Abstract
CRISPR-Cas systems provide bacteria with adaptive immunity against viruses. During spacer adaptation, the Cas1-Cas2 complex selects fragments of foreign DNA, called prespacers, and integrates them into CRISPR arrays in an orientation that provides functional immunity. Cas4 is involved in both the trimming of prespacers and the cleavage of protospacer adjacent motif (PAM) in several type I CRISPR-Cas systems, but how the prespacers are processed in systems lacking Cas4, such as the type I-E and I-F systems, is not understood. In Escherichia coli, which has a type I-E system, Cas1-Cas2 preferentially selects prespacers with 3′ overhangs via specific recognition of a PAM, but how these prespacers are integrated in a functional orientation in the absence of Cas4 is not known. Using a biochemical approach with purified proteins, as well as integration, prespacer protection, sequencing, and quantitative PCR assays, we show here that the bacterial 3′–5′ exonucleases DnaQ and ExoT can trim long 3′ overhangs of prespacers and promote integration in the correct orientation. We found that trimming by these exonucleases results in an asymmetric intermediate, because Cas1-Cas2 protects the PAM sequence, which helps to define spacer orientation. Our findings implicate the E. coli host 3′–5′ exonucleases DnaQ and ExoT in spacer adaptation and reveal a mechanism by which spacer orientation is defined in E. coli.
Keywords: CRISPR/Cas, CRISPR-RNA (crRNA), DNA, deoxyribonuclease (DNase), integrase, 3'-5' exonuclease, bacterial immunity, DnaQ, prespacer processing, primed adaptation, protospacer adjacent motif (PAM)
Introduction
Bacteria and archaea defend against phage infection and invasion by mobile genetic elements using an adaptive immune system composed of CRISPR arrays and Cas (CRISPR-associated) proteins (1). In these systems, during spacer adaptation, short fragments of foreign dsDNA, called prespacers, are incorporated into host CRISPR arrays (2–4). Prespacers are thought to derive from events that result in DNA breaks and free DNA termini (5–9). The CRISPR array is transcribed and processed into CRISPR RNAs (crRNAs),2 which assemble with Cas proteins to form RNA-guided surveillance complexes (10–12). These complexes then recognize and degrade foreign nucleic acids that are complementary to the crRNA, a process termed interference. To defend against divergent viruses that have been previously encountered, type I CRISPR systems use a positive feedback mechanism between interference and adaptation, called priming. During primed adaptation, prespacers are thought to derive from degradation products generated by interference (13–16).
Adaptation relies on two Cas proteins, Cas1 and Cas2, which are almost universally conserved in CRISPR-Cas systems (17). These proteins form a heterohexameric complex (which we call here Cas1-Cas2). Cas1 is an integrase, and Cas2 is a dimeric structural protein that bridges two dimers of Cas1 (16, 18–20). The Cas1-Cas2 complex captures prespacers and catalyzes the direct nucleophilic attack of the 3′-OH ends of each strand of the prespacer into the CRISPR array. The first attack occurs between the leader and the first repeat of the CRISPR array, generating a half-site intermediate. The second attack occurs between the other end of the first repeat and the first spacer, generating a full-site integration product (21, 22). Specificity for this region is enhanced by an integration host factor (IHF), which binds to the leader sequence (23). In vitro, Cas1-Cas2 has been shown to preferentially integrate prespacers with 3′ overhangs. The most efficient prespacer for in vitro integration consists of a 23-bp duplex flanked by 5-nucleotide (nt) 3′ overhangs, which we refer to as the preprocessed prespacer (24, 25).
Escherichia coli Cas1-Cas2 selects prespacers that contain a protospacer adjacent motif, or PAM (25–27). PAM is also essential for recognition of foreign DNA by the surveillance complex during interference (28). The PAM is recognized through specific contacts between the C-terminal tail of Cas1 and the PAM sequence, which is 5′-CTT-3′ in E. coli (25). Cas1-Cas2 selects prespacers with a 3′ overhang that contains a PAM, but because prespacers derive from heterogeneous fragments produced by other cellular processes, the length of the 3′ overhangs will vary. Thus, for integration to occur efficiently, 3′ overhangs that are longer than the optimal length for integration must be trimmed to 5 nt and contain 3′-OH ends. Integration of the prespacer strand containing the PAM, which we refer to as the PAM strand, requires removal of the entire PAM. However, in E. coli, only the 5′-TT-3′ sequence of the PAM is removed, because the last nucleotide of each repeat derives from the last nucleotide of the PAM sequence (13, 25, 29, 30). Recent findings have highlighted the role of Cas4 in both the trimming of prespacers and the cleavage of PAM in several type I CRISPR-Cas systems (31–35), but how prespacers are processed in systems lacking Cas4, such as the type I-E and I-F system, is not understood.
To incorporate a functional spacer that will produce a crRNA complementary to the target strand of an invader, integration must occur in a defined orientation (Fig. S1). In addition to PAM recognition and prespacer trimming, Cas4 has also been shown to play a critical role in ensuring that spacers are integrated in the correct orientation (32). In the type I-E system of E. coli, in vitro integration of the preprocessed prespacer displays minimal bias for the correct orientation (24). In contrast, in vivo, integration of a prespacer that contains a PAM sequence results in a strong bias for the correct orientation (36). Although PAM dictates orientation, the mechanism by which prespacers are integrated in the correct orientation in type I-E systems is not known.
Here, using a biochemical approach, we show that two host 3′–5′ exonucleases, DnaQ and ExoT, can trim unprocessed prespacers for Cas1-Cas2 to integrate into a CRISPR array. As determined by quantitative PCR (qPCR), integration in the presence of these exonucleases displays a strong bias for the correct orientation. DNA protection experiments reveal that these exonucleases differentially trim the non-PAM and PAM strands of an unprocessed prespacer bound by Cas1-Cas2. The result of this differential protection is an asymmetric intermediate that is directed to integrate in the correct orientation. Together, our findings demonstrate how host 3′–5′ exonucleases process prespacers and define spacer orientation in E. coli.
Results
DnaQ and ExoT recover integration of unprocessed prespacers
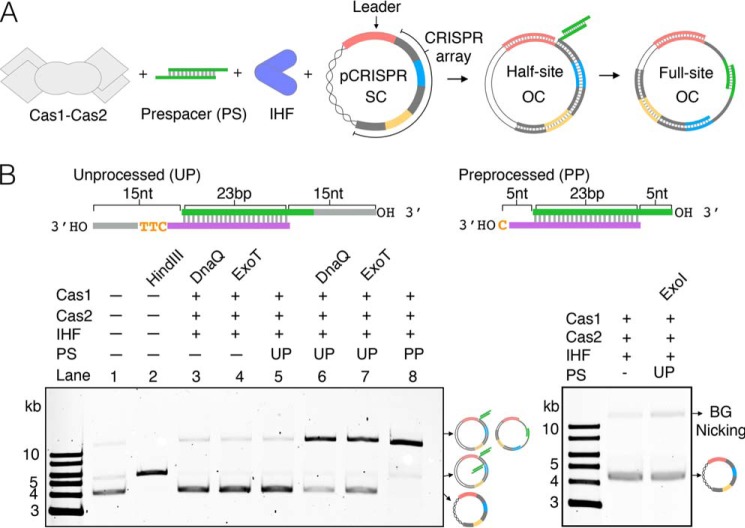
Given that most biochemical studies of spacer adaptation in the type I-E CRISPR system of E. coli have been conducted using the preprocessed prespacer, we wanted to investigate whether Cas1-Cas2 could integrate an unprocessed prespacer, consisting of a 23-bp duplex flanked by two 15-nt 3′ overhangs. We used a plasmid topology assay, whereby integration of a prespacer into plasmid DNA containing a CRISPR array (pCRISPR) results in the conversion of supercoiled plasmid to open circle plasmid (24) (Fig. 1A). The sequence of the unprocessed prespacer derives from the most abundant spacer acquired following M13 bacteriophage infection (13) (Fig. S2). Whereas Cas1-Cas2 integrates the preprocessed prespacer (5-nt 3′ overhangs) efficiently (Fig. 1B, lane 8), we were unable to detect integration of the unprocessed prespacer (Fig. 1B, lanes 3 and 4).
Figure 1.
Integration of unprocessed prespacer recovered by DnaQ and ExoT. A, schematic of plasmid integration assay. SC, supercoiled plasmid; OC, open circle plasmid. Repeats are gray, spacers are blue and yellow, and the leader sequence is pink. The entire array is not represented. B, agarose gels of integration assays using a processed prespacer (PP) or unprocessed prespacer (UP) in the absence or presence of 3′–5′ exonucleases, alongside linearized (HindIII-treated) plasmid, a control reaction lacking prespacer, and untreated plasmid.
The type I-E CRISPR system of Streptococcus thermophilus contains a Cas2 that is fused to a DnaQ-like domain. This DnaQ-like domain functions as a 3′–5′ exonuclease that trims prespacer 3′ overhangs to promote integration (37). In this species, in vitro integration of an unprocessed prespacer by DnaQ-mutant complexes is undetectable (37), which is consistent with our data (Fig. 1B) and suggests that Cas1-Cas2 cannot efficiently integrate unprocessed prespacers. Given these observations and the fact that the majority of type I-E CRISPR-Cas systems do not have a DnaQ-like domain fused to Cas2 (38), we wondered whether host 3′–5′ exonucleases could trim unprocessed prespacers in E. coli. E. coli contains several exonucleases that belong to the DnaQ superfamily, including the proofreading subunit of DNA polymerase III, DnaQ (39), and the repair nucleases ExoT and ExoI (40, 41). We therefore tested whether the addition of one of these 3′–5′ exonucleases could recover integration of the unprocessed prespacer. For DnaQ, we used a construct corresponding to its catalytic domain (residues 1–186), as the full-length protein is insoluble (42) (from here onward, we refer to this construct as DnaQ). We found that the addition of DnaQ or ExoT recovered integration (Fig. 1B, lanes 6 and 7), presumably by trimming the 3′ overhangs of the prespacers, but the addition of ExoI failed to recover integration (Fig. 1B).
Integration in the presence of DnaQ and ExoT displays a strong bias for the correct orientation, which is dictated by the PAM sequence
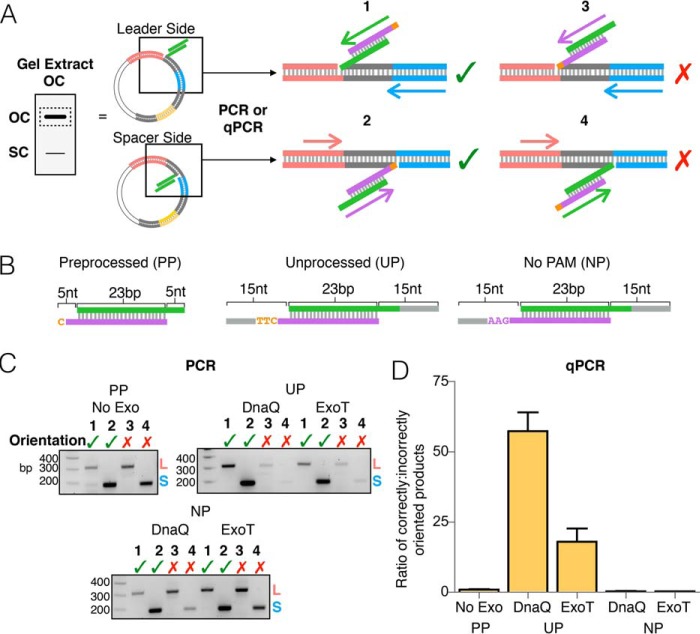
We next wished to determine the orientation of prespacers integrated into pCRISPR in the presence of DnaQ or ExoT. We therefore used PCR with primers specific to the integrated M13 spacer and either the leader (spacer-side integration) or a downstream spacer (leader-side integration) (Fig. 2A and Fig. S3). Integration reactions were first run through agarose gels, and open circle plasmid products were extracted and used as templates for PCR (Fig. 2A). Products from reactions without prespacers and without exonuclease were tested to verify that primers are specific to integration events (Fig. S4). Integration of a prespacer in the correct orientation results in insertion of the PAM strand at the spacer-side junction and the non-PAM strand at the leader-side junction (Fig. 2A). Consistent with previous data (21), integration of a preprocessed prespacer, with 5-nt 3′ overhangs (Fig. 2B), occurs in both orientations at similar frequencies (Fig. 2C). In the presence of DnaQ or ExoT, integration of the unprocessed prespacer, with 15-nt 3′ overhangs (Fig. 2B), occurs with a noticeable bias for the correct orientation (Fig. 2C). However, removing the PAM sequence from the unprocessed prespacer (Fig. 2B) results in integration with no obvious bias in orientation (Fig. 2C).
Figure 2.
Orientation bias displayed by DnaQ- and ExoT-mediated integration. A, schematic depicting PCR amplification of half-site integration events. Products of integration reactions (open circle plasmid) were gel-extracted and used as templates in four different PCRs, using different primer sets to identify the site of integration and the orientation of integrated spacer. Numbers above schematics indicate the four possible integration events. A green checkmark indicates a half-site product in the correct orientation; a red X indicates a product in the incorrect orientation. B, substrates used in the integration reactions. C, agarose gels of all end-point PCRs. Prespacer (processed prespacer (PP), unprocessed prespacer (UP), or no PAM prespacer (NP)) and exonuclease used in the integration reactions are shown above each gel. Numbers refer to the four reactions in A. D, graph depicting the ratio of correctly oriented to incorrectly oriented half-site products determined by qPCR. Ratios represent the combined number of leader- and spacer-side integration events for each orientation, normalized to picogram amounts of the β-lactamase gene. The average of three replicates is shown with error bars representing S.D.
The inclusion of a PAM in prespacers that are electroporated into cells alters the orientation frequency of expanded arrays significantly, resulting in a bias of ∼25 to 1 correctly oriented spacers to incorrectly oriented spacers (36). To quantify the orientation bias displayed by in vitro integration in the presence of DnaQ and ExoT, we used qPCR, again using primers specific to the integrated spacer sequence and either the leader or a downstream spacer sequence (Fig. 2A). Primer efficiencies were calculated using standard curves that were generated from two control templates, which contained the expanded CRISPR array with the new M13 spacer in either the correct or incorrect orientation (Fig. S5). The gel-extracted products of integration were used as templates for qPCR, and relative amounts of each integration product were calculated using the Ct values for each reaction and the standard curves for each primer set. The amounts of integration products were then normalized to the amount of β-lactamase (bla) template (pCRISPR contains the bla gene). Whereas the preprocessed prespacer integrates into the array in both orientations at similar frequencies, integration of the unprocessed prespacer in the presence of DnaQ or ExoT displays a strong bias for the correct orientation, ∼60 to 1 with DnaQ and ∼18 to 1 with ExoT (Fig. 2D). Consistent with the end-point PCR results (Fig. 2C), removing the PAM sequence significantly diminishes the orientation bias (Fig. 2D). Thus, DnaQ and ExoT confer an orientation bias that is consistent with previous in vivo data (36), and as expected, this orientation bias is dictated by the PAM (28, 43).
Cas1-Cas2 differentially protects the PAM and non-PAM strands from cleavage by DnaQ and ExoT
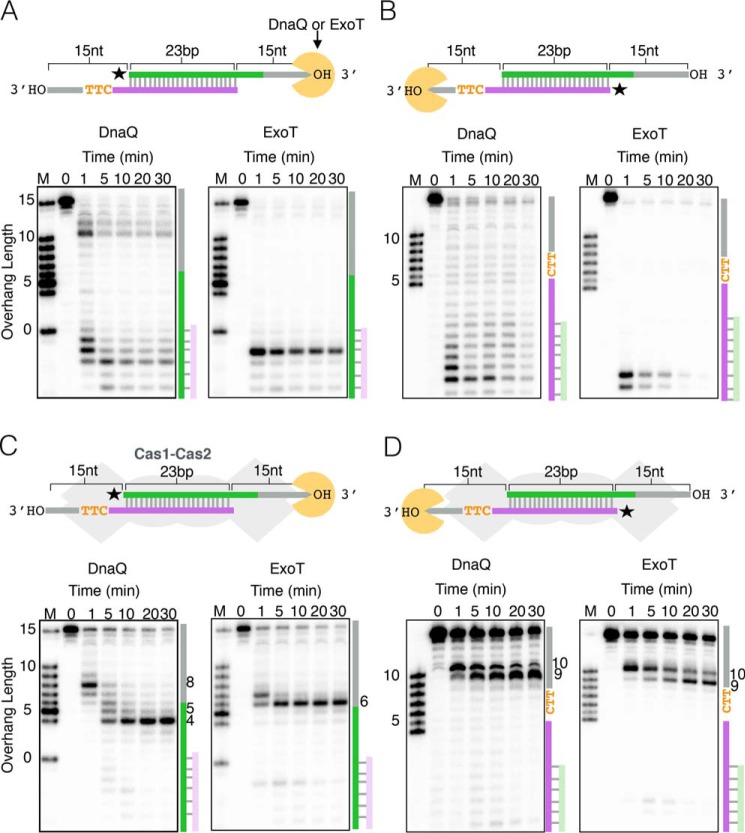
Given that the PAM dictates orientation bias, we wondered whether there are differences in how DnaQ or ExoT trims the PAM and non-PAM strands of the unprocessed prespacer. As such, we monitored cleavage of radiolabeled strands of the unprocessed prespacer over time. In the absence of Cas1-Cas2, both DnaQ and ExoT chew through the entire prespacer, with minimal sequence-dependent differences between strands (Fig. 3, A and B). However, the PAM and non-PAM strands are trimmed differently in the presence of Cas1-Cas2 (Fig. 3, C and D). DnaQ trims the 3′ overhang of the non-PAM strand to 4 nt, generating the optimal 5-nt 3′ overhang at around 5 min (Fig. 3C). However, ExoT trims the non-PAM strand predominantly to 6 nt, generating minimal 5-nt 3′ overhangs (Fig. 3C). In contrast, both exonucleases stall on the PAM strand and generate 3′ overhangs that are 9 or 10 nt long (Fig. 3D). Thus, trimming by DnaQ or ExoT produces asymmetric prespacers with longer 3′ overhangs on the PAM strand, and neither exonuclease removes the PAM sequence.
Figure 3.
Protection of prespacers from 3′–5′ exonuclease cleavage by Cas1-Cas2. Shown are time courses of DnaQ or ExoT cleavage of an unprocessed prespacer. A, radiolabeling of the non-PAM strand in the absence of Cas1-Cas2. B, radiolabeling of the PAM strand in the absence of Cas1-Cas2. C, radiolabeling of the non-PAM strand in the presence of Cas1-Cas2. D, radiolabeling of the PAM strand in the presence of Cas1-Cas2. In all panels, a schematic of the experimental setup is shown above the gel images, with a star indicating which strand is radiolabeled. On the right side of each gel image is a schematic highlighting cleavage of the unprocessed prespacer.
Cas1-Cas2 integration has a greater tolerance of 3′ overhang length on the non-PAM strand than the PAM strand
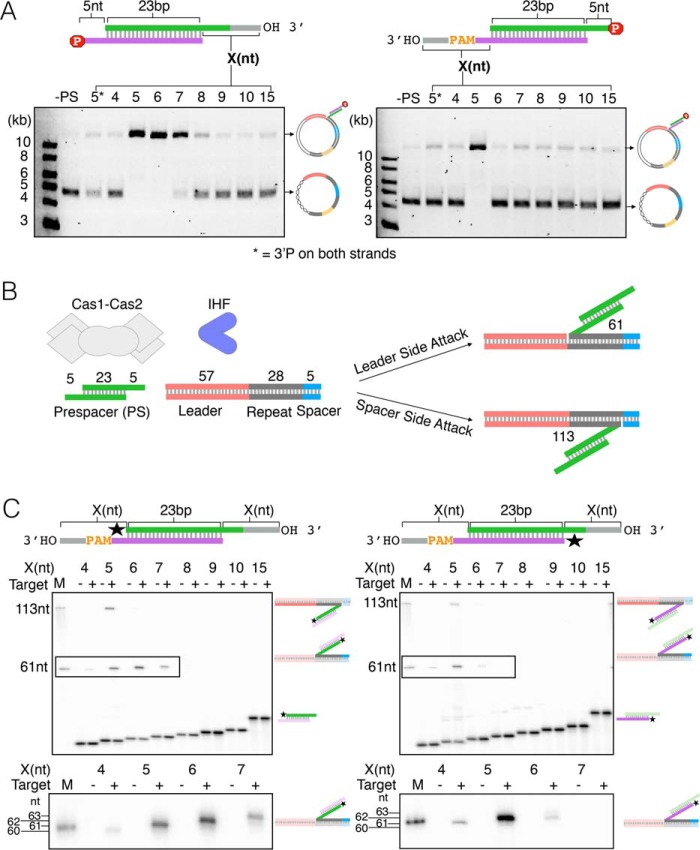
Given the results of our protection experiments (Fig. 3), we next asked how 3′ overhang length affects integration by Cas1-Cas2. To do this, we used the pCRISPR topology assay (Fig. 1A) and a series of prespacers with different length 3′ overhangs, derived from sequences flanking the M13 protospacer (Fig. S2). To distinguish integration of the PAM strand and the non-PAM strand, we selectively blocked the opposite strand for integration with a 3′-phosphate group (Fig. 4A) (21, 22). We found that Cas1-Cas2 efficiently integrates prespacers with 5-, 6-, or 7-nt 3′ overhangs on the non-PAM strand but only integrates prespacers with 5-nt 3′ overhangs on the PAM strand (Fig. 4A). Because DnaQ and ExoT generate 3′ overhangs that are 9 or 10 nt long on the PAM strand (Fig. 3D), this suggests that 4 or 5 additional nucleotides must be subsequently trimmed prior to integration. However, we are unable to resolve whether they are removed by Cas1 or DnaQ/ExoT, which we will discuss in greater detail below.
Figure 4.
Integration tolerance of varied 3′overhangs by Cas1-Cas2. A, plasmid integration assay showing variation in 3′ overhang tolerance between non-PAM strand (left) and PAM (strand). 3′-Phosphate used to block integration of the complementary strand is indicated by a red stop sign symbol containing the letter P. Reactions with varying overhang lengths (X) were run on an agarose gel alongside two control reactions: no prespacer (−PS) and prespacer with 5-nt overhangs containing 3′-phosphate blocks on both strands (5*). B, schematic of an oligonucleotide target integration assay showing reaction components and expected products of leader-side (61 nt) and spacer-side (113 nt) integration of the preprocessed prespacer containing 5-nt 3′ overhangs on both strands. C, denaturing sequencing gel of the oligonucleotide integration assay using prespacers with varied symmetric 3′ overhangs (X). Radiolabel is indicated with a star. Schematics of expected half-site products are shown on the right side of each gel. M, marker; minus or plus symbols indicate the absence or presence of the leader-repeat-spacer oligonucleotide shown in A. For clarity, zoomed-in cutouts of leader-side products, outlined on each gel, are shown below each gel.
To gain more insight into the details of these integration assays, we repeated these experiments with an oligonucleotide target and selectively radiolabeled prespacers that were symmetric, containing the same length 3′ overhangs on the non-PAM and PAM strands. Here, we can distinguish leader-side integration and spacer-side integration because the resulting products differ in length (Fig. 4B). At the leader side, non-PAM strands with 5-, 6-, or 7-nt 3′ overhangs were efficiently integrated into the oligonucleotide target, whereas integration of the PAM strand occurred most efficiently with a 5-nt 3′ overhang (Fig. 4C). At the spacer side, only 5-nt 3′ overhangs were integrated, independent of strand (Fig. 4C). In control experiments, which lacked the oligonucleotide target, we observed no cleavage of the prespacers (Fig. 4C). Together, these data show that leader-side integration of the non-PAM strand is more tolerant of 3′ overhang length than the PAM strand, and efficient integration of the PAM strand only occurs with a 5-nt 3′ overhang. They also suggest that, contrary to a previous report (25), Cas1-Cas2 does not trim prespacers, under the conditions tested.
The presence of PAM blocks spacer-side integration of the non-PAM strand
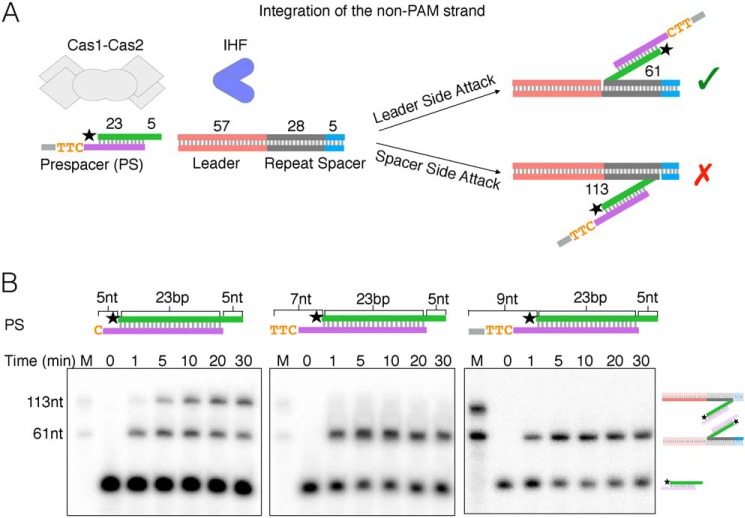
The results of the above experiments suggest that spacer-side integration of symmetric prespacers only occurs with 5-nt overhangs on either the non-PAM or PAM strand. Given that DnaQ and ExoT trim the prespacer non-PAM and PAM strands differently, we wondered how Cas1-Cas2 would integrate prespacers with asymmetric 3′ overhangs. Previous findings have shown that the rate of leader-side integration is faster than the rate of spacer-side integration with a symmetric preprocessed prespacer (5-nt 3′ overhangs), suggesting that leader-side integration occurs first (19, 23, 44). We therefore compared integration of the non-PAM strand (Fig. 5A) in prespacers containing PAM strands with 5-, 7-, and 9-nt 3′ overhangs (Fig. 5B). Consistent with previous results, the symmetric prespacer was integrated more quickly at the leader side. However, if the PAM strand overhang is increased to 7 or 9 nt, both of which contain a PAM, spacer-side integration of the non-PAM strand is not detected (Fig. 5B). Thus, the presence of the PAM ensures that the non-PAM strand is integrated at the leader-repeat junction only, thereby preventing incorrectly oriented spacers that may result from spacer-side integration of the non-PAM strand first (Fig. 5A).
Figure 5.
The presence of PAM blocks spacer-side integration of the non-PAM strand. A, schematic of oligonucleotide target integration assay with asymmetric prespacer, showing expected half-site products of integration of the non-PAM strand in either the correct (green checkmark) or incorrect (red X) orientation. B, denaturing gels of time courses of the oligonucleotide integration assay using three different prespacers (PS), shown by schematics above each gel. The reaction is started with the addition of the CRISPR target. 0, mock reaction without any target. M, marker lane; radiolabels are indicated by a star. Schematics of expected products are to the right of the gel images.
Sequencing of integration products shows PAM cleavage and reveals differences between DnaQ and ExoT
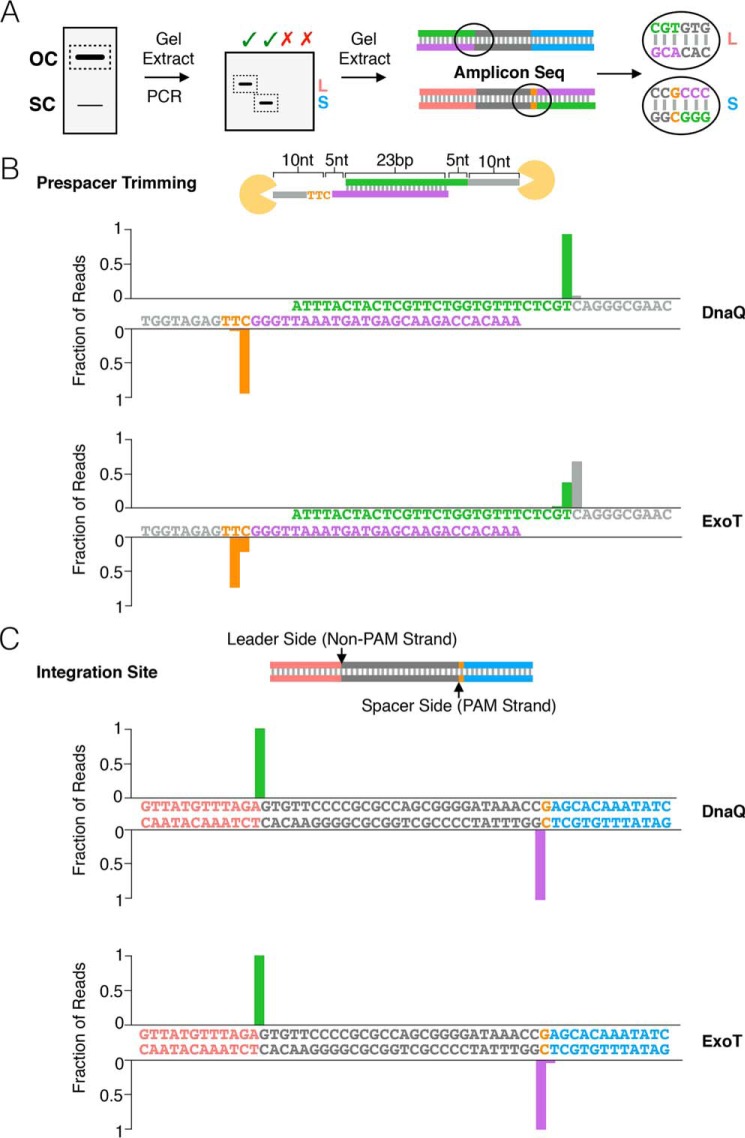
Our results show that trimming by DnaQ or ExoT of the non-PAM strand generates 4- and 6-nt 3′ overhangs, respectively (Fig. 3C), whereas stalling of both exonucleases on the PAM strand produces a 9- or 10-nt overhang (Fig. 3D). However, for integration to occur, both 3′ overhangs should be trimmed to 5 nt, and the 5′-TT-3′ sequence of the PAM should be removed (Fig. 4C) (13, 25, 29, 30). To determine whether integration of an unprocessed prespacer in the presence of DnaQ or ExoT (Fig. 1B) results in appropriate trimming and correct PAM processing, and to map the sites of integration, we Illumina-sequenced the integration products (Fig. 6A). For reactions with DnaQ, the majority of the reads were consistent with trimming of both strands of the unprocessed prespacer to 5-nt 3′ overhangs, with a small proportion resulting in 6-nt 3′ overhangs (Fig. 6B). Trimming of the PAM strand thus removed the 5′-TT-3′ sequence from the PAM. In contrast, reactions with ExoT were more heterogeneous, with most reads consistent with trimming of both strands to 3′ overhangs 6 nt in length (Fig. 6B). Mapping of the integration sites revealed that the majority of integration at the leader and spacer side occurred at the expected sites in the presence of either DnaQ or ExoT (Fig. 6C).
Figure 6.
Sites of DnaQ- and ExoT-mediated prespacer trimming and integration shown by amplicon sequencing. A, schematic of protocol used to analyze sequence of integration products via PCR followed by amplicon deep sequencing. B, graphs depict the sequence of the overhangs produced by DnaQ or ExoT prespacer trimming. Bars on the graphs are aligned above the nucleotide that is left behind upon trimming by the exonuclease. A schematic of unprocessed prespacer used in the integration assay is shown above the graphs. Sequences on the x axes are colored according to the schematic. C, graphs depict sites of integration; bars are aligned at the junction between two nucleotides at which integration occurs in the presence of either DnaQ or ExoT. Sequences are colored according to the schematic of the CRISPR array above. The color of each bar corresponds to the color of the strand (non-PAM or PAM) that is integrated at that junction.
Discussion
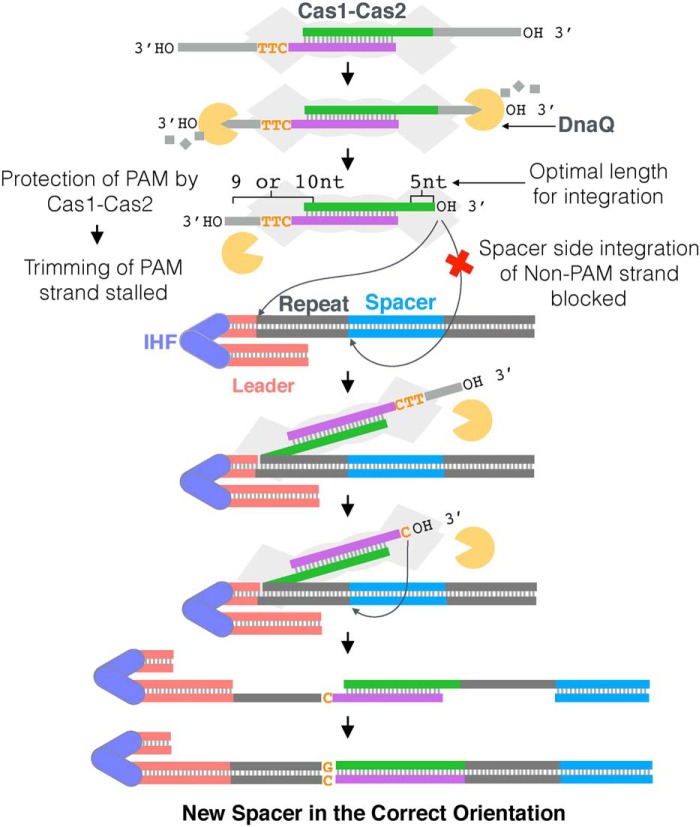
To date, in CRISPR systems that lack Cas4, it is not known how prespacers with long 3′ overhangs are processed and integrated in the functional orientation. Together, our data for the type I-E system from E. coli support a model (Fig. 7) in which DnaQ, or other host 3′–5′ exonucleases, can trim long 3′ overhangs of prespacers bound by Cas1-Cas2. Protection of the PAM by Cas1 leads to differential trimming of the non-PAM and PAM strands (Fig. 3), forming an intermediate that contains asymmetric 3′ overhangs. In the first step of integration, the non-PAM strand of the asymmetric prespacer is directed to the leader side, whereas spacer-side integration is blocked (Fig. 5). We speculate that PAM processing follows formation of a half-side intermediate and that the PAM strand is then integrated at the spacer side. The result is a fully integrated spacer in an orientation that is functional for downstream target recognition and interference (Fig. 7).
Figure 7.
Model for processing and integration mediated by DnaQ in E. coli. Upon binding of a prespacer containing 3′ overhangs longer than the optimal 5 nt by Cas1-Cas2, host exonucleases, such as DnaQ, trim the overhangs to different lengths, depending on the presence of a PAM. Whereas the Non-PAM strand is trimmed to the optimal 5-nt 3′ overhang, trimming of the PAM strand is stalled at a 9- or 10-nt 3′ overhang, due to the protection of PAM by Cas1-Cas2, resulting in an asymmetric intermediate. Spacer-side integration of the non-PAM strand is then blocked, forcing its integration at the leader side and ensuring the formation of a correctly oriented half-site intermediate. Following half-site formation and PAM cleavage, a full-site product is formed, and the repeats are duplicated, resulting in a CRISPR array with a new spacer in the correct orientation.
Previous studies with E. coli Cas1 have suggested that it can cleave 3′ overhangs, but they did not show whether the resulting prespacers could be integrated by Cas1-Cas2 (25). Here, we show that E. coli Cas1-Cas2 does not efficiently integrate unprocessed prespacers with long 3′ overhangs (Fig. 1); nor do we detect any cleavage of these prespacers (Fig. 4), which is consistent with studies of the type I-E system from S. thermophilus (37). The addition of DnaQ or ExoT, but not ExoI, recovers integration of unprocessed prespacers and promotes integration in a functional orientation (Figs. 1 and 2). These results suggest that some, but not all, DnaQ family exonucleases are functional for spacer adaptation in type I-E CRISPR systems.
Host 3′–5′ exonucleases process the two 3′ overhangs of a prespacer differently. DnaQ and ExoT can trim the non-PAM strand of an unprocessed prespacer to overhang lengths of 4 and 6 nt, respectively, but stall on the PAM strand, leaving a 9- or 10-nt overhang (Fig. 3). Based on previous structural studies, we speculate that stalling on the PAM strand is likely due to the protection of the PAM sequence by the C-terminal tail of Cas1, which specifically recognizes the PAM sequence but is disordered when bound to a strand that lacks a PAM (25) (Fig. S6). The result is an asymmetric intermediate, which is consistent with recent evidence that in vivo, spacer precursors detected during primed adaptation in both type I-E and type I-F systems share an asymmetrical structure characterized by a blunt non-PAM end and a 3′ overhang at the PAM end (45).
Although it is clear that PAM dictates spacer orientation (28, 36, 43) (Fig. 2), the mechanism by which it does so in type I-E systems is poorly understood. For prespacers to be incorporated in a functional orientation, their non-PAM strand must be integrated at the leader side of the first repeat and their PAM strand at the spacer side of the first repeat. To achieve this, the processed non-PAM strand of the asymmetric intermediate, generated by host 3′–5′ exonucleases, is directed to integrate at the leader side first. The fidelity of this step is ensured not only by IHF and the intrinsic specificity of the Cas1-Cas2 complex for the leader sequence (22, 23, 44), but also because spacer-side integration is blocked by the PAM sequence in the asymmetric intermediate (Fig. 5). Following formation of the leader-side half-site, the 5′-TT-3′ sequence of the PAM must be removed for subsequent integration of the PAM strand (Figs. 4 and 5). Our sequencing experiments show that in the presence of DnaQ, the majority of integration events do result from PAM processing (Fig. 6). However, our data cannot resolve whether the 5′-TT-3′ sequence of the PAM is removed by Cas1 or DnaQ. Resolving this ambiguity will be an important component of future studies. Together, our data suggest that host 3′–5′ exonucleases dictate spacer orientation in type I-E and possibly type I-F systems (i.e. type I systems that lack Cas4) by differential trimming of unprocessed prespacers.
Recognition of PAM is a critical step during adaptation and is mediated by different CRISPR components, depending on subtype. Cas1 recognizes PAM in the type I-E system of E. coli (25) (Fig. S6). However, in most type I systems, it is thought that Cas4 recognizes PAM (31–34). In the type I-A system, the distinct processing and binding activities of two Cas4 proteins on opposite ends of a prespacer provide the asymmetry that results in integration in a defined orientation (32). In the type I-C system, asymmetrical complexes of Cas4-Cas1-Cas2 have been observed, in which Cas4 only processes the PAM end of the prespacer (33). PAM recognition by Cas1 distinguishes the type I-E systems from type I systems in which Cas4 is thought to recognize PAM and define orientation. Yet, despite this difference in PAM recognition, the asymmetric trimming of prespacer 3′ overhangs by 3′–5′ exonucleases is perhaps a universal mechanism for CRISPR systems to ensure that spacers are functionally oriented for downstream target interference.
Our findings suggest that DnaQ or ExoT promotes integration of unprocessed prespacers via trimming their 3′ overhangs to a length that is optimal for integration. In E. coli, 95% of spacers are 32 bp long, despite spacer length varying between 31 and 34 bp (46). Our sequencing data reveal that the majority of integration events in the presence of DnaQ are consistent with generating 32-bp spacers (Fig. 6B), because the cytosine of the PAM sequence is considered a part of the repeat. We also show that the non-PAM strand can be integrated with 5-, 6-, and 7-nt 3′ overhangs (Fig. 4), which could explain why longer spacers are detected in vivo (46). In contrast to DnaQ, the majority of integration events in the presence of ExoT are consistent with generating 33- or 34-bp spacers (Fig. 6B). This suggests that DnaQ, or possibly other unidentified host 3′–5′ exonucleases, may play a larger role than ExoT in prespacer processing. The 3′–5′ exonucleases that facilitate processing of prespacers add to the growing list of host factors required for spacer adaptation in E. coli (6, 23).
Experimental procedures
Protein purification
The cas1, cas2, IHF, and DnaQ (residues 1–186) genes from E. coli K12 (MG1655) were cloned into expression vectors and purified separately. Cas1 and IHF were cloned into vectors containing a TEV-cleavable N-terminal His6 tag. Cas1 was cloned into pHAT4 (47). IHFα and IHFβ genes were cloned into a pET His6 TEV LIC cloning vector (1B), with the His tag on the N terminus of IHFα; subunits were co-expressed and purified as a heterodimer. Cas2 was cloned into a vector containing a TEV-cleavable N-terminal His6-MBP tag (47), and DnaQ was cloned into a vector with a SENP-cleavable N-terminal His-SUMO tag (pSAT). Constructs were expressed in T7 Express cells (New England Biolabs), grown to 0.4–0.6 A600, and induced overnight at 20 °C with 0.25 mm isopropyl-β-d-thiogalactopyranoside. The cells were harvested and resuspended in buffer A (1 m KCl, 20 mm HEPES-KOH, pH 7.4, 10 mm imidazole, 1 mm TCEP, 10% glycerol) with added protease inhibitors E-64, phenylmethylsulfonyl fluoride, bestatin, and pepstatin A. After lysis by microfluidization, the lysates were cleared by centrifugation and incubated with Ni-NTA affinity resin in batch (Bio-Rad). The resin was washed in buffer A, followed by buffer B (100 mm KCl, 20 mm HEPES-KOH, pH 7.4, 10 mm imidazole, 5% glycerol, and 1 mm TCEP) and then buffer C (500 mm KCl, 20 mm HEPES-KOH, pH 7.4, 10 mm imidazole, 5% glycerol, and 1 mm TCEP). The protein was then eluted with buffer C supplemented with 250 mm imidazole. Samples were loaded onto an immobilized desalting column and exchanged into buffer C to lower the imidazole concentration before further purification via Ni-NTA affinity. To remove the affinity tags, proteins were then treated with His-tagged TEV (for Cas1, Cas2, and IHF) or His-tagged SENP (for DnaQ), with protease concentrations of about one-tenth of the total protein concentration. Samples treated with TEV were incubated overnight at 4 °C, and samples treated with SENP were incubated at 4 °C for 1 h. Samples were then incubated with Ni-NTA affinity resin in batch (Bio-Rad) to remove the proteases or any remaining tagged protein; untagged protein was collected in the flow-through. Cas2 was further purified on an MBPTrap HP (GE Healthcare) column. Proteins were then concentrated and injected onto a HiLoad 26/60 S200 size-exclusion column (GE Healthcare) equilibrated with Buffer B in the absence of imidazole. Purified proteins were stored in 100 mm KCl, 20 mm HEPES-NaOH, 5% glycerol, and 1 mm TCEP at −80 °C before use. SDS-PAGE analysis of the final purified proteins can be found in Fig. S7. ExoT was purchased from New England Biolabs.
DNA preparation
All ssDNA oligonucleotides were purchased from Sigma-Aldrich and purified using urea-PAGE. Sequences of these oligonucleotides are shown in Tables S1–S5. Prespacers and CRISPR target substrates were annealed by mixing the appropriate ssDNA oligonucleotides in 25 mm KCl, 20 mm HEPES-KOH, pH 7.4, and incubating at 95 °C for 5 min, followed by slow-cooling to room temperature. The pCRISPR target plasmid was constructed by cloning the E. coli BL21-AI genomic CRISPR locus via ligation-independent cloning into the pET LIC cloning vector (2A-T). Radiolabeled substrates were prepared by labeling 200 nm ssDNA with 1–2 pmol of [γ-32P]ATP in 1× T4 PNK buffer with 10 units of T4 polynucleotide kinase (New England Biolabs) at 37 °C for 30 min, followed by a 20-min heat inactivation at 65 °C. For prespacer substrates, labeled oligonucleotides were passed through a G25 Sephadex column to remove excess ATP and then annealed with a 1.1-fold excess of complementary unlabeled strands.
Integration assays
Integration assays were performed largely as described previously (21, 44). Reactions were performed in 25 mm KCl, 20 mm HEPES-KOH, pH 7.4, 10 mm MgCl, and 1 mm DTT. Separately purified Cas1 (100 nm) and Cas2 (50 nm) were preincubated for 20 min at 4 °C to allow complex formation. Cas1-Cas2 complex (100 nm) was incubated with 50 nm prespacer DNA, whereas 50 nm IHF was incubated with 5 nm pCRISPR in a separate tube. After a 10–15-min incubation at room temperature, the IHF-pCRISPR mix was added to the Cas1-Cas2-prespacer complex and incubated at 37 °C for 30 min. For reactions containing DnaQ or ExoT, exonuclease (100 nm) was added to the IHF-pCRISPR mix immediately before combining with the Cas1-Cas2-prespacer complex. The reactions were quenched with EDTA and SDS at final concentrations of 28 mm and 0.4%, respectively, and proteins were removed from the reactions via phenol-chloroform extraction. The DNA products were run on a 1% agarose gel post-stained with ethidium bromide.
Integration assays with 5′-radiolabeled prespacer DNA were performed as described above with the exception that the Cas1-Cas2-prespacer complex and the separate IHF-target DNA mix was incubated at 37 °C for 10–15 min instead of room temperature. Reactions contained 200 nm Cas1, 100 nm Cas2, 10 nm radiolabeled prespacer, 200 nm IHF, and 100 nm target DNA. To activate the reaction, the IHF-target DNA mix was added to the Cas1-Cas2-prespacer complex. Reactions were quenched with 2 times the reaction volume of DNA loading buffer containing 95% formamide, 0.01% SDS, 0.01% bromphenol blue, 0.01% xylene cyanol, 1 mm EDTA after 30 min (unless otherwise indicated in time-course experiments) at 37 °C. Products were run on a 6% denaturing polyacrylamide sequencing gel. Gels were dried and visualized by phosphorimaging (FujiFilm FLA-7000).
End-point PCR amplification of integration products
Relaxed, open circle, pCRISPR products were extracted from agarose gels using a GeneJET gel extraction kit (Thermo Scientific) and eluted in 30 μl of water. Samples were then diluted 50-fold, and 1 μl was used as a template in each PCR. 50-μl PCRs were conducted with the appropriate primers and Q5 High-Fidelity Polymerase (New England Biolabs) using the standard protocol and thermocycler conditions provided by New England Biolabs. 10 μl of the PCR products were run on a 2% agarose gel prestained with ethidium bromide.
Quantitative PCR amplification of integration products
Quantitative PCR assays were conducted according to the protocol provided by the Bio-Rad Sso Advanced Universal SYBR Green Supermix kit. Reactions were designed for a final volume of 20 μl. For each integration reaction, 2 μl of the gel-extracted product (eluted in 30 μl of water) was used as the template. Separate reactions for each template were carried out using four separate sets of qPCR primers (Fig. S5). Gel-extracted relaxed pCRISPR from negative control reactions containing Cas1, Cas2, and IHF without any prespacer was used as a control for background amplification.
Positive control pCRISPR plasmids containing the M13 spacer sequence in both orientations were generated by transforming gel-extracted integration reactions, performed with the preprocessed prespacers, into 10β competent E. coli cells (New England Biolabs) and sequencing the colonies. To develop standard curves, positive control pCRISPR plasmids containing the M13 spacer sequence in both orientations were diluted in 10-fold dilution series ranging from 1 pg to 10 ng of total plasmid DNA. Resulting average C(t) values were then plotted with their respective log(pg) amounts of DNA to form a standard curve, which was then used to calculate primer efficiency using the equation, EP = (10 (−1/slope) − 1) × 100 (Fig. S5). For all primer sets, efficiencies varied between 95 and 100%.
Cas1-Cas2 prespacer protection assays
Reactions were performed in the same buffer as integration assays (25 mm KCl, 20 mm HEPES-KOH, pH 7.4, 10 mm MgCl, and 1 mm DTT). Separately purified Cas1 (200 nm) and Cas2 (100 nm) were preincubated for 20 min at 4 °C to allow complex formation. The Cas1-Cas2 complex was incubated with 10 nm 5′-radiolabeled prespacer DNA at 37 °C for 20 min. DnaQ or ExoT was added at 200 nm, and reactions were incubated at 37 °C for 1, 5, 10, 20, and 30 min. Reactions were prepared as master mixes and, upon the addition of exonuclease, were aliquoted into separate tubes and incubated at different time increments. Reactions were quenched with 2 times the reaction volume of DNA loading buffer used in the integration assays above. Products were run on an 8% denaturing polyacrylamide sequencing gel. Gels were dried and visualized by phosphorimaging (FujiFilm FLA-7000).
Sequencing sample preparation
Gel-extracted integration reactions were PCR-amplified using end-point PCR procedure described above. However, instead of 2 μl of a 50-fold dilution, 1 μl of the eluted 30-μl product was used as a template, to generate enough material for amplicon sequencing. 50 μl of the PCR products were run on a 2% agarose gel and gel-extracted using the GeneJET gel extraction kit. Samples were diluted accordingly to 20 ng/μl, and 25 μl was sent for Amplicon-EZ sequencing (GENEWIZ).
Sequencing analysis
Raw paired-end reads obtained from GENEWIZ were trimmed and merged, using the default settings, with the Trimmomatic (48) and PANDAseq (49), respectively. The processed reads were mapped to the sequence of the expanded CRISPR array using BLAST (50), allowing for up to one mismatch. The extent of trimming was determined as the length of sequence between the end of the duplex sequence within the prespacer and the beginning of the repeat sequence with pCRISPR (33). The site of integration was considered to be the position at which the match between the read and pCRISPR began (33).
Author contributions
A. R. and S. B. conceptualization; A. R. and S. B. data curation; A. R. and S. B. formal analysis; A. R., L. S., B. A. L., and L. D. investigation; A. R. writing-original draft; A. R. and S. B. writing-review and editing; S. B. supervision; S. B. funding acquisition.
Supplementary Material
Acknowledgments
We thank Kaitlin Johnson for technical assistance and members of the Bailey laboratory for helpful discussion.
This work was supported by National Institutes of Health Grant GM097330 (to S. B.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article was selected as one of our Editors' Picks.
This article contains Tables S1–S5 and Figs. S1–S7.
- crRNA
- CRISPR RNA
- PAM
- protospacer adjacent motif
- pCRISPR
- plasmid containing a CRISPR array
- IHF
- integration host factor
- nt
- nucleotide(s)
- qPCR
- quantitative PCR
- TEV
- tobacco etch virus
- MBP
- maltose-binding protein
- SUMO
- small ubiquitin-like modifier
- TCEP
- tris(2-carboxyethyl)phosphine
- Ni-NTA
- nickel-nitrilotriacetic acid.
References
- 1. Barrangou R., Fremaux C., Deveau H., Richards M., Boyaval P., Moineau S., Romero D. A., and Horvath P. (2007) CRISPR provides acquired resistance against viruses in prokaryotes. Science 315, 1709–1712 10.1126/science.1138140 [DOI] [PubMed] [Google Scholar]
- 2. Mojica F. J. M., Díez-Villaseñor C. S., García-Martínez J., and Soria E. (2005) Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J. Mol. Evol. 60, 174–182 10.1007/s00239-004-0046-3 [DOI] [PubMed] [Google Scholar]
- 3. Bolotin A., Quinquis B., Sorokin A., and Ehrlich S. D. (2005) Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology 151, 2551–2561 10.1099/mic.0.28048-0 [DOI] [PubMed] [Google Scholar]
- 4. Pourcel C., Salvignol G., and Vergnaud G. (2005) CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA and provide additional tools for evolutionary studies. Microbiology 151, 653–663 10.1099/mic.0.27437-0 [DOI] [PubMed] [Google Scholar]
- 5. Babu M., Beloglazova N., Flick R., Graham C., Skarina T., Nocek B., Gagarinova A., Pogoutse O., Brown G., Binkowski A., Phanse S., Joachimiak A., Koonin E. V., Savchenko A., Emili A., et al. (2011) A dual function of the CRISPR–Cas system in bacterial antivirus immunity and DNA repair. Mol. Microbiol. 79, 484–502 10.1111/j.1365-2958.2010.07465.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ivančić-Baće I., Cass S. D., Wearne S. J., and Bolt E. L. (2015) Different genome stability proteins underpin primed and naïve adaptation in E. coli CRISPR-Cas immunity. Nucleic Acids Res. 43, 10821–10830 10.1093/nar/gkv1213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Levy A., Goren M. G., Yosef I., Auster O., Manor M., Amitai G., Edgar R., Qimron U., and Sorek R. (2015) CRISPR adaptation biases explain preference for acquisition of foreign DNA. Nature 520, 505–510 10.1038/nature14302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Modell J. W., Jiang W., and Marraffini L. A. (2017) CRISPR–Cas systems exploit viral DNA injection to establish and maintain adaptive immunity. Nature 544, 101–104 10.1038/nature21719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Shiimori M., Garrett S. C., Chambers D. P., Glover C. V. C. 3rd, Graveley B. R., and Terns M. P. (2017) Role of free DNA ends and protospacer adjacent motifs for CRISPR DNA uptake in Pyrococcus furiosus. Nucleic Acids Res. 45, 11281–11294 10.1093/nar/gkx839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. van der Oost J., Westra E. R., Jackson R. N., and Wiedenheft B. (2014) Unravelling the structural and mechanistic basis of CRISPR-Cas systems. Nat. Rev. Microbiol. 12, 479–492 10.1038/nrmicro3279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Marraffini L. A. (2015) CRISPR-Cas immunity in prokaryotes. Nature 526, 55–61 10.1038/nature15386 [DOI] [PubMed] [Google Scholar]
- 12. Hille F., Richter H., Wong S. P., Bratovič M., Ressel S., and Charpentier E. (2018) The biology of CRISPR-Cas: backward and forward. Cell 172, 1239–1259 10.1016/j.cell.2017.11.032 [DOI] [PubMed] [Google Scholar]
- 13. Datsenko K. A., Pougach K., Tikhonov A., Wanner B. L., Severinov K., and Semenova E. (2012) Molecular memory of prior infections activates the CRISPR/Cas adaptive bacterial immunity system. Nat. Commun. 3, 945 10.1038/ncomms1937 [DOI] [PubMed] [Google Scholar]
- 14. Künne T., Kieper S. N., Bannenberg J. W., Vogel A. I. M., Miellet W. R., Klein M., Depken M., Suárez-Diez M., and Brouns S. J. J. (2016) Cas3-derived target DNA degradation fragments fuel primed CRISPR adaptation. Mol. Cell. 63, 852–864 10.1016/j.molcel.2016.07.011 [DOI] [PubMed] [Google Scholar]
- 15. Semenova E., Savitskaya E., Musharova O., Strotskaya A., Vorontsova D., Datsenko K. A., Logacheva M. D., and Severinov K. (2016) Highly efficient primed spacer acquisition from targets destroyed by the Escherichia coli type I-E CRISPR-Cas interfering complex. Proc. Natl. Acad. Sci. U.S.A. 113, 7626–7631 10.1073/pnas.1602639113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fagerlund R. D., Wilkinson M. E., Klykov O., Barendregt A., Pearce F. G., Kieper S. N., Maxwell H. W. R., Capolupo A., Heck A. J. R., Krause K. L., Bostina M., Scheltema R. A., Staals R. H. J., and Fineran P. C. (2017) Spacer capture and integration by a type I-F Cas1-Cas2–3 CRISPR adaptation complex. Proc. Natl. Acad. Sci. U.S.A. 114, E5122–E5128 10.1073/pnas.1618421114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yosef I., Goren M. G., and Qimron U. (2012) Proteins and DNA elements essential for the CRISPR adaptation process in Escherichia coli. Nucleic Acids Res. 40, 5569–5576 10.1093/nar/gks216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nuñez J. K., Kranzusch P. J., Noeske J., Wright A. V., Davies C. W., and Doudna J. A. (2014) Cas1–Cas2 complex formation mediates spacer acquisition during CRISPR–Cas adaptive immunity. Nat. Struct. Mol. Biol. 21, 528–534 10.1038/nsmb.2820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Xiao Y., Ng S., Nam K. H., and Ke A. (2017) How type II CRISPR-Cas establish immunity through Cas1-Cas2-mediated spacer integration. Nature 550, 137–141 10.1038/nature24020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rollins M. F., Chowdhury S., Carter J., Golden S. M., Wilkinson R. A., Bondy-Denomy J., Lander G. C., and Wiedenheft B. (2017) Cas1 and the Csy complex are opposing regulators of Cas2/3 nuclease activity. Proc. Natl. Acad. Sci. U.S.A. 114, E5113–E5121 10.1073/pnas.1616395114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nuñez J. K., Lee A. S. Y., Engelman A., and Doudna J. A. (2015) Integrase-mediated spacer acquisition during CRISPR-Cas adaptive immunity. Nature 519, 193–198 10.1038/nature14237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rollie C., Schneider S., Brinkmann A. S., Bolt E. L., White M. F., and Nilsen T. W. (2015) Intrinsic sequence specificity of the Cas1 integrase directs new spacer acquisition. eLife 4, e08716 10.7554/eLife.08716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nuñez J. K., Bai L., Harrington L. B., Hinder T. L., and Doudna J. A. (2016) CRISPR immunological memory requires a host factor for specificity. Mol. Cell. 62, 824–833 10.1016/j.molcel.2016.04.027 [DOI] [PubMed] [Google Scholar]
- 24. Nuñez J. K., Harrington L. B., Kranzusch P. J., Engelman A. N., and Doudna J. A. (2015) Foreign DNA capture during CRISPR-Cas adaptive immunity. Nature 527, 535–538 10.1038/nature15760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang J., Li J., Zhao H., Sheng G., Wang M., Yin M., and Wang Y. (2015) Structural and mechanistic basis of PAM-dependent spacer acquisition in CRISPR-Cas systems. Cell 163, 840–853 10.1016/j.cell.2015.10.008 [DOI] [PubMed] [Google Scholar]
- 26. Díez-Villaseñor C., Guzmán N. M., Almendros C., García-Martínez J., and Mojica F. J. M. (2013) CRISPR-spacer integration reporter plasmids reveal distinct genuine acquisition specificities among CRISPR-Cas I-E variants of Escherichia coli. RNA Biol. 10, 792–802 10.4161/rna.24023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Deveau H., Barrangou R., Garneau J. E., Labonté J., Fremaux C., Boyaval P., Romero D. A., Horvath P., and Moineau S. (2008) Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J. Bacteriol. 190, 1390–1400 10.1128/JB.01412-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shah S. A., Erdmann S., Mojica F. J. M., and Garrett R. A. (2013) Protospacer recognition motifs. RNA Biol. 10, 891–899 10.4161/rna.23764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Goren M. G., Yosef I., Auster O., and Qimron U. (2012) Experimental definition of a clustered regularly interspaced short palindromic duplicon in Escherichia coli. J. Mol. Biol. 423, 14–16 10.1016/j.jmb.2012.06.037 [DOI] [PubMed] [Google Scholar]
- 30. Swarts D. C., Mosterd C., van Passel M. W. J., and Brouns S. J. J. (2012) CRISPR interference directs strand specific spacer acquisition. PLoS One 7, e35888 10.1371/journal.pone.0035888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rollie C., Graham S., Rouillon C., and White M. F. (2018) Prespacer processing and specific integration in a Type I-A CRISPR system. Nucleic Acids Res. 46, 1007–1020 10.1093/nar/gkx1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Shiimori M., Garrett S. C., Graveley B. R., and Terns M. (2018) Cas4 nucleases define the PAM, length, and orientation of DNA fragments integrated at CRISPR loci. Mol. Cell 70, 814–824.e6 10.1016/j.molcel.2018.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lee H., Zhou Y., Taylor D. W., and Sashital D. G. (2018) Cas4-dependent prespacer processing ensures high-fidelity programming of CRISPR arrays. Mol. Cell 70, 48–59.e5 10.1016/j.molcel.2018.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kieper S. N., Almendros C., Behler J., McKenzie R. E., Nobrega F. L., Haagsma A. C., Vink J. N. A., Hess W. R., and Brouns S. J. J. (2018) Cas4 facilitates PAM-compatible spacer selection during CRISPR adaptation. Cell Rep. 22, 3377–3384 10.1016/j.celrep.2018.02.103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lee H., Dhingra Y., and Sashital D. G. (2019) The Cas4-Cas1-Cas2 complex mediates precise prespacer processing during CRISPR adaptation. eLife 8, e44248 10.7554/eLife.44248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shipman S. L., Nivala J., Macklis J. D., and Church G. M. (2016) Molecular recordings by directed CRISPR spacer acquisition. Science 353, aaf1175–aaf1175 10.1126/science.aaf1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Drabavicius G., Sinkunas T., Silanskas A., Gasiunas G., Venclovas Č., and Siksnys V. (2018) DnaQ exonuclease-like domain of Cas2 promotes spacer integration in a type I-E CRISPR-Cas system. EMBO Rep. 19, e45543 10.15252/embr.201745543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Makarova K. S., Wolf Y. I., Alkhnbashi O. S., Costa F., Shah S. A., Saunders S. J., Barrangou R., Brouns S. J. J., Charpentier E., Haft D. H., Horvath P., Moineau S., Mojica F. J. M., Terns R. M., Terns M. P., et al. (2015) An updated evolutionary classification of CRISPR-Cas systems. Nat. Rev. Microbiol. 13, 722–736 10.1038/nrmicro3569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Scheuermann R., Tam S., Burgers P. M., Lu C., and Echols H. (1983) Identification of the ϵ-subunit of Escherichia coli DNA polymerase III holoenzyme as the dnaQ gene product: a fidelity subunit for DNA replication. Proc. Natl. Acad. Sci. U.S.A. 80, 7085–7089 10.1073/pnas.80.23.7085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cooper D. L., Lahue R. S., and Modrich P. (1993) Methyl-directed mismatch repair is bidirectional. J. Biol. Chem. 268, 11823–11829 [PubMed] [Google Scholar]
- 41. Hsiao Y.-Y., Fang W.-H., Lee C.-C., Chen Y.-P., and Yuan H. S. (2014) Structural insights into DNA repair by RNase T—an exonuclease processing 3′ end of structured DNA in repair pathways. PLoS Biol. 12, e1001803 10.1371/journal.pbio.1001803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hamdan S., Brown S. E., Thompson P. R., Yang J. Y., Carr P. D., Ollis D. L., Otting G., and Dixon N. E. (2000) Preliminary X-ray crystallographic and NMR studies on the exonuclease domain of the ϵ subunit of Escherichia coli DNA polymerase III. J. Struct. Biol. 131, 164–169 10.1006/jsbi.2000.4291 [DOI] [PubMed] [Google Scholar]
- 43. Shmakov S., Savitskaya E., Semenova E., Logacheva M. D., Datsenko K. A., and Severinov K. (2014) Pervasive generation of oppositely oriented spacers during CRISPR adaptation. Nucleic Acids Res. 42, 5907–5916 10.1093/nar/gku226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wright A. V., Liu J.-J., Knott G. J., Doxzen K. W., Nogales E., and Doudna J. A. (2017) Structures of the CRISPR genome integration complex. Science 357, 1113–1118 10.1126/science.aao0679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Shiriaeva A. A., Savitskaya E., Datsenko K. A., Vvedenskaya I. O., Fedorova I., Morozova N., Metlitskaya A., Sabantsev A., Nickels B. E., Severinov K., and Semenova E. (2019) Detection of spacer precursors formed in vivo during primed CRISPR adaptation. Nat. Commun. 10, 4603 10.1038/s41467-019-12417-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Savitskaya E., Semenova E., Dedkov V., Metlitskaya A., and Severinov K. (2013) High-throughput analysis of type I-E CRISPR/Cas spacer acquisition in E. coli. RNA Biol. 10, 716–725 10.4161/rna.24325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Peränen J., Rikkonen M., Hyvönen M., and Kääriäinen L. (1996) T7 vectors with a modified T7lac promoter for expression of proteins in Escherichia coli. Anal. Biochem. 236, 371–373 10.1006/abio.1996.0187 [DOI] [PubMed] [Google Scholar]
- 48. Bolger A. M., Lohse M., and Usadel B. (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Masella A. P., Bartram A. K., Truszkowski J. M., Brown D. G., and Neufeld J. D. (2012) PANDAseq: paired-end assembler for illumina sequences. BMC Bioinformatics 13, 31 10.1186/1471-2105-13-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., and Madden T. L. (2009) BLAST+: architecture and applications. BMC Bioinformatics 10, 421 10.1186/1471-2105-10-421 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.