Summary
Fungi have been found in every marine habitat that has been explored, however, the diversity and functions of fungi in the ocean are poorly understood. In this study, fungi were cultured from the marine environment in the vicinity of Woods Hole, MA, USA including from plankton, sponge and coral. Our sampling resulted in 36 unique species across 20 genera. We observed many isolates by time-lapse, differential interference contrast (DIC) microscopy and analyzed modes of growth and division. Several black yeasts displayed highly unconventional cell division cycles compared to those of traditional model yeast systems. Black yeasts have been found in habitats inhospitable to other life and are known for halotolerance, virulence, and stress-resistance. We find that this group of yeasts also shows remarkable plasticity in terms of cell size control, modes of cell division, and cell polarity. Unexpected behaviors include division through a combination of fission and budding, production of multiple simultaneous buds, and cell division by sequential orthogonal septations. These marine-derived yeasts reveal alternative mechanisms for cell division cycles that seem likely to expand the repertoire of rules established from classic model system yeasts.
Introduction
Fungi are critical components of the biosphere with diverse roles in cycling nutrients, shaping microorganism communities, and acting as opportunistic pathogens. These key functions have been extensively explored in terrestrial ecosystems, but the role of fungi in the marine environment are much less appreciated. Large-scale expeditions sampling marine microbiological diversity would have largely missed fungi for technical reasons such as the use of size filtration and the limitations of markers for systematic molecular identification of fungi [1]. Nevertheless, fungi have been found in every part of the marine environment where they have been investigated, associated with marine sediments, invertebrates, marine mammals, algae, driftwood, and throughout the water column [2–13]. Thus, there is a major untapped and unknown biodiversity of this major kingdom of life in the oceans [14].
Phylogenetics studies point to a terrestrial origin of multiple obligate marine fungal lineages [15]. This transition from terrestrial to marine environments may have occurred multiple times, as many of the fungi detected in the marine environments have been previously described in terrestrial habitats [16–18]. However, these marine-derived fungi were detected in samples collected far from shore, suggesting that they are actual inhabitants of the aquatic environment rather than recent arrivals from land. Some of these fungi seem to have a truly amphibious lifestyle, based on gene expression data [19] and strong correlations with abiotic factors [20, 21]. This is indicative of the adaptability of the fungal kingdom and makes it challenging to define what truly constitutes a marine (as opposed to terrestrial) fungus. Suggested criteria include that the fungus was isolated from marine environments on multiple occasions, can grow on marine-origin substrates, forms ecologically-relevant relationships with other marine organisms (pathogen, symbiont, etc.), and has adapted to the marine environment as evident from genetic analyses or metabolic activity [2, 12, 13, 22–24].
Our goal was to identify culturable species of fungi from the sea to assess how fungi in non-terrestrial environments grow and divide. Fungi in these settings face a myriad of potential stresses from temperature, high salinity, buoyancy challenges, UV exposure, and limited organic matter for nutrients. A subgroup of fungi of particular interest are the melanized fungi, also known as black yeasts, that are in the class Dothideomycetes [25]. Black yeasts have attracted the attention of researchers due to their biotechnological potential, high stress tolerance, and ability to cause severe mycosis. Black yeasts have not only been identified in marine environments but also extreme habitats such as salterns, rocks, ice, and desert mats [26]. We speculated that their well-appreciated adaptability might reflect an expanded repertoire of mechanisms regulating growth and division cycles.
Much of our current understanding of the cell division cycle derives from classic studies originating in the 1970s in two model yeast systems: the budding yeast Saccharomyces cerevisiae and the fission yeast Schizosaccharomyces pombe [27–32]. These evolutionarily-distant yeast species were intensively mined for cell division cycle mutations and, together with work from marine invertebrate embryos, gave rise to the molecular framework for understanding the cell cycle that we have today. Cell-cycle progression is regulated so as to maintain a consistent distribution of cell sizes in the population, although the mechanisms underlying such size control remain contested. In addition to size control, budding yeast cells have the added challenge of coordinating bud formation and mitosis so that daughter cells receive a nucleus, and these bud-bound nuclei must navigate the narrow passage of the mother-bud neck. Tight controls have been identified that work to ensure that only a single bud is produced and to sense if a bud has been built [33–35].
Despite sharing many common core components, notable contrasts have emerged between the two model yeast systems. There are substantial differences in how these morphologically distinct yeasts seem to regulate the cell cycle, including how cell size is measured, how nutrient conditions regulate mating and meiosis [36–38], how the substrate specificity of the CDK is controlled [39–41], and how polarity and cytokinesis are regulated by the cell cycle [35]. These variations may arise from essentially different regulatory programs or adaptations of a fundamentally similar regulatory program, or a mixture of the two origins. However, these two systems may still not fully capture the variability in mechanisms controlling division in fungi. In this study, we analyze four different black yeasts isolated from marine environments and find that the division cycles of these yeasts display far more plasticity and behave in unconventional ways not predicted by the studies from model yeasts.
Results
Fungal isolation from the marine environment
Between 2016–2018, we sampled locations around Woods Hole, MA to culture fungi from the marine environment. Fungi were isolated from plankton tows of ocean water in Buzzards Bay, Martha’s Vineyard Sound and Great Harbor, sediment from local beaches, a salt marsh, the sponge Cliona celata and the coral Astrangia poculata (Figure 1, Table 1). Buzzards Bay is separated physically from the other two bodies of water by the southwest corner of mainland Cape Cod and the Elizabeth Islands. Buzzards Bay is fed by the Acushnet River via the New Bedford Harbor, an 18,000-acre Environmental Protection Agency (EPA) superfund site. New Bedford Harbor has been the focus of restoration projects since the EPA found high concentrations of polychlorinated biphenyls (PCBs) and heavy metals such as cadmium and lead in the mid 1970’s [42]. Martha’s Vineyard Sound is encompassed by mainland Cape Cod, Martha’s Vineyard, and Nantucket, but still experiences strong tides and currents.
Figure 1. Culturing marine fungi from marine and coastal environments around Woods Hole, MA.
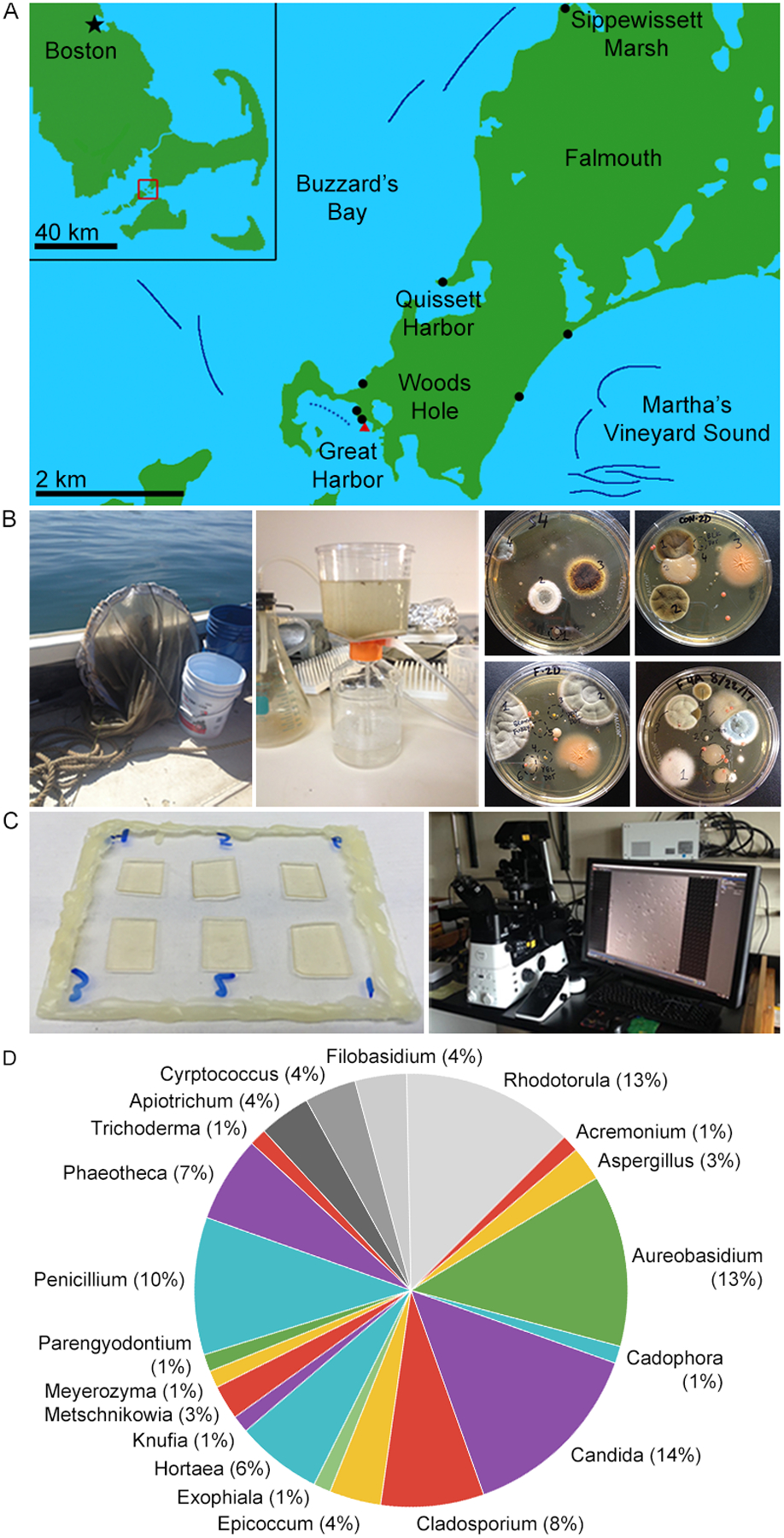
(A) Collection sites. Solid dark blue lines denote cruise tracks of plankton tows. The dotted blue line denotes the approximate cruise track of a Great Harbor tow. Black dots mark beach sediment samples and the red triangle marks where coral and sponge were collected. (B) Collection and culture methodology. Water column samples were collected by conducting plankton tows 1–1.2 km offshore using a 504 μm mesh net (left). 1.0 L water was concentrated using a 0.22 μm filter (center). Cultures were grown on various media for 1–2 weeks at 18–20 °C. (C) DIC imaging methodology. Samples were prepared six per slide on agar pads. Images were collected at 5 min intervals at 18–20 °C. (D) Genus abundance. Numeric labels indicate number of isolates found belonging to each of the 20 different genera we found. We found 16 different genera of Ascomycota (in color) and 4 Basidiomycota (grayscale).
Table 1. Fungal species isolated from marine environments near Woods Hole, Massachusetts.
The four black yeast we focus on in this paper are bolded in the “Species” column. The “Jones List” column denotes species considered marine fungi based on the consensus compiled in Jones et al., 2015 [43]. The columns labeled Marine, Freshwater, and Terrestrial refer to instances where a species was found in a given environment.
| Species | Site | Collection Type | Phylum | Morphotype | Genome | Jones List [43] | Marine | Freshwater | Terrestrial |
|---|---|---|---|---|---|---|---|---|---|
| Acremonium potronii | Buzzards Bay | Plankton tow | Ascomycota | Yeast | Yes | [66] | |||
| Cadophora malorum | Buzzards Bay | Plankton tow | Ascomycota | Hyphal | [67] | Yes | [68, 69] | [70, 71] | |
| Cladosporium cladosporioides | Buzzards Bay | Plankton tow | Ascomycota | Hyphal | Unpub. | Yes | [72, 73] | [74–76] | [71, 77] |
| Cladosporium halotolerans | Buzzards Bay | Plankton tow | Ascomycota | Hyphal | [77] | [77] | [77] | ||
| Cryptococcus fonsecae | Buzzards Bay | Plankton tow | Basidiomycota | Yeast | Yes | [78] | |||
| Cryptococcus mangalensis | Buzzards Bay | Plankton tow | Basidiomycota | Yeast | Yes | [79] | |||
| Exophiala xenobiotica | Buzzards Bay | Plankton tow | Ascomycota | Yeast, Hyphal | [80] | Yes | [81] | [82] | [83] |
| Filobasidium uniguttulatum | Buzzards Bay | Plankton tow | Basidiomycota | Yeast, Hyphal | Yes | [84] | [85, 86] | ||
| Knufia petricola | Buzzards Bay | Plankton tow | Ascomycota | Yeast | [87] | [88] | [49, 87] | ||
| Penicillium thomii | Buzzards Bay | Plankton tow | Ascomycota | Hyphal | Yes | [89] | |||
| Cryptococcus magnum | Buzzards Bay, Great Harbor | Plankton tow (both sites) | Basidiomycota | Hyphal | Yes | [79] | [79] | ||
| Hortaea thailandica | Great Harbor | Coral | Ascomycota | Yeast | [90] | [91] | |||
| Hortaea werneckii | Buzzards Bay, Vineyard Sound | Plankton tow (both sites) | Ascomycota | Yeast | [26, 92] | [93, 94] | [95] | ||
| Phaeotheca salicorniae | Buzzards Bay, Vineyard Sound | Plankton tow (both sites) | Ascomycota | Yeast, Hyphal | [96] | ||||
| Rhodotorula mucilaginosa | Buzzards Bay, Vineyard Sound, Great Harbor |
Plankton tow (Buzzards Bay and Vineyard Sound), Coral |
Basidiomycota | Yeast | [97] | Yes | [98, 99] | [100] | [99, 101] |
| Rhodotorula sphaerocarpa | Buzzards Bay, Vineyard Sound | Plankton tow (both sites) | Basidiomycota | Yeast | [79] | [85, 99, 102] | |||
| Aureobasidium pullulans | Buzzards Bay, Vineyard Sound, Great Harbor |
Plankton tow (all sites), Sediment (Great Harbor), Coral (Great Harbor) |
Ascomycota | Yeast, Hyphal | [52] | Yes | [55, 90] | [79] | [85, 103] |
| Aureobasidium melanogenum | Great Harbor | Coral | Ascomycota | Yeast, Hyphal | [52] | [55, 104] | [104] | [55] | |
| Candida albicans | Great Harbor | Coral | Ascomycota | Yeast, Hyphal, Pseudohyphal | [105] | Yes | [106] | [107] | [108] |
| Candida intermedia | Great Harbor | Coral | Ascomycota | Yeast | [109] | Yes | [79, 98] | [102] | |
| Candida parapsilosis | Great Harbor | Sponge | Ascomycota | Yeast | [110] | Yes | [79, 98, 106, 111] | [71, 102, 111] | |
| Meyerozyma guilliermondii | Great Harbor | Coral | Ascomycota | Yeast | [112] | Yes | [98, 111] | [113] | [111] |
| Phaeotheca triangularis | Great Harbor | Sponge | Ascomycota | Yeast, Hyphal | [114] | ||||
| Rhodotorula diobovata | Great Harbor | Plankton tow, Coral | Basidiomycota | Yeast | Unpub. | [98] | [79] | [102] | |
| Apiotrichum dulcitum | Great Harbor | Sediment | Basidiomycota | Yeast | [62, 63] | ||||
| Apiotrichum porosum | Great Harbor | Sediment | Basidiomycota | Yeast | [62, 63] | ||||
| Cladosporium sphaerospermum | Vineyard Sound | Plankton tow | Ascomycota | Hyphal | [115] | Yes | [81, 116] | [71] | |
| Metschnikowia bicuspidata | Vineyard Sound | Plankton tow | Ascomycota | Yeast | [117] | Yes | [118] | [119] | |
| Trichoderma harzianum | Great Harbor | Coral | Ascomycota | Hyphal | [120] | Yes | [121] | [122] | [123] |
| Epicoccum nigrum | Great Harbor | Coral | Ascomycota | Hyphal | [124] | [125] | [71] | ||
| Aspergillus niger | Great Harbor | Coral | Ascomycota | Hyphal | [126] | Yes | [127, 128] | [74] | [129, 130] |
| Aspergillus sydowii | Great Harbor | Coral | Ascomycota | Hyphal | [131] | Yes | [125] | ||
| Parngyodontium album | Great Harbor | Coral | Ascomycota | Hyphal | Yes | [132] | [76] | ||
| Penicillium chrysogenum | Great Harbor | Coral | Ascomycota | Hyphal | [131] | Yes | [81, 125, 133] | [74, 76] | [134] |
| Penicillium atrovenetum | Great Harbor | Coral | Ascomycota | Hyphal | [135] | [136] |
Samples were plated onto standard fungal media (with yeast extract, malt extract, or potato extract as carbon sources) made with sterile seawater and containing antibiotics to limit bacterial growth. For coral and sponge samples, seawater collected from the vicinity of the specimens was also plated. This was intended as a negative control to see if fungal isolates arising from the coral or sponge plates might just be from the water or contamination from processing pipeline. Individual colonies were then subcultured on individual plates, observed with DIC microscopy, and identified based on amplifying and sequencing the rDNA locus (Figure 1B–C, Table S1). We identified 36 species, including 16 Ascomycetes and 4 Basidiomycetes; 23 of the species have been identified as bona fide marine fungi by Jones et al. [43], although others have also been isolated from marine environments (Figure 1D, Table 1). Multiple species were found uniquely in only one of the habitats sampled; others appeared across several collection sites. The species identified here represent only a subset of the cultured isolates as some were not identified due to challenges isolating DNA and thus this is an underestimate of the diversity and density of fungi in the area.
The isolates exhibit a diversity of colony morphologies and pigmentation and include yeasts and filamentous fungi (Figure 2A, Figure S1). To assess morphogenesis and division patterns at the level of single-cells, a subset of the species was filmed using high-magnification, DIC time-lapse microscopy. This single-cell analysis revealed many modes of growth and division in the Woods Hole marine fungi collection including budding, fission, filamentous growth and meristematic growth. Here, we focus on four black yeasts that are morphologically distinct from each other and from model yeasts (Figure 2B–C). Three of these species display clear black colony growth with a tendency to produce terracotta pigments at the periphery of colonies, and one (A. pullulans) is initially pale pink and slowly melanizes over time under more stressed growth conditions. They each display surprising and unconventional division cycles (Fig 2C–D).
Figure 2. Overview of cultured marine-derived fungal diversity.
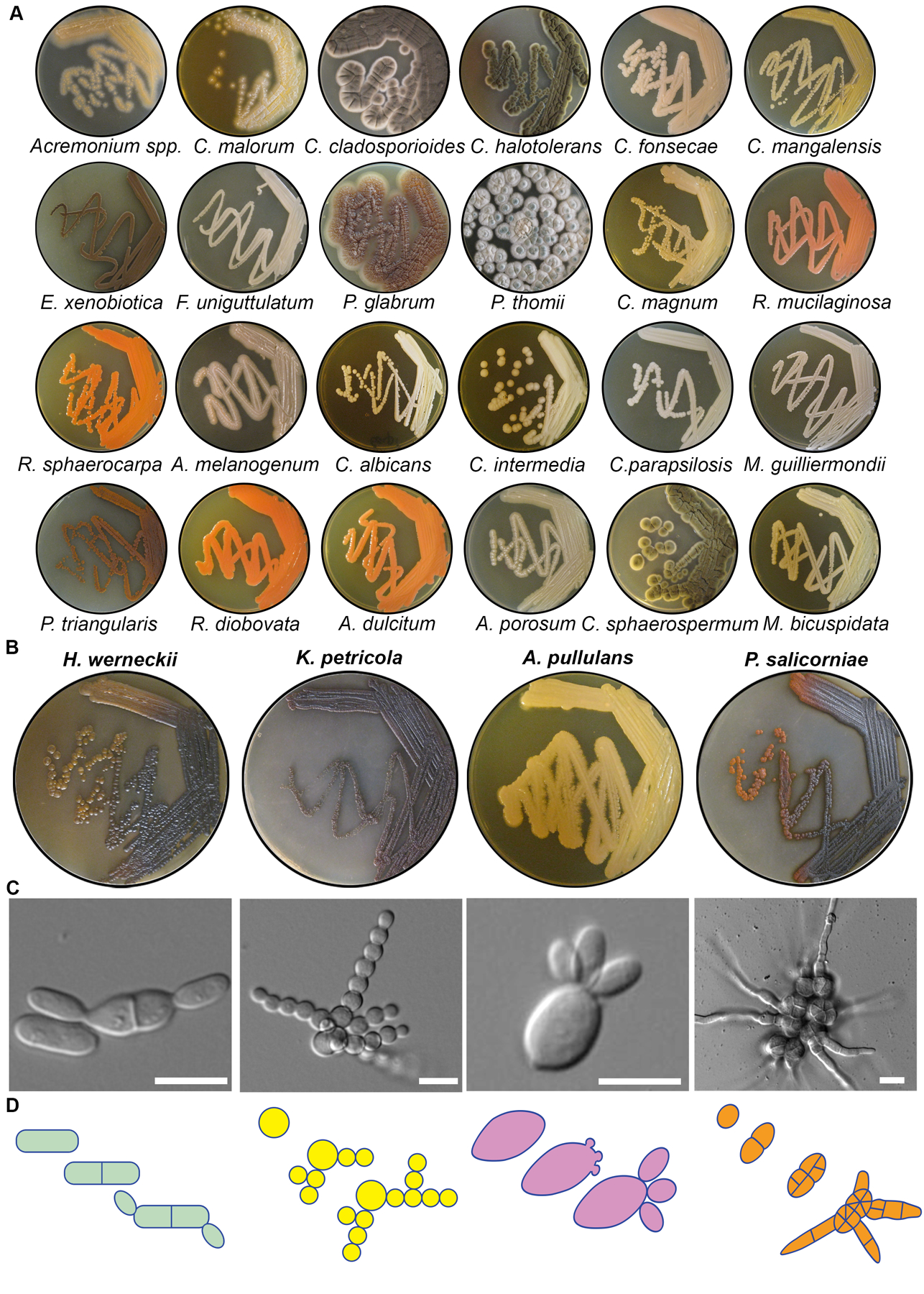
(A) Macroscopic colony morphology of identified marine fungi grown for 10 days on YPD + 0.6M NaCl. Related to Figure S1. (B) Macroscopic colony morphology of the four black yeasts chosen for further characterization. (C) Representative DIC images of each species showing division patterns. These images are used again in Figure 3A (H. werneckii), Figure 4A (K. petricola), Figure 5A (A. pullulans), and Figure 6A (P. salicorniae) and are stills from Video S1, Video S2, Video S3, and Video S6. Scale bar, 10 μm. (D) Schematic diagrams of colony growth for each species.
Alternation between fission and budding in Hortaea werneckii
Hortaea werneckii was identified in multiple locations and has been previously studied due to its extreme halotolerance with growth possible in up to 5 M NaCl [44, 45]. Additionally, H. werneckii is the etiologic agent of tinea nigra, which is an asymptomatic superficial mycosis that is restricted to the palms of the hands and soles of the feet [46]. Remarkably, H. werneckii cells could divide both by fission and budding, with each cell compartment inheriting a single nucleus (Figure 3A–B, Video S1). After placement on a solid medium, cells grew both isotropically and from the cell poles and divided by development of medial septa. This is reminiscent of a typical fission yeast cell cycle with the exception that we did not observe separation of the two daughter cells after the septum was built. Remarkably, 92% of the next division cycles involved the production of a bud, with only 8% of cells displaying a second fission-like process (N = 139) (Figure 3C). Even those cells that performed a second fission then proceeded to bud in the third cell cycle, indicating that all cells switch division modes. There was a strong bias toward alternating between fission and budding rather than consistently dividing by one mode.
Figure 3. Hortaea werneckii cells divide via septation and budding.
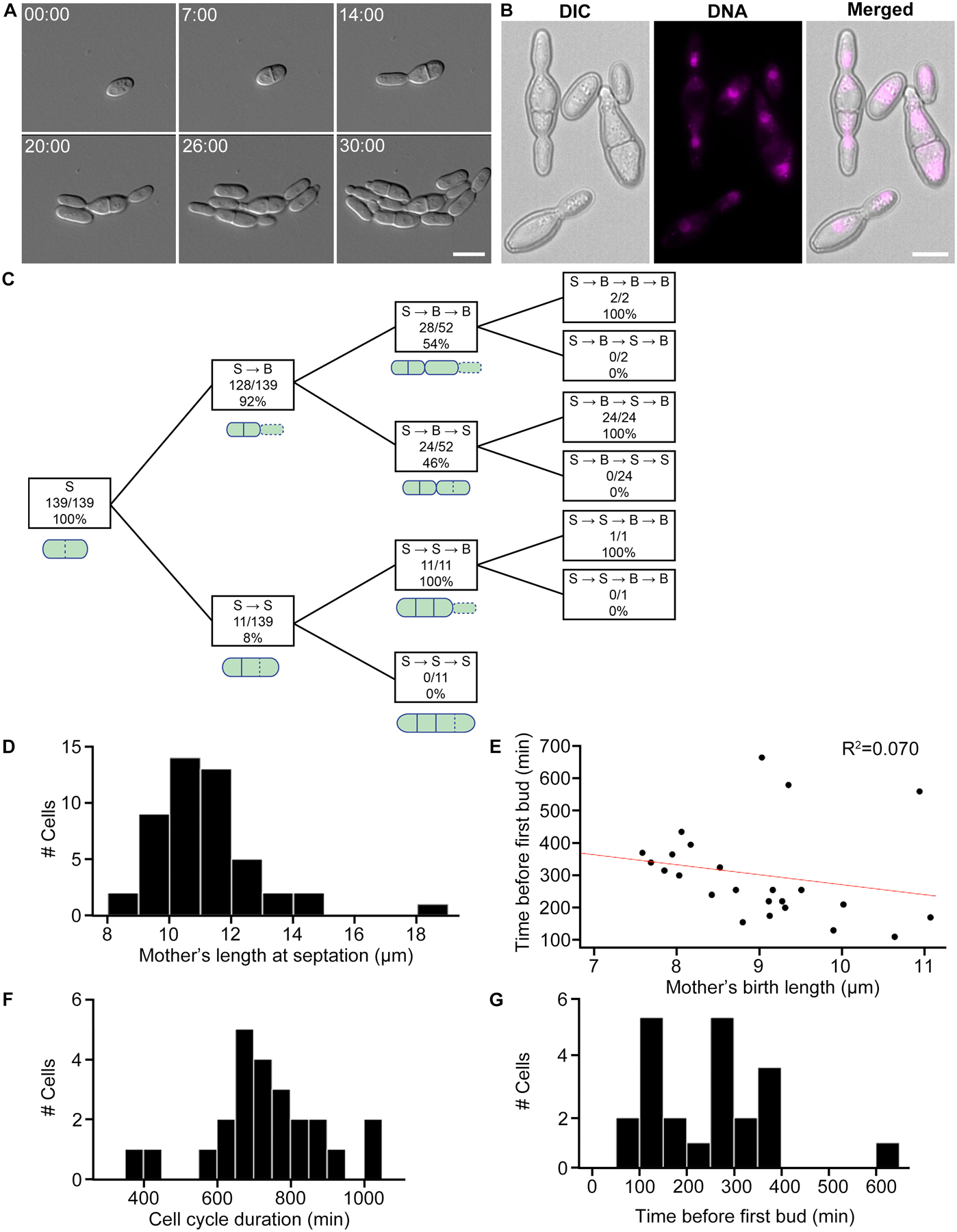
(A) H. werneckii cell growth and development under 40x DIC. These images are stills from Video S1. Scale bar, 10 μm. Time, Hours:Minutes. (B) DNA staining in fixed cells. H. werneckii cells contain one nucleus per compartment. Mitosis seems to occur across the axis of cell division. Nuclei were stained using Hoechst. Scale bar, 10 μm. (C) Quantifying sequence of cell division events. In this strain of H. werneckii we observed two cell division event types, budding and septation. Septation and budding events are represented by “S” and “B”, respectively. Dotted lines in the accompanying schematics indicate the latest division event type in that sequence. The diagram is interpreted as follows: of 139 total cells observed, all underwent septation as a first division event. Of those cells, 128 budded next (S → B) and 11 septated again (S → B). 52 of the 128 S → B cells divided again, 28 of which budded (S → B → B) and 24 septated (S → B → S). (D) Mother’s length at septation. Mothers grew to an average length of 11.2 μm before septating (N = 48, SD = 1.7, CV = 15.2%). (E) Mother’s birth length (μm) vs. cell cycle length (min). Birth length is defined as the length of a cell it breaks off from its mother. Red line denotes linear fit. Larger cells have a shorter division time (N = 24, R2 = 0.070). (F) Cell cycle duration of H. werneckii (min). This is a measurement of the full cell cycle duration (N = 24, mean = 730, SD = 158, CV = 21.6%). (G) Time before first bud (min). Time until a mother cell begins producing its first daughter cell (N = 24, mean = 253, SD = 124, CV = 49.0%).
Cell size and division times were quite variable compared to previous measurements of model systems. The median length at fission was 10.9 μm (N = 48, mean = 11.2 μm, standard deviation [SD] = 1.7 μm, coefficient of variation [CV] = 15.2%, Figure 3D), compared to fission yeasts which are known for their highly robust size control mechanisms, with mean length of 14 μm and CV of 7.7% in S. pombe [38]. Thus, H. werneckii shows substantially greater heterogeneity. The division site was generally centered in the cell (mean ratio of septated halves’ lengths = 1.02, SD = 0.15, CV = 14.2%, N = 48 pairs of septated halves). The median cell cycle duration was 710 min (N = 24, mean = 730 min, SD = 158 min, CV = 21.6%, Figure 3E) for the cells undergoing medial septation. Plotting this time against size at birth revealed some evidence of size control, but only with smaller cell sizes. For all cells, there was a very weak negative correlation with R2 = 0.070 (N = 24, Figure 3E). Subsetting cells of birth length less than 9.5 μm revealed a much stronger, relatively, negative correlation with R2 = 0.520, (N = 18). For cells larger than 9.5 μm, there was a weak positive correlation (R2 = 0.260, N = 6), but this was greatly influenced by an outlier that took over 10 hr to produce its first bud. Median time for a bud to produce a new bud was 265 min (N = 24, mean = 253 min, SD = 124 min, CV = 49.0%, Figure 3F), which is far more variable than for the model budding yeast S. cerevisiae (N = 10, mean generation time = 132 min, SD = 13 min, CV = 9.8%) [47]. Thus, H. werneckii divides by fission and budding, generally alternating between modes of division from cell cycle to cell cycle. The fact that these distinct division patterns co-occur within the same organism indicates that they need not derive from extensive divergent evolution. In both modes, the cell cycle durations and cell sizes were far more variable in H. werneckii than in the model yeasts, and there was not obvious size control, indicating a high degree of plasticity in the growth and division programs.
Spherical non-random budding produces dendritic morphologies in Knufia petricola
We isolated Knufia petricola from a plankton tow in Buzzards Bay. K. petricola is a melanized microcolonial fungus (MCF). MCF exhibit compact colonial structures and protective, melanized cell walls. They are extremotolerant and can grow on bare-rock surfaces. In addition to the Woods Hole marine environment, K. petricola has been found on rocks in extreme conditions from the Antarctic to dry deserts [48, 49] and as part of lichens [50]. The marine strain produces nearly spherical buds that remain connected (Figure 4A–B, Video S2). Given how spherical these cells are, it is unclear how subsequent buds are so precisely positioned at the opposite side from the division site. After multiple cycles, approximately linear chains of spherical cells are produced and, at some frequency, a chain will branch and produce a second chain with the colony taking on a dendritic shape.
Figure 4. K. petricola growth through highly polarized patterning of spherical buds.
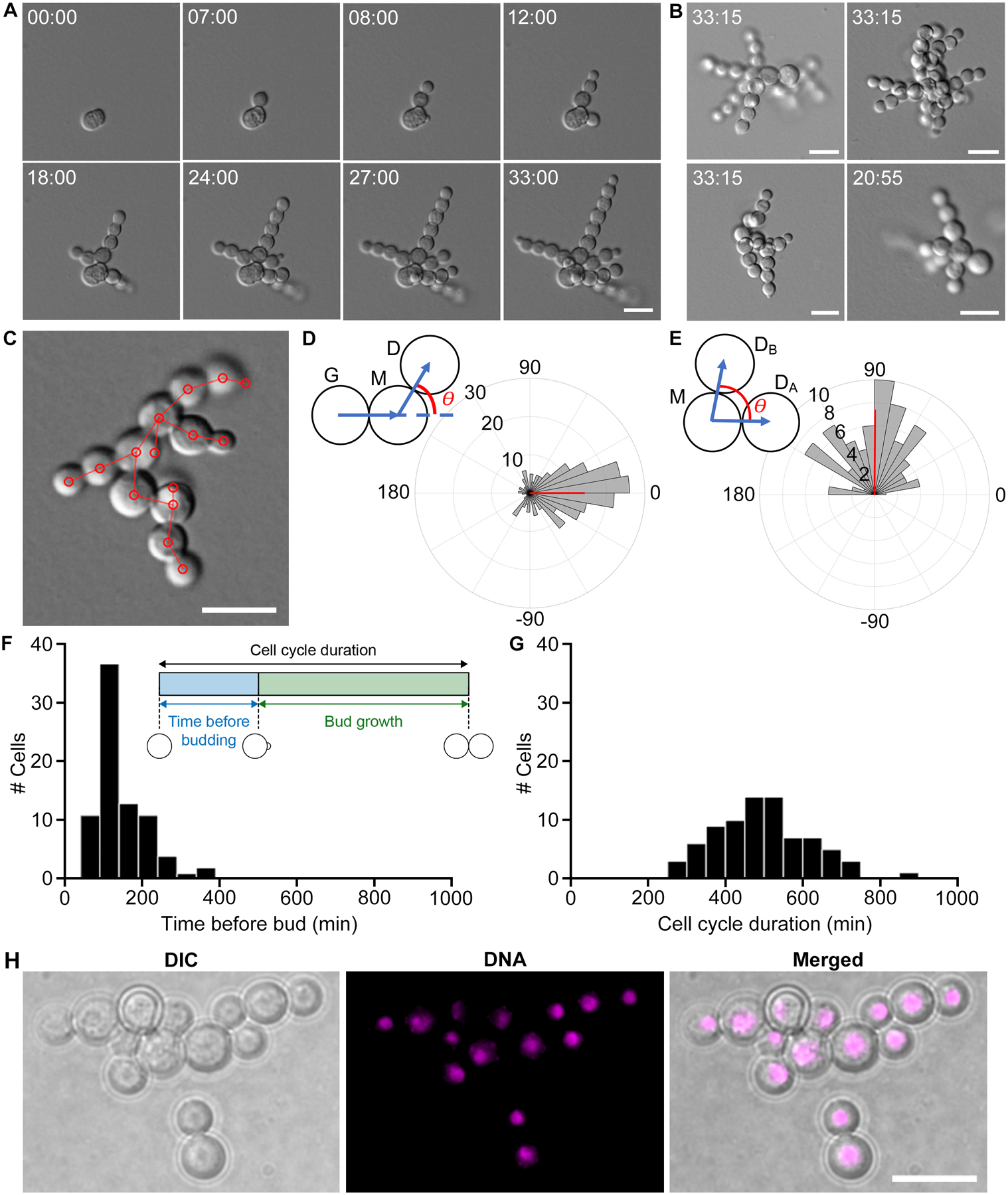
(A) Representative DIC time lapse images of K. petricola colony growth and development (40x). These stills are from Video S2. Scale bar, 10 μm. Time, Hours:Minutes. (B) Final frames from four different colonies. These stills provide more examples of colony growth. Scale bars, 10 μm. Time, Hours:Minutes. (C) Methodological framework of measuring angles between cells. Based on the last frame of each DIC movie we locate the center of each cell (red circles) and define cell lineages (red lines). (D) Angle between daughter (D), mother (M), and grandmother (G) cells. This analysis includes 237 G-M-D lineages, and only considers cases where a cell produces zero or one daughter(s); it excludes cells where branching occurs. The angle θ measures how linear three generations within the same lineage are (see schematic above), where θ = 0° means that the lineage is perfectly linear and θ = 180° means that the daughter buds back in the direction of its grandmother. Positive and negative angles indicate clockwise and counterclockwise growth, respectively. Circular mean of budding angle was 0.07°, indicated by the angle of the red vector, and circular variance was 0.45, indicated by the length of the red vector. (E) Angle between sibling cells. θ is the angle between sibling cells if a mother cell produces multiple daughters, where θ = 0° means that the daughters budded in the same direction and θ = 180° means they budded in opposite directions (see schematic above) (N = 84). Positive and negative angles indicate clockwise and counterclockwise growth, respectively. Circular mean of budding angle was 89.6° and circular variance was 0.26. (F) Time before budding. Cells are capable of a remarkably quick turnaround time of 10 min, but could also take as long as 320 min (N = 28, mean = 110, SD = 64.6, CV = 58.7%). (G) Cell cycle duration (min). Cell cycle duration is highly variable (N = 28, mean = 499, SD = 121, CV = 24.3%). (H) DNA staining in fixed cells. Cells are uninucleate. Scale bar, 10 μm.
To quantify the linearity of the growth pattern, we measured angles between cells of the same lineage. Angles were measured from the centers of cells (Figure 4C). Angles of cell lineages were defined such that 0° means a daughter budded exactly opposite of its grandmother cell, while 180° means a daughter budded in the same direction as its grandmother (Figure 4D) Positive angles corresponded to budding in a clockwise direction and negative angles indicated budding in the opposite direction. From 237 triads analyzed, the average angle was 0.07°, and the circular variance was 0.45. 50% of angles were within ±31°. Thus, many budding events lead to an elongation of a linear chain of cells.
Branching was quantified in a similar way but angles were instead measured between cells produced by the same mother (N = 84). An angle of 0° indicates daughters were produced at the same bud site whereas 180° indicates daughters were produced in opposite directions. Mean angle between daughters was 89.6°, and the circular variance was 0.26 (Figure 4E). Very few of the angles between sibling cells were close to 0° or 180°, which means that if a mother produces multiple daughters, they are unlikely to bud in the same direction. Thus, branching serves as a way to expand the surface covered and start new chains of cells in the same vicinity. We speculate that new bud sites forming maximally far away from all previous bud sites could explain both the linear budding of the first daughter cell, and the perpendicular budding of the second daughter cell, though the molecular mechanism of bud site positioning in this species is unknown.
To quantify cell cycle timing, we considered two phases based on two events observable in the DIC movies: when buds first appeared, and when buds reached their final size (Figure 4F inset). The “time before budding” phase corresponds to the time from when a cell reached its final size to when it produced its first bud, and may be analogous to G1 in S. cerevisiae. The time before budding phase was highly variable, with a mother initiating a bud in as little as 10 min, although the average time to initiate a bud was 110 min (SD = 64.6, CV = 58.7%, N = 28, Figure 4F). The “cell growth” phase corresponds to the time from when a cell first appeared as a bud to when it reached its final size, and it also was quite variable (mean = 193 min, SD = 66.5, CV = 34.5%, N = 138). Together these two phases compose the full cell cycle duration, which also exhibited significant variation (mean = 499 min, SD = 121, CV = 24.3%, N = 28, Figure 4G). Thus, cell cycle timing in K. petricola was highly variable.
It appeared that the linear chains were formed because cells which budded for the first time often budded opposite the previous bud site. The branched morphology developed because cells which budded multiple times often budded in different directions, and such budding events were much less frequent than linear budding events. While the mechanism of cell polarization remains unclear, DAPI staining of nuclei reveals that cells are uninucleate (Figure 4H). Thus, this system is remarkable for using highly spherical cells to produce a polarized colony shape akin to filamentous growth but generated through buds rather than hyphae.
Simultaneous production of multiple buds per cell cycle in Aureobasidium pullulans
Aureobasidium pullulans was isolated from several locations including all plankton tow sites, sponge from the Great Harbor and sediment from Little Sippewissett Marsh. It is a black yeast species known for its potential biotechnological significance as a producer of pullulan (poly-alpha-1,6-maltotriose), a biodegradable, extracellular polysaccharide [51], and it encodes enzymes capable of plastic degradation [52–54].This species is globally ubiquitous and has been isolated from osmotically stressed environments, such as salterns, rocks, and glacial and subglacial ice [55].
A major feature of budding yeast morphogenesis, observed not only in S. cerevisiae but also C. albicans and the distantly related basidiomycetes C. neoformans and U. maydis, is that only a single bud is produced each cell cycle. Singularity in budding ensures that with each mitosis, a daughter and mother each receive a single nucleus. Remarkably, however, the yeast form of A. pullulans produced multiple buds simultaneously, up to 6 from the same vicinity of a mother cell. The number of buds a mother produced could vary each cell cycle, but a single mother displayed a tendency to produce similar numbers of buds in consecutive division cycles, and the sites of bud emergence were often re-used (Figure 5A, Videos S3 and S4). Almost 80% of scored cells produced >1 bud, and of those, 64% produced multiple rounds of simultaneous buds (Figure 5B). Mean cycle length was 159.5 min and was quite variable (N = 82, SD = 80.5 min, CV = 50.5%, Figure 5C) however, cell cycle duration was not obviously correlated with the number of buds a mother produced in a given cell cycle (Figure 5C’).
Figure 5. A. pullulans is multinucleate and is capable of producing multiple buds simultaneously.
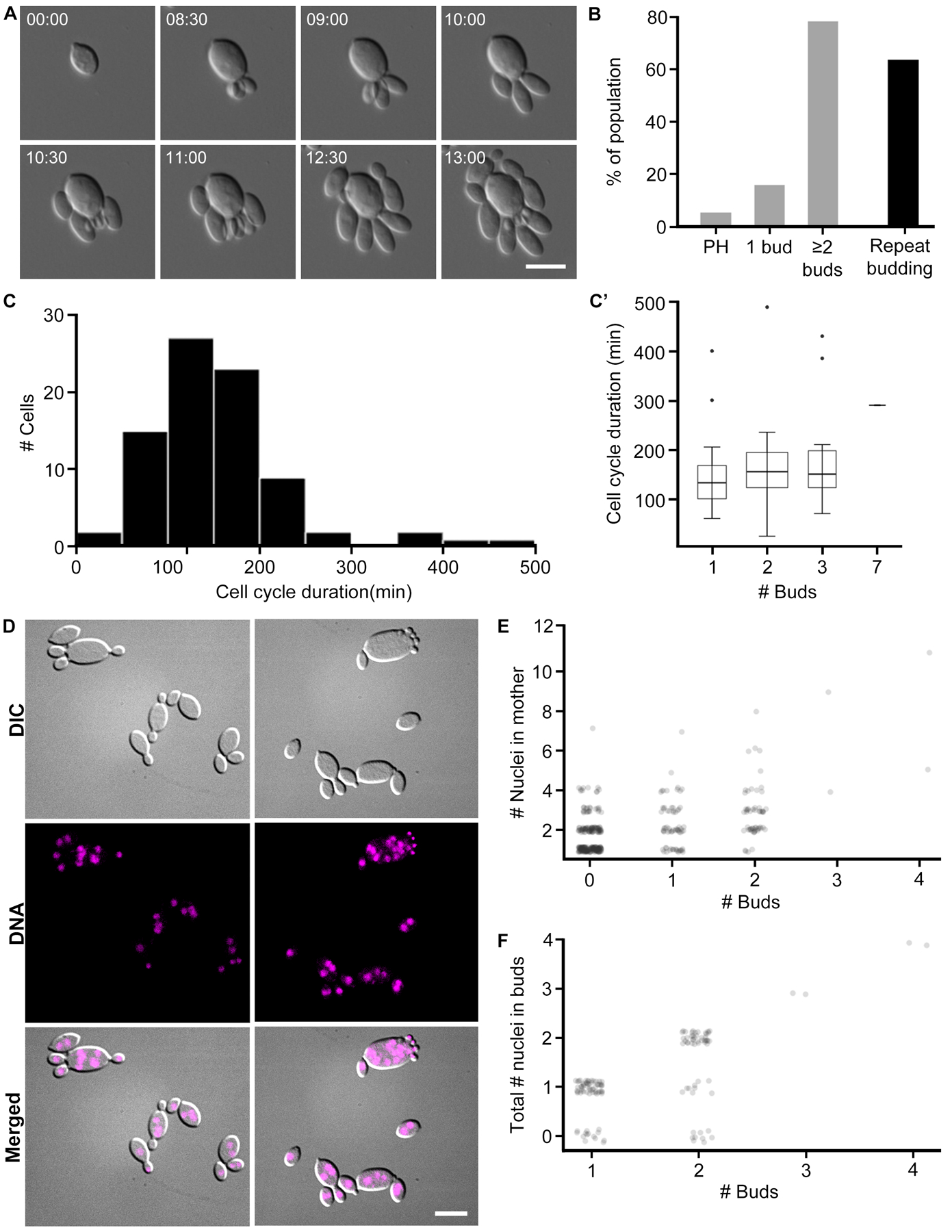
(A) Representative DIC images of A. pullulans cell growth and budding at 40x. Cells may produce multiple buds at once, and sometimes do so in quick succession and from the same site. Scale bar, 10 μm. Time, Hours:Minutes. These stills are from Video S3. Additional images of cells are provided in Video S4. (B) Percentage of population that produce multiple buds. In grey are the percentage of cells that become pseudohyphal (PH), produce one or multiple simultaneous buds (N =75). 59 cells (79%) produce multiple simultaneous buds. Of those, 38 cells (64%) repeatedly produce multiple simultaneous buds, indicated by the black bar. (C) Cell cycle length (min). This is a measurement of the full cell cycle duration (N = 82, mean = 159.5 min, median = 150, SD = 80.5, CV= 50.5%). C’) Cell cycle length based on number of buds that a mother produces. The number of simultaneous daughter cells that a mother produces does not seem to affect the length of its cell cycle. Box widths are representative of sample size. (D) DNA staining in fixed cells reveals that mother cells are multinucleate. Buds are predominantly uninucleate, but some appear to have no nuclei. Scale bar, 10 μm. (E) Number of nuclei in mother cells binned by the number of buds it produces. This counts only the nuclei that are in the mother cells (N = 371). Data were scored from fixed cells. Mothers that produce more buds appear to have more nuclei. (F) Total number of nuclei in buds. This counts the number of nuclei in buds from the same mother (N = 127 family groups). Data were scored from fixed cells. While this does not explicitly reflect the number of nuclei in each bud, we observed no multinucleate buds.
We speculated that the variable numbers of buds might reflect a variable number of nuclei in the mother cell. Mother cells did have highly variable numbers of nuclei based on nuclear staining of fixed cells (Figure 5D). Analysis of fixed yeast indicate that mothers with the most nuclei tended to produce higher numbers of buds in a given division cycle, but the correlation was weak (Figure 5E). Buds generally received no more than one nucleus each, and in some cases the nucleus was deposited even when buds were extremely small (Figure 5D–F). Thus, mothers appear to produce some number of nuclei in the absence of budding, and then segregate nuclei to nascent buds. How nuclear division and budding are coordinated will require analysis of nuclear dynamics in live cells. Thus, this system presents multiple surprises including the ability of cells to host several coexisting polarity sites, and to populate daughter cells with a single nucleus despite a reservoir of many nuclei from which to choose.
Meristematic divisions, filamentation and cellularization in Phaeotheca salicorniae
Phaeotheca salicorniae was isolated twice from plankton tows, once each from Buzzards Bay and the Vineyard Sound. The genus Phaeotheca was first identified in 1981 [56]. P. salicorniae was first identified in 2016 and thus has a very limited literature [57]. We also isolated the very closely related Phaeotheca triangularis [58] from both coral and sponge. We focused on P. salicorniae, which has the most complexity in morphology and division patterns, and is least like any of the well-characterized model yeasts or filamentous fungi in our study. In P. salicorniae we have identified yeast cells and hyphal cells, with cells able to switch bidirectionally between these two types. Colony growth begins from a yeast cell (Figure 6A’, Video S5) which swells and elongates slightly, and then goes through a series of divisions by forming septa (red box in Figure 6C). The first septum is placed in the center of a slightly ovoid cell. Subsequent divisions are perpendicular to the previous division plane, producing a small cluster of cells with shapes reminiscent of sections of a pie (Figure 6A”). After this point, the colony appears to undergo meristematic conversion [58], producing small round cells that seem to be embedded in some sort of extracellular matrix that can be rich in melanin.
Figure 6. P. salicorniae colony growth involves meristematic division and filamentation.
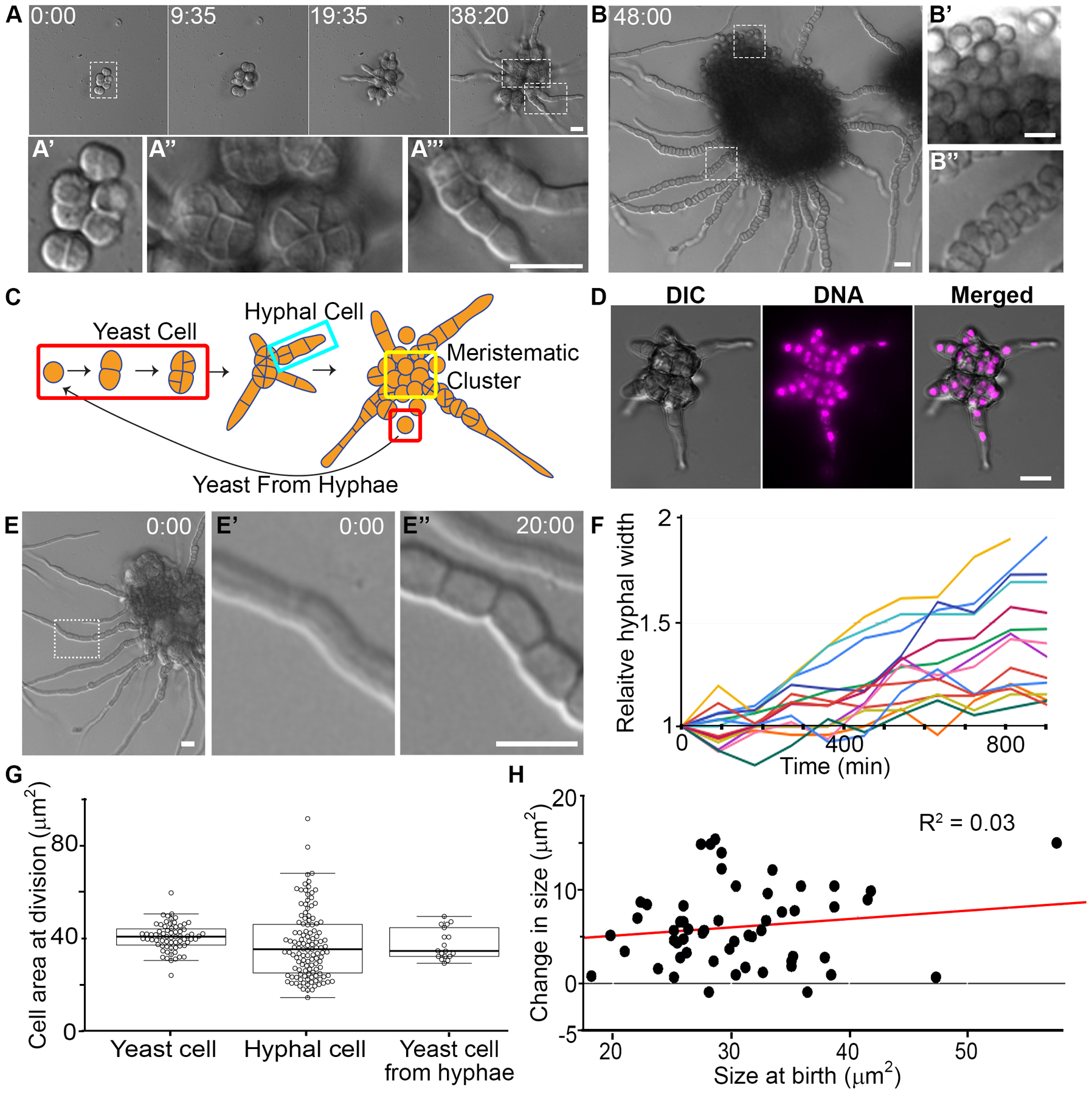
(A) Representative DIC micrographs of P. salicorniae colony growth over the course of 38 hr. Insets show different features of the colony A’ yeast cell, A” wedge shaped cells, A’’’ hyphal cells. These stills are from Video S6. Scale bar, 10 μm. Time, Hours:Minutes. (B) Representative DIC micrograph of a P. salicorniae colony 48 hr after the start of growth. Insets (B’ B”) show different cell types within the colony. These stills are from Video S6. Scale bar, 10 μm. Time, Hours:Minutes. (C) Model for the growth of a P. salicorniae from a single colony. Initial growth begins with a yeast cell (red box). Growth is isotropic and divisions bisect the cell resulting in wedge shaped cells (see A”). At some point some yeast cells begin to grow hyphae (turquoise box), while other cells within the colony divide to form yeast cells (yellow box). Hyphal cells continue to divide until eventually the hyphae burst with cells that are morphologically similar to the initial yeast cell. (D) DNA staining in fixed cells. P. salicorniae compartments contain only a single nucleus. Representative image of a P. salicorniae colony in DIC, stained with DAPI, and in merge. Scale bar, 10 μm. (E) P. salicorniae hyphae continue grow wider while elongating. A representative image of a colony at the start of filming. Inset (E’, E”) showing hyphal width at the start of filming (0:00) and the conclusion of filming (12:00). These stills are from Video S6. Scale bar, 10 μm. Time, Hours:Minutes. (F) Quantification of the change in relative hyphal thickness from the start to end of filming. Each line represents an individual hypha, all hyphae increase in thickness over the length of filming. (G) Quantification of cell size at division in various cell types. The mean size at division is not significantly different between cell types (p > 0.05, t-test) but hyphal divisions are significantly more variable than yeast cells exhibiting both much larger and much smaller cells (p < 0.01, f-test). (H) Quantification of the cell size at birth vs. the cell size at division for hyphal cells and yeast cells.
Two events occur in this meristematic colony. Some cells transition into a second cell type by producing hyphae which extend by polarized growth (Figure 6A’’’ and turquoise box in Figure 6C). At the same time, an abundance of yeast-like cells that are similar in appearance to the original cell (Figure 6B’) are produced and remain associated in a matrix (Video S6). Eventually these yeast cells are released by the breakdown of the matrix around them. In the hyphae, cells proximal to the colony septate, swell, and produce new spherical cells within the hyphae (Figure 6B”, Video S6). These spherical cells appear to be similar to the original yeast cells and can form multiple, orthogonal septa while inside the hyphae. This suggests the ability to switch from hyphal back to yeast-like growth from within a hypha, although it could represent the formation of spores rather than vegetative yeast. As the hyphae grow, the remaining yeast cells continue to divide meristematically and the septa are less clearly visualized (yellow box in Figure 6C).
In addition to alternating between polar, hyphal growth and isotropic, yeast-like growth, we found several other characteristics of this fungus that differ from conventional growth modes. First, the hyphae of P. salicorniae contained only one nucleus per compartment and in the spherical cells in the earliest divisions where it is clear to image single compartments (Figure 6D). Second, P. salicorniae hyphae did not have a fixed width and continued to expand laterally while elongating (Figure 6E–F). We also found that yeast and hyphal cells appeared to have different degrees of cell size control. Both the yeast and the cells within the hyphae divided with a similar mean area of 40.4 μm2 and 37.5 μm2, respectively (not significant, P > 0.05, t-test, N > 62) (Figure 6G). However, the size at division was much more variable in hyphal cells, which had a standard deviation of 15.9 μm2, compared to yeast cells with a standard deviation of just 5.8 μm2 (p < 0.01, F-test, N > 62). Consistently, in hyphal cells, the change in size is virtually uncorrelated with the size at birth, indicating a lack of size control (Figure 6H). Thus, this system shows striking orthogonal divisions unlike those of any model fungal system studied to date for cytokinesis, hyphal growth at the colony periphery, and striking alternation in cell types.
Discussion
Very little is known regarding the distribution, diversity and morphogenesis of marine-derived fungi. Furthermore, there appears to be strong potential for some fungal species to be amphibious as displayed by their highly adaptive nature and their isolation from both terrestrial and marine environments [59–61]. The goal of this study was to identify how fungi found in the marine environment may grow and divide given the different environmental challenges faced in the ocean.
Despite being geographically, temporally and methodologically limited, this study captured substantial fungal diversity. This is surely a significant under sampling of diversity as we used only limited culture conditions, and species which require very specific nutrient requirements were likely missed in our pipeline. A culture-based approach was taken because the goal of this study was to analyze morphogenesis and division of cells. This has the advantage that these systems can now be carefully studied in the lab and empirically analyzed. We are still in the process of identifying other isolates from our collection, but several species, including H. werneckii and A. pullulans, appeared several times and in several places, suggesting that at least some of the species are reasonably abundant. Interestingly, 25% of our 77 sequenced isolates were Basidiomycetes whereas only 10 (2.15%) of the 465 marine isolates identified by Shearer and colleagues [59] were of that phylum. Similar levels of diversity have also been seen in studies of fungi associated with plankton and coral in Hawaii [60, 61]. Metagenomic analysis of samples in the future will give a more comprehensive snapshot at the actual fungal diversity.
Previous studies have suggested there is more overlap between marine and terrestrial species than between marine and freshwater species [59]. 11 of our species had been found, previously or during this study, in both marine and terrestrial environments while only three have been identified in marine and freshwater environments (Table 1). We identified 13 species not listed on Jones et al.’s [43] list of established marine species. These included the three of the black yeasts highlighted in this paper, H. werneckii, K. petricola, and A. pullulans. Two additional species were Apiotrichum dulcitum and A. porosum, which have only been recorded in terrestrial environments [62, 63]. Both Apiotrichum species were isolated only from coastal sediment of Great Harbor during two winter 2018 collections. We identified A. porosum from the first collection and both from the second, which occurred two days after a large storm that produced strong winds and heavy snowfall. It is possible that in the first collection, A. porosum was present because the sand from which it was collected was at a beach near a woodland, and that such transport was then heightened by the storm. The variable timescales of exchange between land and sea complicate the categorization of marine fungi and future work will be critical to resolve exchanges that are by chance and those reflecting adaptation.
There were substantial differences in growth and division amongst the four black yeasts analyzed, both between one another and compared to well-characterized yeast model systems. These differences may support these black yeasts’ flexibility and ability to exist under the challenges associated with marine environments. Size control appeared more limited in the marine yeasts compared to the traditional model systems however this could reflect the significant selection for laboratory growth that the lab strains have experienced. Indeed, there is substantial variation in traits such as the size and shape of the bud and position of nuclei in different wild isolates of S. cerevisiae [64]. However, the variation in programs of morphogenesis in the marine yeasts were particularly striking and unlikely to have been lost in laboratory culturing of the conventional models. Remarkably, the two distinct modes of division in the model yeasts, fission and budding, coexist in the isolates of H. werneckii. In this system, cells systematically alternate between division by fission and budding, readily transitioning between them in a single generation. It is unclear what, if any, adaptive advantage this toggling between forms provides these cells. One possibility is that it represents a form of bet hedging. Depending on specific environmental stress conditions, budding or fission may be more advantageous, and by alternating between these states on a short timeframe, some subset of the population will be ready for any rapid challenge.
A second surprise came from K. petricola which produced spherical cells that appeared to expand isotropically. At the colony level, these bubble-like cells generated linear chains within branched networks. The chains displayed a highly non-random directed budding pattern, and at a lower frequency, the chains branched and sprouted a new chain through a mother producing a second bud at a different orientation than the first bud. This patterning allows these spherical cells to generate dendritic colonies and is reminiscent of the “snowflake” yeast than can arise in S. cerevisiae strains through mutations inactivating cell separation and in turn lead to multicellular chains of cells [65]. We have found that agitation or vigorous pipetting can disrupt the chains of K. petricola, indicating distinction from the evolved snowflake yeasts. We speculate that despite the very isotropic growth of individual cells, there is some mark at the site of bud initiation that persists and biases the site of the next bud. Testing this hypothesis must await the development of molecular tools for this system.
Yet another striking growth pattern that is distinct from the model yeasts was seen in A. pullulans which simultaneously produced up to six buds from a multinucleate mother cell. We also isolated the closely related black yeast, Aureobasidium melanogenum, once from coral and once from sponge from Great Harbor. A. melanogenum also exhibited the pattern of producing multiple buds from the same site, although we did not carry out a detailed analysis of this species. This multi-budding phenotype is in striking contrast to the tightly controlled single bud per cell cycle produced by S. cerevisiae. Singularity in budding is a mechanism to coordinate nuclear division and morphogenesis to ensure that each daughter cell receives only one nucleus. In A. pullulans it is unclear what determines which cells in the population ultimately become mother cells, as the buds produced are generally much smaller and more homogeneous in size that the mothers, which are often highly irregular in shape and size. How the nuclear cycle of the mother cells is coordinated with the multi-budding program, how specific nuclei are selected for deposition into buds, and how it is ensured that each bud receives a nucleus remain fascinating problems for future study in this system.
Finally, the most complex isolate was P. salicorniae, a very recently discovered species with limited descriptions in the literature. The myriad of growth patterns seen in this organism suggest levels of developmental control more complex than in the other black yeasts. The different cell types and meristematic growth resemble other species in the genus such as Phaeotheca triangularis [58], but what stands out as remarkable compared to many traditional fungal models are the series of orthogonal divisions that lead to triangular-shaped compartments and the intercalary growth of hyphae. How division planes are positioned and which cells are fated for hyphal growth from the meristematic colony are fascinating open questions from this new system.
These studies reveal the morphological plasticity present within individual fungal species and the limited view that study of a few model systems have presented as to the rules governing fungal shape and division. Unlike many of the model systems cultivated in lab, there are remarkable variations in patterning in the division cycle of the marine-derived yeasts, with high degrees of variability in cell size for three of the species we examined. This highlights the potential of observing diverse non-model biological systems to test our deeply held models of biological phenomena. We predict many fundamental biological mechanisms have higher degrees of plasticity than appreciated from studies limited to model systems.
STAR METHODS
CONTACT FOR RESOURCE SHARING
Further information and request for resources not already available should be directed to and will be fulfilled by the Lead Contact, Amy S. Gladfelter amyglad@unc.edu.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Collection
Environmental ocean samples.
17 environmental ocean water samples were collected using a plankton net on eight days spanning from summer 2016 to winter 2018 (Figure 1A). The majority of samples were collected in duplicate from two sites (Vineyard Sound and Buzzards Bay) during the summer and fall of 2017 and 2018. One sample was collected during the summer 2016 from Buzzards Bay and one sample from Great Harbor during the winter 2018. The plankton net was 1 m in diameter with a 504 μm mesh. This was trailed for 10 min behind a Boston whaler at an average over-ground speed of 1.2 m/s. The top of the net was kept just under the water surface. Tows were 1–1.5 km offshore and average flow-through volume was 2400 m3. Of the environmental ocean samples described in this paper, the majority were isolated from a set of eight tows conducted on July and August, 2017, four in Buzzards Bay and four in Martha’s Vineyard Sound. Average sea surface temperature at these tow sites was 22.4 °C. We also included several isolates from August 2016 (Buzzards Bay), and March 2018 (Great Harbor). These include both ocean and sediment isolates (see below). Average surface temperatures for ocean samples from August 2016 and March 2018 were 22 °C and 2 °C, respectively.
The Great Harbor Tow was conducted on March 16, 2018. This was two days after a three-day nor’easter that produced heavy winds and several feet of snow.
Plankton tows were also performed in different areas of Little Sippewissett Marsh using a hand-held Lamotte 1063 Plankton net (pore size 153 μm) and wading along the center of water channels. Collection was during the summer of 2017 and at mid-tide.
For all ocean samples, two different concentrations of water were plated in duplicate: 300 μl of seawater with plankton material per plate and 300 μl of concentrated seawater with plankton material per plate. For concentrated samples, 1.0 L of water was passed through a 22 μm filter to a final volume of approximately 50–100 ml- i.e. concentrated 10- to 20-fold. In these concentrated samples, plankton and particles of organic matter were often visible and included in what was plated. Because of heterogeneity in plankton tow collections, we could not systematically plate the same mass for each plate. Additionally, a sample was taken from the filter using sterile cotton tipped applicator and plated or occasionally a piece of the filter was placed on a plate. After filtration various plankton species were present in the concentrate and on the filter so some of our strains are likely parasites or symbionts of plankton.
All samples were plated in duplicate except samples from filters remaining after concentration of ocean samples.
Sediment samples.
Coastal sediment samples were collected nearshore using sterile 50 ml conical tubes (samples always included some amount of water as well). Samples were collected from beaches adjacent to Buzzards Bay, Martha’s Vineyard Sound, and Great Harbor. These sites included Trunk River, Surf Drive Beach, Fay Beach, Garbage Beach, Dog Beach, Quissett Harbor, the Knob, Little Sippewissett Marsh, and by the Woods Hole Town Dock in Great Harbor (Figure 1A). For these samples, conical tubes containing samples were shaken and then 300 μl from each was plated in duplicate.
Sediment samples were collected during summer 2017 and winter 2018. Temperature during summer 2017 was a similar range as the environmental ocean samples, ranging 20–23 °C while temperature during the winter collections was 2–12 °C. One set of winter 2018 sediment samples was collected two days after nor’easter previously mentioned.
Coral and sponge samples.
Coral and sponge samples were collected by SCUBA divers near Garbage Beach in Woods Hole, MA during July and August 2018 and February 2019. Samples were collected 9–13 m below sea level. Corals were rinsed with sea water that had been filtered through a 0.22 μm filter and removed from their substrate of rock or sponge. For each coral sample, an average of 42 polyps of the same color from the same colony were ground up using a mortar and pestle with filtered seawater added with a ratio of 1.0 ml water to 1.0 g of sample. Additionally, some coral and sponge samples were inhabited by small unidentified black annelids. We removed these as much as possible prior to grinding. Average mass per coral colony was 7.74 g. 100 μl of liquid supernatant (the liquid mixture of sample and water produced upon grinding) and 150 μl of slurry (solid ground-up coral) were plated in duplicate on YPD, MEA, and PDA saltwater plates. Sponge samples were prepared in the same manner. Water from the same location was also plated as negative control to ensure isolated fungi were not environmental or pipeline contaminants.
By plating coral samples collected in the summer and winter, we were plating samples enriched with and with limited numbers of symbiont. Polyps with Symbiodinium (symbiotic) are colored [137]. From the two summer collections, we observed a range of colors including white and light pink to darker purple and brown. Some colonies contained polyps of multiple different colors. Polyps without Symbiodinium (aposymbiotic, from the February collection) are much paler in color. Samples from February 2019 samples are still being processed.
Media
Samples were plated on three types of media to offer different carbon sources: 1) Yeast Extract with dextrose (YPD; yeast extract 10 g/L, peptone 10 g/L, dextrose 20 g/L, agar 12 g/L); 2) Malt extract (MEA; malt extract 20 g/L, peptone 6 g/L, dextrose 20 g/L, agar 12 g/L); and 3) Potato Dextrose (PDA; potato dextrose broth 24 g/L, agar 12 g/L). All media were made with seawater, or 36 g/L of Instant Ocean if plates were made inland without easy ocean access. Antibiotics were added to select for fungal isolates (carbenicillin 100 μl/ml, chloramphenicol 25 μl/ml and tetracycline 10 μl/ml).
Culturing
All cultures were incubated at 18–20 °C for 6–14 days. Black yeasts appeared on plates after approximately 7 days. For filming, cells from fresh, isolated streaks were used.
METHOD DETAILS
Filming
Long-term (up to 36 hr) DIC time-lapse data were collected with a Nikon Eclipse Ti2 inverted microscope with a perfect focus system using a 40x dry objective lens using a Nikon DS-Qi2 camera. The microscope was operated using NIS-Elements software. Samples were imaged on agar pads made of the same media type on which they were originally isolated. Up to six isolates (one agar pad per isolate) were filmed per session (four areas of interest per isolate).
Agar pads were made by melting agar media and pipetting the liquid into a mold. The mold was constructed by sandwiching a 1/32 in rubber sheet cut like a rectangular frame missing the top side (so the mold may be filled in) sandwiched between two glass microscope slides and held together with binder clips. The liquid was let to cool and solidify for 10 min. Pads were cut with sterile razor blades to be approximately 1 cm2.
A small amount of sample from a plate was mixed with 50 μl of liquid media, vortexed slowly and rotated for up to 2 hr. In the following order, we placed on a 28×65 mm coverslip: 6 μl of dilute liquid culture containing a fungal strain, the agar pad, 2 μl liquid media, and lastly the microscope slide. The coverslip, agar pad, and microscope slide sandwich are then sealed with VALAP. Several holes were created in the VALAP between the coverslip and slide with a 27-gauge needle to allow oxygen flow. VALAP is Vaseline, Lanolin and Paraffin in a 1:1:1 mass ratio.
Mounting conditions including both solid and liquid media, as well as a solid-air interface along the edges of agar pads to provide multiple options for growth of cells. Multiple species formed visible hyphae or pseudohyphae along the solid-air interface. Long-term imaging of just liquid culture is not feasible due to focus issues. All filming was performed at 18–20 °C.
Molecular identification of isolates
Cells were grown on the same type of media from which they originated. Yeast were harvested from agar plates while filamentous fungi were harvested from liquid culture. For yeast strains, we utilized the DNeasy Plant Mini Kit to extract DNA as per the manufacturer’s protocol. For filamentous isolates for which the DNeasy Plant Mini Kit did not work well, we performed a phenol chloroform extraction (modified from [138]). All samples were amplified using the ITS1 [139], ITS2 [139], D1/D2 [140], and V9 [141] rDNA regions for molecular identification (Table S1). The RPB2 gene[142] was additionally sequenced when rDNA failed to distinguish between closely related species, such as A. pullulans and A. melanogenum.
Isolation of Genomic DNA using Phenol/chloroform
Filamentous fungi were grown in liquid YPD for approximately a week on a rotator at 20 °C. Colonies were removed with a sterile spatula and placed on a stack of sterile paper towels to wick off liquid. Dried colonies were transferred to an Eppendorf tube, with 750 μl of lysis buffer (50 mM Tris-HCl pH 7.4, 50 mM EDTA pH 8, 3% SDS, and 1% 2-mercaptoethanol) and vortexed. Then incubated at 65 °C for 1 hr. Then 750 μl of phenol/chloroform/isoamyl alcohol was added to the tube and then briefly vortexed. The tube was spun at 12,000 × g for 10 min and the aqueous layer was removed to a new tube. A second phenol/chloroform extraction was then performed on the aqueous phase. Following the second extraction, the aqueous phase was transferred to new tube and 20 μl of 3 M NaOAc and 700 μl isopropanol precooled to −20 °C was added. The tube was inverted gently to mix then spun at 12,000 × g for 30 sec to pellet DNA. The supernatant was removed and discarded. The pellet was resuspended in 300 μl of TE (100 mM Tris-HCl pH 7.4, 10 mM EDTA, pH 8.0) at 65 °C for 15 min followed by finger vortexing. DNA was then precipitated by the addition of 10 μl of 3 M NaOAc and 700 μl ethanol at −20 °C. The tube was spun at 12,000 × g for 2 min and the supernatant was removed. The pellet was washed with 500 μl of 70% ethanol at −20°C. The pellet was then spun at 12,000 × g for 30 sec the supernatant was removed and the pellet was allowed to air dry at room temperature for 30 min. The pellet was then resuspended in 100 μl of sterile DI water.
DNA staining
Prior to fixing and subsequent mounting on microscopy slides, cells used for DNA staining were grown in liquid culture. Colony morphology did appear different in liquid culture as compared to when grown on solid media.
H. werneckii was grown in liquid culture for 18 hr at 30 °C in liquid media (YPD + 0.6 M NaCl). Cells were fixed with 3.7% formaldehyde and 25 mM Dithiothreitol (DTT) for 1 hour. Cells were washed with 1x phosphate buffered saline (PBS) 3 times, then stained with Hoechst 33342 in PBS for 30 min.
K. petricola was grown in liquid culture for 92 hr at 22 °C. Cells were fixed in 100% methanol on ice for 20 min, washed with 1x PBS, then stained with NucBlue Live Ready Probe in PBS.
A. pullulans was grown in YPD liquid culture for 24 hr at 24 °C with Complete Supplement Mixture (CSM) and 2% Dextrose + 0.5M NaCl. Cells were fixed in 70% ethanol on ice for 10 min, washed with 1x PBS + 0.5 M NaCl, then stained in 1x PBS + 0.5M NaCl with 50 ng/ml 4′,6-diamidino-2-phenylindole (DAPI).
P. salicorniae was grown affixed to slides coated with Concanavalin A immersed in liquid media (YPD + 0.6M NaCl) for 48 hr at 22 °C. Cells were fixed on the slide with 100% ice cold methanol for 20 min, washed with 1x PBS, then stained with Hoechst 33342 in PBS.
QUANTIFICATION AND STATISTICAL ANALYSIS
Quantifying cell characteristics in H. werneckii and P. salicorniae
Cell size metrics were measured using ImageJ [143]. Area was measured by tracing the outline of a cell with the freehand selection tool. Length and width were measured using the line measurement tool. The time of septation was determined to be when the septum was fully formed line between the two halves of the original cell. To measure the time between divisions, the number of frames between two events was counted and then multiplied by the frame rate.
Measuring angles between cells in K. petricola
Colonies of Knufia petricola appeared as linear chains within branched networks. After budding, cells remained connected and did not move with very few exceptions.
To estimate angles of budding within the colonies, we wrote a script to record the lineage of the cells and their relative positions within the colonies. 237 cells were analyzed over 28 colonies. A user defines the center of each cell by clicking, then assigns the mother of each cell based on the time lapse movie of colony growth. To estimate the error associated with clicking, we wrote a script that presents circles at random positions, then a user tries to click the center of each circle, then the script calculates the displacement between the click and the actual center. The average displacement was 0.6 pixels = 0.1 μm, and since the cell diameter was about 25 pixels = 4.6 μm, this translates to an error in angle of about arctan(0.6/25) = 1.4°. 1.4° is less than the observed standard deviations of 45° for linear chain budding and 44° for branched network budding, so the variation in angles reflects true variation within the colonies. Most of the cells were in a common focal plane between the agar pad and glass cover slip, and the angles were calculated based on the XY positions of the cells.
To quantify the circular variance, each budding event was represented as a unit vector, with an angle corresponding to the direction of budding. The resultant vector was calculated as the mean of the unit vectors. The direction of the resultant vector represents the mean direction of budding. The length of the resultant vector is inversely related to the variance and between 0 and 1. If the unit vectors point in all directions isotropically corresponding to maximal variance, then they add to 0. If the unit vectors all point in the same direction corresponding to minimal variance, then they add to N (where N is the number of budding events), so dividing by N to yield the mean has length 1. The circular variance was defined as 1 - R (where R is the length of the resultant vector).
Analysis of budding pattern and cell cycle duration in A. pullulans
Data from three different days were analyzed: Day 1 7/26/2018 (32 cells), Day 2 8/3/2018 (27 cells), and Day 3 9/26/2018 (26 cells). These dates represent a range of times after the isolate was originally cultured, with Day 1 being 2–3 weeks from initial isolation.
Cells for analysis were chosen at random. Cells that did not bud during the filming period were not scored: Day 1 (7 cells), Day 2 (1 cell) and Day 3 (2 cells), therefore, N = 75 cells. The percentage of cells producing multiple buds was different for each day filmed, with Day 1 the lowest at 68% and Day 3 the highest at 88%. For the analysis in this manuscript, the data from all days were pooled.
The multi-bud phenotype appeared to be specific to a site or sites on the original mother cells. From our fixed analysis, these cells are likely to have multiple nuclei.
For cell cycle duration, only budding events from the initial mother cells were scored, not their daughters.
All statistical analyses were computed using R [144] K. petricola angular data were quantified and analyzed using a custom application in MATLAB [145].
Supplementary Material
Video S1. DIC time lapse of H. werneckii (40x). Scale bar, 10 μm. Time, Hours:Minutes. Related to Figure 3.
Video S2. DIC time lapse of K. petricola (40x). Two examples of colony growth. Scale bar, 10 μm. Time, Hours:Minutes. Related to Figure 4.
Video S3. DIC time lapse of A. pullulans (40x). This mother cell repeatedly produces multiple simultaneous buds from the same site. Scale bar, 10 μm. Time, Hours:Minutes. Related to Figure 5.
Video S4. DIC time lapse of A. pullulans (40x). Three more examples of budding mother cells demonstrating the variability of cell morphology. Many mother cells from Day 3 of filming lyse after producing daughter cells. Scale bar, 10 μm. Time, Hours:Minutes. Related to Figure 5.
Video S5. DIC time lapse of P. salicorniae (40x). Growth of initial yeast cells and sprouting of hyphae which then develop septa. Scale bar, 10 μm. Time, Hours:Minutes. Related to Figure 6.
Video S6. DIC time lapse of P. salicorniae (20x). At left, overview of later stages of colony growth. At right, close-ups of hyphae forming septa and reverting back to yeast cells. Scale bars, 10 μm. Time, Hours:Minutes. Related to Figure 6.
Acknowledgements
CMF, ASG, and LMYM-F thank the MBL for support including Scott Bennett and the MBL Marine Resources Center for help in collecting specimens, Loretta Roberson at MBL for advice on working with coral, and the Physiology and Microbial Diversity courses at MBL for collaborative interactions. CE was a student Physiology course participant. We also thank Nikon at MBL and the Nikon Imaging Center at HMS for microscopes and imaging advice. LMYM-F thanks the Chiara Ricci-Tam in the Springer lab at HMS for initial help with DNA extraction.
Funding sources: ASG, BMS, EJDV, and LMYM-F were supported by an HHMI faculty Scholar award and NSF grant MCB-1615138. JMV-M was supported by NIH Training Grant 5T32AI052080-14. DJL and SVD were supported by R35GM122488. JFP was supported by the Fannie and John Hertz Foundation. CE was supported by the Boehringer Ingelheim Fonds. CMF was supported by NIH grant GM39565.
Footnotes
Declaration of Interests
The authors have no competing interests to declare.
References
- 1.de Vargas C, Audic S, Henry N, Decelle J, Mahé F, Logares, Lara E, Berney C, Le Bescot N, Probert I, et al. (2015). Eukaryotic plankton diversity in the sunlit ocean. Science 348, 1261605.1261601–1261605.1261611. [DOI] [PubMed] [Google Scholar]
- 2.Kohlmeyer J, and Kohlmeyer E (1979). Marine Mycology The Higher Fungi, (Academic Press; ). [Google Scholar]
- 3.Blum LK, Mills AL, Zieman JC, and Zieman RT (1988). Abundance of bacteria and fungi in seagrass and mangrove detritus. Mar Ecol Prog Ser 42, 73–78. [Google Scholar]
- 4.Takami H, Inoue A, Fuji F, and Horikoshi K (1997). Microbial flora in the deepest sea mud of the Mariana Trench. FEMS Microbiol Lett 152, 279–285. [DOI] [PubMed] [Google Scholar]
- 5.Kim K, Harvell CD, Kim PD, Smith GW, and Merkel SM (2000). Fungal disease resistance of Caribbean sea fan corals (Gorgonia spp.). Mar Biol 136, 259–267. [Google Scholar]
- 6.Zuccaro A, Schoch CL, Spatafora JW, Kohlmeyer J, Draeger S, and Mitchell JI (2008). Detection and identification of fungi intimately associated with the brown seaweed Fucus serratus. Appl Environ Microbiol 74, 931–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Le Calvez T, Burgaud G, Mahe S, Barbier G, and Vandenkoornhuyse P (2009). Fungal diversity in deep-sea hydrothermal ecosystems. Appl Environ Microbiol 75, 6415–6421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Q, and Wang G (2009). Diversity of fungal isolates from three Hawaiian marine sponges. Microbiol Res 164, 233–241. [DOI] [PubMed] [Google Scholar]
- 9.Rämä T, Nordén J, Davey ML, Mathiassen GH, Spatafora JW, and Kauserud H (2014). Fungi ahoy! Diversity on marine wooden substrata in the high North. Fungal Ecology 8, 46–58. [Google Scholar]
- 10.Redou V, Navarri M, Meslet-Cladiere L, Barbier G, and Burgaud G (2015). Species richness and adaptation of marine fungi from deep-subseafloor sediments. Appl Environ Microbiol 81, 3571–3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gutierrez MH, Jara AM, and Pantoja S (2016). Fungal parasites infect marine diatoms in the upwelling ecosystem of the Humboldt current system off central Chile. Environ Microbiol 18, 1646–1653. [DOI] [PubMed] [Google Scholar]
- 12.Amend A, Burgaud G, Cunliffe M, Edgcomb VP, Ettinger CL, Gutierrez MH, Heitman J, Hom EFY, Ianiri G, Jones AC, et al. (2019). Fungi in the Marine Environment: Open Questions and Unsolved Problems. MBio 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gladfelter AS, James TY, and Amend AS (2019). Marine fungi. Curr Biol 29, R191–R195. [DOI] [PubMed] [Google Scholar]
- 14.Hawksworth D (1990). The fungal dimension of biodiversity: magnitude, significance, and conservation. Mycol Res 95, 641–655. [Google Scholar]
- 15.Schoch CL, Crous PW, Groenewald JZ, Boehm EW, Burgess TI, de Gruyter J, de Hoog GS, Dixon LJ, Grube M, Gueidan C, et al. (2009). A class-wide phylogenetic assessment of Dothideomycetes. Stud Mycol 64, 1–15S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hibbett DS, and Binder M (2001). Evolution of marine mushrooms. Biol Bull 201, 319–322. [DOI] [PubMed] [Google Scholar]
- 17.Richards TA, Jones MD, Leonard G, and Bass D (2012). Marine fungi: their ecology and molecular diversity. Ann Rev Mar Sci 4, 495–522. [DOI] [PubMed] [Google Scholar]
- 18.Dini-Andreote F, Pylro VS, Baldrian P, van Elsas JD, and Salles JF (2016). Ecological succession reveals potential signatures of marine-terrestrial transition in salt marsh fungal communities. ISME J 10, 1984–1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Velez P, Alejandri-Ramirez ND, Gonzalez MC, Estrada KJ, Sanchez-Flores A, and Dinkova TD (2015). Comparative Transcriptome Analysis of the Cosmopolitan Marine Fungus Corollospora maritima Under Two Physiological Conditions. G3 (Bethesda) 5, 1805–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Orsi WD, Edgcomb VP, Christman GD, and Biddle JF (2013). Gene expression in the deep biosphere. Nature 499, 205–208. [DOI] [PubMed] [Google Scholar]
- 21.Tisthammer KH, Cobian GM, and Amend AS (2016). Global biogeography of marine fungi is shaped by the environment. Fungal Ecology 19, 39–46. [Google Scholar]
- 22.Hyde KD, Jones EBG, Leaño E, Pointing SB, Poonyth AD, and Vrijmoed LLP (1998). Role of fungi in marine ecosystems. Biodiversity Conserv 7, 1147–1161. [Google Scholar]
- 23.Jones EBG (2011). Fifty years of marine mycology. Fungal Diversity 50, 73–112. [Google Scholar]
- 24.Pang K-L, Overy DP, Jones EBG, Calado M.d.L., Burgaud G, Walker AK, Johnson JA, Kerr RG, Cha H-J, and Bills GF (2016). ‘Marine fungi’ and ‘marine-derived fungi’ in natural product chemistry research: Toward a new consensual definition. Fungal Biology Reviews 30, 163–175. [Google Scholar]
- 25.Gostincar C, Muggia L, and Grube M (2012). Polyextremotolerant black fungi: oligotrophism, adaptive potential, and a link to lichen symbioses. Front Microbiol 3, 390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gostincar C, Stajich JE, Zupancic J, Zalar P, and Gunde-Cimerman N (2018). Genomic evidence for intraspecific hybridization in a clonal and extremely halotolerant yeast. BMC Genomics 19, 364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hartwell LH, Culotti J, and Reid B (1970). Genetic control of the cell-division cycle in yeast, I. detection of mutants. Proc Natl Acad Sci 66, 352–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mitchison JM (1972). The Biology of the Cell Cycle, (New York: Cambridge University Press; ). [Google Scholar]
- 29.Hartwell LH (1974). Saccharomyces cerevisiae cell cycle. Bacteriological Reviews 38, 164–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nurse P (1975). Genetic control of cell size at cell division in yeast. Nature 256, 547–551. [DOI] [PubMed] [Google Scholar]
- 31.Nurse P, Thuriaux P, and Nasmyth K (1976). Genetic control of the cell division cycle in the fission yeast Schizosaccharomyces pombe. Molec gen Genet 146, 167–178. [DOI] [PubMed] [Google Scholar]
- 32.Fantes P, and Nurse P (1977). Control of cell size at division in fission yeast by a growth-modulated size control over nuclear division. Exp Cell Res 107, 377–386. [DOI] [PubMed] [Google Scholar]
- 33.Howell AS, Savage NS, Johnson SA, Bose I, Wagner AW, Zyla TR, Nijhout HF, Reed MC, Goryachev AB, and Lew DJ (2009). Singularity in polarization: rewiring yeast cells to make two buds. Cell 139, 731–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kang H, and Lew DJ (2017). How do cells know what shape they are? Curr Genet 63, 75–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chiou JG, Ramirez SA, Elston TC, Witelski TP, Schaeffer DG, and Lew DJ (2018). Principles that govern competition or co-existence in Rho-GTPase driven polarization. PLoS Comput Biol 14, e1006095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Esposito MS, and Esposito RE (1974). GENES CONTROLLING MEIOSIS AND SPORE FORMATION IN YEAST. Genetics 78, 215–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Esposito MS, Esposito RE, Arnaud M, and Halvorson HO (1969). Acetate Utilization and Macromolecular Synthesis During Sporulation of Yeast. Journal of Bacteriology 100, 180–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mitchison JM, and Nurse P (1985). Growth in cell length in the fission yeast Schizosaccharomyces pombe. J Cell Biol 75, 357–376. [DOI] [PubMed] [Google Scholar]
- 39.Loog M, and Morgan DO (2005). Cyclin specificity in the phosphorylation of cyclin-dependent kinase substrates. Nature 434, 104–108. [DOI] [PubMed] [Google Scholar]
- 40.Gutierrez-Escribano P, and Nurse P (2015). A single cyclin-CDK complex is sufficient for both mitotic and meiotic progression in fission yeast. Nat Commun 6, 6871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Swaffer MP, Jones AW, Flynn HR, Snijders AP, and Nurse P (2016). CDK Substrate Phosphorylation and Ordering the Cell Cycle. Cell 167, 1750–1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.EPA (2017). SIXTH EXPLANATION OF SIGNIFICANT DIFFERENCES FOR THE NEW BEDFORD HARBOR SUPERFUND SITE UPPER AND LOWER HARBOR OPERABLE UNIT 1 (OU1) AND THE OUTER HARBOR OPERABLE UNIT 3 (OU3) NEW BEDFORD, MASSACHUSETTS EPA REGION 1., EPA New England, ed. (New Bedford Harbor Superfund Site Administrative Record; ), pp. 1–16. [Google Scholar]
- 43.Jones EBG, Suetrong S, Sakayaroj J, Bahkali AH, Abdel-Wahab MA, Boekhout T, and Pang K-L (2015). Classification of marine Ascomycota, Basidiomycota, Blastocladiomycota and Chytridiomycota. Fungal Diversity 73, 1–72. [Google Scholar]
- 44.Kogej T, Stein M, Volkmann M, Gorbushina AA, Galinski EA, and Gunde-Cimerman N (2007). Osmotic adaptation of the halophilic fungus Hortaea werneckii: role of osmolytes and melanization. Microbiology 153, 4261–4273. [DOI] [PubMed] [Google Scholar]
- 45.Zalar P, Zupančič J, Gostinčar C, Zajc J, de Hoog GS, De Leo F, Azua-Bustos A, and Gunde-Cimerman N (2019). The extremely halotolerant black yeast Hortaea werneckii - a model for intraspecific hybridization in clonal fungi. IMA Fungus 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bonifaz A, Badali H, de Hoog GS, Cruz M, Araiza J, Cruz MA, Fierro L, and Ponce RM (2008). Tinea nigra by Hortaea werneckii, a report of 22 cases from Mexico. Stud Mycol 61, 77–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mitchison JM (1958). The growth of single cells: II. Saccharomyces cerevisiae. Experimental Cell Research 15, 214–221. [DOI] [PubMed] [Google Scholar]
- 48.Wollenzien U, de Hoog GS, Krumbein W, and Uijthof JMJ (1997). Sarcinomyces petricola, a new microcolonial fungus from marble in the Mediterranean basin. Antonie Van Leeuwenhoek 71, 281–288. [DOI] [PubMed] [Google Scholar]
- 49.Nai C, Wong HY, Pannenbecker A, Broughton WJ, Benoit I, de Vries RP, Gueidan C, and Gorbushina AA (2013). Nutritional physiology of a rock-inhabiting, model microcolonial fungus from an ancestral lineage of the Chaetothyriales (Ascomycetes). Fungal Genet Biol 56, 54–66. [DOI] [PubMed] [Google Scholar]
- 50.Noack-Schönmann S, Spagin O, Gründer KP, Breithaupt M, Günter A, Muschik B, and Gorbushina AA (2014). Sub-aerial biofilms as blockers of solar radiation: spectral properties as tools to characterise material-relevant microbial growth. International Biodeterioration & Biodegradation 86, 286–293. [Google Scholar]
- 51.Rekha MR, and Sharma CP (2007). Pullulan as a Promising Biomaterial for Biomedical Applications: A Perspective. Trends Biomater Artif Organs 20. [Google Scholar]
- 52.Gostincar C, Ohm RA, Kogej T, Sonjak S, Turk M, Zajc J, Zalar P, Grube M, Sun H, Han J, et al. (2014). Genome sequencing of four Aureobasidium pullulans varieties: biotechnological potential, stress tolerance, and description of new species. BMC Genomics 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chi Z, Wang F, Chi Z, Yue L, Liu G, and Zhang T (2009). Bioproducts from Aureobasidium pullulans, a biotechnologically important yeast. Appl Microbiol Biotechnol 82, 793–804. [DOI] [PubMed] [Google Scholar]
- 54.Manitchotpisit P, Leathers TD, Peterson SW, Kurtzman CP, Li XL, Eveleigh DE, Lotrakul P, Prasongsuk S, Dunlap CA, Vermillion KE, et al. (2009). Multilocus phylogenetic analyses, pullulan production and xylanase activity of tropical isolates of Aureobasidium pullulans. Mycol Res 113, 1107–1120. [DOI] [PubMed] [Google Scholar]
- 55.Zalar P, Gostincar C, de Hoog GS, Ursic V, Sudhadham M, and Gunde-Cimerman N (2008). Redefinition of Aureobasidium pullulans and its varieties. Stud Mycol 61, 21–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sigler L, Tsuneeda A, and Carmichael JW (1981). Phaeotheca and Phaeosclera, two new genera of dematiaceous Hyphomycetes and a redescription of Sarcinomyces Lindner. Mycotaxon 12, 449–467. [Google Scholar]
- 57.Crous PW, and Roets F (2016). Phaeotheca salicornia. Persoonia 36, 365. [Google Scholar]
- 58.de Hoog GS, Beguin H, and Batenburg-van de Vegte WH (1997). Phaeotheca triangularis, a new meristematic black yeast from a humidifier. Antonie van Leeuwenhoek 71, 289–295. [DOI] [PubMed] [Google Scholar]
- 59.Shearer CA, Descals E, Kohlmeyer B, Kohlmeyer J, Marvanová L, Padgett D, Porter D, Raja HA, Schmit JP, Thorton HA, et al. (2006). Fungal biodiversity in aquatic habitats. Biodiversity and Conservation 16, 49–67. [Google Scholar]
- 60.Gao Z, Li B, Zheng C, and Wang G (2008). Molecular detection of fungal communities in the Hawaiian marine sponges Suberites zeteki and Mycale armata. Appl Environ Microbiol 74, 6091–6101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Amend AS, Barshis DJ, and Oliver TA (2012). Coral-associated marine fungi form novel lineages and heterogeneous assemblages. ISME J 6, 1291–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.James SA, Bond CJ, Stanley R, Ravella SR, Peter G, Dlauchy D, and Roberts IN (2016). Apiotrichum terrigenum sp. nov., a soil-associated yeast found in both the UK and mainland Europe. Int J Syst Evol Microbiol 66, 5046–5050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yurkov AM, Wehde T, Federici J, Schäfer AM, Ebinghaus M, Lotze-Engelhard S, Mittelbach M, Prior R, Richter C, Röhl O, et al. (2016). Yeast diversity and species recovery rates from beech forest soils. Mycological Progress 15, 845–859. [Google Scholar]
- 64.Nogami S, Ohya Y, and Yvert G (2007). Genetic Complexity and Quantitative Trait Loci Mapping of Yeast Morphological Traits. PLOS Genetics 3, e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ratcliff WC, Fankhauser JD, Rogers DW, Greig D, and Travisano M (2015). Origins of multicellular evolvability in snowflake yeast. Nat Commun 6, 6102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zuccaro A, Summerbell R, Gams W, Schroers H, and Mitchell J (2004). A new Acremonium species associated with Fucus spp., and its affinity with a phylogenetically distinct marine Emericellopsis clade. Stud Mycol 50, 283–297. [Google Scholar]
- 67.Rédou V, Kumar A, Hainaut M, Henrissat B, Record E, Barbier G, and Burgaud G (2016). Draft Genome Sequence of the Deep-Sea Ascomycetous Filamentous Fungus Cadophora malorum Mo12 from the Mid-Atlantic Ridge Reveals Its Biotechnological Potential. Genome Announc 4, e00467–00416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Burgaud G, Le Calvez T, Arzur D, Vandenkoornhuyse P, and Barbier G (2009). Diversity of culturable marine filamentous fungi from deep-sea hydrothermal vents. Environ Microbiol 11, 1588–1600. [DOI] [PubMed] [Google Scholar]
- 69.Almeida C, Eguereva E, Kehraus S, Siering C, and König GM (2010). Hydroxylated Sclerosporin Derivatives from the Marine-Derived Fungus Cadophora malorum. Journal of Natural Products 73, 476–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.McColloch LP (1944). A Study of the Apple Rot Fungus Phialophora malorum. Mycologia 36, 576–590. [Google Scholar]
- 71.Arenz BE, Held BW, Jurgens JA, Farrell RL, and Blanchette RA (2006). Fungal diversity in soils and historic wood from the Ross Sea Region of Antarctica. Soil Biology and Biochemistry 38, 3057–3064. [Google Scholar]
- 72.Shao Z, and Sun F (2007). Intracellular sequestration of manganese and phosphorus in a metal-resistant fungus Cladosporium cladosporioides from deep-sea sediment. Extremophiles 11, 435–443. [DOI] [PubMed] [Google Scholar]
- 73.Bonugli-Santos RC, Durrant LR, da Silva M, and Sette LD (2010). Production of laccase, manganese peroxidase and lignin peroxidase by Brazilian marine-derived fungi. Enzyme and Microbial Technology 46, 32–37. [Google Scholar]
- 74.El-Hissy FT, Khallil M, A. R, and A. El-Nagdy M (1990). Fungi associated with Some Aquatic Plants Collected from Freshwater Areas At Assiut (Upper Egypt). Medical Journal of Islamic World Academy of Sciences 3, 298–304. [Google Scholar]
- 75.Buesing N, Filippini M, Burgmann H, and Gessner MO (2009). Microbial communities in contrasting freshwater marsh microhabitats. FEMS Microbiol Ecol 69, 84–97. [DOI] [PubMed] [Google Scholar]
- 76.Perkins AK, Ganzert L, Rojas-Jimenez K, Fonvielle J, Hose GC, and Grossart H-P (2019). Highly diverse fungal communities in carbon-rich aquifers of two contrasting lakes in Northeast Germany. Fungal Ecology 41, 116–125. [Google Scholar]
- 77.Zalar P, de Hoog GS, Schroers HJ, Crous PW, Groenewald JZ, and Gunde-Cimerman N (2007). Phylogeny and ecology of the ubiquitous saprobe Cladosporium sphaerospermum, with descriptions of seven new species from hypersaline environments. Stud Mycol 58, 157–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.de Garcia V, Zalar P, Brizzio S, Gunde-Cimerman N, and van Broock M (2012). Cryptococcus species (Tremellales) from glacial biomes in the southern (Patagonia) and northern (Svalbard) hemispheres. FEMS Microbiol Ecol 82, 523–539. [DOI] [PubMed] [Google Scholar]
- 79.Fell JW, Statzell-Tallman A, Scorzetti G, and Gutierrez MH (2011). Five new species of yeasts from fresh water and marine habitats in the Florida Everglades. Antonie Van Leeuwenhoek 99, 533–549. [DOI] [PubMed] [Google Scholar]
- 80.Teixeira MM, Moreno LF, Stielow BJ, Muszewska A, Hainaut M, Gonzaga L, Abouelleil A, Patane JS, Priest M, Souza R, et al. (2017). Exploring the genomic diversity of black yeasts and relatives (Chaetothyriales, Ascomycota). Stud Mycol 86, 1–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Goncalves VN, Vitoreli GA, de Menezes GCA, Mendes CRB, Secchi ER, Rosa CA, and Rosa LH (2017). Taxonomy, phylogeny and ecology of cultivable fungi present in seawater gradients across the Northern Antarctica Peninsula. Extremophiles 21, 1005–1015. [DOI] [PubMed] [Google Scholar]
- 82.Vaz ABM, Rosa LH, Vieira MLA, de Garcia V, Brandão LR, Teixeira LCRS, Moliné M, Libkind D, van Broock M, and Rosa CA (2011). The diversity, extracellular enzymatic activities and photoprotective compounds of yeasts isolated in Antarctica. Braz J Microbiol 42, 937–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.De Hoog GS, Zeng JS, Harrak MJ, and Sutton DA (2006). Exophiala xenobiotica sp. nov., an opportunistic black yeast inhabiting environments rich in hydrocarbons. Antonie Van Leeuwenhoek 90, 257–268. [DOI] [PubMed] [Google Scholar]
- 84.Volz PA, Jerger DE, Wurzburger AJ, and Hiser JL (1974). A preliminary survey of yeasts isolated from marine habitats at Abaco Island, The Bahamas. Mycopathologia et mycologia applicata 54, 313–316. [DOI] [PubMed] [Google Scholar]
- 85.Sabate J, Cano J, Esteve-Zarzoso B, and Guillamon JM (2002). Isolation and identification of yeasts associated with vineyard and winery by RFLP analysis of ribosomal genes and mitochondrial DNA. Microbiol Res 157, 267–274. [DOI] [PubMed] [Google Scholar]
- 86.Butinar L, Spencer-Martins I, and Gunde-Cimerman N (2007). Yeasts in high Arctic glaciers: the discovery of a new habitat for eukaryotic microorganisms. Antonie Van Leeuwenhoek 91, 277–289. [DOI] [PubMed] [Google Scholar]
- 87.Tesei D, Tafer H, Poyntner C, Piñar G, Lopandic K, and Sterflinger K (2017). Draft Genome Sequences of the Black Rock Fungus Knufia petricola and Its Spontaneous Nonmelanized Mutant. Genome Announc 5, e01242–01217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gnavi G, Garzoli L, Poli A, Prigione V, Burgaud G, and Varese GC (2017). The culturable mycobiota of Flabellia petiolata: First survey of marine fungi associated to a Mediterranean green alga. PLoS One 12, e0175941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Janso JE, Bernan VS, Greenstein M, Bugni TS, and Ireland CM (2005). Penicillium dravuni, a new marine-derived species from an alga in Fiji. Mycologia 97, 444–453. [DOI] [PubMed] [Google Scholar]
- 90.Coutinho ML, Miller AZ, and Macedo MF (2015). Biological colonization and biodeterioration of architectural ceramic materials: An overview. Journal of Cultural Heritage 16, 759–777. [Google Scholar]
- 91.Crous PW, Schoch CL, Hyde KD, Wood AR, Gueidan C, de Hoog GS, and Groenewald JZ (2009). Phylogenetic lineages in the Capnodiales. Stud Mycol 64, 17–47S17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lenassi M, Gostincar C, Jackman S, Turk M, Sadowski I, Nislow C, Jones S, Birol I, Cimerman NG, and Plemenitas A (2013). Whole genome duplication and enrichment of metal cation transporters revealed by de novo genome sequencing of extremely halotolerant black yeast Hortaea werneckii. PLoS One 8, e71328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zalar P, de Hoog GS, and Gunde-Cimerman N (1999). Ecology of halotolerant dothideaceous black yeasts. Studies in Mycology 0, 38–48. [Google Scholar]
- 94.Marchetta A, Gerrits van den Ende B, Al-Hatmi AMS, Hagen F, Zalar P, Sudhadham M, Gunde-Cimerman N, Urzi C, de Hoog S, and De Leo F (2018). Global Molecular Diversity of the Halotolerant Fungus Hortaea werneckii. Life (Basel) 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sterflinger K (1998). Temperature and NaCl- tolerance of rock-inhabiting meristematic fungi. Antonie van Leeuwenhoek 74, 271–281. [DOI] [PubMed] [Google Scholar]
- 96.Crous PW, Wingfield MJ, Richardson DM, Leroux JJ, Strasberg D, Edwards J, Roets F, Hubka V, Taylor PWJ, Heykoop M, et al. (2016). Fungal Planet description sheets: 400–468. Persoonia - Molecular Phylogeny and Evolution of Fungi 36, 316–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gan HM, Thomas BN, Cavanaugh NT, Morales GH, Mayers AN, Savka MA, and Hudson AO (2017). Whole genome sequencing of Rhodotorula mucilaginosa isolated from the chewing stick (Distemonanthus benthamianus): insights into Rhodotorula phylogeny, mitogenome dynamics and carotenoid biosynthesis. PeerJ 5, e4030–e4030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.de Almeida JM (2005). Yeast community survey in the Tagus estuary. FEMS Microbiol Ecol 53, 295–303. [DOI] [PubMed] [Google Scholar]
- 99.Wirth F, and Goldani LZ (2012). Epidemiology of Rhodotorula: an emerging pathogen. Interdiscip Perspect Infect Dis 2012, 465717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Libkind D, Brizzio S, Ruffini A, Gadanho M, van Broock M, and Paulo Sampaio J (2003). Molecular characterization of carotenogenic yeasts from aquatic environments in Patagonia, Argentina. Antonie van Leeuwenhoek 84, 313–322. [DOI] [PubMed] [Google Scholar]
- 101.Connell L, Redman R, Craig S, Scorzetti G, Iszard M, and Rodriguez R (2008). Diversity of soil yeasts isolated from South Victoria Land, Antarctica. Microb Ecol 56, 448–459. [DOI] [PubMed] [Google Scholar]
- 102.Buzzini P, Branda E, Goretti M, and Turchetti B (2012). Psychrophilic yeasts from worldwide glacial habitats: diversity, adaptation strategies and biotechnological potential. FEMS Microbiol Ecol 82, 217–241. [DOI] [PubMed] [Google Scholar]
- 103.Turchetti B, Buzzini P, Goretti M, Branda E, Diolaiuti G, D’Agata C, Smiraglia C, and Vaughan-Martini A (2008). Psychrophilic yeasts in glacial environments of Alpine glaciers. FEMS Microbiol Ecol 63, 73–83. [DOI] [PubMed] [Google Scholar]
- 104.Yanwisetpakdee B, Lotrakul P, Prasongsuk S, Seelanan T, White J, Eveleigh D, Wook Kim S, and Punnapayak H (2016). Associations among halotolerance, osmotolerance and exopolysaccharide production of Aureobasidium melanogenum strains from habitats under salt stress. Pakistan Journal of Botany 48, 1229–1239. [Google Scholar]
- 105.Jones T, Federspiel NA, Chibana H, Dungan J, Kalman S, Magee BB, Newport G, Thorstenson YR, Agabian N, Magee PT, et al. (2004). The diploid genome sequence of Candida albicans. Proc Natl Acad Sci 101, 7329–7334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Buck JD, Bubucis PM, and Combs TJ (1977). Occurrence of human-associated yeasts in bivalve shellfish from Long Island Sound. Applied and Environmental Microbiology 33, 370–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Cook WL, and Schlitzer RL (1981). Isolation of Candida albicans from freshwater and sewage. Applied and environmental microbiology 41, 840–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gow NA, van de Veerdonk FL, Brown AJ, and Netea MG (2011). Candida albicans morphogenesis and host defence: discriminating invasion from colonization. Nat Rev Microbiol 10, 112–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Moreno AD, Tellgren-Roth C, Soler L, Dainat J, Olsson L, and Geijer C (2017). Complete Genome Sequences of the Xylose-Fermenting Candida intermedia Strains CBS 141442 and PYCC 4715. Genome Announc 5, e00138–00117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Logue ME, Wong S, Wolfe KH, and Butler G (2005). A Genome Sequence Survey Shows that the Pathogenic Yeast Candida parapsilosis Has a Defective MTLa1 Allele at Its Mating Type Locus Eukaryot Cell 4, 1009–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Butinar L, Santos S, Spencer-Martins I, Oren A, and Gunde-Cimerman N (2005). Yeast diversity in hypersaline habitats. FEMS Microbiol Lett 244, 229–234. [DOI] [PubMed] [Google Scholar]
- 112.Butler G, Rasmussen MD, Lin MF, Santos MAS, Sakthikumar S, Munro CA, Rheinbay E, Grabherr M, Forche A, Reedy JL, et al. (2009). Evolution of pathogenicity and sexual reproduction in eight Candida genomes. Nature 459, 657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Medeiros AO, Missagia BS, Brandão LR, Callisto M, Barbosa FAR, and Rosa CA (2012). Water quality and diversity of yeasts from tropical lakes and rivers from the Rio Doce basin in Southeastern Brazil. Brazilian Journal of Microbiology 43, 1582–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Burgaud G, Arzur D, Durand L, Cambon-Bonavita MA, and Barbier G (2010). Marine culturable yeasts in deep-sea hydrothermal vents: species richness and association with fauna. FEMS Microbiol Ecol 73, 121–133. [DOI] [PubMed] [Google Scholar]
- 115.Ng KP, Yew SM, Chan CL, Soo-Hoo TS, Na SL, Hassan H, Ngeow YF, Hoh C-C, Lee K-W, and Yee W-Y (2012). Sequencing of Cladosporium sphaerospermum, a Dematiaceous Fungus Isolated from Blood Culture. Eukaryot Cell 11, 705–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Blaylock RB, Overstreet RM, and Klich MA (2001). Mycoses in red snapper (Lutjanus campechanus) caused by two deuteromycete fungi (Penicillium corylophilum and Cladosporium sphaerospermum) In The Ecology and Etiology of Newly Emerging Marine Diseases, Porter JW, ed. (Dordrecht: Springer Netherlands; ), pp. 221–228. [Google Scholar]
- 117.Riley R, Haridas S, Wolfe KH, Lopes MR, Hittinger CT, Göker M, Salamov AA, Wisecaver JH, Long TM, Calvey CH, et al. (2016). Comparative genomics of biotechnologically important yeasts. Proceedings of the National Academy of Sciences 113, 9882–9887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lachance M-A, Miranda M, Miller MW, and Phaff HJ (1976). Dehiscence and active spore release in pathogenic strains of the yeast Metschnikowia bicuspidata var. australis: possible predatory implication. Canadian Journal of Microbiology 22, 1756–1761. [DOI] [PubMed] [Google Scholar]
- 119.Shih-Chu C, Tung-Hung C, Pei-Chi Wang Y-CC, Ju-Ping H, Yu-De L, Hso-Chi C, and Li-Ling L (2003). Metschnikowia bicuspidata and Enterococcus faecium co-infection in the giant freshwater prawn Macrobrachium rosenbergii. Diseases of Aquatic Organisms 55, 161–167. [DOI] [PubMed] [Google Scholar]
- 120.Baroncelli R, Piaggeschi G, Fiorini L, Bertolini E, Zapparata A, Pè ME, Sarrocco S, and Vannacci G (2015). Draft Whole-Genome Sequence of the Biocontrol Agent Trichoderma harzianum T6776. Genome Announc 3, e00647–00615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kobayashi M, Uehara H, Matsunami K, Aoki S, and Kitagawa I (1993). Trichoharzin, a new polyketide produced by the imperfect fungus Trichoderma harzianum separated from the marine sponge Micale cecilia. Tetrahedron Letters 34, 7925–7928. [Google Scholar]
- 122.Khallil A-RM, Bagy MMK, and El-Shimy NA (1991). Mycoflora associated with five species of freshwater leeches. Journal of Basic Microbiology 31, 437–446. [DOI] [PubMed] [Google Scholar]
- 123.Martínez-Medina A, Pascual JA, Lloret E, and Roldán A (2009). Interactions between arbuscular mycorrhizal fungi and Trichoderma harzianum and their effects on Fusarium wilt in melon plants grown in seedling nurseries. Journal of the Science of Food and Agriculture 89, 1843–1850. [Google Scholar]
- 124.Fokin M, Fleetwood D, Weir BS, and Villas-Boas S (2017). Genome Sequence of the Saprophytic Ascomycete Epicoccum nigrum Strain ICMP 19927, Isolated from New Zealand. Genome Announc 5, e00557–00517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Alwakeel SS (2018). Molecular identification of fungi isolated from coastal regions of Red Sea, Jeddah, Saudi Arabia. Journal of the Association of Arab Universities for Basic and Applied Sciences 24, 115–119. [Google Scholar]
- 126.Pel HJ, de Winde JH, Archer DB, Dyer PS, Hofmann G, Schaap PJ, Turner G, de Vries RP, Albang R, Albermann K, et al. (2007). Genome sequencing and analysis of the versatile cell factory Aspergillus niger CBS 513.88. Nature Biotechnol 25, 221–231. [DOI] [PubMed] [Google Scholar]
- 127.Raghukumar C, and Raghukumar S (1998). Barotolerance of fungi isolated from deep-sea sediments of the Indian Ocean. Aquat Microb Ecol 15, 153–163. [Google Scholar]
- 128.Zhang Y, Li XM, Wang CY, and Wang BG (2007). A new naphthoquinoneimine derivative from the marine algal-derived endophytic fungus Aspergillus niger EN-13. Chinese Chemical Letters 18, 951–953. [Google Scholar]
- 129.Wallace GB (1932). Preliminary List of Fungi or Diseases of Economic Plants in Tanganyika Territory. Bulletin of Miscellaneous Information (Royal Botanic Gardens, Kew) 1932, 28–40. [Google Scholar]
- 130.Bleve G, Grieco F, Cozzi G, Logrieco A, and Visconti A (2006). Isolation of epiphytic yeasts with potential for biocontrol of Aspergillus carbonarius and A. niger on grape. Int J Food Microbiol 108, 204–209. [DOI] [PubMed] [Google Scholar]
- 131.de Vries RP, Riley R, Wiebenga A, Aguilar-Osorio G, Amillis S, Uchima CA, Anderluh G, Asadollahi M, Askin M, Barry K, et al. (2017). Comparative genomics reveals high biological diversity and specific adaptations in the industrially and medically important fungal genus Aspergillus. Genome Biol 18, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Khusnullina AI, Bilanenko EN, and Kurakov AV (2018). Microscopic Fungi of White Sea Sediments. Contemporary Problems of Ecology 11, 503–513. [Google Scholar]
- 133.Bringmann G, Lang G, Gulder TAM, Tsuruta H, Mühlbacher J, Maksimenka K, Steffens S, Schaumann K, Stöhr R, Wiese J, et al. (2005). The first sorbicillinoid alkaloids, the antileukemic sorbicillactones A and B, from a sponge-derived Penicillium chrysogenum strain. Tetrahedron 61, 7252–7265. [Google Scholar]
- 134.McRae CF, Hocking AD, and Seppelt RD (1999). Penicillium species from terrestrial habitats in the Windmill Islands, East Antarctica, including a new species, Penicillium antarcticum. Polar Biology 21, 97–111. [Google Scholar]
- 135.Vera J, Gutierrez MH, Palfner G, and Pantoja S (2017). Diversity of culturable filamentous Ascomycetes in the eastern South Pacific Ocean off Chile. World J Microbiol Biotechnol 33, 157. [DOI] [PubMed] [Google Scholar]
- 136.Gonçalves VN, Carvalho CR, Johann S, Mendes G, Alves TMA, Zani CL, Junior PAS, Murta SMF, Romanha AJ, Cantrell CL, et al. (2015). Antibacterial, antifungal and antiprotozoal activities of fungal communities present in different substrates from Antarctica. Polar Biology 38, 1143–1152. [Google Scholar]
- 137.Sharp KH, Pratte ZA, Kerwin AH, Rotjan RD, and Stewart FJ (2017). Season, but not symbiont state, drives microbiome structure in the temperate coral Astrangia poculata. Microbiome 5, 120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Lee SB, Milgroom MG, and Taylor JW (1988). A rapid, high yield mini-prep method for isolation of total genomic DNA from fungi. Fungal Genet Rep 35. [Google Scholar]
- 139.White T, Bruns T, Lee S, and Taylor J (1990). Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols: a guide to methods and applications; PCR protocols: a guide to methods and applications, 315–322. [Google Scholar]
- 140.Kurtzman CP, and Robnett CJ (1998). Identification and phylogeny of ascomycetous yeasts from analysis of nuclear large subunit (26S) ribosomal DNA partial sequences. Antonie van Leeuwenhoek 73, 331–371. [DOI] [PubMed] [Google Scholar]
- 141.Amaral-Zettler LA, McCliment EA, Ducklow HW, and Huse SM (2009). A Method for Studying Protistan Diversity Using Massively Parallel Sequencing of V9 Hypervariable Regions of Small-Subunit Ribosomal RNA Genes. PLOS ONE 4, e6372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Spatafora JW, Sung G-H, Johnson D, Hesse C, O’Rourke B, Serdani M, Spotts R, Lutzoni F, Hofstetter V, Miadlikowska J, et al. (2006). A five-gene phylogeny of Pezizomycotina. Mycologia 98, 1018–1028. [DOI] [PubMed] [Google Scholar]
- 143.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Team RC (2018). R: A Language and Environment for Statistical Computing. (R Foundation for Statistical Computing). [Google Scholar]
- 145.MathWorks (2012b). MATLAB and Statistics Toolbox Release (Natick, Massachusetts, United States: ). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Video S1. DIC time lapse of H. werneckii (40x). Scale bar, 10 μm. Time, Hours:Minutes. Related to Figure 3.
Video S2. DIC time lapse of K. petricola (40x). Two examples of colony growth. Scale bar, 10 μm. Time, Hours:Minutes. Related to Figure 4.
Video S3. DIC time lapse of A. pullulans (40x). This mother cell repeatedly produces multiple simultaneous buds from the same site. Scale bar, 10 μm. Time, Hours:Minutes. Related to Figure 5.
Video S4. DIC time lapse of A. pullulans (40x). Three more examples of budding mother cells demonstrating the variability of cell morphology. Many mother cells from Day 3 of filming lyse after producing daughter cells. Scale bar, 10 μm. Time, Hours:Minutes. Related to Figure 5.
Video S5. DIC time lapse of P. salicorniae (40x). Growth of initial yeast cells and sprouting of hyphae which then develop septa. Scale bar, 10 μm. Time, Hours:Minutes. Related to Figure 6.
Video S6. DIC time lapse of P. salicorniae (20x). At left, overview of later stages of colony growth. At right, close-ups of hyphae forming septa and reverting back to yeast cells. Scale bars, 10 μm. Time, Hours:Minutes. Related to Figure 6.


