Abstract
It has been well-established that the deubiquitinating enzyme ubiquitin-specific peptidase 7 (USP7) supports cancer growth by up-regulating multiple cellular pathways, including Wnt/β-catenin signaling. Therefore, considerable efforts are directed at identifying and developing USP7 inhibitors. Here, we report that sesquiterpene lactone parthenolide (PTL) inhibits USP7 activity, assessed with deubiquitinating enzyme activity assays, including fluorogenic Ub-AMC/Ub-Rho110, Ub-VME/PA labeling, and Di-Ub hydrolysis assays. Further investigations using cellular thermal shift (CETSA), surface plasmon resonance (SPR), and mass spectrum (MS) assays revealed that PTL directly interacts with USP7. Consistent with the role of USP7 in stimulating Wnt signaling and carcinogenesis, PTL treatment inhibited the activity of Wnt signaling partly by destabilizing β-catenin. Moreover, using cell viability assays, we found that PTL suppresses the proliferation of colorectal cancer cells and induces apoptosis in these cells. Additionally, we examined the effects of two other sesquiterpene lactones (costunolide and α-santonin) on USP7 and Wnt signaling and found that α-methylene-γ-butyrolactone may provide a scaffold for future USP7 inhibitors. In summary, our findings reveal that PTL inhibits USP7 activity, identifying a potential mechanism by which PTL suppresses Wnt/β-catenin signaling. We further suggest that sesquiterpene lactones might represent a suitable scaffold for developing USP7 inhibitors and indicate that PTL holds promise as an anticancer agent targeting aberrant USP7/Wnt signaling.
Keywords: small molecule, deubiquitylation (deubiquitination), Wnt signaling, pharmacology, enzyme inhibitor, β-catenin, parthenolide, ubiquitination, USP7, Wnt signaling, ubiquitin-specific peptidase 7, colorectal cancer, proteasome system, protein homeostasis, deubiquitinating activity
Introduction
Ubiquitination mediated by ubiquitin-proteasome system (UPS),3 which consists of E1s, E2s, E3 ligases, and 26S proteasome, plays critical roles in control of the stability, activity, or localization of protein substrates. Oppositely, the process of ubiquitination could be reversed by deubiquitinases (DUBs) that recognize ubiquitylated proteins and remove their ubiquitin tags (1). Over the past decades, numerous studies established that dysregulation of the ubiquitin system has been involved in the pathogenesis of multiple human diseases, including various types of tumors (2). Successful approval of proteasome inhibitors (PIs) for the treatment of multiple myeloma highlights the great potential of developing inhibitors targeting UPS for anti-cancer agents (3). Because of the occurrence of clinical PI resistance and chief roles of E3s and DUBs in governing the specificity of UPS, compounds targeting these two types of enzymes therefore represent novel therapeutics with potentially enhanced specificity and reduced toxicity (4–6).
To date, ∼100 DUBs encoded by human genome have been identified and classified into six families. USPs are the largest subfamily of DUBs, with more than 50 family members reported (7). Among them, USP7 has been initially paid attention because of its regulating effect on oncoprotein MDM2, a major E3 ligase responsible for the proteasomal degradation of the tumor suppressor p53 (8). Beyond the MDM2-p53 axis, subsequent evidence indicates the involvement of USP7 in multiple oncogenic pathways (9–16). For example, our and other studies recently indicated that USP7 positively regulated Wnt signaling through stabilization of the key transcriptional factor β-catenin (10, 11, 17, 18). Several other studies also demonstrated the positive role of USP7 in NF-κB, Hedgehog, and Hippo signaling by deubiquitinating and stabilizing NEK2, Gli, and Yorkie/Yap, respectively (9, 12, 15). Additionally, multiple proteins involved in diverse cellular processes including DNA damage response, transcription, epigenetic control of gene expression, and immune response, such as Chk1, N-Myc, ASXL1, and Foxp3, have been identified as specific substrates of USP7 (19–23). Therefore, it is not surprising that USP7 is often overexpressed and correlates with poor prognosis in diversified human malignancies including breast cancer, hepatocellular carcinoma, leukemia, and on (13, 16, 23–26). The inhibition of the deubiquitinating activity of USP7 thus offers a promising strategy for protein-directed therapies in the treatment of cancer.
Indeed, a number of USP7 inhibitors have been identified, and most of them exhibited in vitro and in vivo anti-cancer activity against various types of tumor (23, 27). Especially, the diversified anti-cancer mechanisms of several inhibitors have been reported, which accorded with the variety of USP7 targets in cells. Two different studies pointed out that P5091, a small molecule inhibitor of USP7, down-regulated the abundance of Yap and up-regulated the level of ARF, which contributed to its cytotoxic effect on hepatocellular carcinoma (15, 25). In addition, our previous study indicated that USP7 inhibition by P5091 attenuated the proliferation of colorectal cancer cells partly through destabilizing β-catenin (28). In addition, P5091 has been reported to accelerate the degradation of N-Myc and Nek2 (12, 20). Notably, pharmacological targeting USP7 by P217564, one derivate of P5091, led to Tip60 degradation and consequent impairment of Treg suppressive function, implicating the potential of USP7 inhibitors in future cancer immunotherapy (29). Overall, these studies demonstrated the rationality for development of USP7 inhibitors as therapeutic agents against cancer.
Natural products harbor structural diversity and are critical for screening of new drug leads. However, small molecule USP7 inhibitors from natural resources have rarely been reported. In this study, we showed that sesquiterpene lactone parthenolide (PTL), first purified from the shoots of the feverfew and used to treat migraine and arthritis for centuries, could inhibit enzymatic activity of USP7 via direct interaction. Also, PTL treatment enhanced the ubiquitination of β-catenin and decreased β-catenin protein levels in colorectal cancer (CRC) cells, which resulted in the inhibition of Wnt signaling and cytotoxity of CRC cells. In sum, our study suggested that sesquiterpene lactones might represent a novel scaffold for designing novel USP7 inhibitors and the potential of PTL in the treatment of cancers driven by dysregulated USP7 and Wnt signaling.
Results
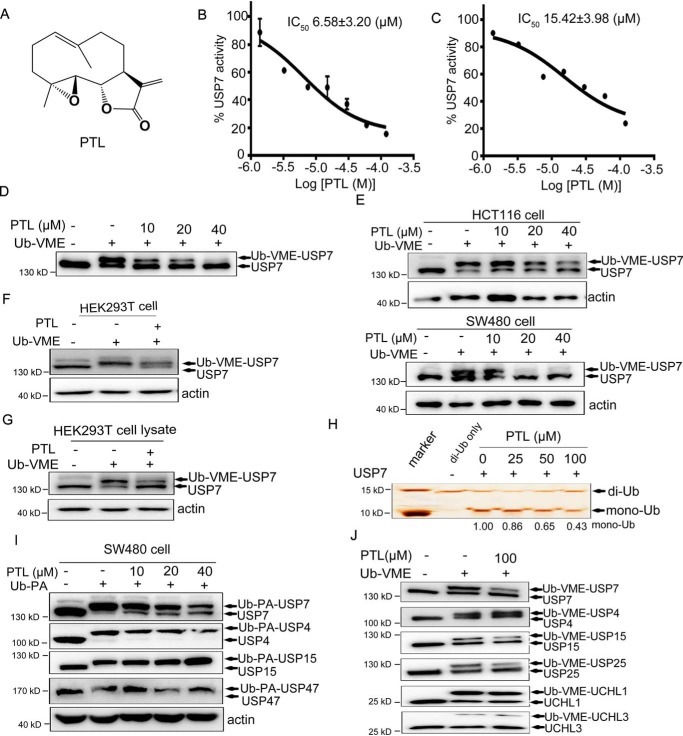
Identification of PTL as a novel USP7 inhibitor
In an effort to identify USP7 inhibitors, we performed an in vitro high-throughput screening assay against a library of natural chemicals using ubiquitin-aminomethylcoumarin (Ub-AMC) as a substrate (30), and the small molecule PTL was discovered (Fig. 1A). Detailed characterization results revealed that PTL inhibited USP7 activity in dose- and time-dependent manner (Fig. 1B and Fig. S1A). Similar inhibitory effects of PTL on USP7 activity were also obtained using an assay based on another ubiquitin precursor, ubiquitin-rhodamine 110 (Ub-Rho110) (Fig. 1C and Fig. S1B).
Figure 1.
Identification and biochemical characterization of PTL as a USP7 inhibitor. A, structure of PTL. B, dose-dependent inhibition of USP7 activity by PTL using the Ub-AMC as substrate. The results are presented as mean ± S.D. (n = 2). C, dose-dependent inhibition of USP7 activity by PTL using the Ub-Rho110 as substrate. The results are presented as mean ± S.D. (n = 2). D, purified FLAG-USP7 were treated with PTL, and the Ub-VME probe was added. Samples were subsequently analyzed by Western blotting using anti-USP7 antibody. E, HCT116 and SW480 cells treated with different doses of PTL were collected and lysed, and Ub-VME probes were added into the cell lysates for 30 min. Samples were subsequently analyzed by Western blotting with anti-USP7 antibody. F, HEK293T cells directly incubated with PTL were then collected and labeled with Ub-VME, followed by immunoblot analysis with anti-USP7 antibody. G, HEK293T cell lysates pretreated with or without PTL were labeled with Ub-VME and analyzed by Western blotting. H, recombinant His-USP7 was pretreated with indicated dose of PTL for 40 min and then incubated with K48-linked di-Ub for 3 h. SDS-PAGE and silver staining were employed to analyze the cleavage of di-Ub by USP7 in the presence or absence of PTL. I, SW480 cells were treated with different doses of PTL for 2 h and then labeled with Ub-PA. Individual DUBs were identified using specific antibodies. J, purified FLAG-USP7 and additional deubiquitinating enzymes (FLAG-USP4, FLAG-USP15, FLAG-USP25, FLAG-UCH-L1, and FLAG-UCH-L3) were treated with PTL for 20 min and then labeled with Ub-VME for another 20 min, followed by SDS-PAGE with FLAG-Tag antibody.
To further confirm the USP7 inhibitory capacity of PTL both in vitro and in the cellular environment, we then performed competition assays between PTL and the Ub active site probe ubiquitin-vinyl methyl ester (Ub-VME). First, purified USP7 was incubated with PTL or equivalent DMSO, followed by the addition of Ub-VME probes. As shown in Fig. 1D, recombinant USP7 incubated with Ub-VME exhibited Ub-USP7 conjugate formation, as reflected by the increase in mass of USP7 of ∼8 kDa. In contrast, Ub-USP7 conjugate formation was dose-dependently inhibited from PTL-treated samples, with a concomitant increase in the unlabeled free form of USP7. In cellular context, HCT116, SW480, and HEK293T cells were treated with or without PTL for 2 h, and cell lysates were labeled with Ub-VME. As expected, PTL also inhibited the covalent binding of Ub-VME with USP7 (Fig. 1, E and F), which is similar to the in vitro results. Besides, we also directly focused on cell lysates. When lysates of HEK293T cells were treated with Ub-VME in the presence or absence of PTL, a strong reduction of the labeled USP7 was observed as well (Fig. 1G). Next, the USP7 activity toward K48-linked diubiquitin (di-Ub) was investigated, and the results showed that PTL could decrease the amount of Ub hydrolyzed from di-Ub in a concentration-dependent manner (Fig. 1H).
To bring insight to the selectivity of PTL for USP7 relative to other DUBs, the Ub-Rho110 hydrolysis test against a panel of 41 DUBs was performed. As the results showed, PTL also exhibited inhibition capacity, to some extent (>25%), against other 6 DUBs including USP8, USP21, USP27X, USP30, USP35, and JOSD2 (Fig. S1C). We then utilized specific antibodies to assess the labeling efficiency for DUBs from SW480 cells. As the results showed, PTL efficiently blocked the labeling of USP7 at 40 μm, but not that of USP4, USP15, and USP47 in SW480 cells (Fig. 1I). The similar findings were observed in HCT116 cells as well (Fig. S1D). We next evaluated the effect of PTL on Ub-VME labeling efficiency against several DUBs individually. For this purpose, seven DUBs, including USP4, USP7, USP15, USP25, UCHL1, and UCHL3, were purified and labeled with Ub-VME in the presence or absence of PTL. As indicated in Fig. 1J, all tested DUBs were labeled with Ub-VME indicated by mobility shift, albeit with different levels of efficiency. Particularly, PTL exhibited major inhibitory activity against USP7 and minor effects against USP25, and negligible effects against USP4, USP15, UCH-L1, and UCH-L3 (Fig. 1J). Taken together, these results confirmed the inhibitory activity of PTL against USP7 and indicated that PTL preferentially inhibited USP7 over a panel of active DUBs.
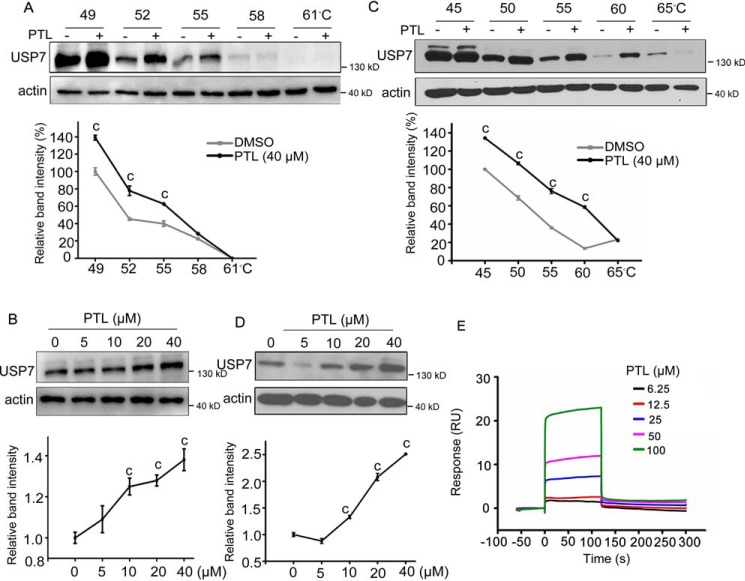
PTL interacts with USP7
Cellular thermal shift assay (CETSA) is a newly developed method to evaluate drug binding to target proteins in cells, which is based on the biophysical principle of ligand-induced thermal stabilization of target proteins (31). To evaluate whether PTL binds to USP7, CETSA was firstly employed in SW480 cell lysates. As shown in Fig. 2A, PTL treatment significantly increased the thermal stability of USP7 in supernatant. Consistent with the results, accumulation of USP7 was also markedly increased by PTL in a concentration manner (Fig. 2B). Next, to test whether PTL interact with USP7 in intact cells, we performed cell-based drug treatment before analysis by temperature shift. To this end, SW480 cells treated with PTL or DMSO were collected, lysed, and then heated. Compared with DMSO treatment, PTL incubation led to an obvious thermostability of USP7 at different temperatures (Fig. 2C) and different doses (Fig. 2D). In short, these results indicated the increased thermostability of USP7 following heat treatment in the presence of PTL, suggesting the interaction between USP7 and PTL. Moreover, we employed surface plasmon resonance (SPR) to further confirm that PTL did bind recombinant USP7 catalytic domain (USP7CD) in a dose-dependent manner (Fig. 2E). Further, we also sought to determine the cysteine(s) in USP7 covalently targeted by PTL. The mass spectrum (MS) data showed that the USP7 protein was identified with 84% sequence coverage (Fig. S2A) and that 13 peptides containing cysteines (Cys) were identified with a mass shift of 248.14 Da that matched the molecular weight of a PTL molecule (Fig. S2B). The identified peptides suggested that PTL covalently modified Cys-90, Cys-315, Cys-334, Cys-478, Cys-488, Cys-510, Cys-702, Cys-721, Cys-799, Cys-896, Cys-917, Cys-925 and Cys-961 of USP7 (Fig. S2B). Taken together, these results demonstrate that PTL directly targets USP7.
Figure 2.
PTL interacts with USP7. A and B, CETSA was performed on SW480 cell lysates at different temperatures (A) and different doses (B). Aliquots of SW480 cell lysates incubated with PTL or DMSO were heated at indicated temperatures. After cooling, samples were centrifuged to separate the soluble fractions from precipitated proteins. The presence of USP7 in the soluble fraction was then analyzed by Western blotting. C and D, for cell-based CETSA, SW480 cells were pre-incubated with PTL for 1 h. Cells were then collected and lysed. The presence of USP7 in the soluble fraction of cell lysates at different temperatures (C) and different doses (D) was analyzed. The intensity of the USP7 bands in (A and B) and (C and D) was quantified by National Institutes of Health ImageJ software. E, SPR analysis of interactions between PTL and recombinant USP7CD. All values were represented as the mean ± S.D. (n = 3). The significance was determined by Student's t test (c, p ≤ 0.001 versus control).
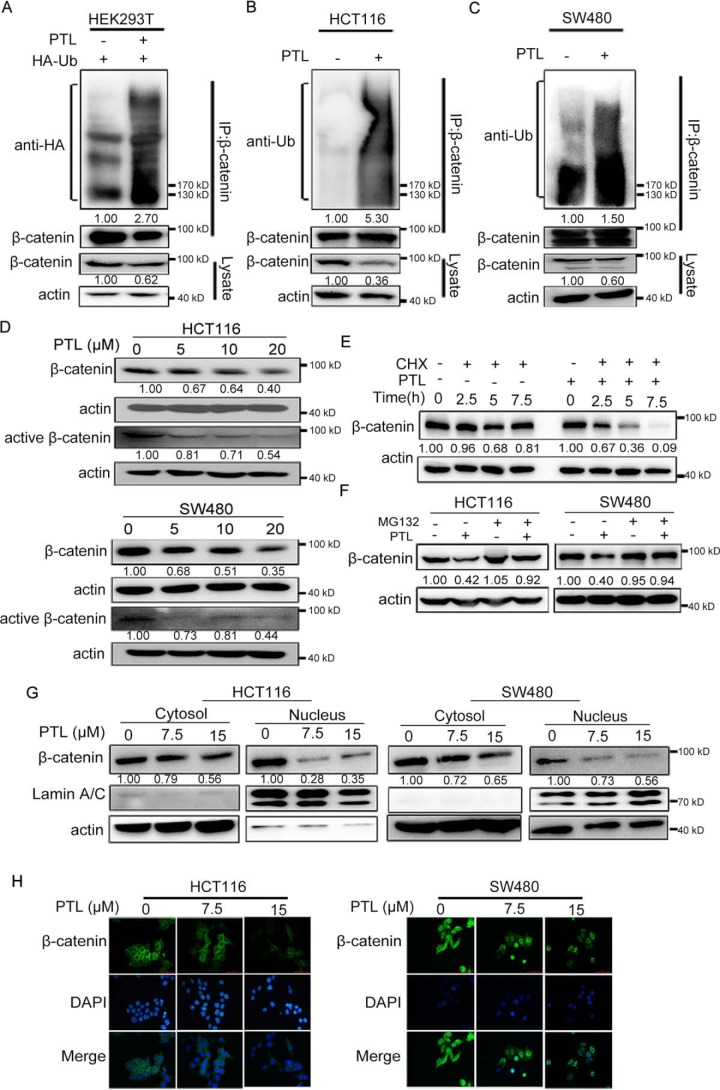
PTL promotes the ubiquitination and degradation of β-catenin
Recent studies indicated that USP7 could deubiquitinate and stabilize β-catenin, the key transcriptional factor of Wnt signaling pathway (10, 11, 17, 18). The effect of PTL on the ubiquitination level of β-catenin was thus explored. HEK293T cells transiently transfected with HA-Ub plasmids were treated with or without PTL, and endogenous β-catenin ubiquitination was analyzed. The results showed that treatment of PTL increased the level of β-catenin ubiquitination (Fig. 3A). The effect of PTL on β-catenin ubiquitination was also tested in HCT116 and SW480 cells without exogenous Ub transfection. Similarly, β-catenin ubiquitination was also enhanced in cells exposed to PTL (Fig. 3, B and C).
Figure 3.
PTL down-regulates β-catenin via promoting its ubiquitination. A, 20 μm for 24 h before collecting. Immunoprecipitation with anti–β-catenin antibody was performed, and ubiquitination of β-catenin was analyzed by anti-HA antibody. B and C, HCT116 (B) and SW480 (C) cells were treated with DMSO or PTL (20 μm) for 12 h, followed by immunoprecipitation with β-catenin antibody, and β-catenin ubiquitination was detected with the antibody against ubiquitin. D, treatment of HCT116 and SW480 cells with PTL for 24 h, total and active forms of β-catenin were determined via Western blotting. E, SW480 cells were pretreated with DMSO or PTL (20 μm) for 4 h, followed by addition of cycloheximide (CHX) (80 μg/ml) for the indicated times. Cell lysates were subjected to immunoblot with antibodies against β-catenin or actin. F, HCT116 and SW480 cells were treated with DMSO or PTL (15 μm) for 20 h, followed by addition of DMSO or MG132 (10 μm) for additional 4 h. Cells were harvested and the protein level of β-catenin and actin were examined. G, PTL reduced β-catenin levels in both cytoplasmic and nuclear fractions. HCT116 and SW480 cells were treated with PTL (7.5 or 15 μm). Cytoplasmic and nuclear fractions were then separated and β-catenin protein levels were analyzed. H, representative immunofluorescence staining images of HCT116 and SW480 cells treated with DMSO or PTL. β-catenin and nucleus was recognized by anti–β-catenin antibody (green) or DAPI (blue), respectively. Scale bar represents 50 μm. Blots for indicated protein expressions were quantified using ImageJ software.
Given that PTL treatment could increase the ubiquitination level of β-catenin, we next determine whether PTL promoted the degradation of β-catenin. As expected, PTL treatment dose-dependently reduced β-catenin levels in HCT116 and SW480 cells (Fig. 3D). Likewise, the presumably transcriptionally active form of β-catenin (nonphosphorylated form of β-catenin) were decreased as well (Fig. 3D). In addition, incubation of SW480 cells with PTL obviously accelerated β-catenin degradation in the presence of cycloheximide, which was used to inhibit protein biosynthesis (Fig. 3E). Especially, the PTL-induced β-catenin degradation was blocked in the presence of proteasome inhibitor MG132 (Fig. 3F), suggesting that β-catenin degradation mediated by PTL is proteasome dependent. In line with the above-mentioned data, the results of cytoplasmic and nuclear fraction and immunofluorescence assays further indicated that PTL treatment down-regulated β-catenin levels of the nuclear and cytoplasmic compartments in both HCT116 and SW480 cells (Fig. 3, G and H). In conclusion, these data suggested that PTL decreased the levels of β-catenin via increasing its ubiquitination.
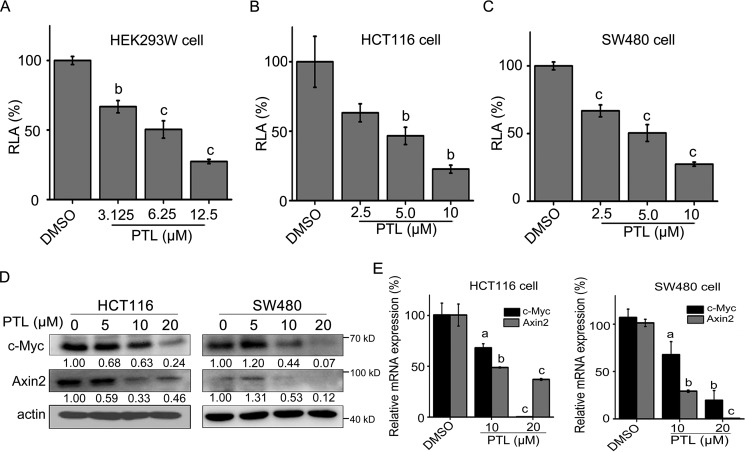
PTL inhibits Wnt signaling in colorectal cancer cells
We then assessed the effect of PTL on Wnt signaling through luciferase activity assay of Super TOPFlash luciferase (ST-Luc), a Wnt/β-catenin signaling reporter. As shown in Fig. 4A, PTL dose-dependently inhibited the activity of ST-Luc reporter in HEK293W cells (HEK293 cells stably co-transfected with Wnt3a, Renilla, and ST-Luc). The inhibition effect of PTL on endogenous Wnt signaling in colon cancer cells was investigated either in line with the results of HEK293W cells, the TOPFlash activity in both HCT116 and SW480 cells treated with PTL was efficiently attenuated in a dose-dependent manner (Fig. 4, B and C). The effects of PTL on the expression of known target genes of Wnt/β-catenin signaling including Axin2 (32) and c-Myc (33) were further monitored. Compared with DMSO-treated HCT116 and SW480 cells, the protein and mRNA levels of Axin2 and c-Myc were both decreased in these two cell lines treated with PTL (Fig. 4, D and E).
Figure 4.
PTL inhibits the ST-Luc activity and Wnt target genes expression in colorectal cancer cells. A) PTL suppressed the Wnt reporter activity in HEK293W cells incubated with indicated doses of PTL for 24 h. The luciferase activity was then measured and normalized to the activity of the Renilla. B-C) HCT116 (B) and SW480 (C) cells were seeded in 96-well plates and transiently co-transfected with SuperTopflash and Renilla. After transfection for 3 h, cells were treated with the indicated doses of PTL for 24 h and lysed. Dual luciferase reporter assay was then undergoing. D, Western blotting was used to detect the protein levels of Wnt target genes (c-Myc and Axin2) in HCT116 and SW480 cells treated with the indicated concentrations of PTL for 24 h; actin was used as the loading control. The actin blots between Fig. 4D and Fig. 3D (upper panel in HCT116 and SW480) were reused. E, the mRNAs levels of c-Myc and Axin2 were determined in HCT116 and SW480 cells treated with PTL for 16 h by real-time PCR. Blots for indicated protein expressions were quantified using ImageJ software. All values were expressed as the mean ± S.D. (n = 3). The significance was determined by Student's t test (a, p < 0.05; b, p ≤ 0.01; and c, p ≤ 0.001 versus control). RLA, relative luciferase activity.
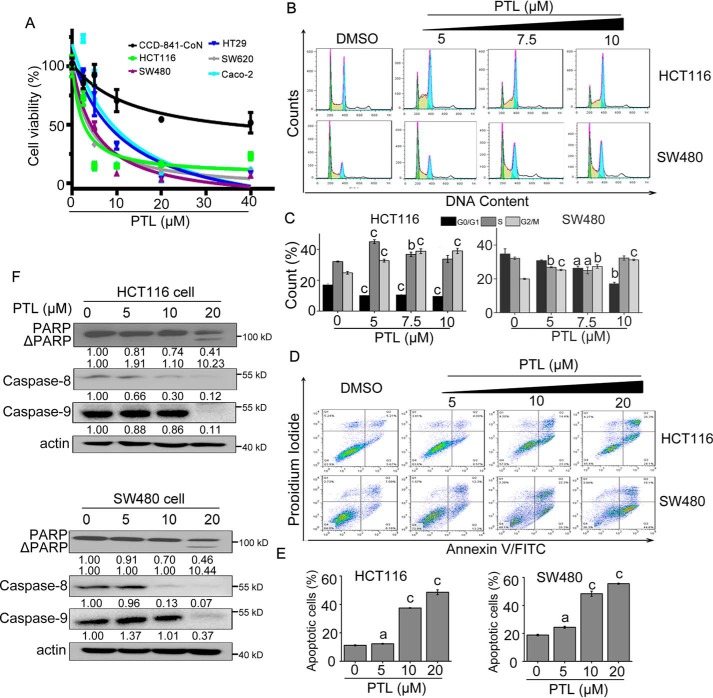
Effect of PTL on proliferation, cell cycle, and apoptosis of colorectal cancer cells
In view of the positive role of USP7 and Wnt/β-catenin signaling in CRC progression, we then employed MTS assay to assess the growth inhibitory effect of PTL in CRC cells. As illustrated in Fig. 5A, PTL exhibited stronger growth inhibition effect on CRC cells than normal colonic epithelial cells. Cell cycle and apoptosis of HCT116 and SW480 cells treated with PTL were determined by flow cytometry. Cell cycle analysis revealed that HCT116 and SW480 cells were efficiently arrested at G2/M phase upon PTL treatment (Fig. 5, B and C). Moreover, we evaluated the ability of PTL to induce apoptosis. Flow cytometry analysis after annexin V/PI double staining established that PTL dose-dependently induced accumulation of cells in early (annexin V+/PI−) and late-stage (annexin V+/PI+) apoptosis (Fig. 5, D and E). To verify the apoptosis observed in PTL-treated cells, apoptosis-related proteins were monitored using Western blotting. As shown in Fig. 5F, treatment of HCT116 and SW480 cells with PTL-activated caspase-8 and caspase-9, accompanied with the cleavage of poly ADP-ribose polymerase in a dose-dependent manner. These results indicated the potential of PTL in the treatment of colorectal cancers mostly driven by deregulated Wnt signaling.
Figure 5.
Activity of PTL on cell proliferation, cell cycle, and apoptosis. A, cells seeded in 96-well plates were cultured overnight and exposed to PTL at various doses for 48 h. Cell viability was determined by MTS assay. B, PTL arrested CRC cells at G2/M phase of cell cycle. HCT116 and SW480 cells seeded in 6-well plates were treated with indicated dose of PTL for 24 h, and cell cycle were analyzed by flow cytometry using PI staining. C, percent of cell cycle phases in (A) were quantified. D, PTL dose-dependently induced apoptosis of CRC cells. HCT116 and SW480 cells were seeded in 6-well plates, after treatment of PTL at indicated concentrations for 48 h, the apoptotic cells were counted by flow cytometry using annexin V/PI double staining. E, flow cytometry analysis of apoptosis in (C) was quantified. F, effect of PTL on the expressions of apoptosis related proteins. HCT116 and SW480 cells were treated with indicated doses of PTL for 24 h, and apoptosis-related proteins were detected by Western blot analysis with indicated antibodies. The actin blots between Fig. 5F and Fig. 3D (lower panel in HCT116 and SW480) were reused. Blots for indicated protein expressions were quantified using ImageJ software. All the results are presented as mean ± S.D. (n = 3). a, p < 0.05; b, p ≤ 0.01; c, p ≤ 0.001, difference versus untreated control.
α-Methylene-γ-butyrolactone of sesquiterpene lactones is responsible for the inhibition toward USP7 and Wnt signaling
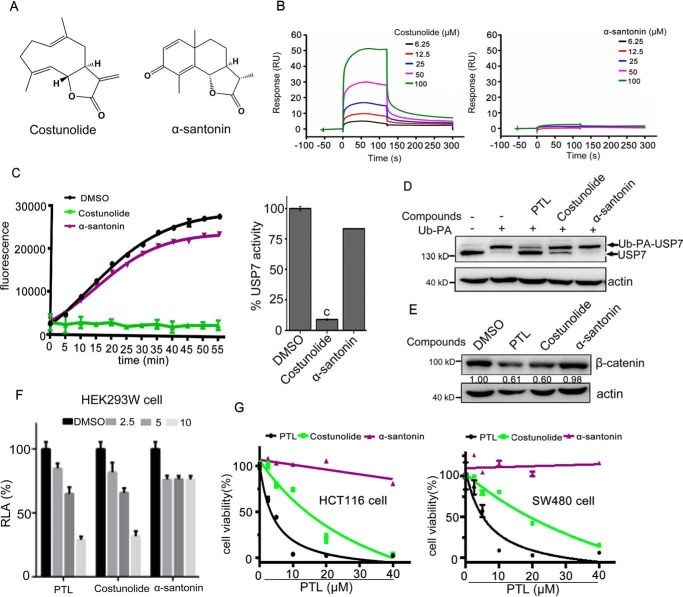
Given that parthenolide is one member of sesquiterpene lactones containing the reactive α-methylene-γ-butyrolactone pharmacophore (34, 35), we sought to determine whether other related sesquiterpene lactones could target USP7 and inhibit Wnt signaling. Costunolide (Fig. 6A), which contains α-methylene-γ-butyrolactone moiety, bound to USP7CD (Fig. 6B). Oppositely, α-santonin (Fig. 6A), which does not contain this reactive functional group, hardly bound to USP7CD (Fig. 6B). Consistent with this result, costunolide, but not α-santonin, inhibited USP7 activity demonstrated by Ub-Rho110–based fluorescent assay (Fig. 6C) and ubiquitin-propargylamid (Ub-PA)–based labeling assay (Fig. 6D). Moreover, parthenolide and costunolide, but not α-santonin, suppressed the activity of Wnt signaling and decreased the levels of β-catenin (Fig. 6, E and F). Consistent with the above results, parthenolide and costunolide impaired the growth of HCT116 and SW480 cells, whereas α-santonin did not attenuate the proliferation of these two cells (Fig. 6G). These data suggested that sesquiterpene lactones containing α-methylene-γ-butyrolactone might be potential scaffolds for the development of USP7 inhibitors.
Figure 6.
α-Methylene-γ-butyrolactone of sesquiterpene lactones is responsible for the inhibition toward USP7 and Wnt signaling. A, structure of costunolide and α-santonin. B, representative SPR sensorgrams of USP7CD incubated with costunolide and α-santonin. Compounds were tested at a series of increasing concentrations. C, inhibition of USP7 activity by costunolide and α-santonin using the Ub-AMC as substrate. The results are presented as mean ± S.D. (n = 2). c, p ≤ 0.001, difference versus DMSO. D, living SW480 cells were treated with PTL, costunolide, and α-santonin (40 μm) for 2 h and then labeled with Ub-PA probe. Samples were subsequently analyzed by Western blotting by anti-USP7 and anti-actin antibodies. E, SW480 cells were treated with PTL, costunolide, and α-santonin for 24 h, β-catenin were determined by Western blotting. Blots for indicated protein expressions were quantified using ImageJ software. F, effects of PTL, costunolide, and α-santonin on the TOPFlash reporter activity. HEK293W cells were incubated with indicated doses of compounds for 24 h. The luciferase activity was then measured and normalized to the activity of the Renilla. G, cells seeded in 96-well plates were cultured overnight and exposed to PTL, costunolide, and α-santonin at various doses for 48 h. Cell viability was determined by MTS assay.
Discussion
DUBs have recently emerged as an attractive drug target class in cancer therapeutics (36). Inhibition of USP7 is of special interest because of its well-established connections to multiple oncogenic pathways (14, 23). In this study, we demonstrate that PTL is a novel inhibitor of USP7 based on the following evidence: (a) PTL could inhibit the USP7-mediated hydrolysis of Ub-AMC/Ub-Rho110 and di-Ub; (b) PTL competed with the binding of Ub-VME/Ub-PA probe to USP7; (c) CETSA, SPR, and MS analyses demonstrated that PTL directly interacted with USP7. According to previous studies, PTL exerted its biological activity through Michael addition reaction, which was based on alkylation of cysteine involving the α-methylene-γ-butyrolactone moiety (34, 35), and the MS data indicated that 13 cysteines were modified by PTL. Interestingly, the catalytic Cys-223 was not modified by PTL. Further experimental data, such as mutation analysis and co-crystal structures, are necessary to confirm the mode of PTL binding to USP7. Comparison of the efficacy of PTL with another well-known USP7 inhibitor P5091 (37) was performed side by side, and the results showed that the inhibitory activity of PTL against USP7 was a bit weaker than that of P5091 in Ub-VME labeling assay and Ub-AMC fluorescent assay (data not shown). Moreover, our study also provided a preliminary selectivity spectrum of PTL against a panel of DUBs and identified that PTL preferentially inhibited USP7. Nevertheless, the molecular basis for PTL preference against USP7 was not defined and warrant further study. As for the specificity, there are both similarities and differences in the specificity of PTL and P5091 toward DUBs. What is similar is that both PTL and P5091 did not inhibit USP2, USP5, USP15, USP20, USP28, UCHL-1, or UCHL-3 (37). However, other DUBs except USP7 targeted by PTL or P5091 were different. Our study indicated that PTL exhibited partial inhibitory activity against USP8, USP21, USP27X, USP30, USP35, and JOSD2. However, only USP47 was reported to be targeted by P5091 (38).
As a major active ingredient from feverfew, PTL exhibits multiple pharmacological activities including anti-cancer and anti-inflammatory activities via modulating various signaling pathways such as the NF-κB, p53-MDM2, and STAT3 pathways. A recent study clarifying that PTL inhibits the activity of JAKs accounts for its inhibitory effect on STAT3 signaling (39). Specially, identification of PTL as a novel USP7 inhibitor here provides a rational explanation for the activation effect of PTL on p53 functions via promoting ubiquitination of MDM2 (40), which is a primary substrate of USP7 (41). In addition, USP7 was reported to interact with and deubiquitinate p65 and NEK2, leading to enhanced activity of NF-κB signaling (12, 42). Although several mechanisms by which PTL inhibited NF-κB have been reported, identification of PTL as a USP7 inhibitor might be a further step for its strong inhibition of NF-κB (43–45). Therefore, the function of PTL suppressing USP7 activity contributes to the illustration of its effect on some signaling pathways.
It should be noted that, although a couple of key signaling pathways were targeted by PTL as mentioned before, poor solubility and bioavailability of PTL leading to its weak in vivo effects are crucial limitations to impede its potential application in clinic (46). Consistently, our results showed that PTL to some extent inhibited tumor growth (∼30.0%) and tumor weight (∼22.7%) of HCT116 xenografts (data not shown). Thus, the chemistry study of PTL deserves further investigation to circumvent these limitations by constructing PTL derivatives with improved efficacy and specificity toward USP7.
Given that USP7 positively regulates Wnt signaling through mediating the deubiquitination of its major transcriptional co-activator β-catenin (10, 11, 17, 18, 28), our further study indicated that PTL suppressed Wnt signaling through accelerating the ubiquitination and subsequent degradation of β-catenin. Recently, Zhu et al. (47) reported that PTL potently inhibited Wnt signaling by blocking TCF4/LEF1 synthesis via targeting RPL10 without affecting β-catenin stability. To be noted, destabilization of β-catenin by PTL has also been validated in SW620 cells, one colorectal cancer cell line, which was consistent with our data (48). The difference in the effect of PTL on β-catenin may be caused by the incubation time and dose treated. In fact, β-catenin level was also decreased in SW480 cells treated with 20 μm PTL in Zhu's results (47). In addition, DNMT1, one validated target of PTL, has been reported to stabilize β-catenin in colorectal cancer cells (49, 50). Whether other targets of PTL, such as IKKβ and FAK (35, 43), were involved in its effect on Wnt signaling remains unclear. Combined with previous studies, we believed that β-catenin destabilization could be one of the mechanisms underlying the Wnt inhibition property of PTL.
Similar with USP7, recent studies validated that USP2a, USP9X, and USP4 positively regulated Wnt signaling through deubiquitinating and stabilizing β-catenin, and pharmacological inhibition with their corresponding inhibitor ML364, WP1130, and Neutral Red led to β-catenin degradation (51–53). Notably, several other DUBs targeting β-catenin including USP5 (54), USP15 (55), USP20 (56), and USP47 (57) were also reported. These studies suggested that targeting the relevant USPs associated with β-catenin might be a novel strategy to promote β-catenin degradation and for the development of Wnt inhibitors.
Collectively, our study demonstrated that USP7 inhibitory activity was a novel bioactivity of PTL and elucidated another potential molecular mechanism of PTL in inhibiting the Wnt signaling pathway. These findings suggested that sesquiterpene lactones containing α-methylene-γ-butyrolactone might represent a novel scaffold for developing USP7 inhibitors and broadened the therapeutic application of PTL in USP7 and Wnt-aberrant cancers.
Experimental procedures
Cell culture
Human colorectal carcinoma cell lines (HCT116, SW480, SW620, Caco-2, and HT-29) and HEK293T cells were purchased from the Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences (Shanghai, China). Normal colonic epithelial cell line CCD-841-CoN was kindly gifted by Professor Lin Li (Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences, Shanghai, China). Cells were cultured in separate medium: High-glucose DMEM medium for HEK293T, SW480, SW620, Caco-2 and CCD-841-CoN cells and RPMI 1640 medium for HCT116 and HT29 cells. All culture media (Biological Industries, Kibbutz Beit Haemek, Israel) were supplemented with 10% FBS (Biological Industries, Kibbutz Beit Haemek, Israel), 100 units/ml penicillin, and 100 μg/ml streptomycin (HyClone, Logan, UT). HEK293W cells were cultured in DMEM, supplemented with 100 μg/ml G418 (Sigma-Aldrich) and hygromycin B (Sigma-Aldrich), in addition to 10% FBS and 1% antibiotics. All the cells were incubated at 37 °C, 5% CO2 in a humidified atmosphere.
Plasmids, cell transfection, protein purification, and luciferase reporter assay
Plasmids were transfected using Lipofectamine 3000 (Invitrogen) according to the manufacturer's instructions. FLAG-USP7 was previously generated in our laboratory. FLAG-USP4/USP15/USP25/UCHL1/UCHL3 constructs were generated using PCR amplification and subcloned into pFLAG-CMV2 vectors. HA-Ub (no. 17608) was purchased from Addgene. Full-length WT human FLAG-USP7 proteins, as well as FLAG-USP4/USP15/USP25/UCHL1/UCHL3 were purified from HEK293T cells. Briefly, HEK293T cells transfected with indicated plasmids were collected and lysed in RIPA buffer (20 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1% Triton X-100, 1 mm EDTA, and 1 mm PMSF) plus protease inhibitors (Roche). After centrifuged, the cell lysates were incubated with Anti-FLAG M2 Affinity Gel (Sigma-Aldrich) at 4 °C for 5 h. The beads were washed four times with the RIPA buffer at 4 °C and then eluted with FLAG Peptide (Sigma-Aldrich) in the buffer (50 mm HEPES, pH 7.5, 100 mm NaCl, 2 mm DTT) at 4 °C. The supernatants were collected after being centrifuged for 15 min at 12,000 rpm.
To confirm PTL (Selleck Chemicals, Houston, TX) as a Wnt signaling inhibitor, HEK293W cells were seeded in 96-well plates with three repeats and treated with PTL for 24 h. HCT116 and SW480 cells were seeded in 96-well plates and then transfected as follows: Wnt/β-catenin signaling responsive Firefly luciferase reporter plasmid SuperTOPFlash (80 ng/well) and Renilla reporter plasmid (8 ng/well). After 3-h transfection, cells were exposed to various concentrations of PTL for 24 h and then lysed. Both Firefly and Renilla luciferase activities were measured using the Dual-Luciferase Reporter Assay kit (Promega, Madison, WI). TOPFlash luciferase activities were normalized to the Renilla activities.
Ub-AMC/Ub-Rho110 assay
Deubiquitinating enzyme activity was monitored in a fluorometric assay using Ub-AMC (Boston Biochem, Cambridge, MA) or Ub-Rho110 (Boston Biochem) as substrates, which could be hydrolyzed by USP7 (27). Both enzyme and substrate were freshly prepared in the Ub-AMC reaction buffer (50 mm Tris-HCl, pH 7.6, 0.5 mm EDTA, 20 mm NaCl, 5 mm MgCl2, 200 μm ATP) or Ub-Rho110 buffer (20 mm Tris-HCl, pH 8.0, 2 mm CaCl2, 2 mm β-mercaptoethanol) for each run. Purified FLAG-USP7 (3.7 nm) was pre-incubated with DMSO or PTL, and the enzymatic reaction was initiated by adding the Ub-AMC/Ub-Rho110 substrate (300 nm). Fluorescence was measured at 5-min intervals using a microplate reader (Perkin-Elmer) (λex = 360 nm, λem = 460 nm for Ub-AMC; λex = 485 nm, λem = 535 nm for Ub-Rho110). The abilities of costunolide and α-santonin to inhibit USP7 were tested using Ub-AMC substrate.
Labeling and competition for deubiquitinating activity with the Ub-VME/Ub-PA activity-based probe
Purified FLAG-USP7, as well as FLAG-USP4, FLAG-USP15, FLAG-USP25, FLAG-UCHL1, and FLAG-UCHL3, was incubated with PTL in 30 μl labeling buffer (50 mm Tris, pH 7.6, 5 mm MgCl2, 0.5 mm EDTA, 2 mm DTT and 250 mm sucrose) at room temperature for 20 min. Ub-VME (to a final concentration of 5 μm) (Boston Biochem, Cambridge, MA) was then added and incubated at 37 °C for an additional 20 min. Samples were boiled in 5× sample buffer at 70 °C for 5 min and then separated by 8% SDS-PAGE.
To evaluate drug occupancy on USP7 in cell lysates, living HCT293T cells were harvested and lysed on ice with TE lysis buffer containing 50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 5 mm MgCl2, 0.5 mm EDTA, 0.5% Nonidet P-40, 10% glycerol, and 2 mm DTT. The clarified cell lysates (40 μg) were labeled with PTL at room temperature for 4 h in labeling buffer described above, and then was added Ub-VME (5 μm) at 37 °C for 20 min. Samples were boiled and then separated by SDS-PAGE. To evaluate drug occupancy on USP7 in intact cells, HEK293T, HCT116, as well as SW480 cells were seeded in 6-well plates with a density of 8 × 105 cells/well and then treated with various concentrations of PTL for 2 h. Cells were harvested and lysed on ice with TE buffer. The ubiquitin-labeling reaction was initiated by adding Ub-VME or Ub-PA (5 μm) in labeling buffer, and the reaction was allowed to proceed in a final volume of 30 μl for 20 min. Samples were boiled and then separated by SDS-PAGE.
Cellular thermal shift assay
For a CETSA in SW480 cell lysates, cells were cultured in 10-cm dishes over 12 h. Cells were harvested and washed with PBS and then diluted in PBS supplemented with complete protease inhibitor mixture. The cell suspensions were freeze-thawed three times with liquid nitrogen. The lysates were separated from the cell debris by centrifugation at 20,000 × g for 20 min at 4 °C. The cell lysates were divided into two aliquots. One aliquot was treated with DMSO, the other was mixed with PTL (40 μm). After 30 min incubation at room temperature, the respective lysates were divided into smaller (50 μl) aliquots and individually heated at the designated temperatures for 3 min, followed by cooling for 3 min at room temperature. For the dose-response analysis, equal amounts of cell lysates were incubated with different concentrations of PTL and DMSO for 30 min at room temperature, followed by heating all samples at 52 °C for 3 min and cooling to room temperature.
For a CETSA in living SW480 cells, cells were seeded in 6-cm dishes and exposed to PTL (40 μm) or DMSO for 1 h. Following incubation, cells were harvested and lysed with PBS supplemented with complete protease inhibitor mixture. The cell lysates were then divided into smaller aliquots and heated. For the dose-response analysis, equal amounts of cells were treated with different doses of PTL for 1 h, followed by heating all samples at 55 °C for 3 min.
All heated lysates were centrifuged at 20,000 × g for 20 min at 4 °C to separate the soluble fractions from the precipitates. All the supernatants were transferred to fresh microtubes before SDS-PAGE and Western blot analysis.
K48-linked diubiquitin gel-based assay
30 nm His6-USP7 (Boston Biochem) and variable concentrations of PTL were preincubated in 30 μl reaction buffer (50 mm Tris-HCl, pH 7.6, 0.5 mm EDTA, 5 mm DTT, 0.01% Triton X-100, and 0.05 mg/ml serum albumin) for 40 min at room temperature. The reaction was initiated by adding 2 μm K48-linked diubiquition and was allowed to proceed for 3 h at 37 °C, and then quenched by the addition of 2× Tricine Sample Buffer (Bio-Rad). The reaction products were separated on a 16.5% denaturing SDS-PAGE gel and stained with Fast Silver Stain Kit (Beyotime Biotechnology).
Deubiquination assays
HEK293T cells were transfected with control (Ctrl) or HA-Ub for 24 h. After that, cells were treated with DMSO or PTL (20 μm) for 12 h and then collected. HCT116 and SW480 cells were treated with DMSO or PTL for 12 h before harvesting. All cells were lysed in lysis buffer (150 mm NaCl, 30 mm Tris, 1 mm EDTA, 1% Triton X-100, 10% glycerol, 0.5 mm DTT, and 1 mm PMSF) plus protease inhibitors (Roche). The cell lysates were incubated with β-catenin antibody at 4 °C overnight, followed by addition of protein A/G beads (Santa Cruz Biotechnology, Dallas, TX) for another 6 h. The beads were washed four times with lysis buffer and then boiled in 2× sample loading buffer for 10 min. Samples were blotted and probed with the indicated antibodies.
Surface plasmon resonance
A Biacore S200 (GE Healthcare) was used for characterization of the interaction between PTL and USP7. USP7CD protein (Boston Biochem), which was diluted to 100 ng/μl in 100 μl coupling buffer (50 mm acetate, pH 4.5, supplemented with 2 mm DTT and 0.5 mm EDTA), was coupled to CM5 sensor chips by amine-coupling in HBS buffer (10 mm HEPES, pH 7.4, and 150 mm NaCl), supplemented with 0.05% Pluronic 127, 2 mm DTT, and 0.5 mm EDTA. CM5 chips were activated with NHS/EDC/ethanolamine injection according to Biacore standard methods. Cell 1 was used as blank reference surface, cell 2 was coupled with USP7CD protein.
A series concentration of PTL solutions with 5% (v/v) DMSO were applied to both cell 1 and cell 2 in HBS buffer supplemented with 0.05% Pluronic 127, 2 mm DTT, 0.5 mm EDTA, and 5% (v/v) DMSO, with a flow rate of 30 μl/min at 25 °C, a contact time of 120 s, and a dissociation time of 180 s. A solvent correction curve from 4.5–5.8% (v/v) DMSO was included. Analysis was performed using the Biacore Evaluation software S200 using an affinity fit and a 1:1 binding model with constant Rmax.
Cell viability assay
Cell viability was determined by MTS assay. Briefly, 5 × 103 cells were seeded in 96-well plates in triplicates and cultured overnight. Cells were next treated with different concentrations of PTL for 48 h. Then, 100 μl culture medium mixed with 20 μl CellTiter 96® Aqueous One Solution Reagent (Promega) was added to each sample. Cells were incubated at 37 °C for 30 min to 2 h. The optical density was measured at a wavelength of 490 nm using a microplate reader (Perkin-Elmer). The IC50 values were calculated by the relative survival curves.
Western blotting assay
Cells were seeded in 6-well plates with a density of 5 × 105 cells/well and treated with various concentrations of PTL at the indicated time. Followed by treatment, cells were harvested and lysed in RIPA buffer (50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1% sodium deoxycholate, 0.1% SDS, 1% Nonidet P-40, 1 mm EDTA, and 1 mm PMSF) containing protease and phosphatase inhibitor mixture (Roche). Lysates were centrifuged, then the supernatants were quantitated and then dissolved with 5× sample loading buffer and boiled for 5 min. Protein extracts were subjected to SDS-PAGE and transferred to PVDF membranes (Millipore). Membranes were blocked with 5% nonfat milk and incubated with the following primary antibodies overnight: anti-USP15, anti-FLAG, anti-Axin2, anti-caspase-9, anti–HA-tag, and anti–active β-catenin (Cell Signaling Technology, Danvers, MA); anti-Ub, anti-c-Myc, anti–caspase-8, and anti–poly ADP-ribose polymerase (Santa Cruz Technology, Dallas, TX); anti-Lamin A/C (Epitomics, Burlingame, CA); anti-β-catenin (BD Biosciences); anti-USP7, anti-USP4, anti-USP47 (Bethyl Laboratories, Montgomery, TX); and anti-β-actin (Sigma-Aldrich). Membranes were then incubated with corresponding secondary antibodies conjugated to horseradish peroxidase. Proteins of interest were incubated with Pierce ECL substrate (Thermo Scientific) and visualized by chemiluminescent detection on an ImageQuant LAS 4000 mini (GE Healthcare).
Cytoplasmic and nuclear fractionation assay
Treated cells were harvested and resuspended in lysis buffer A (10 mm HEPES, pH 7.9, 10 mm KCl, 1.5 mm MgCl2, 0.5 mm DTT, and 1 mm PMSF) containing protease inhibitor mixture, followed by incubation for 10 min on ice. After centrifugation at 3100 rpm, cells were lysed in buffer A with 0.5% Nonidet P-40 for 2 min on ice. The supernatants were collected as cytoplasmic extracts after being centrifuged for 15 min at 6000 rpm. The pellets were then washed with lysis buffer A without Nonidet P-40 three times and resuspended in lysis buffer B (20 mm HEPES, pH 7.9, 400 mm NaCl, 0.5 mm DTT, 0.5 mm EDTA, 25% glycerol, and 1 mm PMSF) with protease inhibitor mixture. After being lysed about 40 min and then centrifuged, the supernatants were collected as nuclear extracts. Samples were separated by SDS-PAGE.
Cell cycle and apoptosis analysis
Cells were seeded in 6-well plates at a density of 2 × 105 cells/well and exposed to PTL for 24 h. Cells were subsequently collected and washed twice with PBS. Cells were fixed with pre-cold 70% ethanol overnight at −20 °C. Fixed cells were washed with PBS and then stained with solution that contained 50 μg/ml propidium iodide (Sigma-Aldrich) and 50 μg/ml RNase A (Sigma-Aldrich) for 30 min in the dark at room temperature. Fluorescence intensity was measured by FACSCalibur flow cytometer (BD Biosciences). The distributions of cells in each phase of the cell cycle were determined using FlowJo 7.6.1 analysis software.
Cell apoptosis was analyzed by the annexin V-FITC/PI Apoptosis kit (BD Biosciences) according to the manufacturer's protocol. Briefly, cells were seeded in 6-well plates at a density of 1 × 105 cells/well and cultured overnight. Cells treated with indicated concentrations of PTL for 48 h, were collected, and washed twice with cold PBS, followed by resuspending in a binding buffer containing annexin V-FITC and PI. After incubation for 15 min at room temperature in dark, the fluorescent intensity was analyzed using the FACSCalibur flow cytometer. The data were analyzed using FlowJo 7.6.1 analysis software.
Immunofluorescence analyzes
Cells seeded in 96-well plates were treated with different doses of PTL for 12 h. 4% paraformaldehyde was used for fixing cells about 20 min and then 0.1% Triton X-100 for permeating about 10 min. Samples were blocked with 3% BSA at 37 °C for 30 min and then incubated with anti–β-catenin antibody at room temperature for 6 h. After washing with PBS three times, cells were incubated with corresponding FITC conjugated secondary antibody (Alexa Fluor® 488) for 1 h at room temperature. Nuclei were stained by DAPI (blue) for 10 min. Cells were then observed and photographed under microscopy (Eclipse, Nikon) at 400×.
RT-PCR assay
HCT116 and SW480 cells were seeded in 6-well plates at a density of 5 × 105 cells/well. After adding PTL for 12 h, total RNA was prepared with TRIzol (Thermo Fisher) according to the manufacturer's protocol. Reverse transcription was performed using RevertAid H Minus First Strand cDNA Synthesis Kit (Thermo Fisher). For qPCR, SYBR Select Master Mix (Thermo Fisher) was used with ABI 7500 Real-Time PCR System. The values of c-Myc and Axin2 were shown against the value of GAPDH that was used as the control. Primer sequences are follows: c-Myc forward, 5′-CTTCTCTCCGTCCTCGGATTCT-3′ and reverse, 5′-GAAGGTGATCCAGACTCTGACCTT-3′; Axin2 forward, 5′-AGCCACACCCTTCTCCAATC-3′ and reverse, 5′-ACCGTCTCATCCTCCCAGAT-3′.
Statistical analysis
Two-tailed Student's t test was used to determine the statistical significance. All the results were expressed as mean ± S.D., and p value of less than 0.05 was considered statistically significant.
Author contributions
X. L. data curation; X. L. formal analysis; X. L., T. A., and Y. L. writing-original draft; X. L., L. K., Q. Y., A. D., X. J., B. C., and L. C. project administration; L. K., T. A., and Y. L. funding acquisition; T. A. and Y. L. conceptualization; Y. L. supervision.
Supplementary Material
Acknowledgments
We thank Dr. Aaron M Zorn (Cincinnati Children's Hospital) for providing the SuperTOPFlash plasmid and Lian Yang (Service Center for Bioactivity Screening, State Key Laboratory of Phytochemistry and Plant Resources in West China, Kunming Institute of Botany) for providing help in SPR assay. We also thank Professor Lin Li (Institute of Biochemistry and Cell Biology) for the gift of normal colonic epithelial cell line (CCD-841-CoN).
This work was supported by the Young Scientists Fund of National Science Foundation of China Grant 81803587 (to T. A.); the National Natural Science Foundation of China Grant 81773783 (to Y. L.); the Young Scientists Fund of National Science Foundation of China Grant 81603161 (to L. K.); the Project of Science and Technology of Yunnan Province Grant 2019FD054 (to T. A.); the West Light Foundation of the Chinese Academy of Sciences (to T. A. and L. K.); and Youth Innovation Promotion Association CAS (to L. K.). The work was also supported by the State Key Laboratory of Phytochemistry and Plant Resources in West China, Kunming Institute of Botany, Chinese Academy of Sciences (to T. A. and L. K.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1 and S2.
- UPS
- ubiquitin-proteasome system
- DUBs
- deubiquitinases
- PIs
- proteasome inhibitors
- PTL
- parthenolide
- CRC
- colorectal cancer
- Ub-AMC
- ubiquitin-aminomethylcoumarin
- Ub-Rho110
- ubiquitin-rhodamine 110
- Ub-VME
- ubiquitin-vinyl methyl ester
- di-Ub
- diubiquitin
- Ub-PA
- ubiquitin-propargylamide
- CETSA
- cellular thermal shift assay
- SPR
- surface plasmon resonance
- MS
- mass spectrum
- ST-Luc
- Super TOPFlash luciferase
- buffer A
- 10 mm HEPES, pH 7.9, 10 mm KCl, 1.5 mm MgCl2, 0.5 mm DTT, and 1 mm PMSF
- PI
- propidium iodide.
References
- 1. Hoeller D., and Dikic I. (2009) Targeting the ubiquitin system in cancer therapy. Nature 458, 438–444 10.1038/nature07960 [DOI] [PubMed] [Google Scholar]
- 2. Huang X., and Dixit V. M. (2016) Drugging the undruggables: Exploring the ubiquitin system for drug development. Cell Res. 26, 484–498 10.1038/cr.2016.31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Merin N. M., and Kelly K. R. (2014) Clinical use of proteasome inhibitors in the treatment of multiple myeloma. Pharmaceuticals 8, 1–20 10.3390/ph8010001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lee M. J., Bhattarai D., Yoo J., Miller Z., Park J. E., Lee S., Lee W., Driscoll J. J., and Kim K. B. (2019) Development of novel epoxyketone-based proteasome inhibitors as a strategy to overcome cancer resistance to carfilzomib and bortezomib. J. Med. Chem. 62, 4444–4455 10.1021/acs.jmedchem.8b01943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gupta I., Varshney N. K., and Khan S. (2018) Emergence of members of TRAF and DUB of ubiquitin proteasome system in the regulation of hypertrophic cardiomyopathy. Front. Genet. 9, 336 10.3389/fgene.2018.00336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sheridan C. (2015) Drug makers target ubiquitin proteasome pathway anew. Nature Biotechnol. 33, 1115–1117 10.1038/nbt1115-1115 [DOI] [PubMed] [Google Scholar]
- 7. Fraile J. M., Quesada V., Rodríguez D., Freije J. M., and López-Otín C. (2012) Deubiquitinases in cancer: New functions and therapeutic options. Oncogene 31, 2373–2388 10.1038/onc.2011.443 [DOI] [PubMed] [Google Scholar]
- 8. Li M., Brooks C. L., Kon N., and Gu W. (2004) A dynamic role of HAUSP in the p53-Mdm2 pathway. Mol. Cell 13, 879–886 10.1016/S1097-2765(04)00157-1 [DOI] [PubMed] [Google Scholar]
- 9. Zhou Z., Yao X., Li S., Xiong Y., Dong X., Zhao Y., Jiang J., and Zhang Q. (2015) Deubiquitination of Ci/Gli by Usp7/HAUSP regulates hedgehog signaling. Dev. Cell 34, 58–72 10.1016/j.devcel.2015.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ma P., Yang X., Kong Q., Li C., Yang S., Li Y., and Mao B. (2014) The ubiquitin ligase RNF220 enhances canonical Wnt signaling through USP7-mediated deubiquitination of beta-catenin. Mol. Cell. Biol. 34, 4355–4366 10.1128/MCB.00731-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Novellasdemunt L., Foglizzo V., Cuadrado L., Antas P., Kucharska A., Encheva V., Snijders A. P., and Li V. S. W. (2017) USP7 is a tumor-specific WNT activator for APC-mutated colorectal cancer by mediating β-catenin deubiquitination. Cell Rep. 21, 612–627 10.1016/j.celrep.2017.09.072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Franqui-Machin R., Hao M., Bai H., Gu Z., Zhan X., Habelhah H., Jethava Y., Qiu L., Frech I., Tricot G., and Zhan F. (2018) Destabilizing NEK2 overcomes resistance to proteasome inhibition in multiple myeloma. J. Clin. Invest. 128, 2877–2893 10.1172/JCI98765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shan H., Li X., Xiao X., Dai Y., Huang J., Song J., Liu M., Yang L., Lei H., Tong Y., Zhou L., Xu H., and Wu Y. (2018) USP7 deubiquitinates and stabilizes NOTCH1 in T-cell acute lymphoblastic leukemia. Signal Transduct. Target. Ther. 3, 29 10.1038/s41392-018-0028-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rawat R., Starczynowski D. T., and Ntziachristos P. (2019) Nuclear deubiquitination in the spotlight: The multifaceted nature of USP7 biology in disease. Curr. Opin. Cell Biol. 58, 85–94 10.1016/j.ceb.2019.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sun X., Ding Y., Zhan M., Li Y., Gao D., Wang G., Gao Y., Li Y., Wu S., Lu L., Liu Q., and Zhou Z. (2019) Usp7 regulates Hippo pathway through deubiquitinating the transcriptional coactivator Yorkie. Nat. Commun. 10, 411 10.1038/s41467-019-08334-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jin Q., Martinez C. A., Arcipowski K. M., Zhu Y., Gutierrez-Diaz B. T., Wang K. K., Johnson M. R., Volk A. G., Wang F., Wu J., Grove C., Wang H., Sokirniy I., Thomas P. M., Goo Y. A., et al. (2019) USP7 cooperates with NOTCH1 to drive the oncogenic transcriptional program in T-cell leukemia. Clin. Cancer Res. 25, 222–239 10.1158/1078-0432.CCR-18-1740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lei A., Chen L., Zhang M., Yang X., Xu L., Cao N., Zhang Z., and Cao Y. (2019) EZH2 regulates protein stability via recruiting USP7 to mediate neuronal gene expression in cancer cells. Front. Genetics 10, 422 10.3389/fgene.2019.00422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhu Y., Gu L., Lin X., Cui K., Liu C., Lu B., Zhou F., Zhao Q., Shen H., and Li Y. (2019) LINC00265 promotes colorectal tumorigenesis via ZMIZ2 and USP7-mediated stabilization of β-catenin. Cell Death Differ. 10.1038/s41418-019-0417-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Alonso-de Vega I., Martín Y., and Smits V. A. (2014) USP7 controls Chk1 protein stability by direct deubiquitination. Cell cycle 13, 3921–3926 10.4161/15384101.2014.973324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tavana O., Li D., Dai C., Lopez G., Banerjee D., Kon N., Chen C., Califano A., Yamashiro D. J., Sun H., and Gu W. (2016) HAUSP deubiquitinates and stabilizes N-Myc in neuroblastoma. Nat. Med. 22, 1180–1186 10.1038/nm.4180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Inoue D., Nishimura K., Kozuka-Hata H., Oyama M., and Kitamura T. (2015) The stability of epigenetic factor ASXL1 is regulated through ubiquitination and USP7-mediated deubiquitination. Leukemia 29, 2257–2260 10.1038/leu.2015.90 [DOI] [PubMed] [Google Scholar]
- 22. van Loosdregt J., Fleskens V., Fu J., Brenkman A. B., Bekker C. P., Pals C. E., Meerding J., Berkers C. R., Barbi J., Gröne A., Sijts A. J., Maurice M. M., Kalkhoven E., Prakken B. J., Ovaa H., Pan F., Zaiss D. M., and Coffer P. J. (2013) Stabilization of the transcription factor Foxp3 by the deubiquitinase USP7 increases Treg-cell-suppressive capacity. Immunity 39, 259–271 10.1016/j.immuni.2013.05.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pozhidaeva A., and Bezsonova I. (2019) USP7: Structure, substrate specificity, and inhibition. DNA Repair 76, 30–39 10.1016/j.dnarep.2019.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang Q., Ma S., Song N., Li X., Liu L., Yang S., Ding X., Shan L., Zhou X., Su D., Wang Y., Zhang Q., Liu X., Yu N., Zhang K., Shang Y., Yao Z., and Shi L. (2016) Stabilization of histone demethylase PHF8 by USP7 promotes breast carcinogenesis. J. Clin. Invest. 126, 2205–2220 10.1172/JCI85747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cai J. B., Shi G. M., Dong Z. R., Ke A. W., Ma H. H., Gao Q., Shen Z. Z., Huang X. Y., Chen H., Yu D. D., Liu L. X., Zhang P. F., Zhang C., Hu M. Y., Yang L. X., et al. (2015) Ubiquitin-specific protease 7 accelerates p14(ARF) degradation by deubiquitinating thyroid hormone receptor-interacting protein 12 and promotes hepatocellular carcinoma progression. Hepatology 61, 1603–1614 10.1002/hep.27682 [DOI] [PubMed] [Google Scholar]
- 26. Agathanggelou A., Smith E., Davies N. J., Kwok M., Zlatanou A., Oldreive C. E., Mao J., Da Costa D., Yadollahi S., Perry T., Kearns P., Skowronska A., Yates E., Parry H., Hillmen P., et al. (2017) USP7 inhibition alters homologous recombination repair and targets CLL cells independently of ATM/p53 functional status. Blood 130, 156–166 10.1182/blood-2016-12-758219 [DOI] [PubMed] [Google Scholar]
- 27. Wu J., Kumar S., Wang F., Wang H., Chen L., Arsenault P., Mattern M., and Weinstock J. (2018) Chemical approaches to intervening in ubiquitin specific protease 7 (USP7) function for oncology and immune oncology therapies. J. Med. Chem. 61, 422–443 10.1021/acs.jmedchem.7b00498 [DOI] [PubMed] [Google Scholar]
- 28. An T., Gong Y., Li X., Kong L., Ma P., Gong L., Zhu H., Yu C., Liu J., Zhou H., Mao B., and Li Y. (2017) USP7 inhibitor P5091 inhibits Wnt signaling and colorectal tumor growth. Biochem. Pharmacol. 131, 29–39 10.1016/j.bcp.2017.02.011 [DOI] [PubMed] [Google Scholar]
- 29. Wang L., Kumar S., Dahiya S., Wang F., Wu J., Newick K., Han R., Samanta A., Beier U. H., Akimova T., Bhatti T. R., Nicholson B., Kodrasov M. P., Agarwal S., Sterner D. E., et al. (2016) Ubiquitin-specific protease-7 inhibition impairs Tip60-dependent Foxp3+ T-regulatory cell function and promotes antitumor immunity. EBioMedicine 13, 99–112 10.1016/j.ebiom.2016.10.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Reverdy C., Conrath S., Lopez R., Planquette C., Atmanene C., Collura V., Harpon J., Battaglia V., Vivat V., Sippl W., and Colland F. (2012) Discovery of specific inhibitors of human USP7/HAUSP deubiquitinating enzyme. Chem. Biol. 19, 467–477 10.1016/j.chembiol.2012.02.007 [DOI] [PubMed] [Google Scholar]
- 31. Jafari R., Almqvist H., Axelsson H., Ignatushchenko M., Lundbäck T., Nordlund P., and Martinez Molina D. (2014) The cellular thermal shift assay for evaluating drug target interactions in cells. Nat. Protoc. 9, 2100–2122 10.1038/nprot.2014.138 [DOI] [PubMed] [Google Scholar]
- 32. Jho E. H., Zhang T., Domon C., Joo C. K., Freund J. N., and Costantini F. (2002) Wnt/β-catenin/Tcf signaling induces the transcription of Axin2, a negative regulator of the signaling pathway. Mol. Cell. Biol. 22, 1172–1183 10.1128/MCB.22.4.1172-1183.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. He T. C., Sparks A. B., Rago C., Hermeking H., Zawel L., da Costa L. T., Morin P. J., Vogelstein B., and Kinzler K. W. (1998) Identification of c-MYC as a target of the APC pathway. Science 281, 1509–1512 10.1126/science.281.5382.1509 [DOI] [PubMed] [Google Scholar]
- 34. Jackson P. A., Widen J. C., Harki D. A., and Brummond K. M. (2017) Covalent modifiers: A chemical perspective on the reactivity of α,β-unsaturated carbonyls with thiols via hetero-Michael addition reactions. J. Med. Chem. 60, 839–885 10.1021/acs.jmedchem.6b00788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Berdan C. A., Ho R., Lehtola H. S., To M., Hu X., Huffman T. R., Petri Y., Altobelli C. R., Demeulenaere S. G., Olzmann J. A., Maimone T. J., and Nomura D. K. (2019) Parthenolide Covalently targets and inhibits focal adhesion kinase in breast cancer cells. Cell Chem. Biol. 26, 1027–1035.e22 10.1016/j.chembiol.2019.03.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Poondla N., Chandrasekaran A. P., Kim K. S., and Ramakrishna S. (2019) Deubiquitinating enzymes as cancer biomarkers: New therapeutic opportunities? BMB Reports 52, 181–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chauhan D., Tian Z., Nicholson B., Kumar K. G., Zhou B., Carrasco R., McDermott J. L., Leach C. A., Fulcinniti M., Kodrasov M. P., Weinstock J., Kingsbury W. D., Hideshima T., Shah P. K., Minvielle S., et al. (2012) A small molecule inhibitor of ubiquitin-specific protease-7 induces apoptosis in multiple myeloma cells and overcomes bortezomib resistance. Cancer Cell 22, 345–358 10.1016/j.ccr.2012.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Weinstock J., Wu J., Cao P., Kingsbury W. D., McDermott J. L., Kodrasov M. P., McKelvey D. M., Suresh Kumar K. G., Goldenberg S. J., Mattern M. R., and Nicholson B. (2012) Selective dual inhibitors of the cancer-related deubiquitylating proteases USP7 and USP47. ACS Med. Chem. Lett. 3, 789–792 10.1021/ml200276j [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Liu M., Xiao C., Sun M., Tan M., Hu L., and Yu Q. (2018) Parthenolide inhibits stat3 signaling by covalently targeting Janus kinases. Molecules 23, E1478 10.3390/molecules23061478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gopal Y. N., Chanchorn E., and Van Dyke M. W. (2009) Parthenolide promotes the ubiquitination of MDM2 and activates p53 cellular functions. Mol. Cancer Ther. 8, 552–562 10.1158/1535-7163.MCT-08-0661 [DOI] [PubMed] [Google Scholar]
- 41. Tavana O., and Gu W. (2016) Modulation of the p53/MDM2 interplay by HAUSP inhibitors. J. Mol. Cell Biol. 9, 45–52 10.1093/jmcb/mjw049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Colleran A., Collins P. E., O'Carroll C., Ahmed A., Mao X., McManus B., Kiely P. A., Burstein E., and Carmody R. J. (2013) Deubiquitination of NF-κB by ubiquitin-specific protease-7 promotes transcription. Proc. Natl. Acad. Sci. U.S.A. 110, 618–623 10.1073/pnas.1208446110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kwok B. H., Koh B., Ndubuisi M. I., Elofsson M., and Crews C. M. (2001) The anti-inflammatory natural product parthenolide from the medicinal herb feverfew directly binds to and inhibits IκB kinase. Chem. Biol. 8, 759–766 10.1016/s1074-5521(01)00049-7 [DOI] [PubMed] [Google Scholar]
- 44. García-Piñeres A. J., Lindenmeyer M. T., and Merfort I. (2004) Role of cysteine residues of p65/NF-κB on the inhibition by the sesquiterpene lactone parthenolide and N-ethyl maleimide, and on its transactivating potential. Life Sci. 75, 841–856 10.1016/j.lfs.2004.01.024 [DOI] [PubMed] [Google Scholar]
- 45. Kong F. C., Zhang J. Q., Zeng C., Chen W. L., Ren W. X., Yan G. X., Wang H. X., Li Q. B., and Chen Z. C. (2015) Inhibitory effects of parthenolide on the activity of NF-κB in multiple myeloma via targeting TRAF6. J. Huazhong Univ. Sci. Technolog. Med. Sci. 35, 343–349 10.1007/s11596-015-1435-0 [DOI] [PubMed] [Google Scholar]
- 46. Ghantous A., Sinjab A., Herceg Z., and Darwiche N. (2013) Parthenolide: From plant shoots to cancer roots. Drug Discov. Today 18, 894–905 10.1016/j.drudis.2013.05.005 [DOI] [PubMed] [Google Scholar]
- 47. Zhu X., Yuan C., Tian C., Li C., Nie F., Song X., Zeng R., Wu D., Hao X., and Li L. (2018) The plant sesquiterpene lactone parthenolide inhibits Wnt/β-catenin signaling by blocking synthesis of the transcriptional regulators TCF4/LEF1. J. Biol. Chem. 293, 5335–5344 10.1074/jbc.M117.819300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Liu Y. C., Kim S. L., Park Y. R., Lee S. T., and Kim S. W. (2017) Parthenolide promotes apoptotic cell death and inhibits the migration and invasion of SW620 cells. Intest. Res. 15, 174–181 10.5217/ir.2017.15.2.174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Liu Z., Liu S., Xie Z., Pavlovicz R. E., Wu J., Chen P., Aimiuwu J., Pang J., Bhasin D., Neviani P., Fuchs J. R., Plass C., Li P. K., Li C., Huang T. H., et al. (2009) Modulation of DNA methylation by a sesquiterpene lactone parthenolide. J. Pharmacol. Exp. Ther. 329, 505–514 10.1124/jpet.108.147934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Song J., Du Z., Ravasz M., Dong B., Wang Z., and Ewing R. M. (2015) A protein interaction between beta-catenin and dnmt1 regulates Wnt signaling and DNA methylation in colorectal cancer cells. Mol. Cancer Res. 13, 969–981 10.1158/1541-7786.MCR-13-0644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kim J., Alavi Naini F., Sun Y., and Ma L. (2018) Ubiquitin-specific peptidase 2a (USP2a) deubiquitinates and stabilizes β-catenin. Am. J. Cancer Res. 8, 1823–1836 [PMC free article] [PubMed] [Google Scholar]
- 52. Nguyen H. H., Kim T., Nguyen T., Hahn M. J., Yun S. I., and Kim K. K. (2019) A selective inhibitor of ubiquitin-specific protease 4 suppresses colorectal cancer progression by regulating β-catenin signaling. Cell. Physiol. Biochem. 53, 157–171 10.33594/000000127 [DOI] [PubMed] [Google Scholar]
- 53. Yang B., Zhang S., Wang Z., Yang C., Ouyang W., Zhou F., Zhou Y., and Xie C. (2016) Deubiquitinase USP9X deubiquitinates β-catenin and promotes high grade glioma cell growth. Oncotarget 7, 79515–79525 10.18632/oncotarget.12819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ma X., Qi W., Pan H., Yang F., and Deng J. (2018) Overexpression of USP5 contributes to tumorigenesis in non-small cell lung cancer via the stabilization of β-catenin protein. Am. J. Cancer Res. 8, 2284–2295 [PMC free article] [PubMed] [Google Scholar]
- 55. Greenblatt M. B., Shin D. Y., Oh H., Lee K. Y., Zhai B., Gygi S. P., Lotinun S., Baron R., Liu D., Su B., Glimcher L. H., and Shim J. H. (2016) MEKK2 mediates an alternative β-catenin pathway that promotes bone formation. Proc. Natl. Acad. Sci. U.S.A. 113, E1226–E1235 10.1073/pnas.1600813113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wu C., Luo K., Zhao F., Yin P., Song Y., Deng M., Huang J., Chen Y., Li L., Lee S., Kim J., Zhou Q., Tu X., Nowsheen S., Luo Q., Gao X., Lou Z., Liu Z., and Yuan J. (2018) USP20 positively regulates tumorigenesis and chemoresistance through β-catenin stabilization. Cell Death Differ. 25, 1855–1869 10.1038/s41418-018-0138-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Shi J., Liu Y., Xu X., Zhang W., Yu T., Jia J., and Liu C. (2015) Deubiquitinase USP47/UBP64E Regulates beta-catenin ubiquitination and degradation and plays a positive role in Wnt signaling. Mol. Cell. Biol. 35, 3301–3311 10.1128/MCB.00373-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.