenteritis but not with in vitro infection. Sci Rep 2018; 8:12961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ströh LJ, Stehle T. Glycan engagement by viruses: receptor switches and specificity. Annu Rev Virol 2014; 1:285–306. [DOI] [PubMed] [Google Scholar]
- 16. Monnier N, Higo-Moriguchi K, Sun ZY, Prasad BV, Taniguchi K, Dormitzer PR. High-resolution molecular and antigen structure of the VP8* core of a sialic acid-independent human rotavirus strain. J Virol 2006; 80:1513–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jiang X, Liu Y, Tan M. Histo-blood group antigens as receptors for rotavirus, new understanding on rotavirus epidemiology and vaccine strategy. Emerg Microbes Infect 2017; 6:e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yu X, Dang VT, Fleming FE, von Itzstein M, Coulson BS, Blanchard H. Structural basis of rotavirus strain preference toward N-acetyl- or N-glycolylneuraminic acid-containing receptors. J Virol 2012; 86:13456–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Koda Y, Soejima M, Kimura H. The polymorphisms of fucosyltransferases. Leg Med (Tokyo) 2001; 3:2–14. [DOI] [PubMed] [Google Scholar]
- 20. Koda Y, Tachida H, Pang H, et al. Contrasting patterns of polymorphisms at the ABO-secretor gene (FUT2) and plasma alpha(1,3)fucosyltransferase gene (FUT6) in human populations. Genetics 2001; 158:747–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nordgren J, Nitiema LW, Ouermi D, Simpore J, Svensson L. Host genetic factors affect susceptibility to norovirus infections in Burkina Faso. PLoS One 2013; 8:e69557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ferrer-Admetlla A, Sikora M, Laayouni H, et al. A natural history of FUT2 polymorphism in humans. Mol Biol Evol 2009; 26:1993–2003. [DOI] [PubMed] [Google Scholar]
- 23. Yu LC, Yang YH, Broadberry RE, Chen YH, Chan YS, Lin M. Correlation of a missense mutation in the human Secretor alpha 1,2-fucosyltransferase gene with the Lewis(a+b+) phenotype: a potential molecular basis for the weak Secretor allele (Sew). Biochem J 1995; 312(Pt 2):329–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Koda Y, Soejima M, Liu Y, Kimura H. Molecular basis for secretor type alpha(1,2)-fucosyltransferase gene deficiency in a Japanese population: a fusion gene generated by unequal crossover responsible for the enzyme deficiency. Am J Hum Genet 1996; 59:343–50. [PMC free article] [PubMed] [Google Scholar]
- 25. Kazi AM, Cortese MM, Yu Y, et al. Secretor and salivary ABO blood group antigen status predict rotavirus vaccine take in infants. J Infect Dis 2017; 215:786–9. [DOI] [PubMed] [Google Scholar]
- 26. Armah GE, Cortese MM, Dennis FE, et al. Rotavirus vaccine take in infants is associated with secretor status. J Infect Dis 2018; 219:756–49. [DOI] [PubMed] [Google Scholar]
- 27. Bucardo F, Nordgren J, Reyes Y, Gonzalez F, Sharma S, Svensson L. The Lewis A phenotype is a restriction factor for Rotateq and Rotarix vaccine-take in Nicaraguan children. Sci Rep 2018; 8:1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pollock L, Bennett A, Jere KC, et al. Non-secretor histo-blood group antigen phenotype is associated with reduced risk of clinical rotavirus vaccine failure in Malawian infants. Clin Infect Dis 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Payne DC, Currier RL, Staat MA, et al. Epidemiologic association between FUT2 secretor status and severe rotavirus gastroenteritis in children in the United States. JAMA Pediatr 2015; 169:1040–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Imbert-Marcille BM, Barbé L, Dupé M, et al. A FUT2 gene common polymorphism determines resistance to rotavirus A of the P[8] genotype. J Infect Dis 2014; 209:1227–30. [DOI] [PubMed] [Google Scholar]
- 31. Ayouni S, Sdiri-Loulizi K, de Rougemont A, et al. Rotavirus P[8] infections in persons with secretor and nonsecretor phenotypes, Tunisia. Emerg Infect Dis 2015; 21:2055–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lee B, Dickson DM, deCamp AC, et al. Histo-blood group antigen phenotype determines susceptibility to genotype-specific rotavirus infections and impacts measures of rotavirus vaccine efficacy. J Infect Dis 2018; 217:1399–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nordgren J, Sharma S, Bucardo F, et al. Both Lewis and secretor status mediate susceptibility to rotavirus infections in a rotavirus genotype-dependent manner. Clin Infect Dis 2014; 59:1567–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Huang P, Xia M, Tan M, et al. Spike protein VP8* of human rotavirus recognizes histo-blood group antigens in a type-specific manner. J Virol 2012; 86:4833–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bishop RF, Barnes GL, Cipriani E, Lund JS. Clinical immunity after neonatal rotavirus infection. A prospective longitudinal study in young children. N Engl J Med 1983; 309:72–6. [DOI] [PubMed] [Google Scholar]
- 36. Bines JE, Danchin M, Jackson P, et al. ; RV3 Rotavirus Vaccine Program Safety and immunogenicity of RV3-BB human neonatal rotavirus vaccine administered at birth or in infancy: a randomised, double-blind, placebo-controlled trial. Lancet Infect Dis 2015; 15:1389–97. [DOI] [PubMed] [Google Scholar]
- 37. Chang JG, Yang TY, Liu TC, et al. Molecular analysis of secretor type alpha(1,2)-fucosyltransferase gene mutations in the Chinese and Thai populations. Transfusion 1999; 39:1013–7. [DOI] [PubMed] [Google Scholar]
- 38. Böhm R, Fleming FE, Maggioni A, et al. Revisiting the role of histo-blood group antigens in rotavirus host-cell invasion. Nat Commun 2015; 6:5907. [DOI] [PubMed] [Google Scholar]
- 39. Hu L, Sankaran B, Laucirica DR, et al. Glycan recognition in globally dominant human rotaviruses. Nat Commun 2018; 9:2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kambhampati A, Payne DC, Costantini V, Lopman BA. Host genetic susceptibility to enteric viruses: a systematic review and metaanalysis. Clin Infect Dis 2016; 62:11–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Günaydın G, Nordgren J, Sharma S, Hammarström L. Association of elevated rotavirus-specific antibody titers with HBGA secretor status in Swedish individuals: the FUT2 gene as a putative susceptibility determinant for infection. Virus Res 2016; 211:64–8. [DOI] [PubMed] [Google Scholar]
- 42. Hong YJ, Hwang SM, Kim TS, et al. Significance of Lewis phenotyping using saliva and gastric tissue: comparison with the Lewis phenotype inferred from Lewis and secretor genotypes. Biomed Res Int 2014; 2014:573652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ameno S, Kimura H, Ameno K, et al. Lewis and secretor gene effects on Lewis antigen and postnatal development of Lewis blood type. Biol Neonate 2001; 79:91–6. [DOI] [PubMed] [Google Scholar]
- 44. Henry S, Oriol R, Samuelsson B. Lewis histo-blood group system and associated secretory phenotypes. Vox Sang 1995; 69:166–82. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.