Abstract
Background
Resident soil microbiota play key roles in sustaining the core ecosystem processes of terrestrial Antarctica, often involving unique taxa with novel functional traits. However, the full scope of biodiversity and the niche-neutral processes underlying these communities remain unclear. In this study, we combine multivariate analyses, co-occurrence networks and fitted species abundance distributions on an extensive set of bacterial, micro-eukaryote and archaeal amplicon sequencing data to unravel soil microbiome patterns of nine sites across two east Antarctic regions, the Vestfold Hills and Windmill Islands. To our knowledge, this is the first microbial biodiversity report on the hyperarid Vestfold Hills soil environment.
Results
Our findings reveal distinct regional differences in phylogenetic composition, abundance and richness amongst microbial taxa. Actinobacteria dominated soils in both regions, yet Bacteroidetes were more abundant in the Vestfold Hills compared to the Windmill Islands, which contained a high abundance of novel phyla. However, intra-region comparisons demonstrate greater homogeneity of soil microbial communities and measured environmental parameters between sites at the Vestfold Hills. Community richness is largely driven by a variable suite of parameters but robust associations between co-existing members highlight potential interactions and sharing of niche space by diverse taxa from all three microbial domains of life examined. Overall, non-neutral processes appear to structure the polar soil microbiomes studied here, with niche partitioning being particularly strong for bacterial communities at the Windmill Islands. Eukaryotic and archaeal communities reveal weaker niche-driven signatures accompanied by multimodality, suggesting the emergence of neutrality.
Conclusion
We provide new information on assemblage patterns, environmental drivers and non-random occurrences for Antarctic soil microbiomes, particularly the Vestfold Hills, where basic diversity, ecology and life history strategies of resident microbiota are largely unknown. Greater understanding of these basic ecological concepts is a pivotal step towards effective conservation management.
Keywords: Antarctica, Soil Microbiome, Bacteria, Eukarya, Archaea, Conservation Ecology
Background
East Antarctica constitutes up to two-thirds of the continent and is home to some of the oldest, coldest and most oligotrophic soils on Earth [1]. Apart from ice-free patches along the coast, most of the sector is covered by a thick layer of permafrost [2]. The Windmill Islands, an ice-free region situated near the Australian Casey research station, consists of five major peninsulas and a number of rock-strewn islands. Approximately 100 km to the north lie the Vestfold Hills, a large expanse of low-lying hilly country deeply indented by sea-inlets and lakes [3, 4]. These diverse edaphic habitats are a legacy of age-involving varied geological and glaciological processes [5]. Soil microbial diversity and functional ecology of the hyperarid Vestfold Hills is virtually unexplored, whilst previous studies at the Windmill Islands have disclosed a relatively high proportion of novel bacterial phyla [6]. However, knowledge on archaea and micro-eukaryotes is still lacking [7]. This is in stark contrast to other regions such as the McMurdo Dry Valleys and Antarctic Peninsula [6, 8, 9]. In our understanding of soil microbiota across the different bioregions of terrestrial Antarctica, addressing these deficiencies will not only improve our understanding of Antarctic microbial biogeography but also guide future conservation planning strategies [10].
Climate and soil age abiotic factors such as pH, moisture and nutrient content exert a strong influence on Antarctic species distribution and life histories [1, 7, 11–13]. These properties may co-vary with local lithology, pedology and aspect—leading to a myriad of edaphic niches [14]. In turn, their microbial occupants are key to establishing and maintaining core ecosystem processes, occasionally involving unique taxa with novel functional traits such as unique biosurfactants and trace gas assimilation as a novel mode of primary production [5, 15]. It is thereby a widely accepted concept that the capacity of microbes to access and utilise resources, as well as tolerate different levels of stress, contributes significantly to the structuring of microbiota dwelling within these oligotrophic soils.
However, our ability to unravel these basic ecological concepts in cold regions has been limited by the small number and depth of studies available [16]. Moreover, the majority of relevant studies have largely been focused on bacteria only. Few micro-eukaryotic and archaeal-specific surveys have been reported on terrestrial Antarctic environments and so their ecological roles remain elusive [17–19]. All three microbial domains are likely to be responsible for the sustainability and evolution of the polar soil microbiome but contemporary dynamics will inevitably change due to the climate-driven emergence of new ice-free areas [2, 20–22]. As consequence, further clarification on their underlying drivers will establish a baseline from which to gauge ecological shifts, which is an important step towards effectively managing microbial biodiversity loss and conserving the key ecosystem functions offered by these assemblages [23–28, 29].
Projected twenty-first century expansion of ice-free habitats across eastern Antarctica means that tools for rapidly assessing soil ecosystem health, such as species abundance distributions (SADs), are gradually becoming more important in managing microbial biodiversity loss, especially in regions where survey data is scarce [2, 30]. In this study, we compiled bacterial 16S (n = 837), eukaryotic 18S (n = 162) and archaeal 16S (n = 144) rRNA gene amplicon sequencing data from soil samples spanning nine east Antarctic sites between the Vestfold Hills (n = 5) and Windmill Islands (n = 4). By taking a multivariate, exploratory network and modelling approach using fitted SADs we aim to (1) elucidate the previously unknown soil microbial biodiversity of the Vestfold Hills, (2) determine the driving processes (i.e. niche or neutral) underlying the microbial communities of east Antarctica and (3) clarify whether they differ between the Vestfold Hills and Windmill Islands.
Results
Amplicon sequencing yield and coverage
We recovered a total of 60,495,244 high-quality bacterial 16S rRNA gene sequences, which clustered into 36,251 operational taxonomic units (OTUs) at 97% identity cut-off. Our micro-eukaryotic and archaeal runs yielded a total of 1,299,519 18S rRNA and 13,373,072 16S rRNA gene sequences after read-quality filtering, which respectively clustered at 97% into 1511 and 589 OTUs (Table S1). Subsampled rarefaction curves of the pooled data revealed that bacterial, micro-eukaryotic and archaeal richness generally approached an asymptote at each site (Fig. S1).
Comparative biodiversity of the east Antarctic polar soil microbiome
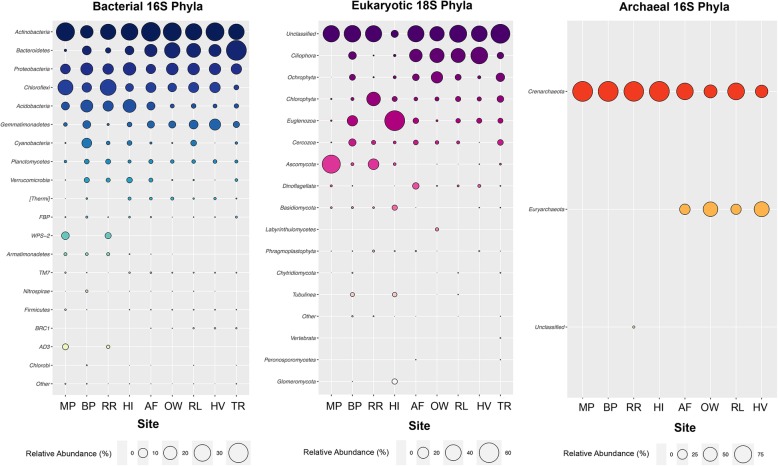
At 97% identity, OTUs were classified into 63 bacterial, 27 micro-eukaryotic and three archaeal phyla. Distributions of phylum abundances for all three domains were uneven, as the majority of sites were dominated by a handful of taxa (Fig. 1). Soil bacterial communities predominantly consisted of the metabolically diverse Actinobacteria (30.5%) and Proteobacteria (14.6%). Bacteroidetes (24.9%) and Gemmatimonadetes (8.0%) were more prevalent at the Vestfold Hills, whereas Chloroflexi (17.8%) and Acidobacteria (13.6%) were present in greater relative abundances throughout the Windmill Islands. With the exception of Rookery Lake (RL = 4.2%), Browning Peninsula (BP = 10.9%) and Herring Island (HI = 3.1 %), Cyanobacteria abundance was relatively low across all sites. At Mitchell Peninsula (MP) and Robinson Ridge (RR), rare candidate phyla namely Candidatus Eremiobacteraeota (WPS-2) and Candidatus Dormibacteraeota (AD3) were present in higher relative abundances (> 4.6%) compared to the other sites. At lower taxonomic levels, bacterial sequences classified into 169 classes, with members largely belonging to Flavobacteria (10.9%) and Actinobacteria (9.0%), followed by similar proportions (6.0%) of Thermoleophilia, Chloracidobacteria, Gamma-proteobacteria and Alpha-proteobacteria (Fig. S2).
Fig. 1.
Bubble plots of relative abundance (%) per site of phyla-level composition of OTUs (97% cut-off), based on bacterial 16S (mean = 490 bp), eukaryotic 18S (mean = 125 bp) and archaeal 16S (mean = 470 bp) SSU rRNA sequences representing > 0.001% of all normalised OTUs sorted by decreasing relative abundance. Greatest phylogenetic diversity is exhibited by bacteria followed by eukarya then archaea. Across all three domains, distribution of phyla abundances is generally uneven as a handful of taxa tend to dominate but strong compositional differences are apparent between the Windmill Islands and Vestfold Hills regions
For micro-eukaryotes, 18S rRNA gene sequences fell into six supergroups consisting of unclassified (46.9%), Chromalveolata (Ciliophora and Dinoflagellata = 20.6%), Archaeplastida (Ochrophyta, Chlorophyta and Phragmoplastophyta = 17.8%), Excavata (Euglenozoa = 5.4%), Opisthokonta (Ascomycota, Basidiomycota, Labyrinthulomycetes, Chytridiomycota, Vertebrata, Peronosporomycetes and Glomermomycota = 4.6%) and Amoebozoa (Cercozoa and Tublinea = 4.4%) (Fig. 1). Unclassified micro-eukarya remained dominant across all taxonomic levels, with moderately higher relative abundance observed throughout the Vestfold Hills (61.3%), compared with the Windmill Island sites (38.8%), particularly at The Ridge (TR). Fungal diversity contributed to a relatively small proportion (10.5%) of the total relative abundance for eukaryotic soil communities, except at MP and RR.
Archaeal diversity was predominantly distributed within the Crenarchaeota phylum (84.5%), whilst members of Euryarchaeota (15.0%) were mainly exclusive to the Vestfold Hills (Fig. 1). In addition, an unusually high proportion (2.3%) of unclassified archaea was observed at RR. At lower taxonomic levels, archaeal sequences belonged to six main families; Nitrososphaeraceae (84.5%) and Halobacteriaceae (15.0%), followed by unclassified, SAGMA-X, Cenarchaeaceae and TMEG families, collectively accounting for 0.01% of total relative archaeal abundance.
Domain-level biotic interactions
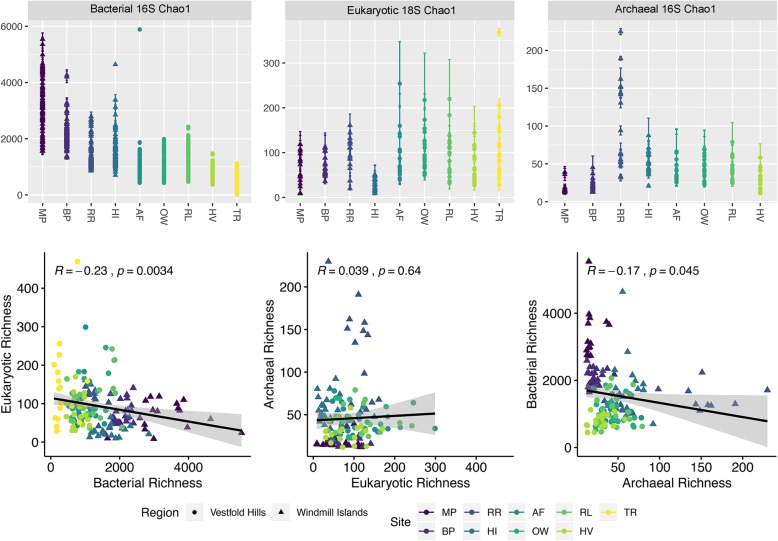
Non-metric multidimensional scaling (NMDS) ordination of microbial OTU communities and corresponding environmental metadata revealed that soils were conserved within sites and broadly by geographic region (Fig. S3). Apart from TR, sites at the Vestfold Hills were more homogenous in terms of community composition and measured soil parameters. Bacterial communities exhibited the greatest overall species richness based on Chao1 estimates (Fig. 2), particularly at the Windmill Islands (mean = 2270.1). In contrast, greater eukaryotic richness was observed throughout the Vestfold Hills (mean = 132.3). Archaeal communities exhibited the lowest overall species richness (mean = 50.9), with RR being an exception (mean = 106.4). Pearson’s correlations between domain-level pooled Chao1 richness estimates revealed weak but significant (α = 0.05) negative relationships of bacterial communities against both micro-eukaryotic (R = − 0.23, P = 0.0034) and archaeal (R = − 0.17, P = 0.045) communities. However, no significant correlation was found between micro-eukaryotic and archaeal richness (R = 0.039, P = 0.64).
Fig. 2.
Chao1 richness estimates and correlations between our soil bacterial, eukaryotic and archaeal communities coloured by site. Polar soil bacterial communities demonstrated highest overall species richness estimates, particularly throughout the Windmill Islands region. Significant (P < 0.05) negative correlations were detected between estimated bacterial species richness against the other two microbial domains
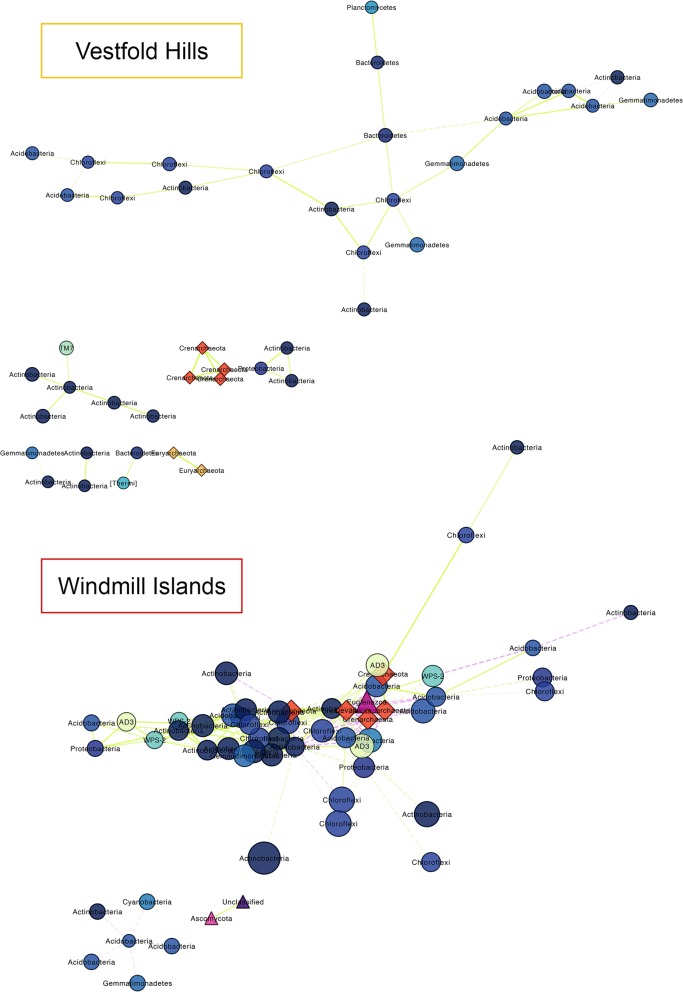
Domain-level networks displaying the co-occurrence of OTUs provided new insights on the potential sharing of niche spaces or interactions between co-existing taxa, many of which are understudied (Fig. 3). The resulting network for the Vestfold Hills consisted of 43 nodes (clustering coefficient = 0.2) and 44 edges (average no. of neighbours = 2.0, characteristic path length = 3.2) across eight connected components with a network diameter of seven edges (Table S2). Whereas, the resulting Windmill Islands network consisted of 58 nodes (clustering coefficient = 0.4) and 201 edges (average no. of neighbours = 6.9, characteristic path length = 2.4) across three connected components with a network diameter of six edges (Table S2).
Fig. 3.
Domain-level OTU co-occurrence network of significant (P < 0.001) and strongly correlated (MIC > 0.8) OTU pairs between the Windmill Islands and Vestfold Hills. Nodes (circles = bacteria, triangles = eukarya, diamonds = archaea) and edges represent individual OTUs and their correlations respectively. Node size is proportional to their degree of connectivity and edge colour is based on linearity (green/solid = positive, purple/dashed = negative). Our soil microbial networks are comprised of moderately connected OTUs, more so at the Windmill Islands, structured amongst multiple components and forming a clustered topology. All three domains of life are present within the Windmill Islands network, most notably Crenarchaeota being strongly embedded and Actinobacteria forming the microbial backbone within these desert soils. In contrast, eukarya are absent from the Vestfold Hills network, suggesting possible competition
Notable associations within the Vestfold Hills network included positive associations between Saccharibacteria (TM7), a parasitic bacterium, and Actinobacteria. Also noted was the lack of co-occurrent micro-eukaryotic species. Crenarchaeota were more strongly embedded within the Windmill Islands network suggesting different life histories or niche preferences between the two regions. Similarly, rare candidate bacterial phyla Candidatus Eremiobacteraeota (WPS-2) and Candidatus Dormibacteraeota (AD3) only formed strong visible associations in this region. The astounding taxonomic diversity of Actinobacteria (Fig. 1 and S2) was reflected in their ability to occupy multiple niches and form the majority of connections to co-existing species, essentially moulding the microbial backbone of these Antarctic desert soils. Overall, microorganisms present within the soil microbial networks tended to co-occur more than expected by chance (P < 0.001).
Correlations between estimated richness and selected environmental predictor variables
Generalised additive models (GAMs) were fitted to test the ability of a range of soil parameters to explain the variation in Chao1 richness of bacteria, eukarya and archaea. A stepwise model selection process (based on the lowest AIC) was used to identify the ‘best model’ and thereby identify the key environmental drivers. These models explained a moderate percentage of variation (45.0–66.8%) in richness for all three microbial communities at the regional scale (Table 1 and Fig. S4, S5,S6). For bacteria, there was a positive relationship between Chao1 richness and copper (Cu), aluminium (Al, Al2O3) and gravel content (Fig. S4). Micro-eukaryote richness exhibited negative relationships with dry matter fraction (DMF), soil pH, nitrite concentrations (NO3) and the amount of mud but displayed a positive relationship with total carbon content (TC) and conductivity (Fig. S5). Archaeal richness had positive relationships with conductivity and total nitrogen content (TN) but displayed a negative relationship with calcium (CECCa) (Fig. S6). Both bacteria and archaea showed a positive relationship with phosphorous (TP, P) and sodium (CECNa) but had a negative relationship with titanium dioxide (TiO2). Only micro-eukaryotes demonstrated a significant (P < 0.05) difference between the two regions.
Table 1.
Summary of best model selection after the removal of co-variates with region as a random effect
| Response | Bacterial Chao1 | Eukaryotic Chao1 | Archaeal Chao1 |
|---|---|---|---|
| Predictor 1 | Total phosphorous | Dry matter fraction | Conductivity |
| Predictor 2 | Phosphorous | Conductivity | Total nitrogen |
| Predictor 3 | Copper | pH | Phosphorous |
| Predictor 4 | Aluminium | Total Carbon | Calcium Cation |
| Predictor 5 | Sodium cation | NO2 | Sodium Cation |
| Predictor 6 | Gravel | Mud | Mud |
| Predictor 7 | TiO2 | Region | TiO2 |
| Predictor 8 | Al2O3 | ||
| Distribution | Negative binomial | Gaussian | Gaussian |
| R2 | 0.601 | 0.34 | 0.611 |
| Deviance Explained | 64.90% | 45% | 66.80% |
Niche or neutral?
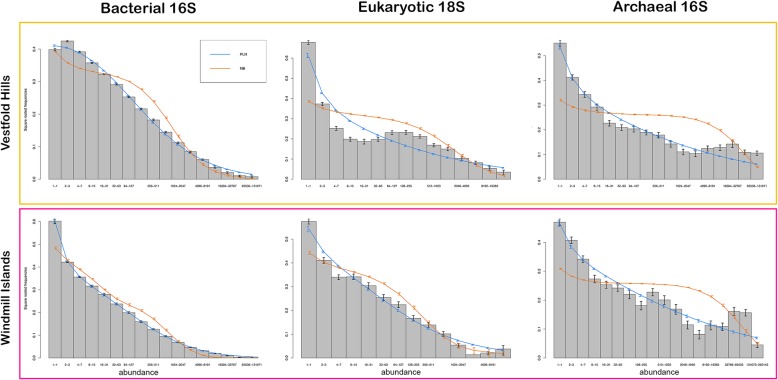
Overall, species abundances were better approximated by Poisson-lognormal (PLN) than negative binomial (NB) distributions (wPLN>wNB), likely attributable to these Antarctic communities being substantially more heterogenous than expected (Fig. 4, Table 2). As is the norm in ecological communities, all distributions were characterised by highly right-skewed patterns, emphasising the disparity between rare and common species. Bacterial communities lacked an internal mode and demonstrated a better PLN-fit (Table 2), particularly at the Windmill Islands (wPLN = 1.000, wNB = < 0.001). By contrast, eukaryotic and archaeal communities demonstrated multimodal distributions accompanied by relatively weaker PLN-fits, particularly for eukaryotic communities at the Vestfold Hills (wPLN = < 0.001, wNB = 1.000). These trends remained consistent at the local scale (Fig. S7, Table S4).
Fig. 4.
Fitted species abundance distribution (SAD) curves of polar soil microbial communities between the Vestfold Hills and Windmill Islands. The bars represent the mean proportion of species at each site in different octave classes of abundance. The blue and orange lines show the mean of fitted values from region-by-region fits of the Poisson-lognormal (PLN) and negative binomial (NB) distributions to the data, respectively. A PLN-fit best explains the overall structure of these communities, particularly for bacterial communities at the Windmill Islands. Eukaryotic and archaeal communities demonstrate slightly weaker PLN-fits and multimodal distributions across both regions, suggesting the emergence of neutrality
Table 2.
Akaike weights (AIC) calculated from regional-scale PLN- and NB-fitted SADs (where a weighted value closer to 1 indicates stronger evidence of one model over the other)
| Dataset | wPLN | wNB | |
|---|---|---|---|
| Bacteria | Windmill Islands | 1.000 | < 0.001 |
| Vestfold Hills | 1.000 | < 0.001 | |
| Eukarya | Windmill Islands | 0.917 | 0.083 |
| Vestfold Hills | < 0.001 | 1.00 | |
| Archaea | Windmill Islands | 1.000 | < 0.001 |
| Vestfold Hills | 0.960 | 0.0405 |
Discussion
Akin to other arid soil environments around the globe (Cowan et al. 2014), this extensive survey of the east Antarctic soil microbiome reveals that whilst bacterial diversity is rich, both micro-eukaryotic and archaeal phylogenies were comparatively low (Figs. 1 and 3). Overall, bacterial communities were dominated by the metabolically and physiologically diverse Actinobacteria phylum. Their ubiquity throughout terrestrial and aquatic ecosystems, including extreme environments like Antarctica, is a direct reflection of their genomic heterogeneity and broad functional capacities [6, 31]. However, regional disparity amongst taxa between the Vestfold Hills and Windmill Islands was observed. The Vestfold Hills, a region comprising of low-lying hilly country indented by lakes, contained a higher prevalence of bacterial members belonging to the Bacteroidetes phylum. This is likely due to its comparatively higher salinity levels than the Windmill Islands (Table S3), manifesting as visible salt crystal encrustations on the soil surface. In contrast, rare bacterial candidate phyla Eremiobacteraeota (WPS-2) and Dormibacteraeota (AD3), previously implicated in a novel mode of primary production using atmospheric energy sources [6], were notably lower in abundance at the Vestfold Hills. Possibly due to the relatively higher proportion of micro-eukaryotic taxa capable of photosynthesis in this region, namely phylum Ochrophyta and Ciliophora (Fig. 1 and S2). In contrast, archaeal communities, being mainly distributed within the Crenarchaeota or Thaumarchaeota phylum, were ubiquitous across both regions. Members in this phylum, more specifically Nitrososphaera, are known for their ammonia oxidising capabilities [32], thereby further highlighting the ecological importance of the vastly understudied polar soil archaea. Although we have only begun to shed light on the hidden complexities of the Antarctic soil microbiome, it is an important step towards achieving an integrated understanding of the basic ecological mechanisms governing these assemblages within such a severely limiting environment.
Strong niche partitioning appear to be driving the establishment and maintenance of contemporary microbiomes of the arid-to-hyperarid east Antarctic soils analysed here (Fig. 4; Table 2). This was particularly evident for bacterial communities at the Windmill Islands, where environmental gradients, such as soil pH and DMF, were generally more pronounced between sites (Fig. S3; Table S3). Whereas, soil parameters between sites at the Vestfold Hills were more similar to one another, with the exception of The Ridge (TR) (Fig. S3). These regional differences are also reflected in their phylogenetic composition, abundance and richness of microbial taxa (Figs. 1 and 3). Reduced niche overlap likely promotes greater biodiversity and long-term species co-existence through the efficient exploitation of resources under the adverse conditions [26, 33]. This may attribute to our observations of high bacterial diversity whilst both eukaryotic and archaeal diversities were relatively low (Figs. 1 and 3). Communities also demonstrated mixed responses to soil environmental predictors such as fertility and grain-related factors as well as metal oxide concentrations, which is likely a reflection of varied life history strategies (Table 1). Most notably, regional effects were only significant in explaining variation in richness for micro-eukarya, suggesting that other influences such as dispersal limitation may come into play for micro-eukaryotic communities between the Vestfold Hills and Windmill Islands [34, 35, 36].
In an era of progressively rapid natural and anthropogenic change, communities demonstrating strong niche-driven responses may have increased susceptibility to disturbance events such as large-scale colonisation, like those observed by Rhizocarpon lichens and invasive grass Poa annua across the Antarctic Peninsula (Chown et al. 2012 [2];; Supp and Ernest 2015). Inevitably, this will alter contemporary ecosystem dynamics and potentially result in the loss of novel polar taxa and associated traits due to the reduced functional insurance of strongly niche-shaped communities [20, 27, 37, 38]. For example, Candidatus Eremiobacteraeota (WPS-2) and Candidatus Dormibacteraeota (AD3) who are comprised of members genetically capable of atmospheric chemosynthesis, a novel process proposed to be contributing to primary production in these nutrient poor desert soils [6].
Neutral processes, however, play larger-than-expected roles within the eukaryotic and archaeal soil communities analysed, particularly throughout the Vestfold Hills (Fig. 4 and S5; Table 2 and S5). Weaker PLN-fits and apparent SAD multimodality suggest the emergence of neutrality for functionally similar groups [39–42] like Nitrososphaera (Fig. S2), a genus of chemotrophic ammonia oxidisers, likely involved in nitrogen cycling within these nutrient-limited Antarctic soils [43]. Interestingly, draft genomes of Thaumarchaeota recovered from Robinson Ridge (RR) soils reported the presence of ammonia monooxygenase [6], the first enzyme in the pathway for nitrification [44], further implicating the functional relevance of archaea in polar soils (Fig. 3). Moreover, members forming metabolic alliances with or competing against co-occurring bacterial taxa, such as Crenarchaeota at the Windmill Islands and micro-eukarya in general at the Vestfold Hills (Fig. 3), are likely critical to the formation of functional microbiomes within these harsh environments ([45]; Bahram et al. 2018 [20];). Unless competition is a major driving force within the relatively species poor eukaryotic and archaeal communities, their emerging neutral status may also promote greater resilience against perturbations due to their ephemeral natures, which is perhaps a cyclic response to seasonal resource availability, such as increased water and nutrient bioavailability during the austral summer [26, 46].
Although there is no current consensus on what drives SAD shape variation [30], a number of studies argue that multimodality occurs quite frequently in nature, and as such it is indeed a characteristic of ecological communities [39, 40, 42]. Emergent neutrality is one hypothesis put forth to explain multimodal SADs, where transient self-organised patterns of functionally similar species coexist within an ecological niche [41, 42]. Other studies claim that multimodality may arise from sampling artefacts [47]. We acknowledge that potential biases may be introduced through amplicon sequencing due to limitations in primer design but multimodality is rarely reported and its implications poorly understood, thus these findings warrant further consideration [39].
Conclusions
Information on biodiversity, assemblage patterns, environmental drivers and non-random co-occurrences are extremely valuable for Antarctic soil ecosystems, particularly the Vestfold Hills, where the basic diversity, ecology and life history strategies of resident microbiota is limited [48, 49]. These findings provide a new understanding of the basic ecological concepts underlying Antarctic species abundance and distribution. Regional disparities between soil communities at the Vestfold Hills and Windmill Islands further support the notion that microbial biogeography exists. Thus, stressing the importance of conserving these unique ecologies in the face of a warming Antarctica. Furthermore, spatial and temporal shifts in the community SAD patterns documented here can potentially be used to infer responses to environmental disturbance, before any local extinctions can occur at the micro-biodiversity scale.
Methods
Study area, soil sampling and physiochemical analysis
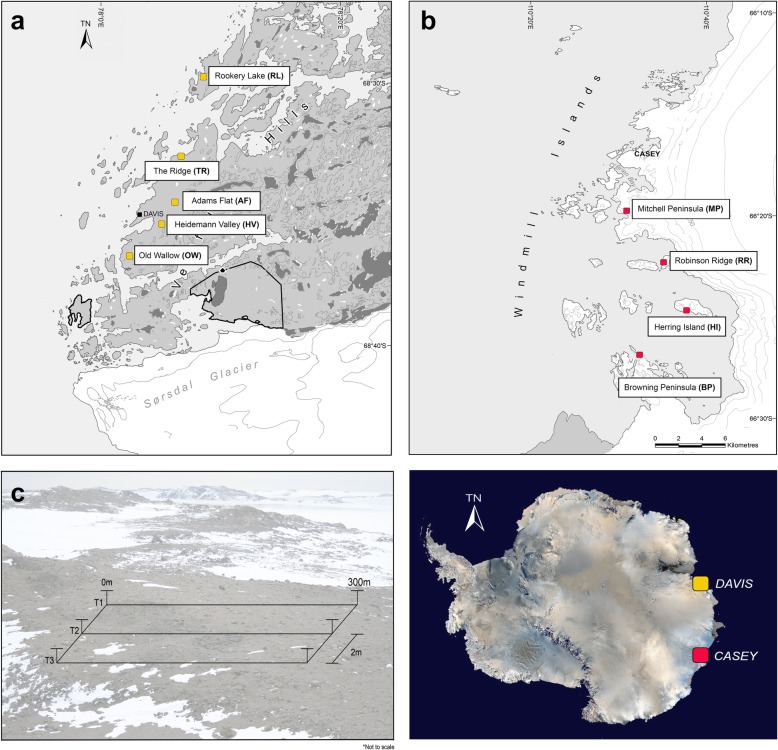
Sampling was performed by expeditioners via the Australian Antarctic Program (AAP) across nine polar desert sites spanning two ice-free regions (the Vestfold Hills and Windmill Islands). All nine sites were within the vicinity of Casey (66° 17′ S, 110° 45′ E) and Davis (68° 35′ S, 77° 58′ E) research stations in Eastern Antarctica (Fig. 4). Five sites were chosen from the Vestfold Hills: Adams Flat (AF: 68° 33′ S, 78° 1′ E); Old Wallow (OW: 68° 36′ S, 77° 58′ E); Rookery Lake (RL: 68° 36′ S, 77° 57′ E); Heidemann Valley (HV: 68° 35′ S, 78° 0′ E); and The Ridge (TR: 68° 54′ S, 78° 07′ E). Four sites were chosen from the Windmill Islands: Mitchell Peninsula (MP: 66° 31′ S, 110° 59′ E); Browning Peninsula (BP: 66° 27′ S, 110° 32′ E); Robinson Ridge (RR: 66° 22′ S, 110° 35′ E); and Herring Island (HI: 66° 24′ S, 110° 39′ E). At each site, soil samples (n = 93) from the top 10 cm were taken along three parallel transects following a geospatially explicit design [7]. All soils (n = 837) included in this study were previously submitted for extensive chemical and physical attributes (Table S2) [7, 50] (Fig. 5).
Fig. 5.
Map of the nine study areas across the a Vestfold Hills (AAD map catalogue No. 14, 499) and b Windmill Islands (No. 14, 179) region of Eastern Antarctica, showing approximate sampling locations and c geospatial transect design. At each site, soil samples (n = 93) were taken at the following distance points along each transect: 0, 0.1, 0.2, 0.5, 1, 2, 5, 10, 20, 50, 100, 100.1, 100.2, 100.5, 101, 102, 105, 110, 120, 150, 200, 200.1, 200.2, 200.5, 201, 202, 205, 210, 220, 250 and 300m. Where underlined distance points refer to a subsample (n = 18) submitted for amplicon sequencing of eukaryotic (18S rRNA) and archaeal (16S rRNA) soil communities
DNA extraction and Illumina amplicon sequencing
DNA was extracted in triplicate from soil samples using the FASTDNA™ SPIN Kit for Soil (MP Biomedicals, Santa Ana, CA, USA) and quantified using the Qubit™ 4 Fluorometer (ThermoFisher Scientific, NSW, Australia) as described in van Dorst et al. 2014. Diluted DNA (1:10 using nuclease-free water) was submitted to the Ramaciotti Centre for Genomics (UNSW, Sydney, Australia) for amplicon paired-end sequencing on the Illumina MiSeq platform (Illumina, California, USA) with negative and positive (mock) controls in accordance to protocols from Bioplatforms Australia (BPA) [50]. All 93 samples from each site were submitted to sequencing for bacteria (n = 837) targeting the 16s rRNA gene using the 27F/519R [51] primer set. As described in Siciliano et al. [7], a non-random subsample (n = 18) at each site was selected for the sequencing of micro-eukarya (n = 162) and archaea (n = 144) targeting the 18S and 16S rRNA genes using the 1391f/EukBr (Amaral-Zettler et al. 2009) and A2F/519R [52] primer sets, respectively. These include distance points at 0, 2, 100, 102, 200 and 202 m along each of the three parallel transects at each site.
Open OTU picking, assignment and classification
We followed the UPARSE OTU algorithm [53] endorsed by BPA through directly employing USEARCH 32-bit v10.0.240 [54] and VSEARCH 64-bit v2.8.0 [55]. Sequences were quality filtered, trimmed and clustered de novo to pick OTUs at 97% identity. Reads were then assigned to separate sample-by-OTU matrices for each amplicon (Table S1). OTUs were taxonomically classified against the SILVA v3.2.1 SSU rRNA database [56]. Where applicable, new OTU matrices were merged with existing ones using the QIIME 2 (https://qiime2.org) feature-table merge option. These were rarefied using the qiime feature-table rarefy function to generate random subsamples (bacterial 16S rRNA gene = 700k reads, micro-eukaryotic 18S rRNA gene = 23k reads, archaeal 16S rRNA gene = 850k reads).
Multivariate and statistical analyses in R
All multivariate and statistical analyses were carried out in the R environment (R Core Team 2018) using the subsampled datasets for bacteria, micro-eukarya and archaea. Subsampled rarefaction curves (q = 0) were generated using the iNEXT package. Non-metric multidimensional scaling (NMDS) ordinations (distance = ‘Euclidean’ for environmental data and distance = ‘Bray-Curtis’ for OTU abundance data) and Chao1 richness estimates were calculated in vegan v2.5-3 [57]. Unless specified otherwise, all plots were visualised using a combination of ggplot2 v3.1.0 [58] and ggpubr v0.2 [59].
Removal of environmental co-variates and model selection
To try and better understand the relationship between a range of environmental co-variates, we first constructed a Pearson correlation matrix to identify co-correlated variables (R > 0.6), and one of each correlated pair was removed from the list of potential predictors. Models were then fitted with all predictors (saturated models) using each of the Chao1 richness variables (bacteria, micro-eukarya and archaea) as the response variable. Starting with this saturated model, the best model (i.e. the most parsimonious—as indicated by the lowest AIC) was then identified using the stepAIC function in the MASS v7.3-51.4 package [60] in R. We also included region (i.e. Windmill Island or Vestfold Hills) as a random effect in the model selection process, to help in understanding the regional effects in explaining variation in richness. We fitted both generalised linear models (GLMs) and generalised additive models (GAMs) with smooth terms as either Gaussian or NB distributions. In addition to AIC, model diagnostic plots (to test normality and heteroscedasticity of variance) were used to help inform final model selection, especially with regard to the distribution used.
Domain-level co-occurrence OTU network from abundance data
OTUs representing > 0.001% of the total relative abundance of the bacterial, eukaryotic and archaeal communities within each region were combined for network analyses. Correlations between the relative abundance of each OTU pair across samples were calculated using the maximal information coefficient (MIC) in the MINE software package [61]. After correction for multiple testing, statistically significant (P < 0.001) co-occurrence relationships between pairs of OTUs were uploaded into the CYTOSCAPE software [62] to generate network diagrams, displaying only very strong associations (MIC > 0.8). Statistical inferences of network topology were calculated using the Network Analyser algorithm (treatment = ‘undirected’) in CYTOSCAPE (Table S2).
PLN- and NB-fitted species abundance distribution curves
As described in [63], PLN and NB models representing niche and neutral distributions, respectively, were fitted to our empirical data using maximum likelihood methods. All available samples for bacteria, micro-eukarya and archaea were included in this analysis. Pooled species abundances were fitted on both regional and local scales then displayed on a logarithmic scale. Akaike weights (wPLN and wNB) were calculated for PLN- and NB-fits on each dataset to provide a measure of the relative goodness for fit [64].
Supplementary information
Additional file 1: Figure S1. Rarefaction curves of subsampled bacterial, eukaryotic and archaeal communities between sites. In all cases, data was approaching asymptote indicating that sufficient sampling depth was achieved. A particularly rich number of bacterial, eukaryotic and archaeal species were observed at MP (Mitchell Peninsula), TR (The Ridge) and RR (Robinson Ridge), respectively. Figure S2. Top 15 most genus of bacterial, eukaryotic and archaeal communities between sites. As taxonomic levels decrease, the number of unclassified taxa increase substantially. Interestingly, archaeal communities were dominated by Nitrososphaera, a genus of ammonia oxidising archaea possibly implicated in nitrogen cycling within these nutrient starved soils. Figure S3. NMDS plots of microbial OTU communities and environmental soil parameters. In all cases, soil samples clustered according to site and broadly by geographic region. Although TR (The Ridge) is more environmentally similar to the Windmill Island sites, it’s soil bacterial and eukaryotic communities cluster more strongly with the Vestfold Hills. Figure S4. GAM model output of negative binomial distributions of best environmental predictor variables against estimated bacterial Chao1 richness based on AIC, where ‘*’ indicates a significant (P<0.05) correlation. A positive relationship is generally observed between bacterial richness and copper (CU), phosphorous (TP, P), aluminium (AL, AL2O3), sodium ion concentrations (CECNA) and the amount of gravel (GRVL) but displayed a negative relationship with titanium dioxide (TIO2). Figure S5. GAM model output of gaussian distributions of best environmental predictor variables against estimated eukaryotic Chao1 richness based on AIC, where ‘*’ indicates a significant (P<0.05) correlation. A negative relationship is generally observed between eukaryotic richness and dry matter fraction (DMF), soil pH, nitrite concentrations (NO2) and mud content but displayed a positive relationship with total carbon (TC) and conductivity (COND). A significant correlation is observed against random regional effects. Figure S6. GAM model output of gaussian distributions of best environmental predictor variables against estimated archaeal Chao1 richness based on AIC, where ‘*’ indicates a significant (P<0.05) correlation. Archaeal richness displayed positive relationships with conductivity (COND), total nitrogen (TN), phosphorous (TP, P) and sodium ion concentrations (CECNA), whilst a negative relationship was observed against titanium dioxide (TIO2). Figure S7. Local scale PLN- (blue) and NB-fitted (orange) SADs of the nine sites studied. These trends remain consistent with those observed for the regional fitted SADs, where bacterial communities display strong niche-driven signatures across all sites whilst eukaryotic and archaeal communities demonstrated weaker PLN-fits and multimodality. Table S1. Summary of amplicon sequencing output and OTU pipeline analysis. Table S2 CYTOSCAPE network topology analysis between regions at the domain-level. Table S3. Environmental soil parameters averaged between sites. Table S4. Akaike weights calculated from local-scale PLN- and NB-fitted SADs. Where NA indicates that the fitting procedure did not converge, which is usual for small datasets.
Acknowledgements
The authors would like to thank the Australian Antarctic Program (AAP), and expeditioners for the successful collection of soil samples between 2005 and 2012 as well as Bioplatforms Australia (BPA) for the provision of the Vestfold Hills biodiversity data.
Authors’ contributions
BCF, AT and EZ designed the study. AT coordinated sample collection and provided the environmental metadata. MR facilitated all the XRF and XRD analysis. JvD, SW and EZ extracted the DNA for sequencing. EZ processed the sequencing data and performed the analyses. LMT provided scripts for the fitted SADs. MMT and SC provided valuable constructive criticism. EZ drafted the manuscript, and all authors read, collaborated and approved the final manuscript.
Funding
We acknowledge funding support through Belinda Ferrari’s ARC Future Fellowship (FT170100341) and the Australian Antarctic Science Grants scheme (AAS-4406).
Availability of data and materials
The datasets generated and analysed during the current study are available through the Australian Antarctic Data (AAD) Centre, [10.4225/15/526F42ADA05B1] and the AusMicrobiome repository, [https://data.bioplatforms.com/organization/about/australian-microbiome]. Please contact author for additional data requests.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s40168-020-00809-w.
References
- 1.Cary SC, McDonald IR, Barrett JE, Cowan DA. On the rocks: the microbiology of Antarctic Dry Valley soils. Nature Reviews. 2010;8:120. doi: 10.1038/nrmicro2281. [DOI] [PubMed] [Google Scholar]
- 2.Lee JR, Raymond B, Bracegirdle TJ, Chadès I, Fuller RA, Shaw JD, Terauds A. Climate change drives expansion of Antarctic ice-free habitat. Nature. 2017;547:49. doi: 10.1038/nature22996. [DOI] [PubMed] [Google Scholar]
- 3.O’Brien PE, Smith J, Stark JS, Johnstone G, Riddle M, Franklin D. Submarine geomorphology and sea floor processes along the cost of Vestfold Hills, East Antarctica, from multibeam bathymetry and video data. Antarctic Science. 2015;27(6):566–586. doi: 10.1017/S0954102015000371. [DOI] [Google Scholar]
- 4.Hirvas H, Nenonen K, Quilty P. Till stratigraphy and glacial history of the Vestfold Hills area, east Antarctica. Quaternary International. 1993;18:81–95. doi: 10.1016/1040-6182(93)90056-L. [DOI] [Google Scholar]
- 5.Anderson JB, Shipp SS, Lowe AL, Wellner JS, Mosola AB. The Antarctic ice sheet during the Last Glacial Maximum and its subsequent retreat history: a review. Quaternary Science Reviews. 2002;21:49–70. doi: 10.1016/S0277-3791(01)00083-X. [DOI] [Google Scholar]
- 6.Ji M, Greening C, Vanwonterghem I, Carere CR, Bay SK, Steen JA, Montgomery K, Lines T, Beardall J, van Dorst J, Snape I, Stott MB, Hugenholtz P, Ferrari BC. Atmospheric trace gases support primary production in Antarctic desert surface soil. Nature. 2017;552:400–403. doi: 10.1038/nature25014. [DOI] [PubMed] [Google Scholar]
- 7.Siciliano SD, Palmer AS, Winsley T, Lamb E, Bissett A, Brown MV, et al. Soil fertility is associated with fungal and bacterial richness, whereas pH is associated with community composition in polar soil microbial communities. Soil Biology & Biochemistry. 2014;78:10–20. doi: 10.1016/j.soilbio.2014.07.005. [DOI] [Google Scholar]
- 8.Chan Y, Van Nostrand JD, Zhou J, Pointing SB, Farrell RL. Functional ecology of an Antarctic Dry Valley. PNAS. 2013;110(22):8990–8995. doi: 10.1073/pnas.1300643110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chong CW, Pearce DA, Convey P. Emerging spatial patterns in Antarctic prokaryotes. Frontiers in Microbiology. 2015;6(1058). 10.3389/fmicb.2015.01058. [DOI] [PMC free article] [PubMed]
- 10.Terauds A, Chown SL, Morgan F, Peat HJ, Watts DJ, Keys H, Convey P, Bergstrom DM. Conservation biogeography of the Antarctic. Diversity and Distributions. 2012;18:726–741. doi: 10.1111/j.1472-4642.2012.00925.x. [DOI] [Google Scholar]
- 11.Aislabie JM, Jordan S, Barker GM. Relation between soil classification and bacterial diversity in soils of the Ross Sea region, Antarctica. Geoderma. 2008;144:9–20. doi: 10.1016/j.geoderma.2007.10.006. [DOI] [Google Scholar]
- 12.Convey P, Chown SL, Clarke A, Barnes DKA, Bokhorst S, Cummings V, et al. The spatial structure of Antarctic biodiversity. Ecological Monographs. 2014;84(2):203–244. doi: 10.1890/12-2216.1. [DOI] [Google Scholar]
- 13.Hogg ID, Cary SC, Convey P, Newsham KK, O’Donnell AG, Adams BJ, et al. Biotic interactions in Antarctic terrestrial ecosystems: Are they a factor? Soil Biology & Biochemistry. 2006;38:3035–3040. doi: 10.1016/j.soilbio.2006.04.026. [DOI] [Google Scholar]
- 14.Chong C.W., Pearce D.A., Convey P., Tan I.K.P. The identification of environmental parameters which could influence soil bacterial community composition on the Antarctic Peninsula - a statistical approach. Antarctic Science. 2012;24(3):249–258. doi: 10.1017/S0954102012000028. [DOI] [Google Scholar]
- 15.Benaud N, Zhang E, van Dorst J, Brown MV, Kalaitzis JA, et al. Harnessing long-read amplicon sequencing to uncover NRPS and Type I PKS gene sequence diversity in polar desert soils. FEMS Microbiology Ecology. 2019. 10.1093/femsec/fiz031. [DOI] [PubMed]
- 16.Cowan D.A., Makhalanyane T.P., Dennis P.G., Hopkins D.W. (2015). Microbial ecology and biogeochemistry of continental Antarctic soils. Frontiers in Microbiology, 5(154), pages 1. [DOI] [PMC free article] [PubMed]
- 17.Pointing SB, Chan Y, Lacap DC, Lau MCY, Jurgens JA, Farrell RL. Highly specialized microbial diversity in hyper-arid polar desert. PNAS. 2009;106(47):19964–19969. doi: 10.1073/pnas.0908274106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rao S, Yuki C, Lacap DC, Hyde KD, Pointing SB, Farrell RL. Low-diversity fungal assemblage in an Antarctic Dry Valley soil. Polar Biology. 2012;35:567–574. doi: 10.1007/s00300-011-1102-2. [DOI] [Google Scholar]
- 19.Richter I, Herbold CW, Lee CK, McDonald IR, Barrett JE, Cary SC. Influence of soil properties on archaeal diversity and distribution in the McMurdo Dry Valleys, Antarctica. FEMS Microbiology Ecology. 2014;89:347–359. doi: 10.1111/1574-6941.12322. [DOI] [PubMed] [Google Scholar]
- 20.Bell TH, Callender KL, Whyte LG, Greer CW. Microbial competition in polar soils: A review of an understudied but potentially important control on productivity. Biology. 2013;2:533–554. doi: 10.3390/biology2020533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Faust K, Raes J. Microbial interactions: from networks to models. Nature Reviews. 2012;10:538–550. doi: 10.1038/nrmicro2832. [DOI] [PubMed] [Google Scholar]
- 22.Fierer N. Embracing the unknown: disentangling the complexities of the soil microbiome. Nature. 2017;15:579–590. doi: 10.1038/nrmicro.2017.87. [DOI] [PubMed] [Google Scholar]
- 23.Dumbrell AJ, Nelson M, Helgason T, Dytham C, Fitter AH. Relative roles of niche and neutral processes in structuring a soil microbial community. The ISME Journal. 2010;4:337–345. doi: 10.1038/ismej.2009.122. [DOI] [PubMed] [Google Scholar]
- 24.Nemergut DR, Schmidt SK, Fukami T, O’Neill SP, Bilinski TM, Stanish LF, et al. Patterns and processes of microbial community assembly. Microbiology and Molecular Biology Reviews. 2013;77(3):342–356. doi: 10.1128/MMBR.00051-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Powell JR, Karunaratne S, Campbell CD, Yao H, Robinson L, Singh BK. Deterministic processes vary during community assembly for ecologically dissimilar taxa. Nature Communications. 2015;6:8444. doi: 10.1038/ncomms9444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scheffer M, van Nes EH, Vergnon R. Toward a unifying theory of biodiversity. PNAS. 2018;115(4):639–641. doi: 10.1073/pnas.1721114115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tylianakis JM, Soper EJ. Vulnerability of ecosystems to climate. Climate Vulnerability. 2013;4:229–237. doi: 10.1016/B978-0-12-384703-4.00432-9. [DOI] [Google Scholar]
- 28.Vellend M. Conceptual synthesis in community ecology. The Quarterly Review of Biology. 2010;85(2):183–206. doi: 10.1086/652373. [DOI] [PubMed] [Google Scholar]
- 29.Verberk WCEP. Explaining general patterns in species abundance and distributions. Nature Education Knowledge. 2011;3(10):38. [Google Scholar]
- 30.Matthews TJ, Whittaker RJ. On the species abundance distribution in applied ecology and biodiversity management. Journal of Applied Ecology. 2015;52:443–454. doi: 10.1111/1365-2664.12380. [DOI] [Google Scholar]
- 31.Ventura M, Canchaya C, Tauch A, Chandra G, Fitzgerald GF, Chater KF, van Sinderen D. Microbiol Mol Biol Rev. 2007;71(3):495–548. doi: 10.1128/MMBR.00005-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schleper C, Nicol GW. Ammonia-Oxidising Archaea – Physiology, Ecology and Evolution. Advances in Microbial Physiology. 2010;57:1–41. doi: 10.1016/B978-0-12-381045-8.00001-1. [DOI] [PubMed] [Google Scholar]
- 33.Finke DL, Snyder WE. Niche partitioning increases resource exploitation by diverse communities. Science. 2008;321:1488–1489. doi: 10.1126/science.1160854. [DOI] [PubMed] [Google Scholar]
- 34.Vyverman W, Verleyen E, Wilmotte A, Hodgson DA, Willems A, Peeters K, Van de Vijver B, De Wever A, Leliaert F, Sabbe K. Evidence for widespread endemism among Antarctic micro-organisms. Polar Science. 2010;4:103–113. doi: 10.1016/j.polar.2010.03.006. [DOI] [Google Scholar]
- 35.Ferrari BC, Bissett A, Snape I, van Dorst J, Palmer AS, Ji M, et al. Geological connectivity drives microbial community structure and connectivity in polar, terrestrial ecosystems. Environmental Microbiology. 2016;18(6):1834–1849. doi: 10.1111/1462-2920.13034. [DOI] [PubMed] [Google Scholar]
- 36.Hubbell SP. The unified neutral theory of biodiversity and biogeography. Monographs in Population Biology, Princeton University Press. 2001;MPB-32:1–448. [Google Scholar]
- 37.Allison SD, Martiny JBH. Resistance, resilience, and redundancy in microbial communities. PNAS. 2008;105:11512–11519. doi: 10.1073/pnas.0801925105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Delgado-Baquerizo M, Giaramida L, Reich PB, Khachane AN, Hamonts K, Edwards C, et al. Lack of functional redundancy in the relationship between microbial diversity ad ecosystem functioning. Journal of Ecology. 2016;104:936–946. doi: 10.1111/1365-2745.12585. [DOI] [Google Scholar]
- 39.Antão LH, Connolly SR, Magurran AE, Soares A, Dornelas M. Prevalence of multimodal species abundance distributions is linked to spatial and taxonomic breadth. Global Ecology and Biogeography. 2016;26:203–215. doi: 10.1111/geb.12532. [DOI] [Google Scholar]
- 40.Dornelas M, Connolly SR. Multiple modes in a coral species abundance distribution. Ecology Letters. 2008;11:1008–1016. doi: 10.1111/j.1461-0248.2008.01208.x. [DOI] [PubMed] [Google Scholar]
- 41.Holt RD. Emergent neutrality. TRENDS in Ecology and Evolution. 2006;21(10):531–533. doi: 10.1016/j.tree.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 42.Vergnon R, van Nes EH, Scheffer M. Emergent neutrality leads to multimodal abundance distributions. Nature Communications. 2012;3:663. doi: 10.1038/ncomms1663. [DOI] [PubMed] [Google Scholar]
- 43.Tourna M, Stieglmeier M, Spang A, Könneke M, Schintlmeister A, Urich T, et al. Nitrososphaera viennensis, an ammonia oxidizing archaeon from soil. PNAS. 2011;108(20):8420–8425. doi: 10.1073/pnas.1013488108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pester M, Rattei T, Flechl S, Gröngröft A, Richter A, Overmann J, Reinhold-Hurek B, Loy A, Wagner M. amoA-based consensus phylogeny of ammonia-oxidizing archaea and deep sequencing of amoA genes from soils of four different geographical regions. Environmental Microbiology. 2012;14(2):525–539. doi: 10.1111/j.1462-2920.2011.02666.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aller JY, Kemp PF. Are archaea inherently less diverse than bacteria in the same environments? FEMS Microbiology Ecology. 2008;65(1):74–87. doi: 10.1111/j.1574-6941.2008.00498.x. [DOI] [PubMed] [Google Scholar]
- 46.Sakavara A, Tsirtsis G, Roelke DL, Mancy R, Spatharis S. Lumpy species coexistence arises robustly in fluctuating resource environments. PNAS. 2018;115(4):738–743. doi: 10.1073/pnas.1705944115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McGill BJ, Etienne RS, Gray JS, Alonso D, Anderson MJ, Benecha HK, et al. Species abundance distributions: moving beyond single prediction theories to integration within an ecological framework. Ecology Letters. 2007;10:995–1015. doi: 10.1111/j.1461-0248.2007.01094.x. [DOI] [PubMed] [Google Scholar]
- 48.Barberán A, Bates ST, Casamayor EO, Fierer N. Using network analysis to explore co-occurrence patterns in soil microbial communities. The ISME Journal. 2012;6:343–351. doi: 10.1038/ismej.2011.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Janssen PH. Identifying the dominant soil bacterial taxa in libraries of 16S rRNA and 16S rRNA genes. Applied Environmental Microbiology. 2006;72(3):1719–1728. doi: 10.1128/AEM.72.3.1719-1728.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bissett A, Fitzgerald A, Meintjes T, Mele PM, Reith F, Dennis PG, et al. Introducing BASE: The Biomes of Australian Soil Environments soil microbial diversity database. Gigascience. 2016;5:21. doi: 10.1186/s13742-016-0126-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lane DJ. 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. New York, NY: John Wiley & Sons; 1991. pp. 115–147. [Google Scholar]
- 52.Reysenbach A.L., Pace N.R., Robb F.T., Place A.R., editors. (1995). Archaea: A laboratory manual—Thermophiles. CSHLP. Protocol, 16, pages 101-107.
- 53.Edgar Robert C. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nature Methods. 2013;10(10):996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 54.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26(19):2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 55.Rognes Torbjørn, Flouri Tomáš, Nichols Ben, Quince Christopher, Mahé Frédéric. VSEARCH: a versatile open source tool for metagenomics. PeerJ. 2016;4:e2584. doi: 10.7717/peerj.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Quast Christian, Pruesse Elmar, Yilmaz Pelin, Gerken Jan, Schweer Timmy, Yarza Pablo, Peplies Jörg, Glöckner Frank Oliver. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Research. 2012;41(D1):D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D, et al. vegan: Community ecology package. R package version. 2018;2:5–2. [Google Scholar]
- 58.Wickham H. ggplot2: Elegant graphics for data analysis. 2016. [Google Scholar]
- 59.Kassambara A (2018). Ggpubr: ‘ggplot2’ Based publication ready plots. R package version 0.1.8. https://CRAN.R-project.org/package=ggpubr.
- 60.Ripley B. MASS. R package version. 2019;7:3–51.4. [Google Scholar]
- 61.Reshef DN, Reshef YA, Finucane HK, Grossman SR, McVean G, Turnbaugh PJ, et al. Detecting novel associations in large datasets. Science. 2011;334(6062):1518–1524. doi: 10.1126/science.1205438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Research. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Connolly SR, MacNeil MA, Caley MJ, Knowlton N, Cripps E, Hisano M, et al. Commonness and rarity in the marine biosphere. PNAS. 2014;111(23):8524–8529. doi: 10.1073/pnas.1406664111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Burnham KP, Anderson DR. Multimodel inference: Understanding AIC and BIC in model selection. Sociological Methods & Research. 2004. https://doi.org/10.117/0049124104268644.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. Rarefaction curves of subsampled bacterial, eukaryotic and archaeal communities between sites. In all cases, data was approaching asymptote indicating that sufficient sampling depth was achieved. A particularly rich number of bacterial, eukaryotic and archaeal species were observed at MP (Mitchell Peninsula), TR (The Ridge) and RR (Robinson Ridge), respectively. Figure S2. Top 15 most genus of bacterial, eukaryotic and archaeal communities between sites. As taxonomic levels decrease, the number of unclassified taxa increase substantially. Interestingly, archaeal communities were dominated by Nitrososphaera, a genus of ammonia oxidising archaea possibly implicated in nitrogen cycling within these nutrient starved soils. Figure S3. NMDS plots of microbial OTU communities and environmental soil parameters. In all cases, soil samples clustered according to site and broadly by geographic region. Although TR (The Ridge) is more environmentally similar to the Windmill Island sites, it’s soil bacterial and eukaryotic communities cluster more strongly with the Vestfold Hills. Figure S4. GAM model output of negative binomial distributions of best environmental predictor variables against estimated bacterial Chao1 richness based on AIC, where ‘*’ indicates a significant (P<0.05) correlation. A positive relationship is generally observed between bacterial richness and copper (CU), phosphorous (TP, P), aluminium (AL, AL2O3), sodium ion concentrations (CECNA) and the amount of gravel (GRVL) but displayed a negative relationship with titanium dioxide (TIO2). Figure S5. GAM model output of gaussian distributions of best environmental predictor variables against estimated eukaryotic Chao1 richness based on AIC, where ‘*’ indicates a significant (P<0.05) correlation. A negative relationship is generally observed between eukaryotic richness and dry matter fraction (DMF), soil pH, nitrite concentrations (NO2) and mud content but displayed a positive relationship with total carbon (TC) and conductivity (COND). A significant correlation is observed against random regional effects. Figure S6. GAM model output of gaussian distributions of best environmental predictor variables against estimated archaeal Chao1 richness based on AIC, where ‘*’ indicates a significant (P<0.05) correlation. Archaeal richness displayed positive relationships with conductivity (COND), total nitrogen (TN), phosphorous (TP, P) and sodium ion concentrations (CECNA), whilst a negative relationship was observed against titanium dioxide (TIO2). Figure S7. Local scale PLN- (blue) and NB-fitted (orange) SADs of the nine sites studied. These trends remain consistent with those observed for the regional fitted SADs, where bacterial communities display strong niche-driven signatures across all sites whilst eukaryotic and archaeal communities demonstrated weaker PLN-fits and multimodality. Table S1. Summary of amplicon sequencing output and OTU pipeline analysis. Table S2 CYTOSCAPE network topology analysis between regions at the domain-level. Table S3. Environmental soil parameters averaged between sites. Table S4. Akaike weights calculated from local-scale PLN- and NB-fitted SADs. Where NA indicates that the fitting procedure did not converge, which is usual for small datasets.
Data Availability Statement
The datasets generated and analysed during the current study are available through the Australian Antarctic Data (AAD) Centre, [10.4225/15/526F42ADA05B1] and the AusMicrobiome repository, [https://data.bioplatforms.com/organization/about/australian-microbiome]. Please contact author for additional data requests.