Abstract
Background
During pregnancy, the vaginal microbiota is relatively stable. However, African women have more diverse vaginal microbiota than their European counterparts, in addition to high human immunodeficiency virus (HIV) prevalence and risk of adverse birth outcomes. Although HIV is associated with alterations in vaginal microbiota and inflammation in nonpregnant women, these relationships are underexplored in pregnant women.
Methods
In this study, we characterize the vaginal microbiota and immune factors in pregnant African women who were HIV-uninfected (n = 314) versus HIV-infected (n = 42). Mucosal samples were collected once at the enrollment visit (between 15 and 35 weeks of gestation) and women were followed until delivery.
Results
Vaginal microbial communities of pregnant women with HIV were significantly more diverse than women without HIV (P = .004), with community structure also differing by HIV status (P = .002, R2 = 0.02). Human immunodeficiency virus infection was also associated with increased risk of preterm birth (PTB) (31% versus 15.3%; P = .066). In a multivariate analysis, HIV infection was independently associated with diverse vaginal community state type (CST)-IVA (P = .005) and CST-IVB (P = .018) as well as PTB (P = .049). No association between HIV status and cytokine concentrations was found.
Conclusions
Longitudinal studies with accurate gestational age assessment would be important to confirm these relationships.
Keywords: cytokines, HIV infection, premature delivery, vaginal microbiota
Pregnant women with HIV have more diverse vaginal communities and altered community structure compared with pregnant uninfected women. However, preterm birth was associated with HIV infections independent of vaginal community state type.
Bacterial communities present in the lower female genital tract play an important role during pregnancy. The vaginal microbiota of most pregnant women is stable with more Lactobacillus species-dominated communities than nonpregnant women [1, 2]. African women and women of African descent often possess vaginal microbiota dominated by Lactobacillus iners or a diverse vaginal community [3, 4], even when pregnant [2]. Vaginal microbial dysbiosis is also more common in women infected with human immunodeficiency virus (HIV) [5], and the prevalence of an altered vaginal microbial environment in HIV-infected pregnant women has been reported to be between 47% and 89% in some African cohorts [6]. Asymptomatic bacterial vaginosis (BV) has been associated with twice the risk of preterm birth (PTB), as well as miscarriage, maternal infection, and low birth weight (LBW) [7]. The increased risk of adverse pregnancy outcome may be due to specific bacterial species, rather than BV itself. Gardnerella vaginalis, Mycoplasma hominis, Mobiluncus, Atopobium vaginae, BV-associated bacterium-1 (BVAB1), Megasphaera, and Sneathia species have all been associated with preterm labor, PTB, and pregnancy loss [8–10]. Although several studies suggest a link between vaginal bacterial diversity and poor pregnancy outcome, others have found no significant link [11, 12]; more important, none have been performed in Africa.
Several studies have demonstrated associations between HIV infection and increased risk of poor pregnancy outcomes [13–16], but whether that is an effect of HIV per se or of associated comorbidities remains unclear [17]. A Spanish study reported a higher risk of both spontaneous and clinically indicated (eg, pre-eclampsia) PTB in HIV-infected women [18]. Past research from the precombination antiretroviral therapy (ART) era suggests that maternal HIV infection is associated with small-for-gestational age infants but not PTB [19]. In France, there was a report of ART-associated increases in PTB [20]. Data from the MmaBana trial in Botswana and earlier work from the Ivory Coast also suggest an association between ART and risk of PTB and/or LBW [21, 22]. In a facility-based cohort in Botswana, women with HIV, regardless of ART use, were at increased risk of adverse pregnancy outcomes, including stillbirth, premature delivery, small-for-gestational age infants, and early neonatal death compared with uninfected women [23]. It is difficult to disentangle the effects of antiretrovirals (ARVs) and HIV itself on adverse birth outcomes. Regardless of the cause, the mechanisms of HIV/ARV-induced PTB need to be investigated so that intervention measures can be identified to mitigate these risks. An important underlying mechanism for the relationship between HIV and poor pregnancy outcomes may be changes in the vaginal microbiota as a result of HIV infection.
Studies in nonpregnant women have demonstrated that HIV infection is associated with increases in proinflammatory cytokine concentrations in the female genital tract [24–26]. In addition, studies conducted in pregnant HIV-uninfected women demonstrated that vaginal inflammatory cytokines were associated with PTB [27, 28]. Despite these facts, the relationship between the composition of vaginal microbial communities, immune factors, and pregnancy outcomes is underexplored in pregnant women with HIV (PWH). Therefore, we assessed this relationship in a cohort of pregnant Zimbabwean women.
METHODS
Women were recruited at Harare and Chitungwiza central hospital antenatal clinics. All pregnant women between 15 and 35 weeks of gestation, above 18 years of age, willing to take part in the study and to provide written consent, with documented HIV testing results in pregnancy were eligible. Additional exclusion criteria included diagnosis of a sexually transmitted infection (STI) in the past month, or antibiotic use in the past month, excluding cotrimoxazole prophylaxis. The study was approved by Harare Central Hospital and Chitungwiza Central hospital ethics committees, the University of Cape Town Human Research Ethics Committee, and the Medical Research Council of Zimbabwe.
After informed consent, a questionnaire was administered, and 2 vaginal swabs were collected; 1 swab was immediately placed into 1.5-mL sterile phosphate-buffered saline, transferred to the laboratory on ice, and stored at −70⁰C within 4 hours of collection. A second vaginal swab was collected, touched on a pH indicator paper to measure vaginal pH, and then used for Gram staining. Diagnosis of BV was done by Gram stain and microscopy using Nugent scoring [29]. A wet prep was done for Trichomonas vaginalis. Birth outcomes were collected from hospital records or maternal health cards.
Samples were thawed, treated with a cocktail of enzymes including lysostaphin, mutanolysin, and lysozyme [30] incubated for 1 hour at 37°C with vortexing every 10–20 minutes. After enzymatic digestion sodium dodecyl sulfate, phenol:chloro:isoamyl were added to the vaginal fluid mixture and subjected to bead beating. Deoxyribonucleic acid (DNA) extraction was done using the phenol chloroform extraction method [3]. Deoxyribonucleic acid was amplified using universal primers (515F/806R) for the hypervariable V4 region of the 16S ribosomal ribonucleic acid (rRNA) gene, and library preparation was performed through 2 amplifications. The first is PCR1: specific primers with overhang adapters were used to amplify the V4 template out of the DNA sample. The purified amplicons were subjected to second PCR: Nextera XT Index Kit (Illumina) was used to add unique sequencing adapters. These adapters were used during the PCR reaction to amplify the insert DNA. Index sequences were added on both ends of the DNA, enabling dual-indexing of pooled libraries. Purified libraries consisting of 96 pooled samples per library were sequenced from both ends using the MiSeq platform (version 3, 2 × 300 base pairs [bp] kit), and the resulting reads were demultiplexed on the MiSeq and analyzed as previously described [31]. In brief, after demultiplexing, raw reads were preprocessed as follows: forward and reverse reads were merged by using usearch7 [32] allowing a maximum of 3 mismatches; merged reads were quality filtered by using usearch7 (reads with E scores of >0.1 were discarded); primer sequences were removed using a custom python script [31]; and merged, filtered reads were truncated at 250 bp. Dereplicated sequences were sorted by abundance (highest to lowest) and clustered de novo into operational taxonomic units (OTUs) at 97% similarity using usearch7. Chimeric sequences were detected (against the Gold database) by using UCHIME [33] and removed. Individual sequences were assigned to the specific identifiers using a 97% similarity threshold. Taxonomic assignment was performed with QIIME 1.8.0 [34] using the RDP classifier (using the default confidence level of 0.5) against the Greengenes 13.8 reference taxonomy for 97% identity. Downstream analysis was done in R using custom scripts [31]. Alpha (α) diversity estimates (Shannon) were calculated using the R package vegan [35]. Beta-diversity was estimated by NMDS of Jensen-Shannon distances matrices [36]. Annotated heat maps were constructed using merged taxa at lowest taxonomic levels for visualization using custom scripts. Differential abundance testing was done using DESeq [37], with significance cutoff of 0.01 after multiple testing correction.
Concentrations of 27 cytokines (fibroblast growth factor [FGF] basic, Eotaxin, granulocyte colony-stimulating factor [CSF], granulocyte macrophage-CSF, interferon-γ, interleukin [IL]-1β, IL-1ra, IL-2, IL-4, IL-5, IL-6, IL-7, IL-8, IL-9, IL-10, IL-12 [p70], IL-13, IL-15, IL-17, interferon-γ-induced protein [IP-10], macrophage chemotactic protein [MCP]-1 [monocyte chemotactic and activating factor], macrophage inflammatory protein [MIP]-1α, MIP-1β, PDGF-BB, RANTES, tumor necrosis factor [TNF]-α, vascular endothelial growth factor [VEGF]) were measured in vaginal fluid using Multiplex bead array. In brief, samples were thawed overnight at 4°C, centrifuged at 13 000 rpm for 10 minutes, and the supernatant was filtered using a Spin-X tube (0.22-μm filter; Corning Costar) at 13 000 rpm for 10 minutes. Cytokine concentrations were measured in the filtrate using a Bio-Plex Pro Human Cytokine 27-Plex Assay (Bio-Rad Laboratories Inc.). Assay plates were read using a Bio-Plex Suspension Array Reader (Bio-Rad Laboratories Inc.). Data were analyzed using Bio-Plex manager software (version 4). Cytokine levels below the lower limit of detection of the assay were reported as the midpoint between zero and the lowest detectable level measured for that given cytokine. As samples were spread across multiple plates, a reference panel of 5 samples was included on each of the plates (interplate controls), in addition to 5 samples being duplicated on each set of plates (intraplate controls) for quality-control measures. Spearman’s rank to measure intra-assay and interassay correlation coefficients was used to determine assay reliability and reproducibility. Cytokines were excluded where interplate or intra-assay correlation coefficients were <0.8 and 0.95, respectively.
Shapiro-Wilk test for normality was performed to determine the distribution of variables within the dataset. Comparison of unpaired non-parametric data was done using the Mann-Whitney U test. The Spearman’s rank test was applied to test for correlation between nonparametric data. Statistical inferences on binary sets of data were performed using the χ 2 test. Non-parametric assessments of variation between groups was carried out through the Kruskal-Wallis analysis of variance (ANOVA), with Dunn’s post hoc test applied to identify which groups were different for the effect of multiple comparisons. Statistical analyses were performed using GraphPad Prism version 7.0 for Mac OS (GraphPad Software, San Diego, CA), R, or Stata version 12. All tests were 2-tailed and P ≤.05 were considered significant. Logistic regression was performed using Stata version 12.
RESULTS
Participant Characteristics
Of the 420 eligible women who consented to the study, we sequenced samples from women without trichomoniasis, diabetes, or other chronic illness unrelated to HIV. Of the 367 samples we sequenced, 356 passed sequencing quality control (≥2000 reads/sample), had HIV status data, and were thus included in downstream analysis. Of these 356 women, 42 (12%) were PWH (Table 1). There were no differences in characteristics such as gestational age at collection (enrollment), gravida, antibiotic use during the past 3 months, vaginal douching practices, and history of previous poor outcomes (including PTB, stillbirth, or miscarriage in prior pregnancies) between PWH and uninfected women. The median CD4+ count of PWH was 495 cells/mm3. Of the women with details on ART use, the majority (36 of 40 [90%]) were on the fixed-dose combination of tenofovir, lamivudine, and efavirenz (Table 1). Of the singleton pregnancies with birth outcomes available, 42 of 244 (17.2%) were preterm, defined as delivery before 37 weeks of gestation. Of those with birth weights available, 35 (15%) were considered LBW (ie, birth weight below 2.5 kg). Of the 42 preterm deliveries in this cohort, a higher prevalence occurred in PWH compared with uninfected women (31% vs 15.3% [P = .066], respectively). Bacterial vaginosis, as diagnosed by Nugent score 7–10, was present in 88 women (24.7%). Pregnant women with HIV were significantly more likely to have BV (P = .01) compared with uninfected women (Table 1). Furthermore, PWH were more likely to be of older age (31 vs 28.7 years, P = .066) and have a partner who was a smoker (7; 16.7% vs 19; 6.1% [P = .03]).
Table 1.
Cohort Characteristics Stratified by HIV Status
| Characteristics | Overall (n = 356) | HIV Uninfected (n = 314) | HIV Infected (n = 42) | P Valuea |
|---|---|---|---|---|
| Maternal age [years], median (IQR) | 29 (24–34) | 28.68 (24.07–33.52) | 31 (25–35) | .066 |
| Gestational age collection [weeks], median (IQR) | 29 (25–33) | 29 (25–33) | 30 (28–32.75) | .754 |
| Partner smoker, n (%) | 26 (7.3) | 19 (6.1) | 7 (16.7) | .030 |
| Blood CD4+ [cell/mm3] (median, IQR) | 495 (398.8–643.5) | NA | 495 (398.8–643.5) | |
| Plasma viral load copies/mL, mean (SD)b | 128.85 (98.49) | NA | 128.85 (98.49) | |
| ART Regimen, n (%)c | NA | |||
| TDF, FTC, EFV | 36 (85.7) | NA | 36 (85.7) | |
| Combivir (AZT + 3TC) | 4 (9.5) | 4 (9.5) | ||
| pH (median, IQR) | 4 (4–4.5) | 4 (4–4.5) | 4 (4–7) | .124 |
| Gravida (median, IQR) | 3 (2–4) | 3 (2–4) | 3. (2–4) | .102 |
| Para (median, IQR) | 1 (0–2) | 1 (0–2) | 2 (1–2) | .041 |
| Antibiotic use past 3 months, n (%)d | 45 (12.6) | 38 (12.1) | 7 (16.7) | .556 |
| Vaginal douching current pregnancy, n (%) | 174 (48.9) | 148 (47.1) | 26 (61.9) | .102 |
| Nugent score (median, IQR) | 2.5 (0–7) | 2 (0–6) | 5 (0–8) | .016 |
| Bacterial vaginosis, n (%) | .010 | |||
| BV positive (Nugent score 7–10) | 88 (24.7) | 70 (22.3) | 18 (42.9) | |
| Intermediate (Nugent score 4–6) | 57 (16.0) | 50 (15.9) | 7 (16.7) | |
| BV negative (Nugent score 0–3) | 211 (59.3) | 194 (61.8) | 17 (40.5) | |
| Pregnancy induced hypertensione, n (%) | 21 (5.9) | 21 (6.7) | 0 (0.0) | .168 |
| Previous poor outcomef, n (%) | 109 (30.6) | 97 (30.9) | 12 (28.6) | .898 |
| Previous preterm birth, n (%) | 53 (14.9) | 48 (15.3) | 5 (11.9) | .728 |
| Outcomes | ||||
| Small for gestational ageg, n (%) | 30 (12.9) | 25 (12.2) | 5 (17.9) | .590 |
| Low birth weightg, n (%) | 35 (15.0) | 28 (13.7) | 7 (25.0) | .196 |
| Pretermh, n (%) | 42 (17.2) | 33 (15.3) | 9 (31.0) | .066 |
| CST, n (%) | .004 | |||
| CST-I | 109 (30.6) | 105 (33.0) | 4 (9.0) | |
| CST-III | 92 (25.8) | 82 (26.0) | 10 (24.0) | |
| CST-IVA | 92 (25.8) | 74 (24.0) | 18 (43.0) | |
| CST-IVB | 55 (15.4) | 45 (14.0) | 10 (24.0) | |
| CST-V | 8 (2.2) | 8 (3.0) | 0 (0.0) | |
| Inflammation (n = 324) n (%) | .565 | |||
| High | 122 (37.65) | 104 (36.9) | 18 (42.9) | |
| Low | 202 (62.35) | 178 (63.1) | 24 (57.1) |
Bolded text signifies significant differences between HIV Uninfected versus Infected.
Abbreviations: ART, antiretroviral treatment; AZT, zidothymidine; BV, bacterial vaginosis; CST, community state type; EFV, efavirenz; FTC, emtricitabine; HIV, human immunodeficiency virus; IQR, interquartile range; NA, not applicable; SD, standard deviation; TDF, tenofovir disoproxil fumarated.
aUnpaired t test was used when assessing frequency associations between groups. χ 2 test was used for BV and Fisher test for CST.
bAnalysis of variance.
cTwo missing values.
dTen were cotrimoxazole prophylaxis.
eCurrent pregnancy.
fStillbirth, miscarriage or preterm birth, prior pregnancy.
gn = 233.
hn = 244.
Vaginal Microbial Composition by 16S Ribosomal Ribonucleic Acid Gene Sequencing
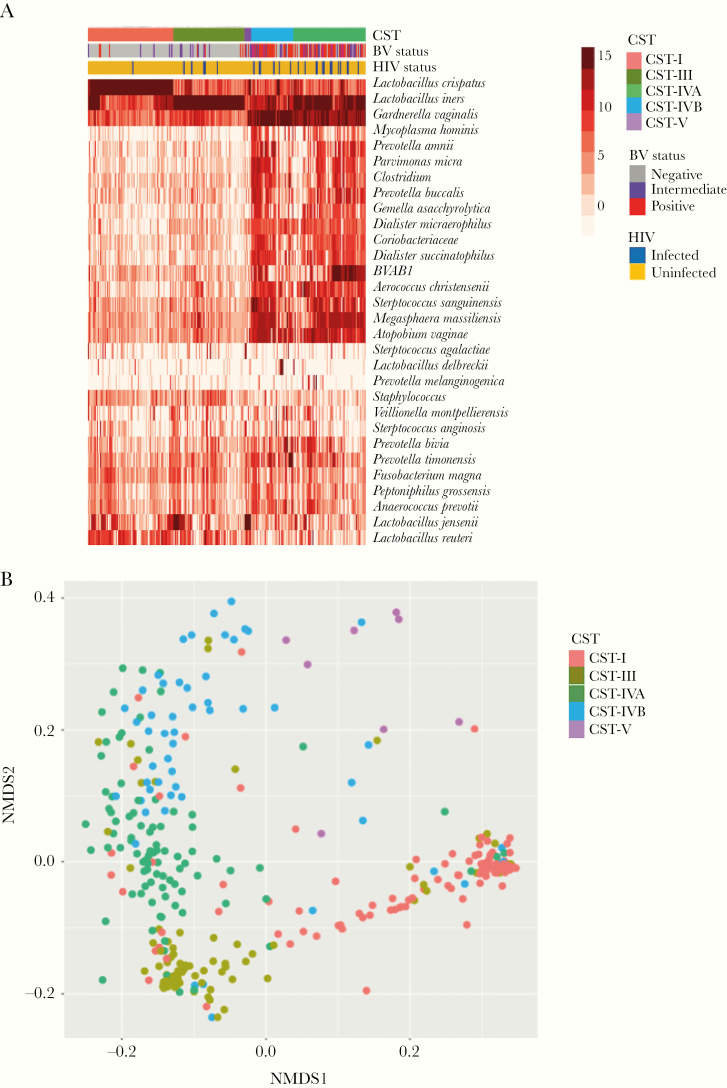
Relative abundance of vaginal microbiota was obtained by amplifying and sequencing the V4 hypervariable regions of the 16S rRNA gene. The median read count was 75 849 (range, 61–352, 921). Samples with fewer than 2000 reads were excluded from downstream analysis. The Greengenes Database [38] was used to assign taxonomy to sequences, and 1185 OTUs were generated. Unsupervised hierarchical clustering of taxa merged at the lowest taxonomic level using Jensen-Shannon distance matrix, and the ward D2 linkage clustering method was performed (Figure 1A). The vaginal microbiota of the women in this cohort clustered into 5 community state types (CST). Using the CST numeric system described by Romero et al [39], these were CST-I (Lactobacillus crispatus dominant), CST-III (L iners dominant), CST-IVA (G vaginalis, BVAB1, and mixed diverse anaerobes), CST-IVB (G vaginalis, L iners, A vaginae, and mixed diverse anaerobes), and CST-V (Lactobacillus jensenii dominant) (Figure 1). The majority of women with CST-IVA or CST-IVB were BV positive by Nugent score (n = 70 of 85; 78.7%). As previously observed in African women, there were no women with vaginal microbiota dominated by L gasseri, and few with L jensenii dominated CST-V. As expected, alpha diversity as measured by Shannon index was significantly lower in CSTs I and III than CST-IV and also lower than CST-V. It is interesting to note that alpha diversity of the L iners-dominant CST-III was lower than CST-I (L crispatus dominant), although the difference was not significant (P = .07) (Figure 1B).
Figure 1.
(A) Unsupervised hierarchical clustering of the 30 most abundant taxa (rows) merged at the lowest taxonomic level as identified by 16S ribosomal ribonucleic acid (rRNA) gene amplicon sequencing using Ward D2 linkage with Jensen-Shannon distances. Red color represents most abundant, whereas blue presents least abundant or absence of log2 transformed operational taxanomic unit counts. (B) Alpha diversity, showing microbial diversities of the 5 community state types (CST). (C) Nonmetric multidimensional scaling (NMDS) using hierarchical clustering with Jensen-Shannon Ward D2 linkage method, showing 5 CSTs: CST-I (coral) dominated by Lactobacillus crispatus, CST-III (olive) dominated by Lactobacillus iners, CST-IVA (green), and CST-IVB (blue)-diverse communities dominated by Gardnerella vaginalis, and other anaerobic bacteria and less Lactobacillus. The CST-V (purple) dominated by Lactobacillus jensenii. BV, bacterial vaginosis.
Relationship Between Vaginal Microbiota and Human Immunodeficiency Virus
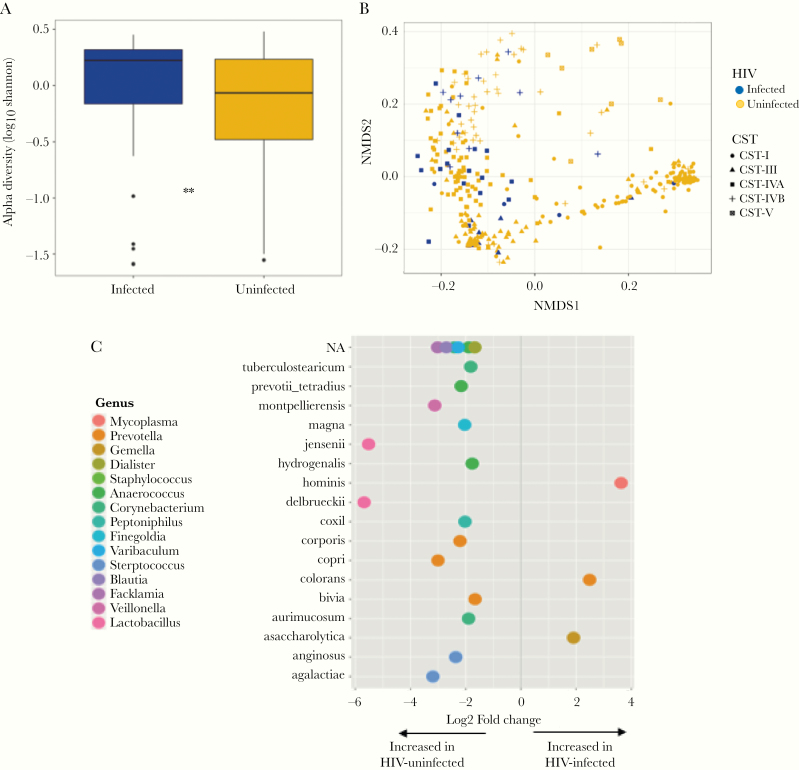
We compared the vaginal microbial composition between PWH and uninfected women. Pregnant women with HIV had significantly more diverse vaginal microbiota than uninfected women (mean log2 Shannon indices 1.66 versus 1.04, P = .004) (Figure 2A). Furthermore, beta diversity using Jensen-Shannon distances using Permutational multivariate ANOVA revealed significant differences between vaginal microbiota of women with and without HIV (P = .002, R2 = 0.02) (Figure 2B). Women infected HIV were less likely to have CST-I and CST-V microbiota and more likely to have CST-IVA and CST-IVB than HIV-negative women (Kruskal-Wallis [ANOVA]; P = .004) (Table 1). The majority of PWH had vaginal microbiota in CST-IVA (18 of 42 [43%]), followed by CST-IVB (10 of 42 [24.0%]) and CST-III (10 of 42 [24.0%]) (Table 1). A multivariate analysis confirmed the significant association between HIV status and diverse CST—specifically CST-IVA (odds ratio [OR], 6.08; 95% confidence interval [CI], 1.87–24.20; P = .004) and CST-IVB (OR, 6.31; 95% CI, 1.55–28.91; P = .01)—after adjusting for possible confounders (Table 2). Using DESeq2, we identified differentially abundant taxa by HIV status at adjusted P < .01. The relative abundance of 26 taxa were significantly differentially abundant with Prevotella colorans, Gemella asaccharolytica, and M hominis most strongly associated with HIV, and L jensenii and Lactobacillus delbrueckii most highly abundant in uninfected women (Figure 2C, Supplementary Table S1).
Figure 2.
(A) Alpha diversity of the vaginal microbiota of human immunodeficiency virus (HIV)-infected and HIV-uninfected women depicted by box plots; (B) beta diversity of vaginal microbiota by Jensen-Shannon distances. (C) Differentially abundant taxa 2-fold change between HIV-infected and HIV-uninfected women differentially abundant taxa using DESeq2. **P < .001. CST, community state types; NA, not applicable; NMDS, nonmetric multidimensional scaling.
Table 2.
Factors Associated With HIV in a Multivariate Analysis
| Characteristic | OR | 95% CI | P Value |
|---|---|---|---|
| Log maternal age (years) | 0.53 | .002–119 | .818 |
| Parity | 1.28 | .88–1.87 | .196 |
| Partner smoker | 4.69 | 1.13–19.55 | .034 |
| Preterm delivery | 2.81 | 1.01–7.83 | .049 |
| Gestational age at collection (weeks) | 1.03 | .94–1.13 | .473 |
| CST-III | 1.38 | .31–6.11 | .668 |
| CST-IVA | 5.89 | 1.70 20.4 | .005 |
| CST-IVB | 5.45 | 1.34–22.19 | .018 |
| CST-V | Omitted |
Bolded values depict signficant associations in the multivarate model.
Abbreviations: CI, confidence interval; CST, community state type; HIV, human immunodeficiency virus; OR, odds ratio.
Vaginal Cytokine Concentrations and Human Immunodeficiency Virus Status
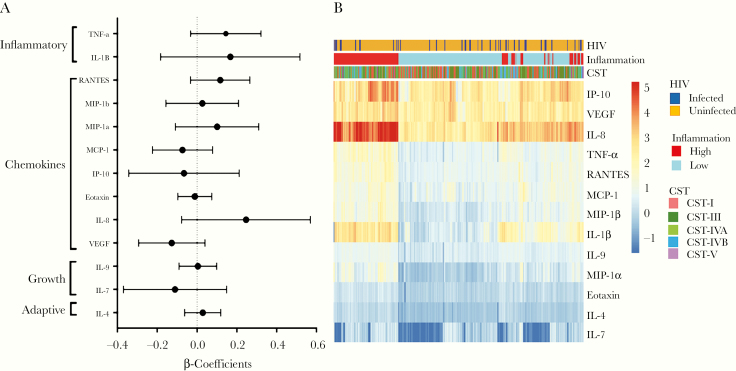
We measured the concentrations of 27 cytokines in vaginal fluid using Luminex. Of these, 13 had interplate and intraplate variation that passed our stringent cutoffs of rho of 0.8 and 0.95, respectively. These included the proinflammatory cytokines IL-1β, TNF-⍺, IL-8, the chemokines MIP-1⍺, MIP-1β, regulated upon activation, normal T-cell expressed, and secreted (RANTES), IP-10 (CXCL-10), MCP-1, Eotaxin, the growth factors VEGF and IL-7, and the Th2 cytokine IL-4. There were no significant differences in the concentrations of these factors according to HIV status (Figure 3A), even after adjusting for CST. Pam clustering of cytokines was used to cluster women into high and low inflammatory groups. The inflammation-high group did not show any clear association with HIV status (Figure 3B).
Figure 3.
(A) Association between human immunodeficiency virus (HIV) status and individual vaginal cytokine levels by univariate logistic regression. Regression β-coefficients are represented by black circles, and 95% confidence intervals are depicted by bars. (B) Heatmap showing cytokine distribution by HIV status, community state type (CST), and inflammation using supervised hierarchical clustering (Jensen-Shannon) of log10-transformed cytokine concentrations. Each column represents a woman, and rows represent individual cytokines. Red color represents higher concentrations (range 5.24 to 1.3 pg/mL), whereas blue represents (ranges 1.29 to −1.7 pg/mL) lower concentrations of cytokines. IL, interleukin; IP-10, interferon-γ-induced protein; MCP, macrophage chemotactic protein; MIP, macrophage inflammatory protein; TNF, tumor necrosis factor; VEGF, vascular endothelial growth factor.
Because vaginal microbiota have been shown to be strongly correlated with cytokine concentrations in nonpregnant women [3, 31], we assessed whether this was also true during pregnancy, a state of relative immune quiescence. There were no correlations between vaginal microbial taxa and cytokine concentrations with an R > 0.5. There were only 6 taxa associated with cytokine concentrations with an R > 0.3 that remained significant after multiple testing correction (Supplementary Figure S1).
Relationship Between Human Immunodeficiency Virus and Pregnancy Outcomes
A multivariate logistic regression analysis was performed, including factors that were associated with HIV in the univariate analysis with P < .1 (Table 1) and factors selected as a priori associated with HIV infection (Table 2). Preterm birth (OR, 3.80; 95% CI, 1.08–12.82; P = .004) and CST-IVA and CST-IVB were independently associated with HIV (OR, 6.08 and 95% CI, 1.87–24.2 and OR, 6.31 and 95% CI, 1.55–28.91, respectively) (Table 2). Because gestational age assessment was not performed via ultrasound and therefore may have been inaccurate, we also performed a multivariate analysis using LBW in a separate model (due to collinearity between LBW and PTB). Human immunodeficiency virus infection was also independently associated with delivery of LBW infants (95% CI, 1.08–12.83; P = .03) (Supplementary Table S2). We also explored predictors of PTB both univariate and in a multivariate linear model, including those factors associated with PTB in the univariate model (maternal age and gravida) or those previously reported to be associated with PTB (pregnancy-induced hypertension, vaginal CST, HIV, and history of prior PTB) (Supplementary Table S3). Only HIV infection remained independently associated with PTB (OR, 2.73; 95% CI, 1.01–7.42). Of the 9 PTBs that occurred in PWH, 7 (77.8%) occurred in women who conceived on ART, but this was not significantly different from the proportion of PWH that conceived on ART and delivered at term (n = 14, 70%).
DISCUSSION
Women with HIV are more likely to have BV and experience high rates of poor pregnancy outcomes, yet the relationship between these is underexplored. In this study, we investigated the relationship between HIV, vaginal microbiota, and pregnancy outcomes in a cohort of African women. As expected, there was a higher prevalence of BV in HIV-infected pregnant women. Consistently, PWH had higher vaginal microbial diversity and significantly different community composition than women without HIV infection. A longitudinal study of nonpregnant women from Chicago found no significant differences in microbial community structure according to HIV status, and the lack of significant differences was attributed to effective ART [40]. In our cohort of Zimbabwean pregnant women, most HIV-infected women were receiving ART yet still had altered vaginal microbiota. Previous studies in African women have reported that high diversity vaginal microbial communities and BV are associated with HIV acquisition [40–42]. Therefore, it remains unclear whether HIV infection is a cause or an effect of the altered vaginal microbiota in these women.
More important, in this cohort of Zimbabwean women, HIV was associated with PTB, and this was independent of vaginal microbiota (as measured by CST). It has long been reported that BV is a risk factor for PTB [10, 43–46] and that HIV-infected women are at risk for delivery of premature infants [18, 47–50]. Whether these factors are related has largely been unexplored. Our data suggest that factors other than nonoptimal vaginal microbiota put women with HIV at risk for adverse birth outcomes. Because all of the HIV-infected women in this cohort were receiving ART, we cannot discern whether HIV infection or the treatment contribute to prematurity in this study; however, in the current era, it is not possible to distinguish the relative contribution of these factors.
Although we found high microbial diversity in HIV-infected pregnant women, few differences in inflammation were observed between HIV-infected and uninfected women. Diverse vaginal microbiota is often associated with genital tract inflammation in nonpregnant women [31]. Because pregnancy is a state of relative immune quiescence, it is possible that factors are in play to dampen inflammation associated with vaginal microbiota. Indeed, we found only weak correlations between vaginal microbiota and cytokine concentrations in this pregnant cohort. In nonpregnant women, Roberts et al [51] found that during early infection, cervicovaginal inflammatory cytokine profiles did not differ between women pre- and post-HIV infection, suggesting that inflammation was a cause, rather than an effect, of HIV acquisition in these high-risk women. It is also possible that the lack of increased vaginal inflammation in the PWH in our cohort was due to the fact that most women were on ART, with median CD4+ counts of 495 cell/mm3. Another potential explanation for the lack of inflammation associated with HIV infection in our cohort is that these women were pregnant.
CONCLUSIONS
Our study was limited by the cross-sectional study design used. Due to this, women were not retested for HIV during pregnancy and therefore could have been misclassified as HIV negative at delivery. We were also limited by lack of STI testing other than wet mount for trichomoniasis. The STIs may alter the relationship between HIV and PTB, although they would likely not influence the relationship between HIV and vaginal microbiota. The study was somewhat underpowered to assess differences by preterm and HIV status because there were relatively low numbers of both. However, we were able to detect associations between HIV and poor pregnancy outcomes independent of vaginal CST. Further research with a longitudinal cohort would be useful to confirm or refute these findings.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Supplementary Figure S1. Significant positive (blue) or negative (red) correlations between standardized read counts of bacteria (merged at lowest taxonomic level) and vaginal concentrations of cytokines using Spearman’s rank correlation, with adjusted P < .05 and R2 > 0.03.
Supplementary Table S1. Differentially abundant taxa by HIV status using DESeq2.
Supplementary Table S2. Factors associated with HIV in a multivariate model including low birth weight.
Supplementary Table S3. Factors associated with preterm birth in univariate and multivariate models.
Notes
Acknowledgments. We express our gratitude to all the pregnant women who volunteered to participate in this study. We further extend our gratitude to Harare central hospital and Chitungwiza central hospital staff for providing their hospitals, the Antenatal Clinic sites for sample collection, and Erica Sinirai Kubvoruno Gudza and Asiyathu Matipuwa for all data collection and data capturing. Computations were performed using facilities provided by the University of Cape Town’s ICTS High Performance Computing team (http://hpc.uct.ac.za). The clinical and technical staff as a whole is gratefully acknowledged. We thank the National Reference Microbiology Laboratory team for all the help given throughout the study.
Disclaimer. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Financial support. This study was funded in part by Letten Foundation Norway. H. B. J. is funded in part by Grants R01HD083040 and R01AI128792 from the US National Institutes of Health.
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest.
References
- 1. MacIntyre DA, Chandiramani M, Lee YS, et al. The vaginal microbiome during pregnancy and the postpartum period in a European population. Sci Rep 2015; 5:8988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Serrano MG, Parikh HI, Brooks JP, et al. Racioethnic diversity in the dynamics of the vaginal microbiome during pregnancy. Nat Med 2019; 25:1001–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Anahtar MN, Bowman BA, Kwon DS. Efficient nucleic acid extraction and 16S rRNA gene sequencing for bacterial community characterization. J Vis Exp 2016. Available at https://www.jove.com/video/53939/efficient-nucleic-acid-extraction-16s-rrna-gene-sequencing-for. Accessed 23 November 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hummelen R, Fernandes AD, Macklaim JM, et al. Deep sequencing of the vaginal microbiota of women with HIV. PLoS One 2010; 5:e12078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mbizvo ME, Musya SE, Stray-Pedersen B, Chirenje Z, Hussain A. Bacterial vaginosis and intravaginal practices: association with HIV. Cent Afr J Med 2004; 50:41–6. [PubMed] [Google Scholar]
- 6. Vallone C, Rigon G, Lucantoni V, Putignani L, Signore F. Pregnancy in HIV-positive patients: effects on vaginal flora. Infect Dis Obstet Gynecol 2012; 2012:287849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Leitich H, Kiss H. Asymptomatic bacterial vaginosis and intermediate flora as risk factors for adverse pregnancy outcome. Best Pract Res Clin Obstet Gynaecol 2007; 21:375–90. [DOI] [PubMed] [Google Scholar]
- 8. Foxman B, Wen A, Srinivasan U, et al. Mycoplasma, bacterial vaginosis-associated bacteria BVAB3, race, and risk of preterm birth in a high-risk cohort. Am J Obstet Gynecol 2014; 210:226.e1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Larsen B, Hwang J. Mycoplasma, Ureaplasma, and adverse pregnancy outcomes: a fresh look. Infect Dis Obstet Gynecol 2010; 2010:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nelson DB, Hanlon A, Nachamkin I, et al. Early pregnancy changes in bacterial vaginosis-associated bacteria and preterm delivery. Paediatr Perinat Epidemiol 2014; 28:88–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Romero R, Hassan SS, Gajer P, et al. The vaginal microbiota of pregnant women who subsequently have spontaneous preterm labor and delivery and those with a normal delivery at term. Microbiome 2014; 2:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Stout MJ, Zhou Y, Wylie KM, Tarr PI, Macones GA, Tuuli MG. Early pregnancy vaginal microbiome trends and preterm birth. Am J Obstet Gynecol 2017; 217:356.e1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gray GE, McIntyre JA. HIV and pregnancy. BMJ 2007; 334:950–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Patil S, Bhosale R, Sambarey P, et al. Impact of maternal human immunodeficiency virus infection on pregnancy and birth outcomes in Pune, India. AIDS Care 2011; 23:1562–9. [DOI] [PubMed] [Google Scholar]
- 15. Petrova MI, van den Broek M, Balzarini J, Vanderleyden J, Lebeer S. Vaginal microbiota and its role in HIV transmission and infection. FEMS Microbiol Rev 2013; 37:762–92. [DOI] [PubMed] [Google Scholar]
- 16. Taha TE, Gray RH, Kumwenda NI, et al. HIV infection and disturbances of vaginal flora during pregnancy. J Acquir Immune Defic Syndr Hum Retrovirol 1999; 20:52–9. [DOI] [PubMed] [Google Scholar]
- 17. Rollins NC, Coovadia HM, Bland RM, et al. Pregnancy outcomes in HIV-infected and uninfected women in rural and urban South Africa. J Acquir Immune Defic Syndr 2007; 44:321–8. [DOI] [PubMed] [Google Scholar]
- 18. Lopez M, Figueras F, Hernandez S, et al. Association of HIV infection with spontaneous and iatrogenic preterm delivery: effect of HAART. AIDS 2012; 26:37–43. [DOI] [PubMed] [Google Scholar]
- 19. Ndirangu J, Newell ML, Bland RM, Thorne C. Maternal HIV infection associated with small-for-gestational age infants but not preterm births: evidence from rural South Africa. Hum Reprod 2012; 27:1846–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Briand N, Mandelbrot L, Le Chenadec J, et al. ; ANRS French Perinatal Cohort No relation between in-utero exposure to HAART and intrauterine growth retardation. AIDS 2009; 23:1235–43. [DOI] [PubMed] [Google Scholar]
- 21. Ekouevi DK, Coffie PA, Ouattara E, et al. ; International Epidemiological Database to Evaluate AIDS West Africa; ANRS 1269 and ANRS 12136 Study Groups in Abidjan Pregnancy outcomes in women exposed to efavirenz and nevirapine: an appraisal of the IeDEA West Africa and ANRS databases, Abidjan, Côte d’Ivoire. J Acquir Immune Defic Syndr 2011; 56:183–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Powis KM, Smeaton L, Ogwu A, et al. Effects of in utero antiretroviral exposure on longitudinal growth of HIV-exposed uninfected infants in Botswana. J Acquir Immune Defic Syndr 2011; 56:131–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chen JY, Ribaudo HJ, Souda S, et al. Highly active antiretroviral therapy and adverse birth outcomes among HIV-infected women in Botswana. J Infect Dis 2012; 206:1695–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Masson L, Barnabas S, Deese J, et al. Inflammatory cytokine biomarkers of asymptomatic sexually transmitted infections and vaginal dysbiosis: a multicentre validation study. Sex Transm Infect 201995:5–12. [DOI] [PubMed] [Google Scholar]
- 25. Masson L, Passmore JA, Liebenberg LJ, et al. Genital inflammation and the risk of HIV acquisition in women. Clin Infect Dis 2015; 61:260–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Passmore JA, Jaspan HB, Masson L. Genital inflammation, immune activation and risk of sexual HIV acquisition. Curr Opin HIV AIDS 2016; 11:156–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gomez-Lopez N, StLouis D, Lehr MA, Sanchez-Rodriguez EN, Arenas-Hernandez M. Immune cells in term and preterm labor. Cell Mol Immunol 2014; 11:571–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Taylor BD, Holzman CB, Fichorova RN, et al. Inflammation biomarkers in vaginal fluid and preterm delivery. Hum Reprod 2013; 28:942–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nugent RP, Krohn MA, Hillier SL. Reliability of diagnosing bacterial vaginosis is improved by a standardized method of Gram stain interpretation. J Clin Microbiol 1991; 29:297–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yuan S, Cohen DB, Ravel J, Abdo Z, Forney LJ. Evaluation of methods for the extraction and purification of DNA from the human microbiome. PLoS One 2012; 7:e33865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lennard K, Dabee S, Barnabas SL, et al. Microbial composition predicts genital tract inflammation and persistent bacterial vaginosis in South African adolescent females. Infect Immun 2017; 86:e00410–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010; 26:2460–1. [DOI] [PubMed] [Google Scholar]
- 33. Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011; 27:2194–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 2010; 7:335–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Dixon P. VEGAN, a package of R functions for community ecology. J Veg Sci 2003; 14:927–30. [Google Scholar]
- 36. Dhariwal A, Chong J, Habib S, King IL, Agellon LB, Xia J. MicrobiomeAnalyst: a web-based tool for comprehensive statistical, visual and meta-analysis of microbiome data. Nucleic Acids Res 2017; 45:W180–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Weiss S, Xu ZZ, Peddada S, et al. Normalization and microbial differential abundance strategies depend upon data characteristics. Microbiome 2017; 5:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. McDonald D, Price MN, Goodrich J, et al. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J 2012; 6:610–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Romero R, Hassan SS, Gajer P, et al. The composition and stability of the vaginal microbiota of normal pregnant women is different from that of non-pregnant women. Microbiome 2014; 2:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chehoud C, Stieh DJ, Bailey AG, et al. Associations of the vaginal microbiota with HIV infection, bacterial vaginosis, and demographic factors. AIDS 2017; 31:895–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Coleman JS, Hitti J, Bukusi EA, et al. Infectious correlates of HIV-1 shedding in the female upper and lower genital tracts. AIDS 2007; 21:755–9. [DOI] [PubMed] [Google Scholar]
- 42. Passmore JS, Jaspan HB. Vaginal microbes, inflammation, and HIV risk in African women. Lancet Infect Dis 2018; 18:483–4. [DOI] [PubMed] [Google Scholar]
- 43.Afolabi BB, Moses OE, Oduyebo OO. Bacterial vaginosis and pregnancy outcome in Lagos, Nigeria. Open Forum Infect Dis 2016; 3:ofw030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hillier SL, Nugent RP, Eschenbach DA, et al. Association between bacterial vaginosis and preterm delivery of a low-birth-weight infant. N Engl J Med 1995; 333:1737–42. [DOI] [PubMed] [Google Scholar]
- 45. Nelson DB, Hanlon A, Hassan S, et al. Preterm labor and bacterial vaginosis-associated bacteria among urban women. J Perinat Med 2009; 37:130–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Svare JA, Schmidt H, Hansen BB, Lose G. Bacterial vaginosis in a cohort of Danish pregnant women: prevalence and relationship with preterm delivery, low birthweight and perinatal infections. BJOG 2006; 113:1419–25. [DOI] [PubMed] [Google Scholar]
- 47. Slyker JA, Patterson J, Ambler G, et al. Correlates and outcomes of preterm birth, low birth weight, and small for gestational age in HIV-exposed uninfected infants. BMC Pregnancy Childbirth 2014; 14:7 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Parekh N, Ribaudo H, Souda S, et al. Risk factors for very preterm delivery and delivery of very-small-for-gestational-age infants among HIV-exposed and HIV-unexposed infants in Botswana. Int J Gynaecol Obstet 2011; 115:20–5. [DOI] [PubMed] [Google Scholar]
- 49. Powis KM, Kitch D, Ogwu A, et al. Increased risk of preterm delivery among HIV-infected women randomized to protease versus nucleoside reverse transcriptase inhibitor-based HAART during pregnancy. J Infect Dis 2011; 204:506–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Townsend CL, Tookey PA, Newell ML, Cortina-Borja M. Antiretroviral therapy in pregnancy: balancing the risk of preterm delivery with prevention of mother-to-child HIV transmission. Antivir Ther 2010; 15:775–83. [DOI] [PubMed] [Google Scholar]
- 51. Roberts L, Passmore JA, Mlisana K, et al. Genital tract inflammation during early HIV-1 infection predicts higher plasma viral load set point in women. J Infect Dis 2012; 205:194–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.