Abstract
Klebsiella pneumoniae is one of the most common hospital-acquired Gram-negative pathogens. During the last decade, the emergence of strains with reduced susceptibility or resistance to carbapenems is becoming a therapeutic challenge. This study takes place after the isolation of 14 strains of carbapenem-resistant K. pneumoniae with similar susceptibility patterns and carriage of OXA-48 and NDM-1 carbapenemases genes. Fourteen patients were found to be colonized (faecal carriage) and/or infected by two different clones of carbapenemase-coproducing K. pneumoniae during a 1-year period of time. Some of the patients had shared a hospital ward and continued to be colonized several months after the outbreak.
Keywords: Antibiotics, carbapenemase, Klebsiella pneumoniae, nosocomial, resistance
Introduction
Carbapenemase production among Enterobacteriaceae is becoming a major problem worldwide. One of the species closely related to the spread of such enzymes is Klebsiella pneumoniae, an important pathogen involved in serious nosocomial infections [1] and whose expansion by means of high-risk clones harbouring plasmid resistance genes has been widely reported [[2], [3], [4], [5]]. OXA-48, NDM-1 and KPC are the most frequent carbapenemases detected nowadays in K. pneumoniae [6]. Their increase favours the emergence of carbapenemase-coproducing isolates. In the present study, we report the outbreak detection of two different clones of K. pneumoniae coproducing NDM-1, OXA-48 and CTX-M-15 enzymes in the same institution in a short period of time.
Consorcio Hospital General Universitario de Valencia is a healthcare facility complex with a reference population of almost 360 000 inhabitants in Valencia, Spain. The first cases of carbapenemase production in Enterobacteriaceae—mainly K. pneumoniae carrying OXA-48—were detected in 2014. At that time no carbapenemase production had been reported in our area, although we had implemented surveillance for carbapenemase production in carbapenem-resistant isolates according to Clinical and Laboratory Standards Institute (CLSI) guidelines [7]. Since then the number of such isolates has raised dramatically in our healthcare setting, especially in 2016 (unpublished data). An imported case of NDM-1 carbapenemase-producing K. pneumoniae in November 2015 marked the emergence of these metallo-β-lactamases in our hospital. In October 2016, coproducing OXA-48, NDM-1 and CTX-M-15 K. pneumoniae was initially isolated in the urine sample from a patient who arrived in the emergency ward from home but who had been discharged from the hospital 10 days before. During the following 10 months, another 13 patients were found to be faecal carriers and/or infected by this carbapenemase-coproducing microorganism.
Since then, no more patients have been found to carry this multidrug-resistant carbapenemase-coproducing K. pneumoniae. Further, interestingly, NDM-1 carbapenemase is at present detected in a very low rate compared to OXA-48 enzyme, which is by far the predominant carbapenemase identified in Enterobacteriaceae in our setting (unpublished data).
The objective of this study was to retrospectively analyse whether the strains coproducing both carbapenemases were clonal and carried related plasmids or not; and to establish a possible epidemiologic relationship between the patients infected and/or colonized by these microorganisms.
Materials and methods
Bacterial identification and broth microdilution susceptibility testing were performed using the commercial platform MicroScan Walkaway Plus system (Beckman Coulter, Brea, CA, USA); when necessary, MIC confirmation was performed by gradient test (Liofilchem, Waltham, MA, USA). The MICs of ampicillin, amoxicillin clavulanate, piperacillin/tazobactam, cefuroxime, cefoxitin, cefotaxime, ceftazidime, cefepime, ertapenem, imipenem, meropenem, norfloxacin, ciprofloxacin, levofloxacin, gentamicin, tobramycin, amikacin, fosfomycin, tigecycline, colistin and trimethoprim/sulfamethoxazole were determined. Some of the antibiotics being tested varied depending on the origin of the sample, such as norfloxacin and fosfomycin in urine samples. Susceptibility breakpoints were interpreted according to the recommendations of the CLSI [7], except for colistin and tigecycline, also tested using MicroScan Walkaway Plus system [8], but interpreted according to the European Committee on Antimicrobial Susceptibility Testing breakpoints [9].
Carbapenemase production and phenotypic characterization were assessed when nonsusceptibility to ertapenem, meropenem or imipenem was detected following CLSI breakpoints [7]. Firstly, screening of carbapenemase with the β-CARBA test (Bio-Rad, Hercules, CA, USA) was performed, followed by the ‘KPC, MBL and OXA-48 confirmation kit: carbapenemases’ (Rosco Diagnostica, Taastrup, Denmark) in case of positive results for the β-CARBA test. Carbapenemase genes were detected by the Xpert-carba-R (Cepheid, Sunnyvale, CA, USA), BDMax CRE (Becton Dickinson, San Diego, CA, USA) or either Eazyplex SuperBug CRE (Amplex Biosystems, Giessen, Germany) and sequenced by in-house Sanger PCR. The primers used in order to obtain PCR products for Sanger sequencing are summarized in Table 1.
Table 1.
Specific primers used for detection and sequencing of OXA-48, NDM-1 and CTX-M β-lactamases
| Gene | Name | Sequence | Reference |
|---|---|---|---|
| blaOXA-48 | OXA-48F | TTGGTGGCATCGATTATCGG | [10] |
| OXA-48R | GAGCACTTCTTTTGTGATGGC | ||
| blaNDM-1 | NDM-1F | CCAATATTATGCACCCGGTCG | |
| NDM-1R | ATGCGGGCCGTATGAGTGATTG | ||
| blaCTX-M-1 | CTX-M-1-SEQ-F | CCCATGGTTAAAAAATCACTGC | [11] |
| CTX-M-1-SEQ-R | CAGCGCTTTTGCCGTCTAAG |
F, forward; R, reverse.
Clonal diversity of isolates was performed by both pulsed-field gel electrophoresis (PFGE) and multilocus sequence typing (MLST). PFGE was performed according to the procedure described by Gautom [12]. Restriction patterns were analysed and interpreted visually by two people according to the criteria of Tenover et al. [13]. Sequence types were determined on the basis of the genetic variation of gapA, infB, mdh, pgi, phoE, rpoB and tonB housekeeping genes, as described online (https://bigsdb.pasteur.fr/klebsiella/klebsiella.html).
Conjugation experiments with azide-resistant Escherichia coli J53 used as recipient and selection on Müeller-Hinton plates containing sodium azide (100 μg/mL) and imipenem (4 μg/mL) were performed in order to characterize the plasmid or plasmids carrying blaOXA-48, blaNDM-1 and/or blaCTX-M-15. Transconjugants were later investigated for extended-spectrum β-lactamase (ESBL) and carbapenemase production by PCR using the Eazyplex SuperBug CRE kit (Amplex Biosystems, Giessen, Germany). Plasmid typing was performed with the PBRT kit (Diatheva, Fano, Italy) in every transconjugant [14].
Results
We found that all the strains harboured OXA-48 and NDM-1 carbapenemases, as well as a CTX-M-15 ESBL, by means of commercial kits followed by sequencing confirmation of the abovementioned bla genes. We obtained two different clones according to results of both PFGE and MLST, which correlated perfectly. Twelve isolates belonged to PFGE pattern A and sequence type (ST) 101 and two strains to PFGE pattern B and ST437. Fig. 1 shows a PFGE gel image of four strains belonging to pattern A.
Fig. 1.
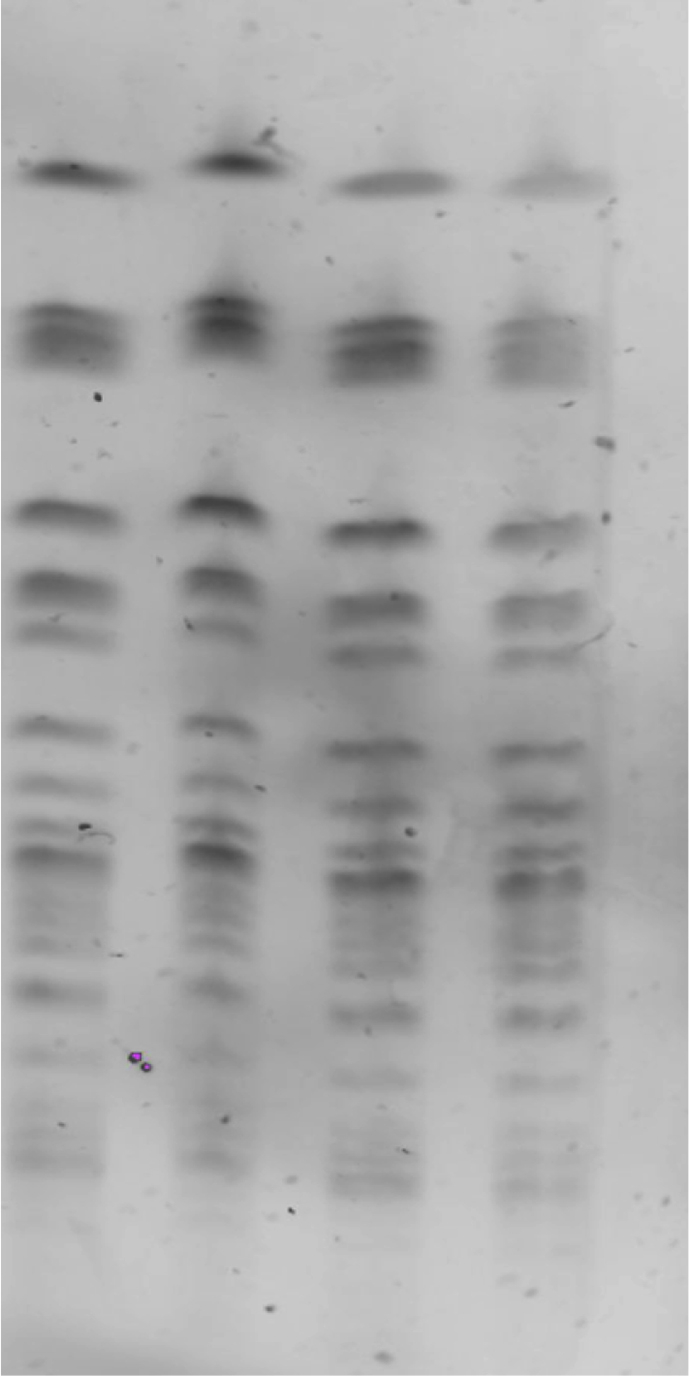
Pulsed-field gel electrophoresis image showing results of four strains belonging to pattern A.
In the conjugation experiment, different transconjugants were detected: one harbouring OXA-48 only, another harbouring NDM-1 and CTX-M-15, and another harbouring OXA-48 and CTX-M-15. Regarding plasmid typing, OXA-48 was found in an IncL/M plasmid without other resistance determinants, and NDM-1 was found in an IncFIB plasmid with other resistance determinants such as CTX-M-15 β-lactamase. Both OXA-48 and NDM-1 carbapenemases were never found together in the same plasmid after conjugation experiments.
Susceptibility was assessed phenotypically: all the strains showed resistance to all β-lactams, including meropenem, imipenem and ertapenem; additionally, they were resistant to ciprofloxacin, gentamicin and tobramycin, but they had different susceptibility patterns to trimethoprim/sulfamethoxazole, fosfomycin, amikacin, tigecycline and colistin. After the detection of the two different clones, the following differences in the susceptibility patterns between both clones were realized: pattern A isolates were always susceptible to trimethoprim/sulfamethoxazole and resistant to amikacin, whereas pattern B isolates were resistant to trimethoprim/sulfamethoxazole and susceptible to amikacin. Table 2 provides a summary of the antimicrobial susceptibility pattern of each clone, its sequence type, its plasmid incompatibility group and the resistance genes detected.
Table 2.
Clones, susceptibility profiles and resistance genes detected
| Strain pattern | ST | Plasmid incompatibility group | Susceptibility profile | Resistance genes detected |
|---|---|---|---|---|
| A | ST101 | IncL/M, IncFIB | S: SXT/TMP,a TIG, FOS, COL R: AK,a AMX/CLV, CPE, PIP/TZB, CTX, CAZ, CFX, ERT, MER, IMI, LEV, CIP, GEN, TOB. |
blaOXA-48, blaNDM-1, blaCTX-M-15 |
| B | ST437 | IncL/M, IncFIB | S: TIG, FOS, COL, AKa SXT/TMP,a AMX/CLV, CPE, PIP/TZB, CTX, CAZ, CFX, ERT, MER, IMI, LEV, CIP, GEN, TOB. |
blaOXA-48, blaNDM-1, blaCTX-M-15 |
AK, amikacin; AMX/CLV, amoxicillin/clavulanate; CIP, ciprofloxacin; COL, colistin; CPE, cefepime; CTX, cefotaxime; ERT, ertapenem; GEN, gentamicin; IMI, imipenem; LEV, levofloxacin; MER, meropenem; PIP/TZB, piperacillin/tazobactam; R, resistant; S, susceptible; ST, sequence type; SXT/TMP, trimethoprim/sulfamethoxazole; TIG, tigecycline; TOB, tobramycin.
Antibiotics with susceptibility that changed depending on ST and pattern.
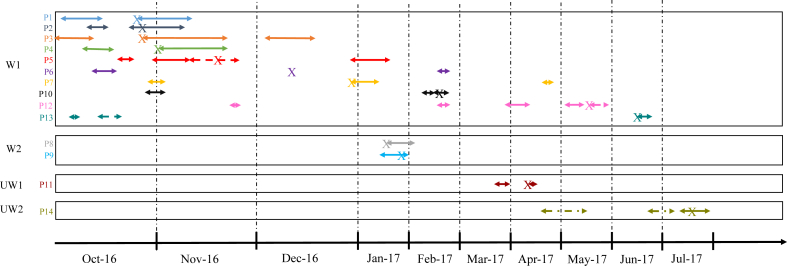
Epidemiologic information showed a possible relationship among ten patients with pattern A strains due to ward sharing (ward 1) in a short period of time (at the same time or within a 2-month difference). It was not possible to find relationship between two of the 12 patients of pattern A, even though the isolates belonged to the same ST and were clonal to the others. The two isolates of pattern B were found in two patients who were hospitalized in another ward (ward 2) than those of pattern A. The first case of pattern B occurred in a patient who had been previously discharged from another hospital in the same city. Time and space sharing between the different patients involved in the outbreaks is shown in Fig. 2.
Fig. 2.
Epidemiologic relationship of patients involved in two outbreaks. Solid arrows indicate periods of hospital stay for each patient; broken arrows, hospital stay in different ward; and X, moment when first isolate was obtained from each patient. P, patient, UW, unrelated ward, W, ward.
Information about clinical, demographic and microbiologic features of the patients and microorganisms is shown in Table 3. It should be noted that during the study period, some patients were discharged from the hospital, then readmitted. It was in the second (or following) hospitalization episode when the coproducing isolate was detected, which complicates the discovery and follow-up of the outbreaks. After recompilation of epidemiologic data, no other relationship but sharing a room during the same period of time could explain the transmission of the two clones among the patients involved in the two outbreaks. Because we could not find any obvious epidemiologic relationship in patients 11 and 14 and they accounted for various admissions before the coproducing K. pneumoniae was isolated from one of their samples, we are not sure of the time these patients had been previously colonized (Table 3).
Table 3.
Clinical, demographic and microbiologic features of 14 patients infected and/or colonized with Klebsiella pneumoniae
| Patient No. | Date of first hospital admission | Date of first isolate | Time elapsed from first admission to carbapenemase detection (days) | Sample source | Hospital ward | ST (pattern) |
|---|---|---|---|---|---|---|
| 1a | 03.10.2016 | 25.10.2016 | 22 | Urine | W1 | 101 (A) |
| 2a | 11.10.2016 | 26.10.2016 | 15 | Blood, urine, faecal carriage | W1 | 101 (A) |
| 3a | 01.10.2016 | 26.10.2016 | 25 | Blood, urine, faecal carriage | W1 | 101 (A) |
| 4a | 09.10.2016 | 26.10.2016 | 15 | Urine, faecal carriage | W1 | 101 (A) |
| 5a | 20.10.2016 | 19.11.2016 | 30 | Blood, urine, faecal carriage | W1 | 101 (A) |
| 6a | 12.10.2016 | 11.12.2016 | 60 | Urine | W1 | 101 (A) |
| 7a | 30.10.2016 | 30.12.2016 | 61 | Urine | W1 | 101 (A) |
| 8 | 17.01.2017 | 17.01.2017 | 0 | Urine | W2 | 437 (B) |
| 9 | 16.01.2017 | 27.01.2017 | 10 | Urine, faecal carriage | W2 | 437 (B) |
| 10a | 28.10.2016 | 17.02.2017 | 112 | Urine | W1 | 101 (A) |
| 11a | 25.03.2017 | 13.04.2017 | 19 | Urine, faecal carriage, blood | UW | 101 (A) |
| 12a | 23.11.2016 or 21.02.2017 or 27.03.2017 | 21.05.2017 | 179 or 89 or 55 | Urine, faecal carriage | W1 | 101 (A) |
| 13a | 05.10.2016 | 19.06.2017 | 257 | Urine | W1 | 101 (A) |
| 14a | 22.04.2017 or 23.06.2017 or 13.07.2017 | 19.07.2017 | 88 or 26 or 6 | Ulcer, faecal carriage | UW | 101 (A) |
UW, unrelated ward; W1, ward 1; W2, ward 2.
These patients were discharged and then readmitted during study period.
Discussion
Dissemination of K. pneumoniae isolates harbouring carbapenem resistance genes continues to increase. NDM-1 or OXA-48 harbouring K. pneumoniae isolates has been identified worldwide [[4], [5], [6]]. However, finding the coproduction of both carbapenemases in the same isolate has not been frequently reported in outbreaks of two or more patients. The first K. pneumoniae strain coproducing NDM-1 and OXA-48 was isolated from the urine sample of an elderly man in Morocco in 2012 [15]. The second was reported in Tunisia, a country where OXA-48 producers are already endemic, as in Turkey [16]. The third was detected in the screening rectal swab of a patient transferred from the intensive care unit of a hospital located in Belgrade, Serbia, to Bern University Hospital in Switzerland, belonging to ST101, as did some of the strains in our study [17]. The present study reveals two outbreaks of carbapenemase-coproducing K. pneumoniae isolates in a tertiary-care hospital at the same time. In our case, the sequence types detected were 101 and 437, which have already been found to harbour NDM-1 and OXA-48 carbapenemases in Switzerland and Slovenia, where they also caused nosocomial outbreaks [17,18]. ST101 has already been identified in Spain as a carbapenemase carrier, and it is one of the sequence types widely distributed across Europe, having been submitted from 15 countries [19]. It is considered to be a high-risk clone that is often associated with outbreaks [20,21]. It probably possesses particular characteristics that increase its tenacity, transmissibility and population size, thereby providing a greater opportunity for the acquisition of antibiotic resistance genes [4]. Regarding ST437, it has already been found to be involved in the interhospital spread of carbapenemase genes in Spain [22], although not coproducing carbapenemases.
In the last few years, K. pneumoniae has developed resistance to several antibiotic classes, leading to an increase in the life-threatening infections and to a major interest in the most appropriate treatments [5,23]. The strains in our study were considered to be multidrug resistant because they were found to be resistant to at least one agent in more than three antimicrobial families [24]. All the strains remained susceptible to colistin, fosfomycin and tigecycline, considered to be last-resort antimicrobial agents used to fight multidrug-resistant K. pneumoniae infections [25].
Regarding plasmid typing, all three types detected in our study have already been reported in the literature as carbapenemase carriers [26]. This shows the high risk of intra- and interspecies horizontal transmission of carbapenemase genes among successful genetic platforms, enhancing dissemination and persistence [27]. IncFIB has been found to carry NDM-1 and CTX-M-15 ESBL in K. pneumoniae [27,28]. Although some other widespread carbapenemase determinants, such as the blaKPC and blaNDM genes, have been shown to disseminate through different plasmid scaffolds [29], the current spread of the blaOXA-48 gene is linked to the wide diffusion of an identical IncL/M plasmid scaffold. Previous studies indicated that plasmids carrying the blaOXA-48 gene from different enterobacterial isolates, different clones and different countries may share very similar features [30].
Regarding epidemiologic relatedness, it is important to mention the fact that some of the patients who were discharged and then readmitted after many months (more than 8 months in patient 13) were still either colonized, or both colonized and infected. It takes a long time for the patient to be uncolonized or for the bacteria to lose the carbapenemase-carrying plasmids, as previously described [31].
The first isolate of pattern B was found in a patient who had been previously discharged from another hospital in the same city, which would explain the coexistence of the two different clones in the same period of time and the lower number of pattern B isolates compared to pattern A. This is consistent with the idea that the co-carriage of different carbapenemases in a same clone is a frequent phenomenon, favoured by the coexistence in similar quantity of strains that carry one enzyme or the other.
It is also remarkable that during the outbreaks there were patients who were infected or colonized by different isolates of K. pneumoniae, some coproducing both carbapenemases and others carrying only OXA-48. This is of great interest because we found isolates carrying OXA-48 on its own, but there were no isolates carrying NDM-1 without OXA-48. This may be because the plasmid carrying OXA-48 confers more stability to the bacteria, as it is a small and self-transferable plasmid that does not carry any additional resistance gene [32]. This would also explain the disappearance of NDM-1 from our hospital and the continuous spread, even in other sequence types, of OXA-48 (unpublished data).
To our knowledge, our description of carbapenemase coproduction is the first to be reported in the area of Valencia. Moreover, the occurrence of two simultaneous different outbreaks of coproducing K. pneumoniae is exceptional in the literature. Further and deeper studies on this subject are needed, as these findings emphasize the importance of systematic detection in order to contain the spread of these multidrug-resistant organisms.
Conflict of Interest
None declared.
References
- 1.Gorrie C.L., Mirceta M., Wick R.R., Edwards D.J., Thomson N.R., Strugnell R.A. Gastrointestinal carriage is a major reservoir of Klebsiella pneumoniae infection in intensive care patients. Clin Infect Dis. 2017;65:208–215. doi: 10.1093/cid/cix270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Navon-Venezia S., Kondratyeva K., Carattoli A. Klebsiella pneumoniae: a major worldwide source and shuttle for antibiotic resistance. FEMS Microbiol Rev. 2017;41:252–275. doi: 10.1093/femsre/fux013. [DOI] [PubMed] [Google Scholar]
- 3.Ferreira Roumayne L., da Silva Brenda C.M., Graziela G.S., Nakamura-Silva R., Pitondo-Silva A., Campanini E.B. High prevalence of multidrug-resistant Klebsiella pneumoniae harboring several virulence and β-lactamase encoding genes in a Brazilian intensive care unit. Front Microbiol. 2019;9:3198. doi: 10.3389/fmicb.2018.03198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mathers A.J., Peirano G., Pitout J.D.D. The role of epidemic resistance plasmids and international high-risk clones in the spread of multidrug-resistant Enterobacteriaceae. Clin Microbiol Rev. 2015;28:565–591. doi: 10.1128/CMR.00116-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Munoz-Price L.S., Poirel L., Bonomo R.A., Schwaber M.J., Daikos G.L., Cormican M. Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect Dis. 2013:13785–13796. doi: 10.1016/S1473-3099(13)70190-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nordmann P., Poirel L. The difficult-to-control spread of carbapenemase producers among Enterobacteriaceae worldwide. Clini Microbiol Infec. 2014;20:821–830. doi: 10.1111/1469-0691.12719. [DOI] [PubMed] [Google Scholar]
- 7.Clinical and Laboratory Standards Institute (CLSI) CLSI guidelines. https://clsi.org/ Available at:
- 8.Pfennigwerth N., Kaminski A., Korte-Berwanger M., Pfeifer Y., Simon M., Werner G. Evaluation of six commercial products for colistin susceptibility testing in Enterobacterales. Clin Microbiol Infect. 2019;25:1385–1389. doi: 10.1016/j.cmi.2019.03.017. [DOI] [PubMed] [Google Scholar]
- 9.European Committee on Antimicrobial Susceptibility Testing (EUCAST) Clinical breakpoints and dosing of antibiotics. http://www.eucast.org/clinical_breakpoints/ Available at:
- 10.Cetinkol Y. The investigation of oxacillinase/metallo-beta-lactamase genes and clonal analysis in carbapenem-resistant Klebsiella pneumoniae. Infez Med Italy. 2016;24:48–53. [PubMed] [Google Scholar]
- 11.Carattoli A. Molecular epidemiology of Escherichia coli producing extended-spectrum beta-lactamases isolated in Rome, Italy. J Clin Microbiol. 2008;46:103–108. doi: 10.1128/JCM.01542-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gautom R.K. Rapid pulsed-field gel electrophoresis protocol for typing of Escherichia coli O157:H7 and other/gram-negative organisms in 1 day. J Clin Microbiol. 1997;35:2977–2980. doi: 10.1128/jcm.35.11.2977-2980.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tenover F.C., Arbeit R.D., Goering R.V., Mickelsen P.A., Murray B.E., Persing D.H., Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.PCR based replicon typing: PBRT KIT–PCR-based replicon typing. https://www.diatheva.com/molecular-biology/pcr-based-replicon-typing/pbrt-2-0-kit-details Available at: [DOI] [PubMed]
- 15.Barguigua A., El Otmani F., Lakbakbi El Yaagoubi F., Talmi M., Zerouali K., Timinouni M. First report of a Klebsiella pneumoniae strain coproducing NDM-1, VIM-1 and OXA-48 carbapenemases isolated in Morocco. APMIS. 2013;121:675–677. doi: 10.1111/apm.12034. [DOI] [PubMed] [Google Scholar]
- 16.Nasr A. Ben, Decré D., Compain F., Genel N., Barguellil F., Arlet G. Emergence of NDM-1 in association with OXA-48 in Klebsiella pneumoniae from Tunisia. Antimicrob Agents Chemother. 2013;57:4089–4090. doi: 10.1128/AAC.00536-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seiffert S.N., Marschall J., Perreten V., Carattoli A., Furrer H., Endimiani A. Emergence of Klebsiella pneumoniae co-producing NDM-1, OXA-48, CTX-M-15, CMY-16, QnrA and ArmA in Switzerland. J Antimicrob Agents. 2014;44:260–262. doi: 10.1016/j.ijantimicag.2014.05.008. [DOI] [PubMed] [Google Scholar]
- 18.Pirš M., Cerar Kišek T., Križan Hergouth V., Seme K., Mueller Premru M., Jeverica S. Successful control of the first OXA-48 and/or NDM carbapenemase-producing Klebsiella pneumoniae outbreak in Slovenia, 2014–2016. J Hosp Infect. 2019;101:142–149. doi: 10.1016/j.jhin.2018.10.022. [DOI] [PubMed] [Google Scholar]
- 19.David S., Reuter S., Harris S.R., Glasner C., Feltwell T., Argimon S. Epidemic of carbapenem-resistant Klebsiella pneumoniae in Europe is driven by nosocomial spread. Nat Microbiol. 2019;4:1919–1929. doi: 10.1038/s41564-019-0492-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pitart C., Solé M., Roca I., Fàbrega A., Vila J., Marco F. First outbreak of a plasmid-mediated carbapenem-hydrolyzing OXA-48 β-lactamase in Klebsiella pneumoniae in Spain. Antimicrob Agents Chemother. 2011;55:4398–4401. doi: 10.1128/AAC.00329-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Snitkin E.S., Zelazny A.M., Thomas P.J., Stock F., NISC Comparative Sequencing Program Group Tracking a hospital outbreak of carbapenem-resistant Klebsiella pneumoniae with whole-genome sequencing. Sci Transl Med. 2012;4:148rr116. doi: 10.1126/scitranslmed.3004129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seara N., Oteo J., Carrillo R., Pérez-Blanco V., Mingorance J., Gómez-Gil R. Interhospital spread of NDM-7–producing Klebsiella pneumoniae belonging to ST437 in Spain. Int J Antimicrob Agents. 2015;46:169–173. doi: 10.1016/j.ijantimicag.2015.04.001. [DOI] [PubMed] [Google Scholar]
- 23.Jacobs D.M., Safir M.C., Huang D., Minhag F., Parker A., Rao G.G. Triple combination antibiotic therapy for carbapenemase-producing Klebsiella pneumoniae: a systematic review. Ann Clin Microbiol Antimicrob. 2017;16:76. doi: 10.1186/s12941-017-0249-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Magiorakos A.P., Srinivasan A., Carey R.B., Carmeli Y., Falagas M.E., Giske C.G. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18:268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 25.Petrosillo N., Taglietti F., Granata G. Treatment options for colistin resistant Klebsiella pneumoniae: present and future. J Clin Med. 2019;8:E934. doi: 10.3390/jcm8070934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carattoli A. Resistance plasmid families in Enterobacteriaceae. Antimicrob Agents Chemother. 2009;53:2227–2238. doi: 10.1128/AAC.01707-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mathers A.J., Cox H.L., Kitchel B., Bonatti H., Brassinga A.K.C., Carroll J. Molecular dissection of an outbreak of carbapenem-resistant Enterobacteriaceae reveals intergenus KPC carbapenemase transmission through a promiscuous plasmid. mBio. 2011;2:e00204–e00211. doi: 10.1128/mBio.00204-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ho P.L., Lo W.U., Yeung M.K., Lin C.H., Chow K.H., Ang I. Complete sequencing of pNDM-HK encoding NDM-1 carbapenemase from a multidrug-resistant Escherichia coli strain isolated in Hong Kong. PLoS One. 2011;21 doi: 10.1371/journal.pone.0017989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Novais A., Cantón R., Moreira R., Peixe L., Baquero F., Coque T.M. Emergence and dissemination of Enterobacteriaceae isolates producing CTX-M-1–like enzymes in Spain are associated with IncFII (CTX-M-15) and broad-host-range (CTX-M-1, -3, and -32) plasmids. Antimicrob Agents Chemother. 2007;51:796–799. doi: 10.1128/AAC.01070-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cuzon G., Ouanich J., Gondret R., Naas T., Nordmann P. Outbreak of OXA-48–positive carbapenem-resistant Klebsiella pneumoniae in France. Antimicrob Agents Chemother. 2011;55:2420–2423. doi: 10.1128/AAC.01452-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mo Y., Hernandez-Koutoucheva A., Musicha P., Bertrand D., Lye D., Ng O.T. Carriage duration of carbapenemase-producing Enterobacteriaceae in a hospital cohort—implications for infection control measures. https://europepmc.org/article/PPR/PPR86839 In press. Available at: [DOI] [PMC free article] [PubMed]
- 32.Poirel L., Potron A., Nordmann P. OXA-48-like carbapenemases: the phantom menace. J Antimicrob Chemother. 2012;67:1597–1606. doi: 10.1093/jac/dks121. [DOI] [PubMed] [Google Scholar]