Abstract
Background:
Platelets are a critical element in coagulation and inflammation, and activated platelets are linked to cancer risk through diverse mechanisms. However, a causal relationship between platelets and risk of lung cancer remains unclear.
Methods:
We performed single and combined multiple instrumental variable Mendelian Randomization (MR) analysis by an inverse-weighted (IVW) method, in addition to a series of sensitivity analyses. Summary data for associations between single nucleotide polymorphisms (SNPs) and platelet count is from a recent publication including 48,666 Caucasian Europeans and International Lung Cancer Consortium and Transdisciplinary Research in Cancer of the Lung data consisting of 29,266 cases and 56,450 controls analyze associations between candidate single nucleotide polymorphisms and lung cancer risk.
Results:
Multiple instrumental variable analysis incorporating six SNPs showed a 62% increased risk of overall NSCLC (OR, 1.62; 95%CI, 1.15–2.27; P = 0.005) and 200% increased risk for small cell lung cancer (OR, 3.00; 95%CI, 1.27–7.06; P = 0.01), respectively. Results showed only a trending association with NSCLC histological subtypes, which may be due to insufficient sample size and/or weak effect size. A series of sensitivity analysis retained these findings.
Conclusion:
Our findings suggest a causal relationship between elevated platelet count and increased risk of lung cancer and provide evidence of possible anti-platelet interventions for lung cancer prevention.
Impact:
Our findings suggest a causal relationship of increased platelet count and risk of lung cancer, which also provide a better understanding of lung cancer etiology and potential evidence for anti-platelet interventions for lung cancer prevention.
Keywords: Lung cancer, Mendelian randomization, Platelet count, Instrumental variable
Introduction
Lung cancer, a highly invasive, rapidly metastasizing cancer, has been the leading cause of cancer deaths worldwide for decades, accounting for more than one million deaths each year [1]. Smoking is a major risk factor for lung cancer and accounts for about 80% of male and 50% of female lung cancer cases [2]. In addition, environmental–occupational exposures [3, 4], lifestyle, and genetic variants [5] have been broadly explored as risks/predisposing factors for lung cancer. However, aspects of lung cancer risk remain largely unexplained and thus warrant further study.
The lung was recently noted to play a major role in platelet biogenesis and act as an ideal bioreactor for production of mature platelets from megakaryocytes, which account for ~50% of total platelet production [6]. Platelets are an important element in coagulation and inflammation, and diverse mechanisms link activated platelets to cancer progression [7, 8]. It has been identified that several variants in those chromosomal regions associated with platelet count (PLT) have associations with myocardial infraction, autoimmune and hematologic disorders. Tumor-educated blood platelets (TEPs) have emerged as promising biomarker sources for non-invasive detection of cancer, and it was demonstrated to discriminates patients with NSCLC from healthy individuals and patients with various non-cancerous inflammatory conditions [9, 10].Indeed, high platelet count (PLT) is associated with increased mortality in a variety of cancers, including malignant mesothelioma [11], gynecological malignancies [12], and breast cancer [13]. In addition, platelet-to-lymphocyte ratio and mean platelet volume also add value in early diagnosis of lung cancer [14] and prognosis prediction [15, 16]. These findings, taken together, indicate that disordered platelet production may be connected to lung carcinogenesis. However, due to potential unmeasured confounders in observational studies, the association between PLT and lung cancer risk remains unclear.
Mendelian Randomization (MR) is based on the principle that an individual’s genotype is randomized at conception[17] and utilizes genetic variants as instrumental variables (IV) for the association between phenotypic exposures and outcomes to eliminate bias due to unmeasured confounders. Genetic variants used as instrumental variables should meet the following assumptions: (1) genetic variants are associated with exposure, (2) genetic variants affect outcome only via the exposure, and (3) genetic variants are not associated with any confounders of the exposure–outcome association.[18] By finding a genetic marker that satisfies instrumental variable assumptions, Mendelian randomization analysis has been broadly used to estimate unconfounded associations between exposure and outcome [19], such as the effect of higher adult height on escalated cancer risk [20–24].
In this study, we performed summary data-based Mendelian randomization (SMR) [25] analysis which is the extension of two sample Mendelian randomization, using curated platelet count-related SNPs as instrumental variables to evaluate the association between platelet count and lung cancer risk by using summary statistics from recent large scale genome-wide association studies (GWAS).
Materials and Methods
Data source and study population
Mendelian Randomization analysis was conducted to estimate the effect of platelet count (X) on risk of lung cancer (Y) using genetic variants (G) as instrumental variables.[26] According to the MR analysis diagram described in Figure 1, we used coefficients of genetic variants on platelet count (bXG) and their standard errors (SEXG) from the recently published study of Gieger et al., which pooled 23 studies and included approximately 48,666 individuals of European descent [27].
Figure 1.
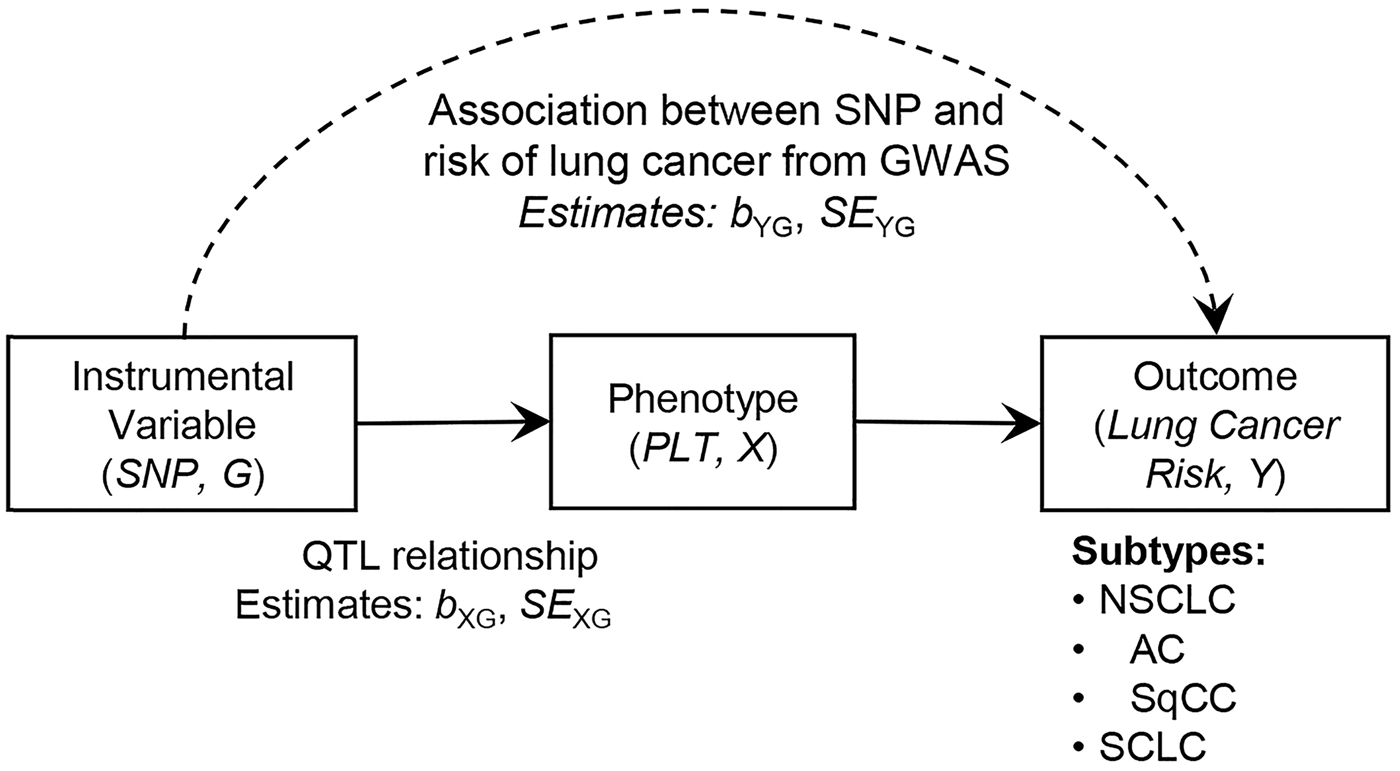
Diagram of Mendelian randomization (MR) analysis. MR aims to estimate the unbiased causal relationship between PLT and lung cancer risk by incorporating genetic variants as instrumental variables (IVs). Dashed line represents the association between instrumental variable (SNP) and outcome (risk of lung cancer), denoted using bYG in log(odds ratio) scale and its standard error (SEYG), which were obtained from GWAS. Estimates of quantitative trait loci relationship between SNP and phenotype (platelet count) were obtained from a recently published article, which were described by bXG and SEXG. Lung cancer risk was assessed for non-small cell lung cancer (NSCLC), adenocarcinoma (AC), squamous cell carcinoma (SqCC), and small cell carcinoma (SCLC).
The 54 genetic variants were identified that were associated with PLT (Table S1). One of the key assumptions underlying Mendelian Randomization is that the genetic variants (SNPs) used as instrumental variables are only related to the outcome of interest through the exposure variable under study. No pleiotropic pathways should exist from platelet-related SNPs to lung cancers through intermediates other than platelet count. Thus, six genetic variants (rs17030845, rs6141, rs3792366, rs210134, rs708382, and rs6065) where further selected as qualified instrumental variables that have prior functional knowledge supporting their association with platelets and no apparent link to cancer through intermediates other than platelets. By the way, the SNP rs6141 in THPO narrowly misses the level required for nominal significance (P<5×10−8) with P=6.18×10−8 in Europeans, but shows genome-wide significance in Japanese [28]. Therefore, it is still included serving as instrument variable for platelet count.
Coefficients (βYG) and corresponding standard errors (SEYG) of the association between genetic variants and lung cancer risk were obtained from meta-analysis of existing Oncoarray and TRICL GWAS studies, which were detailed previously [29]. Briefly, overall non-small cell lung cancer (NSCLC) samples were composed from Oncoarray and TRICL GWASs, including 29,266 cases and 56,450 controls, and subgroup analyses were performed for 11,273 adenocarcinoma (AC), 7,426 squamous cell carcinoma (SqCC), and 2,664 small cell lung cancer (SCLC) cases (Table S2).
Mendelian Randomization (MR) analysis
Mendelian Randomization analysis with multiple instrumental variables was performed using an inverse-variance weighted (IVW) method combining the effect of genetic variants by weighted score. This score was used as an instrumental variable to estimate the effect of PLT on lung cancer risk [26]:
| (1) |
In which N = 6 represents the number of instrumental variables included, and bYX_IVW and SEYX_IVW represent the effect of platelet count on lung cancer risk in log(OR) scale and its corresponding standard error. Associations of platelet count on risk of overall NSCLC and individual subtypes were analyzed. Results are presented as OR for lung cancer risk per 100×109/L increment of platelet count.
Additionally, penalized IVW, robust IVW, MR-Egger, penalized MR-Egger, and robust MR-Egger methods were used for sensitivity analyses to evaluate robustness of the findings [30]. Step forward modeling was used to add an optimal instrumental variable each time from the left 48 SNPs, adding to the 6 curated SNPs for multiple instrumental variable analysis, until there was no improvement of statistical significance (P-value) for the test of causal effect. The modeling process was terminated when no added SNP increased −log10 (P-value) by 20% or 10%. Besides, MR analysis with a single IV (one SNP at a time) was performed as supplementary. Effect of PLT on Lung cancer risk [bYX in log odds ratio (OR) scale] and its standard error (SEYX) were estimated as follows [31]:
| (2) |
All analyses were performed using R Software Version 3.3.1 (The R Foundation). All tests were two-sided, and P ≤ 0.05 was considered statistically significant unless stated otherwise.
Results
Among 48,666 Europeans, 54 SNPs were quantitatively associated with platelet count with P ≤ 5×10−8 (Table S1) [27]. Associations of those 54 SNPs with risk of lung cancer were analyzed among 29,266 cases and 56,450 controls from OncoArray and previous GWAS studies. Demographics and study descriptions were detailed previously [29] and are briefly listed in Table S2 as well. Summarized association results of SNPs and lung cancer risk are listed in Table S3. According to instrumental variable assumptions that had evidence only related to platelets, six SNPs which are relatively independent and situated in different chromosomes were selected for MR analysis (Table 1), and 48 SNPs were excluded (Table S4).
Table 1.
SNPs of specific platelet-related genes
| SNP | Chr:Position (hg19) | Gene | Reference allele | Effect allele | EAF (%) | Function | b (95% CI) | P |
|---|---|---|---|---|---|---|---|---|
| rs17030845 | 2: 43687879 | THADA | C | T | 09.65 | intron | −3.58 (−4.67, −2.49) | 1.27×10−10 |
| rs6141 | 3: 184090266 | THPO | T | C | 47.39 | 3’ UTR | −2.47 (−3.36, −1.57) | 6.18×10−8 |
| rs3792366 | 3: 122839876 | PDIA5 | A | G | 38.68 | intron | 2.153 (1.44, 2.87) | 3.60×10−9 |
| rs210134 | 6: 33540209 | BAK1 | G | A | 29.29 | 500bp downstream | −4.96 (−5.73, −4.18) | 7.11×10−36 |
| rs708382 | 17: 42442344 | FAM171A2-ITGA2B | T | C | 39.66 | 2kb upstream | −2.44 (−3.28, −1.59) | 1.51×10−8 |
| rs6065 | 17: 4836381 | GP1BA | C | T | 08.53 | missense | 4.19 (2.96, 5.43) | 2.92×10−11 |
EAF, effect allele frequency; UTR, untranslated region
In multiple IV analysis combining all six relatively independent SNPs situated in different chromosomes, a significant association between PLT and overall NSCLC risk is revealed, showing that each 100×109/L increment of PLT was associated with a 62% increase in NSCLC risk (95%CI, 1.15–2.27; P = 0.005) (Figure 2A and Figure 3A). In addition, five different methods of sensitivity analysis, including penalized IVW, robust IVW, MR-Egger, penalized MR-Egger, and robust MR-Egger, retained this association (Table 2). In NSCLC subtype analysis, it failed to detected significant associations between PLT and the risk of lung Adenocarcinoma (AC) (OR, 1.51; 95%CI, 0.92–2.48; P = 0.11) (Figure 2B and Figure 3B) and squamous cells carcinomas (SqCC) (OR, 1.59; 95%CI, 0.86–2.92; P = 0.14) (Figure 2C and Figure 3C). On the other hand, it is suggested that PLT is significantly associated with the risk of small cell lung cancer (SCLC) (OR, 3.00; 95%CI, 1.27–7.06; P = 0.01) (Figure 2D and Figure 3D). The results of single IV are presented in supplementary (Table S5). No correction was conducted for them because a single weak instrument will have lower power to reject the null hypothesis [32].
Figure 2.

Causal associations between platelet count and lung cancer risk. Forest plots of causal associations between platelet count (PLT) and risk of lung cancer using Mendelian randomization (MR) analysis incorporating different genetic variants as instrumental variables (IVs). Associations of PLT with risk of (A) non-small cell lung cancer (NSCLC), (B) adenocarcinoma (AC), (C) squamous cell carcinoma (SqCC), and (D) small-cell lung cancer (SCLC) were analyzed based on single IV or multiple IVs using inverse-variance weighted (IVW) analysis.
Figure 3.
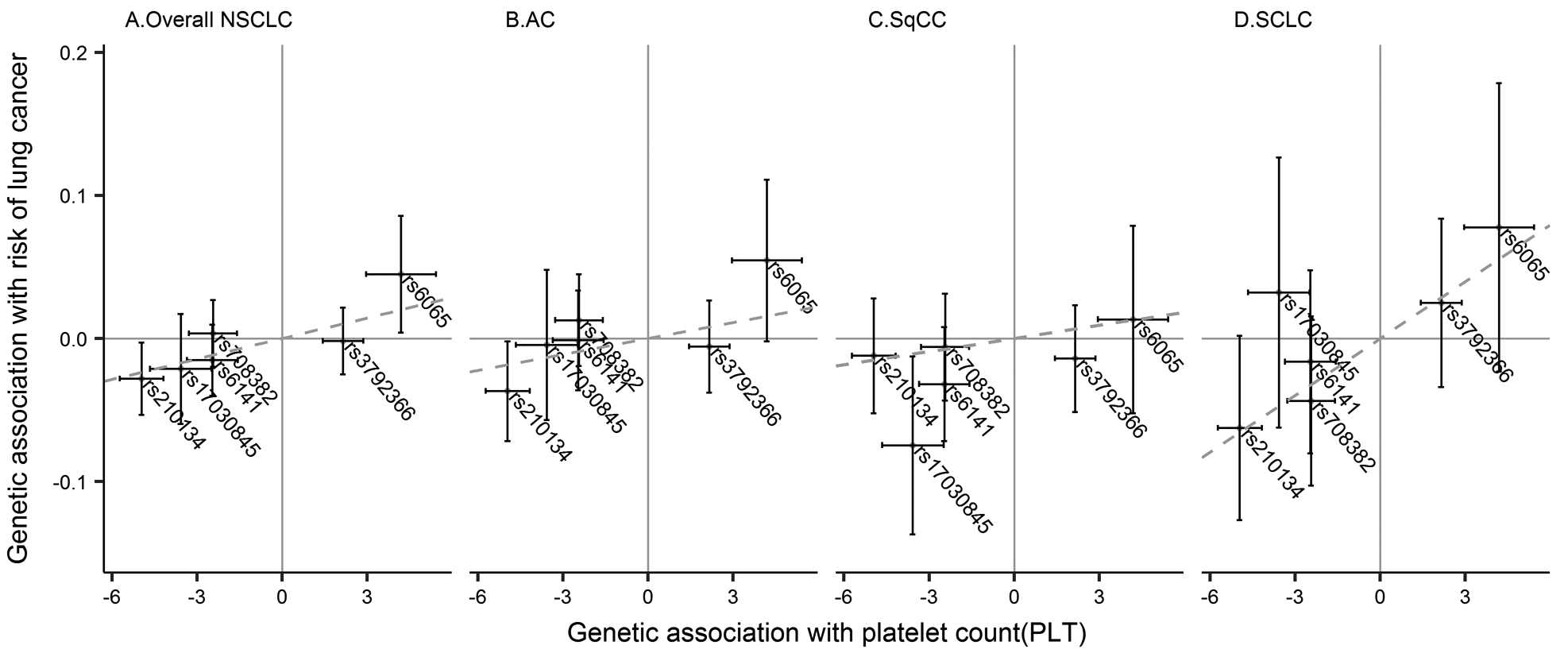
Assocations between SNPs and lung cancer risk. Scatter plots displaying estimates of the association between each SNP and risk of lung cancer against quantitative relationship of each SNP on platelet count (PLT) for (A) non-small cell lung cancer (NSCLC), (B) adenocarcinoma (AC), (C) squamous cell carcinoma (SqCC), and (D) small cell lung cancer (SCLC). Slope of the blue dashed line through the plot represents inverse-variance weighted (IVW) regression estimate for the causal effect of PLT on lung cancer risk.
Table 2.
Association between platelet count and risk of lung cancer using multiple IV analysis
| SNP | Overall NSCLC | AC | SqCC | SCLC | ||||
|---|---|---|---|---|---|---|---|---|
| OR (95%CI) | P | OR (95%CI) | P | OR (95%CI) | P | OR (95%CI) | P | |
| IVW | 1.62 (1.15, 2.27) | 0.005 | 1.51 (0.92, 2.48) | 0.11 | 1.59 (0.86, 2.92) | 0.14 | 3.00 (1.27, 7.06) | 0.01 |
| Penalized IVW | 1.62 (1.15, 2.27) | 0.005 | 1.51 (0.92, 2.48) | 0.11 | 1.59 (0.86, 2.92) | 0.14 | 3.00 (1.27, 7.06) | 0.01 |
| Robust IVW | 1.63 (1.26, 2.11) | <0.001 | 1.51 (0.90, 2.53) | 0.12 | 1.54 (0.93, 2.56) | 0.09 | 3.30 (1.52, 7.15) | 0.003 |
| MR-Egger | 3.25 (1.16, 9.11) | 0.03 | 6.06 (1.45, 25.27) | 0.01 | 1.75 (0.22, 13.84) | 0.59 | 3.29 (0.24, 45.11) | 0.37 |
| Penalized MR-Egger | 3.25 (1.16, 9.11) | 0.03 | 6.06 (1.45, 25.27) | 0.01 | 1.75 (0.22, 13.84) | 0.59 | 3.29 (0.24, 45.11) | 0.37 |
| Robust MR-Egger | 3.23 (1.80, 5.78) | <0.001 | 5.88 (2.74, 12.61) | <0.001 | 1.70 (0.48, 6.08) | 0.41 | 3.56 (1.25, 10.14) | 0.02 |
OR, odds ratio of platelet count (PLT) on lung cancer risk per 100×109/L increment of PLT; NSCLC, non-small cell lung cancer; AC, adenocarcinoma; SqCC, squamous cell carcinoma; SCLC, small-cell carcinoma; IVW, inverse-variance weighted.
We also performed a step forward modeling strategy to include more instrumental SNPs in the multiple instrumental variable model. Including more SNPs as instrumental variables yielded similar, yet more significant, causal estimates (Table S6 and Figure S1).
Discussion
This Mendelian randomization study suggests that each 100×109/L increment in platelets results in a 62% increased risk of non-small cell lung cancer and, notably, a 200% increased risk of small cell lung cancer. However, this study failed to show evidence of a relationship between PLT and risk of AC and SqCC, probably resulting from insufficient sample size. As comparing with SCLC, the effect size of PLT on AC and SqCC are weaker, larger sample size is needed [33].
Platelets have been studied for decades as an important regulator of inflammation and thrombosis [34], which are broadly interrelated with human carcinogenesis [13]. Platelets are also recognized as a stimulator of proangiogenic factors [13] and a major source of vascular endothelial growth factor (VEGF) [35], platelet-derived growth factor (PDGF) [36, 37], and basic fibroblast growth factor (bFGF) [37], which act as promoters of tumor growth in lung [38–44]. New evidence suggests that platelets are relevant to defensive, physiological immune responses of the lungs and to inflammatory lung diseases [45]. Thus, higher platelet count has a potential biological connection to increased risk of lung cancer. Interestingly, p-selectin, an important adhesion molecule expressed on the surface of activated platelets, is more highly expressed in lung adenocarcinomas and squamous cell carcinomas than in healthy populations [46]. These results indicate a considerable role of platelets in lung carcinogenesis.
Intriguingly, a recent study indicates that cancer cells depend on platelets to avoid anoikis and succeed in metastasis [47]. Platelets induce resistance to anoikis in vitro and are critical for metastasis in vivo by activating RhoA-MYPT1-PP1-mediated YAP1 dephosphorylation and promoting its nuclear translocation to inhibit apoptosis. However, the unknown underlying mechanism warrants future well-designed functional experiments to clarify the role of platelets in these cellular processes.
In addition, anti-platelet agents, such as purinergic antagonists, are used clinically because they affect inflammatory pathways [48]. Recent publications demonstrate that platelets suppress T-cell responses against tumors through production and activation of immunosuppressive factors. These results suggest the use of a combination of immunotherapy and platelet inhibitors, such as aspirin [49, 50] and clopidogrel, as a therapeutic strategy against cancer [51, 52]. Therefore, it is possible that anti-platelet therapy could reduce lung cancer risk.
However, we acknowledge some limitations in our study. First, some associations between genetic instrumental variables and phenotype (platelet count) were insufficient and thus may result in a “weak instrument” phenomenon [53]. Second, in some scenarios, inconsistent results were observed between inverse-variance weighted and MR-Egger (or regular and penalized/robust) models. This phenomenon indicates that genetic variants probably have horizontal pleiotropy, and thus MR assumptions are likely violated [54]. Moreover, there is heterogeneity across results incorporating different SNP sets as instrumental variables, which indicates that the instrumental variable should be curated carefully before Mendelian randomization analysis. In this study, all platelet count-related SNPs were curated, and six were retained to better satisfy MR assumptions. Third, a linear association was assumed between PLT and lung cancer risk. However, the shape could be non-linear and thus warrants further study incorporating individual-level data. Fourth, we only evaluated platelet count as a potential causal factor, whereas platelet function plays a comparable causal role in this pathway. More detailed platelet information should be measured in future studies, including immature platelet fractions and function. In addition, we assumed that study populations used for the genetic instrument for platelet count and for risk of lung cancer were representative of the same general Caucasian population, which may not be true. Therefore, additional functional studies are needed to further evaluate the mechanisms that underlie associations between platelets and lung cancer risk.
Nonetheless, our findings do suggest a role of platelet count in risk of lung cancer. The results provide a better understanding of lung cancer etiology and evidence for a possible role of anti-platelet interventions in lung cancer prevention.
Supplementary Material
Acknowledgements
We thank the participants and staff for their important contributions to this study.
Funding
This study was supported by the National Institute of Health (NIH) (CA092824 and CA209414 to D.C. Christiani), National Natural Science Foundation of China (81530088 and 81473070 to F. Chen, and 81373102 to Y. Zhao), State’s Key Project of Research and Development Program (2016YFE0204900 to F. Chen), Key Project of Natural Science Foundation of Jiangsu, China (14JA31002 to F. Chen), A Project Funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD), and Outstanding Young Teachers Training Program of Nanjing Medical University (to Y.Y. Wei). Transdisciplinary Research for Cancer in Lung (TRICL) and Oncoarray funding sources are detailed following. Sponsors had no role in design of the study, collection and analysis of data, or preparation of the manuscript. Transdisciplinary Research for Cancer in Lung (TRICL) of the International Lung Cancer Consortium (ILCCO) was supported by grants U19-CA148127 and CA148127S1. ILCCO data harmonization was supported by the Cancer Care Ontario Research Chair of Population Studies (to R.J. Hung) and the Lunenfeld-Tanenbaum Research Institute, Sinai Health System. The CAPUA study was supported by FIS-FEDER/Spain grants FIS-01/310, FIS-PI03-0365, and FIS-07-BI060604; FICYT/Asturias grants FICYT PB02-67 and FICYT IB09-133; and the University Institute of Oncology (IUOPA) of the University of Oviedo and the Ciber de Epidemiologiay Salud Pública (CIBERESP), Spain. Work performed in the CARET study was supported by the National Institute of Health (NIH)/National Cancer Institute (NCI) UM1 CA167462 (PI: G.E. Goodman), NIH UO1-CA6367307 (PIs: Omen, G.E. Goodman), NIH R01 CA111703 (PI: C. Chen), and NIH 5R01 CA151989-01A1 (PI: J. Doherty). The Liverpool Lung project was supported by the Roy Castle Lung Cancer Foundation. The Harvard Lung Cancer Study was supported by the NIH/NCI grants CA092824, CA090578, and CA074386. The Multiethnic Cohort Study was partially supported by NIH grants CA164973, CA033619, CA63464, and CA148127. Work performed in the MSH-PMH study was supported by the Canadian Cancer Society Research Institute (020214), Ontario Institute of Cancer and Cancer Care Ontario Chair Award (to R.J. Hung and G. Liu), and Alan Brown Chair and Lusi Wong Programs at Princess Margaret Hospital Foundation. NJLCS was funded by the State Key Program of National Natural Science of China (81230067), National Key Basic Research Program (2011CB503805), and Major Program of the National Natural Science Foundation of China (81390543). The Norway study was supported by the Norwegian Cancer Society, Norwegian Research Council. The Shanghai Cohort Study (SCS) was supported by NIH R01 CA144034 (PI: J.M. Yuan) and UM1 CA182876 (PI: J.M. Yuan). The Singapore Chinese Health Study (SCHS) was supported by NIH R01 CA144034 (PI: J.M. Yuan) and UM1 CA182876 (PI: J.M. Yuan). Work in the TLC study has been supported in part the James & Esther King Biomedical Research Program (09KN-15), NIH Specialized Programs of Research Excellence (SPORE) (P50 CA119997), and a Cancer Center Support Grant (CCSG) at the H. Lee Moffitt Cancer Center and Research Institute, an NCI designated Comprehensive Cancer Center (P30-CA76292). The Vanderbilt Lung Cancer Study - BioVU dataset used for the described analyses was obtained from Vanderbilt University Medical Center’s BioVU, which is supported by institutional funding, the 1S10RR025141-01 instrumentation award, and by the Vanderbilt Nature Genetics: doi:10.1038/ng.3892. The Clinical and Translational Science Awards (CTSA) grant UL1TR000445 was from the National Center for Advancing Translational Sciences (NCATS)/NIH. M.C. Aldrich was supported by NIH/NCI K07CA172294 (PI: M.C. Aldrich), and Dr. Bush was supported by National Human Genome Research Institute (NHGRI)/NIH U01HG004798 (PI: Crawford). The Copenhagen General Population Study (CGPS) was supported by the Chief Physician Johan Boserup and Lise Boserup Fund, the Danish Medical Research Council, and Herlev Hospital. The NELCS study was supported by grant P20RR018787 from the National Center for Research Resources (NCRR), a component of the NIH. The Kentucky Lung Cancer Research Initiative was supported by the Department of Defense [Congressionally Directed Medical Research Program, US Army Medical Research and Materiel Command Program] under award 10153006 (W81XWH-11-1-0781). This research was also supported by unrestricted infrastructure funds from the UK Center for Clinical and Translational Science, NIH grant UL1TR000117, and Markey Cancer Center NCI Cancer Center Support Grant (P30 CA177558) Shared Resource Facilities: Cancer Research Informatics, Biospecimen and Tissue Procurement, and Biostatistics and Bioinformatics. The M.D. Anderson Cancer Center study was supported in part NIH grants P50 CA070907 and R01 CA176568 (to X.F. Wu), Cancer Prevention & Research Institute of Texas RP130502 (to X.F. Wu), and University of Texas MD Anderson Cancer Center institutional support for the Center for Translational and Public Health Genomics. The deCODE study of smoking and nicotine dependence was funded in part by grant R01- DA017932 from the National Institutes on Drug Abuse (NIDA). The Lodz center study was partially funded by Nofer Institute of Occupational Medicine under task NIOM 10.13: Predictors of mortality from non-small cell lung cancer - field study. Genetic sharing analysis was funded by NIH grant CA194393. The ResoLuCENT study (Resource for the Study of Lung Cancer Epidemiology in North Trent) is funded by the Sheffield Hospitals Charity, Sheffield Experimental Cancer Medicine Centre and Weston Park Hospital Cancer Charity. F.Taylor was supported by a Cancer Research UK/Yorkshire Cancer Research Clinical Fellowship. B. Zhu’s work was supported by the Intramural Research Program of the Division of Cancer Epidemiology and Genetics, NCI. The Environment And Genetics in Lung cancer Etiology (EAGLE) study (PI: M.T. Landi) was supported by the Intramural Research Program of NIH, NCI, Division of Cancer Epidemiology and Genetics. F.Y. Gu was also supported by Roswell Park Cancer Institute and Cancer Center Supporting Grant P30CA016056. L.L. Marchand was supported by a program project grant from the NCI, NIH: P01 CA168530 and grant U01 CA164973. The Toronto study (PI: J. McLaughlin) was supported by Canadian Cancer Society Research Institute (020214). The Canadian Urban Environmental Health Research Consortium is funded by the Canadian Institutes for Health Research (J. McLaughlin). S.J. Chanock was supported by the Intramural Research Program of the National Institutes of Health NCI’s Division of Cancer Epidemiology, and the American Cancer Society. This work was also supported by Cancer Research UK (C1298/A8362 to R.S. Houlston, C1298/A8780 and C1298/A8362 to J. McLaughlin and C18281/A19169 to R. Carreras-Torres).
Abbreviations:
- SNPs
single nucleotide polymorphisms
- NSCLC
non-small cell lung cancer
- OR
odds ratio
- PLT
platelet count
- MR
Mendelian Randomization
- IV
instrumental variable
- GWAS
genome-wide association studies
- SMR
summary data-based Mendelian randomization
- AC
adenocarcinoma
- SqCC
squamous cell carcinoma
- SCLC
small cell lung cancer
- IVW
inverse-variance weighted
Footnotes
Competing interests
The authors below are claiming a Conflict of Interest.
Geoffrey Liu was on the Speaker’s Bureau and received honoraria from Pfizer, AstraZeneca, Takeda, Roche, Novartis, BMS, Merck.
Erik H.F.M. van der Heijden recieved commercial research support from Philips Medical Systems, and Astra Zeneca Oncology, and was on the Speaker’s Bureau and received honoraria from Pentax Medical.
The other authors declare no potential conflicts of interest.
References
- [1].Siegel R, Ma J Fau - Zou Z, Zou Z Fau - Jemal A, Jemal A. Cancer statistics, 2014. CA Cancer J Clin 2014;64(1):9–29. doi: 10.3322/caac.21208:1542-4863. [DOI] [PubMed] [Google Scholar]
- [2].Jemal A, Bray F Fau - Center MM, Center Mm Fau - Ferlay J, Ferlay J Fau - Ward E, Ward E Fau - Forman D, Forman D. Global cancer statistics. CA Cancer J Clin 2011;65(2):87–108. doi: 10.3322/caac.21262:1542-4863. [DOI] [Google Scholar]
- [3].Katanoda K, Sobue T Fau - Satoh H, Satoh H Fau - Tajima K, Tajima K Fau - Suzuki T, Suzuki T Fau - Nakatsuka H, Nakatsuka H Fau - Takezaki T, et al. An association between long-term exposure to ambient air pollution and mortality from lung cancer and respiratory diseases in Japan. J Epidemiol 2011;21(2):132–43(1349–9092 (Electronic)). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Cohen BL. Testing a BEIR-VI suggestion for explaining the lung cancer vs. radon relationship for U.S. counties. Health Phys 2000;78(5):522–7(0017–9078 (Print)). [DOI] [PubMed] [Google Scholar]
- [5].Kligerman S, White C. Epidemiology of lung cancer in women: risk factors, survival, and screening. AJR Am J Roentgenol 2011;196(2):287–95. doi: 10.2214/AJR.10.5412(1546–3141 (Electronic)). [DOI] [PubMed] [Google Scholar]
- [6].Lefrancais E, Ortiz-Munoz G, Caudrillier A, Mallavia B, Liu F, Sayah DM, et al. The lung is a site of platelet biogenesis and a reservoir for haematopoietic progenitors. Nature 2017;544(7648):105–109. doi: 10.1038/nature21706(1476–4687 (Electronic)). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Gasic Gj Fau - Gasic TB, Gasic Tb Fau - Stewart CC, Stewart CC. Antimetastatic effects associated with platelet reduction. Proc Natl Acad Sci U S A 1968;;61(1):46–52(0027–8424 (Print)). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Ji Y, Sheng L Fau - Du X, Du X Fau - Qiu G, Qiu G Fau - Su D, Su D. Elevated platelet count is a strong predictor of poor prognosis in stage I non-small cell lung cancer patients. Platelets 2015;26(2):138–42. doi: 10.3109/09537104.2014.888547(1369–1635 (Electronic)). [DOI] [PubMed] [Google Scholar]
- [9].Best MG, Sol N, In ‘t Veld S, Vancura A, Muller M, Niemeijer AN, et al. Swarm Intelligence-Enhanced Detection of Non-Small-Cell Lung Cancer Using Tumor-Educated Platelets. Cancer cell 2017;32(2):238–52.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Joosse SA, Pantel K. Tumor-Educated Platelets as Liquid Biopsy in Cancer Patients. Cancer cell 2015;28(5):552–4. [DOI] [PubMed] [Google Scholar]
- [11].Tural Onur S, Sokucu SN, Dalar L, Iliaz S, Kara K, Buyukkale S, et al. Are neutrophil/lymphocyte ratio and platelet/lymphocyte ratio reliable parameters as prognostic indicators in malignant mesothelioma? Ther Clin Risk Manag 2016;12:651–6. doi: 10.2147/TCRM.S104077(1176–6336 (Print)). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Menczer J Preoperative elevated platelet count and thrombocytosis in gynecologic malignancies. Archives of gynecology and obstetrics 2016;295(1):9–15. doi: 10.1007/s00404-016-4212-9(1432–0711 (Electronic)). [DOI] [PubMed] [Google Scholar]
- [13].Franco ATA-Ohoo, Corken A, Ware JA-Ohoo. Platelets at the interface of thrombosis, inflammation, and cancer. Blood 2015;126(5):582–8. doi: 10.1182/blood-2014-08-531582(1528–0020 (Electronic)). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Nikolic I Fau - Kukulj S, Kukulj S Fau - Samarzija M, Samarzija M Fau - Jelec V, Jelec V Fau - Zarak M, Zarak M, Orehovec B Fau - Taradi I, et al. Neutrophil-to-lymphocyte and platelet-to-lymphocyte ratio help identify patients with lung cancer, but do not differentiate between lung cancer subtypes. Croat Med J 2016;57(3):287–92(1332–8166 (Electronic)). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Omar M, Tanriverdi O, Cokmert S, Oktay E, Yersal O, Pilanci KN, et al. Role of increased mean platelet volume (MPV) and decreased MPV/platelet count ratio as poor prognostic factors in lung cancer. LID - 10.1111/crj.12605 [doi]. Clin Respir J 2016; doi: 10.1111/crj.12605(1752–699X (Electronic)). [DOI] [PubMed] [Google Scholar]
- [16].Oncel M, Kiyici A Fau - Oncel M, Oncel M Fau - Sunam GS, Sunam Gs Fau - Sahin E, Sahin E Fau - Adam B, Adam B. Evaluation of Platelet Indices in Lung Cancer Patients. Asian Pac J Cancer Prev 2015;16(17):7599–602(2476–762X (Electronic)). [DOI] [PubMed] [Google Scholar]
- [17].Palmer TM, Sterne Ja Fau - Harbord RM, Harbord Rm Fau - Lawlor DA, Lawlor Da Fau - Sheehan NA, Sheehan Na Fau - Meng S, Meng S Fau - Granell R, et al. Instrumental variable estimation of causal risk ratios and causal odds ratios in Mendelian randomization analyses. Am J Epidemiol 2010;173(12):1392–403. doi: 10.1093/aje/kwr026(1476–6256 (Electronic)). [DOI] [PubMed] [Google Scholar]
- [18].VanderWeele TJ, Tchetgen Tchetgen Ej Fau - Cornelis M, Cornelis M Fau - Kraft P, Kraft P. Methodological challenges in mendelian randomization. Epidemiology 2014;25(3):427–35. doi: 10.1097/EDE.0000000000000081(1531–5487 (Electronic)). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Didelez V, Sheehan N. Mendelian randomization as an instrumental variable approach to causal inference. Stat Methods Med Res 2007;16(4):309–30(0962–2802 (Print)). [DOI] [PubMed] [Google Scholar]
- [20].Thrift AP, Risch HA, Onstad L, Shaheen NJ, Casson AG, Bernstein L, et al. Risk of esophageal adenocarcinoma decreases with height, based on consortium analysis and confirmed by Mendelian randomization. Clin Gastroenterol Hepatol 2014;12(10):1667–76.e1. doi: 10.1016/j.cgh.2014.01.039(1542–7714 (Electronic)). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Thrift AP, Gong J, Peters U, Chang-Claude J, Rudolph A, Slattery ML, et al. Mendelian randomization study of height and risk of colorectal cancer. Int J Epidemiol 2015;44(2):662–72. doi: 10.1093/ije/dyv082(1464–3685 (Electronic)). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Nuesch E, Dale C, Palmer TM, White J, Keating BJ, van Iperen EP, et al. Adult height, coronary heart disease and stroke: a multi-locus Mendelian randomization meta-analysis. Int J Epidemiol 2015;45(6):1927–1937. doi: 10.1093/ije/dyv074(1464–3685 (Electronic)). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Khankari NK, Shu XO, Wen W, Kraft P, Lindstrom S, Peters U, et al. Association between Adult Height and Risk of Colorectal, Lung, and Prostate Cancer: Results from Meta-analyses of Prospective Studies and Mendelian Randomization Analyses. PLoS Med 2016;13(9):e1002118. doi: 10.1371/journal.pmed.1002118(1549–1676 (Electronic)). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Davies NM, Gaunt TR, Lewis SJ, Holly J, Donovan JL, Hamdy FC, et al. The effects of height and BMI on prostate cancer incidence and mortality: a Mendelian randomization study in 20,848 cases and 20,214 controls from the PRACTICAL consortium. Cancer Causes Control 2015;26(11):1603–16. doi: 10.1007/s10552-015-0654-9(1573–7225 (Electronic)). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Zhu Z, Zhang F, Hu H, Bakshi A, Robinson MR, Powell JE, et al. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. 2016;48(5):481–7. [DOI] [PubMed] [Google Scholar]
- [26].Burgess S, Butterworth A Fau - Thompson SG, Thompson SG. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol 2013;37(7):658–65. doi: 10.1002/gepi.21758(1098–2272 (Electronic)). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Gieger C, Radhakrishnan A Fau - Cvejic A, Cvejic A Fau - Tang W, Tang W Fau - Porcu E, Porcu E Fau - Pistis G, Pistis G Fau - Serbanovic-Canic J, et al. New gene functions in megakaryopoiesis and platelet formation. Nature 2011;480(7376):201–8. doi: 10.1038/nature10659(1476–4687 (Electronic)). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Kamatani Y, Matsuda K, Okada Y, Kubo M, Hosono N, Daigo Y, et al. Genome-wide association study of hematological and biochemical traits in a Japanese population. Nature genetics 2010;42(3):210–5. [DOI] [PubMed] [Google Scholar]
- [29].McKay JD, Hung RJ, Han Y, Zong X, Carreras-Torres R, Christiani DC, et al. Large-scale association analysis identifies new lung cancer susceptibility loci and heterogeneity in genetic susceptibility across histological subtypes. Nature genetics 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Bowden J, Del Greco MF, Minelli C, Davey Smith G, Sheehan N, Thompson J. A framework for the investigation of pleiotropy in two-sample summary data Mendelian randomization. Stat Med 2017;36(11):1783–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Burgess S, Small DS, Thompson SG. A review of instrumental variable estimators for Mendelian randomization. Stat Methods Med Res 2016;pii: 0962280215597579(1477–0334 (Electronic)). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Davies NM, von Hinke Kessler Scholder S, Farbmacher H, Burgess S, Windmeijer F, Smith GD. The many weak instruments problem and Mendelian randomization. Stat Med 2015;34(3):454–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Burgess S Sample size and power calculations in Mendelian randomization with a single instrumental variable and a binary outcome. Int J Epidemiol 2014;43(3):922–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Thomas MR, Storey RF. The role of platelets in inflammation. Thromb Haemost 2015;114(3):449–58. doi: 10.1160/TH14-12-1067(0340–6245 (Print)). [DOI] [PubMed] [Google Scholar]
- [35].Verheul HM, Hoekman K Fau - Luykx-de Bakker S, Luykx-de Bakker S Fau - Eekman CA, Eekman Ca Fau - Folman CC, Folman Cc Fau - Broxterman HJ, Broxterman Hj Fau - Pinedo HM, et al. Platelet: transporter of vascular endothelial growth factor. Clin Cancer Res 1997;3(12 Pt 1):2187–90(1078–0432 (Print)). [PubMed] [Google Scholar]
- [36].Pinedo HM, Verheul Hm Fau - D’Amato RJ, D’Amato Rj Fau - Folkman J, Folkman J. Involvement of platelets in tumour angiogenesis? Lancet 1998;352(9142):1775–7(0140–6736 (Print)). [DOI] [PubMed] [Google Scholar]
- [37].Cross MJ, Claesson-Welsh L. FGF and VEGF function in angiogenesis: signalling pathways, biological responses and therapeutic inhibition. Trends Pharmacol Sci 2010;22(4):201–7(0165–6147 (Print)). [DOI] [PubMed] [Google Scholar]
- [38].Ferrara N, Gerber Hp Fau - LeCouter J, LeCouter J. The biology of VEGF and its receptors. Nat Med 2003;9(6):669–76(1078–8956 (Print)). [DOI] [PubMed] [Google Scholar]
- [39].Xiao XY, Lang XP. Correlation Between MMP-7 and bFGF Expressions in Non-small Cell Lung Cancer Tissue and Clinicopathologic Features. Cell Biochem Biophys 2015;73(2):427–432. doi: 10.1007/s12013-015-0656-y(1559–0283 (Electronic)). [DOI] [PubMed] [Google Scholar]
- [40].Otaka Y, Rokudai S, Kaira K, Fujieda M, Horikoshi I, Iwakawa-Kawabata R, et al. STXBP4 Drives Tumor Growth and Is Associated with Poor Prognosis through PDGF Receptor Signaling in Lung Squamous Cell Carcinoma. Clin Cancer Res 2017;doi: 10.1158/1078-0432.CCR-16-1815(1078–0432 (Print)). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Naykoo NA, Dil A, Rasool R, Shah S, Ahangar AG, Bhat IA, et al. Single nucleotide polymorphisms, haplotype association and tumour expression of the vascular endothelial growth factor (VEGF) gene with lung carcinoma. Gene 2017;608:95–102. doi: 10.1016/j.gene.2017.01.007(1879–0038 (Electronic)). [DOI] [PubMed] [Google Scholar]
- [42].Jahanban-Esfahlan R, Seidi K, Monfaredan A, Shafie-Irannejad V, Abbasi MM, Karimian A, et al. The herbal medicine Melissa officinalis extract effects on gene expression of p53, Bcl-2, Her2, VEGF-A and hTERT in human lung, breast and prostate cancer cell lines. (1879–0038 (Electronic)). [DOI] [PubMed]
- [43].Hu M, Hu Y, He J, Li B. Prognostic Value of Basic Fibroblast Growth Factor (bFGF) in Lung Cancer: A Systematic Review with Meta-Analysis. PLoS One 2016;11(1):e0147374. doi: 10.1371/journal.pone.0147374(1932–6203 (Electronic)). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Dadrich M, Nicolay NH, Flechsig P, Bickelhaupt S, Hoeltgen L, Roeder F, et al. Combined inhibition of TGFbeta and PDGF signaling attenuates radiation-induced pulmonary fibrosis. Oncoimmunology 2016;5(5):e1123366. doi: 10.1080/2162402X(2162–4011 (Print)). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Middleton EA, Weyrich AS, Zimmerman GA. Platelets in Pulmonary Immune Responses and Inflammatory Lung Diseases. Physiological reviews 2016;96(4):1211–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Gong L, Cai Y, Zhou X, Yang H. Activated platelets interact with lung cancer cells through P-selectin glycoprotein ligand-1. Pathology oncology research : POR 2012;18(4):989–96. [DOI] [PubMed] [Google Scholar]
- [47].Haemmerle M, Taylor ML, Gutschner T, Pradeep S, Cho MS, Sheng J, et al. Platelets reduce anoikis and promote metastasis by activating YAP1 signaling. 2017;8(1):310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Pitchford SC. Novel uses for anti-platelet agents as anti-inflammatory drugs. Br J Pharmacol 2007;152(7):987–1002(0007–1188 (Print)). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Cao Y, Nishihara R, Wu K, Wang M, Ogino S, Willett WC, et al. Population-wide Impact of Long-term Use of Aspirin and the Risk for Cancer. JAMA Oncol 2016;2(6):762–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Oh SW, Myung SK, Park JY, Lee CM, Kwon HT. Aspirin use and risk for lung cancer: a meta-analysis. Annals of Oncology 2011;22(11):2456–65. [DOI] [PubMed] [Google Scholar]
- [51].Bordon Y Tumour immunology: Platelets - a new target in cancer immunotherapy? Nature reviews Immunology 2017;17(6):348. doi: 10.1038/nri.2017.61(6):348. [DOI] [PubMed] [Google Scholar]
- [52].Saleh Rachidi AM, Riesenberg Brian, Wu Bill X., Nelson Michelle H., Wallace Caroline, Paulos Chrystal M., Rubinstein Mark P., Garrett-Mayer Elizabeth, Hennig Mirko, Bearden Daniel W., Yang Yi, Liu Bei and Li Zihai. Platelets subvert T cell immunity against cancer via GARP-TGFβ axis. Science Immunology 2017;DOI: 10.1126/sciimmunol.aai7911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Burgess S, Thompson SG. Bias in causal estimates from Mendelian randomization studies with weak instruments. Stat Med 2011;30(11):1312–23. doi: 10.1002/sim.4197(1097–0258 (Electronic)). [DOI] [PubMed] [Google Scholar]
- [54].Bowden J, Burgess S, Smith GD. Difficulties in Testing the Instrument Strength Independent of Direct Effect Assumption in Mendelian Randomization. JAMA cardiology 2017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


