Abstract
Objective
Reliable longitudinal risk prediction for hospitalized patients is needed to provide quality care. Our goal is to develop a generalizable model capable of leveraging clinical notes to predict healthcare-associated diseases 24–96 hours in advance.
Methods
We developed a reCurrent Additive Network for Temporal RIsk Prediction (CANTRIP) to predict the risk of hospital acquired (occurring 48 hours after admission) acute kidney injury, pressure injury, or anemia 24 hours before it is implicated by the patient’s chart, labs, or notes. We rely on the MIMIC III critical care database and extract distinct positive and negative cohorts for each disease. We retrospectively determine the date-of-event using structured and unstructured criteria and use it as a form of indirect supervision to train and evaluate CANTRIP to predict disease risk using clinical notes.
Results
Our experiments indicate that CANTRIP, operating on text alone, obtains 74%–87% area under the curve and 77%–85% Specificity. Baseline shallow models showed lower performance on all metrics, while bidirectional long short-term memory obtained the highest Sensitivity at the cost of significantly lower Specificity and Precision.
Discussion
Proper model architecture allows clinical text to be successfully harnessed to predict nosocomial disease, outperforming shallow models and obtaining similar performance to disease-specific models reported in the literature.
Conclusion
Clinical text on its own can provide a competitive alternative to traditional structured features (eg, lab values, vital signs). CANTRIP is able to generalize across nosocomial diseases without disease-specific feature extraction and is available at https://github.com/h4ste/cantrip.
Keywords: deep learning, machine learning, artificial intelligence, natural language processing, medical informatics, decision support systems, clinical
OBJECTIVE
The Centers for Disease Control (CDC) estimates that 1 in every 25 acute care hospitalizations results in a healthcare-associated infection (HAI) and that at least 50% of HAIs are preventable.1,2 Not only are HAIs estimated to cost over $9.8 billion USD annually,2 but they are used to measure quality of care by the Centers for Medicare and Medicaid Services (CMS), with failure to prevent HAIs potentially resulting in financial penalties to the offending hospital. In addition to infections, other types of preventable hospital acquired or associated disease have been reported with high prevalence.3–5 Predicting such nosocomial (ie, hospital acquired) diseases has the potential to reduce costs and improve outcomes.
Predictive modeling is an active area of medical informatics research with over 107 studies published between 2011 and 2017.6 However, the majority of risk prediction frameworks in use today were developed before the adoption of the electronic health record (EHR) and typically rely on a small number of risk factors (eg, signs, social factors, basic measurements) easily assessable by the physician.7 Likewise, automatic approaches typically rely on extracting hand-chosen disease-specific features easily extracted from the structured portions of the EHR (eg, laboratory results, vital signs, and chart information). By contrast, we were interested in discovering whether the information documented in unstructured clinical narratives could supplement or exceed the traditional information contained in structured (ie, tabular) parts of the EHR by enabling more robust prognostication of disease.
Clinical notes typically document or summarize the most important positive and negative observations, potential diagnoses, findings, and treatments about the patient. Moreover, they often provide interpretation of the low-level information present in the patient’s chart or labs. More importantly, unlike structured data, unstructured data can provide important nuanced and contextual information not available in a tabular format including degrees of belief (eg, suggesting possible diagnoses or conditional treatments), relationships (eg, indicating which aspects of the clinical picture are being addressed with specific interventions), and interpretations (eg, indicating that a typically abnormal lab value is effectively normal given patient’s history). This type of rich data is notoriously difficult to incorporate in traditional data-science systems,8 but it is ideally poised for deep learning which can automatically discover and extract significant and meaningful features from raw data.9
To overcome the limitations of structured data and capture the longitudinal information in clinical notes, we present and evaluate a deep learning model harnessing clinical text for temporal risk prediction: reCurrent Additive Network for Temporal RIsk Prediction (CANTRIP). We show how CANTRIP can be trained without direct ground-truth risk labels to predict 3 nosocomial diseases 24–96 hours in advance: hospital acquired acute kidney injury (HAAKI), hospital acquired pressure injury (HAPI), and hospital acquired anemia (HAA). Note: in this study, we are interested only in predicting when and if the patient will develop nosocomial disease—we are not determining causality.
BACKGROUND AND SIGNIFICANCE
Risk prediction from EHR data has received considerable attention over the last decade,6 with the majority of approaches predicting a specific outcome or single disease. In a review of 107 risk prediction studies, Goldstein et al (2017) found that (a) most studies did not fully utilize the depth of information available about patients in the EHR, instead relying on a small predefined list of variables; and (b) most models neglected to consider longitudinal measures.6 Few studies considered clinical text. The use of clinical text was previously explored by Goodwin and Harabagiu to predict congestive heart failure for diabetic patients.10–12 Their methods, too, relied on a small number of predefined features. By contrast, CANTRIP does not rely on any prespecified set of features opting instead to consider all observations documented in each clinical note. This allows CANTRIP to be trained to potentially predict a large variety of diseases without requiring disease-specific feature engineering. To ensure the generalizability of CANTRIP, we apply the model to 3 common nosocomial diseases: hospital acquired acute kidney injury (HAAKI), hospital acquired pressure injury (HAPI), and hospital acquired anemia (HAA)—each with their own cohorts and experimental results.
Hospital acquired acute kidney injury (HAAKI)
Acute kidney injury (AKI) affects as many as 20% of all hospitalizations resulting in an estimated cost of $10 billion annually.5,13 AKI is associated with increased mortality, end-stage renal disease, and chronic kidney disease.13 It has been shown that even small increases in serum creatinine are associated with long-term damage and increased mortality.13 Current criteria for AKI, however, rely on markers of established kidney damage or impaired function, necessitating new approaches for earlier prediction of AKI before significant kidney damage is established.14 Prior work on AKI prediction has largely focused on limited patient populations and a small number of standard features.15–18 Mohamadlou et al (2018) used a Gradient Boosting Machine19 to predict severe AKI using the English National Health Service criteria as their gold standard and relying on vital signs and creatinine values as features.20 Tomašev et al (2019) present a deep learning approach using the KDIGO21 criteria as a gold standard and relying on historical aggregates of 29 numeric structured data elements.22 By contrast, our approach is the first to our knowledge to predict AKI or HAAKI using clinical notes. Moreover, ours is the first approach to predict AKI without relying on extracting AKI-specific features.
Hospital acquired anemia (HAA)
A substantial number of hospital patients with normal HgB on admission become anemic during the course of their hospitalization resulting in increased average length of stay by 10%–88%, hospital charges by 6%–80%, and risk of in-hospital mortality by 51%–228%, depending on HAA severity.3 HAA can result from a large number of factors, such as blood loss (including phlebotomy), erythropoietin deficiencies, nutritional deficiencies, hemolysis, and coagulation abnormalities.23 Thavendiranathan et al (2005) found that phlebotomy is highly associated with changes in HgB and hematocrit noting a mean decrease of 7.0 g/L HgB and 1.9% hematocrit with every 100 mL of blood drawn, while McEvoy et al (2013) indicate that critical care patients average 40–70 mL of blood drawn daily and that every 50 mL of blood drawn increases their risk of moderate to severe HAA by 18%.24,25 Consequently, the ability to automatically predict HAA would enable physicians to switch to small volume phlebotomy tubes, minimizing blood loss from in-dwelling catheters, and reducing blood tests.26 Indeed, Chant et al (2006) found that even small decreases in phlebotomy volume were associated with significantly reduced transfusion requirements in patients with prolonged stays.27 Although there has been some work on predicting anemias such as classifying iron deficiency anemia using artificial neural networks,28 or predicting moderate to severe anemia for patients with ulcerative colitis using logistic regression (LR),29 we were unable to find any prior work on developing automatic methods for predicting hospital acquired anemia whether using structured or unstructured data.
Hospital acquired pressure injury (HAPI)
The development of pressure injuries (ie, pressure ulcers or bed sores) can lead to several complications, including sepsis, cellulitis, osteomyelitis, pain, and depression.30 The mortality rate has been noted to be as high as 60% within 1 year of hospital discharge for older patients who develop a pressure ulcer during their stay.31 The Braden scale is the most widely used risk assessment scale for pressure ulcers.32 However, in an external evaluation, Hyun et al (2013) found that the Braden scale shows “insufficient predictive validity and poor accuracy in discriminating intensive care patients at risk of pressure ulcers developing.”33 Keller et al (2002) reported that, “there are no conclusive studies on the identification of pressure ulcer risk factors. None of the existing risk-assessment scales were developed especially for use in intensive care unit (ICU) patients.”4 Automatic prediction of pressure injuries was explored by Schoonhoven et al (2006),34 wherein LR was applied to a small number of structured features. By contrast, we show that our data-driven deep learning approach can reliably detect pressure ulcer for ICU patients without physician interaction or pre-specified feature extraction, allowing for potentially improved patient outcomes.
METHODS
We first present our cohort selection and data preprocessing approaches and then our proposed model and the evaluation against several baselines.
Cohort selection
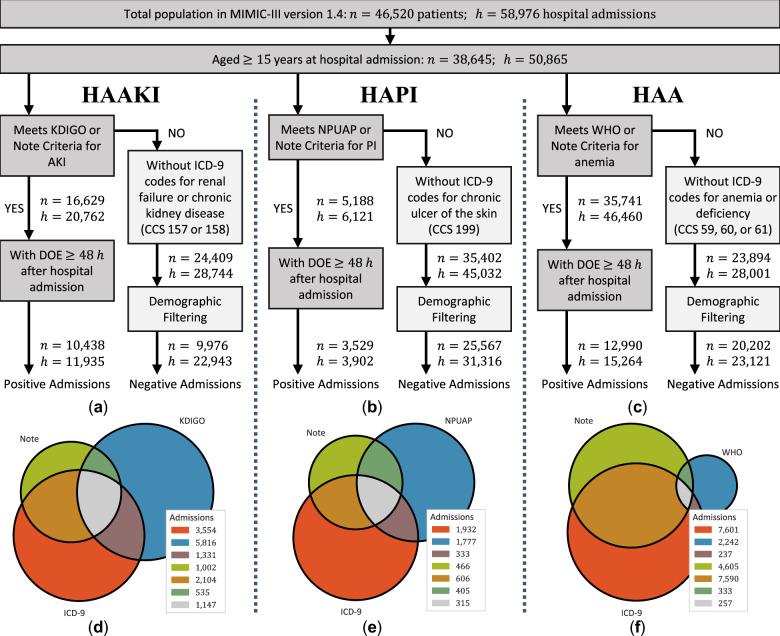
We selected our retrospective cohort, illustrated in Figure 1, from the MIMIC-III critical care database.35,36 MIMIC, developed by the Massachusetts Institute of Technology (MIT) Lab for Computation Physiology to support research in intelligent patient monitoring, is a freely available database containing deidentified health data associated with 46 520 patients. After excluding admissions with fewer than 2 days of notes, our final cohorts consisted of 34 878 hospital admissions for HAAKI (34.2% prevalence); 35 218 for HAPI (11.1% prevalence); and 38 385 for HAA (39.8% prevalence).
Figure 1.
Cohort selection diagram for each experimental cohort where (a), (b), and (c) describe the criteria used to distinguish between the positive and negative admissions for each disease; (d), (e), and (f) present the proportions of positive hospital admissions that met each type of criteria; discharge ICD-9 criteria were based on the Clinical Classifications Software (CCS) diagnosis and procedure categorization scheme provided as part of the Healthcare Cost and Utilization Project (HCUP); and DOE refers to the Date-of-Event for each disease. Admissions that met only ICD-9 criteria were omitted from this study as we were unable to determine their DOE.
Data preparation and preprocessing
To account for irregular gaps in the patient’s hospital visit, we adopt an abstract representation of the patient’s hospital visit which we call their clinical chronology. We represent the chronology as
a discrete, discontiguous sequence of snapshots, , where each snapshot encodes the clinical observations documented in any clinical notes produced on the same (calendar) day, and
a sequence of elapsed times, such that encodes the number of hours between and and encodes the number of hours between hospital admission and the first clinical note.
Natural language preprocessing
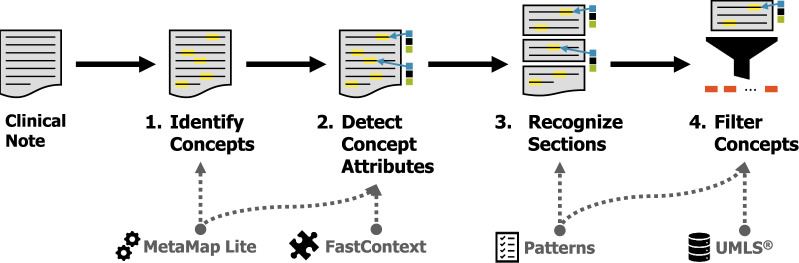
In this work, to evaluate the impact of clinical notes for predicting disease risk, we only considered the clinical observations documented in clinical notes. We extracted the set of observations from each clinical note in 4 steps, illustrated in Figure 2. An initial set of medical concepts corresponding to Unified Medical Language System (UMLS)37 entities was detected using MetaMap Lite.38 In order to account for the physician’s beliefs about each concept, we used FastContext,39 a high-performance reimplementation of ConText,40 to detect the following semantic attributes:
Figure 2.
Natural language preprocessing used to extract observations from clinical notes.
Negation indicating whether the observation was affirmed or negated;
Certainty indicating whether the author was certain or uncertain;
Temporality indicating whether the observation occurred in the present, the past, or is hypothetical; and
Experiencer indicating whether the observation was associated with the patient or someone else (eg, family).
Sections were recognized and normalized using a large number of hand-crafted regular expression rules previously created for InfoBot.41 We then filtered out all observations that (1) were not affirmed, certain, present, and associated with the patient; (2) occurred in a section corresponding to consults, family history, past medical history, or social history; (3) had a UMLS semantic type not corresponding to a medical problem, intervention, drug, or anatomic region; or (4) belonged to InfoBot’s medical stop word list. Semantic types, rules, and stop words are provided in online Supplementary Appendix A.
Determining the Date-of-Event
We determined the Date-of-Event (DOE) as the first date in which the disease is documented in a clinical note, or evidenced by the patient’s labs or chart. Specifically, for each disease, we defined 1 or more (a) seed concepts in the UMLS hierarchy, (b) lexical patterns, and (c) structured criteria using the laboratory, chart, and/or vital sign information in MIMIC. We determined the DOE as the first date in which (1) any observation extracted from a clinical note associated with that date descends from any of the UMLS seed concepts; (2) any observation or any text in the note contains any of the lexical patterns not immediately followed by a colon (to rule out structural matches, eg, “bed sore: none”); or (3) the structured criteria is met. Table 1 provides the seed concepts, lexical patterns, and structured criteria associated with each disease.
Table 1.
Criteria used to detect HAAKI, HAPI, and HAA from clinical notes or structured (eg, chart, laboratory) data, where CUI refers to a concept unique identifier in UMLS
| Disease | UMLS Seed CUI | Lexical pattern(s) | Structured criteria |
|---|---|---|---|
| HAAKI | C0022660 (Kidney Failure, Acute) | kidney failure, renal failure, kidney injury, renal injury, AKI | KDIGO11,12 |
| HAPI | C0011127 (Pressure Ulcer) | bed sore, bed ulcer, pressure sore, pressure ulcer, decub* sore, decub* ulcer | NPUAP13 |
| HAA | C0002871 (Anemia) | anemia, anaemia, HAA | WHO14 |
Abbreviations: AKI, acute kidney injury; CUI, concept unique identifier; HAA, hospital acquired anemia; HAAKI, hospital acquired acute kidney injury; HAPI, hospital acquired pressure injury; KDIGO, Kidney Disease Improving Global Outcomes; NPUAP, National Pressure Uncler Advisory Panel; UMLS, Unified Medical Language System; WHO, World Health Organization.
*represents a regular expression wildcard.
Encoding elapsed times
We encoded elapsed times using the sinusoidal representation proposed in Vaswani et al (2017),42 wherein the number of hours elapsed since the previous note, that is, is represented as a -dimensional vector consisting of pairs of sinusoidal projections with different frequencies:
where is the index into the vector , and is the number of hours between and . This representation was chosen because, for any offset , reduces to a linear function of . We also experimented with other encoding schemes and found the sinusoidal version to be the most effective across all evaluated systems.
Creating positive and negative examples
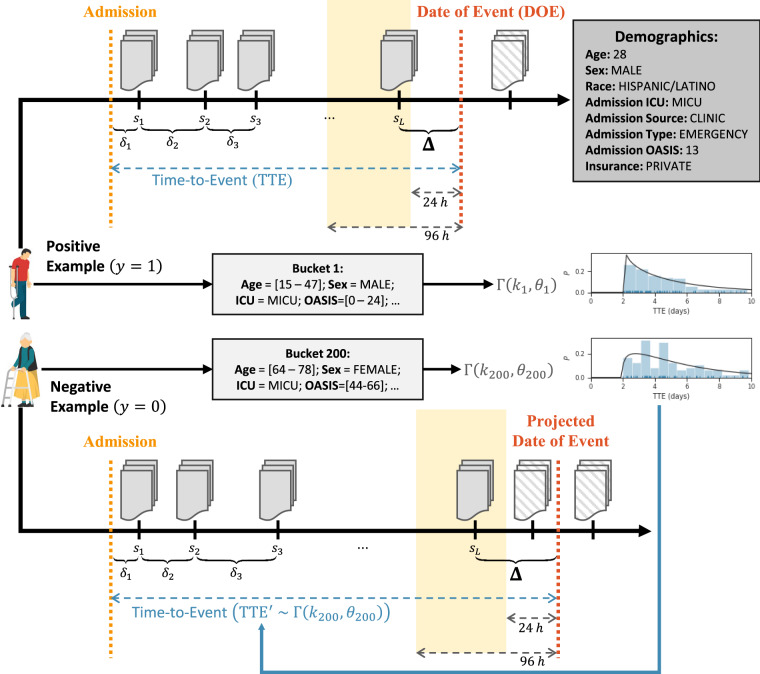
To train and evaluate our model without manually quantifying the risk of disease for each snapshot in each patient’s chronology, we used the DOE as a form of indirect supervision to produce positive and negative examples, as illustrated in Figure 3. Specifically, for each positive admission (ie, admissions with chronologies in which the patient eventually develops the disease) we created a labeled example by:
Figure 3.
How the prediction windows and chronologies are determined for positive and negative examples.
Truncating each chronology to end at the last snapshot occurring 24–96 hours before the DOE;
Defining the prediction window as the elapsed time (in hours) between the final snapshot (after truncation) and the DOE; and
Assigning the label .
To create negative examples, we first grouped positive admissions into buckets based on demographic and admission information including the patient’s age, sex, and race as well as their admitting ICU, source of admission (ie, clinic, physician, transfer, or other), type of admission (ie, elective, emergency, urgent), Oxford Acute Severity of Illness Score,43 and type of insurance (ie, government, private, Medicaid, Medicare, or self pay). For each bucket , we assumed the Time-to-Event (TTE, ie, the number of hours elapsed from hospital admission to DOE) followed a Gamma prior distribution (ie, ) and determined and using maximum likelihood estimates over each positive example in the bucket. This allowed us to create labels for our negative examples by:
Determining which bucket each negative example belonged to;
Sampling ;
Defining the DOE as either (a) the date obtained by projecting from the date of hospital admission or (b) the discharge date, whichever occurred first;
Truncating the chronology to end at the snapshot 24–96 hours before the DOE; and
Defining as the hours elapsed between the final snapshot (after truncation) and the DOE.
Note: negative examples assigned to a bucket without any positive examples were excluded (filtered) from the experiments for that disease (corresponding to “Demographic Filtering” in Figure 1).
Computational approaches
We evaluated 4 computational approaches for predicting nosocomial disease, namely: (1) our proposed CANTRIP, (2) a bidirectional LSTM44 network, and 2 shallow learning systems (3) LR and (4) a support vector machine45 (SVM).
Recurrent additive network for temporal risk prediction
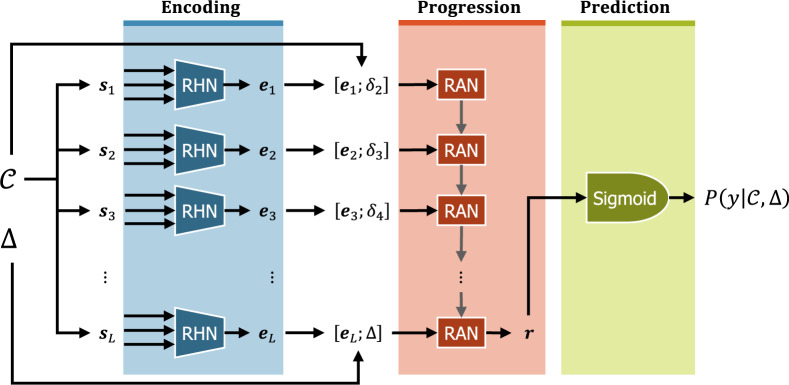
Given a clinical chronology and a prediction window , CANTRIP predicts the probability (ie, risk) that the patient will develop the disease during that window. We factorize this probability into 3 components, each corresponding to 1 of the modules illustrated in Figure 4:
where indicates whether the patient develops the disease (such that if the patient develops the disease, and otherwise), denotes the internal representations of the snapshots learned by Encoding module, is the internal representation clinical picture of the patient produced by the Progression module, and represents the elapsed time between successive snapshots. Details on each module are provided below.
Figure 4.
The reCurrent Additive Network for Temporal RIsk Prediction (CANTRIP).
Encoding clinical snapshots
The goal of the Encoding module in CANTRIP is to learn an optimal encoding of individual clinical snapshots. Formally, we define the vocabulary as the set of all unique clinical observations documented in the positive examples. This allows us to represent each clinical snapshot as a -length binary vector such that the -th element in indicates whether the -th observation in was observed in that snapshot. CANTRIP incorporates a Residual Highway Network46 (RHN) to learn an embedding for each clinical snapshot . Highway networks allow information to flow “around” or across multiple layers, enabling networks with hundreds of layers to be trained efficiently. We used 10 dense layers with batch normalization, regularization, and single-depth residual connections to produce the encoding of the clinical snapshot . All dense layers used Gaussian error linear unit47 activations as in Devlin et al (2019).48
Modeling disease progression
To account for the fact that clinical snapshots provide only an incomplete view of the clinical picture of the patient (ie, an EEG report is unlikely to describe a pressure injury or indicate anemia), we must infer the patient’s clinical picture by combining and accumulating information from each embedded clinical snapshot to model the progression of their disease.
We accomplished this by (1) casting the inferred clinical picture of the patient as the memory of a Recurrent Neural Network (RNN) and (2) training the RNN to accumulate information about the progression of the patient’s disease by processing each snapshot sequentially. Formally, for each encoded snapshot , the RNN is trained to predict the progression of the patient’s disease after elapsed time , such that when considering the final (ie, most recent) encoded snapshot and prediction window , the final output of the RNN, , encodes sufficient information to estimate the probability that the patient will develop the disease within days. We used a recurrent additive network49 (RAN) as our RNN implementation in CANTRIP.
RANs are a substantial simplification of LSTM44 units and gated recurrent units50 with nearly half the number of learnable parameters and have been shown to yield a number of advantages including avoiding the vanishing gradient problem, improving model performance, and significantly reducing model complexity. We believe that these properties make them ideally suited for deep learning with limited datasets.
Predicting disease risk
As all of the heavy lifting is accomplished by the Progression and Encoding modules, the disease risk is calculated by estimating the probability that the patient will develop the disease within days using a logistic sigmoid projection:
where denotes whether the patient develops the disease (ie, if the patient develops the disease, and otherwise), is the encoding of the inferred clinical picture produced by the Progression module, and and denote the learned weight vector and bias value.
Bidirectional LSTM
We also explored the use of a bidirectional LSTM44 network, using a single embedding layer for observations and the final state of the LSTM to predict disease risk as in CANTRIP. The bidirectional LSTM closely resembles CANTRIP if the RHN in the Encoding module were replaced by a single dense layer, and the RAN in the Progression module were replaced with a bidirectional LSTM.
Shallow learning approaches
We evaluated 2 shallow learning approaches: LR and SVMs.45 Both approaches used the set of observations in the final snapshot before the prediction window as their input features.
Evaluation
For each cohort we created training, development, and testing datasets using a stratified 8:1:1 random split based on the demographic and admission criteria illustrated in Figure 3 and, for positive examples, the type of label(s) associated with that chronology as illustrated in Figure 1d–f.51–53 For each system, we report the performance on the test set using the hyperparameters that provided the highest MCC (defined below) on the development set. Hyperparameter optimization is described in online Supplementary Appendix B. We incorporated temperature scaling for probability calibration using the development set.54
Metrics
We report 7 metrics to evaluate the performance of each system for each cohort: Accuracy, Sensitivity (the true positive rate, ie, Recall), Specificity (the true negative rate), Precision (the positive predictive value), the measure (the harmonic mean of Precision and Recall), the area under the Receiver Operating Characteristic (ROC) curve (AUC), and the Mathews correlation coefficient55 (MCC, a balanced measure useful for comparing systems on imbalanced data56). Due to the data imbalance (ie, the low prevalence of HAA, HAPI, and HAAKI) in our dataset, we primarily relied on MCC to compare systems. Additional details on metrics are provided in online Supplementary Appendix C.
RESULTS
Table 2 presents the performance obtained using the best configuration of each system for each disease: HAA, HAPI, and HAAKI. Across all 3 diseases, CANTRIP obtains the highest MCC, , Accuracy, and AUC. Interestingly, bi-LSTM obtains the highest Sensitivity for HAA and HAAKI, at the cost of having the lowest Specificity across the board. By contrast, the SVM exhibited the weakest performance for all 3 diseases. Interestingly, the SVM retained 90% of the positive examples as support vectors when training on all 3 diseases, indicating that nosocomial disease prediction from clinical notes cannot be accomplished focusing only on the most representative examples.
Table 2.
Performance of each evaluated system when predicting HAA, HAPI, and HAAKI 24–96 hours before documented in the clinical notes or directly evidenced by laboratory or chart data
| Disease | System | Accuracy | AUC | Sensitivity | Specificity | Precision | F 1 | MCC |
|---|---|---|---|---|---|---|---|---|
| HAA | LR | 64.58% | 65.55% | 47.55% | 74.51% | 52.09% | 49.72% | 0.2252 |
| SVM | 57.03% | 55.23% | 37.46% | 69.21% | 43.09% | 40.08% | 0.0687 | |
| biLSTM | 62.78% | 70.01% | 72.03% | 57.34% | 49.85% | 58.92% | 0.2844 | |
| CANTRIP | 69.64% | 74.61% | 57.56% | 76.75% | 59.30% | 58.42% | 0.3453 | |
| HAPI | LR | 76.87% | 74.34% | 57.91% | 79.64% | 29.37% | 38.98% | 0.2887 |
| SVM | 78.32% | 62.78% | 24.05% | 86.26% | 20.38% | 22.06% | 0.0961 | |
| biLSTM | 78.77% | 80.84% | 70.57% | 79.99% | 34.41% | 46.27% | 0.3844 | |
| CANTRIP | 83.61% | 87.05% | 71.83% | 85.36% | 42.19% | 53.16% | 0.4632 | |
| HAAKI | LR | 64.84% | 64.49% | 44.99% | 77.20% | 55.12% | 49.55% | 0.2327 |
| SVM | 57.07% | 55.24% | 37.69% | 69.13% | 43.18% | 40.25% | 0.0703 | |
| biLSTM | 67.50% | 74.04% | 71.97% | 64.69% | 56.16% | 63.09% | 0.3569 | |
| CANTRIP | 73.68% | 79.10% | 61.72% | 81.20% | 67.35% | 64.40% | 0.4370 |
Abbreviations: AUC, area under the curve; CANTRIP, reCurrent Additive Network for Temporal RIsk Prediction; biLSTM, bidirectional Long Short-Term Memory network; HAA, hospital acquired anemia; HAAKI, hospital acquired acute kidney injury; HAPI, hospital acquired pressure injury; LR, logistic regression; MCC, Matthews correlation coefficient; SVM, support vector machine.
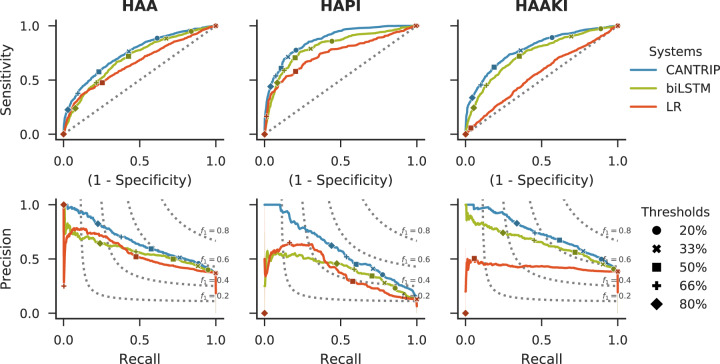
Figure 5 provides insights on system performance as illustrated by receiver operating characteristic and precision-recall curves.
Figure 5.
System performance as illustrated by receiver operating characteristic (ROC) and precision-recall (PR) curves with different decision thresholds indicated. Selecting the decision threshold can be accomplished by (1) determining a clinically actionable level of Precision for the target disease and (2) comparing potential thresholds on ROC and PR curves, with optimal thresholds between 33% and 50% for CANTRIP. In our experiments, we used a threshold of 50%.
DISCUSSION
As shown by Table 2, the highest performance for all systems was obtained when predicting HAPI, followed by HAAKI, and finally by HAA. This is unsurprising given that pressure injury risk factors such as mobility, color, texture, and wound care are often documented in free text. By contrast, anemia is typically defined in terms of HgB and hematocrit, which are only sporadically and inconsistently documented in clinical narratives.
In terms of HAA, Khan et al (2017)29 report an AUC of 69% when predicting moderate to severe anemia based on structured data (including diagnosis of mild anemia) using LR for 789 patients with newly diagnosed ulcerative colitis. CANTRIP obtains similar performance (75% AUC) despite also detecting mild anemias and without considering structured data such as albumin, HgB, or hematocrit.
When predicting HAPI, Schoonhoven et al (2006)34 report an AUC of 70% when using LR and a rule-based classifier on hand-chosen features. A retrospective analysis of the Braden scale reports an AUC of 62%, with 18% Precision and 29% .57 In another retrospective study of the Braden scale, Hyun et al (2013)33 report an AUC of 67%, with 14% Precision and 24% . While these results all use different criteria for pressure injury classification as well as different data sets and study designs, we can see that despite requiring no manual labeling, feature extraction, or physician interaction, CANTRIP obtains similar AUC (87%) and significantly higher Precision (42%) and (53%) compared to both manual and rule-based prediction systems designed specifically for predicting pressure injuries. This suggests that deep learning is able to extract meaningful signals for predicting pressure injury from clinical texts.
Finally, for HAAKI, Mohamadlou et al (2018)20 report an AUC of 67% with 80% Accuracy when predicting moderate to severe AKI up to 72 hours in advance based on structured data. CANTRIP obtains higher AUC (79%) when predicting any type of AKI (including mild) up to 96 hours in advance, based only on clinical notes. DeepMind reports an ROC AUC of 93% with 33% Precision and a Precision-Recall AUC of 29.7% when predicting AKI up to 48 hours in advance using data collected for 703 782 adult patients from all available sites in the US Department of Veterans Affairs and including 620 000 features.22 By contrast, with significantly less data, a more diverse population, and no AKI-specific feature extraction, CANTRIP obtains lower AUC (79%) but more than double Precision (67%) and double Precision-Recall AUC (74%) when predicting up to 96 hours in advance, indicating that clinical text alone provides important cues for predicting HAAKI.
In terms of probability calibration, we measured an Estimated Calibration Error54 of 2.69%, 1.66%, and 2.36%, for HAA, HAPI, and HAAKI, respectively, indicating that CANTRIP under-estimated the empirical probability of each disease and may benefit from site-specific calibration. Likewise, selection of a decision threshold depends on both the disease and the intended use of the model as guided by Figure 5. Reliability plots are provided in online Supplementary Appendix D.
Overall, the relative performance of CANTRIP, when compared to baseline and disease-specific models reported in the literature, suggests that not only can CANTRIP generalize across nosocomial diseases, but that clinical notes provide meaningful information for prognostication of disease.
Limitations
The primary limitation of this study is the fact that all systems relied only on features extracted from clinical notes. This was a deliberate design decision: while our ultimate goal is to combine textual and structured features, we were interested in first examining the power of text alone for predicting nosocomial disease. An additional limitation is that features only indicated the presence or absence of observations, signs, interventions, etc, meaning that values reported in the text, such as “HgB: 7.5,” are not available to the model. Rather than parsing and extracting this information, in future work, we aim to combine both clinical notes and structured data as features. For computational reasons, we chose to exclude hypothetical and negated mentions of observations; we expect that providing explicitly negated information to the model may further improve Specificity and is something we are exploring in future work. In future work, we plan to explore and further validate CANTRIP using patient data from other clinical sites.
CONCLUSION
We presented and evaluated a deep learning model harnessing clinical text to predict nosocomial disease from clinical notes for critical care patients. We showed how CANTRIP can be trained without direct ground-truth risk labels to predict 3 nosocomial diseases 24–96 hours in advance: HAAKI, HAPI, and HAA. Our experimental results indicate that not only does CANTRIP outperform traditional (shallow) learning approaches and a competitive deep learning baseline, but that despite considering only non-disease-specific features extracted from clinical notes, CANTRIP obtains competitive performance to disease-specific systems relying on hand-chosen structured features or hand-crafted rules.
FUNDING
This work was supported by the intramural research program at the US National Library of Medicine, National Institutes of Health.
AUTHOR CONTRIBUTIONS
TG and DDF conceptualized the study. DDF oversaw study design and reviewed and helped analyze the findings. TG designed and implemented the systems, collected and processed the data, performed data analysis, and conducted the experiments and evaluations. TG and DDF contributed to writing and editing of the manuscript.
Supplementary Material
ACKNOWLEDGMENTS
This work utilized the computational resources of the NIH HPC Biowulf cluster (http://hpc.nih.gov). We would like to thank Dr. Clement McDonald for his advice on criteria coding in MIMIC. Figure 2 was designed using assets from MacroVector at Freepik.com.
CONFLICT OF INTEREST STATEMENT
None declared.
REFERENCES
- 1. Magill SS, Edwards JR, Bamberg W, et al. Multistate point-prevalence survey of health care–associated infections. N Engl J Med 2014; 370 (13): 1198–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Schmier JK, Hulme-Lowe CK, Semenova S, et al. Estimated hospital costs associated with preventable health care-associated infections if health care antiseptic products were unavailable. Clincoecon Outcomes Res 2016; 8: 197–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Henderson JM, Blackstone EH, Hixson ED, et al. Hospital-acquired anemia: prevalence, outcomes, and healthcare implications. J Hosp Med 2013; 8: 506–12. [DOI] [PubMed] [Google Scholar]
- 4. Keller B, Wille J, van Ramshorst B, et al. Pressure ulcers in intensive care patients: a review of risks and prevention. Intensive Care Med 2002; 28 (10): 1379–88. [DOI] [PubMed] [Google Scholar]
- 5. Silver SA, Long J, Zheng Y, et al. Cost of acute kidney injury in hospitalized patients. J Hosp Med 2017; 12 (2): 70–6. [DOI] [PubMed] [Google Scholar]
- 6. Goldstein BA, Navar AM, Pencina MJ, et al. Opportunities and challenges in developing risk prediction models with electronic health records data: a systematic review. J Am Med Inform Assoc 2017; 24 (1): 198–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chase HS, Mitrani LR, Lu GG, et al. Early recognition of multiple sclerosis using natural language processing of the electronic health record. BMC Med Inform Decis Mak 2017; 17 (1): 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Demner-Fushman D, Chapman WW, McDonald CJ.. What can natural language processing do for clinical decision support? J Biomed Inform 2009; 42 (5): 760–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Collobert R, Weston J, Bottou L, et al. Natural language processing (almost) from scratch. J Mach Learn Res 2011; 12: 2493–537. [Google Scholar]
- 10. Goodwin T, Harabagiu SM. A probabilistic reasoning method for predicting the progression of clinical findings from electronic medical records. In: proceedings of the American Medical informatics Association (AMIA) Joint Summit on Clinical Research Informatics; March 25–27, 2015; San Francisco, California. [PMC free article] [PubMed]
- 11. Goodwin T, Harabiu SM. A predictive chronological model of multiple clinical observations. In: International Conference on Healthcare Informatics (ICHI); October 21–23, 2015; Dallas, TX.
- 12. Goodwin T, Harabagiu SM.. Inferring the interactions of risk factors from EHRs. AMIA Jt Summits Transl Sci Proc 2016; 2016: 78–87. [PMC free article] [PubMed] [Google Scholar]
- 13. Chertow GM, Burdick E, Honour M, et al. Acute kidney injury, mortality, length of stay, and costs in hospitalized patients. J Am Soc Nephrol 2005; 16 (11): 3365–70. [DOI] [PubMed] [Google Scholar]
- 14. Mehta RL, Pascual MT, Soroko S, et al. Spectrum of acute renal failure in the intensive care unit: the PICARD experience. Kidney Int 2004; 66 (4): 1613–21. [DOI] [PubMed] [Google Scholar]
- 15. Koyner JL, Adhikari R, Edelson DP, et al. Development of a multicenter ward-based AKI prediction model. Clin J Am Soc Nephrol 2016; 11 (11): 1935–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Flechet M, Güiza F, Schetz M, et al. AKIpredictor, an online prognostic calculator for acute kidney injury in adult critically ill patients: development, validation and comparison to serum neutrophil gelatinase-associated lipocalin. Intensive Care Med 2017; 43 (6): 764–73. [DOI] [PubMed] [Google Scholar]
- 17. Kate RJ, Perez RM, Mazumdar D, et al. Prediction and detection models for acute kidney injury in hospitalized older adults. BMC Med Inform Decis Mak 2016; 16 (1): 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Porter CJ, Juurlink I, Bisset LH, et al. A real-time electronic alert to improve detection of acute kidney injury in a large teaching hospital. Nephrol Dial Transplant 2014; 29: 1888–93. [DOI] [PubMed] [Google Scholar]
- 19. Friedman JH. Greedy function approximation: a gradient boosting machine. Ann Stat 2001; 29 (5): 1189–232. [Google Scholar]
- 20. Mohamadlou H, Lynn-Palevsky A, Barton C, et al. Prediction of acute kidney injury with a machine learning algorithm using electronic health record data. Can J Kidney Health Dis 2018;5. doi: 10.1177/2054358118776326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Khwaja A. KDIGO clinical practice guidelines for acute kidney injury. Nephron Clin Pract 2012; 120: c179–84. [DOI] [PubMed] [Google Scholar]
- 22. Tomašev N, Glorot X, Rae JW, et al. A clinically applicable approach to continuous prediction of future acute kidney injury. Nature 2019; 572 (7767): 116–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rawal G, Kumar R, Yadav S, et al. Anemia in intensive care: a review of current concepts. J Crit Care Med 2016; 2 (3): 109–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Thavendiranathan P, Bagai A, Ebidia A, et al. Do blood tests cause anemia in hospitalized patients? The effect of diagnostic phlebotomy on hemoglobin and hematocrit levels. J Gen Intern Med 2005; 20 (6): 520–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. McEvoy MT, Shander A.. Anemia, bleeding, and blood transfusion in the intensive care unit: causes, risks, costs, and new strategies. Am J Crit Care 2013; 22 (6): eS1–13. [DOI] [PubMed] [Google Scholar]
- 26. Harber CR, Sosnowski KJ, Hegde RM.. Highly conservative phlebotomy in adult intensive care: a prospective randomized controlled trial. Anaesth Intensive Care 2006; 34 (4): 434–7. [DOI] [PubMed] [Google Scholar]
- 27. Chant C, Wilson G, Friedrich JO.. Anemia, transfusion, and phlebotomy practices in critically ill patients with prolonged ICU length of stay: a cohort study. Crit Care 2006; 10 (5): R140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Azarkhish I, Raoufy MR, Gharibzadeh S.. Artificial intelligence models for predicting iron deficiency anemia and iron serum level based on accessible laboratory data. J Med Syst 2012; 36 (3): 2057–61. [DOI] [PubMed] [Google Scholar]
- 29. Khan N, Patel D, Shah Y, et al. A novel model for predicting incident moderate to severe anemia and iron deficiency in patients with newly diagnosed ulcerative colitis. Dig Dis Sci 2017; 62 (5): 1295–304. [DOI] [PubMed] [Google Scholar]
- 30. Brem H, Maggi J, Nierman D, et al. High cost of stage IV pressure ulcers. Am J Surg 2010; 200 (4): 473–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Thomas DR, Goode PS, Tarquine PH, et al. Hospital-acquired pressure ulcers and risk of death. J Am Geriatr Soc 1996; 44 (12): 1435–40. [DOI] [PubMed] [Google Scholar]
- 32. Bergstrom N, Demuth PJ, Braden BJ.. A clinical trial of the Braden Scale for Predicting Pressure Sore Risk. Nurs Clin North Am 1987; 22 (2): 417–28. [PubMed] [Google Scholar]
- 33. Hyun S, Vermillion B, Newton C, et al. Predictive validity of the Braden scale for patients in intensive care units. Am J Crit Care 2013; 22: 514–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Schoonhoven L, Grobbee DE, Donders ART, et al. Prediction of pressure ulcer development in hospitalized patients: a tool for risk assessment. Qual Saf Health Care 2006; 15 (1): 65–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Johnson AEW, Pollard TJ, Shen L, et al. MIMIC-III, a freely accessible critical care database. Sci Data 2016; 3 (1): 160035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Goldberger AL, Amaral LA, Glass L, et al. PhysioBank, PhysioToolkit, and PhysioNet: components of a new research resource for complex physiologic signals. Circulation 2000; 101 (23): E215–20. [DOI] [PubMed] [Google Scholar]
- 37. Lindberg DA, Humphreys BL, McCray AT.. The Unified Medical Language System. Methods Inf Med 1993; 32: 281–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Demner-Fushman D, Rogers WJ, Aronson AR.. MetaMap Lite: an evaluation of a new Java implementation of MetaMap. J Am Med Inform Assoc 2017; 24 (4): 841–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Shi J, Hurdle JF.. Trie-based rule processing for clinical NLP: a use-case study of n-trie, making the ConText algorithm more efficient and scalable. J Biomed Inform 2018; 85: 106–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chapman WW, Hillert D, Velupillai S, et al. Extending the NegEx lexicon for multiple languages. Stud Health Technol Inform 2013; 192: 677–81. [PMC free article] [PubMed] [Google Scholar]
- 41. Demner-Fushman D, Seckman C, Fisher C, et al. A prototype system to support evidence-based practice. AMIA Annu Symp Proc 2008; 2008: 151–5. [PMC free article] [PubMed] [Google Scholar]
- 42. Vaswani A, Shazeer N, Parmar N, et al. Attention is all you need In: Guyon I, Luxburg UV, Bengio S, et al. , eds. Advances in Neural Information Processing Systems 30: Annual Conference on Neural Information Processing Systems 2017; December 4–9, 2017: 5998–6008; Long Beach, CA. http://papers.nips.cc/paper/7181-attention-is-all-you-need.pdf Accessed September 18, 2019. [Google Scholar]
- 43. Johnson AEW, Kramer AA, Clifford GD.. A new severity of illness scale using a subset of Acute Physiology and Chronic Health Evaluation data elements shows comparable predictive accuracy. Crit Care Med 2013; 41 (7): 1711–8. [DOI] [PubMed] [Google Scholar]
- 44. Hochreiter S, Schmidhuber J.. Long short-term memory. Neural Comput 1997; 9 (8): 1735–80. [DOI] [PubMed] [Google Scholar]
- 45. Cortes C, Vapnik V.. Support vector machine. Mach Learn 1995; 20 (3): 273–97. [Google Scholar]
- 46. Srivastava RK, Greff K, Schmidhuber J. Training very deep networks. In: proceedings of the 28th International Conference on Neural Information Processing Systems - Volume 2. Cambridge, MA: MIT Press; 2015: 2377–85.
- 47. Hendrycks D, Gimpel K. Gaussian Error Linear Units (GELUs). arXiv preprint arXiv:1606.08415. Published Online First: 27 June 2016. http://arxiv.org/abs/1606.08415 Accessed September 12, 2019.
- 48. Devlin J, Chang M-W, Lee K, et al. BERT: pre-training of deep bidirectional transformers for language understanding. In: proceedings of the 2019 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, Volume 1 (Long and Short Papers); June 2–7, 2019; Minneapolis, MN.
- 49. Lee K, Levy O, Zettlemoyer L. Recurrent Additive Networks. arXiv preprint arXiv:1705.07393. Published Online First: 21 May 2017.
- 50. Glorot X, Bordes A, Bengio Y. Deep sparse rectifier neural networks. In: Gordon G, Dunson D, and Dudík M, eds. proceedings of the 14th International Conference on Artificial Intelligence and Statistics; April 11–13, 2011; PMLR: Fort Lauderdale, FL. http://www.jmlr.org/proceedings/papers/v15/glorot11a/glorot11a.pdf Accessed September 21, 2016.
- 51. Sechidis K, Tsoumakas G, Vlahavas I.. On the stratification of multi-label data In: Gunopulos D, Hofmann T, Malerba D, et al. , eds. Machine Learning and Knowledge Discovery in Databases. Heidelberg: Springer; 2011: 145–58. [Google Scholar]
- 52. Szymański P, Kajdanowicz T.. A network perspective on stratification of multi-label data In: Torgo L, Krawczyk B, Branco P, et al. , eds. Proceedings of the First International Workshop on Learning with Imbalanced Domains: theory and Applications. ECML-PKDD. Skopje, Macedonia: PMLR; 2017: 22–35. [Google Scholar]
- 53. Szymański P, Kajdanowicz T. A scikit-based Python environment for performing multi-label classification. arXiv preprint arXiv:1702.01460. Published Online First: 5 February 2017. http://arxiv.org/abs/1702.01460 Accessed September 16, 2019.
- 54. Guo C, Pleiss G, Sun Y, et al. On calibration of modern neural networks. In: proceedings of the 34th International Conference on Machine Learning - Volume 70. JMLR.org 2017. 1321–1330. http://dl.acm.org/citation.cfm? id=3305381.3305518 Accessed 27 November, 2019.
- 55. Matthews BW. Comparison of the predicted and observed secondary structure of T4 phage lysozyme. Biochim Biophys Acta BBA - Protein Struct 1975; 405 (2): 442–51. [DOI] [PubMed] [Google Scholar]
- 56. Boughorbel S, Jarray F, El-Anbari M.. Optimal classifier for imbalanced data using Matthews Correlation Coefficient metric. PLoS One 2017; 12 (6): e0177678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Cho I, Noh M.. Braden Scale: evaluation of clinical usefulness in an intensive care unit. J Adv Nurs 2010; 66 (2): 293–302. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.