Abstract
Introduction
Hypoplastic left heart syndrome (HLHS) is a rare but devastating congenital heart defect (CHD) accounting for 25% of all infant deaths due to a CHD. The etiology of HLHS remains elusive, but there is increasing evidence to support a genetic cause for HLHS; in particular, this syndrome is associated with abnormalities in genes involved in cardiac development. Consistent with the involvement of heritable genes in structural heart abnormalities, family members of HLHS patients have a higher incidence of both left- and right-sided valve abnormalities, including bicuspid aortic valve (BAV).
Case presentation
We previously described (Am J Med Genet A 173:1848–1857, 2017) a 4-generation family with a 6q25.1 microdeletion encompassing TAB2, a gene known to play an important role in outflow tract and cardiac valve formation during embryonic development. Affected adult family members have short stature, dysmorphic facial features, and multiple valve dysplasia, including BAV. This follow-up report includes previously unpublished details of the cardiac phenotype of affected family members. It also describes a baby recently born into this family who was diagnosed prenatally with short long bones, intrauterine growth restriction (IUGR), and HLHS. He was the second family member to have HLHS; the first died several decades ago. Postnatal genetic testing confirmed the baby had inherited the familial TAB2 deletion.
Conclusions
Our findings suggest TAB2 haploinsufficiency is a risk factor for HLHS and expands the phenotypic spectrum of this microdeletion syndrome. Chromosomal single nucleotide polymorphism (SNP) microarray analysis and molecular testing for a TAB2 loss of function variant should be considered for individuals with HLHS, particularly in those with additional non-cardiac findings such as IUGR, short stature, and/or dysmorphic facial features.
Keywords: Hypoplastic left heart syndrome, TAB2 gene deletion, Bicuspid aortic valve
Introduction
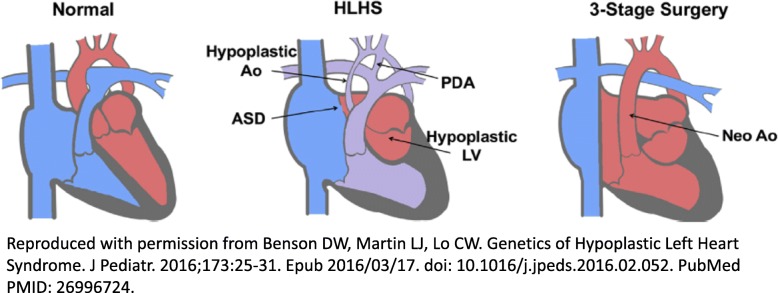
Hypoplastic left heart syndrome (HLHS) is a severe, complex congenital heart defect (CHD) characterized by hypoplasia of the left ventricle and ascending aorta, an atrial septal defect (either large or restrictive), and a patent ductus arteriosus, which provides the only blood flow to the body. It commonly involves atresia or stenosis of the mitral and aortic valves. The prevalence of HLHS is 1.6 per 10,000 live births, and it accounts for 4–8% of all CHD [1]. HLHS is the most severe abnormality in the spectrum of left-sided obstructive CHDs, though it can also be associated with malformation of the tricuspid and pulmonary valves [2]. Although HLHS can be present in a liveborn child, outcomes are universally fatal during infancy without early surgical intervention. Surgical intervention was first implemented in the 1980s, and now involves multiple staged procedures. The end result is that deoxygenated blood is passively directed to the pulmonary circulation via intra-atrial lateral tunnel palliations or more commonly via an extracardiac Fontan circuit (where deoxygenated blood is diverted from the right heart altogether); the right ventricle becomes the systemic ventricle, pumping oxygenated blood through a neo-aorta to the rest of the body (Fig. 1) [1]. Despite surgical advances, HLHS still accounts for 25% of CHD death in infancy, and only 50–70% of affected children live past 5 years of age [2].
Fig. 1.
Hypoplastic left heart syndrome and its surgical repair. Legend: HLHS involves a hypoplastic aorta and left ventricle, a large patent ductus arteriosus (PDA), and an atrial septal defect (ASD). This results in a mixture of oxygenated (from PDA flow) and deoxygenated blood flow to the body. A 3-stage surgical repair involves ligation of the PDA, construction of a neo-Aorta, and a baffle in the right atrium that guides deoxygenated blood into the pulmonary circulation. (Reproduced with permission from Benson DW, Martin LJ, Lo CW. [1])
The pathogenesis of HLHS is unclear, but there is growing literature supporting a genetic etiology. HLHS is highly heritable, with a 500-fold increased incidence among siblings and a 1000-fold increase if a parent has any form of CHD [3]. Approximately 30% of fetuses with HLHS have genetic syndromes or other extra-cardiac abnormalities [4]. Several syndromes caused by chromosomal abnormalities have been associated with HLHS, including Turner syndrome (monosomy X), Edwards syndrome (trisomy 18), DiGeorge syndrome (deletion of 22q11.21), and Jacobsen syndrome (deletion of 11q) [4–6]. Isolated variants in genes involved in cardiac development have been associated with HLHS (Table 1).
Table 1.
Genes Associated with Hypoplastic Left Heart Syndrome
| Gene | Chromosome | Function |
|---|---|---|
| ERBB4 [7] | 2q34 | Encodes a tyrosine protein kinase; required for normal cardiac muscle differentiation during embryonic development, and for postnatal cardiomyocyte proliferation. |
| HAND1 [8] | 5q33.2 | Encodes a basic helix-loop-helix transcription factor important in the formation of the right ventricle and aortic arch arteries |
| NKX2–5 [9–11] | 5q35.1 | Encodes a homeobox-containing transcription factor important in heart formation and development |
| NOTCH1 [12–14] | 9q34.3 | Encodes the Notch 1 protein receptor, which sends signals that are important for normal development of many tissues throughout the body, including the aortic valve |
| GJA1 [15] | 6q22.31 | Encodes a gap junction protein, which play a role in cell-to-cell communication by forming channels, or gap junctions, between cells and are found in many tissues, including the heart |
| TBX5 [16] | 12q24.1 | Encodes T-box protein 5, which plays an important role in the growth and development of the interventricular septum of the heart |
| MYH6 [17] | 14q11.2 | Encodes Myosin-6, found in cardiac muscle cells, where it forms part of a larger protein involved in myocyte contractility |
| FOXC2 [18] | 16q24.1 | Encodes a transcription factor involved in a variety of developmental processes including the cardiovascular system |
Haploinsufficiency or loss of function of TAB2 alone has been shown to be responsible for a multi-system disorder including CHDs. We previously described a 4-generation family (the largest reported to date) with a 6q25.1 microdeletion encompassing TAB2 (TGF-beta activated kinase 1/MAP3K7 binding protein 2) [19]. All affected family members were born with cardiac abnormalities, several with aortic valve malformations, including bicuspid aortic valve (BAV). We now update this family description to include details of the cardiac abnormalities in affected members. We also report the confirmed presence of the TAB2 deletion in a second child in the family to die in infancy from HLHS. These findings suggest that haploinsufficiency of TAB2 is a risk factor for HLHS, expanding the phenotype of the previously reported 6q25.1 microdeletion syndrome [19].
Case presentation
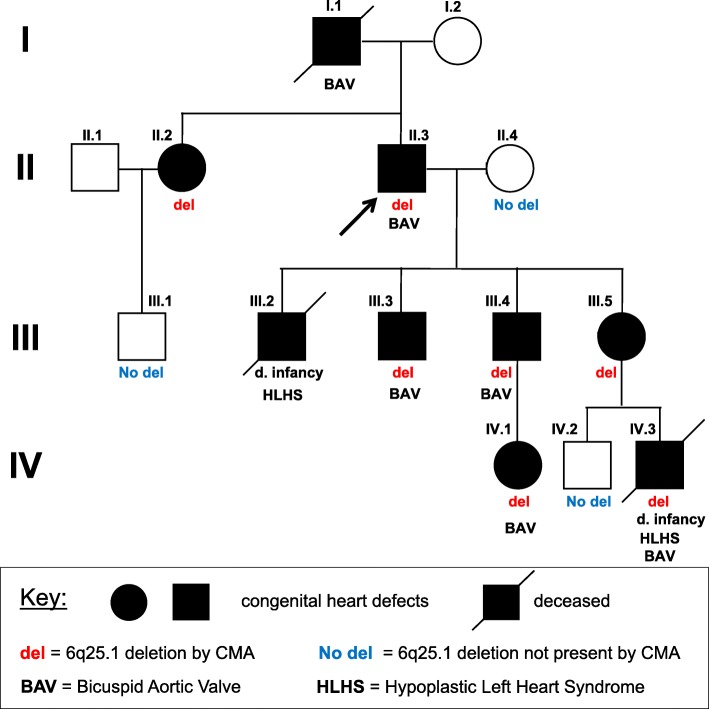
This report focuses on the second member of the family to die during infancy from complications related to HLHS (VI.3; Fig. 2). Except for this newborn baby (VI.3, Fig. 2) this family’s syndromic features, including their extra-cardiac findings, have been previously described [19]. In this report we highlight their echocardiographic findings. For the cardiovascular manifestations in the family, the proband (II.3) was born with BAV and developed progressive aortic dilation, and he ultimately required aortic valve replacement and aortic root repair (Fig. 3). The proband’s father (I.1) also had BAV, along with mitral valve prolapse and a redundant tricuspid valve; he ultimately died from heart failure. The proband had 4 children (III.2-III.5), all born with congenital valve malformations. The first child (III.2) died within a week of birth. He was the first family member to have HLHS, characterized by a hypoplastic/diminutive left ventricle with a large atrial septal defect, dysplastic aortic valve with aortic stenosis, hypoplastic aorta and aortic arch, aortic coarctation, and a large patent ductus arteriosus. He also had redundant atrioventricular valves.
Fig. 2.
Pedigree of the four-generation family showing segregation of the 6q25.1 deletion with congenital heart defects
Fig. 3.

Echocardiogram findings for previously reported family members. Legend: II.3 A: Anterior and posterior aortic leaflets (orange arrows) of a bicuspid aortic valve. II.3B: Calcified aortic valve (red arrow) with a dilated ascending aorta. II.3C: Cardiac MR image showing the ascending aortic aneurysm dilated at 5.8 cm. III.3A: Two leaflets of the bicuspid aortic valve (orange arrows). III.3B: Doming of the aortic valve (blue arrow), consistent with a bicuspid aortic valve, and mitral valve prolapse (red arrow). III.4A: Anterior and posterior leaflets (orange arrows) of a bicuspid aortic valve. III.4B: Thickened mitral valve (red arrow). III.5A: Tri-leaflet aortic valve. III.5B: Thickened mitral valve leaflets (red arrow). IV.1A: Bicuspid aortic valve leaflets (orange arrows). IV.1B-C: Septal defect between the left and right atrium
The second child (III.3) has BAV, bileaflet mitral valve prolapse, and a myxomatous tricuspid valve (Fig. 3). Likewise, the third child (III.4) has BAV and substantial mitral valve thickening (Fig. 3). III.4 has a daughter (IV.1), who was also born with BAV, an atrial septal defect, and a mildly dysplastic pulmonic valve (Fig. 3). Both the fourth child (III.5) and the proband’s sister (II.2) have normal aortic valves but thickened/redundant mitral valve leaflets with mild-to-moderate mitral regurgitation (Fig. 3).
Given the proband’s (II.3) enlarged aorta and bicuspid aortic valve, genetic testing initially focused on genes known to be involved in connective tissue disorders (Marfan, Loeys-Dietz, Ehler-Danlos, Noonan syndromes) and/or BAV (NOTCH1; NKX2–5); no pathogenic variants were identified. Subsequent chromosomal microarray analysis (CMA) performed on the proband (II.3) detected a 1.76 Mb deletion of chromosome 6q24.3-q25.1 ([hg19] chr6:148684028–150,448,233). No other pathogenic copy number variants (CNVs) were present. Testing of the remaining living members of the family showed that the deletion segregates with CHD (Fig. 2). Genetic testing was not possible for III.2, who had died years earlier in the newborn period from complications of HLHS. Thus, while highly probable, it was not definitive that III.2 with HLHS had the familial microdeletion.
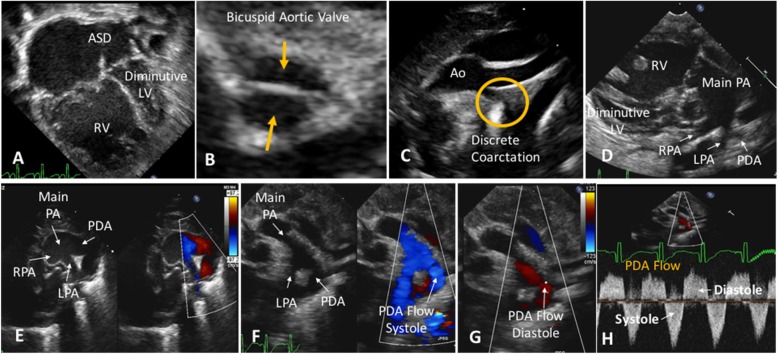
However, III.5 recently had a son (IV.3) also born with HLHS. In addition to HLHS, fetal ultrasound revealed short long bones, intrauterine growth restriction, and a horseshoe kidney. The baby was born at 39 weeks. Birth length was 44.5 cm (< 1%), and birth weight was 3.21 kg (39%). Physical exam on day one of life revealed a sacral dimple and syndromic facies with low-set, posteriorly rotated ears. Despite being born at term, the baby had lung hypoplasia and developed severe respiratory distress. Postnatal echocardiogram showed a diminutive, hypoplastic left ventricle with a parachute mitral valve and BAV. He had a hypoplastic aortic arch with a discrete coarctation. He also had an unrestricted atrial septal defect and a large patent duct arteriosus providing systemic blood flow (Fig. 4). His respiratory distress worsened, and he was too unstable for surgical palliation. The baby died 15 days after birth. Postnatal CMA performed on umbilical cord blood, using the Agilent GGXChip + SNP v1.0 4x180K array platform described previously [19], detected the same microdeletion encompassing TAB2 as seen in the rest of the affected family members.
Fig. 4.
Echocardiogram findings for IV.3. Legend: a Large atrial septal defect (ASD) with a diminutive/hypoplastic left ventricle (LV); RV-right ventricle. b Orange arrows point to bicuspid aortic valve leaflets. c Hypoplastic aorta (Ao) with a discrete coarctation. d and e Two views of the patent ductus arteriosus (PDA); PA -pulmonary artery; RPA - right pulmonary artery; LPA - left pulmonary artery. f-g-h Color and spectral Doppler of the to-and-fro PDA flow from the PA and Ao
Discussion and conclusions
This family’s 6q24.3–25.1 deletion is 1.76 Mb and spans 21 genes (Supplemental Figure 1) [19]. There are multiple lines of evidence implicating TAB2 as the causal gene for structural CHD in this region [19–22], though we cannot definitely exclude involvement of the 20 other genes in this family’s structural heart disease. TAB2 is heavily expressed in the endocardial cushion and plays an important role in outflow tract and valvular formation during human embryonic development. Titrated knockdown of TAB2 in embryonic zebrafish showed dose-sensitive defects in cardiac development [20]. TAB2 was shown to be the only gene within the smallest overlapping region among patients with a 6q25.1 microdeletion and CHD [19], and a balanced translocation that disrupted TAB2 was shown to segregate with familial CHD [20]. Ackerman et al. recently reported a child born with a similar CHD presentation with a sporadic TAB2 nonsense variant (c.1491 T > A; p.Y497X) [21]. TAB2 microdeletions have also been associated with more complex CHD, including tetralogy of Fallot [22]. Our report is the first associating TAB2 haploinsufficiency with HLHS.
Hitz [23] and Carey [24] hypothesize that up to 10% of HLHS is related to chromosomal microdeletions or duplications. In this family with a known chromosomal deletion, two members in differing generations died of HLHS, one of which was verified to have the TAB2 microdeletion. It is unlikely that this is coincidental and unrelated to the deleted gene known to affect cardiac development. Generational skips in phenotype could be related to an autosomal recessive inheritance pattern, but given the rarity of HLHS, the odds of autosomal recessive inheritance are extremely low. The family’s phenotypic and genotypic findings suggest that haploinsufficiency of TAB2 is a risk factor for HLHS. As we collect genetic data on cohorts of individuals with HLHS, it will be worthwhile to see if a 6q25.1 deletion/TAB2 abnormality is more pervasive in this population.
In our 4-generation family, BAV is widely prevalent. A genetic relationship between HLHS and BAV has long been speculated [25]. Approximately 10% of relatives of infants with HLHS have BAV, whereas BAV is present in only 1–2% of the general population [26, 27] . Hinton et al. reported a set of monozygotic twins, one with BAV and the other with HLHS [3]. Pathogenic variants in other genes, such as NOTCH1, cause a spectrum of aortic valve abnormalities, including both BAV [13] and HLHS [28]. Based on observation alone, we cannot definitively prove that BAV and HLHS co-segregate within the family through a common genetic defect. However, like NOTCH1, TAB2 is important in embryonic cardiac development [20]. TAB2 deletions and loss-of-function variants have been shown to cause a variety of left-sided obstructive lesions, so it would not be surprising that BAV and HLHS are both within the phenotypic spectrum of TAB2 haploinsufficiency.
This case report also underscores the complexity of genotype-phenotype predictions. Even within a single family with the identical 6q25.1 microdeletion, there is great variability in the spectrum of CHD, from simple valvular defects to HLHS. Variable expressivity, genetic heterogeneity, and reduced penetrance have been proposed as possible factors contributing to genotype-phenotype differences in CHD [1]. TAB2 appears to be a risk factor for HLHS, but there may be other genetic modifiers and environmental factors critical to the development of this congenital abnormality. Recently, a study using 8 mouse lines with HLHS highlights the genetic heterogeneity of HLHS. Exome sequencing revealed 330 coding or splicing mutations, none which were shared among the mouse lines. In addition, 5 mouse lines had pathogenic variants in 2 or more genes in analogous human chromosomal regions previously associated with HLHS or LV outflow tract obstruction [29]. This discovery favors a multigenic etiology for HLHS. Perhaps the two family members affected with HLHS (III.2 and IV.3) had additional genetic variants predisposing them to more complex CHD.
Although we cannot yet predict who with a TAB2 deletion will have HLHS, our observation that TAB2 haploinsufficiency is associated with HLHS is an important step in further elucidating the genetic underpinnings of this complex congenital heart disease. Thus far, only a handful of gene abnormalities have been linked with this CHD. Given the universal mortality associated with HLHS without early palliation, and the newly recognized association with the 6q25.1 microdeletion, we recommend a fetal echocardiogram in all women carrying an at-risk fetus. Pre-conception genetic counseling is recommended for affected individuals, even those with only a mild phenotype. Furthermore, testing for abnormalities in TAB2 should be considered in patients with HLHS with any non-cardiac abnormalities, including prenatal growth restriction, short stature, and/or dysmorphic facial features. Given the likely genetic heterogeneity of HLHS, chromosomal microarray analysis to evaluate for microdeletions, reflexing to molecular testing for TAB2 loss of function variants should be standard in the genetic work-up of these patients.
Supplementary information
Additional file 1: Supplemental Figure 1. Chromosomal microarray analysis (CMA) results of VI.3 with HLHS using Agilent GGXChip + SNP v1.0 4x180K array. The deletion detected in VI.3 was a 1.76 Mb deletion of chromosome 6q24.3-q25.1 ([hg19] chr6:148684028–150,448,233) and inherited from his mother (III.5). X axis: log2 Ratio. Y axis: genomic location. Non-mosaic one-copy deletions have log2R = − 1. UCSC genes in the deleted region.
Acknowledgements
We thank the patients and their families, without whom this work would not have been possible. We thank Drs. Yu Wu and Yuhua Liu for their technical support.
Abbreviations
- BAV
Bicuspid aortic valve
- CHD
Congenital heart defect
- CMA
Chromosomal microarray analysis
- HLHS
Hypoplastic left heart syndrome
- TAB2
TGF-beta activated kinase 1/MAP3K7 binding protein 2
Authors’ contributions
AC (cardiologist) and PB (medical geneticist) contributed to the clinical examination of the patients in this family. AC interpreted the patient echocardiograms. WNK and YL analyzed and interpreted the cytogenomic microarray data. AC and WNK drafted the manuscript. All authors contributed to, reviewed, and approved the final manuscript.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Availability of data and materials
Chromosomal microarray analysis data for this study is available upon request from the corresponding author.
Ethics approval and consent to participate
This study was approved by the Human Subjects Division and the Institutional Review Board (IRB) at the University of Washington Medical Center (UWMC).
Consent for publication
All the patients/legal guardians provided written informed consent to participate in this study, including review of relevant medical records and consent for publication. A copy of each consent form is available for review by the Editor.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Andrew Cheng, Email: amcheng@uw.edu.
Whitney Neufeld-Kaiser, Email: whitneyn@uw.edu.
Peter H. Byers, Email: pbyers@uw.edu
Yajuan J. Liu, Email: yajuan@uw.edu
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12872-020-01404-5.
References
- 1.Benson DW, Martin LJ, Lo CW. Genetics of Hypoplastic left heart syndrome. J Pediatr. 2016;173:25–31. doi: 10.1016/j.jpeds.2016.02.052. [DOI] [PubMed] [Google Scholar]
- 2.Feinstein JA, Benson DW, Dubin AM, Cohen MS, Maxey DM, Mahle WT, Pahl E, Villafañe J, Bhatt AB, Peng LF, et al. Hypoplastic left heart syndrome: current considerations and expectations. J Am Coll Cardiol. 2012;59(1 Suppl):S1–42. doi: 10.1016/j.jacc.2011.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hinton RB, Martin LJ, Tabangin ME, Mazwi ML, Cripe LH, Benson DW. Hypoplastic left heart syndrome is heritable. J Am Coll Cardiol. 2007;50(16):1590–1595. doi: 10.1016/j.jacc.2007.07.021. [DOI] [PubMed] [Google Scholar]
- 4.Natowicz M, Chatten J, Clancy R, Conard K, Glauser T, Huff D, Lin A, Norwood W, Rorke LB, Uri A. Genetic disorders and major extracardiac anomalies associated with the hypoplastic left heart syndrome. Pediatrics. 1988;82(5):698–706. [PubMed] [Google Scholar]
- 5.Warburton D, Ronemus M, Kline J, Jobanputra V, Williams I, Anyane-Yeboa K, Chung W, Yu L, Wong N, Awad D, et al. The contribution of de novo and rare inherited copy number changes to congenital heart disease in an unselected sample of children with conotruncal defects or hypoplastic left heart disease. Hum Genet. 2014;133(1):11–27. doi: 10.1007/s00439-013-1353-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mattina T, Perrotta CS, Grossfeld P. Jacobsen syndrome. Orphanet J Rare Dis. 2009;4:9. doi: 10.1186/1750-1172-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McBride KL, Zender GA, Fitzgerald-Butt SM, Seagraves NJ, Fernbach SD, Zapata G, Lewin M, Towbin JA, Belmont JW. Association of common variants in ERBB4 with congenital left ventricular outflow tract obstruction defects. Birth Defects Res A Clin Mol Teratol. 2011;91(3):162–168. doi: 10.1002/bdra.20764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reamon-Buettner SM, Ciribilli Y, Inga A, Borlak J. A loss-of-function mutation in the binding domain of HAND1 predicts hypoplasia of the human hearts. Hum Mol Genet. 2008;17(10):1397–1405. doi: 10.1093/hmg/ddn027. [DOI] [PubMed] [Google Scholar]
- 9.McElhinney DB, Geiger E, Blinder J, Benson DW, Goldmuntz E. NKX2.5 mutations in patients with congenital heart disease. J Am Coll Cardiol. 2003;42(9):1650–1655. doi: 10.1016/j.jacc.2003.05.004. [DOI] [PubMed] [Google Scholar]
- 10.Elliott DA, Kirk EP, Yeoh T, Chandar S, McKenzie F, Taylor P, Grossfeld P, Fatkin D, Jones O, Hayes P, et al. Cardiac homeobox gene NKX2-5 mutations and congenital heart disease: associations with atrial septal defect and hypoplastic left heart syndrome. J Am Coll Cardiol. 2003;41(11):2072–2076. doi: 10.1016/S0735-1097(03)00420-0. [DOI] [PubMed] [Google Scholar]
- 11.Stallmeyer B, Fenge H, Nowak-Göttl U, Schulze-Bahr E. Mutational spectrum in the cardiac transcription factor gene NKX2.5 (CSX) associated with congenital heart disease. Clin Genet. 2010;78(6):533–540. doi: 10.1111/j.1399-0004.2010.01422.x. [DOI] [PubMed] [Google Scholar]
- 12.Theis JL, Hrstka SC, Evans JM, O'Byrne MM, de Andrade M, O'Leary PW, Nelson TJ, Olson TM. Compound heterozygous NOTCH1 mutations underlie impaired cardiogenesis in a patient with hypoplastic left heart syndrome. Hum Genet. 2015;134(9):1003–1011. doi: 10.1007/s00439-015-1582-1. [DOI] [PubMed] [Google Scholar]
- 13.Garg V, Muth AN, Ransom JF, Schluterman MK, Barnes R, King IN, Grossfeld PD, Srivastava D. Mutations in NOTCH1 cause aortic valve disease. Nature. 2005;437(7056):270–274. doi: 10.1038/nature03940. [DOI] [PubMed] [Google Scholar]
- 14.McBride KL, Riley MF, Zender GA, Fitzgerald-Butt SM, Towbin JA, Belmont JW, Cole SE. NOTCH1 mutations in individuals with left ventricular outflow tract malformations reduce ligand-induced signaling. Hum Mol Genet. 2008;17(18):2886–2893. doi: 10.1093/hmg/ddn187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dasgupta C, Martinez AM, Zuppan CW, Shah MM, Bailey LL, Fletcher WH. Identification of connexin43 (alpha1) gap junction gene mutations in patients with hypoplastic left heart syndrome by denaturing gradient gel electrophoresis (DGGE) Mutat Res. 2001;479(1–2):173–186. doi: 10.1016/S0027-5107(01)00160-9. [DOI] [PubMed] [Google Scholar]
- 16.Takeuchi JK, Ohgi M, Koshiba-Takeuchi K, Shiratori H, Sakaki I, Ogura K, Saijoh Y, Ogura T. Tbx5 specifies the left/right ventricles and ventricular septum position during cardiogenesis. Development. 2003;130(24):5953–5964. doi: 10.1242/dev.00797. [DOI] [PubMed] [Google Scholar]
- 17.Theis JL, Zimmermann MT, Evans JM, Eckloff BW, Wieben ED, Qureshi MY, O'Leary PW, Olson TM. Recessive MYH6 mutations in Hypoplastic left heart with reduced ejection fraction. Circ Cardiovasc Genet. 2015;8(4):564–571. doi: 10.1161/CIRCGENETICS.115.001070. [DOI] [PubMed] [Google Scholar]
- 18.Stankiewicz P, Sen P, Bhatt SS, Storer M, Xia Z, Bejjani BA, Ou Z, Wiszniewska J, Driscoll DJ, Maisenbacher MK, et al. Genomic and genic deletions of the FOX gene cluster on 16q24.1 and inactivating mutations of FOXF1 cause alveolar capillary dysplasia and other malformations. Am J Hum Genet. 2009;84(6):780–791. doi: 10.1016/j.ajhg.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheng A, Dinulos MBP, Neufeld-Kaiser W, Rosenfeld J, Kyriss M, Madan-Khetarpal S, Risheg H, Byers PH, Liu YJ. 6q25.1 (TAB2) microdeletion syndrome: Congenital heart defects and cardiomyopathy. Am J Med Genet A. 2017;173:1848–1857. doi: 10.1002/ajmg.a.38254. [DOI] [PubMed] [Google Scholar]
- 20.Thienpont B, Zhang L, Postma AV, Breckpot J, Tranchevent LC, Van Loo P, Møllgård K, Tommerup N, Bache I, Tümer Z, et al. Haploinsufficiency of TAB2 causes congenital heart defects in humans. Am J Hum Genet. 2010;86(6):839–849. doi: 10.1016/j.ajhg.2010.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ackerman JP, Smestad JA, Tester DJ, Qureshi MY, Crabb BA, Mendelsohn NJ, Ackerman MJ. Whole exome sequencing, familial genomic triangulation, and systems biology converge to identify a novel nonsense mutation in TAB2-encoded TGF-beta activated kinase 1 in a child with Polyvalvular syndrome. Congenit Heart Dis. 2016;11(5):452–461. doi: 10.1111/chd.12400. [DOI] [PubMed] [Google Scholar]
- 22.Weiss K, Applegate C, Wang T, Batista DA. Familial TAB2 microdeletion and congenital heart defects including unusual valve dysplasia and tetralogy of fallot. Am J Med Genet A. 2015;167A(11):2702–2706. doi: 10.1002/ajmg.a.37210. [DOI] [PubMed] [Google Scholar]
- 23.Hitz MP, Lemieux-Perreault LP, Marshall C, Feroz-Zada Y, Davies R, Yang SW, Lionel AC, D'Amours G, Lemyre E, Cullum R, et al. Rare copy number variants contribute to congenital left-sided heart disease. PLoS Genet. 2012;8(9):e1002903. doi: 10.1371/journal.pgen.1002903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carey AS, Liang L, Edwards J, Brandt T, Mei H, Sharp AJ, Hsu DT, Newburger JW, Ohye RG, Chung WK, et al. Effect of copy number variants on outcomes for infants with single ventricle heart defects. Circ Cardiovasc Genet. 2013;6(5):444–451. doi: 10.1161/CIRCGENETICS.113.000189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hinton RB, Martin LJ, Rame-Gowda S, Tabangin ME, Cripe LH, Benson DW. Hypoplastic left heart syndrome links to chromosomes 10q and 6q and is genetically related to bicuspid aortic valve. J Am Coll Cardiol. 2009;53(12):1065–1071. doi: 10.1016/j.jacc.2008.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loffredo CA, Chokkalingam A, Sill AM, Boughman JA, Clark EB, Scheel J, Brenner JI. Prevalence of congenital cardiovascular malformations among relatives of infants with hypoplastic left heart, coarctation of the aorta, and d-transposition of the great arteries. Am J Med Genet A. 2004;124A(3):225–230. doi: 10.1002/ajmg.a.20366. [DOI] [PubMed] [Google Scholar]
- 27.Braverman AC, Guven H, Beardslee MA, Makan M, Kates AM, Moon MR. The bicuspid aortic valve. Curr Probl Cardiol. 2005;30(9):470–522. doi: 10.1016/j.cpcardiol.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 28.Durbin MD, Cadar AG, Williams CH, Guo Y, Bichell DP, Su YR, Hong CC. Hypoplastic left heart syndrome sequencing reveals a novel NOTCH1 mutation in a family with single ventricle defects. Pediatr Cardiol. 2017;38(6):1232–1240. doi: 10.1007/s00246-017-1650-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu X, Yagi H, Saeed S, Bais AS, Gabriel GC, Chen Z, Peterson KA, Li Y, Schwartz MC, Reynolds WT, et al. The complex genetics of hypoplastic left heart syndrome. Nat Genet. 2017;49(7):1152–1159. doi: 10.1038/ng.3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Supplemental Figure 1. Chromosomal microarray analysis (CMA) results of VI.3 with HLHS using Agilent GGXChip + SNP v1.0 4x180K array. The deletion detected in VI.3 was a 1.76 Mb deletion of chromosome 6q24.3-q25.1 ([hg19] chr6:148684028–150,448,233) and inherited from his mother (III.5). X axis: log2 Ratio. Y axis: genomic location. Non-mosaic one-copy deletions have log2R = − 1. UCSC genes in the deleted region.
Data Availability Statement
Chromosomal microarray analysis data for this study is available upon request from the corresponding author.