Abstract
Breast cancer is a known heterogeneous disease. Current clinically utilized histopathologic biomarkers may undersample tumor heterogeneity, resulting in higher rates of misdiagnosis for breast cancer. MRI can provide a whole-tumor sampling of disease burden and is widely utilized in clinical care. Texture analysis can provide a localized description of breast cancer, with particular emphasis on quantifying breast lesion heterogeneity. The object of this review is to provide an overview of texture analysis applications towards breast cancer diagnosis, prognosis, and treatment response evaluation and review the role of image-based texture features as noninvasive prognostic and predictive biomarkers.
One in eight women are diagnosed with breast cancer in the United States each year.1,2 Breast cancer is a known heterogeneous disease, with inter- and intratumor heterogeneity demonstrated in genomic, histologic, and radiologic analyses.3–5 Intratumor heterogeneity can manifest itself spatially, with microenvironment-specific factors driving the progression and distribution of distinct subpopulations.6 Additionally, the dynamic behavior of breast cancer cells can lead to temporal intratumor heterogeneity, where disease presentation is altered longitudinally due to growth or in response to systemic and local therapies.6 Currently, diagnostic, prognostic, and treatment decisions for breast cancer are made primarily on the basis of established histopathologic biomarkers originating from tissue samples acquired from core biopsy or surgical excision. While advanced treatment and management options have reduced the number of breast cancer-related deaths, 30% of women are either under- or overtreated for breast cancer.7,8 As such, these histopathologic biomarkers may be an undersampling of the heterogeneous tumor, suggesting a clinical need for whole-tumor sampled biomarkers.
Imaging allows for noninvasive sampling of disease burden, with the ability to longitudinally monitor response to treatment.9,10 Magnetic resonance imaging (MRI) is highly sensitive for primary lesion detection, particularly for high-risk women.10 Specific MRI sequences such as diffusion weighted (DW) MRI and dynamic contrast-enhanced (DCE) MRI can provide further insight into tissue architecture and vascularization of and around the tumor.11,12 Current clinical analysis of MR images is largely qualitative, using DCE-MRI to identify tumor regions with contrast uptake or monitor morphologic appearance.13
Recent advances in medical image analysis have highlighted the implementation of computer vision principles and analytic techniques used to quantify and describe medical images.14–16 Image texture has been previously defined as repeating patterns of local variations in gray-level intensities.15,17 Texture analysis has most broadly been used to characterize the spatial distribution of gray-level intensities within an image, capturing image patterns usually unrecognizable or indistinguishable to the human eye. The original utilization of texture analysis can be traced back to computer vision applications for surface inspection and orientation, image and object classification, and shape determination, while current applications extend even beyond medical image analysis.17–19 Within the scope of breast imaging, texture analysis has emerged as a quantitative, surrogate measure for breast parenchymal patterns when applied to images taken during mammographic and tomographic screenings, serving to augment conventional measures of breast percent density in breast cancer risk assessment.20 As developments in MRI have led to improved resolution for 2D and 3D insight into the structure and function of the human body, MRI is now widely utilized for breast cancer screening, diagnosis, and treatment response.9,21 As such, textural information extracted from MR images can have high clinical relevance.22–30 As compared to a global or qualitative report of breast tumor appearance, texture analysis can provide a refined, local description of tumor complexity, heterogeneity, and kinetic behavior as seen in MRI. This quantitative characterization of MR images can have specific applications towards the diagnosis, prognosis, and treatment of breast cancer.
Commonly Extracted Texture Features
Texture analysis aims to extract high-throughput information characterizing image texture within a defined region of interest (ROI). For the analysis of breast lesions as presented in MRI, texture features are often extracted from an ROI selected within a segmented lesion, or from the whole lesion itself. Additionally, recent studies have shown clinical associations of texture within the peritumor region as well, emphasizing the importance of the tumor microenvironment as presented in MRI.31,32
The most commonly used texture features can be stratified by the statistical order of the voxel information encoded within the image. Specifically, first-order texture features include common statistical measures derived from a gray-level histogram, such as mean, median, and skewness. Second-order texture features are often derived from the co-occurrence matrix, as determined by Haralick and Shanmugam,33 and the run-length matrix,34 while higher-order texture features encode structural and frequency-based texture information (Fig. 1).
FIGURE 1:
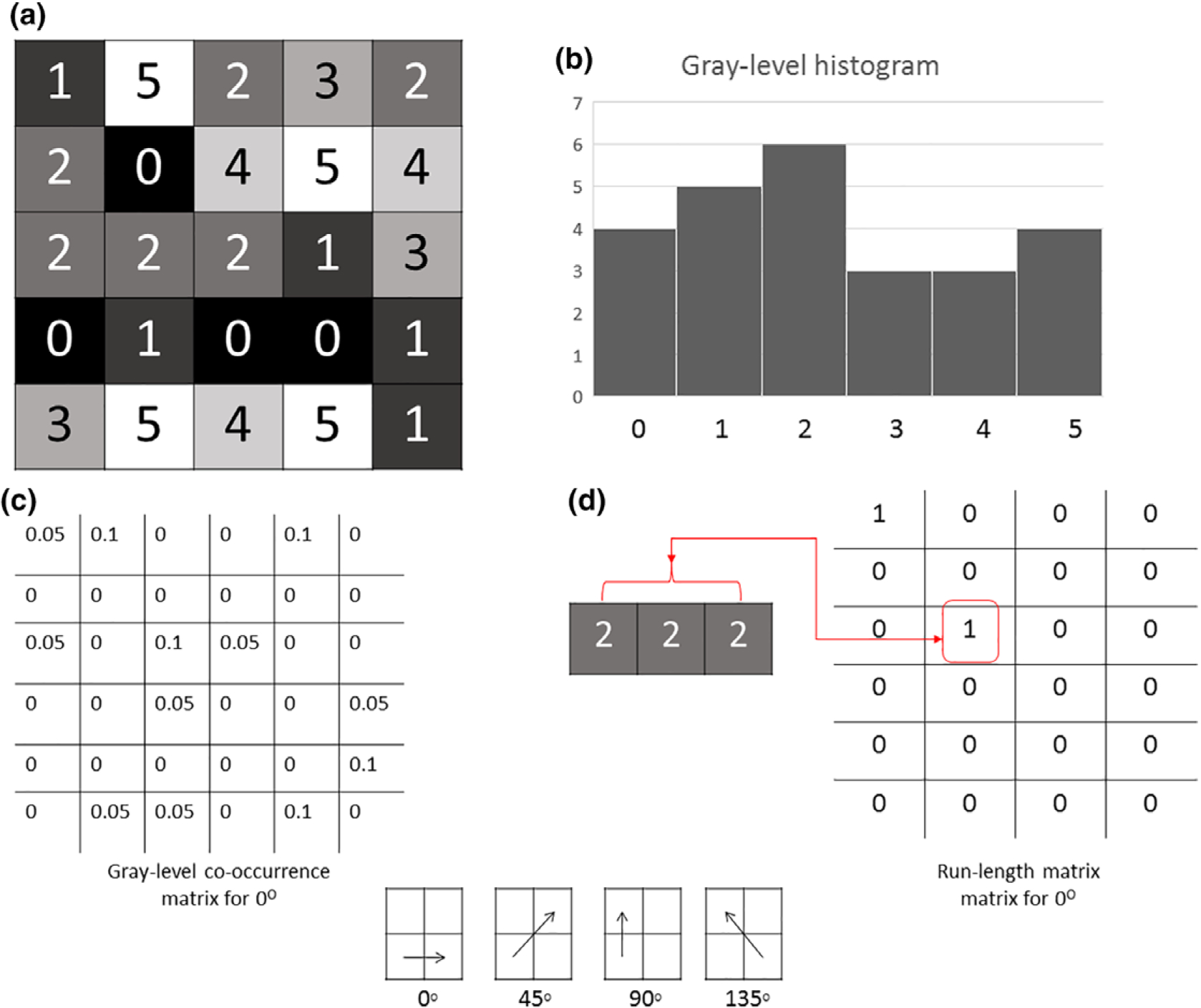
Representative 5 × 5 pixel image with six possible gray levels (0–5) (a). Gray-level histogram generated from representative image (b). Gray-level co-occurrence matrix generated for 0°. The co-occurrence matrix encodes the frequency that two pixels are located a specific distance (1 pixel) away from one another (c). Run-length matrix generated for 0°. Run-length matrix encodes the coarseness of an image in a specified linear direction (d).
First Order: Gray-Level Histogram Features
A gray-level histogram can be generated by calculating a frequency count of the number of voxels of each gray-level intensity value, where the total number of discretized gray-levels is often a user-selected parameter. From the resulting histogram, first-order statistical features may be derived, including the mean, median, and variance. Higher moment features can also be extracted from the histogram including skewness, the measure of the histogram distribution symmetry, and kurtosis, a measure of the histogram distribution shape. While many descriptors can be extracted from the histogram, they often provide cursory insight into the underlying texture, do not account for gray-level intensity spatial relationships within an image, and are dependent on user-selected parameters (Table 1).
TABLE 1.
Gray-Level Histogram Texture Features
| Mean | Mean gray-level value | |
| Min | Min gray-level value | Min(k) |
| Max | Max gray-level value | Max(k) |
| 5th Percentile | Histogram bin that 5% of gray-level values are less than or equal to | k: 5% of values ≤ k |
| Mean 5th | Mean value of gray-level values that 5% of gray-level values are less than or equal to | |
| 95th Percentile | Histogram bin that 95% of gray-level values are greater than or equal to | k: 95% of values ≥ k |
| Mean 95th | Mean value of gray-level values that 95% of gray-level values are greater than or equal to | |
| Sum | Sum of gray-level values | |
| Sigma | Measure of variation of gray-level values around the mean | |
| Entropy | Measure of histogram nonuniformity | |
| Kurtosis | Measure of histogram flatness | |
| Skewness | Measure of histogram symmetry |
Second Order: Gray-Level Co-Occurrence Matrix and Run-Length Features
Gray-level co-occurrence matrix (GLCM) features are the most commonly extracted texture features for MRI quantification. A GLCM encodes the frequency that two voxels of specific gray-level intensities are positioned a specified distance away from each other in a specified image orientation.33 GLCM texture features are most often quantified in the four diagonal image orientations of 0°, 45°, 90°, and 135°. Second-order texture features can then be extracted from the co-occurrence matrix. Examples of such features include contrast, a descriptor of the intensity contrast between a pixel and its neighbor as determined by the distance parameter; correlation, a descriptor of the linear gray-level dependence; and homogeneity, a descriptor of the closeness of distribution in the co-occurrence matrix to the matrix diagonal. Other second-order features such as energy, a descriptor of the certainty of gray-level co-occurrence, and cluster shade, a descriptor of asymmetry in gray-level values, can also be extracted. Entropy, or the randomness of the GLCM, is another commonly extracted feature, often indicating image heterogeneity (Table 2). Run-length features measure the coarseness of an image in specified linear directions34–36 (Table 3).
TABLE 2.
Representative Gray-Level Co-Occurrence Matrix Texture Features
| Contrast | Intensity contrast between pixel and its neighbor | |
| Correlation | Linear gray-level dependence | |
| Homogeneity | Closeness of distribution in co-occurrence matrix to matrix diagonal | |
| Energy | Certainty of gray-level co-occurrence | |
| Entropy | Uncertainty of gray-level co-occurrence | |
| Inverse difference moment (IDM) | Local homogeneity in gray-level co-occurrence | |
| Cluster shade | Asymmetry in gray-level values |
TABLE 3.
Run-Length Texture Features
| Short run emphasis (SRE) | Emphasis on short runs | |
| Long run emphasis (LRE) | Emphasis on long runs | |
| Gray-level nonuniformity (GLN) | Degree of gray-level run dissimilarity | |
| Run length nonuniformity (RLN) | Dissimilarity in length of runs | |
| Run percentage (RP) | Distribution of runs | |
| Low gray-level run emphasis (LGRE) | Emphasis on low gray-level values | |
| High gray-level run emphasis (HGRE) | Emphasis on high gray-level values | |
| Short run low gray-level emphasis (SRLGE) | Emphasis on short runs with low gray-level values | |
| Short run high gray-level emphasis (SRHGE) | Emphasis on short runs with high gray-level values |
nr is the total number of runs, R(i,j) represents the number of runs with pixels of gray-level intensity value, i, and length of run, j. In all, 128 gray levels were used. Estimated by averaging over 0 °, 45°, 90°, and 135 ° orientations.
Higher Order: Structural and Transformation-Based
Structural texture features capture the intensity variations between central voxels and surrounding neighboring voxels37 (Table 4). Extending texture analysis beyond the spatial orientation of gray-level intensity values, transformation-based texture features capture texture information encoded in a different space, such as the frequency space. Transformation-based methods include the Fourier transform, Gabor transform, and Wavelet transform.38–40 The wavelet transform, in particular, is commonly used due to its ability to capture the MR images frequency content both at varying image scales and multiple specified directions.
TABLE 4.
Structural Texture Features
| Structural feature | Qualitative description | Mathematical description |
|---|---|---|
| Local Binary Pattern (LBP) | Intensity variation between a pixel and its neighboring pixels. |
Ic and Ip are gray-level intensity values for pixel (xc,yc) and pixel (xp,yp). q = indicator function, 0 for negative inputs and 1 for non-negative inputs.Q, P = parameters to set pixel neighborhood size, set to 1 and 8, respectively.39,40 |
Applications in Breast Computer-Aided Diagnosis
Computer-aided diagnosis (CAD) of breast tissue was one of the earliest applications of texture analysis in the breast.41 Gibbs and Turnbull42 were one of the first to apply texture analysis toward classifying breast lesions as benign or malignant. The authors reported using 2D DCE-MR images from a cohort of 79 women, of which 45 were diagnosed with breast cancer. An ROI was selected to encompass the entire lesion, within which gray-level intensity values were discretized to 32 levels. Within each lesion ROI, a co-occurrence matrix was determined for adjoining pixels in 0°, 45°, 90°, and 135° directions. The co-occurrence matrices of each direction were averaged and 14 GLCM texture features were extracted. Texture features of variance, sum entropy, and entropy were concluded to be the most significant when discriminating between benign and malignant lesions, as determined by logistic regression performance, suggesting that features quantifying image texture could be a useful tool in lesion delineation. Subsequent studies have followed this preliminary, yet promising, conclusion by utilizing more complex measures of image texture to diagnose breast lesions as benign or malignant.43–49 Nie et al44 investigated the utility of breast lesion texture in parallel with lesion morphology features, using a retrospective study size of 71 women, of which 28 and 42 were diagnosed with benign and malignant lesions, respectively. Ten GLCM texture features were extracted from a co-occurrence matrix summarizing the entire lesion, while eight morphologic features were extracted as well. The study found that the performance of three texture features: GLCM entropy, GLCM Sum Average, and GLCM Homogeneity, and three morphology features resulted in an area under the curve (AUC) of the receiver operating characteristic (ROC) curve of 0.86. Holli et al47 proposed using texture features to distinguish between cancerous and healthy breast tissue within a 15 × 15 pixel ROI, and found that, with 100% accuracy, a combined set of 277 texture features including histogram-based, GLCM, and wavelet features was able to distinguish between healthy and cancerous breast ROIs. Similar conclusions were reported by Nagarajan et al,43 who found that GLCM-based texture features quantifying lesion heterogeneity were particularly accurate when classifying small lesions as benign or malignant. While many of these studies share similar conclusions with regard to specific texture features having high discriminatory capacities, there is a lack of literature for comparative studies performed on the same dataset with defined parameters for extracted features.
A multiparametric CAD approach was suggested by Bhooshan et al46 leveraging T2-weighted and T1-weighted DCE-MRI. GLCM texture features were extracted from breast images from each imaging sequence, while kinetic features were extracted from the DCE-MR images to build a multiparametric feature vector. The study found that the multiparametric feature vector outperformed features extracted from an individual imaging sequence when differentiating between malignant and benign lesions, suggesting that texture analysis, when applied to T2-weighted MR images, provided additional discriminatory information beyond that extracted from T1-weighted imaging.
Traditional applications of texture analysis towards breast diagnosis largely focused on 2D texture features extracted from a 2D slice from 3D images. This idea was extended by Chen et al,48 who extracted 3D GLCM texture features from a 3D breast lesion segmentation. The 3D GLCM features yielded a higher diagnostic accuracy than 2D GLCM features extracted from a 2D ROI, when distinguishing between malignant and benign breast lesions, showing an advantage for 3D breast lesion characterization.
Beyond CAD: Texture Analysis for Histopathologic and Molecular Subtype Classification
The promising conclusions of MRI texture analysis in breast cancer diagnosis suggest an architectural difference between the imaging presentation of benign and malignant lesions that can be quantified using texture features. Recent studies have begun extending this idea, hypothesizing that underlying tumor biological differences can be imaged using MRI and characterized using texture analysis. Consequently, many groups have employed texture analysis to distinguish between the heterogeneous histopathologic32,45,47,50 and molecular51–54 subtypes of breast cancer, with a larger goal of utilizing image texture features to provide a personalized diagnosis (Table 5).
TABLE 5.
Texture Analysis Studies for Breast Cancer Histopathologic and Molecular Subtype Classification
| Holli-Helenius et al51 | 27 | T1-weighted, nonfat-saturated DCE-MRI | GLCM texture features | Sum entropy and sum variance differentiate between luminal A and luminal B subtypes (AUC = 0.88) |
| Waugh et al50 | 200 | DCE-MRI | GLCM texture features | Entropy significantly different between ILC and IDC cancers |
| Sutton et al52 | 178 | T1-weighted fat suppressed MRI | Gray-level histogram, GLCM texture features | Features quantifying heterogeneity were able to classify between molecular subtypes |
| Wang et al32 | 84 | DCE-MRI | Gray-level histogram, GLCM texture features | Adding texture features quantifying tumor microenvironment heterogeneity to model with features quantifying lesion heterogeneity improved classification performance to identify TNBC. |
| Chen et al48 | 121 | T1-weighted DCE-MRI | GLCM texture features | 3D texture features showed better performance than 2D texture features when classifying breast lesions as benign or malignant |
In an attempt to distinguish between invasive lobular carcinoma (ILC) and invasive ductal carcinoma (IDC), Holli et al47 extracted a total of 277 histogram, GLCM, run-length, and wavelet features. The authors found that of these only GLCM-related features characterizing lesion complexity and randomness were significantly different between ILC and IDC lesions. Similar conclusions were reported by Waugh et al,50 who found that entropy, a texture measure of pixel distribution randomness, was significantly different between lobular and ductal lesions, suggesting a difference in underlying growth patterns and tumor heterogeneity.
Increasing the scope of texture analysis to include the surrounding microenvironment in addition to the breast lesion, Wang et al32 investigated the role of kinetic contrast uptake texture in differentiating between histopathologic subtypes of breast cancer. Texture features were extracted from pharmacokinetic parametric maps generated from DCE-MR images of the tumor and surrounding parenchyma. Adding texture features characterizing heterogeneous uptake in the breast parenchyma to a model containing lesion texture features allowed for the identification of triple-negative breast cancers (TNBC). The study concluded that the characterization of heterogeneity, both within the lesion as well as the surrounding parenchyma, could provide noninvasive insight into the heterogeneous tumor behavior associated with more aggressive subtypes. These results were similar to those found in previous studies,55,56 indicating the clinical value of lesion and peritumoral contrast uptake quantification. Texture features quantifying lesion heterogeneity have also been shown to aid in delineating between molecular subtypes of breast cancer. Studies have shown a textural difference between the MR presentation of luminal A and luminal B subtypes, with luminal B lesions having a more quantifiably heterogeneous appearance.51,54
Texture Analysis for Breast Cancer Prognosis and Therapy Response Prediction
Recent studies have shown promising conclusions when exploring the relationship between breast lesion texture and risk of recurrence, and the value of image texture as a noninvasive prognostic biomarker45,57–61 (Table 6). Kim et al57 performed a retrospective analysis of 203 women diagnosed with invasive breast cancer, extracting histogram uniformity and entropy features from both T2-weighted MR images and T1-weighted DCE-MRI. Univariate and multivariate associations between these texture features and disease-free survival determined that increased tumor heterogeneity in T2-weighted MRI could be used to stratify patients more at risk for recurrence. That study suggested that tumor heterogeneity, as quantified by lesion texture, could be used in MRI as an independent prognostic marker. Similar conclusions were drawn by Park et al,59 who generated a multivariate feature vector based on morphologic, histogram texture, and GLCM texture features, from which specifically GLCM cluster tendency, GLCM variance, and GLCM sum variance were selected for in a model stratifying patients at risk for recurrence. Mahrooghy et al60,61 extracted wavelet texture features from within tumor subregions partitioned by pharmacokinetic behavior and concluded that the spatial frequency texture pattern captured using wavelets within the heterogeneous subregions could serve as a strong prognostic biomarker for predicting risk of tumor recurrence (AUC = 0.88).
TABLE 6.
Texture Analysis Studies for Breast Cancer Prognosis
| Kim et al57 | 203 | T2-weighted and T1-weighted DCE-MRI | Histogram uniformity, histogram entropy | Heterogeneous lesions on T2-weighted imaging predicted poor RFS |
| Park et al59 | 294 | DCE-MRI | GLCM texture features | Features characterizing high heterogeneity showed relationship to poor survival outcomes |
| Mahrooghy et al61 | 56 | DCE-MRI | Wavelet features extracted from lesions partitioned by pharmacokinetic uptake patterns | Heterogeneity wavelet features classified tumors by risk of recurrence with AUC = 0.88 |
The potential for texture analysis in breast cancer treatment has been demonstrated in recent studies, showing the potential for MRI-extracted texture features to serve as noninvasive predictive biomarkers31,35,56,57,62–72 (Table 7). In order to predict response to treatment, some studies utilize first-order statistical texture measures extracted from the tumor ROI. Specifically, Johansen et al71 calculated three first-order statistical features of mean, standard deviation, and prediction from a relative signal intensity histogram generated from prechemotherapy DCE-MR scans. Of these, skewness and kurtosis were found to be strongly correlated with complete response to therapy. Similarly, Padhani et al72 conducted a retrospective study of 25 women diagnosed with primary invasive cancer, imaged using DCE-MRI before and after the first cycle of treatment. Leveraging contrast enhancement, the authors generated a histogram from a pharmacokinetic parametric map of the full lesion ROI, and concluded that responsive patients displayed a decrease in pharmacokinetic range, and proposed that this could be attributed to a decrease in heterogeneity after the first cycle of treatment.
TABLE 7.
Texture Analysis Studies for Breast Cancer Treatment Response Prediction
| Teruel et al62 | 58 | T1-weighted DCE-MRI | GLCM texture features | Entropy and sum variance were most significant in predicting stable disease vs. complete responders (AUC = 0.77) and predicting pCR (AUC = 0.69), respectively |
| Thibault et al63 | 38 | DCE-MRI | GLCM, run length features extracted from pharmacokinetic maps | GLCM features most predictive of therapy response |
| Golden et al64 | 60 | DCE-MRI | GLCM texture features extracted from pharmacokinetic maps | Pretherapy features can predict pCR and residual lymph node metastasis |
| Parikh et al65 | 36 | T2-and T1-weighted DCE-MRI | Entropy and uniformity | Responders to NACT showed increase in lesion homogeneity after one round of therapy |
| Michoux et al66 | 69 | T1-weighted DCE-MRI | GLCM, run-length | Model with three texture features and one kinetic feature identified nonresponders to NAC with 84% sensitivity |
| Braman et al31 | 117 | T1-weighted DCE-MRI | Gabor, GLCM, Laws energy measures | Intratumor and peritumor texture features predicted pCR with AUC = 0.78 |
While histogram texture can provide useful information regarding the distribution of gray-level intensity values, it is limited when capturing spatial heterogeneity within a lesion, as it largely ignores the spatial relationships between voxels. Studies extracting higher-order texture features can further quantify the relation between tumor heterogeneity and response to therapy. To this end, Teruel et al62 extracted second-order statistical GLCM features from pharmacokinetic maps generated from DCE-MR images of women diagnosed with locally advanced breast cancer. Eight GLCM features were found to significantly differ between responders and nonresponders, and GLCM sum variance was able to predict response to treatment with an AUC of 0.77. Similar conclusions were drawn by Thibault et al,63 who expanded this 2D analysis by extracting 3D GLCM texture features from DCE-MR pharmacokinetic parametric maps in order to predict response to neoadjuvant chemotherapy (NAC). The 3D GLCM texture features were particularly significant in identifying early responders to NAC, with results showing nonresponders having higher microvascular heterogeneity. In a retrospective study of 36 women who underwent NACT, the change in tumor heterogeneity between pretreatment and mid-treatment, as calculated by entropy and uniformity changes, was predictive of pathologic complete response (pCR) with an AUC of 0.84.65 Comparing this performance to change in tumor size (AUC = 0.66) demonstrates a greater sensitivity for lesion texture in characterizing an early response to pCR.
Current clinical predictions for achieving pCR are based on tumor histopathologic characteristics. As intratumor heterogeneity is associated with adverse clinical outcomes, the limited tissue taken during biopsy may be inadequate for a whole-tumor-based prediction.5,73 Michoux et al66 performed a retrospective analysis on the DCE-MR scans of 69 patients diagnosed with IDC undergoing NAC. For each woman, texture, kinetic, and morphology-based features were extracted from within the pretreatment lesion ROI. The authors concluded that only four parameters—three texture features (GLCM inverse difference moment, gray-level nonuniformity, and long run high gray-level emphasis), and the washin slope kinetic feature—were found to classify nonresponders with 84% sensitivity. Of particular note, clinically utilized histopathologic predictive biomarkers such as estrogen receptor (ER) status, progesterone receptor (PR) status, Ki67 status, and human epidermal growth factor receptor 2 (HER2), along with tumor grade, were not significant when classifying early response, further highlighting the utility of texture analysis. Similarly, Golden et al64 extracted GLCM-based texture features from pharmacokinetic parametric maps generated from DCE-MR images of women diagnosed with TNBC, in order to predict pCR, residual lymph node metastases, and residual tumor with lymph node metastases. The predictive performance of GLCM texture features was compared to “patterns of response,” a qualitative description of lesion appearance before and after chemotherapy, as determined by a radiologist. The GLCM texture features extracted from prechemotherapy MR images could predict pCR and residual lymph node metastasis with a reported AUC of 0.68 when classified in a logistic regression model. In contrast, the radiologist determined “patterns of response” did not predict any of three outcome measures.
The conclusions drawn from studies implementing texture analysis for breast cancer diagnosis, prognosis, and treatment suggest that texture features demonstrating increased lesion heterogeneity are associated with aggressive growth, unfavorable prognosis, and poor treatment outcomes.31,57,61,67,74–76 In addition, they propose a method for noninvasively quantifying the underlying biology of tumor subregions driving recurrence, response, and resistance to therapy (Fig. 2).
FIGURE 2:
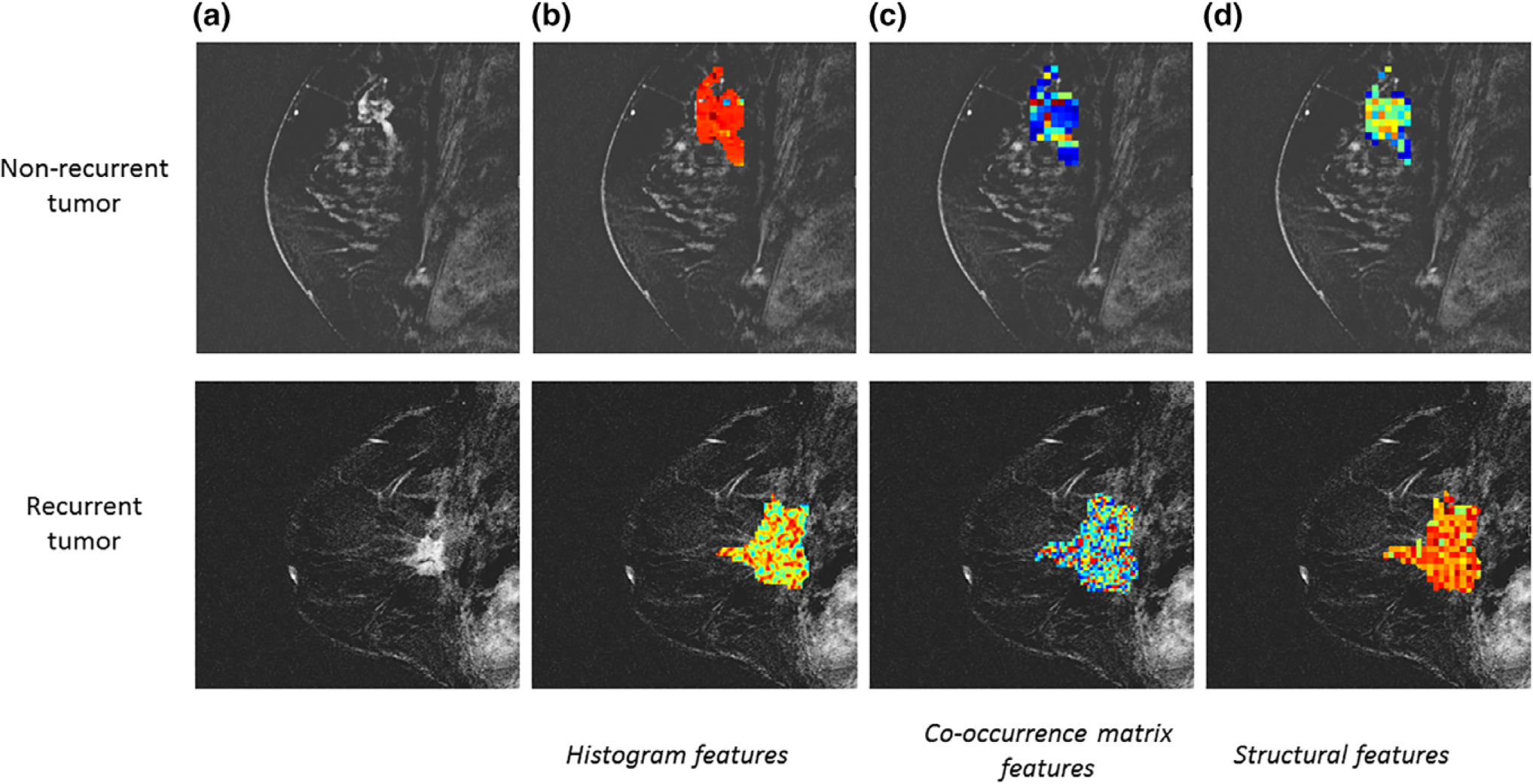
Representative images of a nonrecurrent and recurrent breast tumor (a). Examples of texture feature maps showing distributions of histogram texture features (b), co-occurrence matrix texture features (c), and structural texture features (d).
Texture Analysis Study Designs
Effective use of texture analysis on MR images of breast cancer is highly dependent on appropriate study design and statistical evaluation. There are numerous methods for texture feature extraction, resulting in a myriad of ways to quantify an image’s texture. Consequently, having a high-dimensional texture feature set as compared to a relatively smaller sample size can result in the overfitting of a statistical learning model, resulting in false-positive classification and over-or underestimated statistical associations. Additionally, redundant texture features can often decrease performance accuracy. To alleviate this, methods such as principal component analysis or independent component analysis can be used to reduce the dimensionality of the texture feature set.77 Feature selection methods can also be used to reduce texture feature redundancy and promote relevant texture features for analysis. Statistical correction methods, such as the Benjamini–Hochberg correction,78 can be used to reduce false positives in statistical association conclusions. Ideally, results should be validated using an independent dataset, to ensure the veracity of the texture analysis. While finding comparable independent datasets is not always feasible, splitting the initial dataset into discovery and validation sets is an alternative solution to ensure repeatability. Similarly, crossvalidation can be used to identify robust conclusions. Lastly, utilization of publicly available datasets and detailed methodology of specific texture parameters used during feature extraction can allow for study repeatability.
Future Directions
Texture analysis is limited by user-defined feature parameters, such as selecting the number of discretized gray levels within an image, MR acquisition protocol, and image quality.15,48,79 As such, the literature largely lacks repeated studies performed on the same datasets with standardized texture feature extraction. Additionally, while texture features provide quantitative measures of breast tumor texture, the direct biological interpretation of specific texture feature values remains largely uncertain. Leveraging the specific properties of various functional MRI imaging techniques, texture analysis can be used to quantify different tissue properties. Texture analysis applied to DCE-MRI or DW-MRI could provide insight on the distribution or longitudinal development of tumor vascularization and diffusion, respectively. Similarly, statistical associations between image-derived texture features and histopathologic or genomic expression data could lead to a biological basis for tumor texture. Further work is needed to explore the relationships between specific texture features and underlying biology.
Many studies aiming to analyze MRI presentations of breast lesions for diagnostic, prognostic, and treatment applications have expanded the number and type of features extracted to include morphology, texture, and pharmacokinetic features, allowing for a thorough and quantitative characterization of all tumor properties. This has developed into the new field of “radiomics,” broadly defined as the extraction of high-throughput quantitative features from images obtained from medical imaging modalities.80–82 The promising conclusions from studies employing texture analyses have demonstrated that features defining high lesion heterogeneity have been associated with more aggressive diagnoses and poor response to treatment. Benefited by the whole-tumor sampling and visualization of tumor vasculature afforded by MRI, one of the largest aims in radiomics is to accurately characterize and quantify intratumor heterogeneity.4,5,73 Current advances within radiomics also include quantifying underlying tumor biology by investigating relationships between radiomic features extracted from complementary imaging modalities,83 gene expression pathways,53,84 and existing clinical definitions for breast cancer.85,86 Additionally, leveraging radiomic information from alternative imaging modalities in combination with that acquired from MRI could augment breast tumor characterization.87 Moving forward, radiomic analysis has the potential for use in fully characterizing tumor biology to serve as a noninvasive, quantitative tumor assay, complementing proteomic and genomic tumor analyses for a comprehensive and personalized understanding of breast cancer.
References
- 1.Tao Z, Shi A, Lu C, Song T, Zhang Z, Zhao J. Breast cancer: Epidemiology and etiology. Cell Biochem Biophys 2015;72:333–338. [DOI] [PubMed] [Google Scholar]
- 2.DeSantis C, Ma J, Bryan L, Jemal A. Breast cancer statistics, 2013. CA Cancer J Clin 2014;64:52–62. [DOI] [PubMed] [Google Scholar]
- 3.Marusyk A, Polyak K. Tumor heterogeneity: Causes and consequences. Biochim Biophys Acta Rev Cancer 2010;1805:105–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Polyak K. Heterogeneity in breast cancer. J Clin Invest 2011;121: 3786–3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O’Connor JP, Rose CJ, Waterton JC, Carano RA, Parker GJ, Jackson A. Imaging intratumor heterogeneity: Role in therapy response, resistance, and clinical outcome. Clin Cancer Res 2015;21:249–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dagogo-Jack I, Shaw AT. Tumour heterogeneity and resistance to cancer therapies. Nat Rev Clin Oncol 2018;15:81–94. [DOI] [PubMed] [Google Scholar]
- 7.Bedard PL, Cardoso F. Can some patients avoid adjuvant chemotherapy for early-stage breast cancer? Nat Rev Clin Oncol 2011;8:272. [DOI] [PubMed] [Google Scholar]
- 8.Sabatier R, Gonçalves A, Bertucci F. Personalized medicine: Present and future of breast cancer management. Crit Rev Oncol Hematol 2014;91: 223–233. [DOI] [PubMed] [Google Scholar]
- 9.Gavenonis SC, Roth SO. Role of magnetic resonance imaging in evaluating the extent of disease. Magn Reson Imaging Clin 2010;18:199–206. [DOI] [PubMed] [Google Scholar]
- 10.Weinstein S, Rosen M. Breast MR imaging: Current indications and advanced imaging techniques. Radiol Clin 2010;48:1013–1042. [DOI] [PubMed] [Google Scholar]
- 11.Tse GM, Chaiwun B, Wong KT, et al. Magnetic resonance imaging of breast lesions—A pathologic correlation. Breast Cancer Res Treat 2007; 103:1–10. [DOI] [PubMed] [Google Scholar]
- 12.Hylton NM. Vascularity assessment of breast lesions with gadolinium-enhanced MR imaging. Magn Reson Imaging Clin N Am 2001;9: 321–332, vi. [PubMed] [Google Scholar]
- 13.O’Connor JP, Jackson A, Asselin M-C, Buckley DL, Parker GJ, Jayson GC. Quantitative imaging biomarkers in the clinical development of targeted therapeutics: Current and future perspectives. Lancet Oncol 2008;9:766–776. [DOI] [PubMed] [Google Scholar]
- 14.McInerney T, Terzopoulos D. Deformable models in medical image analysis: A survey. Med Image Anal 1996;1:91–108. [DOI] [PubMed] [Google Scholar]
- 15.Castellano G, Bonilha L, Li L, Cendes F. Texture analysis of medical images. Clin Radiol 2004;59:1061–1069. [DOI] [PubMed] [Google Scholar]
- 16.Ganeshan B, Miles KA. Quantifying tumour heterogeneity with CT. Cancer Imaging 2013;13:140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jain R, Kasturi R, Schunck BG. Machine vision. New York: McGraw-Hill; 1995. [Google Scholar]
- 18.Jain A, Ross A, Prabhakar S. Fingerprint matching using minutiae and texture features In: 2001 Proceedings 2001 International Conference on Image Processing, Vol. 3: IEEE; 2001. p 282–285. [Google Scholar]
- 19.Chen K, Wei H, Hennebert J, Ingold R, Liwicki M. Page segmentation for historical handwritten document images using color and texture features Frontiers in Handwriting Recognition (ICFHR), 2014 14th International Conference on: IEEE; 2014. p 488–493. [Google Scholar]
- 20.Gastounioti A, Conant EF, Kontos D. Beyond breast density: A review on the advancing role of parenchymal texture analysis in breast cancer risk assessment. Breast Cancer Res 2016;18:91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morrow M, Waters J, Morris E. MRI for breast cancer screening, diagnosis, and treatment. Lancet 2011;378:1804–1811. [DOI] [PubMed] [Google Scholar]
- 22.Holli KK, Harrison L, Dastidar P, et al. Texture analysis of MR images of patients with mild traumatic brain injury. BMC Med Imaging 2010;10:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hackmack K, Paul F, Weygandt M, Allefeld C, Haynes J-D, Initiative AsDN. Multi-scale classification of disease using structural MRI and wavelet transform. Neuroimage 2012;62:48–58. [DOI] [PubMed] [Google Scholar]
- 24.Mayerhoefer ME, Breitenseher MJ, Kramer J, Aigner N, Hofmann S, Materka A. Texture analysis for tissue discrimination on T1-weighted MR images of the knee joint in a multicenter study: Transferability of texture features and comparison of feature selection methods and classifiers. J Magn Reson Imaging 2005;22:674–680. [DOI] [PubMed] [Google Scholar]
- 25.Michopoulou S, Costaridou L, Vlychou M, Speller R, Todd-Pokropek A. Texture-based quantification of lumbar intervertebral disc degeneration from conventional T2-weighted MRI. Acta Radiol 2011;52:91–98. [DOI] [PubMed] [Google Scholar]
- 26.Vignati A, Mazzetti S, Giannini V, et al. Texture features on T2-weighted magnetic resonance imaging: New potential biomarkers for prostate cancer aggressiveness. Phys Med Biol 2015;60:2685. [DOI] [PubMed] [Google Scholar]
- 27.Assefa D, Keller H, Ménard C, Laperriere N, Ferrari RJ, Yeung I. Robust texture features for response monitoring of glioblastoma multiforme on-weighted and-FLAIR MR images: A preliminary investigation in terms of identification and segmentation. Med Phys 2010;37:1722–1736. [DOI] [PubMed] [Google Scholar]
- 28.Torheim T, Malinen E, Kvaal K, et al. Classification of dynamic contrast enhanced MR images of cervical cancers using texture analysis and support vector machines. IEEE Trans Med Imaging 2014;33:1648–1656. [DOI] [PubMed] [Google Scholar]
- 29.Harrison L, Dastidar P, Eskola H, et al. Texture analysis on MRI images of non-Hodgkin lymphoma. Comput Biol Med 2008;38:519–524. [DOI] [PubMed] [Google Scholar]
- 30.Zhang Y. MRI texture analysis in multiple sclerosis. J Biomed Imaging 2012;2012:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Braman NM, Etesami M, Prasanna P, et al. Intratumoral and peritumoral radiomics for the pretreatment prediction of pathological complete response to neoadjuvant chemotherapy based on breast DCE-MRI. Breast Cancer Res 2017;19:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang J, Kato F, Oyama-Manabe N, et al. Identifying triple-negative breast cancer using background parenchymal enhancement heterogeneity on dynamic contrast-enhanced MRI: A pilot radiomics study. PLoS One 2015;10:e0143308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haralick RM, Shanmugam K. Textural features for image classification. IEEE Trans Syst Man Cybernet 1973:610–621. [Google Scholar]
- 34.Galloway MM. Texture analysis using grey level run lengths. NASA STI/-Recon Technical Report N 1974;75. [Google Scholar]
- 35.Tang X. Texture information in run-length matrices. IEEE Trans Image Process 1998;7:1602–1609. [DOI] [PubMed] [Google Scholar]
- 36.Chu A, Sehgal CM, Greenleaf JF. Use of gray value distribution of run lengths for texture analysis. Pattern Recogn Lett 1990;11:415–419. [Google Scholar]
- 37.Ojala T, Pietikainen M, Maenpaa T. Multiresolution gray-scale and rotation invariant texture classification with local binary patterns. IEEE Trans Pattern Anal Mach Intell 2002;24:971–987. [Google Scholar]
- 38.Porter R, Canagarajah N. Robust rotation-invariant texture classification: Wavelet, Gabor filter and GMRF based schemes. IEEE Proc Vis Image Sign Process 1997;144:180–188. [Google Scholar]
- 39.Srinivasan G, Shobha G. Statistical texture analysis. In: Proc World Acad Sci Eng Technol Vol. 36; 2008. p 1264–1269. [Google Scholar]
- 40.Livens S, Scheunders P, Van de Wouwer G, Van Dyck D. Wavelets for texture analysis, an overview. 1997. [Google Scholar]
- 41.Cho GY, Moy L, Kim SG, et al. Evaluation of breast cancer using intra-voxel incoherent motion (IVIM) histogram analysis: Comparison with malignant status, histological subtype, and molecular prognostic factors. Eur Radiol 2016;26:2547–2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gibbs P, Turnbull LW. Textural analysis of contrast-enhanced MR images of the breast. Magn Reson Med 2003;50:92–98. [DOI] [PubMed] [Google Scholar]
- 43.Nagarajan MB, Huber MB, Schlossbauer T, Leinsinger G, Krol A, Wismüller A. Classification of small lesions in breast MRI: Evaluating the role of dynamically extracted texture features through feature selection. J Med Biol Eng 2013;33(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nie K, Chen J-H, Hon JY, Chu Y, Nalcioglu O, Su M-Y. Quantitative analysis of lesion morphology and texture features for diagnostic prediction in breast MRI. Acad Radiol 2008;15:1513–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bhooshan N, Giger ML, Jansen SA, Li H, Lan L, Newstead GM. Cancerous breast lesions on dynamic contrast-enhanced MR images: Computerized characterization for image-based prognostic markers. Radiology 2010;254:680–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bhooshan N, Giger M, Lan L, et al. Combined use of T2-weighted MRI and T1-weighted dynamic contrast-enhanced MRI in the automated analysis of breast lesions. Magn Reson Med 2011;66:555–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Holli K, Lääperi A-L, Harrison L, et al. Characterization of breast cancer types by texture analysis of magnetic resonance images. Acad Radiol 2010;17:135–141. [DOI] [PubMed] [Google Scholar]
- 48.Chen W, Giger ML, Li H, Bick U, Newstead GM. Volumetric texture analysis of breast lesions on contrast-enhanced magnetic resonance images. Magn Reson Med 2007;58:562–571. [DOI] [PubMed] [Google Scholar]
- 49.Issa B, Buckley DL, Turnbull LW. Heterogeneity analysis of Gd-DTPA uptake: Improvement in breast lesion differentiation. J Comput Assist Tomogr 1999;23:615–621. [DOI] [PubMed] [Google Scholar]
- 50.Waugh S, Purdie C, Jordan L, et al. Magnetic resonance imaging texture analysis classification of primary breast cancer. Eur Radiol 2016;26: 322–330. [DOI] [PubMed] [Google Scholar]
- 51.Holli-Helenius K, Salminen A, Rinta-Kiikka I, et al. MRI texture analysis in differentiating luminal A and luminal B breast cancer molecular subtypes-a feasibility study. BMC Med Imaging 2017;17:69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sutton EJ, Oh JH, Dashevsky BZ, et al. Breast cancer subtype intertumor heterogeneity: MRI-based features predict results of a genomic assay. J Magn Reson Imaging 2015;42:1398–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li H, Zhu Y, Burnside ES, et al. MR imaging radiomics signatures for predicting the risk of breast cancer recurrence as given by research versions of MammaPrint, Oncotype DX, and PAM50 gene assays. Radiology 2016;281:382–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu J, Sun X, Wang J, et al. Identifying relations between imaging phenotypes and molecular subtypes of breast cancer: Model discovery and external validation. J Magn Reson Imaging 2017;46:1017–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.King V, Brooks JD, Bernstein JL, Reiner AS, Pike MC, Morris EA. Back-ground parenchymal enhancement at breast MR imaging and breast cancer risk. Radiology 2011;260:50–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chamming’s F, Ueno Y, Ferré R, et al. Features from computerized texture analysis of breast cancers at pretreatment MR imaging are associated with response to neoadjuvant chemotherapy. Radiology 2017;286: 412–420. [DOI] [PubMed] [Google Scholar]
- 57.Kim J-H, Ko ES, Lim Y, et al. Breast cancer heterogeneity: MR imaging texture analysis and survival outcomes. Radiology 2016;282:665–675. [DOI] [PubMed] [Google Scholar]
- 58.Pickles MD, Lowry M, Gibbs P. Pretreatment prognostic value of dynamic contrast-enhanced magnetic resonance imaging vascular, texture, shape, and size parameters compared with traditional survival indicators obtained from locally advanced breast cancer patients. Invest Radiol 2016;51:177–185. [DOI] [PubMed] [Google Scholar]
- 59.Park H, Lim Y, Ko ES, et al. Radiomics signature on magnetic resonance imaging: Association with disease-free survival in patients with invasive breast cancer. Clin Cancer Res 2018;37832017. [DOI] [PubMed] [Google Scholar]
- 60.Mahrooghy M, Ashraf AB, Daye D, et al. Heterogeneity wavelet kinetics from DCE-MRI for classifying gene expression based breast cancer recurrence risk In: International Conference on Medical Image Computing and Computer-Assisted Intervention. Berlin: Springer; 2013. p 295–302. [DOI] [PubMed] [Google Scholar]
- 61.Mahrooghy M, Ashraf AB, Daye D, et al. Pharmacokinetic tumor heterogeneity as a prognostic biomarker for classifying breast cancer recurrence risk. IEEE Trans Biomed Eng 2015;62:1585–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Teruel J, Heldahl M, Goa P, et al. Dynamic contrast-enhanced MRI texture analysis for pretreatment prediction of clinical and pathological response to neoadjuvant chemotherapy in patients with locally advanced breast cancer. NMR Biomed 2014;27:887–896. [DOI] [PubMed] [Google Scholar]
- 63.Thibault G, Tudorica A, Afzal A, et al. DCE-MRI texture features for early prediction of breast cancer therapy response. Tomography 2017;3:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Golden DI, Lipson JA, Telli ML, Ford JM, Rubin DL. Dynamic contrast-enhanced MRI-based biomarkers of therapeutic response in triple-negative breast cancer. J Am Med Inform Assoc 2013;20:1059–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Parikh J, Selmi M, Charles-Edwards G, et al. Changes in primary breast cancer heterogeneity may augment midtreatment MR imaging assessment of response to neoadjuvant chemotherapy. Radiology 2014;272: 100–112. [DOI] [PubMed] [Google Scholar]
- 66.Michoux N, Van den Broeck S, Lacoste L, et al. Texture analysis on MR images helps predicting non-response to NAC in breast cancer. BMC Cancer 2015;15:574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wu J, Gong G, Cui Y, Li R. Intratumor partitioning and texture analysis of dynamic contrast-enhanced (DCE)-MRI identifies relevant tumor subregions to predict pathological response of breast cancer to neoadjuvant chemotherapy. J Magn Reson Imaging 2016;44:1107–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ashraf A, Gaonkar B, Mies C, et al. Breast DCE-MRI kinetic heterogeneity tumor markers: Preliminary associations with neoadjuvant chemotherapy response. Transl Oncol 2015;8:154–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ahmed A, Gibbs P, Pickles M, Turnbull L. Texture analysis in assessment and prediction of chemotherapy response in breast cancer. J Magn Reson Imaging 2013;38:89–101. [DOI] [PubMed] [Google Scholar]
- 70.El Adoui M, Drisis S, Larhmam M, Lemort M, Benjelloun M. Breast cancer heterogeneity analysis as index of response to treatment using MRI images: A review. Imaging Med 2017;9:109–119. [Google Scholar]
- 71.Johansen R, Jensen LR, Rydland J, et al. Predicting survival and early clinical response to primary chemotherapy for patients with locally advanced breast cancer using DCE-MRI. J Magn Reson Imaging 2009; 29:1300–1307. [DOI] [PubMed] [Google Scholar]
- 72.Padhani AR, Hayes C, Assersohn L, et al. Prediction of clinicopathologic response of breast cancer to primary chemotherapy at contrast-enhanced MR imaging: Initial clinical results. Radiology 2006;239:361–374. [DOI] [PubMed] [Google Scholar]
- 73.Sala E, Mema E, Himoto Y, et al. Unravelling tumour heterogeneity using next-generation imaging: Radiomics, radiogenomics, and habitat imaging. Clin Radiol 2017;72:3–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Burnside ES, Drukker K, Li H, et al. Using computer-extracted image phenotypes from tumors on breast magnetic resonance imaging to predict breast cancer pathologic stage. Cancer 2016;122:748–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Agner SC, Rosen MA, Englander S, et al. Computerized image analysis for identifying triple-negative breast cancers and differentiating them from other molecular subtypes of breast cancer on dynamic contrast-enhanced MR images: A feasibility study. Radiology 2014;272:91–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhu Y, Li H, Guo W, et al. Deciphering genomic underpinnings of quantitative MRI-based radiomic phenotypes of invasive breast carcinoma. Sci Rep 2015;5:17787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Duda R, Hart PE, Stork DG. Pattern classification and scene analysis. New York: John Wiley & Sons; 2000. [Google Scholar]
- 78.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc B 1995:289–300. [Google Scholar]
- 79.Waugh SA, Lerski RA, Bidaut L, Thompson AM. The influence of field strength and different clinical breast MRI protocols on the outcome of texture analysis using foam phantoms. Med Phys 2011;38:5058–5066. [DOI] [PubMed] [Google Scholar]
- 80.Lambin P, Rios-Velazquez E, Leijenaar R, et al. Radiomics: Extracting more information from medical images using advanced feature analysis. Eur J Cancer 2012;48:441–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gillies RJ, Kinahan PE, Hricak H. Radiomics: Images are more than pictures, they are data. Radiology 2015;278:563–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kumar V, Gu Y, Basu S, et al. Radiomics: The process and the challenges. Magn Reson Imaging 2012;30:1234–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Vallières M, Freeman CR, Skamene SR, El Naqa I. A radiomics model from joint FDG-PET and MRI texture features for the prediction of lung metastases in soft-tissue sarcomas of the extremities. Phys Med Biol 2015;60:5471. [DOI] [PubMed] [Google Scholar]
- 84.Guo W, Li H, Zhu Y, et al. Prediction of clinical phenotypes in invasive breast carcinomas from the integration of radiomics and genomics data. J Med Imaging 2015;2:041007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Koo HR, Cho N, Song IC, et al. Correlation of perfusion parameters on dynamic contrast-enhanced MRI with prognostic factors and subtypes of breast cancers. J Magn Reson Imaging 2012;36:145–151. [DOI] [PubMed] [Google Scholar]
- 86.Li H, Zhu Y, Burnside ES, et al. Quantitative MRI radiomics in the prediction of molecular classifications of breast cancer subtypes in the TCGA/T-CIA data set. NPJ Breast Cancer 2016;2:16012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lubner MG, Smith AD, Sandrasegaran K, Sahani DV, Pickhardt PJ. CT texture analysis: Definitions, applications, biologic correlates, and challenges. RadioGraphics 2017;37:1483–1503. [DOI] [PubMed] [Google Scholar]


