Abstract
Objectives
The homozygous GH receptor (GHR) pseudoexon (6Ψ) mutation leads to growth hormone insensitivity (GHI) with clinical and biochemical heterogeneity. We investigated whether transcript heterogeneity (6Ψ-GHR to WT-GHR transcript ratio) and/or concurrent defects in other short stature (SS) genes contribute to this.
Methods
6Ψ-GHR and WT-GHR mRNA transcripts of four 6Ψ patients (height SDS −4.2 to −3.1) and one control fibroblast were investigated by RT-PCR. Transcripts were quantified by qRT-PCR and delta delta CT analysis and compared using ANOVA with Bonferroni correction. In eleven 6Ψ patients, 40 genes known to cause GHI/SS were analysed by targeted next generation sequencing.
Results
RT-PCR confirmed 6Ψ-GHR transcript in the 6Ψ patients but not in the control. 6Ψ-GHR transcript levels were comparable in patients 1 and 3 but significantly different among all other patients. The mean 6Ψ:WT transcript ratios ranged from 29–71:1 for patients 1–4 and correlated negatively with height SDS (R = −0.85; P < 0.001). Eight deleterious variants in six genes were detected, but the number of gene hits did not correlate with the degree of SS in individual 6Ψ patients.
Conclusion
Variable amounts of 6Ψ- and WT-GHR transcripts were identified in 6Ψ patients but no 6Ψ transcript was present in the control. Higher 6Ψ:WT-GHR transcript ratio correlated with SS severity and may explain the phenotypic variability. Analysis of known SS genes suggested that phenotypic variation is independent of the genetic background. This is the first report of transcript heterogeneity producing a spectrum of clinical phenotypes in different individuals harbouring an identical homozygous genetic mutation.
Keywords: short stature, growth hormone insensitivity, GHR pseudoexon, splicing, gene sequencing
Introduction
Growth hormone insensitivity (GHI) is characterised by growth failure, IGF1 deficiency and normal or elevated GH levels. GHI encompasses a spectrum of genetic, phenotypic and biochemical abnormalities associated with growth failure (1, 2). Monogenic defects of the GH-IGF1 axis leading to GHI have been identified in the GHR (3), STAT5B (4), IGFALS (5), PAPPA2 (6) and IGF1 (7) genes.
Splicing is the process by which introns are precisely identified and excised with the remaining exons united to form a translatable message (8). Intronic DNA frequently encodes potential exonic sequences (9). These ‘pseudoexons’ are sequences between 50 and 300 nucleotides in length with apparently viable 5′ and 3′ splice sites (10, 11). Under normal circumstances, they are actively suppressed and not recognised by the splicing machinery (10, 12, 13, 14, 15). However, point mutations in intronic DNA sequences can lead to the creation of new donor (5′), acceptor (3′) splice or branch sites and activation of ‘pseudoexons’. This often occurs within 100 nucleotide bases of the canonical splice site resulting in the inclusion of intronic sequences immediately flanking the exonic sequence (16). Inclusion of intronic sequences by aberrant splicing is a recognised rare cause of several genetic diseases including neurofibromatosis type 1, cystic fibrosis, Duchenne and Becker muscular dystrophies (8, 17).
The intronic growth hormone receptor (GHR) pseudoexon (6Ψ) mutation was first reported in 2001 in two sets of siblings from a highly consanguineous Pakistani family with growth failure and features of relatively mild GHI (18). In 2007, an additional seven 6Ψ patients were reported with a wider range of short stature (SS) phenotypes (19). In 2013, Walenkamp et al. described two further 6Ψ patients with growth failure followed by partial catch-up growth without treatment (20). We recently reported the spectrum of clinical and biochemical features in 20 6Ψ subjects, which included eleven previously reported individuals and nine additional patients (21).
The GHR 6Ψ mutation (c.618+792A>G) is a homozygous point mutation in the final nucleotide of the pseudoexon, altering the 5′ pseudoexon splice site in intron 6 of the GHR gene. This results in activation of the pseudoexon sequence, efficient splicing and inclusion of an additional 108 bases between exons 6 and 7. This results in the inclusion of 36 amino acids in the extracellular domain of the GHR protein (18), and functional work demonstrated that the mutant GHR protein impaired trafficking rather than signalling (22).
Under normal circumstances, the GHR pseudoexon is disregarded, that is, not spliced into the mature GHR mRNA. Previous work confirmed that binding of heterogenous nuclear ribonucleoprotein E1 (hnRNP E1) and U1 small nuclear ribonucleoprotein (snRNP) to the pre-spliceosomal complex prevented 6Ψ inclusion (8).
GHR 6Ψ mutation patients exhibit a wide spectrum of clinical and biochemical variability, even between individuals within the same kindred (21). The height SDS of 20 GHR 6Ψ patients previously described (21) varied between −1.7 and −5.9 with IGF1 SDS between −1.0 and −6.8. Additionally, 50% patients had ‘classical’ GHI facial features and the remainder had completely normal facial appearance (21).
Splice mutations may not always be 100% efficient in causing aberrant splicing, and normal (WT) and mutant transcripts may exist concurrently due to competitive use of normal and mutant splice sites. Previous data suggested that, alongside the abnormally spliced GHR transcript, a small amount of normally spliced WT GHR mRNA was present in GHR 6Ψ patients (18).
Analysis of cDNA of patients carrying different splice mutations of the same gene have shown that multiple abnormal splicing events occur alongside the production of the normal splice product, leading to a spectrum of phenotypes (23, 24, 25). However, transcript variability has not previously been investigated in individuals with identical splice mutations.
The range of phenotypes observed in patients with the GHR 6Ψ mutation may be related to the presence of transcript heterogeneity, that is, the ratio of abnormal (mutant) to normal (WT) GHR transcript. Genetic and environmental factors may also play a role in defining this ratio (19). We investigated for the first time, whether GHR gene transcript heterogeneity and/or concurrent defects in other known short stature genes contributed to the observed clinical variability of 6Ψ subjects.
Subjects and methods
Subjects
The subjects were diagnosed with homozygous intronic GHR 6Ψ mutations at our centre between 2001 and 2014 (18, 19, 21). The referring physicians completed a proforma detailing the clinical and biochemical details at the time of DNA sampling for genetic analysis (21) and was prior to starting any growth promoting therapy. Height measurements were obtained using a wall-mounted stadiometer. Height was expressed as SDS according to the appropriate UK-WHO growth national standards (26, 27). IGF1 values were expressed as SDS based on the age and sex appropriate ranges provided by the host institution.
Fibroblast culture
Dermal fibroblasts from four GHR 6Ψ subjects (patients 1–4) from two consanguineous Pakistani families were obtained by punch skin biopsies, which were performed according to established protocols (28, 29) after written informed consent was obtained. Control human fibroblasts (normal neonatal male dermal fibroblast cell line) were obtained from American Type Culture Collection (ATCC). Fibroblast cells were cultured in 75 cm2 cell culture flasks (Greiner Bio-One, Germany) in High Glucose-DMEM (Sigma-Aldrich) supplemented with 20% foetal bovine serum (Invitrogen), 50 units/mL penicillin and 50 μg/mL streptomycin.
Reverse-transcriptase PCR (RT-PCR)
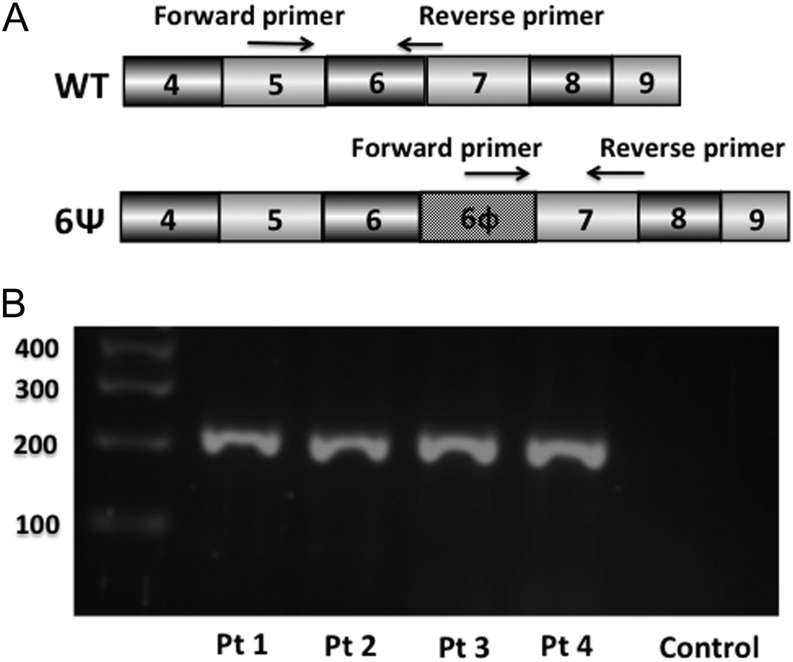
RNA was extracted from fibroblast cell lines using the RNAEasy kit (Qiagen) as per the manufacturer’s instructions. One microgram RNA was reverse transcribed using the Moloney Murine Leukemia Virus Reverse Transcriptase (M-MLV RT) mix (Thermo Fisher Scientific) according to manufacturer’s protocol. cDNA products from four patients and one control subject were amplified using the following intron skipping primers (Fig. 1A):
Figure 1.
Reverse transcriptase PCR (RT-PCR) of WT and mutant transcripts in the 6Ψ and control subjects. (A) Schematic diagram of the GHR gene showing the position of the 6Ψ pseudoexon and intron skipping primers. 6Ψ, mutant pseudoexon transcript; WT, wild type GHR transcript. (B) 2% agarose gel showing products of RT-PCR Reaction 2: 6Ψ transcript (228 bp) in all four 6Ψ patients (patients 1–4) but not in the control subject. Bp, base pairs.
Primers directed to exon 5 of the GHR gene (forward; AGTGCAACCAGATCCACC) and the junction of exons 6 and 7 (reverse; GGAAAATGATGGACCCTATA) to amplify the wild-type (WT-GHR) transcript (Reaction 1).
Primers directed to the GHR 6Ψ (forward; GGCACAGATCACTCCCAG) and the junction of exons 7 and 8 (reverse; GATTTCTACTTTCCATGGCTC) to amplify the mutant (6Ψ-GHR) transcript (Reaction 2).
Thermocycling conditions were: heated lid 110°C; 30 cycles at 95°C for 30 s, 65°C to 55°C for 30 s and 72°C for 30 s followed by 10 cycles at 95°C for 30 s, 60°C for 30 s and 72°C for 30 s. Products were visualised on a 2% agarose gel and verified by Sanger sequencing (GATC Biotech; https://www.gatc-biotech.com).
Quantitative RT-PCR (qRT-PCR)
The relative levels of WT-GHR and 6Ψ-GHR mRNA transcripts were quantified by quantitative RT-PCR using the primer sets previously mentioned (Reactions 1 and 2). Expression levels of the two transcripts were determined quantitatively by the MX3000 real-time PCR system (Stratagene) using SYBR green (Kapabiosystems, USA). Ten nanograms of cDNA were added per 10 μL reaction. Thermocycling conditions were: 1 cycle at 95°C for 3 min, 40 cycles at 95°C for 3 s, 60°C for 20 s and 72°C for 10 s and 1 cycle at 95°C for 1 min, 60°C for 30 s and 72°C for 30 s.
The relative expression levels of the two transcripts were compared in the four patients and one control. Gapdh was used as the internal reference gene, and five technical repeats for each patient were performed. For each experiment, five independent RNA extractions were assayed with three technical replicates. Relative mRNA expression (stated as mean ± s.d.) was calculated by delta delta CT analysis (ΔΔCT), and values in patients and control were compared using one-way ANOVA with Bonferroni correction. P value of < 0.05 was taken as statistical significance.
Genetic analysis by targeted gene sequencing
Genomic DNA was isolated from peripheral blood leukocytes (Qiagen DNeasy Kit) from 11 6Ψ subjects, including patients 1, 2 and 4 from whom skin fibroblasts were obtained. No DNA was available for patient 3. Targeted sequencing of the coding, promotor and intronic regions (2000 bp upstream and 500 bp downstream of each target gene) of 40 short stature genes was undertaken (Table 1). This was processed on an Illumina HiSeq 2500 sequencing platform with paired end of 100 and a designated average coverage of 100× (Otogenetics, Norcross, GA, USA). The raw data from Otogenetics were analysed using DNA Nexus (DNAnexus Inc., Mountain View, CA, USA) by aligning to the H. sapiens GRCh37–b37 (1000 genomes Phase 1) reference genome with BWA-MEM FastQ Readmapper. VCF files were generated by Vendor Human Exome GATK-Lite Variant Caller (Unified Genotyper).
Table 1.
Short stature genes included in the genetic analysis of 11 6Ψ subjects.
| GHR | IGFBP1 | SOS2 | MAP2K1 (MEK1) |
| IGFALS | IGFBP2 | RAF1 | MAP2K2 (MEK2) |
| STAT5B | IGFBP4 | BRAF | A2ML1 |
| IGF1 | IGFBP5 | NRAS | LZTR1 |
| PAPPA-2 | IGF2 | KRAS | SHOC2 |
| IGF1R | OBSL1 | HRAS | ARAF |
| IGFBP3 | CCDC8 | RRAS | NF1 |
| PAPPA | CUL7 | CBL | NPR2 |
| STAT3 | PTPN11 | RIT1 | ACAN |
| JAK2 | SOS1 | RASA2 | SHOX |
Genetic analysis included review of known (GHR, IGFALS, STAT5B, IGF1, PAPPA2 and IGF1R) and putative (IGFBP3, PAPPA, STAT3, JAK2, IGFBP1, IGFBP2, IGFB4 and IGFBP5) monogenic defects of the GH-IGF1 axis leading to GHI and IGF1 resistance phenotypes. We also sought variants in genes associated with overlapping short stature syndromes (2, 34, 35, 36, 37) 3M (OBSL1, CCDC8 and CUL7), Silver–Russell (IGF2) and Noonan (PTPN11, SOS1, SOS2, RAF1, BRAF, NRAS, KRAS, HRAS, RRAS, CBL, RIT1, RASA2, MAP2K1, MAP2K2, A2ML1, LZTR1, SHOC2, ARAF and NF2) syndromes. Other genes associated with short stature (ACAN (61, 62), NPR2 (63) and SHOX (64)) were also included in the analysis.
The resulting VCF files were uploaded to Ingenuity Variant Analysis (Qiagen) and results were analysed with the following filters: call quality ≥20, read depth ≥10 and data outside 5% of most exonically variable 100 base windows in healthy public genomes. Common variants were filtered out by excluding those with a minor allele frequency of ≥0.5% in the 1000 genomes, ExAC and the NHLBI exomes.
Genetic variants were investigated in silico by SIFT (score 0, predicted deleterious to 1, predicted benign), PolyPhen-2 (score 0, predicted benign to 1, predicted deleterious) and CADD (Combined Annotation Dependent Depletion) score to predict the functional outcome. The CADD score assesses the negative effect of single nucleotide variants as well as insertion/deletions variants. A scaled CADD score of 20 represents a variant that is among the top 1% deleterious variants, and a scaled CADD score of 30 means that the variant is in the top 0.1%.
Ethical approval
The study was approved by the Health Research Authority, East of England – Cambridge East Research Ethics Committee (REC reference: 17/EE/0178). Informed written consent for genetic research, skin biopsy and publication of clinical details was obtained from parents/carers and the patients where appropriate.
Results
Clinical phenotypes
The subjects studied had a range of clinical and biochemical heterogeneity as previously described. Patients 1 (height SDS −3.6 and IGF1 SDS −2.0) and 4 (height SDS −3.1 and IGF1 SDS −2.5) were first cousins from a consanguineous Pakistani family with no dysmorphic facial features. Patients 2 (height SDS −4.2 and IGF1 SDS −2.5) and 3 (height SDS −3.8 and IGF1 SDS −2.3) are siblings from another consanguineous Pakistani family. Both had typical facial features of GHI with mid-facial hypoplasia, depressed nasal bridge and prominent forehead. The mean age at presentation of the four patients was 3.2 years (range 2.6–3.8 years) (Table 2).
Table 2.
Phenotypic features and 6Ψ-GHR to WT-GHR transcript ratios in the four 6Ψ subjects.
| Patient | Sex | Age (years) | Height SDS | IGF1 SDS | Facial features | Mean 6Ψ/WT transcript ratio |
|---|---|---|---|---|---|---|
| 1 | M | 3.8 | −3.6 | −2.0 | N | 39.2:1 |
| 2 | F | 3.7 | −4.2 | −2.5 | Y | 70.7:1 |
| 3 | M | 2.6 | −3.8 | −2.3 | Y | 46.9:1 |
| 4 | M | 2.8 | −3.1 | −2.5 | N | 29.4:1 |
6Ψ, mutant pseudoexon transcript; F, female; Facial features, facial features of classical GHI (frontal bossing, mid-facial hypoplasia); M, male; N, no; WT, wild type GHR transcript; Y, yes.
The mean age at presentation of the 11 6Ψ patients who underwent targeted gene sequencing was 4.8 years (range 1.2 to 9.9 years), mean height SDS −4.1 (range −3.0 to −5.1 SDS) and mean IGF1 SDS was −2.6 (range −4.0 to −2.0). 10/11 patients and 1/11 patients were from consanguineous Pakistani families and a non-consanguineous Indian family, respectively. Consistent with our previous report (21), 6/12 (50%) had facial features of GHI, as previously mentioned.
WT and mutant GHR transcript expression
In order to amplify the WT-GHR transcript, a RT-PCR reaction was performed (Reaction 1), using primers directed to exon 5 and the junction of exons 6 and 7 of the GHR gene (Fig. 1A). The WT-GHR transcript (193 bp) was identified in all four 6Ψ subjects and the control. The 6Ψ-GHR transcript was amplified by a RT-PCR reaction (Reaction 2) using primers directed to the GHR 6Ψ and the junction of exons 7 and 8 of the GHR gene (Fig. 1A). The mutant 6Ψ-GHR transcript (228 bp) was identified in all 6Ψ subjects but not the control (Fig. 1B). Sanger sequencing verified all the predicted cDNA sequences.
Quantification of the WT and mutant GHR transcripts
qRT-PCR quantified the relative levels of the WT-GHR and mutant 6Ψ-GHR transcripts, using the primer sets previously mentioned (Reactions 1 and 2).
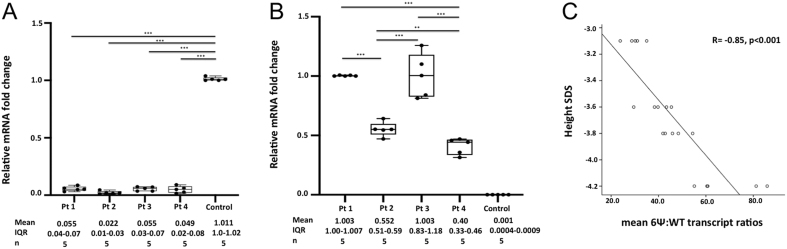
WT-GHR mRNA expression (mean ± s.d.) relative to control was 0.055 ± 0.021, 0.022 ± 0.014, 0.055 ± 0.018 and 0.049 ± 0.034 for patients 1–4, respectively (Fig. 2A). This was significantly lower in all patients compared to control (1.001 ± 0.016); all P values <0.001. This suggests that only small amounts of WT-GHR transcript are present in the GHR 6Ψ patients.
Figure 2.
Quantitative RT-PCR (qRT-PCR). (A) Box and Whisker plot with jitter showing qRT-PCR of WT-GHR mRNA fold change relative to control. Box plots show the mean, upper and lower quartiles and range; IQR = interquartile range; P values calculated by one way ANOVA with Bonferroni correction. *** P value <0.001. (B) Box and Whisker plot with jitter showing qRT-PCR of 6Ψ-GHR mRNA fold change relative to Pt 1. Box plots show the mean, upper and lower quartiles and range; IQR = interquartile range; P values calculated by one-way ANOVA with Bonferroni correction. ***P value <0.001; **P value = 0.017. (C) Scatter plot showing the correlation between the height SDS at presentation and the mean 6Ψ:WT transcript ratios in the four 6Ψ patients. Pt, patient; 6Ψ, pseudoexon; WT, wild type; R, Pearson correlation coefficient.
Mutant 6Ψ-GHR mRNA expression was calculated relative to patient 1, rather than control, as mutant transcript expression was negligible in the control compared to the 6Ψ subjects. Mutant 6Ψ-GHR mRNA expression (mean ± s.d.) relative to patient 1 (1.003 ± 0.004) were 0.552 ± 0.061, 1.003 ± 0.180 and 0.40 ± 0.069 for patients 2–4, respectively, and 0.001 ± 0.0003 for control. There was no significant difference in 6Ψ-GHR transcript levels between patients 1 and 3. However, 6Ψ-GHR transcript levels were significantly different between all the other patients (1 and 2, 1 and 4, 2 and 3 and 3 and 4; P < 0.001 and patients 2 and 4; P = 0.017) (Fig. 2B). This confirms variable amounts of mutant 6Ψ-GHR transcript in the GHR 6Ψ subjects, with negligible levels in control.
The mean 6Ψ:WT transcript ratios for patients 1–4 were 39:1, 71:1, 47:1 and 29:1, respectively (Table 2). These values correlated negatively with height SDS (R = −0.85, P value <0.001) (Fig. 2C). This would suggest that shorter patients have transcript ratios in favour of the mutant GHR.
Genetic analysis
We analysed 11 6Ψ subjects (which included patients 1, 2 and 4) for genetic variants in 40 known human short stature genes. This revealed eight predicted deleterious variants in six genes (IGFALS, OBSL1, CBL, IGF1R, ACAN and CUL7) in eight of the 11 6Ψ subjects (Table 3). Patients 9 and 10 had compound heterozygous missense variants in CUL7 and IGFALS, respectively (Table 3). The remaining 6/8 variants were monoallelic (patients 2, 4, and 5–8). 4/6 variants were missense and 2/6 variants were in-frame insertions. Of the 8 variants, 6 had a CADD score of >20. Patients 4 and 6 had in-frame insertions in CBL and IGF1R genes (CADD score 12.6 and not known, respectively). None of the patients had homozygous variants identified.
Table 3.
Genetic variants identified in known short stature genes in the 6Ψ subjects.
| Pt | Age (years) | Height SDS | IGF-1 SDS | Gene transcript | Translation | SIFT score | Polyphen | CADD score | ACMG/AMP classification |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 3.8 | −3.6 | −2.0 | No deleterious variants detected | N/A | N/A | N/A | N/A | N/A |
| 2 | 3.7 | −4.2 | −2.5 |
ACAN Het c.1223A>G, p.Q532H |
Missense | Damaging | Probably damaging |
23.7 | Uncertain significance |
| 3 | 2.6 | −3.8 | −2.3 | No DNA available | N/A | N/A | N/A | N/A | N/A |
| 4 | 2.8 | −3.1 | −2.5 |
OBSL1 Het c.2671A>G, p.T891A |
Missense | Damaging | Probably damaging |
26.8 | Uncertain significance |
|
CBL Het c.125_127dupACC, p.H42dup |
In-frame | NK | NK | 12.6 | Likely benign | ||||
| 5 | 5.7 | −4.5 | −2.5 |
OBSL1 Het c.2671A>G, p.T891A |
Missense | Damaging | Probably damaging |
26.8 | Uncertain significance |
| 6 | 1.2 | −4.4 | −2.2 |
IGF1R Het c.109_123dupATCGACATCCGCAAC, p.I37_N41dup |
In-frame | NK | NK | NK | Uncertain significance |
| 7 | 7.0 | −4.2 | −2.5 |
CBL Het c.2216C>T, p.S739F |
Missense | Damaging | Benign | 23.1 | Uncertain significance |
| 8 | 5.7 | −3.0 | −2.9 |
ACAN Het c.1596G>C, p.R132C |
Missense | Damaging | Probably damaging |
27.9 | Uncertain significance |
| 9 | 4.3 | −4.1 | −4.0 |
CUL7 Het c.464G>A, p.G155E c.620G>A, p.G207E |
Missense | Damaging | Probably damaging |
26.9 | Uncertain significance |
| 10 | 2.5 | −4.4 | NK |
IGFALS Het c.544C>A, p.L220M c.658C>A, p.L182M |
Missense | Damaging | Probably damaging |
23.5 | Uncertain significance |
| 11 | 5.7 | −4.7 | −3.1 | No deleterious variants detected | N/A | N/A | N/A | N/A | N/A |
| 12 | 9.9 | −5.1 | −2.1 | No deleterious variants detected | N/A | N/A | N/A | N/A | N/A |
Patients 1–4 are the 6Ψ patients analysed for WT-GHR and 6Ψ-GHR transcript ratios (Table 2).
ACMG/AMP classification, classification as per the American College of Medical Genetics and Genomics and the Association for Molecular Pathology’s standards and guidelines for the interpretation of sequence variants; c., coding DNA sequence where nucleotide 1 is the A of the ATG-translation initiation codon (NCBI reference sequences: for ACAN, NM_013227.2; for OBSL1, NM_015311.2; for CBL, NM_005188.3; for IGF1R, NM_000875.3; for CUL7, NM_001168370.1 and for IGFALS, NM_004970.2); CADD, combined annotation dependent depletion; Ht, height; N/A, not applicable; NK, not known; Pt, patient (30).
We classified the pathogenicity of the variants according to the American College of Medical Genetics and Genomics and the Association for Molecular Pathology (ACMG/AMP)’s standards and guidelines for the interpretation of sequence variants (30) (Table 3). Out of the 8 predicted deleterious variants, 7 were variants of ‘uncertain significance’ (30). Although the missense variants are very rare in populations (allele frequency <0.001 in the gnomAD database (31)) and in silico variant prediction tools support a deleterious effect on the gene, they fall into the category of ‘uncertain significance’ as they do not meet the criteria for being either pathogenic or benign (30). Furthermore, although the IGF1R heterozygous c.109_123dupATCGACATCCGCAAC, p.I37_N41dup in-frame insertion variant fulfils two moderate criteria for pathogenicity (30), that is, protein length changes as a result of in-frame deletion in a non-repeat region and is absent from controls as well as large databases (allele frequency 0.002 in the gnomAD database (31)), it does not strictly meet the ACMG/AMP criteria for pathogenicity and hence is classified as ‘uncertain significance’ (30). The CBL heterozygous c.125_127dupACC, p.H42dup variant is ‘likely benign’ (30), as it is an in-frame insertion in a repetitive region without known function and there is 1 homozygote in the gnomAD database (31).
Patients 2 and 8 with heterozygous ACAN variants did not have advanced bone maturation, early onset osteo-arthritis or intervertebral disc disease. Patient 6 (heterozygous in-frame IGF1R insertion) had normal birth weight, normal IGF-1 SDS, no developmental delay and consequently also did not fulfil the published criteria for the identification of patients with suspected IGF1R mutations (total score <3) (32). Patients 4 and 7 with CBL variants had no cardiac defects or developmental delay. Therefore, although these defects could potentially contribute to the short stature, no phenotypic features associated with these heterozygous genetic defects were detected.
In the four patients who underwent transcript heterogeneity analysis, three underwent genetic sequencing of short stature genes. Patient 2 had a heterozygous missense ACAN variant. Patient 4 had a heterozygous missense OBSL1 variant and a heterozygous in-frame CBL variant. Patient 1 had no predicted deleterious variants.
Reviewing all patients assessed on the gene panel (Table 3; n = 11), there was no correlation between the number of predicted deleterious genetic variants (0, 1 or 2) and the degree of short stature (height SDS) in the individual 6Ψ patients, that is, patient 4 had two variants and was not significantly shorter than those with one (n = 7) or no variant(s) (n = 3) (Table 3). Additionally, the mean (±s.d.) height SDS of patients with heterozygous or compound heterozygous variants in autosomal dominant inherited genes (ACAN, CBL and IGF1R) (−3.78 ± 0.67) was not significantly different from that of patients with heterozygous or compound heterozygous variants in autosomal recessive inherited genes (OBSL, CUL7, IGFALS) or patients with no deleterious variants (−4.1 ± 0.56), P = 0.39.
Discussion
GH insensitivity (GHI) is a rare disorder caused by mutations in multiple GH-IGF1 axis genes. Homozygous GHR mutations are the commonest cause of ‘classical’ GHI and are associated with a wide range of clinical and biochemical phenotypes (33). More recently, it has been noted that several other short stature disorders such as 3M, Noonan Syndrome (NS) and Silver–Russell Syndrome (SRS) overlap with GHI (2, 34, 35, 36, 37).
Milder or ‘non-classical’ GHI cases are being increasingly recognised owing to advent of next generation sequencing techniques and an increased awareness of this group of disorders. Other molecular defects of the GH-IGF1 axis that cause ‘non-classical’ or milder GHI phenotypes include dominant negative GHR (38, 39, 40, 41) and STAT5B (42) gene mutations, heterozygous IGF1 (43, 44, 45), IGF2 (46, 47) and IGFALS (48, 49) mutations, homozygous PAPPA2 (6) mutations and the GHR pseudoexon (6Ψ) mutation (18, 19, 20, 21). Non-classical GHI is an important clinical entity and the prevalence may be higher than classical GHI (2). Approximately 70 different GHR gene mutations have been reported in more than 300 patients (1, 2). A recent UK study (37) reporting the genetic diagnoses obtained from candidate gene and whole exome sequencing (WES) in a selected group of 107 patients with GHI showed that the GHR 6Ψ mutation contributed to 25% (8/32) of GHR mutations and 16% (8/51) of all genetic diagnoses.
The GHR 6Ψ mutation is notable, as striking phenotypic variability is observed between different patients harbouring the same homozygous point mutation, even among members of the same kindred (21). We hypothesise that several factors, alone or in combination, could contribute to the phenotypic variability observed between individual patients and include: (1.) differences in the levels of mutant vs WT GHR transcripts, (2.) concurrent genetic variants contributing to the degree of growth failure, (3.) environmental factors and (4.) genetic variability in the cellular processes that regulate the mutant and WT-GHR proteins. These are discussed in detail subsequently.
David et al. (19) suggested that the spectrum of clinical phenotypes observed in the GHR 6Ψ patients may be due to competitive use of both WT and 6Ψ mutant splice sites resulting in different ratios of the two transcripts. The current study tested the hypothesis that transcript heterogeneity could account for the phenotypic differences, particularly the severity of growth failure in the 6Ψ subjects.
We found variable levels of mutant 6Ψ-GHR mRNA expression among all patients except patients 1 and 3, who had similar height deficits of −3.6 and −3.8, respectively. Our work implies that the splicing of the GHR pseudoexon is highly inconsistent, as variable quantities of mutant transcripts are produced in different individuals with the same mutation. Furthermore, consistent with our previous work (18), we confirmed the existence of WT-GHR transcript in the 6Ψ subjects alongside the mutated 6Ψ-GHR transcript. This demonstrates that normal as well as abnormal splicing events co-exist in these subjects. Our results also confirm that a higher WT to mutant transcript ratio correlates negatively with the height SDS of the patients, that is, greater mutant pseudoexon inclusion may lead to a more severe phenotype.
Interestingly, consistent with previously published work (18), no GHR 6Ψ transcript was detected in the control subject. This would indicate that under normal circumstances, the GHR pseudoexon splice site is not recognised by the splicing machinery, thereby preventing its inclusion in the mature mRNA. This corroborates the work by Akker et al. (8) who showed that the heterogenous nuclear ribonucleoprotein E1 (hnRNP E1) and small nuclear ribonucleoprotein U1 (snRNP U1) bind to the relatively weak ‘wild-type’ 5′ GHR pseudoexon splice site in the pre-spliceosomal complex and silence it. The A>G base change at the 5′ splice site in 6Ψ patients does not create a new splice site but increases the bp match of the pre-mRNA with the snRNP U1. Therefore, the spliceosome recognises the GHR pseudoexon sequence and it is included in the mature mRNA of patients carrying the mutation (19).
Genotype-phenotype correlations secondary to variable splicing have been reported in three previous studies. Zhu (23) and Lemahieu et al. (24) investigated the association of different splice mutations of the WASP (Wiskott-Aldrich syndrome protein) gene with the clinical phenotypes of patients with WAS and X-linked thrombocytopaenia. Another study by Gurvich et al. (25) investigated the correlation between the clinical phenotypes of two patients and the splicing efficiency of two different intronic pseudoexon mutations in the DMD (Duchenne muscular dystrophy) gene. All three studies demonstrated that relative amounts of mutant to normal transcripts positively correlated with the severity of the phenotypes of affected individuals. However, these studies examined different splice site mutations of the individual genes. Our work is the first to report the phenotypic impact of transcript heterogeneity in patients with an identical homozygous splice mutation.
The potential underlying mechanism(s) leading to a spectrum of phenotypes in subjects with the same genetic mutation are unclear. Gurvich et al. (25) proposed that the difference in splicing efficiency between the two different DMD gene mutations could be attributed to the strength of the donor splice sites created by the mutation and/or the strength of the cryptic acceptor splice sites in the WT sequence which are further activated by the point mutations. This explanation would not be applicable to our patients, as the 6Ψ intronic point mutation leads to activation of the same 5′ donor splice site in all the subjects.
David et al. (19) also suggested that the genetic background of individual patients and/or environmental factors may play a role in determining the differential use of WT vs mutant splice sites and thus generation of variable quantities of normal vs mutated transcript. Variations in the core spliceosome machinery (comprising hnRNP and snRNP) and variations in genes encoding RNA processing/RNA-binding factors can influence alternative splicing (50, 51, 52). It is possible that differences in these factors between 6Ψ patients regulate the differential use of the mutated and WT splice sites and thus determine the amount of mutant vs WT transcript generated.
It is feasible that the GHI phenotype, particularly the severity of the growth failure observed in the 6Ψ patients, is influenced by genetic variants in different genetic loci. We sought concurrent defects in genes known to cause short stature, GH insensitivity and overlapping phenotypes, for example, Noonan, Silver–Russell and 3M Syndrome in 11 6Ψ patients.
Analysis of 40 genes (including intronic and promoter regions) revealed that a significant proportion (73%) of 6Ψ subjects had predicted deleterious heterozygous or compound heterozygous genetic variants. Of these, three variants in ACAN, IGF1R and CBL genes are recognised to have an autosomal dominant pattern of inheritance leading to short stature (53, 54, 55). It is possible that these variants contributed to the growth failure in patients 2, 4, 6, 7 and 8. However, none of these patients had additional phenotypic features that would be consistent with the identified additional genetic variant. Furthermore, there was no difference in height SDS between the patients with heterozygous/compound heterozygous variants in genes with an autosomal dominant pattern of inheritance compared to patients with heterozygous/compound heterozygous variants in genes with an autosomal recessive inheritance or patients with no deleterious variants. This implies that the ACAN, IGF1R and CBL gene variants identified did not impact significantly on the patient’s phenotypes. Unfortunately, we were unable to perform segregation studies as parental DNA was not available for any of the subjects. However, consistent with our findings, a previous study of two 6Ψ patients also demonstrated that other genetic variants did not contribute to the severity of the phenotypes (20).
Interestingly, given that the majority of patients have consanguineous family structures, no homozygous variants were detected. We were also unable to identify a correlation between the number of predicted deleterious variants and the severity of short stature in the individual GHR 6Ψ patients studied. This indicates that concurrent defects in this subset of known short stature, GHI and overlapping disorders genes do not appear to contribute to the growth phenotype in these patients. We cannot, however, rule out an impact of other known short stature genes or currently undiscovered short stature genes. It is also possible that multiple genes with smaller effect size could account for some or all of the clinical variation.
Genotyping a region spanning 17.10 Mb around the GHR gene on chromosome 5 and analysis of the complex single polymorphic region in intron 9 of the GHR in five GHR 6Ψ families showed the same genotype for all affected members, suggesting the presence of a common ancestor (19). Furthermore, 19/22 of the known GHR pseudoexon patients are of Pakistani ethnicity (18, 19, 20, 21). This implies that the GHR 6Ψ patient cohort share a common genetic background. This is comparable to the p.E180 splice GHR mutation found predominantly in individuals from Ecuador, Brazil and Chile, where a shared genetic background flanking the splice mutation was identified (56, 57). First degree relatives who are heterozygous carriers of the p.E180 mutation are modestly shorter than non-carrier relatives (58). This is in contrast to our cohort, where heterozygous carrier parents of 6Ψ patients had normal stature (19).
It is recognised that a multitude of environmental factors such as nutrition, socio-economic status and adverse environmental conditions affect childhood growth (59, 60). It is possible that any of these factors contribute to the phenotypic variability observed in our patient cohort. However, as there are marked differences between affected individuals within the same kindred, environmental factors are unlikely to be very significant.
It is feasible that differences in processing, trafficking and degradation of the mutant and WT GH receptor proteins account for the clinical differences (19). Maamra et al. (22) confirmed that the mutant GHR 6Ψ protein compromised normal cell surface trafficking but signalling was unimpaired. Therefore, variability in genes regulating these cellular processes might conceivably have a greater influence on the mutant 6Ψ GHR protein than the WT GHR. Further work exploring this hypothesis is required.
It is important to acknowledge that our study has several limitations. First, GHR transcript ratios were studied in only four 6Ψ patients. Furthermore, one of the four patients was not assessed on the gene panel. Our work would not identify variants in other known short stature genes not included in the panel or defects in currently undiscovered short stature genes. Additionally, the phenotypic spectrum of individual genetic defects is expected to broaden as more patients are reported. Finally, we did not explore the mechanisms underlying the observed variable splicing or genetic variability which might affect GHR protein processing, trafficking and degradation. Further work is required to address these.
In conclusion, our work shows that there is a variable ratio of mutated and WT-GHR mRNA transcripts in GHR 6Ψ patients. The mutated GHR 6Ψ transcript is not normally spliced in unaffected subjects. Patients with more severe phenotypes, that is, lower height SDS, have a GHR transcript ratio in favour of the mutant GHR. The reason for the variable splicing in different patients with the same mutation is fascinating. The genetic background of individuals may influence this. However, our preliminary work does not suggest that variants in candidate SS genes contribute significantly to the variability of growth failure. This is the first indication that variable splicing and transcript heterogeneity can lead to a range of short stature phenotypes in subjects harbouring the same genetic mutation.
Declaration of interest
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
Funding
The genetic sequencing service was supported by a large project research grant from Barts charity (H L S). S C was supported by Barts Charity Clinical Research Training Fellowship.
Author contribution statement
S C, S J R, T M and H L S contributed to patient recruitment, data collection and analysis. S C, E C and L A M performed the genetic analysis. S C performed the molecular work and statistical analyses with support from A V M and J W. S C wrote the manuscript with input from L A M, M O S and H L S.
References
- 1.David A, Hwa V, Metherell LA, Netchine I, Camacho-Hubner C, Clark AJ, Rosenfeld RG, Savage MO. Evidence for a continuum of genetic, phenotypic, and biochemical abnormalities in children with growth hormone insensitivity. Endocrine Reviews 2011. 32 472–497. ( 10.1210/er.2010-0023) [DOI] [PubMed] [Google Scholar]
- 2.Storr HL, Chatterjee S, Metherell LA, Foley C, Rosenfeld RG, Backeljauw PF, Dauber A, Savage MO, Hwa V. Nonclassical GH insensitivity: characterization of mild abnormalities of GH action. Endocrine Reviews 2019. 40 476–505. ( 10.1210/er.2018-00146) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amselem S, Duquesnoy P, Attree O, Novelli G, Bousnina S, Postel-Vinay MC, Goossens M. Laron dwarfism and mutations of the growth hormone-receptor gene. New England Journal of Medicine 1989. 321 989–995. ( 10.1056/NEJM198910123211501) [DOI] [PubMed] [Google Scholar]
- 4.Kofoed EM, Hwa V, Little B, Woods KA, Buckway CK, Tsubaki J, Pratt KL, Bezrodnik L, Jasper H, Tepper A, et al Growth hormone insensitivity associated with a STAT5b mutation. New England Journal of Medicine 2003. 349 1139–1147. ( 10.1056/NEJMoa022926) [DOI] [PubMed] [Google Scholar]
- 5.Domené HM, Bengolea SV, Martinez AS, Ropelato MG, Pennisi P, Scaglia P, Heinrich JJ, Jasper HG. Deficiency of the circulating insulin-like growth factor system associated with inactivation of the acid-labile subunit gene. New England Journal of Medicine 2004. 350 570–577. ( 10.1056/NEJMoa013100) [DOI] [PubMed] [Google Scholar]
- 6.Dauber A, Munoz-Calvo MT, Barrios V, Domene HM, Kloverpris S, Serra-Juhe C, Desikan V, Pozo J, Muzumdar R, Martos-Moreno GÁ, et al Mutations in pregnancy-associated plasma protein A2 cause short stature due to low IGF-I availability. EMBO Molecular Medicine 2016. 8 363–374. ( 10.15252/emmm.201506106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Woods KA, Camacho-Hubner C, Savage MO, Clark AJ. Intrauterine growth retardation and postnatal growth failure associated with deletion of the insulin-like growth factor I gene. New England Journal of Medicine 1996. 335 1363–1367. ( 10.1056/NEJM199610313351805) [DOI] [PubMed] [Google Scholar]
- 8.Akker SA, Misra S, Aslam S, Morgan EL, Smith PJ, Khoo B, Chew SL. Pre-spliceosomal binding of U1 small nuclear ribonucleoprotein (RNP) and heterogenous nuclear RNP E1 is associated with suppression of a growth hormone receptor pseudoexon. Molecular Endocrinology 2007. 21 2529–2540. ( 10.1210/me.2007-0038) [DOI] [PubMed] [Google Scholar]
- 9.Dunham I, Shimizu N, Roe BA, Chissoe S, Hunt AR, Collins JE, Bruskiewich R, Beare DM, Clamp M, Smink LJ, et al The DNA sequence of human chromosome 22. Nature 1999. 402 489–495. ( 10.1038/990031) [DOI] [PubMed] [Google Scholar]
- 10.Sun H, Chasin LA. Multiple splicing defects in an intronic false exon. Molecular and Cellular Biology 2000. 20 6414–6425. ( 10.1128/mcb.20.17.6414-6425.2000) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dhir A, Buratti E. Alternative splicing: role of pseudoexons in human disease and potential therapeutic strategies. FEBS Journal 2010. 277 841–855. ( 10.1111/j.1742-4658.2009.07520.x) [DOI] [PubMed] [Google Scholar]
- 12.Fairbrother WG, Chasin LA. Human genomic sequences that inhibit splicing. Molecular and Cellular Biology 2000. 20 6816–6825. ( 10.1128/mcb.20.18.6816-6825.2000) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang XH, Chasin LA. Computational definition of sequence motifs governing constitutive exon splicing. Genes and Development 2004. 18 1241–1250. ( 10.1101/gad.1195304) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pagani F, Buratti E, Stuani C, Bendix R, Dork T, Baralle FE. A new type of mutation causes a splicing defect in ATM. Nature Genetics 2002. 30 426–429. ( 10.1038/ng858) [DOI] [PubMed] [Google Scholar]
- 15.Zhang XH, Leslie CS, Chasin LA. Dichotomous splicing signals in exon flanks. Genome Research 2005. 15 768–779. ( 10.1101/gr.3217705) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakai K, Sakamoto H. Construction of a novel database containing aberrant splicing mutations of mammalian genes. Gene 1994. 141 171–177. ( 10.1016/0378-1119(94)90567-3) [DOI] [PubMed] [Google Scholar]
- 17.Krawczak M, Reiss J, Cooper DN. The mutational spectrum of single base-pair substitutions in mRNA splice junctions of human genes: causes and consequences. Human Genetics 1992. 90 41–54. ( 10.1007/bf00210743) [DOI] [PubMed] [Google Scholar]
- 18.Metherell LA, Akker SA, Munroe PB, Rose SJ, Caulfield M, Savage MO, Chew SL, Clark AJ. Pseudoexon activation as a novel mechanism for disease resulting in atypical growth-hormone insensitivity. American Journal of Human Genetics 2001. 69 641–646. ( 10.1086/323266) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.David A, Camacho-Hubner C, Bhangoo A, Rose SJ, Miraki-Moud F, Akker SA, Butler GE, Ten S, Clayton PE, Clark AJ, et al An intronic growth hormone receptor mutation causing activation of a pseudoexon is associated with a broad spectrum of growth hormone insensitivity phenotypes. Journal of Clinical Endocrinology and Metabolism 2007. 92 655–659. ( 10.1210/jc.2006-1527) [DOI] [PubMed] [Google Scholar]
- 20.Walenkamp MJE, Klammt J, Feigerlova E, Losekoot M, van Duyvenvoorde HA, Hwa V, Pfäffle R, Wit JM. Genetic analysis of GHR should contain sequencing of all coding exons and specific intron sequences, and screening for exon deletions. Hormone Research in Paediatrics 2013. 80 406–412. ( 10.1159/000355928) [DOI] [PubMed] [Google Scholar]
- 21.Chatterjee S, Shapiro L, Rose SJ, Mushtaq T, Clayton PE, Ten SB, Bhangoo A, Kumbattae U, Dias R, Savage MO, et al Phenotypic spectrum and responses to recombinant human igf1 (rhigf1) therapy in patients with homozygous intronic pseudoexon growth hormone receptor mutation. European Journal of Endocrinology 2018. 178 481–489. ( 10.1530/EJE-18-0042) [DOI] [PubMed] [Google Scholar]
- 22.Maamra M, Milward A, Esfahani HZ, Abbott LP, Metherell LA, Savage MO, Clark AJ, Ross RJ. A 36 residues insertion in the dimerization domain of the growth hormone receptor results in defective trafficking rather than impaired signaling. Journal of Endocrinology 2006. 188 251–261. ( 10.1677/joe.1.06252) [DOI] [PubMed] [Google Scholar]
- 23.Zhu Q, Watanabe C, Liu T, Hollenbaugh D, Blaese RM, Kanner SB, Aruffo A, Ochs HD. Wiskott-Aldrich syndrome/X-linked thrombocytopenia: WASP gene mutations, protein expression, and phenotype. Blood 1997. 90 2680–2689. [PubMed] [Google Scholar]
- 24.Lemahieu V, Gastier JM, Francke U. Novel mutations in the Wiskott-Aldrich syndrome protein gene and their effects on transcriptional, translational, and clinical phenotypes. Human Mutation 1999. 14 54–66. () [DOI] [PubMed] [Google Scholar]
- 25.Gurvich OL, Tuohy TM, Howard MT, Finkel RS, Medne L, Anderson CB, Weiss RB, Wilton SD, Flanigan KM. DMD pseudoexon mutations: splicing efficiency, phenotype, and potential therapy. Annals of Neurology 2008. 63 81–89. ( 10.1002/ana.21290) [DOI] [PubMed] [Google Scholar]
- 26.Freeman JV, Cole TJ, Chinn S, Jones PR, White EM, Preece MA. Cross sectional stature and weight reference curves for the UK, 1990. Archives of Disease in Childhood 1995. 73 17–24. ( 10.1136/adc.73.1.17) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.WHO Multicentre Growth Reference Study Group. WHO child growth standards based on length/height, weight and age. Acta Paediatrica: Supplement 2006. 450 76–85. [DOI] [PubMed] [Google Scholar]
- 28.Poliandri A, Miller D, Howard S, Nobles M, Ruiz-Babot G, Harmer S, Tinker A, McKay T, Guasti L, Dunkel L. Generation of kisspeptin-responsive GnRH neurons from human pluripotent stem cells. Molecular and Cellular Endocrinology 2017. 447 12–22. ( 10.1016/j.mce.2017.02.030) [DOI] [PubMed] [Google Scholar]
- 29.Maharaj A, Buonocore F, Meimaridou E, Ruiz-Babot G, Guasti L, Peng HM, Capper CP, Burgos-Tirado N, Prasad R, Hughes CR, et al Predicted benign and synonymous variants in CYP11A1 cause primary adrenal insufficiency through missplicing. Journal of the Endocrine Society 2019. 3 201–221. ( 10.1210/js.2018-00130) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, et al Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genetics in Medicine 2015. 17 405–424. ( 10.1038/gim.2015.30) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, Collins RL, Laricchia KM, Ganna A, Birnbaum DP, et al Variation across 141,456 human exomes and genomes reveals the spectrum of loss-of-function intolerance across human protein-coding genes. bioRxiv 2019. 531210 ( 10.1101/531210) [DOI] [Google Scholar]
- 32.Walenkamp MJE, Robers JML, Wit JM, Zandwijken GRJ, van Duyvenvoorde HA, Oostdijk W, Hokken-Koelega ACS, Kant SG, Losekoot M. Phenotypic features and response to GH treatment of patients with a molecular defect of the IGF-1 receptor. Journal of Clinical Endocrinology and Metabolism 2019. 104 3157–3171. ( 10.1210/jc.2018-02065) [DOI] [PubMed] [Google Scholar]
- 33.Lin S, Li C, Li C, Zhang X. Growth hormone receptor mutations related to individual dwarfism. International Journal of Molecular Sciences 2018. 19 1433 ( 10.3390/ijms19051433) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Akawi NA, Ali BR, Hamamy H, Al-Hadidy A, Al-Gazali L. Is autosomal recessive silver-Russel syndrome a separate entity or is it part of the 3-M syndrome spectrum? American Journal of Medical Genetics. Part A 2011. 155A 1236–1245. ( 10.1002/ajmg.a.34009) [DOI] [PubMed] [Google Scholar]
- 35.Binder G, Neuer K, Ranke MB, Wittekindt NE. PTPN11 mutations are associated with mild growth hormone resistance in individuals with Noonan syndrome. Journal of Clinical Endocrinology and Metabolism 2005. 90 5377–5381. ( 10.1210/jc.2005-0995) [DOI] [PubMed] [Google Scholar]
- 36.Storr HL, Dunkel L, Kowalczyk J, Savage MO, Metherell LA. Genetic characterisation of a cohort of children clinically labelled as GH or IGF1 insensitive: diagnostic value of serum IGF1 and height at presentation. European Journal of Endocrinology 2015. 172 151–161. ( 10.1530/EJE-14-0541) [DOI] [PubMed] [Google Scholar]
- 37.Shapiro L, Chatterjee S, Ramadan DG, Davies KM, Savage MO, Metherell LA, Storr HL. Whole-exome sequencing gives additional benefits compared to candidate gene sequencing in the molecular diagnosis of children with growth hormone or IGF-1 insensitivity. European Journal of Endocrinology 2017. 177 485–501. ( 10.1530/EJE-17-0453) [DOI] [PubMed] [Google Scholar]
- 38.Vairamani K, Merjaneh L, Casano-Sancho P, Sanli ME, David A, Metherell LA, Savage MO, Del Pozo JS, Backeljauw PF, Rosenfeld RG, et al Novel dominant-negative GH receptor mutations expands the spectrum of GHI and IGF-I deficiency. Journal of the Endocrine Society 2017. 1 345–358. ( 10.1210/js.2016-1119) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ayling RM, Ross R, Towner P, Von Laue S, Finidori J, Moutoussamy S, Buchanan CR, Clayton PE, Norman MR. A dominant-negative mutation of the growth hormone receptor causes familial short stature. Nature Genetics 1997. 16 13–14. ( 10.1038/ng0597-13) [DOI] [PubMed] [Google Scholar]
- 40.Iida K, Takahashi Y, Kaji H, Nose O, Okimura Y, Abe H, Chihara K. Growth hormone (GH) insensitivity syndrome with high serum GH-binding protein levels caused by a heterozygous splice site mutation of the GH receptor gene producing a lack of intracellular domain. Journal of Clinical Endocrinology and Metabolism 1998. 83 531–537. ( 10.1210/jcem.83.2.4601) [DOI] [PubMed] [Google Scholar]
- 41.Aisenberg J, Auyeung V, Pedro HF, Sugalski R, Chartoff A, Rothenberg R, Derr MA, Hwa V, Rosenfeld RG. Atypical GH insensitivity syndrome and severe insulin-like growth factor-I deficiency resulting from compound heterozygous mutations of the GH receptor, including a novel frameshift mutation affecting the intracellular domain. Hormone Research in Paediatrics 2010. 74 406–411. ( 10.1159/000314968) [DOI] [PubMed] [Google Scholar]
- 42.Klammt J, Neumann D, Gevers EF, Andrew SF, Schwartz ID, Rockstroh D, Colombo R, Sanchez MA, Vokurkova D, Kowalczyk J, et al Dominant-negative STAT5B mutations cause growth hormone insensitivity with short stature and mild immune dysregulation. Nature Communications 2018. 9 2105 ( 10.1038/s41467-018-04521-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van Duyvenvoorde HA, van Setten PA, Walenkamp MJ, van Doorn J, Koenig J, Gauguin L, Oostdijk W, Ruivenkamp CA, Losekoot M, Wade JD, et al Short stature associated with a novel heterozygous mutation in the insulin-like growth factor 1 gene. Journal of Clinical Endocrinology and Metabolism 2010. 95 E363–E367. ( 10.1210/jc.2010-0511) [DOI] [PubMed] [Google Scholar]
- 44.Batey L, Moon JE, Yu Y, Wu B, Hirschhorn JN, Shen Y, Dauber A. A novel deletion of IGF1 in a patient with idiopathic short stature provides insight into IGF1 haploinsufficiency. Journal of Clinical Endocrinology and Metabolism 2014. 99 E153–E159. ( 10.1210/jc.2013-3106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fuqua JS, Derr M, Rosenfeld RG, Hwa V. Identification of a novel heterozygous IGF1 splicing mutation in a large kindred with familial short stature. Hormone Research in Paediatrics 2012. 78 59–66. ( 10.1159/000337249) [DOI] [PubMed] [Google Scholar]
- 46.Begemann M, Zirn B, Santen G, Wirthgen E, Soellner L, Büttel HM, Schweizer R, van Workum W, Binder G, Eggermann T. Paternally inherited IGF2 mutation and growth restriction. New England Journal of Medicine 2015. 373 349–356. ( 10.1056/NEJMoa1415227) [DOI] [PubMed] [Google Scholar]
- 47.Yamoto K, Saitsu H, Nakagawa N, Nakajima H, Hasegawa T, Fujisawa Y, Kagami M, Fukami M, Ogata T. De novo IGF2 mutation on the paternal allele in a patient with Silver-Russell syndrome and ectrodactyly. Human Mutation 2017. 38 953–958. ( 10.1002/humu.23253) [DOI] [PubMed] [Google Scholar]
- 48.Grandone A, Miraglia del Giudice E, Cirillo G, Abbondanza C, Cioffi M, Romano T, Micillo F, Marzuillo P, Perrone L. Clinical features of a new acid-labile subunit (IGFALS) heterozygous mutation: anthropometric and biochemical characterization and response to growth hormone administration. Hormone Research in Paediatrics 2014. 81 67–72. ( 10.1159/000355017) [DOI] [PubMed] [Google Scholar]
- 49.Fofanova-Gambetti OV, Hwa V, Wit JM, Domene HM, Argente J, Bang P, Högler W, Kirsch S, Pihoker C, Chiu HK, et al Impact of heterozygosity for acid-labile subunit (IGFALS) gene mutations on stature: results from the international acid-labile subunit consortium. Journal of Clinical Endocrinology and Metabolism 2010. 95 4184–4191. ( 10.1210/jc.2010-0489) [DOI] [PubMed] [Google Scholar]
- 50.Chen M, Manley JL. Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nature Reviews: Molecular Cell Biology 2009. 10 741–754. ( 10.1038/nrm2777) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nilsen TW, Graveley BR. Expansion of the eukaryotic proteome by alternative splicing. Nature 2010. 463 457–463. ( 10.1038/nature08909) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Saltzman AL, Pan Q, Blencowe BJ. Regulation of alternative splicing by the core spliceosomal machinery. Genes and Development 2011. 25 373–384. ( 10.1101/gad.2004811) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stattin EL, Wiklund F, Lindblom K, Onnerfjord P, Jonsson BA, Tegner Y, Sasaki T, Struglics A, Lohmander S, Dahl N, et al A missense mutation in the aggrecan C-type lectin domain disrupts extracellular matrix interactions and causes dominant familial osteochondritis dissecans. American Journal of Human Genetics 2010. 86 126–137. ( 10.1016/j.ajhg.2009.12.018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Abuzzahab MJ, Schneider A, Goddard A, Grigorescu F, Lautier C, Keller E, Kiess W, Klammt J, Kratzsch J, Osgood D, et al IGF-I receptor mutations resulting in intrauterine and postnatal growth retardation. New England Journal of Medicine 2003. 349 2211–2222. ( 10.1056/NEJMoa010107) [DOI] [PubMed] [Google Scholar]
- 55.Martinelli S, De Luca A, Stellacci E, Rossi C, Checquolo S, Lepri F, Caputo V, Silvano M, Buscherini F, Consoli F, et al Heterozygous germline mutations in the CBL tumor-suppressor gene cause a Noonan syndrome-like phenotype. American Journal of Human Genetics 2010. 87 250–257. ( 10.1016/j.ajhg.2010.06.015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jorge AA, Menezes Filho HC, Lins TS, Guedes DR, Damiani D, Setian N, Arnhold IJ, Mendonça BB. Founder effect of E180splice mutation in growth hormone receptor gene (GHR) identified in Brazilian patients with GH insensitivity. Arquivos Brasileiros de Endocrinologia e Metabologia 2005. 49 384–389. ( 10.1590/s0004-27302005000300009) [DOI] [PubMed] [Google Scholar]
- 57.Espinosa C, Sjoberg M, Salazar T, Rodriguez A, Cassorla FG, Mericq MV, Carvallo P. E180splice mutation in the growth hormone receptor gene in a Chilean family with growth hormone insensitivity: a probable common Mediterranean ancestor. Journal of Pediatric Endocrinology and Metabolism 2008. 21 1119–1127. ( 10.1515/jpem.2008.21.12.1119) [DOI] [PubMed] [Google Scholar]
- 58.Guevara-Aguirre J, Rosenbloom AL, Guevara-Aguirre M, Yariz K, Saavedra J, Baumbach L, Shuster J. Effects of heterozygosity for the E180 splice mutation causing growth hormone receptor deficiency in Ecuador on IGF-I, IGFBP-3, and stature. Growth Hormone and IGF Research 2007. 17 261–264. ( 10.1016/j.ghir.2007.01.016) [DOI] [PubMed] [Google Scholar]
- 59.Bozzoli C, Deaton A, Quintana-Domeque C. Adult height and childhood disease. Demography 2009. 46 647–669. ( 10.1353/dem.0.0079) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gat-Yablonski G, De Luca F. Effect of nutrition on statural growth. Hormone Research in Paediatrics 2017. 88 46–62. ( 10.1159/000456547) [DOI] [PubMed] [Google Scholar]
- 61.Dateki S, Nakatomi A, Watanabe S, Shimizu H, Inoue Y, Baba H, Yoshiura KI, Moriuchi H. Identification of a novel heterozygous mutation of the aggrecan gene in a family with idiopathic short stature and multiple intervertebral disc herniation. Journal of Human Genetics 2017. 62 717–721. ( 10.1038/jhg.2017.33) [DOI] [PubMed] [Google Scholar]
- 62.Nilsson O, Guo MH, Dunbar N, Popovic J, Flynn D, Jacobsen C, Lui JC, Hirschhorn JN, Baron J, Dauber A. Short stature, accelerated bone maturation, and early growth cessation due to heterozygous aggrecan mutations. Journal of Clinical Endocrinology and Metabolism 2014. 99 E1510–E1518. ( 10.1210/jc.2014-1332) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang SR, Jacobsen CM, Carmichael H, Edmund AB, Robinson JW, Olney RC, Miller TC, Moon JE, Mericq V, Potter LR, et al Heterozygous mutations in natriuretic peptide receptor-B (NPR2) gene as a cause of short stature. Human Mutation 2015. 36 474–481. ( 10.1002/humu.22773) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Binder G. Short stature due to SHOX deficiency: genotype, phenotype, and therapy. Hormone Research in Paediatrics 2011. 75 81–89. ( 10.1159/000324105) [DOI] [PubMed] [Google Scholar]



 This work is licensed under a
This work is licensed under a