Abstract
Polypyrimidine tract-binding protein 1 (PTBP1) plays an essential role in splicing and is expressed in almost all cell types in humans, unlike the other proteins of the PTBP family. PTBP1 mediates several cellular processes in certain types of cells, including the growth and differentiation of neuronal cells and activation of immune cells. Its function is regulated by various molecules, including microRNAs (miRNAs), long non-coding RNAs (lncRNAs), and RNA-binding proteins. PTBP1 plays roles in various diseases, particularly in some cancers, including colorectal cancer, renal cell cancer, breast cancer, and glioma. In cancers, it acts mainly as a regulator of glycolysis, apoptosis, proliferation, tumorigenesis, invasion, and migration. The role of PTBP1 in cancer has become a popular research topic in recent years, and this research has contributed greatly to the formulation of a useful therapeutic strategy for cancer. In this review, we summarize recent findings related to PTBP1 and discuss how it regulates the development of cancer cells.
Keywords: Polypyrimidine tract-binding protein 1 (PTBP1), Alternative splicing, Glycolysis, M2 isoform of pyruvate kinase (PKM2), Cancer
1. Introduction
Polypyrimidine tract-binding protein (PTBP) preferentially binds to polypyrimidine-rich stretches of RNA. It functions mainly in splicing, and can shuttle between the cytoplasm and the nucleus. PTBP1 belongs to the PTB family, which also includes PTBP2 and PTBP3. PTBP1 is expressed in nearly all cell types, PTBP2 is expressed exclusively in the nervous system, and PTBP3 is found predominantly in immune cells (Ghetti et al., 1992; Wagner et al., 1999). These three paralogs share more than 70% sequence homology at the protein level, and all contain four RNA recognition motifs (RRMs) that are used to bind RNA (Oberstrass et al., 2005).
PTBP1 is a known regulator of posttranscriptional gene expression that controls messenger RNA (mRNA) splicing, translation, stability, and localization. It has numerous molecular functions related to RNA metabolism and is a major repressive regulator of alternative splicing, causing exon skipping in many alternatively spliced precursor mRNAs (pre-mRNAs) (Takahashi et al., 2015). Alternative splicing is a regulated process that results in multiple mRNAs and proteins encoded from a single gene. During gene expression, particular exons of the gene can be included or excluded from the final processed mRNA (Black, 2003). Abnormal alternative splicing leads to splicing variants and contributes to the development of cancer (Sveen et al., 2016). PTBP1 has been shown to play roles in modulating splice-site selection and functions as an alternative splicing regulator in mammalian cells (Busch and Hertel, 2012). It crosslinks to the pyrimidine tracts within 3' splice sites and represses exon selection, preventing the exon from splicing (Xue et al., 2009; Llorian et al., 2010). When PTBP1 binds to exonic or/and flanking intronic sequences around the alternative exon, it promotes the inclusion of alternative exons (Corrionero and Valcarcel, 2009; Fu and Ares, 2014). PTBP1 influences pre-mRNA processing and is associated with pre-mRNAs. It is also related to mRNA transport and metabolism (Han et al., 2014). PTBP1 participates in many kinds of activities in different cells, such as the growth and differentiation of neuronal cells, activation of T cells, spermatogenesis, embryonic development, and erythrocyte development. Among its activities in all kinds of cells, the role that PTBP1 plays in the growth and differentiation of neuronal cells is the most important. PTBP1 functions in the entire progression of neuronal cells, including the neuronal transcriptional program, neurogenesis, synapse maturation, and differentiation (Yap et al., 2012; Zheng et al., 2012; Linares et al., 2015).
In cancer cells, glycolysis is the most important process in which PTBP1 is involved (Shinohara et al., 2016). PTBP1 promotes the expression of M2 isoform of pyruvate kinase (PKM2) and decreases the expression of PKM1, which leads mainly to a metabolic shift from oxidative phosphorylation to glycolysis in cancer cells and affects tumorigenesis (He et al., 2014). Apoptosis, proliferation, migration, and invasion are also regulated by PTBP1 via different pathways and molecules (Cui and Placzek, 2016; Jo et al., 2017). It is widely reported that colorectal cancer and glioma are the most common types of cancer involving PTBP1 (Cheung et al., 2009; Takahashi et al., 2015; Zhang et al., 2018). However, the exact mechanisms of PTBP1 function in these and other cancers have not been clearly understood, and more thorough studies may be needed.
2. Overview
2.1. Structure
PTBP1 is an RNA-binding protein that belongs to the subfamily of ubiquitously expressed heterogeneous nuclear ribonucleoproteins (hnRNPs), the gene for which is located on chromosome 19p13.3 in humans. PTBP1 is 531 amino acids long, and has an N-terminal nuclear shuttling domain and four repeats of quasi-RRM domains that participate in the process of RNA-binding (Keppetipola et al., 2012). PTBP1 has a preference for promoters, and coincides with RNA polymerase II (RNA Pol II). It exerts functions in RNA–protein complexes at the initiation of transcription of some genes (Swinburne et al., 2006).
PTBP1 and PTBP2 are both alternative splicing regulators to which many developmentally regulated exons show different sensitivities. The differential targeting arises from different cofactor interactions rather than different RNA interactions (Vuong et al., 2016). In addition, many distinct phosphate modifications are located in the unstructured N-terminal regions of PTBP1 and PTBP2. PTBP1 is ubiquitously expressed, while the expression of PTBP2 is highly tissue-dependent (Licatalosi et al., 2012; Yap et al., 2012). PTBP1 and PTBP2 are non-conserved cryptic exon repressors, and they use CU microsatellites to repress both non-conserved cryptic exons and tissue-specific exons (Ling et al., 2016). The location, structure, and formation of PTBP1 are shown in Fig. 1.
Fig. 1.
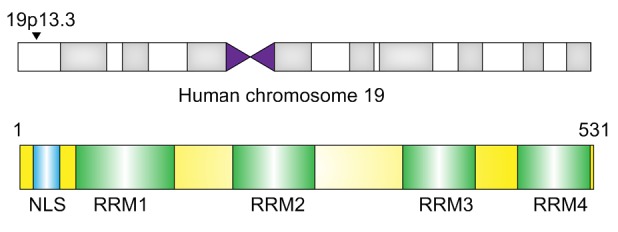
Location, structure, and formation of PTBP1
The PTBP1 gene is located on chromosome 19p13.3 in humans and has one N-terminal nuclear shuttling domain and four repeats of quasi-RNA recognition motif (RRM) domains. NLS: nuclear localization signal
2.2. Regulation in normal cells
The expression of PTBP1 is controlled by other RNA-binding proteins at the transcriptional level. The presence of an alternative exon 11 results in two different splice isoforms of PTBP1 mRNA, and only the isoform that includes exon 11 is able to be transformed to the final product (Méreau et al., 2015). PTBP1 plays a crucial role in the development of the skin of frogs in the Xenopus genus, and downregulation of PTBP1 results in developmental skin defects regulated by the RNA-binding protein Esrp1 (Méreau et al., 2015; Noiret et al., 2017). Esrp1 increases the expression of PTBP1 by promoting the inclusion of exon 11 in the Xenopus epidermis, and high levels of PTBP1 also cause nonsense-mediated decay by skipping exon 11 of PTBP’s own mRNA (Wollerton et al., 2004; Méreau et al., 2015).
Another RNA-binding protein, MATR3, also regulates the function of PTBP1. During RNA processing, long mammalian introns prevent accurate exon identification, and LINE-derived sequences (LINEs) help with the selection by recruiting RNA-binding proteins, such as MATR3, to introns. Then, PTBP1 is recruited by MATR3 to bind to multivalent binding sites within LINEs. These two RNA-binding proteins repress splicing and 3' end processing within LINEs, which contributes to the function of LINEs (Attig et al., 2018).
At the post-transcriptional level, H2O2 treatment decreases the protein level of PTBP1 by inducing a pre-existing protein degradation pathway in breast cells. H2O2 promotes the expression of the oxidation-resistant C-α1 soluble guanylyl cyclase subunit to initiate the formation of PTBP1 dimers, resulting in subsequent protein degradation (Cote et al., 2012).
2.3. Functions of PTBP1 in normal cells
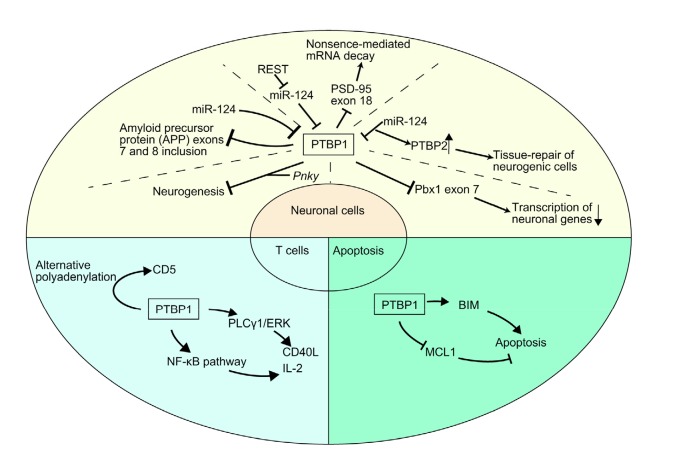
PTBP1 acts as a master regulator of splicing to activate or repress alternative exons depending on the pre-mRNA recruitment position. This regulation plays an important role mainly in the growth and differentiation of neuronal cells (Vuong et al., 2016; Hamid and Makeyev, 2017). PTBP1 blocks up-frameshift 1 (UPF1), a nonsense-mediated mRNA decay (NMD) protein, from binding to 3' untranslated regions (UTRs) to preserve the capacity of long 3' UTRs to regulate gene expression and the ability of NMD to accurately detect aberrant mRNAs (Ge et al., 2016). PTBP1 also acts as a potential dynamic biomarker and is downregulated in the blood of individuals with Parkinson’s disease (PD) (Santiago and Potashkin, 2015b). In addition to its primary role in the nervous system, PTBP1 plays critical roles in the activation of T cells in the immune system (la Porta et al., 2016), the regulation of the cell cycle and apoptosis (Juan et al., 2014), spermatogenesis and Xist induction (Juan et al., 2014; Stork et al., 2018), embryonic development (Dou and Zhang, 2016), and erythrocyte development (Liu JH et al., 2018). All the known or commonly recognized functions and regulatory mechanisms of PTBP1 in normal cells are shown in Fig. 2.
Fig. 2.
Functions and regulation of PTBP1 in normal cells
PTBP1 functions mainly in neuronal cells, in which it interacts with Pnky, miR-124, PSD-95, and Pbx1. PTBP1 regulates neurogenesis, NMD, transcription of neuronal genes, and tissue repair of neurogenic cells. It also regulates CD5 via alternative polyadenylation and CD40L and IL-2 via the PLCγ1/ERK1/2 and NF-κB pathways. PTBP1 regulates apoptosis via BIM and MCL1. miR-124: microRNA-124; PSD-95: postsynaptic density protein 95; NMD: nonsense-mediated mRNA decay; BIM: BCL-2-like 11; MCL1: myeloid cell leukemia 1; REST: repressor element 1 silencing transcription factor; PLCγ1: phospholipase Cγ1; ERK: extracellular signal-regulated kinase; NF-κB: nuclear factor-κB
2.3.1 Growth and differentiation of neuronal cells
PTBP1 functions mainly in the process of neuronal cell growth, in which it reprograms developmental pre-mRNA splicing in neurons (Vuong et al., 2016). PTBP1 suppresses the neuronal transcriptional program by controlling the activity of Pbx1 prior to the induction of neuronal progenitor cell development (Linares et al., 2015). In the mammalian brain, alternative splicing is prevalent, and dynamic control of alternative splicing controls cell fate in the cerebral cortical development of mammals (Zhang XC et al., 2016). PTBP1 also regulates alternative splicing with Pnky, a long non-coding RNA (lncRNA), in neural stem cells (NSCs) (Ramos et al., 2015). Double-knockdown of Pnky and PTBP1 depletes the NSC population and enhances neurogenesis and neuronal differentiation (Ramos et al., 2015), which suggests that the Pnky–PTBP1 complex leads to a splicing program, resulting in suppression during neurogenesis (Grammatikakis and Gorospe, 2016).
2.3.2 T-cell activation and the immune system
PTBP1 functions as a critical regulator of cluster of differentiation 4 (CD4) T-cell activation, which regulates the expression of CD40 ligand (CD40L) and interleukin-2 (IL-2) through the activation of the phospholipase Cγ1 (PLCγ1)/extracellular signal-regulated kinase 1/2 (ERK1/2) and nuclear factor-κB (NF-κB) pathways (la Porta et al., 2016). It also regulates the expression of CD5 via alternative polyadenylation during human T-cell activation (Domingues et al., 2016). PTBP1 also participates in CD46 alternative splicing regulation (Tang et al., 2016). It is also upregulated in B lymphocytes and plays a critical role in B-cell selection in germinal centers (Monzón-Casanova et al., 2018). Furthermore, Sasanuma et al. (2018) have reported that PTBP1 is necessary for B cell receptor (BCR)-mediated antibody production.
2.3.3 Cell cycle and apoptosis
BCL-2-like 11 (BIM), a pro-apoptotic protein, plays a crucial role in impaired or excessive apoptosis, which can contribute to degenerative disorders and tumorigenesis. Researchers have found that the downregulation of PTBP1 can inhibit BIM-mediated apoptosis (Juan et al., 2014). Myeloid cell leukemia 1 (MCL1), a member of the BCL-2 protein family, is a crucial anti-apoptotic protein. A recent study demonstrated that PTBP1 represses MCL1 expression by destabilizing MCL1 mRNA and enhancing the miR-101-guided argonaute 2 (AGO2) interaction with MCL1 (Yang et al., 2018).
2.3.4 Other functions of PTBP1 in cell biology
PTBP1 is a splicing regulatory protein with multiple functions, and acts as a post-transcriptional regulation factor in many processes. It regulates cholesterol biosynthesis by regulating 3-hydroxy-3-methylglutaryl coenzyme A reductase (HMGCR) and low-density lipoprotein (LDL) receptor (LDLR), which are involved in the cholesterol biosynthesis process. These genes undergo alternative splicing, resulting in depletion of the enzyme for cholesterol biosynthesis (Medina et al., 2011). The alternative splicing of dopamine receptor D2 (DRD2) is regulated by PTBP1. DRD2 modulates the intracellular localization of PTBP1 by inhibiting cyclic adenosine monophosphate (cAMP)-dependent protein kinase A. PTBP1 enhances the inclusion of the alternative exon 6 to regulate the expression of the isoforms of DRD2, leading to the autoregulation of DRD2 (Sasabe et al., 2011).
PTBP1 represses the splicing of exons of some genes (Llorian et al., 2016). During the repression of the splicing of the c-src N1 exon, RRM 1 and RRM 2 of PTBP1 directly interact with the pyrimidine-rich internal loop of the U1 small nuclear RNA (snRNA) stem loop 4, which results in the prevention of further spliceosomal component assembly downstream and the U1 small nuclear ribonucleoprotein particle (snRNP) (Sharma et al., 2011). PTBP1 plays a crucial role in the repression of defective splicing caused by ISCU intron mutation in patients who suffer from myopathy, and this repression of defective splicing leads to a drastic reduction in mutant transcripts (Nordin et al., 2012).
2.4. Roles in diseases other than cancer
PTBP1 plays important roles in various diseases. In most of these diseases, it functions mainly as a splicing factor that is regulated by several molecules, including maternally expressed gene 3 (MEG3), H19, miR-124, and glucose. The roles of PTBP1 in diseases other than cancer are shown in Table 1.
Table 1.
Roles of PTBP1 in diseases other than cancer
| Disease | Mechanism | Function/effect | Reference |
| Cholestatic liver injury | MEG3/PTBP1/SHP | Disruption of bile acid (BA) homeostasis, elevation of liver enzymes, dysregulation of BA synthetic enzymes and metabolic genes | Zhang et al., 2017 |
| Pulmonary arterial hypertension | (1) ↓miR-124/PTBP1/PKM1 and PKM2 (2) H19/↓PTBP1/let-7 | (1) Metabolic and proliferative abnormalities (2) Promote the progression of cholestatic liver fibrosis | Kang et al., 2013; Caruso et al., 2017; Zhang et al., 2017; Wang et al., 2019 |
| Fatty liver | H19/↓PTBP1/sterol regulatory element-binding protein 1 | Lipid accumulation; exacerbate the development of fatty liver | Liu C et al., 2018; del Rio-Moreno et al., 2019 |
| Hereditary myopathy with lactic acidosis | PTBP1/ISCU mis-splicing | Substantial decrease in ISCU mis-splicing | Rawcliffe et al., 2018 |
| Idiopathic dilated cardiomyopathy | RBM20 and PTBP1/FHOD3 | Functional cytoskeleton and sarcomere proteins | Lorenzi et al., 2019 |
| Cardiovascular diseases | (1) PTBP1/FHOD3 (2) PTBP1/exons 8/8a L-type CaV1.2 calcium channels | (1) Alteration of CaV1.2 channel (2) Actin filament functional organization | Tang et al., 2011; Lorenzi et al., 2019 |
| Diabetes | Glucose/insulin receptor/PTBP1/insulin | Nuclear retention of PTBP1 and impaired insulin secretion | Ehehalt et al., 2010; Jeong et al., 2018 |
| Alzheimer’s disease | miR-124/PTBP1/amyloid precursor protein | Abnormal neuronal splicing of APP, β-amyloid peptide accumulation | Smith et al., 2011; Vaquero-Garcia et al., 2016 |
| Parkinson’s disease | Unclear | mRNA biomarker, longitudinally dynamic biomarker, aberrant alternative splicing | Santiago and Potashkin, 2015a, 2015b |
MEG3: maternally expressed gene 3; SHP: small heterodimer partner; RBM20: RNA binding motif protein 20; FHOD3: formin homology 2 domain containing 3; APP: amyloid precursor protein
3. Significance in cancers
3.1. Pathways in cancer
PTBP1 functions in many kinds of pathways, such as the epidermal growth factor receptor (EGFR) (Ferrarese et al., 2014; Shi et al., 2018), mitogen-activated protein kinase (MAPK) (Hollander et al., 2016), phosphoinositide 3-kinase (PI3K)/Akt (Ouyang et al., 2018), and hypoxia inducible factor-1α pathways (Shan et al., 2018). It works mainly as a splicing factor and interacts with miR-124 and PKM splicing isoforms in various types of cancer. PTBP1 promotes the progression of glioblastomas and regulates apoptosis during colon tumorigenesis via these pathways. The pathways can be considered as potential targets for treating the corresponding diseases (Zhou et al., 2011; Yao et al., 2015; Wang et al., 2018).
PTBP1 mediates tissue-specific alternative splicing, which is important for the emergence of tissue identity during the process of tissue development. In glioblastomas, the loss of a brain-enriched microRNA (miR-124) leads to PTBP1 amplification. PTBP1 mediates annexin A7 (ANXA7) exon splicing, which eliminates the tumor suppressor functions and promotes glioblastoma progression (Ferrarese et al., 2014). Furthermore, lineage-specific splicing of a brain-enriched cassette exon in ANXA7, a membrane-binding tumor suppressor, decreases endosomal targeting of the EGFR oncoprotein (Ferrarese et al., 2014). To conclude, this mechanism enhances EGFR signaling to promote the progression of glioblastomas (Ferrarese et al., 2014). Oncogenic KRAS mutations increase ELK1 transcription factor levels via the RAS-MAPK pathway, and this increase in ELK1 increases the transcription factors MYC and PTBP1. This process affects the proliferation, migration, and differentiation of colon cancer in a cell-context-specific and cell-type-specific manner. Moreover, PTBP1 promotes major triggers of colon tumorigenesis, including RAC1, NUMB, and PKM splicing isoforms (Hollander et al., 2016). PTBP1 also regulates apoptosis and the cell cycle to promote tumorigenesis (Zhang et al., 2018). It also regulates the PTEN-PI3K/Akt pathway and the hypoxia inducible factor-1α pathway to alter tumor cell growth, migration, and invasion. The latter pathway is specifically involved in renal cell cancer (RCC) (Ouyang et al., 2018; Shan et al., 2018). However, the exact mechanisms of the participation of PTBP1 in the PI3K/Akt pathway and the hypoxia inducible factor-1α pathway have not been completely elucidated.
3.2. Processes regulated by PTBP1 in cancer
3.2.1 Glycolysis
PTBP1 acts as an alternative splicing repressor of the PKM1, leading to the expression of the PKM2 (Shinohara et al., 2016). PTBP1 increases the transformation of PKM1 to PKM2, and the upregulation of PTBP1 is associated with the proliferation, invasion, and migration of human breast cancer, clear-cell renal cell carcinoma (ccRCC), and anaplastic large cell lymphoma (ALCL) (He et al., 2014; Hwang et al., 2017; Jiang et al., 2017). The modulation of PKM alternative splicing and the upregulation of PTBP1 in pancreatic ductal adenocarcinoma (PDAC) cells confers drug resistance, which leads to the transition to drug-resistant PDAC (DR-PDAC) (Calabretta et al., 2016). miR-145 inactivates the MAPK/ERK and PI3K/AKT pathways and impairs cancer-specific energy metabolism by inhibiting the c-Myc/PTBP1/PKMs axis in bladder cancer cells (Takai et al., 2017). The expression of miR-145 downregulates c-Myc expression at the translation level, and c-Myc acts upstream of PTBP1 to positively regulate the expression of PTBP1 (Sachdeva et al., 2009). Moreover, PKM2 is partly upregulated by PTBP1, and PKM2 increases the phosphorylation of Beclin-1 and mediates autophagic activation in acute myeloid leukemia with mutated nucleophosmin (Wang et al., 2019).
Serine and arginine-rich splicing factor 3 (SRSF3) is one of the splicing factors that regulate the alternative splicing of pre-mRNAs and enhance the inclusion of an alternative exon 4 to regulate its own expression (Guo et al., 2015). The silencing of SRSF3 induces marked growth inhibition in human colon cancer cells and causes an increase in the PKM1/PKM2 ratio, which leads to a metabolic shift from glycolysis to oxidative phosphorylation. Subsequently, the cancer cells produce reactive oxygen species (ROS) and induce autophagic cell death (Taniguchi et al., 2018). PTBP1 inhibits the inclusion of an exonic splicing suppressor in exon 4 by binding to it, which leads to the overexpression of full-length functional SRSF3 associated with oncogenesis. Specifically, in oral squamous cell carcinoma (OSCC) cells, SRSF3 exon 4 is damaged (Guo et al., 2015).
Apigenin (AP) and AIC-47 act as anticancer agents in the above diseases. AP ensures a high PKM1/PKM2 ratio in colon cancer cells by blocking the β-catenin/c-Myc/PTBP1 signaling pathway, which reduces PKM2 activity and expression to block cellular glycolysis and induces anti-colon cancer effects (Shan et al., 2017). PTBP1 plays an important role in promoting the growth of chronic myeloid leukemia cells. In the treatment of chronic myeloid leukemia cells, AIC-47 and imatinib decrease the expression of PTBP1 and increase the PKM1 level, which strengthens the attack on cancer energy metabolism (Shinohara et al., 2016). PKM2 alternative splicing may be a promising target for the treatment of ccRCC (Jiang et al., 2017). PTBP1 is recruited to PKM pre-mRNA in DR-PDAC, and the knockdown of PTBP1 reduces its recruitment, which may improve the response of PDAC to chemotherapy if it is considered a new potential therapeutic target (Calabretta et al., 2016). The upregulation of the expression of PKM1 promotes oxidative phosphorylation and reduces tumorigenesis (He et al., 2014).
3.2.2 Apoptosis and proliferation
In cancer cells, PTBP1 regulates apoptosis and proliferation through alternative splicing of MCL1 and adenosine deaminase acting on RNA 1 (ADAR1), respectively. MCL1 is an essential regulator of intrinsic apoptosis and is a member of the anti-apoptotic BCL2 family. PTBP1 regulates MCL1 mRNA by controlling the apoptotic response to tubulin chemotherapeutics, and the knockdown of PTBP1 increases MCL1 expression and MCL1 mRNA accumulation in the cytoplasm of cancer cells (Cui and Placzek, 2016). PTBP1 mediates the translational mode of ADAR1 p110 by an internal ribosomal entry site (IRES)-like element. The protein levels of ADAR1 p110 are upregulated in glioma cells, which is critical for the maintenance of gliomagenesis. The knockdown of ADAR1 p110 significantly decreases the proliferation of glioma cells (Yang et al., 2015).
3.2.3 Tumorigenesis
Cell division control protein 42 homolog (CDC42) and the senescence-associated secretory phenotype (SASP) participate in tumorigenesis via PTBP1. CDC42 has two variants (CDC42-v1 and CDC42-v2) generated by alternative splicing, which act as the main regulators of filopodia formation regulated by PTBP1 in tumorigenesis. Knockdown of PTBP1 promotes the expression of CDC42-v2. CDC42-v2 functions as a tumor suppressor and is expressed in lower levels in ovarian tumor tissues, while CDC42-v1 has an opposite effect (He et al., 2015). Thus PTBP1 plays a vital role in promoting ovarian cancer. PTBP1 controls SASP by regulating exocyst complex exo70 subunit (EXOC7), which is involved in intracellular trafficking. The inhibition of PTBP1 impairs immune surveillance, reducing the risk of tumorigenesis and preventing the protumorigenic effects of SASP. This inhibition functions as a safe and effective therapy against inflammation-driven cancer (Georgilis et al., 2018).
3.2.4 Invasion, migration, and metastasis
PTPB1 interacts with the mRNA of autophagy-related genes (ATGs) directly, and negatively regulates the ATG10 expression level, which plays an important role in colorectal cancer cells (Ouyang et al., 2018). The downregulation of PTBP1 increases ATG10 levels, while the overexpression of PTBP1 decreases ATG10 expression. The knockdown of ATG10 induces the invasion and migration of colorectal cancer cells (Jo et al., 2017). Thus, PTBP1 facilitates metastasis of colorectal cancer cells through downregulation of ATG10.
3.3. Molecules regulating PTBP1 in cancer
3.3.1 LncRNAs
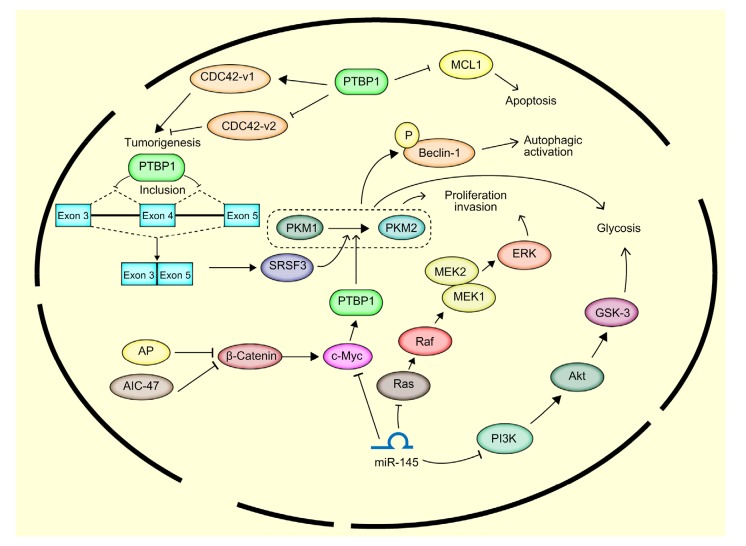
The depletion of ovarian adenocarcinoma-amplified lncRNA (OVAAL), a commonly upregulated lncRNA that confers apoptotic resistance in cancer, triggers cellular senescence via p27 expression. PTBP1-mediated p27 expression is regulated by the competitive binding of p27 mRNA and OVAAL to PTBP1 (Sang et al., 2018). Promotor of CDKN1A antisense DNA damage-activated RNA (PANDAR) is another lncRNA that is involved in the regulation of senescence and proliferation and is transcribed from the CDKN1A promoter. The upregulation of PANDAR results in a decreased level of the short pro-apoptotic BCL-X splice variant (BCL-XS), which is regulated by PTBP1. The overexpression of PTBP1 rescues this effect, which suggests that PANDAR regulates splicing events with its interaction partner PTBP1 (Pospiech et al., 2018). All known or commonly recognized functions and regulatory roles of PTBP1 in cancer cells are shown in Fig. 3.
Fig. 3.
Functions and regulatory roles of PTBP1 in cancer cells
PTBP1 plays an important role in glycolysis, proliferation, invasion, apoptosis, and tumorigenesis. It is regulated by the miR-145/c-Myc and AP/β-catenin/c-Myc pathways. PTBP1 increases the transformation of PKM1 to PKM2, which is a crucial process for glycolysis. PKM2 increases the phosphorylation of Beclin-1 and mediates autophagic activation. PTBP1 inhibits the inclusion of an exonic splicing suppressor in exon 4 of the SRSF3 gene. This inhibition leads to the overexpression of SRSF3 and increases the transformation of PKM1 to PKM2. PTBP1 regulates tumorigenesis via CDC42-v1/v2 and regulates apoptosis via MCL1. CDC42: cell division control protein 42 homolog; MCL1: myeloid cell leukemia 1; SRSF3: serine and arginine-rich splicing factor 3; ERK: extracellular signal-regulated kinase; MEK: mitogen-activated protein kinase kinase; GSK-3: glycogen synthase kinase-3; PI3K: phosphoinositide 3-kinase; P: phosphorylation; AP: apigenin
3.3.2 MicroRNAs
The Warburg effect is a well-known cancer-specific metabolic effect that shifts the metabolism from oxidative phosphorylation to glycolysis. miR-145 disturbs the Warburg effect by inhibiting the Kruppel-like factor 4 (KLF4)/PTBP1/PKMs pathway via specifically binding to the 3' UTR of KLF4 in human bladder cancer cells (Minami et al., 2017). miR-133b functions as a tumor suppressor by negatively regulating the Warburg effect in gastric cancer cells. This regulation reduces PTBP1 expression at the translational level. For the treatment of gastric cancer, knockdown of PTBP1 and ectopic expression of miR-133b switch the PKM isoform expression from PKM2 to PKM1 to inhibit cell proliferation (Sugiyama et al., 2016). The overexpression of linc-ROR (long intergenic non-protein coding RNA, regulator of reprogramming) increases basal autophagy in pancreatic cancer cells via the miR-124/PTBP1/PKM2 axis. miR-124 overexpression negatively correlates with linc-ROR expression and reduces PTBP1 and PKM2 levels to enhance gemcitabine-induced cell apoptosis (Li et al., 2016). miR-124 also regulates a DDX6/c-Myc/PTBP1 positive feedback circuit, which indicates its important role in the maintenance of the Warburg effect in colorectal cancer (Taniguchi et al., 2015).
3.3.3 Other molecules that regulate PTBP1 in cancer
In addition to lncRNAs and microRNAs, other molecules regulate PTBP1, including NOVA1, MYCN, BAF45d, H2O2, and ELK1. PTBP1 coordinates with NOVA1 to promote the splicing of human telomerase reverse transcriptase (hTERT) transcripts and to produce full-length TERT, which promotes telomere maintenance in cancer cells. The knockdown of PTBP1 results in an increased expression of PTBP2, and PTBP2 interacts with NOVA1 to prevent the interaction of NOVA1 with hTERT pre-mRNA (Sayed et al., 2018). MYCN directly binds to the promoter regions of PTBP1 to regulate the expression of this key splicing factor in neuroblastoma (NBL). Knocking down PTBP1 leads to repressed growth of NBL cells, and PTBP1 displays a unique expression pattern in MYCN-amplified tumors and may act as a potential therapeutic target in high-risk NBL with MYCN amplification (Zhang SL et al., 2016). BAF45d, part of the switch non-fermentable complex in glioblastoma, regulates PTBP1. Its function suggests a reciprocal interplay between transcription and RNA splicing regulation (Aldave et al., 2018). H2O2 decreases PTBP1 and hnRNP A2/B1 splice factors at the protein level, which functions as a treatment for breast cancer (Cote et al., 2012). The transcription factor ETS domain-containing protein ELK1 induces the transcription factor MYC and then induces the alternative splicing factor PTBP1 in colon cancer (Hollander et al., 2016).
3.4. Roles in various cancers
PTBP1 participates in various cancers mainly as a splicing factor, thereby playing a critical role in the processes of migration, metastasis, proliferation, and carcinogenesis. It is regulated by various molecules, such as miR-137, miR-206, and miR-133b. The roles of PTBP1 in various cancers are shown in Table 2.
Table 2.
Roles of PTBP1 in various cancers
| Type of cancer | Mechanism | Function/effect | Reference |
| Melanoma | PTBP1/CD44v6 | Migration of melanoma brain metastasis | Marzese et al., 2015 |
| Hepatoma | miR137 and miR206/↓PTBP1/PKM1 | Negative effects related to tissue characteristics and carcinogenesis | Taniguchi et al., 2018 |
| Rhabdomyosarcoma | ↓miR-133b/PAX3-FOXO1/PTBP1 and ↓miR-1 and ↓miR-133b/PTBP1 | Positive regulation of cancer-specific energy metabolism | Sugito et al., 2017 |
| Cervical cancer | HPV infection/PTBP1 | Promotion of DNA replication and cancer cell proliferation | Xu et al., 2019 |
| Acute myeloid leukemia with mutated nucleophosmin | PTBP1/PKM2/phosphorylation of Beclin-1 | Autophagic activation and cancer cell survival | Wang et al., 2019 |
| Anaplastic large cell lymphoma | PTBP1/PKM2/STAT3 | Transcription of genes involved in cell survival and proliferation, oncogenesis | Hwang et al., 2017 |
| Chronic myeloid leukemia | AIC-47 and imatinib/↓PBTP1/↓PKM2 | Perturbation of energy metabolism | Shinohara et al., 2016 |
| Gastric cancer | miR-133b/↓PTBP1 | Negative regulation of the Warburg effect | Sugiyama et al., 2016 |
| Oral squamous cell carcinoma | PTBP1/SRSF3 | Overexpression of SRSF3, oncogenesis | Guo et al., 2015 |
| Ovarian cancer | (1) PTBP1/CDC42 (2) ↓OVAAL/PTBP1/p27 | (1) Filopodia formation, proliferation, invasiveness (2) Cellular senescence | He et al., 2015; Sang et al., 2018 |
| Bladder cancer | (1) miR-145/↓KLF4/↓PTBP1/↓PKMs (2) PTBP1/MEIS2/PKM2 | (1) Perturbation of the Warburg effect and inhibition of carcinogenesis (2) Lymph node metastasis, tumor stage, histological grade, and poor prognosis | Minami et al., 2017; Takai et al., 2017; Xie et al., 2019 |
| Pancreatic ductal adenocarcinoma | (1) Gemcitabine/PTBP1/PKM2 (2) Linc-ROR/↓miR-124/PTBP1/PKM2 | (1) Resistance to gemcitabine (2) Resistance to gemcitabine | Calabretta et al., 2016; Li et al., 2016 |
| Breast cancer | (1) PTBP1/PKM2 (2) PTBP1/PTEN-PI3K/Akt (3) PTBP1/FGFR1α/FGFR1β splicing | (1) Warburg effect, tumorigenesis, and transformation properties (2) Proliferation, invasion, and metastasis (3) Proliferation and migration | He et al., 2014; Ouyang et al., 2018; Zhao et al., 2019 |
| Renal cell cancer | (1) PTBP1/PKM2 (2) ↓PTBP1/HIF-1α pathway | (1) Proliferation, migration, and invasion (2) Inhibit cell migration, invasion, and angiogenesis | Jiang et al., 2017; Shan et al., 2018 |
| Glioblastoma | (1) ↓miR-124/PTBP1/ANXA7/EGFR (2) BAF45d/PTBP1 (3) PTBP1/USP5 (4) ↓CircSMARCA5/SRSF1/SRSF3/PTBP1 | (1) Glioblastoma progression (2) Malignant phenotype and undifferentiated cellular state (3) Oncogenesis (4) Invasion and migration | Izaguirre et al., 2012; Ferrarese et al., 2014; Aldave et al., 2018; Barbagallo et al., 2018 |
| Neuroblastoma | (1) MYCN/PTBP1/PKM2 (2) Hydrogen peroxide/PTBP1/sGC | (1) Proliferation, metastasis, and poor overall survival (2) Cellular adaptation to oxidative stress | Cote et al., 2012; Zhang SL et al., 2016 |
| Glioma | (1) PTBP1/RTN4 (2) PTBP1/IRES-like element/ADAR1 p110 | (1) Proliferation, invasion, and migration (2) Gliomagenesis and proliferation | Cheung et al., 2009; Yang et al., 2015 |
| Colorectal cancer | (1) ↓miR-1 and ↓miR-133b/PTBP1/PKM2 (2) DDX6/c-Myc/PTBP1 (3) PTBP1/ATG10/EMT-associated proteins (4) PTBP1/cortactin | (1) Warburg effect (2) Warburg effect (3) EMT, migration and invasion (4) Migration and invasion | Taniguchi et al., 2015, 2016; Jo et al., 2017; Wang et al., 2017 |
| Colon cancer | (1) ELK1/MYC/PTBP1/RAS-MAPK (2) ↓TPBP1/TRAIL and ↓PTBP1/PKM2→PKM1 (3) Apigenin/↓PTBP1/↓PKM2 | (1) Tumorigenesis (2) Apoptosis and perturbation of the Warburg effect (3) Warburg effect and oncogenesis | Hollander et al., 2016; Kumazaki et al., 2016; Shan et al., 2017 |
PAX3: paired box 3; FOXO1: forkhead box protein O1; HPV: human papillomavirus; FGFR: fibroblast growth factor receptor; HIF-1α: hypoxia-inducible factor-1α; USP5: ubiquitin specific peptidase 5; CircSMARCA5: circular RNA SMARCA5; sGC: soluble guanylyl cyclase; EMT: epithelial-mesenchymal transition; MAPK: mitogen-activated protein kinase; TRAIL: TNF-related apoptosis-inducing ligand
4. Conclusions and perspective
Recently, the most innovative therapy for cancer has been immunotherapy. There are two basic forms of immunotherapy: immune checkpoint therapy and chimeric antigen receptor (CAR) T-cell therapy (Iwai et al., 2017; Miyajima et al., 2017; Finney et al., 2019; Ren et al., 2019). Two of the most important pathways in immune checkpoint therapy are the programmed death 1 (PD-1) and the cytotoxic T-lymphocyte-associated antigen 4 (CTLA4) pathways. The reason why cancer cells escape from the immune system is that cancer cells inhibit the immune system of our body in different ways, and these two pathways reactivate the immune system to kill all cancer cells (Iwai et al., 2017; Miyajima et al., 2017). CAR T-cell therapy targets mainly CD19 and is used to treat B-cell lymphoma (Brown and Mackall, 2019).
Compared with these two main therapies to treat cancer, PTBP1 has the potential to act as another core molecule in immunotherapy. As a splicing factor, PTBP1 regulates many different ligands and receptors of the immune system, including CD40L, CD5, CD46, and IL-2 (Domingues et al., 2016; la Porta et al., 2016; Tang et al., 2016). PTBP1 is also upregulated in B lymphocytes and plays an important role in B-cell selection (Monzón-Casanova et al., 2018). All of these functions can be considered as supporting the immune system, and reveal the potential of PTBP1 in the clinical treatment of cancer.
PKM2 functions as an essential gene for the Warburg effect, a cancer-specific energy metabolism state that is regulated by the alternative splicing of PTBP1 (Hwang et al., 2017; Taniguchi et al., 2018). PTBP1 is a splicer of PKM mRNA and a positive regulator of the Warburg effect. These roles suggest that PTBP1 may be a useful therapeutic target for the Warburg effect (Sugito et al., 2017; Wang et al., 2019). Furthermore, miR-133b can reduce PTBP1 expression and silence the Warburg effect as a tumor suppressor in gastric cancer (Sugiyama et al., 2016). PTBP1 functions in almost all cell types in humans. It mediates growth, differentiation, and proliferation and is regulated by various molecules, including miRNA, lncRNA, RNA-binding protein, H2O2, and other splicing factors. PTBP1 participates in various diseases, including neurodegenerative diseases, cardiovascular diseases, and some cancers, including colon cancer, colorectal cancer, RCC, breast cancer, glioma, NBL, and glioblastoma. In cancers, PTBP1 participates mainly in glycolysis, apoptosis, proliferation, tumorigenesis, invasion, and migration. These findings identify PTBP1 as a novel potential candidate for clinical diagnosis and targeted therapy.
It is interesting that lncRNA PNCTR limits the alternative splicing of PTBP1 and inhibits the apoptosis of cancer cells (Bubenik and Swanson, 2018). In normal cells, PTBP1 promotes apoptosis through the regulation of BIM and MCL1 (Juan et al., 2014; Yang et al., 2018). Moreover, PTBP1 depletion protects cancer cells from antitubulin agent-induced apoptosis via an MCL1-dependent mechanism (Cui and Placzek, 2016). PTBP1 may be a useful therapeutic target for the Warburg effect and tumor cell metabolism reprogramming, but its exact functions in cellular apoptosis and tumor cells are still unclear. The relevant mechanisms are worth exploring in further studies.
Footnotes
Contributors: Cai-ping REN, Wei ZHU, and Bo-lun ZHOU contributed the design of the study. Wei ZHU and Bo-lun ZHOU drafted and critically revised the manuscript. Cai-ping REN, Li-juan RONG, Li YE, Hong-juan XU, Yao ZHOU, Xue-jun YAN, Wei-dong LIU, Bin ZHU, Lei WANG, and Xing-jun JIANG discussed and revised the manuscript. All authors were involved in writing the paper and provided final approval of the submitted and published versions.
Project supported by the National Natural Science Foundation of China (Nos. 81773179, 81272972, and 81472355), the Program for New Century Excellent Talents in University (No. NCET-10-0790), the Hunan Provincial Science and Technology Department (Nos. 2016JC 2049 and 2014FJ6006), the Hunan Provincial Natural Science Foundation of China (No. 2016JJ2172), and the Undergraduate Training Programs for Innovation and Entrepreneurship (Nos. 201810533368, GS201910533474, and GS201910533236), China
Compliance with ethics guidelines: Wei ZHU, Bo-lun ZHOU, Li-juan RONG, Li YE, Hong-juan XU, Yao ZHOU, Xue-jun YAN, Wei-dong LIU, Bin ZHU, Lei WANG, Xing-jun JIANG, and Cai-ping REN declare that they have no conflict of interest.
This article does not contain any studies with human or animal subjects performed by any of the authors.
References
- 1.Aldave G, Gonzalez-Huarriz M, Rubio A, et al. The aberrant splicing of BAF45d links splicing regulation and transcription in glioblastoma. Neuro-Oncology. 2018;20(7):930–941. doi: 10.1093/neuonc/noy007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Attig J, Agostini F, Gooding C, et al. Heteromeric RNP assembly at LINEs controls lineage-specific RNA processing. Cell. 2018;174(5):1067–1081e17. doi: 10.1016/j.cell.2018.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barbagallo D, Caponnetto A, Cirnigliaro M, et al. CircSMARCA5 inhibits migration of glioblastoma multiforme cells by regulating a molecular axis involving splicing factors SRSF1/SRSF3/PTB. Int J Mol Sci. 2018;19(2):480. doi: 10.3390/ijms19020480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- 5.Brown CE, Mackall CL. CAR T cell therapy: inroads to response and resistance. Nat Rev Immunol. 2019;19(2):73–74. doi: 10.1038/s41577-018-0119-y. [DOI] [PubMed] [Google Scholar]
- 6.Bubenik J, Swanson MS. Strring up cancer with lncRNA. Mol Cell. 2018;72(3):399–401. doi: 10.1016/j.molcel.2018.10.026. [DOI] [PubMed] [Google Scholar]
- 7.Busch A, Hertel KJ. Evolution of SR protein and hnRNP splicing regulatory factors. Wiley Interdiscip Rev RNA. 2012;3(1):1–12. doi: 10.1002/wrna.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Calabretta S, Bielli P, Passacantilli I, et al. Modulation of PKM alternative splicing by PTBP1 promotes gemcitabine resistance in pancreatic cancer cells. Oncogene. 2016;35(16):2031–2039. doi: 10.1038/onc.2015.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caruso P, Dunmore BJ, Schlosser K, et al. Identification of microRNA-124 as a major regulator of enhanced endothelial cell glycolysis in pulmonary arterial hypertension via PTBP1 (polypyrimidine tract binding protein) and pyruvate kinase M2. Circulation. 2017;136(25):2451–2467. doi: 10.1161/circulationaha.117.028034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheung HC, Hai T, Zhu W, et al. Splicing factors PTBP1 and PTBP2 promote proliferation and migration of glioma cell lines. Brain. 2009;132(8):2277–2288. doi: 10.1093/brain/awp153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Corrionero A, Valcarcel J. RNA processing: redrawing the map of charted territory. Mol Cell. 2009;36(6):918–919. doi: 10.1016/j.molcel.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 12.Cote GJ, Zhu W, Thomas A, et al. Hydrogen peroxide alters splicing of soluble guanylyl cyclase and selectively modulates expression of splicing regulators in human cancer cells. PLoS ONE. 2012;7(7):e41099. doi: 10.1371/journal.pone.0041099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cui J, Placzek WJ. PTBP1 modulation of MCL1 expression regulates cellular apoptosis induced by antitubulin chemotherapeutics. Cell Death Differ. 2016;23(10):1681–1690. doi: 10.1038/cdd.2016.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.del Rio-Moreno M, Alors-Perez E, Gonzalez-Rubio S, et al. Dysregulation of the splicing machinery is associated to the development of non-alcoholic fatty liver disease. J Clin Endocrinol Metab. 2019;104(8):3389–3402. doi: 10.1210/jc.2019-00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Domingues RG, Lago-Baldaia I, Pereira-Castro I, et al. CD5 expression is regulated during human T-cell activation by alternative polyadenylation, PTBP1, and miR-204. Eur J Immunol. 2016;46(6):1490–1503. doi: 10.1002/eji.201545663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dou XM, Zhang XS. RNA-binding protein PTB in spermatogenesis: progress in studies. Nat J Androl. 2016;22(9):856–860. (in Chinese) [PubMed] [Google Scholar]
- 17.Ehehalt F, Knoch K, Erdmann K, et al. Impaired insulin turnover in islets from type 2 diabetic patients. Islets. 2010;2(1):30–36. doi: 10.4161/isl.2.1.10098. [DOI] [PubMed] [Google Scholar]
- 18.Ferrarese R, Harsh IV GR, Yadav AK, et al. Lineage-specific splicing of a brain-enriched alternative exon promotes glioblastoma progression. J Clin Invest. 2014;124(7):2861–2876. doi: 10.1172/jci68836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Finney OC, Brakke H, Rawlings-Rhea S, et al. CD19 CAR T cell product and disease attributes predict leukemia remission durability. J Clin Invest. 2019;129(5):2123–2132. doi: 10.1172/jci125423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fu XD, Ares M., Jr Context-dependent control of alternative splicing by RNA-binding proteins. Nat Rev Genet. 2014;15(10):689–701. doi: 10.1038/nrg3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ge ZY, Quek BL, Beemon KL, et al. Polypyrimidine tract binding protein 1 protects mRNAs from recognition by the nonsense-mediated mRNA decay pathway. eLife, 5:e11155. 2016 doi: 10.7554/eLife.11155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Georgilis A, Klotz S, Hanley CJ, et al. PTBP1-mediated alternative splicing regulates the inflammatory secretome and the pro-tumorigenic effects of senescent cells. Cancer Cell. 2018;34(1):85–102e9. doi: 10.1016/j.ccell.2018.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ghetti A, Pinol-Roma S, Michael WM, et al. hnRNP 1, the polyprimidine tract-binding protein: distinct nuclear localization and association with hnRNAs. Nucleic Acids Res. 1992;20(14):3671–3678. doi: 10.1093/nar/20.14.3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grammatikakis I, Gorospe M. Identification of neural stem cell differentiation repressor complex Pnky-PTBP1. Stem Cell Investig, 3:10. 2016 doi: 10.21037/sci.2016.03.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo JH, Jia J, Jia R. PTBP1 and PTBP2 impaired autoregulation of SRSF3 in cancer cells. Sci Rep. 2015;5(1):14548. doi: 10.1038/srep14548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hamid FM, Makeyev EV. A mechanism underlying position-specific regulation of alternative splicing. Nucleic Acids Res. 2017;45(21):12455–12468. doi: 10.1093/nar/gkx901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Han W, Wang L, Yin B, et al. Characterization of a novel posttranslational modification in polypyrimidine tract-binding proteins by SUMO1. BMB Rep. 2014;47(4):233–238. doi: 10.5483/bmbrep.2014.47.4.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He X, Arslan AD, Ho TT, et al. Involvement of polypyrimidine tract-binding protein (PTBP1) in maintaining breast cancer cell growth and malignant properties. Oncogenesis. 2014;3(1):e84. doi: 10.1038/oncsis.2013.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.He XL, Yuan CF, Yang JL. Regulation and functional significance of CDC42 alternative splicing in ovarian cancer. Oncotarget. 2015;6(30):29651–29663. doi: 10.18632/oncotarget.4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hollander D, Donyo M, Atias N, et al. A network-based analysis of colon cancer splicing changes reveals a tumorigenesis-favoring regulatory pathway emanating from ELK1. Genome Res. 2016;26(4):541–553. doi: 10.1101/gr.193169.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hwang SR, Murga-Zamalloa C, Brown N, et al. Pyrimidine tract-binding protein 1 mediates pyruvate kinase M2-dependent phosphorylation of signal transducer and activator of transcription 3 and oncogenesis in anaplastic large cell lymphoma. Lab Invest. 2017;97(8):962–970. doi: 10.1038/labinvest.2017.39. [DOI] [PubMed] [Google Scholar]
- 32.Iwai Y, Hamanishi J, Chamoto K, et al. Cancer immunotherapies targeting the PD-1 signaling pathway. J Biomed Sci. 2017;24(1):26. doi: 10.1186/s12929-017-0329-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Izaguirre DI, Zhu W, Hai T, et al. PTBP1-dependent regulation of USP5 alternative RNA splicing plays a role in glioblastoma tumorigenesis. Mol Carcinog. 2012;51(11):895–906. doi: 10.1002/mc.20859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jeong DE, Heo S, Han JH, et al. Glucose controls the expression of polypyrimidine tract-binding protein 1 via the insulin receptor signaling pathway in pancreatic β cells. Mol Cells. 2018;41(10):909–916. doi: 10.14348/molcells.2018.0147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiang JY, Chen X, Liu H, et al. Polypyrimidine tract-binding protein 1 promotes proliferation, migration and invasion in clear-cell renal cell carcinoma by regulating alternative splicing of PKM. Am J Cancer Res. 2017;7(2):245–259. [PMC free article] [PubMed] [Google Scholar]
- 36.Jo YK, Roh SA, Lee H, et al. Polypyrimidine tract-binding protein 1-mediated down-regulation of ATG10 facilitates metastasis of colorectal cancer cells. Cancer Lett. 2017;385:21–27. doi: 10.1016/j.canlet.2016.11.002. [DOI] [PubMed] [Google Scholar]
- 37.Juan WC, Roca X, Ong ST. Identification of cis-acting elements and splicing factors involved in the regulation of BIM pre-mRNA splicing. PLoS ONE. 2014;9(4):e95210. doi: 10.1371/journal.pone.0095210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kang K, Peng X, Zhang X, et al. MicroRNA-124 suppresses the transactivation of nuclear factor of activated T cells by targeting multiple genes and inhibits the proliferation of pulmonary artery smooth muscle cells. J Biol Chem. 2013;288(35):25414–25427. doi: 10.1074/jbc.M113.460287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Keppetipola N, Sharma S, Li Q, et al. Neuronal regulation of pre-mRNA splicing by polypyrimidine tract binding proteins, PTBP1 and PTBP2. Crit Rev Biochem Mol Biol. 2012;47(4):360–378. doi: 10.3109/10409238.2012.691456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kumazaki M, Shinohara H, Taniguchi K, et al. Perturbation of the Warburg effect increases the sensitivity of cancer cells to trail-induced cell death. Exp Cell Res. 2016;347(1):133–142. doi: 10.1016/j.yexcr.2016.07.022. [DOI] [PubMed] [Google Scholar]
- 41.la Porta J, Matus-Nicodemos R, Valentin-Acevedo A, et al. The RNA-binding protein, polypyrimidine tract-binding protein 1 (PTBP1) is a key regulator of CD4 T cell activation. PLoS ONE. 2016;11(8):e0158708. doi: 10.1371/journal.pone.0158708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li CG, Zhao ZM, Zhou ZP, et al. Linc-ROR confers gemcitabine resistance to pancreatic cancer cells via inducing autophagy and modulating the miR-124/PTBP1/PKM2 axis. Cancer Chemother Pharmacol. 2016;78(6):1199–1207. doi: 10.1007/s00280-016-3178-4. [DOI] [PubMed] [Google Scholar]
- 43.Licatalosi DD, Yano M, Fak JJ, et al. Ptbp2 represses adult-specific splicing to regulate the generation of neuronal precursors in the embryonic brain. Genes Dev. 2012;26(14):1626–1642. doi: 10.1101/gad.191338.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Linares AJ, Lin CH, Damianov A, et al. The splicing regulator PTBP1 controls the activity of the transcription factor Pbx1 during neuronal differentiation. eLife, 4: e09268. 2015 doi: 10.7554/eLife.09268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ling JP, Chhabra R, Merran JD, et al. PTBP1 and PTBP2 repress nonconserved cryptic exons. Cell Rep. 2016;17(1):104–113. doi: 10.1016/j.celrep.2016.08.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu C, Yang Z, Wu J, et al. Long noncoding RNA H19 interacts with polypyrimidine tract-binding protein 1 to reprogram hepatic lipid homeostasis. Hepatology. 2018;67(5):1768–1783. doi: 10.1002/hep.29654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu JH, Li YP, Tong JY, et al. Long non-coding RNA-dependent mechanism to regulate heme biosynthesis and erythrocyte development. Nat Commun. 2018;9(1):4386. doi: 10.1038/s41467-018-06883-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Llorian M, Schwartz S, Clark TA, et al. Position-dependent alternative splicing activity revealed by global profiling of alternative splicing events regulated by PTB. Nat Struct Mol Biol. 2010;17(9):1114–1123. doi: 10.1038/nsmb.1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Llorian M, Gooding C, Bellora N, et al. The alternative splicing program of differentiated smooth muscle cells involves concerted non-productive splicing of post-transcriptional regulators. Nucleic Acids Res. 2016;44(18):8933–8950. doi: 10.1093/nar/gkw560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lorenzi P, Sangalli A, Fochi S, et al. RNA-binding proteins RBM20 and PTBP1 regulate the alternative splicing of FHOD3 . Int J Biochem Cell Biol. 2019;106:74–83. doi: 10.1016/j.biocel.2018.11.009. [DOI] [PubMed] [Google Scholar]
- 51.Marzese DM, Liu M, Huynh JL, et al. Brain metastasis is predetermined in early stages of cutaneous melanoma by CD44v6 expression through epigenetic regulation of the spliceosome. Pigment Cell Melanoma Res. 2015;28(1):82–93. doi: 10.1111/pcmr.12307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Medina MW, Gao F, Naidoo D, et al. Coordinately regulated alternative splicing of genes involved in cholesterol biosynthesis and uptake. PLoS ONE. 2011;6(4):e19420. doi: 10.1371/journal.pone.0019420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Méreau A, Anquetil V, Lerivray H, et al. A posttranscriptional mechanism that controls Ptbp1 abundance in the Xenopus epidermis. Mol Cell Biol. 2015;35(4):758–768. doi: 10.1128/mcb.01040-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Minami K, Taniguchi K, Sugito N, et al. MiR-145 negatively regulates Warburg effect by silencing KLF4 and PTBP1 in bladder cancer cells. Oncotarget. 2017;8(20):33064–33077. doi: 10.18632/oncotarget.16524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Miyajima M, Zhang BH, Sugiura Y, et al. Metabolic shift induced by systemic activation of T cells in PD-1-deficient mice perturbs brain monoamines and emotional behavior. Nat Immunol. 2017;18(12):1342–1352. doi: 10.1038/ni.3867. [DOI] [PubMed] [Google Scholar]
- 56.Monzón-Casanova E, Screen M, Díaz-Muñoz MD, et al. The RNA-binding protein PTBP1 is necessary for B cell selection in germinal centers. Nat Immunol. 2018;19(3):267–278. doi: 10.1038/s41590-017-0035-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Noiret M, Méreau A, Angrand G, et al. Robust identification of Ptbp1-dependent splicing events by a junction-centric approach in Xenopus laevis . Dev Biol. 2017;426(2):449–459. doi: 10.1016/j.ydbio.2016.08.021. [DOI] [PubMed] [Google Scholar]
- 58.Nordin A, Larsson E, Holmberg M. The defective splicing caused by the ISCU intron mutation in patients with myopathy with lactic acidosis is repressed by PTBP1 but can be derepressed by IGF2BP1. Hum Mutat. 2012;33(3):467–470. doi: 10.1002/humu.22002. [DOI] [PubMed] [Google Scholar]
- 59.Oberstrass FC, Auweter SD, Erat M, et al. Structure of PTB bound to RNA: specific binding and implications for splicing regulation. Science. 2005;309(5743):2054–2057. doi: 10.1126/science.1114066. [DOI] [PubMed] [Google Scholar]
- 60.Ouyang GQ, Xiong L, Liu ZP, et al. Inhibition of autophagy potentiates the apoptosis-inducing effects of photodynamic therapy on human colon cancer cells. Photodiagn Photodyn Ther. 2018;21:396–403. doi: 10.1016/j.pdpdt.2018.01.010. [DOI] [PubMed] [Google Scholar]
- 61.Pospiech N, Cibis H, Dietrich L, et al. Identification of novel PANDAR protein interaction partners involved in splicing regulation. Sci Rep. 2018;8(1):2798. doi: 10.1038/s41598-018-21105-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ramos AD, Andersen RE, Liu SJ, et al. The long noncoding RNA Pnky regulates neuronal differentiation of embryonic and postnatal neural stem cells. Cell Stem Cell. 2015;16(4):439–447. doi: 10.1016/j.stem.2015.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rawcliffe DFR, Osterman L, Nordin A, et al. PTBP1 acts as a dominant repressor of the aberrant tissue-specific splicing of ISCU in hereditary myopathy with lactic acidosis. Mol Genet Genomic Med. 2018;6(6):887–897. doi: 10.1002/mgg3.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ren SS, Deng JW, Hong M, et al. Ethical considerations of cellular immunotherapy for cancer. J Zhejiang Univ-Sci B (Biomed & Biotechnol) 2019;20(1):23–31. doi: 10.1631/jzus.B1800421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sachdeva M, Zhu SM, Wu FT, et al. p53 represses c-Myc through induction of the tumor suppressor miR-145 . Proc Natl Acad Sci USA. 2009;106(9):3207–3212. doi: 10.1073/pnas.0808042106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sang B, Zhang YY, Guo ST, et al. Dual functions for OVAAL in initiation of RAF/MEK/ERK prosurvival signals and evasion of p27-mediated cellular senescence. Proc Natl Acad Sci USA. 2018;115(50):E11661–E11670. doi: 10.1073/pnas.1805950115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Santiago JA, Potashkin JA. Blood biomarkers associated with cognitive decline in early stage and drug-naive Parkinson’s disease patients. PLoS ONE. 2015;10(11):e0142582. doi: 10.1371/journal.pone.0142582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Santiago JA, Potashkin JA. Network-based metaanalysis identifies HNF4A and PTBP1 as longitudinally dynamic biomarkers for Parkinson’s disease. Proc Natl Acad Sci USA. 2015;112(7):2257–2262. doi: 10.1073/pnas.1423573112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sasabe T, Futai E, Ishiura S. Polypyrimidine tract-binding protein 1 regulates the alternative splicing of dopamine receptor D2 . J Neurochem. 2011;116(1):76–81. doi: 10.1111/j.1471-4159.2010.07086.x. [DOI] [PubMed] [Google Scholar]
- 70.Sasanuma H, Ozawa M, Yoshida N. RNA-binding protein Ptbp1 is essential for BCR-mediated antibody production. Int Immunol. 2018;31(3):157–166. doi: 10.1093/intimm/dxy077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sayed ME, Yuan L, Robin JD, et al. NOVA1 directs PTBP1 to hTERT pre-mRNA and promotes telomerase activity in cancer cells. Oncogene. 2018;38(16):2937–2952. doi: 10.1038/s41388-018-0639-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shan H, Hou P, Zhang M, et al. PTBP1 knockdown in renal cell carcinoma inhibits cell migration, invasion and angiogenesis in vitro and metastasis in vivo via the hypoxia inducible factor-1α pathway. Int J Oncol. 2018;52(5):1613–1622. doi: 10.3892/ijo.2018.4296. [DOI] [PubMed] [Google Scholar]
- 73.Shan SH, Shi JY, Yang P, et al. Apigenin restrains colon cancer cell proliferation via targeted blocking of pyruvate kinase M2-dependent glycolysis. J Agric Food Chem. 2017;65(37):8136–8144. doi: 10.1021/acs.jafc.7b02757. [DOI] [PubMed] [Google Scholar]
- 74.Sharma S, Maris C, Allain FHT, et al. U1 snRNA directly interacts with polypyrimidine tract-binding protein during splicing repression. Mol Cell. 2011;41(5):579–588. doi: 10.1016/j.molcel.2011.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shi Y, Liu N, Lai WW, et al. Nuclear EGFR-PKM2 axis induces cancer stem cell-like characteristics in irradiation-resistant cells. Cancer Lett. 2018;422:81–93. doi: 10.1016/j.canlet.2018.02.028. [DOI] [PubMed] [Google Scholar]
- 76.Shinohara H, Kumazaki M, Minami Y, et al. Perturbation of energy metabolism by fatty-acid derivative AIC-47 and imatinib in BCR-ABL-harboring leukemic cells. Cancer Lett. 2016;371(1):1–11. doi: 10.1016/j.canlet.2015.11.020. [DOI] [PubMed] [Google Scholar]
- 77.Smith P, al Hashimi A, Girard J, et al. In vivo regulation of amyloid precursor protein neuronal splicing by microRNAs. J Neurochem. 2011;116(2):240–247. doi: 10.1111/j.1471-4159.2010.07097.x. [DOI] [PubMed] [Google Scholar]
- 78.Stork C, Li ZL, Lin L, et al. Developmental Xist induction is mediated by enhanced splicing. Nucleic Acids Res. 2018;47(3):1532–1543. doi: 10.1093/nar/gky1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sugito N, Taniguchi K, Kuranaga Y, et al. Cancer-specific energy metabolism in rhabdomyosarcoma cells is regulated by microRNA. Nucleic Acid Ther. 2017;27(6):365–377. doi: 10.1089/nat.2017.0673. [DOI] [PubMed] [Google Scholar]
- 80.Sugiyama T, Taniguchi K, Matsuhashi N, et al. MiR-133b inhibits growth of human gastric cancer cells by silencing pyruvate kinase muscle-splicer polypyrimidine tract-binding protein 1. Cancer Sci. 2016;107(12):1767–1775. doi: 10.1111/cas.13091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sveen A, Kilpinen S, Ruusulehto A, et al. Aberrant RNA splicing in cancer; expression changes and driver mutations of splicing factor genes. Oncogene. 2016;35(19):2413–2427. doi: 10.1038/onc.2015.318. [DOI] [PubMed] [Google Scholar]
- 82.Swinburne IA, Meyer CA, Liu XS, et al. Genomic localization of RNA binding proteins reveals links between pre-mRNA processing and transcription. Genome Res. 2006;16(7):912–921. doi: 10.1101/gr.5211806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Takahashi H, Nishimura J, Kagawa Y, et al. Significance of polypyrimidine tract-binding protein 1 expression in colorectal cancer. Mol Cancer Ther. 2015;14(7):1705–1716. doi: 10.1158/1535-7163.mct-14-0142. [DOI] [PubMed] [Google Scholar]
- 84.Takai T, Yoshikawa Y, Inamoto T, et al. A novel combination RNAi toward Warburg effect by replacement with miR-145 and silencing of PTBP1 induces apoptotic cell death in bladder cancer cells. Int J Mol Sci. 2017;18(1):179. doi: 10.3390/ijms18010179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tang SJ, Luo SF, Ho JXJ, et al. Characterization of the regulation of CD46 RNA alternative splicing. J Biol Chem. 2016;291(27):14311–14323. doi: 10.1074/jbc.M115.710350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tang ZZ, Sharma S, Zheng S, et al. Regulation of the mutually exclusive exons 8a and 8 in the CaV1.2 calcium channel transcript by polypyrimidine tract-binding protein. J Biol Chem. 2011;286(12):10007–10016. doi: 10.1074/jbc.M110.208116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Taniguchi K, Sugito N, Kumazaki M, et al. Positive feedback of DDX6/c-Myc/PTB1 regulated by miR-124 contributes to maintenance of the Warburg effect in colon cancer cells. Biochim Biophys Acta Mol Basis Dis. 2015;1852(9):1971–1980. doi: 10.1016/j.bbadis.2015.06.022. [DOI] [PubMed] [Google Scholar]
- 88.Taniguchi K, Sakai M, Sugito N, et al. PTBP1-associated microRNA-1 and -133b suppress the Warburg effect in colorectal tumors. Oncotarget. 2016;7(14):18940–18952. doi: 10.18632/oncotarget.8005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Taniguchi K, Sugito N, Shinohara H, et al. Organ-specific microRNAs (MIR122, 137, and 206) contribute to tissue characteristics and carcinogenesis by regulating pyruvate kinase M1/2 (PKM) expression. Int J Mol Sci. 2018;19(5):1276. doi: 10.3390/ijms19051276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Vaquero-Garcia J, Barrera A, Gazzara MR, et al. A new view of transcriptome complexity and regulation through the lens of local splicing variations. Elife, 5:e11752. 2016 doi: 10.7554/eLife.11752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Vuong JK, Lin CH, Zhang M, et al. PTBP1 and PTBP2 serve both specific and redundant functions in neuronal pre-mRNA splicing. Cell Rep. 2016;17(10):2766–2775. doi: 10.1016/j.celrep.2016.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wagner EJ, Carstens RP, Garcia-Blanco MA. A novel isoform ratio switch of the polypyrimidine tract binding protein. Electrophoresis. 1999;20(4-5):1082–1086. doi: 10.1002/(SICI)1522-2683(19990101)20:4/5<1082::AID-ELPS1082>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 93.Wang JL, Yang MY, Xiao S, et al. Downregulation of castor zinc finger 1 predicts poor prognosis and facilitates hepatocellular carcinoma progression via MAPK/ERK signaling. J Exp Clin Cancer Res. 2018;37(1):45. doi: 10.1186/s13046-018-0720-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wang L, Yang LY, Yang ZL, et al. Glycolytic enzyme PKM2 mediates autophagic activation to promote cell survival in NPM1-mutated leukemia. Int J Biol Sci. 2019;15(4):882–894. doi: 10.7150/ijbs.30290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wang ZN, Liu D, Yin B, et al. High expression of PTBP1 promote invasion of colorectal cancer by alternative splicing of cortactin. Oncotarget. 2017;8(22):36185–36202. doi: 10.18632/oncotarget.15873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wollerton MC, Gooding C, Wagner EJ, et al. Autoregulation of polypyrimidine tract binding protein by alternative splicing leading to nonsense-mediated decay. Mol Cell. 2004;13(1):91–100. doi: 10.1016/S1097-2765(03)00502-1. [DOI] [PubMed] [Google Scholar]
- 97.Xie R, Chen X, Chen Z, et al. Polypyrimidine tract binding protein 1 promotes lymphatic metastasis and proliferation of bladder cancer via alternative splicing of MEIS2 and PKM. Cancer Lett. 2019;449:31–44. doi: 10.1016/j.canlet.2019.01.041. [DOI] [PubMed] [Google Scholar]
- 98.Xu J, Liu H, Yang Y, et al. Genome-wide profiling of cervical RNA-binding proteins identifies human papillomavirus regulation of RNASEH2A expression by viral E7 and E2F1. mBio. 2019;10(1):e02687–18. doi: 10.1128/mBio.02687-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Xue YC, Zhou Y, Wu TB, et al. Genome-wide analysis of PTB-RNA interactions reveals a strategy used by the general splicing repressor to modulate exon inclusion or skipping. Mol Cell. 2009;36(6):996–1006. doi: 10.1016/j.molcel.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yang B, Hu PS, Lin XH, et al. PTBP1 induces ADAR1 p110 isoform expression through IRES-like dependent translation control and influences cell proliferation in gliomas. Cell Mol Life Sci. 2015;72(22):4383–4397. doi: 10.1007/s00018-015-1938-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yang Y, Wang CF, Zhao KL, et al. TRMP, a p53-inducible long noncoding RNA, regulates G1/S cell cycle progression by modulating IRES-dependent p27 translation. Cell Death Dis. 2018;9(9):886. doi: 10.1038/s41419-018-0884-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Yao WL, Yue P, Zhang GJ, et al. Enhancing therapeutic efficacy of the MEK inhibitor, MEK162, by blocking autophagy or inhibiting PI3K/AKT signaling in human lung cancer cells. Cancer Lett. 2015;364(1):70–78. doi: 10.1016/j.canlet.2015.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yap K, Lim ZQ, Khandelia P, et al. Coordinated regulation of neuronal mRNA steady-state levels through developmentally controlled intron retention. Genes Dev. 2012;26(11):1209–1223. doi: 10.1101/gad.188037.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhang L, Yang Z, Trottier J, et al. Long noncoding RNA MEG3 induces cholestatic liver injury by interaction with PTBP1 to facilitate shp mRNA decay. Hepatology. 2017;65(2):604–615. doi: 10.1002/hep.28882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zhang SL, Wei JS, Li SQ, et al. MYCN controls an alternative RNA splicing program in high-risk metastatic neuroblastoma. Cancer Lett. 2016;371(2):214–224. doi: 10.1016/j.canlet.2015.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zhang T, Li JJ, He Y, et al. A small molecule targeting myoferlin exerts promising anti-tumor effects on breast cancer. Nat Commun. 2018;9(1):3726. doi: 10.1038/s41467-018-06179-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zhang XC, Chen MH, Wu XB, et al. Cell-type-specific alternative splicing governs cell fate in the developing cerebral cortex. Cell. 2016;166(5):1147–1162e15. doi: 10.1016/j.cell.2016.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zhao M, Zhuo ML, Zheng X, et al. FGFR1β is a driver isoform of FGFR1 alternative splicing in breast cancer cells. Oncotarget. 2019;10(1):30–44. doi: 10.18632/oncotarget.26530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zheng SK, Gray EE, Chawla G, et al. PSD-95 is post-transcriptionally repressed during early neural development by PTBP1 and PTBP2. Nat Neurosci. 2012;15(3):381–388. doi: 10.1038/nn.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zhou C, Wu YL, Chen G, et al. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): a multicentre, open-label, randomised, phase 3 study. Lancet Oncol. 2011;12(8):735–742. doi: 10.1016/s1470-2045(11)70184-x. [DOI] [PubMed] [Google Scholar]