Abstract
The Golgi apparatus is a central intracellular membrane-bound organelle with key functions in trafficking, processing, and sorting of newly synthesized membrane and secretory proteins and lipids. To best perform these functions, Golgi membranes form a unique stacked structure. The Golgi structure is dynamic but tightly regulated; it undergoes rapid disassembly and reassembly during the cell cycle of mammalian cells and is disrupted under certain stress and pathological conditions. In the past decade, significant amount of effort has been made to reveal the molecular mechanisms that regulate the Golgi membrane architecture and function. Here we review the major discoveries in the mechanisms of Golgi structure formation, regulation, and alteration in relation to its functions in physiological and pathological conditions to further our understanding of Golgi structure and function in health and diseases.
Golgi Architecture and Its Maintenance
The Golgi apparatus is a central intracellular membrane-bound organelle often located adjacent to the nucleus in mammalian cells. Electron microscope (EM) images revealed its unique feature as stacks of five to seven flattened cisternae overlaying one another, with multiple stacks often lined up and interconnected by tubular structures to form a ribbon (Shorter and Warren 2002; Rabouille and Kondylis 2007; Wei and Seemann 2010). The Golgi stacks are polarized; they receive proteins and lipids from the endoplasmic reticulum (ER) by the cis cisternae and export them from the trans cisternae and the trans-Golgi network (TGN) to other intracellular membranes such as the endosomes, lysosomes, plasma membrane, and outside of the cell (Tang and Wang 2013; Wang and Seemann 2011). While traversing the Golgi stack, cargo molecules are modified and processed. The sub-compartments of the Golgi stacks house a set of glycosidases and glycosyltransferases responsible for the synthesis of glycoproteins and glycolipids. In the TGN, many secretory proteins are proteolytically cleaved by lumenal proteinases (Huang and Wang 2017; Zhang and Wang 2016; Huttner et al. 1995).
Many efforts have been made to understand the mechanism of Golgi structure formation. The formation of the Golgi ribbon depends on Golgi matrix proteins and an intact microtubule organization. Cytosolic dynein moves Golgi membranes along centrosome-derived microtubules toward the (–) end of the microtubules (Matteis et al. 2008; Rabouille and Kondylis 2007; Wei and Seemann 2010). Subsequently, Golgi-oriented microtubules maintain Golgi stacks in the proximity and facilitate tubular connections between them (Zhu and Kaverina 2013). While dynein and microtubules are required for the concentration of Golgi stacks in the pericentrosomal region and Golgi ribbon formation, they are not essential for the generation and maintenance of the stacked structure, as depolymerization of microtubules disrupts the Golgi ribbon but not the stacks (Thyberg and Moskalewski 1999). From the 1960s, morphological studies have shown connections in the space between cisternae that might be involved in the adhesion of cisternae into stacks (Franke et al. 1972; Mollenhauer 1965; Cluett and Brown 1992), which were later identified as Golgi matrix proteins. These include Golgi stacking proteins and membrane tethers, as discussed below.
Golgi Matrix Proteins and Golgi Structure Formation
In 1994, the concept of “Golgi matrix” proteins was first introduced (Slusarewicz et al. 1994). Since then, several Golgi matrix proteins have been identified to be responsible for maintaining the unique architecture and function of the Golgi apparatus. Major components of the Golgi matrix are summarized in Table 19.1. Key proteins involved in Golgi structure formation are discussed below.
Table 19.1.
Major components of the Golgi matrix
| Names | Golgi localization | Membrane anchor | Associated GTPases | Interactions | Functions |
|---|---|---|---|---|---|
| GRASPs | |||||
| GRASP55/GORASP2 | medial/trans | N-myr (Shorter et al. 1999) | Rab2 (Barr 2005; Short et al. 2001) |
– Golgin-45 (Short et al. 2001; Zhao et al. 2017) – p24 (Barr et al. 2001) – TGF-α (Kuo et al. 2000) – LC3, LAMP2 (Zhang et al. 2018) – Sec16 (Piao et al. 2017) – CFTR (Gee et al. 2011) – JAM-B, JAM-C (Cartier-Michaud et al. 2017) – KCTD5 (Dementieva et al. 2009) |
• Golgi stacking (Shorter et al. 1999; Xiang and Wang 2010; Bekier et al. 2017) • Golgi ribbon formation (Feinstein and Linstedt 2008) • Cell cycle control (Duran et al. 2008) • Transport of specific cargo (Kuo et al. 2000; D’Angelo et al. 2009) • p24 cargo receptor retention (Barr et al. 2001) • Autophagy (Zhang et al. 2018; Zhang and Wang 2018a, b) • Unconventional secretion (Dupont et al. 2011; Rabouille and Linstedt 2016; Vinke et al. 2011; Gee et al. 2011; Piao et al. 2017) • Spermatogenesis (Cartier-Michaud et al. 2017) |
| GRASP65/GORASP1 | cis | N-myr (Barr et al. 1997) |
– GM130 (Barr et al. 1998) – Mena (Tang et al. 2016) – DjA1 (Li et al. 2019) – p24 (Barr et al. 2001) – CD8a, Frizzled 4 (D’Angelo et al. 2009) – Bcl-XL (Cheng et al. 2010) |
• Stacking (Barr et al. 1997; Xiang and Wang 2010; Bekier et al. 2017; Shin et al. 2018) • Golgi ribbon formation (Puthenveedu et al. 2006; Tang et al. 2016) • Cell cycle progression (Preisinger et al. 2005; Sutterlin et al. 2005; Yoshimura et al. 2005; Duran et al. 2008; Tang et al. 2010b) • Mitotic spindle formation (Sutterlin et al. 2005) • Transport of specific cargo (D’Angelo et al. 2009) • p24 cargo receptor retention (Barr et al. 2001) • Unconventional secretion (Gee et al. 2011) • Apoptosis (Lane et al. 2002) |
|
| Golgins | |||||
| GM130/GOLGA2 | cis | P | Rab1 (Weide et al. 2001) |
– p115 (Nakamura et al. 1997) – GRASP65 (Hu et al. 2015) – Syntaxin 5 (Diao et al. 2008) – AKAP450 (Rivero et al. 2009) – ZFPL1 (Chiu et al. 2008) – Tuba (Kodani et al. 2009) |
• Golgi ribbon formation (Puthenveedu et al. 2006) • COPII vesicle tethering (Moyer et al. 2001; Alvarez et al. 2001) • Non-centrosomal microtubule organization (Rivero et al. 2009) • Centrosome regulation (Kodani and Sutterlin 2008; Kodani et al. 2009) • Spindle formation (Kodani and Sutterlin 2008) • Cell migration (Preisinger et al. 2004) • Apoptosis (Walker et al. 2004) • Purkinje neuron development (Liu et al. 2017) |
| p115 | cis | P | Rab1 (Allan et al. 2000) |
– GM130 (Nakamura et al. 1997) – Giantin (Sonnichsen et al. 1998) – syntaxin 5, GOS-28 (Shorter et al. 2002) |
• Post-mitotic Golgi reassembly (Shorter and Warren 1999; Brandon et al. 2003; Dirac-Svejstrup et al. 2000) • SNARE assembly (Wang et al. 2015; Shorter et al. 2002) • Membrane tethering (Shorter and Warren 1999; Nakamura et al. 1997; Alvarez et al. 2001; Seemann et al. 2000; Shorter et al. 2002; Allan et al. 2000) • Apoptosis (Chiu et al. 2002) • Nuclear import (Mukherjee and Shields 2009) |
| Giantin/GOLGB1 | cis/medial | TMD | Rab1/6 (Rosing et al. 2007) |
– p115 (Sonnichsen et al. 1998) – GM130 (Sonnichsen et al. 1998) – GCP60/ACBD3 (Sohda et al. 2001) |
• Golgi stack organization (Rosing et al. 2007; Koreishi et al. 2013) • Ribbon organization (Koreishi et al. 2013) • Membrane tethering (Sonnichsen et al. 1998; Linstedt et al. 2000; Lowe et al. 2004; Misumi et al. 2001; Derby and Gleeson 2007; Alvarez et al. 2001; Munro 2011) • ER-to-Golgi trafficking (Sohda et al. 2001) • Apoptosis (Lowe et al. 2004) • Cilia formation (Bergen et al. 2017) |
| Golgin-45/BLZF1 | medial/trans | P | Rab2 (Short et al. 2001) | – GRASP55 (Short et al. 2001, Zhao et al. 2017) |
• Golgi stacking (Short et al. 2001, Zhao et al. 2017) • Membrane tethering (Short et al. 2001) |
| Golgin-67/GOLGA8B | TMDa | • Uncharacterized (Jakymiw et al. 2000, Eystathioy et al. 2000) | |||
| Golgin-84/GOLGA5 | cis | TMD | Rab1(Satoh et al. 2003) |
– CASP (Osterrieder et al. 2017) – COG complex (Sohda et al. 2010) |
• Golgi stacking and reassembly (Satoh et al. 2003) • Golgi ribbon formation (Diao et al. 2003) • ER-Golgi tethering and protein transport (Osterrieder et al. 2017) • Membrane tethering (Malsam et al. 2005) • Intra-Golgi retrograde transport (Sohda et al. 2010) • Proinsulin conversion (Liu et al. 2016) • Tau phosphorylation in Alzheimer’s disease (Jiang et al. 2014) • Bacterial infection (Rejman Lipinski et al. 2009) |
| Golgin-97/GOLGA1 | trans | GRIP |
ARL1/3 (Lu and Hong 2003) Rab6/19/30 (Sinka et al. 2008) |
– TBC1D23 (Shin et al. 2017, 2018) |
• TGN-to-PM trafficking of E-cadherin (Lock et al. 2005) • Endosome-to-TGN trafficking (Lu and Hong 2003; Lu et al. 2004) • Retrograde transport from recycling endosomes to the TGN (Jing et al. 2010) • Poxvirus morphogenesis (Alzhanova and Hruby 2007) |
| Golgin-160/GOLGA3/GCP170 | cis | P |
– GCP60/ACBD3 (Sbodio et al. 2006; Sbodio and Machamer 2007) – GCP16 (Ohta et al. 2003) – β1AR (Gilbert et al. 2014) – ROMK, PIST (Bundis et al. 2006) |
• Post-Golgi trafficking (Bundis et al. 2006; Gilbert et al. 2014) • Apoptosis (Mancini et al. 2000, Maag et al. 2005, Sbodio et al. 2006) |
|
| Golgin-245/GOLGA4/p230 | trans | GRIP |
ARL1/3 (Wu et al. 2004; Sinka et al. 2008) Rab2/6/19/30 (Sinka et al. 2008) |
– TBC1D23 (Shin et al. 2017, 2018) |
• TGN-to-PM traffic (Lu and Hong 2003) • Phagophore formation (Sohda et al. 2015) |
| GCP16 | Acylation | – Golgin-160/GOLGA1/GCP170 (Ohta et al. 2003) | • Golgi-to-PM trafficking (Ohta et al. 2003) | ||
| GCC88 | trans | GRIP | ARL1/3 (Sinka et al. 2008), Rab6/19/30 (Sinka et al. 2008) |
• TGN organization and endosome-to-TGN trafficking (Luke et al. 2003; Lieu et al. 2007) • Golgi ribbon formation (Gosavi et al. 2018) • Actin organization (Makhoul et al. 2019) |
|
| GCC185 | trans | GRIP |
ARL1/3 (Sinka et al. 2008) ARL4A (Lin et al. 2011) Rab1/2/6/9/30 (Burguete et al. 2008, Sinka et al. 2008, Hayes et al. 2009) |
– Syntaxin 16 (Ganley et al. 2008) – CLASP (Efimov et al. 2007) |
• Golgi ribbon formation (Derby et al. 2007) • Golgi integrity (Lin et al. 2011) • Membrane tethering (Derby et al. 2007; Reddy et al. 2006; Ganley et al. 2008) • MPR recycling (Reddy et al. 2006) • Attachment of non-centrosomal microtubules (Efimov et al. 2007) |
| ACBD3/GCP60 | P |
– Golgin-45 (Yue et al. 2017) – giantin (Sohda et al. 2001) – Golgin-160 (Sbodio and Machamer 2007; Sbodio et al. 2006) – PI4KIIIβ (Greninger et al. 2012) – TBC1D22 (Greninger et al. 2013) – Aichi virus (Sasaki et al. 2012) – Poliovirus 3A protein (Greninger et al. 2013) – Numb (Zhou et al. 2007) |
• Golgi integrity and ER-to-Golgi trafficking (Sohda et al. 2001) • Nuclear translocation of caspase-generated Golgin-160 fragments (residues 140-311) (Sbodio and Machamer 2007, Sbodio et al. 2006) • Asymmetric cell division (Zhou et al. 2007) • Aichi virus and poliovirus replication (Greninger et al. 2012) |
||
| Bicaudal-D/BICD2 | P | Rab6 | – Dynactin, p50-dynamitin (Hoogenraad et al. 2001) |
• Recruitment of the dynein-dynactin complex (Hoogenraad et al. 2001) • COPI-independent Golgi-to-ER transport (Matanis et al. 2002) • Endosome-to-Golgi transport (Wanschers et al. 2007) • Autosomal-dominant spinal muscular atrophy (Neveling et al. 2013, Martinez-Carrera and Wirth 2015) |
|
| CASP/Coy1 | cis | TMD |
– Golgin-84 (Osterrieder et al. 2017) – Cog3, Sed5, Sly1, Gos1, Sft1/GS15 (Anderson et al. 2017) |
• ER-Golgi tethering and protein transport (Osterrieder et al. 2017) • Retrograde-directed COPI vesicles (Anderson et al. 2017) |
|
| CG-NAP |
– PKN, RIIα, PP2A, protein phosphatase 1 (Takahashi et al. 1999) – Dynein (Kim et al. 2007) – Cyclin E (Nishimura et al. 2005) – Kendrin, GCP2, γ-tubulin (Takahashi et al. 2002) – CK1δ (Sillibourne et al. 2002) |
• Scaffold protein for kinases/phosphatases (Takahashi et al. 1999) • Centrosome amplification (Nishimura et al. 2005) • Microtubule nucleation (Takahashi et al. 2002) |
|||
| COH1/VPS13B | cis | P | • Golgi ribbon formation (Seifert et al. 2011) | ||
| GCP364 | C-terminal hydrophobic domain | • Golgi ribbon formation and perinuclear localization (Toki et al. 1997) | |||
| GMAP-210/Trip11/Trip230 | cis | GRAB (Gillingham et al. 2004) | ARF1 (Gillingham et al. 2004) |
– ITF20, γ-tubulin (Rios et al. 2004) – Thyroid receptor (Chen et al. 1999) |
• Golgi ribbon formation (Rios et al. 2004) • Membrane tethering (Drin et al. 2008) • ER-to-Golgi trafficking (Gillingham et al. 2004) • γ-tubulin recruitment (Rios et al. 2004) • Sorting to primary cilia (Follit et al. 2008) • Interacts with thyroid hormone receptor beta (Chen et al. 1999) |
| IIGP165 | P | • Anti-apoptosis (Ran et al. 2007) | |||
| Imh1/SYS3 | trans | GRIP | ARL1/Arl1p (Panic et al. 2003) | – Env7 (Rao et al. 2016) | • Endosome-to-Golgi trafficking (Tsukada et al. 1999) |
| Lava lamp/Lva | – Dynein heavy chain, Lrrk (Lin et al. 2015) | • Dynein-based Golgi movements (Papoulas et al. 2005) | |||
| NECC1/2 | TMDa | • Uncharacterized (Cruz-Garcia et al. 2007) | |||
| SCOCO | P | • Uncharacterized (Van Valkenburgh et al. 2001) | |||
| SCYL1BP1/GORAB | P | Rab6 (Hennies et al. 2008) | – Scyl1 |
• Gerodermia osteodysplastica (Hennies et al. 2008; Al-Dosari and Alkuraya 2009) • Suppresses Schwann cell (SC) differentiation and neurite outgrowth by enhancing the Rhoa Pathway (Zhang et al. 2016a) • Type 2 diabetes (Peng et al. 2015) • Suppression of hepatocellular carcinoma development (Hu et al. 2012) |
|
| TMP/ARA160 | P | Rab6 (Fridmann-Sirkis et al. 2004) |
– hSNF2a/b (Mori and Kato 2002) – Fer, AR (Hsiao and Chang 1999) – Stat3 (Perry et al. 2004) |
• Golgi integrity (Fridmann-Sirkis et al. 2004) • Early/recycling endosomes-to-TGN trafficking, Golgi enzyme retention (Yamane et al. 2007, Ramirez and Lowe 2009) |
|
| Other Golgi structural proteins | |||||
| p24/TMED2 | cis | TMD |
– GRASP55 (Barr et al. 2001) – GRASP65 (Barr et al. 2001) |
• Post-Golgi trafficking (Luo et al. 2007) • GPI-anchored proteins export from the ER (Bonnon et al. 2010) |
|
| p28 | cis | • Golgi ribbon formation (Koegler et al. 2010) | |||
| p37 | trans | P | – p97 (Uchiyama et al. 2006) | • Membrane fusion (Uchiyama et al. 2006) | |
| p47 | trans | P |
– VCIP135 (Zhang and Wang 2015a) – p97 (Kondo et al. 1997) |
• Post-mitotic Golgi reassembly (Zhang and Wang 2015a) | |
| p97/VCP | trans | P |
– VCIP135 (Zhang and Wang 2015a) – p47 (Kondo et al. 1997) – p37 (Uchiyama et al. 2006) – YOD1, UBXD1 and PLAA (Papadopoulos et al. 2017) |
• Post-mitotic Golgi reassembly (Zhang and Wang 2015a) • Membrane reassembly (Kondo et al. 1997; Ramadan et al. 2007; Hetzer et al. 2001) • Cell cycle progression (Cao et al. 2003; Fu et al. 2003; Parisi et al. 2018) • Protein aggregation (Ghosh et al. 2018; Ristic et al. 2018; Yi and Yuan 2017; Kobayashi et al. 2007; Song et al. 2007; Nishikori et al. 2008) • Autophagy (Papadopoulos et al. 2017) • DNA repair (Van Den Boom et al. 2016; Fujita et al. 2013; Davis et al. 2012; Meerang et al. 2011; Partridge et al. 2003) • ER-stress-induced gene transcription (Marza et al. 2015) |
|
ACBD Acyl-CoA-binding domain-containing protein, AKAP A-kinase anchor protein, AR adrenergic receptor, ARL ADP-ribosylation factor-like protein, BICD2 protein bicaudal D homolog 2, CFTR cystic fibrosis transmembrane conductance regulator, CG-NAP centrosome- and Golgi-localized PKN-associated protein, CK casein kinase, COG complex conserved oligomeric Golgi complex, COH Cohen syndrome protein, DjA1 DnaJ homolog subfamily A member 1, GCP Golgi complex-associated protein, GMAP Golgi-associated microtubule-binding protein, GORAB RAB6-interacting golgin, GPI glycosylphosphatidylinositol, GRAB GRIP-related ARF-binding domain, GRIP Arl-binding domain, KCTD5 potassium channel tetramerization domain-containing protein 5, Lrrk leucine-rich repeat serine/threonine-protein kinase, Mena mammalian enabled homologue, MPR mannose 6-phosphate receptor, NECC neuroendocrine long coiled-coil protein, N-Myr amino-terminal myristoylation, P peripheral membrane protein, PIST PDZ protein interacting specifically with TC10, PM plasma membrane, ROMK renal outer medullary potassium, SCOCO short coiled-coil protein, SNARE soluble NSF attachment protein receptor, TBC1D23 TBC1 domain family member 23, TGN trans-Golgi network, TGF transforming growth factor, TMD transmembrane domain, TMED transmembrane emp24 domain-containing protein, VCIP valosin-containing protein p97/p47 complex-interacting protein, VCP valosin-containing protein, VPS vacuolar protein sorting-associated protein, ZFPL1 zinc finger protein-like 1
This table is updated from Xiang and Wang (2011)
aPredicted
Golgi ReAssembly Stacking Proteins (GRASPs)
Among all Golgi matrix proteins, the Golgi ReAssembly Stacking Proteins GRASP65 and GRASP55 (GRASPs, also called GORASP1 and GORASP2, respectively) are best characterized for their roles in Golgi structure formation, including stacking (Wang et al. 2003, Shorter et al. 1999, Xiang and Wang 2010, Bekier et al. 2017, Shin et al. 2018, Barr et al. 1997), ribbon-linking (Puthenveedu et al. 2006; Tang et al. 2016; Feinstein and Linstedt 2008), cargo transportation (Kuo et al. 2000; D’Angelo et al. 2009; Barr et al. 2001), unconventional secretion (Dupont et al. 2011; Rabouille and Linstedt 2016; Vinke et al. 2011; Gee et al. 2011; Piao et al. 2017), cell cycle regulation (Preisinger et al. 2005; Sutterlin et al. 2005; Yoshimura et al. 2005; Duran et al. 2008; Tang et al. 2010b), apoptosis (Lane et al. 2002), and autophagy (Zhang et al. 2018; Zhang and Wang 2018a, b), although the mechanisms are less well understood.
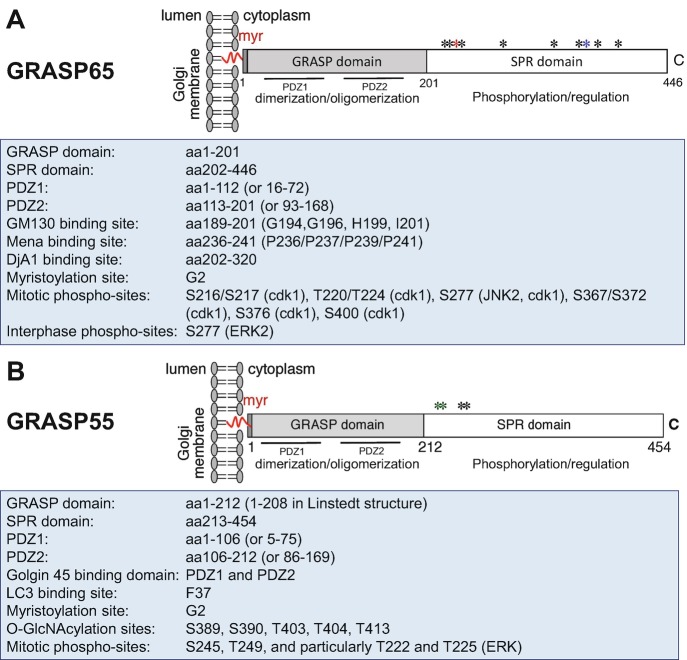
Both GRASPs share a similar structure: a conserved N-terminal GRASP domain consisting of two PDZ domains (PDZ1 and PDZ2) and an intrinsically disordered C-terminal Serine/Proline-Rich (SPR) domain with multiple phosphorylation sites (Zhang and Wang 2015b) (Fig. 19.1). Both GRASP65 and GRASP55 are peripheral membrane proteins that are attached to the Golgi membranes via an N-terminal myristic acid modification and the interaction with their membrane-bound partner proteins (GM130 and Golgin-45, respectively) and therefore are concentrated at the interface between the cisternae where stacking occurs (Short et al. 2001; Barr et al. 1998). GRASP65 is concentrated on the cis-Golgi cisternae, whereas GRASP55 localizes to the medial/trans-Golgi cisternae (Barr et al. 1997; Shorter et al. 1999). Both play complementary roles in Golgi stack formation (Xiang and Wang 2010).
Fig. 19.1.
Structure, modification, and binding sites on GRASP65 (a) and GRASP55 (b). Rat GRASP65 and GRASP55 sequences are used for illustration. Both GRASPs share a similar structure: a conserved N-terminal GRASP domain consisting of two PDZ domains (PDZ1 and PDZ2) and a C-terminal Serine/Proline-Rich (SPR) domain with multiple phosphorylation sites (indicated by asterisks) that are involved in GRASP modulation during the cell cycle. Both GRASP65 and GRASP55 are peripheral membrane proteins attached to the Golgi membranes via N-terminal myristoylation and the interaction with their membrane-bound partner proteins (GM130 and Golgin-45, respectively). GRASP65-binding proteins Mena and DjA1 have been identified to enhance Golgi ribbon linking and stacking, respectively. GRASP55 is regulated by O-GlcNAcylation depending on the glucose level and interacts with LC3 and LAMP2 to facilitate glucose starvation-induced autophagy
Mechanistically, GRASP proteins form homodimers via the N-terminal PDZ domains, and dimers from adjacent Golgi cisternae further oligomerize in trans and function as the “glue” that tethers the cisternae into a stack (Wang et al. 2003, 2005). An in vitro study using modified GRASP domain peptides indicated that insertion of the myristic acid moiety is required for the oriented association to Golgi membranes, which ensures the protein-protein interaction in trans. Furthermore, the conformational change caused by myristoylation affects the tendency of GRASP domain for self-interaction (Heinrich et al. 2014). Depletion of either GRASP65 or GRASP55 reduces the number of cisternae per Golgi stack, whereas depletion of both GRASPs leads to disassembly of the entire Golgi stack (Sutterlin et al. 2005, Xiang and Wang 2010, Bekier et al. 2017). The GRASPs are tightly modulated by a phosphorylation and dephosphorylation cycle during cell division, resulting in mitotic disassembly and post-mitotic reassembly of the Golgi (Feinstein and Linstedt 2008; Cervigni et al. 2015; Lin et al. 2000; Wang et al. 2003; Preisinger et al. 2005; Tang et al. 2012; Xiang and Wang 2010; Tang et al. 2008; Truschel et al. 2012).
Recent studies have identified novel GRASP-binding proteins involved in Golgi biogenesis and morphology modulation. Recent research of the crystal structure of GRASP55 bound to the Golgin-45 C-terminal peptide revealed that Golgin-45 promotes the oligomerization of GRASP55 by forming a new interaction between two neighboring PDZ2 molecules to play an important role in Golgi stacking (Zhao et al. 2017). Meanwhile, using an optimized in vitro system, mammalian enabled homologue (Mena) and DnaJ homolog subfamily A member 1 (DjA1) were identified as GRASP65 binding partners with potential functions on Golgi structure maintenance (Tang et al. 2016; Li et al. 2019). Mena is an actin elongation factor recruited to the Golgi membranes by GRASP65 to facilitate actin polymerization and GRASP65 oligomerization and thus functions with actin as bridging proteins of GRASP65 in Golgi ribbon linking. DjA1 is a co-chaperone of Heat shock cognate 71 kDa protein (Hsc70), but the activity of DjA1 in Golgi structure formation is independent of its co-chaperone activity or Hsc70, rather, through DjA1-GRASP65 interaction to promote GRASP65 oligomerization. Thus, DjA1 facilitates Golgi structure formation through an unconventional Hsc70-independent pathway. These studies further confirmed GRASP65 as a multifaceted protein in Golgi structure formation and indicated that an array of GRASP binding proteins could play important roles in Golgi morphology maintenance (Fig. 19.1). In addition, GRASP55 was reported to be involved in glucose starvation-induced autophagy (Zhang et al. 2018), where it interacts with LC3 on autophagosomes and LAMP2 on lysosomes to facilitate autophagosome maturation, which will be discussed in later sections.
Golgins
Golgins are a family of Golgi-associated coiled-coil proteins that are necessary for vesicle tethering at the Golgi and maintenance of Golgi integrity (Muschalik and Munro 2018; Witkos and Lowe 2015; Gillingham and Munro 2016). Most golgins are peripheral membrane proteins anchored on Golgi membranes via their C-terminus and are associated with small GTPases of Rab, Arf, and Arl families (Table 19.1) (Munro 2011; Sinka et al. 2008). These interactions mediate both membrane attachment and selective localization of a specific golgin to a specific sub-compartment of the Golgi (Witkos and Lowe 2015). Golgins lack significant sequence homology between the family members and localize to different regions of the Golgi to play distinct roles in tethering events, membrane traffic, and Golgi organization. The coiled-coil regions provide the golgins with an extended structure required for the tethering function, while the interactions with Rab GTPases control these molecules in their open (extended) or closed (folded) confirmation (Cheung et al. 2015). In addition, golgins often contain specific sequence and structural features at the N- and C-terminal ends, which allow them to recognize vesicles and Golgi cisternal membranes based on the curvature and lipid composition of the membranes (Drin et al. 2008; Drin et al. 2007; Magdeleine et al. 2016). Detailed information about golgins and their functions are summarized in Table 19.1.
GM130 was the first identified Golgi matrix protein and is predominantly found in the central region of the cis-Golgi (Nakamura et al. 1995), where it forms a stable complex with GRASP65 (Barr et al. 1998). Depletion of GM130 results in the disruption of the Golgi ribbon and causes protein glycosylation defects (Puthenveedu et al. 2006). There are two possible ways GM130 contributes to Golgi ribbon formation. First, GM130 targets GRASP65 to the rim of the cisternae where GRASP65 promotes lateral linking of cisternae via oligomerization (Puthenveedu et al. 2006). Mena and actin cytoskeleton may facilitate GRASP65 in this action (Tang et al. 2016). Second, GM130 recruits A-kinase anchoring protein 450 (AKAP450) onto the cis-Golgi and allows Golgi-associated nucleation of microtubules, which arranges Golgi stacks in close proximity to form a ribbon (Rivero et al. 2009). Similarly, other golgins may also work with the microtubule cytoskeleton in a similar way to facilitate Golgi structure organization. For example, GMAP-210, another cis-Golgi-localized golgin, recruits the γ-tubulin-containing complexes to the Golgi membranes and promotes the formation of tubulin oligomers on Golgi membranes. Depletion of GMAP-210 results in extensive Golgi fragmentation, suggesting a role in Golgi ribbon formation (Rios et al. 2004).
In addition to GRASP65, GM130 also interacts with p115 and giantin to form the GM130-p115-giantin tethering complex (Sonnichsen et al. 1998). The Rab1 effector p115, when recruited to coat protein complex (COP) II vesicles during the budding from the ER, interacts with the soluble ATPases N-ethylmaleimide-sensitive factor (NSF) attachment protein receptors (SNAREs), a specialized set of COPII v-SNAREs, to form a cis-SNARE complex that promotes vesicle targeting to the Golgi apparatus (Allan et al. 2000). Meanwhile, p115 also binds giantin on COPI vesicles and works with GM130 on Golgi membranes to provide a bridging role for vesicle tethering (Sonnichsen et al. 1998). Thus, GM130 and p115 are two major tethering factors in ER-to-Golgi trafficking (Munro 2011).
Other than the well-studied GRASP65-GM130 and GM130-p115-giantin complexes, the GRIP domain containing golgins are another group of proteins associated with the Golgi structure. Most GRIP domain-containing golgins localize to the TGN via their GRIP domains and are involved in Golgi organization (Luke et al. 2005). GCC185 is reported to localize independently of Arl1 on TGN and plays an essential role in Golgi structure formation. Depletion of GCC185 results in fragmentation of both cis- and trans-Golgi that are dispersed throughout the cytoplasm (Derby et al. 2007). On the other hand, another TGN golgin GCC88 is reported to play a role in TGN organization and ribbon-linking. Overexpression of GCC88 causes a loss of the compact Golgi ribbon and dispersal of mini-stacks throughout the cytoplasm, while knockdown of GCC88 results in a longer ribbon structure (Gosavi et al. 2018). A recent report suggests that GCC88-induces Golgi ribbon dispersal via actin and non-muscle myosin IIA. In addition, a novel GCC88-binding partner, the long isoform of intersectin-1 (ITSN-1), a guanine nucleotide exchange factor for Cdc42, is identified to be involved in this process (Makhoul et al. 2019).
Other Golgi Structure-Related Proteins
Besides the Golgi matrix proteins and their cofactors described above, other proteins including SNAREs, kinases, methyltransferases, and GTPases have also been reported to be related to Golgi structural organization and function. A few examples are discussed below.
Vesicle-associated membrane protein 4 (VAMP4), a v-SNARE protein located on the TGN, was first shown to play a role in retrograde trafficking from early endosomes to the TGN (Steegmaier et al. 1999). It was later reported that depletion of VAMP4 led to fragmentation of the Golgi ribbon, although Golgi membranes remained in the juxtanuclear area. EM studies revealed shortened Golgi stacks with a normal arrangement. Depletion of the cognate SNAREs of VAMP4, syntaxin 6, syntaxin 16, and Vti1a also disrupted the Golgi ribbon. These findings suggest that the maintenance of the Golgi ribbon structure requires normal retrograde trafficking, which is likely mediated by the formation of VAMP4-containing SNARE complexes (Shitara et al. 2013).
Serine/threonine-protein kinase H1 (PSKH1) was primarily characterized with multiple intracellular localizations, including Brefeldin A-sensitive Golgi compartment, centrosomes, nucleus, and cytoplasm (Brede et al. 2000). PSKH1 targeting to Golgi depends on dual N-terminal acylation, myristoylation on glycine 2, and palmitoylation on cysteine 3. Expression of palmitoylation site mutant PSKH1 results in the disassembly of the Golgi apparatus to a diffused cytoplasmic pattern without interrupting the microtubule cytoskeleton (Brede et al. 2003). The substrates of this kinase on the Golgi are so far unidentified.
Protein arginine methyltransferase 5 (PRMT5) localizes to the Golgi apparatus and forms complexes with several components, including GM130, which was later identified as a substrate of PRMT5. N-terminal methylation of GM130 does not affect its Golgi localization but is critical for Golgi ribbon formation. Depletion of PRMT5 and expression of methylation-defective GM130 mutants result in fragmentation and dispersal of the Golgi ribbon (Zhou et al. 2010).
In addition to the proteins mentioned above, Rab small GTPases are another group of key regulators of mammalian Golgi organization. Many Rab proteins, including but not limited to Rab1, Rab2 and Rab8 (Aizawa and Fukuda 2015), Rab18 and Rab43 (Dejgaard et al. 2008), Rab6/41 (Goud et al. 1990; Martinez et al. 1997; Liu et al. 2013), and Rab30 (Kelly et al. 2012), have been shown to play a role in Golgi structure organization and reviewed previously in detail (Goud et al. 2018). Considering that Rab proteins switch between inactive GDP-bound and active GTP-bound forms, it has been proposed that Golgi organization-related Rab proteins are divided into two categories. With Class 1 Rabs, the Golgi ribbon is disrupted by Rab inactivation but appears normal with overexpression, whereas with Class 2 Rabs, Rab inactivation has little effect on Golgi ribbon organization, while overexpression leads to the redistribution of Golgi enzymes to the ER (Liu and Storrie 2015). These results indicate that Rabs control the Golgi structure through modulating membrane tethering and trafficking.
Golgi Dynamics in the Mammalian Cell Cycle
The Golgi undergoes a series of sophisticated cell cycle-dependent disassembly and reassembly processes, including the deformation and reformation of Golgi ribbon, stacks, and cisternae. At the onset of mitosis, the Golgi ribbon unlinks into ministacks, which further undergo unstacking and vesiculation. These processes ensure the equal distribution of Golgi compartments into the two daughter cells (Wang 2008; Wei and Seemann 2010). In telophase, the Golgi vesicles fuse into cisternae and form stacks. The new stacks then accumulate in the perinuclear region and further link into a ribbon. The molecular factors that control these processes include Golgi matrix proteins, kinases and phosphatases, ubiquitin ligases and deubiquitinating enzymes, vesicle budding and fusion factors, and actin and microtubule cytoskeleton. An in vitro system has been developed to replicate the Golgi disassembly and reassembly process through sequential treatments of purified Golgi membranes with mitotic (MC) and interphase (IC) cytosol, or with purified proteins (Tang et al. 2008, 2010a). This system provides a powerful tool for testing key proteins in Golgi structure formation, which makes it possible to identify the minimal machinery and key components that control mitotic Golgi disassembly and post-mitotic reassembly (Huang and Wang 2017).
Mechanisms of Golgi Disassembly and Reassembly in the Mammalian Cell Cycle
The first step of Golgi disassembly at the onset of mitosis is Golgi ribbon unlinking. At this step, Golgi stacks in the ribbon are disconnected and dispersed. This step involves disconnecting the tubules between the stacks by the membrane fission protein CtBP/BARS, which is crucial for G2/M transition (Hidalgo Carcedo et al. 2004; Colanzi et al. 2007). Further, GRASPs undergo mitotic phosphorylation which are also required for ribbon-unlinking. The extracellular-signal-regulated kinase (ERK) directly phosphorylates GRASP55 and blocks its activity in both Golgi ribbon formation and trans-oligomerization (Feinstein and Linstedt 2008), while GRASP65 is phosphorylated by c-Jun N-terminal kinase (JNK) on Serine 277 (S277), which causes the separation of the Golgi stacks (Cervigni et al. 2015).
Sequential phosphorylation of GRASP65 on multiple sites by cyclin-dependent kinase 1 (Cdk1) and polo-like kinase 1 (Plk1) results in its conformational changes and subsequent de-oligomerization (Lin et al. 2000; Preisinger et al. 2005; Wang et al. 2003; Tang et al. 2012; Vielemeyer et al. 2009). On the other hand, GRASP55 is phosphorylated by ERK and partially by Cdk1 (Xiang and Wang 2010). In addition to the unique phosphorylation sites in the SPR domains of GRASPs, S189 within the GRASP domain of GRASP65 is modified by Plk1, which causes conformational change and impaired self-association (Sengupta and Linstedt 2010). An in vivo membrane-tethering activity assay using a construct of full-length GRASP55 fused to the C-terminal mitochondrial anchoring sequence shows that the tethering activity is diminished by introducing a phosphomimic S189D mutant. This result suggests that S189 might be a Plk1 target site on both GRASPs, although direct evidence remains to be provided (Truschel et al. 2012). GRASP65 is dephosphorylated by PP2A in late mitosis and the trans-oligomer reformation is therefore rehabilitated to promote cisternae stacking (Tang et al. 2008).
The unstacked cisternae further disassemble into vesicles, which depends on COPI vesicle formation and blockage of vesicle docking and membrane fusion. As mentioned above, the GM130-p115-giantin complex promotes COPII vesicle docking to the Golgi. Cdk1 phosphorylation of GM130 on S25 during mitosis inhibits p115-interaction and therefore blocks vesicle docking (Lowe et al. 1998). Inhibition of Cdk1 causes Golgi vesiculation failure, suggesting an essential role of Cdk1 activity in mitotic Golgi vesiculation. However, expression of the GM130 S25A non-phosphorylatable mutant in GM130-depleted cells causes no apparent defects in Golgi vesiculation and mitotic progression, indicating the existence of GM130 S25 phosphorylation-independent pathways that ensure Golgi vesiculation and mitotic progression in mammalian cells (Sundaramoorthy et al. 2010).
Recently, the recruitment and activation of Aurora kinase family member Aurora A at the centrosomes in M phase was reported to depend on Golgi ribbon unlinking in G2 phase (Barretta et al. 2016). Aurora A functions in centrosome maturation, mitotic entry, and bipolar spindle formation during mitosis (Nikonova et al. 2013; Carmena et al. 2009; Kimura et al. 2013). This finding indicates a potential link between Aurora A activity and cell cycle-associated Golgi structure modulation. Indeed, it was later confirmed that knockdown or inhibition of Aurora A induces Golgi dispersal without affecting the GM130 protein level in interphase. Further investigation revealed that interference of Aurora A causes Golgi dispersal only after mitosis via the dissociation of the Golgi and centrosome (Kimura et al. 2018). These studies revealed a novel relationship between G2 phase Golgi unlinking, M phase Aurora A activation, and interphase Golgi structure formation.
Post-mitotic Golgi Membrane Fusion and Its Regulation
Two AAA ATPases, NSF and p97/VCP, are involved in membrane fusion during post-mitotic Golgi reassembly, and their activities are regulated by phosphorylation during mitosis (Rabouille et al. 1995). For NSF-catalyzed fusion, the p115-GM130 tethering complex is disrupted by GM130 phosphorylation during mitosis (Lowe et al. 1998), while post-mitotic phosphorylation of p115 by a casein kinase II (CKII)-like enzyme is required for cisterna reassembly (Dirac-Svejstrup et al. 2000). Contemporarily, homotypic fusion of Golgi membranes mediated by p97 is also blocked upon phosphorylation of p47 and p37. p97 uses these two distinct cofactors for its membrane fusion function: p47 is essential for the regrowth of Golgi cisternae from mitotic Golgi fragments (Kondo et al. 1997), while p37 is required for the maintenance of the Golgi structure in interphase as well as for its reassembly in late mitosis (Uchiyama et al. 2006). Both pathways are regulated by Cdk1-mediated phosphorylation. Phosphorylation of p47 on S140 abolishes its binding to Golgi membranes, resulting in mitotic inhibition of the p97/p47 pathway (Uchiyama et al. 2003). Phosphorylation on S56 and Threonine 59 (T59) disables p37 from binding to Golgi membranes and consequently blocks p97/p37-mediated Golgi membrane fusion at late mitosis (Kaneko et al. 2010).
In addition to phosphorylation, p97/p47-mediated Golgi membrane fusion is also regulated by ubiquitination (Tang and Wang 2013). Tang et al. discovered that the Homologous to the E6-AP Carboxyl Terminus (HECT) domain containing ubiquitin ligase HACE1 is targeted to the Golgi membrane through the interaction with Rab1 and participates in post-mitotic Golgi biogenesis (Tang et al. 2011). Depletion of HACE1 or expression of an inactive mutant impairs post-mitotic Golgi membrane fusion. The identification of HACE1 as a Golgi-localized ubiquitin ligase provides evidence that ubiquitin has a critical role in Golgi biogenesis during the cell cycle (Tang et al. 2011). Later, the Golgi t-SNARE syntaxin 5 was identified as a ubiquitination substrate (Huang et al. 2016). Syntaxin 5 is monoubiquitinated by HACE1 in early mitosis and deubiquitinated by the de-ubiquitinase VCIP135 in late mitosis (Wang et al. 2004). The monoubiquitination of syntaxin 5 at Lysine 270 (K270) in the SNARE domain impairs the interaction between syntaxin 5 and the cognate v-SNARE Bet1 but increases its binding to the p97 adaptor p47 through the UBA domain of p47, which is required for post-mitotic Golgi membrane fusion (Meyer et al. 2002). Expression of the syntaxin 5 K270R mutant in cells impairs post-mitotic Golgi reassembly. Therefore, monoubiquitinated syntaxin 5 recruits p97/p47 to the mitotic Golgi fragments and promotes post-mitotic Golgi reassembly upon ubiquitin removal by VCIP135 (Huang et al. 2016). VCIP135 was originally identified as a p97 interacting protein (Kano et al. 2005). It was later shown to be a de-ubiquitinating (DUB) enzyme involved in p97/p47-mediated membrane fusion (Wang et al. 2004). VCIP135 DUB activity, as well as its interaction with p97 and association with Golgi membranes, is regulated by phosphorylation (Zhang and Wang 2015a; Zhang et al. 2014). In mitosis, VCIP135 is phosphorylated at S130 by Cdk1 and thus is inactivated, allowing syntaxin 5 to be ubiquitinated by HACE1; in telophase, VCIP135 is dephosphorylated and reactivated, removing ubiquitin from syntaxin 5 to allow p97-mediated membrane fusion (Huang and Wang 2017; Wang 2008). These studies revealed a novel mechanism that monoubiquitination regulates Golgi membrane dynamics during the mammalian cell cycle.
Golgi Stress Response
As stated above, the Golgi apparatus in mammalian cells forms a unique stacked structure under normal growth conditions, which undergoes a regulated disassembly and reassembly process during the cell cycle. However, the Golgi structure and function could be impaired under stress conditions, such as DNA damage, energy and nutrient deprivation, and pro-apoptotic conditions. This could be attributed to perturbation of microtubule organization or phosphorylation, degradation, or cleavage of Golgi structural proteins. Additionally, many signaling molecules have been identified to be associated with the Golgi. Thus, it has been proposed that the Golgi could sense and transduce stress signals and therefore serves as a hub in the cellular signaling network (Farhan and Rabouille 2011; Mayinger 2011; Makhoul et al. 2019).
Apoptotic Stress and Golgi Fragmentation
Apoptosis, also known as programmed cell death, is a cell suicide mechanism carried out by organelle-directed regulators such as the Bcl-2 proteins and ultimately executed by the caspase family proteases (Nicholson and Thornberry 1997). Organellar response to apoptotic initiation includes death receptor endocytosis, mitochondrial and lysosomal permeabilization, ER calcium release, and Golgi fragmentation. The Golgi is one of the first organelles to be affected during apoptosis (Mukherjee et al. 2007; Aslan and Thomas 2009). During apoptosis, several Golgi matrix proteins related to Golgi structure maintenance are cleaved by caspases, leading to Golgi fragmentation (Hicks and Machamer 2005). Apoptotic Golgi fragmentation is one of the most extensively studied Golgi stress responses. Reported caspases-cleaved Golgi proteins include GRASP65, golgin-160, GM130, p115, syntaxin 5, and giantin (Lane et al. 2002; Mancini et al. 2000; Walker et al. 2004; Chiu et al. 2002; Lowe et al. 2004; Machamer 2015), as summarized in Table 19.2 and discussed below.
Table 19.2.
Apoptotic cleavage of Golgi proteins
| Names | Apoptosis inducer | Caspases | Cleavage site | Golgi structural change |
|---|---|---|---|---|
| Golgin-160 | STS; CH11 | Caspase-2, 3 and 7 | D59, D139, D311 | Golgi fragmentation (Mancini et al. 2000; Mukherjee et al. 2007; Nozawa et al. 2002; Hicks and Machamer 2002; Maag et al. 2005) |
| GRASP65 | Anisomycin; STS | Caspase-3 | D320, D375, D393 | Golgi fragmentation (Lane et al. 2002; Cheng et al. 2010) |
| p115 | STS; 4-hydroxytamoxifen; CH11; CA | Caspase-3 and 8 | D757 | Golgi fragmentation (Chiu et al. 2002; How and Shields 2011; Mukherjee et al. 2007; Mukherjee and Shields 2009; Woldemichael et al. 2011) |
| GM130 | CH11 | Caspase-3 | – | Golgi fragmentation (Walker et al. 2004; Lowe et al. 2004; Mukherjee et al. 2007) |
| Syntaxin 5 | STS; anisomycin | Caspase-3 | D1882, D1083 | Secretion inhibition (Lowe et al. 2004) |
| Giantin | STS; anisomycin | Caspase-3 | D263 | Secretion inhibition (Lowe et al. 2004; Nozawa et al. 2002) |
CA carminomycin I, CH11 an anti-Fas monoclonal antibody, D aspartic acid, STS staurosporine
Golgin-160
Golgin-160 is a golgin that plays a role in vesicle tethering and trafficking (Misumi et al. 1997). It is cleaved by caspase-2, caspase-3, and caspase-7 during apoptosis. Under pro-apoptotic conditions stimulated by staurosporine, the Golgi senses and transduces apoptotic signals using a local caspase, caspase-2. Caspase-2 is special in a way that it has both the property of initiator caspases and the substrate specificity of executioner caspases (Mancini et al. 2000). Although it is unclear how caspase-2 is activated by pro-apoptotic signals, in vitro and in vivo caspase cleavage assays showed that caspase-2 cleavage of golgin-160 at aspartate 59 (D59) happens prior to golgin-160 cleavage by caspase-3 and 7 at D139 and D311 (Mancini et al. 2000). Expression of the D59A cleavage-defective mutant of golgin-160 delays Golgi disintegration under staurosporine treatment (Machamer 2003; Hicks and Machamer 2005).
Subsequently, it was shown that an N-terminal 85 amino acid fragment of golgin-160 contains both a Golgi localization signal and a nuclear localization signal (Hicks and Machamer 2002). Expression of a non-cleavable golgin-160 mutant inhibits ER stress or ligation of death receptor-induced apoptosis (Maag et al. 2005). Latterly, yeast two hybrid screening revealed that GCP60 preferentially binds to one of the caspase cleavage products of golgin-160, aa 140-311, to inhibit its nuclear localization (Sbodio et al. 2006). Overexpression of GCP60 sensitizes cells to staurosporine-induced apoptosis, while nuclear localization of a golgin-160 apoptotic cleavage fragment (aa 140-311) protects cells from apoptosis. However, another report indicates that golgin-160 depletion does not affect the Golgi morphology nor constitutive secretion (Williams et al. 2006). Therefore, the mechanism of how golgin-160 transduces apoptotic signals and regulates the apoptotic response needs to be further studied.
GRASP65
GRASP65 is cleaved in apoptosis induced by oxygen- and glucose-deprivation (OGD) as in ischemia-induced cerebral vascular endothelial injury (Yin et al. 2010) and in staurosporine- or Fas ligand-induced apoptosis (Lane et al. 2002, Cheng et al. 2010). In apoptosis, GRASP65 is cleaved by caspase-3 on D320, D375, and D393. Expression of a cleavage-resistant form of GRASP65 delays Golgi fragmentation in apoptosis and protects cells from Fas/CD95-mediated apoptosis, whereas expression of an N-terminal caspase-cleaved fragment dramatically sensitizes cells to Fas/CD95-mediated apoptosis (Lane et al. 2002, Cheng et al. 2010). Further results revealed that the C-terminal fragments of GRASP65 produced by caspase cleavage promotes Fas/CD95-mediated apoptosis via being targeted to mitochondria by binding to Bcl-XL (Cheng et al. 2010). However, the mechanism of how the C-terminal cleavage fragment of GRASP65 regulates apoptosis at the mitochondria and the role of Bcl-XL in this process are still unknown. The Golgi fragmentation phenotype induced by apoptotic GRASP65 cleavage is similar to that of GRASP65 phosphorylation in mitosis (Warren 1995). There is evidence that several kinases involved in mitotic GRASP65 phosphorylation such as Cdk1 and ERK are activated during apoptosis and regulate apoptosis by phosphorylating caspase and Bcl-2 family proteins (Terrano et al. 2010; Yamaguchi et al. 2008; Lu et al. 2011). Whether these kinases directly regulate apoptotic Golgi fragmentation by phosphorylating GRASP65 or other Golgi proteins remains unclear (Ji et al. 2013).
p115
The ER-to-Golgi membrane tether, p115, is cleaved by caspase-3 and caspase-8 during apoptosis. Expression of a caspase-resistant form of p115 delays Golgi fragmentation in apoptosis. Exogenous expression of a p115 C-terminal apoptotic fragment leads to apoptosis and Golgi fragmentation (Chiu et al. 2002). The extreme C-terminal fragment, generated by caspase cleavage during apoptosis, translocates into the nucleus and further activates the apoptosis machinery. Interestingly, translocation of the p115 C-terminal fragment happens prior to major Golgi structural changes, indicating it as an early event (Mukherjee and Shields 2009). The p115 C-terminus is SUMOylated, which regulates its nuclear translocation and amplification of apoptosis signals in a p53-dependent manner (How and Shields 2011; Mukherjee and Shields 2009). In a high-throughput screen, Carminomycin I (CA) was discovered to inhibit cell proliferation of Von Hippel-Lindau (VHL) defective Clear Cell Renal Cell Carcinoma (VHL−/− CCRCC) (Woldemichael et al. 2011). CA activates caspase-2 and caspase-3 to cleave p115, which inhibits CCRCC proliferation (Woldemichael et al. 2011).
Other Proteins
Several other Golgi structural proteins are also involved in apoptosis or cleaved by caspases. The level of GM130 is reduced during Fas-mediated apoptosis but not in staurosporine-induced apoptosis (Walker et al. 2004). However, it is not clear whether the reduction is due to GM130 cleavage or degradation. Syntaxin 5 and giantin are also cleaved by caspase-3 during apoptosis, which inhibits ER-to-Golgi transport (Lowe et al. 2004). Golgin-95 and golgin-97 are cleaved during necrosis but not apoptosis (Nozawa et al. 2002). Cleavage of golgin-95 and golgin-97 during necrosis is also caspase-dependent, since pretreatment of the cells with pan-caspase inhibitor, zVAD-fmk, abolished the cleavage of these two proteins (Nozawa et al. 2002).
Some of the Golgi proteins are also reported to regulate apoptosis. The Golgi SNARE GS28 is involved in cisplatin-induced apoptosis in a p53-dependent manner (Sun et al. 2012). Overexpression of GS28 sensitizes HEK293 cells to the apoptosis-inducer cisplatin via the accumulation of p53 and Bax and the stimulation of p53 pro-apoptotic phosphorylation at S46. It was also shown that GS28 forms a complex with the p53 ubiquitin E3 ligase Murine Double Minute 2 (MDM2) to inhibit its function and consequential p53 ubiquitination and degradation (Sun et al. 2012). Therefore, GS28 promotes cisplatin-induced apoptosis by stabilizing and regulating pro-apoptotic phosphorylation of p53.
A well-studied mediator of intracellular vesicle fusion, NSF attachment protein α (αSNAP), has been reported to have pro-survival functions (Naydenov et al. 2012). Depletion of αSNAP triggers apoptosis in epithelial cells by reducing the anti-apoptotic protein Bcl-2. Depletion of αSNAP in p53 null or Bax null cells still results in apoptosis, indicating that the anti-apoptotic function is independent of p53 and Bax. Interestingly, αSNAP depletion induces apoptosis independent of the cleavage of Golgi proteins such as GRASP65, golgin-160, and p115 but rather by dysregulation of ER-Golgi vesicle cycling and possibly through ER stress (Naydenov et al. 2012). Some other Golgi proteins, including human Golgi anti-apoptotic protein (h-GAAP) (Gubser et al. 2007; Saraiva et al. 2013) and Golgi integral membrane protein 4 (GOLIM4) (Bai et al. 2018), are also reported to have anti-apoptotic functions. GOLIM4 is overexpressed in some head and neck cancers, and depletion of GOLIM4 reduces cell proliferation and cell viability by inducing apoptosis (Bai et al. 2018).
Although microtubule and actin filaments play important roles in Golgi orientation and structure, Golgi fragmentation in apoptosis occurs prior to cytoskeleton disorganization (Mukherjee et al. 2007; Yadav and Linstedt 2011). Furthermore, the level of actin and tubulin did not change during apoptosis, while Golgi structural proteins are cleaved as discussed above. Therefore, Golgi fragmentation in early apoptosis is independent of microtubule and actin filament disorganization.
GOLPH3 and DNA Damage-Induced Golgi Fragmentation
The Golgi phosphoprotein 3 (GOLPH3) is a peripheral membrane protein that regulates vesicle budding and TGN-to-plasma membrane trafficking (Dippold et al. 2009). GOLPH3 is localized to the TGN by binding to phosphatidylinositol 4-phosphate (PI4P). Depletion of PI4P leads to GOLPH3 dissociation from the TGN. GOLPH3 also binds to the actin-based motor protein MYO18A to link Golgi membranes with the actin cytoskeleton. This bridging effect creates a tension required for vesicle budding, trafficking, and maintenance of the Golgi ribbon. Depletion of GOLPH3 or MYO18A leads to the loss of the tensile force, resulting in the shrinkage of the Golgi ribbon and a reduction of vesicles formed at the TGN (Dippold et al. 2009; Bishe et al. 2012; Ng et al. 2013). GOLPH3 is an oncogene known to be overexpressed in some solid tumors, including lung cancer and breast cancer (Scott et al. 2009; Zeng et al. 2012). It is also reported that GOLPH3 increases cell proliferation and cell size by regulating cell proliferation through the interaction with the retromer complex and activation of the mammalian target of rapamycin mTOR (Scott et al. 2009). GOLPH3 induces cell proliferation in breast cancer cells by inhibiting the tumor suppressor transcription factor FOXO1 through activating AKT (Zeng et al. 2012). These findings demonstrate that a trans-Golgi protein can serve as an oncogene (Scott et al. 2009; Buschman et al. 2015; Kuna and Field 2018).
Interestingly, DNA damage causes Golgi dispersal in a GOLPH3-dependent manner. DNA damage activates the DNA-PK kinase to phosphorylate GOLPH3 on T143/T148, which aberrantly increases the tensile force for the Golgi to fragment. Golgi fragmentation in this scenario increases cell survival with an unknown mechanism (Farber-Katz et al. 2014). Depletion of GOLPH3 or MYO18A increases cancer cells’ sensitivity to DNA damage inducing agents, suggesting that GOLPH3 phosphorylation-induced Golgi fragmentation may serve as a protective mechanism (Farber-Katz et al. 2014). Considering that GOLPH3 is overexpressed in many solid tumors, it is reasonable to speculate that this may be a mechanism of how cancer cells escape DNA damage-induced apoptosis (Farber-Katz et al. 2014; Buschman et al. 2015; Li et al. 2016b).
In addition to its high expression level in some cancer cells, GOLPH3 overexpression is also reported in mouse N2A cells under oxygen-glucose deprivation and reoxygenation (OGD/R), a model mimicking severe oxidative injury (Li et al. 2016a). In this OGD/R model, GOLPH3 is overexpressed and forms puncta in the cytosol, which induces the formation of reactive oxygen species (ROS) and lipidation of LC3. Opposed to its anti-apoptotic role in cancer cells, depletion of GOLPH3 in OGD/R desensitizes the cells to apoptosis (Li et al. 2016a).
Golgi in Autophagy Regulation
Most recently, the Golgi stacking protein GRASP55 was reported to regulate autophagy upon energy deprivation (Zhang et al. 2018; Zhang and Wang 2018a, b). Under normal growth condition, GRASP55 is O-GlcNAcylated and localizes in the medial- and trans-Golgi for stacking. However, under glucose starvation, a pool of de-O-GlcNAcylated GRASP55 translocates to the interface between autophagosomes and lysosomes to facilitate autophagosome-lysosome fusion. After a short-term energy deprivation, the Golgi structure is only mildly affected, possibly due to sufficient GRASP55 molecules remaining in the Golgi to maintain its structure. Among over a dozen Golgi proteins tested, only GRASP55, but not GRASP65, GM130, or golgin-45, is O-GlcNAcylated under growth conditions and targets to autophagosomes upon energy deprivation, indicating that GRASP55 serves as an energy sensor on the Golgi to regulate both intracellular trafficking and autophagy. Significantly, the same scenario may be seen in autophagy induced by amino acid starvation and inhibition of mTOR (Zhang et al. 2018).
In addition to Golgi fragmentation, GCC88 overexpression also induces autophagy via reducing the activity of mTOR. A considerable pool of mTOR is localized and activated on the Golgi, which is dependent on the ribbon structure for recruitment but independent of lysosomal mTOR activation (Gosavi et al. 2018). These findings indicate the Golgi ribbon as an important location for the functional regulation of mTOR activity.
Additionally, autophagosomes may directly form on Golgi membranes (Guo et al. 2012) or obtain membranes from the Golgi (Geng et al. 2010; Geng and Klionsky 2010). The only recognized transmembrane ATG protein, ATG9, localizes at the trans-Golgi network and late endosomes and is essential for autophagosome formation (Yamamoto et al. 2012), although the detailed mechanism awaits further investigation (Orsi et al. 2012). Recently, the endoplasmic reticulum-Golgi intermediate compartment (ERGIC) is proposed to serve as a key membrane source for autophagosome formation (Ge et al. 2013). Under normal condition, COPII vesicles are generated from the ER-exit sites (ERES) for ER-Golgi membrane trafficking, while upon starvation, the COPII assembly activator Prolactin Regulatory Element-Binding protein (PREB)/SEC12 relocates to the ERGIC and triggers ERGIC-COPII vesicle formation as membrane templates for LC3 lipidation.
Alteration of Golgi Structure and Function in Diseases
Golgi structure defects and dysfunctions have been observed in many diseases, including pathogen infection, neurodegenerative diseases, and cancer (Aridor and Hannan 2000). Generally, the mechanisms of Golgi fragmentation include imbalanced membrane flux, altered microtubule dynamics, and posttranslational modifications or proteolytic cleavage of Golgi structural proteins (Wei and Seemann 2017). In many cases, the correlation between Golgi defect and disease progression is unclear. A few interesting cases reported recently are discussed below.
Alzheimer’s Disease (AD)
AD is an age-related neurodegenerative disease of the central nerve system characterized by progressive loss of cognition and memory. Golgi fragmentation occurs in neurons of patients with AD since the earliest stages of disease development (Sundaramoorthy et al. 2015). Some cases of early-onset AD are related to mutations in the Amyloid Precursor Protein (APP) or Presenilin 1 and 2 (PSN1 and 2). The amyloid-beta (Aβ) peptide is a proteolytic product of APP, which is considered to be the major inducer of AD (Selkoe and Hardy 2016). Aβ accumulation is likely the direct cause of Golgi fragmentation, as Aβ-treatment causes reversible Golgi fragmentation in cultured neurons (Joshi et al. 2014).
Present research supports that activation of Cyclin-dependent kinase 5 (Cdk5) by Aβ accumulation via the [Ca2+]-calpain-p25 pathway may be the major trigger of Golgi fragmentation in AD (Lee and Linstedt 2000; Joshi et al. 2015; Joshi and Wang 2015; Evin 2015; Ayala and Colanzi 2017). Cdk5 may function in two ways. First, Cdk5 phosphorylates GM130 at S25 and inhibits its interaction with the Golgi tethering protein p115 (Sun et al. 2008). Second, Cdk5 phosphorylates GRASP65 at T220/T224, which inhibits GRASP65 function in Golgi stack formation and ribbon linking (Joshi et al. 2014). Furthermore, inhibiting Cdk5 or expressing non-phosphorylatable GRASP65 mutants both rescued the Golgi structure and reduced Aβ secretion by elevating the α-cleavage of APP (Joshi and Wang 2015; Joshi et al. 2014), indicating that GRASP65 phosphorylation may be the main reason for AD-induced Golgi fragmentation. These studies not only provide molecular mechanisms for Golgi fragmentation but also suggest Golgi as a potential drug target for AD treatment (Joshi et al. 2015; Joshi and Wang 2015; Ayala and Colanzi 2017).
Amyotrophic Lateral Sclerosis (ALS)
ALS is a fatal neurodegenerative disorder specifically targeted to motor neurons. Fragmented Golgi has been observed in numerous models of superoxide dismutase 1 (SOD1), TDP-43, FUS, and optineurin-associated ALS (Fujita et al. 2008; Soo et al. 2015; Van Dis et al. 2014; Wallis et al. 2018). SOD1 inhibits ER-to-Golgi transport and causes Golgi fragmentation (Atkin et al. 2014) by reducing the β-COP protein level, accumulating the ER-Golgi v-SNAREs GS15 and GS28, and destabilizing microtubules by the upregulation of Stathmins 1 and 2 (Bellouze et al. 2016; Atkin et al. 2014). FUS and TDP-43 impair the incorporation of secretory cargo into COPII vesicles (Soo et al. 2015). Furthermore, expression of ALS-related optineurin mutants impairs myosin VI-mediated protein trafficking from Golgi to plasma membrane, which also induces Golgi fragmentation (Sundaramoorthy et al. 2015). Conclusively, impairment of distinct protein trafficking pathways by different ALS-linked proteins are specific triggers for Golgi fragmentation in ALS.
Parkinson’s Disease (PD)
PD is pathologically characterized by the loss of dopamine-containing neurons and by the formation of intracellular protein aggregates known as Lewy bodies in which α-synuclein has been recognized as a major constituent (Forno 1996; Wakabayashi et al. 1998). Golgi fragmentation can be detected in early-stage PD brains (Fujita et al. 2006) and is strongly correlated to the presence of prefibrillar α-synuclein (Gosavi et al. 2002). Since then, emerging studies have provided important insights into the mechanisms of how α-synuclein causes pathological Golgi fragmentation and neuronal degeneration. The primary effect of α-synuclein aggregation is the inhibition of ER-to-Golgi transport (Lashuel and Hirling 2006), which can be rescued by the overexpression of Rab1 and Rab8 and depletion of Rab2 and syntaxin 5 (Rendon et al. 2013; Coune et al. 2011). In a most recent report, mutations in the leucine-rich repeat kinase 2 (LRRK2), a major genetic cause of autosomal-dominantly inherited PD, markedly enhance Rab7L1 phosphorylation on S72, resulting in TGN fragmentation (Fujimoto et al. 2018).
Cancer
Golgi disorganization may be related to cancer progression and metastasis in the following aspects: aberrant glycosylation, abnormal expression of Ras GTPase, dysregulation of kinases, and hyperactivation of myosin motor proteins (Petrosyan 2015). Perturbation of the Golgi morphology in cancers results in an increase of sialylation which is associated with a metastatic cell phenotype (Schultz et al. 2012). Overexpression of sialylated antigens is significantly correlated with tumor progression and therapy resistance due to an anti-apoptotic effect (Lee et al. 2008; Park et al. 2012; Petrosyan et al. 2014). Over-activation of Rabs, which coordinate with golgins in protein transportation and Golgi structure maintenance, has been reported in different types of cancers (Goldenring 2013). Furthermore, Golgi disorganization-related kinases, including Src, ERK9, and P21-activated protein kinase (Pak1), are found elevated in tumor cells (Chia et al. 2014; Ching et al. 2007; Weller et al. 2010). The ubiquitin ligase HACE1, which regulates p97-mediated Golgi membrane fusion as discussed above, is reported as a tumor suppressor downregulated in multiple tumors including Wilms’ tumor (Anglesio et al. 2004), resulting in Golgi fragmentation (Tang et al. 2011; Cui and Wang 2012). Additionally, hyperactivation of Golgi-associated myosins, including MYO18A that directly binds to GOLPH3 to promote Golgi dispersal (Dippold et al. 2009; Allan et al. 2002), is detected in many aggressive cancers. Golgi fragmentation is one of the essential and earliest events in apoptosis, where several golgins and GRASP65 are cleaved by activated caspases, as described in previous sections.
Viral Infection
Several membrane structures including the Golgi are used by viruses as viral factories to replicate, concentrate, and assemble the viral genome and proteins into viral particles (Miller and Krijnse-Locker 2008; Netherton et al. 2007; Salonen et al. 2005). As a highly dynamic organelle, Golgi serves as a membrane scaffold for multiple viruses, including infectious hepatitis C virus, enteroviruses, poliovirus, foot-and-mouth-disease virus, dengue virus, coronavirus, Kunjin virus, tick-borne encephalitis virus, rubella virus, and bunyamwera virus (Miller and Krijnse-Locker 2008; Harak and Lohmann 2015; Risco et al. 2003; Salanueva et al. 2003; Delgui et al. 2013; Westerbeck and Machamer 2015), and is frequently fragmented after infection (Campadelli et al. 1993; Salanueva et al. 2003; Yadav et al. 2016; Avitabile et al. 1995; Lavi et al. 1996; Hansen et al. 2017; Rebmann et al. 2016). Viruses use Golgi membranes directly and/or hijack master controllers of Golgi biogenesis and trafficking to generate vesicles that are used as the site of viral RNA replication (Quiner and Jackson 2010; Hansen et al. 2017; Short et al. 2013), wrapping (Sivan et al. 2016; Alzhanova and Hruby 2007; Alzhanova and Hruby 2006; Nanbo et al. 2018; Lundu et al. 2018; Procter et al. 2018), intracellular transduction (Nonnenmacher et al. 2015), and secretion (Zhang et al. 2016b). Viral infection triggers Golgi fragmentation via diverse mechanisms, ranging from phosphorylating key Golgi structural proteins such as GRASP65 (Rebmann et al. 2016), activating the Src kinase to phosphorylate the Dynamin 2 GTPase (Martin et al. 2017), targeting the immunity-related GTPase M (IRGM) to the Golgi to induce GBF1 phosphorylation (Hansen et al. 2017), modulating vesicular trafficking (Yadav et al. 2016; Johns et al. 2014), to impeding the major histocompatibility complex (MHC) class I trafficking, antigen presentation, and/or cytokine secretion (Moffat et al. 2007; Rohde et al. 2012).
Conclusions and Perspectives
The Golgi is the central hub in the secretory pathway, where proteins and lipids are processed, sorted, and dispatched to distinct destinations. As a well-organized polarized membrane structure, Golgi function is tightly related to its structural integrity. Thus, the first key question in Golgi biology concerns how the stacked Golgi structure forms. During the past decades, proteins with a variety of functions have been identified in the maintenance of Golgi structure and regulation of Golgi function, including but not limited to GRASPs, golgins, kinases, phosphatases, ubiquitin E3 ligases, and deubiquitinases, as summarized above. More detailed investigations need to be done to investigate how Golgi structural proteins and their interacting molecules cooperate together to form the stacked Golgi structure.
Golgi structural and functional defects have been increasingly reported in stress and disease conditions. In addition to its central role in protein sorting and trafficking, the Golgi has been more recently recognized as a hub of signaling pathways, which facilitates Golgi reaction upon stresses and diseases. Thus, the second key question in Golgi biology concerns how the Golgi structure becomes defective in stress and disease conditions. In this regard, much effort and some progress have been made, such as Golgi fragmentation in AD by Cdk5-mediated GRASP65 phosphorylation as discussed above. However, many questions remain. For example, is there an unfolded protein response (UPR)-like mechanism at the Golgi to cope with different stresses? Is there a common stress sensor on the Golgi? How do the signaling pathways on the Golgi sense and transduce stress signals? Apparently, a more systematic analysis of Golgi response to different stressors is necessary. Gene expression profile and posttranslational modification analysis of Golgi structural proteins, Golgi enzymes, and signaling molecules will fast forward the field and shed light on new directions. Considering lipid organization and modification are important for Golgi function and certain lipids can work as signaling molecules, more attention may be put on lipids at the Golgi, in addition to proteins.
The third key question in Golgi biology concerns how Golgi structure alteration affects its function in trafficking, glycosylation, and sorting. The consequence of Golgi fragmentation in different diseases is likely different, but it has been reported that Golgi cisternal unstacking by depleting GRASP proteins enhances protein trafficking. This, however, impairs accurate glycosylation and causes missorting of lysosomal enzymes to the extracellular space (Xiang et al. 2013; Zhang and Wang 2016; Bekier et al. 2017; Wang et al. 2008). Consistently, Golgi fragmentation in AD enhances APP trafficking and Aβ production, while rescue of the Golgi causes APP accumulation in the Golgi and reduces Aβ secretion (Joshi et al. 2014). Most recently, we have obtained evidence that Golgi structure disassembly by GRASP depletion reduces cell attachment and migration while accelerating cell growth and cell cycle progression (Ahat et al. 2019). It will be interesting to investigate how Golgi fragmentation enhances cancer cell proliferation and metastasis in the future. Future efforts may also aim at developing small chemicals or molecular tools to rescue the Golgi structure in diseases, which may delay the disease development.
Acknowledgment
We thank members of the Wang lab for stimulating discussions. This work was supported by the National Institutes of Health (Grants GM112786 and GM105920), MCubed, and the Fast Forward Protein Folding Disease Initiative of the University of Michigan to Y. Wang, and a University of Michigan Rackham Predoctoral fellowship to E. Ahat.
Contributor Information
Malgorzata Kloc, Email: mkloc@houstonmethodist.org.
Jie Li, Email: jieltian@umich.edu.
Erpan Ahat, Email: erpan@umich.edu.
Yanzhuang Wang, Email: yzwang@umich.edu.
References
- Ahat E, Xiang Y, Zhang X, Bekier M, Wang Y. GRASP depletion-mediated Golgi destruction decreases cell adhesion and migration via the reduction of α5β1 integrin. Mol Biol Cell. 2019;30(6):766–777. doi: 10.1091/mbc.E18-07-0462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aizawa M, Fukuda M. Small GTPase Rab2B and its specific binding protein Golgi-associated Rab2B interactor-like 4 (GARI-L4) regulate Golgi morphology. J Biol Chem. 2015;290:22250–22261. doi: 10.1074/jbc.M115.669242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Dosari M, Alkuraya FS. A novel missense mutation in SCYL1BP1 produces geroderma osteodysplastica phenotype indistinguishable from that caused by nullimorphic mutations. Am J Med Genet A. 2009;149A:2093–2098. doi: 10.1002/ajmg.a.32996. [DOI] [PubMed] [Google Scholar]
- Allan BB, Moyer BD, Balch WE. Rab1 recruitment of p115 into a cis-SNARE complex: programming budding COPII vesicles for fusion. Science. 2000;289:444–448. doi: 10.1126/science.289.5478.444. [DOI] [PubMed] [Google Scholar]
- Allan VJ, Thompson HM, Mcniven MA. Motoring around the Golgi. Nat Cell Biol. 2002;4:E236–E242. doi: 10.1038/ncb1002-e236. [DOI] [PubMed] [Google Scholar]
- Alvarez C, Garcia-Mata R, Hauri HP, Sztul E. The p115-interactive proteins GM130 and giantin participate in endoplasmic reticulum-Golgi traffic. J Biol Chem. 2001;276:2693–2700. doi: 10.1074/jbc.M007957200. [DOI] [PubMed] [Google Scholar]
- Alzhanova D, Hruby DE. A trans-Golgi network resident protein, golgin-97, accumulates in viral factories and incorporates into virions during poxvirus infection. J Virol. 2006;80:11520–11527. doi: 10.1128/JVI.00287-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alzhanova D, Hruby DE. A host cell membrane protein, golgin-97, is essential for poxvirus morphogenesis. Virology. 2007;362:421–427. doi: 10.1016/j.virol.2007.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson Nadine S., Mukherjee Indrani, Bentivoglio Christine M., Barlowe Charles. The golgin protein Coy1 functions in intra-Golgi retrograde transport and interacts with the COG complex and Golgi SNAREs. Molecular Biology of the Cell. 2017;28(20):2686–2700. doi: 10.1091/mbc.e17-03-0137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anglesio MS, Evdokimova V, Melnyk N, Zhang L, Fernandez CV, Grundy PE, Leach S, Marra MA, Brooks-Wilson AR, Penninger J, Sorensen PH. Differential expression of a novel ankyrin containing E3 ubiquitin-protein ligase, Hace1, in sporadic Wilms’ tumor versus normal kidney. Hum Mol Genet. 2004;13:2061–2074. doi: 10.1093/hmg/ddh215. [DOI] [PubMed] [Google Scholar]
- Aridor M, Hannan LA. Traffic jam: a compendium of human diseases that affect intracellular transport processes. Traffic. 2000;1:836–851. doi: 10.1034/j.1600-0854.2000.011104.x. [DOI] [PubMed] [Google Scholar]
- Aslan JE, Thomas G. Death by committee: organellar trafficking and communication in apoptosis. Traffic. 2009;10:1390–1404. doi: 10.1111/j.1600-0854.2009.00951.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkin JD, Farg MA, Soo KY, Walker AK, Halloran M, Turner BJ, Nagley P, Horne MK. Mutant SOD1 inhibits ER-Golgi transport in amyotrophic lateral sclerosis. J Neurochem. 2014;129:190–204. doi: 10.1111/jnc.12493. [DOI] [PubMed] [Google Scholar]
- Avitabile E, Di Gaeta S, Torrisi MR, Ward PL, Roizman B, Campadelli-Fiume G. Redistribution of microtubules and Golgi apparatus in herpes simplex virus-infected cells and their role in viral exocytosis. J Virol. 1995;69:7472–7482. doi: 10.1128/jvi.69.12.7472-7482.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayala I, Colanzi A. Alterations of Golgi organization in Alzheimer’s disease: A cause or a consequence? Tissue Cell. 2017;49:133–140. doi: 10.1016/j.tice.2016.11.007. [DOI] [PubMed] [Google Scholar]
- Bai Yunfei, Cui Xiaobo, Gao Dongxue, Wang Yaping, Wang Boqian, Wang Wei. Golgi integral membrane protein 4 manipulates cellular proliferation, apoptosis, and cell cycle in human head and neck cancer. Bioscience Reports. 2018;38(4):BSR20180454. doi: 10.1042/BSR20180454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr FA. Purification and functional interactions of GRASP55 with Rab2. Methods Enzymol. 2005;403:391–401. doi: 10.1016/S0076-6879(05)03034-X. [DOI] [PubMed] [Google Scholar]
- Barr FA, Puype M, Vandekerckhove J, Warren G. GRASP65, a protein involved in the stacking of Golgi cisternae. Cell. 1997;91:253–262. doi: 10.1016/S0092-8674(00)80407-9. [DOI] [PubMed] [Google Scholar]
- Barr FA, Nakamura N, Warren G. Mapping the interaction between GRASP65 and GM130, components of a protein complex involved in the stacking of Golgi cisternae. EMBO J. 1998;17:3258–3268. doi: 10.1093/emboj/17.12.3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr FA, Preisinger C, Kopajtich R, Korner R. Golgi matrix proteins interact with p24 cargo receptors and aid their efficient retention in the Golgi apparatus. J Cell Biol. 2001;155:885–891. doi: 10.1083/jcb.200108102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barretta ML, Spano D, D’Ambrosio C, Cervigni RI, Scaloni A, Corda D, Colanzi A. Aurora-A recruitment and centrosomal maturation are regulated by a Golgi-activated pool of Src during G2. Nat Commun. 2016;7:11727. doi: 10.1038/ncomms11727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekier ME, 2nd, Wang L, Li J, Huang H, Tang D, Zhang X, Wang Y. Knockout of the Golgi stacking proteins GRASP55 and GRASP65 impairs Golgi structure and function. Mol Biol Cell. 2017;28:2833–2842. doi: 10.1091/mbc.e17-02-0112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellouze S, Baillat G, Buttigieg D, De La Grange P, Rabouille C, Haase G. Stathmin 1/2-triggered microtubule loss mediates Golgi fragmentation in mutant SOD1 motor neurons. Mol Neurodegener. 2016;11:43. doi: 10.1186/s13024-016-0111-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergen DJM, Stevenson NL, Skinner REH, Stephens DJ, Hammond CL. The Golgi matrix protein giantin is required for normal cilia function in zebrafish. Biol Open. 2017;6:1180–1189. doi: 10.1242/bio.025502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishe B, Syed GH, Field SJ, Siddiqui A. Role of phosphatidylinositol 4-phosphate (PI4P) and its binding protein GOLPH3 in hepatitis C virus secretion. J Biol Chem. 2012;287:27637–27647. doi: 10.1074/jbc.M112.346569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnon C, Wendeler MW, Paccaud JP, Hauri HP. Selective export of human GPI-anchored proteins from the endoplasmic reticulum. J Cell Sci. 2010;123:1705–1715. doi: 10.1242/jcs.062950. [DOI] [PubMed] [Google Scholar]
- Brandon E, Gao Y, Garcia-Mata R, Alvarez C, Sztul E. Membrane targeting of p115 phosphorylation mutants and their effects on Golgi integrity and secretory traffic. Eur J Cell Biol. 2003;82:411–420. doi: 10.1078/0171-9335-00327. [DOI] [PubMed] [Google Scholar]
- Brede G, Solheim J, Troen G, Prydz H. Characterization of PSKH1, a novel human protein serine kinase with centrosomal, golgi, and nuclear localization. Genomics. 2000;70:82–92. doi: 10.1006/geno.2000.6365. [DOI] [PubMed] [Google Scholar]
- Brede G, Solheim J, Stang E, Prydz H. Mutants of the protein serine kinase PSKH1 disassemble the Golgi apparatus. Exp Cell Res. 2003;291:299–312. doi: 10.1016/j.yexcr.2003.07.009. [DOI] [PubMed] [Google Scholar]
- Bundis F, Neagoe I, Schwappach B, Steinmeyer K. Involvement of Golgin-160 in cell surface transport of renal ROMK channel: co-expression of Golgin-160 increases ROMK currents. Cell Physiol Biochem. 2006;17:1–12. doi: 10.1159/000091454. [DOI] [PubMed] [Google Scholar]
- Burguete AS, Fenn TD, Brunger AT, Pfeffer SR. Rab and Arl GTPase family members cooperate in the localization of the golgin GCC185. Cell. 2008;132:286–298. doi: 10.1016/j.cell.2007.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buschman MD, Xing M, Field SJ. The GOLPH3 pathway regulates Golgi shape and function and is activated by DNA damage. Front Neurosci. 2015;9:362. doi: 10.3389/fnins.2015.00362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campadelli G, Brandimarti R, Di Lazzaro C, Ward PL, Roizman B, Torrisi MR. Fragmentation and dispersal of Golgi proteins and redistribution of glycoproteins and glycolipids processed through the Golgi apparatus after infection with herpes simplex virus 1. Proc Natl Acad Sci USA. 1993;90:2798–2802. doi: 10.1073/pnas.90.7.2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao K, Nakajima R, Meyer HH, Zheng Y. The AAA-ATPase Cdc48/p97 regulates spindle disassembly at the end of mitosis. Cell. 2003;115:355–367. doi: 10.1016/S0092-8674(03)00815-8. [DOI] [PubMed] [Google Scholar]
- Carmena M, Ruchaud S, Earnshaw WC. Making the Auroras glow: regulation of Aurora A and B kinase function by interacting proteins. Curr Opin Cell Biol. 2009;21:796–805. doi: 10.1016/j.ceb.2009.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartier-Michaud A, Bailly AL, Betzi S, Shi X, Lissitzky JC, Zarubica A, Serge A, Roche P, Lugari A, Hamon V, Bardin F, Derviaux C, Lembo F, Audebert S, Marchetto S, Durand B, Borg JP, Shi N, Morelli X, Aurrand-Lions M. Genetic, structural, and chemical insights into the dual function of GRASP55 in germ cell Golgi remodeling and JAM-C polarized localization during spermatogenesis. PLoS Genet. 2017;13:e1006803. doi: 10.1371/journal.pgen.1006803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervigni RI, Bonavita R, Barretta ML, Spano D, Ayala I, Nakamura N, Corda D, Colanzi A. JNK2 controls fragmentation of the Golgi complex and the G2/M transition through phosphorylation of GRASP65. J Cell Sci. 2015;128:2249–2260. doi: 10.1242/jcs.164871. [DOI] [PubMed] [Google Scholar]
- Chen Y, Chen PL, Chen CF, Sharp ZD, Lee WH. Thyroid hormone, T3-dependent phosphorylation and translocation of Trip230 from the Golgi complex to the nucleus. Proc Natl Acad Sci USA. 1999;96:4443–4448. doi: 10.1073/pnas.96.8.4443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng JP, Betin VM, Weir H, Shelmani GM, Moss DK, Lane JD. Caspase cleavage of the Golgi stacking factor GRASP65 is required for Fas/CD95-mediated apoptosis. Cell Death Dis. 2010;1:e82. doi: 10.1038/cddis.2010.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung PY, Limouse C, Mabuchi H, Pfeffer SR (2015) Protein flexibility is required for vesicle tethering at the Golgi. Elife 4. 10.7554/eLife.12790 [DOI] [PMC free article] [PubMed]
- Chia J, Tham KM, Gill DJ, Bard-Chapeau EA, Bard FA. ERK8 is a negative regulator of O-GalNAc glycosylation and cell migration. Elife. 2014;3:e01828. doi: 10.7554/eLife.01828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ching YP, Leong VY, Lee MF, Xu HT, Jin DY, Ng IO. P21-activated protein kinase is overexpressed in hepatocellular carcinoma and enhances cancer metastasis involving c-Jun NH2-terminal kinase activation and paxillin phosphorylation. Cancer Res. 2007;67:3601–3608. doi: 10.1158/0008-5472.CAN-06-3994. [DOI] [PubMed] [Google Scholar]
- Chiu R, Novikov L, Mukherjee S, Shields D. A caspase cleavage fragment of p115 induces fragmentation of the Golgi apparatus and apoptosis. J Cell Biol. 2002;159:637–648. doi: 10.1083/jcb.200208013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu CF, Ghanekar Y, Frost L, Diao A, Morrison D, Mckenzie E, Lowe M. ZFPL1, a novel ring finger protein required for cis-Golgi integrity and efficient ER-to-Golgi transport. EMBO J. 2008;27:934–947. doi: 10.1038/emboj.2008.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cluett EB, Brown WJ. Adhesion of Golgi cisternae by proteinaceous interactions: intercisternal bridges as putative adhesive structures. J Cell Sci. 1992;103:773–784. doi: 10.1242/jcs.103.3.773. [DOI] [PubMed] [Google Scholar]
- Colanzi A, Hidalgo Carcedo C, Persico A, Cericola C, Turacchio G, Bonazzi M, Luini A, Corda D. The Golgi mitotic checkpoint is controlled by BARS-dependent fission of the Golgi ribbon into separate stacks in G2. EMBO J. 2007;26:2465–2476. doi: 10.1038/sj.emboj.7601686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coune PG, Bensadoun JC, Aebischer P, Schneider BL. Rab1A over-expression prevents Golgi apparatus fragmentation and partially corrects motor deficits in an alpha-synuclein based rat model of Parkinson’s disease. J Parkinsons Dis. 2011;1:373–387. doi: 10.3233/JPD-2011-11058. [DOI] [PubMed] [Google Scholar]
- Cruz-Garcia D, Vazquez-Martinez R, Peinado JR, Anouar Y, Tonon MC, Vaudry H, Castano JP, Malagon MM. Identification and characterization of two novel (neuro)endocrine long coiled-coil proteins. FEBS Lett. 2007;581:3149–3156. doi: 10.1016/j.febslet.2007.06.002. [DOI] [PubMed] [Google Scholar]
- Cui F, Wang Y. HACE1 (HECT domain and ankyrin repeat containing E3 ubiquitin protein ligase 1) Atlas Genet Cytogenet Oncol Haematol. 2012;17:333–336. doi: 10.4267/2042/49701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Angelo G, Prencipe L, Iodice L, Beznoussenko G, Savarese M, Marra P, Di Tullio G, Martire G, De Matteis MA, Bonatti S. GRASP65 and GRASP55 sequentially promote the transport of C-terminal valine-bearing cargos to and through the Golgi complex. J Biol Chem. 2009;284:34849–34860. doi: 10.1074/jbc.M109.068403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis EJ, Lachaud C, Appleton P, Macartney TJ, Nathke I, Rouse J. DVC1 (C1orf124) recruits the p97 protein segregase to sites of DNA damage. Nat Struct Mol Biol. 2012;19:1093–1100. doi: 10.1038/nsmb.2394. [DOI] [PubMed] [Google Scholar]
- Dejgaard SY, Murshid A, Erman A, Kizilay O, Verbich D, Lodge R, Dejgaard K, Ly-Hartig TB, Pepperkok R, Simpson JC, Presley JF. Rab18 and Rab43 have key roles in ER-Golgi trafficking. J Cell Sci. 2008;121:2768–2781. doi: 10.1242/jcs.021808. [DOI] [PubMed] [Google Scholar]
- Delgui LR, Rodriguez JF, Colombo MI. The endosomal pathway and the Golgi complex are involved in the infectious bursal disease virus life cycle. J Virol. 2013;87:8993–9007. doi: 10.1128/JVI.03152-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dementieva IS, Tereshko V, McCrossan ZA, Solomaha E, Araki D, Xu C, Grigorieff N, Goldstein SA. Pentameric assembly of potassium channel tetramerization domain-containing protein 5. J Mol Biol. 2009;387(1):175–191. doi: 10.1016/j.jmb.2009.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derby MC, Gleeson PA. New insights into membrane trafficking and protein sorting. Int Rev Cytol. 2007;261:47–116. doi: 10.1016/S0074-7696(07)61002-X. [DOI] [PubMed] [Google Scholar]
- Derby MC, Lieu ZZ, Brown D, Stow JL, Goud B, Gleeson PA. The trans-Golgi network golgin, GCC185, is required for endosome-to-Golgi transport and maintenance of Golgi structure. Traffic. 2007;8:758–773. doi: 10.1111/j.1600-0854.2007.00563.x. [DOI] [PubMed] [Google Scholar]
- Diao A, Rahman D, Pappin DJ, Lucocq J, Lowe M. The coiled-coil membrane protein golgin-84 is a novel rab effector required for Golgi ribbon formation. J Cell Biol. 2003;160:201–212. doi: 10.1083/jcb.200207045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diao A, Frost L, Morohashi Y, Lowe M. Coordination of golgin tethering and SNARE assembly: GM130 binds syntaxin 5 in a p115-regulated manner. J Biol Chem. 2008;283:6957–6967. doi: 10.1074/jbc.M708401200. [DOI] [PubMed] [Google Scholar]
- Dippold HC, Ng MM, Farber-Katz SE, Lee SK, Kerr ML, Peterman MC, Sim R, Wiharto PA, Galbraith KA, Madhavarapu S, Fuchs GJ, Meerloo T, Farquhar MG, Zhou H, Field SJ. GOLPH3 bridges phosphatidylinositol-4-phosphate and actomyosin to stretch and shape the Golgi to promote budding. Cell. 2009;139:337–351. doi: 10.1016/j.cell.2009.07.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirac-Svejstrup AB, Shorter J, Waters MG, Warren G. Phosphorylation of the vesicle-tethering protein p115 by a casein kinase II-like enzyme is required for Golgi reassembly from isolated mitotic fragments. J Cell Biol. 2000;150:475–488. doi: 10.1083/jcb.150.3.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drin G, Casella JF, Gautier R, Boehmer T, Schwartz TU, Antonny B. A general amphipathic alpha-helical motif for sensing membrane curvature. Nat Struct Mol Biol. 2007;14:138–146. doi: 10.1038/nsmb1194. [DOI] [PubMed] [Google Scholar]
- Drin G, Morello V, Casella JF, Gounon P, Antonny B. Asymmetric tethering of flat and curved lipid membranes by a golgin. Science. 2008;320:670–673. doi: 10.1126/science.1155821. [DOI] [PubMed] [Google Scholar]
- Dupont N, Jiang S, Pilli M, Ornatowski W, Bhattacharya D, Deretic V. Autophagy-based unconventional secretory pathway for extracellular delivery of IL-1beta. EMBO J. 2011;30:4701–4711. doi: 10.1038/emboj.2011.398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duran JM, Kinseth M, Bossard C, Rose DW, Polishchuk R, Wu CC, Yates J, Zimmerman T, Malhotra V. The role of GRASP55 in Golgi fragmentation and entry of cells into mitosis. Mol Biol Cell. 2008;19:2579–2587. doi: 10.1091/mbc.e07-10-0998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efimov A, Kharitonov A, Efimova N, Loncarek J, Miller PM, Andreyeva N, Gleeson P, Galjart N, Maia AR, Mcleod IX, Yates JR, 3rd, Maiato H, Khodjakov A, Akhmanova A, Kaverina I. Asymmetric CLASP-dependent nucleation of noncentrosomal microtubules at the trans-Golgi network. Dev Cell. 2007;12:917–930. doi: 10.1016/j.devcel.2007.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evin G. How accelerated Golgi trafficking may drive Alzheimer’s disease (comment on DOI 10.1002/bies.201400116) Bioessays. 2015;37:232–233. doi: 10.1002/bies.201400219. [DOI] [PubMed] [Google Scholar]
- Eystathioy T, Jakymiw A, Fujita DJ, Fritzler MJ, Chan EK. Human autoantibodies to a novel Golgi protein golgin-67: high similarity with golgin-95/gm 130 autoantigen. J Autoimmun. 2000;14:179–187. doi: 10.1006/jaut.1999.0359. [DOI] [PubMed] [Google Scholar]
- Farber-Katz SE, Dippold HC, Buschman MD, Peterman MC, Xing M, Noakes CJ, Tat J, Ng MM, Rahajeng J, Cowan DM, Fuchs GJ, Zhou H, Field SJ. DNA damage triggers Golgi dispersal via DNA-PK and GOLPH3. Cell. 2014;156:413–427. doi: 10.1016/j.cell.2013.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farhan H, Rabouille C. Signalling to and from the secretory pathway. J Cell Sci. 2011;124:171–180. doi: 10.1242/jcs.076455. [DOI] [PubMed] [Google Scholar]
- Feinstein TN, Linstedt AD. GRASP55 regulates Golgi ribbon formation. Mol Biol Cell. 2008;19:2696–2707. doi: 10.1091/mbc.e07-11-1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Follit JA, San Agustin JT, Xu F, Jonassen JA, Samtani R, Lo CW, Pazour GJ. The Golgin GMAP210/TRIP11 anchors IFT20 to the Golgi complex. PLoS Genet. 2008;4:e1000315. doi: 10.1371/journal.pgen.1000315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forno LS. Neuropathology of Parkinson’s disease. J Neuropathol Exp Neurol. 1996;55:259–272. doi: 10.1097/00005072-199603000-00001. [DOI] [PubMed] [Google Scholar]
- Franke WW, Kartenbeck J, Krien S, Vanderwoude WJ, Scheer U, Morre DJ. Inter- and intracisternal elements of the Golgi apparatus. A system of membrane-to-membrane cross-links. Z Zellforsch Mikrosk Anat. 1972;132:365–380. doi: 10.1007/BF02450714. [DOI] [PubMed] [Google Scholar]
- Fridmann-Sirkis Y, Siniossoglou S, Pelham HR. TMF is a golgin that binds Rab6 and influences Golgi morphology. BMC Cell Biol. 2004;5:18. doi: 10.1186/1471-2121-5-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X, Ng C, Feng D, Liang C. Cdc48p is required for the cell cycle commitment point at Start via degradation of the G1-CDK inhibitor Far1p. J Cell Biol. 2003;163:21–26. doi: 10.1083/jcb.200307025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto T, Kuwahara T, Eguchi T, Sakurai M, Komori T, Iwatsubo T. Parkinson’s disease-associated mutant LRRK2 phosphorylates Rab7L1 and modifies trans-Golgi morphology. Biochem Biophys Res Commun. 2018;495:1708–1715. doi: 10.1016/j.bbrc.2017.12.024. [DOI] [PubMed] [Google Scholar]
- Fujita Y, Ohama E, Takatama M, Al-Sarraj S, Okamoto K. Fragmentation of Golgi apparatus of nigral neurons with alpha-synuclein-positive inclusions in patients with Parkinson’s disease. Acta Neuropathol. 2006;112:261–265. doi: 10.1007/s00401-006-0114-4. [DOI] [PubMed] [Google Scholar]
- Fujita Y, Mizuno Y, Takatama M, Okamoto K. Anterior horn cells with abnormal TDP-43 immunoreactivities show fragmentation of the Golgi apparatus in ALS. J Neurol Sci. 2008;269:30–34. doi: 10.1016/j.jns.2007.12.016. [DOI] [PubMed] [Google Scholar]
- Fujita K, Nakamura Y, Oka T, Ito H, Tamura T, Tagawa K, Sasabe T, Katsuta A, Motoki K, Shiwaku H, Sone M, Yoshida C, Katsuno M, Eishi Y, Murata M, Taylor JP, Wanker EE, Kono K, Tashiro S, Sobue G, La Spada AR, Okazawa H. A functional deficiency of TERA/VCP/p97 contributes to impaired DNA repair in multiple polyglutamine diseases. Nat Commun. 2013;4:1816. doi: 10.1038/ncomms2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganley IG, Espinosa E, Pfeffer SR. A syntaxin 10-SNARE complex distinguishes two distinct transport routes from endosomes to the trans-Golgi in human cells. J Cell Biol. 2008;180:159–172. doi: 10.1083/jcb.200707136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge L, Melville D, Zhang M, Schekman R. The ER-Golgi intermediate compartment is a key membrane source for the LC3 lipidation step of autophagosome biogenesis. Elife. 2013;2:e00947. doi: 10.7554/eLife.00947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gee HY, Noh SH, Tang BL, Kim KH, Lee MG. Rescue of DeltaF508-CFTR trafficking via a GRASP-dependent unconventional secretion pathway. Cell. 2011;146:746–760. doi: 10.1016/j.cell.2011.07.021. [DOI] [PubMed] [Google Scholar]
- Geng J, Klionsky DJ. The Golgi as a potential membrane source for autophagy. Autophagy. 2010;6:950–951. doi: 10.4161/auto.6.7.13009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng J, Nair U, Yasumura-Yorimitsu K, Klionsky DJ. Post-Golgi Sec proteins are required for autophagy in Saccharomyces cerevisiae. Mol Biol Cell. 2010;21:2257–2269. doi: 10.1091/mbc.e09-11-0969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh DK, Roy A, Ranjan A. The ATPase VCP/p97 functions as a disaggregase against toxic Huntingtin-exon1 aggregates. FEBS Lett. 2018;592:2680–2692. doi: 10.1002/1873-3468.13213. [DOI] [PubMed] [Google Scholar]
- Gilbert CE, Zuckerman DM, Currier PL, Machamer CE. Three basic residues of intracellular loop 3 of the beta-1 adrenergic receptor are required for golgin-160-dependent trafficking. Int J Mol Sci. 2014;15:2929–2945. doi: 10.3390/ijms15022929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillingham AK, Munro S. Finding the Golgi: Golgin coiled-coil proteins show the way. Trends Cell Biol. 2016;26:399–408. doi: 10.1016/j.tcb.2016.02.005. [DOI] [PubMed] [Google Scholar]
- Gillingham AK, Tong AH, Boone C, Munro S. The GTPase Arf1p and the ER to Golgi cargo receptor Erv14p cooperate to recruit the golgin Rud3p to the cis-Golgi. J Cell Biol. 2004;167:281–292. doi: 10.1083/jcb.200407088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldenring JR. A central role for vesicle trafficking in epithelial neoplasia: intracellular highways to carcinogenesis. Nat Rev Cancer. 2013;13:813–820. doi: 10.1038/nrc3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosavi N, Lee HJ, Lee JS, Patel S, Lee SJ. Golgi fragmentation occurs in the cells with prefibrillar alpha-synuclein aggregates and precedes the formation of fibrillar inclusion. J Biol Chem. 2002;277:48984–48992. doi: 10.1074/jbc.M208194200. [DOI] [PubMed] [Google Scholar]
- Gosavi Prajakta, Houghton Fiona J., McMillan Paul J., Hanssen Eric, Gleeson Paul A. The Golgi ribbon in mammalian cells negatively regulates autophagy by modulating mTOR activity. Journal of Cell Science. 2017;131(3):jcs211987. doi: 10.1242/jcs.211987. [DOI] [PubMed] [Google Scholar]
- Goud B, Zahraoui A, Tavitian A, Saraste J. Small GTP-binding protein associated with Golgi cisternae. Nature. 1990;345:553–556. doi: 10.1038/345553a0. [DOI] [PubMed] [Google Scholar]
- Goud B, Liu S, Storrie B. Rab proteins as major determinants of the Golgi complex structure. Small GTPases. 2018;9:66–75. doi: 10.1080/21541248.2017.1384087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greninger AL, Knudsen GM, Betegon M, Burlingame AL, Derisi JL. The 3A protein from multiple picornaviruses utilizes the golgi adaptor protein ACBD3 to recruit PI4KIIIbeta. J Virol. 2012;86:3605–3616. doi: 10.1128/JVI.06778-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greninger AL, Knudsen GM, Betegon M, Burlingame AL, Derisi JL. ACBD3 interaction with TBC1 domain 22 protein is differentially affected by enteroviral and kobuviral 3A protein binding. MBio. 2013;4:e00098-13. doi: 10.1128/mBio.00098-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubser C, Bergamaschi D, Hollinshead M, Lu X, Van Kuppeveld FJ, Smith GL. A new inhibitor of apoptosis from vaccinia virus and eukaryotes. PLoS Pathog. 2007;3:e17. doi: 10.1371/journal.ppat.0030017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Chang C, Huang R, Liu B, Bao L, Liu W. AP1 is essential for generation of autophagosomes from the trans-Golgi network. J Cell Sci. 2012;125:1706–1715. doi: 10.1242/jcs.093203. [DOI] [PubMed] [Google Scholar]
- Hansen MD, Johnsen IB, Stiberg KA, Sherstova T, Wakita T, Richard GM, Kandasamy RK, Meurs EF, Anthonsen MW. Hepatitis C virus triggers Golgi fragmentation and autophagy through the immunity-related GTPase M. Proc Natl Acad Sci USA. 2017;114:E3462–E3471. doi: 10.1073/pnas.1616683114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Q, Jiao S, Shi Z, Li C, Meng X, Zhang Z, Wang Y, Song X, Wang W, Zhang R, Zhao Y, Wong CC, Zhou Z. A non-canonical role of the p97 complex in RIG-I antiviral signaling. EMBO J. 2015;34:2903–2920. doi: 10.15252/embj.201591888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harak C, Lohmann V. Ultrastructure of the replication sites of positive-strand RNA viruses. Virology. 2015;479–480:418–433. doi: 10.1016/j.virol.2015.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauler F, Mallery DL, Mcewan WA, Bidgood SR, James LC. AAA ATPase p97/VCP is essential for TRIM21-mediated virus neutralization. Proc Natl Acad Sci USA. 2012;109:19733–19738. doi: 10.1073/pnas.1210659109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes GL, Brown FC, Haas AK, Nottingham RM, Barr FA, Pfeffer SR. Multiple Rab GTPase binding sites in GCC185 suggest a model for vesicle tethering at the trans-Golgi. Mol Biol Cell. 2009;20:209–217. doi: 10.1091/mbc.e08-07-0740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich F, Nanda H, Goh HZ, Bachert C, Losche M, Linstedt AD. Myristoylation restricts orientation of the GRASP domain on membranes and promotes membrane tethering. J Biol Chem. 2014;289:9683–9691. doi: 10.1074/jbc.M113.543561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennies HC, Kornak U, Zhang H, Egerer J, Zhang X, Seifert W, Kuhnisch J, Budde B, Natebus M, Brancati F, Wilcox WR, Muller D, Kaplan PB, Rajab A, Zampino G, Fodale V, Dallapiccola B, Newman W, Metcalfe K, Clayton-Smith J, Tassabehji M, Steinmann B, Barr FA, Nurnberg P, Wieacker P, Mundlos S. Gerodermia osteodysplastica is caused by mutations in SCYL1BP1, a Rab-6 interacting golgin. Nat Genet. 2008;40:1410–1412. doi: 10.1038/ng.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetzer M, Meyer HH, Walther TC, Bilbao-Cortes D, Warren G, Mattaj IW. Distinct AAA-ATPase p97 complexes function in discrete steps of nuclear assembly. Nat Cell Biol. 2001;3:1086–1091. doi: 10.1038/ncb1201-1086. [DOI] [PubMed] [Google Scholar]
- Hicks SW, Machamer CE. The NH2-terminal domain of Golgin-160 contains both golgi and nuclear targeting information. J Biol Chem. 2002;277:35833–35839. doi: 10.1074/jbc.M206280200. [DOI] [PubMed] [Google Scholar]
- Hicks SW, Machamer CE. Golgi structure in stress sensing and apoptosis. Biochim Biophys Acta. 2005;1744:406–414. doi: 10.1016/j.bbamcr.2005.03.002. [DOI] [PubMed] [Google Scholar]
- Hidalgo Carcedo C, Bonazzi M, Spano S, Turacchio G, Colanzi A, Luini A, Corda D. Mitotic Golgi partitioning is driven by the membrane-fissioning protein CtBP3/BARS. Science. 2004;305:93–96. doi: 10.1126/science.1097775. [DOI] [PubMed] [Google Scholar]
- Hoogenraad CC, Akhmanova A, Howell SA, Dortland BR, De Zeeuw CI, Willemsen R, Visser P, Grosveld F, Galjart N. Mammalian Golgi-associated Bicaudal-D2 functions in the dynein-dynactin pathway by interacting with these complexes. EMBO J. 2001;20:4041–4054. doi: 10.1093/emboj/20.15.4041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- How PC, Shields D. Tethering function of the caspase cleavage fragment of Golgi protein p115 promotes apoptosis via a p53-dependent pathway. J Biol Chem. 2011;286:8565–8576. doi: 10.1074/jbc.M110.175174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao PW, Chang C. Isolation and characterization of ARA160 as the first androgen receptor N-terminal-associated coactivator in human prostate cells. J Biol Chem. 1999;274:22373–22379. doi: 10.1074/jbc.274.32.22373. [DOI] [PubMed] [Google Scholar]
- Hu L, Liu M, Chen L, Chan TH, Wang J, Huo KK, Zheng BJ, Xie D, Guan XY. SCYL1 binding protein 1 promotes the ubiquitin-dependent degradation of Pirh2 and has tumor-suppressive function in the development of hepatocellular carcinoma. Carcinogenesis. 2012;33:1581–1588. doi: 10.1093/carcin/bgs162. [DOI] [PubMed] [Google Scholar]
- Hu F, Shi X, Li B, Huang X, Morelli X, Shi N. Structural basis for the interaction between the Golgi reassembly-stacking protein GRASP65 and the Golgi matrix protein GM130. J Biol Chem. 2015;290:26373–26382. doi: 10.1074/jbc.M115.657940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Wang Y. Golgi structure formation, function, and post-translational modifications in mammalian cells. F1000Res. 2017;6:2050. doi: 10.12688/f1000research.11900.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Tang D, Wang Y. Monoubiquitination of syntaxin 5 regulates Golgi membrane dynamics during the cell cycle. Dev Cell. 2016;38:73–85. doi: 10.1016/j.devcel.2016.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttner WB, Ohashi M, Kehlenbach RH, Barr FA, Bauerfeind R, Braunling O, Corbeil D, Hannah M, Pasolli HA, Schmidt A, et al. Biogenesis of neurosecretory vesicles. Cold Spring Harb Symp Quant Biol. 1995;60:315–327. doi: 10.1101/SQB.1995.060.01.036. [DOI] [PubMed] [Google Scholar]
- Jakymiw A, Raharjo E, Rattner JB, Eystathioy T, Chan EK, Fujita DJ. Identification and characterization of a novel Golgi protein, golgin-67. J Biol Chem. 2000;275:4137–4144. doi: 10.1074/jbc.275.6.4137. [DOI] [PubMed] [Google Scholar]
- Ji G, Ji H, Mo X, Li T, Yu Y, Hu Z. The role of GRASPs in morphological alterations of Golgi apparatus: mechanisms and effects. Rev Neurosci. 2013;24:485–497. doi: 10.1515/revneuro-2013-0020. [DOI] [PubMed] [Google Scholar]
- Jiang Q, Wang L, Guan Y, Xu H, Niu Y, Han L, Wei YP, Lin L, Chu J, Wang Q, Yang Y, Pei L, Wang JZ, Tian Q. Golgin-84-associated Golgi fragmentation triggers tau hyperphosphorylation by activation of cyclin-dependent kinase-5 and extracellular signal-regulated kinase. Neurobiol Aging. 2014;35:1352–1363. doi: 10.1016/j.neurobiolaging.2013.11.022. [DOI] [PubMed] [Google Scholar]
- Jing J, Junutula JR, Wu C, Burden J, Matern H, Peden AA, Prekeris R. FIP1/RCP binding to Golgin-97 regulates retrograde transport from recycling endosomes to the trans-Golgi network. Mol Biol Cell. 2010;21:3041–3053. doi: 10.1091/mbc.e10-04-0313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johns HL, Gonzalez-Lopez C, Sayers CL, Hollinshead M, Elliott G. Rab6 dependent post-Golgi trafficking of HSV1 envelope proteins to sites of virus envelopment. Traffic. 2014;15:157–178. doi: 10.1111/tra.12134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi G, Wang Y. Golgi defects enhance APP amyloidogenic processing in Alzheimer’s disease. Bioessays. 2015;37:240–247. doi: 10.1002/bies.201400116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi G, Chi Y, Huang Z, Wang Y. Abeta-induced Golgi fragmentation in Alzheimer’s disease enhances Abeta production. Proc Natl Acad Sci USA. 2014;111:E1230–E1239. doi: 10.1073/pnas.1320192111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi G, Bekier ME, 2nd, Wang Y. Golgi fragmentation in Alzheimer’s disease. Front Neurosci. 2015;9:340. doi: 10.3389/fnins.2015.00340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko Y, Tamura K, Totsukawa G, Kondo H. Phosphorylation of p37 is important for Golgi disassembly at mitosis. Biochem Biophys Res Commun. 2010;402:37–41. doi: 10.1016/j.bbrc.2010.09.097. [DOI] [PubMed] [Google Scholar]
- Kano F, Kondo H, Yamamoto A, Kaneko Y, Uchiyama K, Hosokawa N, Nagata K, Murata M. NSF/SNAPs and p97/p47/VCIP135 are sequentially required for cell cycle-dependent reformation of the ER network. Genes Cells. 2005;10:989–999. doi: 10.1111/j.1365-2443.2005.00894.x. [DOI] [PubMed] [Google Scholar]
- Kelly EE, Giordano F, Horgan CP, Jollivet F, Raposo G, Mccaffrey MW. Rab30 is required for the morphological integrity of the Golgi apparatus. Biol Cell. 2012;104:84–101. doi: 10.1111/boc.201100080. [DOI] [PubMed] [Google Scholar]
- Kim HS, Takahashi M, Matsuo K, Ono Y. Recruitment of CG-NAP to the Golgi apparatus through interaction with dynein-dynactin complex. Genes Cells. 2007;12:421–434. doi: 10.1111/j.1365-2443.2007.01055.x. [DOI] [PubMed] [Google Scholar]
- Kimura M, Yoshioka T, Saio M, Banno Y, Nagaoka H, Okano Y. Mitotic catastrophe and cell death induced by depletion of centrosomal proteins. Cell Death Dis. 2013;4:e603. doi: 10.1038/cddis.2013.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M, Takagi S, Nakashima S. Aurora A regulates the architecture of the Golgi apparatus. Exp Cell Res. 2018;367:73–80. doi: 10.1016/j.yexcr.2018.03.024. [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Manno A, Kakizuka A. Involvement of valosin-containing protein (VCP)/p97 in the formation and clearance of abnormal protein aggregates. Genes Cells. 2007;12:889–901. doi: 10.1111/j.1365-2443.2007.01099.x. [DOI] [PubMed] [Google Scholar]
- Kodani A, Sutterlin C. The Golgi protein GM130 regulates centrosome morphology and function. Mol Biol Cell. 2008;19:745–753. doi: 10.1091/mbc.e07-08-0847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodani A, Kristensen I, Huang L, Sutterlin C. GM130-dependent control of Cdc42 activity at the Golgi regulates centrosome organization. Mol Biol Cell. 2009;20:1192–1200. doi: 10.1091/mbc.e08-08-0834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koegler E, Bonnon C, Waldmeier L, Mitrovic S, Halbeisen R, Hauri HP. p28, a novel ERGIC/cis Golgi protein, required for Golgi ribbon formation. Traffic. 2010;11:70–89. doi: 10.1111/j.1600-0854.2009.01009.x. [DOI] [PubMed] [Google Scholar]
- Kondo H, Rabouille C, Newman R, Levine TP, Pappin D, Freemont P, Warren G. p47 is a cofactor for p97-mediated membrane fusion. Nature. 1997;388:75–78. doi: 10.1038/40411. [DOI] [PubMed] [Google Scholar]
- Koreishi M, Gniadek TJ, Yu S, Masuda J, Honjo Y, Satoh A. The golgin tether giantin regulates the secretory pathway by controlling stack organization within Golgi apparatus. PLoS One. 2013;8:e59821. doi: 10.1371/journal.pone.0059821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuna RS, Field SJ. GOLPH3: A Golgi phosphatidylinositol(4)phosphate effector that directs vesicle trafficking and drives cancer. J Lipid Res. 2018;60(2):269–275. doi: 10.1194/jlr.R088328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo A, Zhong C, Lane WS, Derynck R. Transmembrane transforming growth factor-alpha tethers to the PDZ domain-containing, Golgi membrane-associated protein p59/GRASP55. EMBO J. 2000;19:6427–6439. doi: 10.1093/emboj/19.23.6427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane JD, Lucocq J, Pryde J, Barr FA, Woodman PG, Allan VJ, Lowe M. Caspase-mediated cleavage of the stacking protein GRASP65 is required for Golgi fragmentation during apoptosis. J Cell Biol. 2002;156:495–509. doi: 10.1083/jcb.200110007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lashuel HA, Hirling H. Rescuing defective vesicular trafficking protects against alpha-synuclein toxicity in cellular and animal models of Parkinson’s disease. ACS Chem Biol. 2006;1:420–424. doi: 10.1021/cb600331e. [DOI] [PubMed] [Google Scholar]
- Lavi E, Wang Q, Weiss SR, Gonatas NK. Syncytia formation induced by coronavirus infection is associated with fragmentation and rearrangement of the Golgi apparatus. Virology. 1996;221:325–334. doi: 10.1006/viro.1996.0382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TH, Linstedt AD. Potential role for protein kinases in regulation of bidirectional endoplasmic reticulum-to-Golgi transport revealed by protein kinase inhibitor H89. Mol Biol Cell. 2000;11:2577–2590. doi: 10.1091/mbc.11.8.2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M, Lee HJ, Bae S, Lee YS. Protein sialylation by sialyltransferase involves radiation resistance. Mol Cancer Res. 2008;6:1316–1325. doi: 10.1158/1541-7786.MCR-07-2209. [DOI] [PubMed] [Google Scholar]
- Li T, You H, Mo X, He W, Tang X, Jiang Z, Chen S, Chen Y, Zhang J, Hu Z. GOLPH3 mediated Golgi stress response in modulating N2A cell death upon oxygen-glucose deprivation and reoxygenation injury. Mol Neurobiol. 2016;53:1377–1385. doi: 10.1007/s12035-014-9083-0. [DOI] [PubMed] [Google Scholar]
- Li X, Li M, Tian X, Li Q, Lu Q, Jia Q, Zhang L, Yan J, Li X, Li X. Golgi phosphoprotein 3 inhibits the apoptosis of human glioma cells in part by downregulating N-myc downstream regulated gene 1. Med Sci Monit. 2016;22:3535–3543. doi: 10.12659/MSM.900349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Tang D, Ireland SC, Wang Y. DjA1 maintains Golgi integrity via interaction with GRASP65. Mol Biol Cell. 2019;30(4):478–490. doi: 10.1091/mbc.E18-10-0613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieu ZZ, Derby MC, Teasdale RD, Hart C, Gunn P, Gleeson PA. The golgin GCC88 is required for efficient retrograde transport of cargo from the early endosomes to the trans-Golgi network. Mol Biol Cell. 2007;18:4979–4991. doi: 10.1091/mbc.e07-06-0622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CY, Madsen ML, Yarm FR, Jang YJ, Liu X, Erikson RL. Peripheral Golgi protein GRASP65 is a target of mitotic polo-like kinase (Plk) and Cdc2. Proc Natl Acad Sci USA. 2000;97:12589–12594. doi: 10.1073/pnas.220423497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YC, Chiang TC, Liu YT, Tsai YT, Jang LT, Lee FJ. ARL4A acts with GCC185 to modulate Golgi complex organization. J Cell Sci. 2011;124:4014–4026. doi: 10.1242/jcs.086892. [DOI] [PubMed] [Google Scholar]
- Lin CH, Li H, Lee YN, Cheng YJ, Wu RM, Chien CT. Lrrk regulates the dynamic profile of dendritic Golgi outposts through the golgin Lava lamp. J Cell Biol. 2015;210:471–483. doi: 10.1083/jcb.201411033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linstedt AD, Jesch SA, Mehta A, Lee TH, Garcia-Mata R, Nelson DS, Sztul E. Binding relationships of membrane tethering components. The giantin N terminus and the GM130 N terminus compete for binding to the p115 C terminus. J Biol Chem. 2000;275:10196–10201. doi: 10.1074/jbc.275.14.10196. [DOI] [PubMed] [Google Scholar]
- Liu S, Storrie B. How Rab proteins determine Golgi structure. Int Rev Cell Mol Biol. 2015;315:1–22. doi: 10.1016/bs.ircmb.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Hunt L, Storrie B. Rab41 is a novel regulator of Golgi apparatus organization that is needed for ER-to-Golgi trafficking and cell growth. PLoS One. 2013;8:e71886. doi: 10.1371/journal.pone.0071886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Wang Z, Yang Y, Li Q, Zeng R, Kang J, Wu J. Rab1A mediates proinsulin to insulin conversion in beta-cells by maintaining Golgi stability through interactions with golgin-84. Protein Cell. 2016;7:692–696. doi: 10.1007/s13238-016-0298-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Mei M, Li Q, Roboti P, Pang Q, Ying Z, Gao F, Lowe M, Bao S. Loss of the golgin GM130 causes Golgi disruption, Purkinje neuron loss, and ataxia in mice. Proc Natl Acad Sci USA. 2017;114:346–351. doi: 10.1073/pnas.1608576114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lock JG, Hammond LA, Houghton F, Gleeson PA, Stow JL. E-cadherin transport from the trans-Golgi network in tubulovesicular carriers is selectively regulated by golgin-97. Traffic. 2005;6:1142–1156. doi: 10.1111/j.1600-0854.2005.00349.x. [DOI] [PubMed] [Google Scholar]
- Lowe M, Rabouille C, Nakamura N, Watson R, Jackman M, Jamsa E, Rahman D, Pappin DJ, Warren G. Cdc2 kinase directly phosphorylates the cis-Golgi matrix protein GM130 and is required for Golgi fragmentation in mitosis. Cell. 1998;94:783–793. doi: 10.1016/S0092-8674(00)81737-7. [DOI] [PubMed] [Google Scholar]
- Lowe M, Lane JD, Woodman PG, Allan VJ. Caspase-mediated cleavage of syntaxin 5 and giantin accompanies inhibition of secretory traffic during apoptosis. J Cell Sci. 2004;117:1139–1150. doi: 10.1242/jcs.00950. [DOI] [PubMed] [Google Scholar]
- Lu L, Hong W. Interaction of Arl1-GTP with GRIP domains recruits autoantigens Golgin-97 and Golgin-245/p230 onto the Golgi. Mol Biol Cell. 2003;14:3767–3781. doi: 10.1091/mbc.e03-01-0864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L, Tai G, Hong W. Autoantigen Golgin-97, an effector of Arl1 GTPase, participates in traffic from the endosome to the trans-golgi network. Mol Biol Cell. 2004;15:4426–4443. doi: 10.1091/mbc.e03-12-0872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu PH, Yu CC, Chiang PC, Chen YC, Ho YF, Kung FL, Guh JH. Paclitaxel induces apoptosis through activation of nuclear protein kinase C-delta and subsequent activation of Golgi associated Cdk1 in human hormone refractory prostate cancer. J Urol. 2011;186:2434–2441. doi: 10.1016/j.juro.2011.07.088. [DOI] [PubMed] [Google Scholar]
- Luke MR, Kjer-Nielsen L, Brown DL, Stow JL, Gleeson PA. GRIP domain-mediated targeting of two new coiled-coil proteins, GCC88 and GCC185, to subcompartments of the trans-Golgi network. J Biol Chem. 2003;278:4216–4226. doi: 10.1074/jbc.M210387200. [DOI] [PubMed] [Google Scholar]
- Luke MR, Houghton F, Perugini MA, Gleeson PA. The trans-Golgi network GRIP-domain proteins form alpha-helical homodimers. Biochem J. 2005;388:835–841. doi: 10.1042/BJ20041810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundu T, Tsuda Y, Ito R, Shimizu K, Kobayashi S, Yoshii K, Yoshimatsu K, Arikawa J, Kariwa H. Targeting of severe fever with thrombocytopenia syndrome virus structural proteins to the ERGIC (endoplasmic reticulum Golgi intermediate compartment) and Golgi complex. Biomed Res. 2018;39:27–38. doi: 10.2220/biomedres.39.27. [DOI] [PubMed] [Google Scholar]
- Luo W, Wang Y, Reiser G. p24A, a type I transmembrane protein, controls ARF1-dependent resensitization of protease-activated receptor-2 by influence on receptor trafficking. J Biol Chem. 2007;282:30246–30255. doi: 10.1074/jbc.M703205200. [DOI] [PubMed] [Google Scholar]
- Maag RS, Mancini M, Rosen A, Machamer CE. Caspase-resistant Golgin-160 disrupts apoptosis induced by secretory pathway stress and ligation of death receptors. Mol Biol Cell. 2005;16:3019–3027. doi: 10.1091/mbc.e04-11-0971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machamer CE. Golgi disassembly in apoptosis: cause or effect? Trends Cell Biol. 2003;13:279–281. doi: 10.1016/S0962-8924(03)00101-6. [DOI] [PubMed] [Google Scholar]
- Machamer CE. The Golgi complex in stress and death. Front Neurosci. 2015;9:421. doi: 10.3389/fnins.2015.00421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magdeleine M, Gautier R, Gounon P, Barelli H, Vanni S, Antonny B (2016) A filter at the entrance of the Golgi that selects vesicles according to size and bulk lipid composition. Elife 5. 10.7554/eLife.16988 [DOI] [PMC free article] [PubMed]
- Makhoul C, Gosavi P, Duffield R, Delbridge B, Williamson NA, Gleeson PA. Intersectin-1 interacts with the golgin, GCC88, to couple the actin network and Golgi architecture. Mol Biol Cell. 2019;30(3):370–386. doi: 10.1091/mbc.E18-05-0313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malsam J, Satoh A, Pelletier L, Warren G. Golgin tethers define subpopulations of COPI vesicles. Science. 2005;307:1095–1098. doi: 10.1126/science.1108061. [DOI] [PubMed] [Google Scholar]
- Mancini M, Machamer CE, Roy S, Nicholson DW, Thornberry NA, Casciola-Rosen LA, Rosen A. Caspase-2 is localized at the Golgi complex and cleaves golgin-160 during apoptosis. J Cell Biol. 2000;149:603–612. doi: 10.1083/jcb.149.3.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin C, Leyton L, Hott M, Arancibia Y, Spichiger C, Mcniven MA, Court FA, Concha MI, Burgos PV, Otth C. Herpes simplex virus type 1 neuronal infection perturbs Golgi apparatus integrity through activation of Src tyrosine kinase and Dyn-2 GTPase. Front Cell Infect Microbiol. 2017;7:371. doi: 10.3389/fcimb.2017.00371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez O, Antony C, Pehau-Arnaudet G, Berger EG, Salamero J, Goud B. GTP-bound forms of rab6 induce the redistribution of Golgi proteins into the endoplasmic reticulum. Proc Natl Acad Sci USA. 1997;94:1828–1833. doi: 10.1073/pnas.94.5.1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Carrera LA, Wirth B. Dominant spinal muscular atrophy is caused by mutations in BICD2, an important golgin protein. Front Neurosci. 2015;9:401. doi: 10.3389/fnins.2015.00401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marza E, Taouji S, Barroso K, Raymond AA, Guignard L, Bonneu M, Pallares-Lupon N, Dupuy JW, Fernandez-Zapico ME, Rosenbaum J, Palladino F, Dupuy D, Chevet E. Genome-wide screen identifies a novel p97/CDC-48-dependent pathway regulating ER-stress-induced gene transcription. EMBO Rep. 2015;16:332–340. doi: 10.15252/embr.201439123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matanis T, Akhmanova A, Wulf P, Del Nery E, Weide T, Stepanova T, Galjart N, Grosveld F, Goud B, De Zeeuw CI, Barnekow A, Hoogenraad CC. Bicaudal-D regulates COPI-independent Golgi-ER transport by recruiting the dynein-dynactin motor complex. Nat Cell Biol. 2002;4:986–992. doi: 10.1038/ncb891. [DOI] [PubMed] [Google Scholar]
- Matteis M, Mironov A, Beznoussenko G. The Golgi ribbon and the function of the Golgins. In: Mironov A, Pavelka M, editors. The Golgi apparatus. Vienna: Springer; 2008. [Google Scholar]
- Mayinger P. Signaling at the Golgi. Cold Spring Harbor Perspectives in Biology. 2011;3(5):a005314–a005314. doi: 10.1101/cshperspect.a005314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meerang M, Ritz D, Paliwal S, Garajova Z, Bosshard M, Mailand N, Janscak P, Hubscher U, Meyer H, Ramadan K. The ubiquitin-selective segregase VCP/p97 orchestrates the response to DNA double-strand breaks. Nat Cell Biol. 2011;13:1376–1382. doi: 10.1038/ncb2367. [DOI] [PubMed] [Google Scholar]
- Meyer HH, Wang Y, Warren G. Direct binding of ubiquitin conjugates by the mammalian p97 adaptor complexes, p47 and Ufd1-Npl4. EMBO J. 2002;21:5645–5652. doi: 10.1093/emboj/cdf579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller S, Krijnse-Locker J. Modification of intracellular membrane structures for virus replication. Nat Rev Microbiol. 2008;6:363–374. doi: 10.1038/nrmicro1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misumi Y, Sohda M, Yano A, Fujiwara T, Ikehara Y. Molecular characterization of GCP170, a 170-kDa protein associated with the cytoplasmic face of the Golgi membrane. J Biol Chem. 1997;272:23851–23858. doi: 10.1074/jbc.272.38.23851. [DOI] [PubMed] [Google Scholar]
- Misumi Y, Sohda M, Tashiro A, Sato H, Ikehara Y. An essential cytoplasmic domain for the Golgi localization of coiled-coil proteins with a COOH-terminal membrane anchor. J Biol Chem. 2001;276:6867–6873. doi: 10.1074/jbc.M010121200. [DOI] [PubMed] [Google Scholar]
- Moffat K, Knox C, Howell G, Clark SJ, Yang H, Belsham GJ, Ryan M, Wileman T. Inhibition of the secretory pathway by foot-and-mouth disease virus 2BC protein is reproduced by coexpression of 2B with 2C, and the site of inhibition is determined by the subcellular location of 2C. J Virol. 2007;81:1129–1139. doi: 10.1128/JVI.00393-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollenhauer HH. An intercisternal structure in the Golgi apparatus. J Cell Biol. 1965;24:504–511. doi: 10.1083/jcb.24.3.504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori K, Kato H. A putative nuclear receptor coactivator (TMF/ARA160) associates with hbrm/hSNF2 alpha and BRG-1/hSNF2 beta and localizes in the Golgi apparatus. FEBS Lett. 2002;520:127–132. doi: 10.1016/S0014-5793(02)02803-X. [DOI] [PubMed] [Google Scholar]
- Moyer BD, Allan BB, Balch WE. Rab1 interaction with a GM130 effector complex regulates COPII vesicle cis—Golgi tethering. Traffic. 2001;2:268–276. doi: 10.1034/j.1600-0854.2001.1o007.x. [DOI] [PubMed] [Google Scholar]
- Mukherjee S, Shields D. Nuclear import is required for the pro-apoptotic function of the Golgi protein p115. J Biol Chem. 2009;284:1709–1717. doi: 10.1074/jbc.M807263200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S, Chiu R, Leung SM, Shields D. Fragmentation of the Golgi apparatus: an early apoptotic event independent of the cytoskeleton. Traffic. 2007;8:369–378. doi: 10.1111/j.1600-0854.2007.00542.x. [DOI] [PubMed] [Google Scholar]
- Munro S. The golgin coiled-coil proteins of the Golgi apparatus. Cold Spring Harb Perspect Biol. 2011;3:1–14. doi: 10.1101/cshperspect.a005256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muschalik N, Munro S. Golgins. Curr Biol. 2018;28:R374–R376. doi: 10.1016/j.cub.2018.01.006. [DOI] [PubMed] [Google Scholar]
- Nakamura N, Rabouille C, Watson R, Nilsson T, Hui N, Slusarewicz P, Kreis TE, Warren G. Characterization of a cis-Golgi matrix protein, GM130. J Cell Biol. 1995;131:1715–1726. doi: 10.1083/jcb.131.6.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura N, Lowe M, Levine TP, Rabouille C, Warren G. The vesicle docking protein p115 binds GM130, a cis-Golgi matrix protein, in a mitotically regulated manner. Cell. 1997;89:445–455. doi: 10.1016/S0092-8674(00)80225-1. [DOI] [PubMed] [Google Scholar]
- Nanbo A, Noda T, Ohba Y. Epstein-Barr virus acquires its final envelope on intracellular compartments with Golgi markers. Front Microbiol. 2018;9:454. doi: 10.3389/fmicb.2018.00454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naydenov NG, Harris G, Brown B, Schaefer KL, Das SK, Fisher PB, Ivanov AI. Loss of soluble N-ethylmaleimide-sensitive factor attachment protein alpha (alphaSNAP) induces epithelial cell apoptosis via down-regulation of Bcl-2 expression and disruption of the Golgi. J Biol Chem. 2012;287:5928–5941. doi: 10.1074/jbc.M111.278358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netherton C, Moffat K, Brooks E, Wileman T. A guide to viral inclusions, membrane rearrangements, factories, and viroplasm produced during virus replication. Adv Virus Res. 2007;70:101–182. doi: 10.1016/S0065-3527(07)70004-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neveling K, Martinez-Carrera LA, Holker I, Heister A, Verrips A, Hosseini-Barkooie SM, Gilissen C, Vermeer S, Pennings M, Meijer R, Te Riele M, Frijns CJ, Suchowersky O, Maclaren L, Rudnik-Schoneborn S, Sinke RJ, Zerres K, Lowry RB, Lemmink HH, Garbes L, Veltman JA, Schelhaas HJ, Scheffer H, Wirth B. Mutations in BICD2, which encodes a golgin and important motor adaptor, cause congenital autosomal-dominant spinal muscular atrophy. Am J Hum Genet. 2013;92:946–954. doi: 10.1016/j.ajhg.2013.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng MM, Dippold HC, Buschman MD, Noakes CJ, Field SJ. GOLPH3L antagonizes GOLPH3 to determine Golgi morphology. Mol Biol Cell. 2013;24:796–808. doi: 10.1091/mbc.e12-07-0525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson DW, Thornberry NA. Caspases: killer proteases. Trends Biochem Sci. 1997;22:299–306. doi: 10.1016/S0968-0004(97)01085-2. [DOI] [PubMed] [Google Scholar]
- Nikonova AS, Astsaturov I, Serebriiskii IG, Dunbrack RL, Jr, Golemis EA. Aurora A kinase (AURKA) in normal and pathological cell division. Cell Mol Life Sci. 2013;70:661–687. doi: 10.1007/s00018-012-1073-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikori S, Yamanaka K, Sakurai T, Esaki M, Ogura T. p97 Homologs from Caenorhabditis elegans, CDC-48.1 and CDC-48.2, suppress the aggregate formation of huntingtin exon1 containing expanded polyQ repeat. Genes Cells. 2008;13:827–838. doi: 10.1111/j.1365-2443.2008.01214.x. [DOI] [PubMed] [Google Scholar]
- Nishimura T, Takahashi M, Kim HS, Mukai H, Ono Y. Centrosome-targeting region of CG-NAP causes centrosome amplification by recruiting cyclin E-cdk2 complex. Genes Cells. 2005;10:75–86. doi: 10.1111/j.1365-2443.2005.00816.x. [DOI] [PubMed] [Google Scholar]
- Nonnenmacher ME, Cintrat JC, Gillet D, Weber T. Syntaxin 5-dependent retrograde transport to the trans-Golgi network is required for adeno-associated virus transduction. J Virol. 2015;89:1673–1687. doi: 10.1128/JVI.02520-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozawa K, Casiano CA, Hamel JC, Molinaro C, Fritzler MJ, Chan EK. Fragmentation of Golgi complex and Golgi autoantigens during apoptosis and necrosis. Arthritis Res. 2002;4:R3. doi: 10.1186/ar422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta E, Misumi Y, Sohda M, Fujiwara T, Yano A, Ikehara Y. Identification and characterization of GCP16, a novel acylated Golgi protein that interacts with GCP170. J Biol Chem. 2003;278:51957–51967. doi: 10.1074/jbc.M310014200. [DOI] [PubMed] [Google Scholar]
- Orsi A, Razi M, Dooley HC, Robinson D, Weston AE, Collinson LM, Tooze SA. Dynamic and transient interactions of Atg9 with autophagosomes, but not membrane integration, are required for autophagy. Mol Biol Cell. 2012;23:1860–1873. doi: 10.1091/mbc.e11-09-0746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterrieder A, Sparkes IA, Botchway SW, Ward A, Ketelaar T, De Ruijter N, Hawes C. Stacks off tracks: a role for the golgin AtCASP in plant endoplasmic reticulum-Golgi apparatus tethering. J Exp Bot. 2017;68:3339–3350. doi: 10.1093/jxb/erx167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panic B, Whyte JR, Munro S. The ARF-like GTPases Arl1p and Arl3p act in a pathway that interacts with vesicle-tethering factors at the Golgi apparatus. Curr Biol. 2003;13:405–410. doi: 10.1016/S0960-9822(03)00091-5. [DOI] [PubMed] [Google Scholar]
- Papadopoulos C, Kirchner P, Bug M, Grum D, Koerver L, Schulze N, Poehler R, Dressler A, Fengler S, Arhzaouy K, Lux V, Ehrmann M, Weihl CC, Meyer H. VCP/p97 cooperates with YOD1, UBXD1 and PLAA to drive clearance of ruptured lysosomes by autophagy. EMBO J. 2017;36:135–150. doi: 10.15252/embj.201695148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papoulas O, Hays TS, Sisson JC. The golgin Lava lamp mediates dynein-based Golgi movements during Drosophila cellularization. Nat Cell Biol. 2005;7:612–618. doi: 10.1038/ncb1264. [DOI] [PubMed] [Google Scholar]
- Parisi E, Yahya G, Flores A, Aldea M (2018) Cdc48/p97 segregase is modulated by cyclin-dependent kinase to determine cyclin fate during G1 progression. EMBO J 37. 10.15252/embj.201798724 [DOI] [PMC free article] [PubMed]
- Park JJ, Yi JY, Jin YB, Lee YJ, Lee JS, Lee YS, Ko YG, Lee M. Sialylation of epidermal growth factor receptor regulates receptor activity and chemosensitivity to gefitinib in colon cancer cells. Biochem Pharmacol. 2012;83:849–857. doi: 10.1016/j.bcp.2012.01.007. [DOI] [PubMed] [Google Scholar]
- Partridge JJ, Lopreiato JO, Jr, Latterich M, Indig FE. DNA damage modulates nucleolar interaction of the Werner protein with the AAA ATPase p97/VCP. Mol Biol Cell. 2003;14:4221–4229. doi: 10.1091/mbc.e03-02-0111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng D, Wang J, Zhang R, Jiang F, Tang S, Chen M, Yan J, Sun X, Wang S, Wang T, Yan D, Bao Y, Hu C, Jia W. Common variants in or near ZNRF1, COLEC12, SCYL1BP1 and API5 are associated with diabetic retinopathy in Chinese patients with type 2 diabetes. Diabetologia. 2015;58:1231–1238. doi: 10.1007/s00125-015-3569-9. [DOI] [PubMed] [Google Scholar]
- Perry E, Tsruya R, Levitsky P, Pomp O, Taller M, Weisberg S, Parris W, Kulkarni S, Malovani H, Pawson T, Shpungin S, Nir U. TMF/ARA160 is a BC-box-containing protein that mediates the degradation of Stat3. Oncogene. 2004;23:8908–8919. doi: 10.1038/sj.onc.1208149. [DOI] [PubMed] [Google Scholar]
- Petrosyan A. Onco-Golgi: is fragmentation a gate to cancer progression? Biochem Mol Biol J. 2015;1:16. doi: 10.21767/2471-8084.100006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrosyan A, Holzapfel MS, Muirhead DE, Cheng PW. Restoration of compact Golgi morphology in advanced prostate cancer enhances susceptibility to galectin-1-induced apoptosis by modifying mucin O-glycan synthesis. Mol Cancer Res. 2014;12:1704–1716. doi: 10.1158/1541-7786.MCR-14-0291-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piao H, Kim J, Noh SH, Kweon HS, Kim JY, Lee MG. Sec16A is critical for both conventional and unconventional secretion of CFTR. Sci Rep. 2017;7:39887. doi: 10.1038/srep39887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preisinger C, Short B, De Corte V, Bruyneel E, Haas A, Kopajtich R, Gettemans J, Barr FA. YSK1 is activated by the Golgi matrix protein GM130 and plays a role in cell migration through its substrate 14-3-3{zeta} J Cell Biol. 2004;164:1009–1020. doi: 10.1083/jcb.200310061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preisinger C, Korner R, Wind M, Lehmann WD, Kopajtich R, Barr FA. Plk1 docking to GRASP65 phosphorylated by Cdk1 suggests a mechanism for Golgi checkpoint signalling. EMBO J. 2005;24:753–765. doi: 10.1038/sj.emboj.7600569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Procter DJ, Banerjee A, Nukui M, Kruse K, Gaponenko V, Murphy EA, Komarova Y, Walsh D. The HCMV assembly compartment is a dynamic Golgi-derived MTOC that controls nuclear rotation and virus spread. Dev Cell. 2018;45:83–100.e7. doi: 10.1016/j.devcel.2018.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puthenveedu MA, Bachert C, Puri S, Lanni F, Linstedt AD. GM130 and GRASP65-dependent lateral cisternal fusion allows uniform Golgi-enzyme distribution. Nat Cell Biol. 2006;8:238–248. doi: 10.1038/ncb1366. [DOI] [PubMed] [Google Scholar]
- Quiner CA, Jackson WT. Fragmentation of the Golgi apparatus provides replication membranes for human rhinovirus 1A. Virology. 2010;407:185–195. doi: 10.1016/j.virol.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabouille C, Kondylis V. Golgi ribbon unlinking: an organelle-based G2/M checkpoint. Cell Cycle. 2007;6:2723–2729. doi: 10.4161/cc.6.22.4896. [DOI] [PubMed] [Google Scholar]
- Rabouille C, Linstedt AD. GRASP: a multitasking tether. Front Cell Dev Biol. 2016;4:1. doi: 10.3389/fcell.2016.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabouille C, Levine TP, Peters JM, Warren G. An NSF-like ATPase, p97, and NSF mediate cisternal regrowth from mitotic Golgi fragments. Cell. 1995;82:905–914. doi: 10.1016/0092-8674(95)90270-8. [DOI] [PubMed] [Google Scholar]
- Ramadan K, Bruderer R, Spiga FM, Popp O, Baur T, Gotta M, Meyer HH. Cdc48/p97 promotes reformation of the nucleus by extracting the kinase Aurora B from chromatin. Nature. 2007;450:1258–1262. doi: 10.1038/nature06388. [DOI] [PubMed] [Google Scholar]
- Ramirez IB, Lowe M. Golgins and GRASPs: holding the Golgi together. Semin Cell Dev Biol. 2009;20:770–779. doi: 10.1016/j.semcdb.2009.03.011. [DOI] [PubMed] [Google Scholar]
- Ran R, Pan R, Lu A, Xu H, Davis RR, Sharp FR. A novel 165-kDa Golgin protein induced by brain ischemia and phosphorylated by Akt protects against apoptosis. Mol Cell Neurosci. 2007;36:392–407. doi: 10.1016/j.mcn.2007.07.014. [DOI] [PubMed] [Google Scholar]
- Rao KH, Ghosh S, Datta A (2016) Env7p associates with the Golgin protein Imh1 at the trans-Golgi Network in Candida albicans. mSphere 1. 10.1128/mSphere.00080-16 [DOI] [PMC free article] [PubMed]
- Rebmann GM, Grabski R, Sanchez V, Britt WJ (2016) Phosphorylation of Golgi peripheral membrane protein Grasp65 is an integral step in the formation of the human cytomegalovirus cytoplasmic assembly compartment. MBio 7. 10.1128/mBio.01554-16 [DOI] [PMC free article] [PubMed]
- Reddy JV, Burguete AS, Sridevi K, Ganley IG, Nottingham RM, Pfeffer SR. A functional role for the GCC185 golgin in mannose 6-phosphate receptor recycling. Mol Biol Cell. 2006;17:4353–4363. doi: 10.1091/mbc.e06-02-0153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rejman Lipinski A, Heymann J, Meissner C, Karlas A, Brinkmann V, Meyer TF, Heuer D. Rab6 and Rab11 regulate Chlamydia trachomatis development and golgin-84-dependent Golgi fragmentation. PLoS Pathog. 2009;5:e1000615. doi: 10.1371/journal.ppat.1000615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rendon WO, Martinez-Alonso E, Tomas M, Martinez-Martinez N, Martinez-Menarguez JA. Golgi fragmentation is Rab and SNARE dependent in cellular models of Parkinson’s disease. Histochem Cell Biol. 2013;139:671–684. doi: 10.1007/s00418-012-1059-4. [DOI] [PubMed] [Google Scholar]
- Rios RM, Sanchis A, Tassin AM, Fedriani C, Bornens M. GMAP-210 recruits gamma-tubulin complexes to cis-Golgi membranes and is required for Golgi ribbon formation. Cell. 2004;118:323–335. doi: 10.1016/j.cell.2004.07.012. [DOI] [PubMed] [Google Scholar]
- Risco C, Carrascosa JL, Frey TK. Structural maturation of rubella virus in the Golgi complex. Virology. 2003;312:261–269. doi: 10.1016/S0042-6822(03)00384-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ristic G, Sutton JR, Libohova K, Todi SV. Toxicity and aggregation of the polyglutamine disease protein, ataxin-3 is regulated by its binding to VCP/p97 in Drosophila melanogaster. Neurobiol Dis. 2018;116:78–92. doi: 10.1016/j.nbd.2018.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivero S, Cardenas J, Bornens M, Rios RM. Microtubule nucleation at the cis-side of the Golgi apparatus requires AKAP450 and GM130. EMBO J. 2009;28:1016–1028. doi: 10.1038/emboj.2009.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohde J, Emschermann F, Knittler MR, Rziha HJ. Orf virus interferes with MHC class I surface expression by targeting vesicular transport and Golgi. BMC Vet Res. 2012;8:114. doi: 10.1186/1746-6148-8-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosing M, Ossendorf E, Rak A, Barnekow A. Giantin interacts with both the small GTPase Rab6 and Rab1. Exp Cell Res. 2007;313:2318–2325. doi: 10.1016/j.yexcr.2007.03.031. [DOI] [PubMed] [Google Scholar]
- Salanueva IJ, Novoa RR, Cabezas P, Lopez-Iglesias C, Carrascosa JL, Elliott RM, Risco C. Polymorphism and structural maturation of bunyamwera virus in Golgi and post-Golgi compartments. J Virol. 2003;77:1368–1381. doi: 10.1128/JVI.77.2.1368-1381.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salonen A, Ahola T, Kaariainen L. Viral RNA replication in association with cellular membranes. Curr Top Microbiol Immunol. 2005;285:139–173. doi: 10.1007/3-540-26764-6_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saraiva N, Prole DL, Carrara G, Maluquer De Motes C, Johnson BF, Byrne B, Taylor CW, Smith GL. Human and viral Golgi anti-apoptotic proteins (GAAPs) oligomerize via different mechanisms and monomeric GAAP inhibits apoptosis and modulates calcium. J Biol Chem. 2013;288:13057–13067. doi: 10.1074/jbc.M112.414367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki J, Ishikawa K, Arita M, Taniguchi K. ACBD3-mediated recruitment of PI4KB to picornavirus RNA replication sites. EMBO J. 2012;31:754–766. doi: 10.1038/emboj.2011.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh A, Wang Y, Malsam J, Beard MB, Warren G. Golgin-84 is a rab1 binding partner involved in Golgi structure. Traffic. 2003;4:153–161. doi: 10.1034/j.1600-0854.2003.00103.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sbodio JI, Machamer CE. Identification of a redox-sensitive cysteine in GCP60 that regulates its interaction with golgin-160. J Biol Chem. 2007;282:29874–29881. doi: 10.1074/jbc.M705794200. [DOI] [PubMed] [Google Scholar]
- Sbodio JI, Hicks SW, Simon D, Machamer CE. GCP60 preferentially interacts with a caspase-generated golgin-160 fragment. J Biol Chem. 2006;281:27924–27931. doi: 10.1074/jbc.M603276200. [DOI] [PubMed] [Google Scholar]
- Schultz MJ, Swindall AF, Bellis SL. Regulation of the metastatic cell phenotype by sialylated glycans. Cancer Metastasis Rev. 2012;31:501–518. doi: 10.1007/s10555-012-9359-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott KL, Kabbarah O, Liang MC, Ivanova E, Anagnostou V, Wu J, Dhakal S, Wu M, Chen S, Feinberg T, Huang J, Saci A, Widlund HR, Fisher DE, Xiao Y, Rimm DL, Protopopov A, Wong KK, Chin L. GOLPH3 modulates mTOR signalling and rapamycin sensitivity in cancer. Nature. 2009;459:1085–1090. doi: 10.1038/nature08109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seemann J, Jokitalo EJ, Warren G. The role of the tethering proteins p115 and GM130 in transport through the Golgi apparatus in vivo. Mol Biol Cell. 2000;11:635–645. doi: 10.1091/mbc.11.2.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert W, Kuhnisch J, Maritzen T, Horn D, Haucke V, Hennies HC. Cohen syndrome-associated protein, COH1, is a novel, giant Golgi matrix protein required for Golgi integrity. J Biol Chem. 2011;286:37665–37675. doi: 10.1074/jbc.M111.267971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selkoe DJ, Hardy J. The amyloid hypothesis of Alzheimer’s disease at 25 years. EMBO Mol Med. 2016;8:595–608. doi: 10.15252/emmm.201606210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta D, Linstedt AD. Mitotic inhibition of GRASP65 organelle tethering involves Polo-like kinase 1 (PLK1) phosphorylation proximate to an internal PDZ ligand. J Biol Chem. 2010;285:39994–40003. doi: 10.1074/jbc.M110.189449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin JJH, Gillingham AK, Begum F, Chadwick J, Munro S. TBC1D23 is a bridging factor for endosomal vesicle capture by golgins at the trans-Golgi. Nat Cell Biol. 2017;19:1424–1432. doi: 10.1038/ncb3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin JJH, Gillingham AK, Begum F, Chadwick J, Munro S. Author Correction: TBC1D23 is a bridging factor for endosomal vesicle capture by golgins at the trans-Golgi. Nat Cell Biol. 2018;20:222. doi: 10.1038/s41556-017-0026-7. [DOI] [PubMed] [Google Scholar]
- Shitara A, Shibui T, Okayama M, Arakawa T, Mizoguchi I, Sakakura Y, Takuma T. VAMP4 is required to maintain the ribbon structure of the Golgi apparatus. Mol Cell Biochem. 2013;380:11–21. doi: 10.1007/s11010-013-1652-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Short B, Preisinger C, Korner R, Kopajtich R, Byron O, Barr FA. A GRASP55-rab2 effector complex linking Golgi structure to membrane traffic. J Cell Biol. 2001;155:877–883. doi: 10.1083/jcb.200108079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Short JR, Nakayinga R, Hughes GE, Walter CT, Dorrington RA. Providence virus (family: Carmotetraviridae) replicates vRNA in association with the Golgi apparatus and secretory vesicles. J Gen Virol. 2013;94:1073–1078. doi: 10.1099/vir.0.047647-0. [DOI] [PubMed] [Google Scholar]
- Shorter J, Warren G. A role for the vesicle tethering protein, p115, in the post-mitotic stacking of reassembling golgi cisternae in a cell-free system. J Cell Biol. 1999;146:57–70. doi: 10.1083/jcb.146.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorter J, Warren G. Golgi architecture and inheritance. Annu Rev Cell Dev Biol. 2002;18:379–420. doi: 10.1146/annurev.cellbio.18.030602.133733. [DOI] [PubMed] [Google Scholar]
- Shorter J, Watson R, Giannakou ME, Clarke M, Warren G, Barr FA. GRASP55, a second mammalian GRASP protein involved in the stacking of Golgi cisternae in a cell-free system. EMBO J. 1999;18:4949–4960. doi: 10.1093/emboj/18.18.4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorter J, Beard MB, Seemann J, Dirac-Svejstrup AB, Warren G. Sequential tethering of Golgins and catalysis of SNAREpin assembly by the vesicle-tethering protein p115. J Cell Biol. 2002;157:45–62. doi: 10.1083/jcb.200112127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sillibourne JE, Milne DM, Takahashi M, Ono Y, Meek DW. Centrosomal anchoring of the protein kinase CK1delta mediated by attachment to the large, coiled-coil scaffolding protein CG-NAP/AKAP450. J Mol Biol. 2002;322:785–797. doi: 10.1016/S0022-2836(02)00857-4. [DOI] [PubMed] [Google Scholar]
- Sinka R, Gillingham AK, Kondylis V, Munro S. Golgi coiled-coil proteins contain multiple binding sites for Rab family G proteins. J Cell Biol. 2008;183:607–615. doi: 10.1083/jcb.200808018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivan G, Weisberg AS, Americo JL, Moss B. Retrograde transport from early endosomes to the trans-Golgi network enables membrane wrapping and egress of vaccinia virus virions. J Virol. 2016;90:8891–8905. doi: 10.1128/JVI.01114-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slusarewicz P, Nilsson T, Hui N, Watson R, Warren G. Isolation of a matrix that binds medial Golgi enzymes. J Cell Biol. 1994;124:405–413. doi: 10.1083/jcb.124.4.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohda M, Misumi Y, Yamamoto A, Yano A, Nakamura N, Ikehara Y. Identification and characterization of a novel Golgi protein, GCP60, that interacts with the integral membrane protein giantin. J Biol Chem. 2001;276:45298–45306. doi: 10.1074/jbc.M108961200. [DOI] [PubMed] [Google Scholar]
- Sohda M, Misumi Y, Yamamoto A, Nakamura N, Ogata S, Sakisaka S, Hirose S, Ikehara Y, Oda K. Interaction of Golgin-84 with the COG complex mediates the intra-Golgi retrograde transport. Traffic. 2010;11:1552–1566. doi: 10.1111/j.1600-0854.2010.01123.x. [DOI] [PubMed] [Google Scholar]
- Sohda M, Misumi Y, Ogata S, Sakisaka S, Hirose S, Ikehara Y, Oda K. Trans-Golgi protein p230/golgin-245 is involved in phagophore formation. Biochem Biophys Res Commun. 2015;456:275–281. doi: 10.1016/j.bbrc.2014.11.071. [DOI] [PubMed] [Google Scholar]
- Song C, Wang Q, Li CC. Characterization of the aggregation-prevention activity of p97/valosin-containing protein. Biochemistry. 2007;46:14889–14898. doi: 10.1021/bi700499j. [DOI] [PubMed] [Google Scholar]
- Sonnichsen B, Lowe M, Levine T, Jamsa E, Dirac-Svejstrup B, Warren G. A role for giantin in docking COPI vesicles to Golgi membranes. J Cell Biol. 1998;140:1013–1021. doi: 10.1083/jcb.140.5.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soo KY, Halloran M, Sundaramoorthy V, Parakh S, Toth RP, Southam KA, Mclean CA, Lock P, King A, Farg MA, Atkin JD. Rab1-dependent ER-Golgi transport dysfunction is a common pathogenic mechanism in SOD1, TDP-43 and FUS-associated ALS. Acta Neuropathol. 2015;130:679–697. doi: 10.1007/s00401-015-1468-2. [DOI] [PubMed] [Google Scholar]
- Steegmaier M, Klumperman J, Foletti DL, Yoo JS, Scheller RH. Vesicle-associated membrane protein 4 is implicated in trans-Golgi network vesicle trafficking. Mol Biol Cell. 1999;10:1957–1972. doi: 10.1091/mbc.10.6.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun KH, De Pablo Y, Vincent F, Johnson EO, Chavers AK, Shah K. Novel genetic tools reveal Cdk5’s major role in Golgi fragmentation in Alzheimer’s disease. Mol Biol Cell. 2008;19:3052–3069. doi: 10.1091/mbc.e07-11-1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun NK, Huang SL, Chien KY, Chao CC. Golgi-SNARE GS28 potentiates cisplatin-induced apoptosis by forming GS28-MDM2-p53 complexes and by preventing the ubiquitination and degradation of p53. Biochem J. 2012;444:303–314. doi: 10.1042/BJ20112223. [DOI] [PubMed] [Google Scholar]
- Sundaramoorthy S, Goh JB, Rafee S, Murata-Hori M. Mitotic Golgi vesiculation involves mechanisms independent of Ser25 phosphorylation of GM130. Cell Cycle. 2010;9:3100–3105. doi: 10.4161/cc.9.15.12522. [DOI] [PubMed] [Google Scholar]
- Sundaramoorthy V, Sultana JM, Atkin JD. Golgi fragmentation in amyotrophic lateral sclerosis, an overview of possible triggers and consequences. Front Neurosci. 2015;9:400. doi: 10.3389/fnins.2015.00400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutterlin C, Polishchuk R, Pecot M, Malhotra V. The Golgi-associated protein GRASP65 regulates spindle dynamics and is essential for cell division. Mol Biol Cell. 2005;16:3211–3222. doi: 10.1091/mbc.e04-12-1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi M, Shibata H, Shimakawa M, Miyamoto M, Mukai H, Ono Y. Characterization of a novel giant scaffolding protein, CG-NAP, that anchors multiple signaling enzymes to centrosome and the golgi apparatus. J Biol Chem. 1999;274:17267–17274. doi: 10.1074/jbc.274.24.17267. [DOI] [PubMed] [Google Scholar]
- Takahashi M, Yamagiwa A, Nishimura T, Mukai H, Ono Y. Centrosomal proteins CG-NAP and kendrin provide microtubule nucleation sites by anchoring gamma-tubulin ring complex. Mol Biol Cell. 2002;13:3235–3245. doi: 10.1091/mbc.e02-02-0112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D, Wang Y. Cell cycle regulation of Golgi membrane dynamics. Trends Cell Biol. 2013;23:296–304. doi: 10.1016/j.tcb.2013.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D, Mar K, Warren G, Wang Y. Molecular mechanism of mitotic Golgi disassembly and reassembly revealed by a defined reconstitution assay. J Biol Chem. 2008;283:6085–6094. doi: 10.1074/jbc.M707715200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D, Xiang Y, Wang Y. Reconstitution of the cell cycle-regulated Golgi disassembly and reassembly in a cell-free system. Nat Protoc. 2010;5:758–772. doi: 10.1038/nprot.2010.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D, Yuan H, Wang Y. The role of GRASP65 in Golgi cisternal stacking and cell cycle progression. Traffic. 2010;11:827–842. doi: 10.1111/j.1600-0854.2010.01055.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D, Xiang Y, De Renzis S, Rink J, Zheng G, Zerial M, Wang Y. The ubiquitin ligase HACE1 regulates Golgi membrane dynamics during the cell cycle. Nat Commun. 2011;2:501. doi: 10.1038/ncomms1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D, Yuan H, Vielemeyer O, Perez F, Wang Y. Sequential phosphorylation of GRASP65 during mitotic Golgi disassembly. Biol Open. 2012;1:1204–1214. doi: 10.1242/bio.20122659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D, Zhang X, Huang S, Yuan H, Li J, Wang Y. Mena-GRASP65 interaction couples actin polymerization to Golgi ribbon linking. Mol Biol Cell. 2016;27:137–152. doi: 10.1091/mbc.E15-09-0650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terrano DT, Upreti M, Chambers TC. Cyclin-dependent kinase 1-mediated Bcl-xL/Bcl-2 phosphorylation acts as a functional link coupling mitotic arrest and apoptosis. Mol Cell Biol. 2010;30:640–656. doi: 10.1128/MCB.00882-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thyberg J, Moskalewski S. Role of microtubules in the organization of the Golgi complex. Exp Cell Res. 1999;246:263–279. doi: 10.1006/excr.1998.4326. [DOI] [PubMed] [Google Scholar]
- Toki C, Fujiwara T, Sohda M, Hong HS, Misumi Y, Ikehara Y. Identification and characterization of rat 364-kDa Golgi-associated protein recognized by autoantibodies from a patient with rheumatoid arthritis. Cell Struct Funct. 1997;22:565–577. doi: 10.1247/csf.22.565. [DOI] [PubMed] [Google Scholar]
- Truschel ST, Zhang M, Bachert C, Macbeth MR, Linstedt AD. Allosteric regulation of GRASP protein-dependent Golgi membrane tethering by mitotic phosphorylation. J Biol Chem. 2012;287:19870–19875. doi: 10.1074/jbc.M111.326256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukada M, Will E, Gallwitz D. Structural and functional analysis of a novel coiled-coil protein involved in Ypt6 GTPase-regulated protein transport in yeast. Mol Biol Cell. 1999;10:63–75. doi: 10.1091/mbc.10.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchiyama K, Jokitalo E, Lindman M, Jackman M, Kano F, Murata M, Zhang X, Kondo H. The localization and phosphorylation of p47 are important for Golgi disassembly-assembly during the cell cycle. J Cell Biol. 2003;161:1067–1079. doi: 10.1083/jcb.200303048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchiyama K, Totsukawa G, Puhka M, Kaneko Y, Jokitalo E, Dreveny I, Beuron F, Zhang X, Freemont P, Kondo H. p37 is a p97 adaptor required for Golgi and ER biogenesis in interphase and at the end of mitosis. Dev Cell. 2006;11:803–816. doi: 10.1016/j.devcel.2006.10.016. [DOI] [PubMed] [Google Scholar]
- Van Den Boom J, Wolf M, Weimann L, Schulze N, Li F, Kaschani F, Riemer A, Zierhut C, Kaiser M, Iliakis G, Funabiki H, Meyer H. VCP/p97 extracts sterically trapped Ku70/80 rings from DNA in double-strand break repair. Mol Cell. 2016;64:189–198. doi: 10.1016/j.molcel.2016.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dis V, Kuijpers M, Haasdijk ED, Teuling E, Oakes SA, Hoogenraad CC, Jaarsma D. Golgi fragmentation precedes neuromuscular denervation and is associated with endosome abnormalities in SOD1-ALS mouse motor neurons. Acta Neuropathol Commun. 2014;2:38. doi: 10.1186/2051-5960-2-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Valkenburgh H, Shern JF, Sharer JD, Zhu X, Kahn RA. ADP-ribosylation factors (ARFs) and ARF-like 1 (ARL1) have both specific and shared effectors: characterizing ARL1-binding proteins. J Biol Chem. 2001;276:22826–22837. doi: 10.1074/jbc.M102359200. [DOI] [PubMed] [Google Scholar]
- Vielemeyer O, Yuan H, Moutel S, Saint-Fort R, Tang D, Nizak C, Goud B, Wang Y, Perez F. Direct selection of monoclonal phosphospecific antibodies without prior phosphoamino acid mapping. J Biol Chem. 2009;284:20791–20795. doi: 10.1074/jbc.M109.008730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinke FP, Grieve AG, Rabouille C. The multiple facets of the Golgi reassembly stacking proteins. Biochem J. 2011;433:423–433. doi: 10.1042/BJ20101540. [DOI] [PubMed] [Google Scholar]
- Wakabayashi K, Hayashi S, Kakita A, Yamada M, Toyoshima Y, Yoshimoto M, Takahashi H. Accumulation of alpha-synuclein/NACP is a cytopathological feature common to Lewy body disease and multiple system atrophy. Acta Neuropathol. 1998;96:445–452. doi: 10.1007/s004010050918. [DOI] [PubMed] [Google Scholar]
- Walker A, Ward C, Sheldrake TA, Dransfield I, Rossi AG, Pryde JG, Haslett C. Golgi fragmentation during Fas-mediated apoptosis is associated with the rapid loss of GM130. Biochem Biophys Res Commun. 2004;316:6–11. doi: 10.1016/j.bbrc.2004.02.015. [DOI] [PubMed] [Google Scholar]
- Wallis N, Lau CL, Farg MA, Atkin JD, Beart PM, O’Shea RD. SOD1 mutations causing familial amyotrophic lateral sclerosis induce toxicity in astrocytes: evidence for bystander effects in a continuum of astrogliosis. Neurochem Res. 2018;43:157–170. doi: 10.1007/s11064-017-2385-7. [DOI] [PubMed] [Google Scholar]
- Wang Y. Golgi apparatus inheritance. In: Mironov A, Pavelka M, Luini A, editors. The Golgi apparatus. State of the art 110 years after Camillo Golgi’s discovery. New York: Springer; 2008. [Google Scholar]
- Wang Y, Seemann J. Golgi biogenesis. Cold Spring Harb Perspect Biol. 2011;3:a005330. doi: 10.1101/cshperspect.a005330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Seemann J, Pypaert M, Shorter J, Warren G. A direct role for GRASP65 as a mitotically regulated Golgi stacking factor. EMBO J. 2003;22:3279–3290. doi: 10.1093/emboj/cdg317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Satoh A, Warren G, Meyer HH. VCIP135 acts as a deubiquitinating enzyme during p97-p47-mediated reassembly of mitotic Golgi fragments. J Cell Biol. 2004;164:973–978. doi: 10.1083/jcb.200401010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Satoh A, Warren G. Mapping the functional domains of the Golgi stacking factor GRASP65. J Biol Chem. 2005;280:4921–4928. doi: 10.1074/jbc.M412407200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Wei JH, Bisel B, Tang D, Seemann J. Golgi cisternal unstacking stimulates COPI vesicle budding and protein transport. PLoS One. 2008;3:e1647. doi: 10.1371/journal.pone.0001647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Grabski R, Sztul E, Hay JC. p115-SNARE interactions: a dynamic cycle of p115 binding monomeric SNARE motifs and releasing assembled bundles. Traffic. 2015;16:148–171. doi: 10.1111/tra.12242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanschers BF, Van De Vorstenbosch R, Schlager MA, Splinter D, Akhmanova A, Hoogenraad CC, Wieringa B, Fransen JA. A role for the Rab6B Bicaudal-D1 interaction in retrograde transport in neuronal cells. Exp Cell Res. 2007;313:3408–3420. doi: 10.1016/j.yexcr.2007.05.032. [DOI] [PubMed] [Google Scholar]
- Warren G. Intracellular membrane morphology. Philos Trans R Soc Lond B Biol Sci. 1995;349:291–295. doi: 10.1098/rstb.1995.0115. [DOI] [PubMed] [Google Scholar]
- Wei JH, Seemann J. Unraveling the Golgi ribbon. Traffic. 2010;11:1391–1400. doi: 10.1111/j.1600-0854.2010.01114.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei JH, Seemann J. Golgi ribbon disassembly during mitosis, differentiation and disease progression. Curr Opin Cell Biol. 2017;47:43–51. doi: 10.1016/j.ceb.2017.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weide T, Bayer M, Koster M, Siebrasse JP, Peters R, Barnekow A. The Golgi matrix protein GM130: a specific interacting partner of the small GTPase rab1b. EMBO Rep. 2001;2:336–341. doi: 10.1093/embo-reports/kve065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller SG, Capitani M, Cao H, Micaroni M, Luini A, Sallese M, Mcniven MA. Src kinase regulates the integrity and function of the Golgi apparatus via activation of dynamin 2. Proc Natl Acad Sci USA. 2010;107:5863–5868. doi: 10.1073/pnas.0915123107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerbeck JW, Machamer CE. A coronavirus E protein is present in two distinct pools with different effects on assembly and the secretory pathway. J Virol. 2015;89:9313–9323. doi: 10.1128/JVI.01237-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams D, Hicks SW, Machamer CE, Pessin JE. Golgin-160 is required for the Golgi membrane sorting of the insulin-responsive glucose transporter GLUT4 in adipocytes. Mol Biol Cell. 2006;17:5346–5355. doi: 10.1091/mbc.e06-05-0386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witkos TM, Lowe M. The Golgin family of coiled-coil tethering proteins. Front Cell Dev Biol. 2015;3:86. doi: 10.3389/fcell.2015.00086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woldemichael GM, Turbyville TJ, Linehan WM, Mcmahon JB. Carminomycin I is an apoptosis inducer that targets the Golgi complex in clear cell renal carcinoma cells. Cancer Res. 2011;71:134–142. doi: 10.1158/0008-5472.CAN-10-0757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M, Lu L, Hong W, Song H. Structural basis for recruitment of GRIP domain golgin-245 by small GTPase Arl1. Nat Struct Mol Biol. 2004;11:86–94. doi: 10.1038/nsmb714. [DOI] [PubMed] [Google Scholar]
- Xiang Y, Wang Y. GRASP55 and GRASP65 play complementary and essential roles in Golgi cisternal stacking. J Cell Biol. 2010;188:237–251. doi: 10.1083/jcb.200907132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y, Wang Y. New components of the Golgi matrix. Cell Tissue Res. 2011;344:365–379. doi: 10.1007/s00441-011-1166-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y, Zhang X, Nix DB, Katoh T, Aoki K, Tiemeyer M, Wang Y. Regulation of protein glycosylation and sorting by the Golgi matrix proteins GRASP55/65. Nat Commun. 2013;4:1659. doi: 10.1038/ncomms2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav S., Linstedt A. D. Golgi Positioning. Cold Spring Harbor Perspectives in Biology. 2011;3(5):a005322–a005322. doi: 10.1101/cshperspect.a005322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav V, Panganiban AT, Honer Zu Bentrup K, Voss TG. Influenza infection modulates vesicular trafficking and induces Golgi complex disruption. Virusdisease. 2016;27:357–368. doi: 10.1007/s13337-016-0347-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y, Larkin D, Lara-Lemus R, Ramos-Castaneda J, Liu M, Arvan P. Endoplasmic reticulum (ER) chaperone regulation and survival of cells compensating for deficiency in the ER stress response kinase, PERK. J Biol Chem. 2008;283:17020–17029. doi: 10.1074/jbc.M802466200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto H, Kakuta S, Watanabe TM, Kitamura A, Sekito T, Kondo-Kakuta C, Ichikawa R, Kinjo M, Ohsumi Y. Atg9 vesicles are an important membrane source during early steps of autophagosome formation. J Cell Biol. 2012;198:219–233. doi: 10.1083/jcb.201202061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamane J, Kubo A, Nakayama K, Yuba-Kubo A, Katsuno T, Tsukita S. Functional involvement of TMF/ARA160 in Rab6-dependent retrograde membrane traffic. Exp Cell Res. 2007;313:3472–3485. doi: 10.1016/j.yexcr.2007.07.010. [DOI] [PubMed] [Google Scholar]
- Yi Z, Yuan Z. Aggregation of a hepatitis C virus replicase module induced by ablation of p97/VCP. J Gen Virol. 2017;98:1667–1678. doi: 10.1099/jgv.0.000828. [DOI] [PubMed] [Google Scholar]
- Yin KJ, Deng Z, Hamblin M, Xiang Y, Huang H, Zhang J, Jiang X, Wang Y, Chen YE. Peroxisome proliferator-activated receptor delta regulation of miR-15a in ischemia-induced cerebral vascular endothelial injury. J Neurosci. 2010;30:6398–6408. doi: 10.1523/JNEUROSCI.0780-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura S, Yoshioka K, Barr FA, Lowe M, Nakayama K, Ohkuma S, Nakamura N. Convergence of cell cycle regulation and growth factor signals on GRASP65. J Biol Chem. 2005;280:23048–23056. doi: 10.1074/jbc.M502442200. [DOI] [PubMed] [Google Scholar]
- Yue X, Bao M, Christiano R, Li S, Mei J, Zhu L, Mao F, Yue Q, Zhang P, Jing S, Rothman JE, Qian Y, Lee I. ACBD3 functions as a scaffold to organize the Golgi stacking proteins and a Rab33b-GAP. FEBS Lett. 2017;591:2793–2802. doi: 10.1002/1873-3468.12780. [DOI] [PubMed] [Google Scholar]
- Zeng Z, Lin H, Zhao X, Liu G, Wang X, Xu R, Chen K, Li J, Song L. Overexpression of GOLPH3 promotes proliferation and tumorigenicity in breast cancer via suppression of the FOXO1 transcription factor. Clin Cancer Res. 2012;18:4059–4069. doi: 10.1158/1078-0432.CCR-11-3156. [DOI] [PubMed] [Google Scholar]
- Zhang X, Wang Y. Cell cycle regulation of VCIP135 deubiquitinase activity and function in p97/p47-mediated Golgi reassembly. Mol Biol Cell. 2015;26:2242–2251. doi: 10.1091/mbc.E15-01-0041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Wang Y. GRASPs in Golgi structure and function. Front Cell Dev Biol. 2015;3:84. doi: 10.3389/fcell.2015.00084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Wang Y. Glycosylation quality control by the Golgi structure. J Mol Biol. 2016;428:3183–3193. doi: 10.1016/j.jmb.2016.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Wang Y. The Golgi stacking protein GORASP2/GRASP55 serves as an energy sensor to promote autophagosome maturation under glucose starvation. Autophagy. 2018;14:1649–1651. doi: 10.1080/15548627.2018.1491214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Wang Y. GRASP55 facilitates autophagosome maturation under glucose deprivation. Mol Cell Oncol. 2018;5:e1494948. doi: 10.1080/23723556.2018.1494948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Zhang H, Wang Y. Phosphorylation regulates VCIP135 function in Golgi membrane fusion during the cell cycle. J Cell Sci. 2014;127:172–181. doi: 10.1242/jcs.134668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Liu Y, Zhu X, Cao Y, Liu Y, Mao X, Yang H, Zhou Z, Wang Y, Shen A. SCY1-like 1-binding protein 1 (SCYL1BP1) suppressed sciatic nerve regeneration by enhancing the RhoA pathway. Mol Neurobiol. 2016;53:6342–6354. doi: 10.1007/s12035-015-9531-5. [DOI] [PubMed] [Google Scholar]
- Zhang X, Wang T, Dai X, Zhang Y, Jiang H, Zhang Q, Liu F, Wu K, Liu Y, Zhou H, Wu J. Golgi protein 73 facilitates the interaction of hepatitis C virus NS5A with apolipoprotein E to promote viral particle secretion. Biochem Biophys Res Commun. 2016;479:683–689. doi: 10.1016/j.bbrc.2016.09.152. [DOI] [PubMed] [Google Scholar]
- Zhang X, Wang L, Lak B, Li J, Jokitalo E, Wang Y. GRASP55 senses glucose deprivation through O-GlcNAcylation to promote autophagosome-lysosome fusion. Dev Cell. 2018;45:245–261.e6. doi: 10.1016/j.devcel.2018.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Li B, Huang X, Morelli X, Shi N. Structural basis for the interaction between Golgi reassembly-stacking protein GRASP55 and Golgin45. J Biol Chem. 2017;292:2956–2965. doi: 10.1074/jbc.M116.765990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Atkins JB, Rompani SB, Bancescu DL, Petersen PH, Tang H, Zou K, Stewart SB, Zhong W. The mammalian Golgi regulates numb signaling in asymmetric cell division by releasing ACBD3 during mitosis. Cell. 2007;129:163–178. doi: 10.1016/j.cell.2007.02.037. [DOI] [PubMed] [Google Scholar]
- Zhou Z, Sun X, Zou Z, Sun L, Zhang T, Guo S, Wen Y, Liu L, Wang Y, Qin J, Li L, Gong W, Bao S. PRMT5 regulates Golgi apparatus structure through methylation of the golgin GM130. Cell Res. 2010;20:1023–1033. doi: 10.1038/cr.2010.56. [DOI] [PubMed] [Google Scholar]
- Zhu X, Kaverina I. Golgi as an MTOC: making microtubules for its own good. Histochem Cell Biol. 2013;140:361–367. doi: 10.1007/s00418-013-1119-4. [DOI] [PMC free article] [PubMed] [Google Scholar]