Abstract
Cancer is widely considered to be a set of genetic diseases that are currently classified by tissue and cell type of origin and, increasingly, by its molecular characteristics. This latter aspect is based primarily upon oncogene gains, tumor suppressor losses, and associated transcriptional profiles. However, cancers are also characterized by profound alterations in cellular metabolism and epigenetic landscape. It is particularly noteworthy that cancer-causing genomic defects not only activate cell cycle progression, but regulate the opportunistic uptake and utilization of nutrients, effectively enabling tumors to maximize growth and drug resistance in changing tissue and systemic microenvironments. Shifts in chromatin architecture are central to this dynamic behavior. Further, changes in nutrient uptake and utilization directly affect chromatin structure. In this review, we describe a set of recent discoveries of metabolic and epigenetic reprogramming in cancer, and especially focus on the genomically well-characterized brain tumor, glioblastoma. Further, we discuss a new mode of metabolic regulation driven by epigenetic mechanisms, that enables cancer cells to autonomously activate iron metabolism for their survival. Together, these underscore the integration of genetic mutations with metabolic reprogramming and epigenetic shifts in cancer, suggesting a new means to identifying patient subsets suitable for specific precision therapeutics.
Keywords: cancer metabolism, epigenetics, histone acetylation, iron metabolism, glioblastoma
I. Introduction—Epigenetics Is a Driving Force for Cancer Progression
A series of next-generation sequencing approaches have enabled the prognostic and predictive stratification of cancer patients that is framed by the orchestration of genetic aberrations and transcriptome profiles [11, 12, 52, 57]. Large-scale public datasets have provided an infrastructure for this, and the Cancer Genome Atlas (TCGA) project has generated over 2.5 petabytes of genomic, epigenomic, transcriptomic and proteomic data to this end. However, translation of the knowledge into the next clinical stage requires further elucidation of how these genetic abnormalities functionally drive cancer initiation and progression. Importantly, metabolic reprogramming has emerged as a new core hallmark of cancer, and genetic aberrations, in combination with intrinsic and extrinsic molecular signaling, shift intracellular metabolism to support the demands of rapidly proliferating cancer cells [24, 64]. Furthermore, metabolic reprogramming could dynamically shift the epigenetic landscape, as exemplified by DNA and histone modifications, through the production of intermediary metabolites [32, 44].
The genome can effectively be considered from the standpoint of chromatin which is the molecular complex of DNA and histone proteins. A nucleosome is the fundamental unit of chromatin, containing 146 base pairs of DNA, wrapped around a histone octamer (a pair of H2A, H2B, H3 and H4). Notably, the organization and function of the chromatin can be dynamically altered by various modifications of the nucleosomal components (i.e. DNA and histones), at least partly through metabolic reprogramming [1]. Such nucleosomal modifications or epigenetic changes define the state of cellular differentiation, and the Waddington model indicates that differentiating cells proceed downhill along branching canals separated by walls that restrict cell identity in an epigenetic developmental specification [25, 42]. Importantly, cancer cells harness the epigenetic system where aberrant chromatin structures affect the height of the walls between the canals in this epigenetic landscape [19], and genetic, metabolic and environmental stimuli are the driving forces to disrupt chromatin and alter cellular states and responses. This, in turn, can predispose individual cells to a spectrum of cancerous states [66]. In that sense, cancer may be the result of the synergistic combination of genetic, metabolic and epigenetic aberrations, rather than just of one such component.
Here we describe a set of recent discoveries on metabolic and epigenetic reprogramming in cancer, especially focusing on a genomically well-characterized brain tumor, glioblastoma (GBM). We highlight how the common mutations in gliomas promote metabolic and epigenetic reprogramming in the cell. We further use our recent study defining mammalian/mechanistic target of rapamycin (mTOR) complex 2 (mTORC2) as an indicative example of a novel integrator of cancer metabolism and epigenetics and a promising key node which might be therapeutically targeted as a new mode of treatment.
II. Epigenetics as a Key Player in the Biology of Cancer and Its Diagnostic Utility
DNA methylation
DNA methylation is one of the major epigenetic changes that play an important role in regulating chromatin architecture as well as gene expression. Most of the DNA methylation pattern is established by the covalent addition of methyl groups at the 5-carbon of the cytosine rings (5-mC) in short CpG-rich DNA sequences (CpG islands) and large repetitive sequence regions (e.g. centromeric repeats, retrotransposon elements, ribosomal DNA, etc.) [9]. CpG islands preferentially occupy around 60% of human gene promoters, and DNA methylation generally leads to gene silencing in cooperation with methylation regulatory proteins. The DNA methylation patterns are generated and maintained in daughter cells by the cooperation of the replication-independent de novo methyltransferases (DNMT3A and DNMT3B) and the maintenance DNA methyltransferase (DNMT1) which acts during replication [21]. Two additional enzymes (DNMT2 and DNMT3L) may also have more specialized but related functions. In contrast, ten-eleven translocation (TET) family enzymes (TET1, TET2 and TET3) can oxidize 5-methylcytosine (5-mC) to 5-hydroxymethylcytosine (5-hmC) which is a key nexus in demethylation, and further convert 5-hmC to 5-fC (5-formylcytosine), and 5-fC to 5-caC (5-carboxylcytosine) through their hydroxylase activity [35]. Recent studies demonstrated that some types of cancer harbor mutations in the genes of such methyltransferases [21], indicating that the aberrant patterns of DNA methylation might be involved in tumor formation. Of note, for brain tumors, a diagnostic algorithm that integrates histology, conventional molecular tests and DNA methylation arrays has been proposed [28], and certain types of malignant brain tumors are shown to be better subtyped based upon the epigenetic landscape of DNA methylation patterns [29, 74] as compared to traditional histopathology. Further, a subtype of diffuse glioma was associated with DNA demethylation and poor outcome and DNA methylation heterogeneity was demonstrated in a genetically diverse and heterogeneous GBM, suggesting the tight association of DNA methylation with cancer diagnostics and biology [12, 34, 60].
Histone modifications
One type of the essential constituents in the nucleosomal structure is the histone protein class, where their N-terminal tails can undergo a variety of posttranslational covalent modifications including methylation, acetylation, ubiquitylation, sumoylation and phosphorylation on specific residues [7]. These modifications affect chromatin structure and regulate key cellular processes such as transcription, replication and repair, leading to either promotion or suppression of gene expression, depending upon the spatiotemporal patterns of the modification [7]. For example, lysine acetylation is correlated with transcriptional activation, whereas lysine methylation results in transcriptional activation or repression depending upon which residue is modified and the degree of methylation [82]. Furthermore, recent studies demonstrated the presence of bivalent chromatin domains marked by both activating and repressive chromatin modifications which could be associated with a subtype-specific signature in developmental or neoplastic cells [23]. Histone modification patterns are dynamically regulated by enzymes that add and remove covalent modifications to histone proteins. Histone acetyltransferases (HATs) and histone methyltransferases (HMTs) add acetyl and methyl groups, whereas histone deacetylases (HDACs) and histone demethylases (HDMs) remove acetyl and methyl groups, respectively [67]. Aberrant patterns of histone modifications are observed in several types of cancers which could be therapeutically exploitable [6, 37], and the heterogeneity of GBM across the entire age spectrum was demonstrated in terms of histone mutations and subsequent histone modifications on the GBM epigenome [73]. Surprisingly, somatic “oncohistone” mutations occur in approximately 4% of diverse tumor types and in crucial regions of histone proteins [61].
Chromatin remodelers
The innumerable covalent modifications of the nucleosome provides the scaffold and context for dynamic remodeling of the chromatin structures. Based on their biochemical activity and subunit composition, the mammalian chromatin-remodeling complexes can be subclassified into four major families: the switching/sucrose non-fermenting (SWI/SNF) family, the imitation switch (ISWI) family, chromodomain helicase DNA-binding protein (CHD) family, and the inositol requiring 80 (INO80) family [14]. These enzymes are evolutionarily conserved, utilizing ATP as an energy source to mobilize, evict, and exchange histones. Several members from the chromatin-remodeling families are known to be mutated in human malignancies, raising the possibility that abnormal activities of chromatin remodeling may be the driving force for tumor initiation and progression [31, 75]. In brain tumors, genetic defects of the enzymes which are involved in the chromatin remodeling are reported to be the hallmark aberration in some tumor types, notably as driver mutations in histone H3.3 and chromatin remodeling genes in pediatric GBM [58, 70].
Non-coding RNAs
Non-coding RNAs that are not translated into proteins can be divided into housekeeping non-coding RNAs and regulatory non-coding RNAs. Those RNAs with a regulatory role are further divided into two categories based on size [40]: short chain non-coding RNAs (including miRNAs and piRNAs) and long non-coding RNAs (lncRNAs). A newly discovered type of endogenous noncoding RNA, circular RNAs (circRNAs), have also been proposed as part of competing endogenous RNA (ceRNA) regulatory networks [59]. A large number of recent studies have shown that non-encoding RNAs play a significant role in epigenetic modification and can regulate gene expression at the gene and chromosomal levels to control cell differentiation [2]. Importantly, dysregulation of the non-coding RNA networks are the determining factors for human malignancies [4, 33], and recent studies unraveled the role of non-coding RNAs in GBM pathogenesis as well as future application of non-coding RNAs as biomarkers and therapeutics for glioma [46, 68].
III. Metabolic Reprogramming Drives Epigenetic Changes in Gliomas
Mutant IDH-induced epigenetic phenotypes in glioma
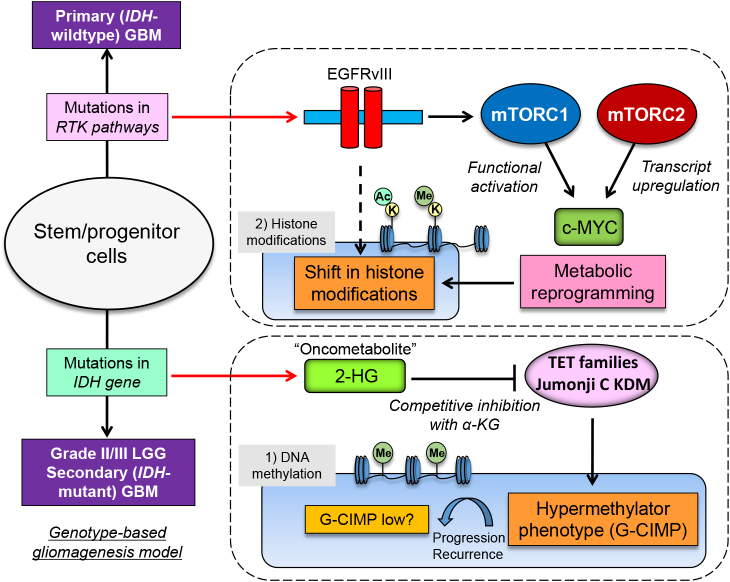
Cancer metabolism is activated by dynamic changes of signaling and transcriptional networks that are induced by activated oncogenes [e.g. epidermal growth factor receptor (EGFR), RAS, MYC] and dysregulated tumor suppressor genes (e.g. TP53, RB1), which are also hallmark genetic aberrations for GBM [51, 55]. Additionally, isocitrate dehydrogenase (IDH) gene mutations (IDH1 and IDH2) were detected in more than 70% of grade II/III diffuse gliomas as well as in a small fraction of GBMs that progress from lower grade gliomas (LGGs). This has caused a paradigm shift in brain tumor diagnostics as well as for the understanding in gliomgenesis through metabolic reprogramming [63, 79]. Such reprogramming results in changes of the levels of intracellular metabolites which can then affect oncogenic signaling by control of epigenetics and, consequently, by globally altering gene transcription [66]. Support for this comes from a series of studies which revealed that IDH mutations could connect genetic mutations, metabolic reprogramming and eventual epigenetic modulation to drive tumor progression. Mutations in IDH produce an enzyme with a neomorphic activity that converts α-ketoglutaric acid (α-KG) to 2-hydroxyglutaric acid (2-HG), which inhibits α-KG-dependent dioxygenases, including Jumonji (JmjC) domain-containing histone demethylases and the TET family of 5'-methlycytosine hydroxylases, which is a dynamic and pathognomonic change in glioma epigenetics (Fig. 1) [78]. The presence of IDH mutations thus leads to a distinct subgroup of glioma with a CpG island methylator phenotype (G-CIMP) and aberrant histone methylation (Fig. 1) [62], which results in differentiation-related genes in an inactive state [45], distortions of chromosomal topology and shifts of the landscape of enhancers [18], eventually contributing to tumorigenesis. In this regard, the findings that the intermediary metabolite 2-HG inhibits α-KG-dependent dioxygenases in glioma has led to the potentially important idea that 2-HG acts as an “oncometabolite” [80]. Furthermore, a shift in the methylation pattern (G-CIMP high to G-CIMP low) in a subtype of IDH-mutant glioma was reported to be linked to recurrence and malignant progression (Fig. 1) [12, 34, 60], further accentuating the importance of IDH-dependent remodeling of metabolism and epigenetics in malignant brain tumors.
Fig. 1.
Metabolic reprogramming modulates epigenetic landscape in IDH-mutated gliomas and GBM with EGFR mutations. Mutations in IDH play an important role in gliomas through its neomorphic activity that converts α-KG to an oncometabolite 2-HG, leading to methylator phenotypes (G-CIMP). Demethylation (G-CIMP low) could be associated with malignant progression of the tumor. Under abnormal EGFR signaling, two mTOR complexes (mTORC1 and mTORC2) could reprogram cellular metabolism for survival through the activation of c-MYC, leading to dynamic shift in the landscape of histone modifications. IDH, isocitrate dehydrogenase; RTK, receptor tyrosine kinase; EGFRvIII, constitutively active mutant of EGFR; LGG, lower grade glioma; α-KG, α-ketoglutaric acid; 2-HG, 2-hydroxyglutaric acid; TET, ten-eleven translocation; KDM, lysine (K)-specific demethylase; G-CIMP, glioma CpG island methylator phenotype; K, lysine residue of the histone protein; Ac, acetyl-group; Me, methyl-group.
EGFR-mTOR pathways: a key player in modulating glioma histone landscapes
In light of the essential contribution of metabolic and epigenetic reprogramming to the pathogenesis of IDH-mutant gliomas, the critical questions could be raised; how metabolism and epigenetics could be remodeled in glioma without IDH mutation, namely the most malignant tumor, GBM. Indeed, in GBM with wildtype IDH, epidermal growth factor receptor (EGFR)-mammalian/mechanistic target of rapamycin (mTOR) pathways are the key regulators of metabolic and epigenetic reprogramming. In addition to reprogramming intracellular metabolic circuits, the ability to appropriately sense and respond to the nutrient in the microenvironment is critical for cancer cells to connect intracellular metabolic changes with cell survival in a timely fashion. One of the mTOR complexes, mTORC1 could respond to a range of amino acids and relevant metabolites, including leucine and arginine [38]. The other mTOR complex, mTORC2 is activated in nutrient-rich (glucose and acetate) conditions by acetylation of Rictor, the main component of mTORC2, suggesting a novel role of mTORC2 as a glucose and acetate sensor in cancer cells [50, 53]. Furthermore, we discovered that mTORC2 could suppress the activity of the cystine-glutamate antiporter, system Xc transporter-related protein (xCT), implicating new roles for mTORC2 as a potential amino acid sensor [22]. Following the appropriate sensing of the nutrients, hyperactivated EGFR signaling increases the activity of a master regulator of metabolism c-MYC through both the phosphoinositide 3-kinase (PI3K)-AKT-mTORC1 pathway, as well as an AKT-independent activity of the mTORC2 complex (Fig. 1) [5, 48].
Metabolic reprogramming, potentially thorough mTOR complexes, could significantly affect the epigenetic status in cancer cells [49]. In fact, a number of enzymes involved in epigenetic gene regulation take use of intermediary metabolites, and multiple intermediary metabolites can be regulated by PI3K-AKT-mTOR signaling (Fig. 2a) [49]. Two representative histone modifications which could be linked with cancer biology are its acetylation and methylation. Acetylation on the N-terminal lysine tail of histones facilitates an open chromatin configuration to promote gene expression, using intermediary metabolite acetyl-CoA as the co-substrate for modification [85]. In addition to DNA methylation, histone methylation is also important in defining the epigenetic status of the cells [85], and the methyl-donor S-adenosylmethionine (SAM) is utilized by methyltransferases for both DNA and histone methylation [71]. In EGFR-mutant GBMs which do not usually possess mutations in IDH or the H3 histone family 3A (e.g. H3K27M) to potentially change the epigenetics [8], aberrant EGFR signaling and downstream PI3K-AKT-mTOR activation could modulate the histone acetylation and methylation to drive tumor progression (Fig. 2b and 2c). Activated EGFR signaling could associate pyruvate kinase isozymes M2 (PKM2) with the phosphorylation of the histone 3 tail which dissociates histone deacetylase 3 (HDAC3) from the chromatin to promote histone acetylation (Fig. 2c) [81]. These eventually promotes transcription of the oncogenic genes including c-MYC and cyclin D1 (CCND1) [81]. Furthermore, the active form of EGFR mutant (EGFRvIII) significantly reprograms the enhancer landscape of GBM (represented by H3K4me1 and H3K27ac), facilitating tumorigenesis through a SOX9 and FOXG1-dependent transcriptional regulatory network (Fig. 2c) [41]. Additionally, our comprehensive metabolome analyses demonstrated that two mTOR complexes (mTORC1 and mTORC2) cooperatively and synergistically drive global histone methylation, which eventually promotes tumor cell survival (Fig. 2c) [Harachi et al. unpublished]. Further studies are necessary to unravel the mechanisms by which glioma cells survive in the various niches through EGFR-mTOR-dependent dynamic shifts in their epigenetic landscapes.
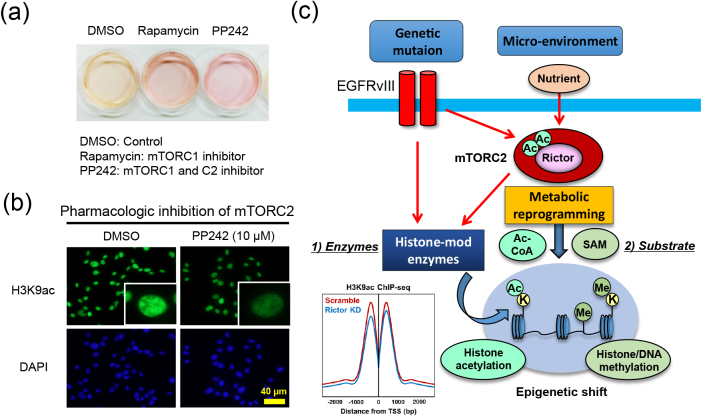
Fig. 2.
mTOR-dependent metabolic reprogramming is a driving force for epigenetic shift in GBM. (a) Treatment of GBM cells (U87 cells) with mTOR inhibitors (Rapamycin and PP242) changes the color of the culture media, indicating a shift in intermediary metabolites. (b) Treatment of GBM cells with dual mTOR inhibitors (PP242) significantly decreases histone acetylation (H3K9ac, green). DAPI staining shows the counter nuclear staining (blue). (c) mTORC2 integrates the information from genetic mutation and the microenvironment into the epigenetic shift in cancer cells through the regulation of both modifying enzymes (histone-mod enzymes) and substrates (Ac-CoA and SAM). ChIP-seq data are taken and modified from ref [54], and H3K9ac peak around TSS (transcriptional start site) was decreased in Rictor (a core component of mTORC2) knockdown GBM cells (blue line) in comparison with control cells (Scramble, red line).
IV. Epigenetic Regulation of Iron Metabolism: Mutual Dependency of Metabolism and Epigenetics in Cancer Cells
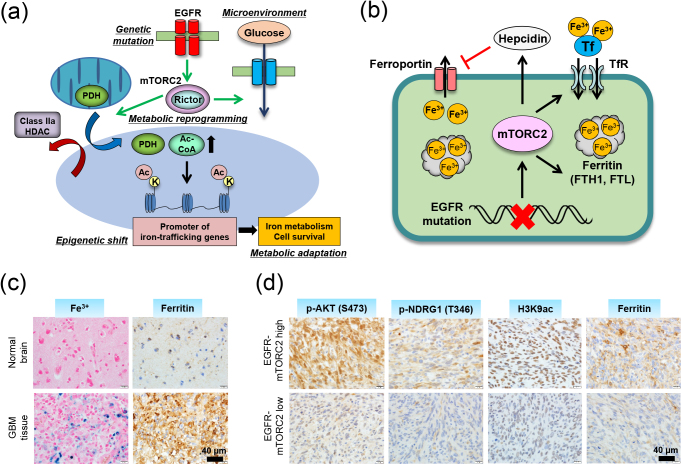
The pathway to epigenetic reprogramming via metabolic change is not a one-way street. Of note, there is a bidirectional relationship between epigenetic modifications and metabolic changes. On one hand, intermediary metabolites and metabolic enzymes regulate epigenetic modifications; on the other hand, epigenetic changes at promoter regions of the metabolic genes modulate the gene expression involved in metabolism, which eventually affects intracellular metabolism [43, 83, 84]. Sensing the microenvironment status with subsequent reprogramming of intracellular metabolism is the driving force to shift the epigenetic landscape, and the timely expression of metabolic genes via epigenetics to respond to the microenvironment would be reasonable, in that the feedback regulation of metabolism could enable cells to respond to changes in microenvironment in a prompt and accurate way. For achieving this, histone modification would seem more appropriate than DNA modification since a change in histone is more sensitive to the nutrient status [20]. We recently unraveled cancer-specific metabolism which is epigenetically driven by global shifts in the histone landscape through metabolic reprogramming [54]. Histone modifications are a dynamic chromatin mark with various important roles in gene regulation [39], and histone H3 acetylation is particularly responsive to shifts in metabolites and highly predictive of gene activity regarding the promoter and enhancer regions of the gene [72]. One of the major marks reported to be found in actively transcribed promoters is acetylation at the ninth lysine residue of histone H3 N-terminal tail (H3K9ac), and we recently demonstrated that GBM cells with activated EGFR-mTORC2 signaling increase H3K9ac through metabolic reprogramming/Warburg effect (hence the production of nuclear acetyl-CoA), in cooperation with histone-modifying enzymes including pyruvate dehydrogenase (PDH) and class IIa subtypes of HDACs (Fig. 3a) [54, 65]. Unexpectedly, comprehensive studies with RNA-seq and ChIP-seq analyses revealed that mTORC2-dependent increase in H3K9ac was uniquely induced at the promoter regions of the components in iron metabolism genes (Fig. 3a) [54]. Upregulated iron metabolism with increased expression of iron-related genes (ferritin, transferrin receptor, divalent metal transporter 1, hepcidin), along with facilitated uptake of iron, enables GBM cells to store more iron in the cell and eventually promotes the survival of GBM cells (Fig. 3b–d). This might be therapeutically exploitable since GBM cells with activated mTORC2 signaling are addicted to iron metabolism for their survival [54]. The mechanisms by which intracellular iron accumulation leads to cell survival awaits further investigation with a novel technique [10, 15, 27, 36], but a recent report demonstrated that GBM stem-like cells epigenetically promote iron trafficking and tumor growth in a ferritin-dependent manner [69]. Normal cells including hepatocytes regulate intracellular iron homeostasis through the post-transcriptional control of iron metabolism genes via the iron responsive element–iron regulatory protein (IRE–IRP) system [10, 26]. In other words, normal cells control iron metabolism according to the amount of iron in the environment while cancer cells regulate iron metabolism autonomously using epigenetic mechanisms (Fig. 3a), and this could ensure cancer cell survival in various niches. Furthermore, we propose a scheme by which cancer cells promote iron metabolism necessary for cell proliferation and growth when they sense appropriate nutrients (glucose) in the microenvironment (Fig. 3a).
Fig. 3.
Co-dependency of metabolism and epigenetics regulates iron metabolism in cancer. (a) Metabolic reprogramming induced by mTORC2 promotes histone acetylation via the regulation of both the substrate and modifying enzymes, leading to the up-regulation of iron-metabolism genes, a key downstream effector of cell survival in GBM. The data are taken and modified from ref [54]. PDH, pyruvated dehydrogenase; HDAC, histone deacetylase. (b) mTORC2-dependent activation of iron metabolism enables cancer cells to store iron in the cell by promoting iron uptake and suppressing its excretion. Tf, transferrin; TfR, transferrin recptor. FTH1, ferritin heavy chain; FTL, ferritin light chain. (c) Histochemical studies revealed that GBM tissue contains more iron (Berlin blue staining) as well as ferritin molecule (immunohistochemical staining) compared with normal brain tissue. (d) The immunoreactivity of histone acetylation (H3K9ac) and iron-related ferritin is well correlated with those of mTORC2 activation markers (p-AKT, p-NDRG1) in human GBM tissue. All methods and experimental protocols related to human subjects were approved by each institutional review board of Ethics Committee, and the procedures related to human subjects were carried out in accordance with each institutional review board-approved protocol and Declaration of Helsinki, 2013.
V. Conclusion and Future Perspective
Cancer is a disease of endogenous somatic mutations, and the traditional phenotypic classifications of neoplasms have been replaced by those based on distinct genetic and epigenetic profiles in each tumor entity. Tumor development, progression and therapy response are profoundly influenced by the intracellular metabolism and the exogenous microenvironment of tumor cells. This potentially shifts the epigenetic landscape, including DNA methylation and histone modifications. Interestingly, metabolism and epigenetics are inherently co-dependent relationships, and their mutual dependence enables tumor cells to appropriately respond to the microenvironment and ensure cell survival. Indeed, the interplay of metabolism and epigenetics plays a role in the development as well as aggressive phenotypes of not only malignant brain tumor GBM, but also other types of cancer; dynamic epigenetic shift is involved in the biology of non-small cell as well as small cell lung cancer [17], and the energy metabolism in lung cancer cells seems to be associated with the epigenetic status of the cell [3]. Further, epigenetic changes, including DNA methylation and histone modifications, have key pathophysiological roles in the initiation and progression of colorectal cancer [30], and metabolic reprogramming regulates the proliferation of colon cancer cells through histone acetylation [16]. Of note, specific subtypes of gastric, colon and lung cancer including their precancerous lesions demonstrate unique epigenetic patterns through the effect of such factors as chronic inflammation and environmental stimuli [17, 30, 56], indicating the deep involvement of metabolism and epigenetics in the biology of a broader range of cancer, the mechanism of which could be exploitable as novel biomarkers and therapeutic/preventive strategies [76]. At the same time, a slight tip in the balance of this regulation is sufficient to result in a cell catastrophe. Further, recent work suggests that tissue context-based cues can shape metabolic dependencies [13], potentially through both genetic and epigenetic mechanisms. This “epigenetic vulnerability” of certain cancer cells recapitulates the notion of “oncogene addiction” [77], the knowledge of which will lead to rational combination of cytotoxic and molecular targeted therapies [47], in order to effectively target the metabolic and epigenetic networks on which glioma cells heavily depend. Future studies are needed to determine precisely how the primary genetic mutations specific for each tumor entity facilitate cancer metabolic reprogramming and epigenetic shifts and how, at the same time, extracellular nutrients modulate oncogenic signaling in order to translate these insights into more effective treatments for cancer patients.
VI. Conflicts of Interest
P.S.M. is a co-founder of Boundless Bio, Inc. He has equity interest and serves as the chair of its scientific advisory board.
VII. Acknowledgments
We thank Department of Neurosurgery, Tokyo Women’s Medical University for biospecimen and biorepository support. This work is supported by a Grant-in-Aid from Takeda Science Foundation (K.M.), Japan Society for the Promotion of Science KAKENHI Grant 19K07649 (K.M.), and National Institutes of Health NS73831 (P.S.M.).
VIII. References
- 1.Allis C. D. and Jenuwein T. (2016) The molecular hallmarks of epigenetic control. Nat. Rev. Genet. 17; 487–500. [DOI] [PubMed] [Google Scholar]
- 2.Amaral P. P. and Mattick J. S. (2008) Noncoding RNA in development. Mamm. Genome 19; 454–492. [DOI] [PubMed] [Google Scholar]
- 3.Amoêdo N. D., Rodrigues M. F., Pezzuto P., Galina A., da Costa R. M., de Almeida F. C., El-Bacha T. and Rumjanek F. D. (2011) Energy metabolism in H460 lung cancer cells: effects of histone deacetylase inhibitors. PLoS One 6; e22264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anastasiadou E., Jacob L. S. and Slack F. J. (2018) Non-coding RNA networks in cancer. Nat. Rev. Cancer 18; 5–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Babic I., Anderson E. S., Tanaka K., Guo D., Masui K., Li B., Zhu S., Gu Y., Villa G. R., Akhavan D., Nathanson D., Gini B., Mareninov S., Li R., Camacho C. E., Kurdistani S. K., Eskin A., Nelson S. F., Yong W. H., Cavenee W. K., Cloughesy T. F., Christofk H. R., Black D. L. and Mischel P. S. (2013) EGFR mutation-induced alternative splicing of Max contributes to growth of glycolytic tumors in brain cancer. Cell Metab. 17; 1000–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Banerjee A., Mahata B., Dhir A., Mandal T. K. and Biswas K. (2019) Elevated histone H3 acetylation and loss of the Sp1-HDAC1 complex de-repress the GM2-synthase gene in renal cell carcinoma. J. Biol. Chem. 294; 1005–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bannister A. J. and Kouzarides T. (2011) Regulation of chromatin by histone modifications. Cell Res. 21; 381–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bechet D., Gielen G. G., Korshunov A., Pfister S. M., Rousso C., Faury D., Fiset P. O., Benlimane N., Lewis P. W., Lu C., David Allis C., Kieran M. W., Ligon K. L., Pietsch T., Ellezam B., Albrecht S. and Jabado N. (2014) Specific detection of methionine 27 mutation in histone 3 variants (H3K27M) in fixed tissue from high-grade astrocytomas. Acta Neuropathol. 128; 733–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bird A. (2002) DNA methylation patterns and epigenetic memory. Genes Dev. 16; 6–21. [DOI] [PubMed] [Google Scholar]
- 10.Bogdan A. R., Miyazawa M., Hashimoto K. and Tsuji Y. (2016) Regulators of iron homeostasis: new players in metabolism, cell death, and disease. Trends Biochem. Sci. 41; 274–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brennan C. W., Verhaak R. G., McKenna A., Campos B., Noushmehr H., Salama S. R., Zheng S., Chakravarty D., Sanborn J. Z., Berman S. H., Beroukhim R., Bernard B., Wu C. J., Genovese G., Shmulevich I., Barnholtz-Sloan J., Zou L., Vegesna R., Shukla S. A., Ciriello G., Yung W. K., Zhang W., Sougnez C., Mikkelsen T., Aldape K., Bigner D. D., Van Meir E. G., Prados M., Sloan A., Black K. L., Eschbacher J., Finocchiaro G., Friedman W., Andrews D. W., Guha A., Iacocca M., O’Neill B. P., Foltz G., Myers J., Weisenberger D. J., Penny R., Kucherlapati R., Perou C. M., Hayes D. N., Gibbs R., Marra M., Mills G. B., Lander E., Spellman P., Wilson R., Sander C., Weinstein J., Meyerson M., Gabriel S., Laird P. W., Haussler D., Getz G. and Chin L.; TCGA Research Network. (2013) The somatic genomic landscape of glioblastoma. Cell 155; 462–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ceccarelli M., Barthel F. P., Malta T. M., Sabedot T. S., Salama S. R., Murray B. A., Morozova O., Newton Y., Radenbaugh A., Pagnotta S. M., Anjum S., Wang J., Manyam G., Zoppoli P., Ling S., Rao A. A., Grifford M., Cherniack A. D., Zhang H., Poisson L., Carlotti C. G. Jr., Tirapelli D. P., Rao A., Mikkelsen T., Lau C. C., Yung W. K., Rabadan R., Huse J., Brat D. J., Lehman N. L., Barnholtz-Sloan J. S., Zheng S., Hess K., Rao G., Meyerson M., Beroukhim R., Cooper L., Akbani R., Wrensch M., Haussler D., Aldape K. D., Laird P. W., Gutmann D. H., Noushmehr H., Iavarone A. and Verhaak R. G.; TCGA Research Network. (2016) Molecular profiling reveals biologically discrete subsets and pathways of progression in diffuse glioma. Cell 164; 550–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chowdhry S., Zanca C., Rajkumar U., Koga T., Diao Y., Raviram R., Liu F., Turner K., Yang H., Brunk E., Bi J., Furnari F., Bafna V., Ren B. and Mischel P. S. (2019) NAD metabolic dependency in cancer is shaped by gene amplification and enhancer remodelling. Nature 569; 570–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clapier C. R., Iwasa J., Cairns B. R. and Peterson C. L. (2017) Mechanisms of action and regulation of ATP-dependent chromatin-remodelling complexes. Nat. Rev. Mol. Cell Biol. 18; 407–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crooks D. R., Maio N., Lane A. N., Jarnik M., Higashi R. M., Haller R. G., Yang Y., Fan T. W., Linehan W. M. and Rouault T. A. (2018) Acute loss of iron-sulfur clusters results in metabolic reprogramming and generation of lipid droplets in mammalian cells. J. Biol. Chem. 293; 8297–8311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Donohoe D. R., Collins L. B., Wali A., Bigler R., Sun W. and Bultman S. J. (2012) The Warburg effect dictates the mechanism of butyrate-mediated histone acetylation and cell proliferation. Mol. Cell 48; 612–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duruisseaux M. and Esteller M. (2018) Lung cancer epigenetics: From knowledge to applications. Semin. Cancer Biol. 51; 116–128. [DOI] [PubMed] [Google Scholar]
- 18.Flavahan W. A., Drier Y., Liau B. B., Gillespie S. M., Venteicher A. S., Stemmer-Rachamimov A. O., Suvà M. L. and Bernstein B. E. (2016) Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature 529; 110–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flavahan W. A., Gaskell E. and Bernstein B. E. (2017) Epigenetic plasticity and the hallmarks of cancer. Science 357; pii: eaal2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao T., Díaz-Hirashi Z. and Verdeguer F. (2018) Metabolic signaling into chromatin modifications in the regulation of gene expression. Int. J. Mol. Sci. 19; pii: E4108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Greenberg M. V. C. and Bourc’his D. (2019) The diverse roles of DNA methylation in mammalian development and disease. Nat. Rev. Mol. Cell Biol. 20; 590–607. [DOI] [PubMed] [Google Scholar]
- 22.Gu Y., Albuquerque C. P., Braas D., Zhang W., Villa G. R., Bi J., Ikegami S., Masui K., Gini B., Yang H., Gahman T. C., Shiau A. K., Cloughesy T. F., Christofk H. R., Zhou H., Guan K. L. and Mischel P. S. (2017) mTORC2 regulates amino acid metabolism in cancer by phosphorylation of the cystine-glutamate antiporter xCT. Mol. Cell 67; 128–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hall A. W., Battenhouse A. M., Shivram H., Morris A. R., Cowperthwaite M. C., Shpak M. and Iyer V. R. (2018) Bivalent chromatin domains in glioblastoma reveal a subtype-specific signature of glioma stem cells. Cancer Res. 78; 2463–2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hanahan D. and Weinberg R. A. (2011) Hallmarks of cancer: the next generation. Cell 144; 646–674. [DOI] [PubMed] [Google Scholar]
- 25.He S., Sun H., Lin L., Zhang Y., Chen J., Liang L., Li Y., Zhang M., Yang X., Wang X., Wang F., Zhu F., Chen J., Pei D. and Zheng H. (2017) Passive DNA demethylation preferentially up-regulates pluripotency-related genes and facilitates the generation of induced pluripotent stem cells. J. Biol. Chem. 292; 18542–18555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hentze M. W., Muckenthaler M. U., Galy B. and Camaschella C. (2010) Two to tango: regulation of mammalian iron metabolism. Cell 142; 24–38. [DOI] [PubMed] [Google Scholar]
- 27.Hirayama T. (2018) Development of chemical tools for imaging of Fe(II) ions in living cells: A Review. Acta Histochem. Cytochem. 51; 137–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jaunmuktane Z., Capper D., Jones D. T. W., Schrimpf D., Sill M., Dutt M., Suraweera N., Pfister S. M., von Deimling A. and Brandner S. (2019) Methylation array profiling of adult brain tumours: diagnostic outcomes in a large, single centre. Acta Neuropathol. Commun. 7; 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johann P. D., Erkek S., Zapatka M., Kerl K., Buchhalter I., Hovestadt V., Jones D. T. W., Sturm D., Hermann C., Segura Wang M., Korshunov A., Rhyzova M., Gröbner S., Brabetz S., Chavez L., Bens S., Gröschel S., Kratochwil F., Wittmann A., Sieber L., Geörg C., Wolf S., Beck K., Oyen F., Capper D., van Sluis P., Volckmann R., Koster J., Versteeg R., von Deimling A., Milde T., Witt O., Kulozik A. E., Ebinger M., Shalaby T., Grotzer M., Sumerauer D., Zamecnik J., Mora J., Jabado N., Taylor M. D., Huang A., Aronica E., Bertoni A., Radlwimmer B., Pietsch T., Schüller U., Schneppenheim R., Northcott P. A., Korbel J. O., Siebert R., Frühwald M. C., Lichter P., Eils R., Gajjar A., Hasselblatt M., Pfister S. M. and Kool M. (2016) Atypical teratoid/rhabdoid tumors are comprised of three epigenetic subgroups with distinct enhancer landscapes. Cancer Cell 29; 379–393. [DOI] [PubMed] [Google Scholar]
- 30.Jung G., Hernández-Illán E., Moreira L., Balaguer F. and Goel A. (2020) Epigenetics of colorectal cancer: biomarker and therapeutic potential. Nat. Rev. Gastroenterol. Hepatol. 17; 111–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kadoch C. (2019) Diverse compositions and functions of chromatin remodeling machines in cancer. Sci. Transl. Med. 11; pii: eaay1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaelin W. G. and McKnight S. L. (2013) Influence of metabolism on epigenetics and disease. Cell 153; 56–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karkhanis V., Alinari L., Ozer H. G., Chung J., Zhang X., Sif S. and Baiocchi R. A. (2020) Protein arginine methyltransferase 5 represses tumor suppressor miRNAs that down-regulate CYCLIN D1 and c-MYC expression in aggressive B-cell lymphoma. J. Biol. Chem. 295; 1165–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klughammer J., Kiesel B., Roetzer T., Fortelny N., Nemc A., Nenning K. H., Furtner J., Sheffield N. C., Datlinger P., Peter N., Nowosielski M., Augustin M., Mischkulnig M., Ströbel T., Alpar D., Ergüner B., Senekowitsch M., Moser P., Freyschlag C. F., Kerschbaumer J., Thomé C., Grams A. E., Stockhammer G., Kitzwoegerer M., Oberndorfer S., Marhold F., Weis S., Trenkler J., Buchroithner J., Pichler J., Haybaeck J., Krassnig S., Mahdy Ali K., von Campe G., Payer F., Sherif C., Preiser J., Hauser T., Winkler P. A., Kleindienst W., Würtz F., Brandner-Kokalj T., Stultschnig M., Schweiger S., Dieckmann K., Preusser M., Langs G., Baumann B., Knosp E., Widhalm G., Marosi C., Hainfellner J. A., Woehrer A. and Bock C. (2018) The DNA methylation landscape of glioblastoma disease progression shows extensive heterogeneity in time and space. Nat. Med. 24; 1611–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kohli R. M. and Zhang Y. (2013) TET enzymes, TDG and the dynamics of DNA demethylation. Nature 502; 472–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kumamoto Y., Harada Y., Takamatsu T. and Tanaka H. (2018) Label-free molecular imaging and analysis by Raman spectroscopy. Acta Histochem. Cytochem. 51; 101–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kyaw M. T. H., Yamaguchi Y., Choijookhuu N., Yano K., Takagi H., Takahashi N., Synn Oo P., Sato K. and Hishikawa Y. (2019) The HDAC inhibitor, SAHA, combined with cisplatin synergistically induces apoptosis in alpha-fetoprotein-producing hepatoid adenocarcinoma cells. Acta Histochem. Cytochem. 52; 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lamb R. F. (2012) Amino acid sensing mechanisms: An achilles heel in cancer? FEBS J. 279; 2624–2631. [DOI] [PubMed] [Google Scholar]
- 39.Lee J. V., Carrer A., Shah S., Snyder N. W., Wei S., Venneti S., Worth A. J., Yuan Z. F., Lim H. W., Liu S., Jackson E., Aiello N. M., Haas N. B., Rebbeck T. R., Judkins A., Won K. J., Chodosh L. A., Garcia B. A., Stanger B. Z., Feldman M. D., Blair I. A. and Wellen K. E. (2014) Akt-dependent metabolic reprogramming regulates tumor cell histone acetylation. Cell Metab. 20; 306–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin C. and He L. (2017) Noncoding RNAs in Cancer Development. Annu. Rev. Cancer Biol. 1; 163–184. [Google Scholar]
- 41.Liu F., Hon G. C., Villa G. R., Turner K. M., Ikegami S., Yang H., Ye Z., Li B., Kuan S., Lee A. Y., Zanca C., Wei B., Lucey G., Jenkins D., Zhang W., Barr C. L., Furnari F. B., Cloughesy T. F., Yong W. H., Gahman T. C., Shiau A. K., Cavenee W. K., Ren B. and Mischel P. S. (2015) EGFR mutation promotes glioblastoma through epigenome and transcription factor network remodeling. Mol. Cell 60; 307–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu J., Zhang W., Wu Z., Dai L. and Koji T. (2018) Changes in DNA methylation of oocytes and granulosa cells assessed by HELMET during folliculogenesis in mouse ovary. Acta Histochem. Cytochem. 51; 93–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu M., Saha N., Gajan A., Saadat N., Gupta S. V. and Pile L. A. (2020) A complex interplay between SAM synthetase and the epigenetic regulator SIN3 controls metabolism and transcription. J. Biol. Chem. 295; 375–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lu C. and Thompson C. B. (2012) Metabolic regulation of epigenetics. Cell Metab. 16; 9–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lu C., Ward P. S., Kapoor G. S., Rohle D., Turcan S., Abdel-Wahab O., Edwards C. R., Khanin R., Figueroa M. E., Melnick A., Wellen K. E., O’Rourke D. M., Berger S. L., Chan T. A., Levine R. L., Mellinghoff I. K. and Thompson C. B. (2012) IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature 483; 474–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Masui K., Cloughesy T. F. and Mischel P. S. (2012) Review: molecular pathology in adult high-grade gliomas: from molecular diagnostics to target therapies. Neuropathol. Appl. Neurobiol. 38; 271–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Masui K., Gini B., Wykosky J., Zanca C., Mischel P. S., Furnari F. B. and Cavenee W. K. (2013) A tale of two approaches: complementary mechanisms of cytotoxic and targeted therapy resistance may inform next-generation cancer treatments. Carcinogenesis 34; 725–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Masui K., Tanaka K., Akhavan D., Babic I., Gini B., Matsutani T., Iwanami A., Liu F., Villa G. R., Gu Y., Campos C., Zhu S., Yang H., Yong W. H., Cloughesy T. F., Mellinghoff I. K., Cavenee W. K., Shaw R. J. and Mischel P. S. (2013) mTOR complex 2 controls glycolytic metabolism in glioblastoma through FoxO acetylation and upregulation of c-Myc. Cell Metab. 18; 726–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Masui K., Cavenee W. K. and Mischel P. S. (2014) mTORC2 in the center of cancer metabolic reprogramming. Trends Endocrinol. Metab. 25; 364–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Masui K., Tanaka K., Ikegami S., Villa G. R., Yang H., Yong W. H., Cloughesy T. F., Yamagata K., Arai N., Cavenee W. K. and Mischel P. S. (2015) Glucose-dependent acetylation of Rictor promotes targeted cancer therapy resistance. Proc. Natl. Acad. Sci. U S A 112; 9406–9411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Masui K., Cavenee W. K. and Mischel P. S. (2016) Cancer metabolism as a central driving force of glioma pathogenesis. Brain Tumor Pathol. 33; 161–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Masui K., Mischel P. S. and Reifenberger G. (2016) Molecular classification of gliomas. Handb. Clin. Neurol. 134; 97–120. [DOI] [PubMed] [Google Scholar]
- 53.Masui K., Shibata N., Cavenee W. K. and Mischel P. S. (2016) mTORC2 activity in brain cancer: Extracellular nutrients are required to maintain oncogenic signaling. Bioessays 38; 839–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Masui K., Harachi M., Ikegami S., Yang H., Onizuka H., Yong W. H., Cloughesy T. F., Muragaki Y., Kawamata T., Arai N., Komori T., Cavenee W. K., Mischel P. S. and Shibata N. (2019) mTORC2 links growth factor signaling with epigenetic regulation of iron metabolism in glioblastoma. J. Biol. Chem. 294; 19740–19751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Masui K., Onizuka H., Cavenee W. K., Mischel P. S. and Shibata N. (2019) Metabolic reprogramming in the pathogenesis of glioma: Update. Neuropathology 39; 3–13. [DOI] [PubMed] [Google Scholar]
- 56.Matsusaka K., Kaneda A., Nagae G., Ushiku T., Kikuchi Y., Hino R., Uozaki H., Seto Y., Takada K., Aburatani H. and Fukayama M. (2011) Classification of Epstein-Barr virus-positive gastric cancers by definition of DNA methylation epigenotypes. Cancer Res. 71; 7187–7197. [DOI] [PubMed] [Google Scholar]
- 57.Matsuzaki I., Iguchi H., Mikasa Y., Morishita H., Okuda K., Nakaguchi K., Mori Y., Iwahashi Y., Warigaya K., Fujimoto M., Kojima F. and Murata S. I. (2017) Novel application of loop-mediated isothermal amplification for rapid detection of gene translocation. Acta Histochem. Cytochem. 50; 169–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Maury E. and Hashizume R. (2017) Epigenetic modification in chromatin machinery and its deregulation in pediatric brain tumors: Insight into epigenetic therapies. Epigenetics 12; 353–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mitra A., Pfeifer K. and Park K. S. (2018) Circular RNAs and competing endogenous RNA (ceRNA) networks. Transl. Cancer Res. 7(Suppl 5); S624–S628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Moure C. J., Diplas B. H., Chen L. H., Yang R., Pirozzi C. J., Wang Z., Spasojevic I., Waitkus M. S., He Y. and Yan H. (2019) CRISPR editing of mutant IDH1 R132H induces a CpG methylation-low state in patient-derived glioma models of G-CIMP. Mol. Cancer Res. 17; 2042–2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nacev B. A., Feng L., Bagert J. D., Lemiesz A. E., Gao J., Soshnev A. A., Kundra R., Schultz N., Muir T. W. and Allis C. D. (2019) The expanding landscape of ‘oncohistone’ mutations in human cancers. Nature 567; 473–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Noushmehr H., Weisenberger D. J., Diefes K., Phillips H. S., Pujara K., Berman B. P., Pan F., Pelloski C. E., Sulman E. P., Bhat K. P., Verhaak R. G., Hoadley K. A., Hayes D. N., Perou C. M., Schmidt H. K., Ding L., Wilson R. K., Van Den Berg D., Shen H., Bengtsson H., Neuvial P., Cope L. M., Buckley J., Herman J. G., Baylin S. B., Laird P. W. and Aldape K.; Cancer Genome Atlas Research Network. (2010) Identification of a CpG Island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell 17; 510–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Parsons D. W., Jones S., Zhang X., Lin J. C., Leary R. J., Angenendt P., Mankoo P., Carter H., Siu I. M., Gallia G. L., Olivi A., McLendon R., Rasheed B. A., Keir S., Nikolskaya T., Nikolsky Y., Busam D. A., Tekleab H., Diaz L. A. Jr., Hartigan J., Smith D. R., Strausberg R. L., Marie S. K., Shinjo S. M., Yan H., Riggins G. J., Bigner D. D., Karchin R., Papadopoulos N., Parmigiani G., Vogelstein B., Velculescu V. E. and Kinzler K. W. (2008) An integrated genomic analysis of human glioblastoma multiforme. Science 321; 1807–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pavlova N. N. and Thompson C. B. (2016) The emerging hallmarks of cancer metabolism. Cell Metab. 23; 27–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Peserico A. and Simone C. (2011) Physical and functional HAT/HDAC interplay regulates protein acetylation balance. J. Biomed. Biotechnol. 2011; 371832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Reid M. A., Dai Z. and Locasale J. W. (2017) The impact of cellular metabolism on chromatin dynamics and epigenetics. Nat. Cell Biol. 19; 1298–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rice J. C. and Allis C. D. (2001) Histone methylation versus histone acetylation: new insights into epigenetic regulation. Curr. Opin. Cell Biol. 13; 263–273. [DOI] [PubMed] [Google Scholar]
- 68.Rynkeviciene R., Simiene J., Strainiene E., Stankevicius V., Usinskiene J., Miseikyte Kaubriene E., Meskinyte I., Cicenas J. and Suziedelis K. (2018) Non-coding RNAs in glioma. Cancers (Basel) 11; pii: E17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schonberg D. L., Miller T. E., Wu Q., Flavahan W. A., Das N. K., Hale J. S., Hubert C. G., Mack S. C., Jarrar A. M., Karl R. T., Rosager A. M., Nixon A. M., Tesar P. J., Hamerlik P., Kristensen B. W., Horbinski C., Connor J. R., Fox P. L., Lathia J. D. and Rich J. N. (2015) Preferential iron trafficking characterizes glioblastoma stem-like cells. Cancer Cell 28; 441–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schwartzentruber J., Korshunov A., Liu X. Y., Jones D. T., Pfaff E., Jacob K., Sturm D., Fontebasso A. M., Quang D. A., Tönjes M., Hovestadt V., Albrecht S., Kool M., Nantel A., Konermann C., Lindroth A., Jäger N., Rausch T., Ryzhova M., Korbel J. O., Hielscher T., Hauser P., Garami M., Klekner A., Bognar L., Ebinger M., Schuhmann M. U., Scheurlen W., Pekrun A., Frühwald M. C., Roggendorf W., Kramm C., Dürken M., Atkinson J., Lepage P., Montpetit A., Zakrzewska M., Zakrzewski K., Liberski P. P., Dong Z., Siegel P., Kulozik A. E., Zapatka M., Guha A., Malkin D., Felsberg J., Reifenberger G., von Deimling A., Ichimura K., Collins V. P., Witt H., Milde T., Witt O., Zhang C., Castelo-Branco P., Lichter P., Faury D., Tabori U., Plass C., Majewski J., Pfister S. M. and Jabado N. (2012) Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature 482; 226–231. [DOI] [PubMed] [Google Scholar]
- 71.Serefidou M., Venkatasubramani A. V. and Imhof A. (2019) The impact of one carbon metabolism on histone methylation. Front. Genet. 10; 764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Strahl B. D. and Allis C. D. (2000) The language of covalent histone modifications. Nature 403; 41–45. [DOI] [PubMed] [Google Scholar]
- 73.Sturm D., Witt H., Hovestadt V., Khuong-Quang D. A., Jones D. T., Konermann C., Pfaff E., Tönjes M., Sill M., Bender S., Kool M., Zapatka M., Becker N., Zucknick M., Hielscher T., Liu X. Y., Fontebasso A. M., Ryzhova M., Albrecht S., Jacob K., Wolter M., Ebinger M., Schuhmann M. U., van Meter T., Frühwald M. C., Hauch H., Pekrun A., Radlwimmer B., Niehues T., von Komorowski G., Dürken M., Kulozik A. E., Madden J., Donson A., Foreman N. K., Drissi R., Fouladi M., Scheurlen W., von Deimling A., Monoranu C., Roggendorf W., Herold-Mende C., Unterberg A., Kramm C. M., Felsberg J., Hartmann C., Wiestler B., Wick W., Milde T., Witt O., Lindroth A. M., Schwartzentruber J., Faury D., Fleming A., Zakrzewska M., Liberski P. P., Zakrzewski K., Hauser P., Garami M., Klekner A., Bognar L., Morrissy S., Cavalli F., Taylor M. D., van Sluis P., Koster J., Versteeg R., Volckmann R., Mikkelsen T., Aldape K., Reifenberger G., Collins V. P., Majewski J., Korshunov A., Lichter P., Plass C., Jabado N. and Pfister S. M. (2012) Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell 22; 425–437. [DOI] [PubMed] [Google Scholar]
- 74.Sturm D., Orr B. A., Toprak U. H., Hovestadt V., Jones D. T. W., Capper D., Sill M., Buchhalter I., Northcott P. A., Leis I., Ryzhova M., Koelsche C., Pfaff E., Allen S. J., Balasubramanian G., Worst B. C., Pajtler K. W., Brabetz S., Johann P. D., Sahm F., Reimand J., Mackay A., Carvalho D. M, Remke M., Phillips J. J., Perry A., Cowdrey C., Drissi R., Fouladi M., Giangaspero F., Łastowska M., Grajkowska W., Scheurlen W., Pietsch T., Hagel C., Gojo J., Lötsch D., Berger W., Slavc I., Haberler C., Jouvet A., Holm S., Hofer S., Prinz M., Keohane C., Fried I., Mawrin C., Scheie D., Mobley B. C., Schniederjan M. J., Santi M., Buccoliero A. M., Dahiya S., Kramm C. M., von Bueren A. O., von Hoff K., Rutkowski S., Herold-Mende C., Frühwald M. C., Milde T., Hasselblatt M., Wesseling P., Rößler J., Schüller U., Ebinger M., Schittenhelm J., Frank S., Grobholz R., Vajtai I., Hans V., Schneppenheim R., Zitterbart K., Collins V. P., Aronica E., Varlet P., Puget S., Dufour C., Grill J., Figarella-Branger D., Wolter M., Schuhmann M. U., Shalaby T., Grotzer M., van Meter T., Monoranu C. M., Felsberg J., Reifenberger G., Snuderl M., Forrester L. A., Koster J., Versteeg R., Volckmann R., van Sluis P., Wolf S., Mikkelsen T., Gajjar A., Aldape K., Moore A. S., Taylor M. D., Jones C., Jabado N., Karajannis M. A., Eils R., Schlesner M., Lichter P., von Deimling A., Pfister S. M., Ellison D. W., Korshunov A. and Kool M. (2016) New brain tumor entities emerge from molecular classification of CNS-PNETs. Cell 164; 1060–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Valencia A. M. and Kadoch C. (2019) Chromatin regulatory mechanisms and therapeutic opportunities in cancer. Nat. Cell Biol. 21; 152–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vander Heiden M. G. and DeBerardinis R. J.. Understanding the intersections between metabolism and cancer biology. Cell 168; 657–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Weinstein I. B. (2002) Cancer. Addiction to oncogenes—the Achilles heel of cancer. Science 297; 63–64. [DOI] [PubMed] [Google Scholar]
- 78.Xu W., Yang H., Liu Y., Yang Y., Wang P., Kim S. H., Ito S., Yang C., Wang P., Xiao M. T., Liu L. X., Jiang W. Q., Liu J., Zhang J. Y., Wang B., Frye S., Zhang Y., Xu Y. H., Lei Q. Y., Guan K. L., Zhao S. M. and Xiong Y. (2011) Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of α-ketoglutarate-dependent dioxygenases. Cancer Cell 19; 17–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yan H., Parsons D. W., Jin G., McLendon R., Rasheed B. A., Yuan W., Kos I., Batinic-Haberle I., Jones S., Riggins G. J., Friedman H., Friedman A., Reardon D., Herndon J., Kinzler K. W., Velculescu V. E., Vogelstein B. and Bigner D. D. (2009) IDH1 and IDH2 mutations in gliomas. N. Engl. J. Med. 360; 765–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yang M., Soga T. and Pollard P. J. (2013) Oncometabolites: linking altered metabolism with cancer. J. Clin. Invest. 123; 3652–3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yang W., Xia Y., Hawke D., Li X., Liang J., Xing D., Aldape K., Hunter T., Alfred Yung W. K. and Lu Z. (2012) PKM2 phosphorylates histone H3 and promotes gene transcription and tumorigenesis. Cell 150: 685–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yen C. Y., Huang H. W., Shu C. W., Hou M. F., Yuan S. S., Wang H. R., Chang Y. T., Farooqi A. A., Tang J. Y. and Chang H. W. (2016) DNA methylation, histone acetylation and methylation of epigenetic modifications as a therapeutic approach for cancers. Cancer Lett. 373; 185–192. [DOI] [PubMed] [Google Scholar]
- 83.Yu X. and Li S. (2017) Non-metabolic functions of glycolytic enzymes in tumorigenesis. Oncogene 36; 2629–2636. [DOI] [PubMed] [Google Scholar]
- 84.Yu X., Ma R., Wu Y., Zhai Y. and Li S. (2018) Reciprocal regulation of metabolic reprogramming and epigenetic modifications in cancer. Front. Genet. 9; 394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhao Z. and Shilatifard A. (2019) Epigenetic modifications of histones in cancer. Genome Biol. 20; 245. [DOI] [PMC free article] [PubMed] [Google Scholar]