Abstract
Mycobacterium bovis is the pathogenic agent responsible for bovine tuberculosis (bTB), a zoonotic disease affecting mostly cattle, but also transmittable to humans and wildlife. Genetic studies on M. bovis allow to detect possible routes of bTB transmission and the identification of genetic reservoirs that may provide an essential framework for public health action.
We used a database with 1235 M. bovis genotypes collected from different regions in Africa with 45 new Mozambican samples. Our analyses, based on phylogeographic and population genetics’ approaches, allowed to identify two clear trends. First, the genetic diversity of M. bovis is geographically clustered across the continent, with the only incidences of long-distance sharing of genotypes, between South Africa and Algeria, likely due to recent European introductions. Second, there is a broad gradient of diversity from Northern to Southern Africa with a diversity focus on the proximity to the Near East, where M. bovis likely emerged with animal domestication in the last 10,000 years.
Diversity indices are higher in Eastern Africa, followed successively by Northern, Central, Southern and Western Africa, roughly correlating with the regional archaeological records of introduction of animal domesticates. Given this scenario M. bovis in Africa was probably established millennia ago following a concomitant spread with cattle, sheep and goat. Such scenario could translate into long-term locally adapted lineages across Africa.
This work describes a novel scenario for the spread of M. bovis in Africa using the available genetic data, opening the field to further studies using higher resolution genomic data.
Author summary
We describe the genetic diversity distribution in Africa of the pathogen Mycobacterium bovis, the responsible for bovine tuberculosis, mostly present in cattle but also transmittable to other animals including humans. This diversity is geographically clustered within the African continent meaning that the genetic diversity was established through independent evolution within different areas. Higher diversity values of M. bovis are found in Eastern and Northern Africa, followed by Central Africa, with Western and Southern Africa displaying the lowest diversity. These levels of diversity correlate well with the introduction of domesticated livestock in the different regions of Africa, following their domestication in the Near East 10,000 ago. We hypothesize that M. bovis emerged in the Near East and it was carried across Africa together with domesticated animals and people that developed herding practices and biological tolerance for digesting milk in adulthood. Such scenario implies a strong evolution and co-evolution of M. bovis across Africa leading to locally adapted strains that could prove a challenge for public health actions.
Introduction
Mycobacterium bovis is the major causal agent of bovine tuberculosis (bTB) in cattle, the world's most neglected zoonotic disease that needs urgent attention, especially in developing countries [1, 2]. The largest disease burden probably occurs in poor, marginalized, rural communities living in close proximity to animals, and with reduced access to safe food and health care. bTB in humans is associated with consumption of unpasteurized dairy products or raw meat products from infected cattle [3–5], or even through inhalation of bacilli when in close proximity to infected cattle or their carcasses [3].
In 2017, Olea-Popelka and co-authors raised awareness for the worldwide importance of bTB, particularly in the African continent, where insufficient surveillance, testing and traditional ways of rearing and consuming cattle may be responsible for under-notification of this zoonotic disease [6]. In fact, an important percentage of cases diagnosed as TB and attributed to Mycobacterium tuberculosis infection may be bTB. The disease also impacts on international trade of animals and animal products [7]. The movement of animals has most probably been the main reason for the spread of M. bovis both within the same country as well as cross-borders [4, 8, 9].
Little is known about the prevalence and epidemiology of bTB in Africa, and its impact on humans, on livestock and wildlife. In sub-Saharan Africa, few studies describe the transmission status of bTB in regions where multiple hosts are present, but it is believed that sharing of water points plays a key role in the transmission and spread of the disease between animals [10]. Indeed, the distribution and epidemiology of bTB varies according to the production system, however, importance in public health and control requires attention of all producers regardless of the system [11].
Different studies have shown that the distribution of bTB in Africa is not uniform, with areas of low and high prevalence [4, 12]. However, It is estimated that 85% of the bovine population and 82% of the human population live in areas where bTB is prevalent and control measures are non-existent or inadequate and the potential for zoonotic transmission is exacerbated by direct coexistence between the breeding/shepherds and cattle [3, 13]. bTB is present in most countries [3, 7] and the political borders do not constitute physical barriers to the movement of animals and their diseases.
In Mozambique, this scenario is common and bTB is assumed to be an important cause of the economic losses in bovine production evidenced by the rejection of carcasses at the slaughterhouse and limitations on national and international trade. However, little is known about the impact of bTB on humans. Mozambique is one of the countries most affected by the TB, TB / HIV and MDR-TB triad with six other African countries, two of which are countries that share borders with Mozambique [14].
The transport of animals might have been a major determinant for the spread of the disease since pre-historic periods. The genetic coalescence of M. bovis have been estimated to at least around 6000 years [15] and domesticated cattle existed along humans in Africa for at least 8–6000 years [16]. The so-called Secondary Product Revolution involved the continuous use of animal products as milk and wool, likely leading to an increase of animals living closely together in herds. Milk consumption left a major mark on human biology and the genetic acquiring of lactose intolerance in human adulthood is arguably one of the clearest signals of positive selection in populations from Europe and Africa [17–19].
Genetic studies on M. bovis allow the possibility to detect possible routes of bTB transmission within and across borders. The main objective of this work is to shed light on the presence of genetic reservoirs of bTB in Africa and to trace historical/prehistoric events in the continent using genetic diversity and phylogeography of M. bovis in Africa.
Methods
Ethics statement
Institutional permission to conduct the study was obtained from the National Directorate of Veterinary Services in Maputo, Mozambique (Nota 162/ MINAG/DNSV/900/2013) and Ethical Principles in Animal Research adopted by Ethic Committee in the Use of Animals (CEUA) of School of Veterinary Medicine and Animal Science, University of São Paulo (Protocol number CEUAx 6755081216). Sampling and culling were performed as part of the Veterinary Services regular activity for disease control, following the procedures determined by the Mozambican Animal Health Regulation (Regulamento de Sanidade Animal decreto 26/2009). The slaughter was done in registered abattoirs according to stipulated procedures. All mycobacterial cultures were performed in the National Tuberculosis Reference Laboratory, Ministry of Health Mozambique.
Mycobacterium bovis’ samples from Mozambican cattle: collection and genotyping
Forty five samples, consisting of tissue/organs containing lesions, were processed in the BSL3 from the National Reference Laboratory of Tuberculosis (LNRT) in Mozambique in duplicate. Cultures grew in Stonebrink (ST) medium with pyruvate and Lowenstein-Jensen (LJ) medium with glycerol. DNA extraction was performed according to the protocol described by Van Soolingen and colleagues [20]. The molecular identification of Mycobacterium tuberculosis complex (MTBC) strains was based on the use of two sets of multiplex PCR for identifying the genus Mycobacterium [21] and for the discrimination of the different MTBC members [22].
Genotyping of the M. bovis samples was performed using three methodologies: spoligotyping, region of difference (RD) analysis and MIRU-VNTR, as described before [23].
Spoligotyping of each sample was determined based on the numerical combination of the presence or absence of the spacers and identified according to mbovis.org database. RD analysis, a PCR-based method to determine the presence or absence of specific regions of difference (RD) was performed to identify four M. bovis groups named Af1, Af2, Eu1 and Eu2. All samples were genotyped using the MIRU-VNTR 24 loci kit (Genoscreen, France) that includes 24 size-variation markers, although only a few have proven to be polymorphic in M. bovis. The genotypic profiles were reported as a series of 24 numbers corresponding to the number of alleles at each of the 24 loci.
Comparative dataset
We collected the largest possible database of genotypes for M. bovis in Africa. A list of sampled locations and numbers is indicated in S1 Table and S2 Table. We included African M. bovis data that encompasses the 43 spoligotyping markers and 5 VNTR markers that were present in most studies (MIRU2165, MIRU2461, MIRU577, MIRU580 and MIRU3192 or ETR-A, -B, -C, -D and -E), that previously displayed enough resolution in the African context [23, 24]. We allowed one marker per sample to be missing, whose allele will be further extrapolated in the phylogenetic analysis below. We included a European Mediterranean dataset for comparison [25]. We also included other members from the M. tuberculosis Complex (MTBC) available at the same genotyping resolution obtained from the reference database of MIRU-VNTR Plus [26] that included genotypes from 31 M africanum, 2 M. canetti, 11 M. caprae, 2 M. pinnipedii, 6 M. microti and 123 M. tuberculosis strains. These genotypes were included in the phylogenetic reconstruction and the dataset of European M. tuberculosis strains were used as an outgroup for the population tree analysis of M. bovis strains.
Phylogenetics reconstruction and statistical analyses of M. bovis genotypes
Reconstruction was performed using a matrix of the 48 markers described above and by applying the reduced median algorithm [27] followed by the median joining algorithm [28]. Both present at the network software (freely available at http://www.fluxus-engineering.com), as we suggested for MIRU-VNTR data in M. tuberculosis [29, 30]. Given the faster evolutionary rate of VNTR markers against binary spoligotyping markers we placed a greater weight of the latter in the analyses against VNTR markers (15 against 10 respectively). Network allows missing genotyping information in the input data that will be extrapolated from the parsimonious phylogenetic analysis.
The diversity of the 48 makers was converted into a binary matrix reflecting genetic distances between each genotype. That binary matrix was used to calculate diversity indices in DNAsp 6 [31]. For each individual population (country) with sample size higher than 20, we estimated the number of haplotypes h, the haplotype diversity Hd (probability that two randomly sampled haplotypes are different), the average number of differences K (average number of differences between all the pairs in a population) and the nucleotide diversity Pi (average number of differences between all the pairs in a population per number of loci).
In order to estimate the coalescence point of M. bovis in each population, we used the previously described outgroups (corresponding to genotypes of six other members of the MTBC) to define the root with Network software. The root was used to obtain the ρ statistic for each dataset. The ρ statistic measures the average number of differences from a root (coalescence point) and each of the samples and represents an unbiased model-free diversity measure and time estimator [32].
The different statistics were plotted geographically using the Kriging algorithm of Surfer 8 software using the geographic points indicated in S1 Fig. A geographically central point of each country was selected as the representative data point instead of the capital where, in many cases, was close to the border with other countries and near the capital of the other country.
Fst distances between pairs of populations were calculated using DNAsp 6 [31] and a population tree (using neighbour-joining) was calculated from the Fst matrix obtained using MEGA7 [33]. We performed the analysis with and without the locations with lower sample size (Burundi and Eritrea), excluding Uganda nevertheless (only 2 genotypes). We included a subset of European M. tuberculosis genotypes to be used as outgroup in the analysis.
Results
To investigate the presence of hypothetical genetic reservoirs of M. bovis across Africa, we performed a detailed phylogeographic and statistical analysis on African diversity of M. bovis. For that we collected 1504 genotypes comprised by 48 markers (43 biallelic and 5 multiallelic) (S1 Table) to which we added 45 new genotypes from Mozambique.
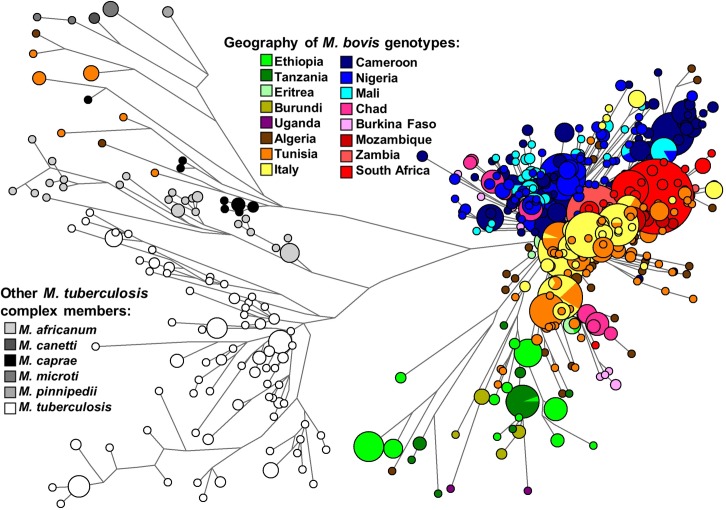
We reconstructed a phylogeny for the genotypes of M. bovis strains in Africa (Fig 1). Incidentally, some strains from Northern Africa clustered with M. caprae instead of M. bovis. The presence of other MTBC strains in the network shows a deep coalescence point on the convergence of M africanum, M. canetti, M. caprae, M. pinnipedii, M. microti and M. tuberculosis genotypes and M. bovis strains’ genotypes. This allows to identify branches that are coalescing at a deeper evolutionary point than most of the M. bovis lineages. These ancient lineages are mostly from Eastern Africa (Ethiopia, Tanzania) and Northern Africa (Algeria, Tunisia), although some haplotypes with minor frequencies from Central Africa are also present. On an overview analysis of the network, Eastern and Northern Africa seem to display a deeper ancestry of M. bovis compared with other African regions.
Fig 1. Median network displaying diversity of Mycobacterium bovis in Africa using 48 markers (43 spoligotype spacers and 5 VNTRs).
The genotypes of M. tuberculosis, M. caprae, M. africanum, M. canetti and M. pinnipedii strains were included as outgroups. Samples were coloured according to the geographic location (in the case of M. bovis strains) or according to the strains (in the case of other M. tuberculosis complex strains). Figure was made using freely available phylogenetic software network (http://www.fluxus-engineering.com).
Most of the M. bovis genotypes radiate into several clades from a single major coalescence point. On that coalescence point it is possible to discern a few longer branches from Ethiopia and Tanzania. There are specific branches from Northern Africa, Mozambique, Southern Africa, Italy and Central Africa, reflecting a strong geographical compartmentalization. Most of the diversity in Central Africa seems present on a single major clade. That clade, defined by the spacer 30, corresponds likely to the common African spoligotype type Af1. Mozambique presents three clades from the main M. bovis radiation point, including one shared with South Africa, but, in general, Mozambique displays, as other locations, a specific local subset of diversity.
In general, the sharing of genotypes across borders is mostly restricted to countries in the vicinity of each other. However, Northern Africa (Algeria) shows common haplotypes with South Africa (Fig 1 and S3 Table). Some of the matching haplotypes could be defined as previously named European clusters Eu1 and Eu2 since they lack spacers 11 and 21 respectively in the spoligotyping data that has have been used to define these clusters [23, 34]. However, spoligotyping data also shows a great level of homoplasy and these definitions might not be exact. It is interesting to notice that, as somewhat expected, there is a great level of sharing between North Africa and the sample from Italy. High levels of gene flow and trading are visible from pre-history until recent times between both regions [35].
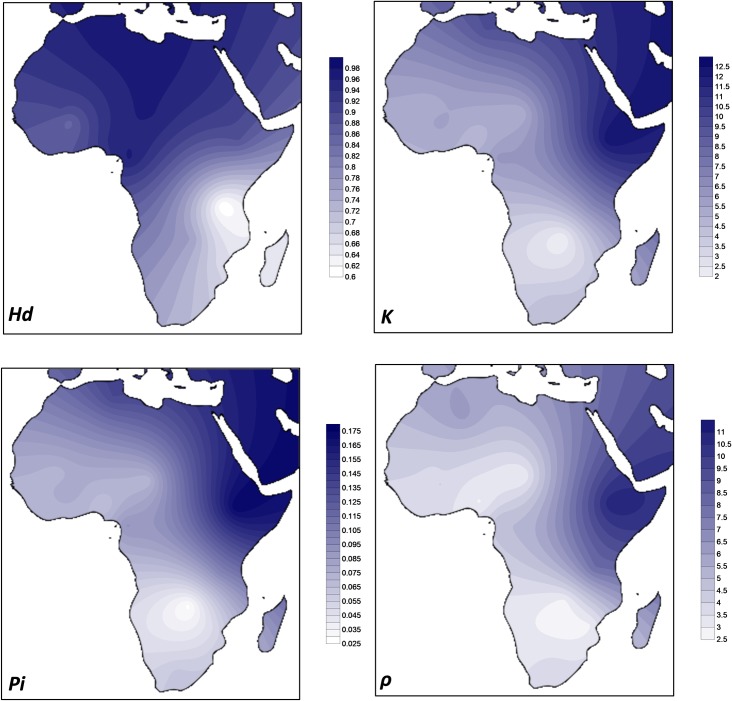
In order to visualize the distribution of the genetic diversity of M. bovis in Africa, we obtained four diversity statistics in all populations with more than 20 samples (Table 1) that were plotted geographically (Fig 2). The measures have different properties. Haplotype diversity Hd reflects the high or low number and frequency of circulating haplotypes independently of the distance between the haplotypes. Recently established reservoirs of diversity would be best detected with this measure than the others. ρ measures depth of diversity in a phylogenetic context and it could establish deep or shallow presence of M. bovis in each region even if concentrated in a small number of haplotypes. K and Pi offers intermediate properties (similar haplotypes have zero distance between them but further genetic distances between other pairs of haplotypes are also accounted).
Table 1. Diversity measures obtained from the sample of Mycobacterium bovis genotypes in different countries.
Diversity indices were not calculated for locations with low sample size (n<20).
| Country | Sample size (n) | Number of haplotypes, h | Haplotype diversity, Hd | Average number of differences, K | Nucleotide diversity, Pi | Rho, ρ |
|---|---|---|---|---|---|---|
| Algeria | 89 | 52 | 0.95812 | 7.56844 | 0.10660 | 6.2410 |
| Burkina Faso | 32 | 11 | 0.85238 | 5.73333 | 0.08075 | 4.0313 |
| Burundi | 10 | 4 | - | - | - | - |
| Cameroon | 269 | 84 | 0.96288 | 6.18543 | 0.09120 | 4.9776 |
| Chad | 67 | 34 | 0.96201 | 5.07825 | 0.07152 | 3.0000 |
| Eritrea | 14 | 5 | - | - | - | - |
| Ethiopia | 67 | 15 | 0.88105 | 12.37268 | 0.17426 | 10.881 |
| Mali | 59 | 11 | 0.90181 | 5.04734 | 0.07109 | 3.8644 |
| Mozambique | 102 | 14 | 0.67618 | 3.62667 | 0.05108 | 2.9314 |
| Nigeria | 178 | 68 | 0.94160 | 4.65454 | 0.06556 | 2.9270 |
| South Africa | 193 | 26 | 0.73343 | 4.38957 | 0.06182 | 3.8083 |
| Tanzania | 27 | 8 | 0.60114 | 7.03704 | 0.09911 | 7.9259 |
| Tunisia | 211 | 89 | 0.97319 | 9.54557 | 0.13499 | 5.5707 |
| Uganda | 2 | 2 | - | - | - | - |
| Zambia | 24 | 6 | 0.75725 | 2.00725 | 0.02827 | 2.8333 |
| Italy | 205 | 33 | 0.87972 | 4.66361 | 0.06565 | 3.0878 |
Fig 2. Geographic distribution of diversity of Mycobacterium bovis in Africa using the Kriging algorithm of Surfer 8 software for four diversity estimators (haplotype diversity Hd, the average number of differences K, the nucleotide diversity Pi and ρ).
Map outline was adapted from https://commons.wikimedia.org/wiki/Atlas_of_the_world.
It is clear that countries in Eastern Africa (Ethiopia and Tanzania) display the higher values of diversity when measures related with deep ancestry are concerned (ρ, K, Pi), mainly in the case of ρ, followed by Northern Africa (Algeria and Tunisia) that display very high values in terms of K and Pi, comparable to Eastern Africa indices. Countries in Central/Western Africa display lower diversity but in general countries in Southern Africa (Mozambique, Zambia and South Africa) display the lowest values. A gradient from North to South is visible in in the maps (Fig 2). It is important to point out that this gradient is disrupted when moving North from Africa. The diversity in the Mediterranean Europe sample is substantially lower than in North Africa, reinforcing the status of Northern and Eastern Africa as regions of deep ancestry of M. bovis diversity.
In terms of Hd, a statistic that could better reflect number of circulating strains, Northern and Central Africa display the highest values. In contrast, neighbour-countries like Mozambique, Tanzania and Zambia display the lowest diversity in terms of this statistics.
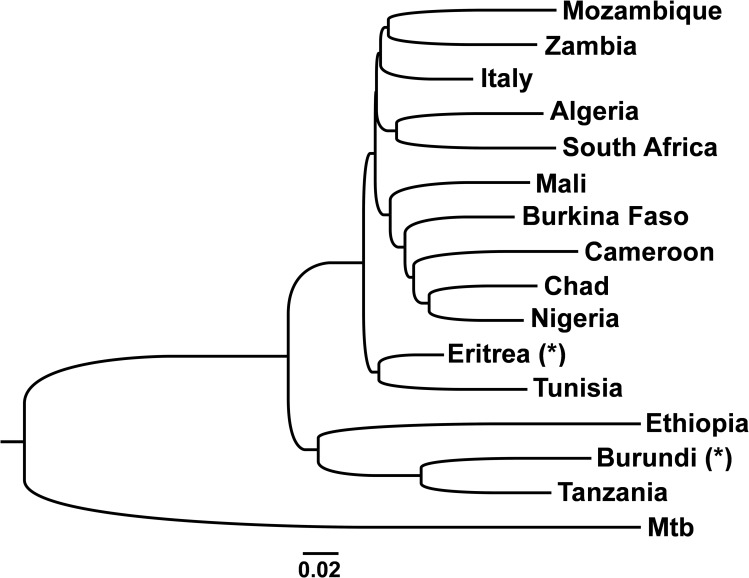
In order to fully evaluate the relationships between countries in terms of general diversity we calculated a population neighbour-joining tree based on Fst values. In one analysis, we included the low sample size locations Burundi and Eritrea in order to check if they fit the general patterns. Their presence did not alter the obtained relationship between the other groups so we present that analysis. The results clearly show Ethiopia and Tanzania as the most divergent populations, branching earlier in the tree (Fig 3). Burundi, on the limit between Eastern and Central Africa clusters with Eastern Africa, despite its low sample size. These were followed by the clustered Eastern Africa Eritrea and North African Tunisia, while the other Northern African population, Algeria, due to the sharing of haplotypes with South Africa was clustered with that population. Southern African Mozambique and Zambia were also clustered. The Italian population is marginally clustered with Mozambique and Zambia but essentially forms a cluster with Algeria and the three Southern African countries in contrast with countries in Central and Western Africa that were also grouped in a separate cluster. Nevertheless, diversity in Central/western Africa and Southern Africa seem more restricted than Eastern and Northern African diversity. In general, the population tree supports the previously performed analyses in terms of phylogeography and general statistics.
Fig 3. Neighbour-joining population tree based on a matrix of Fst values between populations.
A dataset of European M. tuberculosis (Mtb) was used as an outgroup. Sample sets indicated as (*) have a low sample size and their placement in the tree is possibly uncertain.
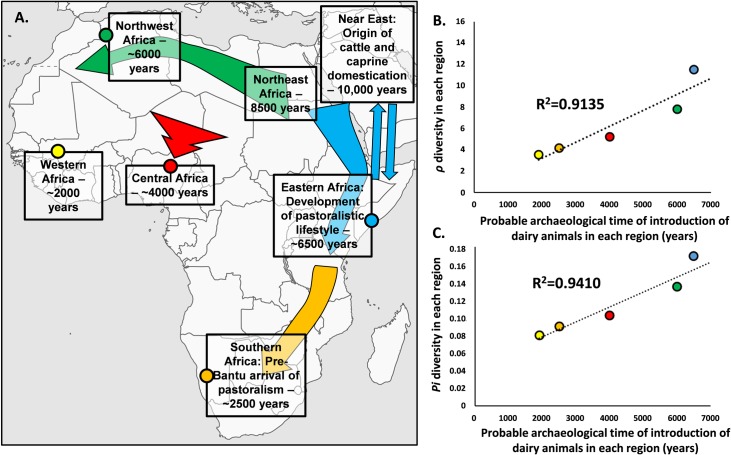
In order to establish general values per region we calculated ρ and Pi for the different general regions and, expectedly, for both measures Eastern Africa was the most diverse (ρ = 11.611, Pi = 0.17113), followed by Northern Africa (ρ = 7.8564, Pi = 0.136349), Central Africa (ρ = 5.3448, Pi = 0.10376), Southern Africa (ρ = 4.2922, Pi = 0.09087) and Western Africa (ρ = 3.978, Pi = 0.08087) (Fig 4).
Fig 4.
Comparison between hypothetical time of introduction of dairy animals across Africa from the archaeological record (A.) and genetic diversity statistics, ρ (B.) and Pi (C.). Colour of data points in (B) and (C) correspond to the regions of the same colour in the map (A). Map was adapted from https://commons.wikimedia.org/wiki/Atlas_of_the_world.
Discussion
M. bovis is the main organism responsible for bTB, a form of TB present in cattle and other domestic and wildlife species, that can be transmitted to humans through zoonosis. bTB represents a high burden in the African continent. In order to understand how the genetic diversity of this pathogen is distributed across Africa, we performed a genetic analysis on its overall diversity. This allows to establish probable regions that can be genetic reservoirs in the continent and where public health actions should focus aiming at bTB eradication. There are two conclusions that erupt from this analysis:
A) One feature is that the transportation of the pathogen across borders is probably more limited than expected–each country displays a subset of diversity that might have been established and maintained for a long period; except for the case of South Africa and Algeria, shared haplotypes seems to occur almost exclusively between neighbour countries. The South African/Algeria sharing of haplotypes can be related with an outside African source. Some of the diversity within these two countries are likely from clades Eu1 and these represent the matches detected, possibly reflecting M. bovis introduced by Europeans in South Africa and Algeria.
B) Considering the haplotype diversity, the number of circulating strains in different regions differs considerably across Africa. In the present analysis Mozambique, Tanzania and Zambia display the lowest diversity in circulating strains and are minor reservoirs of genetic diversity of M. bovis which typically represent more simplistic scenarios for public health than larger genetic reservoirs. However, a real scenario can just be attained by obtaining reliable values of incidence of bTB in different African countries that can be combined with this information. While Mozambique, Tanzania and Zambia show little diversity of circulation genotypes, their estimate of cases that could be related with recent transmission from genetic data is over 70% in the three countries, highlighting probable recent outbreaks.
Nevertheless, the distribution of the diversity of M. bovis in Africa raises questions regarding the processes that shaped that diversity. There has been a wide discussion regarding the origin and spread of the Mycobacterium tuberculosis complex that includes M. tuberculosis, the infectious agent responsible for TB, and M. bovis, the causal infectious agent of bTB that mostly infects animals but that is transmitted to humans. One common hypothesis for the origin of the MTBC relates with the emergence of agriculture in the Fertile Crescent in the Neolithic period, over 10,000 years ago [36], when cattle and other animal were domesticated. While, in the past, an animal-to-human transmission hypothesis was in place for the emergence of M. tuberculosis from M. bovis the sequencing of both genomes clearly showed that the pathogens are not directly related [37] but instead they share a common ancestor. In fact, several studies indicate that the diversity of M. tuberculosis suggests a coalescence of the organism above 40,000 years ago or above, linking its global spread to a probable co-expansion with the modern humans Out-of-Africa [15, 38]. On the other hand, estimates of the coalescence time of M. bovis are as recent as 6000 years placing it well within the Holocene and potentially with the expansion of Neolithic or pastoralist societies in Africa.
It is possible that bTB arose when animals were kept in larger groups in limited areas, that allowed the disease to be more easily transmitted between individuals. Domestication in Africa occurred through introductions from the Near East, where cattle was domesticated about 10,000 years ago, marking also the initiation of dairy practices. None of the native African animals were domesticated. In that sense we can hypothesize that bTB has a Neolithic origin and it was spread across Africa concomitantly with the introduction of cattle. Domestication of cattle and the maintenance of larger groups of animals is also connected with the use of animals, not only as a source of meat and hides, but also as milk producers, part of the so-called Secondary Product Revolution. That change in societies goes beyond a cultural change since it has deep biological implications, as part of the populations was positively selected through time in order to be able to digest lactose in adulthood. Incidentally, the ability to consume milk facilitates zoonotic transfers as it is clear nowadays that milk consumption is the major mode of transmission of bTB to humans. This suggests a co-evolution of humans, cattle and pathogen that allowed the endurance of the disease. Could the data suggest that these genetic patterns are related with anciently established routes? Two points of evidence support this scenario. The first is the strong geographic clustering of the diversity across Africa that provides a direct evidence that this diversity was established in the past and enough time elapsed for local diversity to be established. The second is that the levels of diversity across Africa, that could illustrate different timeframes for diversity to accumulate in each region, correlate well with the estimated arrival of domesticated cattle and pastoralism into those regions (Fig 4).
The two major areas with higher diversity in Africa are Eastern Africa and Northern Africa. These were the first analysed areas where domesticated cattle, goats and sheep were introduced from the Near East [16]. Following an earlier spread of these domesticates into Northeast Africa by 8000 years [16], Eastern Africa, with the higher diversity in our analysis, saw an introduction of people, animals and domestication practices possibly as early as 6500–6000 years ago where a nomadic pastoralistic lifestyle emerged. Specific alleles for lactase persistence, a trait that indicates human co-evolution with cattle, sheep and goat and milk consumption in adulthood, developed and were positively selected in Eastern Africa [19], including the major allele C-14010, dated to around 6–7000 years [19], the time of introduction of domesticated animals. Recent research on ancient DNA suggests a clear admixture of early pastoralistic groups with Eastern Africans by at least 4000 years [39]. Nevertheless, Eastern Africa also display a long history of contact with Arabia/Near East [40, 41] that could have promote multiple transmission events of domesticates and pathogen further increasing the diversity of the M. bovis in Eastern Africa.
As mentioned above, North Africa saw an earlier introduction to the area that is nowadays Egypt (8000 years ago), however in Northwestern Africa (location of our samples) pastoralism was introduced about 6000 years ago [42, 43]. The Sahel Belt was a major travelling corridor both on a West/East axis or North/South [44], where nomadic pastoralistic groups moved. The increased aridity of the Sahara caused pastoralistic groups to migrate South in the Late Holocene. By 4000 years ago, pastoralistic societies and herding practices were archaeologically detected in Nigeria [45] and by 3000 years in Cameroon [46]. Sub-Saharan Western Africa displays low diversity of M. bovis genotypes. While some earlier cases of cattle domestication might exist in this area [47, 48], clear signals of introduction of modern cattle species dates to first millennium AD in Burkina Faso [49].
Populations in Southern Africa, before the massive expansion of Bantu-speaking agriculturists in the last two millennia from Central Africa [50] that replaced part of the indigenous populations, were mostly isolated from the remaining continent [51, 52]. However, frequencies of the previously mentioned lactase persistence allele C-14010 in the indigenous KhoeSan groups suggests a migration that carried that allele, hardly present in Bantu-speaking populations from the South. Such migration probably introduced domesticated species and pastoralistic practices into Southern Africa about 2500 years ago [18], further supported by archaeology [53].
The probable time of introduction of domesticated animals across the continent (Fig 4A) is reflected in the order of diversity values for the ρ and Pi, the statistics that better reflect time and depth of accumulated diversity. More exactly, both set of statistics (Fig 4B and Fig 4C, respectively) correlate well with the hypothetical time of introduction of cattle, sheep and goat. That correlation would be even stronger (>90%) if we consider that the diversity of Eastern Africa could have had later additions from the Near East/Arabia Peninsula, the hypothetical place of origin for M. bovis. In conclusion, using a phylogeographic and population genetics approach, we suggest that the current patterns of diversity of M. bovis in Africa were established in prehistoric times, with the pathogen being carried by introduced domesticated animals across the continent.
It is interesting to highlight that the diversity in Mediterranean Europe was substantially lower than in Mediterranean Africa. If the coalescence estimate for M. bovis is actually below 8000 years [15] the pathogen would not have been carried by the main Neolithic wave from the Near East into Europe, and the Mediterranean low diversity observed could have the result of gene flow from Northern Africa to Southern Europe as observed in human genetics [35, 54].
Spoligotyping and MIRU-VNTR data offer a useful insight into the diversity and transmission of M. bovis and a clear model for the establishment of M. bovis genetic structure in Africa. The current study is limited by the sampling with many locations across Africa without genotyping data. The analysis performed here offers a parsimonious model based on the available data extrapolated for the overall continent. This model can be further tested with more data. In the future, complete genomes of the pathogen across Africa can provide further details to the established scenario, and importantly, it can allow to test the depth of the diversity using a molecular clock and a refined phylogenetic structure.
Understanding the nature of the diversity is of great importance and could dictate the plan of action of public health agents. Strategies should be adjusted for either recently established diversity, possible due to economic trades with other countries, whose public health actions should focus on control of imports and early diagnosis of infected animals; while long–term established diversity can prove more difficult to tackle. In the case where pathogen diversity could have been established for thousands of years, as proposed here, strains could be strongly adapted to local environment and hosts, evolving through strong co-evolution with the domesticates, potentially with wild animals and, zoonotically, with humans.
Supporting information
The colours of the different points correspond to the network colour code in Fig 1. Map outline was adapted from https://commons.wikimedia.org/wiki/Atlas_of_the_world.
(PDF)
(PDF)
(PDF)
(PDF)
Acknowledgments
OI acknowledges Mara Scheffer and Maria Luiza Bazzo for help in spoligotyping techniques and all PGCD students. We thank Direcção Nacional de Veterinária—Mozambique for permits.
Data Availability
The genetic datasets analysed during the current study are available within the manuscripts referenced by us. All new data is available within the manuscript and its supplemental material.
Funding Statement
TR is supported by “PersonalizedNOS”, operation code NORTE-01-0145-FEDER-000013, cofinanced by FEDER through the North Portugal Regional Operational Programme (NORTE 2020). MCN and TR acknowledges project PTDC/SAU-PUB/29521/2017. PS and TR acknowledges PTDC/SOC-ANT/30316/2017. PS acknowledges CBMA strategic programme UID/BIA/04050/2019. OFI is supported by CAPES and IGC through PGCD. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Carruth L, Roess AA, Mekonnen YT, Melaku SK, Nichter M, Salman M. Zoonotic tuberculosis in Africa: challenges and ways forward. The Lancet. 2016;388(10059):2460–1. 10.1016/S0140-6736(16)32186-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.WHO, FAO, OIE. Roadmap for zoonotic tuberculosis. 2017;World Health Organization (WHO), Food and Agriculture Organization of the United Nations (FAO) and World Organisation for Animal Health (OIE)(Available online at: http://apps.who.int/iris/bitstream/handle/10665/259229/9789241513043-eng.pdf;jsessionid=128997632CBEDCBBD8782EC540C238C8?sequence=1).
- 3.Cosivi O, Grange JM, Daborn CJ, Raviglione MC, Fujikura T, Cousins D, et al. Zoonotic tuberculosis due to Mycobacterium bovis in developing countries. Emerging Infectious Diseases. 1998;4(1):59–70. PMC2627667. 10.3201/eid0401.980108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moiane I, Machado A, Santos N, Nhambir A, Inlamea O, Hattendorf J, et al. Prevalence of Bovine Tuberculosis and Risk Factor Assessment in Cattle in Rural Livestock Areas of Govuro District in the Southeast of Mozambique. PLOS ONE. 2014;9(3):e91527 10.1371/journal.pone.0091527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thoen C, LoBue P, de Kantor I. The importance of Mycobacterium bovis as a zoonosis. Veterinary Microbiology. 2006;112(2):339–45. 10.1016/j.vetmic.2005.11.047. [DOI] [PubMed] [Google Scholar]
- 6.Olea-Popelka F, Muwonge A, Perera A, Dean AS, Mumford E, Erlacher-Vindel E, et al. Zoonotic tuberculosis in human beings caused by Mycobacterium bovis-a call for action. The Lancet Infectious Diseases. 2017;17(1):e21–e5. 10.1016/S1473-3099(16)30139-6 [DOI] [PubMed] [Google Scholar]
- 7.Ayele WY, Neill SD, Zinsstag J, Weiss MG, Pavlik I. Bovine tuberculosis: an old disease but a new threat to Africa. The International Journal of Tuberculosis and Lung Disease. 2004;8(8):924–37. [PubMed] [Google Scholar]
- 8.Adesokan HK, Streicher EM, van Helden PD, Warren RM, Cadmus SIB. Genetic diversity of Mycobacterium tuberculosis complex strains isolated from livestock workers and cattle in Nigeria. PLOS ONE. 2019;14(2):e0211637 10.1371/journal.pone.0211637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gilbert M, Mitchell A, Bourn D, Mawdsley J, Clifton-Hadley R, Wint W. Cattle movements and bovine tuberculosis in Great Britain. Nature. 2005;435(7041):491–6. 10.1038/nature03548 [DOI] [PubMed] [Google Scholar]
- 10.Renwick AR, White PCL, Bengis RG. Bovine tuberculosis in southern African wildlife: a multi-species host-pathogen system. Epidemiol Infect. 2007;135(4):529–40. Epub 09/07. 10.1017/S0950268806007205 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Biruk WA. Prevalence of Bovine Cysticercosis at Jijiga Municipal Abattoir, Ethiopia. Journal of Veterinary Science & Technology. 2017;8(3):1–4. [Google Scholar]
- 12.Tanner M, Inlameia O, Michel A, Maxlhuza G, Pondja A, Fafetine J, et al. Bovine Tuberculosis and Brucellosis in Cattle and African Buffalo in the Limpopo National Park, Mozambique. Transboundary and Emerging Diseases. 2015;62(6):632–8. 10.1111/tbed.12210 [DOI] [PubMed] [Google Scholar]
- 13.Michel AL, Bengis RG, Keet DF, Hofmeyr M, Klerk LMd, Cross PC, et al. Wildlife tuberculosis in South African conservation areas: Implications and challenges. Veterinary Microbiology. 2006;112(2):91–100. 10.1016/j.vetmic.2005.11.035. [DOI] [PubMed] [Google Scholar]
- 14.WHO. Global tuberculosis report 2017. Geneva, Switzerland: 2018.
- 15.Wirth T, Hildebrand F, Allix-Béguec C, Wölbeling F, Kubica T, Kremer K, et al. Origin, Spread and Demography of the Mycobacterium tuberculosis Complex. PLOS Pathogens. 2008;4(9):e1000160 10.1371/journal.ppat.1000160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gifford-Gonzalez D, Hanotte O. Domesticating Animals in Africa: Implications of Genetic and Archaeological Findings. Journal of World Prehistory. 2011;24(1):1–23. 10.1007/s10963-010-9042-2 [DOI] [Google Scholar]
- 17.Enattah NS, Jensen TGK, Nielsen M, Lewinski R, Kuokkanen M, Rasinpera H, et al. Independent Introduction of Two Lactase-Persistence Alleles into Human Populations Reflects Different History of Adaptation to Milk Culture. The American Journal of Human Genetics. 2008;82(1):57–72. 10.1016/j.ajhg.2007.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ranciaro A, Campbell Michael C, Hirbo Jibril B, Ko W-Y, Froment A, Anagnostou P, et al. Genetic Origins of Lactase Persistence and the Spread of Pastoralism in Africa. The American Journal of Human Genetics. 2014;94(4):496–510. 10.1016/j.ajhg.2014.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tishkoff SA, Reed FA, Ranciaro A, Voight BF, Babbitt CC, Silverman JS, et al. Convergent adaptation of human lactase persistence in Africa and Europe. Nature Genetics. 2007;39:31–40. 10.1038/ng1946 https://www.nature.com/articles/ng1946#supplementary-information. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Soolingen D, Hermans PW, de Haas PE, Soll DR, van Embden JD. Occurrence and stability of insertion sequences in Mycobacterium tuberculosis complex strains: evaluation of an insertion sequence-dependent DNA polymorphism as a tool in the epidemiology of tuberculosis. Journal of Clinical Microbiology. 1991;29(11):2578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wilton S, Cousins D. Detection and identification of multiple mycobacterial pathogens by DNA amplification in a single tube. Genome Research. 1992;1(4):269–73. [DOI] [PubMed] [Google Scholar]
- 22.Warren RM, Gey van Pittius NC, Barnard M, Hesseling A, Engelke E, de Kock M, et al. Differentiation of Mycobacterium tuberculosis complex by PCR amplification of genomic regions of difference [Short Communication]. The International Journal of Tuberculosis and Lung Disease. 2006;10(7):818–22. [PubMed] [Google Scholar]
- 23.Machado A, Rito T, Ghebremichael S, Muhate N, Maxhuza G, Macuamule C, et al. Genetic diversity and potential routes of transmission of Mycobacterium bovis in Mozambique. PLOS Neglected Tropical Diseases. 2018;12(1):e0006147 10.1371/journal.pntd.0006147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.El-Sayed A, El-Shannat S, Kamel M, Castañeda-Vazquez MA, Castañeda-Vazquez H. Molecular Epidemiology of Mycobacterium bovis in Humans and Cattle. Zoonoses and Public Health. 2016;63(4):251–64. 10.1111/zph.12242 [DOI] [PubMed] [Google Scholar]
- 25.Marianelli C, Amato B, Boniotti MB, Vitale M, Pruiti Ciarello F, Pacciarini ML, et al. Genotype diversity and distribution of Mycobacterium bovis from livestock in a small, high-risk area in northeastern Sicily, Italy. PLOS Neglected Tropical Diseases. 2019;13(7):e0007546 10.1371/journal.pntd.0007546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weniger T, Krawczyk J, Supply P, Niemann S, Harmsen D. MIRU-VNTRplus: a web tool for polyphasic genotyping of Mycobacterium tuberculosis complex bacteria. Nucleic Acids Res. 2010;38(Web Server issue):W326–W31. Epub 2010/05/10. 10.1093/nar/gkq351 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bandelt HJ, Forster P, Sykes BC, Richards MB. Mitochondrial Portraits of Human Populations Using Median Networks. Genetics. 1995;141(2):743–53. PMC1206770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bandelt HJ, Forster P, Röhl A. Median-joining networks for inferring intraspecific phylogenies. Molecular Biology and Evolution. 1999;16(1):37–48. 10.1093/oxfordjournals.molbev.a026036 [DOI] [PubMed] [Google Scholar]
- 29.Rito T, Matos C, Carvalho C, Machado H, Rodrigues G, Oliveira O, et al. A complex scenario of tuberculosis transmission is revealed through genetic and epidemiological surveys in Porto. BMC Infectious Diseases. 2018;18(1):53 10.1186/s12879-018-2968-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oliveira O, Gaio R, Carvalho C, Correia-Neves M, Duarte R, Rito T. A nationwide study of multidrug-resistant tuberculosis in Portugal 2014–2017 using epidemiological and molecular clustering analyses. BMC Infectious Diseases. 2019;19(1):567 10.1186/s12879-019-4189-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rozas J, Ferrer-Mata A, Sánchez-DelBarrio JC, Guirao-Rico S, Librado P, Ramos-Onsins SE, et al. DnaSP 6: DNA Sequence Polymorphism Analysis of Large Data Sets. Molecular Biology and Evolution. 2017;34(12):3299–302. 10.1093/molbev/msx248 [DOI] [PubMed] [Google Scholar]
- 32.Macaulay V, Soares P, Richards MB. Rectifying long-standing misconceptions about the ρ statistic for molecular dating. PLoS One. 2019;14(2):e0212311 10.1371/journal.pone.0212311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Molecular Biology and Evolution. 2016;33(7):1870–4. 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ghebremariam MK, Hlokwe T, Rutten VPMG, Allepuz A, Cadmus S, Muwonge A, et al. Genetic profiling of Mycobacterium bovis strains from slaughtered cattle in Eritrea. PLOS Neglected Tropical Diseases. 2018;12(4):e0006406 10.1371/journal.pntd.0006406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sarno S, Boattini A, Carta M, Ferri G, Alù M, Yao DY, et al. An Ancient Mediterranean Melting Pot: Investigating the Uniparental Genetic Structure and Population History of Sicily and Southern Italy. PLOS ONE. 2014;9(4):e96074 10.1371/journal.pone.0096074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hershkovitz I, Donoghue HD, Minnikin DE, May H, Lee OYC, Feldman M, et al. Tuberculosis origin: The Neolithic scenario. Tuberculosis. 2015;95:S122–S6. 10.1016/j.tube.2015.02.021 [DOI] [PubMed] [Google Scholar]
- 37.Galagan JE. Genomic insights into tuberculosis. Nature Reviews Genetics. 2014;15:307 10.1038/nrg3664 https://www.nature.com/articles/nrg3664#supplementary-information. [DOI] [PubMed] [Google Scholar]
- 38.Comas I, Coscolla M, Luo T, Borrell S, Holt KE, Kato-Maeda M, et al. Out-of-Africa migration and Neolithic coexpansion of Mycobacterium tuberculosis with modern humans. Nature Genetics. 2013;45:1176 10.1038/ng.2744 https://www.nature.com/articles/ng.2744#supplementary-information. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Prendergast ME, Lipson M, Sawchuk EA, Olalde I, Ogola CA, Rohland N, et al. Ancient DNA reveals a multistep spread of the first herders into sub-Saharan Africa. Science. 2019;365(6448):eaaw6275 10.1126/science.aaw6275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gandini F, Achilli A, Pala M, Bodner M, Brandini S, Huber G, et al. Mapping human dispersals into the Horn of Africa from Arabian Ice Age refugia using mitogenomes. Scientific Reports. 2016;6:25472 10.1038/srep25472 https://www.nature.com/articles/srep25472#supplementary-information. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hollfelder N, Schlebusch CM, Günther T, Babiker H, Hassan HY, Jakobsson M. Northeast African genomic variation shaped by the continuity of indigenous groups and Eurasian migrations. PLOS Genetics. 2017;13(8):e1006976 10.1371/journal.pgen.1006976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Savino di L. The Emergence and Spread of Herding in Northern Africa In: Peter M, Paul JL, editors. The Oxford Handbook of African Archaeology Oxford: Oxford University Press; 2013. p. 523–36. [Google Scholar]
- 43.Dunne J, di Lernia S, Chłodnicki M, Kherbouche F, Evershed RP. Timing and pace of dairying inception and animal husbandry practices across Holocene North Africa. Quaternary International. 2018;471:147–59. 10.1016/j.quaint.2017.06.062. [DOI] [Google Scholar]
- 44.Triska P, Soares P, Patin E, Fernandes V, Cerny V, Pereira L. Extensive Admixture and Selective Pressure Across the Sahel Belt. Genome Biology and Evolution. 2015;7(12):3484–95. 10.1093/gbe/evv236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ogundiran A. Four Millennia of Cultural History in Nigeria (ca. 2000 B.C.–A.D. 1900): Archaeological Perspectives. Journal of World Prehistory. 2005;19(2):133–68. 10.1007/s10963-006-9003-y [DOI] [Google Scholar]
- 46.Mbida CM, Van Neer W, Doutrelepont H, Vrydaghs L. Evidence for Banana Cultivation and Animal Husbandry During the First Millennium bc in the Forest of Southern Cameroon. Journal of Archaeological Science. 2000;27(2):151–62. 10.1006/jasc.1999.0447. [DOI] [Google Scholar]
- 47.Linseele V. From the First Stock Keepers to Specialised Pastoralists in the West African Savannah In: Bollig M, Schnegg M, Wotzka H-P, editors. Pastoralism in Africa. Past, Present and Future. 1 ed: Berghahn Books; 2013. p. 145–70. [Google Scholar]
- 48.Watson DJ. Within savanna and forest: A review of the Late Stone Age Kintampo Tradition, Ghana. Azania: Archaeological Research in Africa; 2010;45(2):141–74. 10.1080/0067270X.2010.491361 [DOI] [Google Scholar]
- 49.Dueppen SA. Cattle in the West African savanna: evidence from 1st millennium CE Kirikongo, Burkina Faso. Journal of Archaeological Science. 2012;39(1):92–101.https://doi.org/10.1016/j.jas.2011.09.005. [Google Scholar]
- 50.Rito T, Richards MB, Fernandes V, Alshamali F, Cerny V, Pereira L, et al. The First Modern Human Dispersals across Africa. PLoS One. 2013;8(11):e80031 10.1371/journal.pone.0080031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rito T, Vieira D, Silva M, Conde-Sousa E, Pereira L, Mellars P, et al. A dispersal of Homo sapiens from southern to eastern Africa immediately preceded the out-of-Africa migration. Scientific Reports. 2019;9(1):4728 10.1038/s41598-019-41176-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Soares P, Rito T, Pereira L, Richards MB. A Genetic Perspective on African Prehistory In: Jones SC, Stewart BA, editors. Africa from MIS 6–2: Population Dynamics and Paleoenvironments. Dordrecht: Springer Netherlands; 2016. p. 383–405. [Google Scholar]
- 53.Lander F, Russell T. The archaeological evidence for the appearance of pastoralism and farming in southern Africa. PLoS One. 2018;13(6):e0198941 10.1371/journal.pone.0198941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hernández CL, Soares P, Dugoujon JM, Novelletto A, Rodríguez JN, Rito T, et al. Early Holocenic and Historic mtDNA African Signatures in the Iberian Peninsula: The Andalusian Region as a Paradigm. PLOS ONE. 2015;10(10):e0139784 10.1371/journal.pone.0139784 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The colours of the different points correspond to the network colour code in Fig 1. Map outline was adapted from https://commons.wikimedia.org/wiki/Atlas_of_the_world.
(PDF)
(PDF)
(PDF)
(PDF)
Data Availability Statement
The genetic datasets analysed during the current study are available within the manuscripts referenced by us. All new data is available within the manuscript and its supplemental material.