TAF4b is a subunit of the TFIID complex that is highly expressed in the ovary and testis and required for mouse fertility. TAF4b-deficient male mice undergo a complex series of developmental defects that result in the inability to maintain long-term spermatogenesis. To decipher the transcriptional mechanisms upon which TAF4b functions in spermatogenesis, we used two-hybrid screening to identify a novel TAF4b-interacting transcriptional cofactor, ZFP628. Deletion analysis of both proteins reveals discrete and novel domains of ZFP628 and TAF4b protein that function to bridge their direct interaction in vitro.
KEYWORDS: ZFP628, TAF4b, spermatogenesis, meiosis, spermiogenesis, male fertility, TFIID, transcription
ABSTRACT
TAF4b is a subunit of the TFIID complex that is highly expressed in the ovary and testis and required for mouse fertility. TAF4b-deficient male mice undergo a complex series of developmental defects that result in the inability to maintain long-term spermatogenesis. To decipher the transcriptional mechanisms upon which TAF4b functions in spermatogenesis, we used two-hybrid screening to identify a novel TAF4b-interacting transcriptional cofactor, ZFP628. Deletion analysis of both proteins reveals discrete and novel domains of ZFP628 and TAF4b protein that function to bridge their direct interaction in vitro. Moreover, coimmunoprecipitation of ZFP628 and TAF4b proteins in testis-derived protein extracts supports their endogenous association. Using CRISPR-Cas9, we disrupted the expression of ZFP628 in the mouse and uncovered a postmeiotic germ cell arrest at the round spermatid stage in the seminiferous tubules of the testis in ZFP628-deficient mice that results in male infertility. Coincident with round spermatid arrest, we find reduced mRNA expression of transition protein (Tnp1 and Tnp2) and protamine (Prm1 and Prm2) genes, which are critical for the specialized maturation of haploid male germ cells called spermiogenesis. These data delineate a novel association of two transcription factors, TAF4b and ZFP628, and identify ZFP628 as a novel transcriptional regulator of stage-specific spermiogenesis.
INTRODUCTION
During the course of adult mammalian spermatogenesis, germ cells must transition from a single unipotent and diploid undifferentiated spermatogonial stem cell (SSC) to thousands of highly specialized and haploid mature spermatozoa (1). These abundant and regulated transitions involve rapid cell proliferation (mitotic phase) leading to precise chromosome rearrangements and segregation (meiotic phase). The final phase of differentiation, spermiogenesis, involves remarkable chromatin compaction, cytoplasmic exclusion, and flagellum acquisition events that yield free-swimming spermatozoa (2–5). These processes are all confined within the seminiferous tubules of the testis, an enveloped support structure where somatic Sertoli cells sustain an essential stem/germ cell microenvironment. Although these extensive morphological and cytological features have been appreciated for over a century, significant gaps in our knowledge on the genetic and transcriptional regulation governing spermatogenesis, and more specifically spermiogenesis, remain.
During development, the regulation of RNA polymerase II transcription plays a pivotal role in the commitment toward differentiation in diverse cell lineages. In addition to sequence-specific transcription factors that direct specialized gene expression programs, cell-type-specific or -enriched variant forms of the core transcription machinery are also essential (6). These are required to drive critical gene regulatory programs involved in diverse developmental events associated with multicellularity and differentiation. The diversification of the core transcription machinery, which is required for recruitment of RNA polymerase II to proximal gene promoters, is vast in the germ line (7–9). Germ cell-specific or -enriched variants of the general transcription factors TFIIA and TFIID execute critical and selective functions in regulating reproduction and long-term fertility (9). Such mechanisms of germ cell regulation are conserved between invertebrates and vertebrates and reflect an effective means for evolving germ cell-specific modes of gene regulation. Recent work has demonstrated the germ cell-specific transcriptional requirements of TAF4b, a TFIID subunit and TAF4 paralog, for both male and female fertility (10–13). While the previous studies indicate that TAF4b is required for male germ cell development and long-term male fertility, the transcriptional and gene regulatory mechanisms underlying the requirement for TAF4b remain unclear.
The gonad-enriched TAF4b protein shares extensive sequence similarity within the 300-amino-acid carboxy-terminal histone fold domain (HFD) of its more ubiquitously expressed mouse paralog TAF4a (called TAF4 in humans [14]). Biochemical evidence supports the notion that both TAF4a and TAF4b dimerize with TAF12 via an H2A/H2B-like HFD (15–18). While their well-conserved HFDs direct incorporation of these paralogous TAF4 subunits into the TFIID complex, the amino termini of TAF4a and TAF4b are highly divergent outside of a common TAF4 homology (TAFH) domain. This sequence diversity may facilitate transcriptional regulation through unique interacting transcriptional partners. Structural analysis of the TAFH domain in the ETO protein suggests that it serves as a docking platform for both positive and negative transcriptional regulators (19). While the N-terminal coactivator domain of human TAF4 binds the transcriptional activators Sp1 and CREB, no such specific transcription factor interactions have been identified for TAF4b (20, 21). Therefore, uncovering proteins that interact and function with TAF4b is crucial for the identification of transcriptional cofactors that work together with TAF4b to regulate spermatogenesis.
To further uncover the mechanisms of TAF4b, we assessed the protein-binding properties of an amino-terminal mouse TAF4b coactivator domain using the yeast two-hybrid (Y2H) interaction assay. Our studies reveal the novel association of TAF4b with a sequence-specific transcription factor called, zinc finger protein 628 (ZFP628; also known as ZEC). ZFP628 was known to function as a transcriptional activator and has sequence-specific DNA binding properties in vitro (22). Here, we present the results of diverse protein-protein interaction assays both in vitro and in vivo that support the specific interaction between these two proteins. Phylogenetic analyses of their minimal interacting domains in both ZFP628 and TAF4b suggest that they are well conserved through vertebrate evolution. Loss-of-function genetic analyses reveal an essential role for ZFP628 in the regulation of transcription associated with spermiogenesis and male fertility. Together, these data suggest that TAF4b and ZFP628 cooperate to support stage-specific transcription regulating spermatid development in the mouse testis.
RESULTS
Identification of neonatal testis proteins that interact with the TAF4b N-terminal coactivator domain.
Based on large divergent patches of amino-terminal amino acid sequences between TAF4a and TAF4b paralogs, we hypothesize that they interact with distinct DNA-binding transcription factors through unique coactivator domains. To identify TAF4b-interacting transcription factors, we utilized a Y2H protein-protein interaction screen. Since TAF4b-deficient testes exhibit a late embryonic/early postnatal germ cell defect (12), we generated a prey library from postnatal day 2 (P2) mouse testes and cloned the TAF4b coactivator domain into a bait vector (Fig. 1A). While most of the positive TAF4b-interacting Y2H clones did not map to any known protein-coding genes in the mouse genome, 4 proteins were identified from several clones: scaffolding protein receptor for activated kinase 1 (RACK1), chaperone heat shock protein 90 (HSP90), E3 ubiquitin ligase ring box protein 1 (RBX1), and zinc finger protein 628 (ZFP628) (Fig. 1B). While these proteins exhibit an immensely diverse set of molecular and cellular features, the sequence-specific DNA binding properties of ZFP628 made it a promising TAF4b transcriptional cofactor candidate for further validation (22).
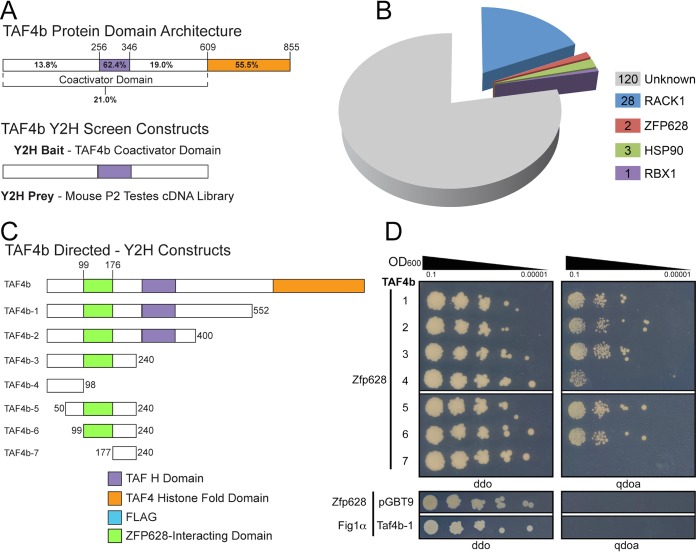
FIG 1.
TAF4b-interacting protein identification by yeast two-hybrid (Y2H) screening assay. (A) TAF4b protein domain architecture featuring comparative domain sequence identity to TAF4a and Y2H screening analysis experimental approach. The mouse TAF4b N-terminal coactivator domain (TAF4b-1) was used as bait to identify interacting proteins from a prey library that was generated from postnatal day 2 testis cDNA. (B) Distribution of positive TAF4b interaction clone frequency identified from 2 independent Y2H screens. The number of positive clones identified for each prey protein is indicated. (C) A schematic depiction for each TAF4b coactivator domain bait construct used in the directed Y2H protein interaction assays. Domain locations are representative of their position within each protein. TAF4b histone fold and TAF H domains were identified using the pfam program (http://pfam.xfam.org/) (44). (D) Directed Y2H analysis. Taf4b bait plasmids (1 to 7) were cotransformed into Y2HGold yeast with the Zfp628 prey plasmid that was identified and isolated from the Y2H screen. Following selective growth in liquid media, 10 μl of the indicated OD600 dilution was plated on agar for either bait/prey plasmid selection (left; ddo) or a stringent selection for bait/prey/protein interaction (right; qdoa). Directed Y2H assays using either the Zfp628 prey plasmid with an empty bait plasmid or a prey plasmid containing Fig1α with the Taf4b-1 bait plasmid were used as negative controls (bottom ddo and qdoa images). The resulting portion of TAF4b necessary for protein interaction with ZFP628 is indicated in panel C.
Our Y2H screens identified the C-terminal 117 amino acids of ZFP628 (922 to 1038) in 2 identical clones from 2 separate screens that specifically interacted with TAF4b (Fig. 1B). To test which portion of the TAF4b coactivator domain ZFP628 interacts with, we generated a series of TAF4b constructs with C-terminal and N-terminal deletions and used these as bait in directed Y2H assays (Fig. 1C; Table 1). As negative controls, we used the empty bait vector along with a prey vector containing Figlα, a germ cell-specific protein not known to interact with the TAF4b coactivator domain. Yeasts were grown on media that selected for the presence of both bait and prey plasmids (Fig. 1D, left) and also selected for the bait-prey interaction (Fig. 1D, right). Robust growth on the bait-prey interaction-selective media by yeasts containing Taf4b-1, -2, -3, -5, and -6 suggests that the ZFP628-TAF4b interaction is highly specific. These data also suggest that TAF4b amino acids 99 to 176 are necessary and amino acids 99 to 240 are sufficient to interact with the C-terminal 117 amino acids of ZFP628 (Fig. 1C and D, ZFP628-interacting domain). We have not yet mapped the minimal domain in ZFP628 (amino acids 922 to 1038) required for interaction with TAF4b.
TABLE 1.
Primers: plasmid construction, Y2H screening, and RT-qPCR
| Construct | Amino acids | Forward primera | Reverse primera | Plasmid |
|---|---|---|---|---|
| Yeast two-hybrid bait constructs | ||||
| Taf4b-1 | 1–552 | TTGGATCCATGCCGGCGGGCCTCAC | TTCTGCAGCTGCCCACAGTTCTGAGTGCTC | pGBKT7 |
| Taf4b-2 | 1–400 | TTGGATCCATGCCGGCGGGCCTCAC | TTCTGCAGGTTTGCTACTCCTGGACCAGCAAG | pGBKT7 |
| Taf4b-3 | 1–240 | TTGGATCCATGCCGGCGGGCCTCAC | TTCTGCAGAGCTTTGAGACGTGCGTCATTTGAG | pGBKT7 |
| Taf4b-4 | 1–98 | TTGGATCCATGCCGGCGGGCCTCAC | TTCTGCAGCGGCGTTTTCACGGTGACTATC | pGBKT7 |
| Taf4b-5 | 50–240 | TTCATATGGTCAGTGTTTGCGTGGAGTCTG | TTGGATCCAGCTTTGAGACGTGCGTCATTTG | pGBKT7 |
| Taf4b-6 | 99–240 | TTCATATGGGCACCACGACAATCCAACTACC | TTGGATCCAGCTTTGAGACGTGCGTCATTTG | pGBKT7 |
| Taf4b-7 | 177–240 | TTCATATGGTTAAAAATTTGACACAGATAGGAACTACTGTG | TTGGATCCAGCTTTGAGACGTGCGTCATTTG | pGBKT7 |
| Yeast two-hybrid library screening primer construct Y2H Hits | CTATTCGATGATGAAGATACCCCACCAAACCC | GTGAACTTGCGGGGTTTTTCAGTATCTACGATTC | pGADT7-Rec | |
| Recombinant ZFP628-TAF4b expression constructs | ||||
| TAF4b-N3 | 1–255 | TTAAGCTTATGCCGGCGGGCCTCAC | GGTACCTAGCACTGTCGGAGGAAGAGCAG | pT7-MAT-FLAG-2 |
| TAF4b-N4 | 1–98 | TTAAGCTTATGCCGGCGGGCCTCAC | TTGGTACCCGGCGTTTTCACGGTGACTATC | pT7-MAT-FLAG-2 |
| TAF4b-N5 | 50–255 | TTAAGCTTGTCAGTGTTTGCGTGGAGTCTG | GGTACCTAGCACTGTCGGAGGAAGAGCAG | pT7-MAT-FLAG-2 |
| TAF4b-N6 | 99–255 | TTAAGCTTGGCACCACGACAATCCAACTACC | GGTACCTAGCACTGTCGGAGGAAGAGCAG | pT7-MAT-FLAG-2 |
| TAF4b-N7 | 177–255 | TTAAGCTTGTTAAAAATTTGACACAGATAGGAACTACTGTG | GGTACCTAGCACTGTCGGAGGAAGAGCAG | pT7-MAT-FLAG-2 |
| TAF4b-N8 | 1–49 | TTAAGCTTATGCCGGCGGGCCTCAC | TTGGTACCAGGAGCCTTAGTCACAGGG | pT7-MAT-FLAG-2 |
| TAF4b-Mid1 | 256–609 | AAGCTTGAAAATGTGAAAAAATGTAAAAACTTCCTTTCAATG | GGTACCGTCATCTTCACCTCTGAAGCATG | pT7-MAT-FLAG-2 |
| TAF4b-C1 | 610–855 | AAGCTTATCAATGATGTGACTTTCATGGCAGAGG | GGTACCCTTAAGAAGGGCAAGGTATAGGGC | pT7-MAT-FLAG-2 |
| GST-ZFP628 | 922–1038 | TTGGATCCATGGTGAGCAGACTAGGCTCTG | TTGAATTCTCAAAACGTGTGTACCAGTTGGACTGC | pGEX-5x-3 |
| 6×His-ZFP628 | 667–1038 | TTGGATCCGCTCAAGATGTTCACGTACTCCCTAAC | TTCTCGAGTAAACGTGTGTACCAGTTGGACTGCAG | pETRP-1B |
| Luciferase assay construct Zfp628-Gal4DBD | 667–1030 | TTTCTAGAGCTCAAGATGTTCACGTACTCCCTAAC | TTGGATCCTAAACGTGTGTACCAGTTGGACTGCAG | pCG-Gal4DBD |
All primers are shown in the 5′-to-3′ direction.
A discrete portion of the TAF4b coactivator domain interacts directly with ZFP628.
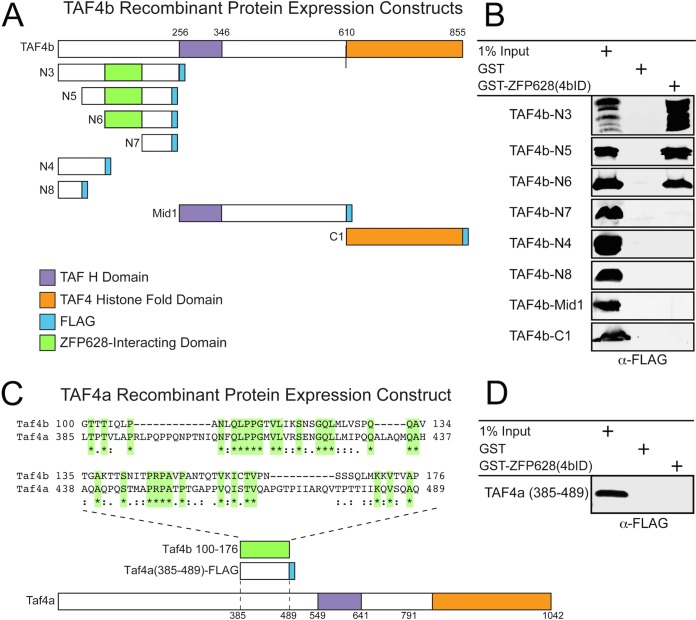
To further test the minimal interaction domains of TAF4b with ZFP628, we generated a glutathione S-transferase (GST)-tagged ZFP628 construct, as well as a series of FLAG-tagged constructs containing different portions of the TAF4b open reading frame (ORF), for recombinant protein expression in Escherichia coli (Fig. 2A; Table 1). By expressing GST-ZFP628 and GST alone and purifying these proteins with glutathione-agarose beads, we tested whether they directly interacted in vitro with the various TAF4b-FLAG proteins. ZFP628 could only pull down TAF4b fragments N3, N5, and N6 (Fig. 2B). These are the only portions of the TAF4b coactivator domain containing amino acids 99 to 176, which is the region identified by the directed Y2H analysis as necessary for interaction with ZFP628. Together, these data indicate that the C-terminal 117 amino acids of ZFP628 are sufficient to directly interact with a small portion of the TAF4b N-terminal coactivator domain (amino acids 99 to 240 [Fig. 1A and 2A]). We also identified a potential ZFP interaction domain in murine TAF4a that is 30% identical to its analogous domain in TAF4b (Fig. 2C) but failed to associate with the TAF4b-interacting domain of ZFP628 (Fig. 2D).
FIG 2.
A discrete portion of the TAF4b coactivator domain is required for its direct interaction with ZFP628. (A) A graphic representation of each TAF4b recombinant protein expression construct containing a C-terminal FLAG epitope, as well as their corresponding TAF H and histone fold domains. The ZFP628-interacting domain identified in Fig. 1 is indicated. The location of each domain is representative of its position within each protein. (B) Defining an interaction domain within TAF4b required for a direct protein-protein interaction with ZFP628. Bacterial soluble protein extracts containing different portions of TAF4b (N3 to N8, Mid1, and C1) were incubated with immobilized GST or GST-ZFP628, and bound proteins were analyzed by immunoblotting with anti-FLAG antibodies. (C) Protein sequence comparisons between mouse TAF4b and TAF4a identified a small domain of TAF4a (amino acids 385 to 389) that displayed limited conservation to the ZFP628-interacting domain of TAF4b. Amino acid identities between these two domains are highlighted in green and indicated by an asterisk, and amino acid conservation is indicated by single and double dots. (D) Bacterial soluble protein extracts containing this domain of TAF4a fused to a C-terminal FLAG epitope were incubated with immobilized GST or GST-ZFP628, and bound proteins were analyzed by immunoblotting with anti-FLAG antibodies.
Comparative phylogenetic analysis of coactivator domain sequence conservation within TAF4b orthologs.
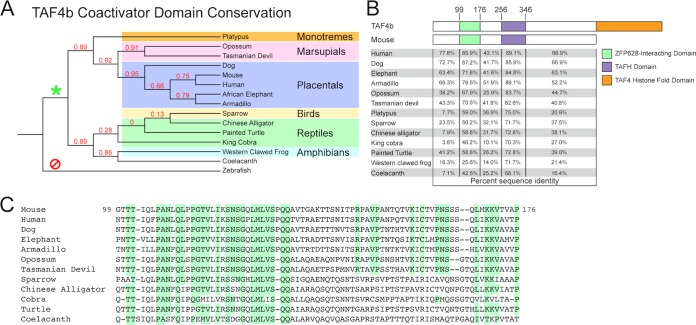
Paralogous TFIID variants are known to function in germ line-specialized transcriptional regulation in both vertebrates and invertebrates through enriched or exclusive gonadal expression (9). Comparative sequence analysis of the testis-specific Taf4 paralog, no hitter (nht), from several Drosophila species indicates that nht arose independently within the Drosophila genus (23). Although TAF4b and the nht product both share a TAF4-like histone fold domain (HFD), the nht product lacks the N-terminal coactivator domain present in the mouse TAF4b ortholog. In light of a direct TAF4b-ZFP628 interaction through a discrete portion of the TAF4b amino-terminal coactivator domain (Fig. 1 and 2), we assessed the extent of this novel coactivator domain throughout metazoans. BLASTP analysis of the mouse N-terminal coactivator domain (amino acids 1 to 609) identified several clear TAF4b orthologs. Phylogenetic analysis places these orthologs among placentals, marsupials, monotremes, amphibians, birds, reptiles, and the lobe-finned fish coelacanth. However, these TAF4b orthologs are not present in ray-finned fishes, cartilaginous fishes, or jawless vertebrates (Fig. 3A). This suggests that the coactivator domain-encoding Taf4b gene originated during the sarcopterygian-osteichthyean evolutionary split approximately 409 to 427 million years ago (24). We found a higher retention of sequence identity within the known TAFH and ZFP628-interacting domains relative to intervening sequences within TAF4b orthologs (Fig. 3B). Furthermore, we also identified several highly conserved residues within the ZFP628-interacting domain, indicating a potential functional and/or structural significance of these sequences (Fig. 3C). As we have highlighted only amino acid sequence identities in these analyses and not conservative substitutions, the extent of conservation is largely underestimated.
FIG 3.
Phylogenetic analysis of the TAF4b coactivator sequence and ZFP628-interacting domain reveals a restricted conservation to vertebrate sarcopterygii and tetrapods. (A) Cladogram of several TAF4b orthologs identified by BLASTP analysis of the mouse TAF4b (NP_001093919) coactivator domain. Sequences from a monotreme (platypus [Ornithorhynchus anatinus] [XP_007666449]), marsupials (opossum [Monodelphis domestica] [XP_007487642] and Tasmanian devil [Sarcophilus harrisii] [XP_012396946]), placentals (dog [Canis lupus familiaris] [XP_547629], human [Homo sapiens] [NP_001280654]), African elephant ([Loxodonta africana] [XP_003406395], and armadillo [Dasypus novemcinctus] [XP_004465202]), a bird (sparrow [Zonotrichia albicollis] [XP_005479722]), reptiles (Chinese alligator [Alligator sinensis] [XP_006016470], painted turtle [Chrysemys picta bellii] [XP_005280060], and king cobra [Ophiophagus hannah] [ETE74136]), an amphibian (western clawed frog [Xenopus tropicalis] [XP_002940973]), and a coelacanth (Latimeria chalumnae [XP_014349716]) are represented. Numbers at the branches represent values obtained after 100 bootstrap replicates. The asterisk and slashed circle indicate the presence and absence, respectively, of an N-terminal coactivator domain within branch-point descendant TAF4b orthologs. (B) Domain structure of the mouse TAF4b coactivator domain and comparative domain sequence identity to TAF4b orthologs. (C) Amino acid sequence alignment of ZFP628-interacting domains from several TAF4b orthologs in panel A. The highlighting corresponds to residues conserved among placentals and marsupials.
Identification and analysis of Zfp628 orthologs.
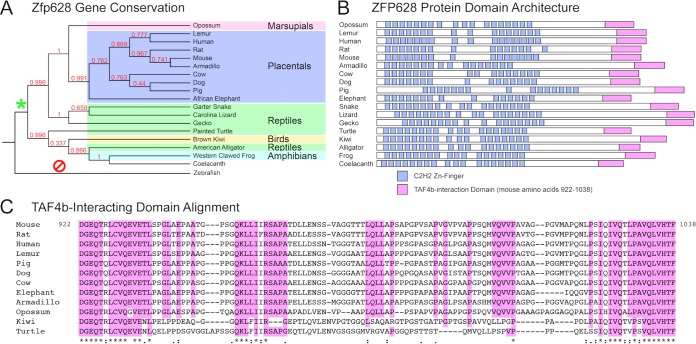
Zinc finger genes represent one of the largest gene classes in the human genome, with 709 identified (3% of all protein-coding genes [25]). Many of these genes have functional orthologs in mice, as well as various other metazoans. Since the ZFP628-interacting domain of TAF4b shares relatively limited conservation, we examined the extent of Zfp628 gene conservation within the animal kingdom. As with Taf4b, Zfp628 orthologs are present in placentals, marsupials, monotremes, amphibians, birds, reptiles, and a lobe-finned fish the coelacanth yet absent in ray-finned fishes, cartilaginous fishes, and jawless vertebrates (Fig. 4A). The 1,038-amino-acid mouse ZFP628 protein contains 16 predicted C2H2 zinc fingers spread over its first 636 amino acids (Fig. 4B). Our Y2H screen identified a TAF4b-interacting clone expressing the C-terminal 117 amino acids of ZFP628 (922 to 1038 [Fig. 1]). To investigate any potential functional conservation within the TAF4b-ZFP628 interaction, we performed a BLASTP analysis of the TAF4b-interacting portion of mouse ZFP628 (922 to 1038). Remarkably, this identified several ZFP628 orthologs that not only contained similar C-terminal TAF4b-interacting sequences but also retained similar N-terminal C2H2 zinc finger domain architectures (Fig. 4B). Phylogenetic analysis reveals a strikingly similar evolutionary conservation profile to the coactivator domain-containing TAF4b orthologs. Regarding the ZFP628-interacting domain in TAF4b, we identified a number of highly conserved residues within the TAF4b-intacting domain of ZFP628 (Fig. 4C). Together, these data demonstrate a clear correlation between Taf4b coactivator domain and Zfp628 gene conservation and also indicate the potential for functional conservation of the ZFP628-TAF4b or related protein-protein interactions during animal evolution.
FIG 4.
Identification and phylogenetic analysis of ZFP628 orthologs: Zfp628 gene conservation correlates with Taf4b coactivator domain conservation. (A) Cladogram of ZFP628 orthologs identified through BLASTP analysis of the mouse ZFP628 (NP_739565). Sequences from a marsupial (opossum [Monodelphis domestica] [XP_016282897]), placentals (gray mouse lemur [Microcebus murinus] [XP_012605717], human [Homo sapiens] [NP_149104], rat [Rattus norvegicus] [XP_008757234], armadillo [Dasypus novemcinctus] [XP_012375274]), cow [Bos taurus] [NP_001179987], dog [Canis lupus familiaris] [XP_541409], pig [Sus scrofa] [XP_013854265], and African elephant [Loxodonta africana] [XP_010585336]), reptiles (garter snake [Thamnophis sirtalis] [XP_013923851], Carolina lizard [Anolis carolinensis] [XP_003224631], gecko [Gekko japonicus] [XP_015277859], painted turtle [Chrysemys picta bellii] [XP_005280811], and American alligator [Alligator mississippiensis] [XP_006275450]), a bird (brown kiwi [Apteryx mantelli] [XP_013794991]), an amphibian (western clawed frog [Xenopus tropicalis] [XP_002941933]), and a coelacanth (Latimeria chalumnae [XP_006010069)]) are represented. Numbers at the branches represent values obtained after 100 bootstrap replicates. The asterisk and slashed circle indicate the presence or absence, respectively, of ZFP628 orthologs within branch-point descendants. (B) Predicted domain architecture of several ZFP628 orthologs. Domain locations are representative of their position within each protein. (C) Amino acid sequence alignment of TAF4b-interacting domains from several ZFP628 orthologs. The highlighting corresponds to residues conserved among placentals and marsupials.
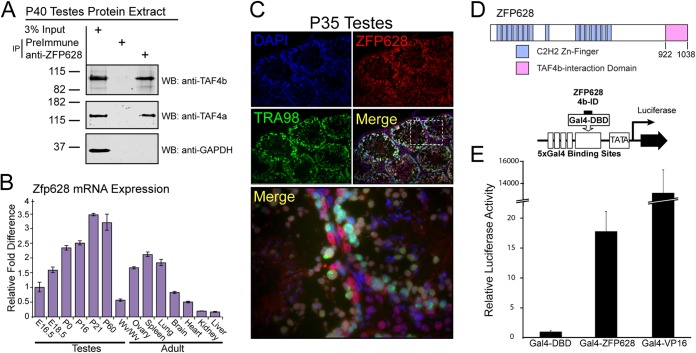
ZFP628 coprecipitates with TAF4b in adult testis protein extracts.
To investigate in vivo ZFP628-TAF4b protein interaction, we generated ZFP628-specific polyclonal antibodies. The ZFP628-specific antiserum was used to coimmunoprecipitate ZFP628-associated proteins from P40 soluble whole testis protein extract. Both TAF4b and TAF4a were detected in ZFP628 immunoprecipitates but not in the negative preimmune serum control. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) did not coimmunoprecipitate with ZFP628 (Fig. 5A). We also examined Zfp628 mRNA expression in adult tissues and during testis development. Zfp628 mRNA was broadly expressed in adult tissues and expression progressively increased during testis development from late embryo to mature adult (Fig. 5B). There is evidence that TAF4a and TAF4b can both associate in the same TFIID complex, and these data suggest that the ZFP628-TAF4b interaction occurs within a TFIID-dependent context. Previous work demonstrated that TAF4b protein is present in both differentiating germ cells and somatic cells within adult testes (12). To test whether there is a similar distribution for ZFP628, we examined ZFP628 protein localization within the adult testis. ZFP628 is found in both TRA98-positive germ cells and TRA98-negative somatic cells (Fig. 5C). Although ZFP628 protein is detectable in most cells, it appears enriched in meiotic spermatocytes, postmeiotic round spermatids, and Sertoli cells. In contrast, ZFP628 was absent from elongating and mature sperm.
FIG 5.
ZFP628 interacts with TAF4b in vivo and has transcriptional activator properties. (A) Endogenous TAF4b and TAF4a were immunoprecipitated from P40 testis extract using polyclonal ZFP628 antisera, whereas GAPDH was not. Rabbit preimmune serum was used as a negative control. WB, Western blotting. (B) Relative mouse Zfp628 mRNA levels during testis development and in adult tissues. (C) P35 testis tissue sections. ZFP628 (red) was detected in both germ cells (TRA98 cells in green) and nongerm cells. Blue, DAPI. ZFP628 colocalization with TRA98 occurs most clearly in round spermatids (yellow merge). (D) Domain architecture of Zfp628 highlighting the TAF4b-interacting portion (pink) and a schematic representation of the luciferase reporter assay using the TAF4b-interacting portion of ZFP628 fused to the Gal4 DNA-binding domain. (E) HEK293T cells were transfected with the negative control Gal4-DBD, positive control Gal4-VP16, or Gal4-ZFP628 and reporter constructs, and transcriptional activity was measured by luminescence.
The TAF4b-interacting portion of ZFP628 can function as a transcriptional activator.
A previous report demonstrated that the entire ZFP628 protein could function as a transcriptional activator (22). To investigate whether the minimal TAF4b-interacting portion of ZFP628 was sufficient for transcriptional activation, we fused it to the yeast GAL4 DNA-binding domain and tested its properties in a human cell-based transcription assay (Fig. 5D). Briefly, we cotransfected a firefly luciferase reporter plasmid containing 5 upstream Gal4 DNA-binding sites into human 293T cells that express very low levels of TAF4b. The transcriptional activity of GAL4-ZFP628 (TAF4b-interacting portion 922 to 1038) regulated by the cytomegalovirus (CMV) promoter was tested compared to GAL4-VP16 as a positive control and the GAL4 DNA-binding domain alone as a negative control. Transfection efficiency was normalized by cotransfection of a CMV promoter-driven Renilla luciferase reporter plasmid. Remarkably, the GAL4-ZFP628 construct yielded an ∼17-fold increase in transcriptional activity, suggesting that on its own the minimal TAF4b-interacting portion of ZFP628 can function as a transcriptional activation domain when fused to the GAL4 DNA-binding domain (Fig. 5E).
Zfp628 is required for spermiogenesis and male fertility.
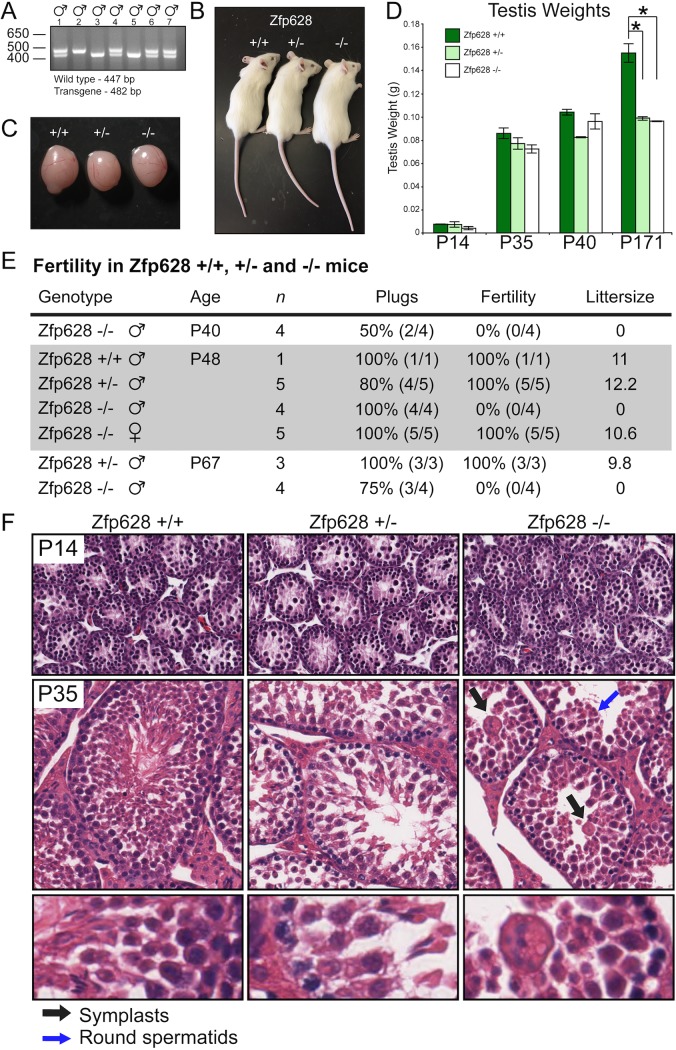
To test the in vivo function of Zfp628 and whether it is functionally required for spermatogenesis, we generated Zfp628-null mice using CRISPR-Cas9 genome editing (26). Following successful production of a male Zfp628+/− founder, we expanded the Zfp628-null transgene by breeding the founder to several Zfp628+/+ females. After obtaining a sufficient number of Zfp628+/− males and females, we conducted several heterozygous intercrosses. We observed normal litter sizes from these crosses compared to +/+ controls. In addition, following offspring genotyping, we observed +/+, +/−, and −/− mice in approximately 1:2:1 ratios (Fig. 6A). These data indicate that −/− mice are viable. In general, Zfp628+/− and Zfp628−/− mice appeared healthy, displayed normal body weights, and did not exhibit any behavioral abnormalities compared to Zfp628+/+ controls (Fig. 6B and C), and testis weights were comparable through P40 mice; at P171, Zfp628+/− and Zfp628−/− mice had significantly smaller testes than those of Zfp628+/+ controls (Fig. 6D). To assess the fertility of Zfp628-null mice, we conducted breeding analysis in both male and female +/+, +/−, and −/− mice. All genotypes had normal mating behaviors, and all crosses generated vaginal plugs. However, no pregnancies or litters were obtained from matings with Zfp628-null males regardless of age (Fig. 6E). In contrast, matings with Zfp628-null females yielded both normal pregnancy rates and litter sizes (Fig. 6E). Together, these data demonstrate that Zfp628 is required for male fertility but is not essential for female fertility.
FIG 6.
Loss of Zfp628 gene function blocks spermiogenesis and causes complete infertility. (A) Genotyping male progeny from a Zfp628+/− mating. (B and C) Size, appearance, and gross morphology of P35 Zfp628 wild-type (WT), heterozygous (Het), and knockout (KO) animals (B) and testes (C) are all normal. (D) Testis weights of Zfp628 WT, Het, and KO mice at the indicated ages. The weights of P171 Zfp628 Het and Zfp628 KO testes were significantly reduced compared to that of WT testes. Data represent means ± SD and are annotated with P values reflecting a two-tailed t test. *, P = 0.01. (E) Summary of breeding studies of Zfp628 transgenic mice. Zfp628−/− mice were infertile, whereas Zfp628+/+, Zfp628+/−, and all female mice were completely fertile (P < 0.01). (F) H&E staining of paraffin-embedded P14 and P35 testes from WT, Het, and Zfp628 KO mice. Black arrows indicate large multinucleated cells (symplasts). Germ cell differentiation is arrested during spermiogenesis in round spermatids (blue arrows) in Zfp628−/− mice.
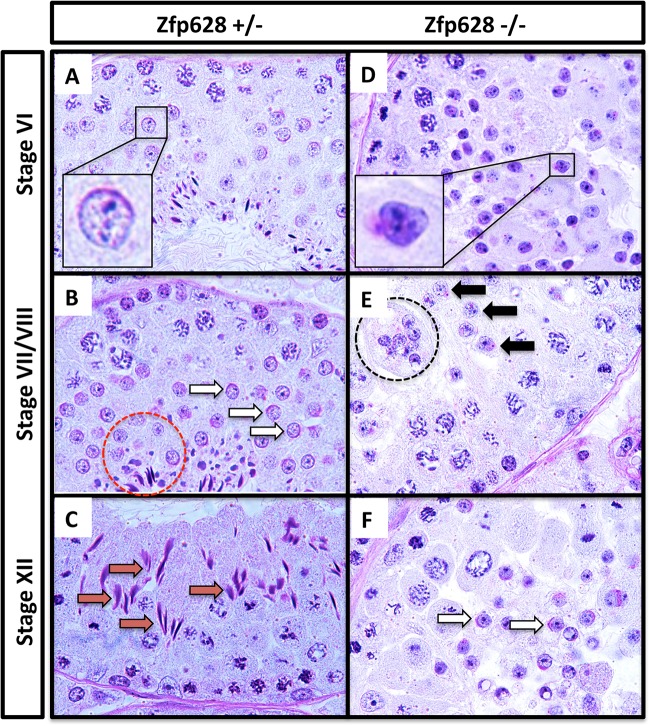
To better understand the developmental nature of the male fertility defect in Zfp628-null mice, we examined testis histology at different ages after hematoxylin and eosin (H&E) staining. At postnatal day 14, testis sections from all genotypes had normal seminiferous tubule, germ cell, and somatic cell morphology (Fig. 6F). In P35 testes, however, several differences appear in the sections derived from Zfp628+/− and Zfp628−/− mice compared to matched Zfp628+/+ controls. Although a full complement of differentiating germ cells (spermatogonia, spermatocytes, spermatids, and mature sperm) was present in Zfp628+/− tubules, a reduced number of germ cells was observed compared to that in Zfp628+/+ controls (Fig. 6F). In P35 Zfp628-null testes, tubules are void of any mature sperm and exhibit a spermiogenesis arrest in round spermatids and several large multinucleated giant cells are present (Fig. 6F). Significant testis weight reductions at P171 in the Zfp628+/− and Zfp628−/− testes also indicate some level of haploinsufficiency (Fig. 6D). To examine the germ cell populations in the testis more closely, we fixed the testes of Zfp628-null mice in Bouin’s solution and embedded and stained paraffin sections with periodic acid-Schiff (PAS) (Fig. 7). Based on the stage-specific comparison of germ cells from the matched fertile Zfp628+/− control, the Zfp628−/− testis displays improperly formed acrosomal caps (Fig. 7A and D, insets), a developmental block at stage VII round spermatids (Fig. 7B, and E) and round spermatid sloughing into the tubule lumen with complete lack of elongating spermatids at stage XII (Fig. 7C and F).
FIG 7.
Stage VII round spermatid block in the Zfp628−/− seminiferous epithelium. Shown is comparative testis histology between Zfp628+/− (A to C) and Zfp628−/− (D to F) testis sections during 3 later stages of the seminiferous epithelium using PAS staining. (A and D) Insets of round spermatids at stage VI show improper acrosomal cap development in Zfp628−/− testis. (B and E) At stage VII/VIII, acrosomal granules (black arrows) are still present in Zfp628−/− testis, while acrosomal caps (white arrows) are properly formed in Zfp628+/− testis. The red circle highlights normal elongated spermatids in the Zfp628+/− testis, whereas the black circle indicates the sloughing of large multinucleated round spermatids in the Zfp628−/− testis that lack these elongated spermatids. (C and F) A stage XII tubule section shows a few round spermatids with acrosomal cap (white arrows) but no elongated spermatids (red arrows) or sperm at this stage of the seminiferous epithelium.
Zfp628 deficiency is associated with elevated apoptosis and downregulation of key spermiogenesis genes.
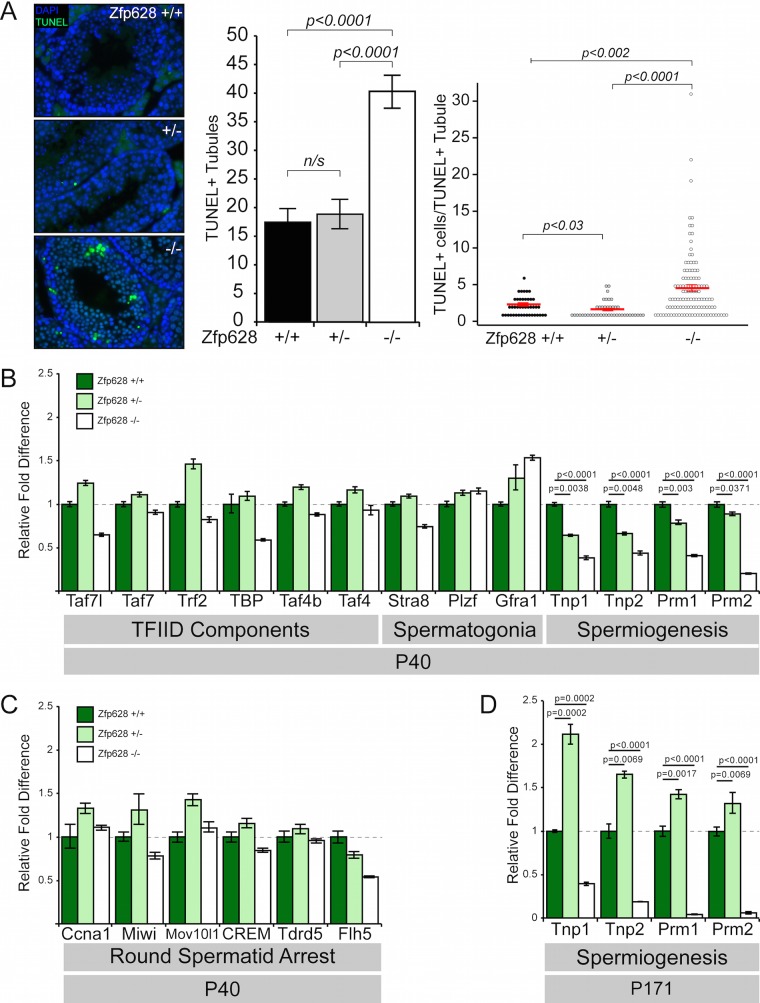
Large multinucleated cells within the Zfp628-null adult testis may correspond to apoptotic germ cells (27). TUNEL labeling in matched +/+, +/−, and −/− testis sections revealed a dramatic and significant increase in both the number of terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL)-positive seminiferous tubules and the number of TUNEL-positive cells within these tubules upon Zfp628 deficiency (Fig. 8A). These results demonstrate that loss of Zfp628 function is accompanied by profound cell death in the adult testis, which likely arises from incomplete spermiogenesis.
FIG 8.
Loss of Zfp628 is associated with extensive apoptosis and downregulation of key spermiogenesis genes. (A) TUNEL staining of P35 testis sections from WT, Het, and Zfp628 KO mice. Quantitative analysis of TUNEL labeling measured both the number of TUNEL-positive tubules and the number of TUNEL-positive cells per TUNEL-positive tubule. Tubules measured: WT, n = 251; Het, n = 238; KO, n = 285. Red bars indicate mean values ± SEM. Data comparisons and P values reflect two-tailed t tests. (B and C) Quantitative RT-PCR (qPCR) analysis of gene expression changes of TFIID component transcripts and transcripts required for spermatogonium maintenance and spermiogenesis (B) and genes whose loss of function phenocopies the Zfp628-null round spermatid arrest (C) in P40 testes from WT, Het, and Zfp628 KO mice. (D) qPCR analysis in P171 testes of genes for spermiogenesis-associated transition proteins 1 and 2 and protamines 1 and 2.
As ZFP628 potentially functions as a sequence-specific transcription factor, we investigated the impact that Zfp628 deficiency has on several potential downstream target genes by quantitative reverse transcription (RT)-PCR (qPCR) in P40 and P171 testes. We first tested whether there are expression changes within genes whose loss of function causes similar germ cell differentiation arrest and increased apoptosis and produces large multinucleated germ cells, many of which were unaffected by the loss of Zfp628 (Fig. 8B). Surprisingly, early spermatocyte gene expression was unaffected (Fig. 8C). In contrast, transition protein and protamine gene expression was compromised in the Zfp628−/− testis at both earlier (Fig. 8B) and later (Fig. 8D) time points. Thus, the developmental block and demise of round spermatids in the absence of Zfp628 may be associated with a block in transcription of the transition and protamine genes required for the timely compaction of the haploid sperm genome.
DISCUSSION
Mechanisms of transcriptional regulation are known to involve specific protein-protein interactions between sequence-specific transcription factors and the general transcription machinery that recruits RNA polymerase II to the start site of transcription. Clearly in development, transcription factors have a dominant role in specifying cell states and fates in comparison to the general transcription machinery. However, throughout evolution, subunits of the general transcription factor TFIID have undergone extensive gene duplication events that allow for these variants to more intimately participate in cell specification events. One such factor is TAF4b, which is a germ cell-enriched subunit of TFIID required for male and female fertility in the mouse. We hypothesized that serving as a coactivator for a testis-specific transcription factor, TAF4b could bridge enhancer-promoter interactions that integrate male germ cell-specific networks of gene expression. To test this hypothesis, we used the yeast two-hybrid method to identify testis-expressed proteins that interact with the amino-terminal coactivator domain of TAF4b. Here we demonstrate the interaction of TAF4b with a putative sequence-specific transcription factor ZFP628 (also called ZEC). A novel amino-terminal domain of TAF4b (amino acids 99 to 240) is necessary for the association with a 117-amino-acid C-terminal domain of ZFP628 (922 to 1038). Moreover, the previously uncharacterized minimal interacting domains of TAF4b and ZFP628 are extensively conserved through vertebrate evolution. The less direct but robust endogenous interaction of TAF4b and ZFP628 in testis-derived protein extracts is supported by coimmunoprecipitation assays. CRISPR/Cas9 targeting of the mouse Zfp628 gene in vivo revealed its potential role in transcription of the transition protein and protamine genes and its essential role in transitioning spermiogenesis in the mouse testes. However, since Taf4b-null males can complete spermiogenesis during the first wave of spermatogenesis, it is still unclear if its interaction with ZFP628 is essential for fertility.
The finding that ZFP628 is required for spermiogenesis is novel and unexpected. Following meiosis, male haploid spermatids undergo extensive chromatin remodeling and compaction as they mature into spermatozoa (28). Crucial to this nuclear condensation process are the replacements of histones first by transition proteins and subsequently by protamines (29–31). In P40 Zfp628−/− mice, we observed a dramatic reduction in transition protein 1 (62%) and transition protein 2 (57%) transcripts, as well as both protamine 1 (59%) and protamine 2 (80%) transcripts; however, we do not yet know if these gene expression changes are direct effects of ZFP628. These expression changes were further exacerbated in P171 Zfp628−/− mice, with reductions of 61% in Tpn1, 82% in Tpn2, 96% in Prm1, and 94% in Prm2 transcript levels, although this may reflect the dramatic loss of round spermatids at this time point. Transcription of transition protein and protamine genes initiate in round spermatids and continue until transcriptional cessation occurs in the condensing nuclei of elongating spermatids (32). Zfp628−/− germ cells do form abundant round spermatids (Fig. 6F) and presumably possess the temporal opportunity for transition protein and protamine transcription. These results suggest that ZFP628 is required for proper Tnp1, Tnp2, Prm1, and Prm2 transcription. Insufficient expression of these critical genes may render round spermatids incapable of differentiating into elongating spermatids. Due to its sequence-specific DNA binding and transcriptional activator properties, it is tempting to speculate that these are direct transcriptional targets for ZFP628 regulation. Indeed, Chen and colleagues identified C/GC/TA/TGGTTGGTTGC as a ZFP628 consensus DNA binding sequence in vitro (22). However, exploring the presence of this DNA binding consensus sequence within promoters of putative target genes and global promoter occupancy and transcriptional studies are needed to address the potential functions of the association between ZFP628 and TAF4b in the future.
Although TAF4b itself has not been previously linked to spermiogenesis, other germ cell-expressed subunits of TFIID, including Trf2 (also called Tbpl1) and Taf7l, are known to be required for spermiogenesis in the mouse. In light of the ZFP628 interaction with TAF4b, as well as phenotypic similarities between ZFP628-, Trf2-, and Taf7l-deficient mice (33, 34), we investigated gene expression differences for several TFIID subunits. In contrast to the dramatic effects on transition protein and protamine mRNA levels, we detected only modest reductions in TAF7, Trf2, Taf4b, Taf4a, Taf7l (35% decrease), and TBP (41% decrease) transcript levels. There is evidence that TBP transcripts accumulate in rodent round spermatids approximately 1,000-fold over somatic cell levels (35, 36). Although the precise function of this transcript elevation is unknown, the decrease in TBP mRNA in Zfp628−/− testes suggests that ZFP628 may also regulate TBP transcription in round spermatids. The germ cell sloughing observed in Zfp628−/− testes resembles symplastic spermatids observed in other mouse models of spermatocyte or spermatid arrest, such as Ccna1-null, Trf2-null, and Rfx2-null mice (33, 37, 38). These symplasts are generally a germ cell degeneration by-product resulting from the arrest in spermiogenesis. Ccna1 is a germ line-specific cell cycle regulator essential for male meiosis (37). Mice lacking the Ccna1 gene lack postmeiotic germ cells, accumulate abundant large multinucleated symplasts, and have elevated apoptosis. MIWI and MOV10L1, involved in Piwi-interacting RNA (piRNA) biogenesis, are required for male fertility, and their loss of function results in a germ cell development arrest at the round spermatid and zygotene spermatocyte stages, respectively (39–41). Miwi exhibited a slight (22%) decrease expression in Zfp628−/− testes, whereas Ccna1 and Mov10l1 were not substantially altered. CREM is a transcriptional master regulator essential for spermiogenesis, and Flh5 is a CREM-binding transcriptional activator that may coregulate CREM target genes (27, 42). Although Zfp628−/− testes had a slight (16%) reduction in CREM expression, they showed a 46% reduction in Flh5 transcript levels. ZFP628 may influence expression of these known spermiogenesis regulators, either directly or indirectly, and these combined effects may lead to the block in spermiogenesis observed in the Zfp628−/− testes.
In addition to ascertaining mechanisms of cell-type-specific gene expression, these studies further implicate Taf4b and Zfp628 in the regulation of fertility in men. While it is not yet known whether their specific interaction occurs during human spermiogensis, the relatively high conservation of the minimal interaction domains of TAF4b and ZFP628 suggests that this interaction may be conserved in men, and its disruption may lead to male infertility. Moreover, the extensive conservation of this interaction domain throughout vertebrate evolution suggests that this protein-protein interaction might be critical for spermatogenesis in many diverse organisms and/or that these domains may reflect novel functional or structural domains involved in other related proteins. Future studies of the potential transcriptional and developmental mechanisms of TAF4b and ZFP628 in mouse spermatogenesis may reveal unanticipated and productive opportunities to address issues of male infertility and contraception in the future.
MATERIALS AND METHODS
Yeast two-hybrid library and bait construction.
Total RNA was purified from postnatal day (P2) testes by TRIzol extraction (Invitrogen, Carlsbad, CA). P2 testis cDNA was synthesized from the poly(A) RNA transcripts, transformed into the Y187 yeast strain, and inserted into the pGADT7-Rec prey vector through in vivo homologous recombination and selective growth on synthetic defined medium lacking Leu with agar (SD/-Leu agar), using the make-your-own Mate & Plate library system (Clontech, Mountainview, CA). Y2H bait constructs were generated by PCR-amplifying portions of the TAF4b N-terminal coactivator domain and cloning into the pGBT9 bait vector following either BamHI and PstI restriction digests of Taf4b-1 to -4 constructs or NdeI and BamHI restriction digests of Taf4b-5 to -7 constructs (Table 1; Fig. 1). Each bait plasmid was transformed into the Y2HGold yeast strain, and positive clones were selected by growth on SD/-Trp agar (Clontech).
Yeast two-hybrid screening.
The bait for the Y2H screen was prepared by growing a positive Taf4b-1 yeast colony overnight in 50 ml of SD/-Trp liquid medium to an optical density at 600 nm (OD600) of 0.8, pelleting the cells by centrifugation, and resuspending the pellet to a cell density of >1 × 108 per ml in SD/-Trp medium. The concentrated Y2HGold bait yeast was combined with 1 ml of the Y187 P2 testis cDNA prey library and 45 ml of 2× yeast extract-peptone-dextrose-adenine (YPDA) medium. Following incubation at 30°C for ∼20 h, the presence of diploid yeast zygotes was confirmed through observation by phase-contrast microscopy. Diploid clones containing candidate TAF4b-interacting prey proteins were selected by growth at 30°C on SD/-Leu/-Trp agar medium containing X-α-Gal (40 μg/ml) and aureobasidin A (125 ng/ml). Positive blue clones were then rigorously tested for a true protein-protein interaction by restreaking on high-stringency SD/-Leu/-Trp/-His/-Ade agar medium containing X-α-Gal and aureobasidin A and growth at 30°C.
TAF4b-interacting-protein identification by colony PCR.
Following growth under high-stringency conditions, a small amount of yeast from positive clones was resuspended in 200-μl PCR tubes containing 2 μl of Zymolyase buffer (50 mM Tris-HCl [pH 7.9], 10 mM MgCl2, 0.02 U/μl of Zymolyase 20T) and incubated for 15 min at 30°C. TAF4b-interacting bait clones were then amplified by PCR using pGADT7-specific primers and identified by sequence analysis (Table 1).
Directed yeast two-hybrid assays.
The prey plasmid identified in the Y2H screen, containing a TAF4b-interacting portion of ZFP628 (amino acids 922 to 1038), was isolated and cotransformed into Y2HGold yeast with each of the Taf4b-1 to -7 bait plasmids. As negative controls, the Zfp628 prey plasmid along with an empty bait plasmid or a prey plasmid expressing the mouse Fig1α coding sequence and the Taf4b-1 bait plasmid were cotransformed into Y2HGold yeast. Following selective growth on SD/-Leu/-Trp agar, colonies containing both bait and prey plasmids were isolated and grown overnight at 30°C in SD/-Leu/-Trp liquid medium. Each culture was diluted in 10-fold serial increments from OD600s of 0.1 to 0.00001, and 10 μl of each dilution was plated on SD/-Leu/-Trp agar (bait/prey plasmid selection) and SD/-Leu/-Trp/-His/-Ade agar containing aureobasidin A (bait/prey interaction selection).
Recombinant TAF4b, TAF4a, and ZFP628 protein production and purification.
The TAF4b-interacting C-terminal 117 amino acids encoded in the Zfp628 prey plasmid were fused to GST by PCR amplification and cloning into pGEX-5x-3 (GE Healthcare Lifesciences, Piscataway, NJ) following BamHI and EcoRI digestion (Table 1). Various portions of the Taf4b open reading frame were generated by PCR and subcloned into pT7-MAT-FLAG-2 (Sigma-Aldrich, St. Louis, MO) following HindIII and KpnI digestion (Table 1; Fig. 2). Recombinant fusion proteins were expressed in E. coli BL21(DE3) Rosetta cells (EMD Millipore, Billerica, MA), and cell pellets were resuspended in 10 ml of protein binding buffer (25 mM Tris [pH 7.5], 150 mM NaCl, 10% glycerol, 0.1% Triton X-100, 0.75% Sarkosyl, and complete protease inhibitor cocktail [Roche, Indianapolis, IN]). The cells were lysed with 20 mg of lysozyme for 30 min at 25°C, freeze-thawed at –80°C and 37°C 3 times, cooled on ice for 15 min, and sonicated for 2 cycles: 1 min each at 100% duty cycle and 1.5 power using a Branson 3000 Sonifier. Insoluble material was removed by centrifugation and the soluble protein lysate was collected and stored at –80°C. GST and GST-ZFP628 fusion protein were purified with glutathione-coupled agarose beads (Sigma-Aldrich, St. Louis, MO) by incubating 70 μl of swollen beads with 300 μl of lysate and 1 ml of protein binding buffer at 25°C for 3 h while rotating. The beads were washed 6 times with 1 ml of protein binding buffer. Purification of recombinant GST-ZFP628 was confirmed by SDS-PAGE followed by Western blotting with anti-GST antibodies (1:5,000) and Coomassie brilliant blue staining (GE Healthcare Lifesciences, Piscataway, NJ).
Recombinant ZFP628-TAF4b protein binding assay.
Recombinant ZFP628-TAF4b binding assays were carried out using methods described by Gustafson et al. (43). Protein samples were prepared by adding 60 μl of 2× sample buffer containing 10 mM dithiothreitol (DTT) to the beads, heated at 100°C for 5 min, and centrifuged for 30 s at 18,000 × g, and denatured protein supernatants were collected. The samples (20 μl) were resolved on a 12% Tris-glycine polyacrylamide gel for immunoblot analysis on a nitrocellulose membrane with mouse monoclonal anti-FLAG primary antibodies (1:5,000; Sigma-Aldrich, St. Louis, MO) and IRDye 800CW goat anti-mouse secondary antibodies (Li-Cor, Lincoln, NE). The TAF4b and TAF4a C-terminal FLAG fusion proteins were detected and imaged with the Odyssey infrared imaging system (Li-Cor).
Zfp628 and Taf4b phylogenetic analysis.
TAF4b orthologs containing coactivator domains were identified using BLASTP analysis of the mouse TAF4b coactivator domain (amino acids 1 to 609) under the default algorithm parameters (https://blast.ncbi.nlm.nih.gov/). Sequence analysis from the Y2H screen identified the C-terminal 117 amino acids from ZFP628 in 2 clones from 2 independent Y2H screens that were positive for a TAF4b interaction. ZFP628 orthologs were identified using BLASTP analysis of the mouse ZFP628 protein sequence. TAF4b-interacting domains were identified by comparison to the mouse ZFP628 C-terminal 117 amino acids. ClustalW analysis was used for amino acid sequence alignments of the ZFP628-interacting domain on TAF4b and the TAF4b-interacting domain on ZFP628 (http://www.ch.embnet.org/software/ClustalW.html). The TAFH and histone fold domains on TAF4b orthologs were identified using the pfam program (http://pfam.xfam.org/) (44). C2H2 domains on ZFP628 orthologs were identified using the MOTIF Search program (http://www.genome.jp/tools/motif/). ZFP628-interacting domain sequence identities in TAF4b orthologs were analyzed using the EMBOSS Needle pairwise alignment algorithm (http://www.ebi.ac.uk/Tools/emboss/align). TAF4b coactivator domain and ZFP628 cladograms were constructed by sequential analyses using MUSCLE for multiple alignment, GBlocks for alignment management, PhyML for tree building, and TreeDyn for illustration through the Phylogeny.fr analysis platform and maximum likelihood analyses (http://phylogeny.fr/) (45–49).
Cell culture, transfection, and luciferase transcription assay.
HEK293T cells were plated at density of 120,000 per well in 12-well plates and cultured in Dulbecco modified Eagle medium (DMEM; Sigma-Aldrich, St. Louis, MO) with 10% fetal bovine serum and 1% penicillin-streptomycin for 24 h. Using the FuGENE6 transfection reagent (Promega, Madison, WI) at a 3:1 reagent-to-DNA ratio, cells were transfected with 150 ng of luciferase reporter plasmid (p5xGal4-E1b-Luciferase), a Renilla luciferase reporter plasmid (pCMV-RL) to control for transfection efficiency, and 40 ng of either pCG-Gal4DBD(1-94) as a negative control for transcriptional activity, pCG-Gal4-VP16 as a positive control for transcriptional activity, or pCG-Gal4-Zfp628 (Table 1; Fig. 5). Each transfection condition was performed in triplicate. The cells were then cultured for an additional 24 to 48 h, and luciferase activity was measured using a Dual-Luciferase reporter assay system (Promega, Madison, WI) and a luminometer.
Quantitative RT-PCR.
Total RNA was purified from several dissected adult tissues (spleen, lung, brain, heart, kidney, liver, ovary, and testes), neonatal testes, and embryonic testes by TRIzol extraction (Invitrogen, Carlsbad, CA). Corresponding template cDNA was generated using the iScript cDNA synthesis kit (Bio-Rad, Hercules, CA). Primer sets were designed by PrimerBank (https://pga.mgh.harvard.edu/primerbank/index.html) (50–52) (Table 2). The ABI 7900H real-time PCR system (Applied Biosystems) and Power SYBR green qPCR master mix with ROX (Invitrogen) were used for quantitative reverse transcription (RT)-PCR (qPCR) data acquisition. The qPCR mixtures were incubated at 50°C for 2 min and 95°C for 10 min, followed by 40 cycles of amplification at 95°C for 15 s and 60°C for 60 seconds and a dissociation curve analysis. Data from each gene were normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA levels and represented as a fold change relative to the indicated mRNA level. Each qPCR was performed in triplicate and averaged. Error bars in figures indicate the standard deviations (SD) resulting from experimental and normalized triplicate qPCRs.
TABLE 2.
RT-qPCR and genotyping primers
| Protein or construct | Forward primera | Reverse primera |
|---|---|---|
| RT-qPCR primers | ||
| ZFP628 | ACCGAGGGAACTGGGGAAAA | GAGATGACCACCTGAAGGACT |
| GAPDH | AGGTCGGTGTGAACGGATTTG | TGTAGACCATGTAGTTGAGGTCA |
| Taf7l | AGCCCCCGTTATTCCTGAAG | CCTCGGGTGTCACAAGTGT |
| Taf7 | AACAAAGACGATGCGCCTCAT | CAGGTTGACATGCCCAGACT |
| Trf2 | TTTGGTGCCAGACGTTTAGC | GCCACAGCCTTTACATTGGG |
| TBP | ATGATGCCTTACGGCACAGG | GTTGCTGAGATGTTGATTGCTG |
| Taf4b | TTGCAGCTATTGGACCAAGGA | GTGGCTGTTAGGCTGGAAGT |
| Taf4 | GAATGGTCCTCGTGCGGAG | CCGTGCAATAATAGGTGTTCCT |
| Stra8 | TTTGACGTGGCAAGTTTCCTG | TAACACAGCCAAGGCTTTTGA |
| Plzf | CTGGGACTTTGTGCGATGTG | CGGTGGAAGAGGATCTCAAACA |
| Gfra1 | CACTCCTGGATTTGCTGATGT | AGTGTGCGGTACTTGGTGC |
| Tnp1 | ACCAGCCGCAAGCTAAAGAC | GCTTCCACCTCTCTTGACGC |
| Tnp2 | TCACACCAGTAACCAGTGCAA | CCCTGAGCTACGCCTCTTAG |
| Prm1 | CCGTCGCAGACGAAGATGTC | CACCTTATGGTGTATGAGCGG |
| Prm2 | ATGGTTCGCTACCGAATGAGG | CTCCGCCTTCTGCATGACC |
| Ccna1 | TGATGCTTGTCAAATGCTCAGC | AGGTCCTCCTGTACTGCTCAT |
| Miwi | GACTCGGAATGGGGAACACG | GGTTTGGAATATCAATCGGGTCA |
| Mov10l1 | CGCTGTGACGAGTACAGTG | CTGACAACCCTTTGCTAGAGTTT |
| CREM | ATGGTTTCTGTAGCTGGATCAGG | GATGTGGTGTCTGAATAACTCCC |
| Tdrd5 | GGAGCCATAAGGTCCGAAACT | AGAACGGGAGACAATGCCAAA |
| Flh5 | TTGTGAGCAGTGTAAAGAACCAA | ACCAAAGAGTGATGGCATTTGTT |
| Zfp628 genotyping primer construct 628sgRNA-F | AGCCTTCAAGGGTTCCTCAG | AGCAGGTTGGAGGAGTGTGT |
All primers are shown in the 5′-to-3′ direction. The sequence for the Zpf628 stop codon cassette is as follows: TGGTCTGGCCTTCAAGTGGTCATCCCATTACCAATACCACTTGAGACAGCATACCGGTGAGTCATGGCGTTTAAACCT*TAAT*TAAGCTGTTGTAGGCGGCCCTACCCGTGCCCTGACTGCCCCAAGGCCTTCAAGAACTCCTCCAGCTTGCGGCG, where bold text indicates the transgene insertion sequence and underlined text indicates the stop codon sequence.
ZFP628-specific antibody production and purification.
The C-terminal ZFP628 amino acids 667 to 1038 were fused to a 6×His tag by PCR amplification and subcloning into the pETRB-1P expression vector (a generous gift from Rebecca Page, Brown University [Table 1]). The 6×His-ZFP628 recombinant protein was expressed in E. coli BL21(DE3) Rosetta cells (EMD Millipore, Billerica, MA), and cell pellets were resuspended in protein purification buffer (8 M urea, 100 mM NaCl, 20 mM HEPES [pH 8.0], and complete protease inhibitor cocktail [Roche, Indianapolis, IN]). The cells were lysed by freeze-thawing at –80°C and 37°C 3 times and sonicated, and insoluble material was removed by centrifugation. The 6×His-ZFP628 and 6×His-TAF4b proteins were purified from the soluble protein lysate using Ni-agarose affinity chromatography and elution with imidazole. Protein purification was assayed by SDS-PAGE and Coomassie staining, as well as immunoblot analysis using mouse monoclonal antipolyhistidine primary antibodies (1:2,000; Sigma-Aldrich, St. Louis, MO) Recombinant proteins were concentrated to 1 to 2 mg/ml by dialysis, lyophilization, and resolubilization in phosphate-buffered saline (PBS) (6×His-ZFP628). Polyclonal antibodies against mouse ZFP628 were then generated by immunizing rabbits and chickens (Cocalico Biologicals Inc., Reamstown, PA). Recombinant 6×His-ZFP628 and 6×His-TAF4b proteins were cross-linked to AminoLink coupling resin (Life Technologies, Grand Island, NY), and the polyclonal antibodies were affinity purified from their respective antisera.
Coimmunoprecipitation of ZFP628-associated proteins.
Several P40 C57/B6 mouse testes were dissected and detunicated. Each testis was homogenized in 500 μl of MAT binding buffer (MBB; 25 mM Tris [pH 7.5], 150 mM NaCl, 10% glycerol, 0.1% Triton X-100, and complete protease inhibitor cocktail (Roche, Indianapolis, IN), and incubated on ice for 1 h. Insoluble material was removed by centrifugation for 15 min at 18,000 × g, and soluble protein extract from each testis was collected and pooled. ZFP628 antiserum and preimmune serum (500 μl each) were bound to a mixture of 30 μl of protein G-Sepharose, 30 μl of protein A-Sepharose, and 500 μl of MBB. Following a 1-h incubation at room temperature under rotation, the different bead mixtures were each washed 4 times with 1 ml of MBB. The anti-ZFP628 and preimmune protein A/G beads were then resuspended in 500 μl of MBB and 500 μl of soluble testis protein extract, incubated overnight at 4°C under rotation, and washed 5 times with 1 ml of MBB. Immunoprecipitated protein samples were prepared by adding 60 μl of 2× sample buffer containing 15 mM DTT to the beads and boiling for 5 min, samples were centrifuged for 30 s at 18,000 × g, and the denatured protein supernatants were collected. An immunoprecipitation input sample was prepared with 25 μl of soluble testis protein and 75 μl of 2× sample buffer containing 15 mM DTT and boiling for 5 min. The samples (20 μl) were resolved on a 10% Tris-glycine polyacrylamide gel and transferred to nitrocellulose membranes for immunoblotting with mouse monoclonal anti-GAPDH (1:1,000; Life Technologies Co., Grand Island, NY), chicken anti-TAF4b (1:1,000 [53]), and mouse monoclonal anti-TAF4 (1:1,000; BD Transduction Laboratories, San Jose, CA) used as primary antibodies. Anti-rabbit IRDye 800CW (1:5,000; Li-Cor Biotechnology, Lincoln, NE), anti-mouse IRDye 800CW (1:5,000; Li-Cor Biotechnology), and anti-mouse IRDye 680CW (1:5,000; Li-Cor Biotechnology) were used as secondary antibodies, and immunoblots were imaged using a Li-Cor Odyssey infrared imaging system.
Genetic ablation of Zfp628.
CRISPR-CAS9 genome editing technology was used to generate Zfp628-null mice.
A Zfp628-specific single guide RNA (sgRNA) was designed and generated (Horizon Discovery, Cambridge, MA). The Zfp628 sgRNA specifically targeted a CAS9-mediated double-strand DNA break within the Zfp628 open reading frame (ORF). A stop codon cassette donor oligonucleotide was designed to facilitate insertion into the Zfp628 ORF by homology-directed repair. The inserted sequence contains stop codons in all three open reading frames to ensure genetic ablation of a functional ZFP628 protein (Table 2). These reagents were injected into single cell zygote pronuclei from CD-1 mice, embryos were implanted at the Brown University Transgenics Facility, and a heterozygous founder male progeny was identified by PCR using Zfp628-specific primers flanking the 35-bp transgene sequence insertion (Table 2). Sequence analysis confirmed the successful incorporation of the stop codon donor oligonucleotide sequence into the Zfp628 gene locus. The Zfp628 knockout (KO) transgene was then bred to homozygosity.
Histology and immunofluorescence.
Testes were dissected from mice of various postnatal ages, fixed, dehydrated, embedded, and sectioned as described previously (12). Sections were deparaffinized and stained with hematoxylin and eosin (H&E) or periodic acid-Schiff (PAS) or prepared for immunofluorescence as described previously (12). Primary antibodies used for immunofluorescence were anti-ZFP628 antiserum (1:100) and anti-TRA98 (1:500; B-Bridge, Cupertino, CA). The secondary antibodies used were Alexa Fluor 488-conjugated anti-rat IgG (H+L) (1:500; Invitrogen, Carlsbad, CA) and Alexa Fluor 594-conjugated anti-rabbit IgG (H+L) (1:500; Invitrogen). DNA was visualized with 4′,6-diamidino-2-phenylindole (DAPI; Vector Laboratories, Burlingame, CA). Fluorescence images were acquired with a Zeiss Axio Imager M1 microscope (Carl Zeiss, Inc., Thornwood, NY). H&E staining images were acquired using the Scanscope CS Aperio (Leica Microsystems, Inc., Buffalo Grove, IL).
TUNEL assay and quantitative analysis.
P35 mouse testis sections were deparaffinized and apoptotic cells were visualized using an in situ cell death detection kit with fluorescein (Roche, Indianapolis, IN). DNA was visualized with DAPI (Vector Laboratories, Burlingame, CA). Fluorescence images were acquired with a Zeiss Axio Imager M1 microscope (Carl Zeiss, Inc., Thornwood, NY). For each Zfp628 genotype, entire testis section composite images were assembled. Seminiferous tubule counts, as well as terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL)-positive cell counts, were then obtained. Bee swarm plots of the TUNEL data were generated using R statistical computing software and the beeswarm package (54).
ACKNOWLEDGMENTS
We thank Kate Grive for her insightful comments on the manuscript.
Author contributions are as follows: E.A.G., conception and design, collection and assembly of data, data analysis and interpretation, and manuscript writing; K.A.S., collection and assembly of data and data analysis and interpretation; K.S., collection and assembly of data; D.G.D.E.R., data analysis and interpretation; and R.N.F., conception and design, manuscript writing, and financial support.
REFERENCES
- 1.De Rooij DG. 1998. Stem cells in the testis. Int J Exp Pathol 79:67–80. doi: 10.1046/j.1365-2613.1998.00057.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Russell LD, Hikim APS, Ettlin RA, Clegg ED. 1990. Histological and histopathological evaluation of the testis. Cache River Press, Clearwater, FL. [Google Scholar]
- 3.De Rooij DG, Russell LD. 2000. All you wanted to know about spermatogonia but were afraid to ask. J Androl 21:776–798. [PubMed] [Google Scholar]
- 4.Handel MA, Schimenti JC. 2010. Genetics of mammalian meiosis: regulation, dynamics and impact on fertility. Nat Rev Genet 11:124–136. doi: 10.1038/nrg2723. [DOI] [PubMed] [Google Scholar]
- 5.O’Donnell L, Nicholls PK, O'Bryan MK, McLachlan RI, Stanton PG. 2011. Spermiation: the process of sperm release. Spermatogenesis 1:14–35. doi: 10.4161/spmg.1.1.14525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Levine M, Cattoglio C, Tjian R. 2014. Looping back to leap forward: transcription enters a new era. Cell 157:13–25. doi: 10.1016/j.cell.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taatjes DJ, Marr MT, Tjian R. 2004. Regulatory diversity among metazoan co-activator complexes. Nat Rev Mol Cell Biol 5:403–410. doi: 10.1038/nrm1369. [DOI] [PubMed] [Google Scholar]
- 8.Thomas MC, Chiang CM. 2006. The general transcription machinery and general cofactors. Crit Rev Biochem Mol Biol 41:105–178. doi: 10.1080/10409230600648736. [DOI] [PubMed] [Google Scholar]
- 9.Freiman RN. 2009. Specific variants of general transcription factors regulate germ cell development in diverse organisms. Biochim Biophys Acta 1789:161–166. doi: 10.1016/j.bbagrm.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grive KJ, Seymour KA, Mehta R, Freiman RN. 2014. TAF4b promotes mouse primordial follicle assembly and oocyte survival. Dev Biol 392:42–51. doi: 10.1016/j.ydbio.2014.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grive KJ, Freiman RN. 2015. The developmental origins of the mammalian ovarian reserve. Development 142:2554–2563. doi: 10.1242/dev.125211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lovasco LA, Gustafson EA, Seymour KA, de Rooij DG, Freiman RN. 2015. TAF4b is required for mouse spermatogonial stem cell development. Stem Cells 33:1267–1276. doi: 10.1002/stem.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lovasco LA, Seymour KA, Zafra K, O’Brien CW, Schorl C, Freiman RN. 2010. Accelerated ovarian aging in the absence of the transcription regulator TAF4B in mice. Biol Reprod 82:23–34. doi: 10.1095/biolreprod.109.077495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tora L. 2002. A unified nomenclature for TATA box binding protein (TBP)-associated factors (TAFs) involved in RNA polymerase II transcription. Genes Dev 16:673–675. doi: 10.1101/gad.976402. [DOI] [PubMed] [Google Scholar]
- 15.Werten S, Mitschler A, Romier C, Gangloff YG, Thuault S, Davidson I, Moras D. 2002. Crystal structure of a subcomplex of human transcription factor TFIID formed by TATA binding protein-associated factors hTAF4 (hTAFII135) and hTAF12 (hTAFII20). J Biol Chem 277:45502–45509. doi: 10.1074/jbc.M206587200. [DOI] [PubMed] [Google Scholar]
- 16.Shao H, Revach M, Moshonov S, Tzuman Y, Gazit K, Albeck S, Unger T, Dikstein R. 2005. Core promoter binding by histone-like TAF complexes. Mol Cell Biol 25:206–219. doi: 10.1128/MCB.25.1.206-219.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hamard PJ, Dalbies-Tran R, Hauss C, Davidson I, Kedinger C, Chatton B. 2005. A functional interaction between ATF7 and TAF12 that is modulated by TAF4. Oncogene 24:3472–3483. doi: 10.1038/sj.onc.1208565. [DOI] [PubMed] [Google Scholar]
- 18.Gazit K, Moshonov S, Elfakess R, Sharon M, Mengus G, Davidson I, Dikstein R. 2009. TAF4/4b·TAF12 displays a unique mode of DNA binding and is required for core promoter function of a subset of genes. J Biol Chem 284:26286–26296. doi: 10.1074/jbc.M109.011486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wei Y, Liu S, Lausen J, Woodrell C, Cho S, Biris N, Kobayashi N, Wei Y, Yokoyama S, Werner MH. 2007. A TAF4-homology domain from the corepressor ETO is a docking platform for positive and negative regulators of transcription. Nat Struct Mol Biol 14:653–661. doi: 10.1038/nsmb1258. [DOI] [PubMed] [Google Scholar]
- 20.Gill G, Pascal E, Tseng ZH, Tjian R. 1994. A glutamine-rich hydrophobic patch in transcription factor Sp1 contacts the dTAFII110 component of the Drosophila TFIID complex and mediates transcriptional activation. Proc Natl Acad Sci U S A 91:192–196. doi: 10.1073/pnas.91.1.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Felinski EA, Quinn PG. 1999. The CREB constitutive activation domain interacts with TATA-binding protein-associated factor 110 (TAF110) through specific hydrophobic residues in one of the three subdomains required for both activation and TAF110 binding. J Biol Chem 274:11672–11678. doi: 10.1074/jbc.274.17.11672. [DOI] [PubMed] [Google Scholar]
- 22.Chen GY, Muramatsu H, Ichihara-Tanaka K, Muramatsu T. 2004. ZEC, a zinc finger protein with novel binding specificity and transcription regulatory activity. Gene 340:71–81. doi: 10.1016/j.gene.2004.06.016. [DOI] [PubMed] [Google Scholar]
- 23.Li VC, Davis JC, Lenkov K, Bolival B, Fuller MT, Petrov DA. 2009. Molecular evolution of the testis TAFs of Drosophila. Mol Biol Evol 26:1103–1116. doi: 10.1093/molbev/msp030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Broughton RE, Betancur RR, Li C, Arratia G, Orti G. 2013. Multi-locus phylogenetic analysis reveals the pattern and tempo of bony fish evolution. PLoS Curr 5:ecurrents.tol.2ca8041495ffafd0c92756e75247483e. doi: 10.1371/currents.tol.2ca8041495ffafd0c92756e75247483e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klug A. 2010. The discovery of zinc fingers and their applications in gene regulation and genome manipulation. Annu Rev Biochem 79:213–231. doi: 10.1146/annurev-biochem-010909-095056. [DOI] [PubMed] [Google Scholar]
- 26.Gagnon JA, Valen E, Thyme SB, Huang P, Akhmetova L, Ahkmetova L, Pauli A, Montague TG, Zimmerman S, Richter C, Schier AF. 2014. Efficient mutagenesis by Cas9 protein-mediated oligonucleotide insertion and large-scale assessment of single-guide RNAs. PLoS One 9:e98186. doi: 10.1371/journal.pone.0098186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nantel F, Sassone-Corsi P. 1996. CREM: a transcriptional master switch during the spermatogenesis differentiation program. Front Biosci 1:d266–d269. doi: 10.2741/a131. [DOI] [PubMed] [Google Scholar]
- 28.Jan SZ, Hamer G, Repping S, de Rooij DG, van Pelt AM, Vormer TL. 2012. Molecular control of rodent spermatogenesis. Biochim Biophys Acta 1822:1838–1850. doi: 10.1016/j.bbadis.2012.02.008. [DOI] [PubMed] [Google Scholar]
- 29.Meistrich ML, Mohapatra B, Shirley CR, Zhao M. 2003. Roles of transition nuclear proteins in spermiogenesis. Chromosoma 111:483–488. doi: 10.1007/s00412-002-0227-z. [DOI] [PubMed] [Google Scholar]
- 30.Rathke C, Baarends WM, Awe S, Renkawitz-Pohl R. 2014. Chromatin dynamics during spermiogenesis. Biochim Biophys Acta 1839:155–168. doi: 10.1016/j.bbagrm.2013.08.004. [DOI] [PubMed] [Google Scholar]
- 31.Bao J, Bedford MT. 2016. Epigenetic regulation of the histone-to-protamine transition during spermiogenesis. Reproduction 151:R55–R70. doi: 10.1530/REP-15-0562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Braun RE. 2000. Temporal control of protein synthesis during spermatogenesis. Int J Androl 23 (Suppl 2):92–94. doi: 10.1046/j.1365-2605.2000.00027.x. [DOI] [PubMed] [Google Scholar]
- 33.Zhang D, Penttila TL, Morris PL, Teichmann M, Roeder RG. 2001. Spermiogenesis deficiency in mice lacking the Trf2 gene. Science 292:1153–1155. doi: 10.1126/science.1059188. [DOI] [PubMed] [Google Scholar]
- 34.Zhou H, Grubisic I, Zheng K, He Y, Wang PJ, Kaplan T, Tjian R. 2013. Taf7l cooperates with Trf2 to regulate spermiogenesis. Proc Natl Acad Sci U S A 110:16886–16891. doi: 10.1073/pnas.1317034110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmidt EE, Schibler U. 1995. High accumulation of components of the RNA polymerase II transcription machinery in rodent spermatids. Development 121:2373–2383. [DOI] [PubMed] [Google Scholar]
- 36.Schmidt EE, Ohbayashi T, Makino Y, Tamura T, Schibler U. 1997. Spermatid-specific overexpression of the TATA-binding protein gene involves recruitment of two potent testis-specific promoters. J Biol Chem 272:5326–5334. doi: 10.1074/jbc.272.8.5326. [DOI] [PubMed] [Google Scholar]
- 37.Liu D, Matzuk MM, Sung WK, Guo Q, Wang P, Wolgemuth DJ. 1998. Cyclin A1 is required for meiosis in the male mouse. Nat Genet 20:377–380. doi: 10.1038/3855. [DOI] [PubMed] [Google Scholar]
- 38.Wu Y, Hu X, Li Z, Wang M, Li S, Wang X, Lin X, Liao S, Zhang Z, Feng X, Wang S, Cui X, Wang Y, Gao F, Hess RA, Han C. 2016. Transcription factor RFX2 is a key regulator of mouse spermiogenesis. Sci Rep 6:20435. doi: 10.1038/srep20435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Deng W, Lin H. 2002. miwi, a murine homolog of piwi, encodes a cytoplasmic protein essential for spermatogenesis. Dev Cell 2:819–830. doi: 10.1016/S1534-5807(02)00165-X. [DOI] [PubMed] [Google Scholar]
- 40.Zheng K, Wang PJ. 2012. Blockade of pachytene piRNA biogenesis reveals a novel requirement for maintaining post-meiotic germline genome integrity. PLoS Genet 8:e1003038. doi: 10.1371/journal.pgen.1003038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weick EM, Miska EA. 2014. piRNAs: from biogenesis to function. Development 141:3458–3471. doi: 10.1242/dev.094037. [DOI] [PubMed] [Google Scholar]
- 42.Lardenois A, Chalmel F, Demougin P, Kotaja N, Sassone-Corsi P, Primig M. 2009. Fhl5/Act, a CREM-binding transcriptional activator required for normal sperm maturation and morphology, is not essential for testicular gene expression. Reprod Biol Endocrinol 7:133. doi: 10.1186/1477-7827-7-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gustafson EA, Yajima M, Juliano CE, Wessel GM. 2011. Post-translational regulation by gustavus contributes to selective Vasa protein accumulation in multipotent cells during embryogenesis. Dev Biol 349:440–450. doi: 10.1016/j.ydbio.2010.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Finn RD, Mistry J, Schuster-Bockler B, Griffiths-Jones S, Hollich V, Lassmann T, Moxon S, Marshall M, Khanna A, Durbin R, Eddy SR, Sonnhammer EL, Bateman A. 2006. Pfam: clans, web tools and services. Nucleic Acids Res 34:D247–D251. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Edgar RC. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 47.Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 48.Chevenet F, Brun C, Banuls AL, Jacq B, Christen R. 2006. TreeDyn: towards dynamic graphics and annotations for analyses of trees. BMC Bioinformatics 7:439. doi: 10.1186/1471-2105-7-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, Claverie JM, Gascuel O. 2008. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res 36:W465–9. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang X, Spandidos A, Wang H, Seed B. 2012. PrimerBank: a PCR primer database for quantitative gene expression analysis, 2012 update. Nucleic Acids Res 40:D1144–D1149. doi: 10.1093/nar/gkr1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Spandidos A, Wang X, Wang H, Dragnev S, Thurber T, Seed B. 2008. A comprehensive collection of experimentally validated primers for polymerase chain reaction quantitation of murine transcript abundance. BMC Genomics 9:633. doi: 10.1186/1471-2164-9-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang X, Seed B. 2003. A PCR primer bank for quantitative gene expression analysis. Nucleic Acids Res 31:e154. doi: 10.1093/nar/gng154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Grive KJ, Gustafson EA, Seymour KA, Baddoo M, Schorl C, Golnoski K, Rajkovic A, Brodsky AS, Freiman RN. 2016. TAF4b regulates oocyte-specific genes essential for meiosis. PLoS Genet 12:e1006128. doi: 10.1371/journal.pgen.1006128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Eklund A. 2015. The Bee Swarm plot, an alternative to stripchart. R package version 0.2.1. http://www.cbs.dtu.dk/~eklund/beeswarm/.