Abstract
Selectins are key to mediating interactions involved in cellular adhesion and migration, underlying processes such as immune responses, metastasis, and transplantation. Selectins are composed of a lectin domain, an epidermal growth factor (EGF)-like domain, multiple short consensus repeats (SCRs), a transmembrane domain, and a cytoplasmic tail. It is well-established that the lectin and EGF domains are required to mediate interactions with ligands; however, the contributions of the other domains in mediating these interactions remain obscure. Using various E-selectin constructs produced in a newly developed silkworm-based expression system and several assays performed under both static and physiological flow conditions, including flow cytometry, glycan array analysis, surface plasmon resonance, and cell-rolling assays, we show here that a reduction in the number of SCR domains is correlated with a decline in functional E-selectin binding to hematopoietic cell E- and/or L-selectin ligand (HCELL) and P-selectin glycoprotein ligand-1 (PSGL-1). Moreover, the binding was significantly improved through E-selectin dimerization and by a substitution (A28H) that mimics an extended conformation of the lectin and EGF domains. Analyses of the association and dissociation rates indicated that the SCR domains, conformational extension, and dimerization collectively contribute to the association rate of E-selectin–ligand binding, whereas just the lectin and EGF domains contribute to the dissociation rate. These findings provide the first evidence of the critical role of the association rate in functional E-selectin–ligand interactions, and they highlight that the SCR domains have an important role that goes beyond the structural extension of the lectin and EGF domains.
Keywords: cell migration, cell adhesion, glycoprotein structure, kinetics, glycobiology, carbohydrate-binding protein, dimerization, binding kinetics, complement regulatory repeats, E-selectin, short consensus repeats, silkworm expression, adhesion, glycoprotein, conformational extension, homing, silkworm expression
Introduction
The multistep paradigm of cell migration outlines a sequence of events that leads cells (e.g. stem cells, leukocytes, or circulating tumor cells) out of the circulating blood and into a target organ/tissue. This sequence of events, called homing, is initiated by the interaction of selectins (specifically, E-, P-, and L-selectins) with their ligands (1–3). Although each step in this process is important, the interactions mediating the first step of homing are crucial to slowing the circulating cells interacting with the endothelium to velocities that are lower than the local flow rate of cells in the blood. The most effective contributors to this critical step are the selectins.
E-selectin is constitutively expressed on bone marrow endothelium, where it recruits circulating hematopoietic stem/progenitor cells (HSPC)2 from the blood and is also important for the maintenance of HSPC within the niche (4, 5). E-selectin shows an affinity toward a prototypic sialylated and fucosylated structure known as sialyl-Lewis X (sLex) (NeuAcα2–3Gal1-4(Fucα1–3)GlcNAc1-R) (2, 6), which is expressed on glycoproteins and glycolipids (7–9). Studies suggest that the primary interaction with these ligands occurs through the carbohydrate-binding lectin domain (10), but less is known about the influence of the remaining structural components of the selectin molecule and how they may contribute to the functional binding activity.
The selectins are structurally composed of five distinct domains: an N-terminal extracellular C-type lectin-like domain, followed by an endothelial growth factor (EGF)–like domain, a defined number of short consensus repeats (SCRs) with 60 amino acids per motif, a transmembrane domain, and a C-terminal cytoplasmic tail that is likely involved in signal transduction regulation (11–15). Although the three selectins share similar structures, they differ in both binding specificity toward their ligands and the number of SCR domains. Although the lectin and EGF domains have a high degree of homology, that of their SCR domains is significantly lower (16). Several studies elucidated the role of the lectin and EGF domains in the interaction of selectins with their ligands (17–19), but the role of the SCR domains in this interaction remains elusive. It is speculated that these domains act as structural spacers that extend the lectin-binding domain beyond the crowded glycocalyx of the cell surface (11). However, studies where the SCR domains were deleted suggest that they may play a role in E-selectin–ligand interactions (20–22). Nonetheless, these studies do not provide information about the functional role of SCRs in the binding of E-selectin, especially under physiological flow conditions.
Studies using P-selectin suggest that it adopts a bent (lower affinity) conformation in the absence of its sulfated ligand but in its presence shifts toward an extended (higher affinity) conformation (22). Moreover, P-selectin is found as both a monomer and a dimer/oligomer on activated platelets. This heterogeneity in P-selectin structure is suggested to contribute to tethering through the low-affinity monomeric structures, whereas, at later stages, the dimerization of extended structures may facilitate stronger binding to stabilize interactions during the rolling process (23). It is unclear if and how dimerization influences the E-selectin binding, or whether the formation of an extended conformer that opens the angle between the lectin and EGF domains influences its binding in a manner analogous to that observed in P-selectin.
In this study, we generated five recombinant E-selectin proteins (see Fig. 1A) secreted by a novel silkworm expression system that included dimeric and monomeric forms harboring varying numbers of SCR domains as well as an enforced extended conformer. These E-selectin proteins allowed us to demonstrate the influence of the various structural domains and conformational extension on the overall ability of E-selectin to bind its ligands. By employing a comprehensive set of assays under static and physiological flow conditions, we showed that SCR domains, the conformational extension of the lectin and EGF domains, and protein dimerization contribute to the efficient binding of E-selectin to its glycosylated ligands. Moreover, these three structural elements collectively contribute to the association rate, whereas the dissociation rate was mainly controlled by the lectin and EGF domains. These findings correlate, for the first time, a very important physiological role for the association rate in the functional binding of the selectin to its ligands.
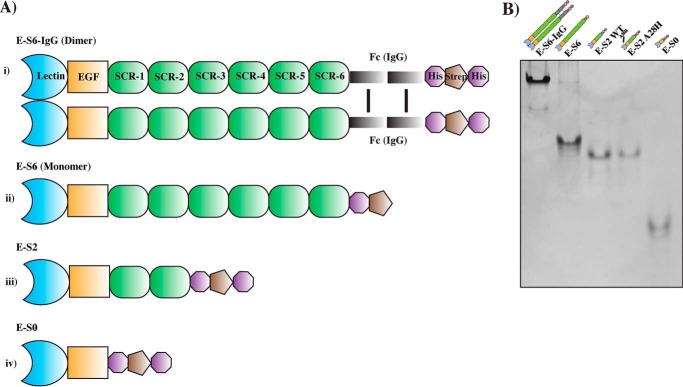
Figure 1.
Expression and purification of functional E-selectin proteins. A, schematic representation of E-selectin constructs. i, E-S6-IgG consisting of the structural domains of a commonly used E-selectin-IgG/Fc (dimer) chimera recombinant protein. ii, monomeric version of full-length E-selectin (E-S6). iii and iv, truncated E-selectin formed by domain deletion of either the last four SCRs, producing constructs with only the first two SCRs (E-S2), or deletion of all of the SCRs, yielding a minimal construct possessing only the main domains for binding (E-S0). Several tags were included at the C terminus of each construct, such as a histidine tag (double His6 or a single His8) and a Strep-tag, to facilitate the purification and subsequent immobilization of E-selectins (note that an Avi tag located after the EGF domain in E-S6-IgG, E-S2, and E-S0 and a TEV tag included before His8 in E-S6 are omitted from this diagram as they were not addressed in this study). B, analytical native PAGE of the recombinant E-selectin proteins. E-S6-IgG, E-S6, E-S2, E-S2-A28H, and E-S0 were diluted in 1× Native PAGE buffer, loaded on a 10% TBE gel and run in 1× Tris-glycine buffer. The majority of molecules in each recombinant protein sample appear as a single species, with a minor amount of oligomerization in case of E-S6; the majority of molecules in E-S6-IgG appear as a dimer through the C terminus Fc region, with some slightly monomeric versions appearing as a faint band of lower molecular weight.
Results
Active E-selectin proteins are expressed and purified from silkworm
To overcome the low level of eukaryotic proteins expressed by the widely used NS0 cells, we chose, for the first time, to use the silkworm expression system for E-selectin protein production (24). This system provides several advantages: it can synthesize and process signal peptides of secreted proteins, it has the machinery to produce proper folding, it can incorporate posttranslational modifications such as glycosylation and phosphorylation, and it is cost-effective (25). It is important to note that the type of N-glycosylation events that take place in insect cells are mainly pauci-mannose whereas in mammalian cells, they are more complex (26, 27) (Fig. S1). The differences in glycosylation patterns that result can affect the activity, stability, and solubility of the protein (28). Although a significant amount of the molecular weight of E-selectin is because of N-linked glycosylation, the protein-binding activity is not affected by the removal of N-glycans (29).
A Western blot analysis comparing the binding of two common E-selectin ligands, CD44/HCELL (hematopoietic cell E- and/or L-selectin ligand) and PSGL-1 (P-selectin glycoprotein ligand-1) (30–33), to dimeric E-selectin produced in silkworm showed similar binding patterns to that produced in a mammalian system (Fig. S2). This observation highlights the suitability of silkworm as an alternative system for expressing E-selectin.
To address the role of SCR domains and protein dimerization in the binding activity of E-selectin, various E-selectin constructs with varying numbers of SCR domains were generated (Fig. 1A): i) E-S6-IgG, which is a full-length dimeric protein that consists of a lectin domain followed by an EGF-like domain; six SCR domains; and an Fc region of IgG that aids in the dimerization of the molecule; ii) E-S6, which is a monomeric version of E-S6-IgG lacking the Fc region; iii) E-S2, which is a truncated protein with only the first two SCR domains; and iv) E-S0, which lacks any SCR domains. These protein constructs were successfully purified and did not appear to aggregate (Fig. 1B). On the other hand, the dimeric E-selectin, E-S6-IgG, was primarily dimerized (as facilitated by the Fc region) with a small amount showing up as a monomer (faint band in Fig. 1B). These observations suggest an absence of aggregation among the E-selectin proteins purified from the silkworm system.
The number of SCR domains, structural extension, and dimerization of E-selectin influence binding to its ligands in both static and flow-based assays
We used flow cytometry to confirm the binding functionality of our recombinant E-selectin proteins on known E-selectin ligands under native conditions. In these experiments, KG1a cells were used to represent HSPC-like (CD34+) cells, which are able to bind E-selectin (30, 31). The E-selectin expressed and purified from the silkworm was incubated with KG1a cells to allow them to interact with the different ligands expressed on these cells. The E-selectin proteins were detected using an antibody toward the Strep-tag located at the C terminus of each E-selectin protein (Fig. 1A), then analyzed by flow cytometry. As illustrated in Fig. 2A, E-S6-IgG, E-S6, E-S2, and E-S0 were all capable of binding the KG1a cells to varying degrees. As shown in Fig. 2B, E-S6-IgG bound to the ligands on 96% ± 0.8 of the live KG1a population, whereas E-S6 and E-S2 bound to significantly fewer (86% ± 2.1 and 76% ± 2.1, respectively; p ≤ 0.05; n = 3, compared with E-S6-IgG). Interestingly, we found that the construct containing only the lectin and EGF domains (E-S0) bound the lowest percentage of KG1a cells (43% ± 6.3; p ≤ 0.05; n = 3, compared with all other constructs). E-S6-IgG displayed the strongest fluorescence signal, followed by E-S6, E-S2, and E-S0 (Fig. 2C). Additionally, samples stained in the presence of EDTA, which removes divalent cations and inhibits the binding of selectins to their ligands, were included as controls to confirm binding specificity (Fig. 2A). Furthermore, we used several increasing concentrations of each construct to test if the binding ability of the weaker constructs would improve upon increasing their concentrations in solution. The results confirmed that the binding trend of each construct remained the same and did not improve for the weaker constructs to the degree that is comparable with the stronger-binding constructs (Fig. S3).
Figure 2.
Flow cytometric and Western blotting analyses comparing the binding functionalities of the various forms of E-selectin proteins to KG1a cells. A, E-S6-IgG, E-S6, E-S2, E-S2-A28H, and E-S0 were tested for their ability to bind to ligands on KG1a cells. Mouse monoclonal anti-Strep antibody (followed by a fluorescently labeled antibody against anti-Strep) was used to detect the E-selectin protein bound to ligands on the surface of KG1a cells in the presence of calcium (green histogram) or EDTA (red histogram). Samples stained with only the secondary antibody are included as a control to determine secondary antibody specificity toward mouse monoclonal anti-Strep antibody (blue histogram). B and C, the percentages of KG1a cells bound to each of the E-selectin proteins and the geometric means of the fluorescence signals were determined from n = 3 independent experiments (n = 3; a, indicates significance compared with E-S6-IgG; b, indicates significance compared with E-S6; c, indicates significance compared with E-S2; and d, indicates significance compared with E-S2-A28H, p ≤ 0.05) and the means are depicted as mean ± S.E.M. in (B) and (C), respectively. D, Western blot analysis of E-selectin protein binding to immunoprecipitated PSGL-1 and CD44. KG1a lysates were prepared, and PSGL-1 and CD44 were immunoprecipitated and subjected to the Western blot analysis. The resulting blots (upper panel, CD44; lower panel, PSGL-1) were stained with 1 μg/ml of E-S6-IgG, E-S6, E-S2, or E-S0, as indicated in the figure, in the presence of calcium. Anti-strep mAb was used against bound E-selectin for subsequent chemiluminescence detection using HRP-conjugated anti-mouse IgG. Blots stained in the presence of EDTA to confirm binding specificity showed no binding activity (Fig. S4).
Western blot analysis was conducted to determine the ability of the recombinant E-selectin protein constructs to bind common E-selectin ligands. To this end, PSGL-1 (33) and CD44/HCELL (32) were each immunoprecipitated from KG1a whole cell lysates and subsequently subjected to a Western blot analysis to measure their binding to the E-selectin protein constructs. E-S6-IgG exhibited the strongest staining for both CD44 (Fig. 2D, upper panel) and PSGL-1 (Fig. 2D, lower panel) compared with the other constructs. E-S6 exhibited visibly stronger staining to both ligands compared with E-S2 or E-S0 (Fig. 2D). Furthermore, identical blots were stained with E-selectin constructs in the presence of EDTA to chelate out the calcium that is essential for binding and binding was abrogated (Fig. S4). Overall, these results suggest that the strongest ligand binding is to the dimeric form of E-selectin and that the SCR domains contribute to this interaction. Because the binding of the E-selectin proteins to PSGL-1 and CD44/HCELL was similar, we chose to focus on the latter for further characterization of the E-selectin proteins.
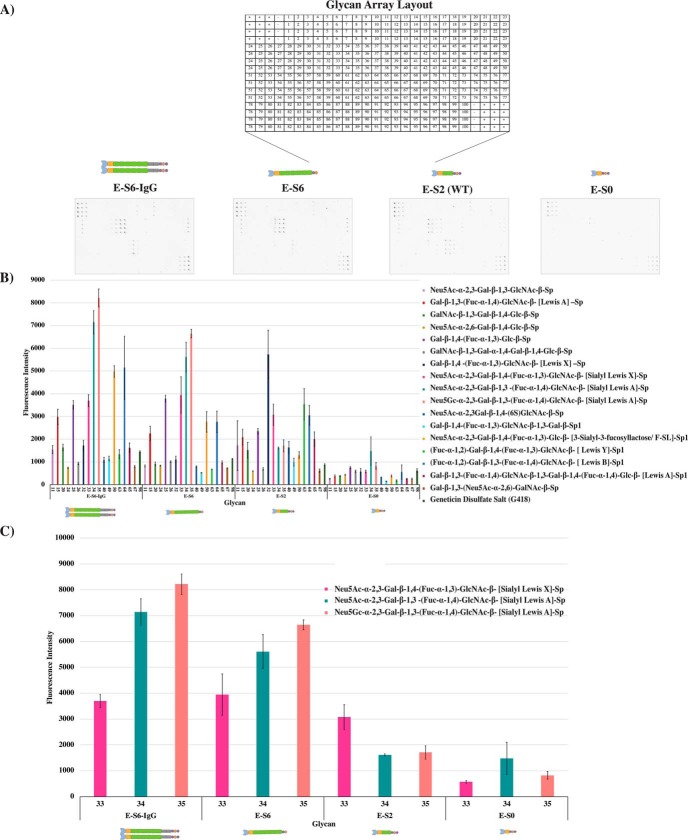
Furthermore, glycan-binding microarray analysis was conducted for each E-selectin construct (Fig. 3A). An overall decrease was observed in the binding of E-selectin to different glycans upon reducing or omitting the SCR domains (Fig. 3B). In particular, shorter E-selectins (E-S2 and E-S0) showed a gradual decrease in the binding to sLex (Neu5Ac-α-2,3-Gal-β-1,4-(Fuc-α-1,3)-GlcNAc-β) and to two sLea structures ((Neu5Ac-α-2,3-Gal-β-1,3-(Fuc-α-1,4)-GlcNAc-β-(sialyl-Lewis A) and Neu5Gc-α-2,3-Gal-β-1,3-(Fuc-α-1,4)-GlcNAc-β)) compared with E-S6-IgG and E-S6 (Fig. 3C). This indicates that the recognition level of each E-selectin construct toward sLex/a structures is similar to that toward intact ligands. A list of all the glycans within a microarray is shown in Fig. S5.
Figure 3.
Glycan microarray analysis. A, upper panel, E-selectins (E-S6-IgG, E-S6, E-S2 (WT), and E-S0) were introduced to 4× glycan subarray slides, and each bound construct was detected with biotin anti-His followed by Cy3 Equivalent Dye-Streptavidin. The layout of each array is shown with the location of each glycan, positive (+) and negative (−) controls. B, fluorescent intensity of each glycan was determined from quadruplicate samples, and a bar chart was generated and the means are depicted as mean ± S.E.M. for each construct. Each glycan is represented as a number and a color on the x-axis. C, a subset of the fluorescent intensity highlighting the differences in binding of each E-selectin to Neu5Ac-α-2,3-Gal-β-1,4-(Fuc-α-1,3)-GlcNAc-β- (sialyl-Lewis X)-Sp (33: magenta), Neu5Ac-α-2,3-Gal-β-1,3-(Fuc-α-1,4)-GlcNAc-β-(sialyl-Lewis A)-Sp (34: green), and Neu5Gc-α-2,3-Gal-β-1,3-(Fuc-α-1,4)-GlcNAc-β-(sialyl-Lewis A)-Sp (35: peach). OCH2CH2Ch2NH2 and NH(CH3)OCH2CH2NH2 linkers are represented as Sp and Sp1, respectively.
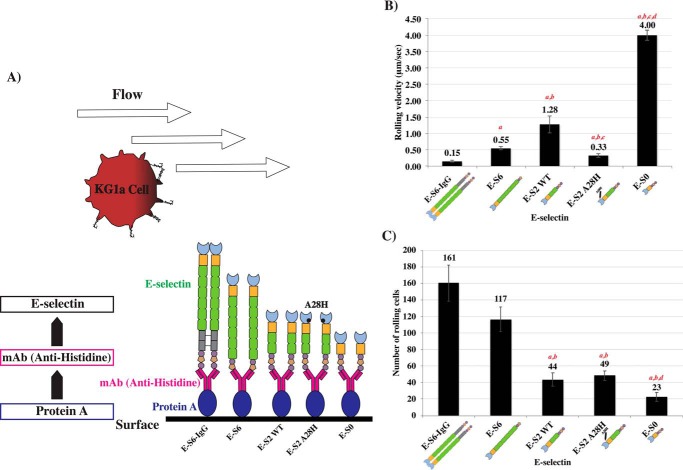
We next characterized the differences in binding among the E-selectin protein constructs using a physiological flow-based binding assay. A modified form of the parallel-plate flow-based binding assay (34–42) was used to achieve a quantitative comparison of the binding and rolling velocities of KG1a cells on various immobilized E-selectin protein constructs. In these experiments, E-selectin constructs were deposited at similar concentrations in each channel of a six-channel microfluidic chamber. To position the E-selectin proteins in the correct orientation, we used the histidine tag located at the C terminus of each E-selectin construct (His) (Fig. 1A). Each channel was coated with equal amounts of protein A, followed by an antibody against the histidine tag (Fig. 4A), which was found to consistently immunoprecipitate each recombinant E-selectin (Fig. S6). The rolling velocities (Fig. 4B) and the numbers of rolling cells (Fig. 4C) were determined for each E-selectin protein and found to be consistent with our observations above. An analysis of the rolling velocities revealed that E-S6-IgG, followed by E-S6, supported the slowest rolling velocities with 0.15 ± 0.03 μm/s and 0.55 ± 0.05 μm/s, respectively, whereas the shorter proteins supported much faster rolling velocities (1.28 ± 0.26 μm/s for E-S2 and 4 ± 0.16 μm/s for E-S0) (Fig. 4B). Longer constructs with six SCR domains supported the highest number of rolling KG1a cells with 161 ± 22 rolling cells on E-S6-IgG and 117 ± 15 on E-S6; significantly fewer cells rolled on the truncated constructs (44 ± 8 cells for E-S2 and 23 ± 5.4 cells for E-S0; p ≤ 0.05; n = 3) (Fig. 4C). Moreover, similar results were obtained when Fab fragments of the anti-histidine antibody were used in place of the intact antibody (Fig. S7). Interestingly, forced dimerization, induced by using the intact antibody, effectively reduced the rolling velocities for the shorter constructs compared with the Fab fragments, further supporting the influence that dimerization of E-selectin has in promoting binding to its ligands.
Figure 4.
Cell rolling analysis of KG1a cells on immobilized E-selectin proteins. A, schematic representation of the experimental steps involved in the microfluidics-based flow assay. To ensure the uniform immobilization of each of the proteins, identical concentrations of protein A were deposited on each channel of an uncoated μ slide VI0.1 microscopy chamber. This was followed by the addition of anti-histidine antibody and, subsequently, E-selectin proteins were introduced and captured through their C-terminal histidine tags (refer to Fig. S6: each E-selectin can be pulled down equivalently using anti-histidine). KG1a cells in perfusion buffer containing 0.5 mm Ca2+ were drawn into the E-selectin–coated channels at various shear stresses (1 dyne/cm2, 2 dyne/cm2, 3 dyne/cm2, 4 dyne/cm2, 5 dyne/cm2, and 6 dyne/cm2) for duration of 30 s each. B, adhesion bar graph of rolling velocities (μm/s) from n = 3 independent experiments, evaluating the rolling of KG1a on histidine-immobilized E-selectins in the presence of 0.5 mm Ca2+, represented as mean ± S.E.M. C, bar graph representing the number of KG1a cells rolling on each construct as an alternative measurement for adhesive strength from n = 3 independent experiments, represented as mean ± S.E.M. (n = 3; a indicates significance compared with E-S6-IgG, b indicates significance compared with E-S6, c indicates significance compared with E-S2, and d indicates significance compared with E-S2-A28H, p ≤ 0.05).
Previous studies of the interaction between selectins and their ligands improved our understanding of the mechanical behavior of selectin-ligand interactions (22, 43–55). A major discovery from these studies was the existence of a higher affinity forms of P-selectin (22) and E-selectin (55), which were achieved by extending the conformation of the lectin and EGF domains. To advance our understanding of the domain contributions to the binding behavior of E-selectin, we asked if the weak binding affinity of E-S2 could be enhanced by an equivalent mutation in the lectin domain (A28H) (54) that mimics the force-free extended form of E-selectin (55). The mutated E-S2 (E-S2-A28H) was tested against the other constructs in the same set of assays described above. In the flow cytometric analysis, the number of KG1a bound to E-S2-A28H increased by ∼1-fold (84% ± 4.2) compared with E-S2 and similar to the number of cells bound by E-S6 (86% ± 2.1) (Fig. 2B). Additionally, the fluorescence signal of E-S2-A28H (1689 ± 352.5) was only ∼2-fold lower than that of E-S6, whereas the signal of E-S2 was ∼3-fold lower than that of E-S6 (Fig. 2C), making the difference between the fluorescence signals of E-S2-A28H and E-S6 insignificant compared with the significant difference observed between E-S2 and E-S6 (p ≤ 0.05; n = 3). Moreover, flow-based experiments evaluating the rolling velocities of KG1a cells on immobilized E-S2-A28H showed a significant reduction in the rolling velocity (0.33 ± 0.06 μm/s) (p ≤ 0.05; n = 3) compared with E-S2 (rolling velocity = 1.28 ± 0.26 μm/s), and even approaches that of E-S6 (Fig. 4B). Together, these data illustrate the importance of the SCR domains, conformational extension, and dimerization in E-selectin binding behavior and in mediating slower rolling velocities.
Binding kinetics reveal SCR domains, extension, and protein dimerization contribute to the association rate
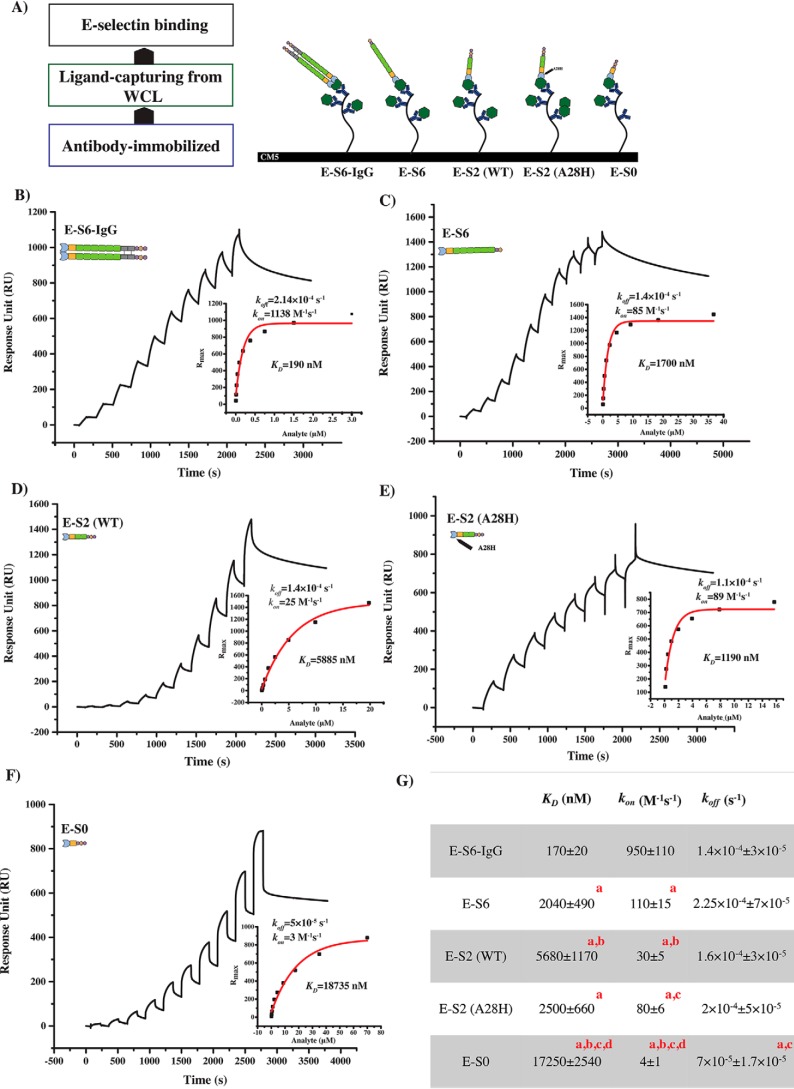
We used our previously described real-time surface plasmon resonance (SPR) binding assay (30) to quantitatively evaluate the interaction of the recombinant E-selectin proteins with the native endogenous selectin ligands expressed on the KG1a cells. Specifically, we captured CD44/HCELL from a KG1a cell lysate via the surface-immobilized monoclonal antibodies (mAb) against CD44 (9) (Fig. 5A). Then, the E-selectin proteins were injected in sequential concentrations at similar flow rates to determine the equilibrium dissociation binding constant (KD) and to estimate the association (kon) and dissociation rate constants (koff). The captured CD44 bound various forms of recombinant E-selectin constructs, with a KD of 170 ± 20 nm for the full-length dimeric E-S6-IgG and significantly higher KD of 2040 ± 490 nm, 5680 ± 1170 nm, and 17250 ± 2540 nm for E-S6, E-S2, and E-S0, respectively (comparing the KD of E-S6-Ig to other constructs, p ≤ 0.05; n = 3) (Fig. 5, B–D and F–G). To better understand the reason for these differences among the KD values of each construct, we determined the kon and koff. At the dissociation phase, all of the E-selectin proteins tested showed similar koff values, demonstrating that neither dimerization nor the SCR domains are required for the stable binding of E-selectin with CD44/HCELL. Strikingly, in contrast to the dissociation rate, the association rate constants revealed different binding kinetics. Full-length E-S6-IgG exhibited a kon (950 ± 110 m−1s−1) that is ∼9-fold higher (p ≤ 0.05; n = 3) than that of monomeric full-length E-S6 (110 ± 15 m−1s−1). Moreover, shorter constructs displayed significantly lower kon values; E-S2 exhibited kon values ∼4-fold lower than E-S6 (30 ± 5 m−1s−1), and E-S0 exhibited kon values ∼28-fold lower than E-S6 (4 ± 1 m−1s−1) (Fig. 5G). Furthermore, we confirmed that the binding of the shortest construct (i.e. E-S0) at high concentration does not seem to be a result of aggregation (Fig. S8).
Figure 5.
Rate constants for E-selectin protein binding to CD44/HCELL. A, experimental schematic diagram of the SPR-based real-time binding assay. Step 1, mAb immobilization on CM5 chip; step 2, KG1a lysate injection to capture CD44/HCELL; step 3, injection of various E-selectin proteins. B, binding of titrated concentrations of E-S6-IgG to captured CD44/HCELL from KG1a lysates via immobilized Hermes-3 (4113 RU); sensorgram representation of sequential injections of E-S6-IgG at 0.005, 0.01, 0.02, 0.05, 0.09, 0.19, 0.38, 0.75, 1.5, and 3 μm at a flow rate of 20 μl/min for 90 s each, spaced by a 60-s washing step. The lysate injection is not shown. The sensorgram profile was corrected for nonspecific interaction by subtracting isotype control (4466 RU). KD and koff values were determined, as described previously (30). kon was calculated using KD and koff values. C, binding of E-S6 (monomer form of the full-length E-selectin) to CD44/HCELL injected at different concentrations, 0.1, 0.19, 0.38, 0.77, 1.53, 3.06, 6.12, 12.25, 24.49, and 48.98 μm, using similar experimental conditions to those described in (B). The RU for Hermes-3 and its isotype control were 8670 and 6770, respectively. D, binding of truncated E-S2 to CD44/HCELL injected consecutively at concentrations 0.04, 0.08, 0.15, 0.31, 0.62, 1.24, 2.48, 4.95, 9.9, and 19.8 μm using similar experimental conditions to (B). The RU for Hermes-3 and its isotype control were 8450 and 5090, respectively. E, binding of titrated concentrations of E-S2-A28H to captured CD44/HCELL at different concentrations, 0.2, 0.3, 0.7, 1.3, 2.6, 5.2, 10.5, and 20.9 μm, using similar experimental conditions to (B). The RU for Hermes-3 and its isotype control were 6680 and 6360, respectively. F, binding of the SCR-deficient E-S0 to CD44/HCELL injected consecutively at concentrations of 0.14, 0.27, 0.56, 1.09, 2.19, 4.38, 8.75, 17.50, 35, and 70 μm using similar experimental conditions to (B). The RU for Hermes-3 and its isotype control were 6200 and 4900, respectively. G, table summarizing the binding constants of the E-selectin proteins to CD44/HCELL from n = 3 independent experiments for each E-selectin (B–F), reported as mean ± S.E.M. (n = 3, a indicates significance compared with E-S6-IgG, b indicates significance compared with E-S6, c indicates significance compared with E-S2, and d indicates significance compared with E-S2-A28H, p ≤ 0.05).
The KD for the binding of E-S2-A28H (Fig. 5, E and G) to immobilized CD44/HCELL was 2500 ± 660 nm. This KD is ∼2-fold higher than E-S2 (KD = 5680 ± 1170 nm). On the other hand, the difference between the affinities of E-S2-A28H and E-S6 (KD = 2040 ± 490 nm) decreased by only ∼1-fold compared with the ∼3-fold difference between E-S2 and E-S6. The kon for E-S2-A28H was ∼3-fold higher (80 ± 6 m−1s−1) than the kon for E-S2 (30 ± 5 m−1s−1) and approached that of E-S6 (kon = 110 ± 15 m−1s−1) (p ≤ 0.05; n = 3) (Fig. 5G). Interestingly, the koff was not influenced by the mutation in E-S2-A28H (Fig. 5, E and G). These findings indicate that binding can be improved by extending the lectin and EGF domains, even under conditions where the binding via the SCR domain is weakened. They also suggest that the extended structure contributes to the on rate of E-selectin binding, similar to the role of dimerization and the SCR domains. Consistent with the physiologically relevant flow data above, the SPR results further support that the SCR domains are indeed intrinsically important to the improved binding affinity of E-selectin. They also reveal the critical roles of the SCR domains, protein extension, and oligomerization in influencing the on rate, but not the off rate, of E-selectin binding to its ligands.
Discussion
Employing a novel silkworm expression system, we were able to purify five different E-selectin proteins in high yield and high purity. We showed that the binding activities of E-selectin to its ligands were improved by increasing the number of SCR domains, by forming an extended lectin and EGF domain structure, and by dimerizing/oligomerizing the protein. These observations were consistently verified by the analysis of these constructs in a number of static and flow-based assays. Our findings further unveiled that all three of these factors contribute to the on rate of E-selectin binding to its ligands, but do not significantly influence its off rate.
The contribution of the SCR domains to ligand binding has been shown previously for P-selectin (56). In that study, similar numbers of neutrophils attached to P-selectin proteins expressing five, six, and nine SCR domains under flow conditions. However, higher concentrations of P-selectin proteins harboring four SCR domains were required for the neutrophils to attach and roll, whereas P-selectin constructs with three or fewer SCR domains failed to bind the neutrophils, even at higher protein densities. Our results from E-selectin are consistent with these findings and established that the SCR domains contribute to the association rate more than to the dissociation rate.
To begin to understand how the SCR domains influence the binding affinity of an E-selectin, it is interesting to note that quite a low conservation of SCR domains exists (only 35%) among the selectins (55), which could potentially have an effect on binding functionality of each selectin toward its ligands. Atomic force microscopy studies revealed that a higher degree of stiffness is associated with the number of SCRs within each selectin implying that these domains act as springs and therefore, L-selectin exhibits the highest level of stiffness (with two SCRs), followed by E-selectin (with six SCRs), and then P-selectin (with nine SCRs) (57). Moreover, the rigidity within the SCR domains is partially provided by the presence of two disulfide bonds on the terminal regions and one central disulfide bond within each SCR domain (11, 55).
Structurally, SCR domains are indeed important for sufficiently extending the lectin domain of E-selectin to access the ligands on cells traveling through the blood. A simple model proposes that the full-length E-selectin tends to be in closer proximity to cells in the blood flow than the truncated constructs, leading to a higher frequency of interactions with their ligands. Additionally, our observations suggest that SCR domains may play a more direct functional role in improving the binding affinity of E-selectin; when E-S6-IgG, E-S6, E-S2, and E-S0 were introduced in solution to their ligands, they each exhibited distinctive binding characteristics. It is likely that the lectin domain is more involved in the actual binding to its sLex-expressing ligand, whereas the SCR domains contribute more to increase the association rate by interacting with other regions of the ligand. It is also possible that the SCR domains might influence the conformation of the selectin to better expose the lectin and EGF domains to facilitate binding to its ligand.
Testing the binding activity of our constructs toward different glycan structures revealed that all the constructs recognize sLex and sLea units (Fig. 3), albeit to varying degrees. This is consistent with the data presented in Figs. 2 and 4, suggesting that binding to E-selectin (and thus sLex/a) varies depending on the dimerization and the number of SCR domains. These findings are more likely to support the role of SCR domains in presenting the lectin and EGF domains in the proper conformation and, therefore, supporting the binding functionality of E-selectin. However, further detailed structural studies are needed to confirm this hypothesis.
The crystal structures of P- and E-selectin containing the lectin and EGF domains exhibited two conformational states: bent (22, 47, 55) and extended (22, 55). In the absence of a ligand, the selectin crystals adopted a bent conformation. When sLex was introduced by soaking it into the bent conformer crystals, it bound to both selectins without any observed changes to the conformation. However, co-crystals formed following the binding of a sulfated fragment of PSGL-1 with P-selectin, or the binding of a glycomimetic of sLex with E-selectin, resulting in an extended conformation of both selectins (22, 55, 58). This conformational shift of the E-selectin molecule toward a higher affinity extended conformer upon binding to a glycomimetic of sLex (58) was not observed previously in crystal structures of either selectin in the presence of soaked sLex (22, 47). Instead of introducing sLex to bent conformer crystals (22), the binding of a mimetic form of sLex to E-selectin was performed in a solution, and the resulting complexed molecules were then co-crystallized (55). The extended conformation of the E-selectin/sLex complex was further confirmed by the observation of a shift in the small-angle X-ray scattering curve, which corresponded to a change in the angle between the lectin and EGF domains.
A mutation in the lectin domain of P-selectin at position A28H was reported to open a split in the lectin domain that triggered a stable version of a higher affinity extended form of the selectin in a force-free fashion (54). Because we observed the functional influence of SCR domains on the binding activity of E-selectin, we tried to improve the binding of the E-selectin molecule bearing only two SCR domains (E-S2) by introducing a similar A28H mutation. The mutated form of this E-S2 (E-S2-A28H) showed improvement in the binding affinity compared with E-S2 and was capable of overcoming the loss of four SCRs by reaching an affinity near that of the full-length monomeric E-selectin molecule (E-S6) with comparable KD values. Furthermore, this observation is supported by the slower rolling velocity of the KG1a cells on E-S2-A28H in rolling assays compared with the cells rolling on E-S2. These results demonstrate that the extended structure of the lectin and EGF domains is another contributor to selectin binding, even under conditions where the contribution of the SCR domain is compromised. Interestingly, the extended structure acted in a similar manner to the SCR domains in that it specifically enhanced the on rate of E-selectin binding. Although the extended conformer decreases the KD to similar -fold in P- and E-selectin, the contribution it plays for the association and dissociation rates appears to be different. The extended conformer in P-selectin decreases the dissociation rate (54, 59), whereas in E-selectin, it only influenced the association rate (current study). These results suggest that the extended conformer may play different roles in the binding kinetics between E- and P-selectin.
Species-specific differences in binding strength indicate that mouse E-selectin possesses stronger binding activity than human E-selectin and that this is correlated to the size of the interdomain angle, which is greater in the mouse than in the human form (60). Consistent with these findings, we reported previously that monomeric human E-selectin binds CD44/HCELL transiently with fast on and off rates (30) whereas the monomeric mouse E-selectin, in the current study, bound CD44/HCELL with slow on and off rates and as tightly as the dimeric form.
A number of studies have focused on the impact of selectin dimerization on binding (61–65). Monomeric E-selectin binds similarly to both the dimeric and monomeric forms of PSGL-1, and dimeric E-selectin binds more strongly to dimeric PSGL-1 (66). Similarly, cells expressing PSGL-1 formed much more stable rolling with dimeric P-selectin compared with monomeric P-selectin (67), suggesting that dimeric versions of P-selectin (and PSGL-1) could support the tethering and rolling stability of cells by increasing their chance of forming a new bond after dissociation, i.e. “rebinding,” as the cell is tethering, thus prolonging the lifetime of the bond (23). Furthermore, kinetic studies comparing soluble monomeric P-selectin to the membrane (most dimeric) P-selectin revealed fast on/fast off binding for monomeric P-selectin and a biphasic on with a slow off rate for membrane P-selectin (68) which is consistent with several binding analyses of the binding kinetics of dimeric and monomeric P-selectin (69, 70). In our current study, both the dimer and monomer bind with slow on and off rates, whereas the dimer primarily contributes to a significantly slower association rate of binding. These findings also support the important role of E-selectin clustering in its interaction with its ligands during physiological cell adhesion processes.
Overall, our results consistently showed that mouse E-selectin binds with slow on and off rates to native ligands. This is in contrast to the fast on and off rate reported for the human monomeric form of P-, L-, and E-selectin (22, 30, 54, 59, 68, 69, 71, 72). It further showed that SCR domains, conformational extension, and dimerization all collectively contribute to the association rate, whereas the lectin and EGF domains are responsible for the dissociation rate. Under physiological conditions of sheer force, the reported differences in the association rate have dramatic effects on the functional binding behavior of E-selectin. Therefore, it is critical to quantitatively characterize the association and dissociation rates of selectin binding to their well-characterized native ligands. We believe that the silkworm expression system will introduce new capabilities to the selectin field at large to help move it toward more quantitative and structural approaches to understanding these interactions.
Experimental procedures
Construction of E-selectins expression vectors
A E-S6-IgG chimera common construct, containing a full-length extracellular region of mouse E-selectin and an Fc region of human IgG1 attached with His6 tag, was designed based on the structure of commercially available materials (575-ES, R&D Systems), except for a single mutation that removed the free cysteine residue (S103C; IgG sequence) to eliminate crosslinking. Then, the construct was synthesized and obtained at the institutional core facility, Bioscience Core Lab (KAUST BCL). The C-terminal His6 tag (73) was replaced with an Avi double-His-Strep II (73, 74) composite tag. Additionally, the monomer E-selectin (E-S6) was synthesized and obtained similarly to E-S6-IgG (described above), with the addition of a TEV cleavage site, and His8 (73) and Strep II tags (74), instead of the double-His-Strep II introduced to E-S6-IgG. E-S6-IgG was also used as the starting template for synthesizing the truncated constructs, E-S2 and E-S0. For the construction of E-S2, the first PCR reaction was completed using the forward primer caccATGAATGCCTCGCGCTTTCTCTCTGC, the overlapping reverse primers CCATCATGCAAAGCTtccggcctgaacgac, tccggcctgaacgacatcttcgaggctcagaaaatcgaatggcacgaacatcatcacc, and aggttaCTCGAGtcaatgatgatggtgatgatgtt for the addition of an Avi tag, a single histidine tag (His6), and an XhoI restriction site. Similarly, E-S0 was produced in a PCR reaction identical to the E-S2 PCR synthesis, with the replacement of the reverse primer CCATCATGCAAAGCTtccggcctgaacgac with CCCAACTGTGAGCAAtccggcctgaacgacatcttcgaggctc. Subsequently, the E-S2 and E-S0 constructs were subjected to a second PCR reaction to add Strep-tag, followed by a second polyhistidine, (His6), with the forward primer ATGAATGCCTCGCGCTTTCTCTCT and the overlapping reverse primers CCAAGCTCTTGAatgatgatggtgatgatgttcgt, catTCAAGAGCTTGGCGTCATCCGCAGTTCGGTGG, tcaGTGATGGTGATGGTGATGACCACCGAACTGCGga, and aggttaCTCGAGtcaGTGATGGTGATGGTGATGAC. The TEV and Avi tags were included for future work and are not utilized in the current study.
Truncated E-selectin constructs were cloned into a pENTR11 (Invitrogen) vector using the GatewayTM cloning system for generating recombinant expression vectors. Briefly, E-selectin amplicons were digested with XhoI (C∧TCGAG), whereas the pENTR11 (Invitrogen) vector was digested with XhoI and XmnI (GAAnn∧nnTTC) (New England Biolabs). The fragments were directionally cloned into pENTR11 by ligation using T4 DNA ligase (Invitrogen). E-S2-A28H was created by subjecting E-S2-pENTR11 recombinant vector to site-directed mutagenesis. A PCR reaction was performed using the forward primer TACACACATCTGGTGcacATTCAGAACAAGGAA and reverse primer TTCCTTGTTCTGAATgtgCACCAGATGTGTGTA. The GatewayTM LR recombination reaction was performed using LR clonase to produce recombinant pDEST8 expression vectors (Fig. S9) containing the different forms of E-selectins under the transcriptional regulation of a PPH Polyhedrin promoter. Design of all constructs and primers was performed using ApE-A Plasmid Editor.
Generation of recombinant BmNPVs and purification of recombinant E-selectins
Recombinant pDEST8 expression vectors, constructed as described above, were used to generate recombinant viral DNAs using a BmNPV/T3 bacmid system, as described previously (75). The viral DNAs were isolated using the FlexiPrep kit (Amersham Biosciences), then transfected into the NIAS-Bm-oyanagi2 (BmO2, kindly provided by Dr. Imanishi) cell line using the FuGENE HD transfection kit (Promega, Madison, WI). Then, 4 days after transfection, the culture medium was gathered as recombinant P1 viruses. High-titer virus (P3) stocks were prepared by serial infection following the protocols recommended in the manufacturer's manual (Invitrogen).
The recombinant E-selectins/BmNPVs were infected into the hemocoel of 2-day-old fifth instar silkworm larvae (NBRP, provided by the Institute of Genetic Resources, Kyushu University Graduate School) using a microliter syringe with a 30-gauge needle (Hamilton Co.). At 4 days post infection, the sera from the silkworm larvae were collected in a 15-ml tube containing 20 mm 1-phenyl-2-thiourea. Following brief centrifugation, they were stored at −80 °C until use.
Purification of the recombinant E-selectins was modified from our previously published protocol (76). Briefly, a two-step purification was performed using standard protocols based on the tandem, terminal His and Strep tags. First, the serum diluted with buffer A (20 mm Tris-HCl, pH 7.4, 0.5 m NaCl, 1 mm PMSF) was purified by nickel affinity chromatography with a HisTrap Excel column (GE Healthcare) and eluted by increasing the concentrations of imidazole solution buffers (100–500 mm imidazole). Then, the eluted solution was concentrated (Amicon Ultra-15 3 or 30K filters, Millipore) and diluted with the binding buffer B (100 mm Tris-HCl, pH 8.4, 150 mm NaCl, 1 mm EDTA, PIC (1 tablet/50 ml)) (Roche). A final 50 ml diluted solution was applied to the StrepTrap HP column (GE Healthcare), followed by elution with buffer B, which contained 2.5 mm desthiobiotin. Finally, the purified recombinant E-selectins were dialyzed against 1× PBS (pH 7.4), separated through 10% SDS-PAGE electrophoresis and quantified by Pierce BCA Protein Assay kit.
Cell lines
Human acute myelogenous leukemia (KG1a) cell line was used as the model for the CD34+ HSPCs (ATCC). The cells were cultured in RPMI 1640 media (Gibco) supplemented with 10% fetal bovine serum (FBS) (Cellgro) and 100 units/ml of HyClone penicillin/streptomycin (Invitrogen). These cell lines were grown and maintained in a suspension culture at a density of 1 × 106 cells/ml, under humid conditions in a 37 °C incubator supplemented with 5% CO2.
Flow cytometry
The KG1a cells (at a concentration of 1 × 106 cells/ml) were stained with 10 μg/ml of each of the E-selectins, E-S6-IgG, E-S6, E-S2, E-S2-A28H, or E-S0, in 100 μl HBSS with 5% FBS, in the presence of either 2 mm CaCl2 or 20 mm EDTA, at 4 °C for 20 min. Next, the stained cell samples were washed with the same HBSS buffer (containing either 2 mm CaCl2 or 20 mm EDTA) one time, and further stained with 10 μg/ml of either Strep-tag II mAb (EMD Millipore), mouse IgG1 PE conjugated isotype control antibody (R&D Systems), or PE rat anti-mouse IgG1 (Clone A85–1) (BD Biosciences) as the secondary control, and then were incubated at 4 °C for 20 min. The samples were then washed two times with the same washing buffer and incubated with 10 μg/ml of PE rat anti-mouse IgG1 (Clone A85–1) for another 20 min at 4 °C. Finally, the stained cells were again washed two times and analyzed using BD FACSCanto (BD Biosciences) and FlowJo software (version 7.6.1).
Cell lysis and immunoprecipitation (IP)
The KG1a cells were lysed using the membrane disruption method with Triton X-100–based lysis buffer (150 mm NaCl, 50 mm Tris-HCl, pH 7.4) (Fisher Scientific), 1× of protease inhibitor (Roche), 1 mm of PMSF (Sigma Aldrich) and 1% of Triton X-100 (Thermo Fisher Scientific) at a density of 1 × 106 cells/16 μl of lysis buffer, and were rotationally mixed for 1 h at 4 °C. The soluble fraction was collected by centrifugation at 16,000 relative centrifugal force for 30 min at 4 °C. PSGL-1 and CD44 were immunoprecipitated separately from the heterogeneous mixture of proteins by incubation with 50 μl of prewashed Protein G Dynabeads (Thermo Fisher Scientific) complexed with either 3 μg of mouse anti-human CD162 (clone KPL-1) (BioLegend) or 3 μg of mouse anti-human CD44 (clone 515) (BD Biosciences) and mixed rotationally overnight at 4 °C. Next, the supernatants were removed, and the beads were washed three times with lysis buffer chilled to 4 °C.
Western blotting and native polyacrylamide agarose gel electrophoresis
Protein G Dynabeads with immunoprecipitated PSGL-1 and CD44 were prepared for PAGE by resuspending the beads in 2× NuPAGE lithium dodecyl sulfate sample buffer (Invitrogen) in PBS (Gibco) and 10% β-mercaptoethanol (Sigma-Aldrich). The sample was then heated at 95 °C for 5 min to assess the denaturation process by releasing the capturing antibody with the immunoprecipitated PSGL-1 or CD44 from the protein G beads and into the solution. For the Western blot analysis, the prepared samples were loaded into a 4–20% SDS-polyacrylamide gradient gel (Bio-Rad) in 1× Tris-Glycine SDS buffer (Sigma) as lysates of 1 × 107 KG1a cells/well and transferred to an immunoblot PVDF membrane (Bio-Rad) in 1× Tris-Glycine buffer (Sigma). The resulting membrane was blocked with 5% of nonfat milk in 1× TBST buffer (Cell Signaling Technology) containing 137 mm sodium chloride, 20 mm Tris, and 0.1% Tween 20 at pH 7.6. The prepared blots were stained individually with 1 μg/ml of E-S6-IgG (either homemade from silkworm or produced from NS0 cells) (R&D Systems), E-S6, E-S2, E-S2-A28H, or E-S0 in 1× TBST buffers containing 2 mm CaCl2 or 20 mm EDTA and were subsequently stained again with 1 μg/ml Strep-tag II mAb, followed by incubation with 0.057 μg/ml of HRP-conjugated goat anti-mouse IgG (Thermo Scientific) or, for staining the dimeric E-S6-IgG proteins from both silkworm and mammalian cells, directly with HRP-goat anti-human IgG (SouthernBiotech) at 1:10,000 (all staining and washing steps were done in 1× TBST buffers containing 2 mm CaCl2 or 20 mm EDTA). Lastly, the membranes were exposed to SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific) for visualization via the chemiluminescence reactions and imaged using ImageQuant LAS 4000 (GE Healthcare).
To characterize the possible aggregation properties of our E-selectins, protein samples were also prepared for native gel electrophoresis at a final volume of 10 μl in 1× of 4× native PAGE sample buffer (Invitrogen). The prepared samples were loaded into a 10% 10 wells TBE gel (Novex), and electrophoresis was conducted in 1× Tris-glycine buffer for 3 h at 200 voltage. Subsequently, the resulting gel was stained using G-250 Coomassie Brilliant Blue stain (Millipore), then destained in water.
Glycan array analysis
Sandwich-based protocol was followed in accordance with the manufacturer's instructions using a 4-array glycan chip (GA-Glycan-100–4; RayBiotech). First, each array was blocked using 400 μl of sample diluent at room temperature for 30 min. Next, the sample diluent was removed and a 135 μg/ml solution of each construct (E-S6-IgG, E-S6, E-S2, and E-S0) was prepared in sample diluent containing 2 mm CaCl2. 400 μl of each construct was then added to one glycan array. Samples were incubated for 3 h at 4 °C with continuous mixing (low shaking speed ∼60 revolutions per minute (rpm); Thermo Fisher). Each array was washed four times with 800 μl of 1× wash buffer I and two times with 800 μl of 1× wash buffer II; each washing step was performed for 5 min at low shaking speed (∼60 rpm). Next, each array was incubated overnight (4 °C) with 400 μl of biotin anti-His6 tag antibody (BioLegend) (prepared in diluent buffer at a final concentration of 1 μg/ml) with low shaking speed (∼60 rpm); the slide was covered with adhesive film to protect it from environment and contamination. The following day, the antibody solution was removed, and each array was washed, as stated above. Next, 400 μl of Cy3 equivalent dye-steptavidin (prepared as 1× in sample diluent buffer) was added to each array and incubated for 1 h at room temperature at low shaking speed (∼60 rpm). Cy3 equivalent dye-steptavidin was then removed, and each array was washed as described above. Subsequently, the gasket defining each array as a separate well was removed, and the slide was soaked consecutively in the following solutions in a slide washer/dryer (RayBiotech): 30 ml 1× wash buffer I, 30 ml 1× wash buffer II, and 30–40 ml milliQ; each wash was performed for 5 min at room temperature. Finally, the residual solution was removed from the surface of the chip and the chip was imaged using the Typhoon Trio variable mode imager (GE Healthcare). Finally, the resulted intensities were measured for each spot using ImageJ software.
Surface plasmon resonance
Affinity and kinetics assessments of the E-selectins' binding were performed using the Biacore T-100 system on a carboxymethylated (CM5) dextran sensor chip (GE Healthcare) at 25 °C, as described previously (30). First, the system and the flow cells of the sensor chip were washed in two priming steps using filtered (through 0.2 μm filter) and degassed 1× HBS-EP buffer (0.01 m HEPES, pH 7.4, 0.15 m NaCl, 3 mm EDTA, 0.005% v/v surfactant P20) (GE Healthcare). Mouse anti-human CD44 Hermes-3 (Abgent) was immobilized on each flow cell of the CM5 surface using an amine-coupling procedure. In this method, each flow cell was activated by injecting a 1:1 ratio of N-hydroxysuccinimide (0.1 m) and 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride (0.4 m) for 8 min at a flow rate of 5 μl/min. Then, 20 μg/ml of the antibody was injected in 10 mm sodium acetate phase (pH 5.0) at a 10 μl/min flow rate, to allow amine-mediated immobilization of the antibody on the CM5 flow cell surface; the resulting amount of immobilized antibody was monitored as indicated in Fig. 5 (4000–9000 RU). Finally, deactivation was achieved by injecting 1 m ethanolamine hydrochloride at a flow rate of 5 μl/min over a total period of 8 min. Using the same procedure, mouse IgG2a κ isotype control (BioLegend) was immobilized on a control flow cell. To prepare for CD44 IP, KG1a cells were lysed at a density of 3 × 107 cells in 300 μl of a lysis buffer containing 1% Triton X-100, 250 mm NaCl, 50 mm Tris base (pH 8.0), and 1 mm CaCl2 mixed on a rotator for 1 h at 4 °C. Subsequently, the clear phase was collected by centrifugation at 16,000 relative centrifugal force for 30 min. The system was primed twice using a filtered and degassed running buffer (50 mm Tris, pH 8.0, 1% Triton X-100, 50 mm NaCl, and 1 mm CaCl2) to exchange the buffer on the flowing cells. CD44 real-time IP was initiated by injecting the lysate at a 20 μl/min flowrate for 13 min, followed by a 15-min washing step. The E-selectins were titrated in running buffer and injected at a sequence of concentrations, as indicated in Fig. 5. Each injection lasted for 1.5 min at a flow rate of 20 μl/min, and 1-min washing steps were included between the injections. The data analysis was conducted using Biacore evaluation software, as described previously (30), and the kinetics and affinity profiles were generated and analyzed using OriginLab software.
Cell rolling assay
The cell rolling assay permitted us to evaluate the incorporated shear stress conditions on the physiological interactions of the E-selectins with their ligands. In this assay, the six channels of an uncoated μ-slide VI0.1 microscopy chamber (ibidi) were coated with protein A (Thermo Fisher) at 10 μg/ml in HBSS (Gibco) for 16 h. Then each channel was washed in one step. After the channels were completely dry, mouse anti-histidine (AbD Serotec) was added as a second coating at a concentration of 10 μg/ml in HBSS for 1 h at 4 °C, then each channel was washed as described above. The E-selectins (E-S6-IgG, E-S6, E-S2, E-S2-A28H, and E-S0) were added to the channels at 5 μg/ml (diluted in HBSS) and incubated at 4 °C for 16 h to uniformly immobilize them by utilizing the histidine tag located at the C terminus of each E-selectin (Fig. 1A). Next, the channels of the chamber were washed and blocked using HBSS containing 1% BSA for 1 h at 4 °C, then washed again. The prepared chamber was placed on the stage of an inverted microscope (Olympus) connected to a CCD camera to visualize and record the rolling events using Cell Sens software. Shear stress within a rectangular channel was calculated as described previously (77) using Equation 1:
| (Eq. 1) |
where τ is the shear stress (dyne/cm2), μ is the viscosity of the fluid (0.00076 Pa.s), Q is flow volume (ml/s), a and b are the height and width of the channel (0.1 mm and 1 mm), respectively.
The inlet of each channel was connected by a tube (0.8 mm silicone tubing from ibidi) to a cell-containing buffer (1.2 × 106 cells in 1 ml of HBSS containing 1% BSA and 0.5 mm CaCl2 or 5 mm EDTA), whereas the outlet was connected to a highly programmable Harvard syringe pump. The suspended KG1a cells were gradually drawn through the channel first at 90 dyne/cm2 for 1 min, and then allowed to attach to the immobilized constructs by decreasing the shear stress rate to 0.3 dyne/cm2 for 5 min. Next, various shear stress rates were introduced sequentially, corresponding to a sequence of 1 dyne/cm2, 2 dyne/cm2, 3 dyne/cm2, 4 dyne/cm2, 5 dyne/cm2, and 6 dyne/cm2 for 30 s each. The recorded rolling incidents of the KG1a cells on the immobilized E-selectins were analyzed using Imaris software to evaluate the number of rolling cells and their rolling velocities on each E-selectin.
Statistical analysis
Data are reported as the means ± S.E.M. Statistically significant differences between the means of each E-selectin were resolved using an unpaired Student's t test. Statistical significance was defined as p ≤ 0.05.
Author contributions
F. A. A., K. S., and J. S. M. conceptualization; F. A. A., K. S., J. M. L., D. B. A., B. A., S. N., M. T., and J. S. M. data curation; F. A. A., K. S., J. M. L., D. B. A., B. A., S. M. H., S. H., T. K., and J. S. M. formal analysis; F. A. A., K. S., J. M. L., D. B. A., and J. S. M. investigation; F. A. A., K. S., J. M. L., D. B. A., B. A., and J. S. M. methodology; F. A. A., K. S., J. M. L., D. B. A., and J. S. M. writing-original draft; F. A. A. and J. S. M. writing-review and editing; J. M. L., T. K., and J. S. M. resources; T. K. and J. S. M. project administration; S. M. H., S. H., T. K., and J. S. M. supervision; S. M. H., S. H., and J. S. M. validation; T. K. and J. S. M. funding acquisition.
Supplementary Material
Acknowledgments
We are grateful to Vlad-Stefan Raducanu for discussion about E-S0 sample aggregates and Maryam Mih, Samar A. Rustum, and Umme Habiba for their support in the management of the laboratory. We also thank Veronica Tremblay from the KAUST Academic Writing Service for editing the manuscript.
This research was supported by a King Abdullah University of Science and Technology (KAUST) Faculty Baseline Research Funding Program (to J. S. M.) and by a Competitive Research Grant (OCRF-2014-CRG3–2276) (to J. S. M.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S9 and supporting methods information.
- HSPC
- hematopoietic stem/progenitor cells
- sLex
- sialyl-Lewis X
- EGF
- epidermal growth factor domain
- SCR
- short consensus repeats
- SPR
- surface plasmon resonance
- TEV
- tobacco etch virus
- HBSS
- Hanks' Balanced Salt Solution
- TBST
- Tris-buffered saline with Tween 20
- IP
- immunoprecipitation
- rpm
- revolutions per minute
- RU
- response units
- KD
- equilibrium dissociation binding constant
- koff
- dissociation rate constant
- kon
- association rate constant.
References
- 1. Butcher E. C. (1991) Leukocyte-endothelial cell recognition: three (or more) steps to specificity and diversity. Cell 67, 1033–1036 10.1016/0092-8674(91)90279-8 [DOI] [PubMed] [Google Scholar]
- 2. Sackstein R. (2005) The lymphocyte homing receptors: Gatekeepers of the multistep paradigm. Curr. Opin. Hematol. 12, 444–450 10.1097/01.moh.0000177827.78280.79 [DOI] [PubMed] [Google Scholar]
- 3. Springer T. A. (1994) Traffic signals for lymphocyte recirculation and leukocyte emigration: The multistep paradigm. Cell 76, 301–314 10.1016/0092-8674(94)90337-9 [DOI] [PubMed] [Google Scholar]
- 4. Schweitzer K. M., Dräger A. M., van der Valk P., Thijsen S. F., Zevenbergen A., Theijsmeijer A. P., van der Schoot C. E., and Langenhuijsen M. M. (1996) Constitutive expression of E-selectin and vascular cell adhesion molecule-1 on endothelial cells of hematopoietic tissues. Am. J. Pathol. 148, 165–175 [PMC free article] [PubMed] [Google Scholar]
- 5. Winkler I. G., Barbier V., Nowlan B., Jacobsen R. N., Forristal C. E., Patton J. T., Magnani J. L., and Lévesque J. P. (2012) Vascular niche E-selectin regulates hematopoietic stem cell dormancy, self renewal and chemoresistance. Nat. Med. 18, 1651–1657 10.1038/nm.2969 [DOI] [PubMed] [Google Scholar]
- 6. Polley M. J., Phillips M. L., Wayner E., Nudelman E., Singhal A. K., Hakomori S., and Paulson J. C. (1991) CD62 and endothelial cell-leukocyte adhesion molecule 1 (ELAM-1) recognize the same carbohydrate ligand, sialyl-Lewis x. Proc. Natl. Acad. Sci. U.S.A. 88, 6224–6228 10.1073/pnas.88.14.6224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Alon R., Feizi T., Yuen C. T., Fuhlbrigge R. C., and Springer T. A. (1995) Glycolipid ligands for selectins support leukocyte tethering and rolling under physiologic flow conditions. J. Immunol. 154, 5356–5366 [PubMed] [Google Scholar]
- 8. Fuhlbrigge R. C., Kieffer J. D., Armerding D., and Kupper T. S. (1997) Cutaneous lymphocyte antigen is a specialized form of PSGL-1 expressed on skin-homing T cells. Nature 389, 978–981 10.1038/40166 [DOI] [PubMed] [Google Scholar]
- 9. Hanley W. D., Burdick M. M., Konstantopoulos K., and Sackstein R. (2005) CD44 on LS174T colon carcinoma cells possesses E-selectin ligand activity. Cancer Res. 65, 5812–5817 10.1158/0008-5472.CAN-04-4557 [DOI] [PubMed] [Google Scholar]
- 10. Geng J. G., A., H. G., and McEver R. P. (1992) Lectin domain peptides from selectins interact with both cell surface ligands and Ca2+ ions. J. Biol. Chem. 267, 19846–19853 [PubMed] [Google Scholar]
- 11. Bevilacqua M. P., Stengelin S., Gimbrone M. A. Jr., and Seed B. (1989) Endothelial leukocyte adhesion molecule 1: An inducible receptor for neutrophils related to complement regulatory proteins and lectins. Science 243, 1160–1165 10.1126/science.2466335 [DOI] [PubMed] [Google Scholar]
- 12. Bevilacqua M. P., Pober J. S., Mendrick D. L., Cotran R. S., and Gimbrone M. A. (1987) Identification of an inducible endothelial-leukocyte adhesion molecule. Proc. Natl. Acad. Sci. U.S.A. 84, 9238–9242 10.1073/pnas.84.24.9238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Collins T., Williams A., Johnston G. I., Kim J., Eddy R., Shows T., Gimbrone M. A. Jr., and Bevilacqua M. P. (1991) Structure and chromosomal location of the gene for endothelial-leukocyte adhesion molecule 1. J. Biol. Chem. 266, 2466–2473 [PubMed] [Google Scholar]
- 14. Johnston G. I., Cook R. G., and McEver R. P. (1989) Cloning of GMP-140, a granule membrane protein of platelets and endothelium: Sequence similarity to proteins involved in cell adhesion and inflammation. Cell 56, 1033–1044 10.1016/0092-8674(89)90636-3 [DOI] [PubMed] [Google Scholar]
- 15. Tedder T. F., Isaacs C. M., Ernst T. J., Demetri G. D., Adler D. A., and Disteche C. M. (1989) Isolation and chromosomal localization of cDNAs encoding a novel human lymphocyte cell surface molecule, LAM-1. Homology with the mouse lymphocyte homing receptor and other human adhesion proteins. J. Exp. Med. 170, 123–133 10.1084/jem.170.1.123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kansas G. S. (1996) Selectins and their ligands: Current concepts and controversies. Blood 88, 3259–3287 10.1182/blood.V88.9.3259.bloodjournal8893259 [DOI] [PubMed] [Google Scholar]
- 17. Kansas G. S., Saunders K. B., Ley K., Zakrzewicz A., Gibson R. M., Furie B. C., Furie B., and Tedder T. F. (1994) A role for the epidermal growth factor-like domain of P-selectin in ligand recognition and cell adhesion. J. Cell Biol. 124, 609–618 10.1083/jcb.124.4.609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Erbe D. V., Watson S. R., Presta L. G., Wolitzky B. A., Foxall C., Brandley B. K., and Lasky L. A. (1993) P- and E-selectin use common sites for carbohydrate ligand recognition and cell adhesion. J. Cell Biol. 120, 1227–1235 10.1083/jcb.120.5.1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gibson R. M., Kansas G. S., Tedder T. F., Furie B., and Furie B. C. (1995) Lectin and epidermal growth factor domains of P-selectin at physiologic density are the recognition unit for leukocyte binding. Blood 85, 151–158 10.1182/blood.V85.1.151.bloodjournal851151 [DOI] [PubMed] [Google Scholar]
- 20. Watson S. R., Imai Y., Fennie C., Geoffrey J., Singer M., Rosen S. D., and Lasky L. A. (1991) The complement binding-like domains of the murine homing receptor facilitate lectin activity. J. Cell Biol. 115, 235–243 10.1083/jcb.115.1.235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li S. H., Burns D. K., Rumberger J. M., Presky D. H., Wilkinson V. L., Anostario M. Jr., Wolitzky B. A., Norton C. R., Familletti P. C., Kim K. J., Goldstein A. L., Cox D. C., and Huang K. S. (1994) Consensus repeat domains of E-selectin enhance ligand binding. J. Biol. Chem. 269, 4431–4437 [PubMed] [Google Scholar]
- 22. Somers W. S., Tang J., Shaw G. D., and Camphausen R. T. (2000) Insights into the molecular basis of leukocyte tethering and rolling revealed by structures of P- and E-selectin bound to SLeX and PSGL-1. Cell 103, 467–479 10.1016/S0092-8674(00)00138-0 [DOI] [PubMed] [Google Scholar]
- 23. Barkalow F. J., Barkalow K. L., and Mayadas T. N. (2000) Dimerization of P-selectin in platelets and endothelial cells. Blood 96, 3070–3077 10.1182/blood.V96.9.3070 [DOI] [PubMed] [Google Scholar]
- 24. Maeda S. F., Kawai T. F., Obinata M. F., Fujiwara H. F., Horiuchi T. F., Saeki Y. F., Sato Y. F., and Furusawa M. (1985) Production of human α-interferon in silkworm using a baculovirus vector. Nature 315, 592–594 10.1038/315592a0 [DOI] [PubMed] [Google Scholar]
- 25. Yin J., Li G., Ren X., and Herrler G. (2007) Select what you need: A comparative evaluation of the advantages and limitations of frequently used expression systems for foreign genes. J. Biotechnol. 127, 335–347 10.1016/j.jbiotec.2006.07.012 [DOI] [PubMed] [Google Scholar]
- 26. Arnold J. N., Wormald M. R., Sim R. B., Rudd P. M., and Dwek R. A. (2007) The Impact of glycosylation on the biological function and structure of human immunoglobulins. Annu. Rev. Immunol. 25, 21–50 10.1146/annurev.immunol.25.022106.141702 [DOI] [PubMed] [Google Scholar]
- 27. Tomiya N., Narang S., Lee Y. C., and Betenbaugh M. J. (2004) Comparing N-glycan processing in mammalian cell lines to native and engineered lepidopteran insect cell lines. Glycoconj. J. 21, 343–360 10.1023/B:GLYC.0000046275.28315.87 [DOI] [PubMed] [Google Scholar]
- 28. Kato T., Kako N., Kikuta K., Miyazaki T., Kondo S., Yagi H., Kato K., and Park E. Y. (2017) N-glycan modification of a recombinant protein via coexpression of human glycosyltransferases in silkworm pupae. Sci. Rep. 7, 1409 10.1038/s41598-017-01630-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Påhlsson P., Strindhall J., Srinivas U., and Lundblad A. (1995) Role of N-linked glycosylation in expression of E-selectin on human endothelial cells. Eur. J. Immunol. 25, 2452–2459 10.1002/eji.1830250907 [DOI] [PubMed] [Google Scholar]
- 30. AbuSamra D. B., Al-Kilani A., Hamdan S. M., Sakashita K., Gadhoum S. Z., and Merzaban J. S. (2015) Quantitative characterization of E-selectin interaction with native CD44 and P-selectin glycoprotein ligand-1 (PSGL-1) using a real time immunoprecipitation-based binding assay. J. Biol. Chem. 290, 21213–21230 10.1074/jbc.M114.629451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Merzaban J. S., Burdick M. M., Gadhoum S. Z., Dagia N. M., Chu J. T., Fuhlbrigge R. C., and Sackstein R. (2011) Analysis of glycoprotein E-selectin ligands on human and mouse marrow cells enriched for hematopoietic stem/progenitor cells. Blood 118, 1774–1783 10.1182/blood-2010-11-320705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sackstein R., and Dimitroff C. J. (2000) A hematopoietic cell L-selectin ligand that is distinct from PSGL-1 and displays N-glycan–dependent binding activity. Blood 96, 2765–2774 [PubMed] [Google Scholar]
- 33. Sako D., Chang X.-J., Barone K. M., Vachino G., White H. M., Shaw G., Veldman G. M., Bean K. M., Ahern T. J., Furie B., Cumming D. A., and Larsen G. R. (1993) Expression cloning of a functional glycoprotein ligand for P-selectin. Cell 75, 1179–1186 10.1016/0092-8674(93)90327-M [DOI] [PubMed] [Google Scholar]
- 34. Fuhlbrigge R. C., King S. L., Dimitroff C. J., Kupper T. S., and Sackstein R. (2002) Direct real-time observation of E- and P-selectin-mediated rolling on cutaneous lymphocyte-associated antigen immobilized on Western blots. J. Immunol. 168, 5645–5651 10.4049/jimmunol.168.11.5645 [DOI] [PubMed] [Google Scholar]
- 35. Sackstein R., and Fuhlbrigge R. (2009) Western blot analysis of adhesive interactions under fluid shear conditions: The blot rolling assay. Methods Mol. Biol. 536, 343–354 10.1007/978-1-59745-542-8_36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Usami S., Chen H.-H., Zhao Y., Chien S., and Skalak R. (1993) Design and construction of a linear shear stress flow chamber. Ann. Biomed. Eng. 21, 77–83 10.1007/BF02368167 [DOI] [PubMed] [Google Scholar]
- 37. Dong C., and Lei X. X. (2000) Biomechanics of cell rolling: Shear flow, cell-surface adhesion, and cell deformability. J. Biomech. 33, 35–43 10.1016/S0021-9290(99)00174-8 [DOI] [PubMed] [Google Scholar]
- 38. Wiese G., Barthel S. R., and Dimitroff C. J. (2009) Analysis of physiologic E-selectin-mediated leukocyte rolling on microvascular endothelium. J. Vis. Exp. 24, e1009 10.3791/1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Levy O., Anandakumaran P., Ngai J., Karnik R., and Karp J. M. (2013) Systematic analysis of in vitro cell rolling using a multi-well plate microfluidic system. J. Vis. Exp. 80, e50866 10.3791/50866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Carlson G. E., Martin E. W., and Burdick M. M. (2013) Simultaneously capturing real-time images in two emission channels using a dual camera emission splitting system: Applications to cell adhesion. J. Vis. Exp. 79, e50604 10.3791/50604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Shetty S., Weston C. J., Adams D. H., and Lalor P. F. (2014) A flow adhesion assay to study leucocyte recruitment to human hepatic sinusoidal endothelium under conditions of shear stress. J. Vis. Exp. 85, e51330 10.3791/51330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ling X., Ye J. F., and Zheng X. X.. Dynamic investigation of leukocyte-endothelial cell adhesion interaction under fluid shear stress in vitro. Acta Biochim. Biophys. Sin. 35, 567–572 [PubMed] [Google Scholar]
- 43. Alon R., Hammer D. A., and Springer T. A. (1995) Lifetime of the P-selectin-carbohydrate bond and its response to tensile force in hydrodynamic flow. Nature 374, 539–542 10.1038/374539a0 [DOI] [PubMed] [Google Scholar]
- 44. Bell G. (1978) Models for the specific adhesion of cells to cells. Science 200, 618–627 10.1126/science.347575 [DOI] [PubMed] [Google Scholar]
- 45. Dembo M., Torney D. C., Saxman K., and Hammer D. (1988) The reaction-limited kinetics of membrane-to-surface adhesion and detachment. Proc. R. Soc. Lond. B Biol. Sci. 234, 55–83 10.1098/rspb.1988.0038 [DOI] [PubMed] [Google Scholar]
- 46. Evans E., Leung A., Heinrich V., and Zhu C. (2004) Mechanical switching and coupling between two dissociation pathways in a P-selectin adhesion bond. Proc. Natl. Acad. Sci. U.S.A. 101, 11281–11286 10.1073/pnas.0401870101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Graves B. J., Crowther R. L., Chandran C., Rumberger J. M., Li S., Huang K. S., Presky D. H., Familletti P. C., Wolitzky B. A., and Burns D. K. (1994) Insight into E-selectin/ligand interaction from the crystal structure and mutagenesis of the lec/EGF domains. Nature 367, 532–538 10.1038/367532a0 [DOI] [PubMed] [Google Scholar]
- 48. Lou J., Yago T., Klopocki A. G., Mehta P., Chen W., Zarnitsyna V. I., Bovin N. V., Zhu C., and McEver R. P. (2006) Flow-enhanced adhesion regulated by a selectin interdomain hinge. J. Cell Biol. 174, 1107–1117 10.1083/jcb.200606056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lou J., and Zhu C. (2007) A structure-based sliding-rebinding mechanism for catch bonds. Biophys. J. 92, 1471–1485 10.1529/biophysj.106.097048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Marshall B. T., Long M., Piper J. W., Yago T., McEver R. P., and Zhu C. (2003) Direct observation of catch bonds involving cell-adhesion molecules. Nature 423, 190–193 10.1038/nature01605 [DOI] [PubMed] [Google Scholar]
- 51. McEver R. P., and Zhu C. (2010) Rolling cell adhesion. Annu. Rev. Cell Dev. Biol. 26, 363–396 10.1146/annurev.cellbio.042308.113238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ramachandran V., Nollert M. U., Qiu H., Liu W.-J., Cummings R. D., Zhu C., and McEver R. P. (1999) Tyrosine replacement in P-selectin glycoprotein ligand-1 affects distinct kinetic and mechanical properties of bonds with P- and L-selectin. Proc. Natl. Acad. Sci. U.S.A. 96, 13771–13776 10.1073/pnas.96.24.13771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sarangapani K. K., Yago T., Klopocki A. G., Lawrence M. B., Fieger C. B., Rosen S. D., McEver R. P., and Zhu C. (2004) Low force decelerates L-selectin dissociation from P-selectin glycoprotein ligand-1 and endoglycan. J. Biol. Chem. 279, 2291–2298 10.1074/jbc.M310396200 [DOI] [PubMed] [Google Scholar]
- 54. Waldron T. T., and Springer T. A. (2009) Transmission of allostery through the lectin domain in selectin-mediated cell adhesion. Proc. Natl. Acad. Sci. U.S.A. 106, 85–90 10.1073/pnas.0810620105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Preston R. C., Jakob R. P., Binder F. P., Sager C. P., Ernst B., and Maier T. (2016) E-selectin ligand complexes adopt an extended high-affinity conformation. J. Mol. Cell Biol. 8, 62–72 10.1093/jmcb/mjv046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Patel K. D., Nollert M. U., and McEver R. P. (1995) P-selectin must extend a sufficient length from the plasma membrane to mediate rolling of neutrophils. J. Cell Biol. 131, 1893–1902 10.1083/jcb.131.6.1893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sarangapani K. K., Marshall B. T., McEver R. P., and Zhu C. (2011) Molecular stiffness of selectins. J. Biol. Chem. 286, 9567–9576 10.1074/jbc.M110.196485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Schwizer D., Patton J. T., Cutting B., Smieško M., Wagner B., Kato A., Weckerle C., Binder F. P., Rabbani S., Schwardt O., Magnani J. L., and Ernst B. (2012) Pre-organization of the core structure of E-selectin antagonists. Chemistry 18, 1342–1351 10.1002/chem.201102884 [DOI] [PubMed] [Google Scholar]
- 59. Phan U. T., Waldron T. T., and Springer T. A. (2006) Remodeling of the lectin–EGF-like domain interface in P- and L-selectin increases adhesiveness and shear resistance under hydrodynamic force. Nat. Immunol. 7, 883–889 10.1038/ni1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Rocheleau A. D., Cao T. M., Takitani T., and King M. R. (2016) Comparison of human and mouse E-selectin binding to sialyl-Lewisx. BMC Struct. Biol. 16, 10 10.1186/s12900-016-0060-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kiely J. M., Hu Y., García-Cardeña G., and Gimbrone M. A. Jr. (2003) Lipid raft localization of cell surface E-selectin is required for ligation-induced activation of phospholipase Cγ. J. Immunol. 171, 3216–3224 10.4049/jimmunol.171.6.3216 [DOI] [PubMed] [Google Scholar]
- 62. Setiadi H., and McEver R. P. (2008) Clustering endothelial E-selectin in clathrin-coated pits and lipid rafts enhances leukocyte adhesion under flow. Blood 111, 1989–1998 10.1182/blood-2007-09-113423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Doyle E. L., Ridger V., Ferraro F., Turmaine M., Saftig P., and Cutler D. F. (2011) CD63 is an essential cofactor to leukocyte recruitment by endothelial P-selectin. Blood 118, 4265–4273 10.1182/blood-2010-11-321489 [DOI] [PubMed] [Google Scholar]
- 64. Poeter M., Brandherm I., Rossaint J., Rosso G., Shahin V., Skryabin B. V., Zarbock A., Gerke V., and Rescher U. (2014) Annexin A8 controls leukocyte recruitment to activated endothelial cells via cell surface delivery of CD63. Nat. Commun. 5, 3738 10.1038/ncomms4738 [DOI] [PubMed] [Google Scholar]
- 65. Yoshida M., Westlin W. F., Wang N., Ingber D. E., Rosenzweig A., Resnick N., and Gimbrone M. A. Jr. (1996) Leukocyte adhesion to vascular endothelium induces E-selectin linkage to the actin cytoskeleton. J. Cell Biol. 133, 445–455 10.1083/jcb.133.2.445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Zhang Y., Jiang N., Zarnitsyna V. I., Klopocki A. G., McEver R. P., and Zhu C. (2013) P-selectin glycoprotein ligand-1 forms dimeric interactions with E-selectin but monomeric interactions with L-selectin on cell surfaces. PLoS One 8, e57202 10.1371/journal.pone.0057202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Ramachandran V., Yago T., Epperson T. K., Kobzdej M. M. A., Nollert M. U., Cummings R. D., Zhu C., and McEver R. P. (2001) Dimerization of a selectin and its ligand stabilizes cell rolling and enhances tether strength in shear flow. Proc. Natl. Acad. Sci. U.S.A. 98, 10166–10171 10.1073/pnas.171248098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Mehta P., Cummings R. D., and McEver R. P. (1998) Affinity and kinetic analysis of P-selectin binding to P-selectin glycoprotein ligand-1. J. Biol. Chem. 273, 32506–32513 10.1074/jbc.273.49.32506 [DOI] [PubMed] [Google Scholar]
- 69. Molenaar T. J. M., Appeldoorn C. C. M., de Haas S. A. M., Michon I. N., Bonnefoy A., Hoylaerts M. F., Pannekoek H., van Berkel T. J. C., Kuiper J., and Biessen E. A. L. (2002) Specific inhibition of P-selectin–mediated cell adhesion by phage display–derived peptide antagonists. Blood 100, 3570–3577 10.1182/blood-2002-02-0641 [DOI] [PubMed] [Google Scholar]
- 70. Revelle B. M., Scott D., Kogan T. P., Zheng J., and Beck P. J. (1996) Structure-function analysis of P-selectin-sialyl Lewis binding interactions: Mutagenic alteration of ligand binding specificity. J. Biol. Chem. 271, 4289–4297 10.1074/jbc.271.8.4289 [DOI] [PubMed] [Google Scholar]
- 71. Deban L., Russo R. C., Sironi M., Moalli F., Scanziani M., Zambelli V., Cuccovillo I., Bastone A., Gobbi M., Valentino S., Doni A., Garlanda C., Danese S., Salvatori G., Sassano M., et al. (2010) Regulation of leukocyte recruitment by the long pentraxin PTX3. Nat. Immunol. 11, 328–334 10.1038/ni.1854 [DOI] [PubMed] [Google Scholar]
- 72. Klopocki A. G., Yago T., Mehta P., Yang J., Wu T., Leppänen A., Bovin N. V., Cummings R. D., Zhu C., and McEver R. P. (2008) Replacing a lectin domain residue in L-selectin enhances binding to P-selectin glycoprotein ligand-1 but not to 6-sulfo-sialyl Lewis x. J. Biol. Chem. 283, 11493–11500 10.1074/jbc.M709785200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Hochuli E., Bannwarth W., Döbeli H., Gentz R., and Stüber D. (1988) Genetic approach to facilitate purification of recombinant proteins with a novel metal chelate adsorbent. Nat. Biotechnol. 6, 1321–1325 10.1038/nbt1188-1321 [DOI] [Google Scholar]
- 74. Schmidt T. G., and Skerra A. (1993) The random peptide library-assisted engineering of a C-terminal affinity peptide, useful for the detection and purification of a functional Ig Fv fragment. Protein Eng. 6, 109–122 10.1093/protein/6.1.109 [DOI] [PubMed] [Google Scholar]
- 75. Ono C., Nakatsukasa T., Nishijima Y., Asano S. I., Sahara K., and Bando H. (2007) Construction of the BmNPV T3 bacmid system and its application to the functional analysis of BmNPV he65. J. Insect Biotechnol. Sericol. 76, 3161–3167 10.11416/jibs.76.3_161 [DOI] [Google Scholar]
- 76. Masuda A., Xu J., Mitsudome T., Morokuma D., Mon H., Banno Y., Kusakabe T., and Lee J. M. (2015) Improvement of Endo-β-N-acetylglucosaminidase H production using silkworm–baculovirus protein expression system. J. Asia-Pacific Entomol. 18, 175–180 10.1016/j.aspen.2015.01.006 [DOI] [Google Scholar]
- 77. Isshiki M., Ando J., Yamamoto K., Fujita T., Ying Y., and Anderson R. G. (2002) Sites of Ca2+ wave initiation move with caveolae to the trailing edge of migrating cells. J. Cell Sci. 115, 475–484 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.