Abstract
Alpinia katsumadai (A. katsumadai), Alpinia oxyphylla (A. oxyphylla) and Alpinia pumila (A. pumila), which belong to the family Zingiberaceae, exhibit multiple medicinal properties. The chloroplast genome of a non-model plant provides valuable information for species identification and phylogenetic analysis. Here, we sequenced three complete chloroplast genomes of A. katsumadai, A. oxyphylla sampled from Guangdong and A. pumila, and analyzed the published chloroplast genomes of Alpinia zerumbet (A. zerumbet) and A. oxyphylla sampled from Hainan to retrieve useful chloroplast molecular resources for Alpinia. The five Alpinia chloroplast genomes possessed typical quadripartite structures comprising of a large single copy (LSC, 87,248–87,667 bp), a small single copy (SSC, 15,306–18,295 bp) and a pair of inverted repeats (IR, 26,917–29,707 bp). They had similar gene contents, gene orders and GC contents, but were slightly different in the numbers of small sequence repeats (SSRs) and long repeats. Interestingly, fifteen highly divergent regions (rpl36, ycf1, rps15, rpl22, infA, psbT-psbN, accD-psaI, petD-rpoA, psaC-ndhE, ccsA-ndhD, ndhF-rpl32, rps11-rpl36, infA-rps8, psbC-psbZ, and rpl32-ccsA), which could be suitable for species identification and phylogenetic studies, were detected in the Alpinia chloroplast genomes. Comparative analyses among the five chloroplast genomes indicated that 1891 mutational events, including 304 single nucleotide polymorphisms (SNPs) and 118 insertion/deletions (indels) between A. pumila and A. katsumadai, 367 SNPs and 122 indels between A. pumila and A. oxyphylla sampled from Guangdong, 331 SNPs and 115 indels between A. pumila and A. zerumbet, 371 SNPs and 120 indels between A. pumila and A. oxyphylla sampled from Hainan, and 20 SNPs and 23 indels between the two accessions of A. oxyphylla, were accurately located. Additionally, phylogenetic relationships based on SNP matrix among 28 whole chloroplast genomes showed that Alpinia was a sister branch to Amomum in the family Zingiberaceae, and that the five Alpinia accessions were divided into three groups, one including A. pumila, another including A. zerumbet and A. katsumadai, and the other including two accessions of A. oxyphylla. In conclusion, the complete chloroplast genomes of the three medicinal Alpinia species in this study provided valuable genomic resources for further phylogeny and species identification in the family Zingiberaceae.
Keywords: Alpinia katsumadai, Alpinia oxyphylla, Alpinia pumila, chloroplast genome, comparative analysis, single nucleotide polymorphisms, indels, phylogenetic relationship
1. Introduction
Alpinia Roxb. is the biggest genus in the family Zingiberaceae, which includes more than 230 species of perennial herb [1,2,3]. Almost all of the Alpinia species center in Southeast Asia, but western outposts of distribution occur in India and extend south into Australia and the islands of the South Pacific [1,2,3]. There are 54 species in China and most of them possess multiple medicinal properties; these species include Alpinia katsumadai K. Schum., Alpinia oxyphylla Miq., Alpinia pumila Hook. f., and others [1,2,4]. Of these species, the dried seeds of A. katsumadai and A. oxyphylla, and dried rhizomes of A. pumila can be used as traditional Chinese medicine and folk medicine, respectively [1,2,4]. A. katsumadai has synonyms Alpinia hainanensis, Alpinia henryi var. densihispida, Alpinia kainantensis, and Alpinia katsumadae [1,3]. A. katsumadai is mainly distributed in forests of Southern China (Guangdong, Guangxi, and Hainan provinces) as well as south into neighboring Vietnam [1,3]. A. pumila is native to shady, humid mountain valleys in Guangdong, Guangxi, Hunan and Yunnan provinces at altitudes of 500–1100 m [1,3]. A. oxyphylla is widely cultivated in Fujian, Guangdong, Guangxi, Hainan, and Yunnan provinces of China [1]. Due to the medicinal value of these three Alpinia species, numerous studies have focused on identifying the effective chemical constituents of these plants, which have many pharmacological effects—for instance, anti-bacterial activity, anti-inflammatory, anti-neoplastic and alzheimer’s disease [4,5,6,7,8,9,10].
Few vegetative characteristics are observed when Alpinia species do not produce bright-colored flowers or fruits, as a result causing difficulty in identifying the species of Alpinia [1,2,3,11]. This genus has also been molecularly studied [12,13]. Results have showed a sufficient resolution among 12 Alpinia species (Alpinia conchigera, Alpinia galanga, Alpinia elegans, Alpinia luteocarpa, Alpinia vittata, Alpinia blepharocalyx, Alpinia intermedia, A. pumila, Alpinia calcarata, Alpinia officinarum, Alpinia foxworthyi and Alpinia carolinensis) and their phylogenetic relationships using nuclear internal transcribed spacer (ITS) and chloroplast genome matK regions [12]. In a recent study, phylogenetic relationships between Alpinia zerumbet and A. oxyphylla sampled from Hainan were evaluated by the whole chloroplast genomes [13]. These publicly available two chloroplast genomes of Alpinia are insufficient to resolve morphological differences at interspecific and intraspecific levels, especially failing to distinguish among three medicinal species, namely, A. katsumadai, A. pumila and A. oxyphylla, and different sources of A. oxyphylla [12,13], because A. katsumadai and different sources of A. oxyphylla were not included and their phylogenetic positions were still unresolved. Therefore, we attempted to report the complete chloroplast genomes of A. katsumadai, A. pumila and A. oxyphylla sampled from Guangdong, as well as to explore their phylogenetic relationships.
The chloroplast is an important organelle that can conduct photosynthesis and produce essential energy in green plants [14,15]. It has its own independent genome, which comprises a closed circular DNA molecule [14,15]. Typical chloroplast genomes of angiosperms have a generally conserved quadripartite circular structure, which consists of a large single copy (LSC) region, a small single copy (SSC) region, and two copies of inverted repeats (IRs) [16,17,18]. With the rapid development of high-throughput sequencing technologies, such as Illumina and PacBio sequencing platforms, it is now convenient to acquire complete chloroplast genome sequences [19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34]. The chloroplast genomes have been widely utilized for species identification, the reconstruction of phylogenetic relationships, the resolution of origin problems, and the development of molecular markers [19,20,21,22,23,24,25,26,27,28,29,30].
In the current study, we firstly sequenced the complete chloroplast genomes of A. katsumadai, A. pumila and A. oxyphylla sampled from Guangdong, using Illumina and PacBio sequencing platforms, respectively. Then, we compared the resulting chloroplast genomes with the published chloroplast genomes of A. zerumbet (JX088668) and A. oxyphylla sampled from Hainan (NC_035895) [13]. Our main objectives were as shown below: (1) to explore the molecular structures of three chloroplast genomes of Alpinia in this study; (2) to examine the variations of simple sequence repeats (SSRs) and long repeats among the five Alpinia chloroplast genomes; (3) to discover highly divergent regions that could be used as specific DNA markers for Alpinia; (4) to deeply understand the interspecific and intraspecific variations among the five Alpinia chloroplast genomes; (5) to reveal phylogenetic relationships among A. katsumadai, A. pumila and A. oxyphylla in family Zingiberaceae.
2. Results and Discussion
2.1. The Chloroplast Genome Features of Alpinia Species
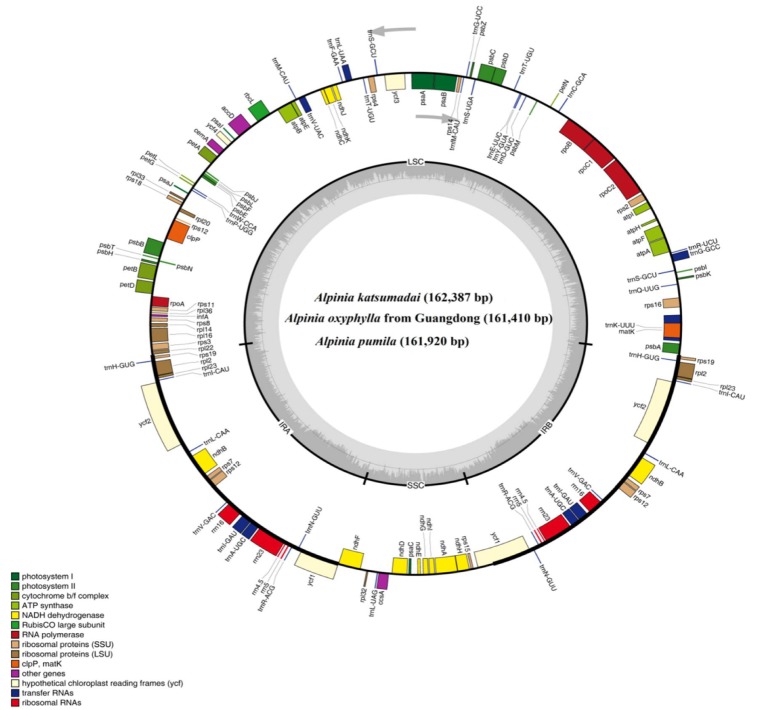
All the three species of Alpinia we sequenced had a typical quadripartite structure, with a single circular molecule ranged from 161,410 bp (A. oxyphylla sampled from Guangdong) to 162,387 bp (A. katsumadai) in length. They had four junction regions: a separate LSC region of 87,261–87,667 bp, an SSC region of 15,306–16,180 bp, and a pair of IRs (IRa and IRb) each 28,964–29,707 bp (Figure 1 and Table 1 and Table S1). The size of the A. katsumadai chloroplast genome (162,387 bp) was the largest among the three sequenced Alpinia species, with 977 bp longer than that of A. oxyphylla sampled from Guangdong and 467 bp longer than that of A. pumila. The GC content of the three species chloroplast genomes varied slightly from 36.15% to 36.17% (Table 1 and Table S1). The AT content at the third codon position (71.35%–71.37%) was higher than that at the first (55.30%–55.44%) and second (62.50%–62.59%) positions in the protein-coding genes of these three Alpinia species (Table S1). Additionally, the AT content was the highest (70.18–70.38%) in the SSC region, the lowest (50.48%–50.79%) in the IR regions, and moderate (66.14%–66.18%) in the LSC region (Table S1). These genomic structures were consistent with most other published chloroplast genomes of family Zingiberaceae, such as two Kaempferia species [23], three Amomum species [24], Zingiber officinale [25], Stahlianthus involucratus [31], Hedychium coronarium [32] and Curcuma longa [33].
Figure 1.
Circular gene map of chloroplast genomes of three Alpinia species. The gray arrowheads indicate the direction of the genes. Genes shown inside the circle are transcribed clockwise and those outside are transcribed counterclockwise. Different genes are color coded. The innermost darker gray corresponds to GC content, whereas the lighter gray corresponds to AT content. IR, inverted repeat; LSC, large single copy region; SSC, small single copy region.
Table 1.
Comparison of the general features of the five Alpinia chloroplast genomes.
| Genome Characteristics | A. katsumadai | A. oxyphylla Guangdong | A. pumila | A. zerumbet | A. oxyphylla Hainan |
|---|---|---|---|---|---|
| GenBank number | MK262728 | MK262729 | MK262731 | JX088668 | NC_035895 |
| Genome size (bp) | 162,387 | 161,410 | 161,920 | 159,773 | 161,351 |
| LSC length (bp) | 87,667 | 87,279 | 87,261 | 87,644 | 87,248 |
| SSC length (bp) | 15,306 | 16,180 | 15,317 | 18,295 | 16,175 |
| IR length (bp) | 29,707 | 28,964/28,987 | 29,671 | 26,917 | 28,964 |
| Total genes | 133 | 133 | 133 | 132 | 132 |
| Protein-coding genes | 87 | 87 | 87 | 86 | 87 |
| tRNA genes | 38 | 38 | 38 | 38 | 37 |
| rRNA genes | 8 | 8 | 8 | 8 | 8 |
| GC content (%) | 36.15 | 36.16 | 36.17 | 36.27 | 36.17 |
A total of 133 predicted genes, consisting of 87 protein-coding genes, 38 tRNA genes, and 8 rRNA genes, were detected in each chloroplast genome of our sequenced three species of Alpinia (Table 1, Table 2 and Table S2). Both of the chloroplast genomes of A. zerumbet and A. oxyphylla sampled from Hainan contained 132 genes (Table 1). As shown in Table 1, the A. zerumbet chloroplast genome had the highest GC content (36.27%), while the A. katsumadai chloroplast genome had the lowest GC content (36.15%). The length of the A. katsumadai chloroplast genome was the longest and the A. zerumbet chloroplast genome (159,773 bp) was the shortest. Interestingly, the SSC region of A. katsumadai (15,306 bp) was the shortest, whereas the SSC region of the A. zerumbet chloroplast genome (18,295 bp) was the longest (Table 1). The complete chloroplast genome of A. oxyphylla sampled from Guangdong was 59 bp longer than that of A. oxyphylla sampled from Hainan (Table 1). The lengths of the IR regions of the five chloroplast genomes, ranging from 26,917 to 29,707 bp (Table 1), were shorter than those of the three species of Amomum, which varied from 29,820 to 29,959 bp [24]. In addition, 86 protein-coding genes were identified in the A. zerumbet chloroplast genome, and 87 were identified in the other four accessions. Eight conserved rRNAs were identified in every chloroplast genome. The chloroplast genome of A. oxyphylla sampled from Hainan encoded 37 types of tRNAs, and the other four accessions encoded 38 (Table 1).
Table 2.
Genes present in the chloroplast genomes of three Alpinia species.
| Category | Gene Names | Amount |
|---|---|---|
| Photosystem Ⅰ | psaA, psaB, psaC, psaI, psaJ | 5 |
| Photosystem Ⅱ | psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbK, psbL, psbM, psbN, psbT, psbZ | 15 |
| Cytochrome b/f complex | petA, petB *, petD *, petG, petL, petN | 6 |
| ATP synthase | atpA, atpB, atpE, atpF *, atpH, atpI | 6 |
| NADH dehydrogenase | ndhA *, ndhB(×2) *, ndhC, ndhD, ndhE, ndhF, ndhG, ndhH, ndhI, ndhJ, ndhK | 12 |
| Rubisco | rbcL | 1 |
| RNA polymerase | rpoA, rpoB, rpoC1 *, rpoC2 | 4 |
| Large subunit ribosomal proteins | rpl2(×2) *, rpl14, rpl16 *, rpl20, rpl22, rpl23(×2), rpl32, rpl33, rpl36 | 11 |
| Small subunit ribosomal proteins | rps2, rps3, rps4, rps7(×2), rps8, rps11, rps12(×2) *, rps14, rps15, rps16 *, rps18, rps19(×2) | 15 |
| Other proteins | accD, ccsA, cemA, clpP **, infA, matK | 6 |
| Proteins of unknown function | ycf1(×2), ycf2(×2), ycf3 **, ycf4 | 6 |
| Ribosomal RNAs | rrn4.5(×2), rrn5(×2), rrn16(×2), rrn23(×2) | 8 |
| Transfer RNAs | trnA-UGC(×2) *, trnC-GCA, trnD-GUC, trnE-UUC, trnF-GAA, trnfM-CAU, trnG-GCC *, trnG-UCC, trnH-GUG(×2), trnI-CAU(×2), trnI-GAU(×2) *, trnK-UUU(×2) *, trnL-CAA(×2), trnL-UAA *, trnL-UAG, trnM-CAU, trnN-GUU(×2), trnP-UGG, trnQ-UUG, trnR-ACG(×2), trnR-UCU, trnS-GCU(×2), trnS-UGA, trnT-UGU, trnV-GAC(×2), trnV-UAC *, trnW-CCA, trnY-GUA | 38 |
* Gene containing one intron; ** gene containing two introns; (×2) gene with two copies.
A total of 20 genes were duplicated in the IR regions, including eight protein-coding genes (ndhB, rpl2, rpl23, rps7, rps12, rps19, ycf1, ycf2), eight tRNA genes (trnH-GUG, trnI-CAU, trnL-CAA, trnV-GAC, trnI-GAU, trnA-UGC, trnR-ACG, trnN-GUU), and all four rRNAs (rrn4.5, rrn5, rrn16 and rrn23) (Figure 1 and Table S2). Sixteen genes (trnA-UGC, trnI-GAU, trnG-GCC, trnK-UUU, trnL-UAA, trnV-UAC, atpF, ndhA, ndhB, rpoC1, petB, petD, rpl2, rpl16, rps12 and rps16) contained one intron, while ycf3 and clpP each contained two introns (Table S3). Among the 18 intron-containing genes, four genes (trnA-UAC, trnI-GAU, rpl2 and ndhB) occurred in the both IRs, 12 genes (trnG-GCC, trnK-UUU, trnL-UAA, trnV-UAC, atpF, rpoC1, petB, petD, rpl16, rps16, ycf3 and clpP) were distributed in the LSC, one gene (ndhA) was in the SSC, and one gene (rps12) was located its first exon in the LSC and the other two exons in both IRs (Figure 1 and Table S3). In addition, the three Alpinia species had the longest introns of trnK-UUU (2,653 bp, 2654 bp, 2626 bp, respectively), all of which were included in the coding region of matK (Tables S2 and S3).
2.2. Codon Usage and Predicted RNA Editing Sites Analyses
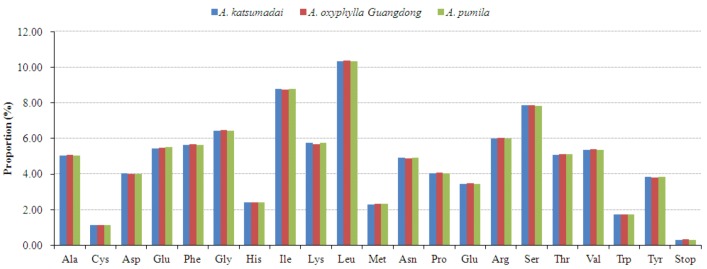
All the protein-coding genes were composed of 27,427–27,669 codons in the three chloroplast genomes of Alpinia species. Of the 27,427–27,669 codons, leucine (Leu) was the most abundant amino acid, with a frequency of 10.34%–10.38%, then isoleucine (Ile) with a frequency of 8.74%–8.80%, while cysteine (Cys) was the least common one with a proportion of 1.12%–1.13% (Figure 2 and Table S4). This phenomenon was consistent with other land plants’ chloroplast genomes, such as Z. officinale [25], Ailanthus altissima [35], Lycium chinense [36], Symplocarpus renifolius [37], and Epipremnum aureum [38]. Due to the value of relative synonymous codon usage (RSCU) >1, thirty codons showed the codon usage bias in the chloroplast genes of all the three Alpinia species (Table S4). Interestingly, out of the above 30 codons, twenty-nine codons were A/T-ending codons. Conversely, the C/G-ending codons had RSCU values of less than one, indicating that they were less common in the chloroplast genes of the three Alpinia species. Stop codon usage was found to be biased toward TAA (RSCU > 1.00). Two amino acids, methionine (Met) and tryptophan (Trp), showed no codon bias both with RSCU values of 1.00 (Table S4).
Figure 2.
Amino acid proportion in three Alpinia species protein-coding sequences.
A total of 56 editing sites in 22 protein-coding genes were identified in A. katsumadai, while similar numbers were found in A. pumila (54 sites) and A. oxyphylla sampled from Guangdong (55 sites) (Table S5). In the three Alpinia species chloroplast genomes we sequenced, the ndhB gene had the highest number of potential editing sites (11), followed by the accD gene (5). Similar to other species, such as Kaempferia galanga [23], Kaempferia elegans [23] and Forsythia suspense [39], the ndhB gene contained the largest number of editing sites. All these editing sites were C-to-T transitions and occurred at the first or second positions of the codons. Interestingly, most conversions at the codon positions changed from serine (S) to leucine (L) and most RNA editing sites led to amino acid changes for hydrophobic products, such as leucine, isoleucine, tryptophan, tyrosine, valine, methionine, and phenylalanine (Table S5). Similar RNA editing features have already been revealed by previous observations [23,39].
2.3. SSRs and Long Repeats Analyses
SSRs were widely dispersed in chloroplast genomes, and have been extensively used in population genetics and molecular phylogenetic researches [40,41]. In this study, 1205 SSRs were identified in five Alpinia chloroplast genomes, including two accessions of A. oxyphylla, A. katsumadai, A. zerumbet, and A. pumila (Figure 3 and Table S6). The most abundant were mononucleotide repeats, located in non-coding regions, and contributed to AT richness (Figure 3). These results are consistent with most reported angiosperms [23,24,25,26,28,29,30]. The total number of SSRs was 237 in A. katsumadai, 244 in A. oxyphylla sampled from Guangdong, 247 in A. pumila, 236 in A. zerumbet, and 241 in A. oxyphylla sampled from Hainan (Figure 3A). In the genomic structure of five chloroplast genomes, the non-coding region had the most abundant SSRs, whereas the coding region had the least SSRs (Figure 3A). The majority of SSRs were located in the LSC regions rather than in the SSC and IR regions of the five chloroplast genomes (Figure 3B). Mono-, di-, tri- and tetranucleotide SSRs were all detected in the five chloroplast genomes (Figure 3C). Additionally, pentanucleotide SSRs were found in two accessions of A. oxyphylla and A. pumila, respectively. Mononucleotide repeats were the largest in a number of these SSRs, with 77.63% and 78.54% found in A. katsumadai and A. pumila, respectively (Figure 3C). A/T repeats were the most common of mononucleotides (70.90%–76.69%), while AT/AT repeats were the majority of dinucleotide repeat sequences (92.30%–94.59%). Interestingly, our results show that AAAT/ATTT repeats were the third abundant SSR types in the five chloroplast genomes (2.96%–3.79%) (Figure 3D).
Figure 3.
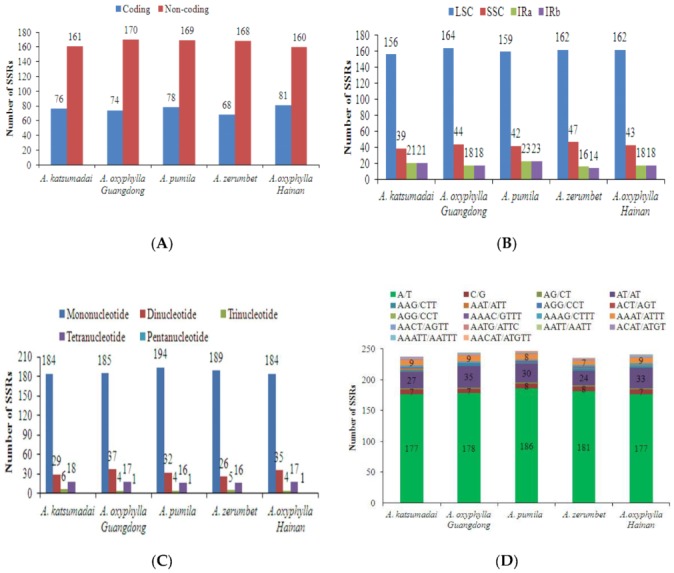
Distribution of small sequence repeats (SSRs) in the chloroplast genomes of five Alpinia accessions. (A) SSR distribution between coding and non-coding regions detected in five Alpinia chloroplast genomes; (B) Frequencies of identified SSRs in the large single copy (LSC), SSC and inverted repeats (IRs) regions; (C) Number of different SSR types detected in five Alpinia chloroplast genomes; (D) Frequency of identified SSRs in different repeat class types.
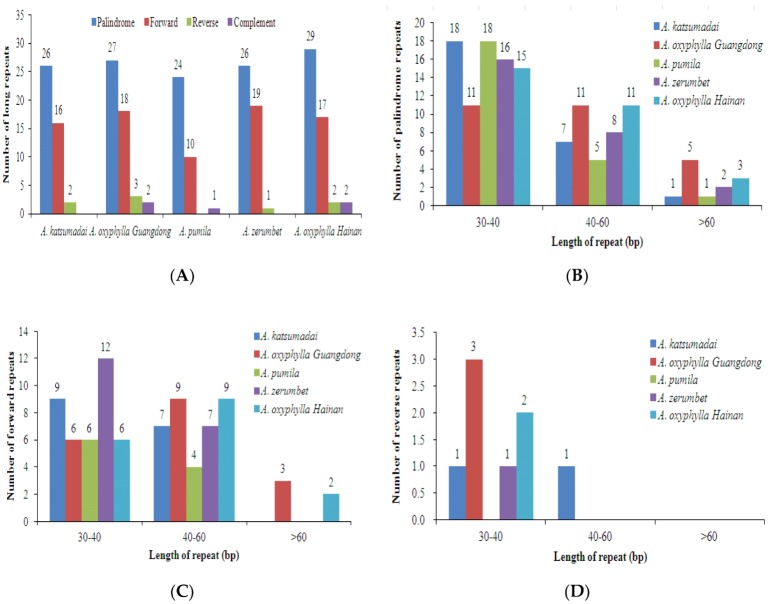
Additionally, we detected long repeats in five chloroplast genomes using REPuter, including forward, complement, reverse, and palindromic repeats. A total of 225 unique long repeats were found from the five chloroplast genomes. In detail, there were 44 (16 forward, 26 palindrome, two reverse), 35 (10 forward, 24 palindrome, one complement), 50 (18 forward, 27 palindrome, three reverse, two complement), 46 (19 forward, 26 palindrome, one reverse) and 50 (17 forward, 29 palindrome, two reverse, two complement) long repeats in A. katsumadai, A. pumila, A. oxyphylla sampled from Guangdong, A. zerumbet and A. oxyphylla sampled from Hainan, respectively (Figure 4A and Table S7). Interestingly, there were no complement repeats in A. katsumadai, similar to A. zerumbet. With 29 palindrome repeats, A. oxyphylla sampled from Hainan contained the highest number of palindrome repeats, while A. zerumbet contained the highest number of forward repeats at 19, and A. oxyphylla sampled from Guangdong contained three reverse repeats, the highest among the compared genomes (Figure 4B–D). The palindrome, forward and reverse repeats with 30–60 bp were found to be the most common in the five chloroplast genomes (Figure 4B–D). Moreover, almost all of the lengths of the reverse repeats were less than 60 bp in the five chloroplast genomes (Figure 4D).
Figure 4.
Analysis of long repeat sequences in the five Alpinia chloroplast genomes. (A) Total of four long repeat types; (B) Frequency of the palindromic repeats by length; (C) Frequency of the forward repeats by length; (D) Frequency of the reverse repeats by length.
2.4. IR Contraction and Expansion Analyses
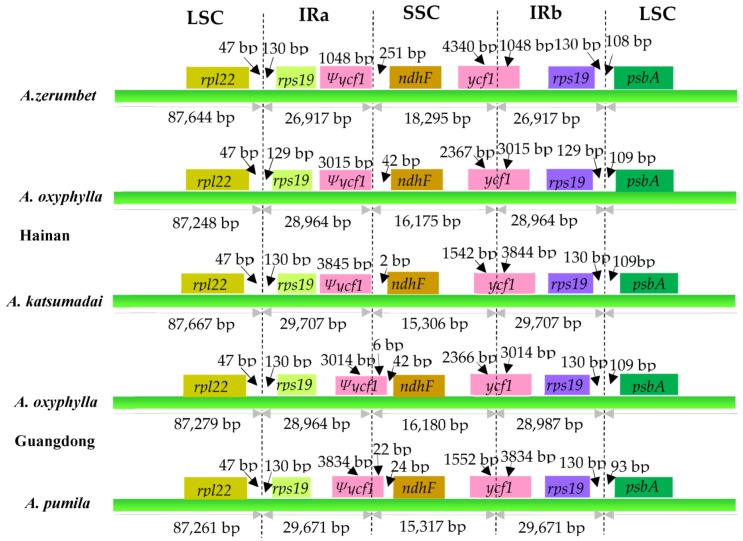
The contractions and expansions at the borders of IR regions were common evolutionary events and may cause size variations of chloroplast genomes [13,23,24,25,26,27,28,29,30]. We compared the IR-SC boundaries information of the five Alpinia chloroplast genomes (Figure 5). The genes rpl22, rps19, pseudogene ycf1 (Ψycf1), ndhF, ycf1 and psbA were present at the junction of the LSC/IRa, IRa/SSC, SSC/IRb, and IRb/LSC borders. As shown in Figure 5, the rpl22-rps19 genes were located in the junctions of the LSC/IRa regions of the five chloroplast genomes. There were 47 bp between rpl22 and the LSC/IRa borders, meanwhile, 129–130 bp between the rps19 and the other LSC/IRa junctions.
Figure 5.
Comparison of the borders of the LSC, SSC, and IR regions among five Alpinia chloroplast genomes. Ψ, pseudogenes. Boxes above the main line indicate the adjacent border genes. The figure is not to scale with respect to sequence length, and only shows relative changes at or near the IR/SC borders.
The Ψycf1-ndhF genes were located at the junctions of the IRa/SSC regions for all five chloroplast genomes; IRa/SSC borders of three species (A. zerumbet, A. oxyphylla sampled from Hainan and A. katsumadai) were all situated just adjacent to the end of Ψycf1; Ψycf1 expanded into the SSC regions for 6 bp in A.oxyphylla sampled from Guangdong and 22 bp in A. pumila, respectively (Figure 5). In the three species (A. zerumbet, A. oxyphylla sampled from Hainan and A. katsumadai), the distances between ndhF and IRa/SSC border were 251 bp, 42 bp, and 2 bp, respectively. There were 24 bp and 42 bp between ndhF and Ψycf1 border in A. pumila and A. oxyphylla sampled from Guangdong, respectively (Figure 5).
The SSC/IRb junction was situated in the ycf1 coding region, which crossed into the IRb region in all five chloroplast genomes. However, the length of ycf1 in the IRb region varied among the five chloroplast genomes from 1048 bp to 3844 bp, which indicated the dynamic variation of the SSC/IRb junctions (Figure 5).
The rps19-psbA genes were located in the junctions of the IRb/LSC regions in five chloroplast genomes (Figure 5). In the five chloroplast genomes, the distances between rps19 and IRb/LSC border were 130 bp, 129 bp, 130 bp, 130 bp and 130 bp, respectively. For all five chloroplast genomes, 93–109 bp distance was found between psbA gene and the IRb/LSC border (Figure 5). Taken together, these data indicated that the contractions and expansions of the IR regions exhibited relatively stable patterns within genus Alpinia, with slight variations.
2.5. Divergence Hotspot Regions Analyses
The whole chloroplast genome sequence of A. pumila (MK262731) was compared to those of A.katsumadai (MK262728), A.oxyphylla sampled from Guangdong (MK262729), A. zerumbet (JX088668) and A.oxyphylla sampled from Hainan (NC_035895) using the mVISTA program (Figure 6). The comparison showed that the two IR regions were less divergent than the LSC and SSC regions and that lower divergence was found in coding regions than in non-coding regions (Figure 6). In the coding regions, most genes were relatively conserved, except for matK, petB, rps15, rpl22 and ycf1. The highly divergent regions were mainly located in the intergenic regions, such as trnH-psbA, atpI-atpH, psbM-petN, trnE-psbD, psbC-psbZ, accD-psaI, psbT-psbN, petD-rpoA, ccsA-ndhD, ndhF-rpl32, rpl32-ccsA, and psaC-ndhE (Figure 6).
Figure 6.
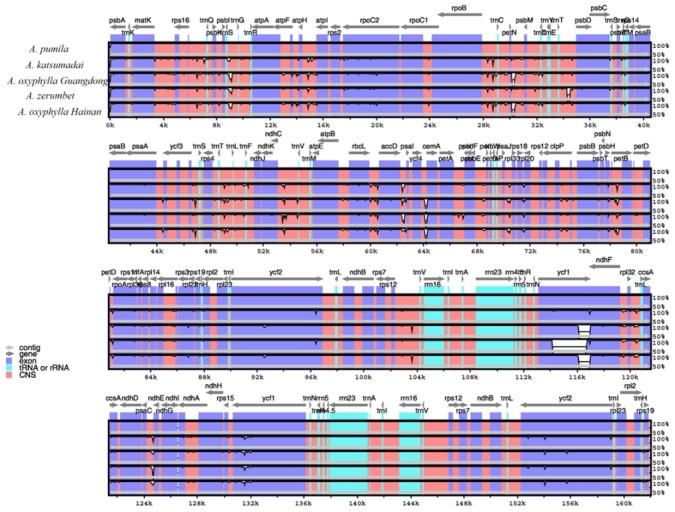
Complete chloroplast genome comparison of five Alpinia accessions using the chloroplast genome of A. pumila as a reference. Gray arrows and thick black lines above the alignment indicate gene orientation. Purple bars represent exons, sky-blue bars represent transfer RNA (tRNA) and ribosomal RNA (rRNA), red bars represent non-coding sequences (CNS), and white peaks represent differences of chloroplast genomes. The y-axis represents the identity percentage ranging from 50 to 100%.
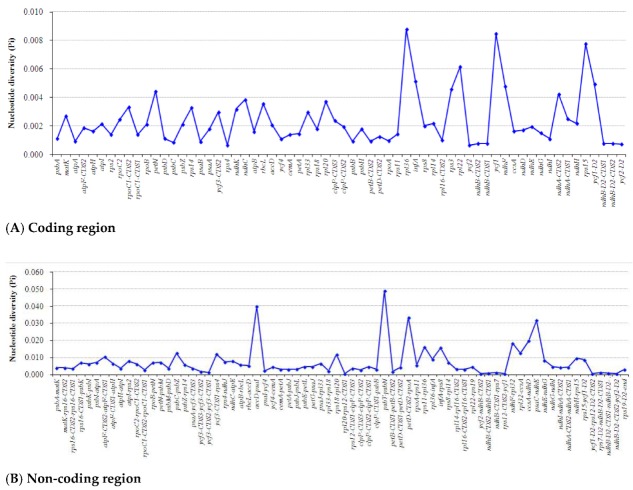
Furthermore, the five Alpinia accessions were observed to have highly variable regions in their chloroplast genomes by sliding window analysis using software DnaSP (Figure 7). Of the 64 protein-coding regions (CDS), nucleotide diversity (Pi) for each locus ranged from 0.0006 (ycf2) to 0.00877 (rpl36) and had the average value of 0.00242. Thereby, five regions (rpl36, ycf1, rps15, rpl22 and infA genes) located at the LSC and SSC regions had remarkably high values (Pi > 0.005; Figure 7A). For the 72 non-coding regions, Pi values ranged from 0.00056 (rpl20-rps12) to 0.04918 (psbT-psbN) and had the average of 0.00746. Ten of these regions also showed remarkably high values (Pi > 0.012), including psbT-psbN, accD-psaI, petD-rpoA, psaC-ndhE, ccsA-ndhD, ndhF-rpl32, rps11-rpl36, infA-rps8, psbC-psbZ, and rpl32-ccsA (Figure 7B). These results also prove that the IR regions were more conserved than the LSC and SSC regions, and the average value of Pi in the non-coding regions was more than three times as much as in the coding regions. Among these regions, ycf1, rps15, rpl22, infA, psbT-psbN, petD-rpoA, psaC-ndhE, ccsA-ndhD, ndhF-rpl32, and rpl32-ccsA have also been reported as highly variable regions in other plant species, such as Kaempferia species [23], Aristolochis species [26], orchid species [42], Lythraceae species [43], Quercus species [44], Lilium species [45], Croomia species [46], Stemona species [46], and Eucommia species [47]. The ndhF-rpl32 and ccsA-ndhD regions had been used as molecular markers for phylogenetic analyses [18,19]. Overall, these highly divergent regions presented abundant information for molecular marker development in plant identification and phylogenetic relationships of Alpinia.
Figure 7.
Sliding window analysis of the whole chloroplast genomes among five Alpinia accessions.Window length: 800 bp; step size: 200 bp. X-axis: position of the midpoint of a window.
2.6. Interspecific Analyses of Alpinia Chloroplast Genomes
Using the A. pumila chloroplast genome sequence as the reference, we compared the SNP/indel loci of the other four chloroplast genomes in the current study. One-hundred-and-twenty-eight and 176 SNP markers were detected between A. pumila and A. katsumadai in protein-coding genes and non-coding regions, respectively (Table S8). One-hundred-and-sixty-two and 205 SNP markers were detected between A. pumila and A. oxyphylla sampled from Guangdong in protein-coding genes and non-coding regions, respectively (Table S8). One-hundred-and-forty-two and 189 SNP markers were detected between A. pumila and A. zerumbet in protein-coding genes and non-coding regions, respectively (Table S8). One-hundred-and-sixty-three and 208 SNP markers were detected between A. pumila and A. oxyphylla sampled from Hainan in protein-coding genes and non-coding regions, respectively (Table S8). The SNPs in the A. katsumadai chloroplast genome were significantly fewer than those in the two accessions of A. oxyphylla. SNPs were detected in 54 protein-coding genes in A. katsumadai, A. zerumbet, and two accessions of A. oxyphylla sampled from Guangdong and Hainan. Nine genes were in the SSC region, one gene was in the IR regions, and 44 genes were in the LSC region (Table 3 and Table S8). These 54 genes were divided into four categories according to their different functions in plant chloroplasts, including photosynthetic apparatus, photosynthetic metabolism, gene expression, and other genes (Table 2). For the 162 and 163 SNP markers in the protein-coding genes of A. oxyphylla sampled from Guangdong and Hainan chloroplast genomes, respectively, 90 and 91 belonged to the synonymous type, and 72 and 72 belonged to the nonsynonymous type (Table 3 and Table S8). Synonymous and nonsynonymous SNP markers in the protein-coding genes shared very similar number in these two chloroplast genomes. There were 67 synonymous SNPs and 61 nonsynonymous SNPs in the protein-coding genes of the A. katsumadai chloroplast genome (Table 3 and Table S8). Seventy-three synonymous and 69 nonsynonymous SNP sites were detected in the chloroplast genome of A. zerumbet (Table 3 and Table S8).
Table 3.
Comparisons of mutation changes, number of synonymous (S) and nonsynonymous (N) substitutions per gene of protein-coding genes among four Alpinia chloroplast genomes.
| Genes | A. katsumadai |
A. oxyphylla Guangdong |
A. zerumbet |
A. oxyphylla Hainan |
Location | ||||
|---|---|---|---|---|---|---|---|---|---|
| S | N | S | N | S | N | S | N | ||
| psbA | 3 | 0 | 3 | 0 | 3 | 0 | 3 | 0 | LSC |
| matK | 2 | 1 | 3 | 3 | 2 | 2 | 3 | 3 | LSC |
| atpA | 1 | 1 | 2 | 1 | 1 | 1 | 2 | 1 | LSC |
| atpF | - | - | - | - | 2 | 0 | - | - | LSC |
| atpH | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | LSC |
| atpI | 1 | 1 | - | - | 2 | 1 | - | - | LSC |
| rps2 | 1 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | LSC |
| rpoC2 | 5 | 7 | 8 | 8 | 6 | 7 | 8 | 8 | LSC |
| rpoC1 | 0 | 2 | 6 | 3 | 0 | 2 | 6 | 3 | LSC |
| rpoB | 7 | 2 | 7 | 3 | 6 | 3 | 7 | 3 | LSC |
| petN | - | - | - | - | 1 | 0 | - | - | LSC |
| psbD | - | - | - | - | 1 | 2 | - | - | LSC |
| psbC | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | LSC |
| psbZ | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | LSC |
| rps14 | - | - | 0 | 1 | 1 | 0 | 0 | 1 | LSC |
| psaB | 2 | 0 | 2 | 1 | 1 | 0 | 2 | 1 | LSC |
| psaA | 6 | 1 | 4 | 1 | 5 | 1 | 4 | 1 | LSC |
| ycf3 | - | - | 1 | 0 | - | - | 1 | 0 | LSC |
| rps4 | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | LSC |
| ndhK | 0 | 2 | 1 | 1 | 1 | 3 | 1 | 1 | LSC |
| ndhC | 1 | 1 | 0 | 2 | 0 | 1 | 0 | 2 | LSC |
| atpB | - | - | 2 | 2 | 1 | 1 | 3 | 2 | LSC |
| rbcL | 3 | 4 | 5 | 1 | 3 | 3 | 5 | 1 | LSC |
| accD | 2 | 1 | 3 | 2 | 2 | 1 | 3 | 2 | LSC |
| ycf4 | - | - | 1 | 0 | - | - | 1 | 0 | LSC |
| cemA | 1 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | LSC |
| petA | 0 | 2 | 1 | 2 | 0 | 2 | 1 | 2 | LSC |
| rpl33 | - | - | 0 | 1 | - | - | 0 | 1 | LSC |
| rps18 | - | - | 1 | 0 | - | - | 1 | 0 | LSC |
| rpl20 | - | - | 2 | 0 | 1 | 0 | 2 | 0 | LSC |
| clpP | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | LSC |
| psbB | 1 | 0 | - | - | 3 | 0 | - | - | LSC |
| psbH | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | LSC |
| petB | 1 | 0 | - | - | 1 | 0 | - | - | LSC |
| petD | - | - | 0 | 1 | - | - | 0 | 1 | LSC |
| rpoA | 1 | 0 | 0 | 1 | - | - | 0 | 1 | LSC |
| rps11 | - | - | 0 | 1 | - | - | 0 | 1 | LSC |
| rpl36 | 1 | 0 | 2 | 0 | 1 | 0 | 2 | 0 | LSC |
| infA | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | LSC |
| rps8 | 2 | 0 | 2 | 0 | 2 | 0 | 2 | 0 | LSC |
| rpl14 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 0 | LSC |
| rpl16 | - | - | - | - | 0 | 1 | - | - | LSC |
| rps3 | 3 | 2 | 2 | 3 | 2 | 3 | 2 | 3 | LSC |
| rpl22 | 0 | 3 | 0 | 1 | 0 | 3 | 0 | 1 | LSC |
| ndhF | 5 | 7 | 8 | 5 | 6 | 6 | 8 | 5 | SSC |
| ccsA | 0 | 3 | 0 | 2 | 1 | 2 | 0 | 2 | SSC |
| ndhD | - | - | 3 | 0 | - | - | 3 | 0 | SSC |
| ndhE | - | - | 1 | 0 | - | - | 1 | 0 | SSC |
| ndhG | 2 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | SSC |
| ndhI | - | - | 1 | 0 | - | - | 1 | 0 | SSC |
| ndhA | 2 | 2 | 2 | 1 | 2 | 2 | 2 | 1 | SSC |
| ndhH | 2 | 2 | 1 | 0 | 2 | 2 | 1 | 0 | SSC |
| rps15 | 1 | 1 | 0 | 3 | 0 | 1 | 0 | 3 | SSC |
| ycf1 | 2 | 15 | 6 | 19 | 3 | 18 | 6 | 19 | IRa/b |
| Total | 67 | 61 | 90 | 72 | 73 | 69 | 91 | 72 | - |
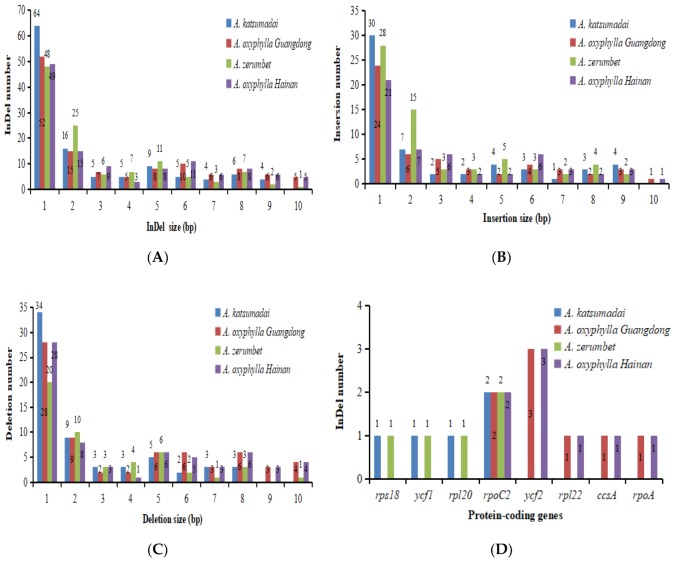
All of the indels were classified into insertions and deletions (Figure 8 and Table S9). Fifty-six insertions and 62 deletions were detected between A. pumila and A. katsumadai chloroplast genomes, respectively (Figure 8A–C). Fifty-three insertions and 69 deletions were detected between A. pumila and A. oxyphylla sampled from Guangdong chloroplast genomes, respectively (Figure 8A–C). Sixty-five insertions and 50 deletions were detected between A. pumila and A. zerumbet chloroplast genomes, respectively (Figure 8A–C). Fifty-three insertions and 67 deletions were detected between A. pumila and A. oxyphylla sampled from Hainan chloroplast genomes, respectively (Figure 8A–C). Eight protein-coding genes from the four Alpinia accessions contained indels (Figure 8D). The gene rpoC2 was a hotspot for indel variation, and all the four Alpinia accessions contained two indels in this gene. Comparison with two Kaempferia species was extremely interesting. The result indicated that SNPs between the four Alpinia species were less common than those between two Kaempferia species, but had more indels than those between two Kaempferia species. There were 536 SNPs and 107 indels between K. galanga and K. elegans [23]. Moreover, the SNPs obtained from chloroplast genomes had been successfully used for phylogenetic studies in several Zingiberaceae species, such as in K. galanga and K. elegans [23], S. involucratus [31], H. coronarium [32] and C. longa [33]. Therefore, these SNPs and indels in Alpinia here would be potential genetic markers to facilitate phylogenetic analysis and species identification in the family Zingiberaceae.
Figure 8.
Indels statistics of four Alpinia chloroplast genomes. The A. pumila chloroplast genome was used as the reference sequence for indels analyses for the other four chloroplast genomes. (A) Total indels statistics. (B) Insertion statistics. (C) Deletion statistics. (D) Indels belonging to different protein-coding genes.
2.7. Intraspecific Analyses of two Chloroplast Genomes of A. oxyphylla
The two chloroplast genomes from A. oxyphylla were found to show a 59-bp difference in length (Table 1). In addition to the total length difference, we obtained indels and SNPs between the two chloroplast genomes of A. oxyphylla in their entirety. Through intraspecific comparison, a total of 23 indels were identified between the two A. oxyphylla accessions (Table S10). Two, one, one, one, and one indels were located in clpP, rpoC1, rps16, rpl16 and trnG-GCC, respectively. The other 17 indels were located in 14 different regions. atpB-rbcL, trnC-GCA-petN and ndhF-rpl32 exhibited the same number indels, of two in total, respectively. There were 20 SNPs identified in the two chloroplast genomes of A. oxyphylla (Table S10). The most frequently occurring mutations were A/C substitutions (4 times), followed by G/T (three times), T/A (three times), and T/G (three times), respectively. ccsA-ndhD contained the highest number of SNPs (6), followed by rps16-trnQ-UUG and trnS-GCU-trnG-GCC, each of which showed three SNPs. Two and two SNPs were located in introns and coding regions, respectively. All of the other four regions contained only one SNP. By contrast, there were more mutational events (SNPs and indels) in Eucommia ulmoides [47] and Scutellaria baicalensis [48]. There were 75 SNPs and 80 indels in two different individuals of E. ulmoides [47], 25 SNPs and 29 indels between the two S. baicalensis genotypes [48]. These 23 indels and 20 SNPs could be used for identification of different sources of A. oxyphylla.
2.8. Phylogenetic Analyses
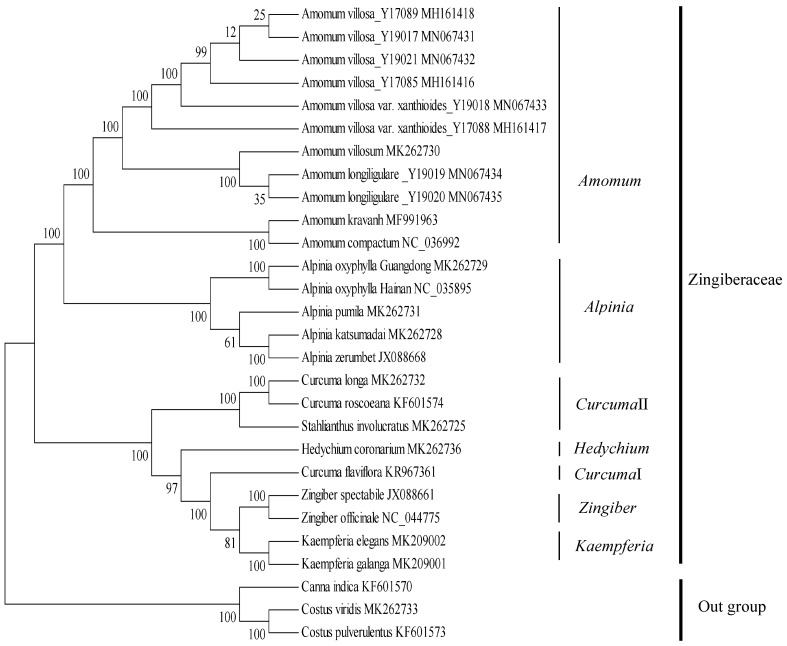
To determine the phylogenetic relationships of five Alpinia accessions in family Zingiberaceae, two phylogenetic trees of 25 representatives from family Zingiberaceae using chloroplast SNP-based matrix, were constructed with Costus pulverulentus, Costus viridis and Canna indica as outgroups (Figure 9 and Figure S1). Both the maximum likelihood (ML) and maximum parsimony (MP) phylogenetic trees strongly indicated that two genera Amomum and Alpinia in the Zingiberaceae clade were strongly supported as a sister monophyletic group, respectively (bootstrap values ≥ 99%). In the genus Alpinia, two accessions of A. oxyphylla clustered together with high statistical support values (bootstrap values ≥ 99%), which was subsequently sister to A. pumila; A. zerumbet and A. katsumadai clustered together (bootstrap values ≥ 99%) was another sister to A. pumila (Figure 9 and Figure S1). Both ML and MP trees confirmed the previously known phylogenetic relationships in the family Zingiberaceae based on earlier studies [12,20,23,24,25,31,32,33,34], while unexpected relationships and positions of certain taxa were also revealed in this study. The reconfirmation of previously known relationships included (1) the monophyletic genera of Amomum, Alpinia, Kaempferia, Zingiber and Hedychium and their relationships to the rest of family Zingiberaceae, (2) the paraphyletic genus Curcuma in family Zingiberaceae, (3) relationships between two genera of Curcuma and Stahlianthus. The most surprising and unexpected findings in this study were the positions and relationships of A. zerumbet and A. oxyphylla. The two accessions of A. oxyphylla formed a group both in the two phylogenetic trees, confirming the validity of the assembled and annotated chloroplast genome of A. oxyphylla in this study. On morphological classifications, A. pumila was placed in subgenus Alpinia, A. zerumbet and A. katsumadai were placed in subgenus Catimbium, and A. oxyphylla was placed in subgenus Probolocalyx [11]. Therefore, our phylogenetic results were congruent with the morphological taxa [11]. On the other hand, several previous studies suggested that, A. oxyphylla was closely related to A. zerumbet based on chloroplast genomes [13], four groups were identified in genus Alpinia based on the ITS and matK trees [12]. We analyzed the reasons for the incongruence between chloroplast SNP-based phylogenetic analyses and chloroplast genomes, ITS and matK phylogenies in the following. Firstly, we did not have the same species samples as the ITS and matK phylogenies [12]. Secondly, we detected further phylogenetic relationships between A. zerumbet and A. oxyphylla with more chloroplast genomes data than the previous study [13]. Thirdly, with more than 230 species, the positions of Alpinia in family Zingiberaceae still remained somewhat uncertain and the future phylogenetic analyses should obtain additional chloroplast genomes of Alpinia species. The current phylogenetic analyses could supply the possibility that chloroplast genomes may be useful for phylogeny and species identification in family Zingiberaceae in the future.
Figure 9.
Phylogenetic tree constructed with SNPs from 28 chloroplast genomes using maximum likelihood method. The bootstrap values were based on 1000 replicates and were indicated next to the branches.
3. Materials and Methods
3.1. Plant Material and DNA Extraction
Fresh leaves were obtained from A. katsumadai (voucher specimen: Ak 2015001), A. oxyphylla sampled from Guangdong (voucher specimen: Ao_Liu 2015002) and A. pumila (voucher specimen: Ap_Liu 2015003) plants, respectively, from the resource garden of the Environmental Horticulture Research Institute, Guangdong Academy of Agricultural Sciences, Guangzhou, China. The total chloroplast DNA was extracted from those leaves using the modified sucrose gradient centrifugation method [49]. The chloroplast DNA sample of good integrity and with both optical density (OD) 260/280 and OD 260/230 ratio greater than 1.8 was used for subsequent experiments.
3.2. Chloroplast Genome Sequencing and Assembly
For each sample, two libraries with insert sizes of 300 bp and 10 kb were constructed after purification, and then sequenced on an Illumina Hiseq X Ten instrument (Biozeron, Shanghai, China) and a PacBio Sequel platform (Biozeron, Shanghai, China), respectively. The resulting Illumina raw data and PacBio raw data were assessed with FastQC. A total of 66.3 M, 80.2 M and 66.4 M clean data of 150 bp Illumina paired-end reads were generated from A. katsumadai, A. oxyphylla sampled from Guangdong and A. pumila, respectively, and 0.62M, 0.56 M and 0.50 M clean data of 8–10 kb subreads were generated from the three species, respectively.
Firstly, the Illumina paired-end clean reads were assembled using SOAPdenova (version 2.04) with default parameters into principal contigs [50], and all contigs were sorted and joined into a single draft sequence using the software Geneious version 11.0.4 [51]. Next, BLASR software was used to compare the PacBio clean data with the single draft sequence and to extract the correction and error correction [52]. Next, the corrected PacBio clean data were assembled using Celera Assembler (version 8.0) with default parameters, thus generating scaffolds [53]. Next, the assembled scaffolds were mapped back to the Illumina clean reads using GapCloser (version 1.12) for gap closing [50]. Finally, the redundant fragments sequences were removed, thus generating the final assembled chloroplast genomic sequence.
3.3. Chloroplast Genome Annotation and Structure Analysis
The assembled chloroplast genome was annotated using the online tool DOGMA (Dual Organellar Genome Annotator) [54] with default parameters and then checked manually. BLASTn searches in the NCBI website were used to identify and confirm both tRNA and rRNA genes. Lastly, further verification of the tRNA genes was carried out using tRNAscanSE with default settings [55]. The circular physical map of the chloroplast genome was drawn using OGDRAWv1.3.1 program with default parameters and subsequent manual editing [56]. The GenBank accession numbers of A. katsumadai, A. oxyphylla sampled from Guangdong and A. pumila are MK262728, MK262729 and MK262731, respectively.
SSRs were identified using MISA (http://pgrc.ipk-gatersleben.de/misa/) [57]. The parameters for SSRs were adjusted for identification of perfect mono-, di-, tri-, tetra-, pena-, and hexanucleotide motifs with a minimum of 8, 5, 4, 3, 3, and 3 repeats, respectively. Furthermore, the size and location of long repeat sequences, including forward, palindrome, reverse and complement repeats, were determined by the online software REPuter [58], with a hamming distance of 3 and a mininal repeat size of 30 bp.
3.4. Codon Usage and Prediction of RNA Editing Sites
Codon usage was determined for all protein-coding genes. To examine the deviation in synonymous codon usage, the relative synonymous codon usage (RSCU) was calculated using MEGA7 software [59]. Amino acid frequency was also calculated and expressed by the percentage of the codons encoding the same amino acid divided by the total codons. To predict the possible RNA editing sites in the three Alpinia species chloroplast genomes, protein-coding genes were used to predict potential RNA editing sites using the online program Predictive RNA Editor for Plants (PREP) suite (http://prep.unl.edu/) with a cutoff value of 0.8 [60].
3.5. Genome Comparison and Divergence Analyses
To compare the chloroplast genome of A. pumila with other four Alpinia accessions (A. katsumadai, A.zerumbet, and two accessions of A. oxyphylla sampled from Guangdong and Hainan), the mVISTA program (http://genome.lbl.gov/vista/mvista/about.shtml) in the Shuffle-LAGAN mode [61] was carried out using the annotation of A. pumila as the reference. To detect the variation in the LSC/IR/SSC boundaries of Alpinia chloroplast genomes, five Alpinia whole genomes were included in comparisons. The nucleotide variability (Pi) among chloroplast genomes was performed using software DnaSP v5 [62], with window length of 600 bp and the step size of 200 bp. The five Alpinia whole genomes were also aligned using MUMmer software [63] and adjusted manually where necessary using Se-Al 2.0 [64], using the annotated A. pumila chloroplast genome as the reference. The SNPs and indels were recorded separately as well as their locations in the chloroplast genome. The two accessions of A. oxyphylla were analyzed to identify SNPs and indels markers that can be selected in subsequent different sources studies.
3.6. Phylogenetic Analyses
In this study, a total of 25 complete chloroplast genome sequences were downloaded from the GenBank (NCBI) database. C. pulverulentus, C. viridis and C. indica were used as outgroups of the family Zingiberaceae. Phylogenetic trees were constructed using SNP matrix from 25 representative chloroplast genomes of the family Zingiberaceae. The reliable SNP sites were obtained by previously described method [23]. For each chloroplast genome, all SNP sites were connected in the same order to obtain a sequence in FASTA format. Multiple FASTA format sequences alignments were carried out by ClustalW in software MEGA7 [59]. To examine the phylogenetic applications of rapidly evolving 11,112 SNP markers (Supplement.SNP matrix.data), maximum likelihood (ML) analysis based on the nucleotide substitution model of Tamura-Nei was conducted using software MEGA7 [59]. The maximum parsimony (MP) method was also employed to construct a phylogenetic tree using the Close-Neighbor-Interchange (CNI) model in software MEGA7 [59]. Both ML and MP analyses were performed with 1000 bootstrap replicates, respectively.
4. Conclusions
In summary, we sequenced and characterized the complete chloroplast genomes of three Alpinia (A. katsumadai, A. pumila and A. oxyphylla sampled from Guangdong) of the family Zingiberaceae. Then, we compared them to the two reported chloroplast genomes of A. zerumbet and A. oxyphylla sampled from Hainan. These five Alpinia chloroplast genomes were highly conserved in terms of gene order, GC content, SSRs, and long repeats, and were typical quadripartite circle molecules consisting of an LSC region, an SSC region, and a pair of separated IRs. The IR expansions and contractions of the border regions resulted in difference of genome size between five Alpinia accessions. Fifteen highly divergent regions (rpl36, ycf1, rps15, rpl22, infA, psbT-psbN, accD-psaI, petD-rpoA, psaC-ndhE, ccsA-ndhD, ndhF-rpl32, rps11-rpl36, infA-rps8, psbC-psbZ, and rpl32-ccsA) were found and could be used as potential markers for Alpinia species on plant identification and phylogenetic studies. Additionally, for the interspecific comparisons, 304, 367, 331 and 371 SNPs were detected in comparisons of A. pumila-A. katsumadai, A. pumila-A. oxyphylla (Guangdong), A. pumila-A. zerumbet, and A. pumila-A. oxyphylla (Hainan), respectively, when the A. pumila chloroplast genome was used as the reference; 118, 122, 115, and 120 indels were accurately located in these four comparisons, respectively. Through the intraspecific comparison, 20 SNPs and 23 indels were identified in the two accessions of A. oxyphylla. The ML and MP trees indicated that the chloroplast SNP-based phylogenetic analyses could be used to identify the five Alpinia accessions. Alpinia showed a close phylogenetic relationship with Amomum species. The molecular data in this study represent a valuable resource for the studies of phylogenetic relationships and species identification in the family Zingiberaceae.
Acknowledgments
A. oxyphylla sampled from Guangdong and A. pumila resources were supplied by professor Nian Liu at department of horticulture and landscape architecture, Zhongkai University of Agriculture and Engineering, Guangzhou, China. The authors are very thankful for the two anonymous reviewers suggestions for improving the manuscript.
Abbreviations
| bp | base pairs |
| BLAST | Basic Local Alignment Search Tool |
| NCBI | National Center for Biotechnology Information |
| IR | Inverted repeat |
| LSC | Large single copy |
| SSC | Small single copy |
| SSRs | Simple Sequence Repeats |
| A | Adenine |
| T | Thymine |
| G | Guanine |
| C | Cytosine |
| DNA | Deoxyribonucleic Acid |
| RNA | Ribonucleic acid |
| tRNA | Transfer RNA |
| rRNA | Ribosomal RNA |
| SNPs | Single-Nucleotide Polymorphisms |
| Indels | Insertion/deletions |
Supplementary Materials
The following are available online at https://www.mdpi.com/2223-7747/9/2/286/s1, Figure S1: Phylogenetic tree constructed with the SNP matrix from 28 chloroplast genomes using maximum parsimony (MP) method, Table S1: Features of the chloroplast genomes of three Alpinia species, Table S2: Gene distribution in the chloroplast genomes of three Alpinia species, Table S3: Genes with introns in the chloroplast genomes of three Alpinia species as well as the exons and introns, Table S4: Codon usages of protein-coding genes in the chloroplast genomes of three Alpinia species, Table S5: RNA editing sites analyses of three Alpinia species, Table S6: SSRs distribution among five Alpinia chloroplast genomes, Table S7: Long repeats distribution among five Alpinia chloroplast genomes, Table S8: SNPs detected in coding and non-coding regions among four Alpinia chloroplast genomes, Table S9: Indels detected among four Alpinia chloroplast genomes, Table S10: Detailed indels and SNPs information detected in the two chloroplast genomes of A. oxyphylla. Supplement.SNP matrix.data.
Author Contributions
D.-M.L. and G.-F.Z. conceived and designed the experiments, generated and analyzed the data, wrote the draft of the manuscript and revised it. Y.-C.X., Y.-J.Y. and J.-M.L. collected plant materials. All authors contributed to the experiments and approved the final draft of the manuscript.
Funding
This work was financially supported by Guangzhou Municipal Science and Technology Project (No.201607010101), National Natural Science Foundation of China (No.31501788), Guangdong Science and Technology Project (No.2015A020209078; No.2015B070701016) and the the Construction, Conservation and Evaluation of Flower Germplasm Resources (Garden) Subproject of Construction, Conservation and Evaluation of Crop Germplasm Resources (Garden) Project in Guangdong Province.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Wu D., Larsen K. Flora of China. Volume 24. Science Press; Beijing, China: 2000. Zingiberaceae; pp. 322–377. [Google Scholar]
- 2.Wu D., Liu N., Ye Y. The Zingiberaceous Resources in China. Volume 1. Huazhong University of Science and Technology University Press; Wuhan, China: 2016. pp. 12, 20, 30, 33. [Google Scholar]
- 3.Branney T.M.E. Hardy Gingers: Including Hedychium, Roscoea and Zingiber. Timber Press, Inc.; Porland, OR, USA: 2005. pp. 41–63. [Google Scholar]
- 4.Zhang J.Q., Li Y.B. Zhongguo Yaoyong Jiangke Zhiwu. Volume 1. China Medical Science Press; Beijing, China: 2015. pp. 133–220. [Google Scholar]
- 5.Pogačar M.Š., Klančnik A., Bucar F., Langerholc T., Možina S.S. Alpinia katsumadai extracts inhibit adhesion and invasion of campylobacter jejuni in animal and human foetal small intestine cell lines. Phytother. Res. 2015;29:1585–1589. doi: 10.1002/ptr.5396. [DOI] [PubMed] [Google Scholar]
- 6.Li Y.Y., Huang S.S., Lee M.M., Deng J.S., Huang G.J. Anti-inflammatory activities of cardamonin from Alpinia katsumadai through heme oxygenase-1 induction and inhibition of NF-κB and MAPK signaling pathway in the carrageenan-induced pawedema. Int. Immunopharmacol. 2015;25:332–339. doi: 10.1016/j.intimp.2015.02.002. [DOI] [PubMed] [Google Scholar]
- 7.Jang H.J., Lee S.J., Lee S., Jung K., Lee S.W., Rho M.C. Acyclic triterpenoids from Alpinia katsumadai inhibit IL-6-induced STAT3 activation. Molecules. 2017;22:1611. doi: 10.3390/molecules22101611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang X.B., Yang C.S., Luo J.G., Zhang C., Luo J., Yang M.H., Kong L.Y. Experimental and theoretical calculation studies on the structure elucidation and absolute configuration of calyxins from Alpinia katsumadai. Fitoterapia. 2017;119:121–129. doi: 10.1016/j.fitote.2017.04.014. [DOI] [PubMed] [Google Scholar]
- 9.Chen F., Li H.L., Tan Y.F., Guan W.W., Zhang J.Q., Li Y.H., Zhao Y.S., Qin Z.M. Different accumulation profiles of multiple components between pericarp and seed of Alpinia oxyphylla capsular fruit as determined by UFLC-MS/MS. Molecules. 2014;19:4510–4523. doi: 10.3390/molecules19044510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qi Y., Cheng X., Jing H., Yan T., Xiao F., Wu B., Bi K., Jia Y. Comparative pharmacokinetic study of the components in Alpinia oxyphylla Miq.-Schisandra chinensis (Turcz.) Baill. herb pair and its single herb between normal and Alzheimer’s disease rats by UPLC-MS/MS. J. Pharm. Biomed. Anal. 2020;177:112874. doi: 10.1016/j.jpba.2019.112874. [DOI] [PubMed] [Google Scholar]
- 11.Flora Reipublicae Popularis Sinicae. Science press; Beijing, China: 1981. Delectis Florae Reipublicae Popularis Sinicae Agendae Academiae Sinicae Edita. [Google Scholar]
- 12.Kress W.J., Prince L.M., Williams K.J. The phylogeny and a new classification of the gingers (Zingiberaceae) evidence from molecular data. Am. J. Bot. 2002;89:1682–1696. doi: 10.3732/ajb.89.10.1682. [DOI] [PubMed] [Google Scholar]
- 13.Gao B., Yuan L., Tang T., Hou J., Pan K., Wei N. The complete chloroplast genome sequence of Alpinia oxyphylla Miq. and comparison analysis within the Zingiberaceae family. PLoS ONE. 2019;14:e0218817. doi: 10.1371/journal.pone.0218817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wicke S., Schneeweiss G.M., DePamphilis C.W., Muller K.F., Quandt D. The evolution of the plastid chromosome in land plants: Gene content, gene order, gene function. Plant Mol. Biol. 2011;76:273–297. doi: 10.1007/s11103-011-9762-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Daniell H., Lin C.S., Yu M., Chang W.J. Chloroplast genomes: Diversity, evolution, and applications in genetic engineering. Genome Biol. 2016;17:134. doi: 10.1186/s13059-016-1004-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eid J., Fehr A., Gray J., Luong K., Lyle J., Otto G., Peluso P., Rank D., Baybayan P., Bettman B., et al. Real-time DNA sequencing from single polymerase molecules. Science. 2009;323:133–138. doi: 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
- 17.Ferrarini M., Moretto M., Ward J.A., Surbanovski N., Stevanovic V., Giongo L., Viola R., Cavalieri D., Velasco R., Cestaro A., et al. An evaluation of the PacBio RS platform for sequencing and de novo assembly of a chloroplast genome. BMC Genom. 2013;14:670. doi: 10.1186/1471-2164-14-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu Z., Gui S., Guan Z., Pan L., Wang S., Ke W., Liang D., Ding Y. A precise chloroplast genome of Nelumbo nucifera (Nelumbonaceae) evaluated with Sanger, Illumina MiSeq, and PacBio RS II sequencing platforms: Insight into the plastid evolution of basal eudicots. BMC Plant Biol. 2014;14:289. doi: 10.1186/s12870-014-0289-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chomicki G., Renner S.S. Watermelon origin solved with molecular phylogenetics including Linnaen material: Another example of museomics. New Phytol. 2015;205:526–532. doi: 10.1111/nph.13163. [DOI] [PubMed] [Google Scholar]
- 20.Wu M., Li Q., Hu Z., Li X., Chen S. The complete Amomum kravanh chloroplast genome sequence and phylogenetic analysis of the commelinids. Molecules. 2017;22:1875. doi: 10.3390/molecules22111875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin M., Qi X., Chen J., Sun L., Zhong Y., Fang J., Hu C. The complete chloroplast genome sequence of Actinidia arguta using the PacBio RSII platform. PLoS ONE. 2018;13:e0197393. doi: 10.1371/journal.pone.0197393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou Y., Nie J., Xiao L., Hu Z., Wang B. Comparative chloroplast genome analysis of rhubarb botanical origins and development of specific identification markers. Molecules. 2018;23:2811. doi: 10.3390/molecules23112811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li D.M., Zhao C.Y., Liu X.F. Complete chloroplast genome sequences of Kaempferia galanga and Kaempferia elegans: Molecular structures and comparative analysis. Molecules. 2019;24:474. doi: 10.3390/molecules24030474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cui Y., Chen X., Nie L., Sun W., Hu H., Lin Y., Li H., Zheng X., Song J., Yao H. Comparison and phylogenetic analysis of chloroplast genomes of three medicinal and edible Amomum species. Int. J. Mol. Sci. 2019;20:4040. doi: 10.3390/ijms20164040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cui Y., Nie L., Sun W., Xu Z., Wang Y., Yu J., Song J., Yao H. Comparative and phylogenetic analyses of ginger (Zingiber officinale) in the family Zingiberaceae based on the complete chloroplast genome. Plants. 2019;8:283. doi: 10.3390/plants8080283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li X., Zuo Y., Zhu X., Liao S., Ma J. Complete chloroplast genomes and comparative analysis of sequences evolution among seven Aristolochia (Aristolochiaceae) medicinal species. Int. J. Mol. Sci. 2019;20:1045. doi: 10.3390/ijms20051045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chiapella J.O., Barfuss M.H.J., Xue Z.Q., Greimler J. The plastid genome of Deschampsia cespitosa (Poaceae) Molecules. 2019;24:216. doi: 10.3390/molecules24020216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li W., Zhang C., Guo X., Liu Q., Wang K. Complete chloroplast genome of Camellia japonica genome structures, comparative and phylogenetic analysis. PLoS ONE. 2019;14:e0216645. doi: 10.1371/journal.pone.0216645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gao C., Deng Y., Wang J. The complete chloroplast genomes of Echinacanthus species (Acanthaceae): Phylogenetic relationships, adaptive evolution, and screening of molecular markers. Front. Plant Sci. 2019;9:1989. doi: 10.3389/fpls.2018.01989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Y., Sylvester S.P., Li M., Zhang C., Li X., Duan Y., Wang X. The complete plastid genome of Magnolia zenii and genetic comparison to Magnoliaceae species. Molecules. 2019;24:261. doi: 10.3390/molecules24020261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li D.M., Xu Y.C., Zhu G.F. Complete Chloroplast genome of the plant Stahlianthus involucratus (Zingiberaceae) Mitochondrial DNA Part B. 2019;4:2702–2703. doi: 10.1080/23802359.2019.1644227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li D.M., Zhao C.Y., Zhu G.F., Xu Y.C. Complete chloroplast genome sequence of Hedychium coronarium. Mitochondrial DNA Part B. 2019;4:2806–2807. doi: 10.1080/23802359.2019.1659114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li D.M., Zhao C.Y., Xu Y.C. Characterization and phylogenetic analysis of the complete chloroplast genome of Curcuma longa (Zingiberaceae) Mitochondrial DNA Part B. 2019;4:2974–2975. doi: 10.1080/23802359.2019.1664343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y., Deng J., Li Y., Gao G., Ding C., Zhang L., Zhou Y., Yang R. The complete chloroplast genome sequence of Curcuma flaviflora (Curcuma) Mitochondrial DNA Part A. 2016;27:3644–3645. doi: 10.3109/19401736.2015.1079836. [DOI] [PubMed] [Google Scholar]
- 35.Saina J.K., Li Z.Z., Gichira A.W., Liao Y.Y. The complete chloroplast genome sequence of tree of heaven (Ailanthus altissima (Mill.)) (Sapindales: Simaroubaceae), an important pantropical tree. Int. J. Mol. Sci. 2018;19:929. doi: 10.3390/ijms19040929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang Z., Huang Y., An W., Zheng X., Huang S., Liang L. Sequencing and structural analysis of the complete chloroplast genome of the medicinal plant Lycium chinense Mill. Plants. 2019;8:87. doi: 10.3390/plants8040087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim S.H., Yang J., Park J., Yamada T., Maki M., Kim S.C. Comparison of whole plastome sequences between thermogenic skunk cabbage Symplocarpus renifolius and nonthermogenic S. nipponicus (Orontioideae; Araceae) in east Asia. Int. J. Mol. Sci. 2019;20:4678. doi: 10.3390/ijms20194678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tian N., Han L., Chen C., Wang Z. The complete chloroplast genome sequence of Epipremnum aureum and its comparative analysis among eight Araceae species. PLoS ONE. 2018;13:e0192956. doi: 10.1371/journal.pone.0192956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang W., Yu H., Wang J., Lei W., Gao J., Qiu X., Wang J. The complete chloroplast genome sequences of the medicinal plant Forsythia suspense (Oleaceae) Int. J. Mol. Sci. 2017;18:2288. doi: 10.3390/ijms18112288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.George B., Bhatt B.S., Awasthi M., George B., Singh A.K. Comparative analysis of microsatellites in chloroplast genomes of lower and higher plants. Curr. Genet. 2015;61:665–677. doi: 10.1007/s00294-015-0495-9. [DOI] [PubMed] [Google Scholar]
- 41.Tong W., Kim T.S., Park Y.J. Rice chloroplast genome variation architecture and phylogenetic dissection in diverse Oryza species assessed by whole-genome resequencing. Rice. 2016;9:57. doi: 10.1186/s12284-016-0129-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dong W.L., Wang R.N., Zhang N.Y., Fan W.B., Fang M.F., Li Z.H. Molecular evolution of chloroplast genomes of orchid species: Insights into phylogenetic relationship and adaptive evolution. Int. J. Mol. Sci. 2018;19:716. doi: 10.3390/ijms19030716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gu C., Ma L., Wu Z., Chen K., Wang Y. Comparative analyses of chloroplast genomes from 22 Lythraceae species: Inferences for phylogenetic relationships and genome evolution within Myrtales. BMC Plant Biol. 2019;19:281. doi: 10.1186/s12870-019-1870-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yin K., Zhang Y., Li Y., Du F.K. Different natural selection pressures on the atpF gene in evergreen sclerophyllous and deciduous oak species: Evidence from comparative analysis of the complete chloroplast genome of Quercus aquifolioides with other oak species. Int. J. Mol. Sci. 2018;19:1042. doi: 10.3390/ijms19041042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Du Y., Bi Y., Yang F., Zhang M., Chen X., Xue J., Zhang X. Complete chloroplast genome sequences of Lilium: Insights into evolutionary dynamics and phylogenetic analyses. Sci. Rep. 2017;7:5751. doi: 10.1038/s41598-017-06210-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lu Q., Ye W., Lu R., Xu W., Qiu Y. Phylogenomic and comparative analyses of complete plastomes of Croomia and Stemona (Stemonaceae) Int. J. Mol. Sci. 2018;19:2383. doi: 10.3390/ijms19082383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang W., Chen S., Zhang X. Whole-genome comparison reveals heterogeneous divergence and mutation hotspots in chloroplast genome of Eucommia ulmoides oliver. Int. J. Mol. Sci. 2018;19:1037. doi: 10.3390/ijms19041037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jiang D., Zhao Z., Zhang T., Zhong W., Liu C., Yuan Q., Huang L. The chloroplast genome sequence of Scutellaria baicalensis provides insight into intraspecific and interspecific chloroplast genome diversity in Scutellaria. Genes. 2017;8:227. doi: 10.3390/genes8090227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li X., Hu Z., Lin X., Li Q., Gao H., Luo G., Chen S. High-throughput pyrosequencing of the complete chloroplast genome of Magnolia officinalis and its application in species identification. Acta Pharm. Sin. 2012;47:124–130. [PubMed] [Google Scholar]
- 50.Luo R., Liu B., Xie Y., Li Z., Huang W., Yuan J., He G., Chen Y., Pan Q., Liu Y. SOAPdenovo2: An empirically improved memory-efficient short-end de novo assembler. Gigascience. 2012;1:18. doi: 10.1186/2047-217X-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kearse M., Moir R., Wilson A., Stoneshavas S., Cheung M., Sturrock S., Buxton S., Cooper A., Markowitz S., Duran C. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chaisson M.J., Tesler G. Mapping single molecule sequencing reads using basic local alignment with successive refinement (BLASR): Application and theory. BMC Bioinform. 2012;13:238. doi: 10.1186/1471-2105-13-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Denisov G., Walenz B., Halpern A.L., Miller J., Axerlrod N., Levy S., Sutton G. Consensus generation and variant detection by celera assembler. Bioinformatics. 2008;24:1035–1040. doi: 10.1093/bioinformatics/btn074. [DOI] [PubMed] [Google Scholar]
- 54.Wyman S.K., Jansen R.K., Boore J.L. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 2004;20:3252–3255. doi: 10.1093/bioinformatics/bth352. [DOI] [PubMed] [Google Scholar]
- 55.Lowe T.M., Chan P.P. tRNAscan-SE On-line: Search and Contextual Analysis of Transfer RNA Genes. Nucleic Acids Res. 2016;44:W54–W57. doi: 10.1093/nar/gkw413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Greiner S., Lehwark P., Bock R. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: Expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 2019;47:W59–W64. doi: 10.1093/nar/gkz238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.MISA-Microsatellite Identification Tool. [(accessed on 20 September 2017)]; Available online: http://pgrc.ipk-gatersleben.de/misa/
- 58.Kurtz S., Choudhuri J.V., Ohlebusch E., Schleiermacher C., Stoye J., Giegerich R. REPuter: The manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001;29:4633–4642. doi: 10.1093/nar/29.22.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kumar S., Stecher G., Tamura K. Mega7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mower J.P. The PREP Suite: Predictive RNA editors for plant mitochondrial genes, chloroplast genes and user-defined alignments. Nucleic Acids Res. 2009;37:W253–W259. doi: 10.1093/nar/gkp337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Frazer K.A., Pachter L., Poliakov A., Rubin E.M., Dubchak I. VISTA: Computational tools for comparative genomics. Nucleic Acids Res. 2004;32:W273–W279. doi: 10.1093/nar/gkh458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Librado P., Rozas J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- 63.Marcais G., Delcher A.L., Phillippy A.M., Coston R., Salzberg S.L., Zimin A. MUMmer4: A fast and versatile genome alignment system. PLoS Comput. Biol. 2018;14:e1005944. doi: 10.1371/journal.pcbi.1005944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rambaut A. Se-Al: Sequence Alignment Editor; Version 2.0. [(accessed on 30 September 2017)]; Available online: http://tree.bio.ed.ac.uk/software.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.