Summary
Genetic influences on psychiatric disorders transcend diagnostic boundaries, suggesting substantial pleiotropy of contributing loci. However, the nature and mechanisms of these pleiotropic effects remain unclear. We performed analyses of 232,964 cases and 494,162 controls from genome-wide studies of anorexia nervosa, attention-deficit/hyperactivity disorder, autism spectrum disorder, bipolar disorder, major depression, obsessive-compulsive disorder, schizophrenia, and Tourette syndrome. Genetic correlation analyses revealed a meaningful structure within the eight disorders, identifying three groups of inter-related disorders. Meta-analysis across these eight disorders detected 109 loci associated with at least two psychiatric disorders, including 23 loci with pleiotropic effects on four or more disorders and 11 loci with antagonistic effects on multiple disorders. The pleiotropic loci are located within genes that show heightened expression in the brain throughout the lifespan, beginning prenatally in the second trimester, and play prominent roles in neurodevelopmental processes. These findings have important implications for psychiatric nosology, drug development, and risk prediction.
Keywords: Psychiatric genetics, cross-disorder genetics, psychiatric disorders, pleiotropy, neurodevelopment, GWAS, genetic correlation, gene expression
INTRODUCTION
Psychiatric disorders affect more than 25% of the population in any given year and are a leading cause of worldwide disability (Global Burden of Disease Injury Incidence Prevalence Collaborators, 2017; Kessler and Wang, 2008). The substantial influence of genetic variation on risk for a broad range of psychiatric disorders has been established by both twin and, more recently, large-scale genomic studies (Smoller et al., 2018). Psychiatric disorders are highly polygenic, with a large proportion of heritability contributed by common variation. Many risk loci have emerged from genome-wide association studies (GWAS) of, among others, schizophrenia (SCZ), bipolar disorder (BIP), major depression (MD), and attention-deficit/hyperactivity disorder (ADHD) from the Psychiatric Genomics Consortium (PGC) and other efforts (Sullivan et al., 2018). These studies have revealed a surprising degree of genetic overlap among psychiatric disorders (Brainstorm Consortium, 2018; Cross-Disorder Group of the Psychiatric Genomics Consoritum, 2013). Elucidating the extent and biological significance of cross-disorder genetic influences has implications for psychiatric nosology, drug development, and risk prediction. In addition, characterizing the functional genomics of cross-phenotype genetic effects may reveal fundamental properties of pleiotropic loci that differentiate them from disorder-specific loci, and help identify targets for diagnostics and therapeutics.
In 2013, analyses by the PGC’s Cross-Disorder Group identified loci with pleiotropic effects across five disorders: autism spectrum disorder (ASD), ADHD, SCZ, BIP, and MD in a sample comprising 33,332 cases and 27,888 controls (Cross-Disorder Group of the Psychiatric Genomics Consortium, 2013). In the current study, we examined pleiotropic effects in a greatly expanded dataset, encompassing 232,964 cases and 494,162 controls, that included three additional psychiatric disorders: Tourette syndrome (TS), obsessive-compulsive disorder (OCD), and anorexia nervosa (AN). We address four major questions regarding the shared genetic basis of these eight disorders: 1) Can we identify a shared genetic structure within the broad range of these clinically distinct psychiatric disorders? 2) Can we detect additional loci associated with risk for multiple disorders (pleiotropic loci)? 3) Do some of these risk loci have opposite allelic effects across disorders? and 4) Can we identify functional features of the pleiotropic loci that could account for their broad effects on psychopathology?
RESULTS
We analyzed genome-wide single nucleotide polymorphism (SNP) data for eight neuropsychiatric disorders using a combined sample of 232,964 cases and 494,162 controls (Table 1; Table S1). The eight disorders included AN (Duncan et al., 2017) ASD (Grove et al., 2019a), ADHD (Demontis et al., 2019), BIP (Stahl et al., 2019), MD (Wray et al., 2018), OCD (International Obsessive Compulsive Disorder Foundation Genetics Collaborative (IOCDF-GC) and OCD Collaborative Genetics Association Studies (OCGAS), 2018), TS (Yu et al., 2019), and SCZ (Schizophrenia Working Group of the Psychiatric Genomics, 2014). All study participants were of self-identified European ancestry, which was supported by principal component analysis of genome-wide data.
Table 1.
Summary of eight neuropsychiatric disorder datasets
| Disorder | # of Cases | # of Controls | # of Total Samples | # of GWAS Loci | Population Prevalence (k) | Liability-based SNP heritability (SE) | References |
|---|---|---|---|---|---|---|---|
| ADHD | 19,099 | 34,194 | 53,293 | 9 | 0.05 | 0.222 (0.014) | Demontis et al. 2019 |
| ANO | 3,495 | 10,983 | 14,478 | 0 | 0.01 | 0.195 (0.029) | Duncan et al. 2017 |
| ASD | 18,381 | 27,969 | 46,350 | 5 | 0.01 | 0.113 (0.010) | Grove et al. 2019 |
| BIP | 20,352 | 31,358 | 51,710 | 17 | 0.01 | 0.182 (0.011) | Stahl et al. 2019 |
| MD | 130,664 | 330,470 | 461,134 | 44 | 0.15 | 0.085 (0.004) | Wray et al. 2018 |
| OCD | 2,688 | 7,037 | 9,725 | 0 | 0.025 | 0.280 (0.041) | IOCDF-GC and OCGAS 2018 |
| SCZ | 33,640 | 43,456 | 77,096 | 108 | 0.01 | 0.222 (0.012) | Ripke et al. 2014 |
| TS | 4,645 | 8,695 | 13,340 | 0 | 0.008 | 0.200 (0.026) | Yu et al. 2019 |
| Total | 232,964 | 494,162 | 727,126 |
Genetic correlations among eight neuropsychiatric disorders indicate three genetic factors
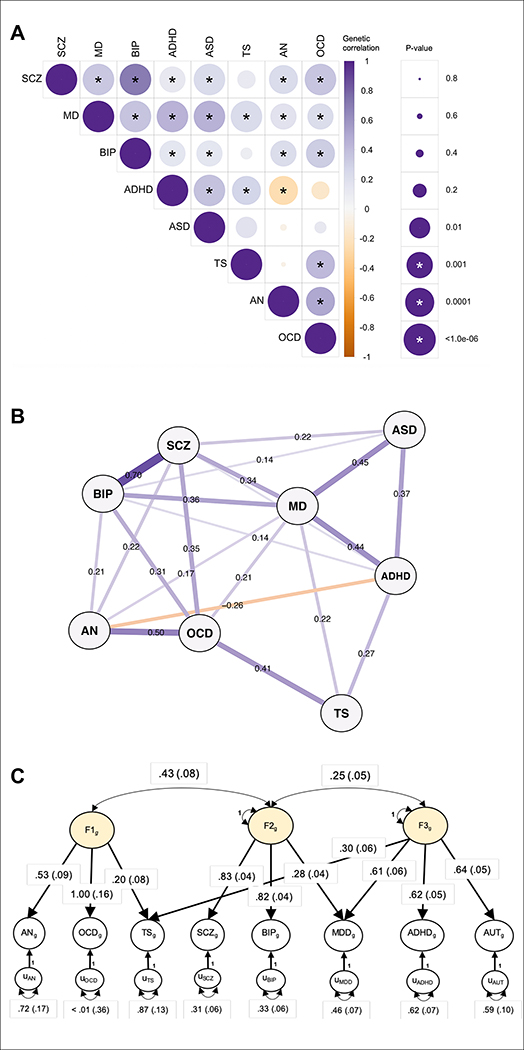
After standardized and uniform quality control, additive logistic regression analyses were performed on individual disorders (Online Methods). 6,786,993 SNPs were common across all datasets and were retained for further study. Using the summary statistics of these SNPs, we first estimated pairwise genetic correlations among the eight disorders using linkage disequilibrium (LD) score regression analyses (Bulik-Sullivan et al., 2015) (Online Methods; Fig. 1a; Table S2.1). The results were broadly concordant with previous estimates (Brainstorm Consortium, 2018; Cross-Disorder Group of the Psychiatric Genomics Consoritum, 2013). The genetic correlation was highest between SCZ and BIP (rg = 0.70 ±0.02), followed by OCD and AN (rg = 0.50 ±0.12). Interestingly, based on genome-wide genetic correlations, MD was closely correlated with ASD (rg=0.45 ±0.04) and ADHD (rg=0.44 ±0.03), two childhood-onset disorders. Despite variation in magnitude, significant genetic correlations were apparent for most pairs of disorders, suggesting a complex, higher-order genetic structure underlying psychopathology (Fig. 1b).
Figure 1. Genetic relationships between eight psychiatric disorders.
A) SNP-based genetic correlations (rg) were estimated between eight neuropsychiatric disorders using LDSC. The size of the circles scales with the significance of the p-values. The darker the color, the larger the magnitude of rg. Star sign (*) indicates statistical significance after Bonferroni correction. (B) SNP-based genetic correlations between eight disorders were depicted using an in-directed graph to reveal complex genetic relationships. Only significant genetic correlations after Bonferroni correction in (A) were displayed. Each node represents a disorder, with edges indicating the strength of the pairwise correlations. The width of the edges increases, while the length decreases, with the absolute values of rg. (C) Based on the results of an exploratory factor analysis of the genetic correlation matrix produced from multivariable LD-score regression, a confirmatory factor model with three correlated genetic factors was specified using Genomic SEM and estimated with the weighted least squares algorithm. In this solution, each common genetic factor (i.e., F1g, F2g, F3g) represents variation in genetic liability that is shared across the disorders that load on it. These common factors are specified so as to account for the genetic covariation among the psychiatric disorders. For example, F1g represents shared genetic liability among disorders characterized by compulsive behaviors (AN, OCD and TS). One-headed arrows connecting the common genetic factors to the individual disorders represent standardized loadings, which can be interpreted as coefficients from a regression of the true genetic liability for the disorder on the common factor. Two-headed arrows connecting the three factors to one another represent their correlations. Two-headed arrows connecting the genetic components of the individual psychiatric disorders to themselves represent residual genetic variances and correspond to the proportion of heritable variation in liability to each individual psychiatric disorder that is unexplained by the three factors. Standardized parameters are depicted with their standard errors in parentheses. Paths labeled 1 with no standard errors reported are fixed parameters, which are used for scaling.
We modeled the genome-wide joint architecture of the eight neuropsychiatric disorders using an exploratory factor analysis (EFA) (Gorsuch, 1988), followed by genomic structural equation modeling (SEM) (Grotzinger et al., 2019) (Online Methods; Fig. 1c). EFA identified three correlated factors, which together explained 51% of the genetic variation in the eight neuropsychiatric disorders (Table S2.2). The first factor consisted primarily of disorders characterized by compulsive/perfectionistic behaviors, specifically AN, OCD, and, more weakly, TS. The second factor was characterized by mood and psychotic disorders (MD, BIP, and SCZ), and the third factor by three early-onset neurodevelopmental disorders (ASD, ADHD, TS) as well as MD. Similar to our EFA results, hierarchical clustering analyses also identified three sub-groups among the eight disorders (Data S1.1). Based on extensive follow-up analyses, this genetic correlational structure does not appear to be biased by sample overlap or sample size differences among the eight disorders (Data S1.2–1.4).
Cross-disorder meta-analysis identifies 109 pleiotropic loci
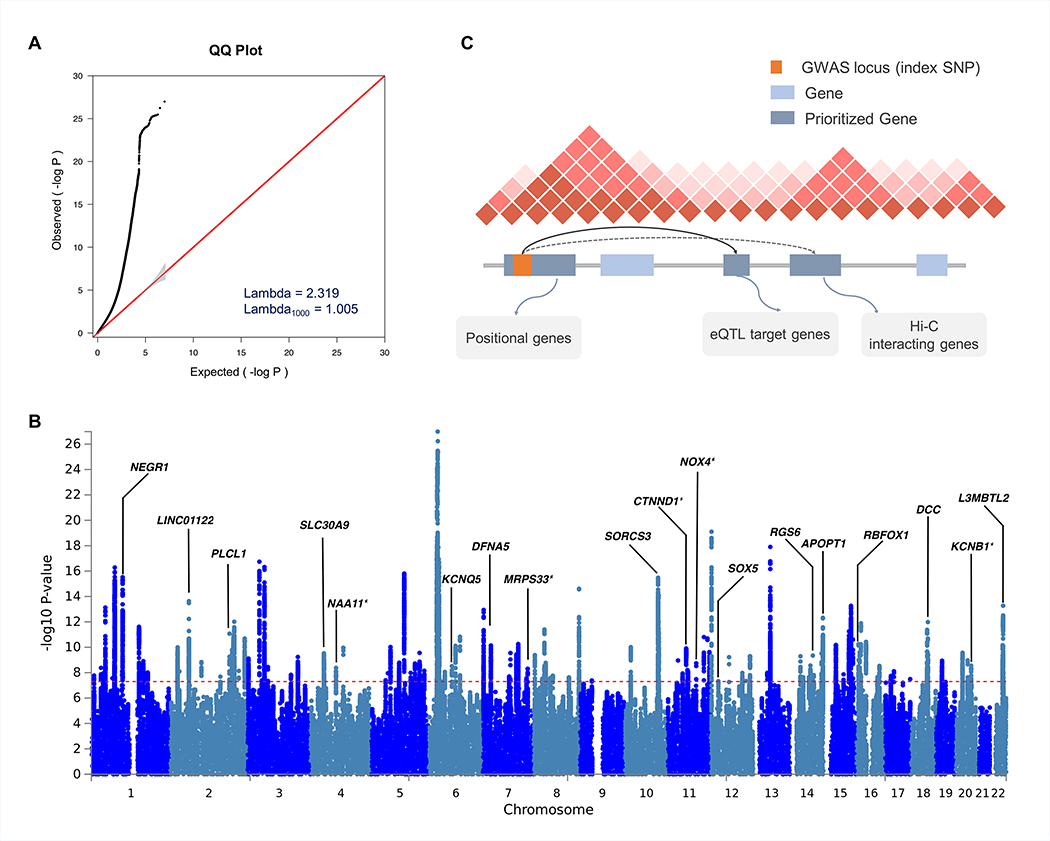
The factor structure described above is based on average effects across the genome, but does not address more fine-grained cross-disorder effects at the level of genomic regions or individual loci. To identify genetic loci with shared risk, we performed a meta-analysis of the eight neuropsychiatric disorders using a fixed-effects-based method (Bhattacharjee et al., 2012) that accounts for the differences in sample sizes, existence of subset-specific effects, and overlapping subjects across datasets (Online Methods). The standardized genomic inflation factor was close to one, suggesting no inflation of test statistics due to confounding (λ1000 = 1.005; Fig. 2a). We identified 136 LD-independent regions with genome-wide significant association (Pmeta ≤ 5×10−8). Due to the extensive LD at the major histocompatibility complex (MHC) region (chromosome 6 region at 25–35 Mb), we considered multiple signals present there as one locus. 101 of the 136 (74.3%) significantly associated regions overlapped with previously reported genome-wide significant regions from at least one individual disorder, while 35 loci (25.7%) represented novel genome-wide significant associations. Simulation analyses confirmed that the number of pleiotropic loci we identified exceeds chance expectation given the sample size and genetic correlations among the eight disorders (p < 9.9×10−3; Data S1.5; for further details, see Online Methods).
Figure 2. Results of cross-disorder meta-analysis and candidate gene mapping.
(A) Quantile-quantile (QQ) plot displaying the observed meta-analysis statistics vs. the expected statistics under the null model of no associations in the -log10(p-value) scale. Although a marked departure is notable between the two statistics, the estimated lambda1000 and the estimated LD Score regression intercept indicate that the observed inflation is mainly due to polygenic signals rather than major confounding factors including population stratification. (B) Gene prioritization strategies for significantly associated loci. Candidate genes were mapped on each locus if the index SNP and credible SNPs reside within a protein-coding gene, are eQTL markers of the gene in the brain tissue, or interact with promoter regions of the gene based on brain Hi-C data. (C) Manhattan plot displaying the cross-disorder meta-analysis results highlighting candidate genes mapped to top pleiotropic regions. When multiple genes were mapped to the same locus, genes encompassing the index SNP or genes with the largest number of evidences were displayed for clarity. Candidate genes that have not previously implicated in individual disorder GWAS are marked with an asterisk.
Within these 136 loci, multi-SNP-based conditional analysis (Yang et al., 2012) identified 10 additional SNPs with independent associations, resulting in a total of 146 independent lead SNPs (Table S3.1). To provide a quantitative estimate of the best fit configuration of cross-disorder genotype-phenotype relationships, we estimated the posterior probability of association (referred to as the m-value) with each disorder using a Bayesian statistical framework (Han and Eskin, 2012) (Online Methods; Table S3.2) As recommended (Han and Eskin, 2012), an m-value threshold of 0.9 was used to predict with high confidence that a particular SNP was associated with a given disorder. Also, m-values of < 0.1 were taken as strong evidence against association. Plots of the SNP p-value vs. m-value for all 146 lead SNPs are shown in Data S2. Nearly 75% (109/146) of the genome-wide significant SNPs were pleiotropic (i.e., associated with more than one disorder). As expected, configurations of disease association reflected the differences in the statistical power and genetic correlations between the samples (Fig. S1). Of the 109 pleiotropic loci, 83% and 72% involved SCZ and BIP, respectively. MD, which had the largest case-control sample, was associated with 48% of the pleiotropic loci (N=52/109). Despite the relatively small sample size, ASD was implicated in 36% of the pleiotropic loci. Most of the ASD associations co-occurred with SCZ and BIP. The other disorders, ADHD, TS, OCD, and AN featured associations in 16%, 14%, 11%, and 7% of the pleiotropic loci, respectively. Of the single-disorder-specific loci, 81% and 16% were associated with SCZ and MD, respectively.
Table 2 summarizes 23 pleiotropic loci associated with at least four of the disorders. Among these loci, heterogeneity of effect sizes was minimal (p-value of Q > 0.1). Eleven of the 23 lead SNPs map to the intron of a protein-coding gene, and seven additional lead SNPs had at least one protein-coding gene within 100 kb. We used an array of functional genomics resources, including brain eQTL and Hi-C data (Wang et al., 2018; Won et al., 2016) to prioritize potential candidate genes to the identified regions (Online Methods; Fig. 2b). The Manhattan plot in Fig. 2c highlights some of the prioritized candidate genes.
Table 2.
Summary of 23 loci with the broadest cross-disorder association
| SNP | CHR | BP | Candidate | ADHD | ANO | ASD | BIP | MD | OCD | SCZ | TS | m |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs8084351 | 18 | 50726559 | DCC(g,q) | 0.961 | 0.905 | 0.97 | 0.965 | 1 | 0.951 | 1 | 0.984 | 8 |
| rs7193263 | 16 | 6315880 | RBFOX1(g) | 0.924 | 0.802 | 0.984 | 0.995 | 1 | 0.902 | 0.901 | 0.932 | 7 |
| rs12658451 | 5 | 103904037 | - | 0.963 | 0.165 | 0.999 | 0.972 | 1 | 0.574 | 1 | 0.963 | 6 |
| rs34215985 | 4 | 42047778 | SLC30A9(g,q) DCAF4L1(tss) | 0.908 | 0.926 | 0.992 | 0.843 | 1 | 0.88 | 0.929 | 0.913 | 6 |
| rs61867293 | 10 | 106563924 | SORCS3(g,ha,hf) | 0.987 | 0.954 | 0.992 | 0.985 | 1 | 0.854 | 1 | 0.886 | 6 |
| rs9360557 | 6 | 73132745 | KCNQ5(ha,hf) | 0.905 | 0.938 | 0.976 | 0.984 | 0.993 | 0.897 | 1 | 0.892 | 6 |
| KCNQ5-IT1(hf) | ||||||||||||
| rs10149470 | 14 | 104017953 | APOPT1(fg) | 0.844 | 0.833 | 0.998 | 0.979 | 1 | 0.868 | 0.997 | 0.97 | 5 |
| C14orf2(ha) | ||||||||||||
| rs11570190 | 11 | 57560452 | CTNND1(g,tss) | 0.927 | 0.79 | 0.97 | 0.58 | 1 | 0.916 | 1 | 0.832 | 5 |
| OR5AK2(q) | ||||||||||||
| rs117956829 | 11 | 89339666 | GRM5(hf) | 0.723 | 0.929 | 0.972 | 0.906 | 1 | 0.66 | 0.997 | 0.789 | 5 |
| NOX4(ha,hf) | ||||||||||||
| rs1484144 | 4 | 80217597 | NAA11(fg) | 0.97 | 0.884 | 0.973 | 0.98 | 1 | 0.84 | 0.998 | 0.85 | 5 |
| rs6969410 | 7 | 110069015 | - | 0.836 | 0.827 | 0.987 | 0.93 | 0.999 | 0.917 | 1 | 0.729 | 5 |
| rs7531118 | 1 | 72837239 | NEGR1(hf) | 0.74 | 0.949 | 0.963 | 0.785 | 1 | 0.858 | 0.973 | 0.921 | 5 |
| rs9787523 | 10 | 106460460 | SORCS3(g) | 0.944 | 0.855 | 0.972 | 0.877 | 1 | 0.853 | 0.999 | 0.963 | 5 |
| rs10265001 | 7 | 140665521 | MRPS33(tss); KDM7A(ha) | 0.716 | 0.772 | 0.986 | 0.999 | 0.783 | 0.921 | 0.988 | 0.692 | 4 |
| rs11688767 | 2 | 57988194 | BCL11A(h) | 0.845 | 0.899 | 0.929 | 0.983 | 1 | 0.849 | 1 | 0.698 | 4 |
| LINC01122(ha,hf) | ||||||||||||
| rs12129573 | 1 | 73768366 | - | 0.929 | 0.835 | 0.894 | 0.948 | 1 | 0.85 | 1 | 0.539 | 4 |
| rs1518367 | 2 | 198807015 | PLCL1(g); SF3B1(ha,q) | 0.897 | 0.783 | 0.913 | 0.991 | 1 | 0.674 | 1 | 0.865 | 4 |
| rs2332700 | 14 | 72417326 | RGS6(g) | 0.755 | 0.884 | 0.951 | 0.948 | 0.999 | 0.885 | 1 | 0.817 | 4 |
| rs5758265 | 22 | 41617897 | L3MBTL2(g) | 0.735 | 0.885 | 0.89 | 0.885 | 1 | 0.913 | 1 | 0.978 | 4 |
| CHADL(g) | ||||||||||||
| rs6125656 | 20 | 48090779 | KCNB1(g) SPATA2(hf) | 0.768 | 0.885 | 0.986 | 0.995 | 0.985 | 0.731 | 0.999 | 0.707 | 4 |
| rs7405404 | 16 | 13749859 | - | 0.763 | 0.765 | 0.99 | 0.939 | 1 | 0.726 | 1 | 0.562 | 4 |
| rs78337797 | 12 | 23987925 | SOX5(g) | 0.849 | 0.797 | 0.97 | 0.954 | 1 | 0.831 | 0.996 | 0.885 | 4 |
| rs79879286 | 7 | 24826589 | DFNA5(fg,tss) MPP6(fg) | 0.865 | 0.854 | 0.966 | 0.999 | 1 | 0.734 | 0.999 | 0.798 | 4 |
SNP ID, location, prioritized candidate gene, disorder-specific m-values for 23 most pleiotropic loci. The number of disorders with high confidence association (m-values ≥0.9) is shown in the last column. Evidence for candidate gene mapping include: g (gene containing index SNP); fg (credible SNP gene); q (brain cis-eQTLs); h (hi-C interacting gene based on FUMA); hf (hi-C-based interaction between associated SNP and target gene in the fetal brain from Won et al. 2016); ha (hi-C-based interaction in the adult brain from Wang et al. 2018); and tss (transcription start sites). Loci were highlighted if the LD-independent regions do not overlap with genome-wide significant associations previously identified in the GWAS of individual disorders. At most two candidate genes are listed here. Full list of associated gene information is available in Table S4.
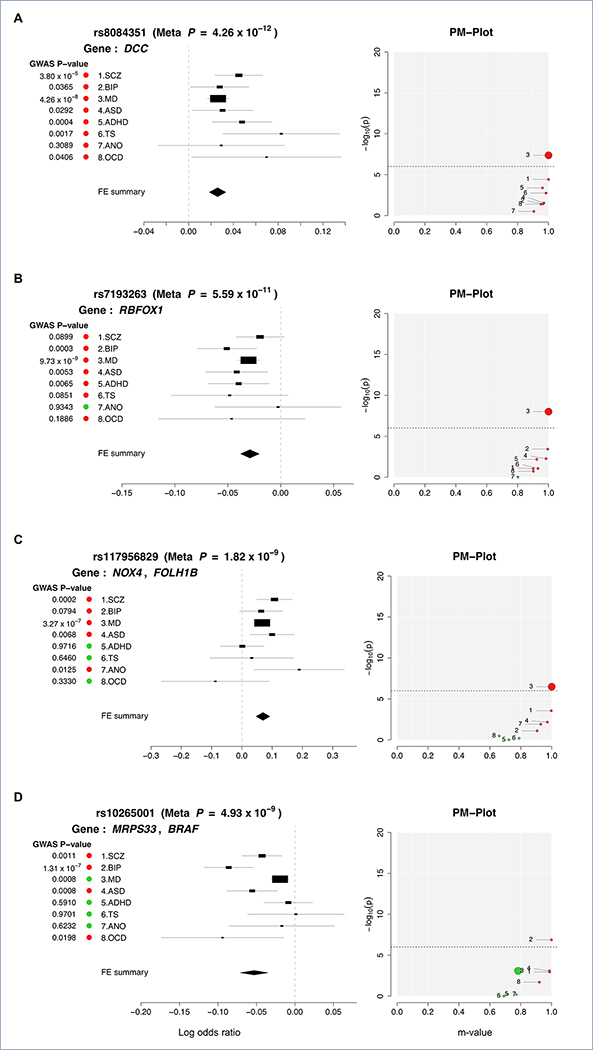
Of the 109 risk loci with shared effects, the 18q21.2 region surrounding SNP rs8084351 at the netrin 1 receptor gene DCC featured the most pleiotropic association (Pmeta = 4.26 × 10−12; Fig. 3a). This region showed association with all eight psychiatric disorders, and has been previously associated with both MD and neuroticism (Turley et al., 2018; Wray et al., 2018). The signal in our meta-analysis colocalizes with brain eQTLs for DCC (eQTL association FDR q = 2.27 × 10−5), supporting DCC as a plausible candidate gene (Fig. S2). The product of DCC plays a key role in guiding axonal growth during neurodevelopment and serves as a master regulator of midline crossing and white matter projections (Bendriem and Ross, 2017). Gene expression data indicate that DCC expression peaks during early prenatal development (Fig. S3).
Figure 3. Profile of disorder associations for illustrative pleiotropic loci: (A) rs8084351 on 18q21.2; (B) rs7193263 on 16p13.3; (C) rs117956829 on 11q14.3; and (D) rs10265001 on 7q34.
For each locus, disorder-specific effects of the index SNP are shown using ForestPMPlot. The first panel is the forest plot, displaying disorder-specific association p-value, log odds ratios (ORs), and standard errors of the SNP. The meta-analysis p-value and the corresponding summary statistic are displayed on the top and the bottom of the forest plot, respectively. The second panel is the PM-plot in which X-axis represents the m-value, the posterior probability that the effect eixsts in each disorder, and the Y-axis represents the disorder-specific association p-value as -log10(p-value). Disorders are depicted as a dot whose size represents the sample size of individual GWAS. Disorders with estimated m-values of at least 0.9 are colored in red, while those with m-values less than 0.9 are marked in green.
The second most pleiotropic locus in our analysis was identified in an intron of RBFOX1 (RNA Binding Fox-1 Homolog 1) on 16p13.3 (lead SNP rs7193263; Pmeta = 5.59 × 10−11). The lead SNP showed association with all of the disorders except AN (Fig. 3b). RBFOX1 (also called A2BP1) encodes a splicing regulator mainly expressed in neurons and known to target several genes important to neuronal development, including NMDA receptor 1 and voltage-gated calcium channels (Gandal, 2018; Gehman, 2011; Hamada et al., 2015). Knock-down and silencing of RBFOX1 during mouse corticogenesis impairs neuronal migration and synapse formation (Hamada et al., 2015; Hamada et al., 2016), implying its pivotal role in early cortical maturation. In contrast to DCC, however, developmental gene-expression of RBFOX1 showed gradually increasing gene expression throughout the prenatal period (Fig. S3). Animal models and association studies have implicated RBFOX1 in aggressive behaviors, a trait observed in several of the disorders in our analysis (Fernandez-Castillo et al., 2017).
Of the 109 pleiotropic loci, 76 were identified in the GWAS of individual disorders, while the remaining 33 are novel. The most pleiotropic among these novel loci was a region downstream of NOX4 (NADPH Oxidase 4) that was associated with SCZ, BIP, MD, ASD, and AN (rs117956829; Pmeta = 1.82 × 10−9; Fig. 3c). Brain Hi-C data (Wang et al., 2018; Won et al., 2016) detected a direct interaction of the cross-disorder association region with NOX4 in both adult and fetal brain (interaction p=3.2×10−16 and 9.3×10−6, respectively). As a member of the family of NOX genes that encode subunits of NADPH oxidase, NOX4 is a major source of superoxide production in human brain and a promoter of neural stem cell growth (Kuroda et al., 2014; Topchiy et al., 2013).
Figure 3d illustrates another novel psychiatric risk locus associated with SCZ, BIP, ASD, and OCD (Pmeta = 3.58 × 10−8). The lead SNP rs10265001 resides between MRPS33 (Mitochondrial Ribosomal Protein S33) and BRAF (B-Raf Proto-Oncogene, Serine/Threonine Kinase) on 7q34. The brain Hi-C data indicated interaction of the associated region with the promoters of two nearby genes: BRAF, which contributes to the MAP kinase signal transduction pathway and plays a role in postsynaptic responses of hippocampal neurons (Grantyn and Grantyn, 1973), and KDM7A (encoding Lysine Demethylase 7A), which plays a central role in the nervous system and midbrain development (Horton et al., 2010; Qi et al., 2010; Tsukada et al., 2010).
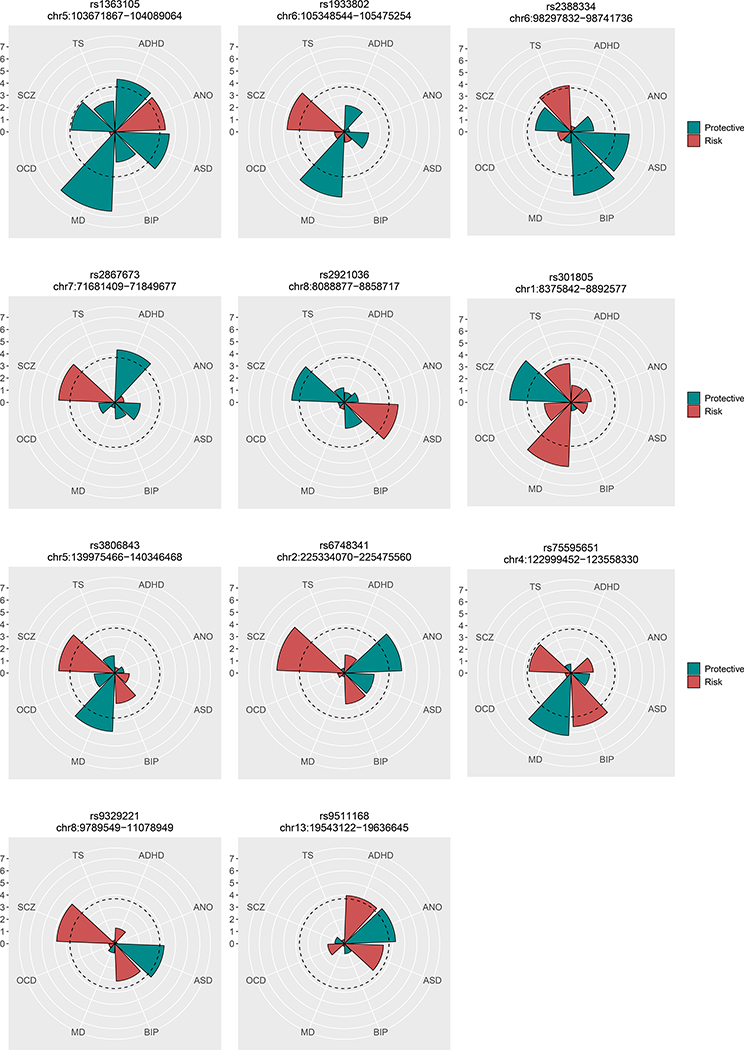
Our prior cross-disorder meta-analysis of five psychiatric disorders (Cross-Disorder Group of the Psychiatric Genomics Consortium, 2013) found no evidence of SNPs with antagonistic effects on two or more disorders. Here, we examined whether any variants with meta-analysis p ≤ 1×10−6 had opposite directional effects between disorders (Online Methods). After adjusting for having examined 206 loci across eight disorders (q < 0.001), we identified 11 loci with evidence of opposite directional effects on two or more disorders (Fig. 4; Table S3.3). The disorder configuration of opposite directional effects varied for the 11 loci, including three loci with opposite directional effects on SCZ and MD (rs301805, rs1933802, rs3806843), two loci between SCZ and ASD (rs9329221, rs2921036), and one locus (rs75595651) with opposite directional effects on the two mood disorders, BIP and MD. Notably, all of the six loci involving SCZ and BIP exhibited the same directional effect on the two disorders (Pbinom < 0.05), in line with their strong genome-wide genetic correlation.
Figure 4. Eleven loci with opposite directional effects.
The radius of each wedge corresponds to the absolute values of the Z-scores (log(Odds ratios)/S.E) obtained from association tests of the SNP for eight disorders. The color indicates whether the examined SNP carries risk (red) or protective effects (green) for each disorder. The dotted line around the center indicates statistically significant SNP effects that account for multiple testing of 206 SNPs the q-value of 0.001.
Functional characterization of pleiotropic risk loci
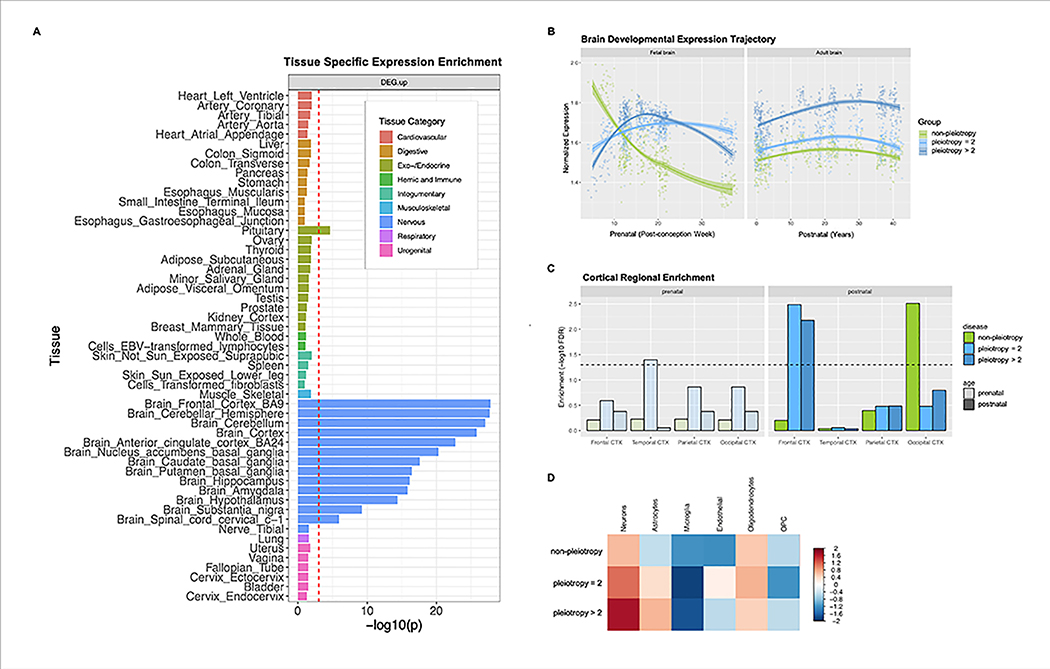
We conducted a series of bioinformatic analyses that examined whether loci with shared risk effects on multiple neuropsychiatric disorders had characteristic features that distinguished them from non-pleiotropic risk loci. First, we annotated the functional characteristics of 146 lead SNPs using various public data sources (Online Methods; Tables S4). Overall, they showed significant enrichment of genes expressed in the brain (beta=0.123, SE=0.0109, enrichment p = 1.22×10−29) and pituitary (beta=0.0916, SE=0.0136, p = 8.74 × 10−12), but not in the other Genotype-Tissue Expression (GTEx) tissues. (Table S5.1; Fig. 5a). A separate analysis of 109 pleiotropic risk loci also showed specific enrichment of genes expressed in multiple brain tissues (p = 1.55 × 10−5; Table S5.2), while disorder-specific loci showed nominally enriched brain gene expression in the cortex (p =2.14 × 10−2; Table S5.3).
Figure 5. Results of functional genomics data analysis for pleiotropic vs. disorder-specific loci.
(A) GTEX tissue-specific enrichment results for 146 risk loci associated with at least one of eight neuropsychiatric disorders. GTEX tissues were classified as 9 distinct categories, of which the brain tissues were colored in blue. The dotted red line indicates a statistically significant p-value after conducting Bonferroni correction for multiple testing. Psychiatric disorder-associated loci show significant enrichment in genes expressed in pituitary and all brain tissues except nerve_tibal. (B) Brain developmental expression trajectory displayed for the three groups of genes based on (Kang et al., 2011) The 146 genome-wide significant loci from the cross-disorder meta analysis were clustered into three groups based on predicted disorder-specific associations: (1) no-pleiotopy; (2) pleiotropy=2; and (3) pleiotropy>2. The “no-pleiotropy” group included 37 loci that showed a single-disorder-specific association, while the “pleiotropy=2” and “pleiotropy>2” groups included 60 and 49 loci that were associated with two and more than two disorders, respectively. (C) In the adult cortex, genes mapped to pleiotropic loci were enriched for frontal cortex specific genes, while genes mapped to non-pleiotropic loci are enriched for occipical cortex specific genes. (D) Genes mapped to 146 risk loci show higher expression values in neurons and oligodendrocytes, with much higher neuronal specificity for pleiotropic loci.
Gene-set enrichment analyses using Gene Ontology data suggested involvement of pleiotropic risk loci in neurodevelopmental processes (Table S6.1). The 109 pleiotropic risk loci were enriched for genes involved in neurogenesis (gene-set enrichment p = 9.67 × 10−6), regulation of nervous system development (p = 3.41 × 10−5), and neuron differentiation (p = 3.30 × 10−5), while enrichment of these gene-sets was not seen for the 37 disorder-specific risk loci (adjusted enrichment p > 0.05; Table S6.2). Pleiotropic risk loci also showed enrichment of genes involved in specific neurotransmitter-related pathways -- glutamate receptor signaling (p = 2.45 × 10−6) and voltage-gated calcium channel complex (p = 5.72 × 10−4) -- while non-pleiotropic risk loci, which were predominantly SCZ-associated, were over-represented among acetylcholine receptor genes (p = 7.25 × 10−8). Analysis of cortical gene expression data also suggested enrichment of pleiotropic risk genes in cortical glutamatergic neurons through layers 2–6 (Table S6.3), further supporting the shared role of glutamate receptor signaling in the pathogenesis of diverse neuropsychiatric disorders.
In contrast to the differences in neuronal development and neuronal signaling pathways, pleiotropic and non-pleiotropic risk loci shared several characteristics related to genomic function. For instance, gene-set enrichment analyses indicated that both pleiotropic and non-pleiotropic risk loci were enriched for genes involved in the regulation of synaptic plasticity, neurotransmission, and synaptic cellular components. More than 41% of the genes associated with our genome-wide significant loci, both pleiotropic and non-pleiotropic, were intolerant of loss of function mutations (pLI score ≥ 0.9); this is highly unlikely to occur by chance (Fisher’s exact p=4.90×10−8). This finding was consistent when examining pleiotropic (p=2.85×10−11) and non-pleiotropic risk loci (p=1.56×10−3) separately.
Next, we compared spatio-temporal gene-expression patterns for the 109 pleiotropic risk loci and the 37 disorder-specific loci using post-mortem brain data. On average, disorder-specific and pleiotropic risk loci showed a similar level of gene expression in both prenatal and postnatal development after multiple testing correction (t-test p > 0.025 ×10−2; Fig. S4). During prenatal development, non-pleiotropic loci (mainly SCZ-associated) showed peak expression in the first trimester, after which expression rapidly decreased, while pleiotropic genes associated with only 2 disorders (“pleiotropy=2”; 60 loci) and those associated with more than 2 (“pleiotropy>2”, 49 loci) showed peak expression around the second trimester (Fig. 5b). After birth, all three groups showed gradually increasing gene expression until adulthood. Expression levels were associated with the degree of pleiotropy, with the pleiotropy>2 group showing higher gene expression than either the pleiotropy=2 group (t-test p < 2.10×10−4) or non-pleiotropic risk loci (t-test p < 2.2×10−16).
Enrichment analyses using the genes preferentially expressed in specific cortical regions suggested that pleiotropic loci were over-represented among genes expressed in the frontal cortex, while non-pleiotropic loci were enriched in the occipital cortex (FDR q<0.05; Fig. 5c). Cell-type-specific analysis indicated that genes implicated in pleiotropic loci were mainly expressed in neurons (FDR q<0.05) but not in glial cell types. Further, enrichment of pleiotropic loci in neuronal cells was also associated with the degree of pleiotropy, as highlighted in Fig. 5d.
Previous studies of model organisms using gene knock-out experiments suggested that pleiotropic risk loci may undergo stronger selection than non-pleiotropic loci (Hill and Zhang, 2012). However, we found no evidence that pleiotropic risk variants are under stronger evolutionary constraints (Table S6.4). Various comparative genomics resources, including PhyloP (Pollard et al., 2010), PhastCons (Siepel et al., 2005), and GERP++ (Davydov et al., 2010), showed our top loci to have similar properties regardless of the extent of pleiotropy. Neither did we find differences between disorder-specific lead SNPs and pleiotropic SNPs with respect to their minor allele frequencies, average heterozygosity, or predicted allele ages (Kiezun et al., 2013). Pleiotropic and non-pleiotropic SNPs also did not differ in terms of the distance to nearest genes, distance to splicing sites, chromosome compositions, and predicted functional consequences of non-coding regulatory elements.
Relationship between cross-disorder genetic risk and other brain-related traits and diseases
To explore the genetic relationship of cross-disorder genetic risk with other traits, we treated this 8-disorder GWAS meta-analysis as a single “cross-disorder phenotype.” We applied LDSC to estimate SNP heritability (h2SNP) and genetic correlations with other phenotypes, using block jackknife-based standard errors to estimate statistical significance. The estimated h2SNP of the cross-disorder phenotype was 0.146 (SE 0.0058; observed scale). Using data for 25 brain-related traits selected from LDHub (Zheng et al., 2017), we found significant genetic correlations of the cross-disorder phenotype with seven traits (at a FDR-corrected p-value threshold 0.002): never/ever smoking status, years of education, neuroticism, subjective well-being, and three sleep-related phenotypes (chronotype, insomnia, and excessive daytime sleepiness) (Table S7.1).
GWAS catalog data for the 109 pleiotropic risk loci showed enrichment of implicated genes in a range of brain-related traits (Table S7.2). As expected, the associated traits included SCZ, BIP, and ASD. In addition, the pleiotropic risk loci were enriched among genes previously associated with neuroticism (corrected enrichment p= 5.28×10−6; GRIK3, CTNND1, DRD2, RGS6, RBFOX1, ZNF804A, L3MBTL2, CHADL, RANGAP1, RSRC1, GRM3), cognitive ability (corrected p= 7.15×10−5; PTPRF, NEGR1, ELOVL3, SORCS3, DCC, CACNA1I), and night sleep phenotypes (corrected p= 1.86×10−2; PBX1, NPAS3, RGS6, GRIN2A, MYO18A, TIAF1, CNTN4, PPP2R2B, TENM2, CSMD1). We also found significant enrichment of pleiotropic risk genes in multiple measures of body mass index (BMI), supporting previous studies suggesting a shared etiologic basis between a range of neuropsychiatric disorders and obesity (Hartwig et al., 2016; Lopresti and Drummond, 2013; Milaneschi et al., 2018).
DISCUSSION
In the largest cross-disorder GWAS meta-analysis of neuropsychiatric disorders to date, comprising more than 725,000 cases and controls across eight disorders, we identified 146 LD-independent lead SNPs associated with at least one disorder, including 35 novel loci. Of these, 109 loci were found to affect two or more disorders, although characterization of this pleiotropy is partly dependent on per-disorder sample size. Our results provide five major insights into the shared genetic basis of psychiatric disorders.
First, modeling of genetic correlations among the eight disorders using two different methods (EFA and hierarchical clustering) identified three groups of disorders based on shared genomics: one comprising disorders characterized by compulsive behaviors (AN, OCD and TS), a second comprising mood and psychotic disorders (MD, BIP and SCZ), and a third comprising two early-onset neurodevelopmental disorders (ASD and ADHD) and one disorder each from the first two factors (TS and MD). The loading of MD on two factors may reflect biological heterogeneity within MD, consistent with recent evidence showing that early-onset depression is associated with genetic risk for ADHD and with neurodevelopmental phenotypes (Rice et al., 2018). Overall, these results indicate a substantial pairwise genetic correlation between multiple disorders along with a higher-level genetic structure that point to broader domains underlying genetic risk to psychopathology. These findings are at odds with the classical, categorical classification of mental illness.
Second, variant-level analyses support the existence of substantial pleiotropy, with nearly 75% of the 146 genome-wide significant SNPs influencing more than one of the eight examined disorders. We also identified a set of 23 loci with particularly extensive pleiotropic profiles, affecting four or more disorders. The most highly pleiotropic locus in our analyses, with evidence of association with all eight disorders, maps within DCC, a gene fundamental to the early development of white matter connections in the brain (Bendriem and Ross, 2017). Prior studies showed that DCC is a master regulator of axon guidance (through its interactions with netrin-1 and draxin (Liu et al., 2018). Loss of function mutations in DCC cause severe neurodevelopmental syndromes involving loss of midline commissural tracts and diffuse disorganization of white matter tracts (Bendriem and Ross, 2017; Jamuar et al., 2017; Marsh et al., 2017). A highly pleiotropic effect of variation in DCC on diverse psychiatric disorders with childhood and adolescent onset would be consistent with its role in both early organization of neuronal circuits and the maturation of mesolimbic dopaminergic connections to the prefrontal cortex during adolescence (Hoops and Flores, 2017; Reynolds et al., 2018; Vosberg et al., 2018).
Third, we identified a set of loci that have opposite effects on risk of psychiatric disorders. Notably, these included loci with opposing effects on pairs of disorders that are genetically correlated and have common clinical features. For example, a SNP within MRSA was associated with opposing effects on two neurodevelopmental disorders (ASD and SCZ), and a variant within KIAA1109 had opposite directional effects on major mood disorders (BIP and MD) (Table S3.3). These results underscore the complexity of genetic relationships among related disorders and suggest that overall genetic correlations may obscure a more complex set of genetic relationships at the level of specific loci and pathways, as seen in immune-mediated diseases (Baurecht et al., 2015; Lettre and Rioux, 2008; Schmitt et al., 2016). This heterogeneity of effects between genetically correlated disorders is also consistent with a recent analysis that revealed loci contributing to biological differences between BIP and SCZ and found polygenic risk score associations with specific symptom dimensions (Bipolar Disorder and Schizophrenia Working Group of the Psychiatric Genomics Consortium, 2018). A complete picture of cross-phenotype genetic relationships will require understanding both same and opposite directional effects. In addition, to the extent that pleiotropic loci may reveal targets for drug discovery, opposite directional effects on psychiatric disorders could help anticipate problematic off-target effects.
Fourth, we found extensive evidence that neurodevelopmental effects underlie the cross-disorder genetics of mental illness. In addition to DCC, a link between pleiotropy and genetic effects on neurodevelopment was also seen for other top loci in our analysis, including RBFOX1, BRAF, and KDM7A, all of which have been shown in prior research to influence aspects of nervous system development. Gene enrichment analyses showed that pleiotropic loci were distinguished from disorder-specific loci by their involvement in neurodevelopmental pathways including neurogenesis, regulation of nervous system development, and neuron differentiation. These results are consistent with those of a smaller recent analysis in the population-based Danish iPSYCH cohort (comprising 46,008 cases and 19,526 controls across six neuropsychiatric disorders) (Schork et al., 2019). In that analysis, consistent with the present findings, functional genomic characterization of cross-disorder loci implicated fetal neurodevelopmental processes, with greater prenatal than postnatal expression. In addition, SORCS3 emerged as a genome-wide significant cross-disorder locus in both studies. However, other specific loci, cell types, and pathways implicated in the iPSYCH analysis differed from those identified in our study. In supplementary analyses, we did not find evidence of significant overrepresentation of genes related to pleiotropic SNPs identified here among previously defined genomic disorder regions or genes associated with neurodevelopmental disorders from rare variant studies (including ASD, intellectual disability, and developmental delay) (Samocha et al., 2017; Satterstrom et al., 2019) (Data S3.1–3.3).
Fifth, our analyses of spatiotemporal gene expression profiles revealed that pleiotropic loci are enriched among genes expressed in neuronal cell types, particularly in frontal or prefrontal regions. They also demonstrated a distinctive feature of genes related to pleiotropic loci: compared with disorder-specific loci, they are on average expressed at higher levels both prenatally and postnatally (Figure 5). More specifically, single-disorder (mainly SCZ) loci were related to genes that were preferentially expressed in the first fetal trimester followed by a decline over the prenatal period and then relatively stable levels postnatally. In contrast, average expression of genes related to pleiotropic loci peaked in the second trimester and remained overexpressed throughout the lifespan. When dividing the pleiotropic loci into bins of those associated with two disorders (mainly SCZ and BIP) vs. three or more disorders, we observed a consistent gradient of greater expression associated with broader pleiotropy. These results are based on average expression profiles, and not all individual gene expression patterns follow this pattern.
Taken together, our results suggest that pleiotropic loci appear to be distinguished by both their differential importance in neurodevelopmental processes and their heightened brain expression after the first trimester. Apart from this, however, pleiotropic loci were similar to non-pleiotropic loci across a range of other functional features, including intolerance to loss-of-function mutations, evidence of selection, minor allele frequencies, and genomic position relative to functional elements.
Overall, our results identify a range of pleiotropic effects among loci associated with psychiatric disorders. Consistent with prior research (Brainstorm Consortium, 2018; Cross-Disorder Group of the Psychiatric Genomics Consoritum, 2013), we found substantial pairwise genetic correlations across child- and adult-onset disorders and extended these findings by demonstrating clusters of genetically-related disorders. These results augment a substantial body of research demonstrating that genetic influences on psychopathology do not map cleanly onto the clinical nosology instantiated in the DSM or ICD (Geschwind and Flint, 2015; Smoller et al., 2019) Using a range of bioinformatic and functional genomic analyses, we find that loci with pleiotropic effects are distinguished by their involvement in early neurodevelopment and increased expression beginning in the second trimester of fetal development and persisting throughout adulthood.
Taken together, the analyses presented here suggest that genetic influences on psychiatric disorders comprise at least two general classes of loci. The first comprises a set of genes that confer relatively broad liability to psychiatric disorders by acting on early neurodevelopment and the establishment of brain circuitry. These pleiotropic genes begin to come online by the second trimester of fetal development and exhibit differentially high expression thereafter. The expression and differentiation of this generalized genetic risk into discrete psychiatric syndromes (e.g., ASD, BIP, AN) may then involve direct and/or interactive effects of additional sets of common and rare loci and environmental factors, possibly mediated by epigenetic effects, that shape phenotypic expression via effects on brain structure/function and behavior. Further research will be needed to clarify the nature of such effects.
Our results should be interpreted in light of several limitations. First, while our dataset is the largest genome-wide cross-disorder analysis to date, data available for individual disorders varied substantially—from a minimum of 9,725 cases and controls for OCD to 461,134 cases and controls for MD. This imbalance of sample size may have limited our power to detect pleiotropic effects on underrepresented disorders. The future availability of larger samples will improve power for detection of cross-disorder effects. Second, it is possible that comorbidity among disorders contributed to apparent pleiotropy; we found, however, that fewer than 2% of cases overlapped between disorder datasets (excluding 23andMe data) and we adjusted for sample overlap in meta-analysis. Third, the method we applied to detect cross-phenotype association, which combines an all-subsets fixed-effects GWAS meta-analysis with a Bayesian method for evaluating the best-fit configuration of genotype-phenotype associations, is one of several approaches (Solovieff et al., 2013). However, we have previously shown that this method outperforms a range of alternatives for detecting pleiotropy under various settings (Zhu et al., 2018). Fourth, our designation of loci as pleiotropic vs. non-pleiotropic loci refers only to their observed effects on the eight target brain disorders. Thus, some of the “non-pleiotropic” loci may have additional effects on psychiatric phenotypes that were not included in our meta-analysis and/or on non-psychiatric phenotypes. Fifth, our functional genomic analyses were constrained by the limitations of existing resources (e.g. spatiotemporal gene expression data resources). Our work underscores the need for more comprehensive functional data including single cell transcriptomic and epigenomic profiles across development and brain tissues. Lastly, we included only individuals of European ancestry to avoid potential confounding due to ancestral heterogeneity across distinct disorder studies. Similar efforts are needed to examine these questions in other populations.
In sum, in a large-scale cross-disorder genome-wide meta-analysis, we identified three genetic factors underlying the genetic basis of eight psychiatric disorders. We also identified 109 genomic loci with pleiotropic effects, of which 33 have not previously been associated with any of the individual disorders. In addition, we identified 11 loci with opposing directional effects on two or more psychiatric disorders. These results highlight disparities between our clinically-defined classification of psychiatric disorders and underlying biology. Future research is warranted to determine whether more genetically-defined influences on cross-diagnostic traits or subtypes of dissect may inform a biologically-informed reconceptualization of psychiatric nosology. Finally, we found that genes associated with multiple psychiatric disorders are disproportionately associated with biological pathways related to neurodevelopment and exhibit distinctive gene expression patterns, with enhanced expression beginning in the second prenatal trimester and persistently elevated expression relative to less pleiotropic genes. Therapeutic modulation of pleiotropic gene products could have broad-spectrum effects on psychopathology.
STAR* METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
Any inquiries about analytical results or other information should be directed to Lead Contact, Jordan W. Smoller (jsmoller@mgh.harvard.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Genotyped sample description
Genotype data from eight studies of genetic associations with psychiatric disorders conducted by the Psychiatric Genomics Consortium were included in this report. A summary of each study is provided below, however, detailed sample descriptions are available in the primary publication. The lead PI of every cohort included across studies certified that their protocol was approved by their local Ethical Committee. Supplementary Table S1 lists for each disorder the number of cases and controls, the number of loci identified in the single disorder genome-wide association study, and SNP-based heritability.
Schizophrenia | Ripke et al., 2014
108 loci were identified as associated with schizophrenia in a case-control meta-analysis including 150,064 individuals. For the current study, the 46 case-control cohorts of European ancestry were retained, totaling 33,640 cases and 43,546 controls. Cases were defined as individuals diagnosed with schizophrenia or schizoaffective disorder, which was determined by research-based assessment or clinician diagnosis depending on the sample.
Bipolar disorder | Stahl et al., 2019
Thirty-two case-control cohorts from Europe, North America, and Australia including 20,352 cases and 31,358 controls of European ancestry were meta-analyzed to identify 30 loci associated with bipolar disorder. Cases met criteria for lifetime diagnosis of bipolar disorder as defined by DSM-IV, ICD-9, or ICD-10, which was established using interview-based structured assessment, clinician-administered checklists, or review of medical records. All subjects in the meta-analysis were included in the current study.
Major depression | Wray et al., 2018
Seven case-control cohorts were combined to identify 44 loci associated with major depression. The first cohort included 29 case-control samples of European descent where lifetime diagnosis of major depressive disorder was ascertained using structured clinical interviews (DSM-V, ICD-9, ICD-10), clinician-administered checklists, or review of medical records. Six additional cohorts of European ancestry, including the Hyde et al study (23andMe, Inc), determined case status using other methods including national or hospital treatment registers, self-reported symptoms or treatment by a medical professional, or direct interviews. Analyses comparing the original cohort with the additional ones indicated strong correlation of common genetic variants and little evidence of heterogeneity. 130,664 cases and 330,470 controls from these cohorts were included in the current analyses.
Attention deficit hyperactive disorder | Demontis et al., 2019
Twelve cohorts of European, North American, and Chinese descent were aggregated in a meta-analysis of attention deficit and hyperactive disorder, revealing 12 associated loci. For the first cohort, cases were ascertained using the Danish Psychiatric Central Research Registrar and diagnoses were confirmed by psychiatrists according to ICD-10. The remaining studies included four parent-offspring trio cohorts and seven case-control cohorts. Cases were recruited from clinics, hospitals or through medical registries and diagnosed using research-based assessments administered by clinicians or trained staff. 19,099 cases and 34,194 controls of European ancestry were included in the current study.
Autism spectrum disorder | Grove et al., 2019
Five family-based cohorts of European descent and a population-based case-control sample from Denmark were combined to discover five loci associated with autism spectrum disorder. In each family study, diagnosis was confirmed for all affected individuals using standard research tools and expert clinical consensus diagnosis. In the population-based cohort, cases were identified using the Danish Psychiatric Central Research Register and were diagnosed with ASD before 2013 by a psychiatrist according to ICD-10. All subjects in this sample were included here (18,381 cases; 27,969 controls).
Obsessive compulsive disorder | IOCDF-GC and OCGAS, 2018
Individuals of European descent from two cohorts were combined in this meta-analysis including 2,688 cases and 7,037 controls; no loci reached genome-wide significance. Case diagnoses were established using DSM-IV criteria and controls were unscreened. All cases and controls were included in the current analyses.
Anorexia nervosa | Duncan et al., 2017
3,495 cases from two consortia and 10,982 matched controls from the Psychiatric Genomics Consortium, all of European descent, were meta-analyzed to identify one locus associated with anorexia nervosa. Cases met criteria as defined by DSM-IV for lifetime diagnosis of anorexia nervosa (restricting or binge-purging subtype), bulimia nervosa, or anorexia nervosa – not otherwise specified, anorexia nervosa subtype. All individuals included in the primary study were included in the current analyses.
Tourette Syndrome | Yu et al., 2019
Three case-control cohorts and one family-based cohort from Europe and North America including 4,819 cases and 9,488 controls of European ancestry were meta-analyzed to identify one locus associated with Tourette Syndrome. All cases met DSM-IV-TR or DSM-5 criteria for Tourette syndrome, except for 12 cases who met DSM-5 criteria for chronic motor or vocal tic disorder. All cases were recruited by Tourette syndrome specialty clinics or by email/online recruitment combined with validated, web-based phenotypic assessments.
Genotype quality control, imputation, and association analysis
All primary studies used the standardized PGC ricopili pipeline for quality control, imputation and association testing. Briefly, for each dataset, poor quality SNPs and samples missing >5% SNPs were removed. Next, pre-phasing and imputation were implemented using IMPUTE2 (Howie et al., 2011) and the 1000 Genomes reference panel. High quality SNPs (INFO > 0.8) with low missingness (<1%) were retained. A subset of these markers (MAF > 0.05; pruned for linkage disequilibrium, r2 > 0.02) were used to assess relatedness and population stratification. Only one of any pair of related individuals was retained. Each imputed dataset was tested for association with the disease outcome of interest using an additive logistic regression model in PLINK (Purcell et al., 2007) with age, sex, and 10 principal components included as covariates. Finally, a meta-analysis within each disease category was done using an inverse-weighted fixed effects model. After extracting SNPs commonly exist in all eight disorder studies, we removed 3,591 SNPs whose alleles were incompatible. For palindromic SNPs, we compared allele frequencies between eight studies to check strand ambiguity. 50 SNPs with frequency difference greater than 15% from the 1KG reference was excluded. As a result, 6,786,993 autosomal SNPs remained for further analysis.
QUANTIFICATION AND STATISTICAL ANALYSIS
Genome-wide SNP-heritability estimation
For each of the eight GWAS disorders, LD Score regression was performed on the summary statistics of individual disease using LDSC to estimate SNP-based heritability in the liability scale and genetic correlation between pairs of disorders (Bulik-Sullivan et al., 2015b). LD scores and weights for European populations were downloaded from the LDSC website (http://www.broadinstitute.org/~bulik/eur_ldscores/). SNPs were removed if the minor allele frequency is smaller than 5% or an imputation quality score is less than 0.9; MHC region was excluded from the analysis. For single-trait LDSC, the slope of the regression estimates the SNP-based heritability, and the intercept greater than one captures the inflation in the summary statistics due to population stratification or other confounding factors. We confirmed that the heritability Z-scores (i.e., a measure of the polygenic signals) are greater than four, and the LDSC intercepts are approximately one and less than. suggesting that the increase in mean χ2 statistics is due to polygenicity and not due to stratification.
Factor analysis and genomic SEM
Genomic SEM’s Multivariable LD score regression method (Grotzinger et al., 2019) was first used to estimate the genetic covariance matrix (S) and sampling covariance matrix (V) for the eight psychiatric traits. Quality control for this step included removing SNPs with an MAF < 1%, information scores < .9, SNPs from the MHC region, and filtering SNPs to HapMap3. All SNP effects were standardized using the sumstats function in Genomic SEM. To examine genome-wide factor structure, models using only the genetic covariance and sampling covariance matrix were fit. Genomic SEM provides indices of model fit—standardized root mean square residual (SRMR), model 2, Akaike Information Criteria (AIC), and Comparative Fit Index (CFI)—that can be used to determine how well the proposed model captures the observed data. Model fit for the common factor model in which the loadings were freely estimated was only fair, (2 (20) = 313.94, AIC = 345.9, CFI = .786, SRMR = .149), suggesting that there were nuances in the genetic architecture not fully captured by a single cross-trait index of genetic risk. An exploratory factor analysis (EFA) of the S matrix with three-factors using the promax rotation in the R package factanal was then used to guide construction of a follow-up model (Table S2.2). A follow-up confirmatory model with three correlated factors was specified in Genomic SEM based on the EFA parameter estimates (positive standardized loadings > .2 were retained; Figure 2b). This model provided good fit to the data (2 (15) = 85.35, AIC = 127.36, CFI = .945, SRMR = .079). Results indicated there was a moderate genetic correlation between the compulsive and mood/psychotic disorders factors (rg = .43, SE = .08), a smaller genetic correlation between the mood/psychotic and early onset factors (rg = .25, SE = .05), and next to no correlation between the compulsive and early onset factors (rg = < .01, SE = .07). A model that included additional negative cross-loadings provided similar fit to the data and highly similar correlations across the genetic factors. Given this consistency in results, the correlated factors model with SNP effects only included positive loadings.
Summary-data-based meta-analysis
To identify genomic loci shared across multiple neuropsychiatric disorders, we performed primary meta-analysis using the subset-based fixed-effects method ASSET (Bhattacharjee et al., 2012). Standard meta-analysis pools the effect of a given SNP across K studies, weighting the effects by the size of the study. By exhaustive investigation of all subset-based effects, the maximum SNP effect was identified as:
where the absolute value of the subset-specific effect [Z(S)] over class S of all possible subsets of K studies is highest. The numbers of shared subjects across eight disorder studies were identified using the PGC checksum algorithm, and Zmeta was standardized so that covariance between the statistics can be accounted for as previously described (Bhattacharjee et al., 2012; Lin and Sullivan, 2009). Tail probabilities for the distribution of the maximum, adjusting for multiple testing of all combination of subsets, were then estimated with the discrete local maxima method, which uses the correlation structure of test statistics across subsets. Based on the derived p-value, standard deviation of the SNP effect was adjusted to reflect the multiple-testing correction. Even when correcting for all subset tests (2K-1), simulations suggest there is a substantial gain in power using this test relative to traditional meta-analysis (Bhattacharjee et al., 2012. Standardized genomic inflation factor (λ1000) for the meta-analysis result was close to one. LDSC intercept was substantially less than λGC (0.79 vs 1.55), suggesting that the increase in mean χ2 statistics in the cross-disorder meta-analysis is mainly due to polygenicity and not due to stratification or other confounding biases.
Once SNPs with genome-wide significant association were identified, we identified LD-independent genomic regions using PLINK clumping (--clump-r2=0.4, --clump-kb=500, --clump-p1=5e-08, --clump-p2=5e-02). Genomic regions were merged if they physically overlap using bedtools. Due to extensive LD, the MHC region was considered as one region (chr6:25–35Mb). To detect secondary signals independent of index SNP in each of the candidate cross-disorder loci, conditional analysis was performed with GCTA-COJO (Yang et al., 2012) using meta-analysis summary statistics from ASSET. 1KG EUR population was used as the reference panel for estimating LD. For each genomic region harboring a cross-disorder signal, we tested the presence of any additional associated SNPs using a stepwise procedure (--cojo-slct), conditioning on the primary significant SNP for model initiation. A conditional p-value for each variant was reported, adjusted for genomic control and collinearity. In each region, additional SNPs were selected as a distinct association signal if having a conditional p-value < 1e-06.
Disease-association modeling
We estimated posterior probabilities for each of the top loci identified from the meta-analysis to quantify disorder-specific effects (Han and Eskin, 2012). This estimation, known as the m-value, relies on two assumptions, 1) effects are either present or absent in studies, and 2) if they are present, they are similarly sized across studies. Assume Xi is the observed effect size of study i, and Ti is a random variable with value 1 if study i has an effect and 0 if not, then the m-value can be estimated using Bayes’ theorem:
which can then be used to predict whether an effect exists in a given study (>.9) or not (<.1) under the binary effects assumption.
Examination of the Impact of Sample Size Imbalance on Genetic Correlations and Genomic SEM Results
We conducted several analyses to examine whether differences in sample size among the 8 disorders influenced the pattern of cross-disorder genomic relationships we observed. First, we note that while sample size will affect the precision of a genetic correlation estimate (ie standard error) it should not affect the magnitude of the estimate itself (Bulik-Sullivan et al. 2015). As shown in Data S1.2, there is no substantial relationship between the estimated genetic correlations and the effective sample sizes of the corresponding disorder pairs (p-value for the slope = 0.055 ). The slightly positive linear relationship appears to be driven by MD and its genetic correlation with the other four major psychiatric disorders (SCZ, BIP, ASD, ADHD), however, these estimates are generally consistent with previously reported ones when sample sizes are much smaller (except for ASD) (Brainstorm et al., 2018) (Cross-Disorder Group of the Psychiatric Genomics et al., 2013) (Data S1.3). Furthermore, the largest among all pairwise comparisons, such as those between SCZ-BIP, AN-OCD, and ADHD-AN, do not scale with sample size.
Next, we investigated the impact of variable sample sizes on the Genomic SEM analysis results by re-running Genomic SEM analysis using a Maximum Likelihood (ML) estimator that does not take into account the differing precisions of the genetic covariance estimates (resulting from, for example, uneven sample sizes across traits) when optimizing parameters. As shown in Data S1.4, the results were consistent with those from the primary analysis reported in the main text that is based on a Weighted Least Squares (WLS) estimator, which does take into account the differing precisions of the genetic covariance estimates. Specifically, the nontrivial standardized factor loadings of MD on two of the three factors is evident in both the WLS and ML solutions and is therefore unlikely to be an artifact of its large N. Note that, in both the WLS and the ML solution, the standard errors are smaller for the loadings involving the better-powered GWAS phenotypes, as we would expect.
To further evaluate whether sample size imbalance across the eight disorders biased the number of pleiotropic signals we observed, we conducted simulation studies of UK Biobank data. In particular, we examine whether the number of pleiotropic loci we identified exceeds chance expectation given the sample size and genetic correlations among the eight disorders. We used the full release of 488,377 UK Biobank (UKBB; (Sudlow et al., 2015)) individual data, imputed with the Haplotype Reference Consortium (HRC), UK 10K, and 1000 Genomes reference panels (under the application number 31063). Data was QC’ed as described in the Neale Lab UK BIOBANK GWAS webpage (http://www.nealelab.is/uk-biobank/), including 361,194 unrelated individuals of Caucasian ancestry and 13.7 million genetic variants (MAF > 0.0001, INFO > 0.8). For the purpose of the simulation, we removed individuals who were in the UKBB interim release to avoid sample overlap with the MD GWAS where these subjects were included (Wray et al., 2018) and restricted the analysis to variants present in both the current study (PGC-CDG2) and the UKBB datasets, resulting in 6,691,733 SNPs.
Because SCZ and MD accounted for the majority of the total sample size in our study as well as the two most statistically powerful studies (estimated by calculating their effective sample size and multiplying that by heritability), we generated simulated datasets similar in size and heritability, as well as cross-correlation to the other datasets, for each of the six smaller studies (BIP, ADHD, ASD, TS, ANO, and OCD); In brief, simulated genetic data was created from the post-QC UKBB imputed data for each of the six disorders by randomly selecting subjects without any overlap given their original sample sizes. In each simulation replicate, we then simulated quantitative phenotypes (Y = ) given true effect sizes, the standardized genotype matrix X, and a non-genetic error term. The true effect sizes of each SNP were drawn from a multivariate normal distribution, where M is the total number of SNPs in the genome, μ is a zero vector of length 6, and ∑ is the covariance matrix that accounts for the genetic correlations (rg) among the six disorders (with disease-specific SNP-heritabilities on the diagonal and hihjrg,ij on the off-diagonals). Individual phenotypes were then generated by calculating the sum of betas weighted by the standardized allele dosages (mean 0 and variance 1) with the --score variance-standardize option in PLINK2 v2.00a2LM (Chang et al., 2015) and a noise term drawn from N(0,) for each disorder. Case-control phenotypes were generated by sorting Y in descending order and assigning the top fcase to be cases, where fcase corresponds to the fraction of cases of each disorder in the original GWAS. Association statistics were estimated using logistic regression, assuming an additive effect of alleles. We then matched the reference and the alternate alleles in UKBB to those in the current study and reversed the sign of the effect sizes when necessary. We then performed meta-analysis using ASSET (Bhattacharjee et al., 2012) and estimated m-values as was done in the original analysis. Finally, we compared the distribution of the number of pleiotropic loci across the 100 simulation replicates against the observed value in the actual study. For this analysis, we focused on chromosome 1 where the largest number of cross-disorder associations were identified in the actual analysis. Data S1.5 displays the distribution of the number of cross-disorder loci identified in meta-analysis of chromosome 1 across 100 simulation replicates. We compared this to the number of pleiotropic loci found in our meta-analysis compared to those seen in the simulations, given the sample size and genetic correlations among the eight disorders to determine whether the observed number of pleiotropic loci exceeds chance expectation.
Functional annotation and gene-mapping of genome-wide significant variants
For the 146 genome-wide significant variants, gene mapping and functional annotation was conducted using various resources, including SNPNexus (Dayem et al., 2018) and FUMA (Watanabe et al., 2017). Nearest genes and functional consequence of each SNP on gene functions were annotated based on ANNOVAR (Wang et al., 2010). Combined Annotation Dependent Depletion (CADD) score (Kircher, 2014) indexes the deleteriousness of variants computed based on 67 annotation resources. SNPs with the CADD score higher than 12 were considered to confer deleterious effects. The RegulomeDB (Boyle, 2012) provides a categorical score that describes how likely a SNP is likely to play a regulatory role based on the integration of high-throughput datasets. The RDB score of 1a suggests the strongest evidence, while the score 7 represents the least support for a regulatory potential. The minChrState and the commonChrState represent the minimum and the most common15-core chromatin state across 127 tissue/cell type predicted by ChrHMM. The chromatin state of less than 8 suggests an open chromatin state. eQTL mapping provides significant cis-SNP-gene pairs (up to 1Mb apart) in brain tissue types from GTEx and BRAINEAC.
For chromatin interaction mapping, we first refined the localization of potential causal variants for top 146 lead SNPs using FINEMAP (Benner et al., 2016). For each region, we considered only SNPs located in the LD region with the lead SNP (r2 > 0.6). We then applied the method to calculate the posterior probability of being causal for each of the remaining SNPs. A 95% credible set of SNPs for each region was constructed by ordering the posterior probability from largest to smallest and selecting in the corresponding SNPs up to a cumulative probability of 95%. Credible SNPs were then grouped into those that are located within the promoter or exons and those that are non-coding/intronic. Promoter/exonal SNPs were directly assigned to their target genes using positional mapping, while non-coding/intronic SNPs were assigned to their target genes based on long range interactions (Hi-C) or expression quantitative trait loci (eQTLs). Two Hi-C datasets originated from the human brain (fetal brain Hi-C (Won et al., 2016) and adult brain Hi-C (Wang et al., 2018)) were used to map credible SNPs to remotely interacting genes as previously described (Wang et al., 2018). A colocalization analysis with the recent eQTL dataset from adult prefrontal cortices (PFC) was also used to map 146 GWS loci into their target genes (Wang et al., 2018). In the end, we obtained two sets of candidate genes, one from fetal brain (positional mapping, fetal brain Hi-C), the other from adult brain (positional mapping, adult brain Hi-C, adult brain eQTLs).
GTEx gene expression enrichment analysis
MAGMA gene-property analysis (de Leeuw et al., 2015) was performed using gene expression data from 83 tissues based on GTEx RNA-seq data (v7). Expression values (RPKM) were log2 transformed with pseudo-count one after winsorization at 50, and average expression values were taken per tissue. Analysis was performed separately for 30 general tissue types and 53 specific tissue types, and Bonferroni-based multiple testing correction was done for the examined tissue types.
Pathway analysis using Gene Ontology
We used FUMA (Watanabe et al., 2017) to map SNPs to genes and then test for enrichment of specific Gene Ontology functions and pathways among genome-wide significant pleiotropic and disorder-specific SNPs separately. Hypergeometric tests identify any statistical over-representation of genes from the input list (mapped from SNPs) in predefined MSigDB Gene Ontology gene sets which describe biological processes, molecular functions, and cellular components. Multiple test correction was applied by category.
Enrichment analysis using brain developmental, regional, and cell-type-specific data
Developmental expression trajectories for candidate genes were plotted using a published transcriptome atlas constructed from post-mortem brain data (Kang et al. 2011). As this dataset contains expression values from multiple brain regions, we selected transcriptomic profiles of cerebral cortex with developmental epochs that span prenatal (6–37 post-conception weeks, PCW) and postnatal (4 months-42 years) periods. Expression values were log-transformed and centered to the mean expression level for each sample using a scale(center=T, scale=F)+1 function in R. This normalization method has been frequently used in other papers to plot developmental expression trajectories (e.g. (Grove et al., 2019b; Li et al., 2018; Mah and Won, 2019; Satterstrom et al., 2019). Instead of measuring the expression values of individual disease associated gene, we measured the average expression values of the entire gene set. To do this, disease risk genes were selected for each sample and their average centered expression values were calculated and plotted (individual dots in the plot denote different samples or individuals, not different genes). It is of note that the average expression values each gene set correspond to representative expression patterns of the disease risk genes, so individual genes may behave differently.
We used candidate genes identified in fetal brain and adult brain to plot prenatal and postnatal gene expression profiles, respectively.
To obtain genes that show cortical regional enrichment (e.g. frontal cortical enrichment), we computed t-statistics for each gene for a specific cortical region (e.g. frontal cortex) versus all other cortical regions (e.g. parietal cortex, temporal cortex, and occipital cortex, Kang et al. 2011). The top 5% of genes that show heightened expression patterns for each cortical region were selected as region-specific genes. These genes were then overlapped with candidate genes by Fisher’s exact test to measure cortex regional enrichment.
Single cell expression profiles from the adult brain (Darmanis et al., 2015) were used to identify cell-type specificity of candidate genes. Single cell expression values were log-transformed and centered using the mean expression values. Average centered expression values for candidate genes were calculated in each cell. Cells were then grouped into cell clusters (neurons, astrocytes, microglia, oligodendrocytes, OPC, and endothelial cells), and a relative expression level for a given cell cluster was calculated by a scale function in R.
Comparison with other brain-related traits and diseases
To explore the genome-wide relationship of our cross-disorder phenotype with other traits and diseases, we estimated pairwise genetic correlations using LD Hub (Zheng et al., 2017). We selected 25 brain-related traits from LD Hub, including phenotypes related to smoking behavior, education, personality, neurological disorders, sleeping, cognitive function, and brain volume (Table S7.1). Summary statistics for different phenotypes were harmonized via the default options provided by LD Hub, and SNPs in the MHC regions were removed before the analysis. For each of the selected traits, a bivariate LDSC analysis was performed to estimate its genetic correlation with our meta-analyzed cross-disorder phenotype. We then applied FDR correction to control for multiple testing and identify significant associations.
For GWAS catalog data, FUMA (Watanabe et al., 2017) GENE2FUNC module was used to test for enrichment of specific GWAS catalog-associated gene sets for genome-wide significant pleiotropic risk loci. Hypergeometric tests identified any statistical over-representation of genes from the input list in predefined GWAS catalog data. Human protein-coding genes were used as background genes. All identified traits with multiple-testing adjusted P < 0.05 were included as results.
Relationship of Lead SNPs from Meta-analysis to Rare CNVs and Mutations Previously Associated with Neurodevelopmental Genomic Disorders
We conducted additional analyses to determine whether our 146 genome-wide significant loci are enriched in CNVs spanning defined genomic disorder (GD) regions or damaging mutations previously shown to be associated with neurodevelopmental disorders (including autism spectrum disorder, intellectual disability, and developmental delay), also known as genomic disorders (GDs). The reference data comprise a curated set of 51 GD loci (encompassing 823 protein-coding genes) with multiple reports of ASD/ID/DD-associated CNVs (Satterstrom et al., 2019). The GD curation process is described in the original publication. Each of our 146 lead SNPs were assigned to its candidate genes using various functional genomics datasets including Hi-C data, overlap with gene and regulatory elements. We examined all SNPs as well as dividing SNPs into groups based on their degree of pleiotropic association and conducted permutation testing to assess significant enrichment. Permutation testing was performed by first assigning each lead (sentinel) SNP to the nearest gene, then randomly sampling 1,000 new genes from the genome with replacement while matching on chromosome and gene length. P-values were derived by comparing the empirically observed number of overlaps to the distribution of expected overlaps based on 1,000 matched permutations (Data S3.1).
We also examined overlap of our 146 genomewide significant loci with genes containing damaging de novo (truncating, highly damaging missense and damaging missense) mutations among children with ASD (data from (Satterstrom et al., 2019)). In this autism dataset, 102 genes had higher frequencies of damaging de novo mutations (DNMs) in cases than controls (FDR q ≤ 0.1) (Satterstrom et al., 2019). Each permutation test consisted of randomly sampling 1,000 new sets of genes with replacement from the genome, where each new set of genes contained the same total number of genes as the observed set of candidate genes for each set of loci. Sampling was also performed while controlling for per-gene mutation rates and brain expression levels using a quantile-based binning approach, as has been described in detail in a recent study (Satterstrom, et al., 2019). P-values were derived by comparing the empirically observed number of genes present in the list of 102 dominant-acting ASD risk genes to the distribution of expected count of dominant-acting ASD risk genes based on 1,000 matched permutations (Data S3.2).
Finally, we examined whether genes linked to our SNPs were enriched for DNMs associated with ASD using the same reference data set. Each permutation test consisted of randomly sampling 1,000 new sets of genes with replacement from the genome, where each new set of genes contained the same total number of genes as the observed set of candidate genes for each set of loci. Sampling was also performed while controlling for per-gene mutation rates and brain expression levels using a quantile-based binning approach, as has been described in detail in a recent study (Satterstrom, et al., 2019). P-values were derived by comparing the empirically observed number of genes present in the list of 102 dominant-acting ASD risk genes to the distribution of expected count of dominant-acting ASD risk genes based on 1,000 matched permutations (Data S3.3).
DATA AND SOFTWARE AVAILABILITY
The Psychiatric Genetics Consortium (PGC)’s policy is to make genome-wide summary results publicly available. Summary statistics for a combined meta-analysis of eight psychiatric disorders without 23andMe data are available on the PGC web site (https://www.med.unc.edu/pgc/results-and-downloads). Results for 10,000 SNPs for eight disorders including 23andMe are also available on the PGC web site. The summary-level GWAS association statistics for PGC individual disorders are available at the website (https://www.med.unc.edu/pgc/results-and-downloads).
GWAS summary statistics for the 23andMe cohort (Hyde, 2016) must be obtained separately. These can be obtained by individual researchers under an agreement with 23andMe that protects the privacy of the 23andMe participants. Contact Aaron Petrakovitz (apetrakovitz@23andme.com) to apply for access to the data.
Supplementary Material
Table S1. Summary of eight neuropsychiatric disorder datasets (Related to Table 1). Disease-specific cases and controls included in the meta-analysis, number of individual GWAS loci, and liability-based SNP heritability estimates are provided. Heritability was estimated from available European summary statistics using LD score regression.
Figure S1. Statistical power and number of cross-disorder associations (Related to Table 2 and Table S3.1). Power to detect associations across pairs of disorders was plotted with the number of cross-disorder associations identified in the current meta-analysis. For each pair of disorders, power was estimated using the number of cases and heritability for each disorder, as well as the genetic correlation between the disorders. In general, as power increased, so did the number of identified SNPs.
Figure S2. Cross-disorder GWAS SNPs in the DCC locus colocalize with eQTLs for DCC (Related to Table 2, Table S4.2, and Figure 3). Dark blue dots refer to SNPs that overlap between brain eQTLs and GWAS plots. LD region for the GWAS locus and TAD boundaries are depicted.
Figure S3. Gene expression of top loci across development (Related to Figure 3). Gene expression trajectories from a transcriptome atlas of post-mortem brain tissue across development are plotted for four top loci, DCC, RBFOX1, NOX4 and BRAF in six different brain tissue types. AMY = amygdala; MD = mediodorsal nucleus of the thalamus; CBC = cerebellar cortex; NCX = neocortex; HIP = hippocampus; STR = striatum.
Figure S4. Gene expression in the brain for pleiotropic and non-pleiotropic loci (Related to Figure 5). Average normalized gene expression in fetal and adult post-mortem brain tissue for pleiotropic (109) and non-pleiotropic (37) loci were plotted. Disorder-specific and pleiotropic risk loci showed a similar level of gene expression in prenatal and postnatal development after multiple testing correction (t-test p > 0.025).
Supplemental Datafile 1: Additional analysis results to assess the robustness of genetic correlation and Genomic SEM study (Related to Figure 1). Data S1.1. displays the hierarchical clustering of genetic architecture across eight neuropsychiatric disorders. Data S 1.2. summarizes the relationship between the estimated genetic correlation and the effective sample sizes for each pair of disorders. Data S1.3. shows the relationship between the estimated genetic correlation and the effective sample sizes for MD and four other disorders, SCZ, BIP, ADHD, and ASD. Data S1.4. depicts the genomic SEM analysis result using a Weighted Least Squares estimator and that based on a Maximum Likelihood estimator. Data S1.5. summarizes the distribution of the number of cross-disorder loci identified in meta-analysis of chromosome 1 across 100 simulation replicates.
Supplementary Datafile 2: P-M plots for 146 LD-independent genome-wide significant loci (Related to Figure 3). P-M plot is displayed on the right side for each SNP locus. The X-axis on a P-M plot represents M-values estimated for each locus for each disorder, while the Y-axis displays the significance of disease-SNP association. Disorders displayed in red dots were predicted to have an effect (M-value 0.9), disorders in blue were predicted to not have an effect (M-value 0.1), and studies in green indicated its effect is uncertain (0.1< M-value < 0.9). The dot size reflects the relative sample size of the studies. On the left side, a forest plot displays log odds ratios for the effect of the SNP on each disorder and a meta-analyzed effect across disorders. As expected, most of the top SNPs (109/146) show pleiotropic effects.
Supplementary Datafile 3: Analyses of rare CNVs and mutations previously associated with neurodevelopmental genomic disorders (Related to STAR Methods section “Relationship of Lead SNPs from Meta-analysis to Rare CNVs and Mutations Previously Associated with Neurodevelopmental Genomic Disorders”) Data S3.1. shows that sentinel SNPs are not overrepresented among genomic disorder loci. Data S3.2. and Data S3.3. indicate that genes linked to sentinel SNPs are not overrepresented among dominant-acting ASD risk genes and for damaging de novo mutations in ASD cases, respectively.
Table S2. Tables Related to STAR Methods, Figure 1 and Figure S1. Table S2.1. displays the genetic correlations estimated by LD score regression, while Table S2.2. shows the results from EFA of genetic covariance matrix.
Table S3. Tables Related to Figure 2, 3, and 4. Table S3.1. displays the list of 146 lead SNPs, and Table S3.2. shows the M-value estimates for quantifying confidence of disorder-specific association for all 146 lead SNPs. Table S3.3. summarizes the loci with opposite directional effects.
Table S5: Tables Related to Figure 5, Gene Expression Enrichment Analyses. Table S5.1. summarizes the GTEx gene enrichment analysis results using MAGMA. Table S5.2. and S5.3 show the tissue enrichment analysis for pleiotropic and disorder-specific risk loci using MAGMA (GTEx v7), respectively.
Table S6: Tables Related to Figure 5. Table S6.1. and S6.2 summarize the Gene ontology analysis for pleiotropic and disease-specific risk loci, respectively. Table S6.3. lists the enrichment analysis results of cortical gene expression data. Table S6.4. shows the comparison of disease-specific vs. pleiotropic risk loci by various functional and genomic features.
Table S4: Tables Related to Table 2, Table S3.1. Table S4.1. summarizes the functional annotation of 146 lead SNPs, while Table S4.2. lists the Brain eQTL and Hi-C data annotation based on FUMA database. Table S4.3. lists the GWAS catalog data for lead SNPs.
Table S7: Tables Related to STAR Methods section “Comparison with other brain-related traits and diseases” - Relationship between Cross-disorder Genetic Risk and other Brain-related Traits and Diseases. Table S7.1. summarizes the genetic correlation analysis of the cross-disorder phenotype with other brain-related traits. Table S7.2. shows the GWAS catalog enrichment analysis for pleiotropic risk loci.
KEY RESOURCES TABLE.
ACKNOWLEDGMENTS
The work of the contributing groups was supported by numerous grants from governmental and charitable bodies as well as philanthropic donation. Specifically, P.H.L. (R00MH101367; R01MH119243), and J.W.S. (R01MH106547; R01MH117599; U01HG008685). The PGC has been supported by the following grants: MH085508, MH085513, MH085518, MH085520, MH094411, MH094421, MH094432, MH096296, MH109499, MH109501, MH109514, MH109528, MH109532, MH109536, MH109539. Funding for the work in Bipolar Disorder was supported by the Research Council of Norway (#223273, 248778, 262656, 273291, 283798, 248828), South East Norway Health Authority (2017-112), and KG Jebsen Stiftelsen. Funding for the work in eating disorders was supported by grants from the Klarman Family Foundation, Swedish Research Council (Vetenskapsrådet: 538-2013-8864), National Institute of Mental Health (K01MH106675, K01 MH109782, K01MH100435, R01MH119084), and NIAAA (K01 AA025113). The iPSYCH project is supported by grants from the Lundbeck Foundation (R165-2013-15320, R102-A9118, R155-2014-1724 and R248-2017-2003) and the universities and university hospitals of Aarhus and Copenhagen. Genotyping of iPSYCH samples was supported by grants from the Lundbeck Foundation, the Stanley Foundation, the Simons Foundation (SFARI 311789 to MJD), and NIMH (5U01MH094432-02 to MJD). The Danish National Biobank resource was supported by the Novo Nordisk Foundation. Data handling and analysis on the GenomeDK HPC facility was supported by NIMH (1U01MH109514-01 to ADB). High-performance computer capacity for handling and statistical analysis of iPSYCH data on the GenomeDK HPC facility was provided by the Center for Genomics and Personalized Medicine and the Centre for Integrative Sequencing, iSEQ, Aarhus University, Denmark (grant to ADB). Funding for the work in Tourette Syndrome/Obsessive Compulsive Disorder was supported by NIH grants U01NS040024, R01NS016648, K02NS085048, R01 MH096767, ARRA grants NS040024-09S1 and NS040024-07S1, P30 NS062691, R01MH092293, R01MH092513, R01MH092289, R01MH071507, R01MH079489, R01MH079487, R01MH079488, R01MH079494, R01MH002930-06, R01MH073250 and MH087748, and grants from the Tourette Association of America and the David Judah Foundation. Funding support for the Study of Addiction: Genetics and Environment (SAGE) was provided through the NIH Genes, Environment, and Health Initiative [GEI] (U01 HG004422); SAGE is one of the genome-wide association studies funded as part of the Gene Environment Association Studies (GENEVA) under the NIH GEI. Assistance with phenotype harmonization and genotype cleaning, as well as with general study coordination, was provided by the GENEVA Coordinating Center (U01 HG004446). Assistance with data cleaning was provided by the National Center for Biotechnology Information. Support for collection of data sets and samples was provided by the Collaborative Study on the Genetics of Alcoholism (U10 AA008401), the Collaborative Genetic Study of Nicotine Dependence (P01 CA089392), and the Family Study of Cocaine Dependence (R01 DA013423). Funding support for genotyping, which was performed at the Johns Hopkins University Center for Inherited Disease Research, was provided by the NIH GEI (U01HG004438), the National Institute on Alcohol Abuse and Alcoholism,NIDA, and the NIHcontract “High Throughput Genotyping for Studying the Genetic Contributions to Human Disease” (HHSN268200782096C). The data sets used for the analyses described here were obtained from dbGaP (http://www.ncbi.nlm.nih.gov/projects/gap/cgibin/study.cgi?study_id=phs000092.v1.p1). All research at Great Ormond Street Hospital NHS Foundation Trust and UCL Great Ormond Street Institute of Child Health is made possible by the NIHR Great Ormond Street Hospital Biomedical Research Centre. The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR or the Department of Health. We thank the research participants and employees of 23andMe, Inc. for their contribution to this study. We are grateful to Emily Madsen for assistance with manuscript preparation.
DECLARATIONS OF INTERESTS
J.W.S. is an unpaid member of the Bipolar/Depression Research Community Advisory Panel of 23andMe. HRK (Henry R. Kranzler) is a member of the American Society of Clinical Psychopharmacology’s Alcohol Clinical Trials Initiative, which was supported in the last three years by AbbVie, Alkermes, Ethypharm, Indivior, Lilly, Lundbeck, Otsuka, Pfizer, Arbor, and Amygdala Neurosciences. HRK and JG (Joel Gelernter) are named as inventors on PCT patent application #15/878,640 entitled: “Genotype-guided dosing of opioid agonists,” filed January 24, 2018. BMN (Benjamin M Neale) is a member of the Deep Genomics Scientific Advisory Board, a consultant for Camp4 Therapeutics Corporation, a consultant for Merck & Co., a consultant for Avanir Pharmaceuticals, Inc, and a consultant for Takeda Pharmaceutical. KMV (Kirsten Müller-Vahl) has nonfinancial competing interests as a member of the TAA medical advisory board, the scientific advisory board of the German Tourette Association TGD, the board of directors of the German (ACM) and the International (IACM) Association for Cannabinoid Medicines, and the committee of experts for narcotic drugs at the federal opium bureau of the Federal Institute for Drugs and Medical Devices (BfArM) in Germany; has received financial or material research support from the EU (FP7-HEALTH-2011 No. 278367, FP7-PEOPLE-2012-ITN No. 316978), the German Research Foundation (DFG: GZ MU 1527/3-1), the German Ministry of Education and Research (BMBF: 01KG1421), the National Institute of Mental Health (NIMH), the Tourette Gesellschaft Deutschland e.V., the Else-Kroner-Fresenius-Stiftung, and GW, Almirall, Abide Therapeutics, and Therapix Biosiences; has served as a guest editor for Frontiers in Neurology on the research topic “The neurobiology and genetics of Gilles de la Tourette syndrome: new avenues through large-scale collaborative projects”, is an associate editor for “Cannabis and Cannabinoid Research” and an Editorial Board Member of “Medical Cannabis and Cannabinoids”; has received consultant’s honoraria from Abide Therapeutics, Fundacion Canna, Therapix Biosiences and Wayland Group, speaker’s fees from Tilray, and royalties from Medizinisch Wissenschaftliche Verlagsgesellschaft Berlin, and is a consultant for Zynerba Pharmaceuticals. JIN has been an investigator for Assurex and is currently an investigator for Janssen. BF has received educational speaking fees from Medice and Shire. The other authors declare no competing interests.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes 13 figures, 7 tables, a list of consortium author affiliations, and can be found with this article online. Related supplementary figures are grouped into “Supplementary Datafiles”.
Note: Related Figures are grouped into “Supplementary Datafiles”
REFERENCES
- Baurecht H, Hotze M, Brand S, Buning C, Cormican P, Corvin A, Ellinghaus D, Ellinghaus E, Esparza-Gordillo J, Folster-Holst R, et al. (2015). Genome-wide comparative analysis of atopic dermatitis and psoriasis gives insight into opposing genetic mechanisms. Am J Hum Genet 96, 104–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendriem RM, and Ross ME (2017). Wiring the Human Brain: A User’s Handbook. Neuron 95, 482–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharjee S, Rajaraman P, Jacobs KB, Wheeler WA, Melin BS, Hartge P, GliomaScan C, Yeager M, Chung CC, Chanock SJ, et al. (2012). A subset-based approach improves power and interpretation for the combined analysis of genetic association studies of heterogeneous traits. Am J Hum Genet 90, 821–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bipolar Disorder and Schizophrenia Working Group of the Psychiatric Genomics Consortium (2018). Genomic dissection of bipolar disorder and schizophrenia, including 28 subphenotypes. Cell 173, 1705–1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brainstorm Consortium (2018). Analysis of shared heritability in common disorders of the brain. Science 360, eaap8757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulik-Sullivan B, Finucane HK, Anttila V, Gusev A, Day FR, Loh PR, ReproGen C, Psychiatric Genomics, C., Genetic Consortium for Anorexia Nervosa of the Wellcome Trust Case Control, C., Duncan L, et al. (2015). An atlas of genetic correlations across human diseases and traits. Nat Genet 47, 1236–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross-Disorder Group of the Psychiatric Genomics Consoritum (2013). Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nat Genet 45, 984–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross-Disorder Group of the Psychiatric Genomics Consortium (2013). Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet 381, 1371–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davydov E, Goode D, Sirota M, Cooper G, Sidow A, and Batzoglou S (2010). Identifying a high fraction of the human genome to be under selective constraint using GERP++. PLoS Comput Biol 6, e1001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demontis D, Walters RK, Martin J, Mattheisen M, Als TD, Agerbo E, Baldursson G, Belliveau R, Bybjerg-Grauholm J, Baekvad-Hansen M, et al. (2019). Discovery of the first genome-wide significant risk loci for attention deficit/hyperactivity disorder. Nat Genet 51, 63–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan L, Yilmaz Z, Gaspar H, Walters R, Goldstein J, Anttila V, Bulik-Sullivan B, Ripke S, Eating Disorders Working Group of the Psychiatric Genomics, C., Thornton L, et al. (2017). Significant Locus and Metabolic Genetic Correlations Revealed in Genome-Wide Association Study of Anorexia Nervosa. Am J Psychiatry 174, 850–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Castillo N, Gan G, van Donkelaar MMJ, Vaht M, Weber H, Retz W, Meyer-Lindenberg A, Franke B, Harro J, Reif A, et al. (2017). RBFOX1, encoding a splicing regulator, is a candidate gene for aggressive behavior. European neuropsychopharmacology : the journal of the European College of Neuropsychopharmacology 2017 Nov 23. pii: S0924–-977X(17)32003–5.. doi: 10.1016/j.euroneuro.2017.11.012 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandal M, Haney JR, Parikshak NN, Leppa V, Ramaswami G, Hartl C, Schork AJ, Appadurai V, Buil A, Werge TM, Liu C, White KP; CommonMind Consortium; PsychENCODE Consortium; iPSYCH-BROAD Working Group, Horvath S, Geschwind DH (2018). Shared molecular neuropathology across major psychiatric disorders parallels polygenic overlap. Science 359, 693–697.29439242 [Google Scholar]
- Gehman L, Stoilov P, Maguire J, Damianov A, Lin CH, Shiue L, Ares M Jr, Mody I, Black DL. (2011). The splicing regulator Rbfox1 (A2BP1) controls neuronal excitation in the mammalian brain. Nat Genet 43, 706–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geschwind DH, and Flint J (2015). Genetics and genomics of psychiatric disease. Science 349, 1489–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Global Burden of Disease Injury Incidence Prevalence Collaborators (2017). Global, regional, and national incidence, prevalence, and years lived with disability for 328 diseases and injuries for 195 countries, 1990–2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet 390, 1211–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorsuch R (1988). Exploratory Factor Analysis. In Handbook of Multivariate Experimental Psychology Perspectives on Individual Differences, Nesselroade JR, and Cattell RB, eds. (Boston, MA: Springer; ), pp. 231–258. [Google Scholar]
- Grantyn R, and Grantyn A (1973). Postsynaptic responses of hippocampal neurons to mesencephalic stimulation: depolarizing potentials and discharge patterns. Brain Res 53, 55–69. [DOI] [PubMed] [Google Scholar]
- Grotzinger AD, Rhemtulla M, de Vlaming R, Ritchie SJ, Mallard TT, Hill WD, Ip HF, Marioni RE, McIntosh AM, Deary IJ, et al. (2019). Genomic structural equation modelling provides insights into the multivariate genetic architecture of complex traits. Nat Hum Behav 3, 513–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grove J, Ripke S, Als TD, Mattheisen M, Walters R, Won H, Pallesen J, Agerbo E, Andreassen OA, Anney R, et al. (2019a). Identification of common genetic risk variants for autism spectrum disorder. Nat Genet 51, 431–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grove J, Ripke S, Als TD, Mattheisen M, Walters RK, Won H, Pallesen J, Agerbo E, Andreassen OA, Anney R, et al. (2019b). Identification of common genetic risk variants for autism spectrum disorder. Nat Genet 51, 431–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada N, Ito H, Iwamoto I, Morishita R, Tabata H, and Nagata K (2015). Role of the cytoplasmic isoform of RBFOX1/A2BP1 in establishing the architecture of the developing cerebral cortex. Mol Autism 6, 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada N, Ito H, Nishijo T, Iwamoto I, Morishita R, Tabata H, Momiyama T, and Nagata K (2016). Essential role of the nuclear isoform of RBFOX1, a candidate gene for autism spectrum disorders, in the brain development. Sci Rep 6, 30805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwig FP, Bowden J, Loret de Mola C, Tovo-Rodrigues L, Davey Smith G, and Horta BL (2016). Body mass index and psychiatric disorders: a Mendelian randomization study. Sci Rep 6, 32730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill W, and Zhang X (2012). Assessing pleiotropy and its evolutionary consequences: pleiotropy is not necessarily limited, nor need it hinder the evolution of complexity. Nat Rev Genet 13, 296. [DOI] [PubMed] [Google Scholar]
- Hoops D, and Flores C (2017). Making Dopamine Connections in Adolescence. Trends Neurosci 40, 709–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton JR, Upadhyay AK, Qi HH, Zhang X, Shi Y, and Cheng X (2010). Enzymatic and structural insights for substrate specificity of a family of jumonji histone lysine demethylases.”. Nat Struct Mol Biol 17, 38–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Obsessive Compulsive Disorder Foundation Genetics Collaborative (IOCDF-GC) and OCD Collaborative Genetics Association Studies (OCGAS) (2018). Revealing the complex genetic architecture of obsessive compulsive disorder using meta-analysis. Mol Psychiatry 23, 1181–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamuar SS, Schmitz-Abe K, D’Gama AM, Drottar M, Chan WM, Peeva M, Servattalab S, Lam AN, Delgado MR, Clegg NJ, et al. (2017). Biallelic mutations in human DCC cause developmental split-brain syndrome. Nat Genet 49, 606–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HJ, Kawasawa YI, Cheng F, Zhu Y, Xu X, Li M, Sousa AM, Pletikos M, Meyer KA, Sedmak G, et al. (2011). Spatio-temporal transcriptome of the human brain. Nature 478, 483–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler RC, and Wang PS (2008). The descriptive epidemiology of commonly occurring mental disorders in the United States. Annual review of public health 29, 115–129. [DOI] [PubMed] [Google Scholar]
- Kiezun A, Pulit S, Francioli L, van Dijk F, Swertz M, Boomsma D, van Duijn C, Slagboom P, van Ommen G, Wijmenga C, et al. (2013). Deleterious alleles in the human genome are on average younger than neutral alleles of the same frequency. PLoS Genet 9, e1003301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda J, Ago T, Nishimura A, Nakamura K, Matsuo R, Wakisaka Y, Kamouchi M, and Kitazono T (2014). Nox4 is a major source of superoxide production in human brain pericytes. J Vasc Res 51, 429–438. [DOI] [PubMed] [Google Scholar]
- Lettre G, and Rioux JD (2008). Autoimmune diseases: insights from genome-wide association studies. Hum Mol Genet 17, R116–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Santpere G, Imamura Kawasawa Y, Evgrafov OV, Gulden FO, Pochareddy S, Sunkin SM, Li Z, Shin Y, Zhu Y, et al. (2018). Integrative functional genomic analysis of human brain development and neuropsychiatric risks. Science 362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Bhowmick T, Liu Y, Gao X, Mertens HDT, Svergun DI, Xiao J, Zhang Y, Wang JH, and Meijers R (2018). Structural Basis for Draxin-Modulated Axon Guidance and Fasciculation by Netrin-1 through DCC. Neuron 97, 1261–1267 e1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopresti AL, and Drummond PD (2013). Obesity and psychiatric disorders: commonalities in dysregulated biological pathways and their implications for treatment. Prog Neuropsychopharmacol Biol Psychiatry 45, 92–99. [DOI] [PubMed] [Google Scholar]
- Mah W, and Won H (2019). The three-dimensional landscape of the genome in human brain tissue unveils regulatory mechanisms leading to schizophrenia risk. Schizophr Res EPub 2019/03/22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh AP, Heron D, Edwards TJ, Quartier A, Galea C, Nava C, Rastetter A, Moutard ML, Anderson V, Bitoun P, et al. (2017). Mutations in DCC cause isolated agenesis of the corpus callosum with incomplete penetrance. Nat Genet 49, 511–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milaneschi Y, Simmons WK, van Rossum EFC, and Penninx BW (2018). Depression and obesity: evidence of shared biological mechanisms. Mol Psychiatry [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Pollard K, Hubisz M, Rosenbloom K, and Siepel A (2010). Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res 20, 110–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi HHS, Hu M, Wang GQ, Bhattacharjee Z, Gordon A, Gonzales DB, Lan M, P.P. FO;Huarte M;Yaghi NK; Lim H;Garcia BA;Brizuela L;Zhao K;Roberts TM,, and Shi Y (2010). Histone H4K20/H3K9 demethylase PHF8 regulates zebrafish brain and craniofacial development. Nature 466, 503–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds LM, Pokinko M, Torres-Berrio A, Cuesta S, Lambert LC, Del Cid Pellitero E, Wodzinski M, Manitt C, Krimpenfort P, Kolb B, et al. (2018). DCC Receptors Drive Prefrontal Cortex Maturation by Determining Dopamine Axon Targeting in Adolescence. Biol Psychiatry 83, 181–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice F, Riglin L, Thapar AK, Heron J, Anney R, O’Donovan MC, and Thapar A (2018). Characterizing Developmental Trajectories and the Role of Neuropsychiatric Genetic Risk Variants in Early-Onset Depression. JAMA Psychiatry. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samocha KE, Kosmicki JA, Karczewski KJ, O’Donnell-Luria AH, Pierce-Hoffman E, MacArthur DG, Neale BM, and Daly MJ (2017). Regional missense constraint improves variant deleteriousness prediction. bioRxiv, 148353. [Google Scholar]
- Satterstrom FK, Kosmicki JA, Wang J, Breen MS, De Rubeis S, An J-Y, Peng M, Collins R, Grove J, Klei L, et al. (2019). Large-scale exome sequencing study implicates both developmental and functional changes in the neurobiology of autism. bioRxiv, 484113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schizophrenia Working Group of the Psychiatric Genomics, C. (2014). Biological insights from 108 schizophrenia-associated genetic loci. Nature 511, 421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt J, Schwarz K, Baurecht H, Hotze M, Folster-Holst R, Rodriguez E, Lee YAE, Franke A, Degenhardt F, Lieb W, et al. (2016). Atopic dermatitis is associated with an increased risk for rheumatoid arthritis and inflammatory bowel disease, and a decreased risk for type 1 diabetes. J Allergy Clin Immunol 137, 130–136. [DOI] [PubMed] [Google Scholar]
- Schork AJ, Won H, Appadurai V, Nudel R, Gandal M, Delaneau O, Revsbech Christiansen M, Hougaard DM, Baekved-Hansen M, Bybjerg-Grauholm J, et al. (2019). A genome-wide association study of shared risk across psychiatric disorders implicates gene regulation during fetal neurodevelopment. Nat Neurosci 22, 353–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel A, Bejerano G, Pedersen J, Hinrichs A, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier L, Richards S, et al. (2005). Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res 15, 1034–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smoller JW, Andreassen OA, Edenberg HJ, Faraone SV, Glatt SJ, and Kendler KS (2018). Psychiatric genetics and the structure of psychopathology. Mol Psychiatry [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smoller JW, Andreassen OA, Edenberg HJ, Faraone SV, Glatt SJ, and Kendler KS (2019). Psychiatric genetics and the structure of psychopathology. Mol Psychiatry 24, 409–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solovieff N, Cotsapas C, Lee PH, Purcell SM, and Smoller JW (2013). Pleiotropy in complex traits: challenges and strategies. Nat Rev Genet 14, 483–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl EA, Breen G, Forstner AJ, McQuillin A, Ripke S, Trubetskoy V, Mattheisen M, Wang Y, Coleman JRI, Gaspar HA, et al. (2019). Genome-wide association study identifies 30 loci associated with bipolar disorder. Nat Genet 51, 793–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan PF, Agrawal A, Bulik CM, Andreassen OA, Borglum AD, Breen G, Cichon S, Edenberg HJ, Faraone SV, Gelernter J, et al. (2018). Psychiatric Genomics: An Update and an Agenda. Am J Psychiatry 175, 15–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topchiy E, Panzhinskiy E, Griffin WS, Barger SW, Das M, and Zawada WM (2013). Nox4-generated superoxide drives angiotensin II-induced neural stem cell proliferation. Developmental neuroscience 35, 293–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukada Y, Ishitani T, and Nakayama KI (2010). KDM7 is a dual demethylase for histone H3 Lys 9 and Lys 27 and functions in brain development. Genes Dev 24, 432–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vosberg DE, Zhang Y, Menegaux A, Chalupa A, Manitt C, Zehntner S, Eng C, DeDuck K, Allard D, Durand F, et al. (2018). Mesocorticolimbic Connectivity and Volumetric Alterations in DCC Mutation Carriers. J Neurosci 38, 4655–4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Liu S, Warrell J, Won H, Shi X, Navarro FCP, Clarke D, Gu M, Emani P, Yang YT, et al. (2018). Comprehensive functional genomic resource and integrative model for the human brain. Science 362, Epub 2018/12/14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won H, de la Torre-Ubieta L, Stein JL, Parikshak NN, Huang J, Opland CK, Gandal MJ, Sutton GJ, Hormozdiari F, Lu D, et al. (2016). Chromosome conformation elucidates regulatory relationships in developing human brain. Nature 538, 523–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray NR, Ripke S, Mattheisen M, Trzaskowski M, Byrne EM, Abdellaoui A, Adams MJ, Agerbo E, Air TM, Andlauer TMF, et al. (2018). Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat Genet 50, 668–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Sul JH, Tsetsos F, Nawaz MS, Huang AY, Zelaya I, Illmann C, Osiecki L, Darrow SM, Hirschtritt ME, et al. (2019). Interrogating the Genetic Determinants of Tourette’s Syndrome and Other Tic Disorders Through Genome-Wide Association Studies. Am J Psychiatry 176, 217–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J, Erzurumluoglu A, Elsworth B, Kemp J, Howe L, Haycock P, Hemani G, Tansey K, Laurin C, Early Genetics and Lifecourse Epidemiology (EAGLE) Eczema Consortium, et al. (2017). LD Hub: a centralized database and web interface to perform LD score regression that maximizes the potential of summary level GWAS data for SNP heritability and genetic correlation analysis. Bioinformatics 33, 272–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Anttila V, Smoller JW, and Lee PH (2018). Statistical power and utility of meta-analysis methods for cross-phenotype genome-wide association studies. PLoS One 13, e0193256. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Summary of eight neuropsychiatric disorder datasets (Related to Table 1). Disease-specific cases and controls included in the meta-analysis, number of individual GWAS loci, and liability-based SNP heritability estimates are provided. Heritability was estimated from available European summary statistics using LD score regression.
Figure S1. Statistical power and number of cross-disorder associations (Related to Table 2 and Table S3.1). Power to detect associations across pairs of disorders was plotted with the number of cross-disorder associations identified in the current meta-analysis. For each pair of disorders, power was estimated using the number of cases and heritability for each disorder, as well as the genetic correlation between the disorders. In general, as power increased, so did the number of identified SNPs.
Figure S2. Cross-disorder GWAS SNPs in the DCC locus colocalize with eQTLs for DCC (Related to Table 2, Table S4.2, and Figure 3). Dark blue dots refer to SNPs that overlap between brain eQTLs and GWAS plots. LD region for the GWAS locus and TAD boundaries are depicted.
Figure S3. Gene expression of top loci across development (Related to Figure 3). Gene expression trajectories from a transcriptome atlas of post-mortem brain tissue across development are plotted for four top loci, DCC, RBFOX1, NOX4 and BRAF in six different brain tissue types. AMY = amygdala; MD = mediodorsal nucleus of the thalamus; CBC = cerebellar cortex; NCX = neocortex; HIP = hippocampus; STR = striatum.
Figure S4. Gene expression in the brain for pleiotropic and non-pleiotropic loci (Related to Figure 5). Average normalized gene expression in fetal and adult post-mortem brain tissue for pleiotropic (109) and non-pleiotropic (37) loci were plotted. Disorder-specific and pleiotropic risk loci showed a similar level of gene expression in prenatal and postnatal development after multiple testing correction (t-test p > 0.025).
Supplemental Datafile 1: Additional analysis results to assess the robustness of genetic correlation and Genomic SEM study (Related to Figure 1). Data S1.1. displays the hierarchical clustering of genetic architecture across eight neuropsychiatric disorders. Data S 1.2. summarizes the relationship between the estimated genetic correlation and the effective sample sizes for each pair of disorders. Data S1.3. shows the relationship between the estimated genetic correlation and the effective sample sizes for MD and four other disorders, SCZ, BIP, ADHD, and ASD. Data S1.4. depicts the genomic SEM analysis result using a Weighted Least Squares estimator and that based on a Maximum Likelihood estimator. Data S1.5. summarizes the distribution of the number of cross-disorder loci identified in meta-analysis of chromosome 1 across 100 simulation replicates.
Supplementary Datafile 2: P-M plots for 146 LD-independent genome-wide significant loci (Related to Figure 3). P-M plot is displayed on the right side for each SNP locus. The X-axis on a P-M plot represents M-values estimated for each locus for each disorder, while the Y-axis displays the significance of disease-SNP association. Disorders displayed in red dots were predicted to have an effect (M-value 0.9), disorders in blue were predicted to not have an effect (M-value 0.1), and studies in green indicated its effect is uncertain (0.1< M-value < 0.9). The dot size reflects the relative sample size of the studies. On the left side, a forest plot displays log odds ratios for the effect of the SNP on each disorder and a meta-analyzed effect across disorders. As expected, most of the top SNPs (109/146) show pleiotropic effects.
Supplementary Datafile 3: Analyses of rare CNVs and mutations previously associated with neurodevelopmental genomic disorders (Related to STAR Methods section “Relationship of Lead SNPs from Meta-analysis to Rare CNVs and Mutations Previously Associated with Neurodevelopmental Genomic Disorders”) Data S3.1. shows that sentinel SNPs are not overrepresented among genomic disorder loci. Data S3.2. and Data S3.3. indicate that genes linked to sentinel SNPs are not overrepresented among dominant-acting ASD risk genes and for damaging de novo mutations in ASD cases, respectively.
Table S2. Tables Related to STAR Methods, Figure 1 and Figure S1. Table S2.1. displays the genetic correlations estimated by LD score regression, while Table S2.2. shows the results from EFA of genetic covariance matrix.
Table S3. Tables Related to Figure 2, 3, and 4. Table S3.1. displays the list of 146 lead SNPs, and Table S3.2. shows the M-value estimates for quantifying confidence of disorder-specific association for all 146 lead SNPs. Table S3.3. summarizes the loci with opposite directional effects.
Table S5: Tables Related to Figure 5, Gene Expression Enrichment Analyses. Table S5.1. summarizes the GTEx gene enrichment analysis results using MAGMA. Table S5.2. and S5.3 show the tissue enrichment analysis for pleiotropic and disorder-specific risk loci using MAGMA (GTEx v7), respectively.
Table S6: Tables Related to Figure 5. Table S6.1. and S6.2 summarize the Gene ontology analysis for pleiotropic and disease-specific risk loci, respectively. Table S6.3. lists the enrichment analysis results of cortical gene expression data. Table S6.4. shows the comparison of disease-specific vs. pleiotropic risk loci by various functional and genomic features.
Table S4: Tables Related to Table 2, Table S3.1. Table S4.1. summarizes the functional annotation of 146 lead SNPs, while Table S4.2. lists the Brain eQTL and Hi-C data annotation based on FUMA database. Table S4.3. lists the GWAS catalog data for lead SNPs.
Table S7: Tables Related to STAR Methods section “Comparison with other brain-related traits and diseases” - Relationship between Cross-disorder Genetic Risk and other Brain-related Traits and Diseases. Table S7.1. summarizes the genetic correlation analysis of the cross-disorder phenotype with other brain-related traits. Table S7.2. shows the GWAS catalog enrichment analysis for pleiotropic risk loci.
Data Availability Statement
The Psychiatric Genetics Consortium (PGC)’s policy is to make genome-wide summary results publicly available. Summary statistics for a combined meta-analysis of eight psychiatric disorders without 23andMe data are available on the PGC web site (https://www.med.unc.edu/pgc/results-and-downloads). Results for 10,000 SNPs for eight disorders including 23andMe are also available on the PGC web site. The summary-level GWAS association statistics for PGC individual disorders are available at the website (https://www.med.unc.edu/pgc/results-and-downloads).
GWAS summary statistics for the 23andMe cohort (Hyde, 2016) must be obtained separately. These can be obtained by individual researchers under an agreement with 23andMe that protects the privacy of the 23andMe participants. Contact Aaron Petrakovitz (apetrakovitz@23andme.com) to apply for access to the data.