Abstract
Background
Rupture of intracranial aneurysms (IA) is associated with high rates of mortality around the world. Use of intestinal probiotics can regulate the pathophysiology of aneurysms, but the details of the mechanism involved have been unclear.
Material/Methods
The GEO2R analysis website was used to detect the DEGs between IAs, AAAs, samples after supplementation with probiotics, and normal samples. The online tool DAVID provides functional classification and annotation analyses of associated genes, including GO and KEGG pathway. PPI of these DEGs was analyzed based on the STRING database, followed by analysis using Cytoscape software.
Results
We found 170 intersecting DEGs (contained in GSE75240 and more than 2 of the 4 aneurysms datasets), 5 intersecting DEGs (contained in all datasets) and 1 intersecting DEG (contained in GSE75240 and all IAs datasets). GO analysis results suggested that the DEGs primarily participate in signal transduction, cell adhesion, immune response, response to drug, extracellular matrix organization, cell-cell signaling, and inflammatory response in the BP terms, and the KEGG pathways are mainly enriched in focal adhesion, cytokine-cytokine receptor interaction, ECM-receptor interaction, amoebiasis, chemokine signaling pathway, proteoglycans, and PI3K-Akt signaling pathway in cancer pathways. Through PPI network analysis, we confirmed 2 candidates for further study: CAV1 and MYH11. These downregulated DEGs are associated with the formation of aneurysms, and the change of these DEGs is the opposite in probiotics-treated animals.
Conclusions
Our study suggests that MYH11 and CAV1 are potential target genes for prevention of aneurysms. Further experiments are needed to verify these findings.
MeSH Keywords: Blood, Intracranial Aneurysm, Probiotics
Background
Intracranial aneurysms (IAs) are the abnormal bulging that occurs on the wall of the artery. It is an important cause of spontaneous subarachnoid hemorrhage (SAH) [1]. In the past few years, hypertension, smoking, atherosclerosis, and other factors were shown to lead to the development and occurrence of aneurysms. Recently, many scholars have come to believe that congenital defects also affect the formation of aneurysms [2]. Pathological conditions and inflammation that alter the morphologies of the vasculature can activate vascular remodeling, including stenosis and vascular weakening [3,4]. Intestinal probiotics are an important group of bacteria that regulate homeostasis of the internal body environment. Recent studies have shown that gut probiotics affect various diseases and impact inflammation, which plays a considerable role in determining the severity and course of disease [5,6]. Findings indicate that intestinal probiotics can affect the pathological condition of aneurysms by regulating vascular inflammation [7]. With the rapid development of gene chip technologies, the Gene Expression Omnibus (GEO) has gradually come to play a significant role in bioinformatic analysis. In this research, we used bioinformatics analysis to discover potential core genes affected by gut probiotics that play a role in the formation of IAs, which may ultimately provide potential therapeutic targets.
Material and Methods
Acquisition of the gene expression datasets
The gene expression datasets were acquired from the GEO database (https://www.ncbi.nlm.nih.gov/geo/). Three gene expression profiles (GSE26969, GSE75436, and GSE54083) of IAs, 1 gene expression profile (GSE7084) of abdominal aortic aneurysm, and 1 gene expression profile (GSE75240) after supplementation with probiotics were selected. Among them, GSE26969 and GSE75436 were based on platform GPL570([HG-U133_Plus_2] Affymetrix Human Genome U133 Plus 2.0 Array), GSE54083 was based on platform GPL4133(Agilent-014850 Whole Human Genome Microarray 4x44K G4112F), GSE7084 was based on platform GPL2507 (Sentrix Human-6 Expression BeadChip), and GSE75240 was based on platform GPL11649 (Agilent-023647 B. Taurus (Bovine) Oligo Microarray v2). The GSE26969 dataset was submitted by Li et al., including 3 IAs samples and 3 normal superficial temporal artery (STAs) samples [8]. The GSE75436 dataset was submitted by Yu et al., including 15 IAs tissues and 15 STAs tissues. The GSE54083 dataset was submitted by Inoue, including 5 unruptured intracranial aneurysms (UIAs) and 10 STAs samples [9]. The microarray data of GSE7084 was submitted by Tromp, which consisted of 8 abdominal aortas and 7 abdominal aortic aneurysms (AAAs) [10]. The microarray data of GSE75240 was submitted by Worku, which consisted of 4 whole-blood samples collected at the beginning (Day 0) and end of the study (Day 60) after supplementation with probiotics [11].
Identification of DEGs
The GEO2R analysis website (https://www.ncbi.nlm.nih.gov/geo/geo2r/) was used to find differentially expressed genes (DEGs) between IAs and normal samples, AAAs and normal samples, or sample after supplementation with probiotics and normal samples. The P-values and Benjamini & Hochberg (false discovery rate) were applied to provide a balance between discovery of statistically significant genes and limitations of false-positives [12]. Probe sets without corresponding gene symbols or genes with more than 1 probe set were removed or we took the highest value of the absolute value, respectively. Subsequently, the |log fold change (FC)| and P-value were calculated [13]. The cut-off criteria for defining DEGs were P-value <0.05 and |logFC| ≥1.5. Statistical analysis was further evaluated for each dataset, and the significative DEGs was exhibited using the Venn diagram online analysis tool (bioinformatics.psb.ugent.be/webtools/Venn/) and the TBtoolsv0.6654 software.
GO and KEGG pathway analysis of DEGs
To study DEGs at a functional level, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis and Gene Ontology (GO) analysis were analyzed and exhibited using the online biological analysis tool (DAVID version 6.8, https://david.ncifcrf.gov/), database for the annotation, visualization, and integrated discovery, which provides functional classification and annotation analyses of DEGs [14]. GO analysis is an available method for large-scale functional enrichment research and can be classified into biological process (BP), cellular component (CC), and molecular function (MF). KEGG pathway enrichment analysis includes biological pathways, diseases, chemical, genomes, and metabolic information. The corrected P-value <0.05 was regarded as a statistically significant difference, and the top 5 counts of GOs and KEGGs were selected.
PPI network construction and module analysis
The Search Tool for the Retrieval of Interacting Genes (STRING) version 11.0 database (http://string-db.org/) is designed to explore protein–protein interactions (PPI) [15]. The DEGs identified previously were first mapped to the STRING database. Only the interactions with a combined score ≥0.4 were considered as significant. Subsequently, the PPI network was visualized using Cytoscape 3.6.1 software, which was used with the maximal clique centrality (MCC) method to explore the PPI network for hub genes. We performed quantitative expression analysis of 30 candidate genes that could better explain the reliability of the results [16].
Results
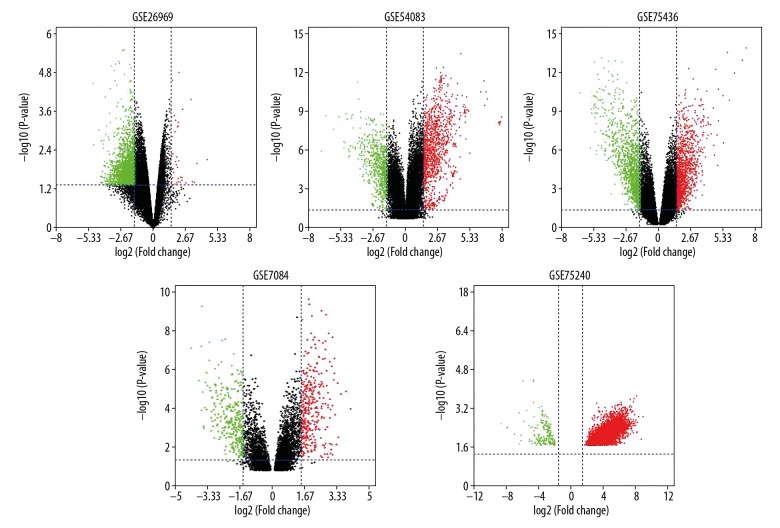
Identification of significantly DEGs (associated with aneurysms or IAs)
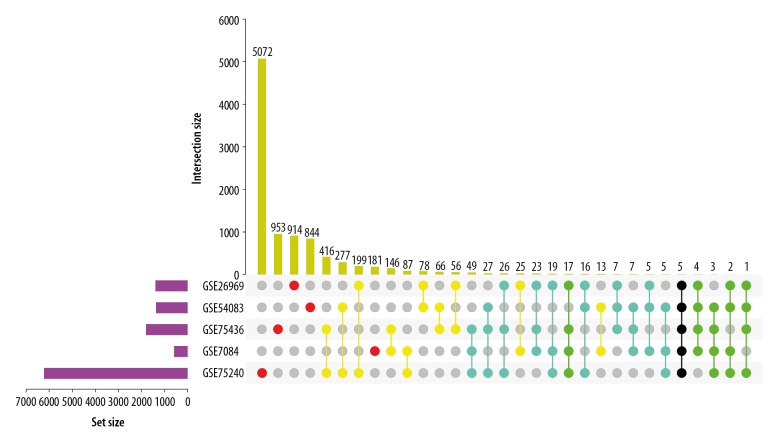
Three datasets of IAs-related gene expression profiles (GSE26969, GSE54083, and GSE75436) were collected from the GEO database via GEO2R. One dataset of AAAs-related gene expression profiles (GSE7084) and probiotics-related gene expression profiles (GSE75240) were also downloaded from the GEO database. We set |log2 fold change (FC)| ≥1.5 and P-value<0.05 as the criterion to identify significantly DEGs. We found 1398, 1372, 1807, 592, and 6233 significantly DEGs in the GSE26969, GSE54083, GSE75436, GSE7084, and GSE75240 datasets, respectively (Figures 1, 2). By using the Venn diagram webtool, we found that some DEGs about probiotics were associated with aneurysms formation, including 170 intersecting (contain GSE75240 and more than 2 of the 4 aneurysms datasets), 28 intersecting DEGs (contain GSE75240 and more than 3 of the 4 aneurysms datasets), 5 intersecting DEGs (contain all datasets), 70 intersecting DEGs (contain GSE75240 and more than 2 of the 3 IAs datasets), and 1 intersecting DEG (contain GSE75240 and all IAs datasets). These different groups were exhibited using the Venn diagram webtool and the TBtools v0.6654 software, including 6, 39, 53, 30, 167 upregulated and 80, 20, 75, 70, 3 downregulated genes in the 170 intersecting DEGs (GSE26969, GSE54083, GSE75436, GSE7084, and GSE75240 datasets) (Table 1, Supplementary Table 1). The 5 intersecting DEGs associated with aneurysms and probiotics were Cofilin 2 (CFL2), LIM domain containing preferred translocation partner in lipoma (LPP), Myosin Heavy Chain 11 (MYH11), Monoamine Oxidase B (MAOB), and MAS-Related GPR Family Member F (MRGPRF). One intersecting DEG associated with IAs and probiotics was caveolin 1 (CAV1). These 6 intersecting DEGs were distributed in different parts of the chromosome (Supplementary Figure 1).
Figure 1.
UpSet Plot analysis of DGEs. Red: in one dataset; yellow: in two datasets; Blue: in three datasets; green: in four datasets; Black: in five datasets
Figure 2.
Volcano plot of DGEs. Red: up-regulation with fold change of more than 1.5; Green: down-regulation with a fold change of more than 1.5; Black: unchanged genes.
Table 1.
Intersecting DEGs.
| Names | Total | Elements |
|---|---|---|
| GSE26969, GSE54083, GSE7084, GSE75240, GSE75436 | 5 | CFL2, LPP, MYH11, MAOB, MRGPRF |
| GSE26969, GSE54083, GSE7084, GSE75436 | 4 | RERG, ACTG2, SBSPON, CPED1 |
| GSE26969, GSE54083, GSE7084, GSE75240 | 2 | SORBS1, FBLN1 |
| GSE26969, GSE7084, GSE75240, GSE75436 | 17 | SYNM, PLN, PDGFD, KCNAB1, FHL5, SORBS2, KANK1, PRUNE2, CSRP2, INPP5A, ZAK, PPP1R14A, CNN1, CSRP1, AOC3, LMOD1, TAGLN |
| GSE54083, GSE7084, GSE75240, GSE75436 | 3 | AIF1L, LTBP2, NUP210 |
| GSE26969, GSE54083, GSE75240, GSE75436 | 1 | CAV1 |
| GSE26969, GSE54083, GSE7084 | 5 | KLHL42, ADH5, MYL9, AIM1, JCHAIN |
| GSE26969, GSE7084, GSE75436 | 23 | MAOA, RERGL, ADIRF, PALLD, HLA-DRA, PTPRC, SMTN, C11orf96, C2orf40, SYNPO2, FAM129A, RNASET2, DSTN, CCDC3, FILIP1L, EGR2, HLA-DMA, MYLK, EVI2B, MFAP4, NEXN, ADH1B, ACTA2 |
| GSE26969, GSE7084, GSE75240 | 19 | PLSCR4, SMOC2, SRPX, CAV2, IGFBP7, TMEM47, GPX3, SERPING1, TGFBR3, TCEAL4, PDLIM3, CPE, HSPB8, RGS5, SPARCL1, IGFBP5, RNASE1, CYBRD1, LAMC1 |
| GSE54083, GSE7084, GSE75436 | 7 | ACTC1, SLC43A2, TREM2, HMOX1, IL1B, HLA-DQB1, COTL1 |
| GSE54083, GSE7084, GSE75240 | 5 | PIK3CD, EFHD1, ERRFI1, MYH10, CXCL5 |
| GSE7084, GSE75240, GSE75436 | 49 | SLCO2B1, ACADL, RBPMS, ALOX5AP, SCARA3, IL10RA, CLEC5A, MPEG1, CKMT2, TIMP4, FLVCR2, MRAP2, KCNA5, NOV, GDF15, CD300A, MMP12, SLC37A2, APOD, IL1RN, FXYD1, RCAN2, ALDH1B1, TSPAN33, AGTR1, THBS2, CD300LF, CCR5, IRF8, KCNMA1, WISP2, APOE, CD8A, IGFBP2, MGP, MMP1, MYOC, CD4, SNX10, LMO3, PNKD, GLDN, CCL4, CD14, DEF6, BCL11A, PLAC8, CXCL16, CP |
| GSE26969, GSE54083, GSE75436 | 7 | SLMAP, LOX, PDLIM5, IGHM, IGHA2///IGHA1///IGH, ENTPD4, TNC |
| GSE26969, GSE54083, GSE75240 | 16 | AKR1B10, RAB14, LRRC40, DIXDC1, MAP3K7, HSP90AA1, RPS17, TIMP3, FGB, PDCD7, PRKAR2B, PNN, CFL1, ANKRD44, PNPLA8, NMD3 |
| GSE26969, GSE75240, GSE75436 | 26 | COL1A1, NEGR1, COL11A1, CHRDL1, PRR11, HNRNPC, SFRP1, TMTC1, MPP7, CLDND1, ADAMTS1, CDH11, PDE5A, PGM5, COL1A2, CXCL14, CD36, GREM1, CRISPLD2, FILIP1, ANGPTL1, ACACB, RASSF3, DES, NUP54, FLNA |
| GSE54083, GSE75240, GSE75436 | 27 | CCL19, ROR2, IGSF11, PPARGC1A, FGF12, RHAG, ZNF280B, GPRIN3, FN1, GNAO1, NGFR, ESR1, CLEC7A, SGSM1, SULT1E1, GABRA4, GALNT3, CHST11, NRAP, TMEM169, PMP2, MSR1, IGSF21, NETO2, SMPDL3A, CTNNA3, STAC |
Functional and pathway enrichment analyses of DEGs
Three terms of GO functional annotation analysis were analyzed and exhibited on these 170 intersecting DEGs mentioned above, including biological process (BP), cellular component (CC), and molecular function (MF). With a threshold of P-value <0.05, the DEGs were enriched in 115 GO terms and 12 KEGG pathways (Figure 3). The enriched GO functions for these DEGs showed the most significant terms were signal transduction, cell adhesion, immune response, extracellular matrix organization, cell–cell signaling, and inflammatory response in the BP term. These DEGs were enriched in plasma membrane, extracellular exosome, extracellular space, extracellular region, and membrane in the CC term, and these DEGs were enriched in protein binding, identical protein binding, calcium ion binding, heparin binding, and actin binding in the MF term. In the above GOs analysis, CFL2 was enriched in extracellular exosome, extracellular space, and protein binding. LPP was enriched in cell adhesion, plasma membrane, and protein binding. MYH11 was enriched in extracellular exosome and protein binding. MAOB was enriched in response to drug and extracellular exosome. MRGPRF was enriched in plasma membrane and extracellular exosome. CAV1 was mainly involved in plasma membrane, membrane, protein binding, and identical protein binding. Subsequently, we conducted KEGG pathway enrichment analysis. The enriched KEGG pathways mainly contained focal adhesion, cytokine-cytokine receptor interaction, amoebiasis, ECM-receptor interaction, chemokine signaling pathway, proteoglycans, and PI3K-Akt signaling pathway. These results demonstrate that CAV1 plays a central role in the focal adhesion and proteoglycans in cancer pathways.
Figure 3.
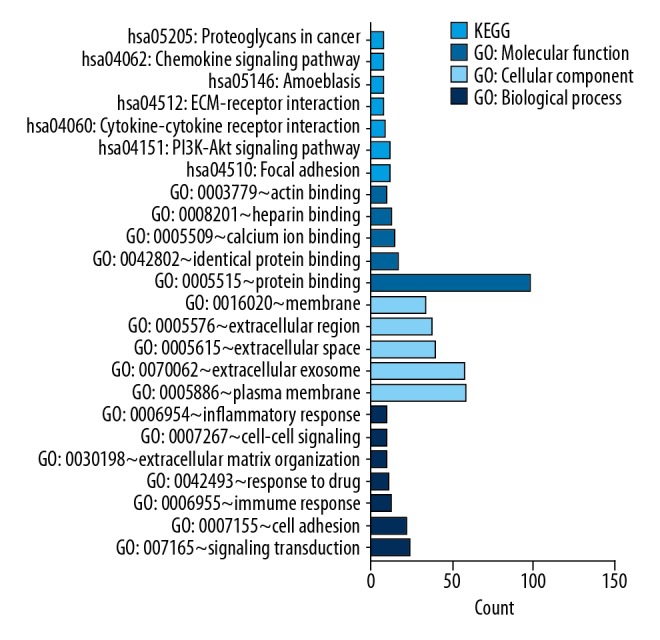
GO and KEGG pathway analyses results of DGEs. GO and KEGG pathway analyses results of DGEs. BP –biological process; CC – cellular component; MF – molecular function; KEGG – Kyoto Encyclopedia of Genes and Genomes. P<0.05.
Construction and analysis of PPI network
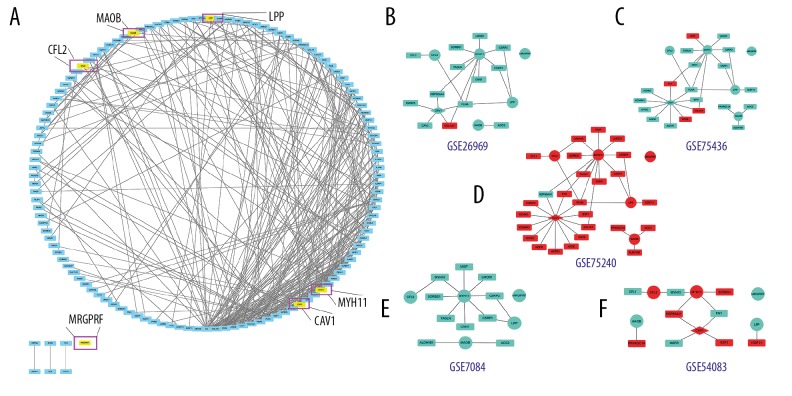
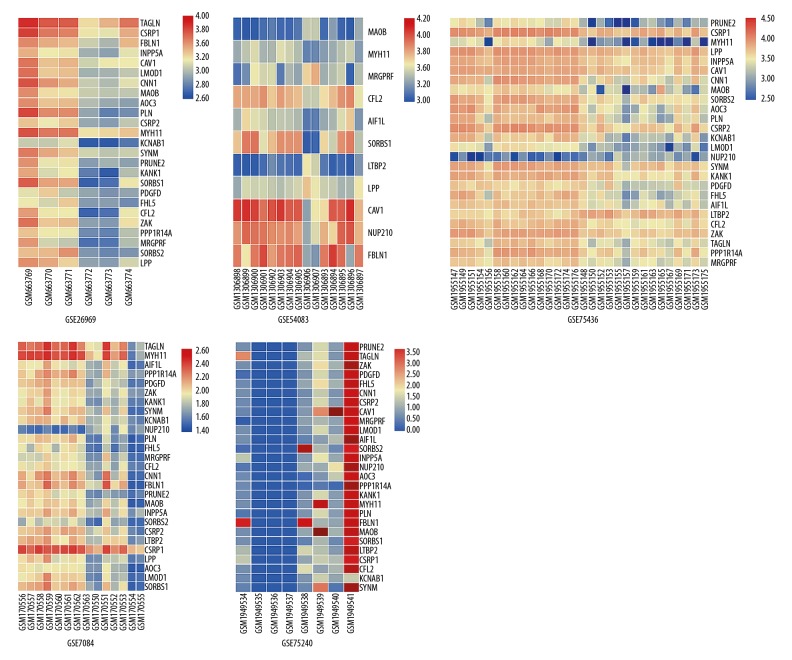
The PPI networks of 170 DEGs were analyzed using the STRING database. The subset of 170 DEGs with 319 interactions was included in the PPI network. In addition, those interactions were constructed by using Cytoscape software. The top 30 DEGs according to the MCC method were identified using the CytoHubba plugin and were ordered as follows: FN1, IGFBP5, APOE, LAMC1, CP, SPARCL1, IGFBP7, CHRDL1, COL1A1, TIMP3, COL1A2, FBLN1, THBS2, COL11A1, CCR5, CCL4, CXCL16, CXCL5, CCL19, CXCL14, TAGLN, CAV1, MYH11, MMP1, ESR1, MGP, FLNA, ADAMTS1, AGTR1, and HSP90AA1 (Table 2). Notably, CAV1 and MYH11 were included in these 30 DEGs. Through PPI network, we found genes that were tightly bound to CFL2, LPP, MYH11, MAOB, MRGPRF, and CAV1 in different GEO datasets (Figure 4). The Heatmap and PPI network clearly display that CFL2 and MYH11 were upregulated in GSE75240 and downregulated in GSE26969, GSE75436, and GSE7084. LPP, MAOB, and MRGPRF were upregulated in GSE75240 and downregulated in GSE26969, GSE75436, GSE7084, and GSE54083. CAV1 was upregulated in GSE75240 and downregulated in GSE26969 and GSE75436 (Table 3, Figures 4, 5). In general, these genes were basically increased in the formation of aneurysms, while the expression was decreased in probiotics. We also found that FLNA was strongly correlated with CAV1, CFL2, LPP, and MYH11 (Table 4).
Table 2.
Top 30 in network, ranked by MCC method.
| Rank | Name | Score |
|---|---|---|
| 1 | FN1 | 6182 |
| 2 | IGFBP5 | 5220 |
| 3 | APOE | 5096 |
| 4 | LAMC1 | 5064 |
| 5 | CP | 5055 |
| 6 | SPARCL1 | 5050 |
| 7 | IGFBP7 | 5043 |
| 8 | CHRDL1 | 5041 |
| 9 | COL1A1 | 1000 |
| 10 | TIMP3 | 940 |
| 11 | COL1A2 | 926 |
| 12 | FBLN1 | 868 |
| 13 | THBS2 | 754 |
| 14 | COL11A1 | 720 |
| 15 | CCR5 | 184 |
| 16 | CCL4 | 166 |
| 17 | CXCL16 | 130 |
| 18 | CXCL5 | 129 |
| 19 | CCL19 | 120 |
| 19 | CXCL14 | 120 |
| 21 | TAGLN | 93 |
| 22 | CAV1 | 68 |
| 23 | MYH11 | 64 |
| 24 | MMP1 | 62 |
| 25 | ESR1 | 55 |
| 26 | MGP | 50 |
| 27 | FLNA | 39 |
| 28 | ADAMTS1 | 36 |
| 29 | AGTR1 | 31 |
| 30 | HSP90AA1 | 25 |
Figure 4.
PPI network. (A) The PPI networks of 170 DGEs. (B–F) Genes related to CFL2, LPP, MYH11, MAOB, MRGPRF, and CAV1 in different GEO datasets. The nodes indicate the genes and the lines represent the corresponding interactions. (B–F) Red: up-regulation; Blue: down-regulation. Circle: 5 intersecting DEGs; Diamond: 1 intersecting DEG; Square: others DGEs related to 6 intersecting DEGs
Table 3.
List of 6 intersecting DEGs (log FC).
| GSE75240 | GSE7084 | GSE26969 | GSE75436 | GSE54083 | |
|---|---|---|---|---|---|
| CFL2 | 3.71922 | −1.75373 | −2.58 | −1.81832 | 2.144287 |
| LPP | 4.870317 | −2.39111 | −2.79 | −1.55623 | −1.92875 |
| MYH11 | 4.312605 | −3.53764 | −2.65 | −4.71788 | 1.595093 |
| MAOB | 5.841989 | −2.05551 | −1.78 | −3.0551 | −2.42214 |
| MRGPRF | 5.190016 | −1.61803 | −2.35 | −2.09833 | −2.37477 |
| CAV1 | 5.990843 | NA | −1.86 | −1.53252 | 3.940476 |
NA – not applicable.
Figure 5.
Heatmap of the top DGEs. Heatmap of the 28 DGEs with a fold change of more than 1.5. Red: up-regulation; blue: down-regulation. Normal superficial temporal artery: GSM663769, GSM663770, GSM663771, GSM306898, GSM306899, GSM306900, GSM306901, GSM306902, GSM306903, GSM306904, GSM306905, GSM306906, GSM306907, GSM1955147, GSM1955149, GSM1955151, GSM1955154, GSM1955156, GSM1955158, GSM1955160, GSM1955162, GSM1955164, GSM1955166, GSM1955168, GSM1955170, GSM1955172, GSM1955174, GSM1955176; Intracranial aneurysm: GSM663772, GSM663773, GSM663774, GSM306893, GSM306894, GSM306895, GSM306896, GSM306897, GSM1955148, GSM1955140, GSM1955153, GSM1955155, GSM1955157, GSM1955159, GSM1955161, GSM1955163, GSM1955165, GSM1955167, GSM1955169, GSM1955171, GSM1955173, GSM1955175; Abdominal aorta: GSM170556, GSM170557, GSM170558, GSM170559, GSM170560, GSM170561, GSM170562, GSM170564; Abdominal aortic aneurysm: GSM170563, GSM170550, GSM170551, GSM170552, GSM170553, GSM170554, GSM170555; Gene expression of initial (0day) cow blood sample before supplementation with probiotics: GSM1949534, GSM1949535, GSM1949536, GSM1949537; Gene expression of end of study (60day) cow blood sample after supplementation with probiotics: GSM1949538, GSM1949539, GSM1949540, GSM1949541.
Table 4.
PPI with 6 intersecting DEGs.
| CFL2 | LPP | MYH11 | MAOB | MRGPRF | CAV1 |
|---|---|---|---|---|---|
| FLNA | FLNA | FLNA | AOC3 | NA | FLNA |
| CFL1 | CSRP1 | CSRP1 | ALDH1B1 | ESR1 | |
| MYH10 | IGSF11 | MYH10 | PPARGC1A | KCNA5 | |
| CSRP2 | CSRP2 | KCNMA1 | |||
| FN1 | FN1 | ||||
| COL1A1 | COL1A1 | ||||
| HSP90AA1 | HSP90AA1 | ||||
| MGP | AGTR1 | ||||
| LMOD1 | IGFBP5 | ||||
| SORBS1 | NGFR | ||||
| CNN1 | APOE | ||||
| TAGLN | MYOC | ||||
| CAV2 |
NA – not applicable.
Discussion
The rupture of IAs can cause serious SAH, which has significant socioeconomic implications. Many factors affect the formation of aneurysms by altering hemodynamics and morphologies of the cerebral vasculature. Intestinal probiotics can regulate homeostasis of the internal body environment. Recent studies showed that gut microbiota affect the pathological condition of aneurysms by modulating the immune system [5,6]. Understanding the molecular mechanism of probiotics in reducing aneurysms formation is of vital clinical significance for diagnosis and treatment.
More than 300 scientists from 25 countries eventually came to the conclusion that the cattle genome contain at least 22 000 protein-coding genes and the genetic similarity between cattle and humans is 80%, which is greater than that between humans and mice or rats [17]. This means that cattle can replace mice or rats as animal models of human disease. Some studies have also used cattle for studying the characteristics of human disease. Nobuhiro Kajihara et al. found that low glucose conditions exacerbated mtROS production through activation of fatty acid oxidation in bovine aortic endothelial cells [18]. Other researchers have also used cattle as their own experimental models [19–24]. We used the GSE75240 dataset to show that gut microbes can change the expression of proteins or genes in the blood. The changes in blood vessels can affected by intestinal probiotics, so we used aneurysm datasets from other sites. In this study, we obtained gene expression data from the GSE26969, GSE75436, GSE54083, GSE7084, and GSE75240 datasets, and selected 170 DEGs (containing a gut probiotic dataset and more than 2 of the 4 aneurysms datasets). To further investigate the interactions of these DEGs, we carried out GO term and KEGG pathway analyses. The GO term analysis results indicated that DEGs were mainly enriched in signal transduction, cell adhesion, immune response, extracellular matrix organization, cell-cell signaling, and inflammatory response in the BP terms. The KEGG pathways analysis suggested that DEGs were significantly involved in focal adhesion, PI3K-Akt signaling pathway, cytokine-cytokine receptor interaction, ECM-receptor interaction, amoebiasis, and chemokine signaling pathway. Previous studies showed that the dysfunction of cell adhesion, immune response, inflammatory response, focal adhesion, PI3K-Akt signaling pathway, chemokine signaling pathway, cytokine-cytokine receptor interaction, and ECM-receptor interaction can cause changes in the vascular structure of aneurysms [25–28].
We constructed a Heatmap that clearly shows that CFL2 and MYH11 were upregulated in GSE75240, GSE54083 and downregulated in GSE26969, GSE75436, GSE7084, while LPP, MAOB, and MRGPRF were upregulated in GSE75240 and downregulated in GSE26969, GSE75436, GSE7084, and GSE54083. In general, probiotics can decrease the formation of aneurysms through the increased expression of CFL2, MYH11, LPP, MAOB, and MRGORF, and by inhibiting the formation of IAs through upregulating the expression of CAV1.
PPI of the 170 intersecting DEGs was analyzed based on the STRING database, followed by use of Cytoscape3.6.1 software. We confirmed 2 candidates for our further study: CAV1 and MYH11. We also found that CFL2, LPP, MYH11, MAOB, and MRGPRF were involved with the association between aneurysm and probiotics, and CAV1 was involved with the associated between IAs and probiotics. CAV1 may be a unique factor in the formation of IAs. CAV1, which is located in cell membrane caveolae, is a member of the caveolin family of proteins [29]. It is generally distributed in smooth-muscle cells, endothelial cells, skeletal myoblasts, and fibroblasts [30]. CAV1 modulates a wide range of cellular events such as proliferation, lipid metabolism, cellular tracking, and signal transduction [29]. MYH11 is a smooth-muscle myosin belonging to the myosin heavy chain family, and has been identified as a major contractile protein that converts chemical energy into mechanical energy by the hydrolysis of ATP [31,32]. Previous research, in which MYH11 mutations were found, showed that a structural defect in MYH11 alters smooth-muscle cell phenotype and myosin contractile function, causing an increase of proliferation and aggravating vascular injury [33].
The PPI network analysis clearly shows that FLNA has a strong correlation with CAV1, CFL2, LPP, and MYH11. Protein filamin A (FLNA) helps build extensive internal network of protein filaments in cells, called the cytoskeleton. FLNA can also bind to many other proteins in the cell to carry out various functions, including cell adhesion, migration, and determination of cell shape [34]. FLNA has been found to play roles in regulating skeletal and brain development, the formation of heart tissue and blood vessels, and blood clotting [34–36]. FLNA was lower in the IAs group than in the normal group (log2 (FC)=−2.14 and −1.83, respectively) and was significantly higher in the probiotics-treated group than in the control group (log2 (FC)=4.10). The incidence of aneurysms was reduced by adjusting the interaction between CAV1, CFL2, LPP, MYH11, and FLNA. Further studies are needed to explore the effects of gut probiotics on FLNA and the impact on aneurysm formation.
The above results promote understanding of the molecular mechanism of the pathogenesis of aneurysms, and identify potential therapeutic targets using gut probiotics. The role of these genes in aneurysms, however, is not clear, and further molecular biological experiments are needed to verify these findings.
Conclusions
This bioinformatics analysis identified DEGs obtained from the GEO databases. Using multiple datasets, 6 genes that are associated with probiotics affecting the formation of intracranial aneurysms were identified. Through further analysis of bioinformatics, 2 high-correlation hub genes were confirmed: MYH11 and CAV1. All of these genes were downregulated in aneurysms, and upregulation of these genes was associated with probiotics. These 7 DEGs may be potential targets for aneurysms prevention.
Supplementary Data
Supplementary Table 1.
List of DGEs considering P<0.05 and |log FC| ≥1.5.
| GSE75240 | GSE26969 | GSE54083 | GSE75436 | GSE7084 | |||||
|---|---|---|---|---|---|---|---|---|---|
| Gene | Log FC | Gene | Log FC | Gene | Log FC | Gene | Log FC | Gene | Log FC |
| ACACB | 5.01 | ACACB | −1.86 | AIF1L | 1.74 | ACACB | −1.68 | ACADL | −1.65 |
| ACADL | 6.19 | ADAMTS1 | −2.58 | AKR1B10 | −4.99 | ACADL | −1.71 | AGTR1 | −1.60 |
| ADAMTS1 | 6.23 | AKR1B10 | 1.67 | ANKRD44 | 1.70 | ADAMTS1 | −1.65 | AIF1L | −1.56 |
| AGTR1 | 6.39 | ANGPTL1 | −2.7 | CAV1 | 3.94 | AGTR1 | −3.63 | ALDH1B1 | −1.56 |
| AIF1L | 6.00 | ANKRD44 | −2.25 | CCL19 | 1.79 | AIF1L | −2.57 | ALOX5AP | 1.53 |
| AKR1B10 | 7.28 | AOC3 | −2.9 | CFL1 | −2.58 | ALDH1B1 | −2.07 | AOC3 | −2.17 |
| ALDH1B1 | 6.35 | CAV1 | −1.86 | CFL2 | 2.14 | ALOX5AP | 2.22 | APOD | −2.31 |
| ALOX5AP | 5.10 | CAV2 | −2.41 | CHST11 | 1.76 | ANGPTL1 | −4.74 | APOE | 2.94 |
| ANGPTL1 | 4.71 | CD36 | −2.66 | CLEC7A | 4.92 | AOC3 | −4.02 | BCL11A | 1.60 |
| ANKRD44 | 6.00 | CDH11 | 1.84 | CTNNA3 | 2.52 | APOD | −2.31 | CAV2 | −1.54 |
| AOC3 | 3.37 | CFL1 | −2.3 | CXCL5 | −5.51 | APOE | 2.67 | CCL4 | 3.16 |
| APOD | 5.50 | CFL2 | −2.58 | DIXDC1 | 4.89 | BCL11A | 2.16 | CCR5 | 1.63 |
| APOE | 4.50 | CHRDL1 | −2.32 | EFHD1 | 3.68 | CAV1 | −1.53 | CD14 | 2.16 |
| BCL11A | 2.30 | CLDND1 | −2.72 | ERRFI1 | 1.68 | CCL19 | −2.11 | CD300A | 1.73 |
| CAV1 | 5.99 | CNN1 | −3.17 | ESR1 | 2.54 | CCL4 | 3.72 | CD300LF | 1.55 |
| CAV2 | 4.61 | COL11A1 | 3.59 | FBLN1 | 4.52 | CCR5 | 2.63 | CD4 | 1.66 |
| CCL19 | 6.29 | COL1A1 | 1.52 | FGB | 2.42 | CD14 | 2.76 | CD8A | 1.52 |
| CCL4 | 2.80 | COL1A2 | 1.51 | FGF12 | 2.39 | CD300A | 2.45 | CFL2 | −1.75 |
| CCR5 | 3.94 | CPE | −3.69 | FN1 | −2.74 | CD300LF | 2.15 | CKMT2 | −1.91 |
| CD14 | 5.19 | CRISPLD2 | −1.86 | GABRA4 | 3.16 | CD36 | −3.14 | CLEC5A | 1.73 |
| CD300A | 2.77 | CSRP1 | −2.74 | GALNT3 | −1.95 | CD4 | 1.63 | CNN1 | −2.77 |
| CD300LF | 3.70 | CSRP2 | −1.96 | GNAO1 | 2.30 | CD8A | 2.30 | CP | −1.84 |
| CD36 | 3.30 | CXCL14 | −2.67 | GPRIN3 | 1.65 | CDH11 | 2.12 | CPE | −2.69 |
| CD4 | 5.48 | CYBRD1 | −3.29 | HSP90AA1 | 7.78 | CFL2 | −1.82 | CSRP1 | −1.94 |
| CD8A | 4.05 | DES | −2.24 | IGSF11 | 2.49 | CHRDL1 | −3.76 | CSRP2 | −2.40 |
| CDH11 | 5.21 | DIXDC1 | −2.37 | IGSF21 | −1.64 | CHST11 | 2.50 | CXCL16 | 1.67 |
| CFL1 | 3.86 | FBLN1 | −1.89 | LPP | −1.93 | CKMT2 | −2.80 | CXCL5 | 2.18 |
| CFL2 | 3.72 | FGB | 1.53 | LRRC40 | 2.88 | CLDND1 | −1.51 | CYBRD1 | −1.95 |
| CHRDL1 | 3.47 | FHL5 | −2.05 | LTBP2 | −2.39 | CLEC5A | 3.31 | DEF6 | 1.94 |
| CHST11 | 4.81 | FILIP1 | −2.5 | MAOB | −2.42 | CLEC7A | 2.43 | EFHD1 | −3.38 |
| CKMT2 | 3.30 | FLNA | −2.14 | MAP3K7 | −1.53 | CNN1 | −3.27 | ERRFI1 | −1.72 |
| CLDND1 | 3.49 | GPX3 | −3.13 | MRGPRF | −2.37 | COL11A1 | 7.24 | FBLN1 | −2.03 |
| CLEC5A | 5.65 | GREM1 | −3.86 | MSR1 | 3.19 | COL1A1 | 3.21 | FHL5 | −2.46 |
| CLEC7A | 5.65 | HNRNPC | −1.51 | MYH10 | −2.70 | COL1A2 | 1.74 | FLVCR2 | 1.65 |
| CNN1 | 2.95 | HSP90AA1 | −3.44 | MYH11 | 1.60 | CP | 2.76 | FXYD1 | −1.74 |
| COL11A1 | 5.32 | HSPB8 | −1.55 | NETO2 | −3.24 | CRISPLD2 | −1.97 | GDF15 | −1.54 |
| COL1A1 | 5.08 | IGFBP5 | −3 | NGFR | −2.12 | CSRP1 | −2.23 | GLDN | −1.84 |
| COL1A2 | 4.10 | IGFBP7 | −2.23 | NMD3 | 1.59 | CSRP2 | −2.68 | GPX3 | −2.87 |
| CP | 5.40 | INPP5A | −1.67 | NRAP | −2.94 | CTNNA3 | −4.40 | HSPB8 | −2.04 |
| CPE | 6.01 | KANK1 | −2.65 | NUP210 | 3.22 | CXCL14 | −4.32 | IGFBP2 | −2.97 |
| CRISPLD2 | 5.12 | KCNAB1 | −2.37 | PDCD7 | −2.95 | CXCL16 | 2.48 | IGFBP5 | −1.53 |
| CSRP1 | 5.08 | LAMC1 | −1.88 | PIK3CD | −1.82 | DEF6 | 1.66 | IGFBP7 | −2.01 |
| CSRP2 | 2.89 | LMOD1 | −2.8 | PMP2 | 2.93 | DES | −5.09 | IL10RA | 1.73 |
| CTNNA3 | 5.82 | LPP | −2.79 | PNN | 2.83 | ESR1 | −2.15 | IL1RN | 2.86 |
| CXCL14 | 3.42 | LRRC40 | −1.9 | PNPLA8 | 1.71 | FGF12 | −1.95 | INPP5A | −1.72 |
| CXCL16 | 5.14 | MAOB | −1.78 | PPARGC1A | 1.84 | FHL5 | −3.39 | IRF8 | 1.63 |
| CXCL5 | 3.60 | MAP3K7 | −1.51 | PRKAR2B | 2.56 | FILIP1 | −2.18 | KANK1 | −2.28 |
| CYBRD1 | 4.78 | MPP7 | −2.33 | RAB14 | −2.66 | FLNA | −1.83 | KCNA5 | −3.29 |
| DEF6 | 5.52 | MRGPRF | −2.35 | RHAG | 3.09 | FLVCR2 | 2.26 | KCNAB1 | −2.38 |
| DES | 6.00 | MYH11 | −2.65 | ROR2 | −1.81 | FN1 | 2.06 | KCNMA1 | −1.93 |
| DIXDC1 | 4.54 | NEGR1 | −2.47 | RPS17 | 2.24 | FXYD1 | −1.50 | LAMC1 | −1.80 |
| EFHD1 | 5.22 | NMD3 | −1.5 | SGSM1 | 2.28 | GABRA4 | −1.87 | LMO3 | −2.50 |
| ERRFI1 | 2.96 | NUP54 | −1.6 | SMPDL3A | 2.56 | GALNT3 | 1.84 | LMOD1 | −2.86 |
| ESR1 | 4.29 | PDCD7 | −1.77 | SORBS1 | 4.45 | GDF15 | 2.31 | LPP | −2.39 |
| FBLN1 | 5.19 | PDE5A | −2.07 | STAC | 1.80 | GLDN | −2.90 | LTBP2 | −2.00 |
| FGB | 5.53 | PDGFD | −1.66 | SULT1E1 | 1.71 | GNAO1 | −2.10 | MAOB | −2.06 |
| FGF12 | 4.16 | PDLIM3 | −2.33 | TIMP3 | 2.56 | GPRIN3 | 2.07 | MGP | −2.51 |
| FHL5 | 5.73 | PGM5 | −2.57 | TMEM169 | −1.59 | GREM1 | 2.80 | MMP1 | 1.59 |
| FILIP1 | 3.32 | PLN | −5.01 | ZNF280B | 1.77 | HNRNPC | 1.55 | MMP12 | 2.70 |
| FLNA | 4.10 | PLSCR4 | −2.8 | IGFBP2 | 2.42 | MPEG1 | 2.51 | ||
| FLVCR2 | 2.48 | PNN | −1.73 | IGSF11 | −3.31 | MRAP2 | −3.66 | ||
| FN1 | 7.15 | PNPLA8 | −2.45 | IGSF21 | 1.75 | MRGPRF | −1.62 | ||
| FXYD1 | 4.48 | PPP1R14A | −2.17 | IL10RA | 1.80 | MYH10 | −3.62 | ||
| GABRA4 | 5.61 | PRKAR2B | −2.32 | IL1RN | 2.31 | MYH11 | −3.54 | ||
| GALNT3 | 4.44 | PRR11 | −2.31 | INPP5A | −1.61 | MYOC | −1.74 | ||
| GDF15 | 4.98 | PRUNE2 | −2.43 | IRF8 | 1.75 | NOV | −3.26 | ||
| GLDN | 5.70 | RAB14 | −1.86 | KANK1 | −2.06 | NUP210 | 1.59 | ||
| GNAO1 | 3.35 | RASSF3 | −3.17 | KCNA5 | −4.42 | PDGFD | −2.78 | ||
| GPRIN3 | 3.90 | RGS5 | −2.35 | KCNAB1 | −3.42 | PDLIM3 | −2.82 | ||
| GPX3 | 5.12 | RNASE1 | −1.72 | KCNMA1 | −2.49 | PIK3CD | 1.99 | ||
| GREM1 | 5.02 | RPS17 | −1.97 | LMO3 | −2.38 | PLAC8 | 1.63 | ||
| HNRNPC | 4.61 | SERPING1 | −2.19 | LMOD1 | −3.19 | PLN | −3.28 | ||
| HSP90AA1 | −3.60 | SFRP1 | −1.99 | LPP | −1.56 | PLSCR4 | −1.62 | ||
| HSPB8 | 5.57 | SMOC2 | −2.48 | LTBP2 | 1.54 | PNKD | 1.58 | ||
| IGFBP2 | 3.58 | SORBS1 | −4.15 | MAOB | −3.06 | PPP1R14A | −2.63 | ||
| IGFBP5 | 4.43 | SORBS2 | −3.51 | MGP | 2.20 | PRUNE2 | −2.61 | ||
| IGFBP7 | 3.50 | SPARCL1 | −2.56 | MMP1 | 2.32 | RBPMS | −2.20 | ||
| IGSF11 | 2.78 | SRPX | −2.67 | MMP12 | 1.98 | RCAN2 | −3.16 | ||
| IGSF21 | 3.24 | SYNM | −2.44 | MPEG1 | 2.14 | RGS5 | −3.42 | ||
| IL10RA | 2.78 | TAGLN | −2.54 | MPP7 | −3.58 | RNASE1 | 1.67 | ||
| IL1RN | 6.21 | TCEAL4 | −2.97 | MRAP2 | −1.56 | SCARA3 | −1.67 | ||
| INPP5A | 3.63 | TGFBR3 | −2.01 | MRGPRF | −2.10 | SERPING1 | −2.03 | ||
| IRF8 | 5.86 | TIMP3 | −2.01 | MSR1 | 3.35 | SLC37A2 | 1.93 | ||
| KANK1 | 3.89 | TMEM47 | −2.52 | MYH11 | −4.72 | SLCO2B1 | 1.67 | ||
| KCNA5 | 4.74 | TMTC1 | −1.67 | MYOC | −4.14 | SMOC2 | −2.86 | ||
| KCNAB1 | 2.95 | ZAK | −2.74 | NEGR1 | −2.23 | SNX10 | 2.22 | ||
| KCNMA1 | 3.39 | NETO2 | 2.28 | SORBS1 | −3.16 | ||||
| LAMC1 | 3.59 | NGFR | −1.57 | SORBS2 | −1.52 | ||||
| LMO3 | 3.12 | NOV | −3.14 | SPARCL1 | −3.06 | ||||
| LMOD1 | 3.06 | NRAP | −2.27 | SRPX | −1.93 | ||||
| LPP | 4.87 | NUP210 | 1.86 | SYNM | −2.49 | ||||
| LRRC40 | 4.39 | NUP54 | −1.61 | TAGLN | −3.01 | ||||
| LTBP2 | 5.15 | PDE5A | −1.58 | TCEAL4 | −1.85 | ||||
| MAOB | 5.84 | PDGFD | −1.91 | TGFBR3 | −1.96 | ||||
| MAP3K7 | 5.05 | PGM5 | −3.02 | THBS2 | −2.57 | ||||
| MGP | 5.97 | PLAC8 | 2.26 | TIMP4 | −1.62 | ||||
| MMP1 | 4.57 | PLN | −3.07 | TMEM47 | −3.01 | ||||
| MMP12 | 6.16 | PMP2 | −3.58 | TSPAN33 | 1.70 | ||||
| MPEG1 | 4.02 | PNKD | 2.24 | WISP2 | −2.09 | ||||
| MPP7 | 4.65 | PPARGC1A | −1.68 | ZAK | −1.85 | ||||
| MRAP2 | 5.45 | PPP1R14A | −2.16 | ||||||
| MRGPRF | 5.19 | PRR11 | 1.71 | ||||||
| MSR1 | 4.34 | PRUNE2 | −3.87 | ||||||
| MYH10 | 4.89 | RASSF3 | −2.12 | ||||||
| MYH11 | 4.31 | RBPMS | −2.21 | ||||||
| MYOC | 3.88 | RCAN2 | −1.72 | ||||||
| NEGR1 | 3.15 | RHAG | −2.59 | ||||||
| NETO2 | 2.77 | ROR2 | 2.05 | ||||||
| NGFR | 4.62 | SCARA3 | −2.17 | ||||||
| NMD3 | 2.96 | SFRP1 | −2.93 | ||||||
| NOV | 6.11 | SGSM1 | −1.75 | ||||||
| NRAP | 4.85 | SLC37A2 | 1.51 | ||||||
| NUP210 | 4.76 | SLCO2B1 | 2.77 | ||||||
| NUP54 | 4.12 | SMPDL3A | 1.54 | ||||||
| PDCD7 | 4.38 | SNX10 | 1.94 | ||||||
| PDE5A | 6.51 | SORBS2 | −2.71 | ||||||
| PDGFD | 3.20 | STAC | −2.66 | ||||||
| PDLIM3 | 3.76 | SULT1E1 | 2.27 | ||||||
| PGM5 | 5.00 | SYNM | −1.69 | ||||||
| PIK3CD | 2.66 | TAGLN | −2.31 | ||||||
| PLAC8 | 6.22 | THBS2 | 1.91 | ||||||
| PLN | 3.39 | TIMP4 | −2.13 | ||||||
| PLSCR4 | 4.04 | TMEM169 | 1.57 | ||||||
| PMP2 | 6.15 | TMTC1 | −1.62 | ||||||
| PNKD | 5.77 | TSPAN33 | 1.59 | ||||||
| PNN | 5.23 | WISP2 | −4.84 | ||||||
| PNPLA8 | 2.97 | ZAK | −1.93 | ||||||
| PPARGC1A | 5.22 | ZNF280B | 1.88 | ||||||
| PPP1R14A | 3.77 | ||||||||
| PRKAR2B | 4.79 | ||||||||
| PRR11 | 2.70 | ||||||||
| PRUNE2 | 5.93 | ||||||||
| RAB14 | −3.28 | ||||||||
| RASSF3 | 4.94 | ||||||||
| RBPMS | 3.96 | ||||||||
| RCAN2 | 3.42 | ||||||||
| RGS5 | 7.11 | ||||||||
| RHAG | 5.03 | ||||||||
| RNASE1 | 4.97 | ||||||||
| ROR2 | 4.97 | ||||||||
| RPS17 | 4.36 | ||||||||
| SCARA3 | 3.58 | ||||||||
| SERPING1 | 5.94 | ||||||||
| SFRP1 | 5.46 | ||||||||
| SGSM1 | 4.44 | ||||||||
| SLC37A2 | 5.00 | ||||||||
| SLCO2B1 | 5.05 | ||||||||
| SMOC2 | 4.96 | ||||||||
| SMPDL3A | 4.21 | ||||||||
| SNX10 | 4.75 | ||||||||
| SORBS1 | 3.91 | ||||||||
| SORBS2 | 4.13 | ||||||||
| SPARCL1 | 3.31 | ||||||||
| SRPX | 5.76 | ||||||||
| STAC | 4.51 | ||||||||
| SULT1E1 | 4.90 | ||||||||
| SYNM | 5.98 | ||||||||
| TAGLN | 4.45 | ||||||||
| TCEAL4 | −2.02 | ||||||||
| TGFBR3 | 3.51 | ||||||||
| THBS2 | 3.75 | ||||||||
| TIMP3 | 5.40 | ||||||||
| TIMP4 | 5.95 | ||||||||
| TMEM169 | 2.84 | ||||||||
| TMEM47 | 4.99 | ||||||||
| TMTC1 | 4.60 | ||||||||
| TSPAN33 | 3.69 | ||||||||
| WISP2 | 4.37 | ||||||||
| ZAK | 2.34 | ||||||||
| ZNF280B | 3.05 | ||||||||
The location of DGEs on the chromosome. 6 intersecting DEGs are located at different positions on the chromosome. Blue: intersection of IAs and probiotics; Purple: intersection of aneurysms and probiotics
Footnotes
Source of support: This work was financed by a grant-in-aid for scientific research from the National Natural Science Foundation of China (No. 81870914)
Conflict of interest
None.
References
- 1.Macdonald RL, Schweizer TA. Spontaneous subarachnoid haemorrhage. Lancet (London, England) 2017;389(10069):655–66. doi: 10.1016/S0140-6736(16)30668-7. [DOI] [PubMed] [Google Scholar]
- 2.Mohan D, Munteanu V, Coman T, Ciurea AV. Genetic factors involves in intracranial aneurysms – actualities. J Med Life. 2015;8(3):336–41. [PMC free article] [PubMed] [Google Scholar]
- 3.Frösen J, Tulamo R, Paetau A, et al. Saccular intracranial aneurysm: pathology and mechanisms. Acta Neuropathol. 2012;123(6):773–86. doi: 10.1007/s00401-011-0939-3. [DOI] [PubMed] [Google Scholar]
- 4.Kanematsu Y, Kanematsu M, Kurihara C, et al. Critical roles of macrophages in the formation of intracranial aneurysm. Stroke. 2011;42(1):173–78. doi: 10.1161/STROKEAHA.110.590976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koeth RA, Wang Z, Levison BS, et al. Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat Med. 2013;19(5):576–85. doi: 10.1038/nm.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Holmes E, Li JV, Marchesi JR, Nicholson JK. Gut microbiota composition and activity in relation to host metabolic phenotype and disease risk. Cell Metab. 2012;16(5):559–64. doi: 10.1016/j.cmet.2012.10.007. [DOI] [PubMed] [Google Scholar]
- 7.Shikata F, Shimada K, Sato H, et al. Potential influences of gut microbiota on the formation of intracranial aneurysm. Hypertension (Dallas, Tex: 1979) 2019;73(2):491–96. doi: 10.1161/HYPERTENSIONAHA.118.11804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li L, Yang X, Jiang F, et al. Transcriptome-wide characterization of gene expression associated with unruptured intracranial aneurysms. Eur Neurol. 2009;62(6):330–37. doi: 10.1159/000236911. [DOI] [PubMed] [Google Scholar]
- 9.Nakaoka H, 1, Tajima A, Yoneyama T, et al. Gene expression profiling reveals distinct molecular signatures associated with the rupture of intracranial aneurysm. Stroke. 2014;45(8):2239–45. doi: 10.1161/STROKEAHA.114.005851. [DOI] [PubMed] [Google Scholar]
- 10.Lenk GM, 1, Tromp G, Weinsheimer S, et al. Whole genome expression profiling reveals a significant role for immune function in human abdominal aortic aneurysms. BMC Genomics. 2007;8:237. doi: 10.1186/1471-2164-8-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Adjei-Fremah S, Ekwemalor K, Asiamah E, et al. Transcriptional profiling of the effect of lipopolysaccharide (LPS) pretreatment in blood from probiotics-treated dairy cows. Genom Data. 2016;10:15–18. doi: 10.1016/j.gdata.2016.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang JZ, Wu ZH, Cheng Q. Screening and identification of key biomarkers in nasopharyngeal carcinoma: Evidence from bioinformatic analysis. Medicine. 2019;98(48):e17997. doi: 10.1097/MD.0000000000017997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davis S, Meltzer PS. GEOquery: A bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics (Oxford, England) 2007;23(14):1846–47. doi: 10.1093/bioinformatics/btm254. [DOI] [PubMed] [Google Scholar]
- 14.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 15.Szklarczyk D, Morris JH, Cook H, et al. The STRING database in 2017: Quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45:D362–68. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shannon P, Markiel A, Ozier O, et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elsik CG, Tellam RL, Worley KC, et al. The genome sequence of taurine cattle: A window to ruminant biology and evolution. Science (New York, NY) 2009;324(5926):522–28. doi: 10.1126/science.1169588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kajihara N, Kukidome D, Sada K, et al. Low glucose induces mitochondrial reactive oxygen species via fatty acid oxidation in bovine aortic endothelial cells. J Diabetes Investig. 2017;8(6):750–61. doi: 10.1111/jdi.12678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fujie T, Murakami M, Yoshida E, et al. Transcriptional induction of metallothionein by Tris(pentafluorophenyl)stibane in cultured bovine aortic endothelial cells. Int J Mol Sci. 2016;17(9) doi: 10.3390/ijms17091381. pii: E1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qu L, Yu L, Wang Y, et al. Inward rectifier K+ currents are regulated by CaMKII in endothelial cells of primarily cultured bovine pulmonary arteries. PLoS One. 2015;10(12):e0145508. doi: 10.1371/journal.pone.0145508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fujie T, Takenaka F, Yoshida E, et al. Possible mechanisms underlying transcriptional induction of metallothionein isoforms by tris(pentafluorophenyl)stibane, tris(pentafluorophenyl)arsane, and tris(pentafluorophenyl)phosphane in cultured bovine aortic endothelial cells. J Toxicol Sci. 2019;44(5):327–33. doi: 10.2131/jts.44.327. [DOI] [PubMed] [Google Scholar]
- 22.Chandra S, Fulton DJR, Caldwell RB. Hyperglycemia-impaired aortic vasorelaxation mediated through arginase elevation: Role of stress kinase pathways. Eur J Pharmacol. 2019;844:26–37. doi: 10.1016/j.ejphar.2018.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chang S, Lanctot AC, McCarter MD. The prediction of radiofrequency ablation zone volume using vascular indices of 3-dimensional volumetric colour Doppler ultrasound in an in vitro blood-perfused bovine liver model. Br J Radiol. 2017;90(1070) doi: 10.1259/bjr.20160661. 20160661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chung KY, Smith SB, Choi SH, Johnson BJ. Oleic acid enhances G protein coupled receptor 43 expression in bovine intramuscular adipocytes but not in subcutaneous adipocytes. J Anim Sci. 2016;94(5):1875–83. doi: 10.2527/jas.2015-0010. [DOI] [PubMed] [Google Scholar]
- 25.Liu P, Shi Y, Fan Z, et al. Inflammatory smooth muscle cells induce endothelial cell alterations to influence cerebral aneurysm progression via regulation of integrin and VEGF expression. Cell Transplant. 2019;28(6):713–22. doi: 10.1177/0963689718815824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Erhart P, Cakmak S, Grond-Ginsbach C, et al. Inflammasome activity in leucocytes decreases with abdominal aortic aneurysm progression. Int J Mol Med. 2019;44(4):1299–308. doi: 10.3892/ijmm.2019.4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Z, Guo J, Han X, et al. Metformin represses the pathophysiology of AAA by suppressing the activation of PI3K/AKT/mTOR/autophagy pathway in ApoE(–/–) mice. Cell biosci. 2019;9:68. doi: 10.1186/s13578-019-0332-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Y, Jia L, Xie Y, et al. Involvement of macrophage-derived exosomes in abdominal aortic aneurysms development. Atherosclerosis. 2019;289:64–72. doi: 10.1016/j.atherosclerosis.2019.08.016. [DOI] [PubMed] [Google Scholar]
- 29.Huang Q, Zhong W, Hu Z, Tang X. A review of the role of cav-1 in neuropathology and neural recovery after ischemic stroke. J neuroinflammation. 2018;15(1):348. doi: 10.1186/s12974-018-1387-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anderson RG. The caveolae membrane system. Ann Rev Biochem. 1998;67:199–225. doi: 10.1146/annurev.biochem.67.1.199. [DOI] [PubMed] [Google Scholar]
- 31.Zhu L, Vranckx R, Khau Van Kien P, et al. Mutations in myosin heavy chain 11 cause a syndrome associating thoracic aortic aneurysm/aortic dissection and patent ductus arteriosus. Nat Genet. 2006;38(3):343–49. doi: 10.1038/ng1721. [DOI] [PubMed] [Google Scholar]
- 32.Milewicz DM, Trybus KM, Guo DC, et al. Altered smooth muscle cell force generation as a driver of thoracic aortic aneurysms and dissections. Arterioscler Thromb Vasc Biol. 2017;37(1):26–34. doi: 10.1161/ATVBAHA.116.303229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuang SQ, Kwartler CS, Byanova KL, et al. Rare, nonsynonymous variant in the smooth muscle-specific isoform of myosin heavy chain, MYH11, R247C, alters force generation in the aorta and phenotype of smooth muscle cells. Circ Res. 2012;110(11):1411–22. doi: 10.1161/CIRCRESAHA.111.261743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Falet H. New insights into the versatile roles of platelet FlnA. Platelets. 2013;24(1):1–5. doi: 10.3109/09537104.2011.654004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hart AW, Morgan JE, Schneider J, et al. Cardiac malformations and midline skeletal defects in mice lacking filamin A. Hum Mol Genet. 2006;15(16):2457–67. doi: 10.1093/hmg/ddl168. [DOI] [PubMed] [Google Scholar]
- 36.Feng Y, Chen MH, Moskowitz IP, et al. Filamin A (FLNA) is required for cell-cell contact in vascular development and cardiac morphogenesis. Proc Natl Acad Sci USA. 2006;103(52):19836–41. doi: 10.1073/pnas.0609628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1.
List of DGEs considering P<0.05 and |log FC| ≥1.5.
| GSE75240 | GSE26969 | GSE54083 | GSE75436 | GSE7084 | |||||
|---|---|---|---|---|---|---|---|---|---|
| Gene | Log FC | Gene | Log FC | Gene | Log FC | Gene | Log FC | Gene | Log FC |
| ACACB | 5.01 | ACACB | −1.86 | AIF1L | 1.74 | ACACB | −1.68 | ACADL | −1.65 |
| ACADL | 6.19 | ADAMTS1 | −2.58 | AKR1B10 | −4.99 | ACADL | −1.71 | AGTR1 | −1.60 |
| ADAMTS1 | 6.23 | AKR1B10 | 1.67 | ANKRD44 | 1.70 | ADAMTS1 | −1.65 | AIF1L | −1.56 |
| AGTR1 | 6.39 | ANGPTL1 | −2.7 | CAV1 | 3.94 | AGTR1 | −3.63 | ALDH1B1 | −1.56 |
| AIF1L | 6.00 | ANKRD44 | −2.25 | CCL19 | 1.79 | AIF1L | −2.57 | ALOX5AP | 1.53 |
| AKR1B10 | 7.28 | AOC3 | −2.9 | CFL1 | −2.58 | ALDH1B1 | −2.07 | AOC3 | −2.17 |
| ALDH1B1 | 6.35 | CAV1 | −1.86 | CFL2 | 2.14 | ALOX5AP | 2.22 | APOD | −2.31 |
| ALOX5AP | 5.10 | CAV2 | −2.41 | CHST11 | 1.76 | ANGPTL1 | −4.74 | APOE | 2.94 |
| ANGPTL1 | 4.71 | CD36 | −2.66 | CLEC7A | 4.92 | AOC3 | −4.02 | BCL11A | 1.60 |
| ANKRD44 | 6.00 | CDH11 | 1.84 | CTNNA3 | 2.52 | APOD | −2.31 | CAV2 | −1.54 |
| AOC3 | 3.37 | CFL1 | −2.3 | CXCL5 | −5.51 | APOE | 2.67 | CCL4 | 3.16 |
| APOD | 5.50 | CFL2 | −2.58 | DIXDC1 | 4.89 | BCL11A | 2.16 | CCR5 | 1.63 |
| APOE | 4.50 | CHRDL1 | −2.32 | EFHD1 | 3.68 | CAV1 | −1.53 | CD14 | 2.16 |
| BCL11A | 2.30 | CLDND1 | −2.72 | ERRFI1 | 1.68 | CCL19 | −2.11 | CD300A | 1.73 |
| CAV1 | 5.99 | CNN1 | −3.17 | ESR1 | 2.54 | CCL4 | 3.72 | CD300LF | 1.55 |
| CAV2 | 4.61 | COL11A1 | 3.59 | FBLN1 | 4.52 | CCR5 | 2.63 | CD4 | 1.66 |
| CCL19 | 6.29 | COL1A1 | 1.52 | FGB | 2.42 | CD14 | 2.76 | CD8A | 1.52 |
| CCL4 | 2.80 | COL1A2 | 1.51 | FGF12 | 2.39 | CD300A | 2.45 | CFL2 | −1.75 |
| CCR5 | 3.94 | CPE | −3.69 | FN1 | −2.74 | CD300LF | 2.15 | CKMT2 | −1.91 |
| CD14 | 5.19 | CRISPLD2 | −1.86 | GABRA4 | 3.16 | CD36 | −3.14 | CLEC5A | 1.73 |
| CD300A | 2.77 | CSRP1 | −2.74 | GALNT3 | −1.95 | CD4 | 1.63 | CNN1 | −2.77 |
| CD300LF | 3.70 | CSRP2 | −1.96 | GNAO1 | 2.30 | CD8A | 2.30 | CP | −1.84 |
| CD36 | 3.30 | CXCL14 | −2.67 | GPRIN3 | 1.65 | CDH11 | 2.12 | CPE | −2.69 |
| CD4 | 5.48 | CYBRD1 | −3.29 | HSP90AA1 | 7.78 | CFL2 | −1.82 | CSRP1 | −1.94 |
| CD8A | 4.05 | DES | −2.24 | IGSF11 | 2.49 | CHRDL1 | −3.76 | CSRP2 | −2.40 |
| CDH11 | 5.21 | DIXDC1 | −2.37 | IGSF21 | −1.64 | CHST11 | 2.50 | CXCL16 | 1.67 |
| CFL1 | 3.86 | FBLN1 | −1.89 | LPP | −1.93 | CKMT2 | −2.80 | CXCL5 | 2.18 |
| CFL2 | 3.72 | FGB | 1.53 | LRRC40 | 2.88 | CLDND1 | −1.51 | CYBRD1 | −1.95 |
| CHRDL1 | 3.47 | FHL5 | −2.05 | LTBP2 | −2.39 | CLEC5A | 3.31 | DEF6 | 1.94 |
| CHST11 | 4.81 | FILIP1 | −2.5 | MAOB | −2.42 | CLEC7A | 2.43 | EFHD1 | −3.38 |
| CKMT2 | 3.30 | FLNA | −2.14 | MAP3K7 | −1.53 | CNN1 | −3.27 | ERRFI1 | −1.72 |
| CLDND1 | 3.49 | GPX3 | −3.13 | MRGPRF | −2.37 | COL11A1 | 7.24 | FBLN1 | −2.03 |
| CLEC5A | 5.65 | GREM1 | −3.86 | MSR1 | 3.19 | COL1A1 | 3.21 | FHL5 | −2.46 |
| CLEC7A | 5.65 | HNRNPC | −1.51 | MYH10 | −2.70 | COL1A2 | 1.74 | FLVCR2 | 1.65 |
| CNN1 | 2.95 | HSP90AA1 | −3.44 | MYH11 | 1.60 | CP | 2.76 | FXYD1 | −1.74 |
| COL11A1 | 5.32 | HSPB8 | −1.55 | NETO2 | −3.24 | CRISPLD2 | −1.97 | GDF15 | −1.54 |
| COL1A1 | 5.08 | IGFBP5 | −3 | NGFR | −2.12 | CSRP1 | −2.23 | GLDN | −1.84 |
| COL1A2 | 4.10 | IGFBP7 | −2.23 | NMD3 | 1.59 | CSRP2 | −2.68 | GPX3 | −2.87 |
| CP | 5.40 | INPP5A | −1.67 | NRAP | −2.94 | CTNNA3 | −4.40 | HSPB8 | −2.04 |
| CPE | 6.01 | KANK1 | −2.65 | NUP210 | 3.22 | CXCL14 | −4.32 | IGFBP2 | −2.97 |
| CRISPLD2 | 5.12 | KCNAB1 | −2.37 | PDCD7 | −2.95 | CXCL16 | 2.48 | IGFBP5 | −1.53 |
| CSRP1 | 5.08 | LAMC1 | −1.88 | PIK3CD | −1.82 | DEF6 | 1.66 | IGFBP7 | −2.01 |
| CSRP2 | 2.89 | LMOD1 | −2.8 | PMP2 | 2.93 | DES | −5.09 | IL10RA | 1.73 |
| CTNNA3 | 5.82 | LPP | −2.79 | PNN | 2.83 | ESR1 | −2.15 | IL1RN | 2.86 |
| CXCL14 | 3.42 | LRRC40 | −1.9 | PNPLA8 | 1.71 | FGF12 | −1.95 | INPP5A | −1.72 |
| CXCL16 | 5.14 | MAOB | −1.78 | PPARGC1A | 1.84 | FHL5 | −3.39 | IRF8 | 1.63 |
| CXCL5 | 3.60 | MAP3K7 | −1.51 | PRKAR2B | 2.56 | FILIP1 | −2.18 | KANK1 | −2.28 |
| CYBRD1 | 4.78 | MPP7 | −2.33 | RAB14 | −2.66 | FLNA | −1.83 | KCNA5 | −3.29 |
| DEF6 | 5.52 | MRGPRF | −2.35 | RHAG | 3.09 | FLVCR2 | 2.26 | KCNAB1 | −2.38 |
| DES | 6.00 | MYH11 | −2.65 | ROR2 | −1.81 | FN1 | 2.06 | KCNMA1 | −1.93 |
| DIXDC1 | 4.54 | NEGR1 | −2.47 | RPS17 | 2.24 | FXYD1 | −1.50 | LAMC1 | −1.80 |
| EFHD1 | 5.22 | NMD3 | −1.5 | SGSM1 | 2.28 | GABRA4 | −1.87 | LMO3 | −2.50 |
| ERRFI1 | 2.96 | NUP54 | −1.6 | SMPDL3A | 2.56 | GALNT3 | 1.84 | LMOD1 | −2.86 |
| ESR1 | 4.29 | PDCD7 | −1.77 | SORBS1 | 4.45 | GDF15 | 2.31 | LPP | −2.39 |
| FBLN1 | 5.19 | PDE5A | −2.07 | STAC | 1.80 | GLDN | −2.90 | LTBP2 | −2.00 |
| FGB | 5.53 | PDGFD | −1.66 | SULT1E1 | 1.71 | GNAO1 | −2.10 | MAOB | −2.06 |
| FGF12 | 4.16 | PDLIM3 | −2.33 | TIMP3 | 2.56 | GPRIN3 | 2.07 | MGP | −2.51 |
| FHL5 | 5.73 | PGM5 | −2.57 | TMEM169 | −1.59 | GREM1 | 2.80 | MMP1 | 1.59 |
| FILIP1 | 3.32 | PLN | −5.01 | ZNF280B | 1.77 | HNRNPC | 1.55 | MMP12 | 2.70 |
| FLNA | 4.10 | PLSCR4 | −2.8 | IGFBP2 | 2.42 | MPEG1 | 2.51 | ||
| FLVCR2 | 2.48 | PNN | −1.73 | IGSF11 | −3.31 | MRAP2 | −3.66 | ||
| FN1 | 7.15 | PNPLA8 | −2.45 | IGSF21 | 1.75 | MRGPRF | −1.62 | ||
| FXYD1 | 4.48 | PPP1R14A | −2.17 | IL10RA | 1.80 | MYH10 | −3.62 | ||
| GABRA4 | 5.61 | PRKAR2B | −2.32 | IL1RN | 2.31 | MYH11 | −3.54 | ||
| GALNT3 | 4.44 | PRR11 | −2.31 | INPP5A | −1.61 | MYOC | −1.74 | ||
| GDF15 | 4.98 | PRUNE2 | −2.43 | IRF8 | 1.75 | NOV | −3.26 | ||
| GLDN | 5.70 | RAB14 | −1.86 | KANK1 | −2.06 | NUP210 | 1.59 | ||
| GNAO1 | 3.35 | RASSF3 | −3.17 | KCNA5 | −4.42 | PDGFD | −2.78 | ||
| GPRIN3 | 3.90 | RGS5 | −2.35 | KCNAB1 | −3.42 | PDLIM3 | −2.82 | ||
| GPX3 | 5.12 | RNASE1 | −1.72 | KCNMA1 | −2.49 | PIK3CD | 1.99 | ||
| GREM1 | 5.02 | RPS17 | −1.97 | LMO3 | −2.38 | PLAC8 | 1.63 | ||
| HNRNPC | 4.61 | SERPING1 | −2.19 | LMOD1 | −3.19 | PLN | −3.28 | ||
| HSP90AA1 | −3.60 | SFRP1 | −1.99 | LPP | −1.56 | PLSCR4 | −1.62 | ||
| HSPB8 | 5.57 | SMOC2 | −2.48 | LTBP2 | 1.54 | PNKD | 1.58 | ||
| IGFBP2 | 3.58 | SORBS1 | −4.15 | MAOB | −3.06 | PPP1R14A | −2.63 | ||
| IGFBP5 | 4.43 | SORBS2 | −3.51 | MGP | 2.20 | PRUNE2 | −2.61 | ||
| IGFBP7 | 3.50 | SPARCL1 | −2.56 | MMP1 | 2.32 | RBPMS | −2.20 | ||
| IGSF11 | 2.78 | SRPX | −2.67 | MMP12 | 1.98 | RCAN2 | −3.16 | ||
| IGSF21 | 3.24 | SYNM | −2.44 | MPEG1 | 2.14 | RGS5 | −3.42 | ||
| IL10RA | 2.78 | TAGLN | −2.54 | MPP7 | −3.58 | RNASE1 | 1.67 | ||
| IL1RN | 6.21 | TCEAL4 | −2.97 | MRAP2 | −1.56 | SCARA3 | −1.67 | ||
| INPP5A | 3.63 | TGFBR3 | −2.01 | MRGPRF | −2.10 | SERPING1 | −2.03 | ||
| IRF8 | 5.86 | TIMP3 | −2.01 | MSR1 | 3.35 | SLC37A2 | 1.93 | ||
| KANK1 | 3.89 | TMEM47 | −2.52 | MYH11 | −4.72 | SLCO2B1 | 1.67 | ||
| KCNA5 | 4.74 | TMTC1 | −1.67 | MYOC | −4.14 | SMOC2 | −2.86 | ||
| KCNAB1 | 2.95 | ZAK | −2.74 | NEGR1 | −2.23 | SNX10 | 2.22 | ||
| KCNMA1 | 3.39 | NETO2 | 2.28 | SORBS1 | −3.16 | ||||
| LAMC1 | 3.59 | NGFR | −1.57 | SORBS2 | −1.52 | ||||
| LMO3 | 3.12 | NOV | −3.14 | SPARCL1 | −3.06 | ||||
| LMOD1 | 3.06 | NRAP | −2.27 | SRPX | −1.93 | ||||
| LPP | 4.87 | NUP210 | 1.86 | SYNM | −2.49 | ||||
| LRRC40 | 4.39 | NUP54 | −1.61 | TAGLN | −3.01 | ||||
| LTBP2 | 5.15 | PDE5A | −1.58 | TCEAL4 | −1.85 | ||||
| MAOB | 5.84 | PDGFD | −1.91 | TGFBR3 | −1.96 | ||||
| MAP3K7 | 5.05 | PGM5 | −3.02 | THBS2 | −2.57 | ||||
| MGP | 5.97 | PLAC8 | 2.26 | TIMP4 | −1.62 | ||||
| MMP1 | 4.57 | PLN | −3.07 | TMEM47 | −3.01 | ||||
| MMP12 | 6.16 | PMP2 | −3.58 | TSPAN33 | 1.70 | ||||
| MPEG1 | 4.02 | PNKD | 2.24 | WISP2 | −2.09 | ||||
| MPP7 | 4.65 | PPARGC1A | −1.68 | ZAK | −1.85 | ||||
| MRAP2 | 5.45 | PPP1R14A | −2.16 | ||||||
| MRGPRF | 5.19 | PRR11 | 1.71 | ||||||
| MSR1 | 4.34 | PRUNE2 | −3.87 | ||||||
| MYH10 | 4.89 | RASSF3 | −2.12 | ||||||
| MYH11 | 4.31 | RBPMS | −2.21 | ||||||
| MYOC | 3.88 | RCAN2 | −1.72 | ||||||
| NEGR1 | 3.15 | RHAG | −2.59 | ||||||
| NETO2 | 2.77 | ROR2 | 2.05 | ||||||
| NGFR | 4.62 | SCARA3 | −2.17 | ||||||
| NMD3 | 2.96 | SFRP1 | −2.93 | ||||||
| NOV | 6.11 | SGSM1 | −1.75 | ||||||
| NRAP | 4.85 | SLC37A2 | 1.51 | ||||||
| NUP210 | 4.76 | SLCO2B1 | 2.77 | ||||||
| NUP54 | 4.12 | SMPDL3A | 1.54 | ||||||
| PDCD7 | 4.38 | SNX10 | 1.94 | ||||||
| PDE5A | 6.51 | SORBS2 | −2.71 | ||||||
| PDGFD | 3.20 | STAC | −2.66 | ||||||
| PDLIM3 | 3.76 | SULT1E1 | 2.27 | ||||||
| PGM5 | 5.00 | SYNM | −1.69 | ||||||
| PIK3CD | 2.66 | TAGLN | −2.31 | ||||||
| PLAC8 | 6.22 | THBS2 | 1.91 | ||||||
| PLN | 3.39 | TIMP4 | −2.13 | ||||||
| PLSCR4 | 4.04 | TMEM169 | 1.57 | ||||||
| PMP2 | 6.15 | TMTC1 | −1.62 | ||||||
| PNKD | 5.77 | TSPAN33 | 1.59 | ||||||
| PNN | 5.23 | WISP2 | −4.84 | ||||||
| PNPLA8 | 2.97 | ZAK | −1.93 | ||||||
| PPARGC1A | 5.22 | ZNF280B | 1.88 | ||||||
| PPP1R14A | 3.77 | ||||||||
| PRKAR2B | 4.79 | ||||||||
| PRR11 | 2.70 | ||||||||
| PRUNE2 | 5.93 | ||||||||
| RAB14 | −3.28 | ||||||||
| RASSF3 | 4.94 | ||||||||
| RBPMS | 3.96 | ||||||||
| RCAN2 | 3.42 | ||||||||
| RGS5 | 7.11 | ||||||||
| RHAG | 5.03 | ||||||||
| RNASE1 | 4.97 | ||||||||
| ROR2 | 4.97 | ||||||||
| RPS17 | 4.36 | ||||||||
| SCARA3 | 3.58 | ||||||||
| SERPING1 | 5.94 | ||||||||
| SFRP1 | 5.46 | ||||||||
| SGSM1 | 4.44 | ||||||||
| SLC37A2 | 5.00 | ||||||||
| SLCO2B1 | 5.05 | ||||||||
| SMOC2 | 4.96 | ||||||||
| SMPDL3A | 4.21 | ||||||||
| SNX10 | 4.75 | ||||||||
| SORBS1 | 3.91 | ||||||||
| SORBS2 | 4.13 | ||||||||
| SPARCL1 | 3.31 | ||||||||
| SRPX | 5.76 | ||||||||
| STAC | 4.51 | ||||||||
| SULT1E1 | 4.90 | ||||||||
| SYNM | 5.98 | ||||||||
| TAGLN | 4.45 | ||||||||
| TCEAL4 | −2.02 | ||||||||
| TGFBR3 | 3.51 | ||||||||
| THBS2 | 3.75 | ||||||||
| TIMP3 | 5.40 | ||||||||
| TIMP4 | 5.95 | ||||||||
| TMEM169 | 2.84 | ||||||||
| TMEM47 | 4.99 | ||||||||
| TMTC1 | 4.60 | ||||||||
| TSPAN33 | 3.69 | ||||||||
| WISP2 | 4.37 | ||||||||
| ZAK | 2.34 | ||||||||
| ZNF280B | 3.05 | ||||||||
The location of DGEs on the chromosome. 6 intersecting DEGs are located at different positions on the chromosome. Blue: intersection of IAs and probiotics; Purple: intersection of aneurysms and probiotics