Abstract
Proper function of many physiological processes requires a robust circadian clock. Disruptions of the circadian clock can result in metabolic diseases, mood disorders, and accelerated aging. Therefore, identifying small molecules that specifically modulate regulatory core clock proteins may potentially enable better management of these disorders. In this study, we applied a structure-based molecular-docking approach to find small molecules that specifically bind to the core circadian regulator, the transcription factor circadian locomotor output cycles kaput (CLOCK). We identified 100 candidate molecules by virtual screening of ∼2 million small molecules for those predicted to bind closely to the interface in CLOCK that interacts with its transcriptional co-regulator, Brain and muscle Arnt-like protein-1 (BMAL1). Using a mammalian two-hybrid system, real-time monitoring of circadian rhythm in U2OS cells, and various biochemical assays, we tested these compounds experimentally and found one, named CLK8, that specifically bound to and interfered with CLOCK activity. We show that CLK8 disrupts the interaction between CLOCK and BMAL1 and interferes with nuclear translocation of CLOCK both in vivo and in vitro. Results from further experiments indicated that CLK8 enhances the amplitude of the cellular circadian rhythm by stabilizing the negative arm of the transcription/translation feedback loop without affecting period length. Our results reveal CLK8 as a tool for further studies of CLOCK's role in circadian rhythm amplitude regulation and as a potential candidate for therapeutic development to manage disorders associated with dampened circadian rhythms.
Keywords: drug design, circadian clock, gene expression, protein expression, drug development, transcription, transcription coregulator, brain and muscle Arnt-like protein-1 (BMAL1), circadian locomotor output cycles kaput (CLOCK), circadian regulation, circadian rhythm amplitude, gene regulation
Introduction
The circadian clock generates a 24-h rhythm through which physiology and behavior adapt to daily changes in the environment (1). Many biological processes such as hormone secretion and sleep-wake cycles are controlled by the circadian clock (2). Therefore, an innate malfunctioning of the circadian clock or a shift between internal circadian rhythm and the external environment can cause various pathologies. Sleep disorders, altered metabolism, obesity, diabetes, mood disorders, cancer, and cardiovascular diseases are all linked to an abnormal circadian rhythm. Also, a decrease in the robustness of the circadian rhythm (amplitude decline) is correlated with different pathologies and chronic diseases such as mood disorders and metabolic diseases. Aging is also associated with dampened amplitude and imperfect timing of the circadian rhythm, and therefore weakening of many clock-controlled physiological processes (3–6).
In mammals, the molecular clock is generated by transcription/translation feedback loop (TTFL).3 In the positive arm of one of the TTFLs, circadian locomotor output cycles caput (CLOCK), and aryl hydrocarbon receptor nuclear translocator-like (BMAL1) form heterodimers and positively regulate the expression of circadian clock-controlled genes including Period (Per) and Cryptochrome (Cry) (7–10). In the negative arm of the loop, the PER:CRY complex along with casein kinase IΔ translocates into the nucleus, interacts with CLOCK:BMAL1, and inhibits the transcriptional activity of CLOCK:BMAL1, thereby repressing the transcription of Per and Cry and other clock-controlled genes. After the PER and CRY proteins are degraded, the transcription repression is relieved, and a new cycle starts. A second feedback consists of the nuclear receptors RORs and REV-ERBs, which are expressed under the control of CLOCK:BMAL1 and activate and repress the transcription of Bmal1, respectively (11). A robust and precise circadian rhythm is generated only if the amounts, localization, and activity of the clock-related proteins are precisely regulated; a process in which several post-translational modifications are involved (12).
Without a robust and well-aligned circadian rhythm, organisms cannot adapt to environmental changes adequately, which defeats the evolutionary purpose of the circadian clock. To modify a disrupted circadian rhythm, small molecules are of great importance because they have reversible and time- and dose-tunable effects on the system (13, 14). So far, unbiased screening based on phenotypic changes in the circadian rhythm of reporter cells has resulted in the discovery of multiple clock-modulating small molecules with various circadian phenotypes (15–17). One such molecule is KL001, which stabilizes and binds to CRY (16). Enhancers of CLOCK:BMAL1 transcriptional activity, and a molecule that augments the repression activity of CRY have been detected by high-throughput screening using mechanistic approaches (18, 19). Targeting nuclear receptors and protein kinases involved in the circadian clock mechanism has resulted in the identification of ROR agonists/antagonists, REV-ERB agonists (20), and kinase inhibitors (21, 22). Structure-based design methods also have been used successfully to identify small molecules that perturb biological systems. Now that the crystal structures of clock proteins are available, this approach has been applied to identify clock-modulating compounds that target clock proteins. The crystal structure of the CLOCK:BMAL1 heterodimer revealed that similar domains in the CLOCK and BMAL1 structures (bHLH, PAS-A, and PAS-B) interacted and provided three protein-protein interfaces (23). The hollows and clefts that were present on the surface of each protein allowed them to interlock to form a stable interaction. PAS domains, as internal sensors of living cells, can bind different cofactors, so proteins that contain PAS domains are good candidate targets for drug design (24).
Amplitude is one of the major circadian rhythm parameters, which is defined as a measure of the mean difference between the peaks and troughs of the rhythm. It is commonly used to characterize the oscillator or stability of the rhythm. Several studies have suggested that the amplitude is reduced in older animals (25). A mouse study, Clock mutation changed the resetting effects of phase-shifting stimuli by reducing pacemaker amplitude (26). Furthermore, the stoichiometric relationships between core clock proteins were found to be critical for the robustness and amplitude of circadian rhythms by affecting both the negative and positive feedback TTFLs (27).
The aim of this study was to find a CLOCK-binding small molecule using a structure-based approach. We started by virtually screening a library of ∼2 million small molecules, then gradually narrowed down the number of selected molecules using different biochemical and molecular evaluation steps. We identified a CLOCK-binding small molecule (CLK8) that decreased the interaction between CLOCK and BMAL1 interfering in the translocation of CLOCK into the nucleus both in vivo and in vitro. Our results show that a decrease in nuclear CLOCK leads to the stabilization of the negative arm of the TTFL and, in turn, enhances the amplitude of the circadian rhythm with no change in period length. Based on its circadian phenotype, CLK8 may be used to improve the low amplitude associated with aging, mood disorders, and metabolic syndrome as well as to improve the effectiveness of cancer treatments.
Results
Molecular dynamics simulation and virtual screening
First, we applied an energy minimization simulation to refine the atom coordinates of the CLOCK structure (PDB ID 4F3L) (23) as described previously (28). After the energy minimization, the temperature of the system was increased to the physiological temperature, 310 K. Then, the simulation was continued with the Langevin piston method for 10 ns. The root mean square deviation (RMSD) analysis of the Cα atoms in the PAS domains showed that the simulation reached equilibration in 1 ns. The final structure of CLOCK at the end of the molecular dynamics simulation was used for the molecular docking. AutoDock Vina (29) was used to perform blind docking of ∼2 million commercially available small molecules filtered by Lipinski's Rule of Five (30) as described previously (31). The structures of the top 500 compounds are listed from the highest to the lowest affinity and visually inspected. Compounds with docking positions far away from the CLOCK:BMAL1 interfaces were eliminated. A final number of 100 compounds with affinities ranging from −7 to −10 kcal/mol were selected by considering features such as favorable shape complementarity and diversity in binding region and chemical properties. These top 100 compounds were tested experimentally.
Cell viability test
The toxicity of the compounds was assessed by the MTT-based assay using human osteosarcoma (U2OS) cells treated with the different compounds at different concentrations. Compounds with a relative cell viability of <80% at 1.25 μm were eliminated. As a result, 72 compounds were statistically identified as nontoxic and used for the next characterization step. For the in vitro experiments, the final concentration for each nontoxic compound was set to the maximum concentration at which the compound had at least 80% relative cell viability.
Effect of the selected compounds on CLOCK:BMAL1 interaction and circadian rhythm
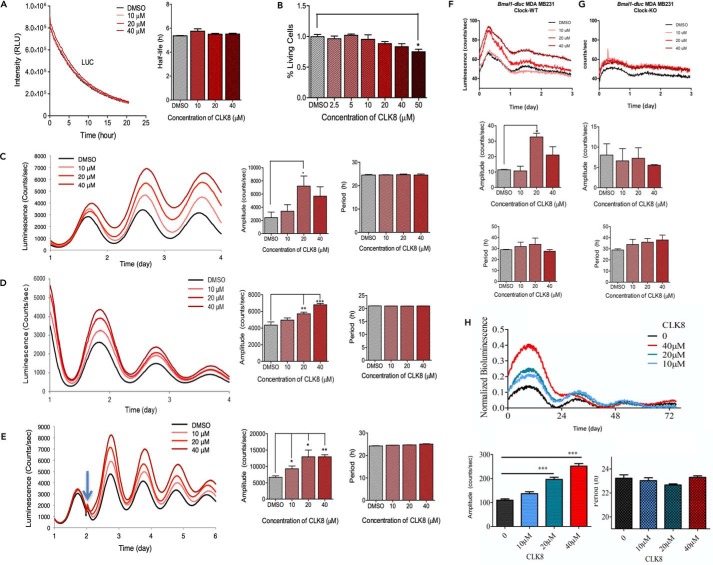
Our expectation was that the selected small molecules would regulate the interaction between CLOCK and BMAL1. Although the mammalian two-hybrid system is not a quantitative assay that is used to measure the degree of interaction between two proteins, we initially used the mammalian two-hybrid system to identify compounds that significantly altered the interaction between CLOCK and BMAL1. We found 24 compounds that significantly altered the CLOCK-BMAL1 interaction without changing the protein level (data are not shown), suggesting that these compounds might modulate the positive arm of the loop at the molecular level. Next, we assessed the effect of these compounds on the circadian rhythm of U2OS cells stably expressing the Bmal1-dLuc luciferase reporter. Each of the small molecules had different effects on the circadian rhythm and were classified as amplitude enhancers, period lengtheners, and amplitude reducers. The subsequent dose-response validation found only four compounds that passed the secondary screening step successfully. To assess the specificity of these four compounds for the circadian rhythm on different genetic backgrounds, NIH 3T3 cells stably expressing Bmal1-dLuc were treated with the four compounds. The amplitude-enhancing compound, CLK8, was selected for further characterization. Until now, none of the reported amplitude-enhancing small molecules directly affected the positive or negative arm of the TTFLs of the circadian clock; indeed, their targets and complex mechanisms of action are still unknown. Some of them caused period shortening in addition to amplitude enhancement, and only one enhanced the amplitude in suprachiasmatic nuclei (SCN) explants from mouse (14). One possible explanation for not observing the same phenotype in the SCN might be the lack of expression of (unknown) target proteins in the SCN (14). Because CLOCK is a core component of the circadian rhythm mechanism, an amplitude-enhancing compound that targets CLOCK may have the potential to consistently affect peripheral clocks and SCN. Molecules that regulate amplitude enhancement and dampened amplitude associated with chronic diseases and aging may be used in their treatment (32, 33). To ensure that CLK8 did not interfere with the luminescence signal itself, we examined the degradation rates of luciferase in HEK293T cells transfected with pcDNA-Luc and treated with CLK8. We found that the half-life of luciferase was unchanged in the cells treated with CLK8, confirming the luminescence signal as an independent and consistent measuring tool (Fig. 1A). This result confirmed the idea that CLK8 likely modulates CLOCK:BMAL1 activity.
Figure 1.
Initial screening for identification of CLK8. A, HEK293T cells were transfected with pcDNA-Luc and the effect of 10, 20, and 40 μm CLK8 on the luciferase (LUC) half-life was measured. The Luc signal was monitored after CLK8 treatment until it reached a plateau. Data are mean ± S.E.; n = 3 independent experiments. B, dose-dependent cytotoxicity of CLK8 in the U2OS cells. All the measurements were normalized to 0.5% DMSO control. Data are mean ± S.E.; n = 3 independent experiments. C and D, continuous monitoring of the luminescence rhythms of Bmal1-dLuc U2OS and Bmal1-dLuc NIH 3T3-transfected cells to determine the dose-dependent effect of CLK8 on circadian rhythm. Amplitude and period parameters are shown in the right panel. E, Bmal1-dLuc U2OS cells were synchronized at time 0. Two days later (blue arrow), cells were treated with 10, 20, or 40 μm CLK8. Cells treated with 0.5% DMSO were used as the control. Luminescence profiles are the means of three independent experiments. Amplitude and period parameters (shown in the right panel) were obtained by fitting first-order polynomial baseline-subtracted data with sin (damped). Day 1 was not included in the analysis. Data are mean ± S.E.; n = 3 independent experiments. *, p value <0.05; **, p value <0.01; ***, p value <0.001. F, WT MDA MB231 cells and G, CLOCK knock-out MDA MB231 cells were transduced with Bmal1-dluc lentiviral particles. At 72 h post-transduction, the luminescence rhythms of the cells were monitored continuously to determine the dose-dependent effect of CLK8 on circadian rhythm. Amplitude and period parameters (shown in the bottom panel) were obtained by fitting first-order polynomial baseline-subtracted data with sin (damped). Luminescence profiles are means of three independent experiments. Data are mean ± S.E.; n = 3 independent experiments. *, p value <0.05. H, primary mouse skin fibroblast cells were transduced with Bmal1-dluc lentiviral particles. Day 1 was not included in the analysis. Data are mean ± S.E.; n = 4 independent experiments. ***, p value <0.001.
Effect of CLK8 on circadian rhythm
The highest concentration of CLK8 used for the in vitro experiments was 40 μm, which was not toxic to the U2OS cells and had cell viability >80% (Fig. 1B). We found that CLK8 enhanced the amplitude of the Bmal1-dLuc signal in U2OS and NIH 3T3 cells in a dose-dependent manner and no period changes were observed in either of the cell lines (Fig. 1, C and D). Subsequently, we monitored the reporter rhythm before and after treatment with CLK8 to better understand the effects of this compound. The Bmal1-dLuc U2OS cells were synchronized and the reporter rhythm was recorded for 2 days (Fig. 1E). Then, different doses of CLK8 were added to the cells without replacing the media to prevent any phase resetting (34). The bioluminescence rhythms were recorded for another 4 days. We found that the luminescence profiles were the same for all the samples until CLK8 was added. Compared with the results for the DMSO control, CLK8 advanced the first trough immediately after it was added, and enhanced the amplitude by >50% at 10 μm (Fig. 1E). We analyzed the effect of CLK8 on the Bmal1-dLuc expressing reporter in CLOCK-deficient and WT MDA MB231 cells. We found that CLK8 enhanced the amplitude in WT MDA MB231 cells (Fig. 1F), and that the CLK8-mediated increase in Bmal1-luc intensity was abolished in the Clock knockout cells at different concentrations of CLK8 (Fig. 1G). Finally, we tested the effect of CLK8 on primary mouse skin fibroblast (MSF) cells and found that CLK8 enhanced the amplitude of the circadian rhythm in a dose-dependent manner with no period change, which was similar to its effects on the other cell lines tested (Fig. 1H). These results suggest that CLK8 specifically binds to CLOCK and results in amplitude enhancement.
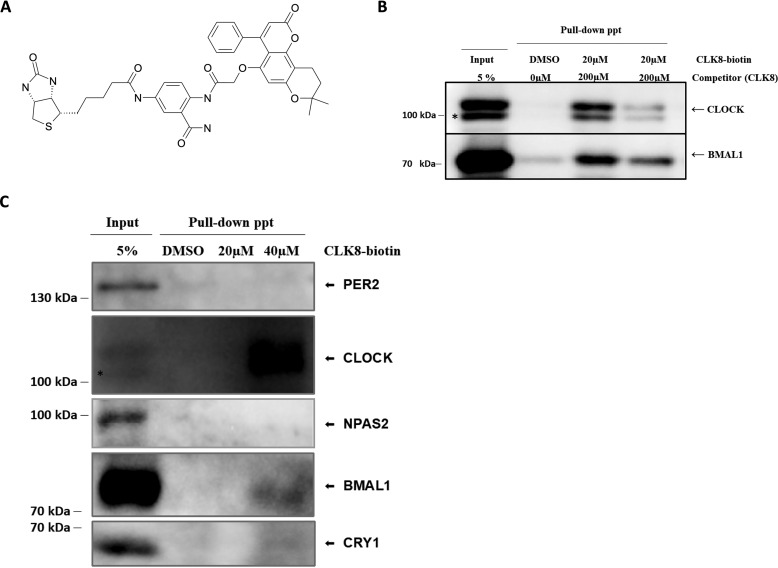
Specific binding of CLK8 to CLOCK
After identifying CLK8 as a promising small molecule for regulating the circadian rhythm amplitude, its ability to physically bind to CLOCK was assessed by pulldown assay using biotinylated CLK8 as bait. The biotinylated CLK8 used in this study was synthesized by Enamine Ltd. (Ukraine) (Fig. 2A). Whole cell lysate of HEK293T cells overexpressing CLOCK and BMAL1 were incubated with biotinylated CLK8 in the absence or presence of the competitor (CLK8). CLOCK was precipitated with the biotinylated CLK8 but not with the unbiotinylated (free) CLK8 (Fig. 2B). The results showed that CLK8 specifically bound to CLOCK. To eliminate any artifact of the overexpressed system, whole cell lysate of U2OS cells expressing endogenous levels of CLOCK (and BMAL1) were used in a pulldown assay. The difference in the amount of CLOCK between the input sample and the sample precipitated with biotinylated CLK8 revealed that CLK8 had high affinity for CLOCK (Fig. 2C). Except for BMAL1, which always was co-precipitated with CLOCK (although in very low amounts), none of the other core clock components were detected as targets for CLK8 (Fig. 2C), including NPAS2, a homologue of CLOCK.
Figure 2.
Specific binding of CLK8 to CLOCK. A, chemical structure of biotinylated CLK8 (bait). Pulldown assay using whole cell lysates of (B) HEK293T cells overexpressing Clock and Bmal1 from pSport6 plasmids and (C) U2OS cells expressing endogenous levels of CLOCK (and BMAL1). Free CLK8 was used as the competitor. Data are representative of three independent experiments. Asterisk indicates nonspecific band.
We next checked for off-targets of CLK8 using LC-tandem MS (LC-MS/MS) to analyze the precipitated proteins. Because the amount of precipitated endogenous CLOCK was insufficient to be detected in LC-MS/MS, we used whole cell lysate of HEK293T cells overexpressing CLOCK and BMAL1 for the LC-MS/MS. We determined proteins that could potentially bind to CLK8 using a minimum threshold for peptide spectrum matches and coverage equal to those used for the main target, CLOCK. We also excluded proteins that were detected in negative controls of the CRAPome database (35), assuming them to be abundant sticky proteins. In the presence of the competitor (CLK8), CLOCK was not detected by MS/MS, which explicitly showed the specificity of the binding. BMAL1 was detected in small amounts when the competitor was present. A decrease in the amount of the neuron navigator NAV2 in the presence of the competitor identified it as a possible off-target of CLK8 (Table 1).
Table 1.
Potential targets of CLK8
Whole cell lysates of HEK293T cells overexpressing Clock and Bmal1 were incubated with 20 μm biotinylated CLK8. When needed, CLK8 was added as a competitor. Precipitated proteins in the pulldown assay were subjected to LC-MS/MS. The peptide spectrum matches and coverage of the main target, CLOCK, were considered as the threshold in data analysis. Proteins with peptide spectrum matches and coverage lower than threshold were eliminated.
| Protein | Peptide spectrum matches |
||
|---|---|---|---|
| Competitor (μm) | |||
| 0 | 200 | ||
| CLOCK | Circadian locomoter output cycles protein kaput | 42 | – |
| BMAL1 | Aryl hydrocarbon receptor nuclear translocator-like protein 1 | 56 | 46 |
| NAV2 | Neuron navigator 2 | 263 | 188 |
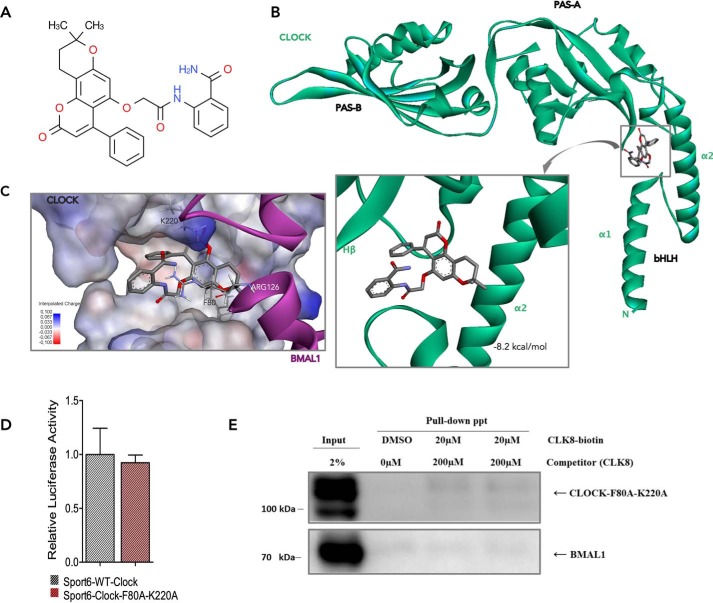
The docking results showed that CLK8 (Fig. 3A) could bind in the hollow created between the α2 helix of the bHLH domain and the Hβ strand of the PAS-A domain of CLOCK (Fig. 3B). The molecular dynamics simulation of the CLOCK:BMAL1 heterodimer indicated that this hollow was where Arg-126 of BMAL1 entered and interacted with Phe-80 of CLOCK, a conserved hydrophobic core residue on the α2 helix of the bHLH domain (Fig. 3, B and C). The docking results indicated that Phe-80 also may be involved in the CLOCK-CLK8 interaction. Lys-220 also was shown to play an important role in the CLOCK-CLK8 interaction through a cation-π interaction. To confirm this hollow as the CLK8-binding site, we replaced Phe-80 and Lys-220 of CLOCK with Ala residues by site-directed mutagenesis using appropriate primers (Table S2). After generating the double mutant (CLOCK-F80A,K220A), we determined the effect of the mutations on the functions of CLOCK by transactivation assay. To evaluate the functional consequences of these mutations on clock function, we monitored the activity of Per1:luc using Clock/Bmal1 and Clock-F80A,K220A/Bmal1 in HEK293T cells. The results showed that both WT and mutant CLOCK had the same degree of activation on the Per1:luc reporter along with BMAL1 (Fig. 3D). Consequently, we used HEK293T cells overexpressing CLOCK-F80A,K220A and BMAL1 to verify the predicted binding site of CLK8. We performed a pulldown assay using biotinylated CLK8 in the absence and presence of the competitor (CLK8). The results clearly indicated that mutant CLOCK did not precipitate with biotinylated CLK8 to the same extent as WT CLOCK (Fig. 3E). Besides, the very small amount of precipitated mutant CLOCK did not disappear in the presence of the competitor (CLK8), indicating that CLK8 bound to the computationally predicted region (Fig. 3B). Notably, BMAL1 did not appear in the Western blotting, as was the case in the pulldown experiment with WT CLOCK (Fig. 3E), suggesting that BMAL1 came along with WT CLOCK rather than because of nonspecific binding to CLK8.
Figure 3.
Docking of CLK8 into the CLOCK-binding site. A, chemical structure of CLK8. B, best binding mode of CLK8 to CLOCK (green) with a predicted binding energy of −8.2 kcal/mol. CLK8 can enter the hollow between the α2 helix of the bHLH domain and the Hβ strand of the PAS-A domain of CLOCK. C, superposition of CLK8 and Arg-126 of BMAL1 (magenta). CLK8 and Arg-126 of BMAL1 share the same binding region in the CLOCK structure, where Phe-80 plays an essential role. The positively charged Lys-220 in the PAS-A domain of CLOCK also contributes to a π-cation interaction with CLK8. D, comparison of WT CLOCK and mutant CLOCK-F80A,K220A. The transcriptional functions of WT and mutant CLOCK were compared by a Per1-Luc assay in HEK293T cells. Data are mean ± S.E.; n = 3 independent experiments. E, pulldown assay using whole cell lysate of HEK293T cells overexpressing CLOCK-F80A,K220A and BMAL1.
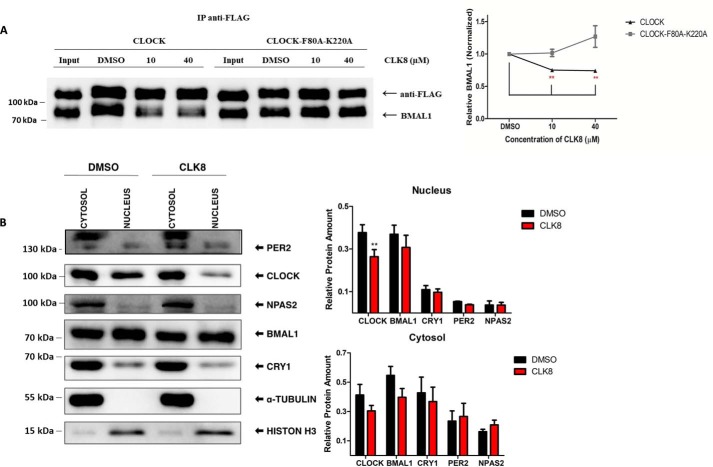
Interaction between CLOCK and BMAL1 in the presence of CLK8
The mammalian two-hybrid results implied that CLK8 reduced the interaction between CLOCK and BMAL1. We used a co-immunoprecipitation assay to verify this finding. HEK293T cells were transfected with plasmids encoding FLAG-tagged Clock and Bmal1. An anti-FLAG resin was used to precipitate FLAG-CLOCK after adding 10 or 40 μm CLK8. Whereas FLAG-CLOCK was precipitated to the same extent in all the samples, the amount of precipitated BMAL1 was reduced at the higher concentration of CLK8 (Fig. 4A). These results suggested that the interaction between CLOCK and BMAL1 decreased at the higher concentration of CLK8 (Fig. 4A). If this is true, the interaction between CLOCK-F80A,K220A and BMAL1 should not be affected in the presence of CLK8. We repeated the pulldown assays side by side using both the mutant and WT CLOCK. The results showed the BMAL1-CLOCK interaction was reduced in the presence of CLK8, whereas the interaction between CLOCK-F80A,K220A and BMAL1 was not affected, even at 40 μm CLK8 (Fig. 4A). These results suggest that CLK8 interfered with the interaction between CLOCK and BMAL1.
Figure 4.
Reduction of CLOCK and BMAL1 interaction and nuclear localization of CLOCK by CLK8. A, effect of CLK8 on the interaction of CLOCK and BMAL1 were evaluated by co-immunoprecipitation. Anti-FLAG affinity gel was used to precipitate FLAG-tagged CLOCK together or FLAG-tagged CLOCK-F80A,K220A mutant with BMAL1. Western blot analysis using anti-BMAL1 and anti-FLAG antibodies was performed to compare the association of CLOCK and the CLOCK-F80A,K220A mutant with BMAL1 in samples with different concentrations of CLK8. Quantification of the Western blotting is shown in the right panel. The vertical axis, which indicates the amount of BMAL1 (normalized by CLOCK) in each sample, was calculated relative to the DMSO control. B, unsynchronized U2OS cells were treated with 20 μm CLK8. Control cells were treated with 0.5% DMSO. Two days later, cells were fractionated and examined by Western blot analysis, where CLOCK, NPAS2, PER2, BMAL1, and CRY1 were normalized by α-tubulin or histone-H3. Quantifications are shown on the right. Data are mean ± S.E.; n = 5 independent experiments. The Western blotting data are representative of five independent experiments. *, p value <0.05; **, p value <0.01.
Effect of CLK8 on subcellular localization of CLOCK
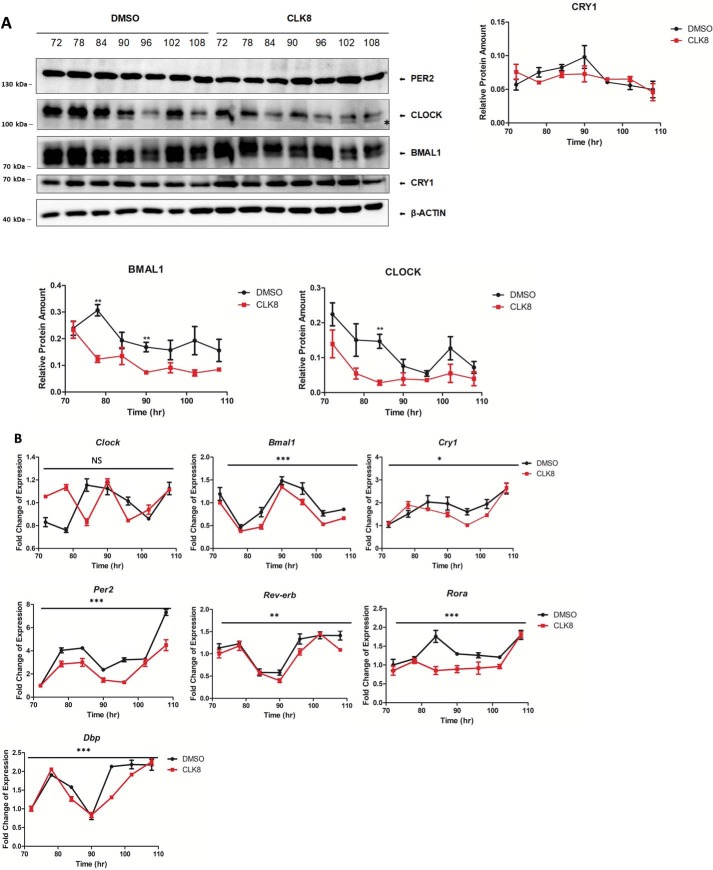
The localization and degradation of CLOCK is regulated by its direct interaction with BMAL1 (36). We hypothesized that the inability of CLOCK to bind to BMAL1 in the presence of CLK8 might alter its subcellular localization. To test this idea, U2OS cells treated with 20 μm CLK8 were fractionated and the cytosolic and nuclear proteins were isolated. Proteins that are specifically localized in the nucleus (histone-H3) and cytoplasm (tubulin) were used as controls to evaluate the purity of the fractions (Fig. 4B). The results indicated that the translocation of CLOCK into the nucleus was affected by the presence of CLK8 compared with the control samples without CLK8. The levels of PER2, NPAS2, BMAL1, and CRY1 were comparable with their levels in the control samples (Fig. 4B). These results suggested that the amount of CLOCK in the nucleus was reduced in the presence of CLK8, and this affected the stoichiometry of the BMAL1:CLOCK dimer in the nucleus. Considering the CRY1 and PER2 levels were unaltered, we expected the negative feedback loop would be more effective on CLOCK/BMAL1 transactivation. To evaluate this possibility, U2OS cells were synchronized with dexamethasone and treated with CLK8. Then, the cells were collected every 6 h to measure the transcriptional and protein levels of core clock components. The oscillatory amplitude of Clock expression was generally lower in the CLK8-treated cells (Fig. 5, A and B). In particular, the protein levels of CLOCK and BMAL1 in the positive arm of the oscillator were reduced (Fig. 5A); however, the overall change in transcriptional level of Clock was not statistically significant (Fig. 5B). Furthermore, CRY1 and PER2, which are components of the negative arm of the oscillator, showed comparable protein abundances in the presence and absence of CLK8 (Fig. 5A) despite their transcript levels being significantly reduced (Fig. 5B). These results suggested that CLK8 caused a reduction in protein levels in the positive arm (BMAL1 and CLOCK) without altering the protein levels in the negative arm (CRY and PER), and therefore caused the reduction in transcriptional levels of clock-controlled genes (Cry1, Per2, Rev-erb, Roar, and Dbp). These results suggested that stabilization of the repression arm by CRY and PER reduced CLOCK/BMAL1 transactivation and led to enhanced overall circadian amplitude in the cell lines.
Figure 5.
CLK8 altered CLOCK protein levels in a time-dependent manner. Confluent U2OS cells were synchronized by 2-h treatment with dexamethasone (0.1 μm) and the medium was replaced with fresh medium containing CLK8 or DMSO. Cells were harvested at the indicated time points. Asterisk indicates nonspecific band. A, time-dependent analysis of core clock proteins by Western blotting. Data are mean ± S.E.; n = 3. B, qPCR analysis of the core clock genes in a time-dependent manner. Data are the mean ± S.E.; n = 3 independent experiments. ***, p < 0.001; **, p < 0.005; *, p < 0.05 versus the DMSO control by two-way ANOVA.
In vivo effect of CLK8 on mice liver
The in vitro studies suggested that CLK8 specifically bound to CLOCK and inhibited its interaction with BMAL1, thereby interfering with nuclear translocation. First, we evaluated the toxicity of CLK8 in vivo by performing a single dose toxicity study. CLK8 was administered intraperitoneally to C57BL/6J mice at doses of 5, 25, 300, and 1000 mg/kg. General toxicity was evaluated by mortality, body weight changes, body temperature, clinical signs, food and water consumption, behavior assessment, and gross pathologies at terminal necropsy. Animals treated with 300 and 1000 mg/kg of CLK8 exhibited the following clinical symptoms: dyspnea, hyporeflexia, reduced locomotor activity, piloerection, hunched posture, and corneal opacity, so these doses were considered as lethal dose and the animals were euthanized for ethical reasons. No mortality or clinical signs were observed at the other doses (5 and 25 mg/kg) of CLK8; therefore, we selected the 25 mg/kg dose for use in our study.
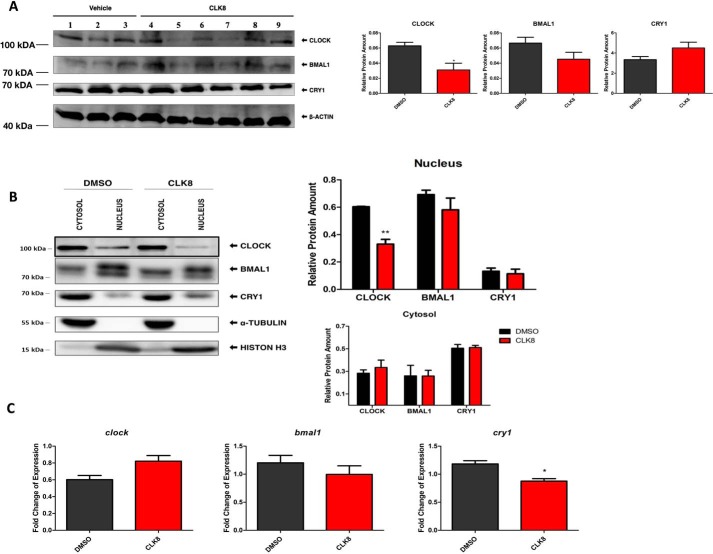
To assess its in vivo effect, CLK8 was administered intraperitoneally into mice at 25 mg/kg. Mice were euthanized 6 h after the injection and all the organs were kept for downstream analysis. A decrease in CLOCK levels was detected in whole cell lysates of the mouse livers (Fig. 6A), whereas the levels of BMAL1 and CRY1 were unaltered (Fig. 6A). To further analyze the CLOCK levels in the cytosolic and nuclear fractions of the mouse livers, we fractionated liver samples from vehicle- and CLK8-treated animals euthanized 6 h after administration and separately isolated the cytosolic and nuclear proteins. Proteins known to be specifically localized in the nucleus (histone-H3) and cytoplasm (tubulin) were used as controls to evaluate the purity of the fractions (Fig. 6B). The abundance of CLOCK in the nucleus was significantly lower in the CLK8-treated mice compared with its abundance in the control animals (Fig. 6B). However, the abundances of cytosolic and nuclear BMAL1 and CRY1 were unaltered in the CLK8-treated mice liver compared with the controls (Fig. 6B). To further explore the influence of CLK8 on circadian transcriptional function in vivo we measured the Bmal1, Clock, and Cry1 transcriptional levels by qPCR. The Cry1 transcriptional level was the only one that was significantly changed in the CLK8-treated mice compared with the transcriptional levels in the untreated mice (Fig. 6C). All the in vivo data were consistent with the in vitro data, which suggested that CLK8 had the same phenotype in mice liver.
Figure 6.
Effect of CLK8 in mice liver. A, the mice were treated with CLK8 or vehicle and then euthanized. Liver protein lysates were prepared and probed with antibodies for different core clock proteins. B, mice livers (n = 3 for vehicle-treated animals; n = 5 for CLK8-treated animals) were fractionated and the samples were subjected to Western blot analysis. Quantifications are shown in the right panel. Data are mean ± S.E.; n = 3 independent experiments for the control samples; n = 5 independent experiments for CLK8-treated samples. **, p value <0.01 by one-way ANOVA. C, transcriptional levels of Clock, Cry1, and Bmal1 were measured by qPCR. Data are mean ± S.E.; n = 3 independent for the control samples; n = 5 for CLK8-treated samples. *, p value <0.05. The lane numbers 1, 2, and 3 indicate the animals used as the controls (treated with vehicle); lanes 4, 5, 6, 7, 8, and 9 indicate the animals treated with CLK8.
Discussion
We identified a novel CLOCK-binding small molecule CLK8 that affected the translocation of CLOCK into the nucleus by modulating the BMAL1-CLOCK interaction. We used a target-directed structure-based design method to disrupt or enhance the circadian clock. Among the core clock components, only the main repressor CRY1 had been considered as a target for small molecule modification, leaving the activators of the feedback loops as novel targets (16, 37). The clock-modulating small molecule CLK8 is the first compound to be identified that binds to CLOCK, a core protein in generating the circadian rhythm. The computationally predicted binding domain was a hollow between the α2 helix of the bHLH domain and the Hβ strand of the PAS-A domain of CLOCK. Although three of the four core clock components contain PAS domains, the pulldown assay and LC-MS/MS results confirmed the specific binding of CLK8 to CLOCK.
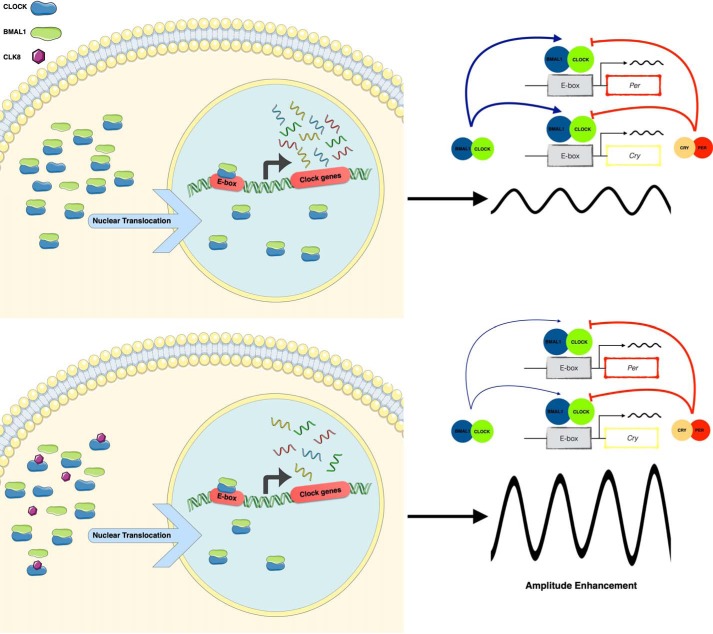
CLK8 enhanced the reporter rhythm amplitude persistently, and also advanced the first trough immediately after treatment. Although the exact molecular mechanisms involved in producing these effects may be complex, our results showed that CLK8 partially inhibited the CLOCK-BMAL1 interaction. This inhibition resulted in a reduction of CLOCK:BMAL1 dimerization and, in turn, a reduction in the translocation of CLOCK into the nucleus. Indeed, it has been shown that phosphorylation of CLOCK upon its dimerization with BMAL1 led to its nuclear localization and degradation (36, 38). Different studies have indicated the importance of the stoichiometric relationships between core clock components in determining the circadian rhythm properties (27, 39). These studies support the possibility of enhancing the amplitude or shortening the period by altering the levels or activities of the positive and negative arms of the TTFL of the circadian clock (13). Our data suggest that CLK8 can affect the amplitude of circadian rhythm by decreasing the amount of CLOCK (Fig. 7), without affecting the protein levels in the negative arm (CRY and PER2). Another small molecule nobiletin, a natural polymethoxylated flavonoid, also has amplitude enhancement and period shortening effects through the ROR nuclear receptors that function in stabilizing the negative arm of the TTFL, the molecular oscillator, by elevating the PER2 level (40). Our data suggested that stabilization of the negative arm of the core clock mechanism by decreasing the PER2 level in the positive arm of the TTFL also enhanced the amplitude.
Figure 7.
Schematic representation of how CLK8 affects CLOCK and the circadian rhythm. CLOCK and BMAL1 interact dynamically. CLK8 binds to CLOCK and reduces the CLOCK-BMAL1 interaction. When CLK8 binds to CLOCK, the translocation of CLOCK into the nucleus is abolished. CLK8 enhances the amplitude of the circadian rhythm at the cellular level, so the role of CLOCK in regulating the circadian clock amplitude can be investigated using CLK8 to transiently modulate CLOCK. The thin line indicates reduction in the strength of the positive arm of the transcription/translation feedback loop (TTFL); the thick line indicates enhanced strength in the negative arm of the TTFL.
The mechanism became more apparent when the expression levels of the clock output genes, DBP and PER2, were found to be reduced in agreement with the decreased interaction between CLOCK and BMAL1 in the presence of CLK8. A possible explanation is that a reduction of CLOCK/BMAL1 entry into the nucleus resulted in the strengthening of the repressor activity of CRY/PER on CLOCK/BMAL-driven transcription.
By taking together our results, we propose CLK8 as a novel CLOCK-binding compound that can be used to enhance circadian rhythm. This clock-enhancing small molecule may be a lead compound for new therapeutics in the prevention and treatment of diseases associated with dampened amplitude. Further pharmacodynamic and pharmacokinetic studies will help to elucidate its exact mechanism of action and possible required chemical modifications of this compound. Understanding the mechanism of action itself will improve our understanding of the circadian clock regulatory mechanism.
Experimental procedures
Molecular dynamics of CLOCK
The initial structure of the CLOCK protein was obtained from the Protein Data Bank (PDB ID 4F3L) (23). Molecular dynamic (MD) simulations were carried out by NAMD (41) software using CHARMM force field. First, the protein structure files for MD simulations were prepared by visual MD (42) after removing crystallographic water molecules and adding hydrogen atoms using AutoDockTools4 (43). Using the psfgen package, atom and residue names were replaced with the ones recognized by NAMD. Then, the structure was dissolved in a water-box and the system was ionized. In the first 10,000 steps of minimization, the backbone was fixed. Further 10,000 steps of minimization were performed on all atoms with no pressure control. Subsequently, the system was brought to physiological temperature (310 K) by 10 K increments with 10-ps simulation for each increment in which α-carbons were restrained. The constraint scaling decreases from 1 to 0.25 kcal/(mol Å2) in 0.25 increments, with each increment being 5,000 steps. Further 90,000 steps with zero constraint scaling were performed as the final part of energy minimization before the RMSD of protein was converged and stabilized. MD simulation for equilibrium was performed using the Langevin dynamics at 310 K with a damping coefficient of 5 ps−1, 1 atm constant pressure, and the Langevin piston period and decay of 100 and 50 fs, respectively. The bonded interactions, the van der Waals interaction with 12 Å cutoff, and the long-range electrostatic interactions with the particle-mesh Ewald were considered in the calculations of the forces acting on the system. At the end, the RMSD of the CLOCK backbone with respect to its initial structure showed that the equilibrium was reached after 3–4 ns. The final structure of the CLOCK after 10 ns of simulation was used as the receptor for docking step.
Docking setup
After MD simulations, water molecules of the system were deleted and the PDBQT file of the receptor, CLOCK, was prepared using AutoDockTools4 (43). The library of commercially available small molecules (by Ambinter) was filtered according to “Lipinski's Rule of Five” (30) considering a maximum limit of one violation. Finally, ∼2 million small molecules were selected for docking. PDBQT files of ligands were prepared by means of an automated script in Python language. We used AutoDock Vina for estimating protein-ligand affinity and predicting the best binding conformations of the compounds (29). The search space was defined to include the whole CLOCK protein to perform blind docking. The exhaustiveness was set as the default. Ultimately, the compounds were ranked based on their binding affinities (kcal/mol). The protein-compound interactions of the top 500 hit compounds were visually examined by the Discovery Studio Visualizer. The threshold value for hydrogen bonding was set as 3.4 Å and the accessible surface area was created considering a radius of 1.4 Å for solvent molecules. In the process of selecting hit molecules, compounds with docking positions far from CLOCK:BMAL1 protein interfaces were eliminated. Diversity in both binding region and chemical structure and shape complementarity as other important factors contributing in protein-ligand interactions were considered.
Cell cytotoxicity test
We measured cell viability with a MTT-based cell cytotoxicity test. U2OS cells were seeded on a 96-well-plate with 4 × 103 cell/well density. Two days later, the cells were treated with an appropriate amount of different small molecules. Control cells were treated with 0.5% vehicle (DMSO (Biofroxx, catalogue number 1264)). After 48 h, the media was replaced with a 20% solution of 5 mg/ml of MTT in DMEM (Gibco, catalogue number 41966-029). After 4 h of incubation, the media was discarded and a 1:1 solution of ethanol and DMSO was used to dissolve the formazan crystals. Cell viability was measured as stated in Sigma-Aldrich product information.
Mammalian two-hybrid assay
The procedure was carried out according to Checkmate, Promega's protocol. Samples were examined in triplicate sets in a 96-well-plate. HEK293T cells with the density of 4 × 104 cell/well were transfected with 50 ng of pG5-luc, 50 ng of pACT-mBmal1, 50 ng of pBind-hsClock plasmids, and 0.5 ng of pRL-TK for normalization. Following the transfection, the cells were treated with different concentrations of small molecules. Control cells were treated with 0.5% vehicle (DMSO). Ultimately, luminescence was measured using Dual Luciferase Reporter Assay System (Promega, catalogue number 6066706) with Fluoroskan Ascent FL plate reader.
Real-time monitoring of circadian rhythm
We used Bmal1-dLuc U2OS and Bmal1-dLuc NIH 3T3 cell lines. We continuously monitored the bioluminescence to record circadian rhythm of these cell lines. This experiment was performed in 96-well-plates (triplicates) and 35-mm dishes (duplicates) in the secondary screen and detailed characterization steps, respectively. Briefly, cells were seeded to reach confluence. A day after, the medium was replaced with DMEM containing 0.1 μm dexamethasone (Sigma, catalogue number D-8893) to synchronize cells. Two hours later, 0.1 mm luciferin (Promega, catalogue number E160C) was freshly added to the recording medium (low glucose DMEM powder (with l-glutamine) supplemented with 10 mm HEPES (Gibco, catalogue number 15630-080), 0.35 mg/ml of sodium bicarbonate, 3.5 mg/ml of d-(+)-glucose powder, 5% fetal bovine serum (Gibco, catalogue number 16000044), 0.1 mm NEAA (Multicell, catalogue number 321-011-EL), 100 units/ml of penicillin and 100 mg/ml of streptomycin (Multicell, catalogue number 450-201-EL). Then, the medium was replaced with recording medium containing an appropriate amount of each compound. Control cells were treated with 0.5% vehicle (DMSO). For a detailed characterization of CLK8, whenever required, and without any further medium change, small molecule treatment was postponed to 2 days after bioluminescence recording started. The 96-well-plates were covered, and bioluminescence was monitored by Synergy H1 Hybrid Multi-Mode Reader every 32 min. Similarly, we sealed the 35-mm plates with vacuum grease prior to mounting them on a LumiCycle luminometer (Actimetrics). Bioluminescence was recorded every 10 min for 4–6 days. Raw data (counts/sec) were plotted against time. The period and amplitude parameters were obtained using LumiCycle Analysis software (Actimetrics). First, the raw data were baseline fitted (n = 1), and then the baseline-subtracted data were fitted to a sine wave (damped). The first day's data were excluded from the analysis due to transient perturbations of luminescence after the medium change.
Pulldown assay
HEK293T cells were transiently transfected with tag-free hsClock and mBmal1. After 48 h of transfection, cells were harvested and whole cell lysate was prepared with ice-cold lysis buffer (50 mm Tris-HCl, pH 7.4, 2 mm EDTA, 1 mm MgCl2, 0.2% Nonidet P-40, 1 mm sodium orthovanadate, 1 mm sodium fluoride and protease inhibitor mixture) followed by centrifugation at 7,000 × g at 4 °C for 10 min. Lysate was diluted with ice-cold 2× binding buffer (100 mm Tris-HCl, pH 7.4, 300 mm NaCl, 0.2% Nonidet P-40, 2 mm sodium orthovanadate, 2 mm sodium fluoride, and protease inhibitor mixture). The lysate was divided into three fractions to be treated with either DMSO, or 20 μm bitoinylated-CLK8 (bait) with or without 200 μm CLK8 (competitor). The lysate-compound samples were incubated and mixed continuously at 4 °C for 1 h. After equilibrating NeutroAvidin-agarose resin (Thermo Scientific, catalogue number 29200) with lysis buffer, the lysate-compound mixture and NeutroAvidin-agarose resin were incubated and mixed continuously at 4 °C overnight. The beads were washed four times with 1× binding buffer. To elute the bound proteins Laemmli buffer was added and samples were heated at 95 °C for 7 min. The pulldown precipitates were subjected to SDS-PAGE and transferred to a polyvinylidene difluoride membrane (Millipore). Anti-CLOCK (Bethyl Lab, Inc., catalogue number A302-618A) and anti-BMAL1 (Santa Cruz Biotechnology, catalogue number sc-365645) antibodies were used to detect CLOCK and BMAL1 proteins, respectively. The LC-MS/MS examination of the pulldown precipitates was performed in Koc University's Proteomics Facility (Istanbul, Turkey). Bitoinylated-CLK8 and CLK8 were synthesized by Enamine, Ukraine.
Co-immunoprecipitation
HEK293T cells co-expressing WT or F80A,K220A mutant FLAG-tagged hsCLOCK together with tag-free mBMAL1 were lysed with ice-cold lysis buffer (50 mm Tris-HCl, pH 7.7, 150 mm NaCl, 0.1% Triton X-100, 1 mm phenylmethylsulfonyl fluoride and protease inhibitor mixture) and centrifuged at 15,000 × g at 4 °C for 20 min. DMSO and different concentrations of CLK8 were added to the whole cell lysate. The mixtures were incubated with anti-FLAG M2 affinity gel (Sigma-Aldrich, catalogue number A2220) at 4 °C overnight with continuous mixing. After three wash steps, bound proteins were eluted with Laemmli buffer heated at 95 °C for 7 min. At the end of the Western blot analysis, CLOCK and BMAL1 were detected by monoclonal anti-FLAG M2 antibody (Sigma-Aldrich, catalogue number F1804) and anti-BMAL1 (Santa Cruz Biotechnology, catalogue number sc-365645), respectively. The amount of co-precipitated BMAL1 was normalized with the amount of precipitated CLOCK.
mRNA and protein analysis
Unsynchronized Bmal1-dLuc U2OS cells were seeded on 6-well dishes as described in Ref. 44. Confluent cells were treated with an appropriate amount of CLK8 for 48 h. Control cells were treated with 0.5% vehicle (DMSO). After harvesting the cells, mRNAs were isolated (RNeasy, Qiagen with catalogue number 74104) and converted to cDNA (Thermo Scientific). Finally, cDNAs were subjected to SYBR Green-based RT-qPCR assay. PRLP0 was used as an internal control. The primers used in RT-qPCR are listed in Table S1. For Western blot analysis RIPA buffer was used to prepare whole cell lysates. The amount of protein for each sample was normalized to the β-ACTIN amount detected by β-Actin antibody (Cell Signaling, catalogue number 3700).
Luciferase degradation assay
The procedure was similar to the one described in a previous study (16). Briefly, HEK293T cells were seeded in triplicates in opaque 96-well-plates 24 h prior to transfection. Cells were transiently transfected with 10 ng of pcDNA-luc plasmid. After 24 h, cells were treated with different concentrations of CLK8 or DMSO. A day later, luciferin and HEPES (pH 7.2) were added to the medium to the final concentrations of 0.4 and 10 mm, respectively, and incubated for 1 h. Subsequently, protein synthesis was ceased by CHX (cycloheximide) treatment and the luminescence signal was recorded by Synergy H1 Hybrid Multi-Mode Reader for 20 h. The results were first normalized and then fitted to one-phase decay to calculate the half-life of Luciferase protein.
Transactivation assay
HEK293T cells with the density of 4 × 104 cell/well were transfected with 50 ng of mPer1-luc, 50 ng of Sport6-Bmal1, and 125 ng of Sport6-Clock (WT) or 125 ng of Sport6-Clock(F80A,K220A) plasmids and 0.5 ng of pRL-TK for normalization. The same experiment was also performed using FLAG-tagged constructs of CLOCK instead. The Dual Luciferase Reporter Assay System (Promega) was used to measure the luminescence by a Fluoroskan Ascent FL plate reader.
Site-directed mutagenesis
Site-directed mutagenesis of Sport6-hsClock and pCMV-Flag-hsClock was performed using platinum Pfx DNA polymerase (Invitrogen) as described in the product's manual. The primer sets are listed in Table S2.
Cytosol-nuclear fractionation
U2OS cells treated with 20 μm CLK8 and 0.5% DMSO were used for fractionation analysis. The 4 × 105 was suspended in 500 μl of cytosolic lysis buffer (10 mm HEPES, pH 7.9, 10 mm KCl, 0,1 mm EDTA, 0,05% Nonidet P-40 with protease inhibitors (Thermo Scientific, catalogue number 79429) and incubated on ice for 10 min. The lysate was centrifuged at 3,000 rpm for 3 min at 4 °C. The resulting supernatant was centrifuged one more time and the supernatant was collected as a cytosolic fraction. On the other hand, the resulting pellet from lysate centrifugation was suspended in nuclear lysis buffer (20 mm HEPES, pH 7.9, 0.4 m NaCl, 1 mm EDTA, 10% glycerol with protease inhibitors) and sonicated 2 times for 10 s with 60% power. After centrifuging at 15,000 × g for 5 min at 4 °C, the supernatant was collected as nuclear fraction. Collected fractions were used for Western blot analysis. Fractionation efficiency was checked with Western blotting using tubulin (Sigma, catalogue number T9026) as cytosolic marker and Histone-H3 (Abcam, catalogue number ab1791) as nuclear marker.
Lentivirus production and transduction
HEK293T cells at 75–85% confluence were transfected with 10 ng of pLV6-Bmal1-dLuc plasmid, 9 ng of pCMV-ΔR8.2dvpr packaging vector, and 1 ng of pCMV-VSVG envelope vector for 16 h. The following day, the medium was replaced with fresh growth medium. Lentiviral particles were harvested at 72- and 96-h post-transfection. Clock-deficient (generated by targeting TCTAGCATTACCAGGAAGCA sequence using CRISPR-Cas9 gene editing technique) or WTMDAMB231, as described in Ref. 45, and primary MSF cells were transduced with Bmal1-dLuc lentiviral particles for 16 h. MSF cells were kindly provided by Dr. Aziz Sancar (University of North Carolina at Chapel Hill, NC) and was described previously (46). Medium was replaced with fresh growth medium and transduction was repeated one more time. At 72-h post-transduction cells were synchronized with 0.1 μm dexamethasone and the circadian rhythms of cells were monitored using Lumicycle as described above.
In vivo studies in mouse liver
Male C57BL/6J mice, 8 weeks of age, weighing 18–24 g, were treated with two different single doses of CLK8 (25 mg/kg, n = 5 per each dose) intraperitoneally. Control mice (n = 3) were only treated with vehicle (DMSO:Cremophor EL:0.9% NaCl; 2.5:15:82.5, v/v, i.p.). Water and food were provided ad libitum throughout the experiments. Six hours after injection, mice were euthanized by cervical dislocation. Liver tissues were removed, frozen in liquid nitrogen, and stored at −80 °C until further processing.
Investigators agreed to abide by Good Animal Practice guidelines and under the guiding principles detailed in the Declaration of Helsinki, and in accordance with any applicable local law(s) and regulation(s). Mice were obtained from Koç University Animal Research Facility (KUTTAM) and experiments were conducted in accordance with the guidelines approved for animal experimental procedures by the Koc University Animal Research Local Ethics Committee (number 2018/011).
Quantification of Western blotting
All Western blots were obtained from Bio-Rad ChemiDoc Imaging System and the amount of each protein was quantified using Image Lab Software applying volume tools.
Statistical analysis
Statistical significance was evaluated using one-way or two-way analysis of variance (ANOVA), followed by a Tukey's multiple comparisons test using Prism software (GraphPad Software). Biological replica was at least n = 3 for all of the experiments (*, p value <0.05; **, p value <0.01; ***, p value <0.001).
Author contributions
Y. U. D., D. Y., Y. K. A., S. G., A. C. T., F. Y., I. B., Nuri Ozturk, M. T., Narin Ozturk, and A. O. data curation; Y. U. D. and Y. K. A. investigation; Y. U. D., D. Y., Nuri Ozturk, M. T., A. O., and I. H. K. methodology; D. Y., A. O., and I. H. K. conceptualization; D. Y. and I. H. K. formal analysis; I. H. K. funding acquisition; Y. U. D., D. Y., and I. H. K. writing-original draft; I. H. K. project administration; I. H. K. writing-review and editing.
Supplementary Material
Acknowledgment
We thank Emma Harris from the University of Edinburgh for critical reading.
This work was supported by The Scientific and Technological Research Council of Turkey (TUBITAK KBAG) Grant 118Z140 (to I. H. K). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Fig. S1 and Tables S1 and S2.
- TTFLs
- transcription/translation feedback loops
- CLOCK
- circadian locomotor output cycles caput
- BMAL1
- Brain and muscle Arnt-like protein-1 (BMAL1), brain and muscle
- RMSD
- root mean square deviation
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- SCN
- suprachiasmatic nuclei
- MSF
- mouse skin fibroblast
- qPCR
- quantitative PCR
- bHLH
- basic helix loop helix
- MD
- molecular dynamics
- DMEM
- Dulbecco's modified Eagle's medium
- CMV
- cytomegalovirus
- ANOVA
- analysis of variance.
References
- 1. Lowrey P. L., and Takahashi J. S. (2011) Genetics of circadian rhythms in mammalian model organisms. Adv. Genet. 74, 175–230 10.1016/B978-0-12-387690-4.00006-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dunlap J. C. (1999) Molecular bases for circadian clocks. Cell 96, 271–290 10.1016/S0092-8674(00)80566-8 [DOI] [PubMed] [Google Scholar]
- 3. Gloston G. F., Yoo S. H., and Chen Z. J. (2017) Clock-enhancing small molecules and potential applications in chronic diseases and aging. Front. Neurol. 8, 100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Manoogian E. N. C., and Panda S. (2017) Circadian rhythms, time-restricted feeding, and healthy aging. Ageing Res. Rev. 39, 59–67 10.1016/j.arr.2016.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Schroeder A. M., and Colwell C. S. (2013) How to fix a broken clock. Trends Pharmacol. Sci. 34, 605–619 10.1016/j.tips.2013.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zee P. C., Attarian H., and Videnovic A. (2013) Circadian rhythm abnormalities. Continuum. (Minneap Minn) 19, 132–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gekakis N., Staknis D., Nguyen H. B., Davis F. C., Wilsbacher L. D., King D. P., Takahashi J. S., and Weitz C. J. (1998) Role of the CLOCK protein in the mammalian circadian mechanism. Science 280, 1564–1569 10.1126/science.280.5369.1564 [DOI] [PubMed] [Google Scholar]
- 8. Hogenesch J. B., Gu Y. Z., Jain S., and Bradfield C. A. (1998) The basic-helix-loop-helix-PAS orphan MOP3 forms transcriptionally active complexes with circadian and hypoxia factors. Proc. Natl. Acad. Sci. U.S.A. 95, 5474–5479 10.1073/pnas.95.10.5474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. King D. P., Zhao Y., Sangoram A. M., Wilsbacher L. D., Tanaka M., Antoch M. P., Steeves T. D., Vitaterna M. H., Kornhauser J. M., Lowrey P. L., Turek F. W., and Takahashi J. S. (1997) Positional cloning of the mouse circadian clock gene. Cell 89, 641–653 10.1016/S0092-8674(00)80245-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kume K., Zylka M. J., Sriram S., Shearman L. P., Weaver D. R., Jin X., Maywood E. S., Hastings M. H., and Reppert S. M. (1999) mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell 98, 193–205 10.1016/S0092-8674(00)81014-4 [DOI] [PubMed] [Google Scholar]
- 11. Takahashi J. S. (2017) Transcriptional architecture of the mammalian circadian clock. Nat. Rev. Genet. 18, 164–179 10.1038/nrg.2016.150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hirano A., Fu Y. H., and Ptáček L. J. (2016) The intricate dance of post-translational modifications in the rhythm of life. Nat. Struct. Mol. Biol. 23, 1053–1060 10.1038/nsmb.3326 [DOI] [PubMed] [Google Scholar]
- 13. Antoch M. P., and Chernov M. V. (2009) Pharmacological modulators of the circadian clock as potential therapeutic drugs. Mutat. Res. Gen. Tox. En. 680, 109–115 10.1016/j.mrgentox.2009.07.015 [DOI] [PubMed] [Google Scholar]
- 14. Chen Z., Yoo S. H., and Takahashi J. S. (2013) Small molecule modifiers of circadian clocks. Cell Mol. Life Sci. 70, 2985–2998 10.1007/s00018-012-1207-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hirota T., Lee J. W., Lewis W. G., Zhang E. E., Breton G., Liu X., Garcia M., Peters E. C., Etchegaray J. P., Traver D., Schultz P. G., and Kay S. A. (2010) High-throughput chemical screen identifies a novel potent modulator of cellular circadian rhythms and reveals CKIα as a clock regulatory kinase. PLos Biol. 8, e1000559 10.1371/journal.pbio.1000559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hirota T., Lee J. W., St John P. C., Sawa M., Iwaisako K., Noguchi T., Pongsawakul P. Y., Sonntag T., Welsh D. K., Brenner D. A., Doyle F. J. 3rd, Schultz P. G., and Kay S. A. (2012) Identification of small molecule activators of cryptochrome. Science 337, 1094–1097 10.1126/science.1223710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Isojima Y., Nakajima M., Ukai H., Fujishima H., Yamada R. G., Masumoto K. H., Kiuchi R., Ishida M., Ukai-Tadenuma M., Minami Y., Kito R., Nakao K., Kishimoto W., Yoo S. H., Shimomura K., et al. (2009) CKI ϵ/δ-dependent phosphorylation is a temperature-insensitive, period-determining process in the mammalian circadian clock. Proc. Natl. Acad. Sci. U.S.A. 106, 15744–15749 10.1073/pnas.0908733106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chun S. K., Jang J., Chung S., Yun H., Kim N. J., Jung J. W., Son G. H., Suh Y. G., and Kim K. (2014) Identification and validation of cryptochrome inhibitors that modulate the molecular circadian clock. ACS Chem. Biol. 9, 1213–1213 10.1021/cb5002872 [DOI] [PubMed] [Google Scholar]
- 19. Hu Y., Spengler M. L., Kuropatwinski K. K., Comas-Soberats M., Jackson M., Chernov M. V., Gleiberman A. S., Fedtsova N., Rustum Y. M., Gudkov A. V., and Antoch M. P. (2011) Selenium is a modulator of circadian clock that protects mice from the toxicity of a chemotherapeutic drug via upregulation of the core clock protein, BMAL1. Oncotarget 2, 1279–1290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kojetin D. J., and Burris T. P. (2014) REV-ERB and ROR nuclear receptors as drug targets. Nat. Rev. Drug Discov. 13, 197–216 10.1038/nrd4100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yagita K., Yamanaka I., Koinuma S., Shigeyoshi Y., and Uchiyama Y. (2009) Mini screening of kinase inhibitors affecting period-length of mammalian cellular circadian clock. Acta Histochem. Cytochem. 42, 89–93 10.1267/ahc.09015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yoshitane H., Honma S., Imamura K., Nakajima H., Nishide S. Y., Ono D., Kiyota H., Shinozaki N., Matsuki H., Wada N., Doi H., Hamada T., Honma K., and Fukada Y. (2012) JNK regulates the photic response of the mammalian circadian clock. EMBO Rep. 13, 455–461 10.1038/embor.2012.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Huang N., Chelliah Y., Shan Y., Taylor C. A., Yoo S. H., Partch C., Green C. B., Zhang H., and Takahashi J. S. (2012) Crystal structure of the heterodimeric CLOCK:BMAL1 transcriptional activator complex. Science 337, 189–194 10.1126/science.1222804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. McIntosh B. E., Hogenesch J. B., and Bradfield C. A. (2010) Mammalian Per-Arnt-Sim proteins in environmental adaptation. Annu. Rev. Physiol. 72, 625–645 10.1146/annurev-physiol-021909-135922 [DOI] [PubMed] [Google Scholar]
- 25. Turek F. W., Penev P., Zhang Y., van Reeth O., and Zee P. (1995) Effects of age on the circadian system. Neurosci. Biobehav. Rev. 19, 53–58 10.1016/0149-7634(94)00030-5 [DOI] [PubMed] [Google Scholar]
- 26. Vitaterna M. H., Ko C. H., Chang A. M., Buhr E. D., Fruechte E. M., Schook A., Antoch M. P., Turek F. W., and Takahashi J. S. (2006) The mouse Clock mutation reduces circadian pacemaker amplitude and enhances efficacy of resetting stimuli and phase-response curve amplitude. Proc. Natl. Acad. Sci. U.S.A. 103, 9327–9332 10.1073/pnas.0603601103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lee Y., Chen R., Lee H. M., and Lee C. (2011) Stoichiometric relationship among Clock proteins determines robustness of circadian rhythms. J. Biol. Chem. 286, 7033–7042 10.1074/jbc.M110.207217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Çakir B., Daǧliyan O., Daǧyildiz E., Bariș İ., Kavakli I. H., Kizilel S., and Türkay M. (2012) Structure based discovery of small molecules to regulate the activity of human insulin degrading enzyme. PLoS ONE 7, e31787 10.1371/journal.pone.0031787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Trott O., and Olson A. J. (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31, 455–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lipinski C. A. (2000) Drug-like properties and the causes of poor solubility and poor permeability. J. Pharmacol. Toxicol. Methods 44, 235–249 10.1016/S1056-8719(00)00107-6 [DOI] [PubMed] [Google Scholar]
- 31. Armutlu P., Ozdemir M. E., Ozdas S., Kavakli I. H., and Turkay M. (2009) Discovery of novel CYP17 inhibitors for the treatment of prostate cancer with structure-based drug design. Lett. Drug Des. Discov. 6, 337–344 10.2174/1570180810906050337 [DOI] [Google Scholar]
- 32. Brown S. A., Pagani L., Cajochen C., and Eckert A. (2011) Systemic and cellular reflections on ageing and the circadian oscillator: a mini-review. Gerontology 57, 427–434 [DOI] [PubMed] [Google Scholar]
- 33. Nohara K., Yoo S. H., and Chen Z. J. (2015) Manipulating the circadian and sleep cycles to protect against metabolic disease. Front. Endocrinol. (Lausanne) 6, 35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nishide S. Y., Honma S., and Honma K. (2008) The circadian pacemaker in the cultured suprachiasmatic nucleus from pup mice is highly sensitive to external perturbation. Eur. J. Neurosci. 27, 2686–2690 10.1111/j.1460-9568.2008.06231.x [DOI] [PubMed] [Google Scholar]
- 35. Mellacheruvu D., Wright Z., Couzens A. L., Lambert J. P., St-Denis N. A., Li T., Miteva Y. V., Hauri S., Sardiu M. E., Low T. Y., Halim V. A., Bagshaw R. D., Hubner N. C., Al-Hakim A., Bouchard A., et al. (2013) The CRAPome: a contaminant repository for affinity purification-mass spectrometry data. Nat. Methods 10, 730–736 10.1038/nmeth.2557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kondratov R. V., Chernov M. V., Kondratova A. A., Gorbacheva V. Y., Gudkov A. V., and Antoch M. P. (2003) BMAL1-dependent circadian oscillation of nuclear CLOCK: posttranslational events induced by dimerization of transcriptional activators of the mammalian clock system. Genes Dev. 17, 1921–1932 10.1101/gad.1099503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kavakli I. H., Baris I., Tardu M., Gül S., Öner H., C̦al S., Bulut S., Yarparvar D., Berkel C̦., Ustaoǧlu P., and Aydin C. (2017) The photolyase/cryptochrome family of proteins as DNA repair enzymes and transcriptional repressors. Photochem. Photobiol. 93, 93–103 10.1111/php.12669 [DOI] [PubMed] [Google Scholar]
- 38. Kwon I., Lee J., Chang S. H., Jung N. C., Lee B. J., Son G. H., Kim K., and Lee K. H. (2006) BMAL1 shuttling controls transactivation and degradation of the CLOCK/BMAL1 heterodimer. Mol. Cell Biol. 26, 7318–7330 10.1128/MCB.00337-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Li Y., Xiong W., and Zhang E. E. (2016) The ratio of intracellular CRY proteins determines the clock period length. Biochem. Biophys. Res. Commun. 472, 531–538 10.1016/j.bbrc.2016.03.010 [DOI] [PubMed] [Google Scholar]
- 40. He B., Nohara K., Park N., Park Y. S., Guillory B., Zhao Z., Garcia J. M., Koike N., Lee C. C., Takahashi J. S., Yoo S. H., and Chen Z. (2016) The small molecule nobiletin targets the molecular oscillator to enhance circadian rhythms and protect against metabolic syndrome. Cell Metab. 23, 610–621 10.1016/j.cmet.2016.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Phillips J. C., Braun R., Wang W., Gumbart J., Tajkhorshid E., Villa E., Chipot C., Skeel R. D., Kalé L., and Schulten K. (2005) Scalable molecular dynamics with NAMD. J. Comput. Chem. 26, 1781–1802 10.1002/jcc.20289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Humphrey W., Dalke A., and Schulten K. (1996) VMD: visual molecular dynamics. J. Mol. Graph 14, 33–38, 27–38 [DOI] [PubMed] [Google Scholar]
- 43. Morris G. M., Huey R., Lindstrom W., Sanner M. F., Belew R. K., Goodsell D. S., and Olson A. J. (2009) AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem. 30, 2785–2791 10.1002/jcc.21256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ozber N., Baris I., Tatlici G., Gur I., Kilinc S., Unal E. B., and Kavakli I. H. (2010) Identification of two amino acids in the C-terminal domain of mouse CRY2 essential for PER2 interaction. BMC Mol. Biol. 11, 69 10.1186/1471-2199-11-69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Korkmaz T., Aygenli F., Emisoglu H., Ozcelik G., Canturk A., Yilmaz S., and Ozturk N. (2018) Opposite carcinogenic effects of circadian clock gene BMAL1. Sci. Rep. 8, 16023 10.1038/s41598-018-34433-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gaddameedhi S., Reardon J. T., Ye R., Ozturk N., and Sancar A. (2012) Effect of circadian clock mutations on DNA damage response in mammalian cells. Cell Cycle 11, 3481–3491 10.4161/cc.21771 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.