Abstract
The pathogenesis of many human diseases has been attributed to the over production of reactive oxygen species (ROS), particularly superoxide (O2●-) and hydrogen peroxide (H2O2), by-products of metabolism that are generated by the premature reaction of electrons with molecular oxygen (O2) before they reach complex IV of the respiratory chain. To date, there are 32 known ROS generators in mammalian cells, 16 of which reside inside mitochondria. Importantly, although these ROS are deleterious at high levels, controlled and temporary bursts in H2O2 production is beneficial to mammalian cells. Mammalian cells use sophisticated systems to take advantage of the second messaging properties of H2O2. This includes controlling its availability using antioxidant systems and negative feedback loops that inhibit the genesis of ROS at sites of production. At its core, ROS production depends on fuel metabolism. Therefore, desensitizing H2O2 signals would also require the temporary inhibition of fuel combustion and fluxes through metabolic pathways that promote ROS production. Additionally, this would also demand the diversion of fuels and nutrients into pathways that generate NADPH and other molecules required to maintain cellular redox buffering capacity. Therefore, fuel selection and metabolic flux plays an integral role in dictating the strength and duration of cellular redox signals. In the present review I provide an updated view on the function of protein S-glutathionylation, a ubiquitous redox sensitive modification involving the formation of a disulfide between the small molecular antioxidant glutathione and a cysteine residue, in the regulation of cellular metabolism on a global scale. To date, these concepts have mostly been reviewed at the level of mitochondrial bioenergetics in the contexts of health and disease. Careful examination of the literature revealed that glutathionylation is a temporary inhibitor of most metabolic pathways including glycolysis, the Krebs cycle, oxidative phosphorylation, amino acid metabolism, and fatty acid combustion, resulting in the diversion of fuels towards NADPH-producing pathways and the inhibition of ROS production. Armed with this information, I propose that protein S-glutathionylation reactions desensitize H2O2 signals emanating from catabolic pathways using a three-pronged regulatory mechanism; 1) inhibition of metabolic flux through pathways that promote ROS production, 2) diversion of metabolites towards pathways that support antioxidant defenses, and 3) direct inhibition of ROS-generating enzymes.
keywords: glutathoinylation, ROS, redox signaling, metabolic regulation
1. Introduction
The regulation of fuel selection and metabolism by mammalian tissues has been an area of great interest in life sciences and medical research for many years due to the relationship of dysfunction in these pathways with the development of a wide array of diseases. For example, selection between carbohydrate, amino acid, and fatty acid utilization by skeletal muscle tissues has been a major focus of study for metabolic transitions in response to feeding and fasting, changing diet compositions, and exercise. Skeletal muscle relies on a high degree of metabolic flexibility for fuel selection and metabolism, which is regulated by the complex interplay of hormones and metabolic factors in response to changes in physiology and energy demands [1]. Fuel selection hinderance due to dysfunctional muscle signaling results in the induction of “metabolic inflexibility”, a hallmark in disorders like obesity, type II diabetes mellitus, liver disease, and many others [1]. The heart relies on a similar degree of flexibility in fuel selection and metabolism to supply enough adenosine triphosphate (ATP) to maintain its hemodynamic function. The heart of an average human male consumes ~430 L of molecular oxygen (O2) a day to support mitochondrial ATP production [2]. Approximately 90% of this ATP is supplied by mitochondria where ~70–90% is produced from the oxidation of fats [2]. The remainder is generated by the combustion of ketone bodies, glucose, amino acids, and lactate [3]. Metabolic gridlock and inflexibility can have dire consequences for heart tissue since it prevents the selection of substrates that are required to supply ATP for cardiomyocyte contraction [3]. This inevitably culminates with the development of heart disease due to reliance on glycolysis, which does not supply enough ATP to maintain the hemodynamic function of the heart. Hepatocytes in the liver also rely on having a high degree of metabolic flexibility to not only sustain intrahepatic ATP levels, but also to maintain glucose, amino acid, and fat homeostasis in response to changes in whole-body energy and nutritional demands. Overall, fuel selection and metabolism and its regulation by the concerted action of complex hormonal circuits and intracellular signals is vital for physiology.
Mammalian cells contain an entire “redoxome” that is required to change protein function in response to fluctuations in redox buffering capacity. These changes are mediated through the redox modification of protein cysteine thiols in response to alterations in the availability of H2O2 and NADPH. Increased H2O2 production oxidizes protein cysteine thiols [4]. NADPH has the opposite effect; restoring the reductive capacity of cell redox buffering networks. These pathways cross communicate with other signaling cascades to modulate a wide array of cell functions including cellular division and growth, wound healing, embryonic development, motility and immune cell function, energy sensing and bioenergetics [[5], [6], [7]]. At its core, nutrient metabolism and fuel combustion relies on redox reactions which produce H2O2 and NADPH. Changes in redox state feedback onto nutrient oxidizing pathways for the regulation of fuel metabolism and ROS production. An excellent example of this concept is the impact of protein S-glutathionylation reactions on mitochondrial bioenergetics where oxidation of glutathione pools results in the inhibition of numerous proteins, decreasing ROS production (reviewed in Refs. [8]). This topic has been reviewed extensively in the context of the inhibition of ROS production in mitochondria. However, the global effects of S-glutathionylation on cellular metabolism and its role in diverting fuels into pathways that promote NADPH production whilst simultaneously desensitizing H2O2 signals has not been discussed. Here, I provide a unifying hypothesis for redox signaling wherein H2O2 signals are not only mediated and amplified through the oxidation of glutathione pools and the subsequent S-glutathionylation of proteins but also desensitized thereafter via a three-pronged mechanism that involves the global regulation of cellular metabolism; deactivation of pathways that generate ROS, promotion of flux through pathways that generate NADPH, and direct inhibition of H2O2-producing enzymes.
2. Protein S-glutathionylation reactions and the inhibition of metabolic flux
Once viewed as a toxic molecule that induces oxidative distress, H2O2 is now considered an important second messenger that, when produced in the cytosol, by mitochondria, or extracellular sources, modulates fuel selection and metabolism in response to changes in the surrounding environment (e.g. nutritional status) [4,9,10]. Hydrogen peroxide is thought by some to fulfill its second messenger function by directly oxidizing a protein cysteine thiol (-SH) to a reactive sulfenic acid (-SOH) intermediate, which activates or deactivates a protein [11]. The caveat associated with H2O2 serving as a direct messenger is that it reacts very slowly with a vast majority of protein and non-protein thiols. This is attributed to several factors; 1) high activation energy of H2O2, 2) reactions are kinetically slow (~20 M−1 s−1 with low molecular weight thiols), which is attributed to the high pKa of most protein and non-protein –SH groups (~8.2 and the spontaneous oxidation of thiols by H2O2 requires deprotonation and thiolate anion formation), 3) concentration of H2O2 normal cells is 10−7-10−9 M [12]. Another important consideration is the formation of –SOH. It has been reported that several enzymes can be directly oxidized by H2O2, resulting in the generation of a sulfenic acid moiety. This occurs in enzyme active sites or areas surrounding these sites where the presence of positively charged amino acids lower the pKa values of catalytic cysteine thiols, promoting their ionization to a nucleophilic thiolate anion that attacks an oxygen in H2O2 [13]. It has been indicated that this plays an integral role in epidermal growth factor signaling through the inhibition of protein tyrosine phosphatase-1B (PTP1B) [14]. However, a report published by Forman et al contended that this reaction would be outcompeted by glutathione because 1) the rate constant for sulfur oxidation in PTP1B is 9–43 M−1s−1, 2) the rate constant for elimination of H2O2 by glutathione peroxidases (GPX; ~107 M−1s−1), and 3) the rapid rate for glutathionylation and deglutathionylation of target proteins by glutathione S-transferase P (GSTP) and glutaredoxin (GRX) enzymes, respectively [12,15]. Additionally, –SOH is unstable and rapidly forms mixed disulfides with glutathione or generates sulfonamides. Generally, a second messenger should be able to elicit a rapid and robust change in cell behavior in response to intra- and extracellular cues and modify target proteins whilst avoiding unwanted side reactions. These properties negate the ability of H2O2 to elicit rapid and robust changes in cell behavior through the direct oxidation of proteins.
The concept that protein S-glutathionylation is essential for amplifying H2O2 signals to change cell behavior in response to stimuli has been reviewed extensively (Fig. 1) [8,12]. Briefly, protein S-glutathionylation is highly responsive to changes in the availability of GSH and GSSG [16]. Oxidation of cellular glutathione pools by H2O2 results in the S-glutathionylation of proteins, which alters their functions. The NADPH-mediated reduction of GSSG and restoration of GSH levels has the opposite effect [16]. The second important feature that allows glutathionylation to interface between the exposome and regulation of cell behavior is that the modification fulfills all the criteria that are required for a post-translational modification (PTM) to serve as a regulatory device [17]. This concept has been reviewed extensively elsewhere [5,17]. In brief, unlike other cysteine redox modifications, the S-glutathionylation/deglutathionylation of proteins is mediated by GSTP and GRX enzymes and are reversible, specific, and respond to changes in the intra-and extracellular environment [12]. Notably, GSTP has been documented to rapidly modify several protein targets by glutathionylation in the cytosol and potentially the nucleus and mitochondria (reviewed in Ref. [12]). The deglutathionylase activities of GRX1 and GRX2 are also well-documented [18] and GRX2 has also been implicated in catalyzing the reverse reaction [16]. However, the capacity of the glutaredoxins to catalyze the S-glutathionylation of proteins remains questionable given that high concentrations of GSSG would be required to do so and the steric and electrostatic restraints associated with this reaction [19]. Another key feature of a second messenger is that it is required to amplify cell signals through rapid and robust changes in its cellular concentration. Unfortunately, H2O2 levels in normal cells occur in the nM range and induce oxidative distress at micromolar concentrations [20]. By contrast, the glutathione concentration in cells is in the millimolar range and the relative concentration of GSH and GSSG can change rapidly in response to H2O2 quenching by GPX and the action of GR. Therefore, the rapid spatiotemporal oxidation and reduction of glutathione pools would serve as an ideal redox signal amplifier for H2O2.
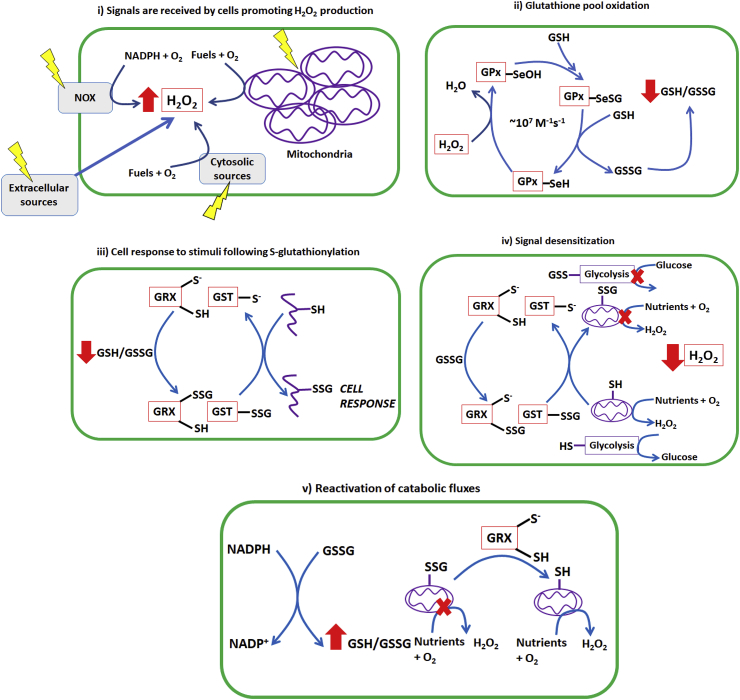
Fig. 1.
A unifying hypothesis for H2O2 second messaging through cellular glutathione pools. I) An intra- or extracellular environmental cue is received by the cell resulting in a temporary burst in H2O2 production. The source of H2O2 can also originate from adjacent cells or extracellular generators. The stimuli can induce the site-specific generation of H2O2 by extracellular, cytosolic, or mitochondrial sources. Additionally, the H2O2 signals can self-amplify one another, either through the temporary inhibition of cellular peroxiredoxins or via the propagation of ROS production through the stimulation of another generator. II) H2O2 is rapidly quenched near its source of production by the rapid action of glutathione peroxidases (GPx). Removal involves an active site selenocysteine, which is oxidized by H2O2 to form a selenol that is resolved by two reduced glutathione (GSH) molecules. This results in production of glutathione disulfide and the oxidation of the cellular glutathione pool (decrease in the ratio of GSH/GSSG). III) Decrease in GSH/GSSG is sensed by glutaredoxins (GRX1; cytosol and intermembrane space, GRX2; matrix) and glutathione S-transferase (GSTP) resulting in the S-glutathionylation of a protein resulting in information transmission. IV) Metabolic enzymes involved in glycolysis, the Krebs cycle, fatty acid and amino acid oxidation, and oxidative phosphorylation are also subjected to S-glutathionylation by GSTP and GRX1/2. This event inhibits catabolic pathways decreasing ROS production and promoting the diversion of metabolites towards pathways that synthesize NADPH. V) Provision of NADPH and inhibition of ROS production results in the reduction of glutathione pools and the restoration of cell redox buffering capacities. This activates the deglutathionylase activities of the glutaredoxins restoring metabolic fluxes through catabolic pathways.
Protein S-glutathionylation is a ubiquitous redox modification that regulates a myriad of cell processes ranging from cell division, migration, phagocytosis, and signaling to calcium homeostasis, and energy sensing [21,22]. Importantly, protein S-glutathionylation is also required to regulate metabolic flux. Most of this work has focused on mitochondria, where S-glutathionylation serves as a prevailing redox sensor for the regulation of bioenergetics, fission/fusion, solute transport and proton leaks, and apoptosis [23]. However, protein S-glutathionylation events in mitochondria have two other important characteristics; 1) these reactions inhibit the activities of most enzymes in the Krebs cycle and electron transport chain and 2) serve as a negative feedback loop for the inhibition of ROS production [24]. Additionally, few articles and studies have focused on the global impact of protein S-glutathionylation on other metabolic pathways. For example, a key word search for the terms “glutathionylation + glycolysis” in PubMed yields mostly articles on the regulation of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and only a handful of articles on how it regulates glycolysis in plants. Few articles have addressed the importance of glutathionylation in the regulation of glycolytic flux in mammals as well as other pathways such as amino acid and fatty acid degradation in mitochondria. However, careful examination of the literature has revealed that like mitochondria, glycolysis is also inhibited by protein S-glutathionylation in mammals which diverts metabolites towards NADPH production. Based on this and in consideration of its capacity to inhibit other metabolic pathways such as amino acid and fatty acid catabolism, protein S-glutathionylation would also play a vital role in redox signal desensitization. This is achieved via a three-pronged approach; 1) by inhibiting metabolic flux through pathways that promote ROS production, 2) redirecting metabolites into pathways that support antioxidant defense systems, and 3) direct inhibition of enzymes that produce ROS (Fig. 1). Additionally, S-glutathionylation has the added benefit of protecting catalytic cysteines and vicinal thiols that are vulnerable towards oxidative deactivation. In the following sections, I survey current literature on the impact of protein S-glutathionylation on glycolysis, glycogen biosynthesis, gluconeogenesis, and pyruvate uptake and oxidation by mitochondria and its role in desensitizing ROS signals and protecting enzymes from oxidative deactivation. I also discuss its role in inhibiting metabolic flux through amino acid and fatty acid catabolism pathways followed by the presentation of a unifying hypothesis wherein protein S-glutathionylation reactions are required to both amplify H2O2 signals and desensitize them thereafter (Fig. 1).
3. Glycolysis
ROS-mediated alteration in glycolytic flux in favor of the diversion of glucose-6-phosphate towards the pentose phosphate pathway is the canonical example of the regulatory control redox signals hold over metabolic fluxes and fuel selection. Glycolysis is a metabolic pathway that involves the conversion of monosaccharides to pyruvate in the cytosol [25]. The pathway is comprised of 10 chemical reactions and produces ATP and reduced β-nicotinamide adenine dinucleotide (NADH). ATP is subsequently used to drive cell processes whereas NADH is oxidized by either cytosolic malate dehydrogenase (MDH2) or cytoplasmic-l-lactate dehydrogenase (c-L-LDH), producing malate and lactate, respectively [26,27]. Malate and lactate are subsequently imported into the matrix of mitochondria and metabolized by mitochondrial malate dehydrogenase (MDH1) and mitochondrial-l-lactate dehydrogenase (m-L-LDH), respectively, reforming NADH which is then oxidized by complex I of the respiratory chain [27]. In anaerobic or hypoxic cells, NADH is oxidized exclusively by c-L-LDH and lactate is exported from the cell to maintain glycolytic flux.
The glycolytic pathway begins with the cellular uptake of the monosaccharide glucose and its ATP-dependent phosphorylation on the C-6 position by hexokinase (or glucokinase in the liver), generating glucose-6-phosphate [25]. Glucose-6-phosphate then enters the “preparatory phase” of the pathway, wherein the hexose ring if phosphorylated again, rearranged and degraded to form glyceraldehyde-3-phosphate (GAP) and dihydroxyacetone phosphate (DHAP) [25]. The two trioses GAP and DHAP then enter the “pay-off phase”, which generates ATP. First, GAP is oxidized and phosphorylated by glyceraldehyde-3-phosphate dehydrogenase (GAPDH), forming the high energy molecule 1,3-biphosphoglycerate [25]. The activity of GAPDH depends on a catalytic cysteine. The cysteine thiol is first ionized by a neighboring histidine residue producing a nucleophilic thiolate anion (-S-) which attacks the C-1 carbonyl in GAP [28]. The resulting covalent adduct is then oxidized in the presence of NAD+ and then subjected to nucleophilic attack by inorganic phosphate, releasing 1,3-bisphosphoglycerate [28]. Phosphoglycerate kinase then transfers the phospho-group from the C-1 position forming 3-phosphoglycerate and ATP. Once produced 3-phosphoglycerate undergoes several rearrangements to generate phosphoenolpyruvate (PEP), which is then used in the final step of glycolysis to produce pyruvate and ATP.
Many other metabolic pathways are strongly reliant on the metabolites formed by glycolysis. For example, glucose-6-phosphate is required to produce glycogen, remove xenobiotics, and is used to generate ribose sugars and NADPH via the pentose phosphate pathway. GAP and DHAP are substrates for glyceroneogenesis and glycerol-3-phosphate oxidation by mitochondria. Finally, pyruvate can generate alanine or lactate or be taken up and fully oxidized by mitochondria for the aerobic production of ATP. Additionally, the metabolism of other monosaccharides (e.g. fructose or galactose) requires their conversion into intermediates of the glycolytic pathway. These factors make glycolysis a primary site for regulation of fuel combustion, selection, and storage in response to environmental cues. This includes maintaining blood glucose homeostasis in response to fasting and fed-state conditions and the production of fat for energy storage in the liver and mobilization of glucose units from glycogen in muscle for energy production during exercise. The regulation of fuel selection and flux through glycolysis is also an important response to oxidative distress [29]. Exposure of cells to high levels of ROS results in a decrease in glycolytic flux and an increase in net flux towards the pentose phosphate pathway for the genesis of NADPH to support antioxidant defenses [29]. This adaptive and rapid redox response has been attributed to H2O2 messaging [30].
3.1. Glyceraldehyde-3-phosphate dehydrogenase
Evidence for the modification of GAPDH by glutathione can be traced back to the early 1990's when it was found that it can undergo “S-thiolation”, now referred to as S-glutathionylation [31]. Modification required the stimulation of respiration, the production of H2O2, and oxidation of cell redox buffers [31]. Glutathionylation of GAPDH lowered its activity by 40%, which was restored with reducing agents [31]. Modification of GAPDH with glutathione first requires oxidation of the active site cysteine thiolate anion to –SOH. Intriguingly, Peralta et al demonstrated that GAPDH contains a shallow H2O2 binding pocket which promotes its SN2-type nucleophilic attack by the catalytic cysteine thiol [32]. Following its formation, –SOH is glutathionylated, yielding a GAPDH mixed protein disulfide (GAPDH-SSG) [33]. It is likely that this mechanism is required to protect GAPDH from irreversible deactivation during a bout of oxidative distress. Once oxidative stress subsides and the reductive capacity of cellular glutathione pools is restored, the glutathionyl moiety is removed by GRX1 reactivating GAPDH [34,35].
The reversible S-glutathionylation of GAPDH in response to oxidative distress fulfills several important cellular functions. First, the sensitivity of GAPDH towards temporary oxidative deactivation makes it an excellent redox sensor. Inhibition of GAPDH by glutathionylation inhibits glycolysis, diverting glucose units towards the pentose phosphate pathway, a critical source of NADPH for antioxidant defenses. This was recently demonstrated by Peralta et al where treatment of yeast cells with H2O2 resulted in GAPDH inhibition and an increase in NADPH/NADP+ [32]. The inhibition of glycolysis through GAPDH glutathionylation has the added benefit of limiting pyruvate production for oxidation by mitochondria, which decreases ROS production and restores cellular redox buffering capacity. In a proteomic study aimed at identifying S-glutathionylation sites following fatiguing exercise, Kramer et al identified GAPDH target for modification [36]. Hydrogen peroxide-mediated oxidation of glutathione pools has been identified as an important mediator for the adaption of muscle towards exercise. In the case of GAPDH, glutathionylation protects it from irreversible oxidation during fatiguing exercise while simultaneously diverting fuels towards the pentose phosphate pathway to desensitize H2O2 signals. It is worth noting, however, that prolonged GAPDH deactivation can be detrimental and has been related to disease progression and drug toxicity [37]. Therefore, like other target proteins, it is vital that the inhibition of GAPDH is temporary and short term to avoid the negative consequences associated with prolonged inhibition of cell metabolism. To this end, recent evidence has shown that GRX1 is required for the rapid deglutathionylation of GAPDH, which restores its activity [38]. Taken together, the temporary inhibition of GAPDH by S-glutathionylation plays several important roles; 1) diverts glucose-6-phosphate towards NADPH production, restoring cell redox capacity for the degradation of H2O2 signals, 2) protects GAPDH from irreversible oxidation during H2O2 signaling, and 3) inhibits carbon flux into pathways that generate ROS.
3.2. Other glycolytic targets for S-glutathionylation
In a recent study using hepatocyte cell lines to model acetoaminophen (APAP) toxicity, Chan and colleagues were able to demonstrate that several glycolytic enzymes are targets for S-glutathionylation [37]. APAP hepatotoxicity is associated with its oxidation by CYP2E1 in the liver to N-acetyl-p-benzoquinone imine (NAPQI), a strong electrophile that forms covalent adducts with protein cysteine thiols [37]. There is a strong correlation between the severity of APAP toxicity and the number of NAPQI adducts in plasma [37]. However, the authors also pointed out that there is a disconnect between adduct formation and APAP toxicity thereafter [37]. For instance, NAPQI adduct formation only moderately affects the activities of some hepatic enzymes like GPX1 [37]. Additionally, other proteins like calcium-dependent ATPases and phosphatases, that exhibit a decrease in activity during APAP toxicity are not modified by NAPQI [37]. Intriguingly, it was shown that glutathionylation patterns in liver sections isolated following APAP toxicity overlapped strongly with NAPQI modifications [37]. Additionally, NAPQI can form adducts with reduced glutathione, creating a good leaving group for nucleophilic attack by a protein cysteine thiol resulting in protein S-glutathionylation [37]. Alternatively, NAPQI can react directly with a protein thiol resulting in attack by GSH [37]. Using isotope-coded affinity tags (ICAT), Chan et al revealed that APAP toxicity was associated with the glutathionylation of fructose bisphosphate aldolase, triose phosphate isomerase, phosphoglycerate kinase, enolase, and pyruvate kinase [37]. These findings are significant for two reasons; first it demonstrates that there are multiple glutathionylation targets in the glycolytic pathway, which would hinder the production of pyruvate from glucose. The second important finding is that it reveals a novel mechanism for drug toxicity. In this case, APAP hepatotoxicity is associated with the non-specific S-glutathionylation of various glycolytic enzymes. It is critical to point out that oxidative distress and disease progression is related to the over oxidation of glutathione pools resulting in an accumulation of GSSG and the spontaneous and non-specific modification of proteins (however; this only occurs across a cell when GSH/GSSG = 1) [18].
Using affinity-base probes, Kehr and colleagues profiled the S-glutathionylated proteome in Plasmodium falciparum and found that several glycolytic enzymes were among the 493 modified proteins [39]. This included GAPDH, LDH, pyruvate kinase (PK), and phosphoglucomutase [39]. Glutathionylation lowered the activity of GAPDH, but had no effect on LDH [39]. Pyruvate kinase was identified as a target for S-glutathionylation in 1983 [40]. The same study also documented that S-glutathionylation was required to protect PK from oxidative deactivation and that its activity could be restored by a thiol oxidoreductase, later identified as GRX1 [40]. This work was highly significant since it was one of the first demonstration that S-glutathionylation served as a cellular defense against oxidative distress [41]. This was followed by the identification of creatine kinase, carbonic anhydrase, and glycogen phosphorylase as glutathionylation targets [42,43]. Cytosolic triose phosphate isomerase (cTPI), which catalyzes the reversible conversion of dihydroxyacetone phosphate to glyceraldehyde-3-phosphate, was also found to be S-glutathionylated in in A. thaliana [44]. Like GAPDH in mammals, cTPI is modified on catalytic Cys127 residue, inhibiting its activity [44]. This results in the diversion of carbon towards the pentose phosphate pathway in response to biotic or abiotic stress [44]. Clearly, more work is required to fully delineate the impact of S-glutathionylation on glycolytic flux. However, the evidence collected so far indicates that S-glutathionylation inhibits several glycolytic enzymes, which occurs through the covalent modification of active site cysteine residues. This leads to the diversion of glycolytic intermediates into pathways that generate NADPH whilst simultaneously preventing their entry into catabolic pathways that produce ROS. Overall, the temporary inhibition of glycolysis by S-glutathionylation plays an important role in the adaptation of cells towards oxidative eustress imposed during H2O2 signaling. Importantly as well, non-specific S-glutathionylation of glycolytic enzymes and their prolonged inhibition occurs during APAP toxicity, resulting in cell death.
3.3. Regulation of 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 3 (PFKFB3)
PFKFB is a bi-functional enzyme that is required for the reciprocal regulation of glycolysis and gluconeogenesis for the provision of blood glucose levels in response to fed, fasting, or exercise conditions. In the fed state, 6-phosphofructo-2-kinase is activated, generating fructose-2,6-bisphosphate (F-2,6-BP), an allosteric activator and inhibitor of PFK-1 and fructose-1,6-bisphosphatase-1 (FBPase-1), respectively. This results in net flux of glucose through glycolysis and the production of ATP, amino acids, and lipids. In response to blood glucose depletion, fructose-2,6-bisphosphatase is activated resulting in the degradation of F-2,6-BP and activation of gluconeogenesis. The reciprocal regulation of glucose production and degradation occurs almost exclusively in hepatocytes and to a lesser extent in the kidney. Cancer cells also rely on this mechanism to control fuel selection and flux but rather invoke it to divert glucose-6-phosphate to the pentose phosphate pathway for the maintenance of antioxidant defenses [45]. In the context of redox signaling, one study found that PFKFB3 undergoes S-glutathionylation on cysteine-205, which is located near the entrance to the catalytic active site [45]. Protein S-glutathionylation was found to be ROS-dependent and sensitive towards the accumulation of GSSG [45]. Furthermore, the authors observed that this modification occurred in cultured HepG2 and Hela cells and resulted in a decrease in kinase activity of the bi-functional enzyme [45]. This resulted in a decrease in F-2,6-BP levels and inhibition of glycolytic flux [45]. Finally, it was observed that this resulted in a net increase in flux through the pentose phosphate pathway and the provision of NADPH for the maintenance of antioxidant defenses [45].
3.4. Glycogen biosynthesis and degradation
The work by Axelsson and Mannervik described above led to an explosion of research aimed at identifying other metabolic targets for S-glutathionylation [40,41,46]. Groups started developing isoelectric focusing techniques to identify novel S-glutathionylation events that resulted from the exposure of purified proteins or tissue lysates to oxidative distress. These new techniques at the time led to the identification of glycogen phosphorylase b as another target for modification [42,47]. Furthermore, the “S-thiolation” event on glycogen phosphorylase b could also be reversed suggesting that it is required for its regulation [47].
Outside of these earlier studies, only two other recent articles have addressed the relationship between protein S-glutathionylation reactions and its impact on glycogen storage. However, based on these initial findings, S-glutathionylation may play a significant role in glycogen homeostasis. In one study, Young et al investigated the impact of deleting the Grx2 gene on the induction of diet-induced obesity and obesity-related disorders [48]. In this study it was observed that wild-type C57BL/6N mice rapidly developed diet-induced obesity which was associated with intrahepatic lipid accumulation and elevated circulating insulin levels [48]. Mice heterozygous for GRX2 (GRX2+/−) were completely protected from diet-induced weight gain, displayed normal circulating insulin and triglyceride levels, and had no signs of fatty liver disease [48]. Of note as well, circulating glucose levels were normal in WT and GRX2 ± fed a high fat diet [48]. A common hallmark for diet-induced obesity and fatty liver disease is the disruption of the glucose-glycogen axis wherein intrahepatic glycogen stores are depleted to maintain blood glucose levels. Young et al found glycogen stores were almost completely depleted in the livers of wild-type mice fed a high fat diet, which is consistent with the concept that “metabolic gridlock” disrupts the glucose-glycogen axis [48]. Intriguingly, GRX2 ± mice fed a high fat diet displayed glycogen levels similar to littermates fed a control-matched diet [48]. GRX2 does play an integral role in preserving and maintaining the efficiency of mitochondrial metabolism and fuel selection and the authors found that loss of GRX2 improved mitochondrial bioenergetics [48]. Therefore, although they did not generate direct evidence that glycogen metabolizing enzymes are S-glutathionylation targets, the authors did show that depleting GRX2 in mitochondria can have profound cellular effects on fatty acid handling and glucose-glycogen homeostasis.
In Dong et al, the authors investigated the function of S-glutathionylation in the regulation of AMP-dependent protein kinase and downstream effects associated with its modification [49]. In this extensive study, it was found that silencing the gene encoding Grx1 or AMPK α2 subunit decreased the expression of glycogen synthase [49]. Although this finding is correlative, it does point to the potential for GRX1 and AMPK cross-talk for control over intrahepatic glycogen homeostasis in response to complex changes in the surrounding nutritional environment.
4. Mitochondrial fuel metabolism
Mitochondria are dynamic double-membraned organelles that serve as central hubs for cellular metabolism. This is by virtue of its superior energy conserving capacity, which is responsible for satisfying the ATP demands of most mammalian cells. The production of ATP by mitochondria depends on the conversion of different fuels into common intermediates that are then combusted by the Krebs cycle [50]. The burning of fuels by the Krebs cycle results in the production of the electron carriers, NADH and FADH2, which are oxidized by complexes I and II, respectively [50]. The disparate metabolic pathways that produce common intermediates for Krebs cycle oxidation also form NADH. Electrons yielded from NADH and FADH2 oxidation then flow through complexes I and II, reducing ubiquinone (UQ) to ubiquinol (UQH2) [51]. Note that several other dehydrogenases that absorb to the outer or inner leaflets of the mitochondrial inner membrane also couple the oxidation of their cognate substrates to the transfer of electrons through an FAD center to UQ [5]. Ubiquinol is then oxidized by complex III and electrons are passed through cytochrome c to complex IV, reducing molecular oxygen (O2) to H2O at the end of the chain [51]. The passage of electrons through the chain establishes an electrochemical gradient of protons across the MIM, which is then used to make ATP by complex V.
Mitochondria are also quantifiably the most important source of ROS in mammalian cells. The proximal ROS generated by mitochondria are superoxide (O2●-) and H2O2 [52]. Mitochondria can contain up to sixteen ROS sources, twelve of which are associated with energy metabolism (reviewed in Ref. [53]). Because of their reactivity, ROS studies initially focused on the contribution of mitochondrial ROS towards the induction of oxidative distress and tissue damage. This led to the development of the “Free Radical Theory of Aging” by Denham Harman, which stipulated that ROS production and the accumulation of oxidative damage are responsible for aging [54]. In addition, mitochondria were often at the center of these studies since high ROS release from these organelles correlated with the induction of oxidative distress and disease. These studies also paved the way for understanding how ROS can deactivate mitochondrial proteins, which eventually led to findings demonstrating that oxyradicals, like H2O2, are required for the reversible regulation of enzymes in response to changes in the environment. For example, in 1983 Hillered and Ernster found that O2●—induced oxidative distress causes respiratory dysfunction in isolated brain mitochondria [55]. Later studies found that several fuel combusting enzymes and respiratory complexes are deactivated directly by ROS [56]. It was eventually found that the oxidative deactivation of these mitochondrial enzymes is reversible and required for protection from damage [57]. Now, it is known that H2O2 signals are mediated through the reversible protein S-glutathionylation, which plays a vital role in regulating several mitochondrial functions including apoptosis, fission and fusion, ROS production, energy production, solute import and proton leaks, and antioxidant defenses (reviewed in Ref. [8]). So far, ~2200 modifiable peptides have been identified, with a significant fraction found in mitochondria [58]. In the following section, the role of glutathionylation in the control of fuel selection will be summarized.
4.1. Entry of pyruvate into the Krebs cycle
Glucose is one of the most important fuels required for energy metabolism and storage. Its high value to mammalian cells is underscored by the hormonal and cellular responses towards small fluctuations in its bioavailability where system wide signals are used to safeguard blood glucose levels for neural function and utilize it for energy metabolism, storage, antioxidant defense, and the biosynthesis of macromolecules. The breakdown of 1 mol of glucose to 2 mol of pyruvate supplies only 2 mol of ATP (ΔG'o ~ −144 kJ mol−1), which is hardly enough energy to meet the demands of mammalian cells. Thus, for glucose metabolism to meet cellular energy demands, pyruvate must be imported into mitochondria where it undergoes further oxidation. The full oxidative degradation of 2 mol of pyruvate by mitochondria yields 36 ATP (2 from the Krebs cycle and 34 from oxidative phosphorylation). Along with the 2 ATP from glycolysis, glucose oxidation by mammalian cells generates 38 ATP (ΔG'o ~ −2840 kJ mol−1).
Pyruvate oxidation by mitochondria begins with its selective import into the matrix by pyruvate carrier [59]. It is then converted to acetyl-CoA by pyruvate dehydrogenase (PDH) through an oxidative decarboxylation reaction that involves coupling the reduction of NAD+ to the evolution of CO2 [60]. PDH is α-keto acid dehydrogenase that is comprised of three catalytic components; pyruvate decarboxylase (E1), dihydrolipoamide transacetylase (E2), and dihydrolipoamide dehydrogenase (E3) [60]. These subunits can form a multi-subunit holoenzyme comprised of multiple copies of each catalytic component, forming high molecular mass oligomers [60]. Since PDH is the major entry point for pyruvate into the Krebs cycle, its activity is subjected to heavy regulation in response to changes in fuel supply and energy demands. PDH is targeted for allosteric induction by its substrates, NAD+ and CoASH, and is also activated by ADP [23]. By contrast, high [acetyl-CoA]/[CoASH], [NADH]/[NAD+], and [ATP]/[ADP] serve as feedback inhibitors of PDH [23]. This is crucial for when ATP demands are low or during fatty acid oxidation since acetyl-CoA is being supplied by the degradation of fats. PDH is also subjected to regulation by phosphorylation/dephosphorylation in response to changes in energy demands and fuel supplies. This mechanism has been reviewed extensively and its importance in modulating pyruvate metabolism following glucose degradation is highlighted by the many studies dedicated to understanding the relationship between dysfunction in this regulatory node and the development of various metabolic diseases.
Redox signals also play an integral role in controlling PDH activity. This is achieved through the direct oxidation of vicinal thiols in PDH. The activity of PDH relies on the several cofactors and prosthetic groups that are associated with the three subunits. The catalytic cycle of PDH begins with the decarboxylation of pyruvate and the acetylation of thiamine pyrophosphate (TPP) [61]. The acetyl group is then transferred to the vicinal thiols of lipoamide in the E2 subunit. The E2 subunit then catalyzes the transfer of the acetyl group to CoASH through a thiol disulfide exchange reaction forming acetyl-CoA and dihydrolipoamide. Next, dihydrolipoamide is oxidized and the hydrides are transferred through a FAD-centered E3 subunit reducing NAD+ [61]. Discussing the catalytic cycle of PDH is important for our purposes here since the vicinal thiols of the lipoamide in the E2 subunit are targets for redox modification. It had been known for some time that other the α-keto acid dehydrogenases and PDH can be deactivated through the oxidation of the vicinal lipoic acid thiols in the E2 subunit. Indeed, in 1996 it was found that purified PDH of porcine heart origin could be deactivated by ROS generated by xanthine/xanthine oxidase, which was speculated by the same authors to contribute to ischemia/reperfusion injury [62]. Around the same period, a second study revealed that PDH is deactivated through the formation of covalent adducts with lipid peroxidation end-product 4-hydroxy-2-nonenal (HNE) [63]. In 2013, evidence generated by Fisher-Wellman et al showed that oxidizing or depleting glutathione pools amplifies ROS production by PDH, leading the authors to speculate that this was associated with protein S-glutathionylation [64]. The same authors later found that this phenomenon contributed towards glucose intolerance and increased tissue fat mass [65].
In 2017, the first evidence was generated demonstrating that PDH is a target for S-glutathionylation. O'Brien et al found that there was a dose dependent relationship between the inhibition of purified PDH and its glutathionylation by diamide [66]. Further studies with isolated liver mitochondria revealed similar trends; both chemical glutathionylation catalysts diamide and disulfiram induced the S-glutathionylation of the E2 subunit of PDH [66]. This modification compromised coupled respiration using pyruvate as a substrate and abolished mitochondrial ROS production [66]. In a separate study published by the same group, it was found that GRX2 was required to catalyze the deglutathionylation of PDH [67]. Indeed, mice containing a deletion for the Grx2 gene displayed increased PDH E2 subunit glutathionylation, which could be reversed using β-mercaptoethanol [67]. In the studies published by O'Brien et al and Chalker et al, it was observed that modification of the E2 subunit of PDH was required to decrease ROS production, leading the authors to conclude that glutathionylation may serve as a negative feedback loop that is required to lower H2O2 generation in response to glutathione pool oxidation [66,67]. This mechanism would also serve as an excellent way of controlling fuel metabolism in response to changes in mitochondrial redox buffering capacities by diminishing substrate oxidation and inhibiting ROS production. For instance, glucose toxicity, which manifests in pancreatic beta-cells due to a deficiency in antioxidant defenses, is related to the over production of ROS by mitochondria [68]. It is feasible that the temporary S-glutathionylation of PDH may protect cells from glucose toxicity and the over production of ROS by stymieing the flow of combustable fuels into the Krebs cycle. This potential mechanism is re-enforced by a third study by the same group showing that S-glutathionylation of pyruvate carrier inhibits pyruvate uptake into the matrix, which substantially decreases mitochondrial ATP production [69]. It is also vital to indicate that PDH is an important mitochondrial ROS generator and over production of H2O2 has been implicated in the pathogenesis of metabolic disorders [65]. Therefore, the modification of PDH with glutathione serves as a negative feedback loop that is required to turn down mitochondrial ROS production in response to cell redox pool oxidation. Overall, this serves as an important mechanism that is required to desensitize H2O2 signals through the deactivation of ROS production and restoration of cell redox buffering capacities.
4.2. Amino acid metabolism
KGDH is comprised of three basic catalytic components that are homologous to the subunits found in PDH (E1; α-ketoglutarate decarboxylase, E2; dihydrolipoamide acyltransferase, E3; dihydrolipoamide dehydrogenase) and thus shares a similar catalytic cycle. Additionally, like PDH, KGDH serves as an important entry point for fuels into the Krebs cycle. Amino acid degradation relies on the transfer of the amino group covalently bound to the α-carbon of an amino acid to α-ketoglutarate, generating glutamate and a carboxy acid-containing carbon skeleton [70]. For example, the degradation of aspartic acid requires the transfer of its amino group to α-ketoglutarate by aspartate aminotransferase, producing oxaloacetate, a Krebs cycle intermediate [70]. It is worth noting that the degradation of all 20 amino acids are intimately linked to the Krebs cycle. The branched chain amino acids give rise to acetyl-CoA and succinyl-CoA. Aromatic amino acids generate fumarate, pyruvate, and acetyl-CoA while histidine, proline, glutamine, and arginine yield glutamate. The degradation of these amino acids first starts with the transamination of α-ketoglutarate forming glutamate [71]. Once formed glutamate can be used as a neurotransmitter, for protein biosynthesis, or in the malate-aspartate shuttle. Glutamate is also subjected to degradation as well, a process that is facilitated by its oxidative deamidation by glutamate dehydrogenase (GDH). This generates α-ketoglutarate which is subjected to oxidative decarboxylation by KGDH.
Because it is a major entry point for carbon derived from amino acid degradation into the Krebs cycle, KGDH is also a major site for the regulation of fuel metabolism. All three subunits are subjected to allosteric activation and inhibition by several metabolites and micronutrients. High [ATP]/[ADP], [NADH]/[NAD+], or [succinyl-CoA]/[CoASH] serve as allosteric inhibitors of the enzyme complex whereas Ca2+, ADP, and Pi have the opposite effect [72]. An intriguing study also found that KGDH activity may be controlled through its full assembly [73]. This is achieved using a novel component, KGD4, which acts as a molecular adaptor for the successful recruitment of the E3 subunit to the E1-E2 core, allowing for the formation of a stable enzyme structure [73]. KGDH is also a well-documented target for redox signaling and is known to undergo irreversible oxidative deactivation in several pathologies. Using intact isolated nerve terminals, Tretter and Adam-Vizi were the first to demonstrate that KGDH can be subjected to oxidative deactivation [74]. In this study, the authors observed that low amounts of H2O2 (<50 μM) deactivated aconitase but KGDH activity remained unchanged allowing mitochondria to maintain NADH production through glutamate oxidation [74]. However, exposure to higher H2O2 amounts led to the oxidative deactivation of KGDH and a depletion in NADH availability [74]. Using rat heart mitochondria, Nulton-Persson and Szweda later discovered that the deactivation of KGDH by H2O2 is reversible [75]. This led to several groups postulating that KGDH serves as a mitochondrial redox sensor that is required to adjust metabolic fluxes and fuel metabolism and selection in response to fluctuations in H2O2 and NADPH availability. The caveat associated with direct oxidation by H2O2 is that the vicinal thiols in the lipoic acid residue remain vulnerable towards further oxidative attack or the formation of irreversible adducts with electrophiles such as HNE. For instance, KGDH has been shown to form irreversible adducts with HNE during ischemia-reperfusion to tissues like the myocardium and as noted above –SOH groups can be irreversibly oxidized [76]. In 2003, Nulton-Persson et al found that H2O2-mediated thiol oxidation results in the S-glutathionylation of the –SOH group thereafter, providing protection from oxidative deactivation [75]. The modification was found to be reversible wherein removal of the glutathionyl moiety restores KGDH activity [75]. Indeed, exposure of mitochondrial preparations to exogenous GRX1 resulted in the reduction of the protein-glutathione mixed disulfide on the E2 subunit [77,78]. The same group also generated evidence demonstrating that S-glutathionylation of a vicinal lipoamide thiol in the E2 subunit also prevents the formation of adducts with HNE [77]. Therefore, S-glutathionylation is vital for protecting KGDH from irreversible deactivation in response to increased H2O2 levels.
It was recently discovered that S-glutathionylation serves as a negative feedback loop for the regulation of ROS production by KGDH using a similar mechanism as described for PDH [66,67]. This observation is significant for several reasons; first, KGDH is a well-documented source of cellular ROS in several tissues and has been shown to produce more H2O2 than conventional generators like complex I [79]. Secondly, mitochondrial H2O2 is an important second messenger that is required to communicate changes in the mitochondrial redox environment with the rest of the cell [4]. This includes adjusting fuel selection and metabolism in response to increased H2O2. For example, mitochondrial H2O2 is integral for fulfilling several physiological and cellular functions from coordinating cell division and growth to facilitating muscle growth and healing in response to exercise [[80], [81], [82]]. Like any second messenger, following the transmission of information, the signal needs to be desensitized thereafter. This can be achieved with this negative feedback loop that decreases ROS production by KGDH. Additionally, S-glutathionylation also decreases ROS production by several other mitochondrial enzymes re-enforcing the concept that it is required to desensitize cellular redox signaling whilst simultaneously promoting the use of amino acids for protein biosynthesis. Two separate studies also found that GRX2 can deglutathionylate KGDH [67,83]. This was found to restore its activity and ROS generating potential [67,83]. The deglutathionylase activity of GRX2 is activated by restoration of the reducing capacity of glutathione pools in the matrix [67,83]. Furthermore, it was recently found that there is a sex dimorphism in the regulation of KGDH by GRX2 [84]. In contrast to male mice where S-glutathionylation inhibits ROS production by KGDH in liver and muscle, which is reversed by GRX2, female mice do not use this mechanism to govern H2O2 generation [84]. This sex difference was attributed to mitochondria from the muscles and liver of female mice having superior redox buffering and ROS handling capacities and were thus less reliant on using S-glutathionylation as a negative feedback loop to desensitize ROS production. Furthermore, a similar effect was observed with PDH wherein only mitochondria collected from the muscle and liver of male mice utilized S-glutathionylation to negatively regulate ROS production [84]. Overall, the modification of KGDH has a three-pronged effect; 1) it protects KGDH from irreversible deactivation, 2) inhibits any further ROS production by KGDH to desensitize mitochondrial redox signals, 3) decreases amino acid catabolism, allowing these fuels to be preserved for protein biosynthesis during cell growth and division while also lowering ROS production by other generators in mitochondria, like the respiratory chain.
Outside of KGDH, only a few other amino acid metabolizing enzymes have been found to undergo S-glutathionylation. GDH is a target for modification in Plasmodium falciparum but its impact on its activity is not known [39]. Another target is cytosolic human branched chain aminotransferase (hBCATc) [85]. In this study, it was found that redox modification of hBCATc lowered its activity by ~45% [85]. This was due to the oxidation of the CXXC motif cysteines, C335 and C338 [85]. Furthermore, oxidative distress was found to induce S-glutathionylation through the initial formation of an –SOH moiety [85]. The glutathionyl moiety could also be removed by exogenous GSH/GRX1 [85]. These observations support the notion that oxidation of cell redox buffering networks lowers fuel oxidation in response to H2O2 signaling, favoring the diversion of amino acids towards protein production during cell growth and division.
4.3. Regulation of Krebs cycle flux
The regulation of Krebs cycle flux by protein S-glutathionylation has been reviewed extensively elsewhere and will thus be only briefly discussed here (reviewed extensively in Ref. [8]). The Krebs cycle is the most central metabolic pathway on the planet. It is an amphibolic pathway that is integral for the catabolism of most fuels and the provision of reducing equivalents for oxidative phosphorylation but is also responsible for supplying basic building blocks for cellular growth, division, and the biosynthesis of macromolecules. As such the Krebs cycle is subjected to heavy regulation and fluxes through this pathway are tightly controlled in response to cellular and whole-body energy demands. The control of PDH and KGDH was discussed in detail above but there are several other important sites for S-glutathionylation in the Krebs cycle. One excellent example in terms of the role of S-glutathionylation in protecting enzymes from oxidative deactivation is aconitase (ACN). Briefly, ACN contains a cubane iron-sulfur cluster that is sensitive to oxidative deactivation through its disassembly by O2●- [86]. Notably, the iron-sulfur cluster becomes less sensitive to oxidative disassembly when the Fea center coordinates citrate. However, when citrate levels decrease this leaves the Fea vulnerable to oxidative attack which could be prevented by S-glutathionylation. Indeed, it was found that the degree of ACN S-glutathionylation was inversely proportional to the availability of citrate [87]. Additionally, although it protects ACN from deactivation, S-glutathionylation also lowers its activity [87]. Other documented targets include isocitrate dehydrogenase-2 (IDH2), succinyl-CoA synthetase (SCS), and malate dehydrogenase (MDH) [37]. Protein S-glutathionylation of IDH2 and MDH also lowers their activities [58,88]. Coupled with the inhibition of PDH and KGDH, the aim of modifying these other Krebs cycle enzymes plays similar roles; 1) to protect Krebs cycle enzyme from oxidative deactivation. This preserves enzyme activity for fuel metabolism once redox buffering capacities are restored and 2) aids in further lowering ROS production to safeguard the cell from oxidative distress while desensitizing mitochondrial redox signaling.
4.4. Oxidative phosphorylation, ATP production, and fatty acid combustion
The regulation of electron flux through the respiratory chain by protein S-glutathionylation and its impact on oxidative phosphorylation has been reviewed extensively (reviewed in Ref. [23]). Here, I will only discuss the role of protein S-glutathionylation in the regulation of fuel selection by the respiratory chain following fluctuations in redox buffering capacity. Complex I is the most heavily studied target for protein S-glutathionylation in mitochondria. It has been found to be regulated by reversible S-glutathionylation in heart and muscle tissue and lens epithelia cells [69,89,90]. GRX2 catalyzes the S-glutathionylation of complex I in response to oxidation of GSH pools, inhibiting the multiprotein enzyme, whereas reduction of the pool has the opposite effect [16]. The temporary inhibition of complex I by S-glutathionylation is integral for limiting ROS production by the electron transport chain [69]. However, prolonged S-glutathionylation of complex I due to loss of GRX2 severely limits ATP production and induces oxidative distress, contributing to the development of heart disease and cataracts [89]. Complex I has approximately fourteen subunits in total that have been shown to undergo glutathionylation, including subunits that form part of the NADH binding site. Another important site for modification is the ND3 subunit which is part of the UQ binding site. The redox modification of Cys39 in the ND3 subunit deactivates complex I by blocking access to the UQ binding site [91]. Overall, S-glutathionylation serves an important mechanism for the regulation of complex I in response to changes in GSH availability.
Unlike complex I, glutathionylation is required to maintain succinate dehydrogenase (complex II) activity. The deglutathionylation of complex II decreases its activity and augments ROS production [92]. Selecting for succinate oxidation during oxidative eustress signaling (e.g. when the glutathione pool is oxidized) would serve as an important adaptive response for the following reasons; 1) it would preserve the integrity of the mitochondrial membrane potential during the temporary deactivation of complex I and the Krebs cycle, thereby preventing permeability transition and the induction of apoptosis and 2) maintain the availability of a transmembrane potential of protons for the production of NADPH by nicotinamide nucleotide transhydrogenase, a major source of matrix reducing equivalents for antioxidant defense. Another important consideration is potential reverse electron transfer from complex II to complex I. It is documented that this pathway is an important source of ROS in mitochondria fueled by succinate [93]. The S-glutathionylation of complex I would severely impede electron flow from the UQH2 pool, limiting ROS production during reverse electron flow from succinate. Therefore, during succinate oxidation when complex I is in an inactivated S-glutathionylated state, it is anticipated that most electrons will feed forward towards complex IV for the maintenance of membrane potential integrity.
Another consideration regarding fuel selection in response to oxidative eustress signaling is whether S-glutathionylation also regulates the oxidation of other enzymes that feed electrons into the respiratory chain. This includes proline dehydrogenase, glycerol-3-phosphate dehydrogenase, dihydroorotate dehydrogenase, sulfide:quinone oxidoreductase, and electron transferring-flavoprotein:quinone oxidoreductase (ETFQO). These five enzymes couple the oxidation of their substrates to the direct reduction of UQ in the electron transport chain. However, out of these UQ-electron donors, only fatty acid oxidation, which involves ETFQO, has been found to be regulated by redox reactions. Fatty acid oxidation (FAO) in mammalian cells depends first on the conversion of acyl-CoA to acyl-carnitine by carnitine acyl-transferase 1 (CAT1). Acyl-carnitine is then imported into the matrix through carnitine:acyl-carnitine carrier (CAC) [1]. It is then oxidized by acyl-CoA dehydrogenase, enoyl-CoA hydratase, 3-hydroxy-acyl-CoA dehydrogenase, and β-ketothiolase [1]. Notably, all four of these fatty acid oxidation enzymes have been found to be subjected to redox modifications. In 2013, Doulias et al observed that all four enzymes can be S-nitrosylated, which the authors indicated could profoundly affect cell metabolism and mitochondrial bioenergetics [94]. Unfortunately, S-nitrosylation suffers from the same drawbacks as sulfenylation. Protein S-nitrosylation is reliant on the formation of S-nitrosylated glutathione (GSNO) or S-nitrosylated-CoA (SNO-CoA), both of which are degraded rapidly by GSNO and SNO-CoA reductases [3]. However, recent work has demonstrated that several FAO enzymes can be subjected to S-glutathionylation. Using an APAP cell culture model and ICAT technology, Chen et al identified long chain and very long chain acyl-CoA dehydrogenases and many other FAO enzymes, including ETFQO, as S-glutathionylation targets [37]. This may result in the inhibition of FAO curtailing ROS production by the electron transport chain. Additionally, even though the respiratory chain is often described as the main source of ROS during FAO, ETFQO and long chain and very long chain acyl-CoA dehydrogenases can also serve as significant sites of production [37]. In addition, induction of protein deglutathionylation in skeletal muscle from mice heterozygous for GRX2 results in an increase mitochondrial fat oxidation [48]. This effect is associated with increased mitochondrial respiration, increased proton leaks through UCP3 and ANT, and an augmentation of ROS production [48]. The same authors also observed that there is a sex dimorphism in this pathway, wherein female mice do not utilize S-glutathionylation to regulate fatty acid oxidation in muscles (unpublished data). It has also been documented that CAC can be subjected to reversible inhibition of S-glutathionylation [95]. This would limit the import of acyl-carnitine into the matrix for FAO. Therefore, in response to a temporary oxidative eustress, several mitochondrial fuel oxidation pathways are inhibited to curtail ROS production and protect mitochondria from irreversible damage.
5. A unifying hypothesis for the amplification of H2O2 signals and their subsequent desensitization through protein S-glutathionylation
Several reviews have addressed the important role of protein S-glutathionylation in the rapid and reversible regulation of various cell functions. Glutathione pool oxidation results in protein S-glutathionylation, culminating with the rapid response of cells towards environmental changes. However, what has not been addressed is the potential function of protein S-glutathionylation in the desensitization of H2O2 signals through the global inhibition of catabolic pathways. Based on the accumulated evidence so far, it is likely that H2O2 signaling and desensitization thereafter occurs in five steps. First, H2O2 production is triggered by stimuli and the origin of its production depends on the cellular location that the stimuli were received (Fig. 1). Second, H2O2 is quenched rapidly by GSH and GPxs resulting in GSSG formation and the oxidation of glutathione pools (Fig. 1). Third, it is documented that GRX1 and 2 harbour both glutathionylase and deglutathionlase activities, which are activated by the oxidation or reduction, respectively, of the glutathione pool. Additionally, GSTP, which is now known to rapidly catalyze the modification of proteins with glutathione, would also catalyze the S-glutathionylation of protein targets. Thus, glutathione pool oxidation triggers S-glutathionylation by GSTP and the glutaredoxins changing cell behavior (e.g. chemotaxis or calcium handling) (Fig. 1). In step fourl, metabolic enzymes are also modified promoting flux through pathways that generate NADPH and the inhibition of the utilization of fuels that produce ROS. Highly vulnerable enzymes that sit at critical metabolic junctions (e.g. GAPDH, PDH, KGDH, complex I) are also modified to prevent irreversible deactivation during H2O2 signaling. Inhibition of ROS production and the provision of NADPH restores the redox buffering capacity of cell glutathione pools resulting in the deglutathionylation of target proteins (Fig. 1). And finally, restoration of the redox buffering capacity of glutathione pools through the provision of NADPH and inhibition of ROS production is sensed by glutaredoxins, triggering their deglutathionylase activities resulting in the removal of glutathionyl moieties from proteins and the re-activation of catabolic fluxes (Fig. 1).
6. Concluding remarks
Research into understanding how cells use redox signals for communication has made substantial progress due to the development and use of novel techniques. This includes the development of intracellular reporters that measure spatiotemporal redox changes in real time and proteomic techniques for the identification of novel S-glutathionylation targets. As such, over ~2200 targets have been identified and use of these approaches have allowed various groups to demonstrate that protein S-glutathionylation reactions are integral for embryonic development, heart physiology, eyesight, immune cell function, and adaptation of muscles to exercise. A number of these targets include metabolic enzymes central to catabolism and H2O2 production. The present review demonstrates that protein S-glutathionylation reactions in response to oxidation of glutathione pools serves a cell-wide inhibitor of fluxes through catabolic pathways including glycolysis, mitochondrial solute import, amino acid metabolism, the Krebs cycle, oxidative phosphorylation, and fatty acid combustion. This leads to a decrease in the flux of metabolites into catabolic pathways that generate ROS and the provision of substrates for NADPH production. Additionally, S-glutathionylation also serves as a negative feedback loop for the deactivation of cellular ROS generators. Collectively, the temporary deactivation of catabolism by protein S-glutathionylation serves as a mechanism for H2O2 signal desensitization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was funded by a start-up grant provided by the Faculty of Agriculture and Environmental Sciences and the School of Human Nutrition at McGill University.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2020.101472.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Muoio D.M. Metabolic inflexibility: when mitochondrial indecision leads to metabolic gridlock. Cell. 2014;159(6):1253–1262. doi: 10.1016/j.cell.2014.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kembro J.M., Aon M.A., Winslow R.L., O'Rourke B., Cortassa S. Integrating mitochondrial energetics, redox and ROS metabolic networks: a two-compartment model. Biophys. J. 2013;104(2):332–343. doi: 10.1016/j.bpj.2012.11.3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mailloux R.J. Application of mitochondria-targeted pharmaceuticals for the treatment of heart disease. Curr. Pharmaceut. Des. 2016;22(31):4763–4779. doi: 10.2174/1381612822666160629070914. [DOI] [PubMed] [Google Scholar]
- 4.Jones D.P., Sies H. The redox code. Antioxidants Redox Signal. 2015;23(9):734–746. doi: 10.1089/ars.2015.6247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mailloux R.J., Treberg J.R. Protein S-glutathionlyation links energy metabolism to redox signaling in mitochondria. Redox Biol. 2016;8:110–118. doi: 10.1016/j.redox.2015.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Checconi P., Limongi D., Baldelli S., Ciriolo M.R., Nencioni L., Palamara A.T. Role of glutathionylation in infection and inflammation. Nutrients. 2019;11(8) doi: 10.3390/nu11081952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Short J.D., Downs K., Tavakoli S., Asmis R. Protein thiol redox signaling in monocytes and macrophages. Antioxidants Redox Signal. 2016;25(15):816–835. doi: 10.1089/ars.2016.6697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Young A., Gill R., Mailloux R.J. Protein S-glutathionylation: the linchpin for the transmission of regulatory information on redox buffering capacity in mitochondria. Chem. Biol. Interact. 2019;299:151–162. doi: 10.1016/j.cbi.2018.12.003. [DOI] [PubMed] [Google Scholar]
- 9.Allister E.M., Robson-Doucette C.A., Prentice K.J., Hardy A.B., Sultan S., Gaisano H.Y., Kong D., Gilon P., Herrera P.L., Lowell B.B., Wheeler M.B. UCP2 regulates the glucagon response to fasting and starvation. Diabetes. 2013;62(5):1623–1633. doi: 10.2337/db12-0981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Robson-Doucette C.A., Sultan S., Allister E.M., Wikstrom J.D., Koshkin V., Bhattacharjee A., Prentice K.J., Sereda S.B., Shirihai O.S., Wheeler M.B. Beta-cell uncoupling protein 2 regulates reactive oxygen species production, which influences both insulin and glucagon secretion. Diabetes. 2011;60(11):2710–2719. doi: 10.2337/db11-0132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heppner D.E., Janssen-Heininger Y.M.W., van der Vliet A. The role of sulfenic acids in cellular redox signaling: reconciling chemical kinetics and molecular detection strategies. Arch. Biochem. Biophys. 2017;616:40–46. doi: 10.1016/j.abb.2017.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang J., Ye Z.W., Singh S., Townsend D.M., Tew K.D. An evolving understanding of the S-glutathionylation cycle in pathways of redox regulation. Free Radic. Biol. Med. 2018;120:204–216. doi: 10.1016/j.freeradbiomed.2018.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gallogly M.M., Starke D.W., Leonberg A.K., Ospina S.M., Mieyal J.J. Kinetic and mechanistic characterization and versatile catalytic properties of mammalian glutaredoxin 2: implications for intracellular roles. Biochemistry. 2008;47(42):11144–11157. doi: 10.1021/bi800966v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sarma B.K., Mugesh G. Redox regulation of protein tyrosine phosphatase 1B (PTP1B): a biomimetic study on the unexpected formation of a sulfenyl amide intermediate. J. Am. Chem. Soc. 2007;129(28):8872–8881. doi: 10.1021/ja070410o. [DOI] [PubMed] [Google Scholar]
- 15.Forman H.J., Davies M.J., Kramer A.C., Miotto G., Zaccarin M., Zhang H., Ursini F. Protein cysteine oxidation in redox signaling: caveats on sulfenic acid detection and quantification. Arch. Biochem. Biophys. 2017;617:26–37. doi: 10.1016/j.abb.2016.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beer S.M., Taylor E.R., Brown S.E., Dahm C.C., Costa N.J., Runswick M.J., Murphy M.P. Glutaredoxin 2 catalyzes the reversible oxidation and glutathionylation of mitochondrial membrane thiol proteins: implications for mitochondrial redox regulation and antioxidant DEFENSE. J. Biol. Chem. 2004;279(46):47939–47951. doi: 10.1074/jbc.M408011200. [DOI] [PubMed] [Google Scholar]
- 17.Mieyal J.J., Chock P.B. Posttranslational modification of cysteine in redox signaling and oxidative stress: focus on s-glutathionylation. Antioxidants Redox Signal. 2012;16(6):471–475. doi: 10.1089/ars.2011.4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gallogly M.M., Mieyal J.J. Mechanisms of reversible protein glutathionylation in redox signaling and oxidative stress. Curr. Opin. Pharmacol. 2007;7(4):381–391. doi: 10.1016/j.coph.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 19.Xiao Z., La Fontaine S., Bush A.I., Wedd A.G. Molecular mechanisms of glutaredoxin enzymes: versatile hubs for thiol-disulfide exchange between protein thiols and glutathione. J. Mol. Biol. 2019;431(2):158–177. doi: 10.1016/j.jmb.2018.12.006. [DOI] [PubMed] [Google Scholar]
- 20.Sies H. Hydrogen peroxide as a central redox signaling molecule in physiological oxidative stress: oxidative eustress. Redox Biol. 2017;11:613–619. doi: 10.1016/j.redox.2016.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mailloux R.J. Cysteine switches and the regulation of mitochondrial bioenergetics and ROS production. Adv. Exp. Med. Biol. 2019;1158:197–216. doi: 10.1007/978-981-13-8367-0_11. [DOI] [PubMed] [Google Scholar]
- 22.Cooper A.J., Pinto J.T., Callery P.S. Reversible and irreversible protein glutathionylation: biological and clinical aspects. Expet Opin. Drug Metabol. Toxicol. 2011;7(7):891–910. doi: 10.1517/17425255.2011.577738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mailloux R.J., Jin X., Willmore W.G. Redox regulation of mitochondrial function with emphasis on cysteine oxidation reactions. Redox Biol. 2014;2:123–139. doi: 10.1016/j.redox.2013.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuksal N., Chalker J., Mailloux R.J. Progress in understanding the molecular oxygen paradox - function of mitochondrial reactive oxygen species in cell signaling. Biol. Chem. 2017;398(11):1209–1227. doi: 10.1515/hsz-2017-0160. [DOI] [PubMed] [Google Scholar]
- 25.Li X.B., Gu J.D., Zhou Q.H. Review of aerobic glycolysis and its key enzymes - new targets for lung cancer therapy. Thorac Cancer. 2015;6(1):17–24. doi: 10.1111/1759-7714.12148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang C., Chen H., Zhang J., Hong Y., Ding X., Ying W. Malate-aspartate shuttle mediates the intracellular ATP levels, antioxidation capacity and survival of differentiated PC12 cells. Int J Physiol Pathophysiol Pharmacol. 2014;6(2):109–114. [PMC free article] [PubMed] [Google Scholar]
- 27.Brooks G.A. The science and translation of lactate shuttle theory. Cell Metabol. 2018;27(4):757–785. doi: 10.1016/j.cmet.2018.03.008. [DOI] [PubMed] [Google Scholar]
- 28.Reis M., Alves C.N., Lameira J., Tunon I., Marti S., Moliner V. The catalytic mechanism of glyceraldehyde 3-phosphate dehydrogenase from Trypanosoma cruzi elucidated via the QM/MM approach. Phys. Chem. Chem. Phys. 2013;15(11):3772–3785. doi: 10.1039/c3cp43968b. [DOI] [PubMed] [Google Scholar]
- 29.Mullarky E., Cantley L.C. Diverting glycolysis to combat oxidative stress. In: Nakao K., Minato N., Uemoto S., editors. Innovative Medicine: Basic Research and Development, Tokyo. 2015. pp. 3–23. [Google Scholar]
- 30.Molavian H.R., Kohandel M., Sivaloganathan S. High concentrations of H2O2 make aerobic glycolysis energetically more favorable for cellular respiration. Front. Physiol. 2016;7:362. doi: 10.3389/fphys.2016.00362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ravichandran V., Seres T., Moriguchi T., Thomas J.A., Johnston R.B., Jr. S-thiolation of glyceraldehyde-3-phosphate dehydrogenase induced by the phagocytosis-associated respiratory burst in blood monocytes. J. Biol. Chem. 1994;269(40):25010–25015. [PubMed] [Google Scholar]
- 32.Peralta D., Bronowska A.K., Morgan B., Doka E., Van Laer K., Nagy P., Grater F., Dick T.P. A proton relay enhances H2O2 sensitivity of GAPDH to facilitate metabolic adaptation. Nat. Chem. Biol. 2015;11(2):156–163. doi: 10.1038/nchembio.1720. [DOI] [PubMed] [Google Scholar]
- 33.Barinova K.V., Serebryakova M.V., Muronetz V.I., Schmalhausen E.V. S-glutathionylation of glyceraldehyde-3-phosphate dehydrogenase induces formation of C150-C154 intrasubunit disulfide bond in the active site of the enzyme. Biochim. Biophys. Acta Gen. Subj. 2017;1861(12):3167–3177. doi: 10.1016/j.bbagen.2017.09.008. [DOI] [PubMed] [Google Scholar]
- 34.Muronetz V.I., Melnikova A.K., Saso L., Schmalhausen E.V. Influence of oxidative stress on catalytic and non-glycolytic functions of glyceraldehyde-3-phosphate dehydrogenase. Curr. Med. Chem. 2018 doi: 10.2174/0929867325666180530101057. [DOI] [PubMed] [Google Scholar]
- 35.Cotgreave I.A., Gerdes R., Schuppe-Koistinen I., Lind C. S-glutathionylation of glyceraldehyde-3-phosphate dehydrogenase: role of thiol oxidation and catalysis by glutaredoxin. Methods Enzymol. 2002;348:175–182. doi: 10.1016/s0076-6879(02)48636-3. [DOI] [PubMed] [Google Scholar]
- 36.Kramer P.A., Duan J., Gaffrey M.J., Shukla A.K., Wang L., Bammler T.K., Qian W.J., Marcinek D.J. Fatiguing contractions increase protein S-glutathionylation occupancy in mouse skeletal muscle. Redox Biol. 2018;17:367–376. doi: 10.1016/j.redox.2018.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chan J.C.Y., Soh A.C.K., Kioh D.Y.Q., Li J., Verma C., Koh S.K., Beuerman R.W., Zhou L., Chan E.C.Y. Reactive metabolite-induced protein glutathionylation: a potentially novel mechanism underlying acetaminophen hepatotoxicity. Mol. Cell. Proteomics. 2018;17(10):2034–2050. doi: 10.1074/mcp.RA118.000875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lopez-Grueso M.J., Gonzalez-Ojeda R., Requejo-Aguilar R., McDonagh B., Fuentes-Almagro C.A., Muntane J., Barcena J.A., Padilla C.A. Thioredoxin and glutaredoxin regulate metabolism through different multiplex thiol switches. Redox Biol. 2019;21:101049. doi: 10.1016/j.redox.2018.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kehr S., Jortzik E., Delahunty C., Yates J.R., 3rd, Rahlfs S., Becker K. Protein S-glutathionylation in malaria parasites. Antioxidants Redox Signal. 2011;15(11):2855–2865. doi: 10.1089/ars.2011.4029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Axelsson K., Mannervik B. An essential role of cytosolic thioltransferase in protection of pyruvate kinase from rabbit liver against oxidative inactivation. FEBS Lett. 1983;152(1):114–118. doi: 10.1016/0014-5793(83)80494-3. [DOI] [PubMed] [Google Scholar]
- 41.Ziegler D.M. Role of reversible oxidation-reduction of enzyme thiols-disulfides in metabolic regulation. Annu. Rev. Biochem. 1985;54:305–329. doi: 10.1146/annurev.bi.54.070185.001513. [DOI] [PubMed] [Google Scholar]
- 42.Park E.M., Thomas J.A. S-thiolation of creatine kinase and glycogen phosphorylase b initiated by partially reduced oxygen species. Biochim. Biophys. Acta. 1988;964(2):151–160. doi: 10.1016/0304-4165(88)90161-4. [DOI] [PubMed] [Google Scholar]
- 43.Chai Y.C., Jung C.H., Lii C.K., Ashraf S.S., Hendrich S., Wolf B., Sies H., Thomas J.A. Identification of an abundant S-thiolated rat liver protein as carbonic anhydrase III; characterization of S-thiolation and dethiolation reactions. Arch. Biochem. Biophys. 1991;284(2):270–278. doi: 10.1016/0003-9861(91)90295-t. [DOI] [PubMed] [Google Scholar]
- 44.Dumont S., Bykova N.V., Pelletier G., Dorion S., Rivoal J. Cytosolic triosephosphate isomerase from Arabidopsis thaliana is reversibly modified by glutathione on cysteines 127 and 218. Front. Plant Sci. 2016;7:1942. doi: 10.3389/fpls.2016.01942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seo M., Lee Y.H. PFKFB3 regulates oxidative stress homeostasis via its S-glutathionylation in cancer. J. Mol. Biol. 2014;426(4):830–842. doi: 10.1016/j.jmb.2013.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Axelsson K., Mannervik B. General specificity of cytoplasmic thioltransferase (thiol:disulfide oxidoreductase) from rat liver for thiol and disulfide substrates. Biochim. Biophys. Acta. 1980;613(2):324–336. doi: 10.1016/0005-2744(80)90087-x. [DOI] [PubMed] [Google Scholar]
- 47.Miller R.M., Sies H., Park E.M., Thomas J.A. Phosphorylase and creatine kinase modification by thiol-disulfide exchange and by xanthine oxidase-initiated S-thiolation. Arch. Biochem. Biophys. 1990;276(2):355–363. doi: 10.1016/0003-9861(90)90732-e. [DOI] [PubMed] [Google Scholar]
- 48.Young A., Gardiner D., Kuksal N., Gill R., O'Brien M., Mailloux R.J. Deletion of the glutaredoxin-2 gene protects mice from diet-induced weight gain, which correlates with increased mitochondrial respiration and proton leaks in skeletal muscle. Antioxidants Redox Signal. 2019;31(17):1272–1288. doi: 10.1089/ars.2018.7715. [DOI] [PubMed] [Google Scholar]
- 49.Dong K., Wu M., Liu X., Huang Y., Zhang D., Wang Y., Yan L.J., Shi D. Glutaredoxins concomitant with optimal ROS activate AMPK through S-glutathionylation to improve glucose metabolism in type 2 diabetes. Free Radic. Biol. Med. 2016;101:334–347. doi: 10.1016/j.freeradbiomed.2016.10.007. [DOI] [PubMed] [Google Scholar]
- 50.Brand M.D., Nicholls D.G. Assessing mitochondrial dysfunction in cells. Biochem. J. 2011;435(2):297–312. doi: 10.1042/BJ20110162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nicholls D.G. Forty years of Mitchell's proton circuit: from little grey books to little grey cells. Biochim. Biophys. Acta. 2008;1777(7–8):550–556. doi: 10.1016/j.bbabio.2008.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mailloux R.J., Gardiner D., O'Brien M. 2-Oxoglutarate dehydrogenase is a more significant source of O2(.-)/H2O2 than pyruvate dehydrogenase in cardiac and liver tissue. Free Radic. Biol. Med. 2016;97:501–512. doi: 10.1016/j.freeradbiomed.2016.06.014. [DOI] [PubMed] [Google Scholar]
- 53.Sies H., Berndt C., Jones D.P. Oxidative stress. Annu. Rev. Biochem. 2017;86:715–748. doi: 10.1146/annurev-biochem-061516-045037. [DOI] [PubMed] [Google Scholar]
- 54.Harman D. Aging: a theory based on free radical and radiation chemistry. J. Gerontol. 1956;11(3):298–300. doi: 10.1093/geronj/11.3.298. [DOI] [PubMed] [Google Scholar]
- 55.Hillered L., Ernster L. Respiratory activity of isolated rat brain mitochondria following in vitro exposure to oxygen radicals. J. Cerebr. Blood Flow Metabol. 1983;3(2):207–214. doi: 10.1038/jcbfm.1983.28. [DOI] [PubMed] [Google Scholar]
- 56.Malis C.D., Bonventre J.V. Susceptibility of mitochondrial membranes to calcium and reactive oxygen species: implications for ischemic and toxic tissue damage. Prog. Clin. Biol. Res. 1988;282:235–259. [PubMed] [Google Scholar]
- 57.Balijepalli S., Annepu J., Boyd M.R., Ravindranath V. Effect of thiol modification on brain mitochondrial complex I activity. Neurosci. Lett. 1999;272(3):203–206. doi: 10.1016/s0304-3940(99)00593-5. [DOI] [PubMed] [Google Scholar]
- 58.Feng S., Chen Y., Yang F., Zhang L., Gong Y., Adilijiang G., Gao Y., Deng H. Development of a clickable probe for profiling of protein glutathionylation in the central cellular metabolism of E. coli and Drosophila. Chem. Biol. 2015;22(11):1461–1469. doi: 10.1016/j.chembiol.2015.09.012. [DOI] [PubMed] [Google Scholar]
- 59.Halestrap A.P. The mitochondrial pyruvate carrier: has it been unearthed at last? Cell Metabol. 2012;16(2):141–143. doi: 10.1016/j.cmet.2012.07.013. [DOI] [PubMed] [Google Scholar]
- 60.Bunik V.I. Redox-driven signaling: 2-oxo acid dehydrogenase complexes as sensors and transmitters of metabolic imbalance. Antioxidants Redox Signal. 2019;30(16):1911–1947. doi: 10.1089/ars.2017.7311. [DOI] [PubMed] [Google Scholar]
- 61.Bunik V.I., Brand M.D. Generation of superoxide and hydrogen peroxide by side reactions of mitochondrial 2-oxoacid dehydrogenase complexes in isolation and in cells. Biol. Chem. 2018;399(5):407–420. doi: 10.1515/hsz-2017-0284. [DOI] [PubMed] [Google Scholar]
- 62.Tabatabaie T., Potts J.D., Floyd R.A. Reactive oxygen species-mediated inactivation of pyruvate dehydrogenase. Arch. Biochem. Biophys. 1996;336(2):290–296. doi: 10.1006/abbi.1996.0560. [DOI] [PubMed] [Google Scholar]
- 63.Humphries K.M., Szweda L.I. Selective inactivation of alpha-ketoglutarate dehydrogenase and pyruvate dehydrogenase: reaction of lipoic acid with 4-hydroxy-2-nonenal. Biochemistry. 1998;37(45):15835–15841. doi: 10.1021/bi981512h. [DOI] [PubMed] [Google Scholar]
- 64.Fisher-Wellman K.H., Gilliam L.A.A., Lin C.T., Cathey B.L., Lark D.S., Darrell Neufer P. Mitochondrial glutathione depletion reveals a novel role for the pyruvate dehydrogenase complex as a key H2O2-emitting source under conditions of nutrient overload. Free Radic. Biol. Med. 2013;65:1201–1208. doi: 10.1016/j.freeradbiomed.2013.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fisher-Wellman K.H., Lin C.T., Ryan T.E., Reese L.R., Gilliam L.A., Cathey B.L., Lark D.S., Smith C.D., Muoio D.M., Neufer P.D. Pyruvate dehydrogenase complex and nicotinamide nucleotide transhydrogenase constitute an energy-consuming redox circuit. Biochem. J. 2015;467(2):271–280. doi: 10.1042/BJ20141447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.O'Brien M., Chalker J., Slade L., Gardiner D., Mailloux R.J. Protein S-glutathionylation alters superoxide/hydrogen peroxide emission from pyruvate dehydrogenase complex. Free Radic. Biol. Med. 2017;106:302–314. doi: 10.1016/j.freeradbiomed.2017.02.046. [DOI] [PubMed] [Google Scholar]
- 67.Chalker J., Gardiner D., Kuksal N., Mailloux R.J. Characterization of the impact of glutaredoxin-2 (GRX2) deficiency on superoxide/hydrogen peroxide release from cardiac and liver mitochondria. Redox Biol. 2018;15:216–227. doi: 10.1016/j.redox.2017.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Robertson R.P., Harmon J., Tran P.O., Tanaka Y., Takahashi H. Glucose toxicity in beta-cells: type 2 diabetes, good radicals gone bad, and the glutathione connection. Diabetes. 2003;52(3):581–587. doi: 10.2337/diabetes.52.3.581. [DOI] [PubMed] [Google Scholar]
- 69.Gill R.M., O'Brien M., Young A., Gardiner D., Mailloux R.J. Protein S-glutathionylation lowers superoxide/hydrogen peroxide release from skeletal muscle mitochondria through modification of complex I and inhibition of pyruvate uptake. PloS One. 2018;13(2) doi: 10.1371/journal.pone.0192801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Haussinger D., Lamers W.H., Moorman A.F. Hepatocyte heterogeneity in the metabolism of amino acids and ammonia. Enzyme. 1992;46(1–3):72–93. doi: 10.1159/000468779. [DOI] [PubMed] [Google Scholar]
- 71.Owen O.E., Kalhan S.C., Hanson R.W. The key role of anaplerosis and cataplerosis for citric acid cycle function. J. Biol. Chem. 2002;277(34):30409–30412. doi: 10.1074/jbc.R200006200. [DOI] [PubMed] [Google Scholar]
- 72.Mailloux R.J., Willmore W.G. S-glutathionylation reactions in mitochondrial function and disease. Front Cell Dev Biol. 2014;2:68. doi: 10.3389/fcell.2014.00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Heublein M., Burguillos M.A., Vogtle F.N., Teixeira P.F., Imhof A., Meisinger C., Ott M. The novel component Kgd4 recruits the E3 subunit to the mitochondrial alpha-ketoglutarate dehydrogenase. Mol. Biol. Cell. 2014;25(21):3342–3349. doi: 10.1091/mbc.E14-07-1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tretter L., Adam-Vizi V. Inhibition of Krebs cycle enzymes by hydrogen peroxide: a key role of [alpha]-ketoglutarate dehydrogenase in limiting NADH production under oxidative stress. J. Neurosci. 2000;20(24):8972–8979. doi: 10.1523/JNEUROSCI.20-24-08972.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nulton-Persson A.C., Starke D.W., Mieyal J.J., Szweda L.I. Reversible inactivation of alpha-ketoglutarate dehydrogenase in response to alterations in the mitochondrial glutathione status. Biochemistry. 2003;42(14):4235–4242. doi: 10.1021/bi027370f. [DOI] [PubMed] [Google Scholar]
- 76.Tretter L., Adam-Vizi V. Alpha-ketoglutarate dehydrogenase: a target and generator of oxidative stress. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2005;360(1464):2335–2345. doi: 10.1098/rstb.2005.1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Applegate M.A., Humphries K.M., Szweda L.I. Reversible inhibition of alpha-ketoglutarate dehydrogenase by hydrogen peroxide: glutathionylation and protection of lipoic acid. Biochemistry. 2008;47(1):473–478. doi: 10.1021/bi7017464. [DOI] [PubMed] [Google Scholar]
- 78.McLain A.L., Cormier P.J., Kinter M., Szweda L.I. Glutathionylation of alpha-ketoglutarate dehydrogenase: the chemical nature and relative susceptibility of the cofactor lipoic acid to modification. Free Radic. Biol. Med. 2013;61:161–169. doi: 10.1016/j.freeradbiomed.2013.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Slade L., Chalker J., Kuksal N., Young A., Gardiner D., Mailloux R.J. Examination of the superoxide/hydrogen peroxide forming and quenching potential of mouse liver mitochondria. Biochim. Biophys. Acta Gen. Subj. 2017;1861(8):1960–1969. doi: 10.1016/j.bbagen.2017.05.010. [DOI] [PubMed] [Google Scholar]
- 80.Deng P., Haynes C.M. The mitokine quest(ion) Cell Res. 2016;26(12):1265–1266. doi: 10.1038/cr.2016.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Horn A., Van der Meulen J.H., Defour A., Hogarth M., Sreetama S.C., Reed A., Scheffer L., Chandel N.S., Jaiswal J.K. Mitochondrial redox signaling enables repair of injured skeletal muscle cells. Sci. Signal. 2017;10(495) doi: 10.1126/scisignal.aaj1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kramer P.A., Duan J., Qian W.J., Marcinek D.J. The measurement of reversible redox dependent post-translational modifications and their regulation of mitochondrial and skeletal muscle function. Front. Physiol. 2015;6:347. doi: 10.3389/fphys.2015.00347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mailloux R.J., Craig Ayre D., Christian S.L. Induction of mitochondrial reactive oxygen species production by GSH mediated S-glutathionylation of 2-oxoglutarate dehydrogenase. Redox Biol. 2016;8:285–297. doi: 10.1016/j.redox.2016.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mallay S., Gill R., Young A., Mailloux R.J. Sex-dependent differences in the bioenergetics of liver and muscle mitochondria from mice containing a deletion for glutaredoxin-2. Antioxidants (Basel) 2019;8(8) doi: 10.3390/antiox8080245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Conway M.E., Coles S.J., Islam M.M., Hutson S.M. Regulatory control of human cytosolic branched-chain aminotransferase by oxidation and S-glutathionylation and its interactions with redox sensitive neuronal proteins. Biochemistry. 2008;47(19):5465–5479. doi: 10.1021/bi800303h. [DOI] [PubMed] [Google Scholar]
- 86.Murphy M.P. How mitochondria produce reactive oxygen species. Biochem. J. 2009;417(1):1–13. doi: 10.1042/BJ20081386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Han D., Canali R., Garcia J., Aguilera R., Gallaher T.K., Cadenas E. Sites and mechanisms of aconitase inactivation by peroxynitrite: modulation by citrate and glutathione. Biochemistry. 2005;44(36):11986–11996. doi: 10.1021/bi0509393. [DOI] [PubMed] [Google Scholar]
- 88.Kil I.S., Park J.W. Regulation of mitochondrial NADP+-dependent isocitrate dehydrogenase activity by glutathionylation. J. Biol. Chem. 2005;280(11):10846–10854. doi: 10.1074/jbc.M411306200. [DOI] [PubMed] [Google Scholar]
- 89.Mailloux R.J., Xuan J.Y., McBride S., Maharsy W., Thorn S., Holterman C.E., Kennedy C.R., Rippstein P., deKemp R., da Silva J., Nemer M., Lou M., Harper M.E. Glutaredoxin-2 is required to control oxidative phosphorylation in cardiac muscle by mediating deglutathionylation reactions. J. Biol. Chem. 2014;289(21):14812–14828. doi: 10.1074/jbc.M114.550574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wu H., Yu Y., David L., Ho Y.S., Lou M.F. Glutaredoxin 2 (Grx2) gene deletion induces early onset of age-dependent cataracts in mice. J. Biol. Chem. 2014;289(52):36125–36139. doi: 10.1074/jbc.M114.620047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Drose S., Stepanova A., Galkin A. Ischemic A/D transition of mitochondrial complex I and its role in ROS generation. Biochim. Biophys. Acta. 2016;1857(7):946–957. doi: 10.1016/j.bbabio.2015.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chen Y.R., Chen C.L., Pfeiffer D.R., Zweier J.L. Mitochondrial complex II in the post-ischemic heart: oxidative injury and the role of protein S-glutathionylation. J. Biol. Chem. 2007;282(45):32640–32654. doi: 10.1074/jbc.M702294200. [DOI] [PubMed] [Google Scholar]
- 93.Chouchani E.T., Pell V.R., Gaude E., Aksentijevic D., Sundier S.Y., Robb E.L., Logan A., Nadtochiy S.M., Ord E.N.J., Smith A.C., Eyassu F., Shirley R., Hu C.H., Dare A.J., James A.M., Rogatti S., Hartley R.C., Eaton S., Costa A.S.H., Brookes P.S., Davidson S.M., Duchen M.R., Saeb-Parsy K., Shattock M.J., Robinson A.J., Work L.M., Frezza C., Krieg T., Murphy M.P. Ischaemic accumulation of succinate controls reperfusion injury through mitochondrial ROS. Nature. 2014;515(7527):431–435. doi: 10.1038/nature13909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Doulias P.T., Tenopoulou M., Greene J.L., Raju K., Ischiropoulos H. Nitric oxide regulates mitochondrial fatty acid metabolism through reversible protein S-nitrosylation. Sci. Signal. 2013;6(256):rs1. doi: 10.1126/scisignal.2003252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Giangregorio N., Palmieri F., Indiveri C. Glutathione controls the redox state of the mitochondrial carnitine/acylcarnitine carrier Cys residues by glutathionylation. Biochim. Biophys. Acta. 2013;1830(11):5299–5304. doi: 10.1016/j.bbagen.2013.08.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.