Abstract
Autologous transplantation of spermatogonial stem cells is a promising new avenue to restore fertility in infertile recipients. Expansion of the initial spermatogonial stem cell pool through cell culturing is a necessary step to obtain enough cells for effective repopulation of the testis after transplantation. Since in vitro propagation can lead to (epi-)genetic mutations and possibly malignant transformation of the starting cell population, we set out to investigate genome-wide DNA methylation status in uncultured and cultured primary testicular ITGA6+ sorted cells and compare them with germ cell tumor samples of the seminoma subtype. Seminomas displayed a severely global hypomethylated profile, including loss of genomic imprinting, which we did not detect in cultured primary testicular ITGA6+ cells. Differential methylation analysis revealed altered regulation of gamete formation and meiotic processes in cultured primary testicular ITGA6+ cells but not in seminomas. The pivotal POU5F1 marker was hypomethylated in seminomas but not in uncultured or cultured primary testicular ITGA6+ cells, which is reflected in the POU5F1 mRNA expression levels. Lastly, seminomas displayed a number of characteristic copy number variations that were not detectable in primary testicular ITGA6+ cells, either before or after culture. Together, the data show a distinct DNA methylation patterns in cultured primary testicular ITGA6+ cells that does not resemble the pattern found in seminomas, but also highlight the need for more sensitive methods to fully exclude the presence of malignant cells after culture and to further study the epigenetic events that take place during in vitro culture.
Introduction
In vitro propagation of cryopreserved spermatogonial stem cells (SSCs) followed by autologous transplantation of cultured SSCs (SSCT) into the testes is viewed as a promising new technique to treat male survivors of childhood cancer for sub- or infertility [1–5]. Theoretically, by utilizing SSCT in this group of otherwise infertile patients, endogenous spermatogenesis can be permanently enhanced or rescued. SSCT has the additional benefit of using the patient’s own cells to rescue fertility, rendering SSCT a preferred option to current clinical alternatives such as the use of donor sperm to achieve pregnancy. The robustness of spermatogenic rescue following SSCT has been demonstrated for various species including mice, cattle and primates [6–10] and SSCT treated animals are capable of producing offspring which appears healthy [11–15] and fertile, at least in rodents [12, 14–16]. The odds of successful testicular colonization after SSCT are predominantly dictated by the number of transplanted SSCs [17]. Since the proportion of true SSCs will be limited in a biopsy from a human prepubertal small testis, in vitro propagation of the initial SSC pool is a necessary step in the SSCT protocol.
A potential risk with SSCT for clinical use is the risk of cancer induction in the recipient originating from the transplanted cell population [18, 19]. This risk is two-fold: either primary cancerous cells (originating from non-solid tumors) that were present in the original biopsy can be re-introduced into the recipient upon transplantation, or normal germ/somatic cells could give rise to a transformed line of cancer cells with malignant properties during in vitro propagation. Several studies have been published that describe the use of different techniques, such as cell culture or FACS, to successfully eliminate malignant cells from contaminated testicular tissue samples [20–22]. The possible secondary risk of testicular cells undergoing culture-induced malignant transformation remains largely unexplored in the context of fertility restoration.
A common hallmark of malignant cells is the occurrence of disturbances in 5-cytosine methylation marks, resulting in an epigenetic landscape that may differ greatly from normal cells [23]. Such alterations in DNA methylation have been found in testicular malignant germ cell tumors of all histological variants, including seminomas, as well its precursor lesion germ cell neoplasia in situ (GCNIS, previously known as CIS) [24–26]. In addition to alterations in their methylome, these malignant cells are often susceptible to and characterized by DNA copy number variations and can in fact even be subclassified based on the occurrence of CNVs in certain loci [27], including TGCTs [28].
Due to the available evidence that in vitro proliferation of primary (stem) cells can affect both the genetic [29, 30] and epigenetic [31–34] integrity of the cell’s genome and possibly trigger malignant transformation, we set out to study whether primary human testicular cells can become malignant during propagation in cell culture. Sporadic genetic aberrations have been observed in long-term cultures of induced pluripotent stem cells (iPS) and embryonic stem cells (ES) [35, 36], displaying recurrent duplications of chromosomal regions associated with increased genetic instability and apoptotic resistance (12q, 17p) also found in various GCNIS-derived TGCTs.
Here, we investigated genome-wide methylation patterns and genomic CNVs in freshly sorted and long-term cultured and sorted integrin alpha-6 (ITGA6) positive human testicular cells (n = 4 cultures) and compared these methylation patterns to primary seminoma tumors (n = 3), focusing on genome-scale DNA patterning as well as known oncogenic regions. Selection of ITGA6 positive cells was based on its localization to the plasma membrane of spermatogonia in the in vivo human testis [37] and previous demonstrations of the testis-repopulating capacity of the ITGA6+ testicular cell subpopulation, both before and after culturing [38–41].
Materials and methods
Ethical approval
This study does not report research involving human participants. We used spare human testicular tissue fragments with written or oral informed consent from prostate cancer patients (n = 4) ranging in age from 49–83, that underwent bilateral orchidectomy as part of their cancer treatment at the Academic Medical Center (AMC), Amsterdam, The Netherlands. In accordance with Dutch law, spare tissues can be used for research with permission of the patients without further permission of an ethical committee since no additional interventions were needed to obtain these samples. Seminoma DNA samples, also used in a previous study [25], were obtained with informed consent from patients in the Netherlands and were handled according to the Code for Proper Secondary Use of Human Tissue as directed by the Dutch Federation of Medical Scientific Societies (https://www.federa.org/codes-conduct).
Patient samples
Adult testicular donor tissues were cryopreserved in 1× MEM, 20% FCS, 8% DMSO upon retrieval and stored for later cell isolations. Haematoxylin-eosin stainings of testicular cross sections were used to confirm the presence of all germ cell stages and revealed normal spermatogenesis in all donor tissues. Seminoma samples were stage I tumors that did not show metastases during follow-up. Seminoma samples were obtained and used directly after surgery. Part of the biopsies were submitted to the pathology department for histological examination and alkaline phosphatase (dAP) reactivity assays [42], the remainder was subjected to DNA isolation as described in previous work [25]. Lymphocyte infiltration scoring showed moderate to high infiltration in two of the tumors (L11-119: 57.5%-66.1% and L11-123: 68.4%-71.1%) and moderate infiltration in the remaining tumor (L10-358: 37.3%-52.5%) with an expected overestimation of 10–20% lymphocyte component.
Primary testicular cell isolation and culture
Biopsies were thawed and the germ cell fraction located on the basal membrane of seminiferous tubules was isolated as described previously [43]. Briefly, testicular biopsies were subjected to a two-step enzyme digestion (1mg/mL type I collagenase (4196, Worthington), 1mg/mL type II hyaluronidase (H2126, Sigma) and 1mg/mL trypsin TRL3 (3707, Worthington), followed by 1mg/mL type I collagenase and 1mg/mL type II hyaluronidase) and the resulting single-cell suspension was plated overnight in 1× MEM containing 20% FCS. The next day, floating cells (containing the germ cell fraction) were separated from the attached cell fraction via gentle pipetting and transferred to a new culture flask. This mixed primary testicular cell (PTC) fraction was then either directly subjected to magnetic-activated cell sorting for ITGA6+ cells (day 0; d0-PTC) to enrich for spermatogonia or first brought into culture and then sorted (long-term; LT-PTC). The medium used for PTC culture contained StemPro-34 SFM medium (10639–011, Life Technologies) supplemented with several key growth factors promoting spermatogonial stem cells self-renewal (GDNF, EGF, PDGF, LIF) as described elsewhere [43]. Cells were cultured at 37°C and 5% CO2 and passaged using trypsin/EDTA at 80–90% confluence, which took on average 13 days for the first passage and from that point on every 7 to 10 days over a period of 50–54 days. Medium was refreshed twice a week.
Magnetic-activated cell sorting
To enrich the initial testicular cell fractions for spermatogonia before and after culture, we performed magnetic assistant cell sorting (MACS) for cells expressing the ITGA6 membrane protein [38–40]. Cells were harvested using 0.25% trypsin/EDTA, filtered using a 40 μm sterile filter and labeled with biotinylated anti-ITGA6 antibody (BioLegend, 1μl ab per 5 × 105 cells) in 100 μl MACS buffer (0.5%BSA, 2mM EDTA in sterile PBS) for 30 minutes at 4°C, followed by incubation with 1:5 (v:v) anti-biotin micro magnetic beads (Miltenyi Biotec) in 100 μl MACS buffer for 15 minutes at 4°C. Cell-bead complexes were then washed 3× in MACS buffer and transferred to a magnetic stand to separate labeled cells from non-labelled cells using a Large Cell separation column (Miltenyi Biotec).
DNA isolation and bisulfite conversion
Genomic DNA was isolated from ITGA6+ PTCs using the QIAamp DNA Mini Kit (Qiagen, 51306) and eluted in low-TE buffer (10 mM Tris-HCl, 0.1 mM EDTA in sterile H20, pH 8.0). Concentration and quality of DNA was measured on a ND-1000 (Thermo Scientific) spectrophotometer and 1 μg of high quality DNA (absorbance 260/230 ≥ 1.8; 260/280 ≥ 2.0) was subjected to bisulfite conversion using the EZ DNA Methylation Gold Kit (Zymo Research). High bisulfite conversion rates were confirmed by single curves using high resolution melt analysis (HRMA) of the H19 imprinting control region 1 (H19-IC1) in sperm control DNA as described elsewhere [44], as well as a two-step verification using internal control probes on the Human Methylation-450k BeadChip microarrays. Primers used for HRMA were as follows: forward ATGTAAGATTTTGGTGGAATAT and reverse ACAAACTCACACATCACAACC.
Human Methylation-450k BeadChip microarray analysis
Methylation profiles of uncultured and cultured ITGA6+ PTCs were generated using HumanMethylation 450k BeadChip microarrays (Illumina), which cover 485,577 genomic CpG sites and capture ≥ 99% of RefSeq genes. Sample preparation and bisulfite quality checks were conducted at the department of Clinical Genetics at the Academic Medical Center (AMC), Amsterdam, the Netherlands. Bisulfite-converted DNA samples were hybridized to the arrays at Service XS, Leiden, the Netherlands according to manufacturer’s protocol. BeadChip images were scanned using an Illumina iScan array scanner and data was extracted into GenomeStudio (v2011.1, Methylation Module v1.9.0) using default analysis settings. Data in this study were further processed using R (v3.1.1, platform: x86_64-w64-mingw32/x64) and Rstudio (v0.98.1091) for Windows 7 (64 bits); packages included lumi (v2.20.1), limma (3.22.7) and ggplot2 (v1.0.0). The data set supporting the results of this article is available in the Gene Expression Omnibus repository, accession number GSE72444.
Data preprocessing
Raw scanner data (.iDat files) were imported into R and preprocessed using lumi [45]; preprocessing steps included removal of non-CpG probes and non-CpG SNP probes, CpG probes not passing the fluorescence detection limit (detectable signal in 95% of samples, p ≤ 0.01), CpG probes known to have multiple targets in the genome and CpG probes with SNPs at or within 10bp of a target CpG (allele frequency ≥ 0.05) [46]. Preprocessing with these criteria resulted in 452,354/485,577 probes suitable for downstream analysis. Fluorescence values of preprocessed CpG probes were subjected to optimized color-bias adjustment and quantile normalization [47] followed by BMIQ-based correction for type I and type II probe bias [48]. Data processing yielded both ß values (in,methylated / (in,unmethylated + in,methylated)), where in is intensity signal of the nth probe and related M-values (log2(ßn / 1–ßn) where ßn is the ß value of the nth probe) for each probe as methylation estimates. We used an extended annotation file (available in the Gene Expression Omnibus repository, accession number GPL18809) and the VariantAnnotation package for CpG annotation, adding a number of functional genomic classes to the standard annotation manifest provided by Illumina.
DMR identification
We used DMRforPairs (v1.2.0) [49] to identify CpG dense regions and define DMRs in the dataset using default settings. For statistical analysis of candidate DMRs we opted to use M-values as opposed to ß-values due to the higher rates of true discovery [50]. A genomic interval was classified as a DMR when it contained at least 4 CpGs within a distance of ≤ 200bp from each other and showed a significant difference in median M-value of ≥ 1.4 (Benjamini-Hochberg adjusted p-value ≤ 0.05, Mann Whitney U test) between two samples.
Copy number variation analysis
Copy number variations were calculated using a previously published approach [51] that we modified to also include CpGs on the X chromosome. CpG probes were divided over 8,949 chromosomal bins and median probe fluorescence was calculated for each bin in a DNA methylation microarray control dataset of 10 female blood samples with stable copy numbers. Based on the fluorescence levels in this control set, a baseline was defined for stable copy numbers of all autosomes (chromosomes 1–22) and chromosome X. Bins where the median probe fluorescence in an experimental sample (PTC, SE) was significantly different from bin fluorescence in the control set were assigned as having a CNV in that sample, designated as +1 for gains and –1 for losses. Cutoff levels for the designation of chromosomal copy number variants provided in S1 File were adapted from [51], distinguishing between homozygous deletions (≤–0.96, brown), hemizygous/mosaic deletions (≤–0.24, red), neutral (between 0.12 and –0.24, grey), duplications (≥ +0.12, green) and high-copy gains (≥ +0.72, blue).
Reverse transcriptase PCR & qPCR
Reverse transcriptase and quantitative polymerase chain reactions (PCR) were carried out using amplified RNA from the same cells that were used for DNA methylation profiling. RNA was isolated using a MagnaPure LC machine (Life Technologies) and amplified using an Ovation RNA-Seq System V2 kit (NuGEN Technologies) prior to analysis. Integrity of the starting RNA was verified by Bio-Analyzer gel electrophoresis analysis (RIN scores ranged from 7.1 to 9.8). Oligonucleotide primer sequences used for the detection of ITGA6, DISL3 (rt-PCR) and POU5F1, EPN2, HeatR6 (qPCR) transcripts are listed in S1 Table. Primer oligonucleotides used in this study were designed to be intron-spanning to avoid amplification of genomic DNA during the reaction. RT-PCR reactions were set up using 1x PCR buffer containing 0.5U Taq polymerase (both Roche), 0.5 μM of forward and reverse primer and 2mM dNTPs in a final volume of 25 μl.
Quantitative PCR reactions were carried out in triplicate 10 μl reactions and reactions were set up using 2.5 ng amplified cDNA, 2x Roche cyber green mix (SY Green Master mix, Roche), 0.2 μM of forward and reverse primer in a final volume 10 μl. Fluorescence per cycle was measured on a LightCycler480 PCR machine (Roche). Specificity of the PCR reactions was ascertained through detection of single bands on agarose gel of the amplified products. We utilized the LinReg method [52] to calculate the starting concentration (N0 value) of POU5F1, EPN2 and HeatR6 transcripts in a given sample based on the fluorescence level per cycle number. Products of PCR were separated on a 3% agarose TBE gel containing ethidium bromide and visualized on a Gel Doc XR system (Bio-Rad).
Results
To study the possible transformation of normal human testicular cells into malignant cells during prolonged cell culture, we generated genome-scale 5-mC profiles of ITGA6+ enriched primary testicular cells (PTC, n = 4) both before culture (d0-PTC) and after long-term culture (LT-PTC), as well as three primary seminoma lesions (SE, n = 3) (Fig 1A). The general consensus in the literature is that spermatogonial stem cells (SSC) represent the fraction of male germ cells that are able to self-renew and differentiate into sperm via the process of spermatogenesis (reviewed in [53]). A distinction is made between SSCs and spermatogonia, the latter of which is an umbrella term for sperm cell precursors including both veritable SSCs as well as more differentiated premeiotic germ cell types that do not possess stem cell potential. We have previously demonstrated that our culture method, either with or without selection of ITGA6+ cells, yields a population of cells that have the ability to migrate to the basal membrane of the seminiferous tubules after xenotransplantation to mouse recipients, as evidenced by positive immunofluorescent staining of COT-1 DNA sequences (a sequence exclusively present in human DNA) in the transplanted tissues [40, 43, 54]. Although human SSCs cannot differentiate in the mouse testis microenvironment due to the large phylogenetic difference between mice and humans [17], these findings form the golden standard to evaluate the presence of spermatogonial stem cells in a transplanted population of cells and confirm that the human ITGA6+ testicular population contains SSCs.
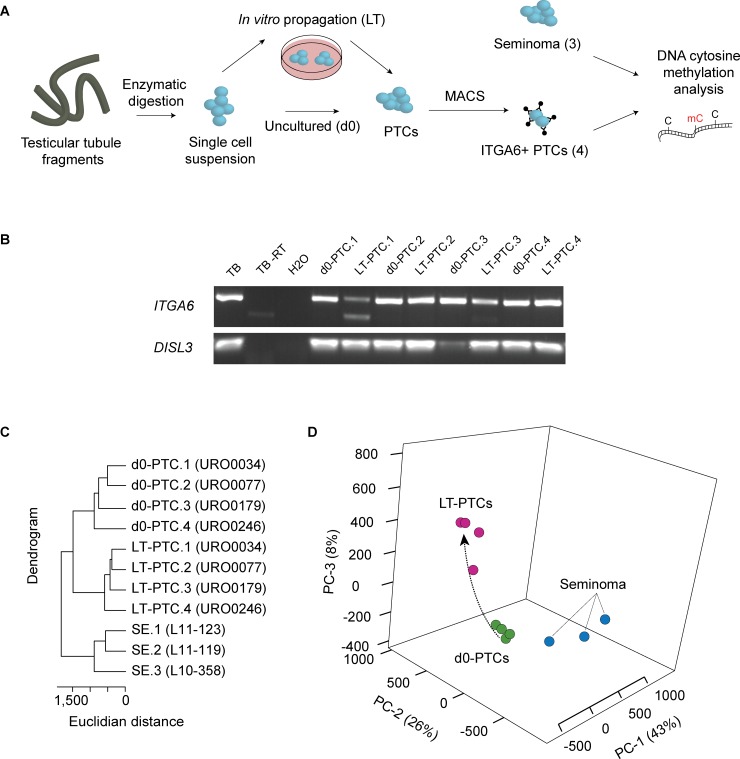
Fig 1. Generation of genome-scale DNA methylation profiles in primary cultured and sorted ITGA6+ testicular cells (PTC) and seminomas tumors (SE).
In total, four PTC cultures and three primary seminoma samples were analyzed using the Illumina HumanMethylation 450k microarray platform. (A) Testicular tubule fragments were dissociated using enzymatic digestions and the resulting single cell suspension was either directly sorted for ITGA6+ cells (d0, day 0) or first cultured for 50–54 days (LT, long-term) and then sorted. Both d0-PTC/LT-PTC sorted fractions and SE were subjected to DNA cytosine methylation analysis. (B) Expression of ITGA6 and DISL3 (reference gene) mRNA transcripts in d0-PTCs and LT-PTCs. Total testicular tissue biopsy (TB) was used as a positive control, a cDNA synthesis using demineralized H2O as input was used as a negative control. (C) Unsupervised hierarchical clustering revealed unique DNA methylation landscapes in LT-PTCs and seminomas as compared to d0-PTCs with a clear segregation of PTCs and seminomas into distinct groups. (D) 3D principal component analysis (PCA) shows that the observed variance in DNA methylation levels in LT-PTCs and SE samples as compared to d0-PTCs occurs at different genomic regions. d0-PTCs samples are displayed in green, LT-PTCs in pink and SE in blue.
As expected, d0-PTCs and LT-PTCs displayed continued expression of the ITGA6 mRNA transcript during culture (Fig 1B). Unsupervised hierarchical clustering revealed clear segregation of the d0-PTC, LT-PTC and seminoma samples into three separate groups, highlighting a distinct DNA methylation profile in seminomas as compared to PTCs both before and after culturing (Fig 1C and 1D).
When analyzing the DNA methylation distribution over all included CpG probes (n = 451,524 CpGs), we found that seminomas display a distinct globally hypomethylated profile compared to cultured and uncultured PTCs (Fig 2A). Global genomic hypomethylation is a well described feature of tumor cells and seminomas in particular [28, 55–57], contrasting with normal cells which have a bimodal distribution of either fully methylated CpGs (100% methylation) or unmethylated CpGs (0% methylation). The extent of hypomethylation in seminomas correlated negatively with the lymphocyte infiltration score, i.e. the tumor possessing the lowest degree of lymphocyte infiltration (L10-358, 37% lymphocyte infiltration) had the most severely hypomethylated profile. This corroborates the findings of Shen et al., who recently studied 137 TGCTs and described the existence of multiple subtypes of seminoma lesions that are distinguishable in terms of lymphocyte infiltration, genetic mutations (KIT locus) and the extent of global DNA hypomethylation [58].
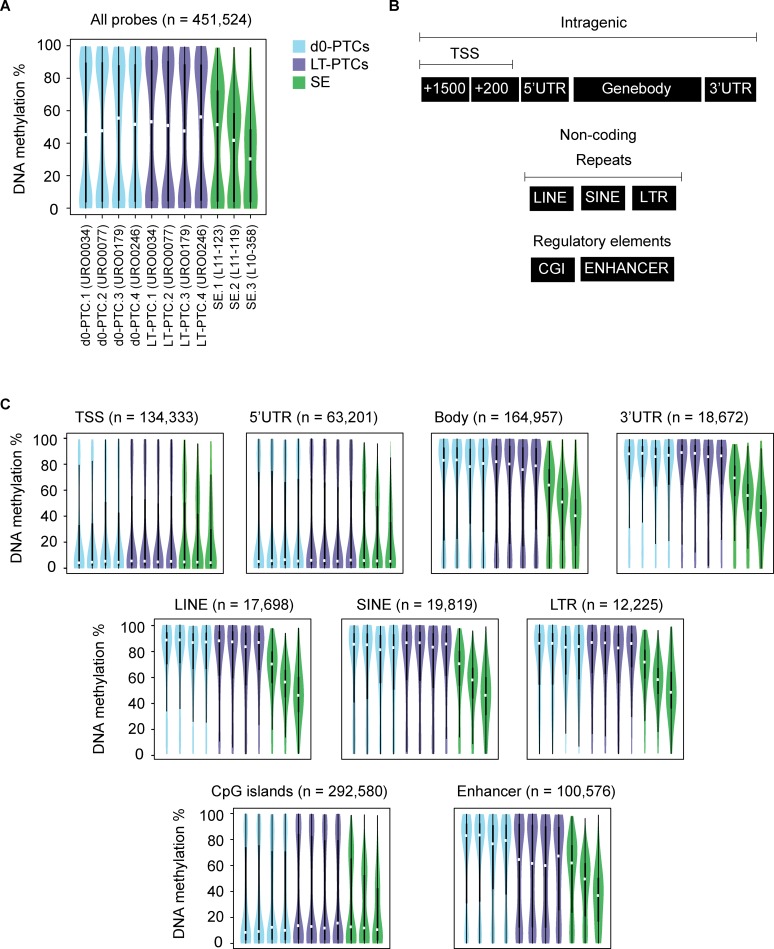
Fig 2. SE samples display a generally hypomethylated DNA methylation profile as compared to d0-PTCs and LT-PTCs.
(A) DNA methylation is deposited in a bipolar fashion in d0-PTCs and LT-PTCS, with most probes displaying a DNA methylation of either 0–10% or 90–100%, while a clear global hypomethylation is observed in SE. (B) Probe DNA methylation data was divided into nine genomic classes according to the genomic annotation of probes to the hg38 human genome reference build. A distinction was made between intragenic sequences (+1500bp of transcription start site (TSS), +200bp of TSS, 5' untranslated region (UTR), first exon, rest of genebody and 3'UTR) and non-coding sequences (long interspersed elements (LINE), short interspersed elements (SINE), long terminal repeats (LTR), CpG islands (CGI) and enhancer regions). (C) Distribution of DNA methylation over the nine genomic classes in d0-PTCs, LT-PTCS and SE.
To further study the extent of DNA hypomethylation in seminomas compared to uncultured and cultured PTCs, we next analyzed CpG methylation levels at nine defined genomic classes; transcription start sites (TSS), 5’-UTR, gene bodies, 3’-UTR, long and short interspersed elements (LINE and SINE, respectively), long terminal repeats (LTR), CpG islands and enhancer regions (Fig 2B). In line with the observed global DNA hypomethylation, six of these nine classes were hypomethylated in seminomas as compared to d0-PTCs and LT-PTCs, namely gene bodies, 3’UTR, LINE, SINE, LTR and enhancer regions (Fig 2C). Interestingly, DNA methylation levels were found to be normally distributed in all groups at the promoter regions (TSS and 5’UTR) and CpG islands. Together, these data suggest that LT-PTCs do not acquire an overall deregulated epigenetic status as is shown in seminoma with a concurrent general loss of DNA methylation, as well as possible decreased genetic stability due to demethylation at retrotransposon sequences.
In order to determine which genomic sequences were differentially methylated between groups, we selected CpG-dense regions represented on the array and tested each region for statistically significant differences in methylation (differentially methylated region, DMR). CpG-dense regions were defined as genomic sequences where a minimum of 4 CpG probes are located within 200bp of each other, resulting in 28,901 regions of interest to be statistically tested. Adhering to significance cut-offs of an adjusted Benjamini-Hochberg corrected p-value ≤ 0.01 and a minimum median methylation difference of 25% between groups, we identified a total of 353, 486 and 550 DMRs between d0-PTCs/LT-PTCs, LT-PTCs/SE and d0-PTCs/SE, respectively.
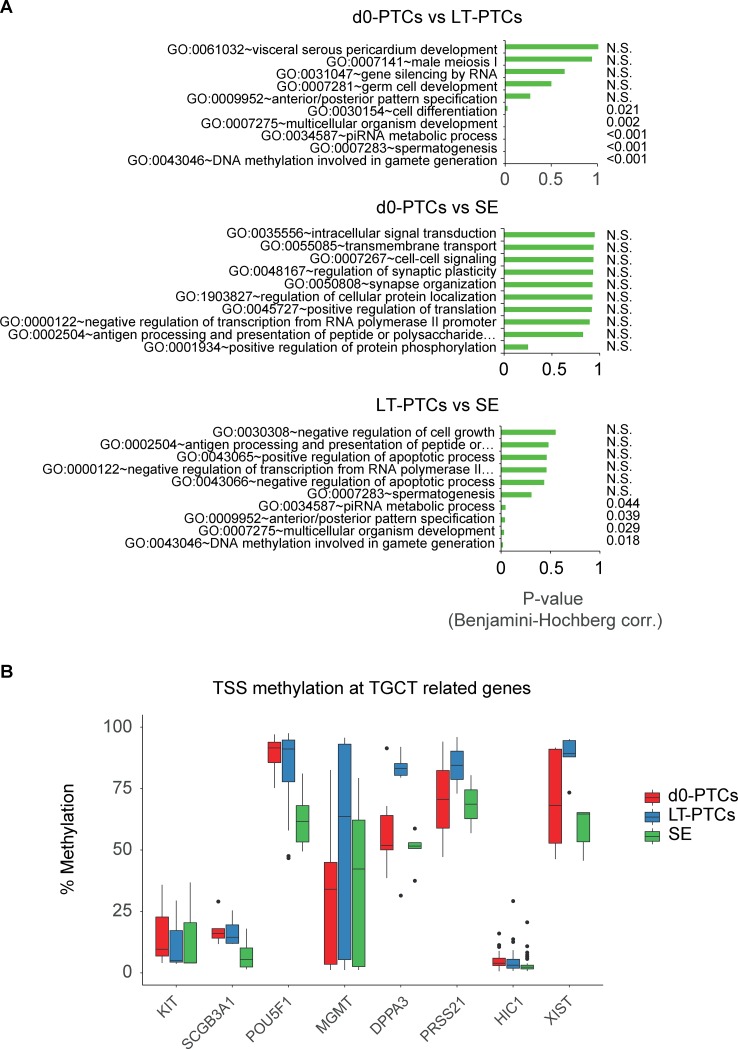
GO-enrichment analysis using the online DAVID tool [59] revealed enrichment of biological processes between d0-PTCs and LT-PTCs related to spermatogenesis and meiosis (Fig 3A). The top enriched GO-term “DNA methylation involved in gamete generation” (GO:0043046) contains several key regulators of male spermatogenesis; PICK1, MAEL, ASZ1, PIWIL2, MOV10L1, DDX4 and TDRD1 were differentially methylated in LT-PTCs as compared to d0-PTCs. All DMRs in this GO-term displayed increased methylation in LT-PTCs (increase of 48% ± 5.7% as compared to d0-PTCs) with the exception of MOV10L, which displayed decreased methylation (54%). In the LT-PTC versus seminoma comparison we found 4 enriched GO-terms that share a high degree of similarity with the terms found in the d0-PTC versus LT-PTC comparison, suggesting that long-term culture results in DNA methylation differences in LT-PTCs as compared to both d0-PTCs and seminomas, seemingly unrelated to malignant progression. There were no GO-terms between d0-PTCs and seminoma samples that reached statistical significance, possibly due to statistical power being too low or p-value cut-off levels.
Fig 3. Detection of differentially methylated regions in d0-PTCs, LT-PTCs and seminomas.
(A) Top 10 most enriched GO-terms (bottom to top) of DMRs between d0-PTC/LT-PTC, d0-PTC/seminoma and LT-PTC/seminoma. N.S. = not significant. (B) Gene-specific DNA methylation levels at tumor suppressor genes (SCGB3A1, MGMT, PRSS21, HIC1), testis-specific genes (KIT, DPPA3), XIST and the pluripotency marker POU5F1.
Seminomas are known to show selective hypermethylation at several tumor suppressors, such as MGMT, SCGB3A1, RASSF1A, HIC1, and PRSS21 [60]. In addition, seminomas can display genetic mutations in the KIT gene and they express the pluripotency marker POU5F1, which is not detectable in normal testis [61]. Analysis of DNA methylation levels at the TSS in specific tumor-related genes revealed a decreased promoter methylation level for POU5F1 in seminomas (approximately 35% reduction in DNA methylation) as compared to both d0-PTCs and LT-PTCs, while we did not detect significant differential methylation in SCGB3A1, PRSS21 or HIC1 (Fig 3B). The DPPA3 and XIST genes were hypermethylated in LT-PTCs as compared to d0-PTCs and seminomas, MGMT displayed a variable DNA methylation pattern between and within groups and the KIT promoter was stably hypomethylated in all groups including seminomas.
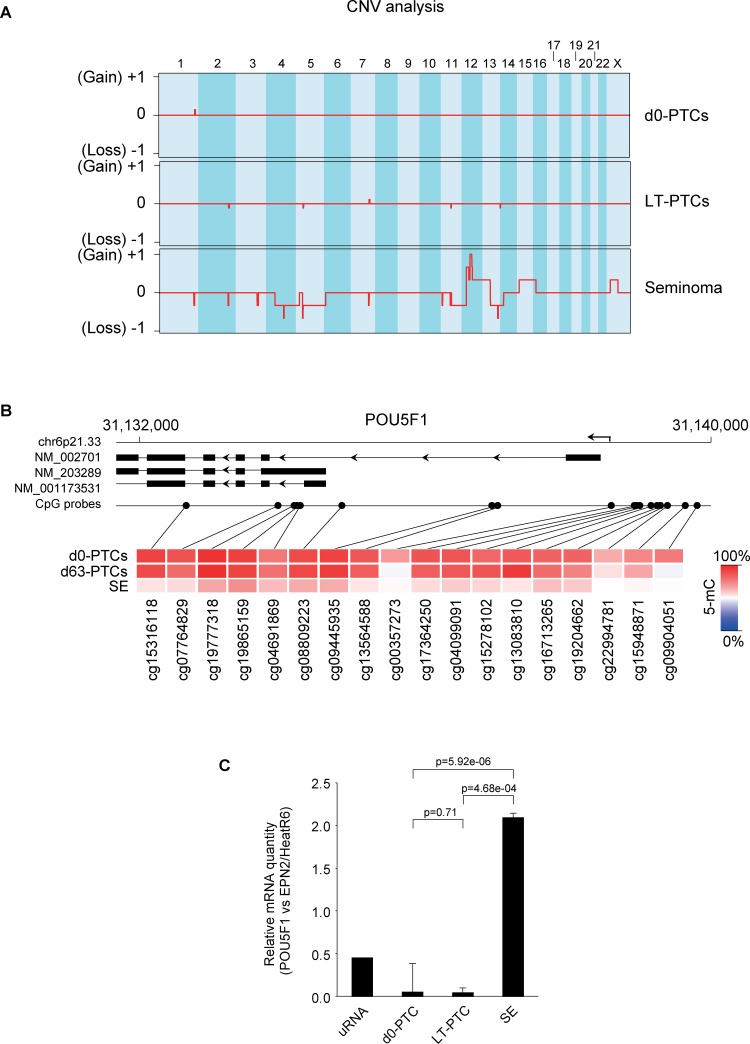
In addition to loss of both global and specific DNA hypermethylation patterning, seminomas are known to acquire structural chromosomal aberrations such as extra p-arms of chromosome 12 (preferentially as isochromosome i(12p)) during progression to invasive growth [62]. Other non-recurrent chromosomal aberrations in seminomas include gains of chromosomes 7, 8, 12p, 21 and X, and loss of chromosomes 1p, 11, 13 and 18 with varying occurrence in individual tumors [63]. To determine whether chromosomal stability in PTC cultures is maintained, we compared CNVs in uncultured and cultured PTCs with that of the seminomas samples using a previously published approach to estimate CNVs from DNA methylation array data [51]. LT-PTCs displayed some small CNVs (chromosomes 2, 5, 7, 11 and 14) that did not reach the significance threshold, corresponding to heterogeneity within individual PTC cultures. Indeed, CNVs occurring in d0-PTCs and LT-PTCs groups were located to different chromosomal regions in each individual culture, suggesting the absence of persistent CNV ‘hotspots’ (S1 File). The seminomas samples showed large-scale losses and gains on several chromosomes including the staple i(12p) duplication, as well as gains of chromosome 15, 16 and X and losses on chromosomes 4, 5, 11 and 13 (Fig 4A).
Fig 4. Copy number variation analysis reveals chromosomal aberrations in seminomas.
Probe fluorescence data was used to calculate average intensity in 8,949 chromosomal bins covering all autosomal chromosomes (1–22) and the X chromosome as aggregates of each sample group (see Methods). Average increases in probe fluorescence are designated as 'Gains' and average decreases as 'Losses' with the y-axis describing the extent of 2 fold amplification or reduction (for example, a gain of +1 signifies a two-fold increase in copy number within that particular bin). (B) The POU5F1 locus on chromosome 6p21.33 (also known as OCT3/4) is an oncogene that is activated in most germ cell cancers. SE samples displayed hypomethylation across the 18 CpG probes localized to this locus. Both d0-PTCs and LT-PTCs show a hypermethylated state of these CpG sites. (C) POU5F1 mRNA expression normalized to the expression of two reference genes (EPN2 and HeatR6) in d0-PTCs, LT-PTCs and seminoma samples as measured by quantitative fluorescence PCR. P-values were calculated using a two-sided student’s t-test. Universal RNA (uRNA) was used as a positive control.
Finally, we investigated the pluripotency marker POU5F1 in more detail due to its importance in TGCT diagnostics. The POU5F1 locus is covered by 18 CpG sites on the array platform that collectively displayed a hypomethylated state in seminomas as compared to d0-PTCs and LT-PTCs, including the genomic region directly upstream of the transcription start site (Fig 4B). To confirm the activated gene expression of POU5F1 in seminomas we performed gene expression analysis of the POU5F1 isoform 1 transcript (NM_002701) by qPCR and found high expression levels in seminomas while it was nearly undetectable in PTCs both before and after culturing (Fig 4C).
Discussion
We here present evidence that in vitro propagated and ITGA6+ enriched primary human testicular cells display a normal global DNA methylation profile. Compared to seminoma, the uncultured and cultured ITGA6+ PTCs do not harbor large-scale CNVs nor show signs of oncogene or retrotransposon activation in terms of regional DNA demethylation. This suggests that, in contrast to the hypomethylated profile and copy number variants commonly observed in GCNIS-derived lesions, including seminomas [64–67], primary testicular ITGA6+ cells remain genetically stable during prolonged cell culture. In contrast, we did observe significant differences in DNA methylation between cultured and uncultured ITGA6+ cells, which did not resemble the epigenetic hallmarks of malignant progression but is potentially relevant for their prospective use in clinical transplantation procedures.
The absence of large-scale chromosomal duplications or losses is in line with an earlier publication where long-term cultured ITGA6+ testicular cells (over 50 days in culture) were screened using single-cell CGH arrays and were reported to be genetically stable (68), which we were able to confirm here. In that same study, epigenetic alterations in cultured ITGA6+ PTCs were reported in a panel of five imprinted loci (H19, H19-DMR, MEG3, KCNQ1OT1 and PEG3) as compared to uncultured ITGA6+ PTCs and spermatozoa. The paternally imprinted H19, H19-DMR and MEG3 loci were reported to show demethylation of 28%-11%, 68%-43% and 26%-18%, respectively, while the maternally imprinted KCNQ1OT1 (13%-50%) and PEG3 (30%-38%) loci were hypermethylated during culture.
The data presented here also support previous efforts (totaling 30–40 recipient mice) where uncultured and cultured human testicular cells were transferred to mouse testes using xenotransplantation and during which tumor formation was never observed or reported after careful histological analyses of the transplanted tissues [17, 43, 54, 68]. Consistent with these data, no increases in tumor incidence or lowered life expectancy after long-term follow up (18 months of age) were reported in recipient mice after transplantation of allogenic in vitro propagated SSCs [69]. It should be noted that the human testicular cell culture system used in this study differs from the mouse culture system in several aspects, including but not limited to the use of feeder cells. More specifically, while mouse spermatogonial cell cultures are established by culturing enriched SSC fractions on a monolayer of STO or MEF feeder cells treated with mitomycin [13, 70, 71], the human system utilizes untreated testicular somatic cells originating from the donor testis tissue itself as outgrowing feeder cells. The manner in which germ cells are established and maintained in vitro is vital to the stability and identity of the cultured germ cell fraction, and as such the results from mouse SSCT experiments do not necessarily reflect the human situation. More research is needed in the characterization of cultured human SSCs before and after (xeno)-transplantation.
Microarray analyses are inherently limited by the resolution of the design of the array and might not be suitable for the detection of low-grade cell mosaics, such as trace amounts of tumor cells in a large population of normal cells. In the CNV analysis, we adhered to the detection limits as suggested by Sturm et al. [51] which does not detect chromosomal copy number aberrations present in under 10% of cells. Similarly, because of the dynamic nature of DNA methylation patterning we screened our samples for differentially methylated sites that displayed a significant increase or decrease in DNA methylation between samples (25% or more) in order to maximally reduce technical noise and identify sites that are plausibly biologically relevant. In contrast to seminomas, the POU5F1 transcript was virtually undetectable by qPCR in PTCs after culture, which would argue against the presence of even a small number of tumorigenic cells, although these data are not enough to rule out the presence of trace amounts of malignant cells completely. Moreover, while the 450k DNA methylation array platform is designed to include the majority of RefSeq genes, imprinting control regions (ICR) are not well covered in the design of the 450k array (less than 300 CpG sites covering ICRs (25)), and due to the low number of CpGs and relatively small sample size in our study we did not investigate these regions here. Based on these considerations, we stress that we are not able to fully exclude the presence of trace numbers of malignant cells in the cultured primary ITGA6+ testicular cell population.
There has been discussion in literature regarding the stability of germ cell DNA methylation patterns and the role of somatic contamination in the analyzed cell fractions. A recent study in marmoset monkey revealed stable DNA methylation patterns of the H19, MEST, DDX4 and MAGEA4 genes in germ cells cultured up to 21 days [72]. Since we detected differential methylation of GO-terms related to germ cell development and in particular DNA methylation regulation of germ cell formation, there could be differences in the functionality or cellular composition of the cultured cell fractions between human and marmoset/rodent germ cell cultures. There is some evidence of the role of epigenetic factors in the regulation of sperm development and quality [73] and the potential for transgenerational inheritance of epigenetic germ line mutations. As such, we strongly feel that more studies are required that focus on transcriptomic and epigenetic measurements in human germ cell cultures, for example through the use of novel emerging single-cell sequencing approaches [74–76], to gain insight in the events that occur during in vitro culture of human testicular cells.
Conclusions
In conclusion, we show that human primary cultured and sorted ITGA6+ testicular cells do not possess large-scale CNVs or epigenetic hallmarks found in malignant cells. Nevertheless, we strongly advise additional studies that focus on assessing the consequences of culture-induced epigenetic alterations on the functionality of in vitro propagated germ cells prior to clinical transplantation, focused on identifying detailed characteristics of human SSCs in vitro and after (xeno)-transplantation, as well as developing more sensitive methods to detect and remove trace malignant cells from human testicular cell cultures prior to transplantation.
Supporting information
(XLSX)
CNV analysis using the total sum of unmethylated and methylated signals of the probes located on the Illumina HumanMethylation 450k beadchip array platform in individual samples. Cutoff values for the designation of copy number variant type is displayed on the y-axis on the right side of the plots, colored dots represent CNV type: homozygous deletion in brown, hemizygous deletion in red, neutral in grey, duplication in green and high-copy gain in blue.
(PDF)
(PDF)
Acknowledgments
We thank dr. A. Meißner for providing testicular tissues and S.K.M. van Daalen and C.M. Korver for technical assistance.
Data Availability
All DNA methylation array data is available from the Gene Expression Omnibus repository (accession number GSE72444).
Funding Statement
This study was funded by the “Stichting Kinderen Kankervrij” (Dutch Children Cancer-Free Foundation, grant number KiKa#86) and ZonMW TAS (grant number 116003002). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Brinster RL. Male germline stem cells: from mice to men. Science. 2007;316(5823):404–5. 10.1126/science.1137741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meistrich ML, Finch M, da Cunha MF, Hacker U, Au WW. Damaging effects of fourteen chemotherapeutic drugs on mouse testis cells. Cancer Res. 1982;42(1):122–31. [PubMed] [Google Scholar]
- 3.Kiserud CE, Fossa A, Bjoro T, Holte H, Cvancarova M, Fossa SD. Gonadal function in male patients after treatment for malignant lymphomas, with emphasis on chemotherapy. Br J Cancer. 2009;100(3):455–63. 10.1038/sj.bjc.6604892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gandini L, Sgro P, Lombardo F, Paoli D, Culasso F, Toselli L, et al. Effect of chemo- or radiotherapy on sperm parameters of testicular cancer patients. Hum Reprod. 2006;21(11):2882–9. 10.1093/humrep/del167 [DOI] [PubMed] [Google Scholar]
- 5.Hwang K, Lamb DJ. New advances on the expansion and storage of human spermatogonial stem cells. Curr Opin Urol. 2010;20(6):510–4. 10.1097/MOU.0b013e32833f1b71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hermann BP, Sukhwani M, Winkler F, Pascarella JN, Peters KA, Sheng Y, et al. Spermatogonial stem cell transplantation into rhesus testes regenerates spermatogenesis producing functional sperm. Cell Stem Cell. 2012;11(5):715–26. 10.1016/j.stem.2012.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schlatt S, Rosiepen G, Weinbauer GF, Rolf C, Brook PF, Nieschlag E. Germ cell transfer into rat, bovine, monkey and human testes. Hum Reprod. 1999;14(1):144–50. 10.1093/humrep/14.1.144 [DOI] [PubMed] [Google Scholar]
- 8.Honaramooz A, Megee SO, Dobrinski I. Germ cell transplantation in pigs. Biol Reprod. 2002;66(1):21–8. 10.1095/biolreprod66.1.21 [DOI] [PubMed] [Google Scholar]
- 9.Kawasaki T, Saito K, Sakai C, Shinya M, Sakai N. Production of zebrafish offspring from cultured spermatogonial stem cells. Genes Cells. 2012;17(4):316–25. 10.1111/j.1365-2443.2012.01589.x [DOI] [PubMed] [Google Scholar]
- 10.Nobrega RH, Greebe CD, van de Kant H, Bogerd J, de Franca LR, Schulz RW. Spermatogonial stem cell niche and spermatogonial stem cell transplantation in zebrafish. PLoS One. 2010;5(9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brinster RL, Avarbock MR. Germline transmission of donor haplotype following spermatogonial transplantation. Proc Natl Acad Sci U S A. 1994;91(24):11303–7. 10.1073/pnas.91.24.11303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goossens E, De Rycke M, Haentjens P, Tournaye H. DNA methylation patterns of spermatozoa and two generations of offspring obtained after murine spermatogonial stem cell transplantation. Hum Reprod. 2009;24(9):2255–63. 10.1093/humrep/dep213 [DOI] [PubMed] [Google Scholar]
- 13.Kanatsu-Shinohara M, Ogonuki N, Inoue K, Miki H, Ogura A, Toyokuni S, et al. Long-term proliferation in culture and germline transmission of mouse male germline stem cells. Biol Reprod. 2003;69(2):612–6. 10.1095/biolreprod.103.017012 [DOI] [PubMed] [Google Scholar]
- 14.Ryu BY, Orwig KE, Oatley JM, Lin CC, Chang LJ, Avarbock MR, et al. Efficient generation of transgenic rats through the male germline using lentiviral transduction and transplantation of spermatogonial stem cells. J Androl. 2007;28(2):353–60. 10.2164/jandrol.106.001511 [DOI] [PubMed] [Google Scholar]
- 15.Wu X, Goodyear SM, Abramowitz LK, Bartolomei MS, Tobias JW, Avarbock MR, et al. Fertile offspring derived from mouse spermatogonial stem cells cryopreserved for more than 14 years. Hum Reprod. 2012;27(5):1249–59. 10.1093/humrep/des077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kubota H, Avarbock MR, Schmidt JA, Brinster RL. Spermatogonial stem cells derived from infertile Wv/Wv mice self-renew in vitro and generate progeny following transplantation. Biol Reprod. 2009;81(2):293–301. 10.1095/biolreprod.109.075960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nagano M, Patrizio P, Brinster RL. Long-term survival of human spermatogonial stem cells in mouse testes. Fertil Steril. 2002;78(6):1225–33. 10.1016/s0015-0282(02)04345-5 [DOI] [PubMed] [Google Scholar]
- 18.Struijk RB, Mulder CL, van der Veen F, van Pelt AM, Repping S. Restoring fertility in sterile childhood cancer survivors by autotransplanting spermatogonial stem cells: are we there yet? Biomed Res Int. 2013;2013:903142 10.1155/2013/903142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jahnukainen K, Hou M, Petersen C, Setchell B, Soder O. Intratesticular transplantation of testicular cells from leukemic rats causes transmission of leukemia. Cancer Res. 2001;61(2):706–10. [PubMed] [Google Scholar]
- 20.Geens M, Van de Velde H, De Block G, Goossens E, Van Steirteghem A, Tournaye H. The efficiency of magnetic-activated cell sorting and fluorescence-activated cell sorting in the decontamination of testicular cell suspensions in cancer patients. Hum Reprod. 2007;22(3):733–42. 10.1093/humrep/del418 [DOI] [PubMed] [Google Scholar]
- 21.Sadri-Ardekani H, Homburg CH, van Capel TM, van den Berg H, van der Veen F, van der Schoot CE, et al. Eliminating acute lymphoblastic leukemia cells from human testicular cell cultures: a pilot study. Fertil Steril. 2014;101(4):1072–8 e1. 10.1016/j.fertnstert.2014.01.014 [DOI] [PubMed] [Google Scholar]
- 22.Dovey SL, Valli H, Hermann BP, Sukhwani M, Donohue J, Castro CA, et al. Eliminating malignant contamination from therapeutic human spermatogonial stem cells. J Clin Invest. 2013;123(4):1833–43. 10.1172/JCI65822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128(4):683–92. 10.1016/j.cell.2007.01.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Van Der Zwan YG, Stoop H, Rossello F, White SJ, Looijenga LH. Role of epigenetics in the etiology of germ cell cancer. Int J Dev Biol. 2013;57(2–4):299–308. 10.1387/ijdb.130017ll [DOI] [PubMed] [Google Scholar]
- 25.Rijlaarsdam MA, Tax DM, Gillis AJ, Dorssers LC, Koestler DC, de Ridder J, et al. Genome wide DNA methylation profiles provide clues to the origin and pathogenesis of germ cell tumors. PLoS One. 2015;10(4):e0122146 10.1371/journal.pone.0122146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Williamson SR, Delahunt B, Magi-Galluzzi C, Algaba F, Egevad L, Ulbright TM, et al. The World Health Organization 2016 classification of testicular germ cell tumours: a review and update from the International Society of Urological Pathology Testis Consultation Panel. Histopathology. 2017;70(3):335–46. 10.1111/his.13102 [DOI] [PubMed] [Google Scholar]
- 27.Zhang N, Wang M, Zhang P, Huang T. Classification of cancers based on copy number variation landscapes. Biochim Biophys Acta. 2016;1860(11 Pt B):2750–5. [DOI] [PubMed] [Google Scholar]
- 28.Killian JK, Dorssers LC, Trabert B, Gillis AJ, Cook MB, Wang Y, et al. Imprints and DPPA3 are bypassed during pluripotency- and differentiation-coupled methylation reprogramming in testicular germ cell tumors. Genome Res. 2016;26(11):1490–504. 10.1101/gr.201293.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maitra A, Arking DE, Shivapurkar N, Ikeda M, Stastny V, Kassauei K, et al. Genomic alterations in cultured human embryonic stem cells. Nat Genet. 2005;37(10):1099–103. 10.1038/ng1631 [DOI] [PubMed] [Google Scholar]
- 30.Foudah D, Redaelli S, Donzelli E, Bentivegna A, Miloso M, Dalpra L, et al. Monitoring the genomic stability of in vitro cultured rat bone-marrow-derived mesenchymal stem cells. Chromosome Res. 2009;17(8):1025–39. 10.1007/s10577-009-9090-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tompkins JD, Hall C, Chen VC, Li AX, Wu X, Hsu D, et al. Epigenetic stability, adaptability, and reversibility in human embryonic stem cells. Proc Natl Acad Sci U S A. 2012;109(31):12544–9. 10.1073/pnas.1209620109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Allegrucci C, Wu YZ, Thurston A, Denning CN, Priddle H, Mummery CL, et al. Restriction landmark genome scanning identifies culture-induced DNA methylation instability in the human embryonic stem cell epigenome. Hum Mol Genet. 2007;16(10):1253–68. 10.1093/hmg/ddm074 [DOI] [PubMed] [Google Scholar]
- 33.Frost J, Monk D, Moschidou D, Guillot PV, Stanier P, Minger SL, et al. The effects of culture on genomic imprinting profiles in human embryonic and fetal mesenchymal stem cells. Epigenetics. 2011;6(1):52–62. 10.4161/epi.6.1.13361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tanasijevic B, Dai B, Ezashi T, Livingston K, Roberts RM, Rasmussen TP. Progressive accumulation of epigenetic heterogeneity during human ES cell culture. Epigenetics. 2009;4(5):330–8. 10.4161/epi.4.5.9275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Amir H, Touboul T, Sabatini K, Chhabra D, Garitaonandia I, Loring JF, et al. Spontaneous Single-Copy Loss of TP53 in Human Embryonic Stem Cells Markedly Increases Cell Proliferation and Survival. Stem Cells. 2017;35(4):872–85. 10.1002/stem.2550 [DOI] [PubMed] [Google Scholar]
- 36.Imreh MP, Gertow K, Cedervall J, Unger C, Holmberg K, Szoke K, et al. In vitro culture conditions favoring selection of chromosomal abnormalities in human ES cells. J Cell Biochem. 2006;99(2):508–16. 10.1002/jcb.20897 [DOI] [PubMed] [Google Scholar]
- 37.He Z, Kokkinaki M, Jiang J, Dobrinski I, Dym M. Isolation, characterization, and culture of human spermatogonia. Biol Reprod. 2010;82(2):363–72. 10.1095/biolreprod.109.078550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Izadyar F, Wong J, Maki C, Pacchiarotti J, Ramos T, Howerton K, et al. Identification and characterization of repopulating spermatogonial stem cells from the adult human testis. Hum Reprod. 2011;26(6):1296–306. 10.1093/humrep/der026 [DOI] [PubMed] [Google Scholar]
- 39.Valli H, Sukhwani M, Dovey SL, Peters KA, Donohue J, Castro CA, et al. Fluorescence- and magnetic-activated cell sorting strategies to isolate and enrich human spermatogonial stem cells. Fertil Steril. 2014;102(2):566–80 e7. 10.1016/j.fertnstert.2014.04.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nickkholgh B, Mizrak SC, Korver CM, van Daalen SK, Meissner A, Repping S, et al. Enrichment of spermatogonial stem cells from long-term cultured human testicular cells. Fertil Steril. 2014;102(2):558–65 e5. 10.1016/j.fertnstert.2014.04.022 [DOI] [PubMed] [Google Scholar]
- 41.Shinohara T, Avarbock MR, Brinster RL. beta1- and alpha6-integrin are surface markers on mouse spermatogonial stem cells. Proc Natl Acad Sci U S A. 1999;96(10):5504–9. 10.1073/pnas.96.10.5504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stoop H, Kirkels W, Dohle GR, Gillis AJ, den Bakker MA, Biermann K, et al. Diagnosis of testicular carcinoma in situ '(intratubular and microinvasive)' seminoma and embryonal carcinoma using direct enzymatic alkaline phosphatase reactivity on frozen histological sections. Histopathology. 2011;58(3):440–6. 10.1111/j.1365-2559.2011.03767.x [DOI] [PubMed] [Google Scholar]
- 43.Sadri-Ardekani H, Mizrak SC, van Daalen SK, Korver CM, Roepers-Gajadien HL, Koruji M, et al. Propagation of human spermatogonial stem cells in vitro. JAMA. 2009;302(19):2127–34. 10.1001/jama.2009.1689 [DOI] [PubMed] [Google Scholar]
- 44.Alders M, Bliek J, vd Lip K, vd Bogaard R, Mannens M. Determination of KCNQ1OT1 and H19 methylation levels in BWS and SRS patients using methylation-sensitive high-resolution melting analysis. Eur J Hum Genet. 2009;17(4):467–73. 10.1038/ejhg.2008.197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Du P, Kibbe WA, Lin SM. lumi: a pipeline for processing Illumina microarray. Bioinformatics. 2008;24(13):1547–8. 10.1093/bioinformatics/btn224 [DOI] [PubMed] [Google Scholar]
- 46.Chen YA, Lemire M, Choufani S, Butcher DT, Grafodatskaya D, Zanke BW, et al. Discovery of cross-reactive probes and polymorphic CpGs in the Illumina Infinium HumanMethylation450 microarray. Epigenetics. 2013;8(2):203–9. 10.4161/epi.23470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marabita F, Almgren M, Lindholm ME, Ruhrmann S, Fagerstrom-Billai F, Jagodic M, et al. An evaluation of analysis pipelines for DNA methylation profiling using the Illumina HumanMethylation450 BeadChip platform. Epigenetics. 2013;8(3):333–46. 10.4161/epi.24008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Teschendorff AE, Marabita F, Lechner M, Bartlett T, Tegner J, Gomez-Cabrero D, et al. A beta-mixture quantile normalization method for correcting probe design bias in Illumina Infinium 450 k DNA methylation data. Bioinformatics. 2013;29(2):189–96. 10.1093/bioinformatics/bts680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rijlaarsdam MA, van der Zwan YG, Dorssers LC, Looijenga LH. DMRforPairs: identifying differentially methylated regions between unique samples using array based methylation profiles. BMC Bioinformatics. 2014;15:141 10.1186/1471-2105-15-141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Du P, Zhang X, Huang CC, Jafari N, Kibbe WA, Hou L, et al. Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinformatics. 2010;11:587 10.1186/1471-2105-11-587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sturm D, Witt H, Hovestadt V, Khuong-Quang DA, Jones DT, Konermann C, et al. Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell. 2012;22(4):425–37. 10.1016/j.ccr.2012.08.024 [DOI] [PubMed] [Google Scholar]
- 52.Ramakers C, Ruijter JM, Deprez RH, Moorman AF. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci Lett. 2003;339(1):62–6. 10.1016/s0304-3940(02)01423-4 [DOI] [PubMed] [Google Scholar]
- 53.de Rooij DG. The nature and dynamics of spermatogonial stem cells. Development. 2017;144(17):3022–30. 10.1242/dev.146571 [DOI] [PubMed] [Google Scholar]
- 54.Sadri-Ardekani H, Akhondi MA, van der Veen F, Repping S, van Pelt AM. In vitro propagation of human prepubertal spermatogonial stem cells. JAMA. 2011;305(23):2416–8. 10.1001/jama.2011.791 [DOI] [PubMed] [Google Scholar]
- 55.Ehrlich M. DNA hypomethylation in cancer cells. Epigenomics. 2009;1(2):239–59. 10.2217/epi.09.33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wermann H, Stoop H, Gillis AJ, Honecker F, van Gurp RJ, Ammerpohl O, et al. Global DNA methylation in fetal human germ cells and germ cell tumours: association with differentiation and cisplatin resistance. J Pathol. 2010;221(4):433–42. 10.1002/path.2725 [DOI] [PubMed] [Google Scholar]
- 57.Netto GJ, Nakai Y, Nakayama M, Jadallah S, Toubaji A, Nonomura N, et al. Global DNA hypomethylation in intratubular germ cell neoplasia and seminoma, but not in nonseminomatous male germ cell tumors. Mod Pathol. 2008;21(11):1337–44. 10.1038/modpathol.2008.127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shen H, Shih J, Hollern DP, Wang L, Bowlby R, Tickoo SK, et al. Integrated Molecular Characterization of Testicular Germ Cell Tumors. Cell Rep. 2018;23(11):3392–406. 10.1016/j.celrep.2018.05.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- 60.Lind GE, Skotheim RI, Lothe RA. The epigenome of testicular germ cell tumors. APMIS. 2007;115(10):1147–60. 10.1111/j.1600-0463.2007.apm_660.xml.x [DOI] [PubMed] [Google Scholar]
- 61.Rijlaarsdam MA, van Herk HA, Gillis AJ, Stoop H, Jenster G, Martens J, et al. Specific detection of OCT3/4 isoform A/B/B1 expression in solid (germ cell) tumours and cell lines: confirmation of OCT3/4 specificity for germ cell tumours. Br J Cancer. 2011;105(6):854–63. 10.1038/bjc.2011.270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rosenberg C, Van Gurp RJ, Geelen E, Oosterhuis JW, Looijenga LH. Overrepresentation of the short arm of chromosome 12 is related to invasive growth of human testicular seminomas and nonseminomas. Oncogene. 2000;19(51):5858–62. 10.1038/sj.onc.1203950 [DOI] [PubMed] [Google Scholar]
- 63.Oosterhuis JW, Looijenga LH. Testicular germ-cell tumours in a broader perspective. Nat Rev Cancer. 2005;5(3):210–22. 10.1038/nrc1568 [DOI] [PubMed] [Google Scholar]
- 64.Suijkerbuijk RF, van de Veen AY, van Echten J, Buys CH, de Jong B, Oosterhuis JW, et al. Demonstration of the genuine iso-12p character of the standard marker chromosome of testicular germ cell tumors and identification of further chromosome 12 aberrations by competitive in situ hybridization. Am J Hum Genet. 1991;48(2):269–73. [PMC free article] [PubMed] [Google Scholar]
- 65.Skotheim RI, Lothe RA. The testicular germ cell tumour genome. APMIS. 2003;111(1):136–50; discussion 50–1. 10.1034/j.1600-0463.2003.11101181.x [DOI] [PubMed] [Google Scholar]
- 66.Sheikine Y, Genega E, Melamed J, Lee P, Reuter VE, Ye H. Molecular genetics of testicular germ cell tumors. Am J Cancer Res. 2012;2(2):153–67. [PMC free article] [PubMed] [Google Scholar]
- 67.Williams LA, Mills L, Hooten AJ, Langer E, Roesler M, Frazier AL, et al. Differences in DNA methylation profiles by histologic subtype of paediatric germ cell tumours: a report from the Children's Oncology Group. Br J Cancer. 2018;119(7):864–72. 10.1038/s41416-018-0277-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nickkholgh B, Mizrak SC, van Daalen SK, Korver CM, Sadri-Ardekani H, Repping S, et al. Genetic and epigenetic stability of human spermatogonial stem cells during long-term culture. Fertil Steril. 2014;102(6):1700–7 e1. 10.1016/j.fertnstert.2014.08.022 [DOI] [PubMed] [Google Scholar]
- 69.Mulder CL, Catsburg LAE, Zheng Y, de Winter-Korver CM, van Daalen SKM, van Wely M, et al. Long-term health in recipients of transplanted in vitro propagated spermatogonial stem cells. Hum Reprod. 2018;33(1):81–90. 10.1093/humrep/dex348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nagano M, Avarbock MR, Leonida EB, Brinster CJ, Brinster RL. Culture of mouse spermatogonial stem cells. Tissue Cell. 1998;30(4):389–97. 10.1016/s0040-8166(98)80053-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kanatsu-Shinohara M, Ogonuki N, Iwano T, Lee J, Kazuki Y, Inoue K, et al. Genetic and epigenetic properties of mouse male germline stem cells during long-term culture. Development. 2005;132(18):4155–63. 10.1242/dev.02004 [DOI] [PubMed] [Google Scholar]
- 72.Langenstroth-Rower D, Gromoll J, Wistuba J, Trondle I, Laurentino S, Schlatt S, et al. De novo methylation in male germ cells of the common marmoset monkey occurs during postnatal development and is maintained in vitro. Epigenetics. 2017;12(7):527–39. 10.1080/15592294.2016.1248007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Laurentino S, Borgmann J, Gromoll J. On the origin of sperm epigenetic heterogeneity. Reproduction. 2016;151(5):R71–8. 10.1530/REP-15-0436 [DOI] [PubMed] [Google Scholar]
- 74.Fend-Guella DL, von Kopylow K, Spiess AN, Schulze W, Salzbrunn A, Diederich S, et al. The DNA methylation profile of human spermatogonia at single-cell- and single-allele-resolution refutes its role in spermatogonial stem cell function and germ cell differentiation. Mol Hum Reprod. 2019. [DOI] [PubMed] [Google Scholar]
- 75.Hermann BP, Cheng K, Singh A, Roa-De La Cruz L, Mutoji KN, Chen IC, et al. The Mammalian Spermatogenesis Single-Cell Transcriptome, from Spermatogonial Stem Cells to Spermatids. Cell Rep. 2018;25(6):1650–67 e8. 10.1016/j.celrep.2018.10.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Neuhaus N, Yoon J, Terwort N, Kliesch S, Seggewiss J, Huge A, et al. Single-cell gene expression analysis reveals diversity among human spermatogonia. Mol Hum Reprod. 2017;23(2):79–90. 10.1093/molehr/gaw079 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
CNV analysis using the total sum of unmethylated and methylated signals of the probes located on the Illumina HumanMethylation 450k beadchip array platform in individual samples. Cutoff values for the designation of copy number variant type is displayed on the y-axis on the right side of the plots, colored dots represent CNV type: homozygous deletion in brown, hemizygous deletion in red, neutral in grey, duplication in green and high-copy gain in blue.
(PDF)
(PDF)
Data Availability Statement
All DNA methylation array data is available from the Gene Expression Omnibus repository (accession number GSE72444).