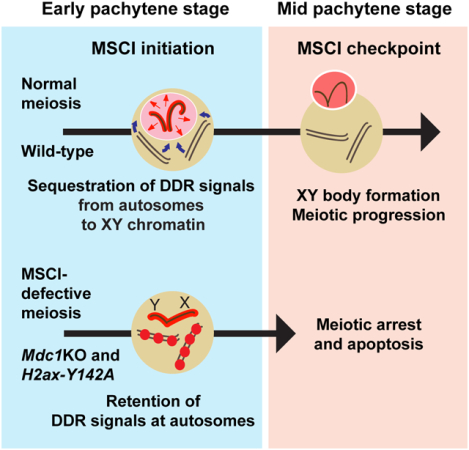
Summary
Meiotic sex chromosome inactivation (MSCI) is an essential event in the mammalian male germline. MSCI is directed by a DNA damage response (DDR) pathway centered on the phosphorylation of histone variant H2AX at serine 139 (termed γH2AX). The failure to initiate MSCI is linked to complete meiotic arrest and elimination of germ cells; however, the mechanisms underlying this arrest and elimination remain unknown. To address this question, we established a new separation-of-function mouse model for H2ax that shows specific and complete defects in MSCI. The genetic change is a point mutation in which another H2AX amino acid residue important in the DDR, tyrosine 142 (Y142), is converted to alanine (H2ax-Y142A). In H2ax-Y142A meiosis, the establishment of DDR signals on the chromosome-wide domain of the sex chromosomes is impaired. The initiation of MSCI is required for stage progression, which enables crossover formation, suggesting that the establishment of MSCI permits the timely progression of male meiosis. Our results further suggest that normal meiotic progression requires the removal of ATR-mediated DDR signaling from autosomes. We propose a novel biological function for MSCI: The initiation of MSCI sequesters DDR factors from autosomes to the sex chromosomes at the onset of the pachytene stage, and the subsequent formation of an isolated XY nuclear compartment—the XY body—sequesters DDR factors to permit meiotic progression from the mid pachytene stage onward.
Keywords: Meiosis, DNA damage signaling, checkpoint, germline, spermatogenesis
Graphical Abstract
eTOC Blurb
Abe et al. demonstrate that tyrosine 142 of histone variant H2AX is required for the initiation of meiotic sex chromosome inactivation (MSCI). Based on new genetic evidence, the study proposes a novel biological function for MSCI: MSCI sequesters DNA damage signaling from autosomes to permit timely progression of male meiosis.
Introduction
Meiosis is a hallmark event in germline development, when paternal and maternal chromosomes undergo synapsis and a reshuffling of the genome prior to producing haploid gametes. During meiosis, the fidelity of meiotic recombination and chromosome synapsis is strictly monitored by checkpoint mechanisms. In coordinating these and other critical events in meiosis, checkpoint mechanisms facilitate timely progression of germ cells through meiosis. Importantly, evolutionarily conserved proteins in DNA damage response (DDR) pathways are implicated in meiotic checkpoint mechanisms of a variety of organisms, from yeast to worms to mammals [1]. Yet despite our understanding of meiotic checkpoints in yeast and worms, their molecular operation remains largely unknown in mammals [2].
In mammalian male meiosis, the X and Y chromosomes are subjected to meiotic sex chromosome inactivation (MSCI) [3, 4]. MSCI is an essential process in the male germline, as failure to initiate MSCI is linked to the complete arrest and timely elimination of male germ cells in the mid pachytene stage of meiotic prophase I [5, 6]. MSCI is a sex chromosome-specific manifestation of a general mechanism for transcriptional silencing in meiosis termed meiotic silencing of unsynapsed chromatin, which operates as a surveillance mechanism for chromosome asynapsis [7–10]. Mechanistically, the initiation of meiotic silencing is directed by a DDR pathway centered on the kinase Ataxia telangiectasia and Rad3 related (ATR) and its phosphorylation of histone H2AX at serine 139 (termed γH2AX) [3, 5, 6, 11–15]. In response to meiotic chromosome asynapsis, a large γH2AX “domain” forms through signal amplification of ATR-mediated γH2AX—first from the unsynapsed axes, then to their protruding loops of chromatin; the signal amplification is directed by Mediator of DNA damage checkpoint protein 1 (MDCI), a γH2AX-binding protein [3, 6]. A major question remains as to how the failure to initiate MSCI is related to complete meiotic arrest and the timely elimination of spermatocytes.
To understand the molecular events that occur in response to MSCI defects, we generated and studied a new separation-of-function mouse model for H2ax. This model contains a point mutation in which another H2AX amino acid residue important in the DDR, tyrosine 142 (Y142) [16, 17], is converted to alanine (H2ax-Y142A). In somatic cells, Y142 is constitutively phosphorylated under physiological conditions; Y142 becomes dephosphorylated upon DNA damage, enabling interactions between γH2AX and other DDR factors, including MDC1, which promote DNA repair [17]. In meiosis, the dephosphorylation of Y142 takes place on the sex chromosomes at the onset of MSCI [18]. Here, we find that H2ax-Y142A mice show specific and complete defects in MSCI, supporting the notion that a common DDR pathway operates in both the somatic DDR and MSCI [3].
In this study, we sought to define the common molecular events that occur in response to defective MSCI. We analyzed the H2ax-Y142A mouse model and an Mdc1 knockout (Mdc1KO) mouse model, both of which exhibit complete impairment of MSCI [6]. Our results suggest that the initiation of MSCI sequesters DDR signaling from autosomes to the sex chromosomes and that the establishment of MSCI permits the timely progression of male germ cells through meiotic prophase I. We propose a novel biological function for MSCI: The initiation of MSCI sequesters DDR factors from autosomes at the onset of the pachytene stage, and the subsequent formation of a distinctly compartmentalized XY body sequesters DDR factors to enable meiotic progression from the mid pachytene stage onward. This model postulates a new mechanism for the meiotic checkpoint: An ATR-mediated DDR pathway that operates in the checkpoint mechanism of the somatic cell cycle has direct regulatory roles in the MSCI checkpoint, which operates to induce cell death if DDR signals remain on autosomes.
Results
An H2AX Y142A point mutation disrupts spermatogenesis
We used CRISPR/Cas9 genome-editing technology to generate a knock-in mouse line harboring a point mutation that changes tyrosine 142 of H2AX into an alanine residue (H2ax-Y142A: Figure 1A). Homozygous males for H2ax-Y142A (hereafter referred to as “H2ax-Y142A”) were infertile, had reduced body weight (Figure 1B), and had significantly smaller testes in comparison to wild-type littermate controls (Figure 1C). We confirmed the abrogation of phosphorylated Y142 (pY142) of H2AX in H2ax-Y142A testes through two methods: western blotting using wild-type and H2ax-Y142A testis lysates (Figure 1D), and immunofluorescence of wild-type and H2ax-Y142A spermatocyte nuclei chromosome spreads (Figure 1E). In wild-type pachytene spermatocytes, pY142 signals were detected throughout nuclei except at XY bodies [18]; pY142 signals were not detected in H2ax-Y142A spermatocytes (Figure 1E). These results confirmed the successful establishment of an H2ax-Y142A mouse line.
Figure 1. H2AX-Y142 is an essential amino residue for completing spermatogenesis.
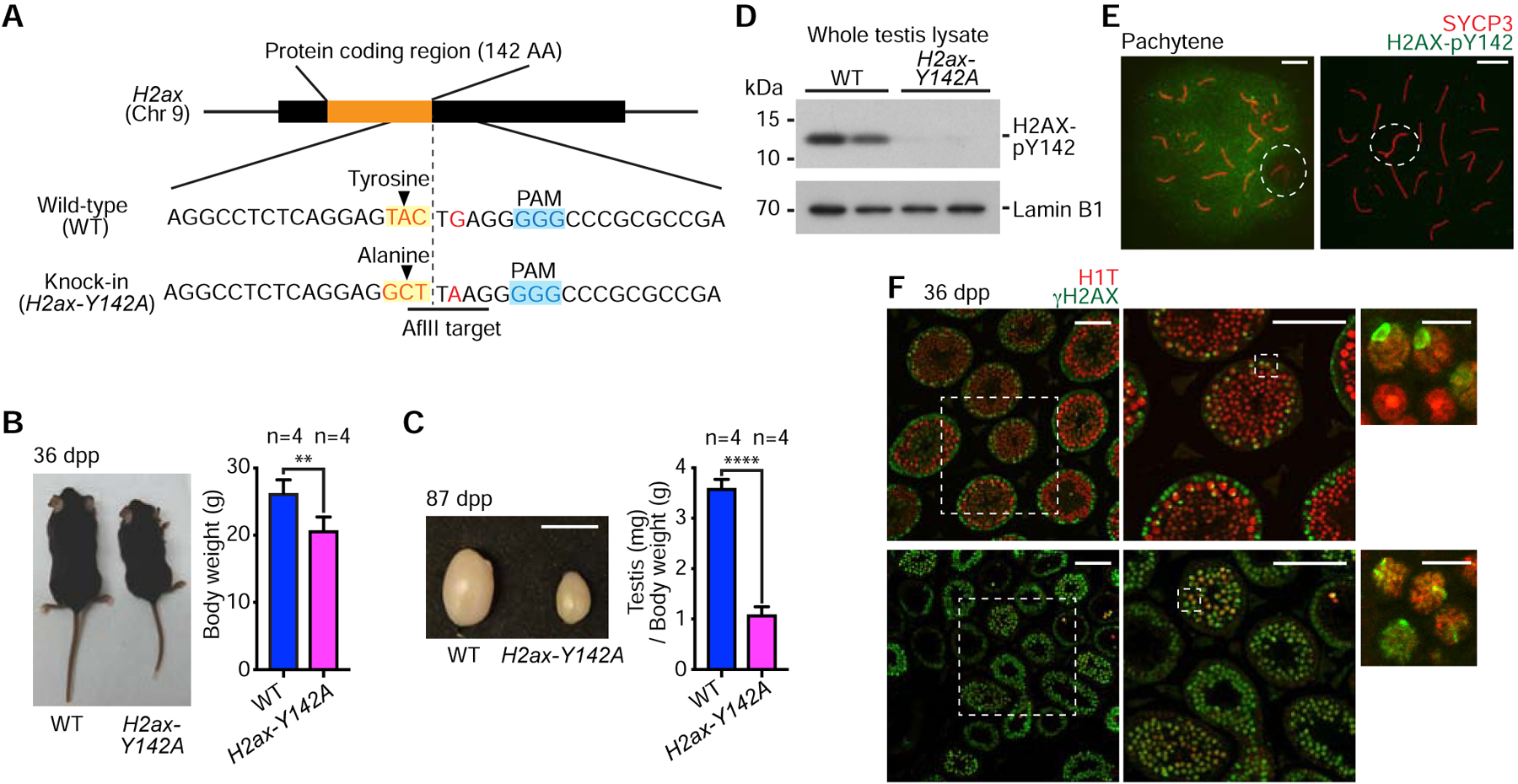
(A) Schematic: Induction of a point mutation and resulting sequence alteration. The introduction of an AflII target site was used to screen mutant mice.
(B) Wild-type (WT) and H2ax-Y142A mice at 36 days post-partum (dpp). Body weights are shown as mean ± s.e.m. from 4 independent pairs of WT and H2ax-Y142A mice. ** p < 0.01, unpaired t test.
(C) Testes from WT and H2ax-Y142A mice at 87 dpp. Scale bar: 1 cm. Ratios of testis weight (mg) to body weight (g) shown as mean ± s.e.m. for 4 independent pairs of WT and H2ax-H142A mice. **** p < 0.0001, unpaired t test.
(D) Western blot of WT and H2ax-Y142A lysates from whole mouse testes with anti-H2AX-pY142 antibody. 20 μg protein samples were loaded in each lane. Two independent samples for WT and H2ax-Y142A are shown. Loading control: Lamin B1.
(E) Chromosome spreads of WT and H2ax-Y142A pachytene spermatocytes immunostained with antibodies raised against SYCP3 and H2AX-pY142. Dashed circles indicate the sex chromosomes. Scale bars: 10 μm.
(F) Testis sections from WT and H2ax-Y142A mice immunostained with antibodies raised against H1T and γH2AX. Dashed squares are magnified in panels to the right. White arrowheads indicate γH2AX signals on sex chromosomes in mid pachytene (H1T-positive) spermatocytes. Scale bars in larger panels: 100 μm; scale bars in smaller panels: 10 μm.
See also Figure S1.
Next, we examined the spermatogenic stage progression of H2ax-Y142A mice using two specific markers: H1T and γH2AX. H1T is a testis-specific histone variant that accumulates in nuclei from the mid-pachytene stage onward [19]. γH2AX is a prominent marker of the DDR at DNA double-strand breaks (DSBs), stalled replication forks, and MSCI [20, 21]. We observed complete spermatogenetic arrest at the mid pachytene stage (H1T-positive) in H2ax-Y142A testes (Figure 1F). Accordingly, the epididymides of H2ax-Y142A mice displayed a complete absence of sperm (Figure S1A). Apoptotic cell death is linked to germ cell loss in H2ax-Y142A testes (Figure S1B), while the point mutation did not noticeably change H2AX protein abundance (Figure S1C). Thus, the Y142A mutation reflects the biological importance of an H2AX residue rather than an effect mediated by destabilization of the H2AX protein. Together, these results suggest that Y142 of H2AX is an amino acid residue essential for the progression of spermatogenesis. In contrast to the severe H2ax-Y142A phenotype observed in males, H2ax-Y142A female mice were fertile (6 independent females were tested; mean litter size: 6.33 ± 0.48, mean ± s.e.m.) similar to H2ax null females [5, 22].
H2AX-Y142 is critical for the establishment of MSCI
In the leptotene stage of meiotic prophase I, the topoisomerase II-like enzyme SPO11 induces programmed DNA double-strand breaks (DSBs) and, in the subsequent zygotene stage, the synapsis of homologs takes places [23–25]. In response to SPO11-dependent DSBs, an initial wave of γH2AX signaling takes place throughout nuclei in the leptotene stage. In chromosome spreads of H2ax-Y142A leptotene spermatocytes, as in wild-type controls, γH2AX displayed apparently normal nuclear accumulation patterns (Figure S2A). Upon homolog synapsis in normal meiosis, γH2AX signals diminish from autosomes, and a chromosome-wide γH2AX domain is established on the sex chromosomes—a hallmark of MSCI (Figure 2A, top panel). In H2ax-Y142A spermatocytes, we detected no sex chromosome-wide accumulation of γH2AX; instead, we observed an intense, linear accumulation of γH2AX along the XY axes (Fig. 2A, bottom panel). This conclusion was confirmed through immunostaining and imaging of testis sections (Fig. 1F, arrows in the upper right panel). Importantly, the linear accumulation of γH2AX along the XY axes comprised a completely penetrant phenotype: We observed no H2ax-Y142A spermatocytes with a chromosome-wide γH2AX domain (>500 nuclei observed from >10 independent H2ax-Y142A mice). Additionally, in staging H2ax-Y142A spermatocytes, we did not observe spermatocytes beyond the mid pachytene stage, neither in our analyses of testis sections nor in our analyses of chromosome spreads of spermatocyte nuclei. In normal male meiosis, a feedforward mechanism mediates the formation of a γH2AX domain on the sex chromosomes; this mechanism sees the accumulation of γH2AX along unsynapsed axes spread to chromatin loops to form a chromosome-wide domain [3, 6] (Figure 2B). The mechanism is centered on MDC1, a γH2AX-binding protein that interacts with TOPBP1 [26]; the binding of MDC1 to γH2AX enables the progressive activation of ATR kinase through its activator protein TOPBP1 [12, 27, 28], which then mediates adjacent γH2AX formation on chromatin loops. MDC1 binds γH2AX again and the mechanism continues onward. In our previous study of the Mdc1KO mouse model, we observed a strikingly similar accumulation of γH2AX signals along sex chromosome axes but not through chromatin loops [6].
Figure 2. H2AX-Y142 is required for the initiation of MSCI.
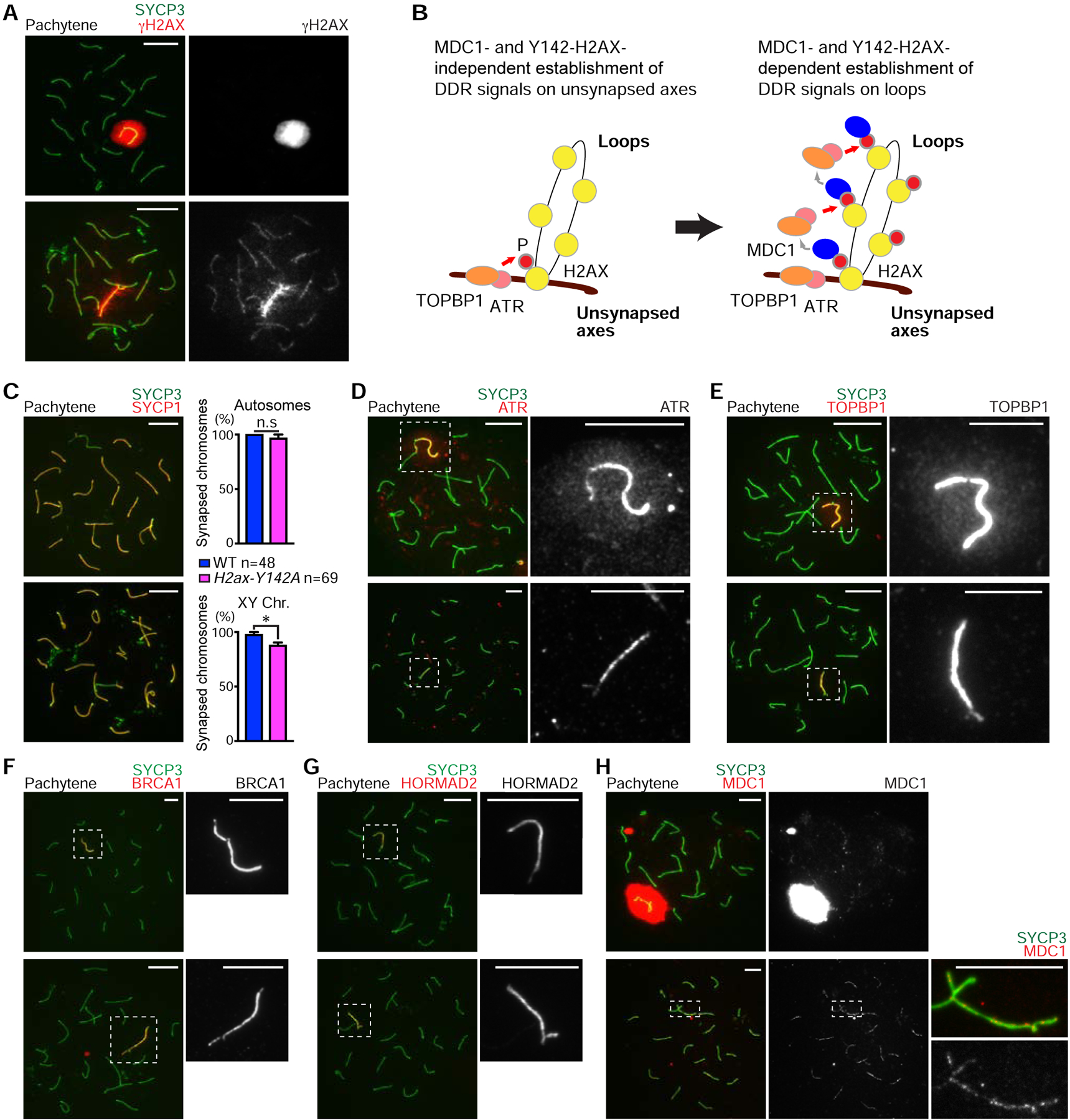
(A, C–H) Chromosome spreads of wild-type (WT) and H2ax-Y142A pachytene spermatocytes immunostained with antibodies raised against the following proteins: SYCP3 (A, C–H), γH2AX (A), SYCP1 (C), ATR (D), TOPBP1 (E), BRCA1 (F), HORMAD2 (G), and MDC1 (H). Dashed squares are magnified in the panels to the right.
(B) Model of the initiation of MSCI. ATR and its activator, TOPBP1, are recruited to unsynapsed axes in an MDC1- and Y142-H2AX-independent manner, resulting in phosphorylation of H2AX to generate γH2AX on axes (left). γH2AX then recruits MDC1, which facilitates progressive recruitment of ATR and TOPBP1, resulting in γH2AX and MDC1 spreading throughout loops (right).
(C) Percentages of mid pachytene chromosome synapsis for autosome and sex chromosomes, shown as mean ± s.e.m.. Total numbers of analyzed nuclei obtained from 3 independent littermate pairs are indicated in the panel. n.s., not significant; * p < 0.05, unpaired t test. Scale bars: 10 μm unless otherwise described in the panels.
See also Figure S2.
Despite a completely penetrant sex chromosome phenotype, autosome synapsis was unimpaired in H2ax-Y142A spermatocytes as mutant chromosome spreads immunostained against SYCP1 (a marker of synapsed chromosome axes) revealed no apparent abnormalities (Figure 2C). We corroborated this observation by immunostaining against another marker of chromosome synapsis, SIX6OS1 [29] (Figure S2B). Together, these data confirm the unimpaired formation of the synaptonemal complex in H2ax-Y142A spermatocytes. Furthermore, the partial synapsis of the X and Y chromosomes at an area of homology known as the pseudoautosomal region was present in 87.86% of observed H2ax-Y142A spermatocytes, a proportion that is slightly lower in comparison to control spermatocytes (Figure 2C). A recent study revealed that autosome synapsis is partially defective in H2ax null mutants, suggesting that H2AX has additional roles in the regulation of autosome synapsis [30]. However, the role of H2AX-Y142 is restricted to the regulation of the sex chromosomes. Thus, we conclude that H2ax-Y142A is a separation-of-function mutation for the H2ax gene.
To determine the function of H2AX-Y142 in the regulation of DDR signals, we examined the localization of ATR and TOPBP1. In H2ax-Y142A spermatocytes, ATR and TOPBP1 are restricted to the axes of sex chromosomes with no apparent spreading to chromatin loops, unlike wild-type spermatocytes (Figures 2D and 2E). These data confirm a function for H2AX-Y142 in the establishment of a chromosome-wide domain. In contrast to these proteins, in H2ax-Y142A spermatocytes, the localization of BRCA1 and HORMAD2 (factors that localize to unsynapsed axes to regulate ATR signaling [13, 15, 31]) was unchanged (Figures 2F and 2G). Therefore, the function of H2AX-Y142 is specific to the XY chromatin domain. Given these findings, we examined the localization of MDC1 on the XY chromatin of H2ax-Y142A spermatocytes. Interestingly, we observed punctate MDC1 foci on H2ax-Y142A sex chromosome axes, unlike the sex chromosome-wide distribution of MDC1 in wild-type spermatocytes (Figure 2H). These data suggest that, while MDC1 was recruited to the axes of H2ax-Y142A sex chromosomes, MDC1 signals were not amplified throughout the XY chromatin. Taken together, we conclude that Y142 of H2AX is required for the MDC1-dependent amplification of DDR signals throughout XY chromatin (Figure 2B).
To further determine the necessity for H2AX-Y142 in MSCI, we examined the status of active transcription on the sex chromosomes. Using slides that preserve the 3D chromatin organization of spermatocytes (3D slides) [32, 33], we judged the localization of RNA polymerase II (RNAPII) signals with respect to the bulbous chromo-nuclear compartment that encompasses XY chromatin in mid pachytene (H1T-positive) spermatocytes. In wild-type mid pachytene spermatocytes, RNAPII signals were largely excluded from the XY body (Figure 3A), consistent with previous reports [6, 32]. However, in stage-matched H2ax-Y142A spermatocytes, RNAPII signals were overtly present throughout XY chromatin, while DAPI-discernible XY bodies were absent (Figure 3A). Likewise, the compaction of the XY axes, another telltale signature of MSCI initiation and XY body formation, was not observed in H2ax-Y142A mid pachytene spermatocytes, neither in our analyses of testis sections (Figure 3B) nor in our analyses of 3D slides (Figure 3C). Interestingly, these observations are essentially the same as those made in studies of other mouse models with complete defects in the initiation of MSCI, including loss-of-function models for H2AX, MDC1, ATR, and TOPBP1 [5, 6, 14, 28]. Therefore, we conclude that H2AX-Y142 is required both for the establishment of MSCI and for formation of the XY body.
Figure 3. Impaired XY body formation in H2ax-Y142A mouse.
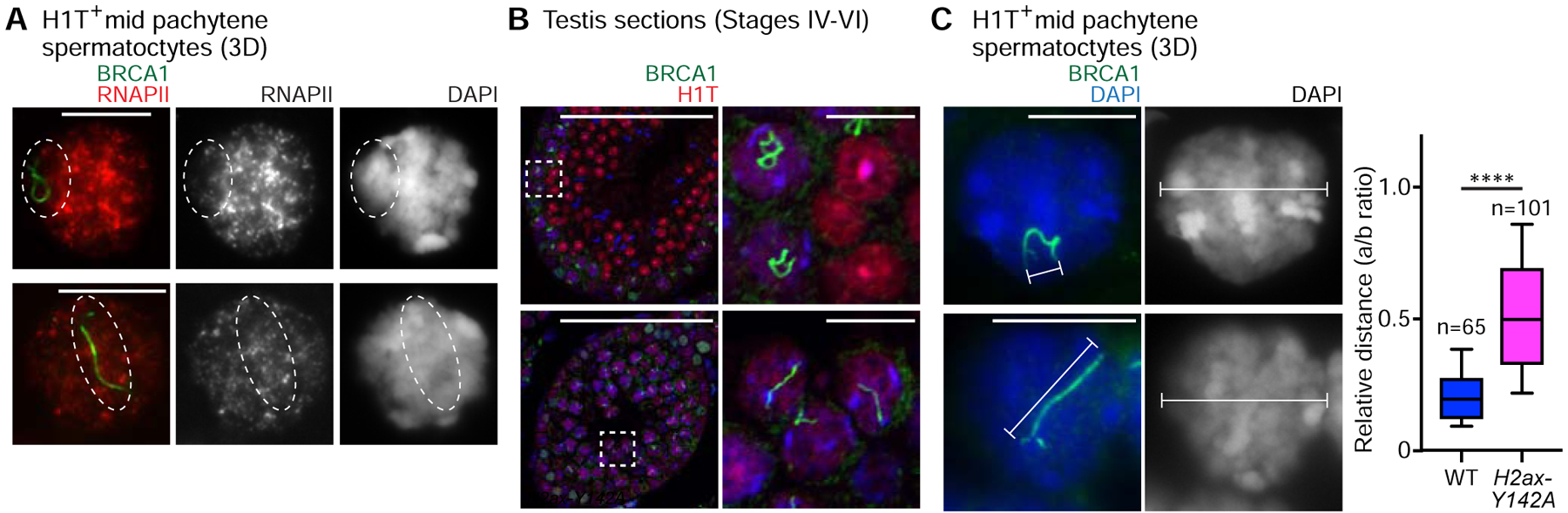
(A) Wild-type (WT) littermate control and H2ax-Y142A pachytene spermatocytes on 3D slides (see STAR METHODS) immunostained with antibodies raised against RNAPII and BRCA1. Although not shown in the panel, the spermatocytes were also immunostained with an anti-H1T antibody to determine their stage; the spermatocytes shown are H1T-positive. Dashed circles indicate the sex chromosomes.
(B) WT littermate control and H2ax-Y142A testis sections immunostained with antibodies raised against BRCA1 and H1T. Dashed squares are magnified in the panels to the right. White arrowheads indicate the axes of sex chromosomes in H1T-positive pachytene spermatocytes. Nuclei were counterstained with DAPI. Scale bars: 100 μm and, in the panels to the right, 10 μm.
(C) WT and H2ax-Y142A pachytene spermatocytes on 3D slides immunostained with an antibody raised against BRCA1. Although not shown in the panel, the spermatocytes were also immunostained with an anti-H1T antibody to determine their stage; the spermatocytes shown are H1T-positive. The relative distances are shown in a box-and-whisker plot: The central line is the median, the bottom edge of the box is the first quartile, the top edge of the box is the third quartile, and the whiskers encompass, from top to bottom, the first to ninth decile. Total numbers of analyzed nuclei, obtained from 3 independent wild-type mice and 4 independent H2ax-Y142A mice, are indicated in the panel. **** p < 0.0001, Mann-Whitney U test. Nuclei were counterstained with DAPI. Scale bars: 10 μm.
The initial steps of autosome DSB repair are normal in MSCI-defective mutants
The results presented here demonstrate the H2ax-Y142A model has a completely penetrant defect in MSCI initiation, a finding that corroborates the essentiality of MSCI in spermatogenesis [5, 6, 13–15, 28, 34]. In turn, this raises the possibility that failure of MSCI is a direct cause of meiotic arrest and germ cell demise. However, if DSB repair in meiotic recombination were defective, that would suggest a recombination-dependent checkpoint operates to trigger germ cell demise in meiotic prophase I [1, 35, 36]. To test the hypothesis that MSCI failure is a direct cause of meiotic arrest in H2ax-Y142A spermatocytes, we evaluated the status of meiotic recombination on H2ax-Y142A autosomes.
To assess the status of DSB repair in H2ax-Y142A spermatocytes, we evaluated RAD51 foci in spermatocyte nuclei. An established surrogate for DSBs, RAD51 foci appear in response to SPO11-induced DSBs in leptotene spermatocytes [37]. As leptotene spermatocytes advance into the zygotene and pachytene stages, there is a progressive decrease in the numbers of RAD51 foci concomitant with the repair of DSBs, which is completed in the pachytene stage [38]. In our analyses of H2ax-Y142A spermatocytes, we observed a normal accumulation of RAD51 foci in the leptotene and zygotene stages (Figure S3A); as well, we observed comparable numbers of RAD51 foci between wild-type and H2ax-Y142A mid pachytene spermatocytes (per nucleus, we observed <10 autosomal foci: Figure 4A). These data indicate that the initial steps of DSB repair are apparently normal on the autosomes of H2ax-Y142A spermatocytes. Interestingly, we observed a slight increase in the numbers of RAD51 foci on the sex chromosome axes of H2ax-Y142A mid pachytene spermatocytes (wild-type mid pachytene spermatocytes: 17.69 ± 0.67, mean ± s.e.m.; H2ax-Y142A mid pachytene spermatocytes: 22.01 ± 0.76, mean ± s.e.m.). This finding further confirms a sex chromosome-specific function for H2AX-Y142—a function apparently extraneous to the formation and resolution of autosomal DSBs.
Figure 4. Initial steps of DSB repair take place normally on autosomes of MSCI defective mutants.
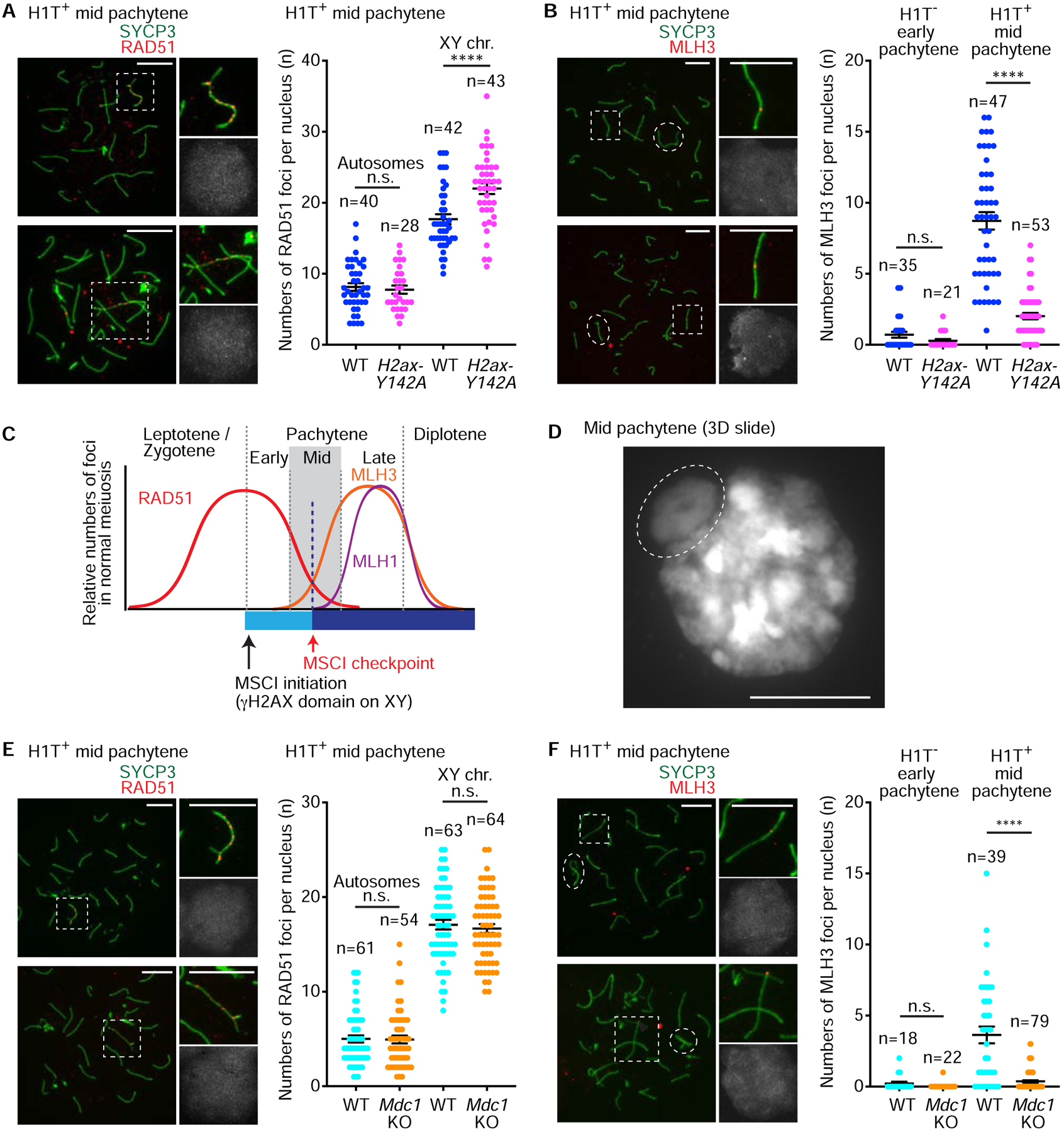
(A, B, E, F) Chromosome spreads of wild-type (WT) littermate control and H2ax-Y142A (A, B) or Mdc1KO (E, F) mid pachytene spermatocytes immunostained with antibodies raised against SYCP3 (A, B, E, F), HIT (A, B, E, F), RAD51 (A, E), or MLH3 (B, F). Sex chromosomes in dashed squares are magnified in the panels to the right (A, E). Autosomes in dashed squares are magnified in the panels to the right, and the dashed circles indicate the sex chromosomes (B, F). Dot plots indicate the numbers of autosome RAD51 foci (A, E), sex chromosome RAD51 foci (A, E), or MLH3 foci (B, F) per mid pachytene (HIT-positive) spermatocyte, shown as mean ± s.e.m. for 3 independent H2ax-Y142A littermate pairs (A, B) and 3 independent Mdc1KO littermate pairs (E, F). Total numbers of analyzed nuclei are indicated in the panels. n.s.: not significant; **** p < 0.0001, unpaired t tests. XY chr.: XY chromosomes. Scale bars: 10 μm.
(C) Model of the MSCI checkpoint and its relationship to meiotic recombination.
(D) DAPI counterstaining of a 3D slide (see STAR METHODS). The dashed circle indicates the XY body. Scale bar: 10 μm.
See also Figures S3 and S4.
While we judged DSB resolution to be grossly normal, a small-but-vitally important proportion of DSBs resolve via a specialized crossover recombination repair pathway that facilitates the reshuffling of genetic information between maternal and paternal chromatids [23–25]. Thus, we sought to evaluate crossover recombination repair in H2ax-Y142A and control spermatocytes. In H2ax-Y142A spermatocytes, we noted a distinct absence of MLH1 foci (Figure S3B), which mark sites of crossover recombination [39]. We infer this is due to the inability of H2ax-Y142A spermatocytes to advance past the mid pachytene stage into the late pachytene stage, when the vast majority of MLH1 focal accumulation takes place (Figure S3B). In a previous report of Mdc1KO spermatocytes, we noted a phenotype that is essentially the same as that in H2ax-Y142A spermatocytes: The absence of MLH1 foci was concomitant with completely defective MSCI [6]. This commonality suggests that the initiation of MSCI is necessary for meiotic progression to the stage of MLH1 focal accumulation.
In a previous report analyzing chromosome spreads of spermatocyte nuclei [40], it was demonstrated that future sites of crossover exhibit focal accumulation of another crossover marker, MLH3, prior to the accumulation of MLH1 foci. To evaluate the initial step of crossover recombination, prior to the arrest of mid pachytene spermatocytes, we scored the numbers of MLH3 foci in control and H2ax-Y142A spermatocytes. In early pachytene (H1T-negative) spermatocytes from both control and H2ax-Y142A mice, MLH3 foci were detected at similarly low levels (Figure 4B). However, in mid pachytene (H1T-positive) spermatocytes, the numbers of MLH3 foci differed between wild-type and H2ax-Y142A: In wild-type samples, MLH3 focal accumulation showed a marked increase (8.72 ± 0.61, mean ± s.e.m.; Figure 4B); in H2ax-Y142A samples, we noted reduced numbers of MLH3 foci (2.01 ± 0.23, mean ± s.e.m.; Figure 4B). In wild-type late pachytene spermatocytes, the numbers of MLH3 foci reached their zenith (>20, with at least one per chromosome; Figure S3C), similar to the focal accumulation of MLH1; this suggests MLH3 focal accumulation grows in and through the mid pachytene stage. Meiotic arrest likely takes place early in the mid pachytene stage, when MLH3 focal accumulation is in its initial growth phase (Figure 4C). Importantly, this timing coincides with the formation of the XY body, a distinct, DAPI-discernible chromo-nuclear compartment at the periphery of the mid pachytene nucleus (Figure 4D) [32]. In the early pachytene stage, the diffuse, indistinct chromosome-wide accumulation of γH2AX on XY chromatin represents the initiation of MSCI, and it is not until the mid pachytene stage that a bulbous, bounded, and distinctly compartmentalized XY body appears at the nuclear periphery [15, 41, 42]. We infer that the complete death of H2ax-Y142A spermatocytes is linked to the failure to form an XY body.
To further define the molecular events that occur in response to defective MSCI, we analyzed meiotic DSB repair in Mdc1KO mice, a separate, independent genetic model exhibiting complete failure of MSCI [6]. We did not examine other mutants for direct regulators of MSCI, such as H2AX, ATR, and TOPBP1, because they are directly involved in meiotic recombination [5, 14, 28, 30]. Similar to our analyses of H2ax-Y142A and control mice, the numbers of RAD51 foci were comparable on the mid pachytene autosomes of Mdc1KO and control mice (Figure 4E). Interestingly, RAD51 foci on Mdc1KO sex chromosomes were comparable with wild-type sex chromosomes (Figure 4E); this is in contrast to the slight increase in RAD51 focal accumulation observed on H2ax-Y142A sex chromosomes (Figure 4A), suggesting that RAD51 foci on the sex chromosomes may be regulated by H2AX-Y142 but not MDC1. Taken together our findings indicate that the initial steps of meiotic DSB repair are grossly normal in Mdc1KO spermatocytes, thereby confirming that MSCI deficiency does not disturb the initial steps of meiotic recombination. Furthermore, reduced numbers of MLH3 foci were observed in mid pachytene spermatocytes of Mdc1KO mice (Figure 4F). Thus, based on findings from two independent mouse models, we conclude that meiotic arrest takes place in the midst of the mid pachytene stage prior to the full accumulation of MLH3 foci (Figure 4C). Importantly, we have excluded the possibility that the meiotic arrest observed in H2ax-Y142A mice is caused by the reduction of MLH3 foci because MLH3-deficient spermatocytes are capable of reaching the metaphase stage of meiosis I [43]. Also, our results further suggest the successful initiation of MSCI is required for stage progression, which enables crossover formation in male meiosis.
Finally, we evaluated meiosis in female H2ax-Y142A mice and confirmed normal meiotic recombination (Figure S4). Consistent with the normal fertility of H2ax-Y142A females, MLH1 foci were observed in H2ax-Y142A pachytene oocytes (Figure S4A). Therefore, Y142 of H2AX is dispensable for the formation of MLH1 foci in female meiosis. This result underscores specific functions for H2AX-Y142 in male-specific MSCI. In support of this notion, regulators of MSCI—such as MDC1, H2AX, BRCA1, and HORMAD2—are not required for female meiosis, and female models for the loss-of-function of these factors are fertile [6, 22, 31, 44, 45].
ATR-associated DDR signals are sequestered from autosomes to the sex chromosomes at the onset of MSCI
Our results suggest the establishment of MSCI permits the timely progression of male germ cells through meiotic prophase. By definition, checkpoints monitor the completion of essential cellular events, enabling cells to progress to their next stages. In meiosis, it is generally thought meiotic checkpoint mechanisms coordinate genetically distinct and/or independent meiotic processes [1]. Since genetic evidence, including the current study, shows that meiotic arrest and subsequent cellular demise ensue when MSCI is abrogated [5, 6, 13–15, 31, 45, 46], we reasoned that the initiation of MSCI functions as a checkpoint to coordinate and pace precise meiotic stage progression during normal meiosis.
A previous study proposed that, in response to defective MSCI, germ cell death results from the ectopic expression of sex chromosome-linked genes [34]. However, DDR signaling—and particularly ATR signaling—is directly involved in the meiotic checkpoints of various organisms, including yeast, worms, and flies [1]. These observations, in combination with our new results, compelled us to hypothesize that DDR signaling itself functions as a checkpoint in MSCI. This is a novel model for the MSCI checkpoint (see Discussion). To test this hypothesis, we sought to identify DDR signaling phenotypes common to both the H2ax-Y142A and Mdc1KO models, which exhibit specific MSCI defects.
In our previous evaluation of mid pachytene Mdc1KO spermatocytes, we observed persistent γH2AX and TOPBP1 foci on autosomes axes when MSCI is abrogated [6]. In H2ax-Y142A spermatocytes in the mid pachytene stage, we observed that γH2AX and TOPBP1 foci were present largely at autosome axes (Figures 2A and 5A). On the other hand, in the transition from the early-to-mid pachytene stages in wild-type spermatocytes, these DDR signals progressively faded from autosomes. This retention of TOPBP1 foci on autosome axes was comparable between H2ax-Y142A and Mdc1KO spermatocytes (Figures 5A–5C: 65.95 ± 4.26, mean ± s.e.m. for H2ax-Y142A; 76.09 ± 2.76, mean ± s.e.m. for Mdc1KO). Importantly, the retention of γH2AX and TOPBP1 foci in H2ax-Y142A and Mdc1KO mice was largely independent of persistent DSBs: The numbers of autosomal γH2AX and TOPBP1 foci were much higher than those of RAD51 foci, and we did not observe differences in the numbers of RAD51 foci between controls and mutants (Figures 4A and 4E). Since TOPBP1 is an ATR activator [47], we also examined the retention of ATR foci. In normal meiosis, γH2AX and ATR foci are confined to the XY body in the mid pachytene stage. In contrast, we confirmed the retention of γH2AX and ATR foci on autosome axes in H2ax-Y142A and Mdc1KO pachytene spermatocytes (Figures 5D and 5E). Furthermore, we found that ATR interacting protein (ATRIP), a binding partner of ATR in checkpoint signaling [48] and a protein that binds unsynapsed meiotic chromosome axes [49], was retained on autosome axes in H2ax-Y142A pachytene spermatocytes (Figure S5). These results raise a compelling possibility: In the early pachytene stage, MDC1-dependent amplification of DDR signals on the XY chromatin sequesters the DDR signaling to the sex chromosomes and away from autosomes (Figure 6). Although another interpretation is possible: MDC1 and H2AX-Y142 are required for the release of DDR signals from autosomes axes. We believe this possibility is unlikely because the release of DDR signals from chromosome axes takes place normally in H2ax-Y142A pachytene oocytes (Figure S4B and S4C).
Figure 5. DDR factors centered on ATR signaling are sequestered from autosomes to the sex chromosomes at the onset of MSCI.
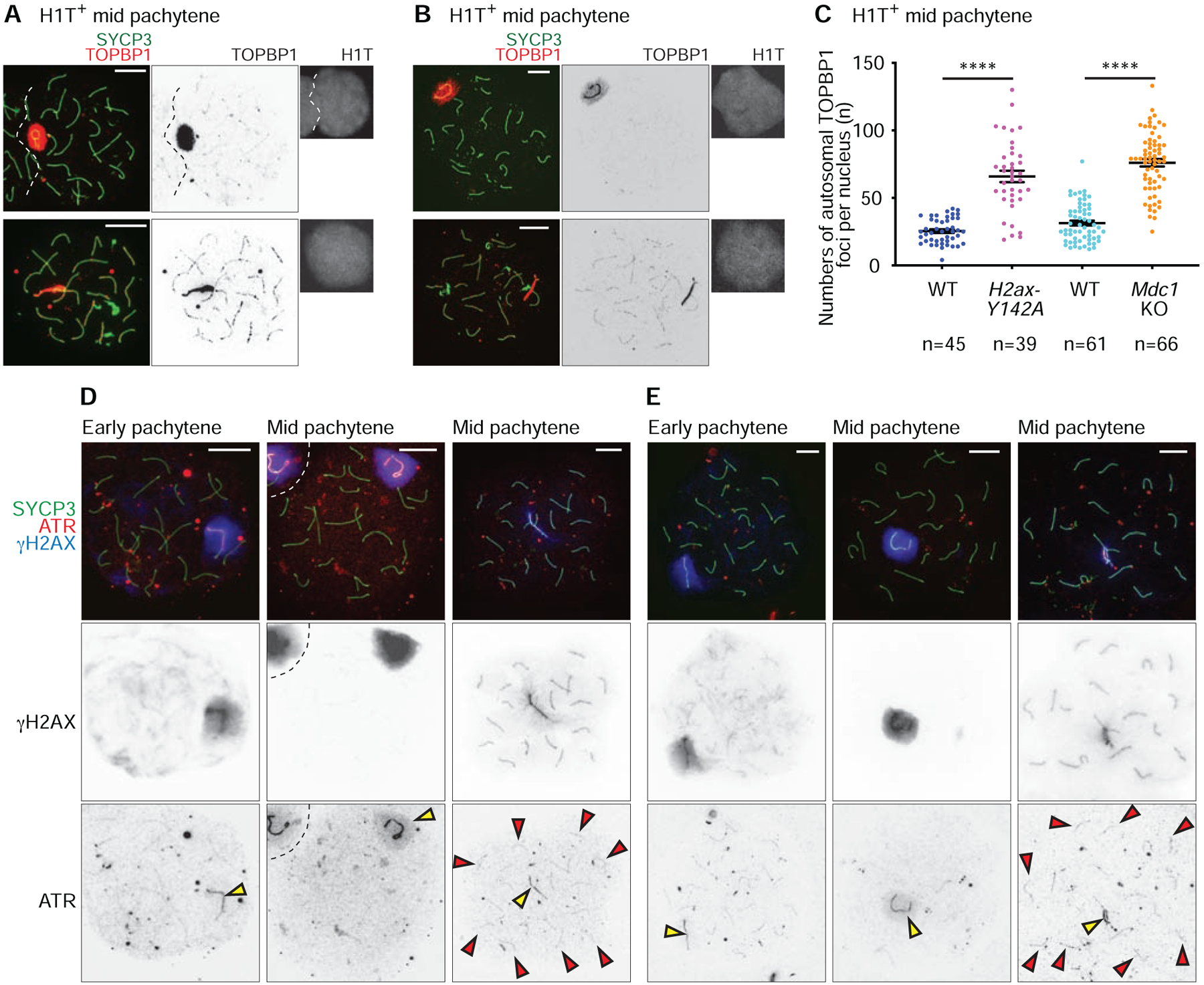
(A, B, D, E) Chromosome spreads of wild-type (WT) littermate control and H2ax-Y142A (A, D) or Mdc1KO (B, E) mid pachytene spermatocytes immunostained with antibodies raised against SYCP3 (A, B, D, E), TOPBP1 (A, B), H1T (A, B), γH2AX (D, E), and ATR (D, E). Yellow arrowheads indicate the sex chromosomes, and red arrowheads indicate ATR foci that persist on H2ax-Y142A autosomes (D, E). Scale bars: 10 μm.
(C) Numbers of TOPBP1 foci on autosomes in mid pachytene (H1T-positive) spermatocytes, shown as mean ± s.e.m. for 3 independent H2ax-Y142A littermate pairs (left) and 3 independent Mdc1KO littermate pairs (right). Total numbers of analyzed nuclei are indicated in the panels. **** p < 0.0001, Mann-Whitney U test.
See also Figure S5.
Figure 6. A model of the MSCI checkpoint: The physical sequestration of DDR factors from autosomes to the XY body is a critical checkpoint in meiosis progression and gamete development.
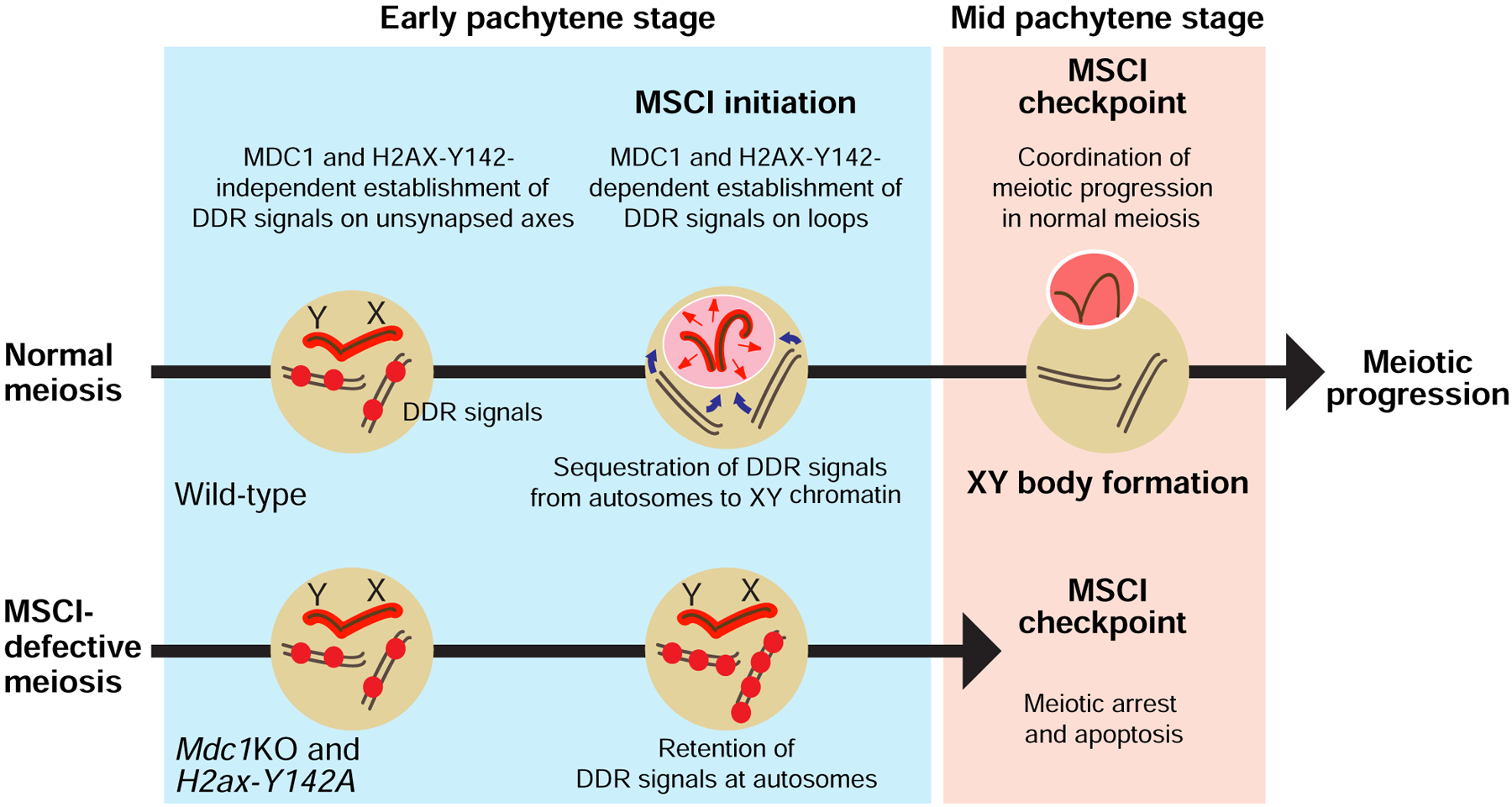
At the onset of MSCI, DDR factors (shown as red spheres) are sequestered from autosomes to the sex chromosomes. The physical sequestration of DDR factors on/at a sex chromosome-specific chromo-nuclear compartment, the XY body, is a critical step in the MSCI checkpoint in the mid pachytene stage of meiotic prophase I. While the MSCI checkpoint ensures meiotic stage progression in normal meiosis, the abolishment of MSCI enables the ectopic retention of DDR signals on/at autosomes; these, in turn, trigger complete meiotic arrest and cell death in response to the checkpoint.
See also Figure S6.
Discussion
In this study, we show that H2ax-Y142A is an H2ax separation-of-function mutation that exhibits specific defects in MSCI. We capitalized on this model to dissect the molecular events that occur in response to defective MSCI. In interpreting our results in the context of the literature, we propose a new model for the MSCI checkpoint wherein MSCI sequesters ATR-mediated DDR signaling from autosomes to permit meiotic progression (Figure 6).
The Y142 residue of H2AX is essential for MSCI
We show that H2AX-Y142 is important for the function of MDC1 in MSCI. In H2ax-Y142A mutants, the localization of MDC1 to the XY axes was significantly reduced (Fig. 2H); this raises the possibility that the binding of MDC1 to H2AX with an A142 point mutation is less than the binding of MDC1 to unphosphorylated H2AX-Y142. This interpretation is in line with results reported for somatic cells [50, 51]: In response to irradiation-induced DNA damage, an H2AX-Y142A point mutation abolishes the binding of MDC1 to γH2AX (i.e., H2AX phosphorylated at S139). However, the PI3-kinases that act in response to irradiation-induced DNA damage in somatic cells differ from those that act in MSCI. At sites of irradiation-induced DSBs, the kinase ATM is thought to amplify γH2AX signals in conjunction with MDC1 [52]; in contrast, ATR-mediated γH2AX is amplified in conjunction with MDC1 in MSCI. Since ATR-mediated γH2AX is linked to stalled replication forks in somatic cells [20], the current study supports a model in which the ATR-mediated DDR pathway that induces MSCI is an adapted version of the ATR-mediated DDR pathway that recognizes replication stress in S phase of somatic cells [3, 6].
Importantly, the initial steps of DSB repair and ATM-mediated γH2AX signaling take place normally in both H2ax-Y142A and Mdc1KO meiosis [6]. Therefore, we conclude H2AX-Y142 and MDC1 are not required for ATM-dependent processes in meiosis, such as the control of DSB numbers [53]. This is in contrast to a recent study postulating H2AX and MDC1 are involved in ATM-dependent processes in meiosis [30]. However, the normal DSB repair found in the current study suggests the ATM-related recombination-dependent checkpoint—triggered by persistent DSBs in early pachytene spermatocytes [35, 36, 54, 55]—may not be activated in H2ax-Y142A and Mdc1KO spermatocytes. Interestingly, H2ax null mice exhibited additional defects beyond those of H2ax-Y142A mice, including chromosome synapsis abnormalities [30]. Such defects may represent H2AX functions genetically distinct from H2AX-Y142.
The numbers of RAD51 foci on the male sex chromosomes in H2ax-Y142A spermatocytes were slightly increased in comparison to wild-type sex chromosomes. In somatic cells, H2AX controls recombination both of homologous chromosomes and sister chromatids [56]. Since sister chromatids comprise the only templates for DSB repair of the male sex chromosomes, we postulate that H2AX-Y142 has a critical role in the control of H2AX-mediated sister chromatid recombination. Alternatively, H2AX may be a key factor in a proposed role for MSCI in the suppression of illegitimate recombination between unsynapsed regions of the X and Y chromosomes [57]; as well, H2AX may be critical for proposed roles of meiotic silencing in suppressing nonhomologous recombination on unsynapsed axes [4]. However, in the above scenarios, the function of H2AX is likely independent of MDC1, since we did not observe increased numbers of RAD51 foci on sex chromosomes in Mdc1KO mice.
A novel model for the MSCI checkpoint
The biological function of MSCI remains a major unsolved question. We propose a novel function for MSCI: Initiation of MSCI sequesters DDR factors from autosomes to the sex chromosomes. According to this model, when MSCI is defective, the retention of ATR-mediated DDR signaling on autosomes directly induces cell arrest and demise (Figure 6). We postulate that this ATR-mediated surveillance mechanism directly functions in the MSCI checkpoint, which, in establishing MSCI, results in an XY body that sequesters ATR-mediated DDR signaling to permit the timely progression of male meiosis (Figure 6). In our reasoning, MSCI can be interpreted as a checkpoint since classical checkpoints function to coordinate essential cell cycle-related events [58, 59]. In support of this, ATR-mediated DDR signaling—including its regulatory mechanisms (e.g., those enabled by BRCA1 and HORMAD1/2)—is tightly linked to the control of meiotic progression, as has been demonstrated in many previous studies [11, 13, 15, 31, 45, 46, 60]. In concert with a possible role for ATR in the MSCI checkpoint, ATR functions to regulate checkpoints in other contexts too, including the intra-S replication checkpoint in somatic cells as well as the G2 DNA damage checkpoint in somatic cells [61, 62]. Importantly, CHEK2, a downstream effector of ATR, was shown to be critical in an oogenic checkpoint [63], further indicating the direct function of DDR signaling in certain checkpoints.
Interestingly, there may exist a specific threshold for the amount of ATR-mediated DDR signaling that triggers mid pachytene cell death. The retention of small amounts of ATR-mediated DDR signaling on autosomes is compatible with meiotic progression through the mid pachytene stage as in, for example, small genomic regions of asynapsis; yet apparently, larger amounts lead to arrest and apoptosis [64, 65]. Furthermore, MSCI is attenuated but not defective in response to a hypomorphic mutation of the Atr gene, and persistent DDR signaling on autosomes is associated with cell death beyond the mid pachytene stage [66]. The current work reveals an intriguing finding: The retention of ATR-mediated DDR signaling on autosomes is largely independent of DSB repair. Therefore, the MSCI checkpoint is functionally separable from the recombination-dependent checkpoint resulting from persistent DSBs [35, 36, 54, 55]. In relation to the MSCI checkpoint model presented here, the depletion of CHEK1, another downstream effector of ATR, results in the accumulation of DSB-independent DDR signaling on autosomes as well as abnormal meiotic progression [41]. A previous study demonstrated that the amounts of DDR factors present in pachytene spermatocytes are fixed [46]; thus, extensive autosome asynapsis may attenuate MSCI by overdrawing from a limited bank of DDR proteins. Thus, our new model provides a logical explanation for why an ATR-mediated DDR pathway recognizes the asynapsis of autosomes and then eliminates germ cells with chromosome abnormalities.
Although the unique characteristics of the MSCI checkpoint do not invalidate other kinds of checkpoints, we need to carefully distinguish between our new model of the MSCI checkpoint and previously described meiotic checkpoint mechanisms. This is because no protein has been identified that, when absent, bypasses the arrest linked to MSCI defects. ATR itself has functions in meiotic recombination [66, 67], and ATR loss-of-function is associated with cell death in the mid pachytene stage [14]. According to our MSCI checkpoint model, sequestration of DDR signaling from autosomes to the XY body, a chromo-nuclear compartment isolated from the rest of the nucleus, permits meiotic progression at the mid pachytene stage. It can be reasonably assumed that, in spermatocytes, a variety of proteins must be sequestered to the XY body at certain thresholds to ensure continued stage progression and gamete development. Notably, in such a scenario, then, the depletion or mutation of a given DDR protein cannot bypass the meiotic checkpoint to permit meiotic progression.
MSCI defects are compatible with cell survival in the early pachytene stage but result in meiotic arrest and death with impressive exactitude in the mid pachytene stage. Indeed, XY bodies are invariably absent in mutants with defective MSCI and/or chromosome synapsis abnormalities [5, 6, 15, 46], indicating a tight, underlying link between the formation of an isolated XY body and meiotic progression. We infer the biological function of the XY body is to sequester specific nuclear components, including DDR signals/checkpoint machinery, from the remainder of the nucleus. The phenotypes of Spo11−/− spermatocytes, which display extensive chromosome asynapsis abnormalities and undergo cell death, could arise from an inability to isolate such nuclear components. While SPO11-deficient spermatocytes manifest one or more diffuse sites of ectopic meiotic silencing—termed “pseudo-XY bodies”—they are not compatible with meiotic progression since pseudo-XY bodies do not constitute distinct, bounded, physically isolated chromo-nuclear compartments like true XY bodies [11, 15]. On the other hand, mutants deficient for XY-body markers that function downstream of the ATR-mediated DDR pathway that initiates MSCI—including, but not limited to, AGO4, RNF8, SCML2, FANCB—are not associated with mid pachytene arrest [68–71]. These XY-body markers may function subsequent to progression through the MSCI checkpoint.
Interestingly, our recent study suggests the 3D chromatin organization of the sex chromosomes during MSCI arises from phase separation [72], a physical process in which membraneless organelles form and behave as liquid droplets [73, 74]. Following on this, it is intriguing to speculate that MDC1-dependent amplification of γH2AX gives rise to nuclear environments in which it is energetically favorable for DDR signals to coalesce, a liquid-liquid condensation that sequesters DDR signals/checkpoint machinery from their initial sites of accumulation on autosome axes and chromatin. In the absence of such a mechanism, DDR signaling is retained on autosomes.
In contrast to our new model, a previous study suggested another potential cause of meiotic cell death in mutants with defective MSCI: Derepression of toxic genes from sex chromosomes such as the Y-linked genes Zfy1 and Zfy2 [34]. Given our growing knowledge of spermatogenesis, and given that both ZFY1 and ZFY2 function in normal spermatogenesis [75–77], derepression of sex-linked genes is unlikely to be the sole mechanism inducing cell death. As well, the extended timespans, noise, and cell-to-cell variability of transcription- and translation-dependent biological processes suggest toxic gene derepression is likely not compatible with the complete, clockwork nature of mid pachytene cell death. In our previous study of Scml2KO mice, the abnormal expression of sex-linked genes in the pachytene stage was found to be compatible with meiotic progression [70], and Zfy1 and Zfy2 were derepressed in Scml2KO pachytene spermatocytes (Figure S6). Furthermore, SCML2 and FANCB are examples of X-linked gene products that function on the XY body [70, 71], indicating that sex-linked gene products can exert physiological functions even after transcriptional silencing via MSCI. Since gene dosage of the sex chromosomes is a critical determinant of fertility [78], it is possible this gene dosage is monitored by quality control mechanisms apart from the MSCI checkpoint. The monitoring of gene dosages and checkpoints are unlikely to be mutually exclusive. Thus, continued investigation will be important to more precisely identify the molecular mechanisms underlying the MSCI checkpoint.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for reagents should be directed to the Lead Contact, Satoshi H. Namekawa (satoshi.namekawa@cchmc.org). H2ax-Y142A mice are available for sharing.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mouse lines
H2AX tyrosine 142-to-alanine 142 point mutation mice (H2ax-Y142A mice or H2ax-Y142A) used in this study were generated via CRISPR-Cas9 gene editing technology in the Transgenic Animal and Genome Editing Core Facility of Cincinnati Children’s Hospital Medical Center. The methods for the design of sgRNAs, the design of donor oligos, and the production of animals have been described [79]. The sgRNA target sequences (5′ GCCTCTCAGGAGTACTGAGG 3′) were cloned through established methods [80] into a modified pX458 vector (Addgene, 48138); the pX458 vector contains an optimized sgRNA scaffold and a high-fidelity Cas9 [81, 82]. Editing activity was validated via T7 Endonuclease 1 mismatch assays in mouse mK4 cells [83] in side-by-side comparisons with Tet2 sgRNA, which is known to work efficiently in mouse embryos [84]. Validated sgRNA was transcribed in vitro using the MEGAshortscript T7 Transcription Kit (ThermoFisher Scientific, AM1354), purified by the MEGAclear Transcription Clean-UP Kit (ThermoFisher Scientific, AM1908), and then stored at −80°C. To prepare the injection mix, we incubated sgRNA and Cas9 protein (ThermoFisher Scientific, A36498) at 37°C for 10 minutes (min) to form ribonucleoproteins; then, we added the single-strand DNA donor oligo (IDT) to the mixture. The final concentrations were as follows: 50 ng/μl sgRNA, 100 ng/μl Cas9 protein, and 100 ng/μl DNA donor oligo. Using a piezoelectric microinjection technique [85], mutant mice were generated via microinjections of the mix into the cytoplasm of fertilized eggs on a C57BL/6 background. On the same day as the microinjections, the eggs were transferred into the oviduct ampulla of pseudopregnant CD-1 female mice.
Pups were born and genotyped by PCR with AflII (NEB, R0520S) enzyme digestion. Animals were housed in a controlled environment with a 12-hour (h) light/12-h dark cycle and with free access to water and a standard chow diet. For genotyping, the wild-type allele and point mutation allele —which codes alanine instead of tyrosine at H2AX residue 142—were distinguished via the following forward primers: wild-type allele, 5′ CGC AGG CCT CTC AGG AGT AC 3′; mutant allele, 5′ CGC AGG CCT CTC AGG AGG CT 3′. The reverse primer was 5′ CTG CGG AGG GAC TAA CCT TC 3′. Heterozygous males and females were bred to produce homozygous mutant males (i.e., H2ax-Y142A mice). The Mdc1 knockout (Mdc1KO) mice used in this study are described in our previous report [6]. For analyses of H2ax-Y142A and Mdc1KO mice, homozygous mice bearing wild-type alleles were used as littermate controls unless otherwise described in the figures and/or figure legends. All mouse tissue samples used in this study were harvested at 5–12 weeks of age unless otherwise noted in the figures and/or figure legends. For analysis of female meiocytes, fetal ovaries were harvested at day 17.5 of pregnancy (where observation of vaginal plug is counted as day 0.5). All subsequent experimental work was performed under protocol no. IACUC2018–0040 approved by the Institutional Animal Care and Use Committee of Cincinnati Children’s Hospital and Medical Center.
METHOD DETAILS
Western blotting
The following western blot experiments were replicated twice; for each experiment, all samples were run on the same gel. To confirm the absence of phosphorylated Y142 of H2AX via western blots, detunicated testis pieces were homogenized in RIPA buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 0.1% SDS, 1% Triton X-100, 1% sodium deoxycholate) containing cOmplete Protease Inhibitor Cocktail (Roche, 11697498001) and Phosphatase Inhibitor Cocktail 2 (Sigma, P5726); then, the homogenate was incubated on ice for 30 min. After DNA fragmentation by sonication and subsequent centrifugation at 10,000×g at 4°C, the supernatant was transferred to a new tube before total protein concentration was quantified via Bradford assays. Volumes of lysates containing 20 μg proteins were separated by electrophoresis on 10% SDS-PAGE gels. Then, the proteins were transferred onto a PVDF membrane (EMD Millipore, IPVH00010). The membranes were blocked with StartingBlockTM T20 (TBS) Blocking Buffer (ThermoFisher Scientific, 37543) at room temperature (RT) for 30 min before incubation with anti-H2AX-pY142 antibody (Millipore, 07–1590), diluted 1/1000 in Tris-buffered saline containing 0.1% Tween 20 detergent (TBST), at 4°C overnight. On the next day, after washing three times in TBST, 5 min per wash, the blot was incubated with VeriBlot for IP Detection Reagent (HRP) (Abcam, ab131366), diluted 1/5000 in TBST at RT, for 1 h. The blot was washed three times in TBST, 5 min per wash, before incubation in Immobilon Western Chemiluminescent HRP Substrate (EMD Millipore, WBKLS0500) at RT for 1 min; then, the blot was imaged using Super RX-N x-ray film (Fujifilm) and a FluorChemQ MultiImage III instrument (Alpha Innotech). To blot loading controls, the initial blot was stripped with Restore Western Blot Stripping Buffer (ThermoFisher Scientific, 21059) at RT for 10 min; then, the stripped blot was washed two times in TBST, 5 min per wash, prior to incubation with anti-Lamin B1 antibody (Abcam, ab16048), diluted 1/2000 in TBST, at RT for 1 h. After washing the blot three times in TBST, 5 min per wash, the blot was incubated with VeriBlot for IP Detection Reagent (HRP), diluted 1/5000 in TBST, at RT for 1 h, and bands were visualized through the procedures described above.
To detect H2AX protein expression in wild-type and H2ax-Y142A mouse heart, liver, and testis tissues, whole tissue lysates were isolated and prepared from mice at two weeks of age. We selected this timepoint because spermatogenesis is in its semi-synchronized “first wave;” thus, the vast majority of testis cells are early pachytene spermatocytes. To prepare tissue lysates, ~20 mg of heart tissue, ~20 mg of liver tissue, and two whole testes were processed as described above. Volumes of lysates containing 20 μg proteins were separated by electrophoresis on a 15% SDS-PAGE gel. After transferal to a PVDF membrane and blocking, the membrane was cut into two pieces and trimmed. One piece was used for the detection of H2AX, and the other piece was used for the detection of a-tubulin, a loading control. Thus, one portion of the split membrane was incubated with anti-H2AX antibody (Cell signaling technology, 2595), diluted 1/2000 in TBST; and the other portion was incubated with anti-α-tubulin antibody (Abcam, ab7291), diluted 1/5000 dilution in TBST. The membranes were incubated at 4°C overnight. The next day, after washing (described above), the membranes were incubated with VeriBlot for IP Detection Reagent (HRP; described above). After additional washing (described above), the membranes were incubated with Immobilon Western Chemiluminescent HRP Substrate (described above); then, the blots were imaged using an Amersham Imager 680 (GE Healthcare).
Preparation and immunofluorescence of meiotic chromosome spreads
Meiotic chromosome spreads from testes were prepared essentially as described [42, 86]; here, we briefly describe our protocol, including a small number of deviations from the cited literature. Testes were excised, detunicated, and placed in 1× phosphate-buffered saline (PBS). Seminiferous tubules were dissociated from whole testes in the following amounts: from wild-type or heterozygous models, approximately one-quarter of an adult testis; from an experimental model—i.e., H2ax-Y142A or Mdc1KO—a whole adult testis. Seminiferous tubules were transferred to a four-well dish (Nunc 4-Well Dishes: ThermoFisher Scientific, 144444) kept on ice. Three of the four wells contained 1 mL PBS, while the fourth well contained 1 mL hypotonic extraction buffer [HEB: 30 mM Tris base, 17 mM trisodium citrate, 5 mM ethylenediaminetetraacetic acid (EDTA), 50 mM sucrose, 5 mM dithiothreitol (DTT), 1× cOmplete Protease Inhibitor Cocktail (Sigma, 11836145001), 1× phosphatase inhibitor cocktail 2 (Sigma, P5726–5ML), pH. 8.2]. In the first well containing 1 mL PBS, seminiferous tubules were gently unraveled into small clumps with fine-point tweezers; care was taken not to tear or mince the tubules. The clumps of seminiferous tubules were transferred to the second and third wells of 1 mL PBS for additional unraveling before transfer to the fourth well containing 1 mL HEB. Once there, fine-point tweezers were used to carefully expose the surface area of tubules to HEB. The seminiferous tubules were incubated in HEB on ice for approximately 2 h with gentle stirring every 30–45 min. After incubation, a small clump of seminiferous tubules—approximately four-to-six seminiferous tubules—was gently pulled and mashed between tweezer tips in 30 μL of sucrose (100 mM) on a plain, uncharged microscope slide (Gold Seal: ThermoFisher Scientific, 3010–002). After approximately 15–25 mashes, a semi-translucent cell suspension was formed. An additional 30 μL of sucrose (100 mM) was added to the suspension, and the suspension was mixed via gentle pipetting up and down several times. The diluted cell suspension was applied to positively charged slides (Probe On Plus: ThermoFisher Scientific, 22-230-900) in 30 μL volumes; before application of the suspension, the slides had been incubating in chilled fixation solution (2% paraformaldehyde, 0.1% Triton X-100, 0.02% sodium monododecyl sulfate, adjusted to pH 9.2 with sodium borate buffer) for a minimum of 2 min. After applying the cell suspension/sucrose mixture, the slide was slowly, gently tilted up and down at slight angles (<10°) to mix the cell suspension/sucrose mixture with remaining fixation solution. The slides were placed in “humid chambers” (closed pipet tip boxes filled to approximately two-thirds volume with water) at RT for a minimum of 1 h (maximum overnight). Then, the slides were washed in a low-concentration surfactant, 0.4% Photo-Flo 200 (Kodak, 146– 4510), at RT two times, 2 min per wash. Slides were dried completely at RT (~30 min) before staining or storage in slide boxes at −80°C.
Chromosome spreads of oocytes at embryonic day 17.5 were prepared as described [42]. Briefly, fetal ovaries at embryonic day 17.5, which contain oocytes in the midst of meiotic prophase, were harvested. Ovaries were in incubated in HEB on ice for 45–60 min with gentle stirring every 15 min. Then, a suspension of oocytes was generated by pulling and mashing an ovary between tweezer tips in 30 μL of sucrose (100 mM) on an uncharged microscope slide. 30 μL of the suspension was applied to positively charged slides and all subsequent steps are the same as those described in the previous paragraph.
For immunostaining experiments, testis and ovary chromosome spreads were incubated in PBS containing 0.1% Tween 20 (PBST) for 5–30 min before blocking in antibody dilution buffer (PBST containing 0.15% BSA) for an additional 30–60 min. Primary and secondary antibodies (described below) were diluted in antibody dilution buffer. Then, chromosome spreads were coated with 100 μL of the antibody/antibody dilution buffer solution, gently covered with Parafilm (Parafilm M All-Purpose Laboratory Film, Bemis Company, Inc.), and stored in “humid chambers” (described in [87]) at RT or 4°C for a minimum timespan of 6 h to a maximum timespan of overnight (~15 h). This study made use of the following primary antibodies at the following dilutions [format: host anti-protein (source or company with product/catalog number if applicable), dilution]: rabbit anti-H2AX-pY142 (Millipore, 07–1590), 1/200; rabbit anti-SYCP3 (Novus, NB300–232), 1/500; mouse anti-H2AX-pS139 (γH2AX: Millipore, 05–636), 1/5000; rabbit anti-SYCP1 (Abcam, ab15090), 1/200; mouse anti-SYCP3 (Abcam, ab97672), 1/5000; rabbit anti-TOPBP1 (gift from Dr. Junjie Chen [88]), 1/2000; rabbit anti-BRCA1 (generated in the Namekawa Lab [6]), 1/500; sheep anti-MDC1 (Bio-Rad, AHP799), 1/500; rabbit anti-RAD51 (Millipore, PC130), 1/50; guinea pig anti-H1T (gift from Dr. Mary Ann Handel [19]), 1/2000; rabbit anti-ATR (Millipore, PC538), 1/2000; rabbit anti-SIX6OS1 (gift from Dr. Alberto M. Pendás [89]), 1/4000; rabbit anti-HORMAD2 (gift from Dr. Attila Tóth [90]), 1/800; rabbit anti-MLH3 (gift from Dr. Paula E. Cohen [43]), 1/1000; rabbit anti-MLH1 (Santa Cruz, sc-11442), 1/500; mouse anti-SYCP3 conjugated with Alexa 488 fluorophore (Abcam, ab205846), 1/500 in Figure 5D, 5E; mouse anti-H2AX-pS139 (γH2AX) conjugated to Alexa 647 fluorophore (Millipore, 05–636-AF647), 1/500 in Figure 5D, 5E; rabbit anti-ATRIP (gift from Dr. Stephen J. Elledge [48]), 1/100. After incubation of the primary antibodies, slides were washed three times in PBST, 5 min per wash. Then, the slides were incubated with appropriate secondary antibodies conjugated to Alexa 488, 555, and/or 647 fluorophores (ThermoFisher Scientific). All secondary antibodies were diluted 1/500 in antibody dilution buffer. Slides were coated with 100 μL of the antibody/antibody dilution buffer solution; then, they were gently covered with Parafilm for 1-h incubation at RT in humid chambers in darkness. After slides were washed three times in PBST in darkness, 5 min per wash, they were counterstained with the DNA-binding chemical 4′,6-diamidino-2-phenylindole (DAPI; Sigma, D9542–5MG) diluted to 1 μg/mL concentration in PBS. Finally, slides were mounted using 20 μL undiluted ProLong Gold Antifade Mountant (ThermoFisher Scientific, P36930). Slides were either imaged immediately or stored at 4°C in darkness. For long-term storage, stained slides were kept at 4°C in darkness.
Images were obtained with an ECLIPSE Ti-E microscope (Nikon) equipped with a Zyla 5.5 sCMOS camera (Andor Technology) and an 60× CFI Apochromat TIRF oil immersion objective NA 1.4 (Nikon), and were processed with NIS-Elements Basic Research (Nikon), Photoshop (Adobe), and Illustrator (Adobe).
3D slide preparation and immunostaining
To conserve the gross conformation of meiotic chromatin, specialized slides that preserve the relative three-dimensional nuclear organization of testis germ cells (3D slides) were prepared as described [33]. Briefly, seminiferous tubules underwent permeabilization in cytoskeleton buffer [CSK buffer: 100 mM sodium chloride, 300 mM sucrose, 10 mM 1,4-Piperazinediethanesulfonic acid (PIPES), 3 mM magnesium chloride, 0.5% Triton X-100], fixation by 4% paraformaldehyde in PBS, and then mechanical dissociation with fine-point tweezers before being cytospun via specialized centrifuge (Shandon, Cytospin 2) onto positively charged slides (Superfrost Plus: ThermoFisher Scientific, 12-550-15). Immunostaining was performed following procedures described above. Briefly, 3D slides were blocked with antibody dilution buffer at RT for 30 min, then the following primary antibodies were diluted in antibody dilution buffer and applied to the slides: rabbit anti-BRCA1 antibody (generated in the Namekawa Lab), diluted 1/2000; mouse anti-RNA Polymerase II antibody (Millipore, 05–952), diluted 1/200. 3D slides were incubated at 4°C overnight in a humid chambers. On the following day, 3D slides were washed three times in PBST, 5 min per wash, before incubation with secondary antibodies conjugated to Alexa 488 or Alexa 555 fluorophores (ThermoFisher Scientific); the secondary antibodies were diluted 1/500 in antibody dilution buffer, then incubated at RT for 1 h in darkness. After washing the 3D slides three times in PBST, 5 min per wash, they were counterstained with DAPI as described above and then mounted using 20 μL undiluted ProLong Gold Antifade Mountant (ThermoFisher Scientific, P36930). Slides were imaged immediately thereafter.
Images were obtained with an ECLIPSE Ti-E microscope (Nikon) equipped with a Zyla 5.5 sCMOS camera (Andor Technology) and an 100× CFI Apochromat TIRF oil immersion objective NA 1.4 (Nikon), and were processed with NIS-Elements Basic Research (Nikon), Photoshop (Adobe), and Illustrator (Adobe).
Immunohistochemistry and TUNEL assay
For the preparation of testis paraffin blocks, excised testes in tunicae albugineae were fixed with 4% paraformaldehyde at 4°C overnight. Testes were dehydrated and embedded in paraffin. For histological analyses, 6 μm-thick paraffin sections were deparaffinized. Standard periodic acid-Schiff staining was performed; for immunostaining, sections were autoclaved in Target Retrieval Solution, Citrate pH 6.1 (DAKO, S-1700), 121°C, 100 kPa (15 psi) for 10 min. The sections were blocked with Blocking One Histo (Nacalai USA, 06349–64) at RT for 10 min; then, the sections were incubated with primary antibodies diluted in PBS at 4°C overnight. The following antibodies were used in the following dilutions [format: host anti-protein (source or company with product/catalog number), dilution]: mouse anti-γH2AX (Millipore, 05–636), 1/2500; guinea pig anti-H1T (gift from Dr. Mary Ann Handel), 1/2500; rabbit anti-BRCA1 (generated in the Namekawa Lab), 1/2000. The resulting signals were detected with appropriate secondary antibodies conjugated to Alexa 488 and 555 (ThermoFisher Scientific), diluted 1/1000 in PBS and incubated at RT for 1 h. TUNEL assays were performed using an In Situ Cell Death Detection Kit (Roche, 11684795910) per the instructions in the manual. Sections were counterstained with DAPI as described above. Images were obtained with an A1RSi Inverted Confocal Microscope (Nikon) and processed with NIS-Elements Basic Research (Nikon), Photoshop (Adobe), and Illustrator (Adobe).
QUANTIFICATION AND STATISTICAL ANALYSIS
The details for statistical analyses performed in this study are described in relevant portions of the Results section, figures, figure legends, supplementary figures, and/or supplementary figure legends. Sample sizes used for analyses are described in relevant portions of the Results section, figures, figure legends, supplementary figures, and/or supplementary figure legends. In predetermining sample sizes, we sought to analyze a minimum of three independent control-mutant littermate pairs except for western blot experiments, in which we analyzed two independent control-mutant littermate pairs; no statistical calculations were used to predetermine sample sizes. No data were excluded from analyses. The experiments were not randomized, and investigators were not blinded to allocation during experiments and outcome assessment. Measurements were recorded in Excel (Microsoft) and Prism 8.0 (GraphPad). Statistical tests were performed with Prism 8.0 (GraphPad).
Quantification of RAD51, TOPBP1, and MLH3 foci in the pachytene stage
To carefully detail the phenotypes of MSCI-deficient mice, we scored foci numbers for the proteins RAD51, TOPBP1, and MLH3 in wild-type and mutant spermatocytes in the initial portion of the mid pachytene stage. In H2ax-Y142A mice, spermatogenesis is arrested in epithelial stages IV, when tubules bear H1T-positive spermatocytes that have just entered the mid pachytene stage of meiotic prophase I. Thus, in our analyses of mutant meiotic chromosome spreads, we judged nuclei to be in the initial portion of the mid pachytene stage based on H1T staining—i.e., H1T-positive H2ax-Y142A nuclei were considered to be mid pachytene spermatocytes. However, we did not rely solely on H1T staining for identification of wild-type mid pachytene spermatocytes: Since all spermatocytes at and after the mid pachytene stage—as well as all spermiogenic cells—are H1T-positive, an H1T-positive status alone is insufficient for this task. To make direct comparisons with H1T-positive H2ax-Y142A spermatocytes, we precisely identified wild-type spermatocytes in the initial portion of the mid pachytene stage drawing on techniques described in our recent chapter in the Methods in Molecular Biology series (SpringerProtocols, Humana Press) [87]. Briefly, we focused on the following chromo-nuclear features to stage spermatocytes in the initial portion of the mid pachytene stage: (1) After immunostaining (described above), spermatocyte nuclei exhibited positive-but-weak H1T signal intensity relative to late pachytene and diplotene spermatocyte nuclei; (2) the pseudoautosomal region (PAR) of the sex chromosomes was clearly visible. After determining the substages of pachytene spermatocytes, we scored clearly visible RAD51, TOPBP1, and/or MLH3 foci overlapping SYCP3-labeled chromosomes axes.
Quantification of XY chromatin compaction
To evaluate chromatin compaction of the sex chromosomes, 3D slides stained with antibodies raised against BRCA1 and H1T, and counterstained with DAPI, were used as described [6]. Wild-type and H2ax-Y142A H1T-positive mid pachytene spermatocytes were evaluated. After obtaining 10 z-sections (0.2 μm interval) for each spermatocyte, we selected a z section containing in-focus XY centromeres for analysis. To quantify chromatin compaction of the sex chromosomes, a straight line was drawn to represent the widest spans of distance between the axial ends of the sex chromosomes as detected via anti-BRCA1 immunostaining (a) in H1T-positive nuclei; then, the widest spans were divided by the diameters of the nuclei (b). Distances were measured using NIS-Elements Basic Research (Nikon). The relative distances (i.e., the a/b distance ratios)in Figure 3 represent distances between XY centromeres with respect to nuclear diameters.
RNA-seq analysis
RNA-seq datasets were obtained from published work (GEO GSE55060) [70]. .fastq files were aligned to the Mus musculus mm10 reference genome via Spliced Transcripts Alignment to a Reference (STAR; version 2.4.2a)[91]; only unique alignments were allowed with a maximum of two errors per alignment. Datasets were processed and visualized through the BioWardrobe Experiment Management Platform [92].
DATA AND CODE AVAILABILITY
The RNA-seq datasets analyzed in this study are available at NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE55060. Software used in this study is described in context under portions of the METHOD DETAILS and/or QUANTIFICATION AND STATISTICAL ANALYSIS headings, and in the Key Resources Table. Software used for this study: NIS-Elements Basic Research (Nikon), Excel (Microsoft), Prism 8.0 (GraphPad), and the BioWardrobe Experiment Management Platform (biowardrobe.com, https://github.com/Barski-lab/biowardrobe) [92]. The manuscript was constructed using Word (Microsoft), Photoshop (Adobe), and Illustrator (Adobe).
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit Anti-Histone H2A.X, phospho (Tyr142) | Millipore | Cat# 07–1590, RRID:AB_1977237 |
| Rabbit Anti-Lamin B1 | Abcam | Cat# ab16048, RRID:AB_443298 |
| Mouse Anti-alpha Tubulin [DM1A] | Abcam | Cat# ab7291, RRID:AB_2241126 |
| Rabbit Anti-Histone H2A.X | Cell Signaling Technology | Cat# 2595, RRID:AB_10694556 |
| Mouse Anti-SYCP3 | Abcam | Cat# ab97672, RRID:AB_10678841 |
| Guinea pig Anti-H1t | [19] | N/A |
| Mouse Anti-Histone H2A.X, phospho (Ser139) | Millipore | Cat# 05–636, RRID:AB_309864 |
| Rabbit Anti-SYCP1 | Abcam | Cat# ab15090, RRID:AB_301636 |
| Rabbit Anti-TOPBP1 | [88] | N/A |
| Rabbit Anti-BRCA1 | [6] | N/A |
| Sheep Anti-Human MDC1 | Bio-Rad | Cat# AHP799, RRID:AB_323725 |
| Mouse Anti-RNA Polymerase II, C-Terminus Domain | Millipore | Cat# 05–952, RRID:AB_492629 |
| Rabbit Anti-Rad51 | Millipore | Cat# PC130, RRID:AB_2238184 |
| Rabbit Anti-ATR | Millipore | Cat# PC538, RRID:AB_2063178 |
| Mouse Anti-SCP3 conjugated with Alexa Fluor 488 | Abcam | Cat# ab205846, RRID: N/A |
| Mouse Anti-phospho Histone H2A.X (Ser139) conjugated with Alexa Fluor 647 | Millipore | Cat# 05–636-AF647, RRID: N/A |
| Rabbit Anti-MLH3 | [43] | N/A |
| Rabbit Anti-MLH1(H-300) | Santa Cruz Biotechnology | Cat# sc-11442, RRID:AB_2145332 |
| Rabbit Anti-ATRIP | [48] | N/A |
| Rabbit Anti- SIX6OS1 | [89] | N/A |
| Rabbit Anti-HORMAD2 | [90] | N/A |
| VeriBlot for IP Detection Reagent (HRP) | Abcam | N/A |
| Donkey Anti-Sheep IgG (H+L) Alexa Fluor 647 | Jackson ImmunoResearch Labs | Cat# 713-606-147, RRID:AB_2340752 |
| Goat Anti-Guinea Pig IgG (H+L) Alexa Fluor 555 | Thermo Fisher Scientific | Cat# A-21435, RRID:AB_2535856 |
| Donkey Anti-Mouse IgG (H+L) Alexa Fluor 555 | Thermo Fisher Scientific | Cat# A-31570, RRID:AB_2536180 |
| Donkey Anti-Mouse IgG (H+L) Alexa Fluor 488 | Thermo Fisher Scientific | Cat# A-21202, RRID:AB_141607 |
| Donkey Anti-Rabbit IgG (H+L) Alexa Fluor 555 | Thermo Fisher Scientific | Cat# A-31572, RRID:AB_162543 |
| Donkey Anti-Rabbit IgG (H+L) Alexa Fluor 488 | Thermo Fisher Scientific | Cat# A-21206, RRID:AB_2535792 |
| Donkey Anti-Sheep IgG (H+L) DyLight 488 | Jackson ImmunoResearch Labs | Cat# 713-486-147, RRID:AB_2340741 |
| Goat Anti-Rabbit IgG (H+L) antibody F(ab’)2 Fragment Cy3 | Jackson ImmunoResearch Labs | Cat# 111-166-003, RRID:AB_2338007 |
| Donkey Anti-Guinea Pig IgG (H+L) antibody F(ab’)2 Fragment Alexa Fluor 647 | Jackson ImmunoResearch Labs | Cat# 706-606-148, RRID:AB_2340477 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Phosphatase inhibitor cocktail 2 | Sigma | Cat# P5726–5ML |
| cOmplete™ Protease Inhibitor Cocktail | Sigma | Cat# 11836145001 |
| StartingBlockTM T20 (TBS) Blocking Buffer | ThermoFisher Scientific | Cat# 37543 |
| Restore™ Western Blot Stripping Buffer | ThermoFisher Scientific | Cat# 21059 |
| Critical Commercial Assays | ||
| In Situ Cell Death Detection Kit, Fluorescein | Sigma | Cat# 11684795910 |
| Deposited Data | ||
| RNA-seq data (Scml2KO) | [70] | GEO: GSE55060 |
| Experimental Models: Organisms/Strains | ||
| Mouse: H2ax-Y142A | This study | N/A |
| Mouse: Mdc1KO | [52] | N/A |
| Oligonucleotides | ||
| Primer: H2ax-Y142A WT Forward: CGC AGG CCT CTC AGG AGT | This study | N/A |
| Primer: H2ax-Y142A KI Forward: CGC AGG CCT CTC AGG AGG CT | This study | N/A |
| Primer: H2ax-Y142A Common Reverse: CTG CGG AGG GAC TAA CCT TC | This study | N/A |
| sgRNA for generating H2ax-Y142A: GCCTCTCAGGAGTACTGAGG | This study | N/A |
| Software and Algorithms | ||
| BioWardrobe Experiment Management Platform | [92] | https://biowardrobe.cchmc.org/frontend/Home |
| Prism 8 | GraphPad software | https://www.graphpad.com/ |
Supplementary Material
Highlights.
The Y142 residue of H2AX is essential for MSCI and XY-body formation.
The initial steps of autosomal DSB repair are normal in MSCI-defective mutants.
ATR-associated DDR signals are sequestered from autosomes at the onset of MSCI.
Novel model for the MSCI checkpoint: MSCI permits meiotic progression.
Acknowledgements
We thank all members of the Namekawa laboratory for discussion and helpful comments. We thank Dr. Junjie Chen for providing the Mdc1KO mice and the anti-TOPBP1 antibody, Dr. Mary Ann Handel for providing the anti-H1T antibody, Dr. Paula E. Cohen for providing the anti-MLH3 antibody, Dr. Stephen J. Elledge for providing the anti- ATRIP antibody, Dr. Alberto M. Pendás for providing the anti-SIX6OS1 antibody, and Dr. Attila Tóth for providing the anti-HORMAD2 antibody. We thank the Transgenic Animal and Genome Editing Core at CCHMC for generating the H2ax-Y142A mice. This work was supported by NIH Grants R01HD089932 to Q.P., R01GM134731 to P.R.A., R01CA207068 to R.S.H., and R01GM098605 to S.H.N.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
The authors declare no competing interests.
References
- 1.Subramanian VV, and Hochwagen A (2014). The meiotic checkpoint network: step-by-step through meiotic prophase. Cold Spring Harb Perspect Biol 6, a016675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Handel MA, and Schimenti JC (2010). Genetics of mammalian meiosis: regulation, dynamics and impact on fertility. Nat Rev Genet 11, 124–136. [DOI] [PubMed] [Google Scholar]
- 3.Ichijima Y, Sin HS, and Namekawa SH (2012). Sex chromosome inactivation in germ cells: emerging roles of DNA damage response pathways. Cell Mol Life Sci 69, 2559–2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Turner JM (2015). Meiotic Silencing in Mammals. Annu Rev Genet 49, 395–412. [DOI] [PubMed] [Google Scholar]
- 5.Fernandez-Capetillo O, Mahadevaiah SK, Celeste A, Romanienko PJ, Camerini-Otero RD, Bonner WM, Manova K, Burgoyne P, and Nussenzweig A (2003). H2AX is required for chromatin remodeling and inactivation of sex chromosomes in male mouse meiosis. Dev Cell 4, 497–508. [DOI] [PubMed] [Google Scholar]
- 6.Ichijima Y, Ichijima M, Lou Z, Nussenzweig A, Camerini-Otero RD, Chen J, Andreassen PR, and Namekawa SH (2011). MDC1 directs chromosome-wide silencing of the sex chromosomes in male germ cells. Genes Dev 25, 959–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Turner JM, Mahadevaiah SK, Fernandez-Capetillo O, Nussenzweig A, Xu X, Deng CX, and Burgoyne PS (2005). Silencing of unsynapsed meiotic chromosomes in the mouse. Nature genetics 37, 41–47. [DOI] [PubMed] [Google Scholar]
- 8.Baarends WM, Wassenaar E, van der Laan R, Hoogerbrugge J, Sleddens-Linkels E, Hoeijmakers JH, de Boer P, and Grootegoed JA (2005). Silencing of unpaired chromatin and histone H2A ubiquitination in mammalian meiosis. Mol Cell Biol 25, 1041–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schimenti J (2005). Synapsis or silence. Nature genetics 37, 11–13. [DOI] [PubMed] [Google Scholar]
- 10.Burgoyne PS, Mahadevaiah SK, and Turner JM (2009). The consequences of asynapsis for mammalian meiosis. Nat Rev Genet 10, 207–216. [DOI] [PubMed] [Google Scholar]
- 11.Bellani MA, Romanienko PJ, Cairatti DA, and Camerini-Otero RD (2005). SPO11 is required for sex-body formation, and Spo11 heterozygosity rescues the prophase arrest of Atm−/− spermatocytes. J Cell Sci 118, 3233–3245. [DOI] [PubMed] [Google Scholar]
- 12.Perera D, Perez-Hidalgo L, Moens PB, Reini K, Lakin N, Syvaoja JE, San-Segundo PA, and Freire R (2004). TopBP1 and ATR colocalization at meiotic chromosomes: role of TopBP1/Cut5 in the meiotic recombination checkpoint. Mol Biol Cell 15, 1568–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Turner JM, Aprelikova O, Xu X, Wang R, Kim S, Chandramouli GV, Barrett JC, Burgoyne PS, and Deng CX (2004). BRCA1, histone H2AX phosphorylation, and male meiotic sex chromosome inactivation. Curr Biol 14, 2135–2142. [DOI] [PubMed] [Google Scholar]
- 14.Royo H, Prosser H, Ruzankina Y, Mahadevaiah SK, Cloutier JM, Baumann M, Fukuda T, Hoog C, Toth A, de Rooij DG, et al. (2013). ATR acts stage specifically to regulate multiple aspects of mammalian meiotic silencing. Genes Dev 27, 1484–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Broering TJ, Alavattam KG, Sadreyev RI, Ichijima Y, Kato Y, Hasegawa K, Camerini-Otero RD, Lee JT, Andreassen PR, and Namekawa SH (2014). BRCA1 establishes DNA damage signaling and pericentric heterochromatin of the X chromosome in male meiosis. The Journal of cell biology 205, 663–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xiao A, Li H, Shechter D, Ahn SH, Fabrizio LA, Erdjument-Bromage H, Ishibe-Murakami S, Wang B, Tempst P, Hofmann K, et al. (2009). WSTF regulates the H2A.X DNA damage response via a novel tyrosine kinase activity. Nature 457, 57–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cook PJ, Ju BG, Telese F, Wang X, Glass CK, and Rosenfeld MG (2009). Tyrosine dephosphorylation of H2AX modulates apoptosis and survival decisions. Nature 458, 591–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Broering TJ, Wang YL, Pandey RN, Hegde RS, Wang SC, and Namekawa SH (2015). BAZ1B is dispensable for H2AX phosphorylation on Tyrosine 142 during spermatogenesis. Biol Open 4, 873–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Inselman A, Eaker S, and Handel MA (2003). Temporal expression of cell cycle-related proteins during spermatogenesis: establishing a timeline for onset of the meiotic divisions. Cytogenet Genome Res 103, 277–284. [DOI] [PubMed] [Google Scholar]
- 20.Sirbu BM, Couch FB, Feigerle JT, Bhaskara S, Hiebert SW, and Cortez D (2011). Analysis of protein dynamics at active, stalled, and collapsed replication forks. Genes Dev 25, 1320–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mahadevaiah SK, Turner JM, Baudat F, Rogakou EP, de Boer P, Blanco-Rodriguez J, Jasin M, Keeney S, Bonner WM, and Burgoyne PS (2001). Recombinational DNA double-strand breaks in mice precede synapsis. Nature genetics 27, 271–276. [DOI] [PubMed] [Google Scholar]
- 22.Celeste A, Petersen S, Romanienko PJ, Fernandez-Capetillo O, Chen HT, Sedelnikova OA, Reina-San-Martin B, Coppola V, Meffre E, Difilippantonio MJ, et al. (2002). Genomic instability in mice lacking histone H2AX. Science 296, 922–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim S, Peterson SE, Jasin M, and Keeney S (2016). Mechanisms of germ line genome instability. Semin Cell Dev Biol 54, 177–187. [DOI] [PubMed] [Google Scholar]
- 24.Hunter N (2015). Meiotic Recombination: The Essence of Heredity. Cold Spring Harb Perspect Biol 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gray S, and Cohen PE (2016). Control of Meiotic Crossovers: From Double-Strand Break Formation to Designation. Annu Rev Genet 50, 175–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang J, Gong Z, and Chen J (2011). MDC1 collaborates with TopBP1 in DNA replication checkpoint control. The Journal of cell biology 193, 267–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reini K, Uitto L, Perera D, Moens PB, Freire R, and Syvaoja JE (2004). TopBP1 localises to centrosomes in mitosis and to chromosome cores in meiosis. Chromosoma 112, 323–330. [DOI] [PubMed] [Google Scholar]
- 28.ElInati E, Russell HR, Ojarikre OA, Sangrithi M, Hirota T, de Rooij DG, McKinnon PJ, and Turner JMA (2017). DNA damage response protein TOPBP1 regulates X chromosome silencing in the mammalian germ line. Proc Natl Acad Sci U S A 114, 12536–12541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gomez HL, Felipe-Medina N, Sanchez-Martin M, Davies OR, Ramos I, Garcia-Tunon I, de Rooij DG, Dereli I, Toth A, Barbero JL, et al. (2016). C14ORF39/SIX6OS1 is a constituent of the synaptonemal complex and is essential for mouse fertility. Nat Commun 7, 13298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Testa E, Nardozi D, Antinozzi C, Faieta M, Di Cecca S, Caggiano C, Fukuda T, Bonanno E, Zhenkun L, Maldonado A, et al. (2018). H2AFX and MDC1 promote maintenance of genomic integrity in male germ cells. J Cell Sci 131 jcs214411. [DOI] [PubMed] [Google Scholar]
- 31.Wojtasz L, Cloutier JM, Baumann M, Daniel K, Varga J, Fu J, Anastassiadis K, Stewart AF, Remenyi A, Turner JM, et al. (2012). Meiotic DNA double-strand breaks and chromosome asynapsis in mice are monitored by distinct HORMAD2-independent and -dependent mechanisms. Genes Dev 26, 958–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Namekawa SH, Park PJ, Zhang LF, Shima JE, McCarrey JR, Griswold MD, and Lee JT (2006). Postmeiotic sex chromatin in the male germline of mice. Curr Biol 16, 660–667. [DOI] [PubMed] [Google Scholar]
- 33.Namekawa SH (2014). Slide preparation method to preserve three-dimensional chromatin architecture of testicular germ cells. J Vis Exp, e50819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Royo H, Polikiewicz G, Mahadevaiah SK, Prosser H, Mitchell M, Bradley A, de Rooij DG, Burgoyne PS, and Turner JMA (2010). Evidence that Meiotic Sex Chromosome Inactivation Is Essential for Male Fertility. Curr Biol 20, 2117–2123. [DOI] [PubMed] [Google Scholar]
- 35.Pacheco S, Marcet-Ortega M, Lange J, Jasin M, Keeney S, and Roig I (2015). The ATM signaling cascade promotes recombination-dependent pachytene arrest in mouse spermatocytes. PLoS genetics 11, e1005017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marcet-Ortega M, Pacheco S, Martinez-Marchal A, Castillo H, Flores E, Jasin M, Keeney S, and Roig I (2017). p53 and TAp63 participate in the recombination-dependent pachytene arrest in mouse spermatocytes. PLoS genetics 13, e1006845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Haaf T, Golub EI, Reddy G, Radding CM, and Ward DC (1995). Nuclear foci of mammalian Rad51 recombination protein in somatic cells after DNA damage and its localization in synaptonemal complexes. Proc Natl Acad Sci U S A 92, 2298–2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Plug AW, Peters AH, Keegan KS, Hoekstra MF, de Boer P, and Ashley T (1998). Changes in protein composition of meiotic nodules during mammalian meiosis. J Cell Sci 111 (Pt 4), 413–423. [DOI] [PubMed] [Google Scholar]
- 39.Baker SM, Plug AW, Prolla TA, Bronner CE, Harris AC, Yao X, Christie DM, Monell C, Arnheim N, Bradley A, et al. (1996). Involvement of mouse Mlh1 in DNA mismatch repair and meiotic crossing over. Nature genetics 13, 336–342. [DOI] [PubMed] [Google Scholar]
- 40.Kolas NK, Svetlanov A, Lenzi ML, Macaluso FP, Lipkin SM, Liskay RM, Greally J, Edelmann W, and Cohen PE (2005). Localization of MMR proteins on meiotic chromosomes in mice indicates distinct functions during prophase I. The Journal of cell biology 171, 447–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Abe H, Alavattam KG, Kato Y, Castrillon DH, Pang QS, Andreassen PR, and Namekawa SH (2018). CHEK1 coordinates DNA damage signaling and meiotic progression in the male germline of mice. Hum Mol Genet 27, 1136–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alavattam KG, Abe H, Sakashita A, and Namekawa SH (2018). Chromosome Spread Analyses of Meiotic Sex Chromosome Inactivation. Methods Mol Biol 1861, 113–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lipkin SM, Moens PB, Wang V, Lenzi M, Shanmugarajah D, Gilgeous A, Thomas J, Cheng J, Touchman JW, Green ED, et al. (2002). Meiotic arrest and aneuploidy in MLH3-deficient mice. Nature genetics 31, 385–390. [DOI] [PubMed] [Google Scholar]
- 44.Bunting SF, Callen E, Kozak ML, Kim JM, Wong N, Lopez-Contreras AJ, Ludwig T, Baer R, Faryabi RB, Malhowski A, et al. (2012). BRCA1 functions independently of homologous recombination in DNA interstrand crosslink repair. Mol Cell 46, 125–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kogo H, Tsutsumi M, Inagaki H, Ohye T, Kiyonari H, and Kurahashi H (2012). HORMAD2 is essential for synapsis surveillance during meiotic prophase via the recruitment of ATR activity. Genes Cells 17, 897–912. [DOI] [PubMed] [Google Scholar]
- 46.Mahadevaiah SK, Bourc’his D, de Rooij DG, Bestor TH, Turner JM, and Burgoyne PS (2008). Extensive meiotic asynapsis in mice antagonises meiotic silencing of unsynapsed chromatin and consequently disrupts meiotic sex chromosome inactivation. The Journal of cell biology 182, 263–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kumagai A, Lee J, Yoo HY, and Dunphy WG (2006). TopBP1 activates the ATR-ATRIP complex. Cell 124, 943–955. [DOI] [PubMed] [Google Scholar]
- 48.Cortez D, Guntuku S, Qin J, and Elledge SJ (2001). ATR and ATRIP: partners in checkpoint signaling. Science 294, 1713–1716. [DOI] [PubMed] [Google Scholar]
- 49.Refolio E, Cavero S, Marcon E, Freire R, and San-Segundo PA (2011). The Ddc2/ATRIP checkpoint protein monitors meiotic recombination intermediates. J Cell Sci 124, 2488–2500. [DOI] [PubMed] [Google Scholar]
- 50.Stucki M, Clapperton JA, Mohammad D, Yaffe MB, Smerdon SJ, and Jackson SP (2005). MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell 123, 1213–1226. [DOI] [PubMed] [Google Scholar]
- 51.Xie A, Odate S, Chandramouly G, and Scully R (2010). H2AX post-translational modifications in the ionizing radiation response and homologous recombination. Cell cycle (Georgetown, Tex.) 9, 3602–3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lou Z, Minter-Dykhouse K, Franco S, Gostissa M, Rivera MA, Celeste A, Manis JP, van Deursen J, Nussenzweig A, Paull TT, et al. (2006). MDC1 maintains genomic stability by participating in the amplification of ATM-dependent DNA damage signals. Mol Cell 21, 187–200. [DOI] [PubMed] [Google Scholar]
- 53.Lange J, Pan J, Cole F, Thelen MP, Jasin M, and Keeney S (2011). ATM controls meiotic double-strand-break formation. Nature 479, 237–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Roig I, Dowdle JA, Toth A, de Rooij DG, Jasin M, and Keeney S (2010). Mouse TRIP13/PCH2 Is Required for Recombination and Normal Higher-Order Chromosome Structure during Meiosis. PLoS genetics 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li XC, and Schimenti JC (2007). Mouse pachytene checkpoint 2 (trip 13) is required for completing meiotic recombination but not synapsis. PLoS genetics 3, e130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xie A, Puget N, Shim I, Odate S, Jarzyna I, Bassing CH, Alt FW, and Scully R (2004). Control of sister chromatid recombination by histone H2AX. Mol Cell 16, 1017–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McKee BD, and Handel MA (1993). Sex chromosomes, recombination, and chromatin conformation. Chromosoma 102, 71–80. [DOI] [PubMed] [Google Scholar]
- 58.van Vugt MA, Bras A, and Medema RH (2005). Restarting the cell cycle when the checkpoint comes to a halt. Cancer Res 65, 7037–7040. [DOI] [PubMed] [Google Scholar]
- 59.Barnum KJ, and O’Connell MJ (2014). Cell cycle regulation by checkpoints. Methods Mol Biol 1170, 29–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Daniel K, Lange J, Hached K, Fu J, Anastassiadis K, Roig I, Cooke HJ, Stewart AF, Wassmann K, Jasin M, et al. (2011). Meiotic homologue alignment and its quality surveillance are controlled by mouse HORMAD1. Nat Cell Biol 13, 599–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Heffernan TP, Simpson DA, Frank AR, Heinloth AN, Paules RS, Cordeiro-Stone M, and Kaufmann WK (2002). An ATR- and Chk1-dependent S checkpoint inhibits replicon initiation following UVC-induced DNA damage. Mol Cell Biol 22, 8552–8561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brown EJ, and Baltimore D (2003). Essential and dispensable roles of ATR in cell cycle arrest and genome maintenance. Genes Dev 17, 615–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bolcun-Filas E, Rinaldi VD, White ME, and Schimenti JC (2014). Reversal of female infertility by Chk2 ablation reveals the oocyte DNA damage checkpoint pathway. Science 343, 533–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Turner JM, Mahadevaiah SK, Ellis PJ, Mitchell MJ, and Burgoyne PS (2006). Pachytene asynapsis drives meiotic sex chromosome inactivation and leads to substantial postmeiotic repression in spermatids. Dev Cell 10, 521–529. [DOI] [PubMed] [Google Scholar]
- 65.Manterola M, Page J, Vasco C, Berrios S, Parra MT, Viera A, Rufas JS, Zuccotti M, Garagna S, and Fernandez-Donoso R (2009). A high incidence of meiotic silencing of unsynapsed chromatin is not associated with substantial pachytene loss in heterozygous male mice carrying multiple simple robertsonian translocations. PLoS genetics 5, e1000625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pacheco S, Maldonado-Linares A, Marcet-Ortega M, Rojas C, Martinez-Marchal A, Fuentes-Lazaro J, Lange J, Jasin M, Keeney S, Fernandez-Capetillo O, et al. (2018). ATR is required to complete meiotic recombination in mice. Nat Commun 9, 2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Widger A, Mahadevaiah SK, Lange J, ElInati E, Zohren J, Hirota T, Pacheco S, Maldonado-Linares A, Stanzione M, Ojarikre O, et al. (2018). ATR is a multifunctional regulator of male mouse meiosis. Nat Commun 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Modzelewski AJ, Holmes RJ, Hilz S, Grimson A, and Cohen PE (2012). AGO4 regulates entry into meiosis and influences silencing of sex chromosomes in the male mouse germline. Dev Cell 23, 251–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sin HS, Barski A, Zhang F, Kartashov AV, Nussenzweig A, Chen J, Andreassen PR, and Namekawa SH (2012). RNF8 regulates active epigenetic modifications and escape gene activation from inactive sex chromosomes in post-meiotic spermatids. Genes Dev 26, 2737–2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hasegawa K, Sin HS, Maezawa S, Broering TJ, Kartashov AV, Alavattam KG, Ichijima Y, Zhang F, Bacon WC, Greis KD, et al. (2015). SCML2 establishes the male germline epigenome through regulation of histone H2A ubiquitination. Dev Cell 32, 574–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kato Y, Alavattam KG, Sin HS, Meetei AR, Pang Q, Andreassen PR, and Namekawa SH (2015). FANCB is essential in the male germline and regulates H3K9 methylation on the sex chromosomes during meiosis. Hum Mol Genet 24, 5234–5249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Alavattam KG, Maezawa S, Sakashita A, Khoury H, Barski A, Kaplan N, and Namekawa SH (2019). Attenuated chromatin compartmentalization in meiosis and its maturation in sperm development. Nat Struct Mol Biol 26, 175–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bergeron-Sandoval LP, Safaee N, and Michnick SW (2016). Mechanisms and Consequences of Macromolecular Phase Separation. Cell 165, 1067–1079. [DOI] [PubMed] [Google Scholar]
- 74.Banani SF, Lee HO, Hyman AA, and Rosen MK (2017). Biomolecular condensates: organizers of cellular biochemistry. Nature reviews. Molecular cell biology 18, 285–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vernet N, Mahadevaiah SK, Yamauchi Y, Decarpentrie F, Mitchell MJ, Ward MA, and Burgoyne PS (2014). Mouse Y-linked Zfy1 and Zfy2 are expressed during the male-specific interphase between meiosis I and meiosis II and promote the 2nd meiotic division. PLoS genetics 10, e1004444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vernet N, Mahadevaiah SK, de Rooij DG, Burgoyne PS, and Ellis PJI (2016). Zfy genes are required for efficient meiotic sex chromosome inactivation (MSCI) in spermatocytes. Hum Mol Genet 25, 5300–5310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nakasuji T, Ogonuki N, Chiba T, Kato T, Shiozawa K, Yamatoya K, Tanaka H, Kondo T, Miyado K, Miyasaka N, et al. (2017). Complementary Critical Functions of Zfy1 and Zfy2 in Mouse Spermatogenesis and Reproduction. PLoS genetics 13, e1006578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Heard E, and Turner J (2011). Function of the sex chromosomes in mammalian fertility. Cold Spring Harb Perspect Biol 3, a002675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yuan CL, and Hu YC (2017). A Transgenic Core Facility’s Experience in Genome Editing Revolution. Adv Exp Med Biol 1016, 75–90. [DOI] [PubMed] [Google Scholar]
- 80.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, and Zhang F (2013). Genome engineering using the CRISPR-Cas9 system. Nat Protoc 8, 2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chen B, Gilbert LA, Cimini BA, Schnitzbauer J, Zhang W, Li GW, Park J, Blackburn EH, Weissman JS, Qi LS, et al. (2013). Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell 155, 1479–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Slaymaker IM, Gao L, Zetsche B, Scott DA, Yan WX, and Zhang F (2016). Rationally engineered Cas9 nucleases with improved specificity. Science 351, 84–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Valerius MT, Patterson LT, Witte DP, and Potter SS (2002). Microarray analysis of novel cell lines representing two stages of metanephric mesenchyme differentiation. Mech Dev 110, 151–164. [DOI] [PubMed] [Google Scholar]
- 84.Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, and Jaenisch R (2013). One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 153, 910–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yang H, Wang H, and Jaenisch R (2014). Generating genetically modified mice using CRISPR/Cas-mediated genome engineering. Nat Protoc 9, 1956–1968. [DOI] [PubMed] [Google Scholar]
- 86.Peters AH, Plug AW, van Vugt MJ, and de Boer P (1997). A drying-down technique for the spreading of mammalian meiocytes from the male and female germline. Chromosome Res 5, 66–68. [DOI] [PubMed] [Google Scholar]
- 87.Alavattam KG, Kato Y, Sin HS, Maezawa S, Kowalski IJ, Zhang F, Pang Q, Andreassen PR, and Namekawa SH (2016). Elucidation of the Fanconi Anemia Protein Network in Meiosis and Its Function in the Regulation of Histone Modifications. Cell Rep 17, 1141–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kim JE, McAvoy SA, Smith DI, and Chen J (2005). Human TopBP1 ensures genome integrity during normal S phase. Mol Cell Biol 25, 10907–10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gomez L, Felipe-Medina N, Sanchez-Martin M, Davies OR, Ramos I, Garcia-Tunon I, de Rooij DG, Dereli I, Toth A, Barbero JL, et al. (2016). C14ORF39/SIX6OS1 is a constituent of the synaptonemal complex and is essential for mouse fertility. Nat Commun 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wojtasz L, Daniel K, Roig I, Bolcun-Filas E, Xu H, Boonsanay V, Eckmann CR, Cooke HJ, Jasin M, Keeney S, et al. (2009). Mouse HORMAD1 and HORMAD2, two conserved meiotic chromosomal proteins, are depleted from synapsed chromosome axes with the help of TRIP13 AAA-ATPase. PLoS genetics 5, e1000702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kartashov AV, and Barski A (2015). BioWardrobe: an integrated platform for analysis of epigenomics and transcriptomics data. Genome biology 16, 158. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq datasets analyzed in this study are available at NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE55060. Software used in this study is described in context under portions of the METHOD DETAILS and/or QUANTIFICATION AND STATISTICAL ANALYSIS headings, and in the Key Resources Table. Software used for this study: NIS-Elements Basic Research (Nikon), Excel (Microsoft), Prism 8.0 (GraphPad), and the BioWardrobe Experiment Management Platform (biowardrobe.com, https://github.com/Barski-lab/biowardrobe) [92]. The manuscript was constructed using Word (Microsoft), Photoshop (Adobe), and Illustrator (Adobe).
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit Anti-Histone H2A.X, phospho (Tyr142) | Millipore | Cat# 07–1590, RRID:AB_1977237 |
| Rabbit Anti-Lamin B1 | Abcam | Cat# ab16048, RRID:AB_443298 |
| Mouse Anti-alpha Tubulin [DM1A] | Abcam | Cat# ab7291, RRID:AB_2241126 |
| Rabbit Anti-Histone H2A.X | Cell Signaling Technology | Cat# 2595, RRID:AB_10694556 |
| Mouse Anti-SYCP3 | Abcam | Cat# ab97672, RRID:AB_10678841 |
| Guinea pig Anti-H1t | [19] | N/A |
| Mouse Anti-Histone H2A.X, phospho (Ser139) | Millipore | Cat# 05–636, RRID:AB_309864 |
| Rabbit Anti-SYCP1 | Abcam | Cat# ab15090, RRID:AB_301636 |
| Rabbit Anti-TOPBP1 | [88] | N/A |
| Rabbit Anti-BRCA1 | [6] | N/A |
| Sheep Anti-Human MDC1 | Bio-Rad | Cat# AHP799, RRID:AB_323725 |
| Mouse Anti-RNA Polymerase II, C-Terminus Domain | Millipore | Cat# 05–952, RRID:AB_492629 |
| Rabbit Anti-Rad51 | Millipore | Cat# PC130, RRID:AB_2238184 |
| Rabbit Anti-ATR | Millipore | Cat# PC538, RRID:AB_2063178 |
| Mouse Anti-SCP3 conjugated with Alexa Fluor 488 | Abcam | Cat# ab205846, RRID: N/A |
| Mouse Anti-phospho Histone H2A.X (Ser139) conjugated with Alexa Fluor 647 | Millipore | Cat# 05–636-AF647, RRID: N/A |
| Rabbit Anti-MLH3 | [43] | N/A |
| Rabbit Anti-MLH1(H-300) | Santa Cruz Biotechnology | Cat# sc-11442, RRID:AB_2145332 |
| Rabbit Anti-ATRIP | [48] | N/A |
| Rabbit Anti- SIX6OS1 | [89] | N/A |
| Rabbit Anti-HORMAD2 | [90] | N/A |
| VeriBlot for IP Detection Reagent (HRP) | Abcam | N/A |
| Donkey Anti-Sheep IgG (H+L) Alexa Fluor 647 | Jackson ImmunoResearch Labs | Cat# 713-606-147, RRID:AB_2340752 |
| Goat Anti-Guinea Pig IgG (H+L) Alexa Fluor 555 | Thermo Fisher Scientific | Cat# A-21435, RRID:AB_2535856 |
| Donkey Anti-Mouse IgG (H+L) Alexa Fluor 555 | Thermo Fisher Scientific | Cat# A-31570, RRID:AB_2536180 |
| Donkey Anti-Mouse IgG (H+L) Alexa Fluor 488 | Thermo Fisher Scientific | Cat# A-21202, RRID:AB_141607 |
| Donkey Anti-Rabbit IgG (H+L) Alexa Fluor 555 | Thermo Fisher Scientific | Cat# A-31572, RRID:AB_162543 |
| Donkey Anti-Rabbit IgG (H+L) Alexa Fluor 488 | Thermo Fisher Scientific | Cat# A-21206, RRID:AB_2535792 |
| Donkey Anti-Sheep IgG (H+L) DyLight 488 | Jackson ImmunoResearch Labs | Cat# 713-486-147, RRID:AB_2340741 |
| Goat Anti-Rabbit IgG (H+L) antibody F(ab’)2 Fragment Cy3 | Jackson ImmunoResearch Labs | Cat# 111-166-003, RRID:AB_2338007 |
| Donkey Anti-Guinea Pig IgG (H+L) antibody F(ab’)2 Fragment Alexa Fluor 647 | Jackson ImmunoResearch Labs | Cat# 706-606-148, RRID:AB_2340477 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Phosphatase inhibitor cocktail 2 | Sigma | Cat# P5726–5ML |
| cOmplete™ Protease Inhibitor Cocktail | Sigma | Cat# 11836145001 |
| StartingBlockTM T20 (TBS) Blocking Buffer | ThermoFisher Scientific | Cat# 37543 |
| Restore™ Western Blot Stripping Buffer | ThermoFisher Scientific | Cat# 21059 |
| Critical Commercial Assays | ||
| In Situ Cell Death Detection Kit, Fluorescein | Sigma | Cat# 11684795910 |
| Deposited Data | ||
| RNA-seq data (Scml2KO) | [70] | GEO: GSE55060 |
| Experimental Models: Organisms/Strains | ||
| Mouse: H2ax-Y142A | This study | N/A |
| Mouse: Mdc1KO | [52] | N/A |
| Oligonucleotides | ||
| Primer: H2ax-Y142A WT Forward: CGC AGG CCT CTC AGG AGT | This study | N/A |
| Primer: H2ax-Y142A KI Forward: CGC AGG CCT CTC AGG AGG CT | This study | N/A |
| Primer: H2ax-Y142A Common Reverse: CTG CGG AGG GAC TAA CCT TC | This study | N/A |
| sgRNA for generating H2ax-Y142A: GCCTCTCAGGAGTACTGAGG | This study | N/A |
| Software and Algorithms | ||
| BioWardrobe Experiment Management Platform | [92] | https://biowardrobe.cchmc.org/frontend/Home |
| Prism 8 | GraphPad software | https://www.graphpad.com/ |


