This study demonstrates that the NGRES quantitative polymerase chain reaction kit can be implemented in routine diagnostic testing to enable rapid prescription of oral ciprofloxacin.
Abstract
Background
Absence of rapid antimicrobial resistance testing of Neisseria gonorrhoeae (Ng) hinders personalized antibiotic treatment. To enable rapid ciprofloxacin prescription, a real-time polymerase chain reaction (PCR) for simultaneous detection of Ng and fluoroquinolone resistance–associated gyrA-S91F mutation was evaluated.
Methods
Analytical NGRES quantitative PCR kit (NYtor BV) performance was assessed on 50 Ng transcription-mediated amplification (TMA)–negative and 100 Ng TMA-positive samples. To assess clinical use, 200 samples were prospectively analyzed, in parallel to routine diagnostic tests. Also, 50 urine, 50 anal, 50 pharyngeal, and 50 vaginal Ng TMA-positive samples were retrospectively analyzed. To assess if patients carried strains with different ciprofloxacin sensitivity at different anatomical locations, 50 urine/anal or vaginal/anal sample pairs collected during a single visit were analyzed.
Results
The NGRES quantitative PCR kit showed 97% sensitivity and 100% specificity for Ng detection and 92% sensitivity and 99% specificity for gyrA-S91F detection. Relative to TMA results, 85% Ng detection sensitivity and 99% specificity were found. Regarding the 200 prospectively analyzed clinical samples, 13 were Ng positive, of which 10 were also tested for antibiotic susceptibility by culture. The kit showed concordance for GyrA-S91F detection in 9 of 10 samples. Ng was detected in 96% and 94% of vaginal and urine TMA-positive samples, in 84% of anal samples and only in 22% of pharyngeal samples. Discordant ciprofloxacin sensitivity was found for 2 of 26 characterized urine/anal sample pairs.
Conclusion
The NGRES quantitative polymerase chain reaction (qPCR) kit can be implemented in diagnostic testing for vaginal, urine, and anal Ng TMA-positive samples to enable rapid prescription of oral ciprofloxacin.
Gonorrhea, the sexually transmitted infection (STI) caused by Neisseria gonorrhoeae (Ng), is one of the most common bacterial STIs worldwide.1 N. gonorrhoeae infection is treated with antimicrobial therapy; however, antimicrobial-resistant Ng is a major global public health concern.2 Treatment recommendations are based on antimicrobial resistance surveillance data and change according to the prevalence of resistant Ng strains. Because of high genomic plasticity of Ng and the exposure to antibiotics without susceptibility testing before treatment, Ng antimicrobial resistance develops continuously.3 The World Health Organization regularly updates their guidelines for the treatment of Ng infection to reduce the spread of multidrug-resistant gonococcal strains. Guidelines of 2016 included the recommendation of single-dose intramuscular ceftriaxone and oral azithromycin as first-choice treatment.4 However, azithromycin- or ceftriaxone-resistant Ng strains have been reported all over the world.2,5–8 Because alternative antimicrobial treatment options are scarce, the current treatment needs to change toward a more personalized approach.9
In the past, fluoroquinolones such as ciprofloxacin were effectively used as treatment of Ng infection. Ciprofloxacin treatment had several advantages over other antimicrobials such as oral administration and relatively few adverse effects. However, fluoroquinolones were eliminated as first-choice treatment once the World Health Organization–defined threshold for the prevalence of fluoroquinolone resistant Ng strains was reached.10 Nevertheless, still large proportions of Ng strains remain susceptible to ciprofloxacin: 70% in the United States, more than 70% in Australia and 53% in Europe.11–13 In the Netherlands, 65% of all Ng strains collected in 2018 were ciprofloxacin sensitive.14 Because ciprofloxacin is highly efficacious in susceptible Ng infections of any anatomic site, the British Association for Sexual Health and HIV guidelines still suggest the use of ciprofloxacin when susceptibility is known before treatment.15 Rapid detection of susceptibility status of clinical Ng isolates could therefore reintroduce treatment with oral ciprofloxacin and provide tailor-made therapy.
The mechanism of ciprofloxacin resistance in Ng is well defined by the presence of mutations in serine codon 91 of the DNA gyrase subunit A (gyrA) gene.16,17 Previous studies have demonstrated the potency of targeting the gyrA gene by using a real-time PCR assay to assess ciprofloxacin susceptibility of clinical Ng isolates.18–20 For even faster analysis, multiplex real-time PCR assays were developed to simultaneously detect Ng and gyrA mutations.21,22 However, most of these assays are not easily available because of in-house development or because extensive validation for use in routine diagnostics has not been performed. In this study, we extensively assessed the performance of the NGRES qPCR kit, developed by the Dutch company NYtor BV, for simultaneous detection of Ng and the ciprofloxacin resistance–associated gyrA-S91F mutation for use in diagnostic routine. The NGRES qPCR kit enables rapid ciprofloxacin susceptibility testing and could allow for the prescription of oral ciprofloxacin based on the results of a single qPCR assay.
METHODS
Sample Selection
NGRES qPCR kit (NYtor BV, Nijmegen, the Netherlands) performance was assessed on clinical samples obtained from patients visiting the STI outpatient clinic of Amsterdam and on samples obtained from general practitioners who requested Ng testing. For samples of STI clinic visitors, routine diagnostic tests consisted of a transcription-mediated amplification (TMA) test on the Aptima Combo 2 CT/NG assay (Hologic, Marlborough, MA). N. gonorrhoeae culture was performed when gram-negative diplococci were present in a gram-stained smear or with material collected at a return visit when the TMA test showed an Ng-positive result. For Ng culture, the clinical sample was inoculated on a plate with BBL GC-Lect Agar (Becton, Dickinson and Company, Franklin Lakes, NJ). A pure Ng colony was picked for growth on a GC agar plate, enriched with 1% IsoVitaleX (BioTRADING Benelux BV, Mijdrecht, the Netherlands), and ciprofloxacin minimum inhibitory concentration (MIC) values were routinely determined with an e-test according to the manufacturer's instructions (bioMérieux SA, Marcy l'Étoile, France). According to the European Committee on Antimicrobial Susceptibility Testing, an MIC ≤0.03 μg/mL is considered as sensitive and an MIC ≥0.06 μg/mL as resistant to ciprofloxacin. Samples obtained from general practitioners were only tested with the TMA according to routine diagnostics because cultures could not be performed on samples from TMA collection tubes and additional culture material was unavailable.
Analytical performance of the NGRES qPCR kit was assessed with 100 Ng TMA-positive and 50 Ng TMA-negative samples from STI clinic visitors that were randomly selected and retrospectively analyzed. Samples were run at the Public Health Laboratory in Amsterdam on the Rotorgene (Qiagen, Holden, Germany) qPCR platform. The same samples were also run on the CFX96 (Bio-Rad, Hercules, CA) by NYtor BV, and results were compared. Successful cultures were performed with additional material from these 100 Ng-positive patients so MIC values were available.
To compare results from the NGRES qPCR kit and routine diagnostic tests, 100 urine and 100 anal samples were selected from male visitors of the STI clinic and prospectively analyzed with the kit in parallel to routine diagnostic tests and culture. Results from the Rotorgene qPCR platform were compared with TMA and culture-based results.
To assess NGRES qPCR kit performance on samples from different anatomical locations, 200 Ng TMA-positive samples (50 urinal, 50 anal, 50 vaginal, 50 pharyngeal) were retrospectively analyzed with the kit on the Rotorgene qPCR platform. All samples were obtained from general practitioners and collected in TMA collection tubes. Because additional culture material was unavailable, MIC values were unknown.
Lastly, we determined the fraction of patients that carried Ng strains with different ciprofloxacin sensitivity at different anatomical locations. From 41 male patients, urine and anal samples collected during a single visit were selected and paired as well as vaginal and anal samples from 9 female patients. All resulting 50 sample pairs were analyzed with the NGRES qPCR kit, and pairwise comparisons were made.
Sample Preparation
Previously collected samples had been stored in Aptima Combo 2 collection buffer (Hologic) at −20°C according to routine storage. Retrospectively analyzed samples were taken from storage, thawed, and vortexed. DNA was extracted from 200 μL of each sample by isopropanol precipitation after lysis with NucliSENS easyMAG Lysis buffer (bioMérieux SA) enriched with glycogen (40 μg/mL). The pellet was washed with 70% ethanol twice, dissolved in 50 μL Tris-HCl at pH 8.0, and stored at −20°C until use in amplification experiments. During each DNA extraction experiment, a negative control was included by adding 200 μL Aptima Combo 2 buffer to the lysis buffer.
NGRES qPCR Kit
The NGRES qPCR kit, developed by NYtor BV, contained a primer/probe mix designed for specific detection of the Ng adenylate kinase (adk) gene and ciprofloxacin resistance–associated gyrA single-nucleotide polymorphism S91F. During analytical validation, an additional mutation A92P in the gyrA probe binding region was detected, which prevented the original probe from binding. Subsequently, NYtor BV provided an adapted NGRES qPCR kit, which effectuated probe binding even in the presence of the gyrA-A92P mutation. The adapted kit was then used for further clinical validation. Positive signals were detected in the FAM (adk) and VIC (gyrA-S91F) channels. The corresponding internal control was detected in the Cy5 channel, and the positive control generated signals in both the FAM and VIC channels.
A qPCR mix was prepared by mixing 10 μL master mix solution and 5 μL primer/probe/internal control solution (provided in NGRES qPCR kit), and 5 μL DNA extract was added. Polymerase chain reactions were performed on the CFX96 and/or the Rotorgene with the following program: 1 cycle 3 minutes 95°C and 45 cycles 15 seconds 95°C and 60 seconds 60°C. Raw data were manually checked and analyzed with Bio-Rad CFX Manager software version 3.1 or Rotorgene software version 1.7.0.75, respectively. According to diagnostic routine guidelines of the Public Health Laboratory, detection of Ng and/or gyrA-S91F was confirmed in case of well-defined sigmoidal curves and Ct values <36 for both samples and controls. Samples that showed sigmoidal curves with Ct values >36 and <40 or badly shaped sigmoidal curves were repeated, and a Ct value <40 was then determined positive.
RESULTS
Concordance Between Results of CFX96 and Rotorgene qPCR Platforms
To assess the analytical performance of the NGRES qPCR kit, 100 Ng TMA-positive and 50 Ng TMA-negative samples were randomly selected for retrospective analysis. One Ng-positive sample could not be traced back and was excluded from analysis. For all 99 Ng TMA-positive samples, ciprofloxacin MICs were determined on routine cultures of additional material from the same patient. All 149 samples were analyzed on both the CFX96 and the Rotorgene qPCR platforms. One sample was inhibited on both qPCR platforms. The NGRES qPCR kit detected Ng in 96 (98%) of 98 and 94 (96%) of 98 Ng TMA-positive samples on the CFX96 and Rotorgene qPCR platform, respectively. Regarding the Ng TMA-negative samples, 50 (100%) of 50 were also negative on both platforms (Table 1).
TABLE 1.
Comparison of Results From Diagnostic Tests and the NGRES qPCR Kit
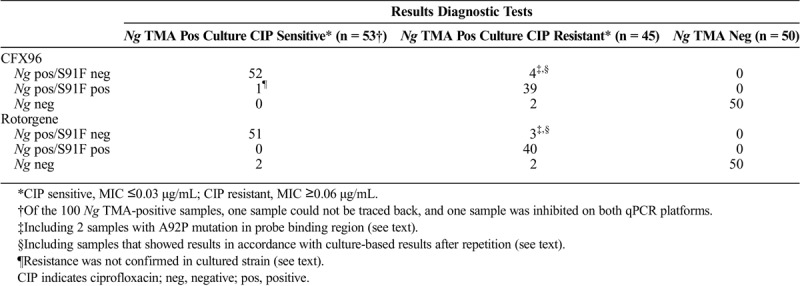
Ciprofloxacin sensitivity of the Ng strains in the samples was determined for all 100 Ng TMA-positive samples on routine cultures of additional material from the same patient. Of all clinical samples that contained a ciprofloxacin-sensitive Ng strain and that were Ng positive with the NGRES qPCR kit (CFX96: n = 53, Rotorgene: n = 51), 52 (98%) of 53 and 51 (100%) of 51 were also gyrA-S91F negative, so ciprofloxacin sensitive, on the CFX96 or Rotorgene qPCR platform. One Ng sample was gyrA-S91F positive on the CFX96 but negative on the Rotorgene (Table 1). Cultured urethra material of the same patient was subsequently tested as ciprofloxacin sensitive, indicating gyrA-S91F absence. Of all 43 clinical samples that contained a resistant Ng strain and were Ng positive with the NGRES qPCR kit, gyrA-S91F was detected in 39 (91%) of 43 and 40 (93%) of 43 samples on the CFX96 or Rotorgene qPCR platform, respectively. The gyrA-S91F mutation was thus not detected in 4 samples on the CFX96 and in the same 3 samples on the Rotorgene (Table 1). For the 4 samples that had shown discordant genotype results, DNA was again extracted from fresh cultures and analyzed by NYtor BV on the CFX96 platform. The gyrA-S91F mutation was detected in 2 of 4 samples after repetition, which was in accordance with the culture-based results. For the other 2 samples, the NGRES qPCR kit was still unable to detect the gyrA-S91F mutation, although these samples contained ciprofloxacin-resistant Ng strains with MIC values ≥0.06 μg/mL. The gyrA genes from the 4 discordant samples were also sequenced, and it seemed that the 2 samples that remained discordant after repetition carried an additional mutation, as described hereinafter.
GyrA-A92P Mutation Prevented Probe Binding
For 2 samples, no gyrA-S91F mutation was detected by using the NGRES qPCR kit on both qPCR platforms (Table 1), although the MIC values obtained from concomitant cultures were >32.0, which indicates ciprofloxacin resistance. These samples were collected from the anus and vagina of the same patient. The gyrA genes of these samples were partly sequenced, and an additional gyrA-A92P mutation was found, which prevented probe binding during the qPCR reaction (Fig. 1). NYtor BV provided an adapted NGRES qPCR kit, which effectuated probe binding even in the presence of a gyrA-A92P mutation. The adapted kit was then used for further clinical validation.
Figure 1.
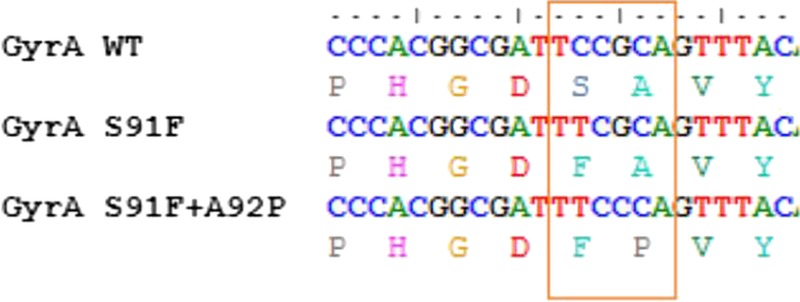
Sequences of the gyrA probe region without mutation (WT), with single S91F or with both S91F and A92P mutations. WT indicates wild type.
Comparison of Results From NGRES qPCR Kit and Routine Diagnostic Tests
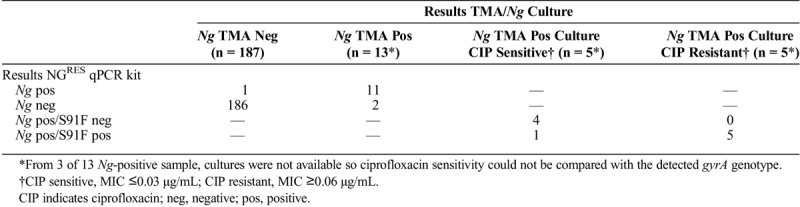
NGRES qPCR kit performance was compared with routine diagnostic test results with 100 urine and 100 anal samples. These were selected from male visitors of the STI clinic and prospectively analyzed with the kit in parallel to routine diagnostic tests, which included culture and MIC assessment. Regarding the total number of samples, 186 (99%) of 187 were negative and 11 (85%) of 13 were positive with the kit compared with TMA results (Table 2). For 3 of 13 Ng-positive samples, cultures were not available so the ciprofloxacin MIC values could not be compared with the gyrA genotype. For the other 10 samples, ciprofloxacin sensitivity was assessed on routine cultures of additional material from the same patient. The kit detected gyrA-S91F in 5 (100%) of 5 samples that contained a ciprofloxacin-resistant Ng strain, and no mutation was detected in 4 (80%) of 5 samples that contained a ciprofloxacin-sensitive Ng strain. For one sample, the kit detected the gyrA-S91F mutation, indicating ciprofloxacin resistance, although the Ng strain had been assessed as ciprofloxacin sensitive on concomitant culture (Table 2).
TABLE 2.
Comparison of Results From NGRES qPCR Kit and Routine Diagnostic Tests: TMA Test and Culture-Based Antibiotic Susceptibility Testing
High NGRES qPCR Kit Sensitivity in Urine and Vaginal Samples
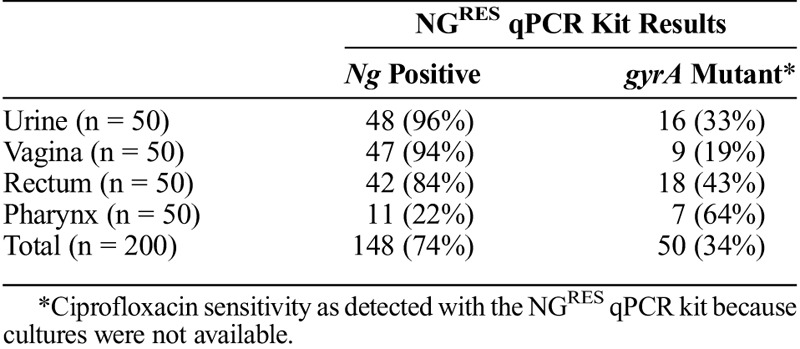
To assess NGRES qPCR kit performance on samples from various anatomical locations, 200 Ng TMA-positive samples were retrospectively analyzed with the kit. Samples were collected from urine (n = 50), vagina (n = 50), anus (n = 50), or pharynx (n = 50). Ng was detected in 48 (96%) of 50 urine samples, 47 (94%) of 50 vaginal samples, and 42 (84%) of 50 anal samples. Only 11 (22%) of 50 pharyngeal samples were Ng positive with the kit. The gyrA-S91F mutation was detected in 50 (34%) of 148 samples that were Ng positive (Table 3). Additional material from these patients was not available for culture so gyrA-S91F detection could not be validated.
TABLE 3.
NGRES qPCR Kit Analysis Results of 200 Ng TMA-Positive Samples Originating From Different Anatomical Locations
Patients Mostly Carried Similar Ng Strains at 2 Anatomical Locations
To determine the fraction of patients that carried Ng strains with discordant ciprofloxacin sensitivity at different anatomical locations, urine/anal or vaginal/anal samples that were collected during a single clinic visit were selected from 41 men and 9 women. All 50 sample pairs were analyzed with the NGRES qPCR kit. For 15 of 41 men, the kit could not detect Ng in the anal sample so these sample pairs could not be further characterized. For the other 26 men, 14 men (54%) carried gyrA-S91F mutant Ng strains and 10 men (38%) carried gyrA wild-type Ng strains in the urine and anus. In the other 2 men, strains with different gyrA types were found in the urine and anus: 1 man carried a gyrA-S91F mutant strain in the urine and a gyrA wild-type strain in the anus, and 1 man carried a gyrA-S91F mutant strain in the anus and a gyrA wild-type strain in the urine.
For 2 of 9 women, the kit could not detect Ng in the anal sample so these sample pairs could not be further characterized. The other 7 women carried gyrA wild-type Ng strains in both the vagina and the anus. Regarding ciprofloxacin susceptibility, 10 (38%) of 26 men carried only ciprofloxacin-sensitive Ng strains in contrast to 7 (100%) of 7 women.
DISCUSSION
We assessed the performance of the NGRES qPCR kit for simultaneous detection of Ng and the ciprofloxacin resistance–associated gyrA-S91F mutation. Use of this kit may allow for the prescription of oral ciprofloxacin based on the results of a single qPCR assay, which takes 4 hours including DNA extraction from the clinical sample. Implementation of the kit in diagnostic screening would increase the turnaround time with about half a day. However, this facilitates more rapid antibiotic sensitivity analysis compared with the current susceptibility testing based on bacterial cultures, which require at least 2 days. A detailed cost-benefit analysis for implementation is hard to perform because both costs and benefits, such as the use of an oral instead of an injectable antibiotic, are very much depending on the local situation.
The NGRES qPCR kit showed good performance compared with our current daily routine tests, which include the highly sensitive TMA test for Ng detection and culture for antimicrobial sensitivity testing. Only 5 samples showed discordant results between culture-based determination of ciprofloxacin sensitivity and detection of the gyrA-S91F mutation with the NGRES qPCR kit (Tables 1, 2). These samples either showed concordant results after repeated culture, DNA extraction, and analysis, or the discrepancy was caused by an additional gyrA-A92P mutation, which prevented probe binding in the initial, nonoptimized kit format. Assessing the type of errors in the discrepant results is important to determine its clinical impact. When typing is impossible, treatment recommendation remains ceftriaxone. False-positive gyrA-S91F detection would result in not using ciprofloxacin, which is not of clinical concern. However, false-negative gyrA-S91F detection would result in using ciprofloxacin to treat-resistant infections, which certainly is of clinical concern. In our study, only 2 samples were false negative for gyrA-S91F (Table 1). For these samples, the Ct value for Ng detection was high (>36), which indicates a low bacterial load. When using the NGRES qPCR kit in clinical practice, samples with such high Ct values should be repeated or assessed as nontypable. Because only 2 samples gave false-negative results, we are confident that the kit performance is sufficient and will increase if a certain cutoff Ct value is established by further validation in clinical practice.
SpeeDx recently developed the ResistancePlus assay for dual detection of Ng and gyrA-S91F mutation, which is already approved and commercially available.22 Ebeyan et al22 also reported 2 samples that gave discordant results when comparing culture and genotyping with this ResistancePlus assay. The results of one sample became concordant after repetition and one sample remained discrepant. A possible explanation for the discrepancies observed by Ebeyan et al and these in our study could be that patients are infected with multiple Ng strains at the same anatomical location, and that strains with discrepant ciprofloxacin sensitivity were taken for culturing and for genotyping with the NGRES qPCR kit. Such mixed infections have been reported to occur in high-risk populations, albeit at low prevalence.23,24 When implementing the kit in clinical practice, it should be further validated to what extent mixed infections influence the genotyping results.
When assessing the performance of the NGRES qPCR kit on samples from different anatomical locations, the NGRES qPCR kit showed excellent results for urine and vaginal samples. N. gonorrhoeae detection sensitivity was lower in anal samples, but for pharyngeal samples, the sensitivity was clearly insufficient, suggesting a low bacterial load in throat swabs as described previously.25,26 Compared with the previously published real-time PCR assay developed by Hemarajata et al,18 the NGRES qPCR kit demonstrated higher ability to characterize the genotype. Compared with the assay developed by Ellis et al,20 higher genotyping sensitivity was found for urine and rectal samples but lower sensitivity for pharyngeal samples. The ResistancePlus assay reported by Ebeyan et al22 showed better gyrA-S91F detection sensitivity and specificity, especially in pharyngeal samples. However, anal samples were missing in the validation of this assay, which seemed difficult to characterize in our study and could have an effect on the proportion of characterized samples. Also, initial Ng screening was not done with the Aptima Combo 2 but with other commercial assays, so direct comparison is not possible. Recently, Allan-Blitz et al27 showed that the ResistancePlus assay was able to genotype some of the samples that were nontypable in the method described by Ellis et al. Comparing the NGRES qPCR kit with the ResistancePlus assay would be valuable. Although the NGRES qPCR kit could not characterize a small proportion of the anal samples, by analyzing urine/anal and vaginal/anal sample pairs that were obtained from one patient during a single visit, we showed that only 2 of 33 patients carried Ng strains that differed in ciprofloxacin sensitivity, whereas the other 31 patients had strains with similar ciprofloxacin sensitivity. This indicated that analysis of only the urine or vaginal sample without the anal sample would be suitable for guided ciprofloxacin treatment for most cases.
When assessing the performance of the kit on samples from different anatomical locations, 148 of 200 samples could be characterized. In 66% of these samples, a gyrA wild-type Ng strain was detected, which is in accordance with the previously published fraction of 65% of ciprofloxacin-sensitive strains in the Netherlands.14 The presence of this significant fraction of ciprofloxacin-sensitive Ng strains shows that ciprofloxacin can still be used for treatment of Ng detection if rapid ciprofloxacin sensitivity detection is part of routine diagnostics. We suggest to implement the NGRES qPCR kit in addition to a highly sensitive assay. In our setting, we used the TMA test, which proved to be very suitable for first screening of Ng-suspected samples, followed by analysis of Ng TMA-positive samples with the NGRES qPCR kit. Based on the genotype determined by the NGRES qPCR kit, physicians could be advised if treatment with ciprofloxacin is indicated. Effectiveness of the assay should be carefully validated by comparing results with current diagnostic test results, and ciprofloxacin use could be monitored by a test-of-cure study.
Results of the analysis of sample pairs from 26 men and 7 women showed that 38% of the men and 100% of the women carried gyrA wild-type Ng strains. Although the number of men included in the study was much higher than the number of women, this result suggests that men are more often carrying resistant Ng strains. It has been reported that pharyngeal Ng infections are particularly important in transmission networks of men who have sex with men and that Neisseria species could exchange resistance determinants at the pharyngeal site.28 Because 80% of the STI clinic visitors are men who have sex with men, this could explain the high percentage of men carrying ciprofloxacin-resistant strains. However, further research on transmission networks of ciprofloxacin-resistant strains among general populations is needed.
In conclusion, our results show that the NGRES qPCR kit is suitable for concomitant detection of Ng and gyrA-S91F, especially in urine and vaginal clinical samples, to enable rapid prescription of ciprofloxacin.
Footnotes
Conflict of Interest and Sources of Funding: The authors declare that they have no competing interests.
The NGRES quantitative polymerase chain reaction kits used in this study were partly financed by NYtor BV. All other costs were financed by the Public Health Laboratory, GGD Amsterdam, the Netherlands (R&D 7572.2172).
REFERENCES
- 1.Rowley J, Vander Hoorn S, Korenromp E, et al. Chlamydia, gonorrhoea, trichomoniasis and syphilis: Global prevalence and incidence estimates, 2016. Bull World Health Organ 2019; 97:548–62P. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wi T, Lahra MM, Ndowa F, et al. Antimicrobial resistance in Neisseria gonorrhoeae: Global surveillance and a call for international collaborative action. PLoS Med 2017; 14:e1002344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buono SA, Watson TD, Borenstein LA, et al. Stemming the tide of drug-resistant Neisseria gonorrhoeae: The need for an individualized approach to treatment. J Antimicrob Chemother 2015; 70:374–381. [DOI] [PubMed] [Google Scholar]
- 4.World Health Organization. WHO guidelines for the treatment of Neisseria gonorrhoeae. Available at: http://www.ncbi.nlm.nih.gov/books/NBK379221/. Accessed August 5, 2019. [PubMed]
- 5.Shimuta K, Unemo M, Nakayama S, et al. Antimicrobial resistance and molecular typing of Neisseria gonorrhoeae isolates in Kyoto and Osaka, Japan, 2010 to 2012: Intensified surveillance after identification of the first strain (H041) with high-level ceftriaxone resistance. Antimicrob Agents Chemother 2013; 57:5225–5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ko KKK, Chio MTW, Goh SS, et al. First case of ceftriaxone-resistant multidrug-resistant Neisseria gonorrhoeae in Singapore. Antimicrob Agents Chemother 2019; 63 pii: e02624-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Unemo M, Golparian D, Nicholas R, et al. High-level cefixime- and ceftriaxone-resistant Neisseria gonorrhoeae in France: Novel penA mosaic allele in a successful international clone causes treatment failure. Antimicrob Agents Chemother 2012; 56:1273–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eyre DW, Town K, Street T, et al. Detection in the United Kingdom of the Neisseria gonorrhoeae FC428 clone, with ceftriaxone resistance and intermediate resistance to azithromycin, October to December 2018. Euro Surveill 2019; 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Trembizki E, Guy R, Donovan B, et al. , GRAND study investigators. Further evidence to support the individualised treatment of gonorrhoea with ciprofloxacin. Lancet Infect Dis 2016; 16:1005–1006. [DOI] [PubMed] [Google Scholar]
- 10.Centers for Disease Control and Prevention (CDC). Update to CDC's sexually transmitted diseases treatment guidelines, 2006: fluoroquinolones no longer recommended for treatment of gonococcal infections. MMWR Morb Mortal Wkly Rep 2007; 56:332–336. [PubMed] [Google Scholar]
- 11.Harris SR, Cole MJ, Spiteri G, et al. Public health surveillance of multidrug-resistant clones of Neisseria gonorrhoeae in Europe: A genomic survey. Lancet Infect Dis 2018; 18:758–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lahra MM, Enriquez R, George CRR. Australian gonococcal surveillance programme annual report, 2017. Commun Dis Intell (2018) 2019; 43. [PubMed] [Google Scholar]
- 13.Centers for Disease Control and Prevention (CDC). Sexually Transmitted Disease Surveillance 2018. Atlanta: U.S. Department of Health and Human Services, 2018. Available at: www.cdc.gov/std/stats18/gonorrhea.htm. Accessed December 16, 2019. [Google Scholar]
- 14.National Institute for Public Health and the Environment. Sexually transmitted infections in the Netherlands in 2018. National Institute for Public Health and the Environment. 2019:189 Report No: 2019-0007. Available at: wwwrivmnl/bibliotheek/rapporten/2019-0007pdf. Accessed August 5, 2019.
- 15.British Association for Sexual Health and HIV. BASHH guidelines. Available at: https://bashh.org/guidelines/. Accessed December 16, 2019.
- 16.Tanaka M, Otsuki M, Nishino T, et al. Mutation in DNA gyrase of norfloxacin-resistant clinical isolates of Neisseria gonorrhoeae. Sex Transm Infect 1996; 72:295–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Allan-Blitz LT, Wang X, Klausner JD. Wild-type gyrase A genotype of Neisseria gonorrhoeae predicts in vitro susceptibility to ciprofloxacin: A systematic review of the literature and meta-analysis. Sex Transm Dis 2017; 44:261–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hemarajata P, Yang S, Soge OO, et al. Performance and verification of a real-time PCR assay targeting the gyrA gene for prediction of ciprofloxacin resistance in Neisseria gonorrhoeae. J Clin Microbiol 2016; 54:805–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Siedner MJ, Pandori M, Castro L, et al. Real-time PCR assay for detection of quinolone-resistant Neisseria gonorrhoeae in urine samples. J Clin Microbiol 2007; 45:1250–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ellis O, Hemarajata P, Shahkolahi A, et al. A multisite implementation of a real-time polymerase chain reaction assay to predict ciprofloxacin susceptibility in Neisseria gonorrhoeae. Diagn Microbiol Infect Dis 2019; 94:213–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perera SR, Khan NH, Martin I, et al. Multiplex real-time PCR assay for simultaneous identification of Neisseria gonorrhoeae and its ciprofloxacin susceptibility status. J Clin Microbiol 2017; 55:3201–3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ebeyan S, Windsor M, Bordin A, et al. Evaluation of the ResistancePlus GC (beta) assay: A commercial diagnostic test for the direct detection of ciprofloxacin susceptibility or resistance in Neisseria gonorrhoeae. J Antimicrob Chemother 2019; 74:1820–1824. [DOI] [PubMed] [Google Scholar]
- 23.Goire N, Kundu R, Trembizki E, et al. Mixed gonococcal infections in a high-risk population, Sydney, Australia 2015: Implications for antimicrobial resistance surveillance? J Antimicrob Chemother 2017; 72:407–409. [DOI] [PubMed] [Google Scholar]
- 24.Martin IMC, Ison CA. Detection of mixed infection of Neisseria gonorrhoeae. Sex Transm Infect 2003; 79:56–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alexander S. The challenges of detecting gonorrhoea and chlamydia in rectal and pharyngeal sites: Could we, should we, be doing more? Sex Transm Infect 2009; 85:159–160. [DOI] [PubMed] [Google Scholar]
- 26.Hananta IPY, De Vries HJC, van Dam AP, et al. Persistence after treatment of pharyngeal gonococcal infections in patients of the STI clinic, Amsterdam, the Netherlands, 2012–2015: A retrospective cohort study. Sex Transm Infect 2017; 93:467–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Allan-Blitz LT, Ellis OL, Wee R, et al. Improved determination of Neisseria gonorrhoeae gyrase A genotype results in clinical specimens. J Antimicrob Chemother 2019; 74:2913–2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Whittles LK, White PJ, Paul J, et al. Epidemiological trends of antibiotic resistant gonorrhoea in the United Kingdom. Antibiotics (Basel) 2018; 7 pii: E60. [DOI] [PMC free article] [PubMed] [Google Scholar]


