Abstract
Motivation
Recent advances in computational methods have facilitated large-scale sampling of protein structures, leading to breakthroughs in protein structural prediction and enabling de novo protein design. Establishing methods to identify candidate structures that can lead to native folds or designable structures remains a challenge, since few existing metrics capture high-level structural features such as architectures, folds and conformity to conserved structural motifs. Convolutional Neural Networks (CNNs) have been successfully used in semantic segmentation—a subfield of image classification in which a class label is predicted for every pixel. Here, we apply semantic segmentation to protein structures as a novel strategy for fold identification and structure quality assessment.
Results
We train a CNN that assigns each residue in a multi-domain protein to one of 38 architecture classes designated by the CATH database. Our model achieves a high per-residue accuracy of 90.8% on the test set (95.0% average per-class accuracy; 87.8% average per-structure accuracy). We demonstrate that individual class probabilities can be used as a metric that indicates the degree to which a randomly generated structure assumes a specific fold, as well as a metric that highlights non-conformative regions of a protein belonging to a known class. These capabilities yield a powerful tool for guiding structural sampling for both structural prediction and design.
Availability and implementation
The trained classifier network, parser network, and entropy calculation scripts are available for download at https://git.io/fp6bd, with detailed usage instructions provided at the download page. A step-by-step tutorial for setup is provided at https://goo.gl/e8GB2S. All Rosetta commands, RosettaRemodel blueprints, and predictions for all datasets used in the study are available in the Supplementary Information.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
While the 3D structures of proteins arise as a consequence of their amino acid sequences, computational design methods operate by creating sequences that minimize the energy of a pre-defined protein backbone. This approach has provided solutions to challenging design problems ranging from enzyme catalysis to viral inhibition (Bialas et al., 2016; Fleishman et al., 2011; Jiang et al., 2008; Joh et al., 2017; Pejchal et al., 2011; Röthlisberger et al., 2008; Smadbeck et al., 2014; Tinberg et al., 2013). Computational design begins with defining a set of constraints that constitute the design problem, followed by either acquiring a natural backbone [from the Protein Data Bank (PDB)] or building one from scratch (de novo design). Once a backbone is defined, amino acid sequences are designed onto it (Leaver-Fay et al., 2011).
Obtaining a suitable backbone is the most challenging step in computational protein design. Highly evolved native backbones tolerate only minimal deviations from their original sequences, leading to a restricted set of solutions for a given design problem. De novo design, which uses physical and chemical principles to build proteins from scratch, offers at least three advantages over native-based methods (Huang, et al., 2016b). First, because the role of each residue in a de novo structure is precisely defined, the range of tolerated modifications is well understood, allowing for more controlled customization. Second, de novo proteins often have greater thermodynamic stability, which facilitates the introduction of function (Bloom et al., 2006; Tokuriki et al., 2008). Lastly, by not being restricted to existing structures, de novo backbones offer solutions otherwise unattainable from native scaffolds, and are likely more adaptable to a wider range tasks (Huang et al., 2016b).
Despite these advantages, de novo design is challenging because it requires the construction of a protein with neither a known structure nor sequence. Since the vastness of the protein torsion space (φ, ψ, χ) prohibits an exhaustive search, current de novo design protocols generate backbones by combining small, continuous segments of φ-ψ torsions (‘fragments’) collected from the PDB. While this allows for a significant reduction in search space, an enormous amount of sampling is still required to find the lengths, types (e.g. helix, beta-sheet) and order of secondary structure elements (here on, ‘topologies’) that result in viable structures. The process of identifying successful models and topologies currently relies on a combination of scoring functions, hydrogen-bonding patterns, and other discernible regularities to screen for models that satisfy the desired criteria (Brunette et al., 2015; Dou et al., 2018; Huang et al., 2016a; Koga et al., 2012; Marcos et al., 2017, 2018; Rocklin et al., 2017). However, such heuristics are often subjective and chosen ad hoc, and there are currently no unbiased, generalizable methods that can perform automated structure selection based on the overall organization of a protein.
In the field of computer vision, convolutional neural networks (CNNs) have revolutionized pattern-recognition tasks ranging from facial recognition (Schroff et al., 2015) to object detection (Redmon et al., 2015). In applications to proteins, several groups have used 1D and 3D CNNs to process protein sequence and structure data, performing tasks such as domain prediction (Hou et al., 2018) and mutation stability evaluation (Torng and Altman, 2017). Nonetheless, several limitations have prevented the use of these models in protein design. In the case of 1D CNNs, low-dimensionality often results in loss of important features needed to describe realistic structures. In the case of 3D CNNs, model sizes (i.e. the number of weights in a network) scale quickly with input size, resulting in significant hardware requirements for efficient processing of full-length protein structures. Most 3D representations also lack rotational and translational invariance, thus requiring large quantities of data to train deep models. Surprisingly, few reported studies have used 2D CNNs to perform protein structure analysis, despite the fact that 2D CNNs are the most well-studied and widely implemented class of neural network.
In this study, we demonstrate how CNNs intended for 2D image processing can be used as powerful 3D structure analysis tools to guide computational protein design. A number of recently reported 2D CNN architectures have enabled advanced forms of image classification, namely instance (Tokuoka et al., 2018) and semantic (Long et al., 2014) segmentation, which predict class labels for individual pixels as opposed to entire images—segmentation provides information about where exactly an object is in an image, rather than simply indicating whether or not an object is present. We speculated that this capability is pertinent to protein structure analysis and hypothesized that image segmentation networks could be adapted to create a model that quantifies structural features at various scales within a protein (e.g. per-residue, per-domain).
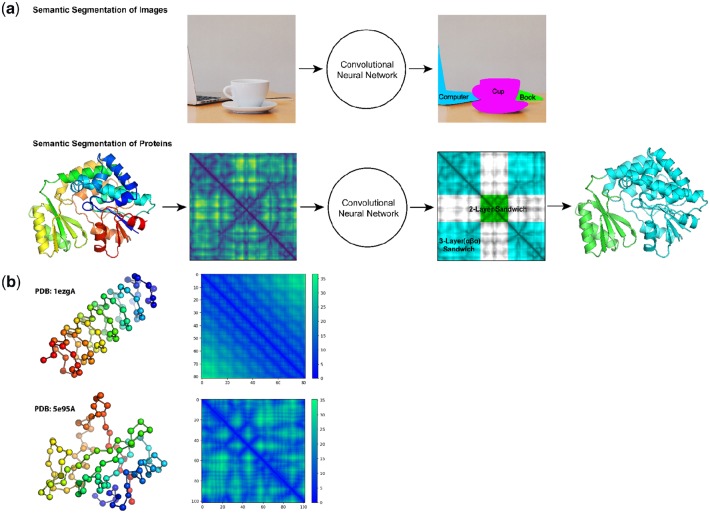
We trained a 2D CNN that classifies each residue in a protein into one of 38 architecture classes designated by the CATH (Dawson et al., 2017) protein database (Fig. 1a). We represent a protein using the pairwise distance matrix (here on, ‘contact map’) between all α-carbons in its structure (Anand and Huang, 2018; Wang et al., 2017) (Fig. 1b). This representation is rotationally and translationally invariant, and provides an image-like rendering of 3D protein structures in 2D. In the same way that image segmentation predicts a class for the basic unit of an image—a pixel, our model predicts classes for the basic unit of a protein—a residue.
Fig. 1.
Semantic segmentation of protein structures using contact maps. (a) A comparison of semantic segmentation for objects in images, and domains in proteins. The two-domain protein shown (PDB ID: 1mlaA) contains a 3-layer-αβα-sandwich (cyan), and a 2-layer sandwich (green). Multiple distal regions in a contact map can correspond to a single domain in a protein. The example segmentation and contact maps are real inputs and outputs obtained from our model. (b) Two examples of single-domain proteins colored as chainbows (left), with α-carbons shown as spheres. Red indicates the beginning, and blue indicates the end of the chain. The corresponding contact maps are shown to the right of each protein with units in Å. Axes correspond to amino acid index
While the most obvious function of our network is in protein classification, its true significance lies in its utility as a de novo design tool. On the single-residue level, Shannon entropies (Shannon and Weaver, 1964) of the CNN-predicted probability distributions can be used as indicators of local structure quality, allowing for identification of low-confidence regions that require further refinement or reconstruction. On the full-protein level, per-residue class probabilities predicted by our classifier can be averaged across an entire protein to provide a quantitative measure of the degree to which a structure assumes a fold. This function is useful for quickly searching large design trajectories for proteins that adopt specific architectures.
2 Materials and methods
2.1 Dataset
Training a CNN for semantic segmentation requires large amounts of densely labeled data, which are often expensive and or difficult to obtain. In the case of proteins, this corresponds to a large collection of proteins with a class assignment for each residue in every protein. Databases such as CATH (Dawson et al., 2017) and SCOPe (Fox et al., 2014) provide this information in the form of residue-level domain boundary assignments and classifications. The dataset used in this study was obtained from CATH v4.2, which is comprised of 132 380 unique, fully annotated protein chains (42 architecture classes, 202 506 domains, 3.3 × 107 residues). From this set, chains shorter than 520 residues that did not contain domains belonging to classes with fewer than 10 members were selected for use. Structures larger than 512 were center-cropped, and smaller structures were zero-padded to 512. The resulting dataset contains 126 069 chains (38 architecture classes, 181 753 domains, 2.9 × 107 residues) spanning 95% of all unique chain data. Selection did not drastically alter the overall structure of the data (Supplementary Figure S1a–c). Of the selected chains, 8000 were reserved for each the test and development sets and the remaining 110 069 were used in training. The dataset is highly imbalanced with the largest class containing nearly 7 million residues and the smallest containing less than 3500. To address this, the split was performed in a stratified manner, but stochastically adjusted so that all sets had at least 650 residues from each class present (Supplementary Fig. S1d–f). During training, examples were weighted to ensure that each class had equal influence.
2.2 Model architecture and implementation
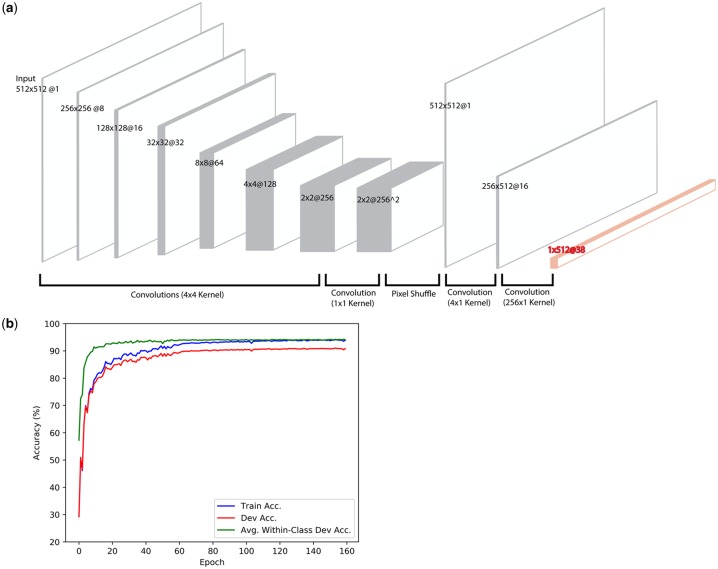
Our architecture is comprised of six convolutional layers (‘encoding’), followed by a ‘pixel-shuffle’ step (Shi et al., 2016) for upsampling to a 512×512 feature map (‘decoding’). Rectangular convolutions are then used to reshape the map into a 1×512@38 tensor (Fig. 2a, orange) that is passed to a Softmax function (Goodfellow et al., 2016). Each convolutional layer in the encoding phase is followed by a BatchNorm (Goodfellow et al., 2016) and a LeakyReLu (Goodfellow et al., 2016). The 4×1 convolution is followed only by a LeakyReLu, and the final layer passes directly into the Softmax function.
Fig. 2.
Model architecture and training. (a) The model is comprised of six down-sampling convolutional layers (encoding), followed by a ‘pixel shuffle’ step for up-sampling to a 512 × 512 map (decoding). Rectangular convolutions are used to reshape the feature map into a 1 × 512@38 which corresponds to a probability distribution over the 38 CATH architecture classes for each residue (orange). (b) Accuracy profiles of the training and development sets during training. The model converges within 100 epochs, achieving a residue-wise training accuracy above 90%. The performance generalizes to the test set, where the model achieves a per-residue accuracy of 90.8%, a per-class accuracy of 95.0% and a per-structure accuracy of 87.8%
The model was implemented in the PyTorch deep learning framework (Paszke et al., 2017). Training was performed for a total of 160 epochs with a mini-batch (Goodfellow et al., 2016) size of 64 using the Adam optimization algorithm. A learning rate of 0.001 was used for the first 60 epochs, 0.0001 for an additional 100 epochs. The loss function is a cross entropy loss (Goodfellow et al., 2016) averaged across every residue in the input chain. Importantly, the loss was weighted so that training examples from under-represented classes had increased influence on training. Weights were chosen so that each class, in total, had equal influence. Dropout regularization (Goodfellow et al., 2016) was used throughout the convolutional layers in the encoding phase with a zeroing-probability of 0.1. All weights were initialized using Xavier initialization (Glorot and Bengio, 2010). Model inputs were scaled by −100 and not normalized.
2.3 A note on accuracy metrics
In our formulation of the residue classification problem, the job of the network is to predict the correct class given the residue’s environment, where each residue is treated as an independent classification problem. Importantly, our model does not provide any information about domain boundaries because the network assigns a structural class to each residue, but does not group the residues into domains. Our accuracy metric is thus the proportion of residues that the model has correctly classified. Per-structure and per-class accuracy are more stringent versions of this metric—the model may be able to achieve high accuracies over all residues, but a good classifier should be accurate within each class, and should also have a high accuracy within each structure. To compute the average per-class accuracy, we compute the accuracy within each class and then average over all classes. Assuming that 90% of all beta-barrel residues, and 70% of all 2-layer sandwich residues are predicted correctly, this would yield an average per-class accuracy of 80%. However, if there are many more beta-barrel residues than 2-layer sandwich residues, the accuracy over all residues might be significantly higher than 80%, which is why these distinctions are necessary. Per-structure accuracy is computed similarly. The domain parser model (described in Supplementary Information), is characterized using an analogous accuracy metric, but instead of architecture classes, the domain parser is required to assign each residue to the correct domain.
3 Results
Our motivation for using a per-residue classifier as a protein evaluation tool is based on the assumption that the environment of each residue is rich in structural information (Mackenzie et al., 2016), which can be aggregated to describe higher level features such as domain architecture, secondary structure organization and structure quality. Importantly, the goal of our study is not to create a protein classifier, but rather to show how a classifier can be used to quantify residue-level environments in the form of probability distributions, which can then be averaged over regions of a protein to obtain information at the desired size scales. Our classifier differs from past protein classification methods (Dawson et al., 2017; Fox et al., 2014) in two ways. First, it operates on protein backbone distance information alone and is completely sequence agnostic. Second, because each residue is treated as an independent classification problem, our network does not explicitly predict which domain each residue belongs to. In the Supplementary Information, we describe how our network can be adapted to provide conventional domain predictions, and compare its performance to prior domain-parsing models.
3.1 Performance as a per-residue classifier
Class predictions for a row (or column) in a contact map are effectively a prediction in 3D space, because each row can be mapped to the location of a residue in the original structure. Using this fact, we trained a CNN to perform semantic segmentation on proteins, which entails predicting one of 38 architectural classes for each residue. In the same way that image segmentation models assess the context of individual pixels in an image, our CNN evaluates local features in the contact map, which encode structural features centered around a residue in 3D space.
A diagram of our CNN is shown in Figure 2a, with additional technical details provided in Section 2. The network converged within 100 epochs of training (Fig. 2b) and achieves average test accuracies of 90.8% per residue, 95.0% per architecture class and 87.8% per structure. Because the proportion of the dataset comprised by each class is highly variable, one concern is that the model may have simply learned to predict frequently occurring classes. Figure 3a suggests otherwise, as there is no correlation between the frequency of a class in the training set and class accuracy in the test set. The majority of class accuracies are above 90% even for the rarest classes (Fig. 3a and b). The class with the highest error rate was the unassigned/loop class at ∼40%. This is largely due to the fact that in CATH, unassigned regions of proteins include not only loops, but also peptide fragments that are not fully resolved in the original structures. The unassigned class thus contains a large number of anomalies that can be difficult for our model to predict. A noticeable trend in Figure 3a is that architectures with features similar to those of other classes (e.g. 4,5,6,7-propellers, αβα, ββα, βαβ-3-layer sandwiches) tended have slightly higher error rates, suggesting that the model may have some difficulty distinguishing between these architectures. While classes with highly similar architectures tended to be confused with one another, this occurs at a very low rate (Fig. 3b). Additionally, the unassigned/loop class tended to be confused with multiple classes, which was expected for reasons described above. Architectures such as aligned prism and trefoil which have distinct structural features, tended to have lower error rates.
Fig. 3.
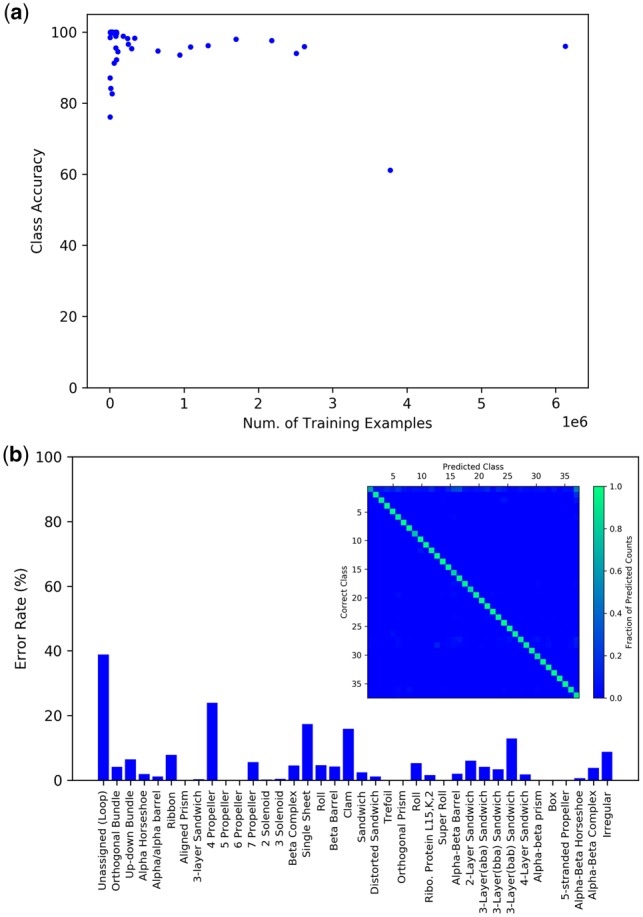
Analysis of classifier performance. (a) A plot of the number of residues in the training set belonging to a specific class, versus the class accuracy in the test set. Each point in the plot represents a single class. No obvious correlation between the number of examples in the training set and class accuracy on the test set is observed. The majority of class accuracies are above 90% even for the rarest classes. (b) Error rates for each architecture class. The confusion matrix is shown as an insert with architectures indexed in the same order as on the horizontal axis of the bar graph. The vertical axis of the confusion matrix indicates the correct class. The horizontal axis indicates the class predicted by the model. Each column is normalized to the total number of predicted counts for each class. An enlarged version of the confusion matrix is provided in Supplementary Figure 8
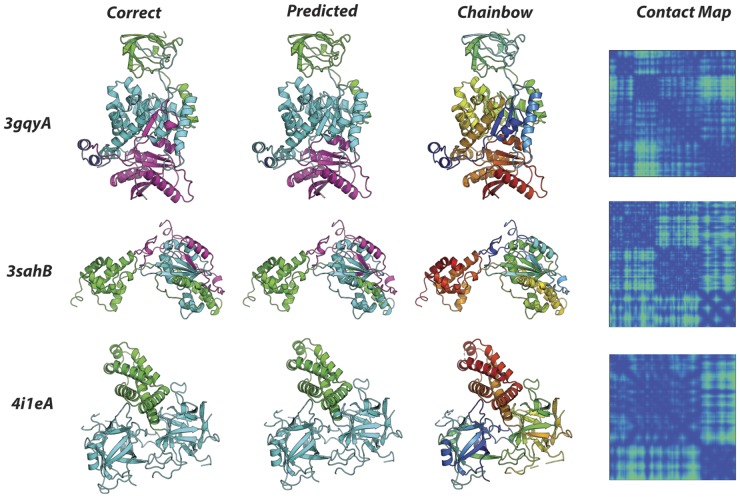
Example outputs (PDB IDs: 3gqyA, 3sahB, 4i1eA) are shown in Figure 4. Overall the model is able to closely reproduce per-residue architecture classifications provided by the CATH database. Inspection of outputs suggests that our model is able to accurately classify individual residues, even if they constitute fragments of a domain that are isolated in sequence space. This is illustrated in the case of the central αβ-barrel in 3gqyA (Fig. 4, 3gqyA classifications, cyan), which is comprised of multiple fragments from distal regions in primary sequence (Fig. 4, 3gqyA, chainbow). This observation suggests that the network is able to correlate delocalized features in the 2D contact map corresponding to the same domain in the 3D structure. Additionally, our network is able to recognize differences in secondary structure organization. For example, both the orthogonal-bundle in 3sahB (Fig. 4, 3sahB, green) and the α-horseshoe in 4i1eA (Fig. 4, 4i1eA, green) are purely alpha-helical in content, and both are comprised of residues in contiguous sequence. Although these architectures differ only in their secondary structure organization, the model is able to successfully differentiate between the two.
Fig. 4.
Classifications and Corresponding Contact Maps. Segmented examples from the test set. From the left, each column depicts: the correctly segmented structure, the model prediction, protein chain colored in residue order (rainbow order), and the input contact map. PDB ID's for each protein are shown on the left. 3gqyA: 86.9% within-structure accuracy: beta-barrel (green), alpha-beta barrel (cyan), loop (dark blue), 3-layer-αβα(magenta). 3sahB: 89.0% within-structure accuracy: orthogonal-bundle (green), 2-layer sandwich (cyan), loop (magenta). 4i1eA: 99.2% within-structure accuracy: alpha-horseshoe (green), trefoil (cyan)
Our model performs well at domain boundaries and provides accurate predictions even at complicated boundaries such as those in Figure 4, 4i1eA. In cases such as the central αβ-barrel in Figure 4, 3gqyA our model arguably provides a more intuitive classifications than CATH’s—while CATH predicts one of the central β-strands and one of the outer α-helices as belonging to the αβα-3-layer sandwich class, our model reasonably assigns both secondary elements to the central barrel. This phenomenon is likely due to the fact that CATH classifications are guided by sequence, while our predictions are informed by structure information alone.
A characteristic of our model that distinguishes it from most conventional structure classification algorithms (Dawson et al., 2017; Fox et al., 2014) is that it is sequence-agnostic. This is particularly important for de novo proteins, which have sequences that lack homology with naturally occurring families. The de novo designed TIM barrel structure, sTIM-11 (PDB ID: 5bvl), is one such example (Huang et al., 2016a). sTIM-11 belongs unequivocally to the αβ-barrel architecture class; however, it remains unclassified in both CATH (Dawson et al., 2017) and SCOPe (Fox et al., 2014), the two largest protein classification database, likely due to its lack of sequence homology to native TIM barrels. Our model is able to correctly classify this structure.
3.2 Class probabilities encode generalized structural features
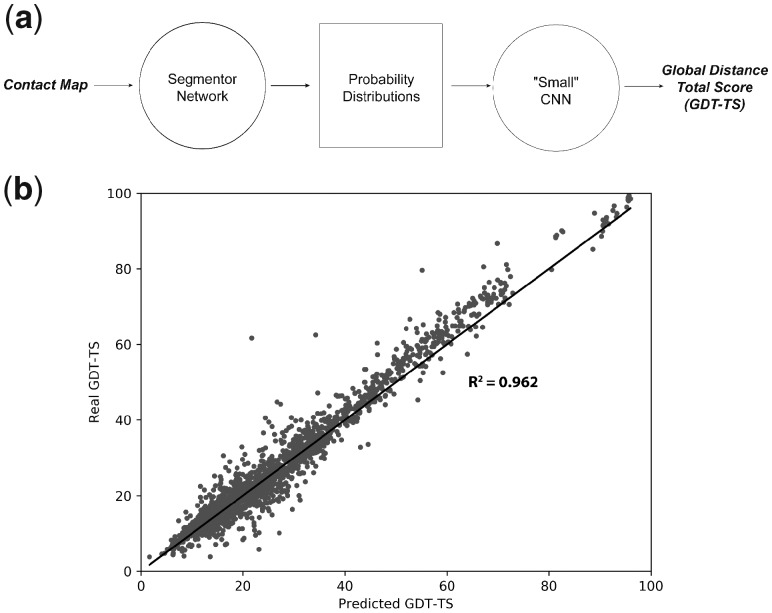
Although our model is able to achieve high classification accuracies, accuracy metrics alone provide little information about the generalizability of the features it has learned. Given that neural networks are highly flexible, it is possible that our classifier is fitting to features such as protein length, which may correlate with architecture class but provide little structural information. To control for such possibilities, we tested whether the features encoded by our classifier could generalize to other structure evaluation tasks (viz. transfer-learning, Goodfellow et al., 2016). Specifically, we trained a ‘small’ 6-layer CNN and assessed its ability to predict the Global Distance Test Total Score (Zemla et al., 1999) (here on, GDT) of CASP submissions (Moult et al., 2018). The input to the small network is the 1×512@38 layer of the classifier network, which encodes the probability distributions over the 38 architecture classes at each residue (Fig. 2a, orange)—this is effectively a compressed representation of a protein, encoded by our classifier (Fig. 5a). During training, the architecture and weights of the original model were not modified, and the small network was used only to process and extract structural information from the probability distributions.
Fig. 5.
GDT-TS predictions for 1960 submissions for 98 CASP target domains. (a) A schematic of the GDT-TS prediction scheme. (b) A plot of real GDT-TS versus the GDT-TS predicted by the small model. The identity function is shown as a black line. Each point corresponds to a single decoy from the test set, which consists of 20 randomly chosen submissions for each of the 98 targets (1960 total decoys). The small model was able to accurately predict the GDT-TS of the CASP submissions, yielding an R-squared value of 0.962 between the real and predicted values. The 98 selected targets were exclusively in the free-modeling categories of CASP10–12, and were not in the training set of the classifier model
The small network was trained on 41 029 CASP submissions for 98 target domains exclusively in the free-modeling categories of CASP 10–12, and that were not contained in the training set of the classifier network. Twenty randomly chosen submissions from each of the 98 targets were reserved for use as a test set comprised of 1960 structures. The small CNN model was able to accurately predict the GDT of the CASP submissions, yielding an R-squared value of 0.962 between the real and predicted GDT values (Fig. 5b). Plots of the real versus predicted GDT values for each target are provided in the Supplementary Figure S7.
While the predictive power of the small model is limited to the 98 targets, these results suggest that the representation learned by the classifier is able to encode diverse structural information at high resolution. The accurate predictions despite many CASP targets not falling into any CATH architecture classes suggests that the structural features encoded by our model can be used to describe a wide variety of proteins, not just those belonging to the predefined classes.
3.3 Selecting valid design topologies
A significant challenge in de novo design is that it is necessary to find a viable set of lengths, types and orders of secondary structure elements that can adopt a target fold. ‘12-residue α-helix, 3-residue loop, 5-residue β-strand’ is one such example, and is referred to as a ‘topology.’ In shorthand, we denote this topology as ‘H12-E5’, where ‘H’ and ‘E’ denote helix and sheet, while numbers denote element lengths.
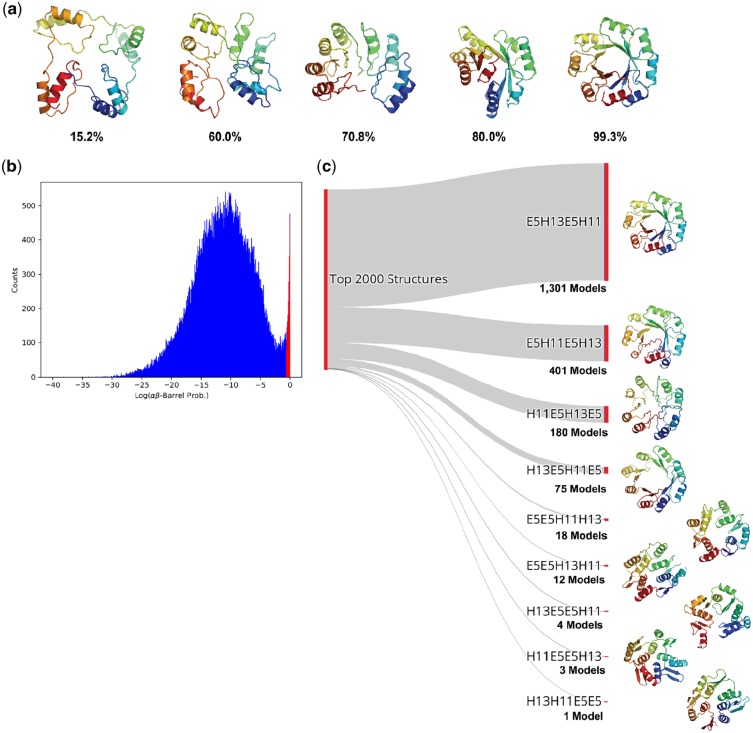
To test whether our CNN can be used to perform topology selection, we generated a series of design trajectories targeting the TIM-barrel fold (αβ-barrel architecture). Our previous study discovered that a 4-fold repeat of E5-H13-E5-H11, with 3-residue loops interspersed between each secondary structure element, constitutes a viable TIM-barrel topology (Huang, et al., 2016a). In order to test whether our model could distinguish this validated topology from a pool of random ones, we generated backbones for every permutation of the secondary structure elements in the 4-fold E5-H13-E5-H11 repeat while maintaining the 3-residue loops between each of the helix or sheet elements; this resulted in 12 unique topologies. We used RosettaRemodel (Huang et al., 2011), which was used in the original study, to generate 7000 backbone designs for each of the topologies, producing a total of 84 000 structures. For each candidate structure, we used our network to predict the average probability of the αβ-barrel architecture over all residues ( where ) denotes the probability of class c at residue i).
We observed that the predicted probabilities correlated with the degree to which a structure adopts a TIM-barrel fold (Fig. 6a). The distribution of probabilities for the generated structures exhibits a bimodal distribution with only ∼2% of structures receiving a probability prediction greater than 50% (Fig. 6b). Collecting the top 2000 designs corresponding to the high probability peak, we observed that the majority of the recovered structures (1301 models, 65%) belonged to 5E-13H-5B-11H topology, suggesting that our model was able to identify the experimentally validated topology as being the most promising. The proportions of the topologies found in the top 2000 structures are shown in Figure 6c along with the highest probability structures from each. Designs from highly represented topologies tended to have fewer structural irregularities relative to under-represented ones. The top four topologies shared the common feature of having two helical elements separated by a single E5 element, likely due to the fact that the having the second E5 element at either the beginning or end of the repeating unit results in near-equivalent structures that are circular permutations of one another. It has been experimentally confirmed that circularly permuted TIM-barrel variants fold correctly (Huang et al., 2016a). These results suggest that our model can be used in combination with established protein backbone generation protocols, such as RosettaRemodel, to automatically select viable protein topologies.
Fig. 6.
Identification of viable TIM barrel topologies. (a) Models generated in the de novo design trajectory and their predicted αβ-barrel probabilities. (b) The distribution of αβ-barrel architecture probabilities for 84 000 structures spanning 12 topologies. The region corresponding to the 2000 structures described in c is shown in red. (c) The proportions of the topologies found in the top 2000 highest αβ-barrel probability structures. The experimentally validated E5-H13-E5-H11 topology constituted 1, 301 models (65%) of the top scoring structures. The highest probability structure from each topology is shown to the right
3.4 Precision and local structure evaluation
While the result above suggests that our model may be useful for design, it does not provide insight into the sensitivity of our metric. Because unviable topologies largely produce models that differ significantly from their target architecture, the task of identifying promising topologies is a coarse-grained problem. To stringently test whether our classifier can quantify the degree to which a structure adopts a particular fold, we assessed our network’s ability to differentiate between highly similar models, using predicted class probabilities and the Shannon entropies of the per-residue probability distributions. We test our classifier on structure prediction data because these trajectories generate a diverse ensemble of structures that converge towards a known structure—a subset of these structures are highly similar and therefore provide a good test of model performance.
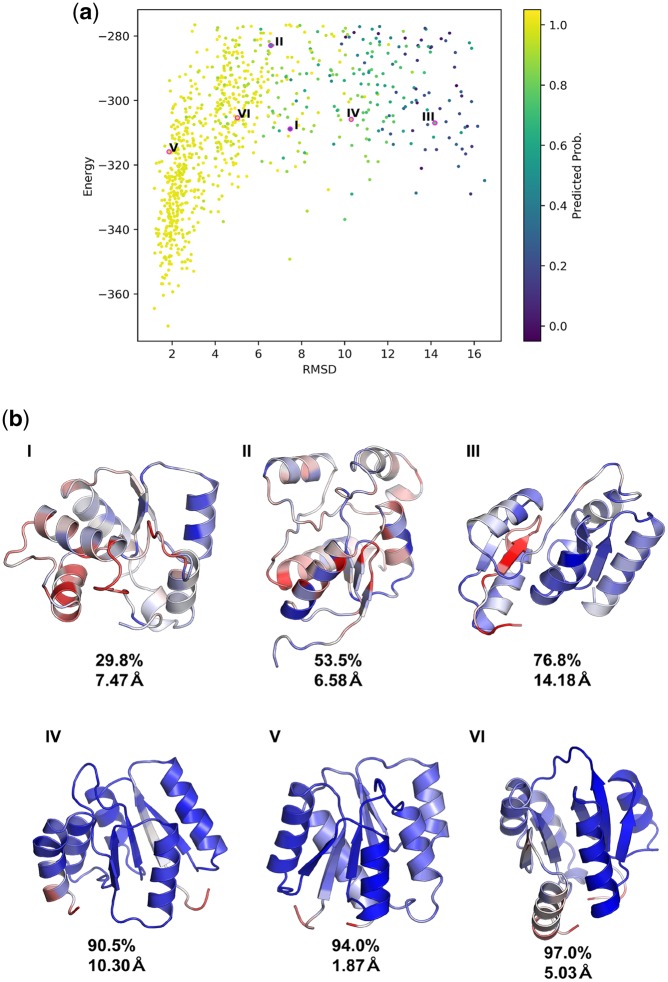
We generated a structure prediction trajectory for the PDB structure 2chfA using the Ab Initio protocol (Bradley, 2005) of Rosetta. 2chfA was not included in the training set of the classifier, and has an αβα-3-layer-sandwich architecture. For each generated decoy we computed the probability of the αβα-3-layer-sandwich class averaged over all residues. With increasing RMSD from the native structure, the model predicted a decreasing probability of belonging to the αβα-3-layer-sandwich class (Fig. 7a). Importantly, the predicted probability of a class was indicative of how consistent the features of a decoy were with the features of a target fold, independent of its RMSD to the native structure (Fig. 7b). Predicted αβα-3-layer-sandwich probabilities correlated with the formation of a well-defined central 5-stranded β-sheet, suggesting that our model correctly identifies this β-core as a feature of this fold. These results suggest that the predicted class probabilities of our network can be used as continuous metrics for selecting decoys assuming a specific fold.
Fig. 7.
Analysis of structure prediction data. (a) 850 randomly selected structures from a structure prediction calculation generated by Rosetta Ab Initio. The vertical axis indicates the energy of each structure as predicted by Rosetta. The horizontal axis indicates the RMSD of each decoy relative to the native structure. Each point is colored by probability for the αβα-3-layer-sandwich architecture, averaged over all residues in the decoy, as predicted by the classifier model. The model generally predicts a decreasing probability of belonging to the αβα-3-layer-sandwich class with increasing RMSD. Roman numerals correspond to decoys shown in b. (b) Six examples from the 2chfA structure prediction calculation. The average predicted probability of the αβα-3-layer-sandwich class, and RMSD relative to the native 2chfA structure is shown below each decoy. The predicted probability of a class was indicative of how closely a decoy resembled the target fold, independently of native RMSD. The structures are colored by the exponential of the entropies of the predicted probability distributions at each residue. The coloring is normalized within each structure; for each decoy, red indicates the highest entropy point, and blue indicates the lowest entropy point
The entropy H of the class probabilities at each residue (), is a measure of the certainty inherent to each prediction. We observed that the high entropy regions of decoys tended to be structurally ambiguous or deviant from the target fold. This effect is shown in Figure 7b, where the normalized spread of entropy values within each structure is shown with a blue-to-red color spectrum. In the high probability models (Fig. 7b, IV–VI), high entropy regions corresponded to loops, and in Figure 7b, III the network correctly identifies the interface of unpaired beta strands failing to form a properly assembled core as a low confidence region. In Figure 8, we show two other examples in which entropy appeared to be indicative of local model quality. The de novo structure in Figure 8a was generated using a αβ-2-layer-sandwich topology described by Koga et al. (2012), and the authors report that atom-pair constraints between the two C-terminal β-strands are required to obtain properly annealed designs. In an unconstrained trajectory, our model identifies the interface of the unpaired strands as being low confidence (Fig. 8a, red), agreeing with this statement. The structure shown in Figure 8b was generated in a structure prediction calculation for the gene NHLRC3, which has an unknown structure thought to be a 6-blade-β-propeller. In the model shown, each of the six blades has formed, however one of the blades has failed to anneal to the rest; our model identifies this sixth blade as being a low-confidence region (Fig. 8b, red).
Fig. 8.
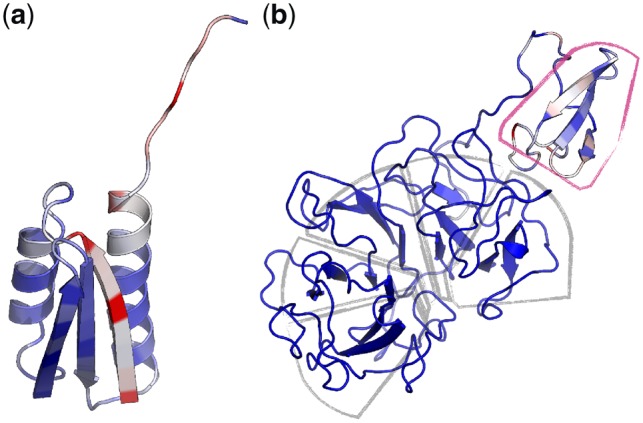
Entropy as an indicator of local model quality. Two structural models colored by the exponential of the entropies of the predicted probability distributions at each residue. The coloring is normalized within each structure; for each decoy, red indicates the highest entropy point, and blue indicates the lowest entropy point. (a) A de novo structural model generated using an αβ-2-layer-sandwich topology described by Koga et al. (2012). The interface of an the unannealed β-strands is identified as a high-entropy region. (b) A structural model generated in a structure prediction calculation for the gene NHLRC3, which has an unknown structure thought to be a 6-blade-β-propeller. Five of the propeller blades have assembled (grey outline), and the un-assembled sixth blade is identified as a high-entropy region (magenta outline)
4 Discussion
In recent years, new computational tools have allowed sampling of protein structural conformations at unprecedented scales, leading to breakthroughs in structure prediction (Das et al., 2007; Ovchinnikov et al., 2014) and the creation of de novo designed structures—an achievement that was considered impossible just 30 years ago (Huang et al., 2016b). Nonetheless, for both prediction and design, finding a target structure requires large-scale sampling of conformational space before a subset of models with the desired features can be found. For structure prediction calculations, promising models can be selected based on criteria such as degree of convergence (Dill et al., 2008) and score function evaluations (Bradley, 2005) because native structures fold into a minimal-energy state dictated by their sequences (Anfinsen, 1973). For de novo design, however, the objectives of a sampling calculation are far less clear as there is no longer a sequence to guide the search process. To identify valid structures, designers have relied heavily on custom-made heuristics based on hydrogen-bonding patterns, geometric parameterizations, modified scoring functions, and other artificially defined constraints. Despite these efforts, a generalizable model selection strategy that captures high-level features such as topology, and secondary structure organization has yet to be developed.
Previously reported methods have shown that recurring structural features (motifs) can be used to accelerate conformational searches and assist in structure evaluation (King et al., 2012; Mackenzie et al., 2016; Marze et al., 2018). We have developed a novel approach that uses a CNN to capture and quantify such features. While our model solves a classification problem that is distinct from domain parsing or domain classification, the architecture can be adapted to perform conventional protein classification, and performs comparably to state-of-the-art protein parser algorithms (Supplementary Information).
Our CNN can be implemented in established protocols to significantly improve sampling efficiency in both structure prediction and protein design: first, the Shannon entropies of per-residue probabilities can be used as a measure of local model quality. Methods such as Iterative Hybridize (Park et al., 2018), which iteratively recombines candidate models to obtain improved structures, could benefit greatly from using this type of method to identify low quality regions requiring rebuilding and refinement. Second, full-structure probabilities can be used to quantify the degree to which a structure assumes a target fold; this can be used to efficiently screen for models adopting a desired architecture in large sampling pools. Finding satisfactory models is comparable to finding a needle in a haystack, and is a daunting task in de novo design. Our novel model selection method can be used to improve the sampling efficiency by focusing searches around viable candidates in structural space. In contrast to the custom heuristics described above, our classifier is unbiased in that it is informed by 95% of all unique structural data, and generalizes to the 38 architecture classes that comprise this dataset. Our CNN is also fast, and can compute both entropies and full-structure probabilities in milliseconds, efficiently processing the millions of candidate structures generated during both prediction and design.
There are two minor limitations to our network. First, albeit by choice, inputs to our current model are limited to single chains that are 512 residues or less (95% of all unique chains in the PDB). For analysis of longer chains, structures need to be fragmented or truncated. Nonetheless, this limitation can be easily remedied by training a new model with a larger input layer. Second, our network is trained on the set of architectures defined by CATH. While CATH classifications account for the vast majority of observed protein structures, this implies that protein designers seeking to build unclassified architectures cannot currently benefit from our model.
Nonetheless, the novel way in which we apply image segmentation methods to protein structure allows us to quantify abstract structural patterns constituting the ‘environment’ of each residue. Our results suggest that by combining per-residue information in different ways, it is possible to create any number of metrics that describe various characteristics of a protein; this approach is powerful in its generalizability and can be used to encode features of proteins that are often difficult to assess when building structural models.
Supplementary Material
Acknowledgements
All data used for model training was obtained from the CATH protein database. A list of unique protein chains was kindly provided by Ian Sillitoe who works with the CATH group at University College London. Namrata Anand provided much technical advice and support through the whole project. Guillaume Postic kindly provided model outputs for SWORD, DDOMAIN, PDP and DP. We also thank Yuexu Jiang for providing DeepDom model outputs.
Funding
This project was supported by startup funds from the Stanford Schools of Engineering and Medicine, the Stanford ChEM-H Chemistry/Biology Interface Predoctoral Training Program and the National Institute of General Medical Sciences of the National Institutes of Health under Award Number [T32GM120007]. Additionally, this project was supported by the U.S. Department of Energy, Office of Science, Office of Advanced Scientific Computing Research, Scientific Discovery through Advanced Computing (SciDAC) program.
References
- Anand N., Huang P. (2018) Generative modeling for protein structures. In: Bengio,S. et al (eds) Advances in Neural Information Processing Systems 31. Curran Associates, Inc, San Jose, CA, USA, pp. 7494–7505. [Google Scholar]
- Anfinsen C.B. (1973) Principles that govern the folding of protein chains. Science, 181, 223–230. [DOI] [PubMed] [Google Scholar]
- Bialas C. et al. (2016) Engineering an artificial flavoprotein magnetosensor. J. Am. Chem. Soc., 138, 16584–16587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom J.D. et al. (2006) Protein stability promotes evolvability. Proc. Natl. Acad. Sci. USA, 103, 5869–5874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley P. et al. (2005) Toward high-resolution de novo structure prediction for small proteins. Science, 309, 1868–1871. [DOI] [PubMed] [Google Scholar]
- Brunette T. et al. (2015) Exploring the repeat protein universe through computational protein design. Nature, 528, 580–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das R. et al. (2007) Structure prediction for CASP7 targets using extensive all-atom refinement with Rosetta@home. Proteins Struct. Funct. Bioinformatics, 69, 118–128. [DOI] [PubMed] [Google Scholar]
- Dawson N.L. et al. (2017) CATH: an expanded resource to predict protein function through structure and sequence. Nucleic Acids Res., 45, D289–D295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill K.A. et al. (2008) The protein folding problem. Annu. Rev. Biophys., 37, 289–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou J. et al. (2018) De novo design of a fluorescence-activating β-barrel. Nature, 561, 485–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleishman S.J. et al. (2011) Computational design of proteins targeting the conserved stem region of influenza hemagglutinin. Science, 332, 816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox N.K. et al. (2014) SCOPe: Structural Classification of Proteins—extended, integrating SCOP and ASTRAL data and classification of new structures. Nucleic Acids Res., 42, D304–D309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glorot X., Bengio Y. (2010) Understanding the difficulty of training deep feedforward neural networks In: Teh Y.W., Titterington M. (eds) Proceedings of the Thirteenth International Conference on Artificial Intelligence and Statistics. pp. 249–256,PMLR. [Google Scholar]
- Goodfellow I. et al. (2016) Deep Learning. MIT Press, Cambridge, MA, USA. [Google Scholar]
- Hou J. et al. (2018) DeepSF: deep convolutional neural network for mapping protein sequences to folds. Bioinformatics, 34, 1295–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P.-S. et al. (2016a) De novo design of a four-fold symmetric TIM-barrel protein with atomic-level accuracy. Nat. Chem. Biol., 12, 29–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P.-S. et al. (2011) RosettaRemodel: a generalized framework for flexible backbone protein design. PLoS One, 6, e24109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P.-S. et al. (2016b) The coming of age of de novo protein design. Nature, 537, 320–327. [DOI] [PubMed] [Google Scholar]
- Jiang L. et al. (2008) De novo computational design of retro-aldol enzymes. Science, 319, 1387–1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joh N.H. et al. (2017) Design of self-assembling transmembrane helical bundles to elucidate principles required for membrane protein folding and ion transport. Philos. Trans. R. Soc. B Biol. Sci., 372, 20160214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King N.P. et al. (2012) Computational design of self-assembling protein nanomaterials with atomic level accuracy. Science, 336, 1171–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga N. et al. (2012) Principles for designing ideal protein structures. Nature, 491, 222–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leaver-Fay A. et al. (2011) Rosetta3 In: Johnson,M.L. and Brand,L. (eds) Methods in Enzymology. Elsevier, Cambridge, MA, USA, pp. 545–574. [Google Scholar]
- Long J. et al. (2014) Fully convolutional networks for semantic segmentation. CoRR, abs/1411.4038. https://arxiv.org/abs/1411.4038. [DOI] [PubMed]
- Mackenzie C.O. et al. (2016) Tertiary alphabet for the observable protein structural universe. Proc. Natl. Acad. Sci. USA, 113, E7438–E7447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcos E. et al. (2018) De novo design of a non-local β-sheet protein with high stability and accuracy. Nat. Struct. Mol. Biol., 25, 1028–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcos E. et al. (2017) Principles for designing proteins with cavities formed by curved b sheets. Science, 355, 201–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marze N.A. et al. (2018) Efficient flexible backbone protein–protein docking for challenging targets. Bioinformatics, 34, 3461–3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moult J. et al. (2018) Critical assessment of methods of protein structure prediction (CASP)-Round XII. Proteins Struct. Funct. Bioinformatics, 86, 7–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovchinnikov S. et al. (2014) Robust and accurate prediction of residue–residue interactions across protein interfaces using evolutionary information. eLife, 3, e02030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H. et al. (2018) Protein homology model refinement by large-scale energy optimization. Proc. Natl. Acad. Sci. USA, 115, 3054–3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paszke A. et al. (2017) Automatic differentiation in PyTorch. NIPS Autodiff Workshop. https://github.com/pytorch/pytorch/issues/4126.
- Pejchal R. et al. (2011) A potent and broad neutralizing antibody recognizes and penetrates the HIV glycan shield. Science, 334, 1097–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redmon J. et al. (2015) You only look once: unified, real-time object detection. ArXiv150602640 Cs https://arxiv.org/abs/1506.02640.
- Rocklin G.J. et al. (2017) Global analysis of protein folding using massively parallel design, synthesis, and testing. Science, 357, 168–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Röthlisberger D. et al. (2008) Kemp elimination catalysts by computational enzyme design. Nature, 453, 190–195. [DOI] [PubMed] [Google Scholar]
- Schroff F. et al. (2015) FaceNet: a unified embedding for face recognition and clustering. In: 2015 IEEE Conference on Computer Vision Pattern Recognition CVPR, IEEE publishing, New York, NY, USA, pp. 815–823. [Google Scholar]
- Shannon C., Weaver W. (1964) The Mathematical Theory of Communication. The University of Illinois Press, Hoboken, NJ, USA.
- Shi W. et al. (2016) Real-time single image and video super-resolution using an efficient sub-pixel convolutional neural network. ArXiv160905158 Cs Stat.https://arxiv.org/abs/1609.05158v2.
- Smadbeck J. et al. (2014) De novo design and experimental characterization of ultrashort self-associating peptides. PLoS Comput. Biol., 10, e1003718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinberg C.E. et al. (2013) Computational design of ligand-binding proteins with high affinity and selectivity. Nature, 501, 212–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuoka Y. et al. (2018) Convolutional neural network-based instance segmentation algorithm to acquire quantitative criteria of early mouse development. bioRxiv, https://www.biorxiv.org/content/10.1101/324186v2.
- Tokuriki N. et al. (2008) How protein stability and new functions trade off. PLoS Comput. Biol., 4, e1000002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torng W., Altman R.B. (2017) 3D deep convolutional neural networks for amino acid environment similarity analysis. BMC Bioinformatics, 18, 302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S. et al. (2017) Accurate de novo prediction of protein contact map by ultra-deep learning model. PLoS Comput. Biol., 13, e1005324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemla A. et al. (1999) Processing and analysis of CASP3 protein structure predictions. Proteins, 37(Suppl 3), 22–29. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.