Peroxisome proliferator-activated receptor γ (PPARγ) is a nuclear receptor essential for adipocyte development and the maintenance of the alternatively polarized macrophage phenotype. Biochemical studies have established that as an obligate heterodimer with retinoid X receptor (RXR), PPARγ binds directly repeated nuclear receptor half sites spaced by one nucleotide (direct repeat 1 [DR1]). However, it has not been analyzed systematically and genome-wide how cis factors such as the sequences of DR1s and adjacent sequences and trans factors such as cobinding lineage-determining transcription factors (LDTFs) contribute to the direct binding of PPARγ in different cellular contexts.
KEYWORDS: PPARγ, RXR, adipocytes, direct repeat 1, lineage-determining factors, macrophages, nuclear receptors
ABSTRACT
Peroxisome proliferator-activated receptor γ (PPARγ) is a nuclear receptor essential for adipocyte development and the maintenance of the alternatively polarized macrophage phenotype. Biochemical studies have established that as an obligate heterodimer with retinoid X receptor (RXR), PPARγ binds directly repeated nuclear receptor half sites spaced by one nucleotide (direct repeat 1 [DR1]). However, it has not been analyzed systematically and genome-wide how cis factors such as the sequences of DR1s and adjacent sequences and trans factors such as cobinding lineage-determining transcription factors (LDTFs) contribute to the direct binding of PPARγ in different cellular contexts. We developed a novel motif optimization approach using sequence composition and chromatin immunoprecipitation with high-throughput sequencing (ChIP-seq) densities from macrophages and adipocytes to complement de novo motif enrichment analysis and to define and classify high-affinity binding sites. We found that approximately half of the PPARγ cistrome represents direct DNA binding; both half sites can be extended upstream, and these are typically not of equal strength within a DR1. Strategically positioned LDTFs have greater impact on PPARγ binding than the quality of DR1, and the presence of the extension of DR1 provides a remarkable synergy with LDTFs. This approach of considering not only nucleotide frequencies but also their contribution to protein binding in a cellular context is applicable to other transcription factors.
INTRODUCTION
Nuclear receptors (NRs), in principle, work as lipid sensor-linked transcription factors (TFs). Most of them form homo- or heterodimers and recognize the direct or inverted repeat (DR or IR) of their common recognition sequence (5′-AGGTCA-3′), the so-called NR half site (1). Class II and class III NRs bind DR elements in which the different lengths of the spacer between the adjacent NR half sites determine which dimer can interact with a given element. Depending on spacer length, DR elements are called DR1, DR2, and DR3, etc. (2, 3). Class II NRs such as the peroxisome proliferator (PP)-activated receptors (PPARs) form heterodimers with the retinoid X receptors (RXRs), while class III “dimeric orphan” receptors, including RXRs, generally form homodimers. However, there is an additional level of specificity with respect to NR binding sites, which is provided by the 5′ extensions of the NR half sites.
During the course of identification of the first PP response elements (PPREs), primarily by transfection-based reporter assays, the importance of any sequences beyond DR1 was not revealed (4–7). Subsequently, based on close to 30 PPREs, a 5′-(A)AACT-3′ sequence directly upstream of DR1s was identified (8, 9). These sporadically identified extended DR1s proved later to be the strongest PPREs in biochemical assays (7, 10, 11). More recently, the importance of the 5′ extension was also confirmed by biotin-labeled double-stranded DNA pulldown assays (12) and microfluidics-based dimer-DNA interaction assays (13). These methods suggested that the length of the 5′ extension is 5 nucleotides. Accordingly, the crystal structure of the PPARγ/RXRα heterodimer was also determined on the 5′-AAACTAGGTCAAAGGTCA-3′ consensus sequence (boldface nucleotides highlight the 5' extension/spacer of NR [AGGTCA] half sites) (14). Based on the obtained structure, the DNA-binding domain (DBD) of the PPARγ sequence specifically interacts with the upstream half site, and the carboxy-terminal extension (CTE) of the DBD directly interacts with the 5′ extension within the minor groove. Similarly, the CTE of RXR is also located within the minor groove, and thus the heterodimer wraps around the DNA in the middle of the PPREs (14).
However, all these analyses were carried out either on very few response elements or in highly artificial model systems, often using nonchromatinized DNA, including transient transfections and irrelevant cell types or in vitro (8, 9, 12, 13). In contrast, chromatin immunoprecipitation (ChIP) with high-throughput sequencing (ChIP-seq) allowed the genome-wide identification of thousands of binding sites from live cells, facilitating the better characterization of PPARγ-chromatin interactions (15–17). Utilizing the knowledge of mammalian genomes and cistromic (ChIP-seq) results, it became clear that there are millions of potential binding sites throughout the genome (e.g., one per kilobase), but only a fraction of these are occupied and functional within a given cell at a given point in time and even fewer represent high-affinity binding sites. A key question is what supports cell-type-specific binding and activity at these sites. In order to answer this, one needs to consider the contribution of context-relevant TFs, such as lineage-determining transcription factors (LDTFs).
Interestingly, PPARγ is an LDTF of both white and brown adipocytes, and its cistromes in the two cell types overlap significantly (15). The other LDTF of adipocytes with which PPARγ works closely, for example, to synergistically activate metabolic genes is CCAAT/enhancer-binding protein alpha (C/EBPα) (18). In the PPARγ cistrome of adipocytes, these two TFs show the highest motif enrichment. In contrast to previous results and assumptions drawn based on reporter assays and high-throughput DNA-protein interaction assays, a conserved, up to 4-nucleotide-long 5′ extension of PPARγ binding sites could be observed in both human and mouse cells by using ChIP-seq-based motif enrichment analyses (18–20). However, the approaches resulting in position weight matrices (PWMs) of the motifs also have major technical limitations. Per-position nucleotide frequencies cannot describe the substantial variety of motifs and are unable to reflect the significance of each nucleotide in DNA-protein interactions, especially the importance of di- or trinucleotides within response elements (21).
We have been studying PPARγ in macrophages, a cell type in which PPARγ does not have a lineage-determining role per se. However, its cistrome is very dynamic, and its expression largely increases during alternative macrophage polarization upon interleukin-4 (IL-4) stimulation in both human and mouse models (17, 22, 23). IL-4 treatment of mouse bone marrow-derived macrophages (BMDMs) results in a significant extension of the PPARγ/RXR cistrome, which is coupled with the commissioning of thousands of previously unoccupied binding sites (17, 24). Although more than half of the PPARγ/RXR cistrome shows some activity, e.g., enhancer transcription, only a small fraction of these sites shows responsiveness to the synthetic PPARγ ligand rosiglitazone (RSG). Importantly, the composition of PPREs differs at PPARγ ligand-responsive and nonresponsive sites. Ligand-responsive DR1s are significantly more frequently extended, and, in addition, higher PPARγ ChIP-seq densities can be observed at these sites (17, 24).
Despite a significant amount of work on the contribution of cis- and trans-acting factors to the nature of PPREs, there is no clear understanding of the significance, exact frequency, required length, composition, or variants of the 5′ and potential 3′ extensions of PPREs in vivo and genome wide. Furthermore, experiments testing sequence requirements of PPAR/RXR heterodimers could not explain how collaborating LDTFs contribute to NR binding, although one can surmise their pivotal roles in this aspect as well.
In order to unravel the contributing molecular interactions within a hierarchy of factors that determine chromatinized DNA binding by PPARγ, we characterized the composition and extension of PPREs of alternatively polarized macrophages and adipocytes and then examined their contribution to DNA/protein contacts in combination with the presence of cell-type-specific LDTFs, PU.1 and C/EBP, respectively. Although we did not systematically connect PPARγ binding events with gene regulation in this work, for simplicity, we refer to PPARγ-specific binding sites as PPREs throughout this paper. We found that the greatest impact on PPARγ binding is exerted by LDTFs, and the second most important contributor is a PPRE containing an exactly 4-nucleotide-long (5′-AACT-3′) 5′ extension. Importantly, the presence of both of these factors had a synergistic effect on PPARγ binding. Furthermore, we observed that the half sites of most PPREs had different, uneven strengths and that the half site of RXR could also be extended. Finally, we uncovered a general shift of LDTF binding upstream to high-affinity PPREs and a so-far unidentified C/EBP:DR1 composite element.
RESULTS
The macrophage PPARγ cistrome is partially independent from the RXR cistrome and highly enriched for PU.1 and AP-1 motifs.
We determined the PPARγ and RXR cistromes of alternatively polarized macrophages exposed to IL-4 for 24 h using ChIP-seq (Fig. 1A, left). Approximately 44,000 PPARγ binding sites could be predicted in the two biological replicates, each by using MACS2 (25), and ∼73% of these overlapped (see Fig. S1A in the supplemental material). This overlap contained 32,180 binding sites, which we termed the “consensus set.” The nonoverlapping peaks showed significantly lower ChIP-seq densities measured by HOMER (26) than the top peaks but displayed motif enrichments similar to those of the top peaks (Fig. S1B and C). Regarding the high quality of these ChIP-seq data (the IP efficiency was over 19 for both replicates according to HOMER), we used the broader range of binding strengths, with any detectable peaks also considered to be part of the PPARγ cistrome. This had the following several advantages to support statistical analyses: (i) we could map a higher number of putative DR1 elements, (ii) these DR1 elements included extremely strong and weak ones, (iii) together, the sequences and (nonbasal) ChIP-seq densities of these DR1 elements could be used to determine their contribution to DNA-protein interactions, and (iv) by using the binding density of other factors, the significance of the presence and collaboration of the DR1 elements could also be better assessed.
FIG 1.
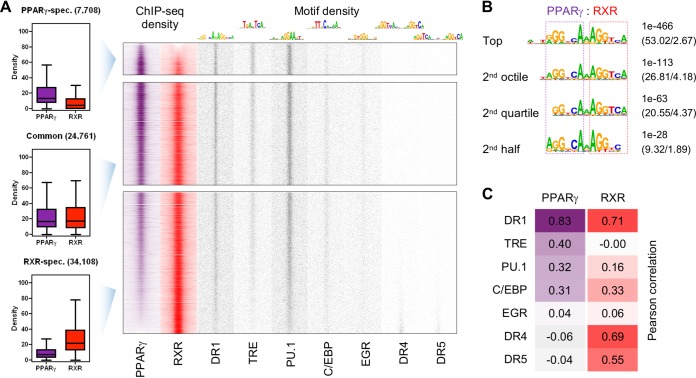
Relationship of the PPARγ and RXR cistromes and enriched motifs in alternatively polarized macrophages. (A) Genome-wide representation of PPARγ (purple) and RXR (red) ChIP-seq densities and motif enrichments (gray) at the PPARγ-specific (n = 7,708), common (n = 24,761), and RXR-specific (n = 34,108) sites. Genomic regions were ordered based on the ratio of PPARγ and RXR ChIP-seq densities and separated at a 1.5-fold difference. Read distribution heat maps are depicted on 1.5-kb regions in 30-bp resolution, relative to the summit position of primarily PPARγ, or RXR, peaks. Motif distribution heat maps represent the densities of the indicated motifs (top), depicted in 10-bp resolution (GEO accession no. GSE115083) (17, 24). (B) De novo DR1 motifs enriched in the top 1,000 peaks of the entire set and the 2nd octile, the 2nd quartile, and the bottom half of the consensus PPARγ peak set. P values and target/background percentages are indicated, and PPAR and RXR half sites are highlighted with purple and red dashed lines, respectively. (C) Correlation between PPARγ (purple) or RXR (red) enrichment and motif density. Genomic regions ranked by PPARγ or RXR ChIP-seq density were split into 1,000 bins (n = 66). The average ChIP-seq density and the cumulative number of the indicated motifs were determined for each bin. Pearson correlation coefficients were calculated by using the per-bin values.
In the next step, we compared the PPARγ and RXR cistromes. In order to make PPARγ and RXR signals comparable, they were normalized based on the median ChIP-seq density of the overlapping fraction of the two cistromes (Fig. 1A, left). We then clustered them into three sets of binding sites: fewer than 12% of the sites showed stronger PPARγ than RXR ChIP-seq density (Fig. 1A, top), ∼37% showed similar (<1.5-fold difference) enrichment for the two proteins (Fig. 1A, middle), and more than half of the sites were more enriched for RXR (Fig. 1A, bottom). These data suggest that the relationship between the two components of the heterodimer is complex even if one considers that RXR can bind other receptors.
We then aimed to map the direct binding events by PPARγ, in order to cover as many PPREs as possible. The DR1 motif derived from the top 1,000 PPARγ peaks covered more than half of this peak set (Fig. S1C) and 18.8% of the entire cistrome. Assuming that the larger the ChIP-seq density the higher the motif specificity, we looked for additional de novo motifs of different compositions (Fig. 1B). For this, we used the top 1,000 PPARγ binding sites of the second octile, second quartile, and bottom half of the consensus peak set. Mapping the position weight matrices (PWMs) of the obtained three DR1 motifs together with that derived from the highest peaks showed a 71% higher DR1 enrichment correlating with PPARγ binding (Fig. 1A, right) and covering close to a third of the cistrome. This means that this portion of the cistrome is almost certainly the result of direct DNA binding. Of note, genome wide, we identified 1.73 million putative DR1s by these four PWMs. Cellular and molecular mechanisms are required to select binding sites mostly from this pool (26).
The additional de novo motifs were determined around the summit of the 1,000 highest peaks that lacked putative DR1s and were dominated by PPARγ or RXR or showed similar protein binding (Fig. 1A, right). 12-O-Tetradecanoylphorbol-13-acetate (TPA) response element (TRE), which is specific for activator protein 1 (AP-1) heterodimers, purine-rich (PU-box or PU.1) response element, and early growth response (EGR) motifs appeared to be specific for PPARγ binding, while C/EBP motifs were enriched at those sites with similar PPARγ and RXR ChIP-seq densities. Sites with frequent RXR binding and no PPARγ gave very specific DR4 and DR5 motif enrichments, showing that other heterodimerizing partners, probably liver X receptor (LXR) and retinoic acid receptor (RAR), respectively, are also readily detectable in alternatively polarized macrophages (Fig. 1A).
In order to verify the trends observed in motif distribution, we applied a correlation analysis (Fig. 1C). The entire PPARγ/RXR cistrome was ranked by ChIP-seq density and split into 1,000 bins (n = 66 for each), and the cumulative numbers of putative elements and average ChIP-seq densities per bin were compared. As expected, PPARγ binding was highly dependent on DR1s, and RXR enrichment showed correlation with the presence of all three DR motifs tested. In contrast to PPARγ binding, low or no correlation could be observed between RXR binding and PU.1 or TRE motif enrichments, respectively (Fig. 1C). Although we had only indirect evidence for the collaboration of PPARγ/RXR with macrophage-specific factors, these results suggested that PPARγ binding correlated with apparent PU.1 and AP-1 binding and with RXR binding.
Notably, in the case of the most robust ChIP-seq peaks, we observed two noteworthy phenomena: (i) there was a bias toward the presence of an upstream 5′-(A/T)NT-3′ sequence extension, and (ii) the first nucleotide of the PPAR half site was preferentially G instead of A (Fig. 1B). In order to sort out the contributions of the different variants and the significance of the different parts of PPREs, we applied novel motif composition analyses coupled with PPARγ binding data.
High-affinity PPREs are extended by 4 nucleotides upstream.
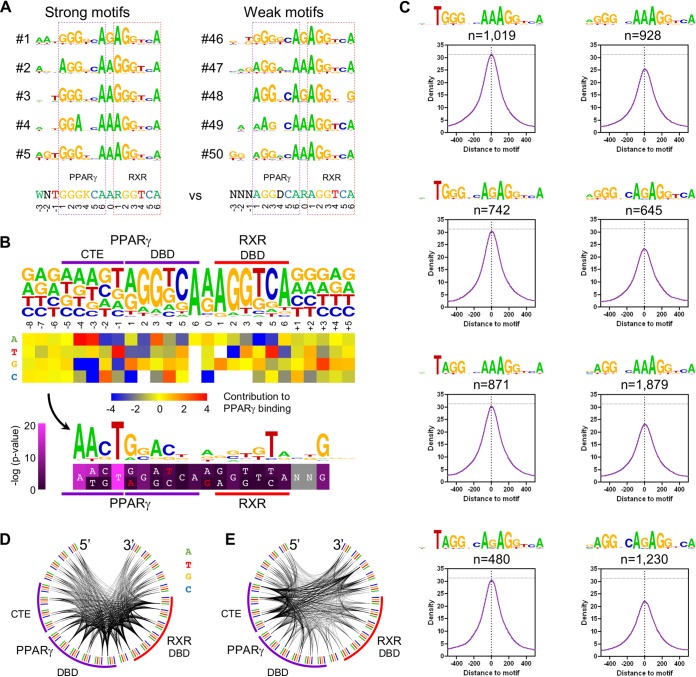
We determined ∼20,000 putative DR1s within 200 nucleotides around PPARγ peak summits by using the four PWMs described above (Fig. 1B). Regions with two or three DR1 motifs showed significantly higher PPARγ ChIP-seq densities than those with one or multiple sites, so to avoid additive effects due to nearby PPREs, only single DR1 binding events (n = 14,924) were included in the downstream analyses (Fig. S2A). In order to discriminate PPREs based on their nucleotide composition, we applied k-means clustering (k = 50) based on the sequence of mapped motifs (Fig. S2B). The composition of the 5′ half site was fairly diverse, with the dominance of 5′-(A/G)GGNCA-3′ sequences: the spacer was A or G (R), and the RXR-bound part showed a high frequency of the canonical 5′-AGGTCA-3′ half sites (Fig. S2B and C). In the 5′ extension, A or W (A/T) at position −3, G or S (C/G) at position −2, and T or W at position −1 showed enrichment, and out of all possible combinations, 5′-ANT-3′ sequences showed the highest frequency. A rank order of motif clusters was established according to the upper quartile of PPARγ coverage values, as indicated in Fig. S2B. Comparing the top and bottom five motif clusters, we found that an initial 5′-GGG-3′ trinucleotide of the PPAR half site was dominant at the highest peaks and were not typical at the smallest ones; in line with the motif enrichment analysis, directly upstream of the top sequences, at least a single A (−3) or T (−1) was enriched (Fig. 2A; Fig. S2B). These results were consistent with the de novo motif hits (Fig. 1B) and also the newest protein-binding microarray results (27) but were in contrast to results for the classical PPREs.
FIG 2.
Identification of the extensions of PPREs. (A) The top and bottom 5 DR1 motifs, determined by k-means clustering (i = 50) of the single DR1 sequences per-PPARγ binding site (n = 14,924) and ranked by per-cluster PPARγ ChIP-seq density. Consensus sequences are included (bottom), and the PPAR and RXR half sites are highlighted with purple and red dashed lines, respectively. (B) The process of nucleotide contribution analysis. Nucleotide frequencies of the single DR1s are depicted (top). The heat map represents the average PPARγ ChIP-seq density of all DR1s harboring the indicated nucleotide (y axis) at the indicated position (x axis), relative to the average PPARγ ChIP-seq density of all single DR1s (middle). The obtained “contribution” values were transformed to a motif logo highlighting those nucleotides improving PPREs. The heat map represents the P values determined between the ChIP-seq densities of those sequences having or lacking the indicated, preferred or canonical, nucleotides (unpaired t test). The upper line represents the higher −log (P value). White characters indicate the improving nucleotides, and red characters indicate the ones being impaired (bottom). Purple and red lines represent the PPAR and RXR binding sites, respectively. (C) Contribution of single nucleotides to direct PPRE binding. Read distribution histograms represent the average PPARγ ChIP-seq coverage of the indicated motifs, depicted in 10-bp resolution. (D and E) Frequency of nucleotide pairs of the single DR1s (D) or the contribution of these nucleotide pairs to PPARγ binding (E), depicted as Circos plots. The color-coded outer arc represents the nucleotides of DR1 and its flanking regions; the cooccurring nucleotides are represented by the inner arcs, whose thickness is proportional to the frequency of the involved nucleotide pairs (D) or the average PPARγ ChIP-seq density calculated on the DR1s harboring the involved nucleotide pairs (E). The purple and red arcs represent the PPARγ and RXR binding sites, respectively.
Previously, the involvement of 4- to 6-nucleotide-long regions upstream of DR1s was suggested for more extensive PPARγ binding (8, 9, 12, 13). Indeed, the initial nucleotide frequencies upstream of DR1s showed some enrichment of 5'-AAAGT-3' (5'-WRWST-3') nucleotides (Fig. 2B, top). In order to test which nucleotides may contribute to frequent PPARγ binding, we used a motif optimization approach, termed nucleotide contribution analysis. For this, we calculated the average PPARγ ChIP-seq density of each DR1 harboring a given nucleotide at a given position (Fig. S2D) and compared these to the average ChIP-seq density of all regions (Fig. 2B, middle). We then translated nucleotide contributions to a motif logo, highlighting nucleotides that had the greatest impact on PPARγ binding, and calculated the P values between the ChIP-seq densities of those sequences having or lacking the preferred or canonical nucleotides (Fig. 2B, bottom; red characters indicate nucleotides being impaired). In addition to the previously found 5′-AST-3′ trinucleotide sequence, an upstream A appeared. Furthermore, nucleotides within the NR half sites also showed some preferences. G, A, and C at positions 1, 3, and 4, respectively, of the upstream half site and T at position 5 of the downstream half site significantly facilitated PPARγ binding. Significantly, within the downstream sequences, G nucleotides showed some preference, especially at position +3.
We were interested whether the different PPAR half sites showed distinct sequence preferences within the 5′ extension, so according to the results of motif clustering, we discriminated the six most frequent variants based on their first three nucleotides (5′-AGG-3′, 5′-GGG-3′, 5′-AAG-3′, 5′-AGA-3′, 5′-GGA-3′, and 5′-GAG-3′) (Fig. S2B and C). Half sites starting with nucleotide A showed a higher frequency of G at position −2, while those starting with G cooccurred instead with C at that position. Regardless, both kinds of half sites showed the most frequent binding in the presence of the 5′-AACT-3′ extension (Fig. S2E).
Within this nucleotide tetramer, the last T was the most frequent and most effective in supporting PPARγ binding, especially in front of G nucleotides (Fig. S2E). In order to demonstrate its effect, we plotted the average PPARγ ChIP-seq densities at DR1s of different compositions and having or lacking this T (Fig. 2C). In this analysis, DR1s were used from the clusters with a 5′-RGGNSARAGGNSA-3′ consensus sequence and thus might contain mismatches at any position. Importantly, neither the first nucleotide of the 5′ half site nor the spacer nucleotide affected PPARγ ChIP-seq density significantly, while the presence of a single upstream T largely improved any of the examined groups of DR1 sequences (Fig. 2C).
By analyzing the contribution of single nucleotides to PPARγ binding frequency, we identified important ones both upstream and downstream of DR1s (Fig. 2B). Importantly, we also found that sequences beyond these sequences do not contribute to binding. In order to test nucleotide concomitances and their effect on DNA-protein interactions, we did the same analysis for cooccurring nucleotides at different positions. Within the Circos plot shown in Fig. 2D, each nucleotide at each position of the DR1 and its flanking sequences is represented on the outer arc; the inner arcs represent the linked nucleotide pairs, and the thickness of these arcs is proportional to the frequency of nucleotide pairs. Although we could not discriminate outstanding nucleotide concomitances, pairwise nucleotide frequencies clearly separated the extended PPREs from the neighboring random sequences—one (or two) thick arc(s) versus four thin arcs arriving at a position, respectively (Fig. 2D). Testing pairwise nucleotide contributions to PPARγ recruitment and visualizing this also as a Circos plot, where thickness is proportional to the contribution of a given pair of nucleotides, a strong preference of the 5′ extension and its cooccurrence (positions −4, −3, and −1) with the downstream G at position +3 are outlined (Fig. 2E). Furthermore, several important pairs within the PPARγ binding site are clearly visible. For instance, A at position −4 together with A at position 3 led to very frequent PPARγ binding, which might indicate even a specific conformation of PPARγ on the DNA.
Since this analysis reinforced that the highest-affinity PPAR half sites started with 5'-GGA-3' nucleotides, we compared PPARγ ChIP-seq densities of the top 6 variants of PPREs (Fig. S2F). This analysis indeed resulted in the finding that G instead of A at position 1 and A instead of G at position 3 improved DR1s, while A instead of G at position 2 impaired the motif. However, median and low PPARγ ChIP-seq densities were essentially unaffected by the composition of DR1s (Fig. S2F).
The LDTF PU.1 synergistically facilitates PPARγ binding at extended PPREs.
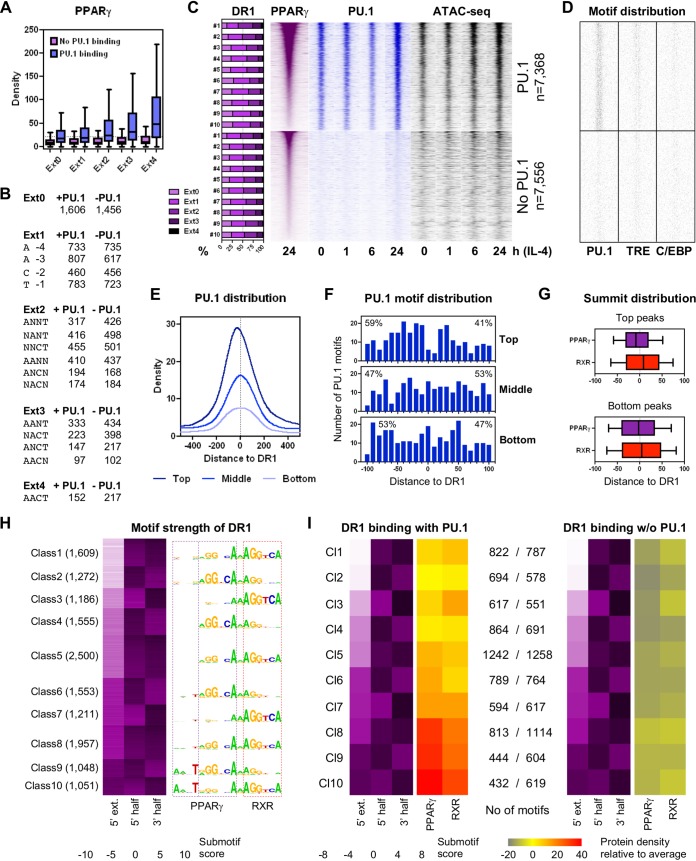
In order to uncover the reasons for the scattered distribution of PPARγ ChIP-seq densities observed even at elements with similar strengths, we utilized PU.1 ChIP-seq data (Fig. 3A to C). We classified PPREs by the ratio of the present CTE-specific nucleotides (Ext0 to Ext4) and the presence or absence of PU.1 binding; this resulted in a striking PPARγ peak size distribution (Fig. 3A and B). In the absence of PU.1, the existence of the perfect 5′ extension had less than a 1.5-fold effect on PPARγ ChIP-seq density, while in the presence of PU.1, this ratio was almost 3-fold (Fig. 3A). The presence of PU.1 caused an ∼2- to ∼5-fold enrichment of PPARγ occupancy, depending on the strength of PPRE extension, and the existence of the extension and the presence of PU.1 together caused the highest, ∼6-fold difference between the extreme binding sites. The enrichment of PPARγ led to PU.1 enrichment at these sites. The higher the ratio of strong DR1s, the higher the PU.1 ChIP-seq density and chromatin openness (detected by assay for transposase-accessible chromatin with high-throughput sequencing, ATAC-seq [28]) could be reached (Fig. 3C), but only in the presence of a greater number of PU.1 motifs (Fig. 3D). In the absence of PU.1, only a small fraction of binding sites showed high PPARγ ChIP-seq densities, and this was coupled with elevated but still moderate chromatin openness (Fig. 3C, bottom). This relatively high PPARγ ChIP-seq density might be explained by the higher ratio of strong DR1 extensions, because no other motifs were enriched at these sites (Fig. 3C and D). During the time course of IL-4 treatment, we observed that the mostly steady-state PU.1-bound and accessible chromatin regions recruited TFs, while those PPREs lacking the pioneer factor PU.1 were typically not efficient in recruiting further TFs and opening chromatin.
FIG 3.
Effects of PPRE extensions on the collaborative binding of PPARγ, RXR, and PU.1. (A) Dependence of PPARγ binding on the presence (blue) or absence (purple) of PU.1 and the strength of the 5′ extension (Ext0 to Ext4). Extensions were discriminated by the number of nucleotides matching the 5′-AACT-3′ sequence. (B) Number of the DR1s with different 5′ extensions in the presence or absence of PU.1. (C) Ratio of the 5′ extensions of different strengths (Ext0 to Ext4) relative to the PPARγ enrichment in the presence (n = 7,368) or absence (n = 7,556) of PU.1. Bars represent the percentages of different DR1s (Ext0 to Ext4) in 2 × 10 deciles (n = 736 and 755, respectively) (left). Read distribution heat maps showing PPARγ (purple) and PU.1 ChIP-seq (blue) and ATAC-seq (black) densities are depicted on 1.5-kb regions in 30-bp resolution, relative to DR1s (right). Stages of the time course upon IL-4 treatment are indicated (GEO accession no. GSE115083 and GSE107456) (17, 24). (D) Distribution of the indicated motifs around DR1s in the presence or absence of PU.1, depicted on 1.5-kb regions in 10-bp resolution. (E and F) Average PU.1 ChIP-seq coverage (E) and PU.1 motif distribution (F) around the top, middle, and bottom 500 DR1s according to PPARγ ChIP-seq densities, depicted in 10-bp resolution. (G) Summit distribution of PPARγ (purple) and RXR (red) peaks relative to extended (Ext4) and nonextended (Ext0) DR1s. (H) Classification of PPREs based on the score of the indicated three submotifs (5′ extension, 5′ half, and 3′ half) by k-means clustering (k = 10). PPARγ and RXR submotifs are highlighted with purple and red dashed lines, respectively. (I) Correlation between the average submotif score and the PPARγ/RXR ChIP-seq density of the 10 motif classes in the presence (left) or absence (right) of PU.1.
By carrying out these analyses, we noticed a shift between the top PPARγ and PU.1 peaks (Fig. 3C, top), and also the PU.1 motif distributions were unequal at these sites (Fig. 3D, top). Plotting PU.1 ChIP-seq densities (Fig. 3E) and motif numbers around PPREs of the top, middle, and bottom 500 PU.1-bound regions (Fig. 3F) indeed showed a shift upstream to DR1s. Notably, both the TRE and C/EBP motif distributions were even more unequal at the top peaks (Fig. S3A), and C/EBP motifs showed a discrete enrichment directly upstream of DR1s, although their number was very low (n = 15 within 10 bp). These results indicated that not only the presence of the motifs of collaborating TFs but also their locations on DNA relative to each other are important, which might be responsible for a more effective interaction between the associating proteins.
PPARγ/RXR heterodimers bind uneven PPREs.
Upon identifying the two major factors, PU.1 binding and extended PPREs, determining robust PPARγ binding in macrophages, we aimed to find out how these lead to asymmetrical PPARγ/RXR binding (Fig. 1A). We examined the DR1 sequences first. Plotting the average PPARγ and RXR ChIP-seq densities calculated at PPREs having or lacking the entire 5′ extension in the presence of PU.1, we obtained similar results as earlier. At the top sites, RXR showed lower ChIP-seq density than PPARγ, while weak PPARγ binding was coupled with more frequent RXR recruitment on average (Fig. S3B). PPARγ and RXR peak summits showed the expected relative locations of the two NRs, although this was more apparent at sites having the 5′ extension (Fig. 3G; Fig. S3B). Next, we examined whether this phenomenon was due also to the NR half sites of different strengths. We calculated scores for three submotifs, 5′ extension, 5′ half site, and 3′ half site, for each putative PPRE and generated 10 motif classes by k-means clustering (Fig. 3H). Ordering the classes by the score of the 5′ extension resulted in a checkered pattern of the strength of NR half sites, indicating that strong half sites typically associated with weak ones (Fig. 3H, left). Generating motif logos indeed mirrored that usually there was only one complete half site within the examined sequences (Fig. 3H, right).
In order to measure the effects of submotif strength on NR binding, we assigned the average ChIP-seq densities to the average submotif scores of the classes in the presence or absence of PU.1 (Fig. 3I). Interestingly, ChIP-seq densities did not always follow the pattern of submotif strength. While the presence of strong 3′ half sites was associated with frequent RXR binding (classes 1, 3, 7, 8, and 10), in the absence of its CTE-binding site, PPARγ associated with these high-affinity RXR sites independently of the quality of its own sites (classes 1, 3, and 5 versus classes 2 and 4). In contrast, the extended PPAR half sites let PPARγ facilitate RXR binding (class 9). This analysis demonstrated that the existence of the 5′ extension is a key feature to specific DNA binding of PPARγ, even in the absence of a strong PPAR half site, as the highest-affinity DR1s carried the best CTE-binding sites and strong RXR half sites but relatively weak PPAR half sites (class 10) (Fig. 3I, left). Notably, RXR preferred 5′-AA(/G)AGGTCA-3′ sequences over DR1s in the lack of strong 5′ extension (class 3 versus classes 1 and 5), suggesting its direct, perhaps monomeric DNA binding. In the absence of PU.1, we observed similar tendencies, even though ChIP-seq density was always below average, and the effect of CTE binding was substantially milder (Fig. 3I, right).
Using the PWM of the 10 DR1 classes resulted in ∼1.62-times more (33,533) putative elements than the initial number (∼20,000) (Fig. S3C, top), and most of the newly identified sites seemed functional if we considered how much they covered the PPARγ cistrome (Fig. S3C, bottom). In this way, DR1s could be assigned to 49% of the entire cistrome and over 90% of the top peaks, suggesting that high PPARγ enrichment is difficult to imagine without its specific elements. Testing the indirectly bound part of the PPARγ cistrome, we observed very low levels of PPARγ enrichment without PU.1, but importantly, PU.1 could anchor PPARγ slightly more efficiently than its own element itself (Fig. S3D). By focusing on the top motifs according to our classes, we found that within the top 100 peaks harboring a class 10 motif, 10 could be annotated to gene promoters within 10 kb, and two of them could not be mapped with any of the initial four PWMs (Fig. S3E). Most of these genes play important roles in lipid metabolism, e.g., beta-oxidation, and the PPRE of the Acaa2 gene is one of the first described DR1s (6, 29).
In order to examine whether there is a difference between the gene expressional outcome of extended DR1s (classes 6 to 10) and that of nonextended DR1s (classes 1 to 5), we thoroughly filtered the PPARγ cistrome to get to functional enhancers. Previously, we described that enhancers show RNA polymerase activities (enhancer transcription) similar to those of their target genes, and this phenomenon may be used for annotating the enhancers to the regulated genes (17, 30). For this reason, we examined the enrichment of the elongation-specific RNA polymerase II (RNAPII-pS2) in the presence or absence of RSG, a synthetic and selective agonist of PPARγ. Our first goal was to identify those extended or nonextended DR1s that are coupled to strong PPARγ binding (top 10% of the consensus peak set) as well as ligand responsiveness (1.5-fold difference, more than 0.5 read per kilobase per million mapped reads [RPKM] in treated samples). We found approximately twice as many extended DR1s as nonextended ones meeting our strict criteria, and there was a small, nonsignificant difference in polymerase II enrichment in favor of extended DR1s (Fig. S3F). There were only 33 and 11 genes with elevated polymerase II levels that could be assigned to the previously determined extended or nonextended DR1s, respectively, within 100 kb of their transcription start sites (TSS). Of these, only 13 and 3, respectively, showed an occupancy level higher than 5 RPKM following ligand treatment (Fig. S3G). Importantly, the top genes with an extended DR1(s) showed higher levels and induction of polymerase II than those with nonextended ones.
We determined the mRNA levels of the annotated genes and found the top ligand-inducible ones already highly expressed and induced at the end of polarization (Fig. S3H), suggesting both ligand-independent and -dependent actions of PPARγ. We observed a difference between the top extended and nonextended DR1-regulated genes, as the adipocyte-specific Plin2, Ucp2, and Fabp4 genes showed much higher expression than the nuclear receptor gene Ear2 (Fig. S3H). Notably, three of four of the RSG-induced genes could be annotated to both extended and nonextended or less frequently bound DR1s, or to DR1s with low polymerase II coverage, suggesting the importance of collaboration between adjacent regulatory sites and protein-protein interactions.
PPREs show similar features in macrophages and adipocytes.
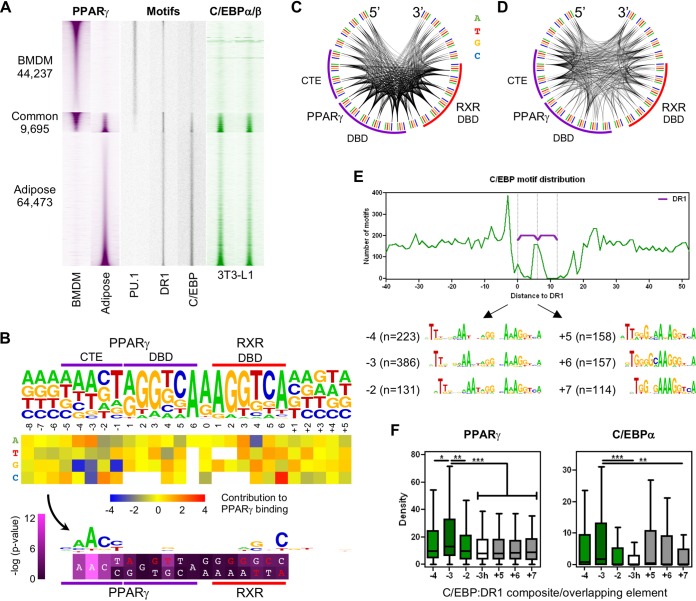
We were interested whether the same principles apply to adipocytes that also highly express PPARγ (31, 32). In adipocytes, ∼37.5% more PPARγ binding sites could be predicted than in alternatively polarized macrophages, and a relatively small fraction (∼13 and 18%, respectively) of their cistromes overlapped, although these appeared to be significant sites regarding the PPARγ ChIP-seq densities (Fig. 4A). The only motif showing high enrichment beyond DR1 was that of the binding sites of C/EBP proteins (Fig. S4A), and its distribution was similarly dense in adipocytes as the PU.1 motif distribution was in macrophages (Fig. 4A). PU.1 motifs did not show any enrichment in adipocytes, except for a fraction common to both cell types, thus showing its exact cell type specificity. In spite of the significant presence of DR1s at common sites, PPARγ followed the motif enrichment of its collaborating factors, and its ChIP-seq density showed a clear negative correlation between the two cell types (Fig. 4A). The presence of C/EBP motifs did not show major effects on macrophage PPARγ, which might be due to the lower expression level of Cebp genes. By aligning C/EBPα and C/EBPβ ChIP-seq densities of differentiated 3T3-L1 cells with the macrophage/adipocyte cistrome, we observed a high correlation with adipocyte PPARγ enrichment (Fig. 4A, right). This result allowed us to more thoroughly compare these ChIP-seq data even if they were derived from different adipocyte models.
FIG 4.
Characterization of the factors determining the adipocyte PPARγ cistrome. (A) Genome-wide representation of the PPARγ cistrome (purple) of alternatively polarized bone marrow-derived macrophages (BMDMs) and adipocytes (left), cell-type-specific motif distribution (gray, middle), and C/EBPα and C/EBPβ ChIP-seq enrichments derived from 3T3-L1 cells (green, right). Genomic regions were ordered based on the cell type specificity of PPARγ ChIP-seq densities. Read distribution heat maps are depicted on 1.5-kb regions in 30-bp resolution, relative to the summit position of primarily the macrophage, or the adipocyte, PPARγ peaks. Motif distribution heat maps represent the densities of the indicated motifs depicted in 10-bp resolution (GEO accession no. GSE74189 and GSE107456) (58). (B) Nucleotide contribution analysis of the adipocyte PPREs. Nucleotide frequencies of the single DR1 sequences per PPARγ binding site (n = 28,116) are depicted (top). The heat map represents the average PPARγ ChIP-seq density of all DR1s harboring the indicated nucleotide (y axis) at the indicated position (x axis) relative to the average PPARγ ChIP-seq density of all single DR1s (middle). The obtained “contribution” values were transformed to a motif logo highlighting those nucleotides improving PPREs. The heat map represents the P values determined between the ChIP-seq densities of those sequences having or lacking the indicated, preferred or canonical, nucleotides (unpaired t test). The upper line represents the higher −log (P value). White characters indicate the improving nucleotides, and red characters indicate the ones being impaired (bottom). The purple and red lines represent PPAR and RXR binding sites, respectively. (C and D) Frequency of nucleotide pairs of the single DR1s (C) or contribution of these nucleotide pairs to PPARγ binding (D), depicted as Circos plots. The color-coded outer arc represents the nucleotides of DR1 and its flanking regions; the cooccurring nucleotides are represented by the inner arcs, whose thickness is proportional to the frequency of the involved nucleotide pairs (C) or the average PPARγ ChIP-seq density calculated on the DR1s harboring the involved nucleotide pairs (D). The purple and red arcs represent the PPARγ and RXR binding sites, respectively. (E) The per-nucleotide frequency of C/EBP elements relative to DR1s (top), resulting in overlapping (C/EBP elements end at positions +5, +6, and +7) and C/EBP:DR1 composite elements (C/EBP elements end at positions −4, −3, and −2) (bottom). (F) PPARγ and C/EBPα ChIP-seq densities at overlapping and composite elements as defined above. A composite element containing a downstream C/EBP half site and a DR1 spaced by two spacer nucleotides is marked as -3h. Differences were considered significant at P values of <0.05 (*), <0.01 (**), and <0.001 (***) according to two-tailed unpaired t tests.
By following the same approach as that applied to macrophages, we did four de novo motif searches for the 1,000 highest and then lower and lower PPARγ peaks of adipocytes. This gave stronger PPRE motif enrichments, e.g., regarding CTE binding sites, than in the case of macrophages (Fig. 1B; Fig. S4A). Moreover, another putative PPRE (also called RAR-related orphan receptor [ROR] response element [RORE]) with a 5′-AACTGGGNSA-3′ sequence was also enriched, reinforcing the significance of the extended PPAR half sites (Fig. S4A). By using the four adipocyte-specific DR1 PWMs, ∼28,000 quasi-single DR1s out of ∼39,000 in total could be mapped, showing a proportion of direct binding events similar to that observed in macrophages. Importantly, adipocyte DR1s showed higher frequencies of nucleotides important for PPARγ binding than those of macrophages (Fig. 4B, top; Fig. S4B). Especially, nucleotide T at position −1 showed strong enrichment, but the entire extension and the following G were also more frequent in adipocytes than in macrophages.
Nucleotide contribution analysis gave similar results for adipocytes as for macrophages, but because of the “better assembly” of adipocyte DR1s, we got “smaller characters” within the resulting motif logo (Fig. 4B, bottom). Unexpectedly, a C at position −1 showed up, which was followed by DR1s starting with A, but these were not typical upstream of G nucleotides (Fig. S4C). Analysis of DR1s separated according to the first nucleotide (A or G) of the 5′ half site also showed that a 5′-AACC-3′ extension of 5′-AGGKSA-3′ half sites promotes PPARγ recruitment (Fig. S4D), but this represented a smaller fraction of PPREs (Fig. S4C). Strikingly, 75% of 5′-GGGKSA-3′ half sites followed a T (at position −1), which appeared to be the most important part of the extension or even the integral part of the first half site (Fig. S4C). Pairwise nucleotide frequencies also showed the importance of the 5′ extension, as it did not separate from the DR1 itself (Fig. 4C). In contrast, the effects of nucleotide pairs on PPARγ binding were not as pronounced as in macrophages, although the 5′-AAC(T/C)-3′ sequence was clearly outlined (Fig. 4D). Notably, we also observed some very weak, downstream sequence preferences in adipocytes that were different from those of macrophages.
To examine whether C/EBP plays a role in adipocytes similar to that of PU.1 in macrophages regarding PPARγ binding, we discriminated PPREs based on the ratio of the present CTE-specific nucleotides (Ext0 to Ext4) and the presence or absence of C/EBP binding (the latter data were derived from 3T3-L1 cells) (Fig. S4E). The observed tendencies in PPARγ ChIP-seq density perfectly followed what could be seen for macrophages, indicating that both the existence of the 5′ extension and the presence of collaborating LDTF were critical for PPARγ binding in adipocytes as well. Furthermore, we detected a shift of C/EBPα binding upstream of DR1s at the top PPARγ peaks (Fig. S4F).
The application of clustering of submotif scores gave a checkered pattern similar to the macrophage-specific one, although only ∼59% of DR1s appeared uneven, and the most frequent RXR binding events were associated with roughly coequal half sites (Fig. S4G). The most frequent PPARγ binding, however, was coupled with frequent RXR binding or the presence of strong 5′ extension, as we had also seen for macrophages. Notably, while in macrophages approximately half of the examined DR1s overlapped with PU.1 binding, in adipocytes only ∼l8% showed C/EBP binding, although this could also be caused by the differences between our adipocyte and the 3T3-L1 models used.
PPARγ and C/EBP have common composite elements.
Due to the great numbers of putative DR1 and C/EBP elements, we were able to determine their single-nucleotide resolution distance distributions relative to each other (Fig. 4E, top). C/EBP elements were highly enriched around DR1s, especially at a certain distance, ending at position −3 (Fig. 4E). In order to examine whether these were functional composite elements, we determined PPARγ ChIP-seq densities at these sites and used other putative composite and overlapping elements as controls. As an additional negative control, we generated a PWM containing a single downstream C/EBP half site ending at position −3 relative to the DR1 (Fig. S4H). PPARγ indeed showed significantly higher ChIP-seq density at the enriched C/EBP:DR1 composite elements than the overlapping ones or those containing only one C/EBP half site, and those sites having a 3- or 1-nucleotide-long spacer between the C/EBP and DR1s also showed elevated PPARγ recruitment (Fig. 4F, left). In order to test whether C/EBP was also recruited to these sites, so there was potential interaction between C/EBP and PPARγ, we used the 3T3-L1 ChIP-seq data (Fig. 4F, right). Despite the potential differences between the two cell types, C/EBP:DR1 composite elements with two spacer nucleotides showed the highest C/EBPα ChIP-seq densities out of all elements, while those sites lacking half of the C/EBP motif were less effective in recruiting the protein.
DISCUSSION
Gene regulatory complexes within the chromatin are assembled at specific anchor points of the DNA and held together by specific molecular interactions. TFs, including NRs, typically form dimers to be able to interact with longer, more specific DNA motifs, but multiunit complexes can also be formed with multiple anchor points (21). Another way to enhance motif specificity is not to duplicate or multiply but to extend motifs. However, the principles of chromatinized DNA binding and the contribution of the various factors can be determined only using systematic genome-wide approaches.
In this study, we aimed to examine the major cis and trans determinants of chromatinized DNA binding by PPARγ, specifically how DNA sequences and LDTFs contribute to PPARγ distribution throughout the genome of alternatively polarized macrophages. We discerned direct and indirect DNA binding events. By mapping 10 DR1 motif classes harboring half sites and 5′ extensions of differing strengths, approximately 49% of the PPARγ cistrome contained at least one putative PPRE, and 51% represented PPARγ recruitment through other TFs, primarily PU.1, and probably AP-1 and C/EBP dimers. In the absence of PU.1 and PPRE, we detected low PPARγ enrichment, indirect recruitment through PU.1 resulted in a slightly higher PPARγ density than direct PPRE binding in the absence of PU.1, and collaborative binding of the two TFs had an additive effect on PPARγ enrichment, suggesting a similar significance of DNA-protein and protein-protein interactions (see Fig. S3D in the supplemental material). In adipocytes, similar ratios of direct/indirect DNA binding could be observed according to the presence of DR1 elements, and based on motif enrichment analyses, no TFs other than C/EBP appeared to have a significant impact on PPARγ binding (Fig. S4A).
Despite their apparent limitations, de novo motifs provided a large number (14,924) of quasi-single DR1 sequences within the macrophage PPARγ cistrome to be tested. By clustering these sequences, we observed that unlike the RXR half sites, the PPAR half sites could greatly differ from the canonical 5′-AGGTCA-3′ sequence; e.g., nucleotide 4 could be any of the four possibilities, and G at position 1 was four times more frequent than in the RXR half sites (Fig. S2C). On the other hand, a fraction of the RXR half sites showed G at position 6, while at this position nucleotide A occurred in the vast majority of the PPAR half sites. Regarding the 5′ extension, we observed tendencies similar to those based on de novo motif analyses—it was specific for higher peaks and missing from smaller ones. Importantly, the initial 5′-GGG-3′ trinucleotide not only was frequent but also resulted in typically stronger PPARγ enrichment than the canonical one (Fig. 2A).
However, like motif enrichment analyses, clustering ultimately also used nucleotide frequencies and the contribution of nucleotides to DNA-protein interactions could not be considered. In order to overcome this limitation, we applied a novel approach, termed nucleotide contribution analysis, which assigned a value to each nucleotide at each position of PPREs according to ChIP-seq density. This value mirrored whether a given nucleotide improved or impaired PPREs, and the large number of DR1 sequences extending in both directions and showing a wide range of binding frequency allowed an unbiased analysis. By using this approach, an exactly 4-nucleotide-long 5′ extension of which a 5′-AACT-3′ sequence was sharply demarked from the further upstream sequences could be identified (Fig. 2B and E). As in the case of ROREs and the Rev-DR2 elements of REV-ERB proteins, this nucleotide tetramer was frequently followed by 5′-GGGNCA-3′ sequences beyond the canonical NR half site (13, 33). In contrast, upstream of canonical 5′-AGGTCA-3′ half sites, the 5′-AAGT-3′ extension was more frequent and almost as effective in anchoring PPARγ; in addition, a 5′-AACC-3′ extension also appeared to be an efficient variant in the adipocyte cistrome. The latter finding was described previously in RORE/Rev-DR2 elements (33, 34), and based on our results, it is specific for canonical half sites (Fig. S4C and D).
Besides the 5′-A/GGGNCA-3′ PPAR half sites, we observed further variants in significant numbers (Fig. S2 and S4C). Of these, the 5′-A/GGANCA-3′ sequences showed ChIP-seq densities that were higher than those measured on canonical elements, suggesting that A instead of G at position 3 improves the motif (Fig. S2F). Furthermore, within the 5′-AACTGGANCA-3′ sequence, both of the underlined A nucleotides and their cooccurrence largely promoted PPARγ binding in macrophages, suggesting that the different DR1 variants might result in different DNA/PPARγ/RXR conformations (Fig. 2B and E). This needs to be validated by further experiments.
Regarding the relationship between PPARγ and RXR in alternatively polarized macrophages, we observed an unusual phenomenon, which contradicted, at least in part, the obligate heterodimer partnership theory. Although in the absence of RXR low PPARγ enrichment could be detected, the most frequent PPARγ binding was associated with low or moderate RXR enrichment. The other extreme, namely, the high ratio of RXR binding sites with low or no PPARγ binding, could easily be explained by the presence of other class II NRs, but PPARγ dominance over RXR at any sites was unexpected (Fig. 1A). There are two simple explanations for unequal PPARγ/RXR recruitment: (i) there is inequality already at the level of PPREs, for instance, in the composition of the NR half sites; and (ii) there are collaborating factors that are specific for PPARγ, not for RXR, and these facilitate PPARγ binding even in the absence of RXR.
In order to test the first hypothesis, we classified PPREs based on the strength of their three submotifs, the 5′ extension and the two NR half sites (Fig. 3H). Using this approach, we identified a fourth submotif, which can be considered the 5′ extension of the RXR half site in the middle of PPREs (classes 3 and 7). The last A of the PPAR half site, showing the highest conservation within PPREs, together with the highly enriched A spacer nucleotide results in a 5′-AAAGGTCA-3′ sequence (5'-AA-3' is the 5' extension of the 5'-AGGTCA-3' half site), which is known to be the specific motif of NR4A proteins (35), and according to the newest protein-binding microarray results, it can be bound also by RXR (27). As the CTE of RXR is capable of minor groove binding (14, 36), we suggest that there is a more extensive interaction between RXR and its extended half site in live cells as well. Submotif-based classification also demonstrated that in line with unequal protein binding, most PPREs were uneven, meaning that one of their half sites was significantly stronger than the other one, and the 5′ extensions, even belonging to RXR, further improved half sites. However, less than 20% of the half sites had strong extension. In addition, both NRs were able to facilitate the recruitment of their heterodimerizing partner (Fig. 3I; Fig. S4G). These results are consistent with the findings of Penvose et al., who detected RXR heterodimers interacting with single half sites beyond the direct binding of entire DR elements (27).
In line with our second hypothesis, PU.1 and TRE motif enrichments were more specific for PPARγ binding than for RXR binding (Fig. 1C). The presence of PU.1 together with the presence of the 5′ extension of DR1s exerted a synergistic effect on PPARγ binding, and PPARγ was also able to facilitate PU.1 recruitment, which ultimately led to chromatin opening (Fig. 3C). While PU.1 motifs showed a broadly invariant enrichment around PPREs that was independent of PPARγ density, TRE motifs showed enrichment together with PPARγ (Fig. 3D). Regardless, both motifs were located more frequently upstream of the highest-affinity PPREs (Fig. 3F; Fig. S3A). A shift between PU.1 and PPARγ/RXR on the surface of DNA could indeed be observed, suggesting a preferred orientation of these TFs, which may promote protein-protein interactions (Fig. 3E and G). In contrast, in the absence of PU.1, a significantly smaller amount of PPARγ could be reached on its elements even if they had strong extensions (Fig. 3A, C, and I).
Regarding the relationship between the two LDTFs of adipocytes, we got results that were very similar to those derived from macrophages. In the absence of C/EBP, the presence of the 5′ extension had minor effects on PPARγ enrichment, while the presence of C/EBP and the 5′ extension of DR1 had a synergistic effect on PPARγ recruitment (Fig. S4E). The location of C/EBP, similarly to the macrophage-specific TFs, was shifted upstream to PPREs (Fig. S4F); more importantly, we identified a composite element in which two nucleotides separated the C/EBP and DR1 elements (Fig. 4E). A composite element of an NR and an LDTF is not unprecedented, as several steroid hormone receptors interact with Forkhead box (Fox) A1, and joint direct DNA binding by androgen receptor and FoxA1 has also been observed (37). Moreover, the other two PPAR family members interact with the non-NR protein BCL6 (38, 39). The C/EBP:DR1 composite element not only enriched was but also showed elevated levels of both the C/EBP and PPARγ proteins, suggesting that the 5′ extension can be bound from both the major and minor grooves (Fig. 4F). The need for DNA binding at both sides may provide an elevated evolutionary pressure on composite elements, and these interactions may lead to a tighter collaboration between C/EBP and PPARγ proteins (40). This type of evolutionary pressure may have resulted in the conserved nucleotide A at the overlapping part of the PPAR half site and the 5′ extension of the RXR half site within DR1s, and a similar process may stabilize the interferon-stimulated response elements (ISREs) that are bound both in the major and minor grooves by IRF dimers (41).
Despite the very extensive cistrome, PPARγ has major effects only on a small set of genes in macrophages (17). Most target genes have multiple enhancers, typically with several DR1s of different compositions, but the number and expression of those genes that are regulated by extended DR1s exclusively are higher than the number and expression of the genes that have only nonextended DR1s (Fig. S3G and H). Among the identified genes with extended DR1s, there were several well-known PPARγ targets, such as Angpl4, Fabp4, and Plin2 (Fig. S3E, G, and H).
Since nucleotide contribution analysis was highly applicable for an unbiased analysis of PPREs, any other cistrome may be tested with it. By using this approach, we were able to characterize and differentiate weak and strong PPREs without using any preliminary data or data other than ChIP-seq data. We achieved similar but more robust and more detailed results essentially from a few ChIP-seq experiments than could be achieved by decades of molecular biology work. Our approach may be applied to the dissection of any element having different variants or specific extensions. For instance, insulator sequences can include satellite elements both upstream and downstream (42) of which cooccurrence and significance could be further explored by using our pairwise nucleotide contribution analysis. Our approach also allows comparison of the contribution of PPREs of different compositions with the contribution of the present LDTFs. We assigned a greater predictive force to this latter factor, although the direct DNA binding by both the DBD and CTE was indispensable for the most frequent binding events.
DNA binding by PPARγ depends on its obligate heterodimerizing partner, RXR. However, there are additional cis and trans factors that greatly determine its interactions with the DNA. In this study, we demonstrate that the affinity of PPARγ and RXR to DNA depends on the strength of their associated half sites, and any of these half sites may have a characteristic 5′ extension that provides even more frequent binding by the involvement of additional interaction surfaces. For PPARγ and RXR, the 5′-AACT-3′ tetranucleotide and the 5′-AA-3′ dinucleotide, respectively, appeared to be the most specific 5′ extension, and the CTE of both NRs is responsible for binding these sequences. We also show that in the absence of collaborative LDTFs, even perfect PPREs are not frequently occupied, but the presence of LDTF and extended PPRE has a synergistic effect on PPARγ binding. Sequential binding of LDTF, PPARγ, and RXR, using even a composite element, further enhances PPARγ binding. In addition to collaborative binding, LDTFs are able to recruit PPARγ, which accounts for approximately half of the entire cistrome.
MATERIALS AND METHODS
Alternative polarization of bone marrow-derived macrophages.
Isolation and differentiation of bone marrow cells derived from 3-month-old male C57BL/6 mice were completed as described previously (30, 43). Bone marrow-derived cells were differentiated for 6 days in the presence of L929 supernatant. Cells then were polarized with IL-4 (20 ng/ml; Peprotech) for 1, 3, 6, or 24 h.
Adipocyte differentiation.
E14 mouse embryonic stem cells (ESCs) were differentiated into adipocytes as described previously (31). ESCs were differentiated by the hanging drop method (2,000 cells/drop) after three passages without feeder culture. Embryonic bodies were collected at day 2 and plated in gelatin-coated dishes. One day later, the cells were treated with retinoic acid (1 μM) and ascorbic acid (AsA) (12.5 μg/ml) for 3 days. From day 7 to day 11, the cells were treated with an adipogenic cocktail containing RSG (0.5 μM), insulin (0.5 μg/ml), triiodothyronine (3 nM), and AsA (12.5 μg/ml). At day 12 of the differentiation, the cells were replated using Accutase and treated with the previously mentioned adipogenic cocktail. The cells were treated with a second adipogenic cocktail from day 15 to day 20: 3-isobutyl-1-methylxanthine (IBMX; 0.5 μM), dexamethasone (0.1 μM), insulin (20 μg/ml), RSG (0.5 μg/ml), indomethacin (0.06 mM), and AsA (25 μg/ml). From day 21, the medium was supplemented with insulin (10 μg/ml), RSG (0.5 μg/ml), AsA (25 μg/ml), and T3 (3 nM). Sampling took place at day 21 for PPARγ ChIP-seq and day 30 for RXR ChIP-seq.
ChIP.
Chromatin immunoprecipitation was performed as described previously (30, 44). Libraries were prepared either with Ovation ultralow library systems (Nugen) or TruSeq ChIP library systems (Illumina) according to the manufacturer’s instructions. Antibodies to the following were used: PPARγ (Perseus no. PP-A3409A), RXR (sc-774), and PU.1 (sc-352).
ChIP-seq analyses.
The primary analysis of raw sequence reads has been carried out using the updated version of our ChIP-seq analysis command line pipeline (45). Alignment to the mm10 mouse genome assembly was done by the BWA tool (46). BAM files were created by SAMTools (47). Peaks were predicted by MACS2 (25), and artifacts were removed according to the blacklist of ENCODE (48). The overlap of different sets of peaks and DR1s was determined by intersectBed (bedtools) (49) and visualized as a Venn diagram by VennMaster (50). Genome coverage (bedgraph) files were generated by makeUCSCfile.pl (HOMER) (26) and visualized with IGV2 (see Fig. S3E in the supplemental material) (51). ChIP-seq densities of peak summits or DR1s for read distribution (RD) histograms (bin size = 10 nucleotides) or heat maps (bin size = 30 nucleotides) were calculated by annotatePeaks.pl (HOMER) and visualized by GraphPad Prism or Java TreeView (52), respectively. RXR signal was normalized to that of PPARγ by using the median read density of PPARγ peak summits (middle bins of the RD heat maps) based on the overlapping part of the two cistromes (Fig. 1A; Fig. S3B and E). The read densities of peak summits or DR1s (middle bins of the RD heat maps) were visualized as box plots by GraphPad Prism. For peak summit distribution, the 1-nucleotide resolution read density of each DR1 was calculated by annotatePeaks.pl (HOMER), and the positions with the highest densities, relative to DR1s, were visualized as a box plot by GraphPad Prism (Fig. 3G). Whiskers were plotted according to the Tukey method in all box plots, and P values were determined by two-tailed unpaired t tests. In the case of PPARγ and RNAPII-pS2 two replicates and in the case of PU.1 and C/EBP a single time course and two isoforms, respectively, as pseudoreplicates were used.
Motif enrichment analysis.
The 1,000 highest peaks of the entire PPARγ cistrome or smaller sets—the 2nd octile, the 2nd quartile, or the bottom half of those peaks predictable from both replicates (consensus set), as well as those peaks predictable from one of the replicates—were determined based on MACS2 peak scores. Peak summit ± 50-nucleotide-long regions and 10-, 12-, 14-, and 16-nucleotide long sequences were used for the de novo motif enrichment analyses, performed by findMotifsGenome.pl (HOMER). P values were calculated by comparing the motif number of the “target” regions (search set) with the motif number of a set of ∼50,000 random (background) regions generated by HOMER.
The top 1,000 peaks of the PPARγ/RXR cistrome, lacking DR1s and showing PPARγ, RXR, or shared dominance based on the obtained read density of peak summits, were used for further de novo motif enrichment analyses with the same parameters as described above (Fig. 1A, top).
The selected PWMs were mapped by using annotatePeaks.pl (HOMER). Motif distribution heat maps (bin size = 10 nucleotides) were visualized by Java TreeView. Correlation analysis between read and motif density was carried out by using the average read density and the cumulative motif number of the peak summit ± 100-nucleotide-long regions calculated for ∼1,000 groups of regions (n = 66); previously, individual regions were ranked based on read density and then grouped (Fig. 1C). Motif distribution relative to DR1s was visualized as a bar chart (bin size = 10 nucleotides) or line plot (bin size = 1 nucleotide) by GraphPad Prism.
Motif clustering.
The four DR1 PWMs derived from the consensus peak set were mapped to the summit ± 100-nucleotide-long regions of the entire PPARγ cistrome by annotatePeaks.pl (HOMER). DR1s shorter than 16 nucleotides were extended to make them overlap the top motif. The sequence of the single DR1s that could be mapped uniquely within a region (14,924 DR1s in macrophages and 28,116 in adipocytes) was recovered by homerTools extract. Each nucleotide was changed to a number (−1, 0, or 1) or “x” (not applicable), and then the sequences of numbers were clustered (k-means, k = 50) by Cluster 3 (53). Per cluster, if a single nucleotide showed at least a 2-fold higher frequency than the other three at a given position, it was indicated as A, T, G, or C; if two nucleotides showed at least a 2-fold higher frequency than the other two but less difference than each other at a given position, these were indicated as R (A/G), Y (C/T), S (G/C), W (A/T), K (G/T), or M (A/C); if three nucleotides showed at least a 2-fold higher frequency than the fourth but less difference than each other at a given position, these were indicated as B (C/G/T), D (A/G/T), H (A/C/T), or V (A/C/G); and in further cases, nucleotides were indicated as N. The nucleotide frequencies of the clusters or the other groups of DR1 sequences were converted to Homer motif matrices and then were visualized by motif2Logo.pl (HOMER).
Nucleotide contribution analysis.
The previously identified 16-nucleotide-long single DR1s were extended by 5 nucleotides in both directions. Sequences were recovered by homerTools extract. The average PPARγ ChIP-seq densities of all sequences harboring a given nucleotide at a given position (4 × 26) and the difference between these values and the average density of all single DR1s (per-nucleotide contribution to PPARγ binding) were visualized as heat maps by Java TreeView. Nucleotides with a frequency below 2% at any position were excluded (marked as not applicable). Unpaired t tests were carried out between the ChIP-seq densities of those sequences having or lacking the preferred or canonical nucleotides, and −log (P values) were visualized by Java TreeView. The per-nucleotide contribution was expressed as the power of two and then as a percentage calculated for each position and visualized as a motif logo by motif2Logo.pl (HOMER). The same analysis was carried out with different sets of sequences with a fixed nucleotide(s).
Filtering DR1 sequences based on the presence of certain nucleotides at certain positions and calculating their number and ratio in different sets of regions were done by using command line tools, including intersectBed (bedtools). The percentage of DR1s harboring different numbers of the present CTE-specific nucleotides (5′-AACT-3′) was plotted as a bar chart by GraphPad Prism (Fig. 3C, left).
Pairwise nucleotide frequencies within the extended DR1 sequences were visualized as Circos plots, in which the 26 × 4 nucleotides were color-coded in the outer arc (ticks) and the thickness of the middle arcs (links) was proportional to the frequencies (54). Pairwise nucleotide contributions as the power of two were visualized as Circos plots, in which the thickness of the links was proportional to the contribution of nucleotide pairs.
Classification by submotif strength.
The PWMs of the submotifs were generated from the resulting PWMs of nucleotide contribution analysis of the macrophage PPREs (5′ extension) as well as the PWMs generated from all single macrophage DR1s (5′ and 3′ NR half sites). Submotif scores for all DR1s were calculated by annotatePeaks.pl (HOMER). DR1 sequences were classified based on these scores by k-means clustering (k = 10) using Cluster 3. DR1 motif classes were visualized as a heat map by Java TreeView and as motif logos by using motif2Logo.pl (HOMER). The average submotif scores of the classes and the difference between the per-class average read density and the average read density of all DR1s (for PPARγ and RXR) in the presence or absence of PU.1 or C/EBP were visualized as heat maps by Java TreeView.
A motif score threshold of DR1 classes was set to a value allowing one motif hit for every 10th random kilobase on average.
Identification of C/EBP:DR1 composite elements.
A 1-nucleotide resolution distance distribution between DR1 and C/EBP elements of adipocytes was visualized as a line plot by using GraphPad Prism (Fig. 4E). The enriched composite and overlapping elements were used for ChIP-seq density analyses. Composite elements containing a downstream C/EBP half site and a DR1 separated by two spacer nucleotides were recovered with a “shortened” C/EBP:DR1 PWM, and then those hits matching the entire PWM were excluded (Fig. S4H). Annotation to genes was carried out by annotatePeaks.pl (HOMER), and GO analysis was performed by ShinyGO (Fig. S4I) (55).
ATAC-seq.
Assay for transposase-accessible chromatin with high-throughput sequencing (ATAC-seq) was carried out as described earlier (28). The primary analysis of raw sequence reads was carried out as described above for ChIP-seq analysis.
RNA sequencing.
RNA-seq was carried out with BMDMs derived from four individual mice as described earlier (56). TopHat and Cufflinks toolkits were used for mapping spliced reads to the mm10 mouse genome assembly with default parameters, obtaining gene expression data in FPKM (fragments per kilobase per million mapped fragments) (57).
Enhancer transcription and gene expression analyses.
PPARγ peaks harboring exclusively extended (classes 6 to 10) or nonextended (classes 1 to 5) DR1s from the top 10% of the consensus peak set of BMDMs were extended to ±250 bp relative to their summits. The RNAPII-pS2 ChIP-seq density, as expressed in RPKM (reads per kilobase per million mapped reads), of two replicates of control and RSG-treated BMDM samples was calculated at the extended PPARγ peaks, gene bodies, and randomly selected intergenic regions used as background. The coverage of the overlapping gene or the average background was subtracted from the coverage of intronic or intergenic peaks, respectively. Peaks with at least 1.5-fold RNAPII-pS2 enrichment upon ligand treatment (comparing any replicates) and more than 0.5 RPKM on average in the treated samples were considered ligand-responsive sites and were plotted as a box plot (Fig. S3F). Genes showing at least 1.5-fold induction upon ligand treatment (comparing any replicates), more than 1 RPKM on average in the treated samples, and of which the transcription start site (TSS) was closer than 100 kb to ligand-responsive sites harboring exclusively extended or nonextended DR1s were considered the direct target of PPARγ and plotted as line plots (Fig. S3G). The FPKM values of the identified genes during the time course upon IL-4 treatment as determined by RNA-seq were visualized as profile plots (Fig. S3H). In figures representing gene expression data (Fig. S3G and H), average values are shown.
Data availability.
The data sets supporting the conclusions of this article are available in the SRA and GEO repositories under the following accession numbers: PRJNA318630, GSE115083, GSE107456, GSE106706, GSE142249, PRJNA299295, and GSE74189, as detailed in Table S1 in the supplemental material.
Supplementary Material
ACKNOWLEDGMENTS
We thank Zsolt Czimmerer, Petros Tzerpos, Lajos Szeles, and other members of the Nagy laboratory for discussions and comments on the manuscript.
This work was supported by grants from the National Institutes of Health (R01DK115924 to L.N.) and the Hungarian Scientific Research Fund (OTKA grant no. K124298, K126885, and K116855 to L.N. and OTKA grant no. PD124843 to G.N.). G.N. is supported by the János Bolyai Research Scholarship of the Hungarian Academy of Sciences and by the ÚNKP-19-4-DE-173 New National Excellence Program of the Ministry of Human Capacities. B.D. is supported by an American Heart Association postdoctoral fellowship (17POST33660450). Library preparation and bioinformatics analysis were performed at the Center of Clinical Genomics and Personalized Medicine of the University of Debrecen. Next-generation sequencing was performed at the Centre National de Genotypage Evry by Steven McGinn, Anne Boland, Doris Lechner, and Marie Thérèse Bihoreau and supported by the European Sequencing and Genotyping Infrastructure (funded by the European Commission; FP7/2007-2013) under grant agreement no. 26205 (École Supérieure de Génie Informatique), as part of the ADIPOMACTX transnational access program and also at the Analytical Genomics Core Facility at the Sanford Burnham Prebys Medical Discovery Institute.
We declare that we have no competing interests.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Mangelsdorf DJ, Thummel C, Beato M, Herrlich P, Schütz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, Evans RM. 1995. The nuclear receptor superfamily: the second decade. Cell 83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Umesono K, Murakami KK, Thompson CC, Evans RM. 1991. Direct repeats as selective response elements for the thyroid hormone, retinoic acid, and vitamin D3 receptors. Cell 65:1255–1266. doi: 10.1016/0092-8674(91)90020-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Evans RM. 2005. The nuclear receptor superfamily: a Rosetta Stone for physiology. Mol Endocrinol 19:1429–1438. doi: 10.1210/me.2005-0046. [DOI] [PubMed] [Google Scholar]
- 4.Osumi T, Wen J-K, Hashimoto T. 1991. Two cis-acting regulatory sequences in the peroxisome proliferator-responsive enhancer region of rat acyl-CoA oxidase gene. Biochem Biophys Res Commun 175:866–871. doi: 10.1016/0006-291x(91)91645-s. [DOI] [PubMed] [Google Scholar]
- 5.Tugwood JD, Issemann I, Anderson RG, Bundell KR, McPheat WL, Green S. 1992. The mouse peroxisome proliferator activated receptor recognizes a response element in the 5′ flanking sequence of the rat acyl CoA oxidase gene. EMBO J 11:433–439. doi: 10.1002/j.1460-2075.1992.tb05072.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kliewer SA, Umesono K, Noonan DJ, Heyman RA, Evans RM. 1992. Convergence of 9-cis retinoic acid and peroxisome proliferator signalling pathways through heterodimer formation of their receptors. Nature 358:771–774. doi: 10.1038/358771a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Issemann I, Prince R, Tugwood J, Green S. 1992. A role for fatty acids and liver fatty acid binding protein in peroxisome proliferation? Biochem Soc Trans 20:824–827. doi: 10.1042/bst0200824. [DOI] [PubMed] [Google Scholar]
- 8.IJpenberg A, Jeannin E, Wahli W, Desvergne B. 1997. Polarity and specific sequence requirements of peroxisome proliferator-activated receptor (PPAR)/retinoid X receptor heterodimer binding to DNA. A functional analysis of the malic enzyme gene PPAR response element. J Biol Chem 272:20108–20117. doi: 10.1074/jbc.272.32.20108. [DOI] [PubMed] [Google Scholar]
- 9.Juge-Aubry C, Pernin A, Favez T, Burger AG, Wahli W, Meier CA, Desvergne B. 1997. DNA binding properties of peroxisome proliferator-activated receptor subtypes on various natural peroxisome proliferator response elements. J Biol Chem 272:25252–25259. doi: 10.1074/jbc.272.40.25252. [DOI] [PubMed] [Google Scholar]
- 10.Rodríguez JC, Gil-Gómez G, Hegardt FG, Haro D. 1994. Peroxisome proliferator-activated receptor mediates induction of the mitochondrial 3-hydroxy-3-methylglutaryl-CoA synthase gene by fatty acids. J Biol Chem 269:18767–18772. [PubMed] [Google Scholar]
- 11.Aldridge TC, Tugwood JD, Green S. 1995. Identification and characterization of DNA elements implicated in the regulation of CYP4A1 transcription. Biochem J 306:473–479. doi: 10.1042/bj3060473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tzeng J, Byun J, Park JY, Yamamoto T, Schesing K, Tian B, Sadoshima J, Oka S-I. 2015. An ideal PPAR response element bound to and activated by PPARα. PLoS One 10:e0134996. doi: 10.1371/journal.pone.0134996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Isakova A, Berset Y, Hatzimanikatis V, Deplancke B. 2016. Quantification of cooperativity in heterodimer-DNA binding improves the accuracy of binding specificity models. J Biol Chem 291:10293–10306. doi: 10.1074/jbc.M115.691154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chandra V, Huang P, Hamuro Y, Raghuram S, Wang Y, Burris TP, Rastinejad F. 2008. Structure of the intact PPAR-gamma-RXR- nuclear receptor complex on DNA. Nature 456:350–356. doi: 10.1038/nature07413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rajakumari S, Wu J, Ishibashi J, Lim H-W, Giang A-H, Won K-J, Reed RR, Seale P. 2013. EBF2 determines and maintains brown adipocyte identity. Cell Metab 17:562–574. doi: 10.1016/j.cmet.2013.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Soccio RE, Chen ER, Rajapurkar SR, Safabakhsh P, Marinis JM, Dispirito JR, Emmett MJ, Briggs ER, Fang B, Everett LJ, Lim H-W, Won K-J, Steger DJ, Wu Y, Civelek M, Voight BF, Lazar MA. 2015. Genetic variation determines PPARγ function and anti-diabetic drug response in vivo. Cell 162:33–44. doi: 10.1016/j.cell.2015.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Daniel B, Nagy G, Czimmerer Z, Horvath A, Hammers DW, Cuaranta-Monroy I, Poliska S, Tzerpos P, Kolostyak Z, Hays TT, Patsalos A, Houtman R, Sauer S, Francois-Deleuze J, Rastinejad F, Balint BL, Sweeney HL, Nagy L. 2018. The nuclear receptor PPARγ controls progressive macrophage polarization as a ligand-insensitive epigenomic ratchet of transcriptional memory. Immunity 49:615–626.e6. doi: 10.1016/j.immuni.2018.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Madsen MS, Siersbæk R, Boergesen M, Nielsen R, Mandrup S. 2014. Peroxisome proliferator-activated receptor and C/EBP synergistically activate key metabolic adipocyte genes by assisted loading. Mol Cell Biol 34:939–954. doi: 10.1128/MCB.01344-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Savic D, Ramaker RC, Roberts BS, Dean EC, Burwell TC, Meadows SK, Cooper SJ, Garabedian MJ, Gertz J, Myers RM. 2016. Distinct gene regulatory programs define the inhibitory effects of liver X receptors and PPARG on cancer cell proliferation. Genome Med 8:74. doi: 10.1186/s13073-016-0328-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sanchez DJ, Steger DJ, Skuli N, Bansal A, Simon MC. 2018. PPARγ is dispensable for clear cell renal cell carcinoma progression. Mol Metab 14:139–149. doi: 10.1016/j.molmet.2018.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Siggers T, Gordân R. 2014. Protein-DNA binding: complexities and multi-protein codes. Nucleic Acids Res 42:2099–2111. doi: 10.1093/nar/gkt1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ricote M, Li AC, Willson TM, Kelly CJ, Glass CK. 1998. The peroxisome proliferator-activated receptor-γ is a negative regulator of macrophage activation. Nature 391:79–82. doi: 10.1038/34178. [DOI] [PubMed] [Google Scholar]
- 23.Szanto A, Balint BL, Nagy ZS, Barta E, Dezso B, Pap A, Szeles L, Poliska S, Oros M, Evans RM, Barak Y, Schwabe J, Nagy L. 2010. STAT6 transcription factor is a facilitator of the nuclear receptor PPARγ-regulated gene expression in macrophages and dendritic cells. Immunity 33:699–712. doi: 10.1016/j.immuni.2010.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Daniel B, Nagy G, Horvath A, Czimmerer Z, Cuaranta-Monroy I, Poliska S, Hays TT, Sauer S, Francois-Deleuze J, Nagy L. 2018. The IL-4/STAT6/PPARγ signaling axis is driving the expansion of the RXR heterodimer cistrome, providing complex ligand responsiveness in macrophages. Nucleic Acids Res 46:4425–4439. doi: 10.1093/nar/gky157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, Liu XS. 2008. Model-based analysis of ChIP-Seq (MACS). Genome Biol 9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Penvose A, Keenan JL, Bray D, Ramlall V, Siggers T. 2019. Comprehensive study of nuclear receptor DNA binding provides a revised framework for understanding receptor specificity. Nat Commun 10:2514. doi: 10.1038/s41467-019-10264-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. 2013. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods 10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hijikata M, Wen JK, Osumi T, Hashimoto T. 1990. Rat peroxisomal 3-ketoacyl-CoA thiolase gene. Occurrence of two closely related but differentially regulated genes. J Biol Chem 265:4600–4606. [PubMed] [Google Scholar]
- 30.Daniel B, Nagy G, Hah N, Horvath A, Czimmerer Z, Poliska S, Gyuris T, Keirsse J, Gysemans C, Van Ginderachter JA, Balint BL, Evans RM, Barta E, Nagy L. 2014. The active enhancer network operated by liganded RXR supports angiogenic activity in macrophages. Genes Dev 28:1562–1577. doi: 10.1101/gad.242685.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cuaranta-Monroy I, Simandi Z, Kolostyak Z, Doan-Xuan Q-M, Poliska S, Horvath A, Nagy G, Bacso Z, Nagy L. 2014. Highly efficient differentiation of embryonic stem cells into adipocytes by ascorbic acid. Stem Cell Res 13:88–97. doi: 10.1016/j.scr.2014.04.015. [DOI] [PubMed] [Google Scholar]
- 32.Chawla A, Schwarz EJ, Dimaculangan DD, Lazar MA. 1994. Peroxisome proliferator-activated receptor (PPAR) gamma: adipose-predominant expression and induction early in adipocyte differentiation. Endocrinology 135:798–800. doi: 10.1210/endo.135.2.8033830. [DOI] [PubMed] [Google Scholar]
- 33.Zhang Y, Fang B, Emmett MJ, Damle M, Sun Z, Feng D, Armour SM, Remsberg JR, Jager J, Soccio RE, Steger DJ, Lazar MA. 2015. Discrete functions of nuclear receptor Rev-erb couple metabolism to the clock. Science 348:1488–1492. doi: 10.1126/science.aab3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Crumbley C, Wang Y, Banerjee S, Burris TP. 2012. Regulation of expression of citrate synthase by the retinoic acid receptor-related orphan receptor α (RORα). PLoS One 7:e33804. doi: 10.1371/journal.pone.0033804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wilson T, Paulsen R, Padgett K, Milbrandt J. 1992. Participation of non-zinc finger residues in DNA binding by two nuclear orphan receptors. Science 256:107–110. doi: 10.1126/science.1314418. [DOI] [PubMed] [Google Scholar]
- 36.Lee MS, Kliewer SA, Provencal J, Wright PE, Evans RM. 1993. Structure of the retinoid X receptor alpha DNA binding domain: a helix required for homodimeric DNA binding. Science 260:1117–1121. doi: 10.1126/science.8388124. [DOI] [PubMed] [Google Scholar]
- 37.Sahu B, Laakso M, Pihlajamaa P, Ovaska K, Sinielnikov I, Hautaniemi S, Jänne OA. 2013. FoxA1 specifies unique androgen and glucocorticoid receptor binding events in prostate cancer cells. Cancer Res 73:1570–1580. doi: 10.1158/0008-5472.CAN-12-2350. [DOI] [PubMed] [Google Scholar]
- 38.Lee C-H, Chawla A, Urbiztondo N, Liao D, Boisvert WA, Evans RM, Curtiss LK. 2003. Transcriptional repression of atherogenic inflammation: modulation by PPARdelta. Science 302:453–457. doi: 10.1126/science.1087344. [DOI] [PubMed] [Google Scholar]
- 39.Sommars MA, Ramachandran K, Senagolage MD, Futtner CR, Germain DM, Allred AL, Omura Y, Bederman IR, Barish GD. 2019. Dynamic repression by BCL6 controls the genome-wide liver response to fasting and steatosis. Elife 8:e43922. doi: 10.7554/eLife.43922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mouthiers A, Baillet A, Deloménie C, Porquet D, Mejdoubi-Charef N. 2005. Peroxisome proliferator-activated receptor alpha physically interacts with CCAAT/enhancer binding protein (C/EBPbeta) to inhibit C/EBPbeta-responsive alpha1-acid glycoprotein gene expression. Mol Endocrinol 19:1135–1146. doi: 10.1210/me.2004-0188. [DOI] [PubMed] [Google Scholar]
- 41.Fujii Y, Shimizu T, Kusumoto M, Kyogoku Y, Taniguchi T, Hakoshima T. 1999. Crystal structure of an IRF-DNA complex reveals novel DNA recognition and cooperative binding to a tandem repeat of core sequences. EMBO J 18:5028–5041. doi: 10.1093/emboj/18.18.5028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li Y, Huang W, Niu L, Umbach DM, Covo S, Li L. 2013. Characterization of constitutive CTCF/cohesin loci: a possible role in establishing topological domains in mammalian genomes. BMC Genomics 14:553. doi: 10.1186/1471-2164-14-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barish GD, Downes M, Alaynick WA, Yu RT, Ocampo CB, Bookout AL, Mangelsdorf DJ, Evans RM. 2005. A nuclear receptor atlas: macrophage activation. Mol Endocrinol 19:2466–2477. doi: 10.1210/me.2004-0529. [DOI] [PubMed] [Google Scholar]
- 44.Barish GD, Yu RT, Karunasiri M, Ocampo CB, Dixon J, Benner C, Dent AL, Tangirala RK, Evans RM. 2010. Bcl-6 and NF- B cistromes mediate opposing regulation of the innate immune response. Genes Dev 24:2760–2765. doi: 10.1101/gad.1998010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barta E. 2011. Command line analysis of ChIP-seq results. Embnet J 17:13. doi: 10.14806/ej.17.1.209. [DOI] [Google Scholar]
- 46.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup . 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.ENCODE Project Consortium. 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kestler HA, Muller A, Gress TM, Buchholz M. 2005. Generalized Venn diagrams: a new method of visualizing complex genetic set relations. Bioinformatics 21:1592–1595. doi: 10.1093/bioinformatics/bti169. [DOI] [PubMed] [Google Scholar]
- 51.Thorvaldsdóttir H, Robinson JT, Mesirov JP. 2013. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform 14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Saldanha AJ. 2004. Java Treeview—extensible visualization of microarray data. Bioinformatics 20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- 53.de Hoon MJL, Imoto S, Nolan J, Miyano S. 2004. Open source clustering software. Bioinformatics 20:1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- 54.Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. 2009. Circos: an information aesthetic for comparative genomics. Genome Res 19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ge SX, Jung D. 2018. ShinyGO: a graphical enrichment tool for animals and plants. bioRxiv doi: 10.1101/315150. [DOI] [PMC free article] [PubMed]
- 56.Czimmerer Z, Daniel B, Horvath A, Rückerl D, Nagy G, Kiss M, Peloquin M, Budai MM, Cuaranta-Monroy I, Simandi Z, Steiner L, Nagy B, Poliska S, Banko C, Bacso Z, Schulman IG, Sauer S, Deleuze J-F, Allen JE, Benko S, Nagy L. 2018. The transcription factor STAT6 mediates direct repression of inflammatory enhancers and limits activation of alternatively polarized macrophages. Immunity 48:75–90.e6. doi: 10.1016/j.immuni.2017.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L. 2013. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol 31:46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lai B, Lee J-E, Jang Y, Wang L, Peng W, Ge K. 2017. MLL3/MLL4 are required for CBP/p300 binding on enhancers and super-enhancer formation in brown adipogenesis. Nucleic Acids Res 45:6388–6403. doi: 10.1093/nar/gkx234. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data sets supporting the conclusions of this article are available in the SRA and GEO repositories under the following accession numbers: PRJNA318630, GSE115083, GSE107456, GSE106706, GSE142249, PRJNA299295, and GSE74189, as detailed in Table S1 in the supplemental material.