Abstract
Pancreatic ductal adenocarcinoma (PDAC) is associated with metaplastic changes in the pancreas but the transcriptional program underlying these changes is incompletely understood. The zinc finger transcription factor, PRDM3, is lowly expressed in normal pancreatic acini and its expression increases during tumorigenesis. Although PRDM3 promotes proliferation and migration of PDAC cell lines, the role of PRDM3 during tumor initiation from pancreatic acinar cells in vivo is unclear. In this study, we showed that high levels of PRDM3 expression in human pancreas was associated with pancreatitis, and well-differentiated but not poorly differentiated carcinoma. We examined PRDM3 function in pancreatic acinar cells during tumor formation and pancreatitis by inactivating Prdm3 using a conditional allele (Ptf1aCreER;Prdm3flox/flox mice) in the context of oncogenic Kras expression and supraphysiological cerulein injections, respectively. In Prdm3-deficient mice, KrasG12D-driven preneoplastic lesions were more abundant and progressed to high-grade precancerous lesions more rapidly. This is consistent with our observations that low levels of PRDM3 in human PDAC was correlated significantly with poorer survival in patient. Moreover, loss of Prdm3 in acinar cells elevated exocrine injury, enhanced immune cell activation and infiltration, and greatly increased acinar-to-ductal cell reprogramming upon cerulein-induced pancreatitis. Whole transcriptome analyses of Prdm3 knockout acini revealed that pathways involved in inflammatory response and Hif-1 signaling were significantly upregulated in Prdm3-depleted acinar cells. Taken together, our results suggest that Prdm3 favors the maintenance of acinar cell homeostasis through modulation of their response to inflammation and oncogenic Kras activation, and thus plays a previously unexpected suppressive role during PDAC initiation.
Subject terms: Pancreatic cancer, Pancreatitis
Introduction
Pancreatic ductal adenocarcinoma (PDAC) is the most common malignancy in pancreas and the third leading cause of cancer-related deaths in US1. Emerging evidence suggests that pancreatic acinar cells can acquire ductal cell-like characteristics and downregulate genes maintaining acinar cell identify, also known as acinar-to-ductal metaplasia (ADM). This process is reversible because once the injury is resolved the acinar cell-derived ductal-like cells can revert back to acinar cells2,3. However, in the presence of additional stresses, such as a KrasG12D mutation, ADM cannot be reversed and cells are “locked” into a transdifferentiated state before converting to precancerous pancreatic intraepithelial neoplasia (PanIN) lesions and subsequently invasive PDAC4–6. In mice, pancreatic tumorigenesis is dramatically hastened by the presence of pancreatitis7,8, while, in humans, induction of chronic inflammation is a common character in many known risk factors for pancreatic cancer including diabetes, pancreatitis, alcohol consumption and tobacco use9. However, the complete transcriptional program that regulates the interconversion of acinar cells to ductal-like cells and vice versa, and the role of these events in the context of tumorigenesis are still unclear.
PRDM3 is a nuclear transcription factor involved in many biological processes including hematopoiesis, development, cell differentiation and apoptosis10. PRDM3 belongs to the positive regulatory domain (PRDM) family proteins, which are characterized by an N-terminal PR (PRDI-BF1-RIZ1 homologous) domain followed by an array of C2H2 zinc finger motifs for sequence-specific DNA binding and a C-terminal binding protein (CtBP)-binding domain for protein-protein interactions11. PRDM3 is necessary for the maintenance of hematopoietic stem cells12,13. A recent study has reported that PRDM3 is weakly expressed in normal pancreatic acinar cells and upregulated in many PDAC precursor lesions and PDAC14. Using siRNA-mediated knockdown of PRDM3 in PK-8 pancreatic cancer cells, Tanaka and colleagues also showed that PRDM3 promotes pancreatic cancer cell proliferation and migration through the inhibition of a KRAS suppressor miR-9614. Despite this characterization of the effects of PRDM3 inhibition in pancreatic tumor cells ex vivo, the role of PRDM3 during tumor initiation from acinar cells in vivo is unclear.
Here, we used a CreER-inducible mouse model to genetically delete Prdm3 specifically in adult acinar cells to examine the functional role of PRDM3 in pancreatic carcinogenesis. Our results show that Prdm3 deficiency potentiates inflammation, promotes tumor initiation and dramatically accelerates malignant progression, which is consistent with our findings indicating that PRDM3 loss is significantly associated with poorer survival in patients with PDAC. We further demonstrate that PRDM3 is important to suppress the expression of genes involved in inflammatory response in pancreatic acinar cells. These findings suggest an inhibitory role of PRDM3 in pancreatic tumorigenesis. Future development of drugs that target PRDM3 might yield novel approaches to benefit the treatment of PDAC.
Results
PRDM3 is upregulated in pancreatitis, as well as well-differentiated PDAC and its high expression is associated with better survival in patients with PDAC
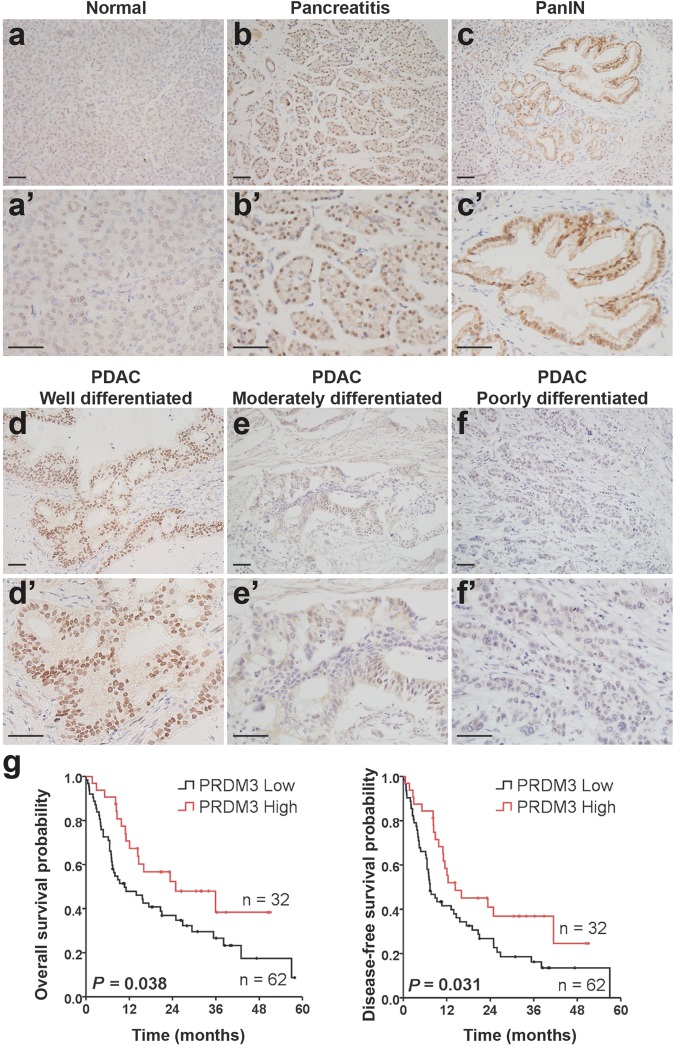
We first characterized the expression of PRDM3 and its relevance to pancreatic cancer prognosis by analyzing a cohort of 94 patients who were diagnosed with PDAC and received surgical resection without preoperative chemotherapy. We found that PRDM3 was strongly expressed in precancerous PanIN lesions (Fig. 1c–c’) and well-differentiated PDAC from patients (Fig. 1d–d’), while moderately to poorly differentiated cancer cells showed little to no staining of PRDM3 (Fig. 1e–e’, f–f’, Table 1). Our observation of heterogeneous expression of PRDM3 in PDAC was supported by a recent study demonstrating that PRDM3 is selectively expressed in low-grade PDAC cells featuring differentiated epithelia, but not high-grade cells showing fibroblastoid morphology15. We performed subsequent overall survival and disease-free survival analyses with these 94 PDAC patients and found that patients with high levels of PRDM3 lived significantly longer than those with low levels of PRDM3 (Overall survival: 16.0 months vs. 9.3 months; Disease-free survival: 12.4 months vs. 7.4 months) (Fig. 1g). Our clinical relevance analysis clearly revealed that a better survival in patient with PDAC was associated with high levels of PRDM3 expression, but not with age, gender, tumor size, location, TNM (tumor-node-metastasis), or CA19-9 (Table 1). Given that pancreatitis is a well-described risk factor for PDAC development, we also analyzed pancreatic tissue from 22 patients with chronic pancreatitis. We found a dramatic increase of PRDM3 protein levels in inflamed tissues compared with normal pancreas (Fig. 1a–a’, b–b’; Supplementary Table 1). Similarly, administration of supraphysiologic concentrations of a cholecystokinin ortholog, cerulein, in mice resulted in acute pancreatitis and Prdm3 upregulation in murine acinar cells (Supplementary Fig. 1a). Consistent with findings from previous reports14, we also observed strong expression of Prdm3 in the precursor lesions of PDAC including ADM, low-grade PanINs, and high-grade PanINs found in pancreata from mice expressing oncogenic Kras in pancreatic acinar cells (Ptf1aCreER; KrasG12D) (Supplementary Fig. 1b). Together, our results demonstrated that elevated levels of PRDM3 are associated with inflamed pancreatic epithelia and well-differentiated pancreatic lesions, while low levels of PRDM3 are associated with poorly differentiated carcinoma and a worse prognostic outcome in patients with PDAC.
Fig. 1. Weak PRDM3 expression is associated with poorly differentiated tumors and a worse prognostic outcome in patients with PDAC.
Prdm3 immunostaining in normal pancreatic tissue (a and a’), pancreatitis (b and b’), PanIN (c and c’), well-differentiated (d and d’), moderately differentiated (e and e’), and poorly differentiated (f and f’) PDAC. g The overall survival probability and disease-free survival probability were compared between low (n = 32, staining index ≤ 6) and high (n = 62, staining index > 6) levels of PRDM3 expression in a cohort of 94 PDAC patients after surgical resection. p-values were calculated based on log-rank test. Scale: 50 μm.
Table 1.
Association between expression levels of PRDM3 and clinical relevance in patients with PDAC (n = 94).
| Characteristics | Low expression (n = 62) | High expression (n = 32) | p-valuea |
|---|---|---|---|
| Histological grade | <0.001 | ||
| Poorly differentiated | 19 | 1 | |
| Moderately differentiated | 43 | 14 | |
| Well differentiated | 0 | 17 | |
| Survivalb | 0.030 | ||
| Alive | 17 | 16 | |
| Death | 45 | 16 | |
| Age (years) | 0.057 | ||
| >60 | 24 | 19 | |
| ≤60 | 38 | 13 | |
| Gender | 0.174 | ||
| Female | 22 | 16 | |
| Male | 40 | 16 | |
| Tumor size (cm) | 0.300 | ||
| ≤2.0 | 10 | 8 | |
| >2.0 | 52 | 24 | |
| Location | 0.333 | ||
| Head | 50 | 23 | |
| Body and tail | 12 | 9 | |
| TNMc | 0.717 | ||
| I, II | 54 | 27 | |
| III, IV | 8 | 5 | |
| CA19-9 | 0.526 | ||
| ≤35 | 12 | 8 | |
| >35 | 50 | 24 |
ap-values were based on t-test (two-sided). p < 0.05 was considered as statistically significant.
bAll PDAC patients received surgical resection without preoperative chemotherapy; median follow-up was 12.1 months after surgical resection; median survival time for patients deceased was 8.0 months; median follow-up for patients alive was 26.4 months.
cThe tumor-node-metastasis (TNM) stages were determined according to the 7th edition TNM classification of the American Joint Committee on Cancer.
Bold values indicate a statistically significant difference with a p-value less than 0.05.
Ablation of Prdm3 enhances KrasG12D-stimulated PDAC initiation and progression
To determine whether Prdm3 is functionally important for pancreatic carcinogenesis in vivo, we applied a genetic strategy to induce expression of oncogenic KrasG12D and deletion of Prdm3 in adult acinar cells, simultaneously. Cre-mediated recombination was induced in pancreatic acinar cells using the tamoxifen-inducible Ptf1aCreER allele16. The Mds1 and Evi1 complex locus (Mecom) encodes a full-length isoform of Prdm3, and a shorter isoform lacking the N-terminus PR domain. We therefore used a Prdm3flox mouse which harbors two LoxP sites flanking exon 4, the first shared exon in the long- and short-isoform of Prdm312, to completely eliminate Prdm3 in pancreatic acinar cells upon tamoxifen induced recombination. By combining the KrasLSL-G12D allele17 with the Ptf1aCreER allele with/without the Prdm3flox allele, we generated control Ptf1aCreER; KrasG12D (KrasG12D) mice, as well as Ptf1aCreER; KrasG12D; Prdm3flox/flox (KrasG12D-Prdm3ΔAcinar) mice (Supplementary Fig. 2a).
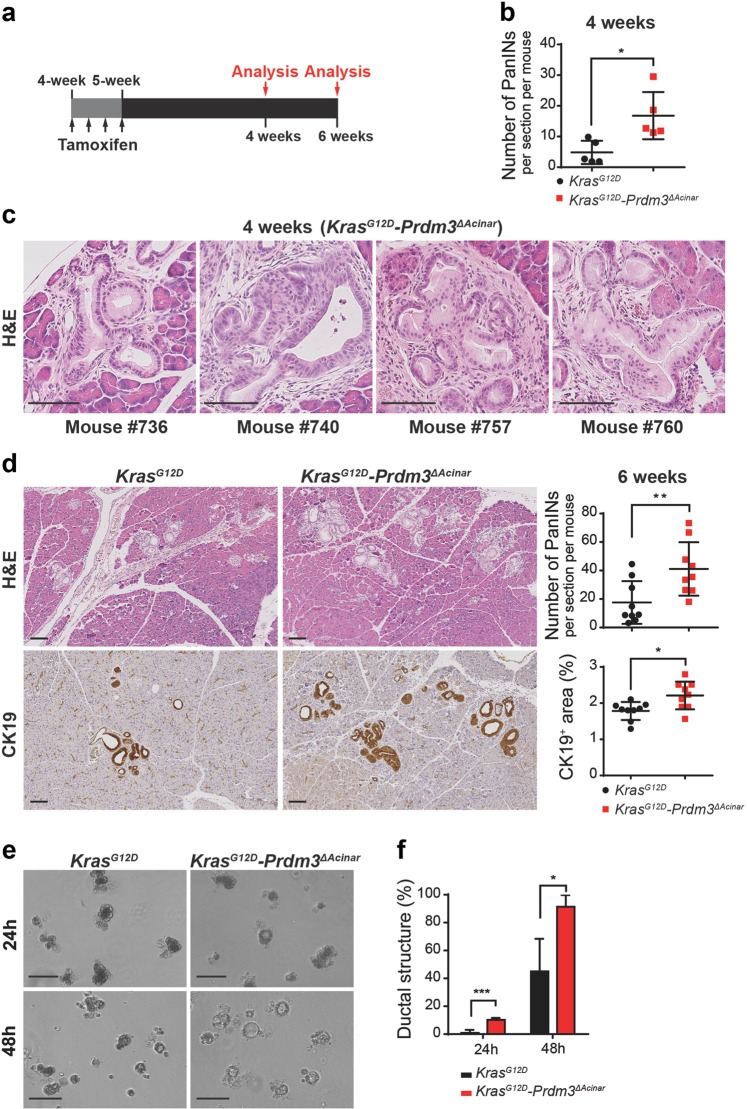
To initiate recombination, we injected mice with tamoxifen at 4 to 5 weeks of age and analyzed pancreata at 4 weeks and 6 weeks post-injection (Fig. 2a). Comparison of Prdm3 expression in KrasG12D-Prdm3ΔAcinar and KrasG12D mice after tamoxifen administration showed an almost complete loss of Prdm3 protein in KrasG12D-Prdm3ΔAcinar acinar cells (Supplementary Fig. 2b, top panel). Four weeks after tamoxifen-mediated recombination, small areas of ADM and occasional low-grade PanINs were observed in the control KrasG12D mice. In contrast, KrasG12D-Prdm3ΔAcinar mice, with loss of Prdm3 in pancreatic acinar cells, exhibited more cuboidal to columnar duct-like structures with enlarged lumens (Supplementary Fig. 2b, bottom panel). Quantification of the number of PanINs revealed that preneoplastic lesions arising in KrasG12D-Prdm3ΔAcinar mice increased significantly compared to KrasG12D mice (Fig. 2b). More intriguingly, we observed 4 out of 5 KrasG12D-Prdm3ΔAcinar mice developed high-grade PanINs at 4 weeks post-tamoxifen injection (Fig. 2c), while no high-grade lesions were found in KrasG12D mice even at 3 months of age (data not shown). Consistent with these findings, at 6 weeks post-tamoxifen injection KrasG12D-Prdm3ΔAcinar mice exhibited higher number of lesions with histological and molecular characteristics of PanINs indicated by the expression of Cytokeratin 19 (CK19) (Fig. 2d) and the present of acidic mucin content indicated by Mucin 5AC (Muc5AC) and Alcian blue staining (Supplementary Fig. 2c). Altogether, these data suggest that deletion of Prdm3 promotes PanIN formation in the presence of oncogenic Kras expression.
Fig. 2. Loss of prdm3 promotes acinar-to-ductal metaplasia and PanIN lesions formation.
a Ptf1aCreER;KrasG12D and Ptf1aCreER;KrasG12D;Prdm3flox/flox mice at 4–5 weeks of age were injected 4 times on alternating days with tamoxifen. Recombined mice were analyzed at 4 and 6 weeks post tamoxifen injection. b The number of PanINs per section for KrasG12D (n = 5) vs. KrasG12D-Prdm3ΔAcinar (n = 5) mice at 4 weeks post tamoxifen injection. c Representative images of high-grade PanINs in KrasG12D-Prdm3ΔAcinar mice at 4 weeks after tamoxifen injection. d Hematoxylin-eosin staining (H&E) and immunohistochemistry staining for the ductal marker Cytokeratin 19 (CK19). Quantification of the number of PanINs, as well as the percent of pancreatic area that is CK19+ in KrasG12D (n = 9) vs. KrasG12D-Prdm3ΔAcinar (n = 9) mice 6 weeks post-tamoxifen injection. e Images of acinar cell explants embedded in Matrigel at 24 and 48 h. f Quantification of the percent of ductal-like structures in explants derived from KrasG12D (n = 3) and KrasG12D-Prdm3ΔAcinar (n = 3) mice at 4 weeks post-tamoxifen injection. Data show mean ± SD. Statistical analysis: Two-tailed t-test. *p < 0.05, **p < 0.01, ***p < 0.001. Scale: 100 μm.
To further examine whether loss of Prdm3 promotes acinar to ductal transformation in the presence of mutant Kras, we isolated primary acinar cell clusters from 8-week-old KrasG12D-Prdm3ΔAcinar and KrasG12D mice, respectively, and performed 3D Matrigel explant culture. In agreement with the histologic observations, Prdm3 deficiency led to a significant increase in earlier ADM evens within 48 h in culture, as quantified by counting the number of duct-like structures vs. the total number of cell clusters (Fig. 2e, f). Previous studies have demonstrated that Tgf-α is upregulated in KrasG12D pancreata18 and acts as a potent inducer for the transdifferentiation of acinar to ductal cells19,20. Therefore, we also tested if Prdm3 depletion impacts TGF-α-induced in vitro ADM. Acinar cell explants derived from Prdm3ΔAcinar mice transformed into duct-like structures within 30 h in culture with addition of TGF-α (Supplementary Fig. 2d, e). This acinar to ductal cyst conversion was almost undetected in explants derived from control (Ptf1aCreER) mice until 48 h in culture. Collectively, our data support that loss of Prdm3 accelerates ductal metaplasia.
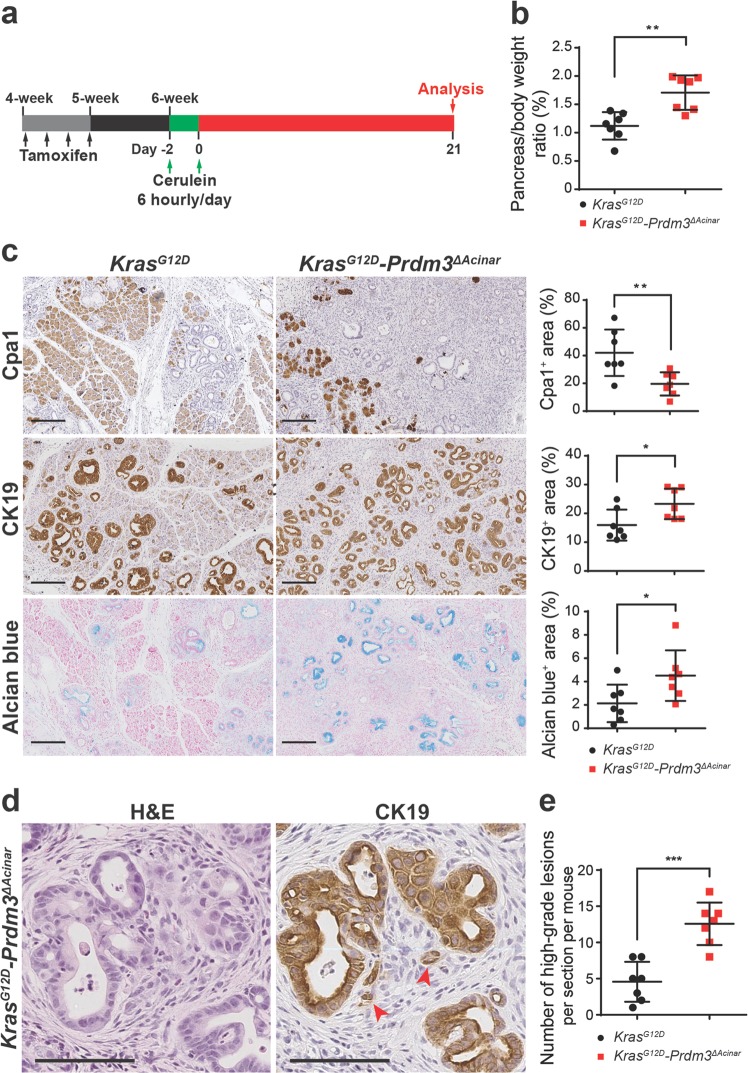
We next examined whether loss of Prdm3 promotes neoplastic progression. To accelerate the formation of invasive lesions, we induced cerulein-mediated acute pancreatitis in cooperation with acinar-cell-specific activation of oncogenic Kras as previously described4. One week after tamoxifen administration, KrasG12D-Prdm3ΔAcinar and KrasG12D mice were injected hourly with 50 μg/kg cerulein over 6 h on alternating days. The pancreata were harvested at 21 days post-cerulein injection (Fig. 3a). Pancreata from KrasG12D-Prdm3ΔAcinar mice had full spectrum of precursor lesions including low-grade PanINs, high-grade PanINs and ductal carcinoma in situ (Supplementary Fig. 3a), which had lost Prdm3 staining (Supplementary Fig. 3b). The control KrasG12D mice had less evidence of tumorigenesis compared with KrasG12D-Prdm3ΔAcinar mice. Specifically, in KrasG12D-Prdm3ΔAcinar mice, the ratio of pancreas-to-body weight increased (Fig. 3b); the number of Cpa1+acinar cells decreased and CK19+ duct-like cells increased dramatically (Fig. 3c); a higher percentage of the pancreas was replaced by acidic mucin content indicated by Alcian blue staining (Fig. 3c); and the number of high-grade PanIN significantly elevated (Fig. 3e). Moreover, we consistently observed tumor budding associated with many high-grade neoplastic lesions in KrasG12D-Prdm3ΔAcinar mice at 21 days post-cerulein injection (Fig. 3d). Tumor budding is a strong prognostic indicator of aggressive tumor behavior, which is defined as the presence of single cells or clusters of less than five tumor cells scattered in the stroma21. In contrast to KrasG12D-Prdm3ΔAcinar mice, tumor budding was rarely observed in KrasG12D mice, suggesting that loss of Prdm3 accelerates pancreatic cancer formation in KrasG12D-expressing mice. Together, our data suggest that progression of low-grade precursor lesions to high-grade PanIN is more rapid in the absence of Prdm3.
Fig. 3. Inhibition of Prdm3 accelerates KrasG12D-driven neoplastic transformation in response to pancreatitis.
a Schematic illustration showing experimental design of cerulein-induced acute pancreatitis in cooperation with activation of oncogenic Kras in Ptf1a-expressing cells. Ptf1aCreER; KrasG12D (n = 7) and Ptf1aCreER; KrasG12D; Prdm3flox/flox (n = 7) mice at 4 to 5 weeks of age were injected 4 times on alternating days with tamoxifen. One week after the last tamoxifen injection, mice were subjected to cerulein (50 μg/kg) injection at hourly intervals over 6 h on alternating days separated by 24 h and analyzed at 21 days post cerulein injection. b Quantification of relative pancreas mass measured as percent of pancreas weight over body weight in KrasG12D vs. KrasG12D-Prdm3ΔAcinar mice. c Hematoxylin-eosin staining (H&E) and immunohistochemistry for Cpa1, Cytokeratin 19 (CK19) and Alcian blue of pancreata from KrasG12D and KrasG12D-Prdm3ΔAcinar mice. Quantification of the percent of pancreatic area that is Cpa1+, CK19+ or Alcian blue+ in KrasG12D vs. KrasG12D-Prdm3ΔAcinar mice. d Representative images of tumor budding in KrasG12D-Prdm3ΔAcinar mice indicated by red arrowheads. Immunohistochemistry staining of CK19 strongly suggests invasive high-grade neoplasia. e Number of high-grade PanINs per section for each genotype. Data show mean ± SD. Statistical analysis: Two-tailed t-test. *p < 0.05, **p < 0.01, ***p < 0.001. Scale: 200 μm (c) and 100 μm (d).
Loss of Prdm3 in acinar cells enhances pancreatitis
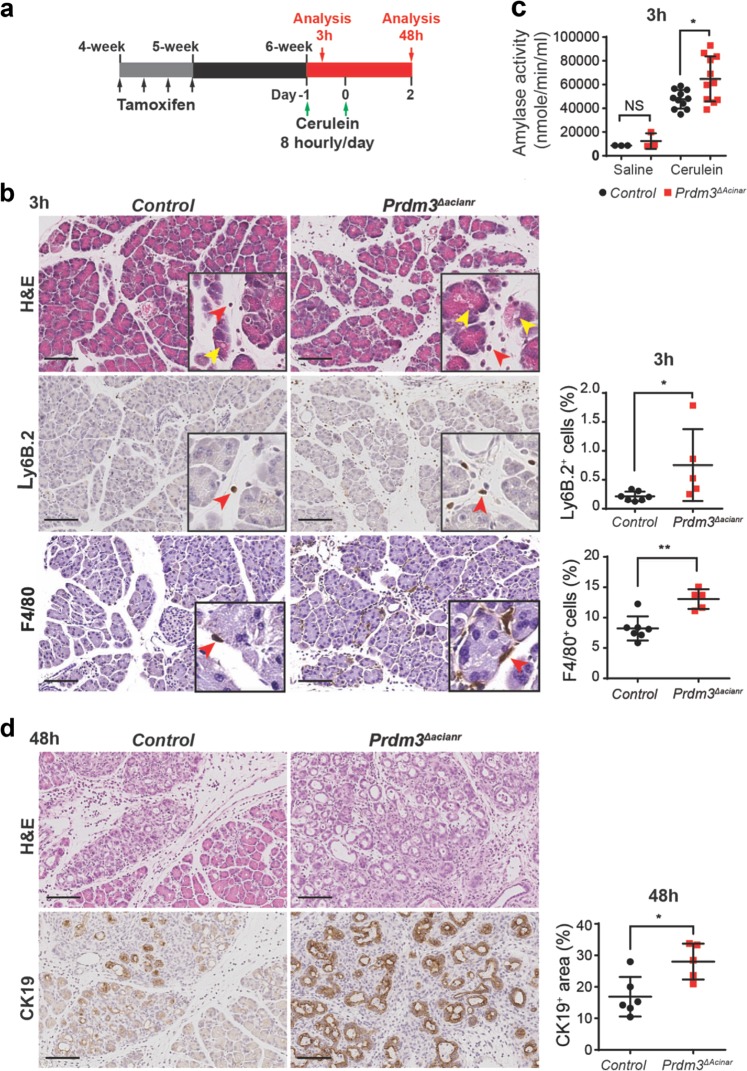
Given that inflammation promotes cancer formation and Prdm3 was significantly increased in humans with pancreatitis, we further determined whether Prdm3 modulated the inflammatory response of the pancreas in Ptf1aCreER;Prdm3flox/flox mice. Mice injected with tamoxifen, but lacking the Prdm3flox allele, were used as controls (Ptf1aCreER mice). Seven days after tamoxifen administration Prdm3 protein was absent in more than 80% of acinar cells in Prdm3ΔAcinar mice, indicating efficient and specific deletion of Prdm3 (Supplementary Fig. 4a). Acute pancreatitis was induced with intraperitoneal injection of cerulein, as described previously22 (Fig. 4a). Specifically, 7 days after the last tamoxifen injection, Prdm3ΔAcinar and control (Ptf1aCreER) mice were injected with 50 μg/kg cerulein at hourly intervals for 8 h. Histological assessment of pancreata from mice 3 h after the last cerulein injection demonstrated exaggerated interstitial edema, cytoplasmic vacuolization and immune cell infiltration in Prdm3ΔAcinar mice compared with control mice (Fig. 4b). Consistently, examination of immune cell infiltration demonstrated a substantial increase in the number of F4/80+ macrophages, as well as Ly6B.2+ neutrophils in cerulein-treated Prdm3ΔAcinar pancreata (Fig. 4b). Elevated blood amylase levels are an indicator of pancreatitis. Consistent with the increased inflammatory infiltrates in the pancreas, we observed significantly higher levels of serum amylase in Prdm3ΔAcinar mice compared to controls (Fig. 4c). To determine whether the expression of inflammatory cytokines is increased, we harvested RNA from pancreata of Prdm3ΔAcinar and control mice to perform quantitative qRT-PCR. As expected, expression of inflammatory cytokines, including Il-6, Cxcl-1, Cxcl-10, Ccl2, and Ccl20, increased significantly in Prdm3-deleted pancreata compared to control pancreata (Supplementary Fig. 4b). We further examined pancreata harvested from Prdm3ΔAcinar and control mice 48 h after two series of 8-hourly injection of cerulein (Fig. 4a). Characterization by immunohistochemistry for the ductal marker CK19 confirmed that acini undergoing acinar-to-ductal metaplasia increased dramatically in Prdm3ΔAcinar mice (Fig. 4d). These results illustrate that acinar-cell-specific ablation of Prdm3 augments the severity of pancreatitis, suggesting a specific role of Prdm3 as a modulator of inflammatory response in the pancreas.
Fig. 4. Prdm3 deletion in pancreatic acinar cells exaggerates cerulein-induced pancreatitis.
a Schematic illustration of experimental design. Ptf1aCreER;Prdm3flox/flox mice at 4 to 5 weeks of age were injected 4 times on alternating days with tamoxifen. One week after the last tamoxifen injection, mice were subjected to cerulein (50 μg/kg) injection at hourly intervals for 8 h per day for two consecutive days and analyzed at the indicated time points. b Histologic characterization was determined with hematoxylin-eosin staining (H&E). Immunohistochemistry staining for neutrophil marker Ly6B.2 and macrophage marker F4/80 with quantitation of the percent of all cells that are Ly6B.2+ or F4/80+ in Ptf1aCreER (control, n = 7) and Prdm3ΔAcinar (n = 5) pancreata. Vacuole is indicated by yellow arrowheads, and infiltrated immune cells are indicated by red arrowheads. c Serum amylase levels of cerulein-treated control (n = 11) vs. Prdm3ΔAcinar (n = 11) mice and saline-treated control (n = 3) vs. Prdm3ΔAcinar (n = 3). d Representative images of control and Prdm3ΔAcinar pancreata stained with H&E and the ductal marker Cytokeratin 19 (CK19). Quantification of the respective percent of all cells that are CK19+ from control (n = 6) vs. Prdm3ΔAcinar (n = 5) pancreata. Data show mean ± SD. Statistical analysis: Two-tailed t-test. *p < 0.05. Scale: 100 μm.
Loss of Prdm3 activates inflammatory response and Hif-1 signaling pathways
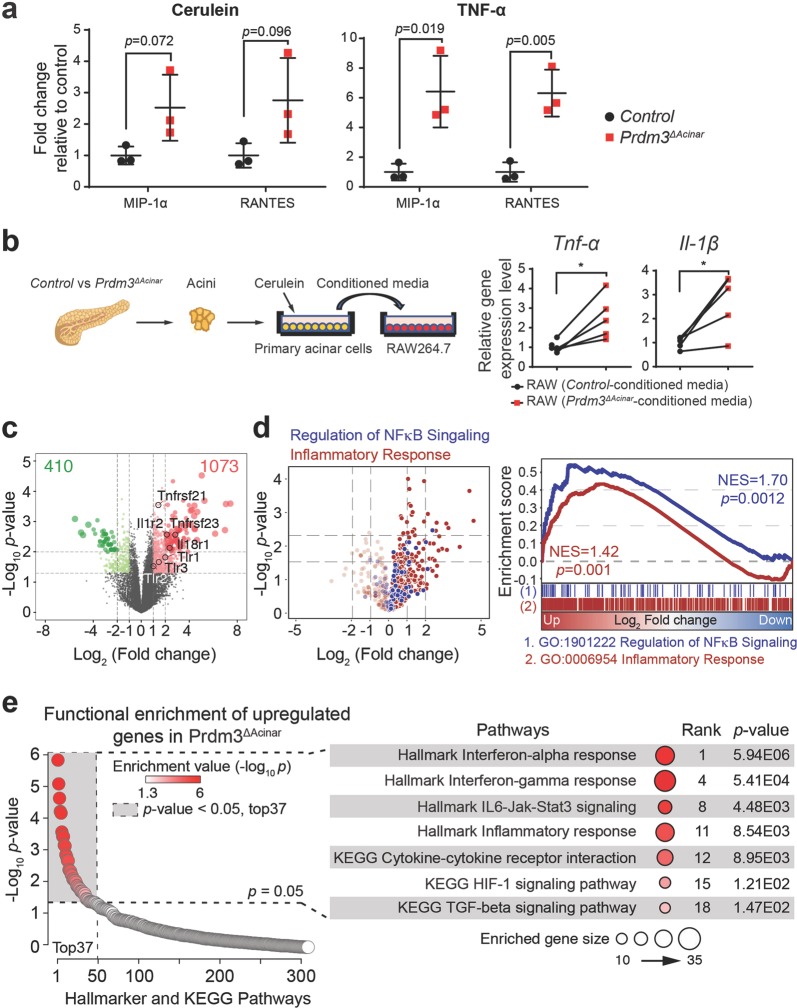
Injured acinar cells initiate inflammatory responses by releasing digestive enzymes and proinflammatory mediators such as a variety of cytokines. These cytokines in turn attract and activate macrophages to produce excessive inflammatory cytokines, like tumor necrosis factor-alpha (TNF-α) and interleukin-1 beta (IL-1β), to exacerbate tissue injury23,24. To investigate whether loss of Prdm3 alters cytokines secretion by pancreatic acinar cells, we performed a cytokine array assay on primary acinar cells in response to stimuli. Briefly, acinar cell clusters were isolated from a cohort of Prdm3ΔAcinar and control (Ptf1aCreER) mice, respectively, and subsequently cultured in the presence of cerulein or TNF-α for 6 h. We found that among 40 cytokines detected in a mouse inflammatory array, macrophage inflammatory protein (MIP)-1α and regulated upon activation normal T expressed and secreted (RANTES) were remarkably increased in Prdm3ΔAcinar incubation medium (Fig. 5a). As MIP-1α and RANTES are potent chemotactic agents for monocytes, we further examined macrophage activation in response to conditioned media from cerulein-stimulated cell cultures. The murine macrophages, Raw246.7 cells, were incubated in conditioned media collected from Prdm3ΔAcinar or control cell culture exposed to cerulein for 6 h (Fig. 5b). After 16-hour incubation, the activation of macrophages was determined by the expression of inflammatory cytokines. We found that the relative expression of inflammatory cytokines Tnf-α and Il-1β was significantly higher in Raw246.7 incubated with Prdm3ΔAcinar conditioned media (Fig. 5b), supporting that more proinflammatory factors were released from Prdm3-deficient cells to stimulate macrophage infiltration (Fig. 4b) and subsequent activation. These results indicate that Prdm3ΔAcinar mice are more susceptible to cerulein-induced injury, at least, in part by contributing to increased macrophage activation.
Fig. 5. Inactivation of Prdm3 enhances inflammatory response pathways.
a Cytokine secretion was determined using a Raybiotech mouse inflammation array. Primary acinar cells, isolated from control (n = 3) and Prdm3ΔAcinar (n = 3) mice, were exposed to cerulein (10 nM) or TNF-α (100 ng/ml) for 6 h. Levels of cytokines (MIP-1α and RANTES) in culture medium were expressed relative to control. p-values were calculated based on two-tailed t-test. b Primary acinar cells were isolated from Prf1aCreER (n = 5) vs. Prdm3ΔAcinar (n = 5) mice and cultured in Waymouth complete media in the presence of 10 nM cerulein for 6 h. Raw264.7 cells were treated with the above acinar-cell-conditioned media for 16 h and collected for quantitative real-time PCR for the indicated cytokines. Each set of connected dots in b depicts an independent biological replicate (n = 5 experiments). Data show mean ± SD. Statistical analysis: Two-tailed t-test. *p < 0.05. c Volcano plot showing a total of 1483 differentially expressed genes in primary acinar cells isolated from Prdm3ΔAcinar mice, relative to control Ptf1aCreER. Individual genes are labeled and circled in black. d Volcano plot showing differentially expressed genes belonging to inflammatory responses and NF-κB signaling are highlighted in red and blue, respectively. Gene Set Enrichment Analysis (GSEA) of differentially expressed genes (Ptf1aCreER vs. Prdm3ΔAcinar) identified enrichment of immune responses and regulation of NF-κB signaling. Normalized enrichment score (NES) and p-values are shown. e Functional annotation on 1073 upregulated genes in primary acinar cells isolated from Prdm3ΔAcinar mice, relative to control. Significant KEGG and Hallmark terms, p-values and ranks are shown.
To further examine the effect of Prdm3 deletion on acinar cell homeostasis, we performed transcriptome analyses of primary acinar cells isolated from Prdm3ΔAcinar and control mice. Ptf1aCreER; Prdm3flox/flox and control Ptf1aCreER mice were injected with four doses of tamoxifen at 4 to 5 weeks of age to induce recombination prior to acini isolation. One week after the last tamoxifen injection, primary acinar cells were isolated as described previously25 and then allowed to recover in oxygenated medium for 2 h. Total RNA was extracted from these isolated acini and subjected to RNA-seq analysis. The extent of Prdm3 deletion was confirmed in Prdm3ΔAcinar pancreata (Supplementary Fig. 5a). Analysis of RNA-seq data sets was performed by DESeq2. Setting a p-value threshold of 0.05, we identified 1483 genes that were significantly differentially expressed (log2|Fold change| > 1) in Prdm3-deficient acini compared with control (Supplementary Table 2). Moreover, 1073 out of 1483 differentially expressed genes (DEGs) were upregulated, while only 410 of them were downregulated (Fig. 5c). We found that the expression of many adhesion molecules involved in the inflammatory response, such as the TNF receptor superfamily Tnfrsf21 and Tnfrsf23, interleukin receptors Il18r1 and Il1r2, and toll like receptors Tlr1, Tlr2 and Tlr3 were significantly altered in Prdm3-deficient cells. Gene Set Enrichment Analysis (GSEA) further demonstrated that genes involved in inflammatory response and regulation of NF-κB signaling were significantly enriched in Prdm3-deficient acinar cells (Fig. 5d). These findings support that loss of Prdm3 changes cellular homeostasis and biases acinar cells toward a proinflammatory state possibly by decreasing the inflammatory response threshold in Prdm3ΔAcinar cells.
To further identify additional biological pathways that were activated upon Prdm3 deletion, we performed functional annotation on the 1703 upregulated genes with KEGG and Hallmark data sets. Loss of Prdm3 significantly affected over 30 pathways (Supplementary Table 3) including the Hif-1 signaling pathways (Fig. 5e). Intriguingly, recent studies demonstrated that mice with acinar cell-specific deletion of Hif1α were less susceptible to cerulein-induced pancreatitis26. As our transcriptome analysis revealed a significant elevation of Hif1a in Prdm3-deficient cells, we compared the protein level of Hif1a in control and Prdm3ΔAcinar pancreata. We confirmed that the expression level of Hif1a was significantly upregulated in Prdm3-depleted pancreatic tissue by immunoblotting and immunohistochemical staining (Supplementary Fig. 5b, c). Our findings are supported by a previous study in which shRNA knockdown of Prdm3 upregulated Hif1a in DA-1 and NFS-60 leukemic cells27. As loss of Prdm3 exaggerated inflammation, we speculate that dysregulation of Hif1a expression might contribute to the dramatic effects of Prdm3 depletion on pancreatitis.
Discussion
In the present study, we investigated the role of Prdm3 in pancreatitis and pancreatic tumorigenesis using mouse models to indelibly delete Prdm3 in adult acinar cells. We demonstrated that PRDM3 was substantially upregulated in pancreatic acinar cells of patients with pancreatitis, as well as well-differentiated PDAC, but not poorly differentiated PDAC. Interestingly, our clinical relevance analysis suggests a prognostic relevance of low PRDM3 expression in PDAC patients with surgical resection. Consistent herewith, we further demonstrated that loss of Prdm3 not only increased the severity of cerulein-induced pancreatitis, but also accelerated cellular atypia and tumorigenic potential in the pancreas, as Prdm3-deficient mice undergo robust formation of precursor lesions in the presence of oncogenic Kras. These findings implicate a potentially protective mechanism of Prdm3 in pancreatic exocrine cells, which is different from the pro-tumor role of PRDM3 in several aggressive forms of cancer including colon, breast and ovarian cancer28–30. A number of alternatively spliced variants, including the long and short forms of PRDM3, are expressed in pancreatic cancer cells14. Our studies cannot distinguish which forms of PRDM3 are necessary for its anti-tumor effect in acinar cells, therefore, more work will be needed to examine the effects of different PRDM3 isoforms on pancreatitis and pancreatic cancer initiation.
Previous studies demonstrated that Prdm3 promotes proliferation and migration in established pancreatic cancer cell lines14. However, when Prdm3 was knocked out in pancreatic acinar cells, we observed that Prdm3 depletion potentiated pancreatic cancer initiation and progression to high-grade lesions including ductal carcinoma in situ, suggesting that Prdm3 plays a suppressive role in acinar-to-ductal transformation. At a molecular level, we found that over 70% of the differentially expressed genes between normal and Prdm3-deficient acinar cells were upregulated in Prdm3ΔAcinar mice, which is consistent with previous studies demonstrating that Prdm3 acts as a transcriptional repressor through interaction with a variety of co-repressors, such as CtBP, histone methyltransferase SUV39H1 and deacetylase HDAC1/210. We therefore postulate that increased Prdm3 expression in transformed ductal-like cells plays an inhibitory role in pancreatic tumorigenesis through the ability of Prdm3 to suppress a series of signaling cascades important for malignant transformation in exocrine pancreas.
Here, we showed that Prdm3-deficient acinar cells were much more susceptible to cerulein-induced injury. Loss of Prdm3 enhanced ADM formation, as well as macrophage and neutrophil infiltration. Our transcriptome analysis suggests that deletion of Prdm3 in adult pancreatic acini induces significant alteration in the expression profiles of cytokine/chemokine receptors, possibly accounting for greater responses to inflammatory stimuli. We speculate that accumulation of Prdm3 is beneficial to limit local inflammation by increasing the threshold for acinar cells to respond to cellular stress. Expression of oncogenic Kras in acinar cells triggers microinflammation and chemoattraction of macrophages31. Infiltrated macrophages secrete matrix-metalloproteinases and inflammatory cytokines, such as TNF-α, to facilitate acinar-to-ductal transdifferentiation through a NF-κB-mediated signaling cascase32. Moreover, several lines of evidence suggest that inflammation leads to an increase of Ras activity and amplification of oncogenic Ras signaling, which is necessary for pancreatic cancer progression18,33,34. Activation of Kras alone in mice leads to PDAC formation at a low frequency and takes over one year35. In contrast, when pancreatitis was induced in KrasG12D mice, tumorigenesis occurred within a few months8,36 suggesting that precancerous lesions can arise from acinar cells through a process dramatically hastened by inflammation32. In this study, we demonstrated that loss of Prdm3 accelerated KrasG12D-induced PanIN initiation and promoted rapid progression of pre-neoplastic lesions to invasive lesions. Given that activation of inflammatory response elevates constitutive Ras activity, we propose that, loss of Prdm3 upregulates the expression of genes involved in inflammatory response in pancreatic acinar cells, which modulates acinar cell homeostasis to lower the threshold of acinar cells to inflammatory stimuli and promote widespread formation of precancerous lesions from Kras mutated acinar cells.
In addition, we found that hypoxia inducible factor Hif1a was significantly upregulated in Prdm3-deficent acinar cells at both mRNA and protein levels. Our findings are consistent with the previous study in which knockdown of Prdm3 upregulated Hif1a in DA-1 and NFS-60 leukemic cells27. It has also recently been established that HIF-1 signaling plays important roles in both pancreatitis and pancreatic cancer. Hif1a is overexpressed in chronic pancreatitis37 and its high expression is associated with poor prognosis in PDAC38,39. Acinar-cell-specific deletion of Hif1a prevented intrapancreatic coagulation of fibrinogen and protected mice from cerulein-induced acute pancreatitis26, suggesting a functional role of Hif1a in the development of pancreatitis. Given that loss of Prdm3 exaggerated inflammation, we speculate that dysregulation of Hif1a expression might contribute to the dramatic effects of Prdm3 depletion on pancreatitis.
Taken together, our data demonstrated that loss of Prdm3 not only increased the severity of cerulein-induced pancreatitis, but also accelerated cellular atypia and tumorigenic potential in the pancreas, as Prdm3-deficient mice undergo robust formation of precursor lesions in the presence of oncogenic Kras. We uncovered a previously unappreciated role for Prdm3 as a suppressor of both pancreatitis and pancreatic tumorigenesis presumably through regulating inflammatory and Hif-1 signaling pathways in the pancreatic acinar cells.
Materials and methods
Human samples
A total of 94 patients diagnosed with PDAC and 22 patients diagnosed with chronic pancreatitis between 2003–2011 were included in this study in accordance with institutional guidelines and approved by the Clinical Research Ethics Committee of the First Affiliated Hospital at Sun Yat-sen University. Written informed consent was received from participants prior to inclusion in this study.
Mice
All animal experiments were approved by the University of Macau Animal Ethics Committees and carried out in accordance to recommendations stated in the Guide for the Care and Use of Laboratory Animals for the National Institutes of Health (US Department of Health, Education, and Welfare). Prdm3flox/flox, KrasLSL-G12D, and Ptf1aCreER mice have previously been described. Recombination was induced by four subcutaneous injection of tamoxifen every other day at 125 mg/kg body weight on animals at 4 to 5 weeks of age.
Cerulein-induced pancreatitis
To induce acute pancreatitis, experimental mice were fasted overnight before administration of cerulein as described previously4. Cerulein (American Peptide) was dissolved in saline and administrated intraperitoneally at 50 μg/kg body weight hourly for 8 h. Mice were sacrificed after 3 h recovery. Alternatively, mice were injected with cerulein (50 μg/kg body weight) at hourly intervals for 8 h per day for two consecutive days and sacrificed at 48 h after the last injection. To accelerate tumorigenesis, mice were given hourly injections of cerulein (50 μg/kg body weight) for 6 h per day on alternating days separated by 24 h and sacrificed after 21 days.
Histology and immunohistochemical analyses
Paraffin-embedded sections were prepared and subjected to hematoxylin, eosin Y, Alcian blue or immunohistochemical staining. H&E staining and IHC followed our established procedures5, including antigen retrieval with citrate buffer (pH 6.0) prior to staining paraffin sections. For Alcian blue staining, paraffin sections were incubated in 3% acetic acid for 3 min, followed by staining in 1% Alcian blue staining solution for 30 min, and subsequently in Nuclear Fast Red for 5 min. All slides were scanned with a 20× objective using a 2D glass slide digital scanner (Leica Biosystems) and examined at high magnification using the Aperio ImageScope software (Leica Biosystems). The Aperio positive pixel Algorithm was used to quantify area with positive staining and Aperio nuclear V9 algorithm was used to quantify the number of nuclei. The percentages of Cpa1-positive, CK19-positive, Muc5AC-positive, and Alcian blue-positive area were calculated by positive pixels divided by the total pixels in selected tissue areas. The percentages of Ly6B.2-positive, and Prdm3-positive cells were calculated by positive number of nuclei divided by the total number of nuclei in selected tissue areas. Six sections, which displayed maximal pancreatic cross-sectional area, from each animal were used for quantification. The number of PanINs were counted based on the characteristics of every gland with cuboidal to columnar duct-like structures and enlarged lumen in six sections per mouse. For the number of high-grade lesions, including PanINs and ductal carcinoma in situ, five 1600 × 920 μm squares in one section per mouse were analyzed and the squares were randomly distributed in the pancreas. Primary and secondary antibodies used for staining is provided in Supplementary Table 4.
Evaluation of immunostaining of PRDM3 in patient specimens
PRDM3 expression was evaluated according to the staining intensity and proportion of positively stained tumor cells. Staining intensity was graded as 0 (negative), 1 (weakly positive), 2 (moderately positive), and 3 (strong positively). The proportion of positively stained tumor cells was scored as 0 (no positive cells), 1 (<10% positive cells), 2 (10–25% positive cells), 3 (25–50% positive cells), and 4 (>50% positive cells). The immunostaining of PRDM3 was determined by staining index (SI) through multiplying the staining intensity by the proportion of positively stained tumor cells as previously described40. The expression levels of PRDM3 was regarded as high if the SI score is > 6, or low if the SI score if ≤ 6. The immunohistochemical specimens were evaluated by two independently pathologists who were blinded to clinical diagnosis.
RNA isolation and quantitative real-time PCR
Mice were sacrificed with CO2 asphyxiation followed by cervical dislocation. Pancreata were immediately harvested and cut into small pieces in RNALater (Qiagen). To detect mRNAs, 20 mg of tissue was homogenized in 1 ml of Trizol using a T 10 basic ULTRA-TURRAX® homogenizer. RNA was extracted from Trizol according to the manufacturer’s instructions (Thermo Fisher Scientific) and subsequently subjected to reverse transcription using the PrimeScript™ RT reagent Kit (TaKara). Quantitative real-time PCR was performed with the Premix Ex Taq (TaKara) on a CFX96 qPCR system (BioRad). Results were normalized to Gapdh for mRNA detection. The quantitative real-time PCR primer sequences are list in Supplementary Table 5.
Western blotting
Pancreata were harvested and snap frozen in liquid nitrogen. The frozen tissue was homogenized in lysis buffer containing 20 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1 mM Na2EDTA, 2 mM Na2VO4, 1% Triton X-100, 5 mM 4-nitrophenyl phosphate, 0.5% sodium deoxylcholate, 1 mM phenylmethanesulfonylfluoride, and protease inhibitor cocktail (Sigma). 20 μg of protein was separated on a 10% sodium dodecyl sulfate-polyacrylamide gel, transferred onto a polyvinylidene difluoride membrane, and probed with antibodies. The membranes were visualized using the ECL™ Western Blotting Detection System (GE Healthcare) and ChemiDoc™ Imaging Systems (BioRad).
Serum amylase assay
Blood was collected by cardiac puncture and placed at room temperature for 30 min. Serum was separated from red blood cells by centrifugation at 2500 × g for 15 min. The top layer which contained serum was transferred to a new tube for further analysis. Amylase activity was measured using the Amylase Colorimetric Assay Kit (Sigma) following manufacturer’s instructions.
Isolation of primary acinar cells
Primary acinar cells were isolated as described in detail previously25 with a small modification. In brief, pancreas was harvested and transferred into ice-cold Hank’s balanced salt solution (HBSS). Lymph nodes, fat and mesenteric tissues were carefully removed. Pancreas was minced into 2-mm to 3-mm pieces and digested with 0.2 mg/ml collagenase P (Roche) at 37 °C for 10–12 min. Cell clusters were washed 3 times with ice-cold HBSS containing 5% FBS and filtered through 100-μm cell strainer (BD Biosciences). The cell suspension containing acini was carefully layered on top of HBSS containing 30% FBS. Primary acinar cells were pelleted (80 × g, 2 min, at 4 °C) and resuspended in Waymouth media (Sigma).
RAW246.7 cell culture and activation
Raw264.7 macrophages were obtained from American Type Culture Collection (ATCC) and maintained in DMEM (Gibco) containing 10% FBS and 100 U/ml penicillin/streptomycin in a 37 °C humified incubator supplemented with 5% CO2. To stimulate macrophage activation, 5 × 105 Raw264.7 cells per well of 24-well plate were culture in DMEM (10% FBS and 100 U/ml penicillin/streptomycin) overnight and then incubated in acinar-cell-conditioned media. To obtain acinar-cell-conditioned media, primary acinar cell clusters isolated from an equal mass of Prdm3ΔAcinar or control pancreata were incubated in Waymouth complete media (10% FBS, 10 mM HEPES, 100 U/ml penicillin/streptomycin, 100 μg/ml Soybean Trypsin Inhibitor, and 1 μg/ml Dexamethasone) containing 10 nM cerulein for 6 h. 0.5 ml cell-free supernatants were collected and applied to Raw264.7 cells for 16 h. RNA from Raw264.7 macrophages was extracted for quantitative real-time PCR. Data were normalized to Gapdh, as well as the amount of proteins extracted from primary acinar cells used to prepare conditioned media.
Mouse cytokine array
Six-week-old recombinant Ptf1aCreER;Prdm3flox/flox and Ptf1aCreER mice were sacrificed one week after the last tamoxifen injection. Primary acinar cells were isolated from an equal mass of Prdm3ΔAcinar (n = 3) and control (n = 3) pancreata as described above. To stimulate cytokine release, acinar cell explants were incubated in 0.5 ml Waymouth complete medium in the presence of cerulein (10 nM) or TNFα (100 ng/ml) for 6 h. 100 μl of supernatants was collected to examine the concentration of 40 cytokines using the Quantibody® Mouse Inflammatory Array Kit (RayBiotech, Catalog number QAM-INF-1) according to the manufacturer’s instructions. The fluorescent signal was detected using an InnoScan® 300 Microarray Scanner and analyzed by the GenePix® Microarray Analysis Software.
3D acinar cell explant culture
Three-dimensional Matrigel explant culture of acinar cells was performed as described previously41. In brief, freshly isolated primary acinar cell clusters were prepared as above, embedded in growth factor reduced Matrigel (BD Biosciences), cultured in Waymouth complete medium, and maintained at 37 °C in 5% CO2 atmosphere. To stimulate acinar to ductal transdifferentiation in the absence of mutant Kras, control and Prdm3ΔAcinar acinar explants were treated with 50 ng/ml TGF-α. The ratio of acinar to ductal conversion was determined with an average of ten random 10× fields using the EVOS FL Imaging System (Thermo Fisher Scientific).
RNA-seq analyses
Total RNA was extracted from primary acinar cells freshly isolated from 6-week-old Ptf1aCreER;Prdm3flox/flox mice (n = 2) and their corollary controls (Ptf1aCreER, n = 2), one week after the last tamoxifen injection (125 mg/kg, 4 times, every other day). RNA concentration and integrity were measured using the Agilent 2100 Bioanalyzer (Agilent Technologies). PolyA tailed RNA were selected using Dynabeads® oligo(dT) (Thermo Fisher Scientific). cDNA libraries were prepared using the NEBNext® Ultra™ RNA Library Prep Kit for Illumina (New England Biolabs) according to the manufacturer’s instructions. Libraries were sequenced at Novogene (Tianjin, China) with 100× coverage and 150 bp paired end reads on an Illumina HiSeq 2500 instrument.
The quality of the sequencing data was analyzed by using FastQC (version 0.11.5), and raw reads with low quality were removed using Trim Galore (version 0.4.4) prior to analysis of the data. All the trimmed reads were mapped to reference mouse genome (mm10, GRCm38) by using STAR (version 020201), and the mapped counts were extracted using feature count from Subread package (version 1.5.3). Subsequently, read count data containing 49,492 quantified transcripts with raw reads was preprocessed by filtering out genes with zero read count across different samples, and 20,069 genes remained after filtering. The read count data were normalized by DESeq2, which can be used for downstream differential expression analysis. Differentially expressed genes (DEG) in Prdm3ΔAcinar vs. control were filtered by |log2 fold change| > 1. P-values were determined using moderated t-statistics implemented in the Limma package. Gene set enrichment analysis (GSEA) was performed using Bioconductor R package clusterProfiler42,43. The gene sets implemented were derived from Cancer hallmark, Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO), which was collected in the Molecular Signatures Database (MSigDB; version 6.2). Functional enrichment was performed on genes upregulated Prdm3-deficient cells. RNA-seq raw data generated for this manuscript is available in the NCBI Sequence Read Archive (SRA) accession number PRJNA605571.
Statistical analysis
All data are presented as mean ± SD from at least three mice in each experimental group. Statistical analysis between two groups was acquired using two-tailed Student’s t-test. Statistical analyses of patient samples were performed with SPSS software using two-sided t-test. Kaplan-Meier survival plots were generated using the log-rank test. Statistical analysis for RNA-seq data is described in the corresponding sections. P-value < 0.05 was considered statically significant.
Supplementary information
Acknowledgements
We thank the University of Macau, Faculty of Health Sciences, Animal Research Facility for animal housing. We thank Christopher Wright (Vanderbilt University) for Ptf1aCreER mice and David Tuveson (Cold Spring Harbor Laboratory) for LSL-KrasG12D mice. This work was supported by the National Natural Science Foundation of China (NSFC 31701276) and the Science and Technology Development Fund of Macau SAR (FDCT 170/2017/A3 and FDCT 0033/2019/A1) to R.X., Canadian Institutes of Health Research Operating Grant (mop-142216) and New Investigator Award to J.L.K., National Natural Science Foundation of China (NSFC 81672417) to W.L., and National Institutes of Health (R01-DK078803) to M.S.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Edited by A. Stephanou
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Jie Ye, Anpei Huang
Contributor Information
Wen Li, Email: liwen@mail.sysu.edu.cn.
Janel L. Kopp, Email: Janel.kopp@ubc.ca
Ruiyu Xie, Email: ruiyuxie@um.edu.mo.
Supplementary information
Supplementary Information accompanies this paper at (10.1038/s41419-020-2371-x).
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J. Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 2.Murtaugh LC, Keefe MD. Regeneration and repair of the exocrine pancreas. Annu. Rev. Physiol. 2015;77:229–249. doi: 10.1146/annurev-physiol-021014-071727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Storz P. Acinar cell plasticity and development of pancreatic ductal adenocarcinoma. Nat. Rev. Gastroenterol. Hepatol. 2017;14:296–304. doi: 10.1038/nrgastro.2017.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morris JPt, Cano DA, Sekine S, Wang SC, Hebrok M. Beta-catenin blocks Kras-dependent reprogramming of acini into pancreatic cancer precursor lesions in mice. J. Clin. Invest. 2010;120:508–520. doi: 10.1172/JCI40045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kopp JL, et al. Identification of Sox9-dependent acinar-to-ductal reprogramming as the principal mechanism for initiation of pancreatic ductal adenocarcinoma. Cancer Cell. 2012;22:737–750. doi: 10.1016/j.ccr.2012.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Habbe N, et al. Spontaneous induction of murine pancreatic intraepithelial neoplasia (mPanIN) by acinar cell targeting of oncogenic Kras in adult mice. Proc. Natl Acad. Sci. USA. 2008;105:18913–18918. doi: 10.1073/pnas.0810097105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McAllister F, et al. Oncogenic Kras activates a hematopoietic-to-epithelial IL-17 signaling axis in preinvasive pancreatic neoplasia. Cancer Cell. 2014;25:621–637. doi: 10.1016/j.ccr.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guerra C, et al. Chronic pancreatitis is essential for induction of pancreatic ductal adenocarcinoma by K-Ras oncogenes in adult mice. Cancer Cell. 2007;11:291–302. doi: 10.1016/j.ccr.2007.01.012. [DOI] [PubMed] [Google Scholar]
- 9.Hassan MM, et al. Risk factors for pancreatic cancer: case-control study. Am. J. Gastroenterol. 2007;102:2696–2707. doi: 10.1111/j.1572-0241.2007.01510.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Glass C, Wilson M, Gonzalez R, Zhang Y, Perkins AS. The role of EVI1 in myeloid malignancies. Blood Cells Mol. Dis. 2014;53:67–76. doi: 10.1016/j.bcmd.2014.01.002. [DOI] [PubMed] [Google Scholar]
- 11.Hohenauer T, Moore AW. The Prdm family: expanding roles in stem cells and development. Development. 2012;139:2267–2282. doi: 10.1242/dev.070110. [DOI] [PubMed] [Google Scholar]
- 12.Goyama S, et al. Evi-1 is a critical regulator for hematopoietic stem cells and transformed leukemic cells. Cell Stem Cell. 2008;3:207–220. doi: 10.1016/j.stem.2008.06.002. [DOI] [PubMed] [Google Scholar]
- 13.Zhang Y, et al. PR-domain-containing Mds1-Evi1 is critical for long-term hematopoietic stem cell function. Blood. 2011;118:3853–3861. doi: 10.1182/blood-2011-02-334680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tanaka M, et al. EVI1 oncogene promotes KRAS pathway through suppression of microRNA-96 in pancreatic carcinogenesis. Oncogene. 2014;33:2454–2463. doi: 10.1038/onc.2013.204. [DOI] [PubMed] [Google Scholar]
- 15.Diaferia GR, et al. Dissection of transcriptional and cis-regulatory control of differentiation in human pancreatic cancer. EMBO J. 2016;35:595–617. doi: 10.15252/embj.201592404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kopinke D, et al. Ongoing Notch signaling maintains phenotypic fidelity in the adult exocrine pancreas. Dev. Biol. 2012;362:57–64. doi: 10.1016/j.ydbio.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jackson EL, et al. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev. 2001;15:3243–3248. doi: 10.1101/gad.943001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ardito CM, et al. EGF receptor is required for KRAS-induced pancreatic tumorigenesis. Cancer Cell. 2012;22:304–317. doi: 10.1016/j.ccr.2012.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Means AL, et al. Pancreatic epithelial plasticity mediated by acinar cell transdifferentiation and generation of nestin-positive intermediates. Development. 2005;132:3767–3776. doi: 10.1242/dev.01925. [DOI] [PubMed] [Google Scholar]
- 20.Sandgren EP, Luetteke NC, Palmiter RD, Brinster RL, Lee DC. Overexpression of TGF alpha in transgenic mice: induction of epithelial hyperplasia, pancreatic metaplasia, and carcinoma of the breast. Cell. 1990;61:1121–1135. doi: 10.1016/0092-8674(90)90075-P. [DOI] [PubMed] [Google Scholar]
- 21.Berg KB, Schaeffer DF. Tumor budding as a standardized parameter in gastrointestinal carcinomas: more than just the colon. Mod. Pathol. 2018;31:862–872. doi: 10.1038/s41379-018-0028-4. [DOI] [PubMed] [Google Scholar]
- 22.Jensen JN, et al. Recapitulation of elements of embryonic development in adult mouse pancreatic regeneration. Gastroenterology. 2005;128:728–741. doi: 10.1053/j.gastro.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 23.Shrivastava P, Bhatia M. Essential role of monocytes and macrophages in the progression of acute pancreatitis. World J. Gastroenterol. 2010;16:3995–4002. doi: 10.3748/wjg.v16.i32.3995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lugea A, et al. Human pancreatic acinar cells: proteomic characterization, physiologic responses, and organellar disorders in ex vivo pancreatitis. Am. J. Pathol. 2017;187:2726–2743. doi: 10.1016/j.ajpath.2017.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gruber R, et al. YAP1 and TAZ control pancreatic cancer initiation in mice by direct up-regulation of JAK-STAT3 signaling. Gastroenterology. 2016;151:526–539. doi: 10.1053/j.gastro.2016.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park MJ, et al. HIF1-alpha regulates acinar cell function and response to injury in mouse pancreas. Gastroenterology. 2018;154:1630–1634. doi: 10.1053/j.gastro.2018.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Glass C, et al. Global identification of EVI1 target genes in acute myeloid leukemia. PLoS ONE. 2013;8:e67134. doi: 10.1371/journal.pone.0067134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang H, et al. Prominent oncogenic roles of EVI1 in breast carcinoma. Cancer Res. 2017;77:2148–2160. doi: 10.1158/0008-5472.CAN-16-0593. [DOI] [PubMed] [Google Scholar]
- 29.Nanjundan M, et al. Amplification of MDS1/EVI1 and EVI1, located in the 3q26.2 amplicon, is associated with favorable patient prognosis in ovarian cancer. Cancer Res. 2007;67:3074–3084. doi: 10.1158/0008-5472.CAN-06-2366. [DOI] [PubMed] [Google Scholar]
- 30.Deng X, et al. Overexpression of Evi-1 oncoprotein represses TGF-beta signaling in colorectal cancer. Mol. Carcinog. 2013;52:255–264. doi: 10.1002/mc.21852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liou GY, et al. Mutant KRAS-induced expression of ICAM-1 in pancreatic acinar cells causes attraction of macrophages to expedite the formation of precancerous lesions. Cancer Disco. 2015;5:52–63. doi: 10.1158/2159-8290.CD-14-0474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liou GY, et al. Macrophage-secreted cytokines drive pancreatic acinar-to-ductal metaplasia through NF-kappaB and MMPs. J. Cell Biol. 2013;202:563–577. doi: 10.1083/jcb.201301001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ji B, et al. Ras activity levels control the development of pancreatic diseases. Gastroenterology. 2009;137:1072–1082. doi: 10.1053/j.gastro.2009.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guerra C, et al. Pancreatitis-induced inflammation contributes to pancreatic cancer by inhibiting oncogene-induced senescence. Cancer Cell. 2011;19:728–739. doi: 10.1016/j.ccr.2011.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hingorani SR, et al. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell. 2003;4:437–450. doi: 10.1016/S1535-6108(03)00309-X. [DOI] [PubMed] [Google Scholar]
- 36.Carriere C, Young AL, Gunn JR, Longnecker DS, Korc M. Acute pancreatitis markedly accelerates pancreatic cancer progression in mice expressing oncogenic Kras. Biochem. Biophys. Res. Commun. 2009;382:561–565. doi: 10.1016/j.bbrc.2009.03.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Crnogorac-Jurcevic T, et al. Proteomic analysis of chronic pancreatitis and pancreatic adenocarcinoma. Gastroenterology. 2005;129:1454–1463. doi: 10.1053/j.gastro.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 38.Sun HC, et al. Expression of hypoxia-inducible factor-1 alpha and associated proteins in pancreatic ductal adenocarcinoma and their impact on prognosis. Int. J. Oncol. 2007;30:1359–1367. [PubMed] [Google Scholar]
- 39.Shibaji T, et al. Prognostic significance of HIF-1 alpha overexpression in human pancreatic cancer. Anticancer Res. 2003;23:4721–4727. [PubMed] [Google Scholar]
- 40.Li W, et al. Sphingosine kinase 1 is associated with gastric cancer progression and poor survival of patients. Clin. Cancer Res. 2009;15:1393–1399. doi: 10.1158/1078-0432.CCR-08-1158. [DOI] [PubMed] [Google Scholar]
- 41.Fleming Martinez AK, Storz P. Mimicking and manipulating pancreatic acinar-to-ductal metaplasia in 3-dimensional cell culture. J. Vis. Exp. 2019;144:e59096. doi: 10.3791/59096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.