Abstract
The Millettioid/Phaseoloid (MP) clade from the subfamily Papilionoideae (Leguminosae) consists of six tribes and ca. 3,000 species. Previous studies have revealed some plastome structural variations (PSVs) within this clade. However, many deep evolutionary relationships within the clade remain unresolved. Due to limited taxon sampling and few genetic markers in previous studies, our understanding of the evolutionary history of this clade is limited. To address this issue, we sampled 43 plastomes (35 newly sequenced) representing all the six tribes of the MP clade to examine genomic structural variations and phylogenetic relationships. Plastomes of the species from the MP clade were typically quadripartite (size ranged from 140,029 to 160,040 bp) and contained 109–111 unique genes. We revealed four independent gene losses (ndhF, psbI, rps16, and trnS-GCU), multiple IR-SC boundary shifts, and six inversions in the tribes Desmodieae, Millettieae, and Phaseoleae. Plastomes of the species from the MP clade have experienced significant variations which provide valuable information on the evolution of the clade. Plastid phylogenomic analyses using Maximum Likelihood and Bayesian methods yielded a well-resolved phylogeny at the tribal and generic levels within the MP clade. This result indicates that plastome data is useful and reliable data for resolving the evolutionary relationships of the MP clade. This study provides new insights into the phylogenetic relationships and PSVs within this clade.
Keywords: evolutionary relationships, inversion, IR expansion/contraction, Leguminosae, Plastome, the Millettioid/Phaseoloid clade
Introduction
The plastid genome (plastome) usually shows a quadripartite structure including a large-single-copy (LSC, 60–90 kb) region, a small-single-copy (SSC, 7–27 kb) region, and a pair of inverted repeats (IR, 20–76 kb) (Raubeson and Jansen, 2005; Zhang et al., 2018). As a circular genome of about 108–218 kb in size, the plastome contains ca. 90–130 unique genes including 80–90 protein-coding genes (PCGs), 30–31 transfer RNA (tRNAs) and 4 ribosomal RNAs (rRNAs) (Bock, 2007; Wu et al., 2009; Green, 2011). The plastome structure of autotrophic plants is usually conserved (Kim and Jansen, 2005; Lin et al., 2015). However, significant structural variations including IR loss, IR contraction/expansion, inversion, pseudogenization, gene duplication, and gene loss have been reported in some gymnosperms (Wu and Chaw, 2016) and angiosperm families such as Campanulaceae (Cosner et al., 1997; Haberle et al., 2008), Geraniaceae (Chumley et al., 2006; Guisinger et al., 2011; Weng et al., 2014), Oleaceae (Lee et al., 2007), Petrosaviaceae (Logacheva et al., 2014), and Leguminosae (Lavin et al., 1990; Wojciechowski, 2003; Luo et al., 2016; Choi and Choi, 2017; Wang et al., 2017).
Some species of Leguminosae, especially those of the subfamily Papilionoideae, have acquired significant plastome structural variations (PSVs) during their evolution. These PSVs includes loss of IR (e.g., Lavin et al., 1990; Doyle et al., 1996), gene or plastome segment inversion (Choi and Choi, 2017), IR expansion, and/or contraction (Choi and Choi, 2017), and gene loss (Jansen et al., 2007; Sabir et al., 2014; Asaf et al., 2017). Most members of papilionoids, with the exception of a few early diverging lineages, share a 50-kb inversion in the LSC (Doyle et al., 1996). Previous studies have reported multiple inversions of 23, 24, or 36-kb in the Genistoid clade (Martin et al., 2014; Choi and Choi, 2017; Feng et al., 2017; Keller et al., 2017), a 39-kb inversion in Robinia (Schwarz et al., 2015), and a large 78-kb inversion in the subtribe Phaseolinae of tribe Phaseoleae (Bruneau et al., 1990). However, only a few studies have examined PSV in the Millettioid/Phaseoloid clade (hereafter referred as the MP clade), one of the most species-rich clades within subfamily Papilionoideae.
The MP clade consists of more than 3,000 extant species with a global distribution (Schrire, 2005a; Schrire, 2005b; Schrire, 2005c and Schrire, 2005d; Schrire et al., 2009). Many species of this clade are economically important (Simpson and Ogorzaly, 2001; Baker, 2004), as edible seeds [Glycine max (L.) Merr (soybean), Cajanus cajan (L.) Millsp. (pigeon pea), Phaseolus vulgaris L. (kidney bean), Vigna unguiculata (L.) Walp. (cowpea), and Pachyrhizus erosus (L.) Urb (Mexican yam bean)], medicines [Abrus precatorius L. (crab eye)], ornamentals [Canavalia gladiata (Jacq.) DC. (Sword beans pea) and Millettia pinnata (L.) Panigrahi (Indian beech)], forages [Pueraria phaseoloides Benth (tropical kudzu)], and woods [M. laurentii De Wild. (African rosewood)].
Some previous studies based on nuclear ribosomal ITS (Hu et al., 2002) and a few plastid loci (Hu et al., 2000; Kajita et al., 2001; Pennington et al., 2001; Wojciechowski et al., 2004; Cardoso et al., 2013; LPWG, 2017) have made progress in clarifying evolutionary relationships of the MP clade. However, some deep relationships, particularly at tribe level, have not been fully resolved, perhaps due to limited phylogenetic signals in these gene loci. Whole plastome sequences have been successfully applied to resolve plant evolutionary relationships (Jansen et al., 2007), and therefore they might be of use for clarifying unresolved relationships in the MP clade. A few recent studies using limited samples have detected multiple types of PSV in this clade, such as a 78-kb inversion in Vigna radiata (L.) R. Wilczek and P. vulgaris, a 36-kb inversion in Lupinus luteus L. (Martin et al., 2014), the loss of rps16 gene in Cajanus Adans. (Guo et al., 2007; Schwarz et al., 2015), the loss of rpl2 and clpP introns (Kaila et al., 2016), and IR contraction/expansion in G. max (Saski et al., 2005; Kim et al., 2015). Investigation of the plastome of more taxa of this clade is essential for a better understanding of PSVs across this clade. In this study, we analyzed plastomes of 43 species (35 newly sequenced) representing all the six tribes of the MP clade. We investigated plastome structural diversification, and conducted phylogenetic reconstruction of the clade using plastome sequences. Deep phylogenetic relationships of the MP clade were investigated using the coding genes (CDs), noncoding regions (NCDs) and complete plastomes (CP). Our study provides important new insights into both phylogenetic relationships and PSVs within the MP clade.
Materials and Methods
Taxon Sampling, DNA Extraction, and Genome Sequencing
For this study, we used a total of plastomes of 43 species from the MP clade including one plastome from NCBI, seven plastomes from Zhang et al. (2020)’s phylogenetic study of the whole family, and newly sequenced plastomes of 35 species from 35 genera ( Supplementary Table S1 ). These species were selected based on the availability of tissues for sampling and their representation of previously recognized tribes in the clade (LPWG, 2013). Total genomic DNA (gDNA) was extracted from either fresh or silica-gel dried leaves using the modified CTAB method (Doyle and Doyle, 1987). The genome skimming method was used to obtain the plastome data (Zeng et al., 2018). The gDNA was fragmented and libraries size were selected for 350 bp inserts. Sequencing with 2 × 150-bp paired-end (PE) reads was performed on the Illumina Hiseq 2500/X-Ten at the Novogene (Tianjin, China) or Illumina Hiseq 2000/2500/4000/X-Ten at the Beijing Genomics Institute (BGI) in Shenzhen, China.
Plastome Assembly and Annotation
The clean-up and quality control checks of the raw reads were performed using the Next Generation Sequencing (NGS) QC Tool Kit with default settings (Patel and Jain, 2012). Then, we assembled contigs from the PE reads via de novo assembly using GetOrganelle (Jin et al., 2019) with K-mer values 21, 45, 65, 85, 105, and 127 calling SPAdes version 3.10 (Bankevich et al., 2012), using a reference genome from subfamily Papilionoideae (Arachis hypogaea L., NC_026676). Bandage v.0.80 (Wick et al., 2015) was used to visualize and filter the assembled contigs to generate a complete circular plastome. For incomplete plastomes, we filled the gaps between the contigs with consensus sequences of raw reads that were initially mapped to the reference plastome in order to obtain the complete plastome. The number of the mapped PE reads and the coverage depth were determined by mapping the paired reads against the plastome using Bowtie2 (Langmead and Salzberg, 2012) incorporated in Geneious v. 8.1.4 (Kearse et al., 2012).
The locations of the single copy (SC) and IR boundaries in the newly sequenced plastomes were determined using the same methods as Qu et al. (2019). The ‘find repeat’ function in Geneious was used to flank the IR regions. Then, the paired reads were remapped to the assembled plastomes to validate the SC/IR regions using Bowtie2. Finally, we visualized the read stacks of the newly assembled plastomes and compared the marked SC/IR boundaries in Geneious. The new plastomes were annotated using Dual Organellar Genome Annotator (DOGMA) web-interface (Wyman et al., 2004). We manually checked the consistency of start/stop codons and intron/exon boundaries in Geneious. The ‘Find ORFs’ function in Geneious was used to re-confirm the PCGs annotations, while tRNAscan-SE web service was applied to determine the tRNA genes (Schattner et al., 2005). The OrganellarGenomeDRAW [web server, (Lohse et al., 2013)] was used to draw the physical genomic map ( Supplementary Figure S1 ). Finally, the complete newly assembled plastomes (35 in the MP clade and four outgroup species) were deposited in GenBank ( Supplementary Table S1 ).
Plastome Structural Analysis
To investigate the patterns of genomic evolution, we analyzed and compared the structural characteristics of the 43 annotated plastomes. We examined structural characteristics such as plastome size (bp), LSC length (bp), SSC length (bp), IR length (bp), GC content (%), and gene distributions of all studied plastomes (Li et al., 2013; Supplementary Table S2 ; Table 1 ). For the contraction and expansion analysis, we compared the newly sequenced plastomes of the species from the MP clade with the A. hypogaea plastome. Afterward, we examined the variation of the genes located at the plastome termini and the boundary shifts (IR-SC) in the four junctions (JLB –LSC/IRB, JSB –IRB/SSC, JSA –SSC/IRA, and JLA –IRA/LSC) ( Supplementary Figure S2 ). To confirm inversions, we aligned the 43 plastomes of species from the MP clade with the A. hypogaea plastome using the progressiveMauve algorithm (Wang et al., 2017). We used default settings to automatically calculate the seed weight (15), and calculated Locally Collinear Blocks (LCBs) with the minimum LCB score of 30,000 (Darling et al., 2004). The detected inversions were illustrated in Figure 3 and Supplementary Figure S3 .
Table 1.
Plastome content and their functions in the MP clade.
| Function | Gene group | Gene |
|---|---|---|
| Biosynthesis of fatty acids | Acetyl-CoA carboxylase | accD |
| Genetic apparatus | Large subunit of ribosomal protein | rpl2 a,b, rpl14 c, rpl16 c,e,f, rpl23, rpl32, rpl33, rpl36 c |
| Small subunit of ribosomal protein | rps2, rps3 c, rps4, rps7, rps8 c, rps11 c, rps12 g , rps14, rps15, rps16, rps18, rps19 h | |
| Subunits of RNA polymerase | rpoA, rpoB, rpoC1, rpoC2 | |
| Photosynthesis pathway | Photosystem I | psaA, psaB, psaI, psaJ |
| Photosystem I assembly | ycf3, ycf4 | |
| Photosystem II | psbA-N, psbT, psbZ | |
| F-type ATP synthase | atpA, atpB, atpE, atpF, atpH, atpI | |
| NADH-plastoquinone oxidoreductase | ndhA, ndhB i , ndhC, ndhD, ndhE, ndhF, ndhG, ndhH, ndhI, ndhJ, ndhK | |
| Component of cytochrome b6/f Complex | petA, petB, petD, petG, petL, petN | |
| Carbon metabolism | cemA | |
| Cytochrome c biogenesis protein | ccsA | |
| Large subunit of Rubisco | rbcL | |
| Protein-modifying | ATP-dependent protease proteolytic subunit | clpP |
| Structural RNAs | Transfer RNAs | trRNA-Ala (trnA-UGC), -Arg (trnR-ACG , trnR-UCU), Asn (trnN-GUU) j , -Cys (GCA), -Gln (trnQ-UUG),-Glu (trnE-UUC), -fMet (trnfM-CAU), - Gly (trnG- GCC, trnG-UCC), -His (trnH-GUG)d , -Ile (trnI-CAU, trnI-GAU), -Lys (trnK-UUU), -Leu (trnL-CAA , trnL-UAA, trnL-UAG), -Met (trnM-CAU), -Phe (trnF-GAA), -Pro (trnP-UGG), -Ser (trnS-GCU, trnS-GGA, trnS-UGA), -Thr (trnT-GGU, trnT-UGU), -Trp (trnW-CCA), -Tyr (trnY-GUA), -Val (trnV-GAC , trnV-UAC), |
| Ribosomal RNAs | rrn16, rrn23, rrn4.5, rrn5 | |
| Post-transcriptional modification | Maturase | matK |
| Protein of unknown function | Other genes | ycf1 k , ycf2 |
Boldface for genes duplicated in the IR regions,
Exon not duplicated in H. ormocarpioides.
Intron 1 duplicated in S. macrobotrys.
Duplicated in the IR expansion of C. cathartica and S. macrobotrys respectively.
Duplicated exon 1 and 2 in C. cathartica.
Duplicated intron 1 in C. cathartica.
trans-spliced gene.
Duplicated in the IR of all species except D. araripensis, L. domingensis, O. pinnata, P. violacea, X. stuhlmannii, I. linifolia and tinctoria.
Duplicated in the IR of C. pubescens and D. falciformis.
Duplicated in the IR of all species except L. domingensis.
Duplicated in C. carthatica and L. cuneata.
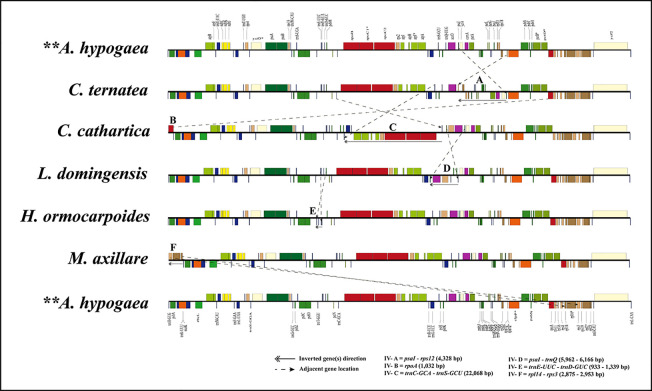
Figure 3.
Plastome inversions in representative species of the MP clade. Gene arrangement as the reference plastome of Arachis hypogaea.
Phylogenetic Analysis
A total of 49 plastomes (including 43 species of the MP clade and six outgroups) were used for the phylogenetic analysis. The outgroups included two loosely related species of the subfamily Caesalpinioideae (Tamarindus indica L., NC026685, and Ceratonia siliqua L., NC026678) with plastome data downloaded from GenBank, and four more closely related species (newly sequenced) of the subfamily Papilionoideae [Parochetus communis Buch.-Ham. ex D.Don, Kotschya aeschynomenoides (Welw. ex Baker) Dewit & P.A.Duvign., Pterocarpus violaceus Vogel, and Podalyria calyptrata Willd.]. We could not perform whole plastome alignment due to high PSVs in the legume plastomes. For this reason, we used the python script “get_annotated_regions_from_gb” (https://github.com/Kinggerm/PersonalUtilities) to extract the CDs and NCDs from the plastomes. We performed individual gene/region alignment in MAFFT v.7.4.0 (Katoh and Standley, 2013) with LINSI algorithm. All alignments were visualized and manually adjusted in Geneious. To reduce systematic error, we excluded noncoding loci with less than 70% taxon occupancy or alignment lengths less than 100 bp. We generated three data matrices for the phylogenetic analyses that included the CDs (81 genes for all species), NCDs (113 loci for all species), and CP (concatenated CDs and NCDs for all species).
The substitution models for the three data matrices were determined using PartitionFinder2 v.2.1.1 (Lanfear et al., 2017). The evolutionary best fit models and data partitioning schemes ( Supplementary Table S3 ) were selected using the corrected Akaike Information Criterion (AICc). Phylogenetic relationships were reconstructed using Maximum Likelihood (ML) and Bayesian Inference (BI). The ML analysis was performed using the IQ-TREE (Nguyen et al., 2015; Chernomor et al., 2016). We used the best partitioning schemes, -spp option (allowing partition-specific rates), and the ultrafast bootstrap replicates at 1000 for the analyses. The BI was performed using MrBayes v.3.2.6 (Ronquist and Huelsenbeck, 2003). The Bayesian posterior probability (PP) was estimated with two independent Markov Chain Monte Carlo (MCMC) runs, which included one cold chain and three hot chains for 10,000,000 generations and the tree sampling frequency at every 1,000 generations. The MCMC convergence was determined, and the first 20% were discarded as burn-in using TRACER v.1.6 (Rambaut and Drummond, 2004). Each parameter for each run obtained a sufficient effective sample size (ESS > 250). The majority-rule consensus tree was generated from the post burn-in trees. The resulting trees (ML and BI) were viewed and edited in FigTree v.1. 3.1 software (Rambaut, 2009).
Results
Plastome Organization and Size
The mean plastome coverage ranged between 162.0 × (Philenoptera violacea (Klotzsch) Schrire, Millettieae) and 1,536.4 × [Cajanus crassus (Prain ex King) Maesen, Phaseoleae]. The plastomes of the 43 species from the MP clade exhibited a typical quadripartite structure ( Figure 1 ; Supplementary Figure S1 ). The plastome size ranged from 148,889 bp in Lonchocarpus domingensis DC. of Millettieae to 160,040 bp in Indigofera linifolia (L.f.) Retz. of Indigofereae. Substantial length variation was evident in the LSC, ranging from 77,970 bp in Canavalia cathartica Thouars. of Phaseoleae to 90,459 bp in I. linifolia of Indigofereae. The SSC length ranged from 14,869 bp in Strongylodon macrobotrys A.Gray of Phaseoleae to 18,965 bp in C. cathartica. Finally, the IR ranged from 24,111 bp in Desmodium renifolium Schindl. of Desmodieae to 30,644 bp in C. cathartica ( Supplementary Table S2 ). We observed only marginal variation in the GC content, which ranged from 34.2% in Dolichos falciformis E.Mey. of Phaseoleae to 35.8% in Indigofera spp. of Indigofereae ( Supplementary Table S2 ).
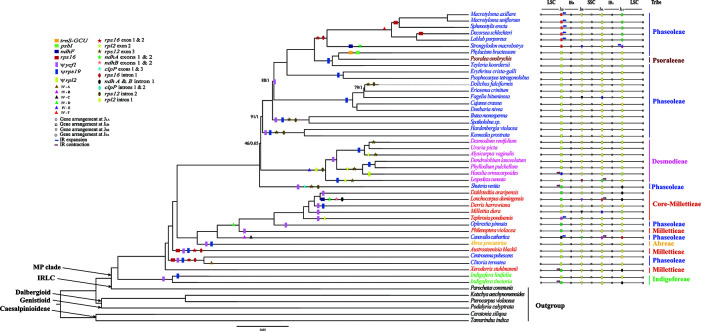
Figure 1.
The ML tree of the MP clade reconstructed based on the CP and the variation of IR/SC junctions. Numbers at nodes correspond to ML bootstrap percentages (only values <100% are shown) and Bayesian inference (BI) posterior probabilities (only probabilities <1.0 are shown). Genes loss, pseudogenes, inversions (IV), exon and intron loss, in the plastome, are indicated on the branches using coloured squares, rectangles, triangles, stars and oval shapes, respectively. The IR expansion and contraction are shown by blue and red arrow, respectively.
Each plastome contained 109–111 unique genes, including 73–90 PCGs, 30 tRNAs, and four rRNAs ( Table 1 ). Nine genes (atpF, ndhA, ndhB, petB, petD, rpl2, rpl16, rpoC1, and rps16) had one intron, while two genes (clpP and ycf3) had two introns ( Table 1 ). The rps12 gene of most species was trans-spliced into three exons (exon 1 in the LSC, and exons 2 and 3 in the IR). Four genes were absent from some species and lineages: the rps16 gene from Austrosteenisia blackii (F.Muell.) R.Geesink, Centrosema pubescens Benth., Clitoria ternatea L., Decorsea schlechteri (Harms) Verdc., Macrotyloma axillare (E.Mey.) Verdc., Macrotyloma uniflorum (Lam.) Verdc., and Sphenostylis erecta Hutch. ex Baker f. (Phaseoleae); the psbI and trnS-GCU genes from Phylacium bracteosum Benn. (Phaseoleae); and the ndhF gene from L. domingensis (Millettieae) and S. macrobotrys (Phaseoleae). We detected pseudogenization of ycf1, rpl2, and rps19 in one to multiple species ( Figure 1 ; Supplementary Figure S2 ).
Plastome Structural Variations in the MP Clade
The locations of IR-SC junctions in many species of the MP clade have experienced significant variations in some species ( Figures 1 and 2 ; Supplementary Figure S2 ). Mostly, the SSC/IRB (JSB) border lies within the ndhF gene, with the duplication of 3’-ends of this gene (from 1 bp in Psophocarpus tetragonolobus DC. to 53 bp in C. cathartica) at the boundary of the IRA/SSC junction (JSA). However, some species contracted their IRs following the shift of the JSB into the IGS region. The JSB lies within the IGS region between trnN and rpl32 in S. macrobotrys because of the loss of ndhF. Instead, the JSB lies within the IGS region between trnR and rpl32 in L. domingensis because of the loss of ndhF and the translocation of trnN into the SSC region. The JSA lies within the ycf1 gene in most species, with the duplication of 3’-ends of this gene (from 374 bp in Shuteria vestita Wight & Arn. to 1,240 bp in Erythrina crista-galli L.) at the boundary of the JSB. The IR is contracted at this boundary following the shift of JSA into the IGS region between ycf1 and trnN in Lespedeza cuneata G.Don and C. cathartica, between trnN and trnR in L. domingenesis.
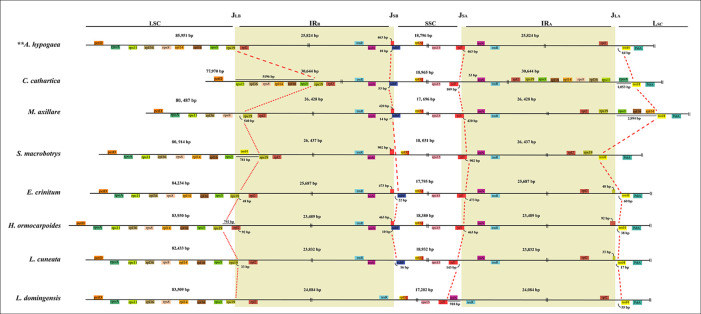
Figure 2.
Comparison of LSC, IRs, and SSC junction positions among plastomes of the MP clade. JLB, JSB, JSA, JLA refer to junctions of LSC/IRB, SSC/IRB, SSC/IRA, LSC/IRA, respectively.
Typically, the LSC/IRB junction (JLB) lies within the rps19 gene, resulting in the duplication of the 5’-ends of this gene (from 2 bp in A. blackii to 68 bp in E. crista-galli) at the boundary of the IRA/LSC junction (JLA). The JLB has experienced expansion into the LSC by 5,196-bp in C. cathartica to include the intact rps3, rps8, rps11, rpl36, rps14, rps16, and rps19 genes. The JLB lies between petD and rps11 in C. cathartica and between rps19 and rps8 in M. axillare, M. uniflorum, S. erecta, D. schlechteri, and Lablab purpureus (L.) Sweet. Also, the JLB lies between rps3 and trnH in S. macrobotrys, and between rps3 and rps19 in Tephrosia pondoensis (Codd) Schrire. Likewise, the JLB has experienced contraction into rpl2 in Hanslia ormocarpoides (DC.) H. Ohashi, and the IGS region between rps19 and rpl2 in S. vestita, Dahlstedtia araripensis (Benth.) M.J.Silva & A.M.G.Azevedo, L. domingensis, Ophrestia pinnata (Merr.) Verdc., P. violacea, Xeroderris stuhlmannii (Taub.) Mendonça & E.P.Sousa, Indigofera tinctoria Gouan and Millettia dura Dunn. The JLA is mostly between rps19 and rpl2 in the IR and trnH in the LSC. However, the JLA lies between rps11 and rpoA in C. cathartica, between rps19 and rps3 in M. axillare, M. uniflorum, S. erecta, D. schlechteri and L. purpureus, and between trnH and psbA in S. macrobotrys.
Multiple inversions (IVs A to F) and intragenomic relocations were detected in the LSC region of some species in the MP clade ( Figure 3 ; Supplementary Figure S3 ), including a 4,328-kb inversion (IV A) from psaI to rps12 in C. ternatea; a 1,032-kb inversion (IV B) of the rpoA gene; a 22,060-kb inversion (IV C) from trnC-GCA to trnS-GCU in C. cathartica; a 5,962- to 6,166-bp inversion (IV D) from trnQ-UUG to psaI in the P. violacea + O. pinnata + T. pondoensis + M. dura + Derris harrowiana + L. domingensis + D. araripensis clade; a 933- to 1,339-bp inversion (IV E) from trnE-UUC to trnD-GUC in the L. cuneata + H. ormocarpoides + Phyllodium pulchellum (L.) Desv. + Dendrolobium lanceolatum (Dunn) Schindl. + Alysicarpus vaginalis (Schumach.) J.Léonard + Uraria picta (Jacq.) Desv. ex DC. + D. renifolium clade; and a 2,875- to 2,952-kb inversion (IV F) from rpl14 to rps3 in the L. purpureus + D. schlechteri + S. erecta + M. uniflorum + M. axillare clade. Interestingly, the rpoA gene translocated from one end of the LSC near the JLB to another end of the LSC near the JLA in C. cathartica. Additionally, the segment comprising the genes rpl14, rpl16 and rps3 was translocated from one end of the LSC near the JLB to another end of the LSC near JLA in a subclade of the tribe Phaseoleae (L. purpureus + D. schlechteri + S. erecta + M. uniflorum + M. axillare).
Phylogenetic Relationships of the MP Clade
The phylogenies of the MP clade inferred from the three data matrices and two methods (ML and BI) yielded largely similar topologies, including well-resolved deep relationships of the MP clade ( Figure 4 ). Our phylogenetic analyses strongly supported (BS ≥ 95 %, and PP = 1.0) the monophyly of the MP clade and most lineages. However, the lineage consisting of Butea monosperma (Lam.) Kuntze and Spatholobus Hassk sp. has different phylogenetic position in trees of CP and NCDs, and that of CDs, but both relationships were weakly supported. Also, the tribe Desmodieae was weakly supported to bemonophyletic in CP and NCDs datamatrices whereas strongly supported by CDs data. The tribe Indigofereae was strongly supported as sister to the remainder of the MP clade (BS = 100%, and PP = 1.0). Based on the current sampling, it is not sure if the tribe Desmodieae is monophyletic, while the tribes Millettieae and Phaseoleae appear non-monophyletic. Psoralea onobrychis Nutt. of the tribe Psoraleeae was nested within a big clade of the tribe Phaseoleae.
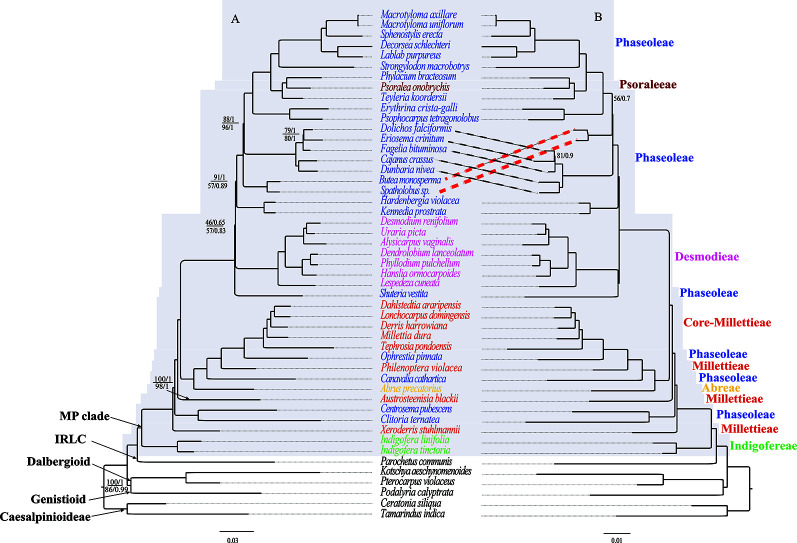
Figure 4.
The ML and BI phylogenetic relationships reconstructed for the MP clade. (A) CP and NCDs, and (B) CDs. Numbers at nodes correspond to ML bootstrap percentages (only values <100% are shown) and Bayesian inference (BI) posterior probabilities (only probabilities <1.0 are shown). For (A), the values above and below the line represents support values for the CP and NCDs, respectively. The thick dotted lines indicate topology differences. The scale bar represents the mean nucleotide substitutions per site along the branch.
Discussion
Evolutionary Pattern of PSV in the MP Clade
Gene Loss and Pseudogenization Events
Previous studies documented the loss of the genes rpl22 and infA in Lotus japonicus (Regel) K.Larsen of the Robinioid clade (Kato et al., 2000), Trifolium subterraneum L. of the IRLC (Cai et al., 2008), and G. max of the MP clade (Saski et al., 2005); this study confirmed the loss of both genes in all studied species of this clade. These two genes (rpl22 and infA) were reported lost in all the previously studied legume species (Saski et al., 2005) and almost all rosids (Millen et al., 2001). The functional copies of rpl22 and infA might have been transferred into nuclear genome [e.g., Pisum sativum L., (Gantt et al., 1991); Lupine L. species, (Martin et al., 2014)]. Previous studies suggested the loss of the ycf4 gene in Cicer L. sp., Glycine Wild. sp., and Medicago L. sp. (Magee et al., 2010; Kaila et al., 2016), or as pseudogene in P. sativum (Smith et al., 1991). Interestingly, we found ycf4 to be a normal gene in all newly sequenced plastomes of the species from the MP clade. We therefore attribute the absence of this gene in previous studies to inaccurate genome annotation, as the ycf4 gene is highly divergent (Kaila et al., 2016). The loss of the rps16 gene has been reported in some legumes (Doyle et al., 1995). Again, we detected the loss of this gene in C. pubescens, C. ternatea, D. schlechteri, S. erecta, M. uniflorum and M. axillare of the tribe Phaseoleae of the MP clade ( Figure 1 ).
The loss of introns (e.g., rpl2 intron 1) has occurred frequently in the plastomes of some angiosperm families as Convolvulaceae, Menyanthaceae, and Saxifragaceae (Downie et al., 1991), Leguminosae (Lee and Hymowitz, 2001; Jansen et al., 2008), and Lythraceae (Gu et al., 2016). Introns, especially those located at specific regions, are momentous in the transformational functionality and regulation of gene expressions (Xu et al., 2003). According to this study, with the exception of the loss of the clpP introns 1 and 2 in a single species of S. vestita (Phaseoleae) and the loss of ndh A and ndh B intron 1 in a single species of L. domingensis (Millettieae), two other introns (rps16 and rps12) have experienced multiple independent loss during the plastome evolution of the species from the MP clade. This finding agrees with the previous studies on the independent loss of rpsl2, rps16, and clpP introns in the MP clade (Guo et al., 2007; Schwarz et al., 2015; Kaila et al., 2016).
Consistent with previous studies in legumes, we observed the rps12 gene to have been trans-spliced (located in LSC region and the duplicated end in IRA) during the plastome evolution of the species from the MP clade (Fonseca and Lohmann, 2017; Wang et al., 2017). Our results showed the expression of two distinct transcripts from a single gene. Previously, the rps12 gene ligation between exon 1 and 2 had been affirmed through complementary DNA sequencing of rps12 messenger RNA (mRNA) (Sharp, 1985). Thus, this evidence suggests that the rps12 gene was trans-spliced (exon 1 and exons 2–3) because of separate transcription. Trans-spliced events of a single gene during evolution are linked with two distinct transcripts encoding protein structural domains (Sharp, 1985) and reverse transcription of the trans-spliced, sequel to the insertion in the plastome (Baltimore, 1985). The exon-rearrangement paradigm during gene evolution propounds that gene fragments coding for protein structural domains (exon) are affected by reorganization into other genes (Gilbert et al., 1986). Also, RNA trans-splicing coding for rpsl2 exon 1 with transcripts from other genes may yield polypeptide variations in the plastome. These may be the underlying factor responsible for the rps12 gene trans-splicing event in the plastomes of the species from the MP clade.
Previous studies have documented pseudogenes in some species of the MP clade, for example rps16 and rpl33 in P. vulgaris (Guo et al., 2007); ycf15, rpl33, rps16, ycf68 and ycf1 in Cajanus scarabaeoides (L.) Thouars (Kaila et al., 2016); and rps16 in Lupinus (Keller et al., 2017). Our study identified rpl2, rps19, and ycf1 as pseudogenes (based on the presence premature stop codons and their reduced length) in most species of the MP clade ( Figure 3 ; Table 1 ), while the rps16 and rpl33 genes were detected as normal genes in the species of the MP clade. The pseudogenization of these genes has been reported in other species, e.g. Melianthus villosus Bolus in Melianthaceae (Weng et al., 2014), Phalaenopsis aphrodite Rchb.f. in Orchidaceae (Chang et al., 2006), and Tylosema spp. in Mimosoideae (Wang et al., 2017). Pseudogenization of some genes is common in the plastomes of some plant taxa (Kim et al., 2015; Naumann et al., 2016; Keller et al., 2017). In previous studies, gene loss/pseudogenization in the plastome is attributed to rate of sequence evolution, gene transfer to the nucleus, or substitution by a nuclear-encoded protein for a plastid gene product (Ueda et al., 2008; Magee et al., 2010; Jansen and Ruhlman, 2012; Williams et al., 2015).
IR Contraction and Expansion
IR-SC boundary shifts played a significant role in the plastome size variation of the species from the MP clade ( Figure 1 ; Supplementary Figure S2 ). Significantly, a substantial expansion of the IR to include six ribosomal protein genes (rps3, rps8, rps11, rpl14, rpl16 and rpl36) resulted in the large plastome of C. cathartica (Phaseoleae) ( Figure 2 ; Supplementary Figure S2 ). In contrast, in L. domingensis (Millettieae), the trnN and ycf1 genes have been relocated into the SSC following IR contraction, resulting in the smallest plastome studied of the MP clade. Additionally, the contraction/expansion of IR regions in the MP clade accounts for new positions of JLA between rps11 and rpoA; rps19 and rps3, and trnH and psbA.
The IR contraction/expansions are frequent evolutionary events in angiosperm lineages, resulting in dramatic differences in the plastome length variations (e.g., Guisinger et al., 2011; Zeng et al., 2017). The rate of gene conversion during cell division/evolution and high content of short repeats (AT-rich) have also been noted as explanations for IR boundary shifts among several angiosperm lineages (Wang et al., 2008; Dugas et al., 2015; Wang et al., 2017). The same mechanisms might explain IR boundary shifts in plastomes of the species from the MP clade. The IR expansion to include the whole rps19 gene is a synapomorphic character for the M. axillare + M. uniflorum + S. erecta + D. schlechteri + L. purpureus clade. Most other IR contractions/expansions occurred independently across the MP clade.
Gene relocation within plastome has been reported in multiple previous studies (e.g., Lee et al., 2007; Kaila et al., 2016; Mower et al., 2019). For instance, the intragenomic transfer of ycf2 from the LSC region to the SSC region in lycophytes (Mower et al., 2019), the relocations of ycf3 and ycf4 within the LSC region of Menodora longiflora Engelm. ex A.Gray (Oleaceae, Lee et al., 2007), and the transfer of a block of ribosomal protein genes (rps19–rps8) from one end of the LSC region to the other end in the legumes— e.g. Vigna Savi (Perry et al., 2002), Phaseolus L. (Bruneau et al., 1990) and Cajanus spp. (Kaila et al., 2016). Similarly, our study detected translocation of genes within the LSC region in the plastomes of multiple species from the MP clade ( Figure 3 ). Additionally, we documented the relocation of a single gene (rpoA) in C. cathartica, and three ribosomal protein genes (rpl14, rpl16 and rps3) in a clade of Phaseoleae from one end of the LSC region to the other. Gene relocation can be associated with the subsequent contraction and expansion of the IR as observed in Pelargonium L’Hér. ex Aiton (Bruneau et al., 1990; Chumley et al., 2006). Alternatively, overlapping inversions and IR direction have been applied to explain the relocation of genes in the plastome of Oleaceae (Lee et al., 2007) and lycophytes (Mower et al., 2019), respectively. The IR expansion to include these genes is followed by the IR contraction at another end to relocate these genes into the SSC region. This appears to represent a more parsimonious explanation for the relocation of the rpoA gene and the segment comprising the genes rpl14, rpl16 and rps3.
Inversions
Several inversions including a 421-bp inversion in the mimosoid species (Wang et al., 2017), a 7.5-kb inversion in the Cercioideae (Kim and Cullis, 2017), and a large inversion of 50-kb in the subfamily Papilionoideae (Guo et al., 2007; Cai et al., 2008; Keller et al., 2017) occur in legumes. A few studies have documented the presence of inversions in species of the MP clade, such as V. radiata (Jansen et al., 2007), L. luteus (Martin et al., 2014), and P. vulgaris (Bruneau et al., 1990). Importantly, an early molecular investigation (Bruneau et al., 1990) on plastome DNA inversions in Papilionoideae detected a large inversion (78-kb in size) between the psbA and rps11 genes in nine species of the tribe Phaseoleae. Also, prior studies documented a 50-kb inversion that spans the genes rbcL and rps16 in the plastomes of C. cajan and C. scarabaeoides (Kaila et al., 2016) and Cyamopsis tetragonoloba (L.) Taub. (Kaila et al., 2017) in the MP clade. By analyzing additional taxa of the MP clade, we discovered six new inversions in three tribes (Desmodieae, Milletieae, and Phaseoleae) of the MP clade ( Figure 1 ; Supplementary Figure S3 ), with the largest size being 22-kb (IV-C, Figure 3 ). These newly discovered inversions significantly increase the number of documented plastome rearrangements in Leguminosae.
Inversions might be linked with IR contraction/expansion (Bruneau et al., 1990), as shown by IV-A, B, and F in the study. The regions flanking three inversions (IV-C, D, and E) contain tRNA genes, which is consistent with the assumption that tRNA activity may influence inversion in plastome (Walker et al., 2014). Also, recombination through repeated sequences can induce inversions in plastome (Rogalski et al., 2006). We failed to detect any repeats in the breakpoint regions of these six inversions. Rearrangements such as inversions in plastid genomes of land plants are considered a useful marker to infer evolutionary relationships (Doyle et al., 1992). Large inversions have been considered informative for defining clades in legumes (Bruneau et al., 1990; Doyle et al., 1996; Dugas et al., 2015). For example, the inversion (IV-E) is synapomorphy of the monophyletic tribe Desmodieae excluding S. vestita. The IV-D occurs multiple times in tribes Millettieae and Phaseoleae. The other four inversions (IV-A, B, C, and F) occur in multiple separate lineages of Phaseoleae.
Phylogenetic Relationships in the MP Clade
Appropriate data partitioning is important for achieving accurate phylogenetic result in simultaneous utilization of multiple genes (Li et al., 2013; Saarela et al., 2018; He et al., 2019), a way may greatly abate the erroneous phylogenetic inferences caused by unequal rates and patterns of nucleotide substitutions in plastomes (Li et al., 2008). Our results indicated that ML and BI analyses with multiple genes partitioned models (CDs, NCDs, and CP) presents well-resolved evolutionary relationships of the MP clade. This study underscores the utility of plastid phylogenomics for resolving intertribal and intergeneric relationships within the MP clade ( Figure 4 ). Evolutionary relationships among the major lineages, tribes, and genera were resolved with high support values. Consisted with previous studies (Hu et al., 2000; Wojciechowski et al., 2004; Cardoso et al., 2013; de Queiroz et al., 2015; LPWG, 2017), our analyses supported the tribe Indigofereae as sister to the remaining members of the MP clade. Desmodieae was supported as monophyletic group in previous studies (Bruneau et al., 1994; Doyle et al., 1997; Kajita et al., 2001; Stefanovic et al., 2009; Cardoso et al., 2013; de Queiroz et al., 2015; Egan et al., 2016), however this tribe was strongly supported as monophyletic by CDs but weakly supported by CP and NCDs ( Figure 4 ). Our phylogenetic analyses suggested the polyphyly of the tribes Millettieae and Phaseoleae, which are consistent with previous studies (Hu et al., 2000; Wojciechowski et al., 2004; de Queiroz et al., 2015; Vatanparast et al., 2018). Previous studies (Wojciechowski et al., 2004; Cardoso et al., 2013; de Queiroz et al., 2015; LPWG, 2017) included multiple genera and supported the monophyly Psoraleeae. The phylogenetic analysis of Stefanovic et al. (2009) based on eight plastid genes supported the tribe Psoraleeae as sister to Phaseoleae, whereas it is nested within the Phaseoleae in this study and several other studies (e.g., Hu et al., 2000; de Queiroz et al., 2015; Vatanparast et al., 2018).
Our study benefits from having a more comprehensive taxon sampling and involving whole plastome sequences for phylogenetic analysis; thus, it marks the beginning of a better understanding of evolutionary relationships in the MP Clade. For instance, our study highly supported the relationships of (1) C. ternatea + C. pubescens (BS = 100/PP = 1) and (2) A. blackii + C. ternatea + C. pubescens (BS = 100/PP = 1); these relationships were only weakly supported in previous studies (Kajita et al., 2001; Vatanparast et al., 2018). Notably, our multi-locus plastome data suggested (BS = 100%, PP = 1) the evolutionary position of S. vestita as sister to the tribe Desmodieae, in contrast with previous placement close to the subtribe Kennediinae of the tribe Phaseoleae (e.g., de Queiroz et al., 2015). Formerly, the genus Shuteria was included in the tribe Phaseoleae based on flower structures shared with core Phaseoleae species (e.g., Amphicarpaea Elliott ex Nutt., Cologania Kunth, and Dumasia DC., Lackey et al., 1981). It is noteworthy that a similar phylogenetic placement in the MP clade has been shown from analysis based on the single plastid region matK (de Queiroz et al., 2015). Therefore, our phylogeny supports the placement of S. vestita as sister to the tribe Desmodieae. Nevertheless, we expect that future phylogenetic studies would improve the understanding of the phylogenetic relationships of the genus Shuteria within the clade. Collectively our results provide important insights on the backbone relationships of the MP clade. However, additional phylogenetic study, perhaps integrating additional molecular data with morphological traits, will be necessary to fully clarify the evolutionary relationships of this clade.
Insights Into the Plastomic Evolution of the MP Clade
Some large inversions in the MP clade seem to have phylogenetic signal for the MP clade ( Figure 1 ). The IV-A was only found in C. ternatea, IV-B and IV-C only in C. cathartica, and IV-F in the clade of L. purpureus + M. axillare + M. uniflorum + S. erecta + D. schlechteri. The IV-E was only detected in the tribe Desmodieae, which supports the monophyly of the tribe. Of note, the IV-D is a synapomorphy of one subclade of the tribes Phaseoleae and core-Millettieae, which is congruent with their closely related evolutionary relationships. Consistent with some previous studies (Martin et al., 2014; Dugas et al., 2015; Choi and Choi, 2017), our results suggest that significant plastome structural rearrangements such as inversion may provide useful information about phylogenetic relationships. However, some previous studies have suggested caution in using inversions in phylogenetic analysis. For example, a 36-kb inversion has been documented in distantly related lineages of papilionoids (Schwarz et al., 2015). Also, a 29-kb inversion has been reported from distantly related species of Ranunculaceae (Anemone L. and Clematis L., Hoot and Palmer, 1994). Additional sampling is necessary to better evaluate the utility of large PSVs for phylogenetic reconstruction in the MP clade. The independent loss of genes, exons, and introns was observed across different lineages of the MP clade. These results are consistent with previous studies that have shown multiple independent losses of specific genes in plastomes of different plant groups (e.g., Gu et al., 2016; Kaila et al., 2016). These kinds of PSV therefore seem to have low phylogenetic signal. Similarly, pseudogenization events have occurred independently across the MP lineages, indicating that these as well are likely not useful for inferences of phylogenetic relationships. Many observed PSVs in the MP clade plastomes suggest significant structural variation following the diversification of this lineage. In total, this study provides new insights into the phylogenetic relationships and PSVs within the MP clade.
Data Availability Statement
The datasets generated for this study can be found in Genbank; the list of accession can be found in Supplementary Table 1 .
Author Contributions
T-SY, OO, and RZ designed the research. OO and RZ performed the experiments and assembled the plastomes. OO, RZ, and S-YC conducted the analysis. OO and T-SY wrote the manuscript. All authors revised the manuscript and approved the final manuscript.
Funding
This study was supported by grants from the Large-scale Scientific Facilities of the Chinese Academy of Sciences (No. 2017-LSF-GBOWS-02), the Strategic Priority Research Program of Chinese Academy of Sciences (XDB31010000), the National Natural Science Foundation of China [key international (regional) cooperative research project No. 31720103903].
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We appreciate the financial support from the aforementioned funding organizations. We gratefully appreciate everyone that participated during the plant collections. We thank the Germplasm Bank of Wild Species at the Kunming Institute of Botany (KIB) for facilitating this study; the curators and staff of the Beijing Botanical Garden (BG), Brisbane BG, Kunming BG, Missouri BG, Royal BG Edinburgh, RBG Kew, RBG Sydney, RBG Victoria (both Melbourne and Cranbourne), San Francisco BG, UC Berkeley BG, Xishuangbanna Tropical BG, and O. Maurin (Johannesburg, now Kew), J. R. Shevock (California), Y.-M. Shui (Kunming), and N. Zamora (Costa Rica) for samples; and S. R. Manchester (Florida) for critical discussion on fossil selection and calibration. We would like to acknowledge the huge assistance rendered by Jian-Jun Jin, Shu-Dong Zhang, and Xiao-Jian Qu during the data analysis.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.00151/full#supplementary-material
References
- Asaf S., Khan A. L., Khan M. A., Imran M. Q., Kang S. M., Al-Hosni K., et al. (2017). Comparative analysis of complete plastid genomes from wild soybean (Glycine soja) and nine other Glycine species. PloS One 12, e0182281. 10.1371/journal.pone.0182281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker M. (2004). Wood for Woodturners (Sussex: Guild of Master Craftsmen Publications; ). [Google Scholar]
- Baltimore D. (1985). Retroviruses and retrotransposons: the role of reverse transcription in shaping the eukaryotic genome. Cell 40, 481–482. 10.1016/0092-8674(85)90190-4 [DOI] [PubMed] [Google Scholar]
- Bankevich A., Nurk S., Antipov D., Gurevich A. A., Dvorkin M., Kulikov A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. 10.1089/cmb.2012.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock R. (2007). “Structure, function, and inheritance of plastid genomes,” in Cell and molecular biology of plastids, vol. 29–63 . Ed. Bock R. (Berlin, Heidelberg: Springer Berlin Heidelberg; ). [Google Scholar]
- Bruneau A., Doyle J. J., Palmer J. D. (1990). A chloroplast DNA Inversion as a subtribal character in the phaseoleae (Leguminosae). Syst . Botany 15, 378–386. 10.2307/2419351 [DOI] [Google Scholar]
- Bruneau A., Doyle J. J., Doyle J. A. (1994). “Phylogenetic relationships in Phaseoleae: evidence from chloroplast DNA restriction site characters,” in Advances in legume systematics, Part 7. Eds. Crisp M., Doyle J. J. (Richmond, Surrey, UK: Royal Botanic Gardens, Kew; ), 309– 330. [Google Scholar]
- Cai Z., Guisinger M., Kim H. G., Ruck E., Blazier J. C., McMurtry V., et al. (2008). Extensive reorganization of the plastid genome of Trifolium subterraneum (Fabaceae) is associated with numerous repeated sequences and novel DNA insertions. J. Mol. Evol. 67, 696–704. 10.1007/s00239-008-9180-7 [DOI] [PubMed] [Google Scholar]
- Cardoso D., Pennington R. T., de Queiroz L. P., Boatwright J. S., Van Wyk B. E., Wojciechowski M. F., et al. (2013). Reconstructing the deep-branching relationships of the papilionoid legumes. S. Afr. J. Bot. 89, 58–75. 10.1016/j.sajb.2013.05.001 [DOI] [Google Scholar]
- Chang C. C., Lin H. C., Lin I. P., Chow T. Y., Chen H. H., Chen W. H., et al. (2006). The chloroplast genome of Phalaenopsis aphrodite (Orchidaceae): comparative analysis of evolutionary rate with that of grasses and its phylogenetic implications. Mol. Biol. Evol. 23, 279–291. 10.1093/molbev/msj029 [DOI] [PubMed] [Google Scholar]
- Chernomor O., von Haeseler A., Minh B. Q. (2016). Terrace aware data structure for phylogenomic inference from supermatrices. Syst. Bot. 65, 997–1008. 10.1093/sysbio/syw037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi I. S., Choi B. H. (2017). The distinct plastid genome structure of Maackia fauriei (Fabaceae: Papilionoideae) and its systematic implications for Genistoids and tribe Sophoreae. PloS One 12, e0173766. 10.1371/journal.pone.0173766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chumley T. W., Palmer J. D., Mower J. P., Fourcade H. M., Calie P. J., Boore J. L., et al. (2006). The complete chloroplast genome sequence of Pelargonium x hortorum: organization and evolution of the largest and most highly rearranged chloroplast genome of land plants. Mol. Biol. Evol. 23, 2175–2190. 10.1093/molbev/msl089 [DOI] [PubMed] [Google Scholar]
- Cosner M. E., Jansen R. K., Palmer J. D., Downie S. R. (1997). The highly rearranged chloroplast genome of Trachelium caeruleum (Campanulaceae): multiple inversions, inverted repeat expansion and contraction, transposition, insertions/deletions, and several repeat families. Curr. Genet. 31, 419–429. 10.1007/s002940050225 [DOI] [PubMed] [Google Scholar]
- Darling A. C. E., Mau B., Blattner F. R., Perna N. T. (2004). Mauve: Multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14, 1394–1403. 10.1101/gr.2289704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Queiroz L. P., Pastore J. F., Cardoso D., Snak C., de C Lima A. L., Gagnon E., et al. (2015). A multilocus phylogenetic analysis reveals the monophyly of a recircumscribed papilionoid legume tribe diocleae with well-supported generic relationships. Mol. Phylogenet. Evol. 90, 1–19. 10.1016/j.ympev.2015.04.016 [DOI] [PubMed] [Google Scholar]
- Downie S. R., Olmstead R. G., Zurawski G., Soltis D. E., Soltis P. S., Watson J. C., et al. (1991). Six independent losses of the chloroplast DNA rpl2 intron in dicotyledons: molecular and phylogenetic implications. Evolution 45, 1245–1259. 10.1111/j.1558-5646.1991.tb04390.x [DOI] [PubMed] [Google Scholar]
- Doyle J. J., Doyle J. L. (1987). A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bull. 19, 11–15. [Google Scholar]
- Doyle J. J., Davis J. I., Soreng R. J., Garvin D., Anderson M. J. (1992). Chloroplast DNA inversions and the origin of the grass family (Poaceae). Proc. Natl. Acad. Sci. U. S. A. 89, 7722–7726. 10.1073/pnas.89.16.7722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle J. J., Doyle J. L., Palmer J. D. (1995). Multiple independent losses of two genes and one intron from legume chloroplast genomes. Syst. Bot. 20, 272–294. 10.2307/2419496 [DOI] [Google Scholar]
- Doyle J. J., Doyle J. L., Ballenger J. A., Palmer J. D. (1996). The distribution and phylogenetic significance of a 50-kb chloroplast DNA inversion in the flowering plant family leguminosae. Mol. Phylogenet. Evol. 5, 429–438. 10.1006/mpev.1996.0038 [DOI] [PubMed] [Google Scholar]
- Doyle J. J., Doyle J. L., Ballenge J. A., Dickson E. E., Kajita T., Ohashi H. (1997). A phylogeny of the chloroplast gene rbcL in the Leguminosae: taxonomic correlations and insights into the evolution of nodulation. Am. J. Bot. 84, 541–554. 10.2307/2446030 [DOI] [PubMed] [Google Scholar]
- Dugas D. V., Hernandez D., Koenen E. J. M., Schwarz E., Straub S., Hughes C. E., et al. (2015). Mimosoid legume plastome evolution: IR expansion, tandem repeat expansions, and accelerated rate of evolution in clpP . Sci. Rep. 5, 16958. 10.1038/srep16958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan A. N., Vatanparast M., Cagle W. (2016). Parsing polyphyletic Pueraria: delimiting distinct evolutionary lineages through phylogeny. Mol. Phylogenet. Evol. 104, 44–59. 10.1016/j.ympev.2016.08.001 [DOI] [PubMed] [Google Scholar]
- Feng L., Gu L. F., Luo J., Fu A. S., Ding Q., Yiu S. M., et al. (2017). Complete plastid genomes of the genus Ammopiptanthus and identification of a novel 23-kb rearrangement. Conserv. Genet. Resour. 9, 647–650. 10.1007/s12686-017-0747-8 [DOI] [Google Scholar]
- Fonseca L. H. M., Lohmann L. G. (2017). Plastome rearrangements in the “Adenocalymma-Neojobertia“ clade (Bignonieae, Bignoniaceae) and its phylogenetic implications. Front. Plant Sci. 8, 1875. 10.3389/fpls.2017.01875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gantt J. S., Baldauf S. L., Calie P. J., Weeden N. F., Palmer J. D. (1991). Transfer of rpl22 to the nucleus greatly preceded its loss from the chloroplast and involved the gain of an intron. EMBO J. 10, 3073–3078. 10.1002/j.1460-2075.1991.tb07859.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert W., Marchionni M., McKnight G. (1986). On the antiquity of introns. Cell 46, 151–153. 10.1016/0092-8674(86)90730-0 [DOI] [PubMed] [Google Scholar]
- Givnish T. J., Spalink D., Ames M., Lyon S. P., Hunter S. J., Zuluaga A., et al. (2015). Orchid phylogenomics and multiple drivers of their extraordinary diversification. Proc. R. Soc B. 282, 1–10. 10.1098/rspb.2015.1553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green B. R. (2011). Chloroplast genomes of photosynthetic eukaryotes. Plant J. 66, 34–44. 10.1111/j.1365-313X.2011.04541.x [DOI] [PubMed] [Google Scholar]
- Gu C., Tembrock L. R., Johnson N. G., Simmons M. P., Wu Z. (2016). The complete plastid genome of Lagerstroemia fauriei and loss of rpl2 intron from Lagerstroemia (Lythraceae). PloS One 11, e0150752. 10.1371/journal.pone.0150752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guisinger M. M., Kuehl J. V., Boore J. L., Jansen R. K. (2011). Extreme reconfiguration of plastid genomes in the angiosperm family Geraniaceae: rearrangements, repeats, and codon usage. Mol. Biol. Evol. 28, 583–600. 10.1093/molbev/msq229 [DOI] [PubMed] [Google Scholar]
- Guo X., Castillo-Ramírez S., González V., Bustos P., Fernández-Vázquez J. L., Santamaría R. I., et al. (2007). Rapid evolutionary change of common bean (Phaseolus vulgaris L.) plastome, and the genomic diversification of legume chloroplasts. BMC Genom. 8, 228. 10.1186/1471-2164-8-228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberle R. C., Fourcade H. M., Boore J. L., Jansen R. K. (2008). Extensive rearrangements in the chloroplast genome of Trachelium caeruleum are associated with repeats and tRNA genes. J. Mol. Evol. 66, 350–361. 10.1007/s00239-008-9086-4 [DOI] [PubMed] [Google Scholar]
- He J., Yao M., Lyu R. D., Lin L. L., Liu H. J., Pei L. Y., et al. (2019). Structural variation of the complete chloroplast genome and plastid phylogenomics of the genus Asteropyrum (Ranunculaceae). Sci. Rep. 9, 15285. 10.1038/s41598-019-51601-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoot S. B., Palmer J. D. (1994). Structural rearrangements, including parallel inversions, within the chloroplast genome of Anemone and related genera. J. Mol. Evol. 38, 274–281. 10.1007/bf00176089 [DOI] [PubMed] [Google Scholar]
- Hu J. M., Lavin M., Wojciechowski M. F., Sanderson M. J. (2000). Phylogenetic systematics of the tribe Millettieae (Leguminosae) based on trnK/matK sequences, and implications for evolutionary patterns in Papilionoideae. Am. J. Bot. 87, 418–430. 10.2307/2656638 [DOI] [PubMed] [Google Scholar]
- Hu J. M., Lavin M., Wojciechowski M. F., Sanderson M. J. (2002). Phylogenetic analysis of nuclear ribosomal ITS/5.8 S sequences in the tribe Millettieae (Fabaceae): Poecilanthe-Cyclolobium, the core Millettieae, and the Callerya group. Sys. Bot. 27, 722–733. [Google Scholar]
- Jansen R. K., Ruhlman T. A. (2012). “Plastid genomes of seed plants,” in Genomics of Chloroplasts and Mitochondria. Eds. Bock R., Knoop V. (Dordrecht: Springer; ), 103–126. [Google Scholar]
- Jansen R. K., Cai Z., Raubeson L. A., Daniell H., Depamphilis C. W., Leebens-Mack J., et al. (2007). Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc. Natl. Acad. Sci. U. S. A. 104, 19369–19374. 10.1073/pnas.0709121104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen R. K., Wojciechowski M. F., Sanniyasi E., Lee S. B., Daniell H. (2008). Complete plastid genome sequence of the chickpea (Cicer arietinum) and the phylogenetic distribution of rps12 and clpP intron losses among legumes (Leguminosae). Mol. Phylogenet. Evol. 48, 1204–1217. 10.1016/j.ympev.2008.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J.-J., Yu W.-B., Yang J.-B., Song Y., dePamphilis C.W., Yi T.-S., Li D.-Z. (2018). GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of oraganelle genomes. bioRxiv, 256479. 10.1101/256479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaila T., Chaduvla P. K., Saxena S., Bahadur K., Gahukar S. J., Chaudhury A., et al. (2016). Chloroplast Genome Sequence of Pigeonpea (Cajanus cajan (L.) Millspaugh) and Cajanus scarabaeoides (L.) Thouars: Genome organization and comparison with other legumes. Front. Plant Sci. 7, 1847. 10.3389/fpls.2016.01847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaila T., Chaduvla P. K., Rawal H. C., Saxena S., Tyagi A., Mithra S. V. A., et al. (2017). Chloroplast genome sequence of cluster bean (Cyamopsis tetragonoloba L.): Genome structure and comparative analysis. Genes 8, E212. 10.3390/genes8090212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajita T., Ohashi H., Tateishi. Y., Bailey C. D., Doyle J. J. (2001). rbcL and legume phylogeny, with particular reference to Phaseoleae, Millettieae, and allies. Syst. Bot. 26, 15–536. 10.1043/0363-6445-26.3.515 [DOI] [Google Scholar]
- Kato T., Kaneko T., Sato S., Nakamura Y., Tabata S. (2000). Complete structure of the chloroplast genome of a legume, Lotus japonicus . DNA Res. 7, 323–330. 10.1093/dnares/7.6.323 [DOI] [PubMed] [Google Scholar]
- Katoh K., Standley D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M., Moir R., Wilson A., Stones-Havas S., Cheung M., Sturrock S., et al. (2012). Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649. 10.1093/bioinformatics/bts199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller J., Rousseau-Gueutin M., Martin G. E., Morice J., Boutte J., Coissac E., et al. (2017). The evolutionary fate of the chloroplast and nuclear rps16 genes as revealed through the sequencing and comparative analyses of four novel legume chloroplast genomes from Lupinus . DNA Res. 24, 343–358. 10.1093/dnares/dsx006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Cullis C. (2017). A novel inversion in the chloroplast genome of marama (Tylosema esculentum). J. Exp. Bot. 68, 2065–2072. 10.1093/jxb/erw500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K. J., Jansen R. K. (2005). Two chloroplast DNA inversions originated simultaneously during the early evolution of the sunflower family (Asteraceae). Mol. Biol. Evol. 22, 1783–1792. 10.1093/jxb/erw500 [DOI] [PubMed] [Google Scholar]
- Kim H. T., Kim J. S., Moore M. J., Neubig K. M., Williams N. H., Whitten W. M., et al. (2015). Seven new complete plastome sequences reveal rampant independent loss of the ndh gene family across orchids and associated instability of the inverted repeat/small single-copy region boundaries. PloS One 10, e0142215. 10.1371/journal.pone.0142215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackey J. A., Polhill R. M., Raven P. H. (1981). “Phaseoleae,” in Advances in Legume Systematics, part 1 (UK: Royal Botanic Gardens, Kew; ), 301– 327. [Google Scholar]
- Lanfear R., Frandsen P. B., Wright A. M., Senfeld T., Calcott B. (2017). PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol. Biol. Evol. 34, 772–773. 10.1093/molbev/msw260 [DOI] [PubMed] [Google Scholar]
- Langmead B., Salzberg S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavin M., Doyle J. J., Palmer J. D. (1990). Evolutionary significance of the loss of the Chloroplast-DNA inverted repeat in the Leguminosae subfamily Papilionoideae. Evolution 44, 390–402. 10.2307/2409416 [DOI] [PubMed] [Google Scholar]
- Lee J., Hymowitz T. (2001). A molecular phylogenetic study of the subtribe Glycininae (Leguminosae) derived from the chloroplast DNA rps16 intron sequences. Am. J. Bot. 88, 2064–2073. 10.2307/3558432 [DOI] [PubMed] [Google Scholar]
- Lee H. L., Jansen R. K., Chumley T. W., Kim K. J. (2007). Gene relocations within chloroplast genomes of Jasminum and Menodora (Oleaceae) are due to multiple, overlapping inversions. Mol. Biol. Evol. 24, 1161–1180. 10.1093/molbev/msm036 [DOI] [PubMed] [Google Scholar]
- Li C. H., Lu G. Q., Orti G. (2008). Optimal data partitioning and a test case for ray-finned fishes (Actinopterygii) based on ten nuclear loci. Syst. Biol. 57, 519–539. 10.1080/10635150802206883 [DOI] [PubMed] [Google Scholar]
- Li R., Ma P. F., Wen J., Yi T. S. (2013). Complete sequencing of five araliaceae chloroplast genomes and the phylogenetic implications. PloS One 8, e78568. 10.1371/journal.pone.0078568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C. S., Chen J. J. W., Huang Y. T., Chan M. T., Daniell H., Chang W. J., et al. (2015). The location and translocation of ndh genes of chloroplast origin in the Orchidaceae family. Sci. Rep. 5, 1–10. 10.1038/srep09040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logacheva M. D., Schelkunov M. I., Nuraliev M. S., Samigullin T. H., Penin A. A. (2014). The plastid genome of mycoheterotrophic monocot Petrosavia stellaris exhibits both gene losses and multiple rearrangements. Genome Biol. Evol. 6, 238–246. 10.1093/gbe/evu001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse M., Drechsel O., Kahlau S., Bock R. (2013). OrganellarGenomeDRAW - a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 41, W575–W581. 10.1093/nar/gkt289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LPWG (2013). Legume phylogeny and classification in the 21st century: progress, prospects and lessons for other species-rich clades. Taxon 62, 217–248. 10.5167/uzh-78167 [DOI] [Google Scholar]
- LPWG (2017). A new subfamily classification of the Leguminosae based on a taxonomically comprehensive phylogeny. Taxon 66, 44–77. 10.12705/661.3 [DOI] [Google Scholar]
- Luo Y., Ma P. F., Li H. T., Yang J. B., Wang H., Li D. Z. (2016). Plastid phylogenomic analyses resolve Tofieldiaceae as the root of the early diverging monocot Order Alismatales. Genome Biol. Evol. 8, 932–945. 10.1093/gbe/evv260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma P. F., Zhang Y. X., Zeng C. X., Guo Z. H., Li D. Z. (2014). Chloroplast phylogenomic analyses resolve deep-level relationships of an intractable bamboo tribe Arundinarieae (Poaceae). Syst. Biol. 63, 933–950. 10.1093/sysbio/syu054 [DOI] [PubMed] [Google Scholar]
- Magee A. M., Aspinall S., Rice D. W., Cusack B. P., Sémon M., Perry A. S., et al. (2010). Localized hypermutation and associated gene losses in legume chloroplast genomes. Genome Res. 20, 1700–1710. 10.1101/gr.111955.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G. E., Rousseau-Gueutin M., Cordonnier S., Lima O., Michon-Coudouel S., Naquin D., et al. (2014). The first complete chloroplast genome of the Genistoid legume Lupinus luteus: evidence for a novel major lineage-specific rearrangement and new insights regarding plastome evolution in the legume family. Ann. Bot. 113, 1197–1210. 10.1093/aob/mcu050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millen R. S., Olmstead R. G., Adams K. L., Palmer J. D., Lao N. T., Heggie L., et al. (2001). Many parallel losses of infA from chloroplast DNA during angiosperm evolution with multiple independent transfers to the nucleus. Plant Cell 13, 645–658. 10.1105/tpc.13.3.645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mower J. P., Ma P. F., Grewe F., Taylor A., Michael T. P., VanBuren R., et al. (2019). Lycophyte plastid genomics: extreme variation in GC, gene and intron content and multiple inversions between a direct and inverted orientation of the rRNA repeat. New Phytol. 222, 1061–1075. 10.1111/nph.15650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumann J., Der J. P., Wafula E. K., Jones S. S., Wagner S. T., Honaas L. A., et al. (2016). Detecting and characterizing the highly divergent plastid genome of the nonphotosynthetic parasitic plant Hydnora visserim (Hydnoraceae). Genome Biol. Evol. 8, 345–363. 10.1093/gbe/evv256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen L. T., Schmidt H. A., von Haeseler A., Minh B. Q. (2015). IQTREE: A fast and effective stochastic algorithm for estimating maximum likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. 10.1093/molbev/msu300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel R. K., Jain M. (2012). NGS QC toolkit: a toolkit for quality control of next generation sequencing data. PloS One 7, e30619. 10.1371/journal.pone.0030619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennington R. T., Lavin M., Ireland H., Klitgaard B. B., Preston J. (2001). Phylogenetic relationships of basal papilionoid legumes based upon sequences of the chloroplast trnL intron. Syst. Bot. 26, 537–566. Retrieved from http://www.jstor.org/stable/3093980. [Google Scholar]
- Perry A. S., Brennan S., Murphy D. J., Wolfe K. H. (2002). Evolutionary re-organization of a large operon in Adzuki bean chloroplast DNA caused by inverted repeat movement. DNA Res. 9, 157–162. 10.1093/dnares/9.5.157 [DOI] [PubMed] [Google Scholar]
- Qu X. J., Fan S. J., Wicke S., Yi T. S. (2019). Plastome reduction in the only parasitic gymnosperm Parasitaxus is due to losses of photosynthesis but not housekeeping genes and apparently involves the secondary gain of a large inverted repeat. Genome Biol. Evol. 11, 2789–2796. 10.1093/gbe/evz187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A. (2009). FigTree version 1.3.1 [computer program] http://tree.bio.ed.ac.uk.
- Rambaut A., Drummond A. J. (2004). Tracer version 1.5 [computer program] http://beast.bio.ed.ac.uk
- Raubeson L. A., Jansen R. K. (2005). “Chloroplast Genomes of Plants,” in Plant Diversity and Evolution: Genotypic and Phenotypic Variation in Higher Plants. Ed. Henry R. J. (Cambridge, MA: CABI Press; ), 45–68. [Google Scholar]
- Rogalski M., Ruf S., Bock R. (2006). Tobacco plastid ribosomal protein S18 is essential for cell survival. Nucleic Acids Res. 34, 4537–4545. 10.1093/nar/gkl634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F., Huelsenbeck J. P. (2003). MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574. 10.1093/bioinformatics/btg180 [DOI] [PubMed] [Google Scholar]
- Saarela J. M., Burke S. V., Wysocki W. P., Barrett M. D., Clark L. G., Craine J. M. (2018). A 250 plastome phylogeny of the grass family (Poaceae): topological support under different data partitions. PeerJ 6, e4299. 10.7717/peerj.4299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabir J., Schwarz E., Ellison N., Zhang J., Baeshen N. A., Mutwakil M., et al. (2014). Evolutionary and biotechnology implications of plastid genome variation in the inverted-repeat-lacking clade of legumes. Plant Biotechnol. J. 12, 743–754. 10.1111/pbi.12179 [DOI] [PubMed] [Google Scholar]
- Saski C., Lee S. B., Daniell H., Wood T. C., Tomkins J., Kim H. G., et al. (2005). Complete chloroplast genome sequence of Glycine max and comparative analyses with other legume genomes. Plant Mol. Biol. 59, 309–322. 10.1007/s11103-005-8882-0 [DOI] [PubMed] [Google Scholar]
- Schattner P., Brooks A. N., Lowe T. M. (2005). The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33, W686–W689. 10.1093/nar/gki366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrire B. D., Lavin M., Barker N. P., Forest F. (2009). Phylogeny of the tribe Indigofereae (Leguminosae- Papilionoideae): geographically structured more in succulent-rich and temperate settings than in grass-rich environments. Am. J. Bot. 96, 816–852. 10.3732/ajb.0800185 [DOI] [PubMed] [Google Scholar]
- Schrire B. D. (2005. a). “Indigofereae,” in Legumes of the world. Ed. Lewis (Kew, UK: Royal Botanic Gardens; ), 361–365. [Google Scholar]
- Schrire B. D. (2005. b). “Millettieae,” in Legumes of the world. Ed. Lewis (Kew, UK: Royal Botanic Gardens; ), 367–387. [Google Scholar]
- Schrire B. D. (2005. c). “Abreae,” in Legumes of the world. Ed. Lewis (Kew, UK: Royal Botanic Gardens; ), 389–391. [Google Scholar]
- Schrire B. D. (2005. d). “Phaseoleae,” in Legumes of the world. Ed. Lewis (Kew, UK: Royal Botanic Gardens; ), 393–431. [Google Scholar]
- Schwarz E. N., Ruhlman T. A., Sabir J. S. M., Hajrah N. H., Alharbi N. S., Al-Malki A. L., et al. (2015). Plastid genome sequences of legumes reveal parallel inversions and multiple losses of rps16 in papilionoids. J. Syst. Evol. 53, 458–468. 10.1111/jse.12179 [DOI] [Google Scholar]
- Sharp P. A. (1985). On the origin of RNA splicing and introns. Cell 42, 397–400. 10.1016/0092-8674(85)90092-3 [DOI] [PubMed] [Google Scholar]
- Simpson B. B., Ogorzaly M. C. (2001). Economic botany: plants in our world. 3rd ed. (New York: McGraw-Hill, Inc.), 544. [Google Scholar]
- Smith A. G., Wilson R. M., Kaethner T. M., Willey D. L., Gray J. C. (1991). Pea chloroplast genes encoding a 4 kDa polypeptide of photosystem I and a putative enzyme of C1 metabolism. Curr. Genet. 19:403–10. 10.1007/bf00309603 [DOI] [PubMed] [Google Scholar]
- Stefanovic S., Pfeil B. E., Palmer J. D., Doyle J. J. (2009). Relationships among phaseoloid legumes based on sequences from eight chloroplast regions. Syst. Bot. 34, 115–128. 10.1600/036364409787602221 [DOI] [Google Scholar]
- Ueda M., Fujimoto M., Takanashi H., Arimura S. I., Tsutsumi N., Kadowaki K. I. (2008). Substitution of the gene for chloroplast rps16 was assisted by generation of dual targeting signal. Mol. Biol. Evol. 25, 1566–1575. 10.1093/molbev/msn102 [DOI] [PubMed] [Google Scholar]
- Vatanparast M., Powell A., Doyle J. J., Egan A. N. (2018). Targeting legume loci: a comparison of three methods for target enrichment bait design in Leguminosae phylogenomics. Appl. Plant Sci. 6, e1036. 10.1002/aps3.1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker J. F., Zanis M. J., Emery N. C. (2014). Comparative analysis of complete chloroplast genome sequence and inversion variation in Lasthenia burkei (Madieae, Asteraceae). Am. J. Bot. 101, 722–729. 10.3732/ajb.1400049 [DOI] [PubMed] [Google Scholar]
- Wang R. J., Cheng C. L., Chang C. C., Wu C. L., Su T. M., Chaw S. M. (2008). Dynamics and evolution of the inverted repeat-large single copy junctions in the chloroplast genomes of monocots. BMC Evol. Biol. 8, 36. 10.1186/1471-2148-8-36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. H., Qu X. J., Chen S. Y., Li D. Z., Yi T. S. (2017). Plastomes of Mimosoideae: structural and size variation, sequence divergence, and phylogenetic implication. Tree Genet. Genomes 13, 41. 10.1007/s11295-017-1124-1 [DOI] [Google Scholar]
- Wang Y. H., Wicke S., Wang H., Jin J. J., Chen S. Y., Zhang S. D., et al. (2018). Plastid genome evolution in the early-diverging legume subfamily Cercidoideae (Fabaceae). Front. Plant Sci. 9, 138. 10.3389/fpls.2018.00138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng M. L., Blazier J. C., Govindu M., Jansen R. K. (2014). Reconstruction of the ancestral plastid genome in Geraniaceae reveals a correlation between genome rearrangements, repeats, and nucleotide substitution rates. Mol. Biol. Evol. 31, 645–659. 10.1093/molbev/mst257 [DOI] [PubMed] [Google Scholar]
- Wick R. R., Schultz M. B., Zobel J., Holt K. E. (2015). Bandage: interactive visualization of de novo genome assemblies. Bioinformatics 31, 3350–3352. 10.1093/bioinformatics/btv383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams A. V., Boykin L. M., Howell K. A., Nevill P. G., Small L. (2015). The complete sequence of the Acacia ligulata chloroplast genome reveals a highly divergent clpP1 gene. PloS One 10, e0138367. 10.1371/journal.pone.0125768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojciechowski M. F., Lavin M., Sanderson M. J. (2004). A phylogeny of legumes (Leguminosae) based on analysis of the plastid matK gene resolves many well-supported subclades within the family. Am. J. Bot. 91, 1846–1862. 10.3732/ajb.91.11.1846 [DOI] [PubMed] [Google Scholar]
- Wojciechowski M. F. (2003). “Reconstructing the phylogeny of legumes (Leguminosae): An early 21st century perspective,” in Advances in Legume Systematics 10. Eds. Klitgaard B. B., Bruneau A. (Kew: Royal Botanic Gardens; ), 5–35. [Google Scholar]
- Wu C. S., Chaw S. M. (2016). Large-Scale comparative analysis reveals the mechanisms driving plastomic compaction, reduction, and inversions in Conifers II (Cupressophytes). Genome Biol. Evol. 8, 740–3750. 10.1093/gbe/evw278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C. S., Lai Y. T., Lin C. P., Wang Y. N., Chaw S. M. (2009). Evolution of reduced and compact chloroplast genomes (cpDNAs) in gnetophytes: selection toward a lower-cost strategy. Mol. Phylogenet. Evol. 52, 115–124. 10.1016/j.ympev.2008.12.026 [DOI] [PubMed] [Google Scholar]
- Wyman S. K., Jansen R. K., Boore J. L. (2004). Automatic annotation of organellar genomes with DOGMA. Bioinformatics 20, 3252–3255. 10.1093/bioinformatics/bth352 [DOI] [PubMed] [Google Scholar]
- Xu J., Feng D., Song G., Wei X., Chen L., Wu X., et al. (2003). The first intron of rice EPSP synthase enhances expression of foreign gene. Sci. China Ser. C. Life. Sci. 46, 561–569. 10.1360/02yc012 [DOI] [PubMed] [Google Scholar]
- Zeng S., Zhou T., Han K., Yang Y., Zhao J., Liu Z. L. (2017). The complete chloroplast genome sequences of six rehmannia species. Genes 8, 103. 10.3390/genes8030103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng C., Hollingsworth P. M., Yang J., He Z. S., Zhang Z. R., Li D. Z., et al. (2018). Genome skimming herbarium specimens for DNA barcoding and phylogenomics. Plant Methods 14, 1–14. 10.1186/s13007-018-0300-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S. D., Jin J. J., Chen S. Y., Chase M. W., Soltis D. E., Li H. T., et al. (2017). Diversification of rosaceae since the late cretaceous based on plastid phylogenomics. New Phytol. 214, 1355–1367. 10.1111/nph.14461 [DOI] [PubMed] [Google Scholar]
- Zhang H., Jin J., Moore M. J., Yi T. S., Li D. (2018). Plastome characteristics of Cannabaceae. Plant Divers. 40, 127–137. 10.1016/j.pld.2018.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., Wang Y. -H., Jin J. -J., Stull G. W., Bruneau A., Cordoso D., et al. (2020). Exploration of plastid phylogenomic conflict yields new insights into the deep relationships of Leguminosae. Syst. Biol. 10.1093/sysbio/syaa013 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated for this study can be found in Genbank; the list of accession can be found in Supplementary Table 1 .