Highlights
-
•
First report of Mesonivirus detection and isolation in Senegal, West Africa.
-
•
Genetic and morphological characterizations were performed.
-
•
Species demarcation criteria allowed to conclude to a new species named Dianke virus.
-
•
Dianke virus found in 21 species of arthropods, among them male individuals.
-
•
Temperature was shown to be important in Dianke virus host restriction.
Keywords: Mesonivirus, Insect-Specific virus, Mosquito, Eastern Senegal
Abstract
An increasing number of insect-specific viruses are found around the world. Very recently, a new group of insect-specific viruses, the Mesoniviridae family, was discovered in Africa, Asia, North America and Australia. Here we report the first detection and isolation of a new virus belonging to Mesonivirus genus in Senegal, West Africa. The so-called Dianke virus was detected in 21 species of arthropods trapped in the eastern part of the country. Male individuals were also infected, supporting vertical transmission assertion of insect specific viruses. As described for other mesoniviruses, no viral replication was observed after inoculation of mammalian cells. Viral replication in mosquito cells was blocked at a temperature of 37 °C, highlighting the importance of thermal conditions in Mesonivirus host restriction. Similar to our study, where a diverse range of arthropod vectors were found infected by the new virus, several studies have detected mesonivirus infection in mosquitoes with concerns for human health. It has been shown that dual infections in mosquito can alter viral infectivity. Due to their extensive geographic distribution and host range, as well as their use as potential disease control agents in vector populations, more studies should be done for a better knowledge of arthropod-restricted viruses prevalence and diversity.
1. Introduction
Since the discovery of the first members of the Mesoniviridae family, order Nidovirales (Junglen et al., 2009; Zirkel et al., 2011; Nga et al., 2011), a growing number of new species has been found in different parts of the world (Africa, Asia, North America and Australia) (Lauber et al., 2012; Kuwata et al., 2013; Zirkel et al., 2013; Vasilakis et al., 2014; Warrilow et al., 2014; Hang et al., 2016). Mesoniviruses are enveloped, positive-sense, single stranded RNA ([+]ssRNA) viruses and constitute the only insect-associated genus of nidoviruses. The 20 kb nidovirus genome organization is generally ORF1a-ORF1b-ORF2a-ORF2b-ORF3a-ORF3b, except the Alphamesonivirus1 species, which encodes a seventh open reading frame (ORF4) (Vasilakis et al., 2014). ORF1a and ORF1b are predicted to encode two polymerase polyproteins (pp) while the other ORFs encode structural proteins (Zirkel et al., 2011; Hang et al., 2016). Viral particles are enveloped, spherical in shape and 60–80 nm in diameter with club-shaped surface spikes (Zirkel et al., 2013). Like insect-specific flaviviruses (Hoshino et al., 2007; Huhtamo et al., 2009) and other mosquito-associated bunyaviruses (Marklewitz et al., 2013) and orbiviruses (Harrison et al., 2016), mesoniviruses are considered insect specific viruses (ISVs) as the totality of detection has come from mosquito pools and isolations only from insect cells lines, while no detection or replication has been reported in mammalian models (Nga et al., 2011; Zirkel et al., 2011; Vasilakis et al., 2014).
Here, we report the detection and isolation of a new closely related mesonivirus from mosquitoes trapped in eastern Senegal. Phylogenetic analyses of the complete genome of the strain showed a new circulating mesonivirus species in sympatric mosquitoes from eastern Senegal. The discovery of this new virus confirms that an increasing number of arthropod-specific viruses (ASVs) are discovered throughout the world with a potential impact on vectors and viruses of public health interest.
2. Material and methods
2.1. Study sites
The study consisted in mosquitoes trapping sessions done in the middle and at the end of the rainy season in 2012 and 2013 in several villages located along the two main roads of eastern Senegal, Africa. Permission to work within the different villages and household was provided by the CNERS (National Ethical Committee for Health Research) under the number 0000167 MSAS/DPRS/CNERS (see CNERS_authorization in supplemental material).
2.2. Mosquitoes trapping
In order to study the arboviruses circulation among domestic and peridomestic mosquitoes in eastern Senegal, the sampling protocol was as described by Diallo et al. (2012). Briefly, trapping was done in different human dwellings of villages of eastern Senegal. In each village, at least 10 houses were selected following a transect going from the peripheral borders to the center in order to cover all the ecological profiles. The different trapping sessions were performed during twilight, where most of arboviruses vectors such as Aedes species are active. Three methods were used: (1) human landing catches, the most appropriate method for determining the risk of human infection (Diallo et al., 2014), (2) CDC light traps with or without CO2 bait, shown to have relatively high sampling efficiency (Okumu et al., 2008) but to be less specific to particular anthropophilic species, (3) indoor residual spraying for endophilic species collection. Adhesive trap catches were also performed for sandflies. After morphological identification, collected insects were pooled by species in tubes and stored at −80 °C until viral detection processing. Each pool was triturated in L-15 medium and RNA was extracted as described below.
2.3. Arboviruses screening
Each pool was triturated in Leibovitz-15 (L-15) medium (GibcoBRL, Grand Island, NY, USA) containing penicillin and streptomycin (Sigma, GmBh, Germany) and 10 % FBS (Gibco BRL, Grand Island, NY, USA) and centrifuged in order to collect the suspension.
RNA was extracted from these different suspensions using the QIAamp RNA Viral Kit (Qiagen, Hilden, Germany) according to the manufacturer's recommendations. RNA was eluted in 50 μl of AVE buffer and stored at −80 °C until use. For the arbovirus screening, conventional RT-PCR systems targeting different genera were used (table in supplement).
Virus isolation attempts were also performed from 150 μl of suspensions of each specimen that were inoculated separately onto monolayers Aedes albopictus C6/36 cells in 25-cm2 tissue-culture flasks. After incubation at 28 °C for a maximum of 7 days post infection (pi), supernatant was collected and an aliquot of 200 μL was used for 4 serial passages, or until the observation of a cytopathic effect (CPE). Supernatants were also tested for several arboviruses by conventional reverse transcription (RT)-PCRs (Table S1 in supplement). Immunofluorescence assay (IFA) was performed as previously described to assess cell infection (Digoutte et al., 1992).
2.4. Next generation sequencing (NGS)
Supernatants obtained from some CPE-inducing samples but without virus identification by IFA or RT-PCR were processed for NGS. Library construction, sequence and genomic analysis were performed as described previously (Vasilakis et al., 2014). Briefly, viral RNA was fragmented using fragmentation buffer (Illumina 15016648) and the Illumina TruSeq RNA Sample Preparation kit was used for strand synthesis, adapter ligation and library amplification under conditions prescribed by the manufacturer (Illumina). Samples were tracked using the “index tags” incorporated into the adapters as defined by the manufacturer. Cluster formation of the library DNA templates was performed using the TruSeq PE Cluster Kit v3 (Illumina) and the Illumina cBot workstation using conditions recommended by the manufacturer. Paired end 50 base sequencing by synthesis was performed using TruSeq SBS kit v3 (Illumina) on an Illumina HiSeq 1000 using protocols defined by the manufacturer. Sequences were assembled into contigs with the SeqMan and NextGen suites of the DNAStar Lasergene 7 program (Bioinformatics Pioneer DNAStar, Inc., Madison, WI) and their relationships to other viruses were determined by a BLAST search. The open reading frames were identified using “SnapGene software (from GSL Biotech; available at snapgene.com)”. The MUSCLE algorithm was used for multiple alignments and maximum likelihood (ML) tree constructions were performed in MEGA software version 7 (Kumar et al., 2016). Calculation of the pairwise evolutionary distances (PED) between the highly conserved protein domains of ORF1ab (3CLpro, RdRp and ZnHel1) of the mesoniviruses was performed using the Jones-Taylor-Thornton matrix based-model in MEGA 7 with complete deletion gap and 1000 bootstrap replicates.
2.5. Transmission electron microscopy
Ultrathin sections were prepared for ultrastructural observation as previously described (Vasilakis et al., 2014). Briefly, after fixation of infected cells for at least 1 h in a mixture of 2.5 % formaldehyde and 0.1 % glutaraldehyde in 0.05 M cacodylate buffer pH 7.3, the monolayers were washed in 0.1 M cacodylate buffer and cells were scraped off. The pellets were post-fixed in 1 % OsO4 in 0.1 M cacodylate buffer pH 7.3 for 1 h, washed with distilled water and en bloc stained with 2 % aqueous uranyl acetate for 20 min at 60 °C. The pellets were dehydrated in ethanol, processed through propylene oxide and embedded in Poly/Bed 812 (Polysciences, Warrington, PA). Ultrathin sections were cut on Leica EM UC7 ultramicrotome (Leica Microsystems, Buffalo Grove, IL), stained with lead citrate and examined in a Philips 201 transmission electron microscope at 60 kV.
2.6. Conventional pan-Mesonivirus RT-PCR
Confirmation of NGS results was done using conventional RT-PCRs. For cDNA synthesis, 10 μl of RNA extract was mixed with 1 μl of random primers (pdN6) following the manufacturer's instructions (2 pmol) and the mixture was heated at 95 °C for 2 min. Reverse transcription was performed in 20 μl mixture containing 2.5 U RNasin (Promega, Madison, USA), 1 μl of deoxynucleotide triphosphate (dNTP) (10 mM each DNTP), 5 U of AMV reverse transcriptase (Promega, Madison, USA) and incubated at 42 °C for 1 h. A pan Mesonivirus nested PCR system was then performed as previously described (Zirkel et al., 2013) using primer pair MeniV-F1/MeniV-R1 followed by MeniV-F2/MeniV-R2 mixed with 10X buffer, 5 μl of dNTPs 10 mM, 3 μl of MgCl2, and 0.5 μl of Taq polymerase (Promega, Madison, USA).
2.7. Development of a pan Mesonivirus qRT-PCR
Multiple alignments of 11 mesonivirus sequences previously available in Genbank (Table 1 ) was made using MEGA 7 (Kumar et al., 2016). Based on a conserved region of 167bp between ORF3b and the 3′poly (A) tail, the primers and probe were then designed using Primer3 software (Untergasser et al., 2012). RT-qPCRs were performed in an ABI Prism 7500 SDS Real-Time cycler (Applied Biosystems, Foster City, USA) using Quantitect One-Step RT-PCR kit (Qiagen, Hilden, Germany) according to manufacturer’s recommendations with 5 μl of RNA template.
Table 1.
Sequences used for primers and probe design of the pan Mesonivirus qRT-PCR assay.
| Strains | Collection place | Collection date | Accession number |
|---|---|---|---|
| Bontang virus strain JKT7774 | Indonesia | 1981 | KC807166.1 |
| Karang Sari virus strain JKT10701 | Indonesia | 1981 | KC807171.1 |
| Ngewotan virus strain JKT9982 | Indonesia | 1981 | KC807170.1 |
| Kamphang Phet virus strain KP84-0156 | Thailand | 1984 | KC807172.1 |
| Cavally virus isolate C79 | Cote d'Ivoire | 2004 | HM746600.1 |
| Hana virus strain A4/CI/2004 | Cote d'Ivoire | 2004 | JQ957872.1 |
| Houston virus strain V3872 | USA: Houston | 2004 | KC807175.1 |
| Nse virus strain F24/CI/2004 | Cote d'Ivoire | 2004 | JQ957874.1 |
| Dak Nong virus | Viet Nam | 2007 | AB753015.2 |
| Nam Dinh virus isolate NDiV-NJ8-09 | China | 2009 | KF771866.1 |
| Casuarina virus isolate 0071 | Australia | 2010 | KJ125489.1 |
2.8. Mesonivirus in vitro infection assays
Viral stock was prepared from C6/36-passage 2 and the same concentration was inoculated into four different cell lines: mosquito C6/36 cells, Cercopithecus aethiops Vero cells, Homo sapiens Hep G2 and human rhabdomyosarcoma RD cells. Each infection was performed in 25-cm2tissue-culture flasks for incubation at 28 °C, 33 °C or 37 °C. Monitoring was done until strong CPE observation or at day 3, day 7 and day 10 p.i. Supernatant were then collected and the viral RNA quantification was done using the pan Mesonivirus qRT-PCR.
Continuous cell lines were originally provided by the ATCC (American Type Culture Collection, Manassas, Va.) and were cultured in Leibovitz-15 (L-15) medium (GibcoBRL, Grand Island, NY, USA) supplemented with 10 % FBS (Gibco BRL, Grand Island, NY, USA), penicillin-streptomycin (Sigma, GmBh, Germany). For C6/36 cells culture, Leibovitz-15 (L-15) medium was also supplemented with additional 10 % of Bacto™ Tryptose Phosphate Broth (Becton, USA).
The experiment was performed in duplicate for each cell line and temperature condition.
3. Results
3.1. Mesonivirus isolation from mosquitoes trapped in Eastern Senegal
During the arbovirus screening, CPE was observed 4 days p.i on C6/36 cells inoculated with extract from seven (7) mosquito pool homogenates from Sabodala, one (1) specimen from Fadiga, six (6) from Kedougou town and two (2) others in Dianke Makhan, with negative results both by PCR and IFA targeting numerous arboviruses (Table S1 in supplement). One of the C6/36 cell supernatants was randomly chosen for Next Generation Sequencing (NGS) as described above, yielding 20,118 bp apart from the extreme 3′-poly (A) tail - termini. The sequence shared 90 % nucleotide identity with some previously described mesoniviruses. This result was confirmed using a conventional pan Mesonivirus RT-nested PCR, which was positive both for corresponding supernatants and original samples.
3.2. Phylogeny and genome analysis
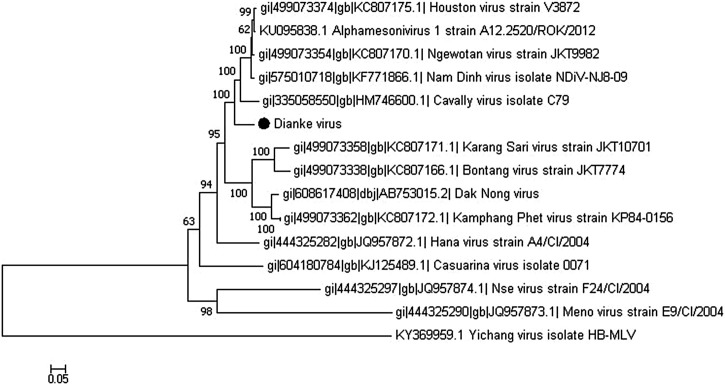
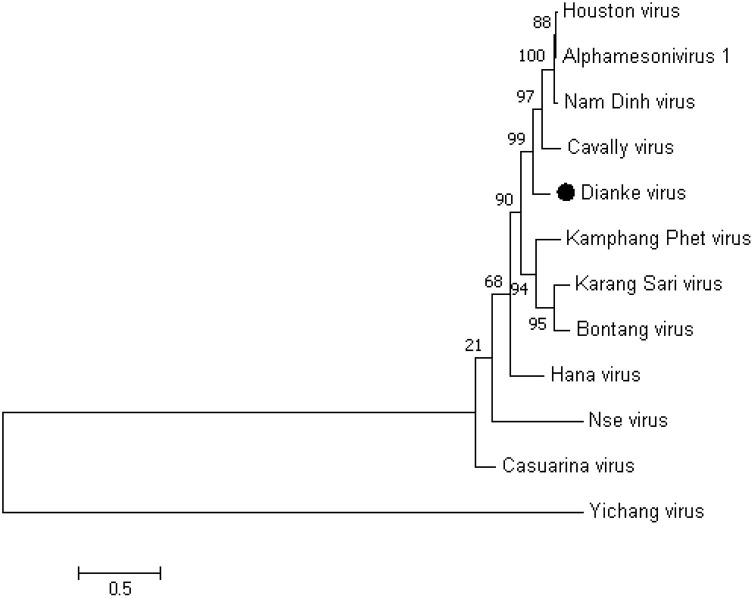
Comparative analysis of the genome showed that the strain is clearly distinct from the other mesoniviruses previously characterized, with 10 % nucleotide sequence difference compared to Houston and Nam Dinh viruses, and forming a separate cluster (Fig. 1 ). This observation was confirmed by a parallel analysis using a neighbor-Joining approach for which one no discrepancy was noted, as shown in figure S1 in supplemental material. The classical mesonivirus genome organization was found, with the typical ORFs and the putative ribosomal frame-shift (RFS) element in ORF1a and ORF1b, the 3C-like serine protease (3CLP), RNA-dependent RNA polymerase (RdRp), zinc-binding (Z), helicase (Hel), exoribonuclease (ExoN), N7-methyltransferase (NMT) and 2′-O-methyltransferase (OMT) domains. Block insertions observed after the first 1300 nt of the ORF1a of some Asian strains (Vasilakis et al., 2014) were absent in the Senegal sequence. Protein sequence identity of ORF 1, ORF 2 and ORF 3 were 91 %, 94 % and 91 %, respectively (Table 4). Pairwise evolutionary distance (PED) is the reference species demarcation criterion in the Mesonivirus genus (Lauber et al., 2012). Using the conserved protein domains of ORF1ab (3CLpro, RdRp and Hel) (Vasilakis et al., 2014), we calculated PED between the virus and some previously described mesoniviruses selected from the blast output using a protein alignment. We considered the same consensus threshold of 0.037 as minimum distance for the demarcation of nidovirus species based on the concatenated protein domains (Lauber et al., 2012). In accordance with a previous study (Warrilow et al., 2014), a PED value of 0.031 was obtained between CavV and NDiV isolates (Table 5), which constitutes the species Alphamesonivirus 1 as previously described (Lauber et al., 2012). The Senegal isolate was closest to Alphamesonivirus 1 with distances of 0.042, 0.044 and 0.053 separating it from Houston virus (HOUV), Nam Dinh virus (NDiV) and Cavally virus (CavV), respectively. The highest distance was observed with Nse (NseV), Casuarina (CASV) and Hana viruses (HAV) with PED values of 0.243, 0.123 and 0.117, respectively (Table 5). These results are reflected by the tree obtained with the different sequences involved in the maximum likelihood analysis (Fig. 2 ), as well as a neighbor-joining approach (figure S2 in supplemental material). Our isolates represent a new species in the Mesonivirus genus. We proposed Dianke virus (DKV) (Accession number: MN622133) as a name for this virus, referring to one of the sampling localities.
Fig. 1.
Maximum likelihood tree of the full-length genome of Dianke virus and other mesoniviruses. Scale bar indicate nucleotide substitutions/site.
Table 4.
Blast results summary of Dianke virus open reading frames (ORFs): HOUV: Houston virus; NGEV: Ngewotan virus; NDiv: Nam Dinh virus; CAVV: Cavally virus; HAV: Hana virus; DkNV: Dak Nong virus; KSV: Karang Sari virus; KPV: Kamphang Phet; BV: Bontang virus; CASV: Casuarina virus; NseV: Nse virus; MeV: Meno virus.
| ORF1 |
ORF2 |
ORF3 |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Nucleotides |
Protein |
Nucleotides |
Protein |
Nucleotides |
Protein |
||||||||||||
| Id | Accession | Id | Accession | Id | Accession | Id | Accession | Id | Accession | Id | Accession | ||||||
| HOUV | 88% | KC807178.1 | NGEV | 91% | AGL73185.1 | HOUV | 91% | KC807177.1 | NGEV | 94% | ASA47342.1 | NGEV | 92% | MF176279.1 | NDiV | 91% | AII17280.1 |
| NGEV | 88% | MF176279.1 | NDiV | 91% | YP_004767306.1 | NGEV | 91% | MF176279.1 | NDiV | 94% | AHG56128.1 | NDiV | 92% | KF522691.1 | NGEV | 91% | ASA47344.1 |
| NDiV | 88% | DQ458789.2 | HOUV | 91% | AGL73209.1 | NDiV | 91% | KF771866.1 | HOUV | 94% | AGL73201.1 | HOUV | 92% | KC807178.1 | HOUV | 91% | AGL73202.1 |
| CAVV | 87% | HM746600.1 | CAVV | 90% | YP_004598982.1 | CAVV | 90% | HM746600.1 | CAVV | 93% | AEH26445.1 | CAVV | 89% | HM746600.1 | CAVV | 86% | YP_004598983.1 |
| HAV | 80% | JQ957872.1 | HAV | 81% | YP_007697630.1 | DkNV | 85% | AB753015.2 | DkNV | 88% | BAN58308.1 | KPV | 89% | KC807174.1 | DkNV | 85% | BAN58309.1 |
| DkNV | 78% | AB753015.2 | DkNV | 78% | BAN58307.2 | KSV | 85% | KC807171.1 | BV | 87% | AGL73177.1 | DkNV | 88% | AB753015.2 | KPV | 85% | AGL73193.1 |
| KSV | 77% | KC807171.1 | KPV | 72% | AGL73197.1 | KPV | 85% | KC807174.1 | KSV | 88% | AGL73189.1 | HAV | 86% | JQ957872.1 | HAV | 82% | YP_007697631.1 |
| KPV | 77% | KC807173.1 | CASV | 71% | YP_009026379.1 | BV | 85% | KC807169.1 | KPV | 87% | AGL73192.1 | BV | 79% | KC807166.1 | KSV | 73% | AGL73190.1 |
| BV | 77% | KC807169.1 | NseV | 59% | YP_007697643.1 | HAV | 85% | JQ957872.1 | CASV | 85% | YP_009026378.1 | CASV | 79% | KJ125489.1 | BV | 72% | AGL73178.1 |
| CASV | 78% | KJ125489.1 | MeV | 50% | YP_007697637.1 | CASV | 82% | KJ125489.1 | HAV | 85% | YP_007697629.1 | CASV | 70% | YP_009026380.1 | |||
| MOUV | 77% | KC768950.1 | NseV | 75% | YP_007697642.1 | NseV | 63% | YP_007697644.1 | |||||||||
| NseV | 76% | JQ957874.1 | MOUV | 76% | AGI52414.1 | MeV | 60% | YP_007697638.1 | |||||||||
Table 5.
Estimates of evolutionary distances between ORF1a conserved domain (3CL-RdRp-HEL) of Dianke virus and other mesoniviruses.
| Nse virus | ||||||||||
| Hana virus | 0.249 | |||||||||
| Casuarina virus | 0.247 | 0.160 | ||||||||
| Houston virus | 0.238 | 0.112 | 0.108 | |||||||
| Kamphang Phet virus | 0.236 | 0.121 | 0.123 | 0.080 | ||||||
| Karang Sari virus | 0.250 | 0.122 | 0.133 | 0.082 | 0.044 | |||||
| Bontang virus | 0.252 | 0.128 | 0.137 | 0.092 | 0.051 | 0.039 | ||||
| Nam Dinh virus | 0.241 | 0.114 | 0.109 | 0.006 | 0.080 | 0.082 | 0.093 | |||
| Cavally virus | 0.249 | 0.117 | 0.112 | 0.028 | 0.085 | 0.090 | 0.102 | 0.031 | ||
| Alphamesonivirus 1 | 0.239 | 0.114 | 0.106 | 0.002 | 0.082 | 0.084 | 0.094 | 0.008 | 0.028 | |
| Dianke virus | 0.243 | 0.117 | 0.123 | 0.042 | 0.086 | 0.085 | 0.092 | 0.044 | 0.053 | 0.044 |
Fig. 2.
Maximum-likelihood tree obtained from amino acid analysis of concatenated conserved protein domains of ORF1ab: 3CLpro-RdRp-HEL1. (Position on Dianke virus genome: 3CLpro: 4167nt-5072nt; RdRp: 9023nt-10387nt; HEL1: 11825nt-12853nt.) Scale bar indicate amino acid substitutions/site.
3.3. Dianke virus morphological characterization
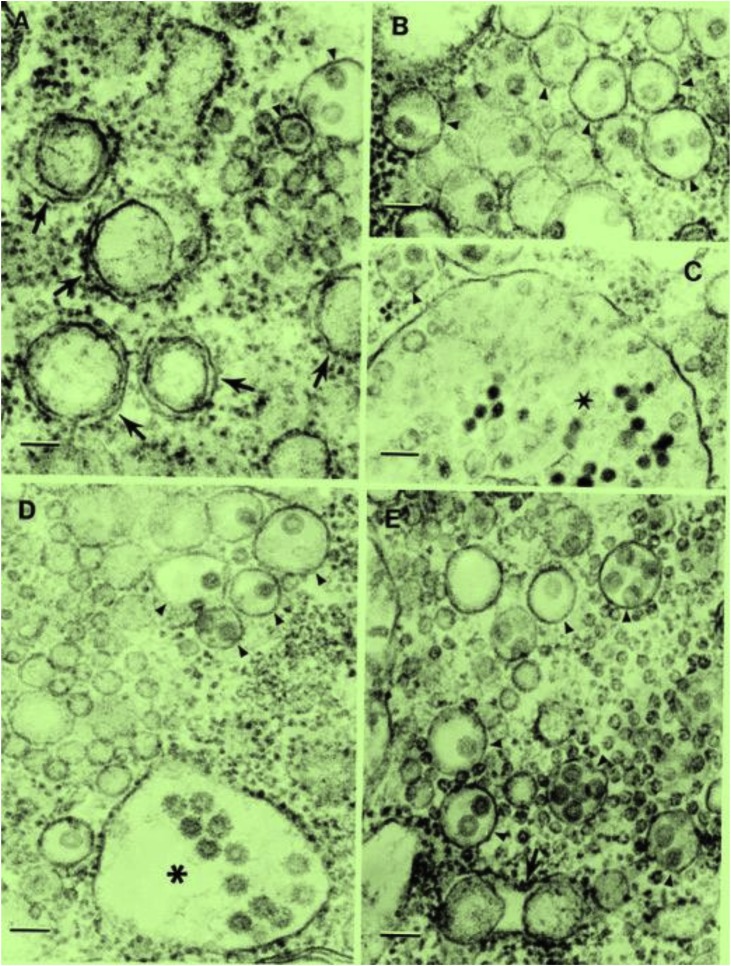
In ultrathin sections of DKV-infected C6/36 cells, three types of virus particles were observed: enveloped spherical particles 55–60 nm in diameter with smooth surface located within smooth membrane-limited vacuoles containing from 1 to 5 particles (Fig. 3 A-E); enveloped spherical particles ∼65 nm in diameter with short (∼5 nm) surface projections located mostly within enlarged cisterns of granular endoplasmic reticulum (Fig. 3D) and smaller spherical enveloped particles with dense core (40–45 nm in diameter) within large vacuoles (Fig. 3C). Characteristically, viruses produced the same kind of CPE – large vesicles 120–250 nm in diameter located mostly individually (Fig.3A) or in pairs (Fig. 3E) inside tight vacuoles, presumably of granular endoplasmic reticulum origin because some of them had ribosomes at their outer surface. Morphologically these vesicles resembled smooth membrane structures (SMS) regularly observed with replicating flaviviruses.
Fig. 3.
Ultrastructure of Dianke virus in C6/36 cells.
Bars in all pictures =100 nm.
A- Virus particles with smooth surface 55–60 nm in diameter inside cytoplasmic vacuoles (arrowheads). Arrows indicate vesicles 120–250 nm in diameter inside vacuoles, presumably of granular endoplasmic reticulum origin.
B- Virus particles with smooth surface 55–60 nm in diameter inside multiple cytoplasmic vacuoles (arrowheads).
C- Three particles with smooth surface 55–60 nm in diameter inside small cytoplasmic vacuole (arrowhead) and smaller (40–45 nm in diameter) enveloped particles with dark core inside huge vacuole (star).
D- Two types of virus particles inside vacuoles: smooth surfaced 55–60 nm in diameter inside small smooth surfaced vacuole s (arrowheads) and particles ∼ 65 nm in diameter with spikes ∼5 nm long inside an expanded cistern of granular endoplasmic reticulum (asterisk).
E- Smooth surfaced virus particles 55–60 nm in diameter inside smooth surfaced cytoplasmic vacuoles (arrowheads) and vesicles 140–160 nm in diameter inside a cistern of granular endoplasmic reticulum (arrow). Cytoplasm contains multiple vesicles 25–30 nm in diameter.
3.4. Pan mesonivirus qRT-PCR validation
A pan Mesonivirus qRT-PCR was designed using an alignment of partial sequences of eleven known mesonivirus strains (see above). The TaqMan probe was flanked by a 6-carboxyfluorescein (FAM) fluorescent reporter dye at 5′end and a black hole quencher 1 (BHQ-1) at the 3′-end. Primers and probe characteristics are shown in Table 2 . A specificity test was performed with several viruses found in mosquito pools from Senegal (Table 3 ). The pan-Mesonivirus qRT-PCR was validated by a specificity test including some strains obtained during this study (Table 3) and showed good results with amplification only for mesonivirus samples. The qRT-PCR was also more sensitive than conventional RT-PCR.
Table 2.
Nucleotide sequences of primers and probe used for qPCR pan Mesonivirus assay (based on Cavally virus isolate C79 strain, HM746600.1).
| Sequences 5’-3’ | Nucleotide position | |
|---|---|---|
| Meso F (Forward primer) | CATGGACDNAACACAACAGCAG | 19877-19898 |
| Meso P (Probe) | FAM-AGGYGTACTGAAYTCYRAGGAGACG—BHQ1 | 19940-19964 |
| Meso R (Reverse primer) | AATGYGTCTCTCRCAAYGTA | 20022-20041 |
FAM: 6-carboxyfluorescein; BHQ1: 6-black hole quencher 1; D: A or G or T; N: A or C or G or T; Y: C or T; R: A or G.
Table 3.
Specificity test for pan Mesonivirus qRT-PCR assay.
| Virus | Strains | Mesonivirus qRT-PCR | Pan Mesonivirus (RT)-PCR | ct value |
|---|---|---|---|---|
| Chikungunya | S27 African prototype (AF369024.2) | Negative | Negative | – |
| Zika | ArD 165,522 (KF383090.1) | Negative | Negative | – |
| West Nile | Eg101 (AF260968.1) | Negative | Negative | – |
| Usutu | SAAR-1776 (AY453412.1) | Negative | Negative | – |
| Rift valley fever | MP-12 (DQ380154.1) | Negative | Negative | – |
| Yellow fever | 17 D (X03700.1) | Negative | Negative | – |
| Dengue 2 | Dak Ar 141,070 (EF105390.1) | Negative | Negative | – |
| Wesselsbron | ArB 4177 | Negative | Negative | – |
| Bagaza | ArB 209 (AY632545.2) | Negative | Negative | – |
| Barkedji | ArD86177 (EU078325.1) | Negative | Negative | – |
| Mesonivirus | Culex species (original) | Positive | Negative | 34.1 |
| Mesonivirus | Aedes species (original) | Positive | Negative | 33.5 |
| Mesonivirus | Culex species (C6/36 passage) | Positive | Positive | 24.3 |
| Mesonivirus | Aedes species (C6/36 passage) | Positive | Positive | 24.87 |
In order to obtain a pan Mesonivirus qRT-PCR standard curve, the linear dynamic range of the qPCR assay was initially assessed using 8-log10 serial dilutions of a synthetic standard RNA. The assay was shown to be linear over the entire range of 108 to 10 genomic copies. The different end-point dilutions of standard RNA were tested in triplicate. A typical standard curve amplification plot and linear regression analysis of these data are shown in the figure S3 in supplemental material. This qRT-PCR system was used for a screening of mesoniviruses in other mosquito pools (MPs) caught in the same areas as the positive ones.
3.5. Evidence of DKV circulation in mosquitoes
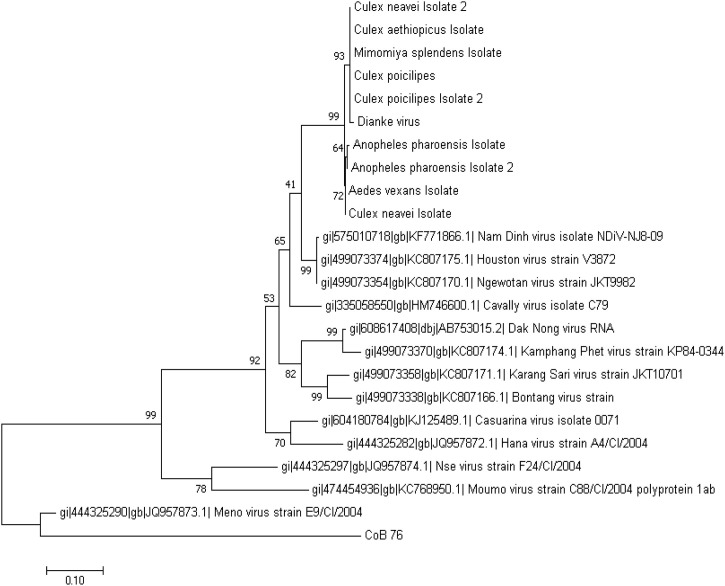
After molecular screening for mesonivirus RNA (qRT-PCR), DKV was detected in 21 species of arthropods (mainly mosquitoes) trapped in the same areas. The most representative genus was Aedes with 7 species: Ae. furcifer (5MPs), Ae. aegypti (2), Ae. cumminsii (1), Ae. fowleri (1), Ae. mcintoshi (1), Ae. minutus (1), and Ae. vittatus (1), followed by Culex species such as Cx. antennatus (5), Cx. decens (3), Cx. neavei (3), Cx. nebulosus (1), Cx. poicilipes (1) and 1 pool of males Cx. quinquefasciatus. Species from the genus Anopheles were also found positive with An. funestus (2), An. gambiae (1), An. pharoensis (1) or An. rufipes (1). A mesonivirus was also detected from 8 pools of Mansonia uniformis (including 1 pool of males) and 2 pools from Ma. africana. The virus was also detected on 1 pool of Uranotaenia sp and another one of biting midges (ceratopogonids). A conventional, specific RT-PCR assay targeting a small part of the ORF1a was performed on all positive samples and the sequences obtained confirmed that the same virus strain was circulating in these different arthropods (99 % identity) in eastern Senegal (Fig. 4 ). The Genbank accession numbers of the different sequences generated during this study are available in the table S2 of the supplement.
Fig. 4.
Maximum likelihood tree of 303bp of the ORF1ab of Dianke virus, the mesoniviruses detected in mosquito pools from the same areas and others published ones. The tree is rooted with CoB76 (Bovine Coronavirus). The sequences from Dianke virus and mosquito isolates from Senegal form a common cluster (99%–100% nucleotide identity) different to the others strains.
3.6. Dianke virus: vertebrate and insect cells infections
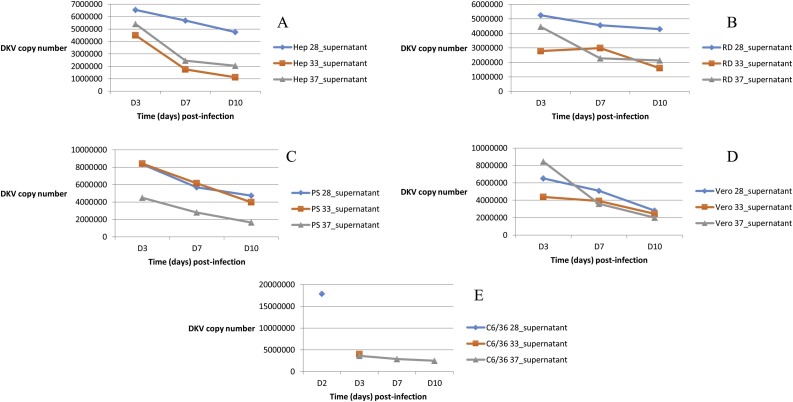
The same concentration of DKV was inoculated both in mosquito and mammalian cells in order to access replication under different conditions of temperature. C6/36 cells exhibited CPE 2 days pi at 28 °C and 3 days p.i at 33 °C, while no CPE was observed at 37 °C (Fig. 5 A). Moreover, a decrease of DKV RNA in C6/36 at 37 °C was observed until day 10. Mammalian cell monolayers were unaltered until day 10 pi with also a constant reduction of the viral quantity over time (Fig. 5B-C-D-E). However, this reduction was less important at 28 °C than the other thermal conditions with, for instance, a difference on the order of twice as pronounced for the Hep-G2 cells at day 10 pi.
Fig. 5.
Dianke virus (DKV) in vitro infections in different thermal conditions (blue: 28 °C, orange: 33 °C and grey (37 °C) on Hep-G2 cells (A), RD cells (B), PS cells (C), Vero cells (D) and C6/6 cells (E). The kinetics were prematurely stopped for C6/36 cells at 28 °C and 33 °C as a strong depletion was observed at day 2 and day 3 post-infection respectively.
4. Discussion
In this study, we reported the isolation of a new viral strain phylogenetically belonging to an insect-specific virus (ISV) group in Senegal. The isolates, obtained from different mosquito pools trapped in the Kedougou and Tambacounda areas (eastern Senegal), clearly belong to the Mesoniviridae family with 90 % nucleotide identity compared to other members. Genome organization showed typical mesonivirus features. The species demarcation based on previously employed criteria (Lauber et al., 2012) allowed us to conclude that this mesonivirus constitute a new species that we named Dianke virus (DKV), referring to one of the places where the strain have been found. Molecular screening undertaken in the same areas resulted in detection of the same virus in 43 mosquito pools from 21 species. These results are similar to previous observations where a broad host species range in mosquitoes suggested a worldwide mesoniviruses distribution (Vasilakis et al., 2014), while Colmant et al. (2017) described a species-specific host restriction for some insect-specific-flaviviruses.
Numerous papers indicate vertical transmission for some ISVs (Sang et al., 2003; Cook et al., 2006; Boiling et al., 2011; Haddow et al., 2013). In this study, DKV detection in some male mosquitoes supports this assertion.
Like our study where a large diversity of vectors was infected by the new strain, several studies have detected mesonivirus infection of mosquitoes with potentially significant health impacts (Zirkel et al., 2011; Nga et al., 2011; Vasilakis et al., 2014). Indeed, it has been shown that dual viral infections in mosquito can alter viral infectivity (Fujita et al., 2018) and ISVs are more and more considered as potential disease control agents in vector populations (Guzman et al., 2018; Öhlund et al., 2019). Virus phenotypic restriction could be more complex as Parry and Asgari (2018) highlighted that restriction of Dengue virus replication in Ae.aegypti mosquitoes could be hindered by interactions between a novel ISV and the endosymbiotic bacterium Wolbachia pipientis. In contrast, Zhang et al. (2017) pointed out that Cell fusing agent virus, another ISV, favor Dengue virus replication in Ae.aegypti cell lines. All together, the current studies highlight gaps in the understanding of interactions between ISVs and other pathogenic viruses. It has been shown that the host range restriction of insect-specific flaviviruses and alphaviruses occurs at different levels of viral replication cycle (Nasar et al., 2015; Junglen et al., 2017). Among intrinsic and extrinsic factors affecting viral replication and transmissibility, temperature was shown to be of an extreme importance (Samuel et al., 2016). The experiments undertaken in this study tends to confirm this assessment as DKV was unable to replicate in permissive mosquito C6/36 cells at 37 °C. However, the virus did not grow in different mammal cells at the mosquito temperature of 28 °C. The host restriction phenomenon for mesoniviruses and the other ISVs remain to be further studied.
An increasing number of arthropod-specific viruses (ASVs) are being discovered in hematophagous arthropods throughout the world (Calisher and Higgs, 2018). Due to their potential impact on the vectors fitness, more studies are needed to improve the knowledge about their prevalence and diversity.
Financial support
This study was supported in part by grants ANR-11-CEPL-0010 and ANR-11-JSV7-0006 (AAS, OF, MMD, AG, YB, MD) from the Agence Nationale de la Recherche, from Institut Pasteur de Dakar funds and grant R24 AT120992 from the National Institutes of Health (RBT, SCW, VP, HG, NV).
Disclosure of competing interest
No competing interests in this scientific work.
Acknowledgements
We thank Dr. Laurent Granjon and Pr. Pascal Handschumacher as well as the CBGP teams of IRD (Institut de Recherche pour le Développement) from France and Dakar, Senegal for their outstanding collaboration for CHANCIRA project.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.virusres.2019.197802.
Appendix A. Supplementary data
The following are Supplementary data to this article:
References
- Boiling B.G., Eisen L., Moore C.G., Blair C.D. Insect-specific flaviviruses from Culex mosquitoes in Colorado, with evidence of vertical transmission. Am. J. Trop. Med. Hyg. 2011;85:169–177. doi: 10.4269/ajtmh.2011.10-0474. 10.4269 %2Fajtmh.2011.10-0474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calisher C.H., Higgs S. The Discovery of Arthropod-Specific Viruses in Hematophagous Arthropods: An Open Door to Understanding the Mechanisms of Arbovirus and Arthropod Evolution? Annu. Rev. Entomol. 2018;7(Jan (63)):87–103. doi: 10.1146/annurev-ento-020117-043033. [DOI] [PubMed] [Google Scholar]
- Colmant A.M.G., Hobson-Peters J., Bielefeldt-Ohmann H. A new clade of insect-specific flaviviruses from australian Anopheles mosquitoes displays species-specific host restriction. mSphere. 2017;2(4) doi: 10.1128/mSphere.00262-17. Published 2017 Jul 12. 10.1128/mSphere.00262-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook S., Bennett S.N., Holmes E.C., De Chesse R., Moureau G., de Lamballerie X. Isolation of a new strain of the flavivirus cell fusing agent virus in a natural mosquito population from Puerto Rico. J. Gen. Virol. 2006;87:735–748. doi: 10.1099/vir.0.81475-0. [DOI] [PubMed] [Google Scholar]
- Diallo D., Sall A.A., Buenemann M. Landscape ecology of sylvatic chikungunya virus and mosquito vectors in southeastern Senegal. PLoS Negl. Trop. Dis. 2012;6:e1649. doi: 10.1371/journal.pntd.0001649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diallo D., Sall A.A., Diagne C.T. Patterns of a sylvatic yellow fever virus amplification in southeastern Senegal, 2010. Am. J. Trop. Med. Hyg. 2014;90(6):1003–1013. doi: 10.4269/ajtmh.13-0404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Digoutte J.P., Calvo-Wilson M.A., Mondo M., Traore-Lamizana M., Adam F. Continuous cell lines and immune ascitic fluid pools in arbovirus detection. Res. Virol. 1992;143(Nov-Dec 96):417–422. doi: 10.1016/s0923-2516(06)80135-4. [DOI] [PubMed] [Google Scholar]
- Fujita R., Kato F., Kobayashi D., Murota K. Persistent viruses in mosquito cultured cell line suppress multiplication of flaviviruses. Heliyon. 2018;4(Aug (8)) doi: 10.1016/j.heliyon.2018.e00736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman H., Contreras-Gutierrez M.A., Travassos da Rosa A.P.A. Characterization of three new insect-specific flaviviruses: their relationship to the mosquito-borne flavivirus pathogens. Am. J. Trop. Med. Hyg. 2018;98(Feb (2)):410–419. doi: 10.4269/ajtmh.17-0350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddow A.D., Guzman H., Popov V.L. First isolation of Aedes flavivirus in the Western Hemisphere and evidence of vertical transmission in the mosquito Aedes (stegomyia) albopictus (Diptera: culicidae) Virology. 2013;440:134–139. doi: 10.1016/J.virol.2012.12.008. [DOI] [PubMed] [Google Scholar]
- Hang J., Klein T.A., Kim H.C. Genome sequences of five arboviruses in field-captured mosquitoes in a unique rural environment of South Korea. Genome Announc. 2016;28(4(1)) doi: 10.1128/genomeA.01644-15. pii: e01644-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison J.J., Warrilow D., McLean B.J. A new orbivirus isolated from mosquitoes in North-Western Australia shows antigenic and genetic similarity to corriparta virus but does not replicate in vertebrate cells. Viruses. 2016;20(May 8(5)) doi: 10.3390/v8050141. pii: E141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino K., Isawa H., Tsuda Y. Genetic characterization of a new insect flavivirus isolated from Culex pipiens mosquito in Japan. Virology. 2007;359:405–414. doi: 10.1016/j.virol.2006.09.039. [DOI] [PubMed] [Google Scholar]
- Huhtamo E., Putkuri N., Kurkela S. Characterization of a novel flavivirus from mosquitoes in northern europe that is related to mosquito-borne flaviviruses of the tropics. J. Virol. 2009;83:9532–9540. doi: 10.1128/JVI.00529-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junglen S., Kurth A., Kuehl H. Examining landscape factors influencing relative distribution of mosquito genera and frequency of virus infection. Ecohealth. 2009;6:239–249. doi: 10.1007/s10393-009-0260-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junglen S., Korries M., Grasse W. Host range restriction of insect-specific flaviviruses occurs at several levels of the viral life cycle. Randall G, ed. mSphere. 2017;2(1) doi: 10.1128/mSphere.00375-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33(Jul (7)):1870–1874. doi: 10.1093/molbev/msw054. Epub 2016 Mar 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwata R., Satho T., Isawa H. Characterization of Dak Nong virus, an insect nidovirus isolated from Culex mosquitoes in Vietnam. Arch. Virol. 2013;158:2273–2284. doi: 10.1007/s00705-013-1741-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauber C., Ziebuhr J., Junglen S. Mesoniviridae: a proposed new family in the order Nidovirales formed by a single species of mosquito-borne viruses. Arch. Virol. 2012;157(8):1623–1628. doi: 10.1007/s00705-012-1295-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marklewitz M., Zirkel F., Rwego I.B. Discovery of a unique novel clade of mosquito-associated bunyaviruses. J. Virol. 2013;87(Dec (23)):12850–12865. doi: 10.1128/JVI.01862-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nga P.T., Parquet M.D.C., Lauber C. Discovery of the first insect nidovirus, a missing evolutionary link in the emergence of the largest RNA virus genomes. PLoS Pathog. 2011;7 doi: 10.1371/journal.ppat.1002215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasar F., Gorchakov R.V., Tesh R.B., Weaver S.C. Eilat virus host range restriction is present at multiple levels of the virus life cycle. J. Virol. 2015;89:1404–1418. doi: 10.1128/JVI.01856-14. PMCID: PMC4300653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Öhlund P., Lundén H., Blomström A.L. Insect-specific virus evolution and potential effects on vector competence. Virus Genes. 2019;55(2):127–137. doi: 10.1007/s11262-018-01629-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumu F.O., Kotas M.E., Kihonda J. Comparative evaluation of methods used for sampling malaria vectors in the Kilombero Valley, South Eastern Tanzania. Open Trop. Med. J. 2008;1:51–55. doi: 10.2174/1874315300801010051. [DOI] [Google Scholar]
- Parry R., Asgari S. Aedes anphevirus (AeAV): an insect-specific virus distributed worldwide in Aedes aegypti mosquitoes that has complex interplays with Wolbachia and dengue virus infection in cells. J. Virol. 2018;92(17) doi: 10.1128/JVI.00224-18. pii: e00224-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel G.H., Adelman Z.N., Myles K.M. Temperature-dependent effects on the replication and transmission of arthropod-borne viruses in their insect hosts. Curr. Opin. Insect Sci. 2016;16(Aug):108–113. doi: 10.1016/j.cois.2016.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang R.C., Gichogo A., Gachoya J. Isolation of a new flavivirus related to cell fusing agent virus (CFAV) from field-collected flood-water Aedes mosquitoes sampled from a dambo in central Kenya. Arch. Virol. 2003;148:1085–1093. doi: 10.1007/s00705-003-0018-8. [DOI] [PubMed] [Google Scholar]
- Untergasser A., Cutcutache I., Koressaar T. Primer3-new capabilities and interfaces. Nucleic Acids Res. 2012;40(15):e115. doi: 10.1093/nar/gks596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasilakis N., Guzman H., Firth C. Mesoniviruses are mosquito-specific viruses with extensive geographic distribution and host range. Virol. J. 2014;11 doi: 10.1186/1743-422X-11-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warrilow D., Watterson D., Hall R.A., Davis S.S., Weir R., Kurucz N. A new species of mesonivirus from the Northern Territory, Australia. PLoS One. 2014;9(3) doi: 10.1371/journal.pone.0091103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G., Asad S., Khromykh A.A., Asgari S. Cell fusing agent virus and dengue virus mutually interact in Aedes aegypti cell lines. Sci. Rep. 2017;7:6935. doi: 10.1038/s41598-017-07279-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zirkel F., Kurth A., Quan P.L., Briese T., Ellerbrok H., Pauli G., Leendertz F.H., Lipkin W.I., Ziebuhr J., Drosten C., Junglen S. An insect nidovirus emerging from a primary tropical rainforest. mBio. 2011;2:e00077–00011. doi: 10.1128/mBio.00077-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zirkel F., Roth H., Kurth A. Identification and characterization of genetically divergent members of the newly established family Mesoniviridae. J. Virol. 2013;87(11):6346–6358. doi: 10.1128/JVI.00416-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.