Abstract
Euphorbia is one of the largest genera in the Euphorbiaceae family, comprising 2000 species possessing commercial, medicinal, and ornamental importance. However, there are very little data available on their molecular phylogeny and genomics, and uncertainties still exist at a taxonomic level. Herein, we sequence the complete chloroplast (cp) genomes of two species, E. larica and E. smithii, of the genus Euphorbia through next-generation sequencing and perform a comparative analysis with nine related genomes in the family. The results revealed that the cp genomes had similar quadripartite structure, gene content, and genome organization with previously reported genomes from the same family. The size of cp genomes ranged from 162,172 to 162,358 bp with 132 and 133 genes, 8 rRNAs, 39 tRNA in E. smithii and E. larica, respectively. The numbers of protein-coding genes were 85 and 86, with each containing 19 introns. The four-junction regions were studied and results reveal that rps19 was present at JLB (large single copy region and inverted repeat b junction) in E. larica where its complete presence was located in the IRb (inverted repeat b) region in E. smithii. The sequence comparison revealed that highly divergent regions in rpoC1, rpocB, ycf3, clpP, petD, ycf1, and ndhF of the cp genomes might provide better understanding of phylogenetic inferences in the Euphorbiaceae and order Malpighiales. Phylogenetic analyses of this study illustrate sister clades of E. smithii with E. tricullii and these species form a monophyletic clade with E. larica. The current study might help us to understand the genome architecture, genetic diversity among populations, and evolutionary depiction in the genera.
Keywords: chloroplast genomics, genus Euphoria, comparative analyses, Euphorbiaceae
1. Introduction
Plants have chloroplasts (cp) that help in photosynthesis [1]. The genomic component of cp is composed of circular and double-stranded DNA molecules [2]. Moreover, it is very essential for fatty acids, starch, and pigments biosynthesis [3]. The chloroplast contains its own independent genomic component, which is highly conserved in angiosperms. The chloroplast genome possesses certain characteristics such as small single copies, multiple copies, and a simple structure [4]. Unlike the other genomes, such as the nuclear genome, which has more repetitive sequences, the mitochondrial genome in which frequent rearrangements of nucleotide occur, the chloroplast genome is conservative [5]. The chloroplast genome is maternally inherited in angiosperm, having its own independent evolutionary route [6]. The chloroplast genome shows collinearity among the plant kingdom, which is why phylogenetic trees are constructed on the basis of chloroplast data, and the genome structure of chloroplast provides information regarding the specie origin, evolution, and also the differences between closely related and other species [7]. In recent years, with the advent of advanced sequencing technology, more chloroplast genomes have been sequenced [4]. In this study, we sequenced the complete chloroplast genome of ecologically endangered species E. smithii and E. larica and performed a comparative analysis with other genomes from the Euphorbiaceae family.
The Euphorbiaceae (Spurge family) is one of the largest families in angiosperm and comprises 300 genera and almost 7500 species [8]. Euphorbia larica Boiss. and Euphorbia smithii S. Carter belong to the genus Euphorbia, which is the largest genus in the Euphorbiaceae family, comprising almost 2000 identified species, which mostly produce latex and possess a unique flower structure [9]. The genus is estimated to have originated in Africa approximately 48 million years ago and expanded to the American continents through two single long-distance dispersal events, i.e., 30 and 25 million years ago [10,11,12,13]. Euphorbia is an ecologically, medicinally, and commercially important genus in the Euphorbiaceae family, and various indigenous based traditional folk recipes are utilized as medicines for curing skin diseases, intestinal parasites, gonorrhea, warts, and migraines [14]. The Euphorbia species constitutes monocyclic diterpenoids that possess anti-bacterial, anti-cancer, anti-HCV, and analgesic activities [15,16,17]. Some plants of this genus secrete a sap which prevents the growth of other species and shows their habitat dominance feature [18]. In addition, some plants from this genus (for example, E. pulcherrima) is used for ornamental purposes [19,20].
In the case of E. larica and E. smithii, E. larica is native and widely found in northern regions of Oman [21], whilst E. smithii (near-threatened) was once considered endemic to Oman but has also been found in Yemen [22]. E. larica is a woody species with a self-supporting habitat, whereas E. smithii is a shrub [23]. The species are rich in flavonoids [23], alkaloids [9], and terpenoids [23]. The latex of E. smithii is used for veterinary medicines at a local level [24]. Similarly, the latex derived from E. larica is used to treat the camel parasite [25]. There are several examples of understanding the genetic diversity of the Euphorbia species, such as E. telephioides [26] and E. pulcherrima [27]. However, no study has been performed on E. larica and E. smithii due to the lack of genome or related sequence data. Understanding the genetic diversity is essential to ensure increased conservation efforts for the decline of such endemic or native species in the world. Looking at the importance of these species, we sequenced the complete chloroplast genome of two important species, E. larica and E. smithii, and performed comparative analysis with related species (E. esula, E. tirucalli, M. esculanta, J. curcas, H. brasiliensis, R. communis, V. fordii, C. tiglium, and D. tonkinensis). In our study, the sequencing of complete chloroplast genomes of E. larica and E. smithii encourages and provides a basis for a more detailed study of chloroplast molecular biology and also helps in the genetic breeding and molecular evolution of this threatened species. This study also provides details of evolutionary analysis and helps in the classification of this morphologically diverse species. Some previous studies have suggested that this group has been difficult to discern mainly due to homoplasious morphological characters and inadequate taxon sampling in previous phylogenetic studies [28].
2. Results
2.1. Comparative Characteristics of Chloroplast genomes
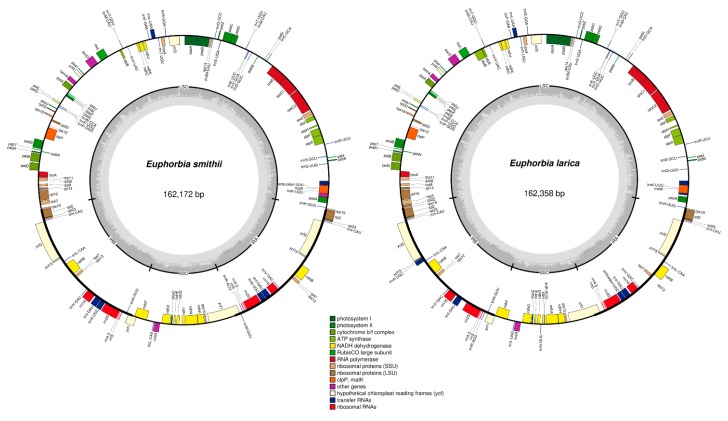
The chloroplast genome of the Euphoria species showed a typical tetrad quadripartite structure comprising (i) small single copy, (ii) large single copy, (iii) inverted repeat A, and (iv) inverted repeat B that are mirror images of each other (Figure 1A,B). The complete chloroplast genome of E. smithii was 162,172 bp, which is 186 bp less than E. larica (162,358 bp). The two sequenced genomes were compared with two from the Euphorbia genus (E. esula and E. tirucalli) and seven (M. esculanta, J. curcas, H. brasiliensis, R. communis, V. fordii, C. tiglium, and D. tonkinensis) other cp genomes from the Euphorbiaceae family. The large single-copy region (LSC) of E. smithii was observed with a length of 91,158, while the LSC of E. larica was 91,537 bp in length. The length of the small single copy region of E. smithii (18,603 bp) was 364 bp larger than E. larica (18,239 bp). The smallest IR region length was 10,100 bp in C. tiglium and the highest was observed in the R. communis (27,347 bp). Overall, there were little differences among the two sequenced genomes, and the main differences were in the LSC and SSC regions. The complete chloroplast genome ranges from 150,021 bp in C. tiglium to 163,856 bp in J. curcas (Table 1).
Figure 1.
Genome circular map of the E. smithii and E. larica. Thick lines indicate the extent of the inverted repeat regions (IRa and IRb), which separate the genome into small (SSC) and large (LSC) single copy regions. Genes drawn inside the circle are transcribed clockwise, while those outside of the circle are transcribed counter-clockwise. Genes belonging to different functional groups are color-coded. The dark gray in the inner circle corresponds to the GC content, while the light gray corresponds to the AT content.
Table 1.
Summary of complete chloroplast genomes of E. laica and E. smithii.
| E. smithii | E. larica | E. esula | E. tirucalli | M. esculanta | J. curcas | H. brasiliensis | R. communis | V. fordii | C. tiglium | D. tonkinensis | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Size (bp) | 162,172 | 162,358 | 160,512 | 163,091 | 161,453 | 163,856 | 161,191 | 163,161 | 161,528 | 150,021 | 163,481 |
| Overall GC contents | 35.8 | 35.6 | 35.6 | 35.6 | 35.9 | 35.4 | 35.7 | 35.7 | 36.0 | 35.4 | 35.7 |
| LSC size in bp | 91,158 | 91,537 | 90,309 | 91,259 | 89,275 | 91,846 | 89,209 | 89,650 | 89,132 | 111,654 | 91,453 |
| SSC size in bp | 18,603 | 18,239 | 17,023 | 18,168 | 18,250 | 17,849 | 18,362 | 18,816 | 18,758 | 18,167 | 18,476 |
| IR size in bp | 26,206 | 26,291 | 26,590 | 26,832 | 26,954 | 27,023 | 26,810 | 27,347 | 26,819 | 10,100 | 26,776 |
| Protein coding regions size in bp | 79,173 | 80,458 | 80,274 | 74,289 | 72,108 | 79,206 | 78,852 | 79,494 | 80,283 | 68,601 | 79,857 |
| tRNA size in bp | 2885 | 2887 | 2925 | 2885 | 2814 | 2797 | 2812 | 2802 | 2742 | 2560 | 2740 |
| rRNA size in bp | 9050 | 9049 | 9049 | 9049 | 6252 | 9047 | 9050 | 9050 | 9048 | 9050 | 9050 |
| Number of genes | 132 | 133 | 132 | 130 | 128 | 129 | 129 | 131 | 129 | 122 | 134 |
| Number of protein coding genes | 85 | 86 | 85 | 82 | 83 | 84 | 84 | 86 | 85 | 78 | 84 |
| Number of rRNA | 8 | 8 | 8 | 8 | 7 | 8 | 8 | 8 | 8 | 8 | 8 |
| Number of tRNA | 39 | 39 | 39 | 39 | 38 | 37 | 37 | 37 | 36 | 34 | 36 |
| Genes with introns | 19 | 19 | 18 | 13 | 20 | 21 | 21 | 19 | 21 | 20 | 20 |
The total numbers of genes annotated in E. smithii and E. larica were 132 and 133, respectively, including 85 and 86 protein coding genes (PCGs), 8 rRNAs, 39 tRNAs, and 19 intron-containing genes (Table 1). The number of protein-coding genes varied among these genomes, the highest of which were recorded in E. larica and R. communis, whilst the lowest were 78 in C. tiglium. The highest number of introns containing genes among the compared genomes was 21, noted in J. curcas, H. brasiliensis, and V. fordii, and the lowest were 13 in E. tirucalli. GC content of E. smithii was found higher than GC content of E. larica; the highest GC content found in the compared genome was 36% found in V. fordii, while the lowest was observed in J. curcas and C. tiglium with 35.4%. Relative conservation of the genome structure and gene contents were observed among all the eleven chloroplast genomes with no specific gene organization and rearrangement observed, though some differences were still found in the number of genes, intron losses, and contraction and expansion in the IR regions. The chloroplast genome contains some of the important genes responsible for the vital process of life, i.e., photosynthesis and self-replication of chloroplast being a self-replicating organelle in the plant cell (Supplementary Table S1). The self-replication of chloroplast includes the gene responsible for the large subunit of ribosomal proteins, the small subunit of ribosome, DNA dependent RNA polymerase, rRNA genes, and tRNA genes. The genes responsible for photosynthesis further include photosystem I and II, and 33 genes are responsible for carrying out photosynthesis in the chloroplast genome (Supplementary Table S1).
The chloroplast genome of E. smithii, E. larica, E. esula, and E. tirucalli contains 18 introns containing genes. Out of these 18 introns containing genes, five are tRNAs, and three genes, ycf3, clpP, and rps12, contain double introns (Table 2). The base composition analyzed in sequenced and compared genomes reveal that adenine (A) at the first position was 31.0%, 31.2%, 31.0%, and 30.9% and on second position 29.7%, 29.8%, 29.8%, and 29.9%, while at third position 13.8%, 32.1%, 28.4%, and 32.4% in the E. larica, E. smithii, and E. tirucalli, respectively. Likewise, the composition of base T (thymine) at the first position was 24.3%, 25.9%, 24.1%, and 23.9%; at second position 32.6%, 32.7%, 28.4%, and 38.4%; and at third position 38.2%, 38.2%, 38.6%, and 38.4%, respectively. Furthermore, the abundance of “G” and “C” at the first, second, and third positions were observed less than the abundance of “A” and “T” (Table 3).
Table 2.
The genes with introns in the Euphorbia species chloroplast genome and the length of exons and introns.
| Gene | Exon I (bp) | Intron 1 (bp) | Exon II (bp) | Intron II (bp) | Exon III (bp) | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| E.l | E.s | E.e | E.t | E.l | E.s | E.e | E.t | E.l | E.s | E.e | E.t | E.l | E.s | E.e | E.t | E.l | E.s | E.e | E.t | |
| atpF | 145 | 145 | 145 | 145 | 670 | 671 | 666 | 667 | 470 | 470 | 470 | 470 | ||||||||
| petB | 6 | 6 | 773 | 779 | 642 | 642 | ||||||||||||||
| PetD | 8 | 8 | 779 | 780 | 496 | 496 | ||||||||||||||
| rpl2* | 400 | 396 | 396 | 396 | 634 | 629 | 624 | 629 | 464 | 468 | 468 | 468 | ||||||||
| rpl16 | 9 | 9 | 1395 | 1395 | 399 | 399 | ||||||||||||||
| rpoC1 | 430 | 432 | 432 | 767 | 769 | 775 | 1613 | 1618 | 1617 | |||||||||||
| rps12* | 114 | 114 | 114 | 114 | 232 | 232 | 232 | 232 | 541 | 536 | 536 | 536 | 26 | 26 | 26 | 26 | ||||
| clpP | 71 | 71 | 71 | 71 | 825 | 831 | 821 | 827 | 291 | 291 | 291 | 291 | 650 | 648 | 653 | 651 | 229 | 229 | 229 | 229 |
| ndhA | 553 | 554 | 553 | 553 | 1138 | 1111 | 1116 | 1137 | 539 | 541 | 539 | 539 | ||||||||
| ndhB* | 777 | 777 | 777 | 682 | 682 | 678 | 756 | 756 | 756 | |||||||||||
| ycf3 | 124 | 124 | 124 | 124 | 733 | 747 | 747 | 733 | 230 | 230 | 230 | 230 | 669 | 676 | 675 | 677 | 153 | 153 | 153 | 153 |
| trnA-UGC* | 38 | 38 | 38 | 38 | 813 | 803 | 813 | 803 | 35 | 35 | 35 | 35 | ||||||||
| trnI-GAU* | 42 | 42 | 42 | 42 | 945 | 945 | 945 | 945 | 35 | 35 | 35 | 35 | ||||||||
| trnL-UAA | 37 | 37 | 37 | 37 | 587 | 583 | 619 | 590 | 50 | 50 | 50 | 50 | ||||||||
| trnK-UUU | 37 | 37 | 37 | 37 | 2551 | 2560 | 2555 | 2563 | 28 | 29 | 29 | 29 | ||||||||
| trnV-UAC | 39 | 582 | 42 | |||||||||||||||||
Euphorbia larica = E.l, Euphorbia smithii = E.s, Euphorbia esula = E.e, Euphorbia tirucalli = E.t.
Table 3.
Base composition of the Euphorbia species in the chloroplast genome.
| T/U | C | A | G | Length (bp) | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| E.l | E.s | E.e | E.t | E.l | E.s | E.e | E.t | E.l | E.s | E.e | E.t | E.l | E.s | E.e | E.t | E.l | E.s | E.e | E.t | |
| Genome | 32.7 | 32.5 | 32.7 | 32.6 | 18.1 | 18.1 | 18 | 18.1 | 31.7 | 31.7 | 31.8 | 31.8 | 17.5 | 17.6 | 17.6 | 17.5 | 162,258 | 162,172 | 160,512 | 163,091 |
| LSC | 34.5 | 34.3 | 34.5 | 34.3 | 16.7 | 16.8 | 16.6 | 16.7 | 32.8 | 32.7 | 32.9 | 32.9 | 16.0 | 16.1 | 16.1 | 16.1 | 91,537 | 91,158 | 90,309 | 91,259 |
| SSC | 35.0 | 34.6 | 34.8 | 35 | 15.7 | 15.8 | 15.9 | 15.8 | 34.9 | 34.9 | 35 | 34.9 | 14.3 | 14.7 | 14.3 | 14.3 | 18,239 | 18,603 | 17,023 | 1818 |
| IR | 28.9 | 29 | 28.7 | 29.1 | 20.5 | 20.5 | 21.9 | 20.4 | 28.6 | 28.5 | 29 | 28.6 | 22.0 | 22.1 | 20.4 | 21.9 | 26,291 | 26,206 | 26,590 | 26,832 |
| tRNA | 26.5 | 25.5 | 25.4 | 25.5 | 21.6 | 23.2 | 23.2 | 23.3 | 24.7 | 22.2 | 22.3 | 22.1 | 27.2 | 29.2 | 29.1 | 29.1 | 2887 | 2885 | 2925 | 2885 |
| rRNA | 18.8 | 18.8 | 18.8 | 18.8 | 23.7 | 23.7 | 23.7 | 23.7 | 25.8 | 25.8 | 25.7 | 25.7 | 31.8 | 31.8 | 31.8 | 31.8 | 2049 | 9050 | 9049 | 9049 |
| Protein Coding genes | 31.8 | 31.8 | 31.8 | 31.6 | 17.4 | 17.4 | 17.3 | 17.3 | 31.1 | 31.1 | 31.2 | 31.1 | 19.8 | 19.8 | 19.6 | 19.9 | 80,458 | 79,173 | 80,274 | 74,289 |
| 1st position | 24.3 | 25.9 | 24.1 | 23.9 | 18.3 | 18.4 | 18.4 | 18.5 | 31.0 | 31.2 | 31.0 | 30.9 | 26.1 | 24.3 | 26.3 | 26.6 | 26,817 | 26,390 | 26,758 | 24,763 |
| 2nd position | 32.6 | 32.7 | 28.4 | 38.4 | 19.9 | 19.8 | 19.9 | 19.9 | 29.7 | 29.8 | 29.8 | 29.9 | 17.5 | 17.5 | 17.4 | 17.5 | 26,817 | 26,390 | 26,758 | 24,763 |
| 3rd position | 38.2 | 38.2 | 38.6 | 38.4 | 32.2 | 13.8 | 16.6 | 13.4 | 13.8 | 32.1 | 28.4 | 32.4 | 15.5 | 18.1 | 13.8 | 15.7 | 26,817 | 26,390 | 26,758 | 24,763 |
Euphorbia larica = E.l, Euphorbia smithii = E.s, Euphorbia esula = E.e, Euphorbia tirucalli = E.t.
2.2. Analysis of Repetitive Sequences in the Genomes
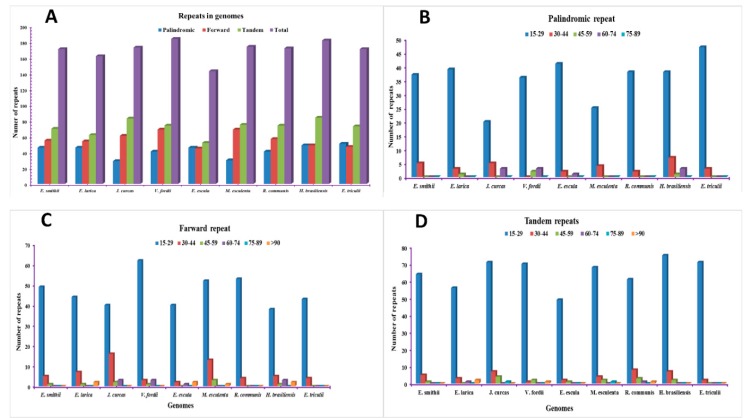
Repeat analyses of the two sequenced Euphoria species and seven other chloroplast genomes were conducted. The result of the repeats shows the total number of repeats present in E. smithii (171) and E. larica (162). Among the compared genomes, the highest number of repeats was found in V. fordii with 184, followed by H. brasilliensis with 182. Furthermore, the lowest repeats were found in E. esula, comprising 143 repeats (Figure 2). Among the repeats, the tandem repeats were found highest, followed by the forward and palindromic repeats. The repeats of different sizes were also studied. In E. smithii, the palindromic repeats were found to be 46 among which the 15–29 size repeats were found to be 37, and the 30–44 size repeats, as well as >90, were 4. The forward repeats were 55 in which 15–29 were found to be 49, and 30–44 were found to be 5. Among the 70 tandem repeats, 15–29 were 64, 30–44 were 5, 45–59 was 1. In E. larica, among the 46 palindromic repeats, the 15–29 were 39, 30–44 were 3, 45–59 were 1 and >90 were 3. The forward repeats were 54, among the 15–29 were 44, 30–44 were 7, 45–59 were 1, and >90 were 2. The tandem repeats were analyzed to be 62 in which 15–29 were 56, 30–44 were 3, 60–74 were 1, and >90 were 2 (Figure 2D).
Figure 2.
Analysis of repetitive sequences in E. smithii and E. larica. (A) Total number of repeats present in the genome. (B) Number of palindromic repeats in the genome. (C) Number of forward repeats present in the genome. (D) Number of tandem repeats present in the genome.
2.3. SSRs Polymorphism Analysis
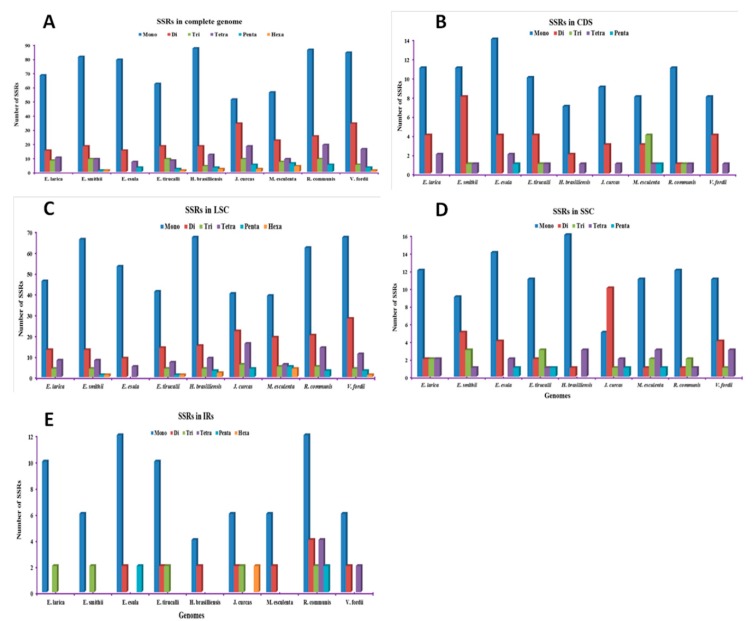
Simple sequence repeats (SSRs) are the microsatellites present in the chloroplast genomes, which play an important role in the cp genome. They are usually varying from one to six base pairs and present in all genome. In our current study, we determined the SSRs in the sequenced and compared cp genomes. Our result for SSRs analysis reveals that there are 101, 119,104, 100, 126, 119, 104, 144, and 143 SSRs found in the E. larica, E. smithii, E. esula, E. tirucalli, H. brasilliensis, J. curcas, M. esculenta, R. communis, and V. fordii genomes, respectively (Figure 3A). The highest number of SSRs was found in R. communis, while the lowest was observed in E. larica. Moreover, the mono nucleotide in E. larica and E. smithii were highest with 68 and 81, while the lowest were tri in E. larica and hexa in E. smithii, which were found and were observed to be absent in the E. larica cp genome (Figure 3B). The SSRs present in the CDS region were 17 and 21, LSC region comprised 71 and 93, SSC region comprised an equal number, which is 18. In In inverted repeat regions, the number of SSRs was 12 and 8 in the E. larica and E. smithii, respectively (Figure 3C–E). In both E. larica and E. smithii, most mononucleotide SSRs were T (61.7%, 46.1%) motifs, with the majority of dinucleotide SSRs being A/T (8, 13) motifs (Supplementary Figure S1).
Figure 3.
Analysis of simple sequence repeats (SSRs) in chloroplast genomes of E. smithii and E. larica. (A) Total number of SSRs present in complete genomes. (B) Total number of SSRs present in the CDS of the genome. (C) Total number of SSRs present in the LSC of genome. (D) Total number of SSRs present in SSC of the genome. (E) Total number of SSRs present in IRs of the genome.
2.4. Compression and Augmentation of IR Region
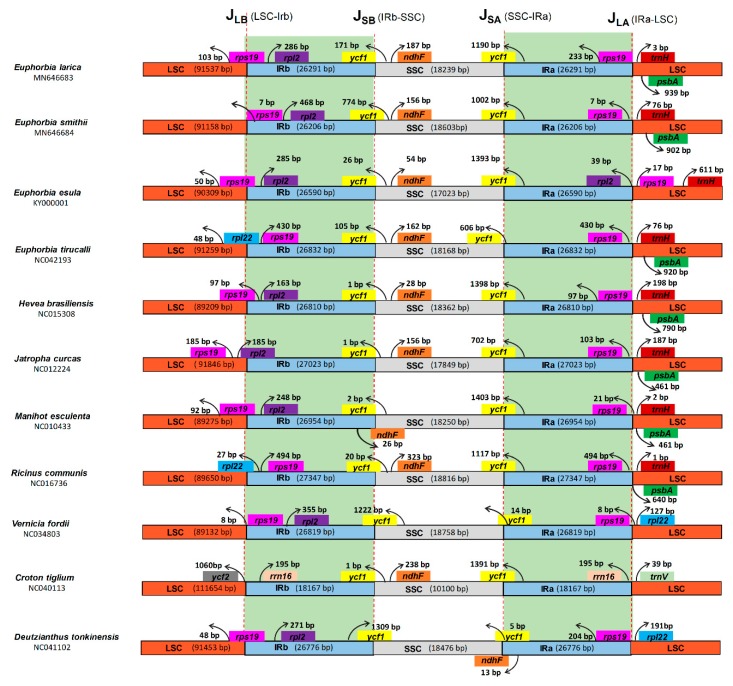
The expansion and contraction of the inverted repeats at the border region in the chloroplast genome is commonly observed and mainly responsible for the size variation in the chloroplast genome. Therefore, for the complete study of inverted repeat regions in the sequenced genomes of E. larica and E. smithii, we compared the IR border regions and the genes present within these junctions with the other nine chloroplasts genomes. Critically analyzing the junctions of E. larica revealed that the length of LSC was found to be 91,537 bp and the rps19 gene was located on the junction of LSC/IRb (JLB). The gene rpl2 was located 286 bp from the JLB in the IRb region. The ycf1 gene was located at JSB and JSA junctions while the ndhF gene was located in the SSC region, 187 bp away from the JSB junction. The JLA junction includes the rps19 gene and the trnH gene located in the IRa and LSC region, respectively. The E. smithii junctions contained the same genes present in the E. larica, with small differences in the location from the junction region like the rps 19 gene that is located in the IRb region 7 bp away from the JLB region. In all the compared genomes, the location of rps19 at the JLB junction shows a similar pattern in E. larica, E. esula, H. brasilliensis, M. esulenta, and D. tonkinesis, while the complete location of rps19 in the IRb region was observed in the E. smithii, E. tirucalli, R. communis, and V. fordii. Surprisingly, the rps19 was found completely in the LSC region in J. curcas. The rps19 gene, like ycf1, is present at two locations in the junction region, while in some genomes, among the compared genome like C. tiglium, it was found completely absent at JLB, while at JLA it was present in the IRa region (Figure 4).
Figure 4.
Distances between adjacent genes and junctions of the small single-copy (SSC), large single-copy (LSC), and two inverted repeat (IR) regions among plastid genomes E. smithii and E. larica and related species within the Euphorbiaceae family. Boxes above and below the primary line indicate the adjacent border genes. The figure is not to scale with regards to sequence length and only shows relative changes at or near the IR/SC borders.
The ycf1 gene was present at both the JSB and JSA junction in all compared genomes. The ndhF gene was found in all compared genomes at the SSC region near the JSB junction, except the few genomes like in M. esulenta and D. tonkinensis. It is present at the JSB while absent in the V. fordii genome. The trnH gene was present in the LSC region near the JLA junction in all genomes, except V. fordii, C. tiglium, and D. tonkinesis, where it was missing and was replaced by rpl22, trnV, and rpl22 in these genomes, respectively. Surprisingly, the rrn16 gene was found in the IRa and IRb regions. The C. tiglium genome was absent in all other sequenced and compared genomes (Figure 4).
2.5. Comparison of the Hotspot Region in the cp Genome
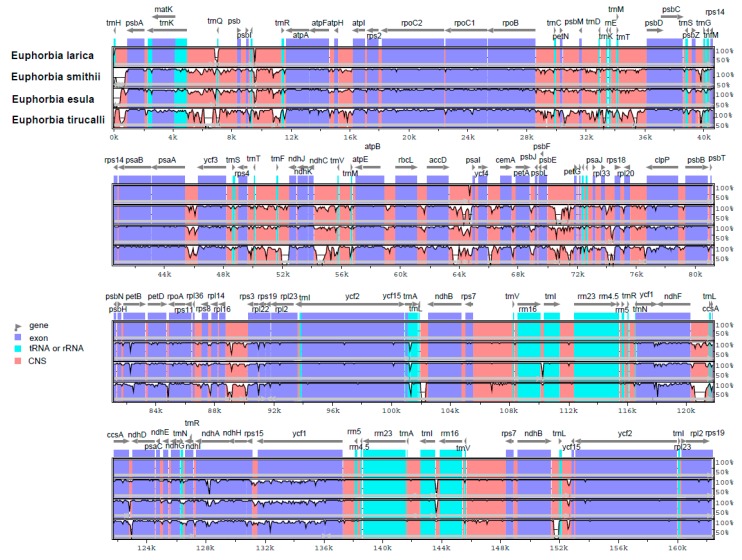
Chloroplast genomes present in most of the higher plants are relatively conserved and stable in terms of their structure and gene content. Despite the conserved structure, some variation in plant groups like genome size, gene content, and genome structure still occur due to the different evolutionary histories and genetic backgrounds. The E. larica cp genome was taken as a reference for detecting a divergence hotspot in E. smithii, E. esula, and E. tirucalli. The divergence in protein-coding genes was also analyzed and 65 genes were studied for the pairwise distance among these genomes. Sequence divergence analysis of the Euphoria species and compared genomes revealed a high conservative degree of the coding region as compared to non-coding regions. Furthermore, it was found that sequence divergence in the single copy region was higher than in inverted repeats regions. Further analysis of genes revealed that some of the divergent regions in these genomes were rpoC1, rpocB, ycf3, clpP, petD, ycf1, and ndhF. These regions were divergent but less divergent than non-coding regions (Figure 5).
Figure 5.
Visual alignment of plastid genomes with the previously reported cp genomes. VISTA-based identity plot showing sequence identities among eight species, using E. larica as a reference.
2.6. Phylogenomic Analysis of E. larica and E. smithii and Its Comparison with Related Species
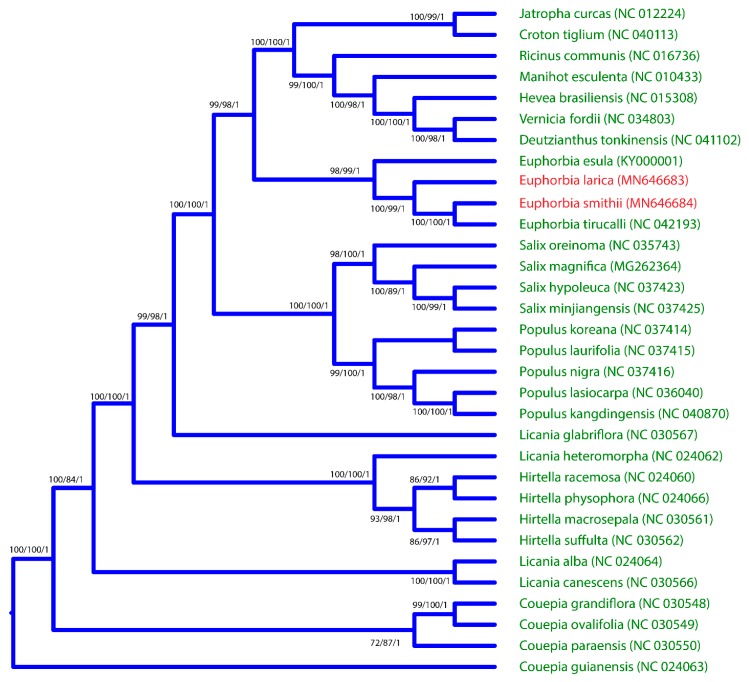
In this study, a dataset of 32 complete chloroplast genomes was used to construct the phylogenetic tree of E. larica and E. smithii. The Couepia paraensis chloroplast genome was used as an out group in this study. The phylogenetic tree was constructed using MP (maximum parsimony), ML (maximum likelihood), and BI (Bayesian interference). The result of the phylogenetic tree based on the complete chloroplast genome shows that E. smithii and E. larica share the same clade, which further makes a sister clade with E. esula with high bootstrap values (Figure 6).
Figure 6.
Phylogenetic trees of E. smithii and E. larica. The entire genome dataset was analyzed using three different methods: Bayesian inference (BI), maximum parsimony (MP), and maximum likelihood (ML). Numbers above the branches represent bootstrap values in the ML and MP, and posterior probabilities in the BI trees. Red color represents the positions of E. smithii and E. larica.
3. Discussion
With the advancement of next-generation sequencing technologies, the number of sequenced genomes has increased rapidly in the NCBI database. The availability of this data provides new insight into the chloroplast genomics, phylogenetic studies, rearrangement of genomes, sequence divergence, simple sequence repeats analysis, and the study of nucleotide substitution in these genomes. Euphorbiaceae is a large family and the number of the sequenced chloroplast genome is very limited [29]. The two sequenced cp genomes are comparatively analyzed with another cp genome to study the various parameters of these genomes. The chloroplast genome structure and gene order of these two chloroplast genomes are highly conserved with no specific genome inversion reported, and the gene order was the same and found consistent with previously reported genomes [30]. In the present study, we compared eleven chloroplast genomes. All of them were assembled into a single chloroplast genome presenting a typical quadripartite structure. Analysis of two sequenced Euphorbia genomes revealed that, like most of the higher angiosperm genomes, they comprised the tetrahedral structure containing two pairs of inverted repeats, one large single-copy region and one small single-copy region [4,31]. There was a 186 bp difference observed between the two sequenced chloroplast genomes, and the size was also comparable with other compared Euphorbia species, as well as the Vernicia fordii chloroplast genome, which is 161,528 bp in length [32], suggesting that chloroplast genomes are conserved. The result was consistent with previously reported studies [30]. The total number of genes presented in the chloroplast genome is divided into three main categories. The first is related to chloroplast gene expression and its self-replication. This includes the majority of rRNA, tRNA, and genes for RNA polymerase synthesis. The second category of genes is related to the vital process of life, i.e., genes responsible for photosynthesis, which includes photosystem I and photosynthesis II. The third category of genes is responsible for other biosynthesis genes and some genes of unknown function, such as matK and ycf1 [33,34], similar to sequenced Euphorbia chloroplast genomes. During evolution, some genomes are liable to gain or lose introns, and this process plays a key role in expression and gene regulation [35]. In the Euphorbia species, there are 12 genes and 6 tRNA which contain introns and were found to be similar in the previously reported V. fordii cp genome belonging to Euphorbiaceae [32]. Some of the genes in the chloroplast genome contain double introns, such as rps12, clpP, and ycf3, and in some genes like rpl2 and rpl16, the second intron is absent, which is consistent with previously reported genomes of Manihot esculenta [35] and Oresitrophe [36]. This phenomenon was absent in H. micrantha cp genomes (EF207446). The GC content of these sequenced genomes was consistent with a previously reported genome from this genus [37,38]. The number of repeats, including forward, tandem, and palindromic repeats, were studied in the chloroplast genomes sequenced and compared, and were found in a larger amount than in the previously reported cp genome of V. fordii (49 repeats) [32]. Among these repeats, tandem repeats were found several times more than palindromic and forward repeats, which are consistent with the Teucrium and Commiphora species [30,31], as well as S. miltiorrhiza [39], as previously reported.
SSRs (simple sequence repeats) are repeats that play an important role in genome stabilization and rearrangement of genome sequences, and these SSRs make the cp genome favorable because of its use as a molecular maker and phylogenetic analysis [40,41]. In our study 101 and 119 total SSRs were found in E. larica and E. smithii respectively, which is higher than the Euphorbiaceae family members [32]. However, it is similar to the previously reported B. sacra cp genome [42]. Among the dinucleotide SSRs, AT was found to be the most abundant in the sequenced and compared cp genomes, similar to the previously reported genomes [30]. Another and important characteristic of the chloroplast genome, which is useful for evolutionary studies, is the location of the boundaries among the four chloroplast regions. Evaluating their contraction and expansion can shed some light on the evolution of some taxa [43]. From our results, we noticed that the length variation in the IR regions created some pseudogenes, like the ycf1Ψ or rps19Ψ. The ycf1 pseudogene is present in all studied species, whereas the rps19 pseudogene is only present in C. icaco, H. racemose (Chrysobalanaceae), V. seoulensis (Violaceae) [44], and M. esculenta (Euphorbiaceae) [35]. Inverted repeats are the most conserved region in the chloroplast genome and the construction and expansion of these IR regions are the common evolutionary events that lead to the differences in the size of chloroplast genomes [45]. In most of the plants, the border and junctions of the quadripartite structure of the genome structure is conserved but some species show inversion at the junction, as previously reported in [46], and loss of genes reported in [47], as well as contraction and expansion, which is a common event observed in the cp genomes of angiosperms [48]. Some angiosperm also show the loss of inverted repeats, such as geranium [49] and fabaceae [50]. Our study analysis of junction regions shows that the rps19 gene is present at the JLB junction in E. larica, while other genes like ndhF also show a pattern that is similar in Violaceae, as previously reported [44]. The rps19 gene present in the IRb region near the JLB junction was found in the present study of the Byrsonima species reported by Alison et al. [51]. Previously, it was identified that the alignment of many genomes contributes and identifies mutational hotspots, which are widely used for interspecies discrimination and species-level phylogenetic studies [52]. The coding region in many previous studies has been proven to play an important role in species-level phylogenetic analyses like some of the genes, such as ycf1 in Anemopaegma [53] and rps16, psaI, psbT, psbH, petB, rpoA, and rps11 in Notopterygium [54], which were more divergent than non-coding regions. However, a number of studies have confirmed that there is more variation in the non-coding region comprising the intergenic spacer regions and introns. For species identification in some previous studies, the clpP, rps16, rpoB-trnC, rbcL-accD, and ccsA-ndhD regions were used as markers [55] and trnH-psbA, trnG-trnM, trnT-trnL, rpl32-trnL, rps15-ycf1, ycf4-cemA, and petD-rpoA were the divergence hotspot regions in Veroniceae and Veronica [48].
In our study, the four Euphorbia species were compared through mVISTA and multiple alignment analyses. It was revealed that some of the regions were found more divergent and consisted of non-coding regions as compared to coding regions. Some regions, like rpoC1, rpocB, ycf3, clpP, petD, ycf1, and ndhF, were larger in number. These results are consistent with previously reported cp genomes [23,28]. Furthermore, we screened the four most mutational hotspots, ndhF, ycf1, ndhA, and rpl32-trnL, which can be used as genetic markers for species delimitation and phylogenetic studies of the genus Euphorbia. However, our study finds that more hotspot regions were present in the SSC region while the IR region was conserved, similar to the previously reported [56]. The phylogenetic position of genus euphorbia and our sequenced species were not identified on the basis of the complete chloroplast genome. Previously, some phylogenetic study was carried out on the basis of ITS regions and the plastid ndhF gene [57]. Based on the previous studies, it was not possible to understand the position of these two sequenced E. smithii and E. larica and the compared genome in this genus. Our study, on basis of complete cp genome sequences, provides a detail of the phylogenetic position of genus Euphorbia species. The current study reported for the first time sequence datasets of the two species, and it might help us to understand the genome architecture, genetic diversity amongst populations, and evolutionary depiction in the genera.
4. Material and Methods
4.1. Chloroplast DNA Extraction and Sequencing
Young fresh healthy green leaves of E. larica and E. smithii were collected from the Nizwa governorate (57°31′59.99″ E) and placed immediately in liquid nitrogen. The contamination-free chloroplast DNA was extracted according to a modified protocol of Shi et al. [58]. An ion torrent sequencing platform was used for the sequencing of these samples using the Ion Torrent S5 sequencer with an ion torrent server (Life Technologies, Carlsbad, CA, USA). Genomic libraries were prepared according to the manufacturer’s instructions (Life Technologies, Carlsbad, CA, USA). Total chloroplast DNA of each sample was sheared enzymatically for 400 bp using the Ion Shear™ Plus Reagents kit, and libraries were prepared using the Ion Xpress™ Plus gDNA Fragment Library kit. Prepared libraries were quantified and qualified on a Qubit 3.0 fluorometer and bioanalyzer (Agilent 2100 Bioanalyzer system, Palo Alto, CA, USA). Libraries preparation was followed by template amplification (Ion OneTouch 2 instrument, Life Technologies, Carlsbad, CA, USA) and enrichment of the amplified template (Ion OneTouch™ ES enrichment system, Life Technologies, Carlsbad, CA, USA) by using Ion 520 and 530 OT2 reagents. The sample was loaded onto the Ion S5 sequencing chip and sequencing was performed according to the protocol of Ion Torrent S5 (Life Technologies, Carlsbad, CA, USA).
4.2. Genome Assembly
A total of 1,018,614 and 1,396,422 raw reads were generated for E. larica and E. smithii, respectively. The obtained reads of the genomes were mapped to the selected reference genome of E. esula using Bowtie2 (v.2.2.3) [59] in Geneious Pro (v.10.2.3) [60] software. The mean coverage of the assemblies for E. larica and E. smithii were 186X and 256X, respectively. The IR junction regions were identified using the already published genome of E. esula, and an iteration method using the MITObim (v.1.8) software [61] was utilized to adjust the sequence length. After sequencing, FastQC (v0.11.6) [61] was performed to check the read quality. To reduce biases in the analysis, an in-house script was used to filter out reads if less than 90% of the bases that made up the read were below Q20. Trimmomatic (v0.36) [62] was used to remove adapter sequences. Only high-quality reads were mapped using Bowtie2 in Geneious Pro (v.10.2.3) [60] as previously performed in cp genome of Vachellia nilotica [63].
4.3. Genome Annotation
Chloroplast genomes were annotated by using Dual Organellar Genome Annotator (DOGMA) [64], and BLASTX and BLASTN were used to identify the positions of ribosomal RNAs, transfer RNAs, and coding genes. The tRNAscan-SE version 1.21 [65] software was used to annotate tRNA genes. Additionally, for manual adjustment, Geneious and tRNAscan-SE [65] were used to compare it with previously reported genomes. Correspondingly, the start and stop codon and intron boundaries were also manually adjusted compared with a pre-published E. esula cp genome. In addition, the structural features of both Euphorbia species cp genomes were illustrated using OGDRAW [66]. Correspondingly, MEGA6 software [67] was used to determine the relative synonymous codon usage and deviations in synonymous codon usage by avoiding the influence of amino acid composition. The divergence of these three Euphorbia species taxa genomes with other related species (Figure 5) was determined by using mVISTA [68] in Shuffle-LAGAN mode, using E. esula as a reference genome.
4.4. Repeat Identification
REPuter software [69] was used for the identification of palindromic, forward, and tandem repeats present in the genome. The criterion was a minimum of >15 base pairs with a sequence identity of 90%. Furthermore, SSRs were determined using Phobos version 3.3.12 [70] with the search parameters set for mononucleotide repeats at ≥10 repeat units, for dinucleotide repeats at ≥ 8repeat units, for tri nucleotide and tetra nucleotide repeats at ≥4 repeat units, and for penta nucleotide and hexa nucleotide repeats at ≥3 repeat units. Tandem Repeats Finder version 4.07 b [71] with default settings was used to determine tandem repeats.
4.5. Sequence Divergence and Phylogenetic Analysis
The average pairwise sequence divergence of the complete cp genomes of Euphorbia species with related species was determined. Comparative sequence analysis after comparing gene order and multiple sequence alignment was used to identify missing and ambiguous gene annotations. MAFFT version 7.222 [72] with default parameters was used for the alignment of complete genomes, and pairwise sequence divergence was calculated by selected Kimura’s two-parameter (K2P) model [73]. To resolve the phylogenetic position of E. larica and E. smithii within the Euphorbiaceae family, cp genomes were downloaded from the NCBI database. Alignment of the complete cp genomes was constructed on the basis of conserved gene order and structure of the cp genome, and three different methods were applied to infer phylogenetic analysis: Bayesian inference (BI), implemented using Mr Bayes 3.1.2 [74,75]; maximum parsimony (MP), implemented using PAUP 4.0 [76]; and both maximum likelihood (ML), implemented using MEGA 6 [60], employing previously described settings [77,78]. For ML analysis, parameters were adjusted with a BIONJ tree with 1000 bootstrap replicates using the Kimura 2-parameter model with gamma-distributed rate heterogeneity and invariant sites. A heuristic search for MP analysis was run with 1000 random addition sequence replicates with the tree-bisection-reconnection (TBR) branch-swapping tree search criterion. The best substitution model, GTR + G model, was used according to the Akaike information criterion (AIC) by jModelTest version 2102 for Bayesian posterior probabilities (PP) in the BI analyses. The Markov Chain Monte Carlo (MCMC) method was run with four incrementally heated chains for 1,000,000 generations, starting from random trees, and sampling one out of every 100 generations. The first 25% of trees were discarded as burn-in to estimate the value of posterior probabilities.
Acknowledgments
We are thankful to Oman Animal and Plant Genetic Resources Center (OAPGRC) for providing instrumental support (Ion Torrent S5 sequencer).
Supplementary Materials
The following are available online at https://www.mdpi.com/2223-7747/9/2/199/s1, Figure S1: Frequency of identified SSR motifs in different repeat class types, Table S1: Genes in the sequenced E. smithii and E. larica chloroplast genomes.
Author Contributions
A.L.K., S.A., and A.K. conceived and designed the experiments. A.K. and S.A. analyzed the sequence data and drafted the manuscript. A.K. participated in data analysis and manuscript writing. T.S., A.L.K., A.A.-H., and A.A.-R. revised the manuscript. All authors have read and agreed to the published version of the manuscript.
Conflicts of Interest
All authors declare that they have no competing interests.
References
- 1.Bauer J., Chen K., Hiltbunner A., Wehrli E., Eugster M., Schnell D., Kessler F. The major protein import receptor of plastids is essential for chloroplast biogenesis. Nature. 2000;403:203. doi: 10.1038/35003214. [DOI] [PubMed] [Google Scholar]
- 2.Sugiura M. The chloroplast genome. Essays Biochem. 1995;30:49–57. [PubMed] [Google Scholar]
- 3.Neuhaus H., Emes M. Nonphotosynthetic metabolism in plastids. Annu. Rev. Plant Biol. 2000;51:111–140. doi: 10.1146/annurev.arplant.51.1.111. [DOI] [PubMed] [Google Scholar]
- 4.Daniell H., Lin C.-S., Yu M., Chang W.-J. Chloroplast genomes: Diversity, evolution, and applications in genetic engineering. Genome Biol. 2016;17:134. doi: 10.1186/s13059-016-1004-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clifton S.W., Minx P., Fauron C.M.-R., Gibson M., Allen J.O., Sun H., Thompson M., Barbazuk W.B., Kanuganti S., Tayloe C. Sequence and comparative analysis of the maize NB mitochondrial genome. Plant Physiol. 2004;136:3486–3503. doi: 10.1104/pp.104.044602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dumolin S., Demesure B., Petit R. Inheritance of chloroplast and mitochondrial genomes in pedunculate oak investigated with an efficient PCR method. Theor. Appl. Genet. 1995;91:1253–1256. doi: 10.1007/BF00220937. [DOI] [PubMed] [Google Scholar]
- 7.Schmidt L., Fischer M., Oja T. Two closely related species differ in their regional genetic differentiation despite admixing. AoB Plants. 2018;10:ply007. doi: 10.1093/aobpla/ply007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Horn J.W., van Ee B.W., Morawetz J.J., Riina R., Steinmann V.W., Berry P.E., Wurdack K.J. Phylogenetics and the evolution of major structural characters in the giant genus Euphorbia, L.(Euphorbiaceae) Mol. Phylogenet. Evol. 2012;63:305–326. doi: 10.1016/j.ympev.2011.12.022. [DOI] [PubMed] [Google Scholar]
- 9.Jassbi A.R. Chemistry and biological activity of secondary metabolites in Euphorbia from Iran. Phytochemistry. 2006;67:1977–1984. doi: 10.1016/j.phytochem.2006.06.030. [DOI] [PubMed] [Google Scholar]
- 10.Zimmermann N., Ritz C.M., Hellwig F. Further support for the phylogenetic relationships within Euphorbia L. (Euphorbiaceae) from nrITS and trnL–trnF IGS sequence data. Plant Syst. Evol. 2010;286:39–58. [Google Scholar]
- 11.Yang Y., Berry P.E. Phylogenetics of the Chamaesyce clade (Euphorbia, Euphorbiaceae): Reticulate evolution and long-distance dispersal in a prominent C4 lineage. Am. J. Bot. 2011;98:1486–1503. doi: 10.3732/ajb.1000496. [DOI] [PubMed] [Google Scholar]
- 12.Genc I., Kültür Ş. Euphorbia akmanii (Euphorbiaceae), a new species from Turkey. Phytotaxa. 2016;265:112–120. doi: 10.11646/phytotaxa.265.2.2. [DOI] [Google Scholar]
- 13.Ernst M., Nothias L.-F., van der Hooft J.J., Silva R.R., Saslis-Lagoudakis C.H., Grace O.M., Martinez-Swatson K., Hassemer G., Funez L.A., Simonsen H.T. Assessing specialized metabolite diversity in the cosmopolitan plant genus Euphorbia, L. Front. Plant Sci. 2019;10:846. doi: 10.3389/fpls.2019.00846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Singla A., Kamla P. Phytoconstituents of Euphorbia species. Fitoterapia. 1990;41:483–516. [Google Scholar]
- 15.Abdelgaleil S.A., Kassem S.M., Doe M., Baba M., Nakatani M. Diterpenoids from Euphorbia paralias. Phytochemistry. 2001;58:1135–1139. doi: 10.1016/S0031-9422(01)00393-4. [DOI] [PubMed] [Google Scholar]
- 16.Ravikanth V., Reddy V.N., Rao T.P., Diwan P., Ramakrishna S., Venkateswarlu Y. Macrocyclic diterpenes from Euphorbia nivulia. Phytochemistry. 2002;59:331–335. doi: 10.1016/S0031-9422(01)00461-7. [DOI] [PubMed] [Google Scholar]
- 17.Ernst M., Grace O.M., Saslis-Lagoudakis C.H., Nilsson N., Simonsen H.T., Rønsted N. Global medicinal uses of Euphorbia, L.(Euphorbiaceae) J. Ethnopharmacol. 2015;176:90–101. doi: 10.1016/j.jep.2015.10.025. [DOI] [PubMed] [Google Scholar]
- 18.Shaaban M., Ali M., Tala M.F., Hamed A., Hassan A.Z. Ecological and Phytochemical Studies on Euphorbia retusa (Forssk.) from Egyptian Habitat. J. Anal. Methods Chem. 2018;2018:9143683. doi: 10.1155/2018/9143683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rahman A.H.M.M., Akter M. Taxonomy and Medicinal Uses of Euphorbiaceae (Spurge) Family of Rajshahi, Bangladesh. Res. Plant Sci. 2013;1:74–80. [Google Scholar]
- 20.Kumar S., Malhotra R., Kumar D. Euphorbia hirta: Its chemistry, traditional and medicinal uses, and pharmacological activities. Pharmacogn. Rev. 2010;4:58. doi: 10.4103/0973-7847.65327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Al-Mahmooli I., Al-Bahri Y., Al-Sadi A., Deadman M. First report of Euphorbia larica dieback caused by Fusarium brachygibbosum in Oman. Plant Dis. 2013;97:687. doi: 10.1094/PDIS-09-12-0828-PDN. [DOI] [PubMed] [Google Scholar]
- 22.Miller A.G., Morris M. Plants of Dhofar: The Southern Region of Oman, Traditional, Economic and Medicinal Uses. Oman: Office of the Adviser for Conservation of the Environment; Diwan of Royal Court; Sultanate, Oman: 1988. [Google Scholar]
- 23.Noori M., Chehreghani A., Kaveh M. Flavonoids of 17 species of Euphorbia (Euphorbiaceae) in Iran. Toxicol. Environ. Chem. 2009;91:631–641. doi: 10.1080/02772240802389936. [DOI] [Google Scholar]
- 24.Patzelt A. Oman Plant: Red Data Book. Oman Botanic Garden; Muscat, Oman: 2015. [Google Scholar]
- 25.Pickering H., Patzelt A. Field Guide to the Wild Plants of Oman. Royal Botanic Gardens; Muscat, Oman: 2008. [Google Scholar]
- 26.Trapnell D.W., Hamrick J., Negrón-Ortiz V. Genetic diversity within a threatened, endemic North American species, Euphorbia telephioides (Euphorbiaceae) Conserv. Genet. 2012;13:743–751. doi: 10.1007/s10592-012-0323-4. [DOI] [Google Scholar]
- 27.Trejo L., Briones-Dumas E., Gómez-Bermejo R., Olson M.E. Molecular evidence for repeated recruitment of wild Christmas poinsettia (Euphorbia pulcherrima) into traditional horticulture in Mexico. Genet. Resour. Crop Evol. 2019;66:481–490. doi: 10.1007/s10722-018-0727-1. [DOI] [Google Scholar]
- 28.Dorsey B.L., Haevermans T., Aubriot X., Morawetz J.J., Riina R., Steinmann V.W., Berry P.E. Phylogenetics, morphological evolution, and classification of Euphorbia subgenus Euphorbia. Taxon. 2013;62:291–315. doi: 10.12705/622.1. [DOI] [Google Scholar]
- 29.Asif M.H., Mantri S.S., Sharma A., Srivastava A., Trivedi I., Gupta P., Mohanty C.S., Sawant S.V., Tuli R. Complete sequence and organisation of the Jatropha curcas (Euphorbiaceae) chloroplast genome. Tree Genet. Genomes. 2010;6:941–952. doi: 10.1007/s11295-010-0303-0. [DOI] [Google Scholar]
- 30.Khan A., Asaf S., Khan A.L., Al-Harrasi A., Al-Sudairy O., AbdulKareem N.M., Khan A., Shehzad T., Alsaady N., Al-Lawati A., et al. First complete chloroplast genomics and comparative phylogenetic analysis of Commiphora gileadensis and C. foliacea: Myrrh producing trees. PLoS ONE. 2019;14:e0208511. doi: 10.1371/journal.pone.0208511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khan A., Asaf S., Khan A.L., Khan A., Al-Harrasi A., Al-Sudairy O., AbdulKareem N.M., Al-Saady N., Al-Rawahi A. Complete chloroplast genomes of medicinally important Teucrium species and comparative analyses with related species from Lamiaceae. PeerJ. 2019;7:e7260. doi: 10.7717/peerj.7260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Z., Long H., Zhang L., Liu Z., Cao H., Shi M., Tan X. The complete chloroplast genome sequence of tung tree (Vernicia fordii): Organization and phylogenetic relationships with other angiosperms. Sci. Rep. 2017;7:1869. doi: 10.1038/s41598-017-02076-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu Y., Zhou H. Research progress of sugarcane chloroplast genome. Agric. Sci. Technol. 2013;14:1693. [Google Scholar]
- 34.Asaf S., Waqas M., Khan A.L., Khan M.A., Kang S.-M., Imran Q.M., Shahzad R., Bilal S., Yun B.-W., Lee I.-J. The complete chloroplast genome of wild rice (Oryza minuta) and its comparison to related species. Front. Plant Sci. 2017;8:304. doi: 10.3389/fpls.2017.00304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Daniell H., Wurdack K.J., Kanagaraj A., Lee S.-B., Saski C., Jansen R.K. The complete nucleotide sequence of the cassava (Manihot esculenta) chloroplast genome and the evolution of atpF in Malpighiales: RNA editing and multiple losses of a group II intron. Theor. Appl. Genet. 2008;116:723. doi: 10.1007/s00122-007-0706-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu L., Wang Y., He P., Li P., Lee J., Soltis D.E., Fu C. Chloroplast genome analyses and genomic resource development for epilithic sister genera Oresitrophe and Mukdenia (Saxifragaceae), using genome skimming data. BMC Genom. 2018;19:235. doi: 10.1186/s12864-018-4633-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Q., Qu Z., Tian X. Complete chloroplast genome of an endangered oil tree, Deutzianthus tonkinensis (Euphorbiaceae) Mitochondrial DNA Part B. 2019;4:299–300. doi: 10.1080/23802359.2018.1542991. [DOI] [Google Scholar]
- 38.Zhang J.-F., Zhao L., Duan N., Guo H.-X., Wang C.-Y., Liu B.-B. Complete chloroplast genome of Euphorbia hainanensis (Euphorbiaceae), a rare cliff top boskage endemic to China. Mitochondrial DNA Part B. 2019;4:1325–1326. doi: 10.1080/23802359.2019.1596761. [DOI] [Google Scholar]
- 39.Qian J., Song J., Gao H., Zhu Y., Xu J., Pang X., Yao H., Sun C., Li X.e., Li C. The complete chloroplast genome sequence of the medicinal plant Salvia miltiorrhiza. PLoS ONE. 2013;8:e57607. doi: 10.1371/journal.pone.0057607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang J.-B., Tang M., Li H.-T., Zhang Z.-R., Li D.-Z. Complete chloroplast genome of the genus Cymbidium: Lights into the species identification, phylogenetic implications and population genetic analyses. BMC Evol. Biol. 2013;13:84. doi: 10.1186/1471-2148-13-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Do Nascimento Vieira L., Faoro H., Rogalski M., de Freitas Fraga H.P., Cardoso R.L.A., de Souza E.M., de Oliveira Pedrosa F., Nodari R.O., Guerra M.P. The complete chloroplast genome sequence of Podocarpus lambertii: Genome structure, evolutionary aspects, gene content and SSR detection. PLoS ONE. 2014;9:e90618. doi: 10.1371/journal.pone.0090618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Khan A.L., Al-Harrasi A., Asaf S., Park C.E., Park G.-S., Khan A.R., Lee I.-J., Al-Rawahi A., Shin J.-H. The first chloroplast genome sequence of Boswellia sacra, a resin-producing plant in Oman. PLoS ONE. 2017;12:e0169794. doi: 10.1371/journal.pone.0169794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nazareno A.G., Carlsen M., Lohmann L.G. Complete chloroplast genome of Tanaecium tetragonolobum: The first Bignoniaceae plastome. PLoS ONE. 2015;10:e0129930. doi: 10.1371/journal.pone.0129930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cheon K.-S., Kim K.-A., Kwak M., Lee B., Yoo K.-O. The complete chloroplast genome sequences of four Viola species (Violaceae) and comparative analyses with its congeneric species. PLoS ONE. 2019;14:e0214162. doi: 10.1371/journal.pone.0214162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shen X., Wu M., Liao B., Liu Z., Bai R., Xiao S., Li X., Zhang B., Xu J., Chen S. Complete chloroplast genome sequence and phylogenetic analysis of the medicinal plant Artemisia annua. Molecules. 2017;22:1330. doi: 10.3390/molecules22081330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cho K.-S., Yun B.-K., Yoon Y.-H., Hong S.-Y., Mekapogu M., Kim K.-H., Yang T.-J. Complete chloroplast genome sequence of tartary buckwheat (Fagopyrum tataricum) and comparative analysis with common buckwheat (F. esculentum) PLoS ONE. 2015;10:e0125332. doi: 10.1371/journal.pone.0125332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fu P.-C., Zhang Y.-Z., Geng H.-M., Chen S.-L. The complete chloroplast genome sequence of Gentiana lawrencei var. farreri (Gentianaceae) and comparative analysis with its congeneric species. PeerJ. 2016;4:e2540. doi: 10.7717/peerj.2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Choi K.S., Chung M.G., Park S. The complete chloroplast genome sequences of three Veroniceae species (Plantaginaceae): Comparative analysis and highly divergent regions. Front. Plant Sci. 2016;7:355. doi: 10.3389/fpls.2016.00355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Blazier J.C., Jansen R.K., Mower J.P., Govindu M., Zhang J., Weng M.-L., Ruhlman T.A. Variable presence of the inverted repeat and plastome stability in Erodium. Ann. Bot. 2016;117:1209–1220. doi: 10.1093/aob/mcw065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang Y.-H., Wicke S., Wang H., Jin J.-J., Chen S.-Y., Zhang S.-D., Li D.-Z., Yi T.-S. Plastid genome evolution in the early-diverging legume subfamily Cercidoideae (Fabaceae) Front. Plant Sci. 2018;9:138. doi: 10.3389/fpls.2018.00138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Menezes A.P.A., Resende-Moreira L.C., Buzatti R.S.O., Nazareno A.G., Carlsen M., Lobo F.P., Kalapothakis E., Lovato M.B. Chloroplast genomes of Byrsonima species (Malpighiaceae): Comparative analysis and screening of high divergence sequences. Sci. Rep. 2018;8:2210. doi: 10.1038/s41598-018-20189-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ahmed I., Matthews P.J., Biggs P.J., Naeem M., McLenachan P.A., Lockhart P.J. Identification of chloroplast genome loci suitable for high-resolution phylogeographic studies of C olocasia esculenta (L.) S chott (A raceae) and closely related taxa. Mol. Ecol. Resour. 2013;13:929–937. doi: 10.1111/1755-0998.12128. [DOI] [PubMed] [Google Scholar]
- 53.Firetti F., Zuntini A.R., Gaiarsa J.W., Oliveira R.S., Lohmann L.G., Van Sluys M.A. Complete chloroplast genome sequences contribute to plant species delimitation: A case study of the Anemopaegma species complex. Am. J. Bot. 2017;104:1493–1509. doi: 10.3732/ajb.1700302. [DOI] [PubMed] [Google Scholar]
- 54.Yang J., Yue M., Niu C., Ma X.-F., Li Z.-H. Comparative analysis of the complete chloroplast genome of four endangered herbals of Notopterygium. Genes. 2017;8:124. doi: 10.3390/genes8040124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shaw J., Shafer H.L., Leonard O.R., Kovach M.J., Schorr M., Morris A.B. Chloroplast DNA sequence utility for the lowest phylogenetic and phylogeographic inferences in angiosperms: The tortoise and the hare IV. Am. J. Bot. 2014;101:1987–2004. doi: 10.3732/ajb.1400398. [DOI] [PubMed] [Google Scholar]
- 56.Wu C.-S., Chaw S.-M. Evolutionary stasis in cycad plastomes and the first case of plastome GC-biased gene conversion. Genome Biol. Evol. 2015;7:2000–2009. doi: 10.1093/gbe/evv125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Peirson J.A., Bruyns P.V., Riina R., Morawetz J.J., Berry P.E. A molecular phylogeny and classification of the largely succulent and mainly African Euphorbia subg. Athymalus (Euphorbiaceae) Taxon. 2013;62:1178–1199. doi: 10.12705/626.12. [DOI] [Google Scholar]
- 58.Shi C., Hu N., Huang H., Gao J., Zhao Y.-J., Gao L.-Z. An improved chloroplast DNA extraction procedure for whole plastid genome sequencing. PLoS ONE. 2012;7:e31468. doi: 10.1371/journal.pone.0031468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kearse M., Moir R., Wilson A., Stones-Havas S., Cheung M., Sturrock S., Buxton S., Cooper A., Markowitz S., Duran C. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hahn C., Bachmann L., Chevreux B. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads—A baiting and iterative mapping approach. Nucleic Acids Res. 2013;41:e129. doi: 10.1093/nar/gkt371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bolger A.M., Lohse M., Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Asaf S., Khan A., Khan A.L., Al-Harrasi A., Al-Rawahi A. Complete Chloroplast Genomes of Vachellia nilotica and Senegalia senegal: Comparative Genomics and Phylogenomic Placement in a New Generic System. PloS oNE. 2019;14 doi: 10.1371/journal.pone.0225469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wyman S.K., Jansen R.K., Boore J.L. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 2004;20:3252–3255. doi: 10.1093/bioinformatics/bth352. [DOI] [PubMed] [Google Scholar]
- 65.Schattner P., Brooks A.N., Lowe T.M. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 2005;33:W686–W689. doi: 10.1093/nar/gki366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lohse M., Drechsel O., Bock R. OrganellarGenomeDRAW (OGDRAW): A tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr. Genet. 2007;52:267–274. doi: 10.1007/s00294-007-0161-y. [DOI] [PubMed] [Google Scholar]
- 67.Kumar S., Nei M., Dudley J., Tamura K. MEGA: A biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief. Bioinform. 2008;9:299–306. doi: 10.1093/bib/bbn017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Frazer K.A., Pachter L., Poliakov A., Rubin E.M., Dubchak I. VISTA: Computational tools for comparative genomics. Nucleic Acids Res. 2004;32:W273–W279. doi: 10.1093/nar/gkh458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kurtz S., Choudhuri J.V., Ohlebusch E., Schleiermacher C., Stoye J., Giegerich R. REPuter: The manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001;29:4633–4642. doi: 10.1093/nar/29.22.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kraemer L., Beszteri B., Gäbler-Schwarz S., Held C., Leese F., Mayer C., Pöhlmann K., Frickenhaus S. S TAMP: Extensions to the S TADEN sequence analysis package for high throughput interactive microsatellite marker design. BMC Bioinform. 2009;10:41. doi: 10.1186/1471-2105-10-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Benson G. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 1999;27:573. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 74.Librado P., Rozas J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- 75.Ronquist F., Huelsenbeck J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 76.Swofford D. PAUP-a computer-program for phylogenetic inference using maximum parsimony. J. Gen. Physiol. 1993;102:A9. [Google Scholar]
- 77.Wu Z., Tembrock L.R., Ge S. Are differences in genomic data sets due to true biological variants or errors in genome assembly: An example from two chloroplast genomes. PLoS ONE. 2015;10:e0118019. doi: 10.1371/journal.pone.0118019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Asaf S., Khan A.L., Khan A.R., Waqas M., Kang S.-M., Khan M.A., Lee S.-M., Lee I.-J. Complete chloroplast genome of Nicotiana otophora and its comparison with related species. Front. Plant Sci. 2016;7:843. doi: 10.3389/fpls.2016.00843. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.