Abstract
Dialister massiliensis strain Marseille-P5638T (= CSUR P5638) is a new species from the genus Dialister and family Veillonellaceae which was isolated from the gut microbiota of a healthy individual.
Keywords: Culturomics, Dialister massiliensis sp. nov., Firmicutes, gut microbiota, taxonogenomics
The genus Dialister belongs to the order Firmicutes and includes anaerobic, nonmotile and Gram-negative bacilli [1]. To date, there are only five bacterial species with standing in nomenclature (https://www.bacterio.net/genus/dialister), all exclusively isolated from human samples. Here we report the isolation of a new Dialister species from a stool sample of a faecal transplant donor. This strain, Marseille-P5638, was obtained using the culturomics approach [[2], [3], [4]].
Isolation and growth conditions
In November 2017, we collected a fresh stool specimen from a 30-year-old Frenchman, who was a faecal transplant donor. The stool was decontaminated with 100% ethanol (v/v) [5]. The stool was preincubated for 5 days in an anaerobic blood culture bottle (Becton Dickinson, Le Pont de Claix, France) containing 2 mL sheep's blood and 2 mL filter-sterilized rumen. Subsequently, culture suspension was inoculated on 5% sheep's blood–enriched Columbia agar (bioMérieux, Marcy l’Etoile, France) and incubated for 72 hours at 37°C in anaerobic atmosphere (anaeroGEN; Oxoid, Dardilly, France). Identification of isolated bacterial colonies was attempted by MALDI-TOF MS with a Microflex LT spectrometer (Bruker Daltonics, Bremen, Germany) and the Biotyper 3.0 software against the Bruker database that was continually incremented with the MEPHI database (https://www.mediterranee-infection.com/urms-data-base/), as previously reported [6]. Among these, the bacterial strain Marseille-P5638 could not be identified (Fig. 1).
Fig. 1.
MALDI-TOF MS reference spectrum of Dialister massiliensis sp. nov. strain Marseille-P5638T. Reference spectrum was generated by comparison of spectra from 12 individual colonies.
The study was approved by the ethics committee of the Institut Mediterranee-Infection under reference 2016-010. The faecal transplant donor provided written informed consent for participation in this study.
Phenotypic characteristics
Colonies from strain Marseille-P5638 were small, transparent and smooth with a mean diameter of 0.1 to 0.2 mm. Bacterial cells were Gram-negative coccobacilli occurring in pairs or in sets of 4, ranging in length from 0.83 to 1.20 μm and in width from 0.70 to 0.80 μm (Fig. 2). Strain Marseille-P5638 exhibited catalase- and oxidase-negative activities. This strain is non–spore forming. The survival of several nonsporulated bacterial species to stool disinfection with ethanol has previously been described [5,7,8]. Characteristics of the strain are summarized in Table 1. Strain Marseille-P5638 differed from closely related species with validly published names in terms of cell length, growth temperature, major fatty acid methyl ester composition, DNA G + C content, alkaline phosphatase and glutamic acid decarboxylase activities (Supplementary Table S1).
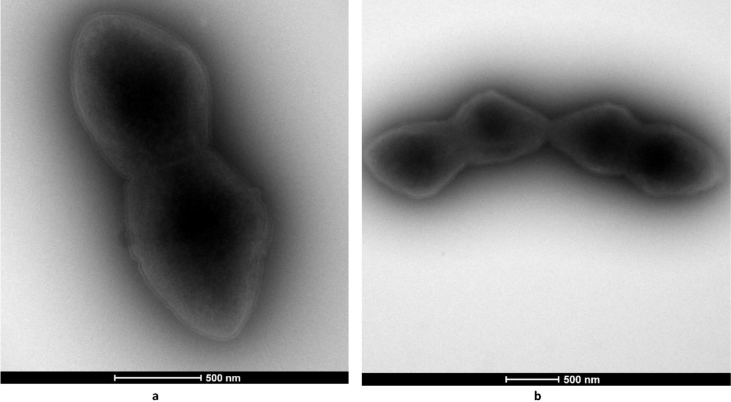
Fig. 2.
Transmission electron micrograph of Dialister massiliensis sp. nov. strain Marseille-P5638T (a) Bacterial cells in pairs. (b) Bacterial cells in quadruplet. Colony was collected from agar and fixed with 2.5% glutaraldehyde in 0.1 M cacodylate buffer for at least 1 hour at 4°C. Drop of cell suspension was deposited for approximately 5 minutes on glow-discharged formvar carbon film with 400 mesh nickel grids (FCF400–Ni, EMS). Grids were dried on blotting paper and cells were negatively stained for 10 seconds with 1% ammonium molybdate solution in filtered water at room temperature. Electron micrographs were acquired with a Tecnai G20 Cryo (FEI Company, Limeil-Brévannes, France) transmission electron microscope operated at 200 keV. Scale bar represents 500 nm.
Table 1.
Description of Dialister massiliensis sp. nov. according to digital protologue TA00779
| Characteristic | Value |
|---|---|
| Taxonumber | TA00779 |
| Date of entry | 31 October 2018 |
| Draft number/date | 001 |
| Version | Draft |
| Species name | Dialister massiliensis |
| Genus name | Dialister |
| Specific epithet | massiliensis |
| Species status | sp. nov. |
| Species etymology | massiliensis (mas.si.li.en'sis, L. masc. adj. massiliensis, ‘of Massilia,’ ancient Roman name for Marseille, where strain was isolated) |
| Submitter | AFOUDA Pamela |
| E-mail of submitter | afoudapamela@yahoo.fr |
| Designation of type strain | Strain Marseille-P5638 |
| Strain collection number | CSUR P5638 |
| 16S rRNA gene accession number | LT996173 |
| Genome accession number (EMBL) | LT996885 |
| Genome status | Draft |
| Genome size | 2 320 000 bp |
| GC mol% | 48.4 |
| Data on origin of sample from which strain had been isolated | |
| Country of origin | France |
| Region of origin | Marseille |
| Date of isolation | 6 November 2017 |
| Source of isolation | Human gut |
| Sampling date | 20 October 2017 |
| Growth medium, incubation conditions (temperature, pH and further information) used for standard cultivation | Columbia agar supplemented with 5% sheep's blood, 37°C for 72 hours of incubation |
| Gram stain | Negative |
| Cell shape | Rod |
| Cell size (length or diameter) | 0.83–1.20 × 0.70–0.80 μm |
| Motility | Nonmotile |
| Colony morphology | Transparent, smooth |
| Temperature range | 28–45°C |
| Temperature optimum | 37°C |
| Lowest pH for growth | 6 |
| Highest pH for growth | 7.5 |
| Highest NaCl concentration for growth | 0.5 |
| Relationship to O2 | Anaerobe |
| O2 conditions for strain testing | Aerobiosis, anaerobiosis, microaerophilic |
| Oxidase | Negative |
| Catalase | Negative |
Digital prologue available at http://imedea.uib-csic.es/dprotologue/.
Strain identification
To identify strain Marseille-P5638, the 16S rRNA gene was amplified using the primer pair fD1 and rP2 (Eurogentec, Angers, France), as previously described [9], and sequenced using the Big Dye Terminator v1.1 Cycle Sequencing Kit and a 3500xLGenetic Analyzer capillary sequencer (Thermo-Fisher, Saint-Aubin, France). The 16S rRNA nucleotide sequence was assembled and corrected using the CodonCode Aligner software (https://www.codoncode.com/).
Strain Marseille-P5638 exhibited a 95.99% sequence identity with Dialister succinatiphilus strain YIT 11850T (GenBank accession no. AB370249), the closest phylogenetically related species with standing in nomenclature (Fig. 3). We consequently classified this strain as type strain of a new species within the genus Dialister (family Veillonellaceae, phylum Firmicutes).
Fig. 3.
Phylogenetic tree showing position of Dialister massiliensis sp. nov., strain Marseille-P5638T, relative to other phylogenetically close neighbours. GenBank accession numbers of 16S ribosomal RNA are indicated in parentheses. Sequences were aligned using MUSCLE with default parameters; phylogenetic inferences were obtained by maximum composite likelihood method and MEGA 6 software. Bootstrap values obtained by repeating analysis 1000 times to generate majority consensus tree are indicated at nodes. Only bootstrap values ≥ 90% were retained. Scale bar indicates 0.5% nucleotide sequence divergence.
Genome sequencing
Genomic DNA was extracted using the EZ1 biorobot (Qiagen, Hilden, Germany) with the EZ1 DNA tissue kit (Qiagen) and then sequenced on a MiSeq sequencer (Illumina, San Diego, CA, USA) with the Nextera Mate Pair sample prep and Nextera XT Paired End kits (Illumina), as previously described [10]. To improve the sequence assembly, a second genomic sequencing was performed using the MinIon sequencer using the SQK-LSK108 kit (Oxford Nanopore Technologies, Oxford, UK). The combination of these two technologies enabled us to obtain a single scaffold as the assembly. The assembly was performed with a pipeline incorporating several software packages (Spades [11] and Trimmomatic [12] for trimmed data). GapCloser [13] was used to reduce gaps.
The genome of strain Marseille-P5638 is 2 320 000 bp long with a 48.4 mol% G + C content. The degree of genomic similarity of strain Marseille-P5638T with closely related species was estimated by OrthoANI [14]. OrthoANI values among closely related species (Fig. 4) ranged from 63.32% between Dialister succinatiphilus and Veillonella tobetsuensis to 87.52% between Veillonella atypica and Veillonella tobetsuensis. When Dialister massiliensis was compared to these closely related species, values ranged from 63.12% with Veillonella tobetsuensis to 73.96% with Dialister succinatiphilus.
Fig. 4.
Heat map generated by OrthoANI values calculated by OAT software between Dialister massiliensis sp. nov. strain Marseille-P5638T and other closely related species with standing in nomenclature.
Conclusion
Strain Marseille-P5638T, exhibiting several phenotypic and genomic differences, as well as a 16S ribosomal RNA sequence divergence of >1.3% and OrthoANI value of <95% with the closest phylogenetically related species with standing in nomenclature, is consequently proposed as the type strain of the new species Dialister massiliensis sp. nov.
Description of Dialister massiliensis sp. nov.
Dialister massiliensis (mas.si.li.en'sis, L. masc. adj. massiliensis from Massilia, the ancient Roman name for Marseille, where the strain was isolated).
Cells are anaerobic, Gram negative, oxidase and catalase negative, and are nonmotile rods. Colonies are small, transparent and smooth, with a diameter ranging from 0.1 to 0.2 mm on 5% sheep's blood–enriched Columbia agar. The growth temperature ranges from 28 to 45°C after 72 hours of incubation, with an optimal growth temperature at 37°C. Cells range in size from 0.83 to 1.20 μm in length and 0.70 to 0.80 μm in width.
Using API 20NE, Rapid ID 32A API and API ZYM galleries, positive reactions were observed for l-arginine, glutamic acid, alkaline phosphatase, esterase (C4), esterase lipase (C8), leucine arylamidase, acid phosphatase and naphthol-AS-BI-phosphohydrolase and negative reactions were observed for β-galactosidase, potassium nitrate (nitrate reductase), l-tryptophan (indole formation), d-glucose (fermentation and assimilation), urease, esculin ferric citrate, gelatin hydrolysis, l-arabinose (assimilation), d-mannose (assimilation), d-mannitol (assimilation), N-acetylglucosamine (assimilation), d-maltose (assimilation), potassium gluconate (assimilation), capric acid (assimilation), adipic acid (assimilation), malic acid (assimilation), trisodium citrate (assimilation), phenylacetic acid (assimilation), 4-nitrophenyl-αd-galactopyranoside, 4-nitrophenyl-βd-galactopyranoside, 4-nitrophenyl-βd-galactopyranoside-6-phosphate-2CHA, 4-nitrophenyl-αd-glucopyranoside, 4-nitrophenyl-βd-glucopyranoside, 4-nitrophenyl-αl-arabinofuropyranoside, 4-nitrophenyl-βd-glucuronide, 4-nitrophenyl-N-acetyl-βd-glucosaminide, d-mannose (fermentation), d-raffinose (fermentation), 4-nitrophenyl-αl-fucopyranoside, 2-naphtyl-phosphate, l-arginine-β-naphthylamide, l-proline-β-naphthylamide, l-leucyl-l-glycine-β-naphthylamide, l-phenylalanine-β-naphthylamide, l-leucine-β-naphthylamide, pyroglutamic β-naphthylamide acid, l-tyrosine-β-naphthylamide, l-alanyl-l-alanine-β-naphthylamide, l-glycine-β-naphthylamide, l-histidine-β-naphthylamide, l-glutamyl-l-glutamic β naphthylamide acid, l-serine-β-naphthylamide, lipase (C14), valine arylamidase, cystine arylamidase, trypsin, α-chymotrypsin, α-galactosidase, β-galactosidase, β-glucuronidase, α-glucosidase, β-glucosidase, N-acetyl-β-glucosaminidase, α-mannosidase and α-fucosidase.
The major fatty acids are C18:1n9, C16:0, and C18:0. The G + C content of the genome is 48.4%. The type strain Marseille-P5638T (= CSUR P5638) was isolated from the stool specimen of a healthy 30-year-old Frenchman.
Nucleotide sequence accession number
The 16S rRNA gene and genome sequences were deposited in GenBank under accession numbers LT996173 and LT996885, respectively.
Deposit in a culture collection
Strain Marseille-P5638T was deposited in Collection de Souches de l’Unité des Rickettsies collection under number CSUR P5638.
Conflict of Interest
None declared.
Acknowledgements
Funded by the Mediterranee-Infection foundation and the programme ‘Investissements d’Avenir,’ managed by the Agence Nationale de la Recherche (reference ANR-10IAHU-03). The authors thank A. Caputo for submitting the genomic sequence to GenBank, the electron microscopy platform of IHU-Mediterranée Infection for the electron micrographs; and M. Lardiere for English-language editorial work.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.nmni.2020.100657.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Moore L.V.H., Moore W.E.C. Oribaculum catoniae gen. nov., sp. nov.; Catonella morbi gen. nov., sp. nov.; Hallella seregens gen. nov., sp. nov.; Johnsonella ignava gen. nov., sp. nov.; and Dialister pneumosintes gen. nov., comb. nov., nom. rev., anaerobic Gram-negative bacilli from the human gingival crevice. Int J Syst Evol Microbiol. 1994;44:187–192. doi: 10.1099/00207713-44-2-187. [DOI] [PubMed] [Google Scholar]
- 2.Lagier J.C., Armougom F., Million M., Hugon P., Pagnier I., Robert C. Microbial culturomics: paradigm shift in the human gut microbiome study. Clin Microbiol Infect. 2012;18:1185–1193. doi: 10.1111/1469-0691.12023. [DOI] [PubMed] [Google Scholar]
- 3.Fournier P.E., Lagier J.C., Dubourg G., Raoult D. From culturomics to taxonomogenomics: a need to change the taxonomy of prokaryotes in clinical microbiology. Anaerobe. 2015;36:73–78. doi: 10.1016/j.anaerobe.2015.10.011. [DOI] [PubMed] [Google Scholar]
- 4.Lagier J.C., Khelaifia S., Alou M.T., Ndongo S., Dione N., Hugon P. Culture of previously uncultured members of the human gut microbiota by culturomics. Nat Microbiol. 2016;1:16203. doi: 10.1038/nmicrobiol.2016.203. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 5.Khanna S., Pardi D.S., Kelly C.R., Kraft C.S., Dhere T., Henn M.R. A novel microbiome therapeutic increases gut microbial diversity and prevents recurrent Clostridium difficile infection. J Infect Dis. 2016;214:173–181. doi: 10.1093/infdis/jiv766. [DOI] [PubMed] [Google Scholar]
- 6.Seng P., Drancourt M., Gouriet F., La Scola B., Fournier P.E., Rolain J.M. Ongoing revolution in bacteriology: routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin Infect Dis. 2009;49:543–551. doi: 10.1086/600885. [DOI] [PubMed] [Google Scholar]
- 7.Koransky J.R., Allen S.D., Dowell V.R. Use of ethanol for selective isolation of sporeforming microorganisms. Appl Environ Microbiol. 1978;35:762–765. doi: 10.1128/aem.35.4.762-765.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Browne H.P., Forster S.C., Anonye B.O., Kumar N., Neville B.A., Stares M.D. Culturing of ‘unculturable’ human microbiota reveals novel taxa and extensive sporulation. Nature. 2016;533:543–546. doi: 10.1038/nature17645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morel A.S., Dubourg G., Prudent E., Edouard S., Gouriet F., Casalta J.P. Complementarity between targeted real-time specific PCR and conventional broad-range 16S rDNA PCR in the syndrome-driven diagnosis of infectious diseases. Eur J Clin Microbiol Infect Dis. 2015;34:561–570. doi: 10.1007/s10096-014-2263-z. [DOI] [PubMed] [Google Scholar]
- 10.Diop A., Khelaifia S., Armstrong N., Labas N., Fournier P.E., Raoult D. Microbial culturomics unravels the halophilic microbiota repertoire of table salt: description of Gracilibacillus massiliensis sp. nov. Microb Ecol Health Dis. 2016;27:32049. doi: 10.3402/mehd.v27.32049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leplae R., Hebrant A., Wodak S.J., Toussaint A. ACLAME: a classification of mobile genetic elements. Nucleic Acids Res. 2004;32:D45–D49. doi: 10.1093/nar/gkh084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grissa I., Vergnaud G., Pourcel C. The CRISPRdb database and tools to display CRISPRs and to generate dictionaries of spacers and repeats. BMC Bioinform. 2007;8:172. doi: 10.1186/1471-2105-8-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Llorens C., Futami R., Covelli L., Domínguez-Escribá L., Viu J.M., Tamarit D. The Gypsy Database (GyDB) of mobile genetic elements: release 2.0. Nucleic Acids Res. 2011;39:D70–D74. doi: 10.1093/nar/gkq1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee I., Ouk Kim Y., Park S.C., Chun J. OrthoANI: an improved algorithm and software for calculating average nucleotide identity. Int J Syst Evol Microbiol. 2016;66:1100–1103. doi: 10.1099/ijsem.0.000760. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.