Abstract
Our primary objective is to provide the clinical informatics community with an introductory tutorial on calibration measurements and calibration models for predictive models using existing R packages and custom implemented code in R on real and simulated data. Clinical predictive model performance is commonly published based on discrimination measures, but use of models for individualized predictions requires adequate model calibration. This tutorial is intended for clinical researchers who want to evaluate predictive models in terms of their applicability to a particular population. It is also for informaticians and for software engineers who want to understand the role that calibration plays in the evaluation of a clinical predictive model, and to provide them with a solid starting point to consider incorporating calibration evaluation and calibration models in their work.
Covered topics include (1) an introduction to the importance of calibration in the clinical setting, (2) an illustration of the distinct roles that discrimination and calibration play in the assessment of clinical predictive models, (3) a tutorial and demonstration of selected calibration measurements, (4) a tutorial and demonstration of selected calibration models, and (5) a brief discussion of limitations of these methods and practical suggestions on how to use them in practice.
Keywords: tutorial, calibration, predictive models
INTRODUCTION
Most clinicians can recall seeing that inpatient who was listed as 50 years old but appeared decades older due to the effects of chronic illness, a physical exam finding commonly described as “Appearing older than stated age.” And yet, that patient’s stated age is used to dictate much of their care, including the calculation of glomerular filtration rate for medication dosing, risk of developing illnesses, and risk of inpatient mortality. Experienced clinicians can “calibrate” their mental models to account for the patients’ appearance, but predictive models cannot do this without further instructions. When predictive models are built based on a population that differs from the population in which they will be used, blind application of these models could result in large “residuals” (ie, a large difference between a model’s estimate and the true outcome) because of factors that are difficult to account for. This deficiency could lead to catastrophic decisions for a single patient, even when the average residual for the overall population is very low. The analysis of such residuals can serve as a proxy for measuring the “calibration” of the model. While calibration-in-the-large is concerned with gross measurements of calibration, such as whether the model’s overall expected number of cases exceeds the observed number, or whether the proportion of expected over observed cases departs significantly from “1,” other measurements of calibration are based on population stratifications, which can include anything from analyzing residuals on a few large subgroups, all the way to analyzing residuals for each individual. Calibration is an essential component of the evaluation of computational models for medical decision making, diagnosis, and prognosis.1,2 In contrast to discrimination, which refers to the ability of a model to rank patients according to risk, calibration refers to the agreement between the estimated and the “true” risk of an outcome.3 A well-calibrated model is one that minimizes residuals, which is equivalent to saying that the model fits the test data well. Note that observing small residuals on the training set does not necessarily mean that it is a good model, as “overfit” models are known not to generalize well to previously unseen data.
There are a number of cases that illustrate the omnipresence and importance of calibration and its critical role in model evaluation. If individualized predictions are used for clinical decision making, well-calibrated estimates are paramount. Take the case of dementia, a neurodegenerative disorder that affects at least 14% of Americans and recently cost the U.S. healthcare system over $150 billion/y.4 One recent study evaluated the calibration-in-the-large of several models for predicting the risk of developing dementia in the general community and found that models drastically overestimated the expected incidence of dementia.5 At a predicted risk of 40%, the observed incidence was still only 10%, so the test overestimated incidence by 30%. For an individual patient, and for the healthcare provider, an overestimation of this magnitude could lead to different decisions. For example, it is recommended by the American College of Cardiology and the American Heart Association that patients with a cardiovascular risk over 7.5% be prescribed statins, and those between 5% and 7.5% be considered.6 Even risk calculators that are not based on percentages may benefit from calibration. An example is the Model for End-Stage Liver Disease,7 which provides a risk score that is used to prioritize cases for liver transplantation. When score thresholds are used (eg, to determine the frequency in which a patient’s score is recalculated for sorting the waiting list), calibration becomes critical. Accurate, well-calibrated estimates are necessary to allocate resources appropriately. Additionally, even when the models are well calibrated-in-the-large (eg, the average predicted risk was 40% and the observed incidence was also 40%), there could be severe discrepancies to particular groups of individuals. Thus, it is critical to understand how a model will be employed in order to emphasize certain performance measures.
This tutorial covers some techniques to assess and correct model calibration in the context of employing clinical predictive models to estimate individualized risk. It is by no means comprehensive and is not intended to replace the extensive body of literature on the topic of calibration. We present issues with measures of calibration that go beyond calibration-in-the-large, and we include examples of some calibration models that have been recently used in the biomedical literature. This tutorial does not include all available measurement methods and calibration models, is complementary to book chapters and articles that serve as references to this topic, and will be of interest to those who want to deepen their understanding of model calibration.3,8–10 We provide here some simple and interpretable calibration measures and calibration models that can illustrate the concepts and have appeared in the recent biomedical predictive modeling literature so that clinical researchers and informaticians may familiarize themselves with this topic. Despite its importance to understanding the utility of a model, calibration is vastly underreported: one systematic review noted that although 63% of published models included a measure of discrimination, only 36% of models provided a measure of calibration.11 Precision medicine involves individualized prevention, diagnosis, and treatment. Thus, it needs to rely on predictive models that are well calibrated. We describe, in a didactic manner, key steps for measuring calibration and applying calibration models to a predictive model.
RANKING PATIENTS VS ESTIMATING INDIVIDUAL RISK
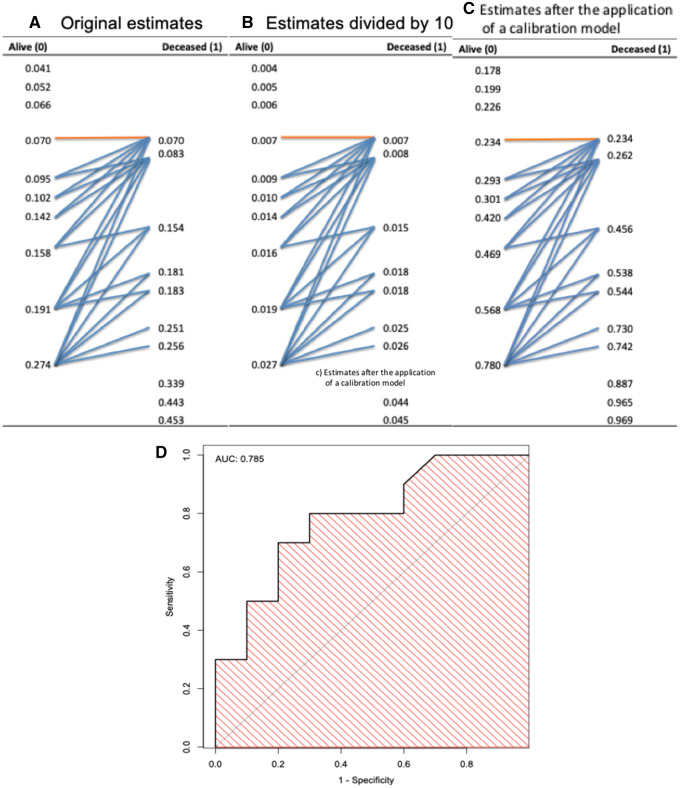
Initial comparison and selection of appropriate models are often done through the evaluation of discrimination, which is measured with the area under the receiver-operating characteristic curve (AUROC), but the AUROC says nothing about the calibration of the model. Figure 1 shows how relying on AUROC overlooks calibration. Figures 1A–1C contain 3 models’ predicted estimates, sorted in ascending order, for 2 groups (Alive = “0” and Deceased = “1”). The 3 models’ estimates are
Figure 1.
Illustration of a receiver-operating characteristic curve and corresponding area under the receiver-operating characteristic curve (AUC) derived from predictive model estimates. (A) Original estimates. (B) Original estimates divided by 10. (C) Original estimates after the application of a calibration model to have the estimates be closer to the actual outcomes. The blue arrows, showing discordant pairs, indicate pairs of estimates in which the estimates for the Alive patients (coded as 0) are greater than the estimates for the Deceased patients (coded as 1), while the orange arrows indicate pairs of estimates where estimates for Alive and Deceased patients are equal (ties). The AUC is equivalent to the concordance index, which can be calculated here by the number of concordant pairs (ie, total number of pairs minus the discordant and half of the tied pairs) over the total number of pairs, shown in the text as equation 1. (D) Identical receiver-operating characteristic curve and AUC (0.785) for the 3 models.
original estimates,
original estimates divided by 10, and
original estimates after applying calibration model to have the estimates be closer to the actual outcomes.
The AUROC, which is equivalent to the concordance index,12,13 can be easily calculated by counting the arrows in Figure 1.
The nonparametric AUROC can be calculated by the concordance index as follows:
| (1) |
where total # pairs is the number of pairs of Alive and Deceased estimates; # discordant pairs is the number of pairs composed of 1 Alive and 1 Deceased patient, in which the estimate for the Deceased (coded as 1) is lower than the estimate for the Alive (coded as 0) patient; and the # ties is for such a pair in which the estimates are equal. As illustrated in Figure 1, there is no need for the actual values of the estimates: only the ranks (ie, the order of the estimates) are necessary to calculate the concordance index or the AUROC. Therefore, when equation 1 is applied to estimates in Figures 1A–1C, the resulting receiver-operating characteristic (ROC) curves for all 3 models are the same, and their corresponding AUROCs are also identical, as shown in Figure 1D. However, the 3 models’ estimates are vastly different. Such differences are reflected in the absolute errors between the estimates and actual outcomes, as well as in the average estimates for Alive and Deceased (Table 1). These values help give an idea of the models’ calibration but still do not reveal whether there is gross under- or overestimation of the probability of Deceased for particular groups or individuals. Without a means for measuring calibration and for choosing appropriate models accordingly, erroneous decisions could be made in processes that rely on the values of the estimates. For example, if a clinical practice guideline recommends that all individuals at >20% risk receive a certain intervention, a noncalibrated model such as the one in Figure 1B could result in no one receiving the intervention. In this tutorial, we illustrate methods that measure calibration in different ways. For models that have improper calibration, we show how calibration models can mitigate the problem of poorly calibrated models.
Table 1.
Average estimates and observed outcomes
| Figure 1A | Figure 1B | Figure 1C | |
|---|---|---|---|
| AUROC | 0.785 | 0.785 | 0.785 |
| Average true outcomes | 0.5 | 0.5 | 0.5 |
| Average absolute error | 0.439 | 0.499 | 0.367 |
| Average estimates: | 0.180 | 0.002 | 0.500 |
| Average estimates (alive) | 0.115 | 0.001 | 0.349 |
| Average estimates (deceased) | 0.241 | 0.002 | 0.633 |
AUROC: area under the receiver-operating characteristic curve.
SIMULATED DATA
We used simulated data to demonstrate the forthcoming calibration measures and calibration models. The code can be found on GitHub (https://github.com/easonfg/cali_tutorial). To create artificial data, we utilized the method from Zimmerman et al.14 Twenty-three artificial features were constructed with 20 binary and 3 continuous independent variables. A uniform distribution was then used to decide the dependent variable, mortality, where 0 indicates Alive and 1 indicates Deceased. The observed mortality frequency was 15%. To compare models, a logistic regression (LR) model and a support vector machine (SVM) model with a linear kernel were built using the 23 variables. SVM uses the hinge loss function as its objective function, so it often produces improperly calibrated estimates.15 We present different measures of calibration and the results of calibration models.
Five thousand samples were created. The entire simulated data were separated into 3 parts: (1) training set part 1 (2500 samples), (2) training set part 2 (1250 samples), and (3) test set (1250 samples). We used hold-out validation: we train the classifier on training set part 1, we train the calibration models on training set part 2, and we use the test set to “validate” (ie, to assess performance when the model is used in previously unseen cases). A comparison of sampling strategies for model building and evaluation (eg, cross-validation, bootstrap) is beyond the scope of this tutorial on calibration. The interested reader is referred to Zou et al16 for an introduction to (re)sampling techniques.
For this tutorial, 2 classification models (LR and SVM) were trained on the training set part 1, and a calibration model was built based on the resulting model applied to training set part 2. Then, the classifier and calibration models were applied to the test set and evaluation of calibration and discrimination were calculated on the test set’s predicted estimates. We compared discrimination and calibration on the original test set estimates (ie, preapplication of the calibration model) and on the calibrated estimates. By separating the entire data into 3 parts and training the classification model and calibration model on different datasets, we aimed to avoid overfitting.
MEASURING CALIBRATION
There is no best method to measure the calibration of predictive models. While some methods are frequently used and have specific strengths, all have limitations. Here, we present these calibration assessment methods and the scenarios in which each can be used appropriately. Additionally, some methods combine implicit measures of calibration with other components such as discrimination, which may be difficult to separate.
BRIER SCORE AND SPIEGELHALTER’S z TEST
The Brier score is the mean squared error between the actual outcome and the estimated probabilities, as shown in equation 2:
| (2) |
where N is the number of patients, is the predicted estimate for patient i, and is the actual outcome for patient i. The Brier score should be interpreted carefully. Without understanding whether the error is caused by a relatively small number of estimates with high error or a large number of estimates with a smaller error, it is difficult to say whether this model could be used in practice. Note that, by squaring errors that are in the [0,1] range, large errors “count less” to the overall score, when compared numerically with smaller errors. The Brier score includes components of discrimination and calibration, so a lower Brier score does not necessarily imply higher calibration.17 However, it can be shown that, from the decomposition of Brier score, a formal measurement that can serve as a proxy for calibration can be calculated: the Spiegelhalter z test.18 The z statistic can be calculated with equation 3.
| (3) |
If , where is the -quantile of the standard normal distribution (0.05), the result is significant, suggesting an improperly calibrated model.
The discrimination (AUROC), Brier scores, and Spiegelhalter’s z-test results for the LR and SVM models are shown in Table 2, as are other measures described in subsequent sections of this article. The AUROCs of the 2 models are the same, but there is a difference in Brier scores. Also, P values for the Spiegelhalter’s z test indicate that the SVM classifier is not well calibrated.
Table 2.
Discrimination and calibration results of the LR and SVM models applied to the test set
| LR | LR Platt scaling | LR isotonic regression | LR BBQ | SVM | SVM Platt scaling | SVM isotonic regression | SVM BBQ | |
|---|---|---|---|---|---|---|---|---|
| AUROC | 0.870 | 0.870 | 0.870 | 0.867 | 0.870 | 0.870 | 0.870 | 0.862 |
| Brier score | 0.087 | 0.088 | 0.088 | 0.089 | 0.111 | 0.086 | 0.088 | 0.090 |
| Spiegelhalter z score | 0.762 | 0.417 | 0.087 | 0.748 | 2.21 | 0.826 | 0.693 | 0.731 |
| Spiegelhalter P value | .223 | .338 | .465 | .227 | .013a | .204 | .244 | .232 |
| Average absolute error | 0.177 | 0.177 | 0.177 | 0.182 | 0.236 | 0.177 | 0.177 | 0.185 |
| H-L C-statistics | 5.88 | 24.6 | 11.7 | 16.0 | 176 | 4.75 | 12.7 | 28.0 |
| H-L C-statistic P value | .661 | .002a | .167 | .042a | <1 × 10–22a | .784 | .122 | 4.71 × 10–4a |
| H-L H-statistics | 9.18 | 16.6 | 10.1 | 11.5 | 160 | 11.2 | 8.15 | 1.86 |
| H-L H-statistic P value | .327 | .030a | .259 | .174 | <1 × 10–22a | .188 | .419 | .984 |
| MCE | 0.038 | 0.072 | 0.033 | 0.042 | 0.403 | 0.028 | 0.034 | 0.052 |
| ECE | 0.014 | 0.035 | 0.012 | 0.022 | 0.109 | 0.011 | 0.018 | 0.027 |
| Cox’s slope | 1.070 | 1.074 | 0.953 | 1.020 | 5.014a | 1.087 | 1.023 | 1.008 |
| Cox’s intercept | 0.080 | 0.072 | –0.092 | –0.007 | 6.193a | 0.081 | –0.001 | –0.02 |
| ICI | 0.010 | 0.034 | 0.012 | 0.012 | 0.104 | 0.008 | 0.013 | 0.020 |
Discrimination is measured by the AUROC. The Brier score is a combined measure of discrimination and calibration. Calibration is measured by the Spiegelhalter z test, average absolute error, H-L test, MCE, ECE, Cox slope and intercept, and ICI. SVM estimates for the test set produced were improperly calibrated. Application of Platt scaling, isotonic regression, or BBQ was performed.
AUROC: area under the receiver-operating characteristic curve; BBQ: Bayesian Binning into Quantiles; ECE: expected calibration error; H-L, Hosmer-Lemeshow; ICI: integrated calibration index; LR: logistic regression; MCE: maximum calibration error; NIS: Nationwide Inpatient Sample; SVM: support vector machine.
shows significance.
Calculation of the Brier score is relatively simple. A line of code is sufficient or use of packages that calculate the Brier score in R (R Foundation for Statistical Computing, Vienna, Austria), such as the “rms” package with the function “val.prob” and the “DescTools” package with the function “BrierScore” (packages that implement the Brier scores and all subsequent calibration methods conducted with the simulated data are listed in Supplementary Table). Spiegelhalter’s z statistic can also be calculated with function “val.prob” from the “rms” package.
AVERAGE ABSOLUTE ERROR
Another easily implemented measure is the average absolute error, which is calculated in equation 4.
| (4) |
The average absolute error is very similar to the Brier score, but small and large errors contribute in the same way to the sum (ie, unlike the Brier score, both contribute in the same way to the sum). The results for LR and the SVM are shown in Table 2. Examples are given in the GitHub folder.19
HOSMER-LEMESHOW TEST
The Hosmer-Lemeshow (H-L) test has been a popular measure of goodness of fit for predictive models of binary outcomes, and is sometimes used as a proxy for calibration20 despite its shortcomings, which we describe in the Discussion section. A PubMed search returns hundreds of articles per year mentioning the H-L goodness-of-fit test. Despite its age and known shortcomings, this test has been frequently used in health sciences research. Because the way in which it groups observations is also used in reliability diagrams and calibration curves, we explain some details here.
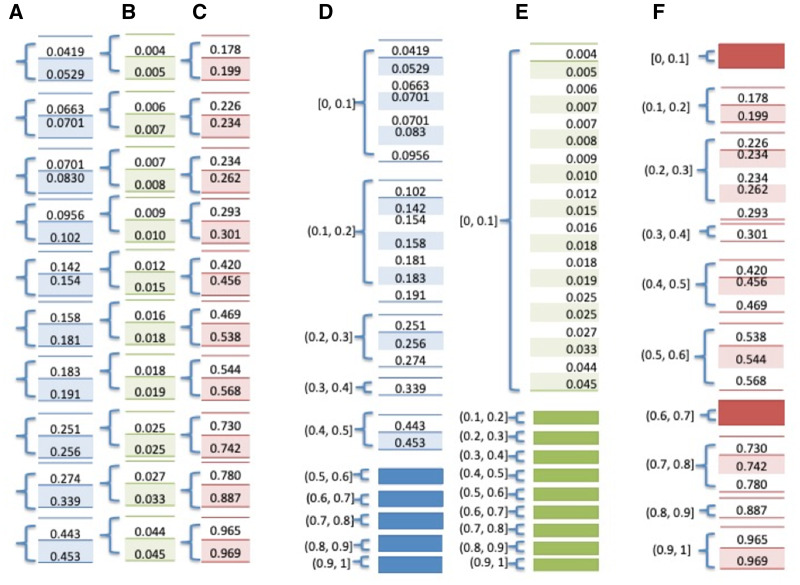
Grouping of Observations to Calculate a Proxy for the “Gold Standard”. There is no ideal method to assess calibration of models. A calibration measure could ideally compare the predicted estimate with the “true” probability for each patient, but the measurement of actual probability for a single individual is challenging. That is, in the data, we can only ascertain the binary outcome, and not actual or “true” probabilities; therefore, a proxy for this probability is used. Forming groups of individuals and calculating the proportion of positive outcomes is a way to achieve such proxy. For example, we can say that the actual probability of death is 10% for a patient if 10 of 100 patients “just like” this patient died. For the H-L test and some other calibration methods, patients who are “just like” each other are patients whose predictive model’s estimates belong to the same group (ie, patients who received similar estimates once a model is applied), and the ratio of event and nonevent within each group is the proxy for the “true” probability for the patients in that group. There are 2 ways by which the H-L test assigns individuals to the same group, resulting in H-L C- or H-statistics, as shown in Figure 2.
Figure 2.
Grouping methods for Hosmer-Lemeshow (H-L) C- and H-statistics. The small number of observations would not warrant a test, but serves to illustrate the contrast between 2 different ways of forming groups for the H-L test: (1) for H-L C-statistics, patients are divided into g groups, where each group contains approximately the same number of patients, typically grouped by deciles of risk (g = 10); and (2) for H-L H-statistics, groups are divided based on equal increment thresholds for the estimates (eg, if there are 10 groups, estimates in the interval [0, 0.1] belong to one group, estimates in the interval [0.1, 0.2] belong to the second group, and so on). Numbers shown in a blue background correspond to Figure 1A estimates, green corresponds to Figure 1B, and red corresponds to Figure 1C estimates. (A–C) Groups of estimates using deciles of samples utilized for the H-L C-statistics. (D–F) Groups of estimates using equal interval groups utilized for the H-L H-statistics. As the degrees of freedom are equal to (g – 2), the degrees of freedom for groups in panels A–F are 8, 8, 8, 3, 0, and 6, respectively.
H-L test statistic and P value. The total estimates of the “similar” patients are then compared with total observed outcomes within each group. The H-L C-statistic or H-L H-statistic can be calculated using equation 5:
| (5) |
where is the number of patients with outcome “1” within each group and is the sum of the estimates of patients with outcome “1” within each group, is the number of patients with outcome “0” within each group, and is the sum of the estimates of patients with outcome “0” within each group. Finally, g is the number of groups. The distribution of the test statistics follows a chi-square distribution with (g – 2) degrees of freedom. The P value can be subsequently calculated. A P value of .1 or higher is considered appropriate, a P value <.1 and >.05 indicates that the model is neither well calibrated nor grossly miscalibrated, and a P value <.05 indicates miscalibrated estimates.21
The ideal number of groups and the method to determine membership in a group are often points of contention when performing an H-L test. With H-L C-statistics, where an approximate equal number of samples are in each group, the range of the estimates could differ wildly, as shown in Figure 2.
H-L statistics and P values results are calculated and shown in Table 2. Results shown in Table 2 used 10 groups or “bins.” The P values for the SVM model (P = 0) are significant, indicating improper calibration, while LR is properly calibrated (P > .1). Packages and functions that implement the H-L test are listed in the Supplementary Table. The package “ResourceSelection” from R with function “hoslem.test” and the package “generalhoslem” with function “logitgof” both provide the same calculation of the H-L test.22 However, these packages are only capable of calculating the H-L C-statistics. A modified method is presented in the GitHub file that allows calculations for both H-L C- and H-statistics.19
RELIABILITY DIAGRAM
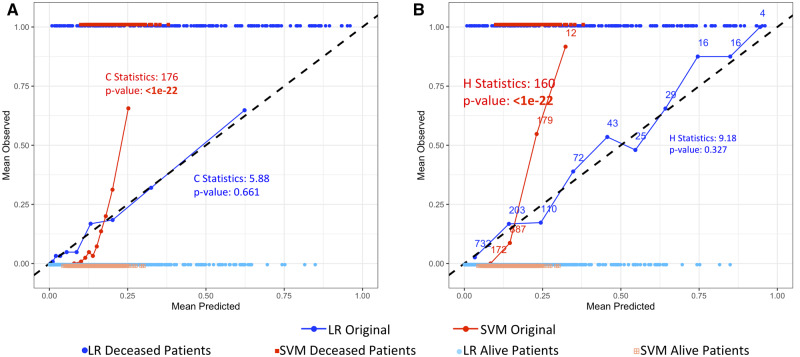
The reliability diagram is a visualization technique that uses observation groupings such as the ones formed for the H-L test.15 However, instead of the sum, the mean of actual outcomes of each group is plotted against the mean of estimates of each group. While the points are typically connected to help with visualization, it is obviously not a true curve. No information can be derived between the points on the diagram. A perfectly calibrated model would result in a 45-degree line. Figure 3 shows the reliability diagrams for the LR and SVM models using the H-L C- and H-statistics in Figures 3A and 3B, respectively. The actual data are also plotted for reference. While the reliability diagram of LR follows the diagonal line, we can see that the reliability diagram for the SVM model deviates from the diagonal, trending upward. This indicates improper calibration and underestimation of actual number of Deceased (outcome = 1) in some groups.
Figure 3.
Reliability diagrams of test set estimates produced by logistic regression (LR) and support vector machine (SVM) models grouped for the Hosmer-Lemeshow (H-L) C-statistics and the H-L H-statistics. Data points of estimates produced by the models and their actual binary outcomes are plotted to show the distribution of the actual data. The Alive outcome is indicated as 0 and the Deceased outcome is indicated as 1. Corresponding H-L statistics and P values are shown in the graphs. (A) LR and SVM estimates grouped for the calculation of the H-L C-statistic. The number of patients within each bin is the same (n = 125). (B) Estimates grouped for the calculation of the H-L H-statistic. The number of patients in each group is shown in the graph. Both graphs show that the SVM underestimates the actual number of deaths, as shown by the red line deviating from the diagonal line. LR is relatively well calibrated (blue line).
The Supplementary Table shows R packages and commands for the calibration measures discussed in this article. A package from “PresenceAbsence” in R is able to draw reliability diagram with the function “calibration.plot.”23 This method groups the estimates according to the H-L H-statistics and plots the average of the actual number of positive outcomes against the midpoint of each group’s interval. R packages for the H-L test use the H-L C-statistics grouping method, whereas the reliability diagram typically uses the H-L H-statistics grouping method. We implemented the H-L test and reliability diagrams in both grouping methods for completeness (see GitHub for the code).19
EXPECTED CALIBRATION ERROR AND MAXIMUM CALIBRATION ERROR
Aside from the H-L test and the reliability diagram, binning was also used in recent papers to calculate the expected calibration error (ECE) and the maximum calibration error (MCE).24–26 To compute the ECE and the MCE, predictions or estimates are sorted and divided into K bins with an approximately equal number of patients in each bin. The ECE calculates the average calibration error over the bins, whereas the MCE calculates the maximum calibration error for the bins:
| (6) |
where is the fraction of all patients who fall into bin i, is the fraction of positive instances in group i, and is the average of the probabilities for the instances in group. The number of groups used is 10. Results for simulated data are shown in Table 2: LR has smaller ECEs and MCEs than the SVM does, confirming what we already knew via the H-L C and H-L H tests.
COX INTERCEPT AND SLOPE
Unlike the previous methods, Cox’s intercept and slope do not group estimates into bins. The Cox method assesses calibration by regressing the observed binary outcome to the log odds of the estimates with a general linear model, as shown in equation 727:
| (7) |
where b is the regression slope and a is the intercept. The estimated regression slope dictates the direction of miscalibration, where 1 denotes perfect calibration (usually achieved by overfitting the model), >1 denotes underestimation of high risk and overestimation of low risk, and <1 denotes underestimation of low risk and overestimation of high risk. The estimated regression intercept represents the overall miscalibration, where 0 indicates good calibration, >0 denotes an average underestimation, and <0 denotes an average overestimation. An example is given in the GitHub folder.19 Results of Cox’s slope and intercept are shown in Table 2. The slope and intercept for LR are close to 1 and 0, respectively, indicating a proper calibration. The SVM, on the other hand, exhibits an underestimation of high risk and overestimation of low risk given its slope >1, and exhibits overall underestimation given its intercept >0.
INTEGRATED CALIBRATION INDEX
Similar to Cox’s method, the integrated calibration index (ICI) assessed calibration by first regressing the binary response to the estimates. However, the ICI uses a locally weighted least squares regression smoother (ie, the Loess algorithm).29 Cox’s slope and intercept can equal to 1 and 0, respectively, while deviations from perfect calibration can still occur (eg, when these deviations “cancel” each other in terms of the linear regression). However, these deviations can be captured by the Loess smoother, and a subsequent numerical summary is the ICI. Other functions such as splines and polynomials can also be used. ICI takes the average of the absolute difference between the estimates and the predicted estimates based on the Loess calibration curve. The results for LR and SVM are shown in Table 2. The ICI of the SVM is higher than the ICI of LR, and particularly for estimates before application of calibration models, as expected.
CALIBRATING MODELS
Calibration models can be applied to improve calibration performance, and there are 2 ways to attempt to obtain calibrated estimates. The first approach is to include measures and terms in the objective function that specifically cater to calibration during model development.30 When retraining a model to emphasize that calibration is not feasible, it is sensible to improve calibration by applying calibration models to the estimates produced by the classifiers. The advantage of applying calibration models to estimates is that the method can be used in addition to any existing classification method and adjusted to the local patient population.
In a relatively recent article from the biomedical informatics literature, Walsh et al31,32 re-emphasize the calls from Van Calster et al33 and Riley et al34 on the importance of calibration in addition to discrimination when evaluating predictive models. Walsh et al31,32 select the following calibration models for their experiments: logistic calibration, Platt scaling, and prevalence adjustment. We utilize the Platt scaling,35 isotonic regression,36 and the Bayesian Binning into Quantiles (BBQ)26 calibration models to illustrate differences in calibration.
PLATT SCALING
Platt scaling transforms model estimates by passing the estimates through a trained sigmoid function.35 The sigmoid function is shown in equation 8:
| (8) |
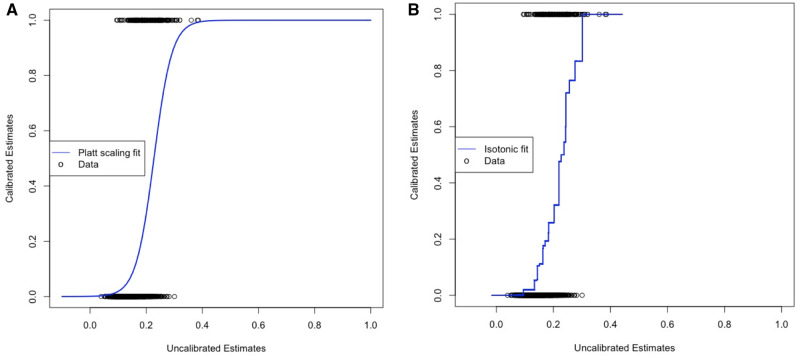
where f is the predicted estimate and parameters A and B are derived using gradient descent. Figure 4A shows the fitted sigmoid function derived using training set part 2. Platt scaling trains a sigmoid function with the codomain constrained to the interval [0,1], using the built-in function “glm” in R, with link function “logit.” It is a univariate LR model that uses the model estimates as independent variables and the binary outcomes as dependent variables. An example is shown in the GitHub folder.28
Figure 4.
Calibration models functions. (A) Example of fitted sigmoid function on support vector machine training set part 2 estimates (Platt scaling). (B) Example of a fitted isotonic regression on the training set part 2 estimates.
ISOTONIC REGRESSION
Isotonic regression uses a step function with monotonically increasing values on the estimates.36 There are 2 algorithms to find the stepwise function. One is the pair-adjacent violator algorithm and the second is the active set algorithm.36 Both minimize residuals under the assumption that there is no change in the ranking of estimates derived from equation 9:
| (9) |
where is the sorted actual outcome and is the fitted value (ie, precalibration estimate). Figure 4B shows an example of a fitted isotonic curve derived using training set part 2. R has a built-in function, “isoreg,” that fits the best monotonically increasing step function using model estimates as independent variable and the binary outcomes as dependent variable. An example is shown in the GitHub folder.19
BAYESIAN BINNING into QUANTILES
In addition to Platt scaling and isotonic regression, an ensemble method called BBQ has been recently proposed to improve calibration.26 BBQ is based on quantile binning developed by Zadrozny and Elkan.37 In quantile binning, estimates are partitioned into K bins of equal numbers of patients. For every estimate within each bin, an estimate is calibrated to be equal to the fraction of positive samples in that bin. One drawback of quantile binning is the arbitrariness of K. BBQ takes an ensemble approach, calculating multiple binning size models and combining them. Individual calibration functions calculated with different-sized bins are combined with a weighted sum.38 The MATLAB (The MathWorks, Natick, MA) implementation of BBQ can be accessed in the original article. Note that this approach, unlike monotonic (ie, order-preserving) transformations such as Platt scaling and isotonic regression, does not require or guarantee that the order of estimates to remain the same after the application of the calibration model, so a decrease or increase in discrimination after the application of such calibration models can occur.
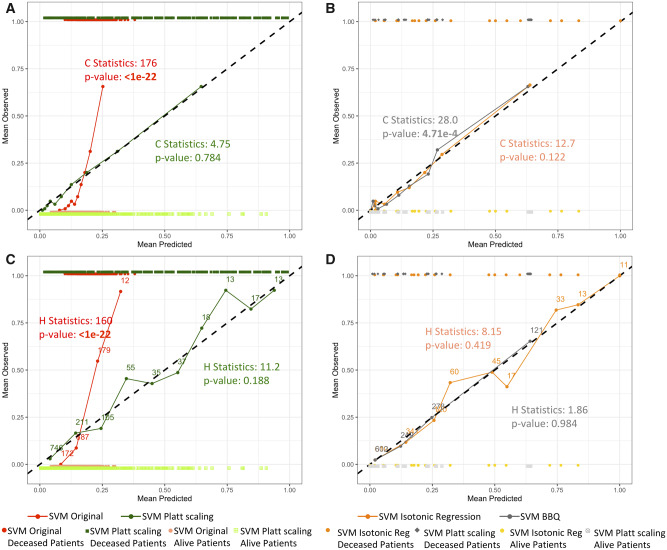
By applying Platt scaling, isotonic regression, and BBQ to test set estimates produced by the SVM model, the estimates became better calibrated, as shown by the Spiegelhalter’s z test and H-L test results in Table 2. The corresponding reliability diagrams are shown in Figure 5. Application of the calibration models also showed lowering of the ECE, MCE, and ICI. Furthermore, Cox’s slope and intercept became closer to 1 and 0, respectively. That is, there was consistency among most calibration measures, in that estimates obtained by calibration models resulted in smaller errors than the ones calculated for the original SVM estimates.
Figure 5.
Reliability diagrams of test set estimates produced by support vector machine (SVM) models after application of Platt scaling, isotonic regression, or Bayesian Binning into Quantiles (BBQ), grouped for the Hosmer-Lemeshow (H-L) C-statistics and the H-L H-statistics. Estimates produced by the models and their actual binary outcomes are plotted to show the distribution of the actual data. The Alive outcome is indicated as 0 and Deceased outcome is indicated as 1. Corresponding H-L statistics and P values are shown in the graphs. (A) Platt scaling and (B) isotonic regression and BBQ are grouped for the calculation of the H-L C-statistic. The number of patients within each bin is the same (n = 125). Panels C and D are grouped for the calculation of the H-L H-statistic. After applying calibration models, the SVM estimates show proper calibration.
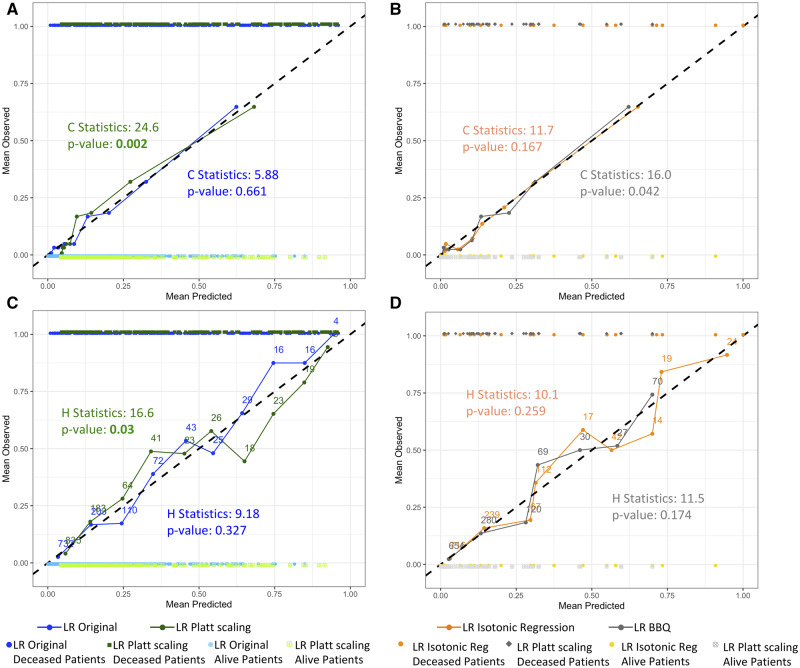
An unintended consequence of applying calibration models can be the worsening of calibration for models that are already well calibrated. Table 2 also shows results after applying calibration models to the test set estimates produced by the LR model, which were already well calibrated, as shown previously in Figures 3A and 3B. After applying Platt scaling, H-L C- or H-statistics returned significant P values, while the Spiegelhalter z test did not. Looking at the corresponding reliability diagrams in Figure 6, the lines with applications of Platt scaling show more deviation from the diagonal line than the original LR line. Such phenomenon sometimes happens with Platt scaling, as its underlying assumption is that the estimates’ distribution is sigmoidal in shape.39 When the logistic parametric assumptions are not met, properly calibrated estimates could suffer and become improperly calibrated. Application of isotonic regression and BBQ on the LR model also raised the H-L C- and H-statistics, indicating worsened calibration. This result is consistent with the increase in ECE, MCE, and ICI for LR calibrated with BBQ, and consistent with the increase in ICI for LR calibrated with isotonic regression.
Figure 6.
Reliability diagrams of test set estimates produced by logistic regression (LR) models after application of Platt scaling, isotonic regression, or Bayesian Binning into Quantiles (BBQ), grouped for the Hosmer-Lemeshow (H-L) C-statistics and the H-L H-statistics. Data points of estimates produced by the models and their actual binary outcomes are plotted to show the distribution of the actual data. The Alive outcome is indicated as 0 and Deceased outcome is indicated as 1. Corresponding H-L statistics and P values are shown in the graphs. Panels A and B are grouped for the calculation of the H-L C-statistic. The number of patients within each bin is the same (n = 125). Panels C and D are grouped for the calculation of the H-L H-statistic.
REAL CLINICAL DATA
The simulated experiments were repeated with real data set from the Nationwide Inpatient Sample.40 We picked 10 000 random patients from Nationwide Inpatient Sample 2014 dataset and predicted whether patients would need a major therapeutic procedure during their stay (20% did). The predictors were preadmission features (age, sex, race, admission month, elective or nonelective admission, expected primary payer, median household income quartile range, and presence or absence of 30 chronic conditions). Experiments were done with LR and SVM with radial kernel.
The results are shown in Table 3. H-L C- and H-statistics and the Spiegelhalter z test showed significance for both LR and SVM. For LR, Platt scaling was not able to produce calibrated estimates, as shown by the H-L test, ECE, MCE, and ICI. However, Cox’s slope and intercept suggest that calibration improved. For the SVM, Platt scaling produced statistically significant H-L C-statistics and H-L H-statistics, and elevated ECE, MCE, and ICI. Cox’s slope and intercept also suggest worse calibration. Isotonic regression and BBQ were able to improve calibration in both LR and SVM, resulting in nonsignificant P values for the Spiegelhalter z and H-L tests. They also lowered the MCE, ECE, and ICI. Cox’s slope and intercept became closer to 1 and 0, respectively, for both LR and SVM. All related reliability diagrams are shown in the Supplementary Figures.
Table 3.
Discrimination and calibration results of the LR and SVM models applied to the Nationwide Inpatient Sample test dataset
| LR | LR Platt scaling | LR isotonic regression | LR BBQ | SVM | SVM Platt scaling | SVM isotonic regression | SVM BBQ | |
|---|---|---|---|---|---|---|---|---|
| AUROC | 0.785 | 0.785 | 0.785 | 0.787 | 0.817 | 0.817 | 0.817 | 0.817 |
| Brier score | 0.119 | 0.121 | 0.118 | 0.120 | 0.109 | 0.110 | 0.104 | 0.105 |
| Spiegelhalter z score | 1.895 | –0.081 | 0.316 | –0.246 | –1.698 | –0.064 | 0.175 | –0.383 |
| Spiegelhalter P value | .029a | .468 | .376 | .402 | .044a | .542 | .351 | .313 |
| Average absolute error | 0.230 | 0.241 | 0.234 | 0.240 | 0.221 | 0.227 | 0.213 | 0.217 |
| H-L C-statistics | 39.473 | 43.439 | 13.744 | 12.351 | 50.540 | 67.228 | 10.760 | 10.865 |
| H-L C-statistic P value | 4.01 × 10–6a | 7.26 × 10–7a | .111 | .136 | 3.21 × 10–8a | 1.746 × 10–11a | .215 | .209 |
| H-L H-statistics | 38.084 | 61.353 | 10.456 | 6.683 | 146.129 | 69.947 | 9.556 | 8.804 |
| H-L H-statistic P value | 7.26 x 10–6a | 2.527 × 10–10a | .234 | .571 | <1 × 10–22a | 5.036 × 10–12a | .297 | .359 |
| MCE | 0.119 | 0.124 | 0.061 | 0.069 | 0.096 | 0.133 | 0.061 | 0.060 |
| ECE | 0.031 | 0.043 | 0.025 | 0.022 | 0.044 | 0.050 | 0.016 | 0.017 |
| Cox’s slope | 0.560 | 0.946 | 0.889 | 0.923 | 0.863 | 1.001 | 0.902 | 1.019 |
| Cox’s intercept | –0.601 | –0.177 | –0.230 | –0.203 | –0.374 | –0.149 | –0.257 | –0.123 |
| ICI | 0.027 | 0.038 | 0.018 | 0.025 | 0.052 | 0.061 | 0.015 | 0.015 |
Discrimination is measured by the AUROC, while calibration is measured by the Spiegelhalter z test, H-L test, MCE, ECE, Cox slope and intercept, and ICI. Estimates of the test set produced by both LR and SVM were improperly calibrated. Application of Platt scaling, isotonic regression, or BBQ was performed.
AUROC: area under the receiver-operating characteristic curve; BBQ: Bayesian Binning into Quantiles; ECE: expected calibration error; H-L, Hosmer-Lemeshow; ICI: integrated calibration index; LR: logistic regression; MCE: maximum calibration error; NIS: Nationwide Inpatient Sample; SVM: support vector machine.
shows significance.
DISCUSSION
Calibration and discrimination measurements are just a part of what needs to be considered when evaluating a model: the specific development-validation strategy is equally important and deserves its own tutorial. In our demonstrations we utilized hold-out validation, but results could differ if other validation techniques were used, such as 10-fold cross-validation, bootstrap techniques, jack-knife, etc.
The H-L test continues to be a very well-known proxy for a calibration measure. In the past 5 years, there have been more than 874 mentions on PubMed. The H-L test is a starting point to measuring calibration and needs to be considered. It has shortcomings, however, including the susceptibility to increase in power as sample size increases and the arbitrariness of number of bins to use. The H-L test’s probability of rejecting a poorly fitted model increases as the sample size increases. To remedy the problem, Paul et al41 created a function to calculate the number of bins according to sample size. The formula was able to keep the power consistent as sample size increased, but it could only handle sample sizes <25 000. For larger sample sizes, more complex techniques have been proposed.42,43 As for the arbitrariness of the number of bins, it is a problem shared by other measures. The MCE, ECE, and reliability diagrams all require binning. The Loess function in the ICI also requires an adjustable window parameter in order to calculate calibration. In model comparison, this is not a huge problem, as one can use the same test set and the same number of bins when comparing calibration of different models. However, it is a problem when asserting whether model estimates are well calibrated, as changing the number of bins can alter the P value.
Calibration measurements can be categorized into 2 groups in this tutorial. The H-L test, ECE, and MCE take an approach that requires binning based on estimates. The Cox intercept and slope and ICI regress the estimates to the true outcomes. In terms of interpretability, the binning methods are more readily understandable by the medical community. More recent methods to measure calibration are increasingly being used, and new guidelines on how to assess whether they are adequate for a particular use case will develop over time. We summarize the pros and cons of the methods we have presented in this article in Table 4. The references refer to studies that used these measures.
Table 4.
Summary of advantages and disadvantages of calibration measurement methods presented in this tutorial
| Calibration measure (examples of studies in which the measure was used) | Pros | Cons |
|---|---|---|
| Brier score44–46 | < id="1124" data-dummy="list" list-type="none">
|
The contribution of each component (discrimination, calibration) is not easy to calculate or interpret. |
| Spiegelhalter’s z test47,48 | Extension of Brier score that measures calibration only. P value can serve as a guide for how calibrated a model is. | Not intuitive. |
| Average absolute error | Easy calculation. Intuitive. | Same problems as Brier score. Rarely used. |
| H-L test28,49 | Widely used in the biomedical literature. P value can serve as a guide for how calibrated a model is. | < id="1155" data-dummy="list" list-type="none">
|
| Reliability diagram25,26,50,51 | Allows for visualization of regions of miscalibration and the “direction” of miscalibration (ie, underestimation, overestimation) | Not a continuous graph. Hard to see when estimates are clustered in certain regions (zoom into a portion of the graph may be needed). |
| Expected calibration error and maximum calibration error25,52,53 | Intuitive. | No statistical test to help determine whether a model is adequately calibrated or not. |
| Cox’s slope and intercept54–56 | Summarizes direction of miscalibration (ie, overall underestimation or overestimation). | Can still result in perfect calibration of 0 and 1 even if regions are miscalibrated. |
| Integrated calibration index | Can capture regions of miscalibration that Cox’s slope and intercept cannot. | Requires Loess to build calibration model. Not intuitive. |
H-L: Hosmer-Lemeshow.
In our simple example, application of Platt scaling and isotonic regression on the SVM-derived estimates had good results. While there are other methods that were built on such techniques, and more are being created for modern deep learning,24–26,39,57–59 Platt scaling and isotonic regression are relatively easy to understand and implement. They can act as the benchmark that subsequent calibration models are compared with. With ease of interpretability as the main advantage of these 2 techniques, they are not without faults. As illustrated in our example, Platt scaling may fail when model is already well calibrated. It performs best under the assumption that the estimates are close to the midpoint and away from the extremes.39 Therefore, Platt scaling may not be suitable for estimates produced by naive Bayes or AdaBoost models, which tend to produce extreme estimates close to 0 and 1. In terms of isotonic regression, the criticism is that it lacks continuousness. Because the fitted regression function is a piecewise function, a slight change in the uncalibrated estimates can result in dramatic difference in the estimates (ie, a change in step). Also, owing to the stepwise nature of the function, uncalibrated estimates that fall on the same “step” result in having the same calibrated value, eliminating any distinction between those patient estimates (similarly to quantile binning). However, there are smoothing techniques to make the estimates continuous.60
Finally, the available packages currently used to measure calibration are sparse and missing some key documentation. There is a need for better descriptions of how such techniques were implemented.
CONCLUSION
While discrimination is the most commonly used measure of how well a predictive model performs, calibration of estimates is also important. With the help of R packages, it is not difficult to measure calibration alongside discrimination when reporting on a model’s predictive performance. Also, there are simple techniques that can improve calibration without the need to retrain a model. To improve discrimination, parameters will need to be tuned or a completely different model may be required, whereas to improve calibration, there are techniques that do not require retraining. In this tutorial, we raise the awareness of the importance and meaning of calibration in clinical predictive modeling by providing simple and readily reproducible examples.
FUNDING
This work was supported by National Institutes of Health grants R01GM118609 (to YH and LO-M) and R01HL136835 (to YH and LO-M).
AUTHOR CONTRIBUTIONS
YH mainly drafted the manuscript. WL conducted experiments on real clinical data and provided guidance to machine learning discussions. FM contributed introductory paragraphs. YH wrote the sample code. RG contributed clinical data and consulted on related questions. LO-M was principal investigator: she provided the original idea, wrote parts of the introduction, oversaw the process, steered the direction of the article, and provided critical editing.
Supplementary Material
ACKNOWLEDGMENTS
We thank 3 anonymous reviewers for insightful suggestions.
CONFLICT OF INTEREST STATEMENT
None declared.
REFERENCES
- 1. Steyerberg EW, Vickers AJ, Cook NR, et al. Assessing the performance of prediction models: a framework for traditional and novel measures. Epidemiology 2010; 21 (1): 128–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Alba AC, Agoritsas T, Walsh M, et al. Discrimination and calibration of clinical prediction models: users’ guides to the medical literature. JAMA 2017; 318 (14): 1377–84. [DOI] [PubMed] [Google Scholar]
- 3. Steyerberg EW. Clinical Prediction Models. New York, NY: Springer; 2009. [Google Scholar]
- 4. Hurd MD, Martorell P, Delavande A, Mullen KJ, Langa KM.. Monetary costs of dementia in the United States. N Engl J Med 2013; 368 (14): 1326–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Licher S, Yilmaz P, Leening MJG, et al. External validation of four dementia prediction models for use in the general community-dwelling population: a comparative analysis from the Rotterdam Study. Eur J Epidemiol 2018; 33 (7): 645–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Firnhaber JM. Estimating cardiovascular risk. Am Fam Physician 2017; 95 (9): 580–1. [PubMed] [Google Scholar]
- 7.MELD Calculator - OPTN. https://optn.transplant.hrsa.gov/resources/allocation-calculators/meld-calculator/ Accessed October 29, 2019.
- 8. Fenlon C, O’Grady L, Doherty ML, Dunnion J.. A discussion of calibration techniques for evaluating binary and categorical predictive models. Prev Vet Med 2018; 149: 107–14. [DOI] [PubMed] [Google Scholar]
- 9. Walsh CG, Sharman K, Hripcsak G.. Beyond discrimination: a comparison of calibration methods and clinical usefulness of predictive models of readmission risk. J Biomed Inform 2017; 76: 9–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Steyerberg EW, Vergouwe Y.. Towards better clinical prediction models: seven steps for development and an ABCD for validation. Eur Heart J 2014; 35 (29): 1925–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wessler BS, Lai Yh L, Kramer W, et al. Clinical prediction models for cardiovascular disease: tufts predictive analytics and comparative effectiveness clinical prediction model database. Circ Cardiovasc Qual Outcomes 2015; 8 (4): 368–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Harrell FE, Lee KL, Califf RM, Pryor DB, Rosati RA.. Regression modelling strategies for improved prognostic prediction. Stat Med 1984; 3 (2): 143–52. [DOI] [PubMed] [Google Scholar]
- 13. Harrell FE., Jr. Evaluating the yield of medical tests. JAMA 1982; 247 (18): 2543–6. [PubMed] [Google Scholar]
- 14. Kramer AA, Zimmerman JE.. Assessing the calibration of mortality benchmarks in critical care: The Hosmer-Lemeshow test revisited. Crit Care Med 2007; 35 (9): 2052–6. [DOI] [PubMed] [Google Scholar]
- 15. Niculescu-Mizil A. Caruana Predicting good probabilities with supervised learning. In: Proceedings of the 22nd International Conference on Machine Learning - ICML ‘05 New York, NY: ACM Press; 2005: 625–32.
- 16. Zou KH, Liu A, Bandos AI, Ohno-Machado L, Rockette HE.. Statistical Evaluation of Diagnostic Performance: Topics in ROC Analysis. Boca Raton, FL: CRC Press; 2016. [Google Scholar]
- 17. Rufibach K. Use of Brier score to assess binary predictions. J Clin Epidemiol 2010; 63 (8): 938–9; author reply 939. [DOI] [PubMed] [Google Scholar]
- 18. Brier GW. Verification of forecasts expressed in terms of probability. Mon Weather Rev 1950; 78 (1): 1–3. [Google Scholar]
- 19.GitHub - easonfg/cali_tutorial. https://github.com/easonfg/cali_tutorial Accessed August 2, 2019.
- 20. Hosmer DW, Lemesbow S.. Goodness of fit tests for the multiple logistic regression model. Commun Stat Theory Methods 1980; 9 (10): 1043–69. [Google Scholar]
- 21. Hosmer DW. Applied Logistic Regression. 2nd ed.Hoboken, NJ: Wiley-Interscience; 2000. [Google Scholar]
- 22. Lele SR. A new method for estimation of resource selection probability function. J Wildl Manag 2009; 73 (1): 122–7. [Google Scholar]
- 23. Freeman EA, Moisen G.. PresenceAbsence: an R package for presence absence analysis. J Stat Softw 2008; 23 (11). doi : 10.18637/jss.v023.i11. [Google Scholar]
- 24. Wang Y, Li L, Dang C.. Calibrating classification probabilities with shape-restricted polynomial regression. IEEE Trans Pattern Anal Mach Intell 2019; 41 (8): 1823–27. doi : 10.1109/TPAMI.2019.2895794. [DOI] [PubMed] [Google Scholar]
- 25. Guo C, Pleiss G, Sun Y, Weinberger K. On calibration of modern neural networks. In: Proceedings of the 34th International Conference on Machine Learning 2017; Sydney, Australia.
- 26. Naeini MP, Cooper GF, Hauskrecht M.. Obtaining well calibrated probabilities using Bayesian binning. Proc Conf AAAI Artif Intell 2015; 2015: 2901–7. [PMC free article] [PubMed] [Google Scholar]
- 27.Cox DR. Two further applications of a model for binary regression. Biometrika 1958; 45 (3–4): 592–65. [Google Scholar]
- 28. Nascimento MS, Rebello CM, Vale L, Santos É, Prado CD.. Spontaneous breathing test in the prediction of extubation failure in the pediatric population. Einstein (Sao Paulo) 2017; 15 (2): 162–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Austin PC, Steyerberg EW.. The Integrated Calibration Index (ICI) and related metrics for quantifying the calibration of logistic regression models. Stat Med 2019; 38 (21): 4051–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jiang X, Menon A, Wang S, Kim J, Ohno-Machado L.. Doubly Optimized Calibrated Support Vector Machine (DOC-SVM): an algorithm for joint optimization of discrimination and calibration. PLoS One 2012; 7 (11): e48823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Walsh CG, Ribeiro JD, Franklin JC.. Predicting risk of suicide attempts over time through machine learning. Clin Psychol Sci 2017; 5 (3): 457–69. [Google Scholar]
- 32. Walsh CG, Ribeiro JD, Franklin JC.. Predicting suicide attempts in adolescents with longitudinal clinical data and machine learning. J Child Psychol Psychiatry 2018; 59 (12): 1261–70. [DOI] [PubMed] [Google Scholar]
- 33. Van Calster B, Nieboer D, Vergouwe Y, De Cock B, Pencina MJ, Steyerberg EW.. A calibration hierarchy for risk models was defined: from utopia to empirical data. J Clin Epidemiol 2016; 74: 167–76. [DOI] [PubMed] [Google Scholar]
- 34. Riley RD, Ensor J, Snell KIE, et al. External validation of clinical prediction models using big datasets from e-health records or IPD meta-analysis: opportunities and challenges. BMJ 2016; 353: i3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Platt JC. Probabilistic outputs for support vector machines and comparisons to regularized likelihood methods In: Smola A, Bartlett P, Schölkopf B, Schuurmans D, eds. Advances in Large Margin Classifiers. Cambridge, MA: MIT Press; 2000. [Google Scholar]
- 36. Leeuw J. D, Hornik K, Mair P.. Isotone optimization inR: Pool-Adjacent-Violators Algorithm (PAVA) and active set methods. J Stat Softw 2009; 32 (5). doi : 10.18637/jss.v032.i05. [Google Scholar]
- 37. Zadrozny B, Elkan C. Obtaining calibrated probability estimates from decision trees and naive Bayesian classifiers. In: Proceedings of the Eighteenth International Conference on Machine Learning; 2001: 609–16.
- 38. Heckerman D, Geiger D, Chickering DM.. Learning Bayesian networks: the combination of knowledge and statistical data. Mach Learn 1995; 20 (3): 197–243. [Google Scholar]
- 39. Kull M, Filho T S, Flach P.. Beta calibration: a well-founded and easily implemented improvement on logistic calibration for binary classifiers. Proc Int Conf Artif Intell Stat 2017; 54: 623–31. [Google Scholar]
- 40.Healthcare Cost and Utilization Project. HCUP Nationwide Inpatient Sample. Rockville, MD: Agency for Healthcare Research and Quality; 2012. [PubMed] [Google Scholar]
- 41. Paul P, Pennell ML, Lemeshow S.. Standardizing the power of the Hosmer-Lemeshow goodness of fit test in large data sets. Stat Med 2013; 32 (1): 67–80. [DOI] [PubMed] [Google Scholar]
- 42. Yu W, Xu W, Zhu L.. A modified Hosmer–Lemeshow test for large data sets. Commun Stat Theory Methods 2017; 46 (23): 11813–25. [Google Scholar]
- 43. Lai X, Liu L.. A simple test procedure in standardizing the power of Hosmer–Lemeshow test in large data sets. J Stat Comput Simul 2018; 88 (13): 2463–72. [Google Scholar]
- 44. Ambale-Venkatesh B, Yang X, Wu CO, et al. Cardiovascular event prediction by machine learning: the multi-ethnic study of atherosclerosis. Circ Res 2017; 121 (9): 1092–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sahm F, Schrimpf D, Stichel D, et al. DNA methylation-based classification and grading system for meningioma: a multicentre, retrospective analysis. Lancet Oncol 2017; 18 (5): 682–94. [DOI] [PubMed] [Google Scholar]
- 46. Bendapudi PK, Hurwitz S, Fry A, et al. Derivation and external validation of the PLASMIC score for rapid assessment of adults with thrombotic microangiopathies: a cohort study. Lancet Haematol 2017; 4 (4): e157–64. [DOI] [PubMed] [Google Scholar]
- 47. Manktelow BN, Draper ES, Field DJ.. Predicting neonatal mortality among very preterm infants: a comparison of three versions of the CRIB score. Arch Dis Child Fetal Neonatal Ed 2010; 95 (1): F9–13. [DOI] [PubMed] [Google Scholar]
- 48. Spiegelhalter DJ. Probabilistic prediction in patient management and clinical trials. Stat Med 1986; 5 (5): 421–33. [DOI] [PubMed] [Google Scholar]
- 49. Khavanin N, Qiu CS, Mlodinow AS, et al. External validation of the breast reconstruction risk assessment calculator. J Plast Reconstr Aesthet Surg 2017; 70 (7): 876–83. [DOI] [PubMed] [Google Scholar]
- 50. Bröcker J, Smith LA.. Increasing the reliability of reliability diagrams. Weather Forecast 2007; 22 (3): 651–661. [Google Scholar]
- 51. Yao S, Zhao Y, Zhang A, et al. Deep learning for the internet of things. Computer 2018; 51 (5): 32–41. [Google Scholar]
- 52. Lee K, Lee K, Lee H, Shin J. Training confidence-calibrated classifiers for detecting out-of-distribution samples. Int: proceedings of the Sixth International Conference on Learning Representations; April 30, 2018 to May 3, 2018; Vancouver, Canada.
- 53. Maddox W, Garipov T, Izmailov P, Vetrov D, Wilson AG. A simple baseline for Bayesian uncertainty in deep learning. arXiv 2019. Dec 31 [E-pub ahead of print].
- 54. Steyerberg EW, Nieboer D, Debray TPA, van Houwelingen HC.. Assessment of heterogeneity in an individual participant data meta-analysis of prediction models: An overview and illustration. Stat Med 2019; 38 (22): 4290–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Norvell DC, Thompson ML, Boyko EJ, et al. Mortality prediction following non-traumatic amputation of the lower extremity. Br J Surg 2019; 106 (7): 879–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Nelson BB, Dudovitz RN, Coker TR, et al. Predictors of poor school readiness in children without developmental delay at age 2. Pediatrics 2016; 138 (2): e20154477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zadrozny B. Elkan Transforming classifier scores into accurate multiclass probability estimates. In: Proceedings of the Eighth ACM SIGKDD International Conference on Knowledge Discovery and Data Mining - KDD ‘02 New York, NY: ACM Press; 2002: 694–9.
- 58. Jiang X, Osl M, Kim J, Ohno-Machado L.. Calibrating predictive model estimates to support personalized medicine. J Am Med Inform Assoc 2012; 19 (2): 263–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Demler OV, Paynter NP, Cook NR.. Tests of calibration and goodness-of-fit in the survival setting. Stat Med 2015; 34 (10): 1659–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Jiang X, Osl M, Kim J, Ohno-Machado L.. Smooth isotonic regression: a new method to calibrate predictive models. AMIA Jt Summits Transl Sci Proc 2011; 2011: 16–20. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.