Abstract
Polarized epithelial cells have functionally distinct apical and basolateral membranes through which they communicate with external and internal bodily environments, respectively. The establishment and maintenance of this asymmetric structure depend on polarized trafficking of specific cargos, but the precise molecular mechanism is incompletely understood. We previously showed that Rab35, a member of the Rab family small GTPases, differentially regulates the trafficking of an apical cargo, podocalyxin (PODXL), in two-dimensional (2D) and three-dimensional (3D) Madin–Darby canine kidney (MDCK) II cell cultures through specific interactions with two distinct effectors, OCRL inositol polyphosphate-5-phosphatase (OCRL) and ArfGAP with coiled-coil, ankyrin repeat and pleckstrin homology domains 2 (ACAP2), respectively. However, whether the upstream regulators of Rab35 also differ depending on the culture conditions remains completely unknown. Here, we investigated four known guanine nucleotide exchange factors (GEFs) of Rab35, namely DENN domain–containing 1A (DENND1A), DENND1B, DENND1C, and folliculin (FLCN), and demonstrate that DENND1A and FLCN exhibit distinct requirements for Rab35-dependent PODXL trafficking under the two culture conditions. In 3D cell cultures, only DENDN1A-knockout cysts exhibited the inverted localization of PODXL similar to that of Rab35-knockout cysts. Moreover, the DENN domain, harboring GEF activity toward Rab35, was required for proper PODXL trafficking to the apical membrane. By contrast, FLCN-knockdown cells specifically accumulated PODXL in actin-rich structures similar to the Rab35-knockdown cells in 2D cell cultures. Our findings indicate that two distinct functional cascades of Rab35, the FLCN-Rab35-OCRL and the DENND1A-Rab35-ACAP2 axes, regulate PODXL trafficking in 2D and 3D MDCK II cell cultures, respectively.
Keywords: cell polarity, epithelial cell, guanine nucleotide exchange factor (GEF), membrane trafficking, Rab, small GTPase, 3D cell culture, podocalyxin, tissue development
Introduction
Polarized epithelial cells have morphologically and functionally distinct apical and basolateral membranes, each of which contains specific proteins and lipids. However, the precise molecular mechanism of polarized cargo trafficking to the apical or basolateral membrane remains largely unknown (1). Rab small GTPases are critical regulators of intracellular membrane trafficking in eukaryotic cells (2–5). An active form of Rab localizes to specific membranes or subcellular compartments and regulates various types (or steps) of membrane trafficking, such as polarized trafficking, by recruiting specific effector(s). We previously performed a comprehensive screening for Rabs that regulate the trafficking of podocalyxin (PODXL),3 an apical cargo protein, and epithelial cell polarity formation in two-dimensional (2D) and three-dimensional (3D) Madin–Darby canine kidney (MDCK) II cell cultures (6, 7) and found that various Rab isoforms regulate PODXL transcytosis at different trafficking steps. Of particular interest was that whereas the route of PODXL transcytosis is seemingly the same between the two culture conditions, Rab35 differentially regulated PODXL trafficking, depending on the culture conditions. In 3D cysts, Rab35 knockout (KO) or knockdown (KD) caused an “inverted cyst” phenotype, in which PODXL remained on the outer membrane (6, 8), whereas in 2D cell cultures, Rab35 depletion caused PODXL accumulation in actin-rich structures during the early stage of polarization (6). Intriguingly, however, these Rab35-KO phenotypes were observed in the early stage of epithelial polarization, and PODXL was transported to the apical membrane in fully polarized cells, suggesting the existence of an alternative Rab35-independent mechanism of PODXL trafficking (6). Moreover, two distinct Rab35 effectors, OCRL inositol polyphosphate-5-phosphatase (OCRL) and ArfGAP with coiled-coil, ankyrin repeat and pleckstrin homology domains 2 (ACAP2; also called centaurin-β2), were found to be required for PODXL trafficking under 2D and 3D culture conditions, respectively (6). These results raised the possibility that the upstream regulators of Rab35 are also different between the two culture conditions.
The activation of Rab is thought to be spatiotemporally regulated by its specific guanine nucleotide exchange factors (GEFs), and four Rab35–GEFs have been reported so far: three DENN domain–containing 1 (DENND1) family proteins and folliculin (FLCN). The DENND1 family consists of three paralogs, DENND1A–C/connecdenn 1–3, all of which have a DENN domain that harbors GEF activity toward Rab35 in their N-terminal regions (9). The C-terminal regions are relatively less conserved and contain several unique domains and motifs, suggesting the functional diversity of the DENND1 family members. Both DENND1A and DENND1B contain clathrin– and AP-2–binding motifs and have been shown to be involved in clathrin-mediated endocytosis, endosome-to-trans-Golgi network trafficking, and the recycling of certain receptors (10–16). By contrast, DENND1C has a unique actin-binding motif and is required for the regulation of actin dynamics (17). Another Rab35–GEF, FLCN (18), also contains a DENN-like domain that harbors GEF activity toward Rab35 (19, 20), but FLCN is also known to act as a GTPase-activating protein (GAP) for other small GTPases, such as RagC/D (21) and Rab7A (22). Despite the presence of four different types of Rab35–GEFs in terms of their domain organization, their involvement in polarized trafficking in epithelial cells is completely unknown.
In this study, we investigated which Rab35–GEFs, specifically DENND1 proteins, are required for the transcytosis of PODXL in 2D and 3D MDCK II cell cultures by analyzing the phenotypes of each Rab35–GEF depletion. We found that only DENND1A-KO causes an inverted cyst phenotype similar to that of Rab35-KO cells. By contrast, none of the DENND1 family proteins are involved in PODXL trafficking in 2D cell cultures; instead, FLCN-KD phenocopied Rab35 deficiency (i.e. PODXL accumulation in actin-rich structures) in 2D cell cultures. Our findings suggest that Rab35-dependent PODXL transcytosis is differentially regulated by its upstream GEFs between 2D and 3D cell cultures.
Results
Active Rab35 regulates PODXL trafficking
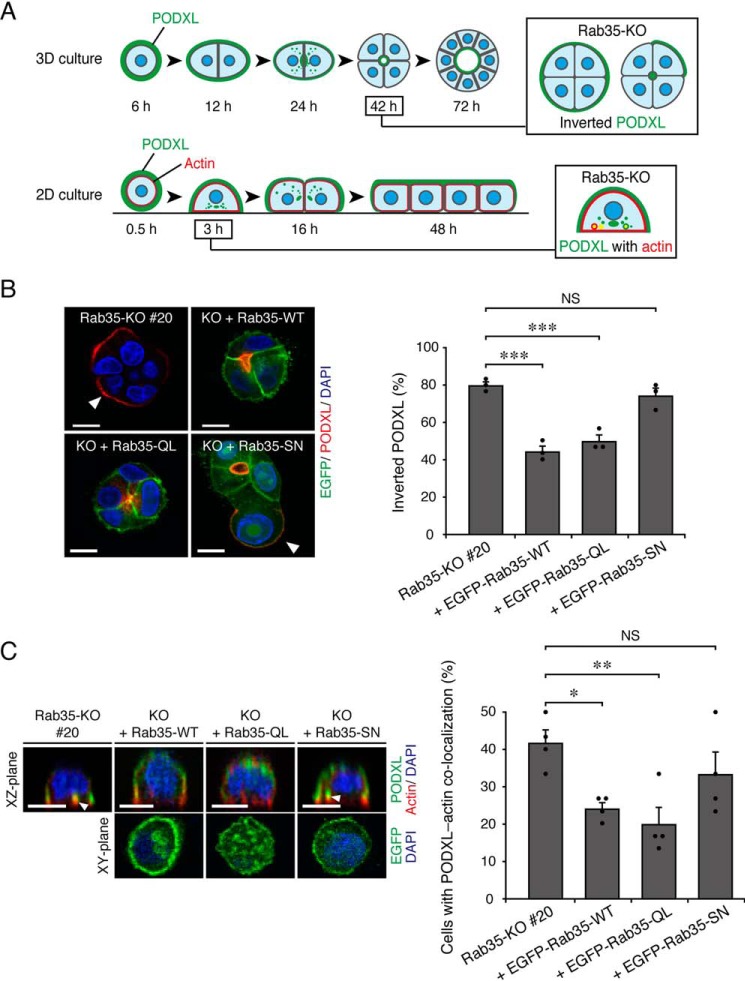
PODXL is a highly glycosylated transmembrane protein that is widely used as an apical membrane marker (23, 24). During the polarization of MDCK II cells, PODXL is actively transcytosed toward the nascent apical membrane (Fig. 1A; modified from Ref. 6). In a previous study, we showed that Rab35 differentially regulates PODXL trafficking, depending on the culture conditions. When Rab35-depleted MDCK II cells were plated on Matrigel (i.e. a 3D culture), they showed an “inverted cyst” phenotype, in which PODXL remained on the outer membrane during the 3D cyst formation (Fig. 1A, top row) (6, 8). On the other hand, when Rab35-depleted cells were plated on uncoated glass-bottom dishes (i.e. a 2D culture), they showed PODXL accumulation in actin-rich structures during the early stage of 2D monolayer formation (Fig. 1A, bottom row) (6). To confirm whether active Rab35 is necessary for the regulation of PODXL trafficking, we first performed KO-rescue experiments using constitutively active and negative mutants of Rab35 (Rab35-Q67L (QL) and -S22N (SN), respectively). Rab35-KO cells and those stably expressing enhanced GFP (EGFP)-Rab35 (WT, QL, or SN) were cultured under 2D and 3D culture conditions and were fixed once the Rab35-KO phenotypes became observable (42 h after plating for the 3D cultures and 3 h after plating for the 2D cultures) (Fig. 1A), followed by the immunostaining of PODXL. The results showed that the expression of Rab35-QL clearly rescued the Rab35-KO phenotypes under both culture conditions, similar to the results of Rab35-WT expression, whereas the expression of Rab35-SN failed to restore PODXL localization (Fig. 1, B and C). We thus concluded that Rab35 regulates PODXL trafficking when it is in an active state regardless of the culture conditions.
Figure 1.
Active Rab35 is required for PODXL trafficking under 2D and 3D culture conditions. A, schematic representation of PODXL localization during 3D cyst and 2D monolayer formation of MDCK II cells (modified from Ref. 6). In a 3D Rab35-KO cell culture, PODXL (green) remained on the outer membrane at 42 h after plating (Inverted PODXL) (top row, 3D culture). In a 2D Rab35-KO cell culture, endocytosed PODXL from the membrane attaching to the bottom plane of the dish localized and accumulated on actin-rich structures (red) at 3 h after plating (PODXL with actin) (bottom row, 2D culture). B, Rab35-KO (#20) and its rescued cells (+EGFP-Rab35 (WT, QL, or SN; shown in green)) were plated on Matrigel and fixed at 42 h after plating. The cells were stained for PODXL (red) and DAPI (blue), followed by counting of the inverted PODXL (30 cysts/condition). The arrowheads show PODXL localized on the outer membrane. Scale bars, 10 μm. The graph shows the means and S.E. (error bars) of three independent experiments. ***, p < 0.001; NS, not significant (Dunnett's test). C, Rab35-KO (#20) and its rescued cells (+EGFP-Rab35 (WT, QL, or SN)) were plated on glass-bottom dishes and fixed at 3 h after plating. The cells were stained for PODXL (green), actin (red), and DAPI (blue), followed by counting of cells with co-localized PODXL and actin (30 cells/condition). The confocal xz-plane (top) and the xy-plane (bottom) are shown. The arrowheads show PODXL co-localizing with actin. Scale bars, 10 μm. The graph shows the means and S.E. (error bars) of four independent experiments. *, p < 0.05; **, p < 0.01; NS, not significant (Dunnett's test).
DENND1A, but not DENND1B or DENND1C, is specifically required for PODXL trafficking in 3D cysts
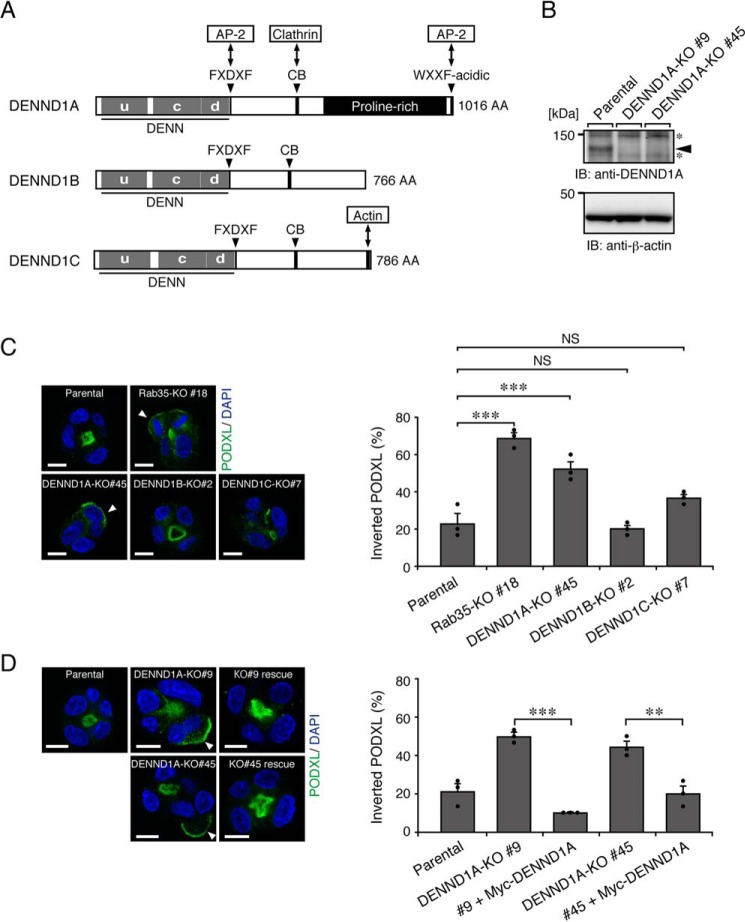
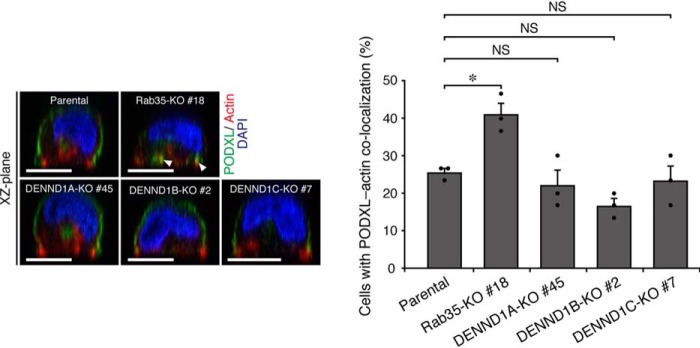
The activation of Rabs is generally thought to be regulated by their specific GEFs (25, 26). Because Rab35 differentially regulates PODXL trafficking in 2D and 3D cell cultures, we hypothesized that distinct Rab35–GEFs are required for Rab35 activation, depending on the culture conditions. We first focused on the well-known Rab35–GEFs, the DENND1 family proteins (9–11), because their C-terminal regions are relatively divergent in addition to the conserved N-terminal DENN domain, which exerts Rab35–GEF activity in vitro (Fig. 2A). Actually, the localization of EGFP-tagged DENND1 family proteins was different in both 2D and 3D MDCK II cell cultures (Fig. S1). The results of a quantitative RT-PCR analysis indicated that all of the DENND1 family genes were expressed in MDCK II cells (the relative mRNA expression level was DENND1B (1.755 ± 0.151, mean ± S.E.) > DENND1A (1.0) > DENND1C (0.358 ± 0.179); Fig. S2A). To investigate the possible involvement of DENND1A–C in PODXL trafficking, we used a genome-editing technology to establish each DENND1-KO MDCK II cell line. We confirmed that all indels occurred within the DENN domain–coding sequence to disrupt the Rab35–GEF function of DENND1 proteins (Fig. S2, B and C). The loss of DENND1A protein in DENND1A-KO cells was also confirmed by immunoblotting with anti-DENND1A antibody (Fig. 2B). When these KO cells together with the parental and Rab35-KO cells were cultured under 3D culture conditions, only the DENND1A-KO cysts showed the inverted localization of PODXL, similar to the Rab35-KO cysts (Fig. 2C). By contrast, DENND1B-KO (#2 and #11) and DENND1C-KO cells (#7 and #20; two independent clones) formed 3D cysts with normal PODXL localization, similar to parental MDCK II cells (Fig. 2C) (data not shown). Because the inverted PODXL phenotype of the DENND1A-KO clone (#45) was also observed in another independent DENND1A-KO clone (#9) and because the phenotypes of both clones were clearly rescued by the stable expression of mouse DENND1A (Fig. 2D), these results represent a bona fide phenotype induced by the specific loss of the DENND1A gene that is not caused by an off-target effect of guide RNA or by clonal variations among cell lines. These results strongly suggest that only DENND1A is required for PODXL trafficking in 3D cell cultures.
Figure 2.
DENND1A-KO, but not DENND1B-KO or DENND1C-KO, induces the inverted localization of PODXL in 3D cysts. A, schematic representation of mouse DENND1 family proteins. The domains and motifs are depicted according to previous reports (15, 17) and the UniProtKB (DENND1A, Q8K382; DENND1B, Q3U1T9; DENND1C, Q8CFK6). uDENN, upstream DENN; cDENN, central/core DENN; dDENN, downstream DENN; FXDXF, AP-2 α-ear platform subdomain–binding motif; CB, clathrin heavy chain–binding motif; WXXF-acidic, AP-2 α-ear sandwich subdomain–binding motif; AA, amino acids. B, lysates of parental cells and two DENND1A-KO cells (#9 and #45) were analyzed by immunoblotting (IB) with anti-DENND1A and anti-β-actin antibodies. The arrowhead indicates the position of endogenous DENND1A. The asterisks indicate nonspecific bands of the primary antibody. C, parental, Rab35-KO, DENND1A-KO, DENND1B-KO, and DENND1C-KO cells were plated on Matrigel and fixed at 42 h after plating. The cells were stained for PODXL (green) and DAPI (blue), followed by counting of the inverted PODXL (30 cysts/condition). The arrowheads show PODXL localizing on the outer membrane. Scale bars, 10 μm. The graph shows the means and S.E. (error bars) of three independent experiments. ***, p < 0.001; NS, not significant (Dunnett's test). D, parental cells, DENND1A-KO (#9 and #45) cells, and their rescued cells (+Myc-DENND1A) were plated on Matrigel and fixed at 42 h after plating, followed by counting of the inverted PODXL (30 cysts/condition). The arrowheads show PODXL localizing on the outer membrane. Scale bars, 10 μm. The graph shows the means and S.E.(error bars) of three independent experiments. ***, p < 0.001; **, p < 0.01 (Tukey's test).
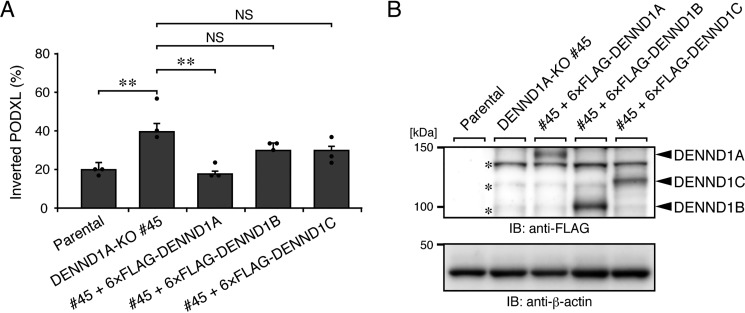
To further confirm the specific involvement of DENND1A in PODXL trafficking in 3D cysts, we next asked whether DENND1B or DENND1C can rescue the phenotype of DENND1A-KO cells. Consistent with the result of the DENND1B/1C-KO analysis described above, neither the stable expression of human DENND1B nor DENND1C in DENND1A-KO cells rescued the inverted PODXL phenotype (Fig. 3A and Fig. S3A), even though equivalent amounts of exogenous DENND1 family proteins were expressed in DENND1A-KO cells (Fig. 3B). These results allowed us to conclude that DENND1A is specifically required for PODXL trafficking in 3D cysts.
Figure 3.
DENND1A is specifically required for PODXL trafficking in 3D cysts. A, parental cells, DENND1A-KO (#45) cells, and its rescued cells (+6×FLAG-tagged DENND1A, DENND1B, or DENND1C) were plated on Matrigel and fixed at 42 h after plating, followed by counting of the inverted PODXL (30 cysts/condition). The graph shows the means and S.E. (error bars) of three independent experiments. **, p < 0.01; NS, not significant (Tukey's test). Representative images are shown in Fig. S3A. B, lysates of the cells used in A were analyzed by immunoblotting (IB) with anti-FLAG and anti-β-actin antibodies. The asterisks show the predicted 3×FLAG-tagged SpCas9 degradation products, because an ∼160-kDa protein was detected by immunoblotting with anti-FLAG antibody in DENND1A-KO #45 cells, but not in parental cells (data not shown).
The N-terminal DENN domain of DENND1A is required for PODXL trafficking in 3D cysts
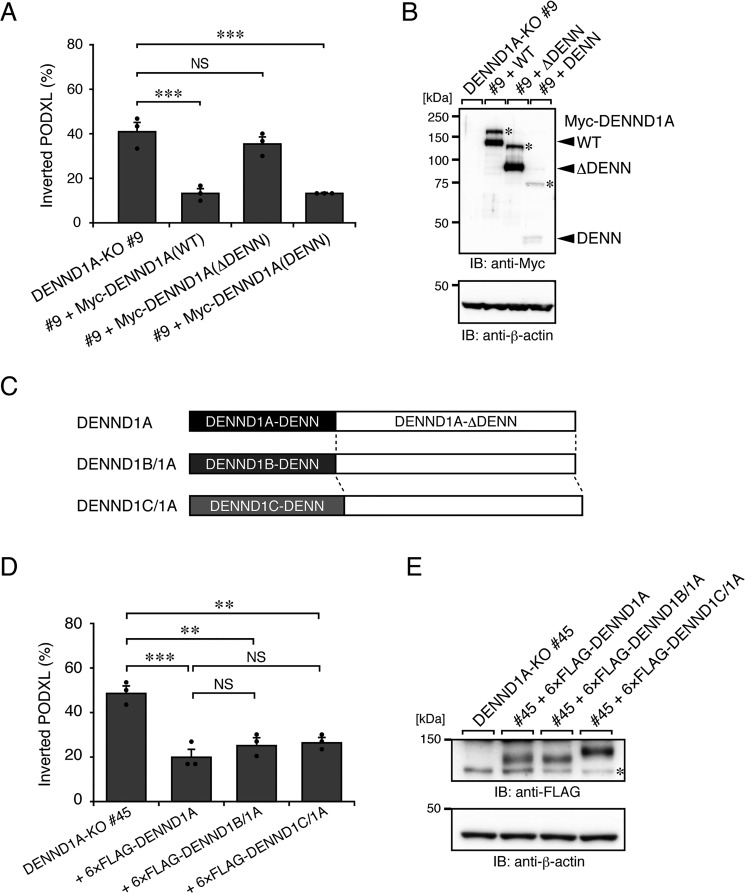
To clarify whether DENND1A regulates PODXL trafficking through the activation of Rab35 in 3D cysts, we performed a rescue experiment using a DENN domain–truncated mutant (named DENND1A(ΔDENN)) of DENND1A. As expected, the stable expression of the ΔDENN mutant did not rescue the inverted PODXL localization in DENND1A-KO cysts (Fig. 4 (A and B) and Fig. S3B). By contrast, the DENN domain of DENND1A (named DENND1A(DENN)) alone was able to rescue the inverted PODXL localization despite its cytosolic localization (Fig. 4 and Fig. S4). These results suggest that the activation of Rab35 by the DENN domain of DENND1A is responsible for PODXL trafficking under 3D culture conditions.
Figure 4.
The N-terminal DENN domain of DENND1A is required for PODXL trafficking in 3D cysts. A, DENND1A-KO (#9) and its rescued cells (+Myc-DENND1A (WT, ΔDENN, or DENN)) were plated on Matrigel and fixed at 42 h after plating, followed by counting of the inverted cysts (30 cysts/condition). The graph shows the means and S.E. (error bars) of three independent experiments. ***, p < 0.001; NS, not significant (Dunnett's test). Representative images are shown in Fig. S3B. B, lysates of the cells used in A were analyzed by immunoblotting (IB) with anti-Myc and anti-β-actin antibodies. The additional higher bands (asterisks) in the blots presumably result from post-translational modifications, such as phosphorylation (30). Alternatively, the DENN domain of DENND1A could form an SDS-insensitive dimer. C, schematic representation of DENND1B/1A and DENND1C/1A chimera proteins. The DENND1B/1A (or DENND1C/1A) chimera protein consists of the DENN domain of human DENND1B (DENND1B-DENN) (or DENND1C (DENND1C-DENN)) fused with the C-terminal part of mouse DENND1A (DENND1A-ΔDENN). D, DENND1A-KO (#45) and its rescued cells (+6×FLAG-tagged DENND1A, DENND1B/1A, or DENND1C/1A) were plated on Matrigel and fixed at 42 h after plating, followed by counting of the inverted PODXL (30 cysts/condition). The graph shows the means and S.E. (error bars) of three independent experiments. ***, p < 0.001; **, p < 0.01; NS, not significant (Tukey's test). Representative images are shown in Fig. S3C. E, lysates of the cells used in D were analyzed by immunoblotting with anti-FLAG and anti-β-actin antibodies. The asterisk shows the predicted 3×FLAG-tagged SpCas9 degradation products.
Because the DENN domain of all DENND1 family proteins basically has Rab35–GEF activity in vitro (15), we hypothesized that the C-terminal domain of DENND1A is also required for efficient activation of Rab35 during PODXL trafficking in 3D cysts. To test this hypothesis, we further prepared DENND1 chimera proteins, named DENND1B/1A (DENND1B-DENN + DENND1A-ΔDENN) and DENND1C/1A (Fig. 4C), and performed a rescue experiment. The results showed that the inverted PODXL phenotype in DENND1A-KO cysts was clearly rescued by the expression of either DENND1B/1A or DENND1C/1A, the same as the DENND1A expression (Fig. 4, D and E). Thus, the C-terminal domain of DENND1A also contributes to the Rab35-dependent PODXL trafficking in 3D cysts.
DENND1 family proteins are not involved in PODXL trafficking under 2D culture conditions
To determine the functional diversity of the DENND1 family proteins in PODXL trafficking even in 2D monolayer formation, we further investigated whether DENND1A–C are required for PODXL trafficking under 2D culture conditions. To our surprise, however, none of the DENND1-KO cells showed an accumulation of PODXL in actin-rich structures, which is a characteristic phenotype of Rab35-KO cells in 2D cell cultures (Fig. 5). Therefore, another Rab35–GEF protein other than the DENND1 family proteins is probably required for PODXL trafficking under 2D culture conditions.
Figure 5.
DENND1 family proteins are not required for PODXL trafficking under 2D culture conditions. Parental, Rab35-KO, DENND1A-KO, DENND1B-KO, and DENND1C-KO cells were plated on glass-bottom dishes and fixed at 3 h after plating. The cells were stained for PODXL (green), actin (red), and DAPI (blue), followed by counting of cells with co-localized PODXL and actin. The arrowheads show PODXL co-localized with actin. Scale bars, 10 μm. The graph shows the means and S.E. (error bars) of three independent experiments. *, p < 0.05; NS, not significant (Dunnett's test).
FLCN regulates PODXL trafficking under 2D culture conditions
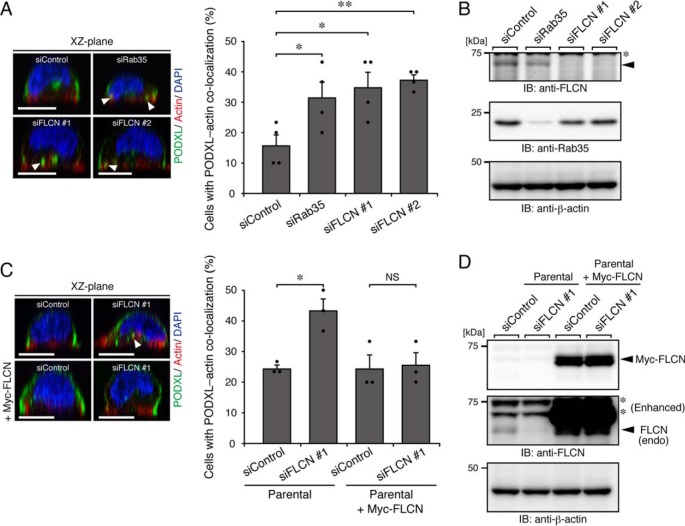
Finally, we focused on FLCN, another type of Rab35–GEF, which also contains a DENN-like domain (19, 20), and investigated its involvement in PODXL trafficking in 2D cell cultures. To this end, we knocked down endogenous FLCN using two independent siRNAs (#1 and #2). Parental MDCK II cells that had been transfected with control siRNA or siRNAs against FLCN (or Rab35 as a positive control) were fixed at 3 h after replating on glass-bottom dishes, followed by immunostaining for PODXL. As a result, FLCN-KD cells, but not control cells, showed PODXL accumulation in actin-rich structures, similar to the Rab35-KD cells (Fig. 6, A and B). The observed effect was clearly rescued by the stable expression of mouse FLCN (Fig. 6, C and D), ruling out the possibility of an off-target effect of siRNAs. By contrast, no apparent phenotype was observed in FLCN-KD cells for 3D cell cultures (Fig. S5). We thus concluded that FLCN is required for Rab35-dependent PODXL trafficking under 2D culture conditions.
Figure 6.
FLCN regulates PODXL trafficking under 2D culture conditions. A, parental cells that had been transfected with control siRNA (siControl), siRNAs against Rab35 (siRab35), or FLCN (two independent target sites: #1 and #2) (siFLCN) were plated on glass-bottom dishes and fixed at 3 h after plating, followed by counting of cells with co-localized PODXL and actin (30 cells/condition). The arrowheads show PODXL co-localizing with actin. Scale bars, 10 μm. The graph shows the means and S.E. (error bars) of four independent experiments. *, p < 0.05; **, p < 0.01 (Dunnett's test). B, KD efficiency of Rab35 and FLCN as revealed by immunoblotting (IB) with anti-FLCN, anti-Rab35, and anti-β-actin antibodies. The arrowhead indicates the position of endogenous FLCN. The asterisk indicates a nonspecific band of the primary antibody. C, parental cells and Myc-FLCN–expressing cells (+Myc-FLCN) that had been transfected with control siRNA or siRNA against FLCN (#1) were plated on glass-bottom dishes and fixed at 3 h after plating, followed by counting of cells with co-localized PODXL and actin (30 cells/condition). The graph shows the means and S.E. (error bars) of three independent experiments. *, p < 0.05; NS, not significant (Tukey's test). The arrowhead shows PODXL co-localizing with actin. D, lysates of the cells used in C were analyzed by immunoblotting with anti-FLCN and anti-β-actin antibodies. Note that endogenous (endo) FLCN, but not exogenous mouse FLCN, were efficiently knocked down by the siFLCN #1. Because knockdown of endogenous FLCN in Myc-FLCN–expressing cells was not observed, presumably because of the existence of the degradation product of Myc-FLCN, the decrease of endogenous FLCN mRNA in parental and Myc-FLCN–expressing MDCK II cells that had been transfected with siFLCN#1 was also confirmed by real-time PCR analysis (data not shown). The asterisks indicate nonspecific bands of the primary antibody.
Discussion
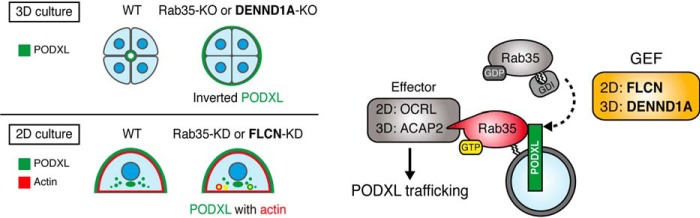
We previously found that Rab35 differentially regulates PODXL trafficking in 2D and 3D MDCK cell cultures through its interactions with two downstream effectors, OCRL and ACAP2, respectively (6). In the present study, we presented new evidence indicating that distinct Rab35–GEFs (upstream regulators of Rab35) regulate PODXL trafficking under two culture conditions. Namely, DENND1A is specifically required for PODXL trafficking in 3D cysts (Figs. 2–5; DENND1A deficiency caused inverted PODXL localization), whereas FLCN is required for PODXL trafficking in 2D cell cultures (Fig. 6; FLCN deficiency caused PODXL accumulation in actin-rich structures). Our findings indicate that two functional GEF-Rab-effector cascades (i.e. the FLCN-Rab35-OCRL axis and the DENND1A-Rab35-ACAP2 axis) regulate PODXL trafficking during epithelial polarization under 2D and 3D culture conditions, respectively (Fig. 7).
Figure 7.
A proposed model of Rab35 functional cascades in PODXL trafficking under 2D and 3D culture conditions. Rab35 activation during PODXL trafficking is regulated by distinct Rab35–GEF proteins, depending on the culture conditions (i.e. DENND1A in 3D and FLCN in 2D culture conditions, respectively). DENND1A activates Rab35 and enables it to recruit a Rab35 effector, ACAP2 (Arf6-GAP), in 3D cell cultures. On the other hand, FLCN activates Rab35 and enables it to recruit another Rab35 effector, OCRL (phosphatidylinositol 4,5-bisphosphate phosphatase), in 2D cell cultures, followed by PODXL apical trafficking (6).
How does DENND1A specifically regulate PODXL trafficking in 3D cell cultures? One possible mechanism is the difference in the gene expression between 2D and 3D cell cultures (27). However, this mechanism is unlikely because the mRNA expression levels of DENND1A–C and FLCN are reportedly unaltered between 2D and 3D cell cultures (27). Moreover, the DENND1A–C mRNA expression levels were comparable in MDCK II cells (Fig. S2A). Thus, a high gene expression level would not simply explain the specific involvement of DENND1A in PODXL trafficking in 3D cell cultures (or FLCN in 2D cell cultures).
Because it has been proposed that the activation and localization of Rabs are generally defined by their upstream GEFs (28, 29), another possible mechanism is the specific localization of Rab35–GEFs during the polarization of MDCK II cells. Actually, different subcellular localizations of DENND1 family proteins have previously been reported in nonepithelial cells (10, 11, 15, 17). We especially noted the fact that only DENND1A was specifically localized near the apical membrane, where PODXL (and Rab35) is localized in 3D cysts (Fig. S1A) (6, 8). By contrast, DENND1C seemed to become localized in the basolateral membrane, and DENND1B mostly showed a cytosolic localization in addition to its weak association with membranes. Moreover, FLCN showed a cytosolic localization (Fig. S1A). Thus, it seems likely that the specific localization of DENND1A at the apical membrane of 3D cysts specifically regulates PODXL trafficking. Actually, DENND1B/1A and DENND1C/1A chimera proteins, but not DENND1B or DENND1C, can rescue the inverted PODXL phenotype in DENND1A-KO cells (Fig. 4D), strongly supporting the importance of the C-terminal domain of DENND1A in PODXL trafficking.
The apical localization of DENND1A should be mediated by its C-terminal domain, because the DENND1A(ΔDENN) mutant still has an ability to localize near the apical membrane (Fig. S4), although the ΔDENN mutant itself did not rescue the inverted PODXL phenotype (Fig. 4A). Consistent with our observation, DENND1A is known to localize to clathrin-coated vesicles via its several clathrin– and AP-2–binding motifs (Fig. 2A) (10, 15), whereas DENND1B only partially localizes to clathrin-coated vesicles despite having clathrin– and AP-2–binding motifs in its C-terminal region (15). Actually, EGFP-tagged DENND1A, but not DENND1B or DENND1C, was strongly co-localized with monomeric Strawberry (mStr)-tagged clathrin light chain in 2D and 3D MDCK cell cultures (Fig. S1, C and D). Whether or not DENND1A is recruited near the apical membrane by means of clathrin/AP-2 binding awaits further investigation.
Although we favored the above possibility that the apical membrane localization of DENND1A is a prerequisite for the activation of Rab35 and PODXL trafficking, our other finding that the cytosolically expressed DENN domain alone can rescue the inverted PODXL phenotype in DENND1A-KO cells (Fig. 4) seemed to be inconsistent with this possibility. However, the apparent discrepancy can be explained by a previously reported autoinhibition mechanism of DENND1A on Rab35–GEF activity: the Rab35–GEF activity of DENND1A is autoinhibited by an intramolecular interaction between the DENN domain and its downstream region, and the isolated DENN domain shows a 2–3 times greater Rab35–GEF activity than the full-length protein does in vitro (30). We thus speculated that such a hyperactive GEF fragment would promiscuously activate Rab35 throughout the cytoplasm. Because it has been proposed that the phosphorylation of DENND1A regulates its Rab35–GEF activity through an intramolecular interaction (30), the extracellular matrix–dependent phosphorylation of DENND1A might locally activate Rab35 near the apical membrane. Further extensive research is necessary to determine the regulatory mechanism for the Rab35–GEF activity of DENND1A.
As noted above, because the DENN domain of DENND1B and DENND1C possesses Rab35–GEF activity, it is possible that an excessively high expression level of DENND1B or DENND1C could support PODXL trafficking in DENND1A-KO 3D cysts. As expected, forced overexpression of DENND1B or DENND1C (>30 times higher than endogenous DENND1A) was able to rescue DENND1A-KO cells (Fig. S6), although such a condition is physiologically irrelevant. We speculate that some portions of overexpressed DENND1B (or DENND1C) localize in cytosol, which would activate Rab35 just near the apical region to support apical localization of PODXL.
In contrast to 3D cysts, FLCN was found to be specifically required for the Rab35-dependent PODXL trafficking in 2D cell cultures (Fig. 6). Although the role of FLCN in the endocytic/recycling pathway in mammals is poorly understood, a previous study on Caenorhabditis elegans development showed that both Rab35 and FLCN-1 (C. elegans FLCN) are required for the maturation and clearance of phagosomes that contain apoptotic cells (31). Both Rab35 and FLCN-1 transiently localize to nascent phagosomes, and the loss of Rab35 delays PI(4,5)P2 removal from phagosomal membranes. Notably, the loss of OCRL-1 in C. elegans also results in the persistent accumulation of PI(4,5)P2 on phagosomal membranes (32), suggesting that the FLCN-1-Rab35-OCRL-1 axis regulates phosphoinositide metabolism during phagosome maturation. Under our experimental conditions, EGFP/mStr-tagged FLCN seemed to be diffusely present throughout the cytoplasm and did not show any robust vesicular localization in MDCK II cells (Fig. S1); however, it is still possible that FLCN very weakly and transiently associates with PODXL-containing endosomes as with phagosomes in C. elegans. Further investigation is needed to specify the exact time points and places at which FLCN functions during 2D epithelial polarization.
The main differences between 2D and 3D culture conditions are the existence of the extracellular matrix and substrate stiffness (i.e. hard coverslips versus soft gels). Because epithelial tissues generally develop in the presence of the soft extracellular matrix, the 3D cell culture is thought to be more suitable to mimic the normal physiological conditions. However, because tumors are known to be much stiffer than normal tissues, the phenotypes observed in the 2D cell culture may reflect the cellular behaviors in such pathological environments. Therefore, our findings suggest the physiological roles of DENND1A and FLCN in normal tissue development and tumor progression/suppression, respectively, and further research will be needed to determine the physiological roles of these Rab35–GEFs.
In summary, we have investigated the regulation of PODXL trafficking by Rab35–GEFs in 2D and 3D epithelial cell cultures, and we discovered that FLCN and DENND1A differentially regulate PODXL trafficking in 2D monolayer formation and 3D cyst formation, respectively. Our findings indicate that two distinct Rab35-dependent PODXL-trafficking pathways are present in polarizing epithelial cells, depending on the extracellular environments.
Experimental procedures
Materials
Anti-PODXL rabbit polyclonal antibody was prepared as described previously (6). Anti-DENND1A rabbit polyclonal antibody was raised against a synthetic peptide, CVEQLRRQWETFE, which corresponds to the C terminus of mouse/human/canine DENND1A (underlined), with an artificial cysteine being added to its N terminus. The following antibodies were obtained commercially: anti-β-actin mouse mAb (Applied Biological Materials, #G043), anti-FLAG (M2) mouse mAb (Sigma–Aldrich, #F1804), anti-Myc (9E10) mouse mAb (Santa Cruz Biotechnology, Inc., #sc-40), anti-FLCN rabbit polyclonal antibody (OriGene, #TA309661), anti-Rab35 rabbit polyclonal antibody (Proteintech, #11329-2-AP), horseradish peroxidase (HRP)-conjugated anti-mouse IgG goat polyclonal antibody (SouthernBiotech, #1031-05), HRP-conjugated anti-rabbit IgG donkey antibody (GE Healthcare, #NA934-1ML), Alexa Fluor 488–conjugated anti-rabbit IgG goat polyclonal antibody (Thermo Fisher Scientific, #A11034), and Alexa Fluor Plus 555–conjugated anti-rabbit IgG goat polyclonal antibody (Thermo Fisher Scientific, #A32732). Alexa Fluor 568– and 633–conjugated phalloidin were purchased from Thermo Fisher Scientific (#A12380 and #A22284, respectively).
Plasmid construction
The single guide RNA (sgRNA)- and Cas9-encoding vector was constructed by using the pSpCas9(BB)-2A-Puro vector (33). The target sequences against canine DENND1A, DENND1B, and DENND1C were chosen using the web tool CRISPRdirect (Table S1) (34).
The standard plasmid used in the real-time PCR analysis was constructed from the pGEM-T-EASY vector (Promega) as follows. Partial cDNA fragments of DENND1A, DENND1B, and DENND1C were amplified from the cDNA library of MDCK II cells (see “RNA isolation, reverse transcription, and real-time PCR”) using specific primers (Table S1), and each cDNA was subcloned into the pGEM-T-EASY vector. The DENND1B and DENND1C cDNA fragments were then excised by PstI/SphI and SphI/BstXI, respectively, and were together ligated to the PstI/BstXI-digested pGEM-T-DENND1A vector to obtain the standard plasmid (named pGEM-T-DENND1A-1B-1C).
The pMRX-IRES-puro retrovirus vector was kindly donated by Dr. Shoji Yamaoka (Tokyo Medical and Dental University, Tokyo, Japan) (35), and Myc, EGFP, or mStr tag was inserted and/or a puromycin resistance gene (puro) was replaced by a blasticidin S resistance gene (bsr) to obtain pMRX-bsr-Myc, pMRX-puro-EGFP, and pMRX-bsr-mStr vectors (7). cDNAs encoding mouse Rab35 (NM_198163.1; WT, QL, and SN) (36–38), mouse DENND1A (NM_001362973.1) (39), human DENND1B (XM_005244931.2), human DENND1C (NM_024898.4) (40), and mouse FLCN (NM_001271356.1) were subcloned into the pMRX-puro-EGFP vector. cDNAs encoding DENND1A (WT, ΔDENN (lacking amino acids 1–378), and DENN (amino acids 1–378)) and FLCN were subcloned into the pMRX-bsr-Myc vector. cDNAs encoding DENND1B/1A chimera (amino acids 1–372 of human DENND1B + amino acids 376–1059 of mouse DENND1A) and DENND1C/1A chimera (amino acids 1–393 of human DENND1C + amino acids 374–1059 of mouse DENND1A) were prepared by two-step PCR techniques (41) using specific primers (Table S1). cDNAs encoding mouse FLCN and mouse CLC (clathrin light chain) (NM_001080384.3) were subcloned into the pMRX-bsr-mStr vector.
To achieve low expression levels of exogenous DENND1 proteins, which were comparable with those of endogenous ones, 6×FLAG-tagged DENND1A–C (or DENND1 chimera) were fused downstream of the bsr and internal ribosome entry site (IRES) sequences. In addition, the WT IRES sequence was replaced by a largely truncated IRES sequence (from 620 to 832 of the encephalomyocarditis virus genome (NC_001479.1)), resulting in the pMRX-bsr-IRES(620)-6×FLAG-DENND1A–C (or DENND1 chimera) vectors.
Cell cultures
MDCK II cells were grown in culture medium (Dulbecco's modified Eagle's medium; Fujifilm Wako Pure Chemical, Osaka, Japan) supplemented with 10% fetal bovine serum, penicillin G (100 units/ml), and streptomycin (100 μg/ml) at 37 °C under 5% CO2. For the 2D cell culture observation, the cells were trypsinized to create a single-cell suspension, 1 × 105 cells were plated on an uncoated glass-bottom dish (35-mm dish; MatTek), and the cells were incubated for 3 h. For the 3D cell culture observation, the cells were trypsinized to create a single-cell suspension, 2 × 104 cells were plated on a Matrigel (growth factor–reduced; BD Biosciences)-coated coverglass in a well of a 24-well plate, and the cells were incubated in a culture medium containing 2% Matrigel for 42 h. The Plat-E cells were kindly donated by Dr. Toshio Kitamura (University of Tokyo) and were grown in the same culture medium and then used for retrovirus production.
RNA isolation, reverse transcription, and real-time PCR
Total RNA was isolated from parental MDCK II cells (100% confluent in a 60-mm dish) with TRI reagent (Sigma-Aldrich), followed by chloroform extraction and 2-propanol precipitation. Reverse transcription was performed using ReverTra Ace (Toyobo, Osaka, Japan) with 5 μg of the total RNA as a template and oligo(dT) primer. Real-time PCR was performed with the LightCycler 480 system (Roche, Basel, Switzerland) and TB GreenTM Premix Ex TaqTM II (Tli RNaseH plus) kit (Takara Bio Inc., Shiga, Japan). The reaction mixture (final volume: 20 μl including 1 μl of template DNA (the standard plasmid or the reverse transcription product)) was prepared according to the manufacturer's instructions. The primers for canine DENND1A, DENND1B, and DENND1C were designed using LightCycler Probe Design Software 2.0 (Idaho Technology Inc.) (Table S1). The real-time PCR was performed using a shuttle PCR standard protocol, including 40 cycles of denaturation at 95 °C for 5 s and annealing/extension at 60 °C for 30 s, according to the manufacturer's instructions. Experiments were performed by using three independent cDNA samples, each of which was measured twice and averaged. The relative expression level of each mRNA was calculated using the known amount of the standard plasmid (pGEM-T-DENND1A-1B-1C) described above.
Establishment of KO cells
MDCK II cells (1 × 105 cells/6-cm dish) were cultured overnight and were transfected with the sgRNA/Cas9-encoding plasmid using Lipofectamine 2000 (Thermo Fisher Scientific). In the case of DENND1A-KO, two distinct sgRNAs were used simultaneously to induce deletions between the two target sites. One day after transfection, the medium was changed and a fresh one was applied, and the cells were cultured for one additional day. The transfected cells were selected with 2 μg/ml puromycin (Merck) for 24 h. The cells were then cloned by limiting dilution, and each clone was checked for target gene disruption using genomic PCR followed by the sequencing of its product (42) (see Fig. S2C). The loss of DENND1A protein expression in DENND1A-KO cells was also checked using immunoblotting. Rab35-KO cell lines (#18 and #20) were prepared as described previously (6).
Genomic PCR and direct sequencing
Cells were lysed with a digesting buffer (0.5% SDS, 100 mm NaCl, 10 mm Tris-HCl, pH 8.0, 25 mm EDTA, pH 8.0, and 0.1 mg/ml proteinase K) and gently agitated overnight at 50 °C. An equal volume of phenol/chloroform was added to the lysate, followed by vortexing and then centrifugation at 1,700 × g for 5 min at room temperature. Genomic DNA in the supernatant was ethanol-precipitated and subjected to a PCR. A genomic region containing the target sequence was amplified with LA Taq or Ex Taq (Takara Bio Inc.) and appropriate pairs of primers (Table S1), and the products were directly sequenced using either of the primers.
Retrovirus production and establishment of stable expression cell lines
For retrovirus production, Plat-E cells (43) (5 × 105 cells/35-mm dish coated with poly-l-lysine) were cultured overnight and were transfected with 2 μg of pMRX plasmids together with 1 μg of pLP-VSVG (Thermo Fisher Scientific) using Lipofectamine 2000. One day after transfection, the medium was changed with a fresh one, and the cells were cultured for one additional day. The medium was collected and centrifuged at 17,800 × g for 2 min to remove debris. The virus-containing supernatant was used for subsequent viral infection. To establish stable cell lines, MDCK II cells (1.5 × 104 cells/35-mm dish) were cultured overnight, and a retrovirus-containing medium was added to the culture medium in the presence of 8 μg/ml Polybrene. One day after the addition of the virus, the medium was changed, and a fresh one without the virus was applied; the cells were then cultured for one additional day. The infected cells were selected using 3–6 μg/ml puromycin for 24 h or 13 μg/ml blasticidin S (Fujifilm Wako Pure Chemical) for 48 h.
Knockdown experiments
MDCK II cells (2 × 104 cells/well of a 6-well plate) were cultured overnight and transfected with 12.5 nm control siRNA or siRNA against Rab35 or FLCN (#1 or #2) using Lipofectamine RNAiMAX (Thermo Fisher Scientific). Three days after transfection, the cells for 2D cell cultures were replated for immunofluorescence analysis or were lysed for immunoblotting. For 3D cell cultures, the cells were cultured in the Matrigel-containing medium for 42 h as described above, and 12.5 nm each of siRNAs and Lipofectamine RNAiMAX were also added to the medium (6). The siRNAs used in this study are listed in Table S1.
Immunofluorescence
Immunofluorescence staining of 2D and 3D MDCK II cell cultures was performed essentially as described previously (6, 44). In brief, the cells were fixed with 4% paraformaldehyde for 10 min (2D) or 30 min (3D), permeabilized with 0.3% Triton X-100 in PBS for 2 min (2D) or 30 min (3D), and blocked with a blocking buffer (1% BSA and 0.1% Triton X-100 in PBS) for 15 min (2D) or 30 min (3D). The cells were incubated for 1 h (2D) or overnight (3D) in a blocking buffer containing the primary antibody against PODXL (1:2,000 dilution), followed by incubation with a secondary antibody and DAPI for 1 h. For actin staining, the cells were incubated with Alexa Fluor–conjugated phalloidin (1:1,000 dilution) together with the secondary antibody and DAPI. All of the steps were performed at room temperature.
Quantification of KO phenotypes in 2D and 3D MDCK cell cultures
For the 2D cell cultures, 30 cells at the one-cell stage in xz images were observed in each experiment. Cells in which endosomal PODXL was co-localized with actin were manually counted (see Fig. 1A, PODXL with actin). For the 3D cell cultures, 30 cysts containing 3–6 cells/cyst in xy-confocal images were observed in each experiment. Cysts in which PODXL was localized to the outer membrane were also manually counted. Cysts containing at least one cell that showed PODXL in the outer membrane were counted as “inverted PODXL” (see Fig. 1A, Inverted PODXL).
Immunoblotting
Denatured proteins in an SDS sample buffer (62.5 mm Tris-HCl, pH 6.8, 2% 2-mercaptoethanol, 2% SDS, 10% glycerol, and 0.02% bromphenol blue) were separated using SDS-PAGE and were transferred to polyvinylidene difluoride membranes (Merck Millipore). The membranes were blocked with a blocking buffer (1% skim milk and 0.1% Tween 20 in PBS) for 15 min and were incubated for 1 h at room temperature or overnight at 4 °C in the blocking buffer containing appropriate primary antibodies. The membranes were washed three times with 0.1% Tween 20 containing PBS and were subsequently incubated in the blocking buffer containing appropriate HRP-conjugated secondary antibodies. Chemiluminescence signals were detected using an ECL substrate (Bio-Rad) and a chemiluminescence imager (ChemiDoc Touch, Bio-Rad).
Statistical analysis
The statistical analysis was performed using Tukey's test or Dunnett's test on R software, and the following significance levels were used: *, p < 0.05; **, p < 0.01; and ***, p < 0.001.
Author contributions
R. K. and Y. H. conceptualization; R. K. and Y. H. data curation; R. K. and Y. H. formal analysis; R. K. and Y. H. investigation; R. K., Y. H., and M. F. writing-original draft; R. K., Y. H., and M. F. writing-review and editing; Y. H. and M. F. funding acquisition; Y. H. and M. F. project administration; M. F. supervision.
Supplementary Material
Acknowledgments
We thank Dr. Toshio Kitamura and Dr. Shoji Yamaoka for kindly donating materials, Megumi Takada-Aizawa and Kazuyasu Shoji for technical assistance, Kan Etoh for preparing DENND1 materials, Paulina S. Wawro and Miki Matsui for initial experiments, and members of the Fukuda laboratory for valuable discussions.
This work was supported in part by Grant-in-Aid for Young Scientists 18K14692 from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan (to Y. H.); Grant-in-Aid for Scientific Research (B) 19H03220 from MEXT (to M. F.); and Japan Science and Technology Agency (JST) CREST Grant JPMJCR17H4 (to M. F.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Table S1 and Figs. S1–S6.
- PODXL
- podocalyxin
- ACAP2
- ArfGAP with coiled-coil, ankyrin repeat and pleckstrin homology domains 2
- bsr
- blasticidin S resistance gene
- DAPI
- 4',6-diamidino-2-phenylindole
- DENND1
- DENN domain–containing 1
- EGFP
- enhanced green fluorescent protein
- FLCN
- folliculin
- GAP
- GTPase-activating protein
- GEF
- guanine nucleotide exchange factor
- HRP
- horseradish peroxidase
- IRES
- internal ribosome entry site
- KD
- knockdown
- KO
- knockout
- MDCK
- Madin–Darby canine kidney
- mStr
- monomeric Strawberry
- OCRL
- OCRL inositol polyphosphate-5-phosphatase
- puro
- puromycin resistance gene
- QL
- Q67L
- sgRNA
- single guide RNA
- SN
- S22N
- 2D
- two-dimensional
- 3D
- three-dimensional.
References
- 1. Román-Fernández A., and Bryant D. M. (2016) Complex polarity: building multicellular tissues through apical membrane traffic. Traffic 17, 1244–1261 10.1111/tra.12417 [DOI] [PubMed] [Google Scholar]
- 2. Fukuda M. (2008) Regulation of secretory vesicle traffic by Rab small GTPases. Cell Mol. Life Sci. 65, 2801–2813 10.1007/s00018-008-8351-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Stenmark H. (2009) Rab GTPases as coordinators of vesicle traffic. Nat. Rev. Mol. Cell Biol. 10, 513–525 10.1038/nrm2728 [DOI] [PubMed] [Google Scholar]
- 4. Hutagalung A. H., and Novick P. J. (2011) Role of Rab GTPases in membrane traffic and cell physiology. Physiol. Rev. 91, 119–149 10.1152/physrev.00059.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pfeffer S. R. (2013) Rab GTPase regulation of membrane identity. Curr. Opin. Cell Biol. 25, 414–419 10.1016/j.ceb.2013.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mrozowska P. S., and Fukuda M. (2016) Regulation of podocalyxin trafficking by Rab small GTPases in 2D and 3D epithelial cell cultures. J. Cell Biol. 213, 355–369 10.1083/jcb.201512024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Homma Y., Kinoshita R., Kuchitsu Y., Wawro P. S., Marubashi S., Oguchi M. E., Ishida M., Fujita N., and Fukuda M. (2019) Comprehensive knockout analysis of the Rab family GTPases in epithelial cells. J. Cell Biol. 218, 2035–2050 10.1083/jcb.201810134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Klinkert K., Rocancourt M., Houdusse A., and Echard A. (2016) Rab35 GTPase couples cell division with initiation of epithelial apico-basal polarity and lumen opening. Nat. Commun. 7, 11166 10.1038/ncomms11166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chaineau M., Ioannou M. S., and McPherson P. S. (2013) Rab35: GEFs, GAPs and effectors. Traffic 14, 1109–1117 10.1111/tra.12096 [DOI] [PubMed] [Google Scholar]
- 10. Allaire P. D., Ritter B., Thomas S., Burman J. L., Denisov A. Y., Legendre-Guillemin V., Harper S. Q., Davidson B. L., Gehring K., and McPherson P. S. (2006) Connecdenn, a novel DENN domain-containing protein of neuronal clathrin-coated vesicles functioning in synaptic vesicle endocytosis. J. Neurosci. 26, 13202–13212 10.1523/JNEUROSCI.4608-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yoshimura S., Gerondopoulos A., Linford A., Rigden D. J., and Barr F. A. (2010) Family-wide characterization of the DENN domain Rab GDP-GTP exchange factors. J. Cell Biol. 191, 367–381 10.1083/jcb.201008051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sato M., Sato K., Liou W., Pant S., Harada A., and Grant B. D. (2008) Regulation of endocytic recycling by C. elegans Rab35 and its regulator RME-4, a coated-pit protein. EMBO J. 27, 1183–1196 10.1038/emboj.2008.54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Allaire P. D., Marat A. L., Dall'Armi C., Di Paolo G., McPherson P. S., and Ritter B. (2010) The connecdenn DENN domain: a GEF for Rab35 mediating cargo-specific exit from early endosomes. Mol. Cell 37, 370–382 10.1016/j.molcel.2009.12.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cauvin C., Rosendale M., Gupta-Rossi N., Rocancourt M., Larraufie P., Salomon R., Perrais D., and Echard A. (2016) Rab35 GTPase triggers switch-like recruitment of the Lowe syndrome lipid phosphatase OCRL on newborn endosomes. Curr. Biol. 26, 120–128 10.1016/j.cub.2015.11.040 [DOI] [PubMed] [Google Scholar]
- 15. Marat A. L., and McPherson P. S. (2010) The connecdenn family, Rab35 guanine nucleotide exchange factors interfacing with the clathrin machinery. J. Biol. Chem. 285, 10627–10637 10.1074/jbc.M109.050930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yang C. W., Hojer C. D., Zhou M., Wu X., Wuster A., Lee W. P., Yaspan B. L., and Chan A. C. (2016) Regulation of T cell receptor signaling by DENND1B in TH2 cells and allergic disease. Cell 164, 141–155 10.1016/j.cell.2015.11.052 [DOI] [PubMed] [Google Scholar]
- 17. Marat A. L., Ioannou M. S., and McPherson P. S. (2012) Connecdenn 3/DENND1C binds actin linking Rab35 activation to the actin cytoskeleton. Mol. Biol. Cell 23, 163–175 10.1091/mbc.e11-05-0474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nickerson M. L., Warren M. B., Toro J. R., Matrosova V., Glenn G., Turner M. L., Duray P., Merino M., Choyke P., Pavlovich C. P., Sharma N., Walther M., Munroe D., Hill R., Maher E., et al. (2002) Mutations in a novel gene lead to kidney tumors, lung wall defects, and benign tumors of the hair follicle in patients with the Birt-Hogg-Dubé syndrome. Cancer Cell 2, 157–164 10.1016/S1535-6108(02)00104-6 [DOI] [PubMed] [Google Scholar]
- 19. Nookala R. K., Langemeyer L., Pacitto A., Ochoa-Montaño B., Donaldson J. C., Blaszczyk B. K., Chirgadze D. Y., Barr F. A., Bazan J. F., and Blundell T. L. (2012) Crystal structure of folliculin reveals a hidDENN function in genetically inherited renal cancer. Open Biol. 2, 120071 10.1098/rsob.120071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zheng J., Duan B., Sun S., Cui J., Du J., and Zhang Y. (2017) Folliculin interacts with Rab35 to regulate EGF-induced EGFR degradation. Front. Pharmacol. 8, 688 10.3389/fphar.2017.00688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tsun Z. Y., Bar-Peled L., Chantranupong L., Zoncu R., Wang T., Kim C., Spooner E., and Sabatini D. M. (2013) The folliculin tumor suppressor is a GAP for the RagC/D GTPases that signal amino acid levels to mTORC1. Mol. Cell 52, 495–505 10.1016/j.molcel.2013.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Laviolette L. A., Mermoud J., Calvo I. A., Olson N., Boukhali M., Steinlein O. K., Roider E., Sattler E. C., Huang D., Teh B. T., Motamedi M., Haas W., and Iliopoulos O. (2017) Negative regulation of EGFR signalling by the human folliculin tumour suppressor protein. Nat. Commun. 8, 15866 10.1038/ncomms15866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mrozowska P. S., and Fukuda M. (2016) Regulation of podocalyxin trafficking by Rab small GTPases in epithelial cells. Small GTPases 7, 231–238 10.1080/21541248.2016.1211068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ojakian G. K., and Schwimmer R. (1988) The polarized distribution of an apical cell surface glycoprotein is maintained by interactions with the cytoskeleton of Madin-Darby canine kidney cells. J. Cell Biol. 107, 2377–2387 10.1083/jcb.107.6.2377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ishida M., Oguchi M. E., and Fukuda M. (2016) Multiple types of guanine nucleotide exchange factors (GEFs) for Rab small GTPases. Cell Struct. Funct. 41, 61–79 10.1247/csf.16008 [DOI] [PubMed] [Google Scholar]
- 26. Lamber E. P., Siedenburg A. C., and Barr F. A. (2019) Rab regulation by GEFs and GAPs during membrane traffic. Curr. Opin. Cell Biol. 59, 34–39 10.1016/j.ceb.2019.03.004 [DOI] [PubMed] [Google Scholar]
- 27. Gálvez-Santisteban M., Rodriguez-Fraticelli A. E., Bryant D. M., Vergarajauregui S., Yasuda T., Bañón-Rodríguez I., Bernascone I., Datta A., Spivak N., Young K., Slim C. L., Brakeman P. R., Fukuda M., Mostov K. E., and Martín-Belmonte F. (2012) Synaptotagmin-like proteins control the formation of a single apical membrane domain in epithelial cells. Nat. Cell Biol. 14, 838–849 10.1038/ncb2541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Blümer J., Rey J., Dehmelt L., Mazel T., Wu Y. W., Bastiaens P., Goody R. S., and Itzen A. (2013) RabGEFs are a major determinant for specific Rab membrane targeting. J. Cell Biol. 200, 287–300 10.1083/jcb.201209113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gerondopoulos A., Langemeyer L., Liang J. R., Linford A., and Barr F. A. (2012) BLOC-3 mutated in Hermansky-Pudlak syndrome is a Rab32/38 guanine nucleotide exchange factor. Curr. Biol. 22, 2135–2139 10.1016/j.cub.2012.09.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kulasekaran G., Nossova N., Marat A. L., Lund I., Cremer C., Ioannou M. S., and McPherson P. S. (2015) Phosphorylation-dependent regulation of connecdenn/DENND1 guanine nucleotide exchange factors. J. Biol. Chem. 290, 17999–18008 10.1074/jbc.M115.636712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Haley R., Wang Y., and Zhou Z. (2018) The small GTPase RAB-35 defines a third pathway that is required for the recognition and degradation of apoptotic cells. PLoS Genet. 14, e1007558 10.1371/journal.pgen.1007558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cheng S., Wang K., Zou W., Miao R., Huang Y., Wang H., and Wang X. (2015) PtdIns(4,5)P2 and PtdIns3P coordinate to regulate phagosomal sealing for apoptotic cell clearance. J. Cell Biol. 210, 485–502 10.1083/jcb.201501038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ran F. A., Hsu P. D., Wright J., Agarwala V., Scott D. A., and Zhang F. (2013) Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281–2308 10.1038/nprot.2013.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Naito Y., Hino K., Bono H., and Ui-Tei K. (2015) CRISPRdirect: software for designing CRISPR/Cas guide RNA with reduced off-target sites. Bioinformatics 31, 1120–1123 10.1093/bioinformatics/btu743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Saitoh T., Nakayama M., Nakano H., Yagita H., Yamamoto N., and Yamaoka S. (2003) TWEAK induces NF-κB2 p100 processing and long lasting NF-κB activation. J. Biol. Chem. 278, 36005–36012 10.1074/jbc.M304266200 [DOI] [PubMed] [Google Scholar]
- 36. Etoh K., and Fukuda M. (2019) Rab10 regulates tubular endosome formation through KIF13A and KIF13B motors. J. Cell Sci. 132, jcs226977 10.1242/jcs.226977 [DOI] [PubMed] [Google Scholar]
- 37. Itoh T., Satoh M., Kanno E., and Fukuda M. (2006) Screening for target Rabs of TBC (Tre-2/Bub2/Cdc16) domain-containing proteins based on their Rab-binding activity. Genes Cells 11, 1023–1037 10.1111/j.1365-2443.2006.00997.x [DOI] [PubMed] [Google Scholar]
- 38. Tamura K., Ohbayashi N., Maruta Y., Kanno E., Itoh T., and Fukuda M. (2009) Varp is a novel Rab32/38-binding protein that regulates Tyrp1 trafficking in melanocytes. Mol. Biol. Cell 20, 2900–2908 10.1091/mbc.e08-12-1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fukuda M., Kobayashi H., Ishibashi K., and Ohbayashi N. (2011) Genome-wide investigation of the Rab binding activity of RUN domains: development of a novel tool that specifically traps GTP-Rab35. Cell Struct. Funct. 36, 155–170 10.1247/csf.11001 [DOI] [PubMed] [Google Scholar]
- 40. Nishikimi A., Ishihara S., Ozawa M., Etoh K., Fukuda M., Kinashi T., and Katagiri K. (2014) Rab13 acts downstream of the kinase Mst1 to deliver the integrin LFA-1 to the cell surface for lymphocyte trafficking. Sci. Signal. 7, ra72 10.1126/scisignal.2005199 [DOI] [PubMed] [Google Scholar]
- 41. Fukuda M., Kojima T., Aruga J., Niinobe M., and Mikoshiba K. (1995) Functional diversity of C2 domains of synaptotagmin family: mutational analysis of inositol high polyphosphate binding domain. J. Biol. Chem. 270, 26523–26527 10.1074/jbc.270.44.26523 [DOI] [PubMed] [Google Scholar]
- 42. Kinoshita R., Homma Y., and Fukuda M. (2020) Methods of establishing Rab knockout MDCK cells. Methods Mol. Biol., in press [DOI] [PubMed] [Google Scholar]
- 43. Morita S., Kojima T., and Kitamura T. (2000) Plat-E: an efficient and stable system for transient packaging of retroviruses. Gene Ther. 7, 1063–1066 10.1038/sj.gt.3301206 [DOI] [PubMed] [Google Scholar]
- 44. Yasuda T., Mrozowska P. S., and Fukuda M. (2015) Functional analysis of Rab27A and its effector Slp2-a in renal epithelial cells. Methods Mol. Biol. 1298, 127–139 10.1007/978-1-4939-2569-8_11 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.