Abstract
Aim
The loss of insulin-secreting β-cells, ultimately characterizing most diabetes forms, demands the development of cell replacement therapies. The common endpoint for all ex vivo strategies is transplantation into diabetic patients. However, the effects of hyperglycemia environment on the transplanted cells were not yet properly assessed. Thus, the main goal of this study was to characterize global effect of brief and prolonged in vivo hyperglycemia exposure on the cell fate acquisition and maintenance of transplanted human pancreatic progenitors.
Methods
To rigorously study the effect of hyperglycemia, in vitro differentiated human induced pluripotent stem cells (hiPSC)-derived pancreatic progenitors were xenotransplanted in normoglycemic and diabetic NSG RIP-DTR mice. The transplants were retrieved after one-week or one-month exposure to overt hyperglycemia and analyzed by large-scale microscopy or global proteomics. For this study we pioneer the use of the NSG RIP-DTR system in the transplantation of hiPSC, making use of its highly reproducible specific and absolute β-cell ablation property in the absence of inflammation or other organ toxicity.
Results
Here we show for the first time that besides the presence of an induced oxidative stress signature, the cell fate and proteome landscape response to hyperglycemia was different, involving largely different mechanisms, according to the period spent in the hyperglycemic environment. Surprisingly, brief hyperglycemia exposure increased the bihormonal cell number by impeding the activity of specific islet lineage determinants. Moreover it activated antioxidant and inflammation protection mechanisms signatures in the transplanted cells. In contrast, the prolonged exposure was characterized by decreased numbers of hormone+ cells, low/absent detoxification signature, augmented production of oxygen reactive species and increased apoptosis.
Conclusion
Hyperglycemia exposure induced distinct, period-dependent, negative effects on xenotransplanted human pancreatic progenitor, affecting their energy homeostasis, cell fate acquisition and survival.
Keywords: differentiation, endocrine progenitors, human iPSCs, hyperglycemia, RIP-DTR, xenotransplantation
Introduction
Diabetes mellitus is a group of metabolic diseases characterized by chronic increase in blood sugar levels (hyperglycemia) triggered by the inability of the body to produce or use sufficient insulin1. Insulin is produced by the β-cells, which reside in the pancreatic islets together with other cell types secreting hormones involved in glucose homeostasis: the α-cells (glucagon), δ-cells (somatostatin), γ-cells (pancreatic polypeptide) and ε-cells (ghrelin). Most forms of diabetes are ultimately characterized by the loss of functional insulin-secreting β-cells, thus a stable cure will require their replacement.
Human induced pluripotent stem cells (hiPSCs) can be differentiated towards insulin-producing cells by stepwise addition of factors mimicking pancreas development2–4. The outcome of most current protocols is a heterogeneous population comprising of cells expressing different islet hormones, including polyhormonal entities5–7. In vivo transplantation of these cells at the last stages of guided differentiation improves β-cell maturation2,8, normalizing the glycemia in diabetic mice9,10, 11–13.
Although the above examples prove that transplanted cells are able to differentiate and function in hyperglycemic conditions, it remains an open question whether hyperglycemia impedes or promotes the differentiation potential of the transplanted cells. To our knowledge, there are no rigorous studies addressing the in vivo impact of hyperglycemia exposure on the transplanted cells differentiation as compared to a normoglycemic environment. Addressing this problem has both fundamental science and clinical implications.
Results
NSG RIP-DTR as an ideal model system for the in vivo study of the hyperglycemia effect on human islet cell differentiation.
To investigate the in vivo impact of hyperglycemia on islet cells differentiation, we xenotransplanted (TX) alginate-encapsulated hiPSC-derived pancreatic progenitor cells (here forth S5-cells) in either normal or diabetic humanized NSG RIP-DTR mice (Figure 1A). For β-cell differentiation we used the protocol designed by Rezania et al.9 with slight modifications as previously described5,14. The encapsulated cells were exposed to either a brief (1 week, here on 1w-postTX) or an extended period of time (4 weeks, here on 4w-postTX) in the in vivo hyperglycemic environment. These time points were selected to allow both the examination of the initial changes in the cells’ regulatory landscape, known to occur rapidly in response to altered cell signaling or environmental cues15,16, and the assessment of the long-term effect of hyperglycemia on the cells.
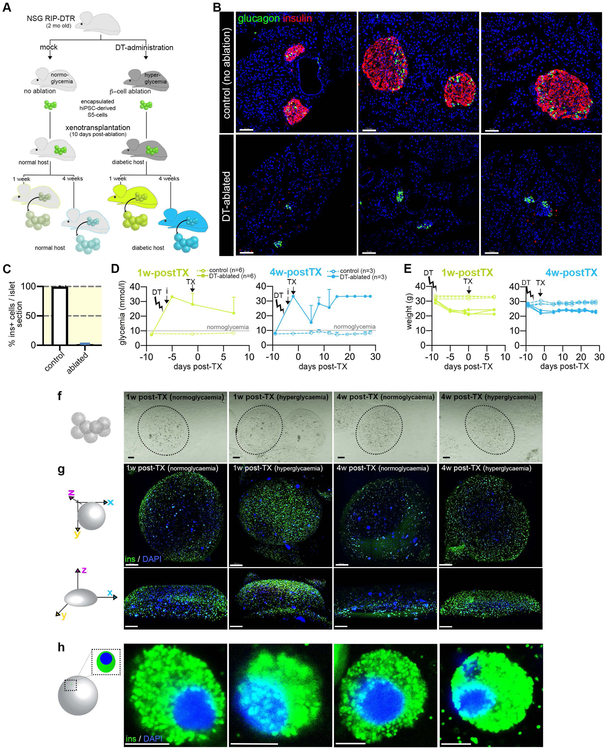
Figure 1. Transplantation setup.
(A) Experimental design (B) Confocal imaging of control and diphtheria toxin (DT)-ablated islet sections (insulin –red, glucagon –green, scale bar: 70μm), (C) Efficiency of DT-induced β-cell ablation in RIP-DTR NSG strain expressed as % insulin cells / islet section. (D) Glycemia and (E) Weight evolution in ablated and control mice (chartreuse – 1w-posTX, blue – 4w-postTX, TX – transplant, DT – DT-induced ablation), (F) Binocular imaging of the mounted alginate capsules and (G) 3D reconstructions of the immunofluorescence on whole alginate beads of the 4 conditions analyzed of the 4 conditions analyzed following the retrieval procedure (insulin – green, and DAPI – blue, scale bar: 200μm) (H) High magnification confocal imaging of insulin+ cells (scale bar: 5μm). Data are shown as mean ± SD.
To achieve highly reproducible hyperglycemia conditions, we employed the RIP-DTR genetic system, allowing the rapid and specific ablation of the insulin-producing β-cells in vivo, in the absence of autoimmunity or inflammation17,18. In these mice, the diphtheria toxin receptor (DTR) is specifically overexpressed on the surface of the β-cells, under the control of the rat insulin promoter (RIP). As previously described, upon diphtheria toxin (DT) administration, the β-cell compartment is specifically ablated with extremely high and reproducible efficiency (97.88% in NSG RIP-DTR, Figure 1B, C), similar to the one reported for SCID RIP-DTR (97.8%)19. Ablated animals developed hyperglycemia and displayed lower weight compared to their normoglycemic counterparts (Figure 1D, E). In contrast to the available chemical models of β-cell ablation, DT does not affect other organs or systems, due to the absent DTR expression in murine tissues, explaining the high animal survival rate following the transplant procedure (93.3%, 1/15).
Diabetic animals were xenotransplanted intraperitoneally with ~5 mil hiPSC-derived S5-cells (pancreatic progenitor stage), expressing typical markers of pancreatic progenitors (Supp. Figure 1A) obtained by guided in vitro differentiation, 10 days following DT-administration (Figure 1A, D). The cells were encapsulated in alginate20 just before the transplantation as alginate (i) allows easy sample recovery, (ii) protects against the immune system attack and (iii) promotes the islet cell differentiation potential probably through the mechanical forces elicited by encapsulation21.
Cell viability was over 94% (97.65%±0.35 for 1w-postTX and 95.5%±1.7 for 4w-postTX). Higher levels of human insulin were detected in the blood of the xenotransplanted hyperglycemic mice as compared to normoglycemic hosts, probably due to chronic high glucose stimulation (Supp. Figure 1B) and potentially contributing to the slight drop in glycemia values observed following transplant (Figure 1D). Capsules’ sizes remained unchanged in the beads recovered 1w- or 4w-postTX, regardless of the glycemic status (Figure 1 F, G). Following capsule retrieval, the encapsulated cells presented spheroidal morphology, typical of cells in suspension in all conditions analyzed (Figure 1H, Figure 2A).
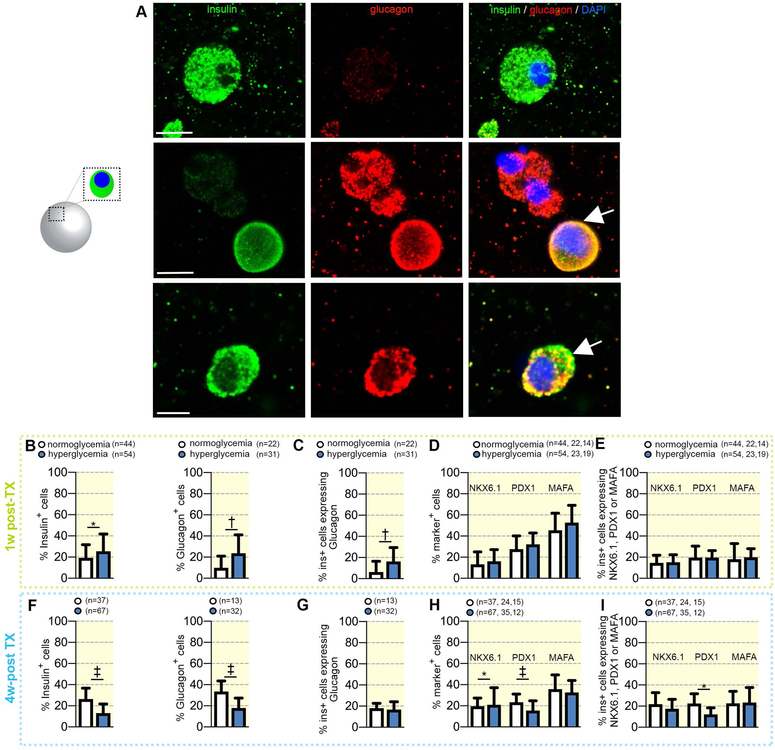
Figure 2. Comparison of hormone expression in encapsulated transplanted cells retrieved from either normoglycemic or diabetic mice.
(A) High magnification confocal imaging of representative insulin+ (green), glucagon+ (red) and bihormonal cells (arrows) cells (scale bar: 5μm). (B) Proportion of transplanted cells expressing insulin or glucagon in 1w_postTX conditions, (C) Fraction of insulin cells co-expressing glucagon (bihormonal cells) in 1w_postTX conditions, (D) Proportion of transplanted cells expressing NKX6.1, PDX1 or MAFA in 1w_postTX conditions. (e) Fraction of insulin+ cells co-expressing NKX6.1, PDX1 or MAFA in 1w-postTX conditions. (f) Proportion of transplanted cells expressing insulin or glucagon in 4w_postTX conditions, (g) Fraction of insulin cells co-expressing glucagon (bihormonal cells) and (h) Proportion of transplanted cells expressing NKX6.1, PDX1 or MAFA in 4w_postTX conditions,. (i) Fraction of insulin+ cells co-expressing NKX6.1, PDX1 or MAFA in 4w-postTX conditions. Data are shown as mean ± SD
Short in vivo exposure to hyperglycemia increases the number of bihormonal glucagon+insulin+ cells
To assess the impact of hyperglycemia on the differentiation outcome of the transplanted cells, we performed whole mount immunofluorescence staining on the retrieved capsules and quantified the numbers of hormone positive cells (Figure 2A). To allow a good coverage of the capsule volume, 9 fields of view (FoV) were acquired over 100 different focal planes, followed by 3D reconstruction and automatic quantification with manual supervision on Imaris 9.1.2.
Following the short in vivo exposure to hyperglycemia, a slight increase was observed in the percentage of ins+-cells (~6%, 19.23% to 25.47%) and, to a higher extent, glu+-cells (~13%, 9.84% to 23.69%) as compared to normoglycemic controls (Figure 2B). Nevertheless, most of these cells were bihormonal (10%, 6.31% to 16.3%), coexpressing both insulin and glucagon, suggestive of their immature or defective status (Figure 2A, Figure 2B). Moreover, there was no significant increase in the fraction of cells expressing PDX1, NKX6.1 or MAFA, three vital β-cell markers, neither in the total transplanted cell population (Figure 2D), nor in the insulin+ cell subpopulation (Figure 2E).
These data suggest that despite apparently incrementing the numbers of hormone+ cells, short exposure to hyperglycemia does not promote the desired differentiation outcome.
Prolonged hyperglycemia exposure hinders islet hormone expression
In contrast, the cells exposed to prolonged hyperglycemia exhibited a marked decline of both the proportion of ins+-cells (~−13%, 26.34% to 13.04%) and glu+-cells (~−15%, 33.57% to 17.99%) (Figure 2F), however displaying no significant impact on the bihormonal cell fraction (Figure 2G). In addition, the number of NKX6.1+ and PDX+ cells, but not MAFA, was also significantly decreased (~−4.5%, 21.9% to 17.46% and ~−10%, 22.44% to 11.96%, respectively) when compared to cells exposed for 4 weeks to a normoglycemic environment (Figure 2H). Consistently, the fraction of insulin cells coexpressing Pdx1+ declined by ~−8%, from 23.36% to 15.46% (Figure 2I).
These results indicate that differentiation or maintenance of islet cell phenotypes is hindered by long exposure to a hyperglycemic environment.
Global proteomics revealed changes in the energy metabolism and redox homeostasis following short hyperglycemia exposure
To thoroughly assess the hyperglycemia effect on the transplanted cells differentiation outcome, we compared by global proteomics (using tandem mass tag (TMT) 10-plex proteomics) the encapsulated cells retrieved from diabetic (D) and normoglycemic mice (N), at either 1 or 4 weeks following xenotransplantation (Figure 3A, 4A). Only the proteins detected in at least one sample of each compared condition (2183 for 1w-postTX) were further considered. About one fifth (22.35%) represented differentially expressed proteins (DEPs) between the cells xenotransplanted for 1 week into diabetic mice (1w-postTX_(D)) and the ones introduced into normoglycemic mice for an equivalent period of time (1w-postTX_(N)) (Figure 3A, Supp. Table 2). To identify the canonical pathways, upstream regulators and protein networks defining this landscape, we performed pathway analysis on this DEP set by using the Ingenuity Pathway Analysis (IPA) software.
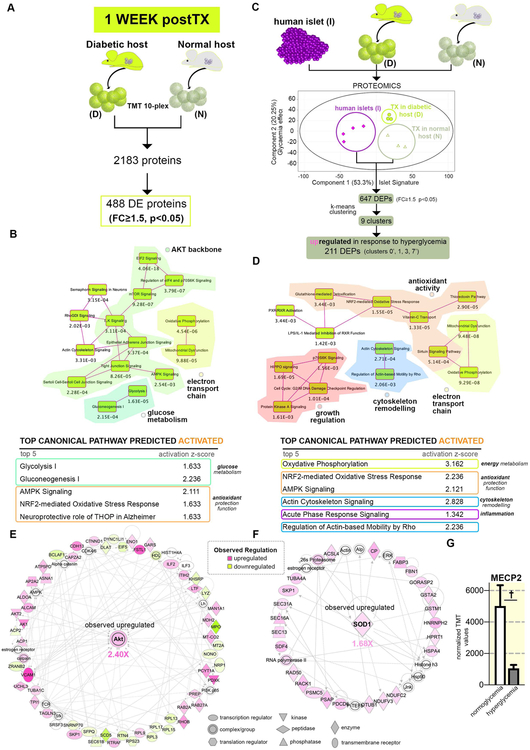
Figure 3. Pathway analysis of the proteome landscape following short-term hyperglycemia exposure.
(A) Experimental design of the comparison analyzed. (B) overlapping canonical pathways map and tables depicting the top canonical pathways predicted activated for the proteins differentially expressed between hyperglycemia and normoglycemia samples retrieved after 1 week. (C) Selected top radial network centered on Akt. (D) PCA plot of the conditions analyzed and analysis workflow depicting the k-means clustering and clusters selected in the upregulated proteins class. (E) Overlapping canonical pathways map and tables depicting the top canonical pathways predicted activated of the 1-week hyperglycemia-exposure upregulated protein class (F) Selected top radial network centered on SOD1. (G) Graph of the observed MECP2 downregulation in short-term hyperglycemia exposed samples. Data are shown as mean ± SD
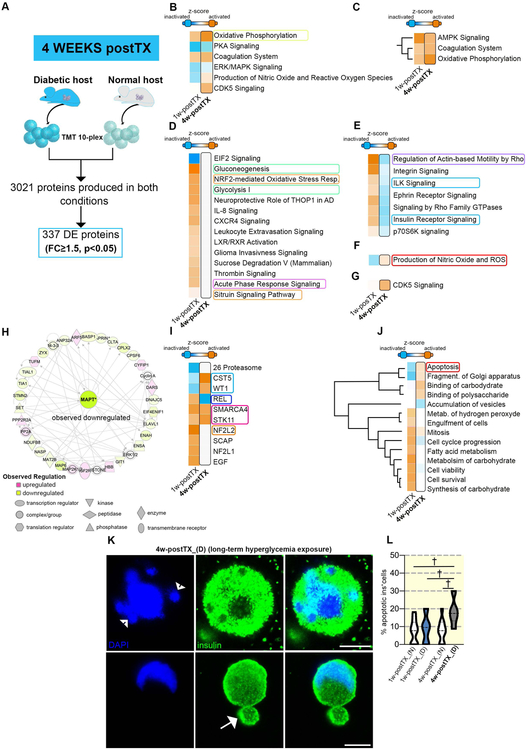
Figure 4. Comparative pathway analysis of the proteome landscapes following brief and long-term hyperglycemia exposure.
(A) Experimental design of the comparison analyzed. (B) Comparative analyses of the canonical pathways between hyperglycemia exposed and normoglycemia samples at 1w-postTX (left column) and 4w-postTX (right column, bordered) (C) the 3 pathways predicted activated in both conditions compared. (D) The canonical pathways predicted activated only in the samples exposed for short-term to hyperglycemia as compared to normoglycemic samples. (E) The canonical pathways exhibiting opposing activation patterns between the two conditions analyzed. (F) Production of Nitric Oxide and ROS pathway contrasting activation status is predicted activated in the 4w-postTX condition. (G) CDK5 Signaling predicted activation solely in the 4w-postTX condition. (H) Selected top IPA radial network centered on MAPT. (I) Comparative IPA activation predictions between the 1w-postTx and 4w-postTX upstream regulators. (J) IPA generated hierarchical clustering of the predicted disease and function processes for the conditions compared. Apoptosis and Fragmentation of Golgi apparatus exhibit different predicted activation pattern, with predicted activation in the 4w-postTX condition. (K) high magnification confocal images of apoptotic insulin+ cells in the long-term hyperglycemia exposed samples illustrating apoptosis-specific nuclear fragmentation (arrowheads) and membrane blebbing (arrows) (scale bar: 5μm). (L) Quantification of apoptotic insulin+ cells (n=9–11 bead confocal fields).
The analysis identified pathways involved in oxidative phosphorylation (OXPHOS) and glucose metabolism in the top canonical pathways defining the set (Figure 3B, yellow and dark-green islands, Supp. Figure 2A). Based on their components observed regulation these pathways were predicted as activated (Figure 3B table, Supp. Figure 2A), indicating an increased energy metabolism of the hyperglycemia-exposed cells, a regulation pattern suggestive of increased reactive oxygen species (ROS) production22. In response to ROS overproduction the cells usually deploy detoxifying mechanisms23. Several pathways with known antioxidant protection function, such as NRF2-mediated Oxidative Stress Response (Figure 3B, orange border), were found amongst the top 5 activated pathways. Moreover, NF2L2 (NRF2), the major key regulator of antioxidant gene expression regulation, protecting against oxidative stress24 and NUPR1, a transcription regulator involved in cell cycle control that confers resistance to microenvironment changes stress, were predicted as the leading two activated transcription factors accountable for the observed proteome landscape regulation (Supp. Figure 2B).
These observations are consistent with hyperglycemia exposure increasing the glycolysis and consequently the respiratory activity and ROS production of the transplanted differentiating cells, which respond by activating key detox mechanisms.
Furthermore, several top pathways shared as common denominator the AKT2 signaling cascade (Figure 3B, light-green island)(Supp. Figure 2A). In accordance with AKT2 observed upregulation, regulatory hubs like RICTOR and ADORA2A were predicted as top upstream regulators (Supp. Figure 2C). Interestingly, the AKT-based signaling was linked to the energy metabolism pathways by our network analysis (Figure 3C). A large body of literature involved AKT2 either in a ROS-stimulated positive feedback loop, promoting the glucose flux or in ROS signaling transduction, facilitating diverse ROS-mediated changes in gene expression25–28.
Last, sectors of the observed proteome landscape reflected the inactivation of chromatin binders involved in differentiation regulation, such as the epigenetic modifier DNMT3B (methylase) and the estrogen receptor alpha inhibitors SAFB2 and SAFB, involved in cell cycle/differentiation modulation in response to stress29 (Supp. Figure 2D). In contrast, SMARCA4, an ATP-dependent chromatin remodeler was predicted activated (Supp. Figure 2B). These data point at subtle epigenetic changes in gene expression as result of hyperglycemia exposure.
Altogether, these results suggest that the response of the transplanted differentiating cells to short-term hyperglycemia revolves around actively attempting to maintain the redox homeostasis. Nevertheless, the inferred changes in the epigenetic landscape regulation suggest the tilting of balance towards ROS accumulation.
Brief hyperglycemia exposure promotes proteins involved in redox balance
To investigate the impact of the hyperglycemia exposure on islet cells signature, we focused on proteins normally found in bonafide islet cells. For this we compared by TMT 10-plex-based quantitative proteomics the pancreatic progenitors transplanted for 1 week in diabetic/normoglycemic mice and human islets isolated from apparently healthy cadaveric donors. Both the Principal Component Analysis (PCA) plot (Figure 3D) and hierarchical clustering (Supp. Figure 2E) displayed a clear separation of the conditions analyzed. As expected, the transplanted cells conditions cluster close to each other and at distance from islets samples.
We then addressed the effect of hyperglycemia on the proteins differentially expressed between normoglycemia-exposed transplanted samples and human islets. Briefly, 647 DEPs (FC≥1.5, p<0.05, Supp. Table 3) were filtered between 1w-postTX_(N) and human islets samples, representing the difference in the proteome landscape between the standard (normoglycemia differentiating pancreatic progenitors) and the bonafide islets cells. These were k-means clustered based on their islet-normalized regulation pattern following brief hyperglycemia exposure (Figure 3D). Nine clusters were obtained, manually curated for errors and distributed into two classes according to their profile dynamic: (1) upregulated and (2) downregulated in response to hyperglycemia.
The upregulated class represented one third of the total DEP set (~33% 211/647). The pathway analysis revealed that the energy metabolism pathways identified above are transduced by proteins belonging in this group (Figure 3E, chartreuse islands, Supp. Figure 2F), with signaling cascades performing an antioxidant function found in the top canonical pathways (orange islands, Figure 3E, Supp. Figure 2F) and predicted activated (table Figure 3E). This was further corroborated by the network analysis that identified upregulated SOD1 (superoxide dismutase), one of the two key antioxidant enzymes involved in destroying reactive oxygen species in the body30,31, as a node of a leading network defining the analyzed proteome landscape (Figure 3F). Accordingly, NRF2 was predicted as the top activated upstream regulator (Supp. Figure 2G).
In addition, the program inferred that proteins of this group are also involved in inflammation and actin cytoskeleton remodeling, two processes linked to oxidative stress32–34 (Figure 3E, magenta, blue rectangles). Accordingly, NFkB, the lead molecule linking oxidative stress to inflammation35–38, was predicted as center of the lead network (Supp. Figure 2H), despite not being significantly regulated in the actual assay.
These results suggest that this class consists of proteins involved in redox balance regulation and inflammation in response to hyperglycemia.
Proteins downregulated in response to brief-hyperglycemia exposure are involved in protein synthesis and differentiation regulation
We subsequently focused on the proteins downregulated following the short-term exposure to hyperglycemia. The analysis of the resulted 129 DEPs revealed that most are involved in protein and amino acid metabolism (Supp. Figure 2I,J, Supp. Table 3). Moreover, JNK, NFkB and P38 MAPK, well-known ROS signaling transducers24,39–43, were central nodes of a lead protein network (Supp. Figure 2K), further corroborating the potential involvement of NfkB in signal transduction in response to brief hyperglycemia. Of note, the proteins used to infer the above-mentioned DNMT3b inactivation, resided in this class (Supp. Figure 2L). These results suggest that short hyperglycemia exposure decreases the abundance of proteins involved in the epigenetic regulation of the transcriptional landscape, amongst others.
Short-term hyperglycemia exposure impedes the activity of specific epigenetic modifiers with key role in β-cells identity maintenance
To test a possible connection between the predicted decrease in methylase activity and the increased generation of bihormonal glu+ins+ cells observed one week after in vivo hyperglycemia exposure (Figure 2A, C), we mined the dataset for epigenetic modifiers described to play a role in islet endocrine cell fate decisions44. We identified the significant downregulation of MECP2 (Figure 3G), an essential enzyme involved in maintaining and propagating the repression of α-cell fate in β-cells. This methyl-binding protein is recruited in β-cells to the methylated locus of the main α-cell lineage determinant gene Arx (aristaless-related homeobox), actively repressing its transcription and thus maintaining β-cell fate. ARX affects α-cell differentiation, but not directly glucagon gene transcription, nevertheless it has a critical role in suppressing the β-cell phenotype. Loss of MECP2 in β-cells was shown to result in a steep increase of Arx expression, coupled with the decrease of the main β-cell markers PDX1, PAX4 and Insulin, and the upregulation of key α-cell markers, such as Glucagon and MafB45.
Unfortunately, due to the unavoidable sensitivity limitation caused by performing proteomics on encapsulated cells, we were unable to detect proteins with very low abundance, such as ARX, hence we could not confirm its upregulation following MECP2 loss. Moreover, to our knowledge there is no study describing the ARX target genes in the pancreatic islet, hence their activation could not be assessed.
Thus, to partially validate in a different setup the role of increased oxidative stress on the loss of cell identity and Arx regulation, we analyzed Matrigel-cultured differentiated cells treated with WNT5a, a previously reported inducer of oxidative stress46. This in vitro setup grants a much better coverage of the proteome, allowing mining for ARX expression.
The global proteome analysis of the differentially expressed proteins between the treated and untreated cells confirmed an increase in the energy metabolism and antioxidant protection signatures (Supp. Figure 3A). Interestingly, the data mining of the set revealed the downregulation of MECP2 and the subsequent ARX upregulation (Supp. Figure 3B). Furthermore, there was a significant increase in the fraction of insulin+ cells also expressing glucagon+ (bihormonal cells) in this context as compared to untreated control (from 24.29% to 43.23%, Supp. Figure 3C, D).
Globally, the above series of analyses advance a model in which short-term in vivo exposure to hyperglycemia induces oxidative stress, caused by increased glucose flux, promoting increased OXPHOS and accumulation of ROS. Moreover, our data suggest that AKT2-mediated signaling pathways contribute to the oxidative stress response mechanism. The transplanted cells respond to the ROS overproduction by activating detox mechanisms. Nevertheless, these seem to be inefficient in maintaining the redox balance as processes like inflammation, cytoskeleton remodeling and changes in the epigenetic landscape regulation (such as cells with mixed identity) were detected, probably transduced via the NFkB hub.
Cells exposed in vivo to long-term hyperglycemia exhibit minimal antioxidant protection response to increased oxidative stress
When compared to brief hyperglycemia exposure, surprisingly fewer proteins (11.18%) were differentially expressed between the 4w-postTX_(D) and 4w-postTX_(N) samples. Moreover, only 54 DEPs were regulated by both hyperglycemia exposures periods, representing 11.27% and 16.27% of the respective DEP sets, suggesting that exposure length impacts differently the proteome landscape.
We further characterized the proteome changes in 4w-postTX_(D) as compared to their control counterparts (4w-postTX_(N)) (Supp. Table 2). The analysis revealed in the top toxicity list (i) the Mitochondrial Disorder and (ii) Increases Transmembrane Potential of Mitochondria and Mitochondrial Membrane. As for the short-term hypoglycemia exposure, the strong activation of OXPHOS pathway was predicted with a high activation score, suggesting it as a common response to hyperglycemia regardless of the exposure period (Figure 4B). Similar regulation patterns (activation) were also exhibited by AMPK signaling and Coagulation system pathway (Figure 4C).
Despite these similarities, the comparison identified substantial differences in the proteome fingerprint between the two contexts. A large group of pathways were predicted to display an absent or even an opposing pattern of regulation between the two hyperglycemia exposure periods. Specifically, glucose metabolism (such as Glycolysis I, Gluconeogenesis I) and Antioxidant protection signaling (such as NRF2-mediated Oxidative Stress Response or Sirtuin Signaling), that govern the response to short-term hyperglycemia were not predicted as regulated in this context (Figure 4D). Moreover, pathways like Insulin Receptor and ILK Signaling displayed opposing activation patterns, being predicted as inactivated in the long-term condition (Figure 4E). Importantly, the sole pathway exhibiting an inactive-to-active between brief and prolonged hyperglycemia exposure was Production of Nitric Oxide and Reactive Oxygen Species (Figure 4F), indicating changes in the proteome landscape caused by high generation of oxidants. In addition, CDK5 Signaling is one of only three entities to be predicted as activated exclusively as a result of extended hyperglycemia exposure (Figure 4G). CDK5 Signaling is known to be active in β cells, inhibiting insulin exocytosis and thus reducing insulin secretion47.
Furthermore, the network analysis identified downregulated MAPT (microtubule associate protein tau), as the main node of the Energy Production network of this proteome landscape (Figure 4H). Of relevance, this regulator was recently correlated with mitochondrial hyperpolarization and oxidative stress in hiPSC-derived neurons from patients bearing mutations in the Mapt gene48.
Several regulators shared similar activation profiles between the hyperglycemia periods, such as SMARCA4 and STK11 (Figure 4I). Nevertheless, as revealed also by the canonical pathways analysis, many were predicted to exhibit opposing regulations, thus disclosing radically different target proteome landscapes in response to the different hyperglycemia exposure periods. One of these, REL (mammalian NF-kB subunit), is a major player in regulating ROS amounts in the cell by increasing the expression of antioxidant proteins and protecting against oxidative damage. REL was predicted as activated following brief hyperglycemia exposure, while the analysis of the long-term effect revealed it as inactivated. Similarly, NF2L2 (NFR2), the key antioxidant protection regulator, confidently predicted as activated based on the one week transplanted samples signature, could not be inferred as regulated based on the four weeks proteome landscape. In contrast, WT1, levels were directly correlated with oxidative stress by previous research49 was predicted activated following long- but not short-term hyperglycemia exposure.
Moreover, the comparative diseases and functions analysis also confirmed a cohort of processes displaying opposite regulation. Active cell proliferation and cell survival on a background of inflammatory processes characterized the short-term hyperglycemia exposure landscape, whereas the long-term exposure seemed to be defined by low inflammation, cell cycle inhibition and increased apoptosis (Figure 4J). Indeed, a brief high resolution assessment of the retrieved encapsulated cells 4w-postTX_(D) hyperglycemic exposure revealed that at this stage significantly more insulin+ cells displayed typical signs of apoptosis, such as apoptotic blebbing and nuclei fragmentation, which were not present in 4w-postTX_(N) normoglycemic samples (Figure 4K,L).
Thus, based on the above series of analyses, a model can be proposed where prolonged in vivo exposure to hyperglycemia will overcome the antioxidative protection of the cells, leading to increased ROS accumulation. Excessive ROS will promote apoptotic cell death in susceptible cells.
Discussion
To the best of our knowledge, this is the first study reporting the in vivo impact of hyperglycemia on the islet cell fate acquisition and identity of xenotransplanted human pancreatic progenitors. Although hyperglycemia increased the oxidative stress signature regardless of exposure period, the cellular and proteome landscape response to the ROS overproduction was different, relying on largely distinct mechanisms according to the period spent in the hyperglycemic environment. Brief hyperglycemia exposure increased the bihormonal cell number and activated antioxidant protection mechanisms, while prolonged exposure was characterized by decreased hormone+ cells, low/absent detoxification signature, activation of oxidative stress pathways (Production of Nitric Oxide and ROS) and increased apoptosis.
We believe we are the first to report xenotransplantation of differentiating hiPSC-derived cells into the RIP-DTR NSG mouse model. By using this approach we eliminated many of the variables inherently occurring when employing classical chemical models of β-cells destruction, which are characterized by high individual variations in β-cell ablation efficiency, inefficient ablation or overall toxicity issues, such as renal failure.
Of note, this study was strictly focused on characterizing the global proteome landscape changes in response to in vivo hyperglycemia exposure, limited to the period immediately following the transplant. Consequently, it provides only a broad picture of the molecular events governing this process. Undoubtedly, the complex network of regulation uncovered needs further experimental dissection and validation in future studies.
Although a totally new finding in regard to transplanted hiPSC-derived pancreatic progenitor cells differentiating towards islet cell fates, the ability of hyperglycemia to drive ROS overproduction was previously described in other contexts42,50,51. These studies showed that hyperglycemia is increasing the glucose flux and TCA cycle (Krebs cycle), consequently raising the levels of electron transfer donors (NADH, FADH2) that, in turn, enhance OXPHOS and ROS production23,52,53. Our analysis was consistent with this scenario revealing a pattern of regulation reflecting the consistent activation of both Glycolysis and OXPHOS in the top activated pathways, regardless of the hyperglycemia exposure period. Moreover, the TCA cycle activation was further inferred in the prolonged-exposure samples.
Several studies proposed hyperglycemia-induced oxidative stress as a main driver for multiple diabetes complications54–57. Our results contribute to this list, by exposing the harmful effect of hyperglycemia exposure on islet cell fate acquisition. This is especially relevant in the context of future hiPSC-based therapies for diabetes, as it highlights the importance of patient glycemia control, especially during the initial period following the transplant. Of course, we cannot exclude that further matured islet cells might cope better with sustained hyperglycemia exposure, nevertheless in the light of increasing amount of studies linking hyperglycemia to oxidative stress in mature cell types, it is expected that they will also be affected. Interestingly, a previous study based on the transplantation of mouse bonafide pancreatic islets (mature islet cells) underneath the kidney capsule of streptozocin-induced hyperglycemic mice with or without insulin treatment also supports this hypothesis58. Despite employing a different experimental setup, the authors observed that transplanted islets exposed to hyperglycemia showed increased β-cell mass 10-days after transplant, followed by a decrease at 30-days due to β-cell apoptosis. These results are comparable with our observations, suggesting that finally matured cells are also negatively affected by long-term hyperglycemia. Nevertheless, it should be considered that parameters such as the difference in oxygen exposures between cells in suspension and bonafide islets as well as distinct transplantation sites are expected to influence the raw readouts between the studies. Of note, the authors also observed a steep decrease in the β-cell numbers 3-days following transplantation regardless of glycemia condition58, suggesting that the decrease in the islet cell numbers is a result of the transplant (necrosis) and not hyperglycemia.
In addition, we observed the downregulation of MECP2 methyl-binding protein, a critical repressor of α-cell fate in β-cells45. This provides a potential explanation for the increase in bihormonal cells expressing both glucagon and insulin. To our knowledge, this is the first study proposing a link connecting increased hyperglycemia induced oxidative stress and the loss in cell identity manifested by occurrence of bihormonal cell entities. If proven true by future studies, this will have important repercussions for the interpretation of the origin of bihormonal cells in human diabetic patients59,60.
Finally, we propose a working model in which the transplanted cells activate antioxidant protection mechanisms to cope with hyperglycemia-induced ROS production. However, sustained hyperglycemia exposure will in time exceed these mechanisms, causing ROS accumulation, leading to altered proteome signatures and impacting islet cell fate acquisition. When further prolonged, ROS overproduction will lead to increased cellular damage and cell death61–63.
Materials and methods
Cells and mice sources
The Norwegian Regional Committee of Medical and Health Research Ethics approved the reported experimental protocols used for hiPSCs (REK 2010/2295) and for human islets (REK 2011/426). All methods were carried out in accordance with the Helsinki Declaration. Informed consent was obtained from the healthy donor (skin fibroblasts) or from the relatives (organ donations). We used human induced pluripotent stem cells line previously generated via episomal reprogramming from skin fibroblasts collected from a healthy donor5. Human islets were obtained as previously described64, here from 8 donors, men and women between 41–63 years old (Supp. Table 1). Briefly, human pancreas were obtained from brain-dead donors, and digested by collagenase solution, and islets were separated by a Ficoll gradient. In this study, one of the human islet samples was excluded due to technical issues. We used the following transgenic mouse line NOD.Cg-PrkdcscidIl2rgtm1Wjl Tg(Ins2-HBEGF)6832Ugfm/Sz mice65, referred to as NSG RIP-DTR mice. All animal procedures were performed in accordance with the EU Directive 2010/63/EU for animal experiments. The breeding strategy and experimental protocols were approved by The Norwegian Animal Research Authority (FOTS ID 8329 and 8423).
In vitro differentiation
The hiPSCs line (passage >20) was characterized and confirmed to have normal karyotype and tested negative for mycoplasma on MycoAlert Mycoplasm Detection Kit (Lonza, LT07–418). The hiPSC line was enriched for SSEA4+ cells using magnetic beads (cat.#130097855 MACS Miltenyi Biotec) before in vitro differentiation. Two million hiPS cells per condition were differentiated following a seven-stage differentiation protocol9 in planar culture conditions on Matrigel-coated plates up to stage S5 (pancreatic endocrine precursors), which involves adding different mixtures of chemicals to mimic the stages of mouse pancreatic development for a duration of two weeks as described previously5.
Alginate encapsulation
S5 cells were collected using TrypLE Select Enzyme (cat.#12563011, Thermo Fisher), after viability check and counting on NucleoCounter NC-200, were resuspended in MCDB131 medium and mixed to a total concentration of 1.8 % alginate in 0.3 M mannitol. We used ultra-pure LVG (70 % G and 198 mPas) sodium alginate (batch #BP-0907–02, FMC BioPolymer AS NovaMatrix, Norway). Gel beads were created by using an electrostatic bead machine (Nisco Engineering AG, Switzerland) having a potential difference of 7 kV at a flow of 50 mL/h, with a standard nozzle with flat cut tip with an inner diameter of 0.5 mm. Alginate beads were incubated for less than 10 min in gelling solution (50mM CaCl2, 1mM BaCl2 in 0.15M mannitol, 10mM HEPES, pH 7.2)20, and rinsed three times in MCDB131 medium.
Transplantation
Males RIP-DTR NSG mice (8 to 12 weeks old) were separated into two groups, six mice per group. The mice were housed in individually ventilated cages (IVC) enriched with wooden bedding, nesting material, in a temperature-controlled environment at 22°C under a 12-h light-dark cycle. The mice were given ad libitum access to water and standard diet RM1A (SDS). First group received three intraperitoneally (ip) diphtheria toxin (Sigma, cat.# D0564–1MG) injections to induce diabetes as previously described18,19. Blood glucose was monitored twice a week at 4–5pm with a Contour XT glucometer (Bayer) to confirm diabetes (blood glucose >300 mg/dL on 2 consecutive measurements). Diabetic mice received an insulin implant (Linbit) 5 days after DT injections to ensure survival. The diabetic mice remained hyperglycemic after implant. Each diabetic and normoglycemic mouse received intraperitoneally alginate beads containing in total 5 million pancreatic progenitor cells differentiated in vitro. For transplantation, the mice were anesthetized using inhalable sevoflurane administered via Datex-Ohmeda Sevotec 5. Transplanted mice were given 0.5 mg/ml paracetamol in the drinking water for 5 days after transplantation. After 1 or 4 weeks, blood was collected from the tail and the mice were euthanized by cervical dislocation and the alginate beads were collected from the intra peritoneal cavity and prepared for either immunofluorescence or proteomics analyses. Three mice from each group were analyzed at each timepoint. The pancreas was also collected and processed for immunofluorescence.
Human insulin measurement
Blood from the mice were collected in Lithium Heparin tubes and centrifuged at 14 000 g for 10 min, and plasma were collected. Human insulin quantities were measured using the Mercodia Ultrasensitive Insulin ELISA (10-1132-01, Mercodia).
Immunofluorescence staining
Alginate beads were fixed in 4% paraformaldehyde for 1 hour at room temperature and stored in PBS at 4 °C until use. The NSG RIPDTR pancreas was fixed in 4% PFA for 2 hours at RT, then dehydrated in a sucrose gradient of 10, 20, 30% sucrose and embedded in Tissue Tek OCT compound (Sakura JP). 10 μm sections were obtained by using a cryotome (Leica CM 1950, Leica, DE) and added on SuperFrost® Plus slides (Thermo Scientific). The immunofluorescence staining was performed conformed indications provided by the supplier. The following primary antibodies were used: mouse anti-insulin IgG1 (1/500, I2018, Sigma-Aldrich), guinea-pig anti-porcine insulin (1/400, A056401–2, Dako), mouse anti-porcine glucagon (1/1000, G2654, Sigma-Aldrich), guinea-pig anti-PDX1 (1/500, ab47308, Abcam), rabbit anti-NKX6.1 (1/100, NBP1–82553, Novus), rabbit anti-MAFA (1/200, ab98859, Abcam), and rabbit anti-NEUROD1 (1/200, ab16508, Abcam). The following secondary antibodies were used at dilution 1/500: goat anti-mouse IgG1 A488, goat anti-guinea-pig A488, goat anti-mouse IgG1 A647, goat anti-guinea-pig A546, donkey anti-rabbit A647, and donkey anti-rabbit A647 (Molecular Probes). DAPI (1/1000, D1306, Molecular Probes) was used to stain the nuclei. The samples were mounted in Prolong Diamond Antifade Mountant Media (P36970, Life technologies). Image acquisition was performed on Andor Dragonfly confocal microscope. Antibody validations: endocrine markers were tested on different tissues, for example mouse pancreas cryosections, transplanted and in vitro hiPS-derived differentiating hormone expressing cells. A known positive slide of pancreatic tissue was used for staining validation.
Confocal Imaging
Whole mount beads were imaged using the Andor Dragonfly 5050 (Andor Technologies, Inc) confocal microscope with 20x dry objective (CFI Plan Apochromat Lambda 20x). Each bead was imaged with 3×3 fields of view, which covered the entire bead. The z-stacks were acquired from the top of each bead, and 100 steps of 4 μm with a total of 400 μm depth, which corresponded to the imaging depth. Each image was taken with high-speed iXon 888 Life EMCCD camera with 1024×1024 resolution. For nuclear imaging we used 405 nm laser with intensity of 30–50 % and an exposure time of 40–100 ms. For detecting proteins, laser 488, 546, and 647 were used with laser intensity ranging from 5–30 % and exposure time of 30–50, depending on the antibody and sample. High magnification images were acquired on Leica TCS SP5 confocal (Leica Microsystems CMS GmbH).
Automated cell counting
Imaris 9.1.2. (Bitplane AG) was used to analyze the immunofluorescence pictures. For counting each bead was considered one unit. A surface mask was used on the DAPI signal, with filters on absolute intensity from 600 or 1200 to max, quality minimum between 30 and 100, and size between 100 μm and 10 000 μm with separation of nuclei in clusters of 8 μm in diameter. For the different proteins spot masking was used with quality minimum ranging from 30 to 1500 depending on antibody and intensity of staining. The same settings were used for all beads of same staining from the same mouse. The MatLab plugin “Find spots close to surface” within 1 μm was used to analyze only spots belonging to a nucleus, which removed the unspecific staining from the analysis. The MatLab plugin “Find colocalizing spots” within 1 μm was used to identify the cells co-expressing two proteins.
Global proteomics analysis
Encapsulated cells were lysed directly in the alginate beads, in a buffer containing 8M Urea, 200 mM EPPS pH8.5 and protease inhibitors (Roche complete with EDTA) and sonicated (30 seconds × 3 times at 30% power). Human islets were processed as previously described5,14, is short by boiling in 4 % SDS buffer before sonication Chloroform-Methanol precipitation was performed on alginate beads as previously described66, involving adding methanol, chloroform and water before centrifugation thereafter the alginate were removed as best as possible. All peptides were desalted and dried. Protein processing and Tandem Mass Tag (TMT) 11-plex labeling was performed as described previously5,14 apart from halved volume of TMT reagents. In short, the desalted peptides were re-suspended in 200 mM HEPES before addition of CAN and TMT reagents for 1,5 h before quenching of the reaction by 5 % hydroxylamine. TMT labeled samples were combined, concentrated to near dryness and desalted with C18 solid phase extraction. We fractionated the pooled TMT-labelled peptide sample using the Pierce High pH Reversed-Phase Peptide Fractionation Kit (cat. # 84868). Twelve fractions were collected using: 7.5%, 10%, 12.5%, 15%, 17.5%, 20%, 22.5%, 25%, 27.5%, 30%, 35%, and 60% acetonitrile. Every sixth fraction was combined to yield a total of 6 fractions. Samples were subsequently acidified with 1% formic acid and vacuum centrifuged to near dryness. Each fraction was desalted via StageTip, dried again via vacuum centrifugation, and reconstituted for LC-MS/MS processing.
LC-MS3 analysis
From each of the 6 fractions, ~5 μg was dissolved in 15% aqueous formic acid/5% acetonitrile prior to LC-MS/MS analysis on an Orbitrap Fusion Lumos mass spectrometer (Thermo Fisher Scientific, San Jose, CA) coupled to a Proxeon EASY-nLC 1200 liquid chromatography (LC) pump (Thermo Fisher Scientific). Peptides were fractionated on a 100-μm inner diameter microcapillary column packed with ~35 cm of Accucore resin (2.6 μm, 150 Å, ThermoFisher Scientific). For each analysis, we loaded ~1 μg onto the column. Subsequent separation and acquisition were performed as previously described5,14. Samples were analyzed in duplicate, one with advanced peak determination (ADP) activated and a second run with this option off. Both analyses used the real-time search (RTS) algorithm67. Data was searched against the UniProt human database (downloaded: October, 2016).
Data analysis
The mass spectrometry data were analyzed as previously described5,14. Briefly, the mass spectra were processed using a Sequest-based in-house software pipeline66, database searching from the human Uniprot database, linear discriminant analysis for peptide-spectrum matches, before summing reporter ion counts to quantify proteins. Protein profiles from each mouse or human islet were treated as one unit. Protein quantitation values were exported for further analysis in Microsoft Excel and GraphPad Prism (version 8). The dataset was uploaded to ProteomeXchange via the PRIDE (http://www.proteomexchange.org) partner repository with the dataset identifier PXD015071.
The hierarchical clustering was performed with GeneSpring 14.9.1 GX software (Agilent), with clustering on both entities and conditions by using Squared Euclidian distance metric and Ward’s linkage rule. The same program was used to generate 9 and 12 k-mean clusters for the 1W and 4W time points respectively. Clusters 0 and for 1W and 0, 5, and 8 for 4W were manually curated. The pathway analyses were generated by QIAGEN’s Ingenuity Pathway Analysis program (IPA®, QIAGEN Redwood City, www.qiagen.com/ingenuity)68 as previously described5,14, here using 35 molecules/network; 25 networks/analysis for generating the interaction networks.
RNA extraction and Sequencing
RNA was purified from stage 5 cells (S5-cells) and human islets using the Qiagen RNeasy Micro Kit (ID74004, Qiagen, DE). The amount and quality of the purified RNA was measured using a NanodropOne (Thermo Scientific, US), before the samples were frozen and shipped to Qiagen Next-Generation Sequencing Center (Hilden, Germany) for sample sequencing and analysis. Second quality control, library preparation, sequencing, mapping and analysis up to normalized counts files were done at Qiagen Genomic Facility. We mined the normalized (fpkm) datasets for Neurod1 and Neurog3 expression levels; the full datasets were deposited to the NCBI Gene Expression Omnibus repository, accession number GSE141309.
Wnt5a treatments
In vitro differentiated cells were stimulated with Wnt5A (645-WN, R&D Systems) (400 ng/mL) for 4 h and further cultured for 48 h in differentiation media before sample collection. Wnt5A-stimulated cells and non-treated controls were washed in DPBS 1x and incubated with TrypLE™ Select Enzyme (1X) (cat.#12563011, Thermo Fisher Scientific) for 5 min at 37°C, followed by centrifugation (250g for 5 min). Cell pellets were processed for proteomics as described above. The raw mass spectrometry proteomics data containing these samples were deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD012081.
Statistical analysis
The group assignment of the host mice was randomized. Due to the difference in glycemia the groups could not be blinded during the collection of the blood glucose or weight measurements. Data are expressed as mean ± SD unless otherwise specified. Statistical analysis on the proteomics data was tested using unpaired two-tailed Student’s t-test and a p-value of ≤0.05 was considered significant. Two-way ANOVA with Tukey’s multiple comparisons test were used to compare between the groups for number of cells positive for the different markers by IF staining. This analysis was performed using GraphPad Prism v8.1.2.
Supplementary Material
Acknowledgements
We thank A. Altankhuyag and E. Stordal for animal care, A. H. Knudsen for performing the ELISA, and H.A. Dale for Imaris assistance. The confocal imaging was performed at the Molecular Imaging Center (MIC), Department of Biomedicine, University of Bergen. The NSG RIP-DTR mice were kindly provided by Pedro Herrera. Human islets were provided through the Islet Distribution Program at University of Oslo to H.S. The authors are grateful to the human islet isolation team in Oslo, Norway; to the Gygi Lab and Taplin Facility at Harvard Medical School, particularly Dr. Steven P. Gygi for the use of his mass spectrometers. This work was supported by funds from the Research Council of Norway (NFR 247577 and 251041) and Novo Nordic Foundation (NNF15OC0015054) to S.C. J.A.P. is funded by NIH/NIDDK grant DK098285; H.R. is supported by Bergen Forskningsstiftelse (BFS2014REK02) and the Western Norway Regional Health Authority (Bergen Stem Cell Consortium); L.G. is supported by Diabetesforbundet.
The funding sources had no role in the study design, its execution, analyzes, interpretation of the data, nor the decision to publish these results.
Footnotes
Conflict of Interest
The authors declare that they have no competing financial interests.
References
- 1.Aye MM, Atkin SL. Patient safety and minimizing risk with insulin administration - role of insulin degludec. Drug, healthcare and patient safety. 2014;6:55–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rezania A, Bruin JE, Riedel MJ, et al. Maturation of human embryonic stem cell-derived pancreatic progenitors into functional islets capable of treating pre-existing diabetes in mice. Diabetes. 2012;61(8):2016–2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang D, Jiang W, Liu M, et al. Highly efficient differentiation of human ES cells and iPS cells into mature pancreatic insulin-producing cells. Cell research. 2009;19(4):429–438. [DOI] [PubMed] [Google Scholar]
- 4.Pagliuca FW, Millman JR, Gurtler M, et al. Generation of functional human pancreatic beta cells in vitro. Cell. 2014;159(2):428–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vethe H, Bjørlykke Y, Ghila LM, et al. Probing the missing mature β-cell proteomic landscape in differentiating patient iPSC-derived cells. Scientific Reports. 2017;7(1):4780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bruin JE, Erener S, Vela J, et al. Characterization of polyhormonal insulin-producing cells derived in vitro from human embryonic stem cells. Stem Cell Research. 2014;12(1):194–208. [DOI] [PubMed] [Google Scholar]
- 7.Petersen MBK, Azad A, Ingvorsen C, et al. Single-Cell Gene Expression Analysis of a Human ESC Model of Pancreatic Endocrine Development Reveals Different Paths to beta-Cell Differentiation. Stem Cell Reports. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kroon E, Martinson LA, Kadoya K, et al. Pancreatic endoderm derived from human embryonic stem cells generates glucose-responsive insulin-secreting cells in vivo. Nature biotechnology. 2008;26:443. [DOI] [PubMed] [Google Scholar]
- 9.Rezania A, Bruin JE, Arora P, et al. Reversal of diabetes with insulin-producing cells derived in vitro from human pluripotent stem cells. Nat Biotechnol. 2014;32(11):1121–1133. [DOI] [PubMed] [Google Scholar]
- 10.Pagliuca FW, Millman JR, Gürtler M, et al. Generation of functional human pancreatic β cells in vitro. Cell. 2014;159(2):428–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Agulnick AD, Ambruzs DM, Moorman MA, et al. Insulin-Producing Endocrine Cells Differentiated In Vitro From Human Embryonic Stem Cells Function in Macroencapsulation Devices In Vivo. Stem Cells Transl Med. 2015;4(10):1214–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bochenek MA, Veiseh O, Vegas AJ, et al. Alginate encapsulation as long-term immune protection of allogeneic pancreatic islet cells transplanted into the omental bursa of macaques. Nat Biomed Eng. 2018;2(11):810–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vegas AJ, Veiseh O, Gurtler M, et al. Long-term glycemic control using polymer-encapsulated human stem cell-derived beta cells in immune-competent mice. Nat Med. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vethe H, Ghila L, Berle M, et al. The Effect of Wnt Pathway Modulators on Human iPSC-Derived Pancreatic Beta Cell Maturation. Frontiers in Endocrinology. 2019;10(293). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.López-Maury L, Marguerat S, Bähler J. Tuning gene expression to changing environments: from rapid responses to evolutionary adaptation. Nature Reviews Genetics. 2008;9:583. [DOI] [PubMed] [Google Scholar]
- 16.Hah N, Danko CG, Core L, et al. A rapid, extensive, and transient transcriptional response to estrogen signaling in breast cancer cells. Cell. 2011;145(4):622–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chera S, Baronnier D, Ghila L, et al. Diabetes recovery by age-dependent conversion of pancreatic δ-cells into insulin producers. Nature. 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thorel F, Nepote V, Avril I, et al. Conversion of adult pancreatic alpha-cells to beta-cells after extreme beta-cell loss. Nature. 2010;464(7292):1149–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cigliola V, Ghila L, Thorel F, et al. Pancreatic islet-autonomous insulin and smoothened-mediated signalling modulate identity changes of glucagon+ α-cells. Nature Cell Biology. 2018;20(11):1267–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Formo K, Cho CH, Vallier L, Strand BL. Culture of hESC-derived pancreatic progenitors in alginate-based scaffolds. J Biomed Mater Res A. 2015;103(12):3717–3726. [DOI] [PubMed] [Google Scholar]
- 21.Vethe H, Legøy TA, Abadpour S, et al. Encapsulation boosts islet-cell signature in differentiating human induced pluripotent stem cells via integrin signalling. bioRxiv. 2019:791442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ott M, Gogvadze V, Orrenius S, Zhivotovsky B. Mitochondria, oxidative stress and cell death. Apoptosis. 2007;12(5):913–922. [DOI] [PubMed] [Google Scholar]
- 23.Liemburg-Apers DC, Willems PH, Koopman WJ, Grefte S. Interactions between mitochondrial reactive oxygen species and cellular glucose metabolism. Arch Toxicol. 2015;89(8):1209–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schieber M, Chandel NS. ROS function in redox signaling and oxidative stress. Curr Biol. 2014;24(10):R453–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Niwa K, Inanami O, Yamamori T, Ohta T, Hamasu T, Kuwabara M. Redox regulation of PI3K/Akt and p53 in bovine aortic endothelial cells exposed to hydrogen peroxide. Antioxid Redox Signal. 2003;5(6):713–722. [DOI] [PubMed] [Google Scholar]
- 26.Zhang J, Wang X, Vikash V, et al. ROS and ROS-Mediated Cellular Signaling. Oxid Med Cell Longev. 2016;2016:4350965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koundouros N, Poulogiannis G. Phosphoinositide 3-Kinase/Akt Signaling and Redox Metabolism in Cancer. Front Oncol. 2018;8:160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Afanas’ev I Mechanisms of superoxide signaling in epigenetic processes: relation to aging and cancer. Aging Dis. 2015;6(3):216–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Townson SM, Dobrzycka KM, Lee AV, et al. SAFB2, a new scaffold attachment factor homolog and estrogen receptor corepressor. J Biol Chem. 2003;278(22):20059–20068. [DOI] [PubMed] [Google Scholar]
- 30.Fridovich I Superoxide anion radical (O2-.), superoxide dismutases, and related matters. J Biol Chem. 1997;272(30):18515–18517. [DOI] [PubMed] [Google Scholar]
- 31.Murphy MP. How mitochondria produce reactive oxygen species. Biochem J. 2009;417(1):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huot J, Houle F, Marceau F, Landry J. Oxidative stress-induced actin reorganization mediated by the p38 mitogen-activated protein kinase/heat shock protein 27 pathway in vascular endothelial cells. Circ Res. 1997;80(3):383–392. [DOI] [PubMed] [Google Scholar]
- 33.Wilson C, Gonzalez-Billault C. Regulation of cytoskeletal dynamics by redox signaling and oxidative stress: implications for neuronal development and trafficking. Front Cell Neurosci. 2015;9:381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stanley A, Thompson K, Hynes A, Brakebusch C, Quondamatteo F. NADPH oxidase complex-derived reactive oxygen species, the actin cytoskeleton, and Rho GTPases in cell migration. Antioxid Redox Signal. 2014;20(13):2026–2042. [DOI] [PubMed] [Google Scholar]
- 35.Mauro C, Leow SC, Anso E, et al. NF-kappaB controls energy homeostasis and metabolic adaptation by upregulating mitochondrial respiration. Nat Cell Biol. 2011;13(10):1272–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baker RG, Hayden MS, Ghosh S. NF-kappaB, inflammation, and metabolic disease. Cell Metab. 2011;13(1):11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kabe Y, Ando K, Hirao S, Yoshida M, Handa H. Redox regulation of NF-kappaB activation: distinct redox regulation between the cytoplasm and the nucleus. Antioxid Redox Signal. 2005;7(3–4):395–403. [DOI] [PubMed] [Google Scholar]
- 38.Takada Y, Mukhopadhyay A, Kundu GC, Mahabeleshwar GH, Singh S, Aggarwal BB. Hydrogen peroxide activates NF-kappa B through tyrosine phosphorylation of I kappa B alpha and serine phosphorylation of p65: evidence for the involvement of I kappa B alpha kinase and Syk protein-tyrosine kinase. J Biol Chem. 2003;278(26):24233–24241. [DOI] [PubMed] [Google Scholar]
- 39.Kaneto H, Matsuoka TA, Nakatani Y, Kawamori D, Matsuhisa M, Yamasaki Y. Oxidative stress and the JNK pathway in diabetes. Curr Diabetes Rev. 2005;1(1):65–72. [DOI] [PubMed] [Google Scholar]
- 40.Ben-Neriah Y, Karin M. Inflammation meets cancer, with NF-kappaB as the matchmaker. Nat Immunol. 2011;12(8):715–723. [DOI] [PubMed] [Google Scholar]
- 41.Kaneto H, Kawamori D, Nakatani Y, Gorogawa S, Matsuoka TA. Oxidative stress and the JNK pathway as a potential therapeutic target for diabetes. Drug News Perspect. 2004;17(7):447–453. [DOI] [PubMed] [Google Scholar]
- 42.Evans JL, Goldfine ID, Maddux BA, Grodsky GM. Oxidative stress and stress-activated signaling pathways: a unifying hypothesis of type 2 diabetes. Endocrine reviews. 2002;23(5):599–622. [DOI] [PubMed] [Google Scholar]
- 43.Son Y, Cheong YK, Kim NH, Chung HT, Kang DG, Pae HO. Mitogen-Activated Protein Kinases and Reactive Oxygen Species: How Can ROS Activate MAPK Pathways? J Signal Transduct. 2011;2011:792639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chakravarthy H, Gu X, Enge M, et al. Converting Adult Pancreatic Islet alpha Cells into beta Cells by Targeting Both Dnmt1 and Arx. Cell Metab. 2017;25(3):622–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dhawan S, Georgia S, Tschen SI, Fan G, Bhushan A. Pancreatic beta cell identity is maintained by DNA methylation-mediated repression of Arx. Dev Cell. 2011;20(4):419–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yoon JC, Ng A, Kim BH, Bianco A, Xavier RJ, Elledge SJ. Wnt signaling regulates mitochondrial physiology and insulin sensitivity. Genes Dev. 2010;24(14):1507–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wei FY, Nagashima K, Ohshima T, et al. Cdk5-dependent regulation of glucose-stimulated insulin secretion. Nat Med. 2005;11(10):1104–1108. [DOI] [PubMed] [Google Scholar]
- 48.Esteras N, Rohrer JD, Hardy J, Wray S, Abramov AY. Mitochondrial hyperpolarization in iPSC-derived neurons from patients of FTDP-17 with 10+16 MAPT mutation leads to oxidative stress and neurodegeneration. Redox Biol. 2017;12:410–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shimizu N, Hasunuma H, Watanabe Y, et al. The Simultaneous Elevation of Oxidative Stress Markers and Wilms’ Tumor 1 Gene during the Progression of Myelodysplastic Syndrome. Intern Med. 2016;55(24):3661–3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stefano GB, Challenger S, Kream RM. Hyperglycemia-associated alterations in cellular signaling and dysregulated mitochondrial bioenergetics in human metabolic disorders. Eur J Nutr. 2016;55(8):2339–2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wisneski JA, Stanley WC, Neese RA, Gertz EW. Effects of acute hyperglycemia on myocardial glycolytic activity in humans. J Clin Invest. 1990;85(5):1648–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rolo AP, Palmeira CM. Diabetes and mitochondrial function: Role of hyperglycemia and oxidative stress. Toxicology and Applied Pharmacology. 2006;212(2):167–178. [DOI] [PubMed] [Google Scholar]
- 53.Du XL, Edelstein D, Dimmeler S, Ju Q, Sui C, Brownlee M. Hyperglycemia inhibits endothelial nitric oxide synthase activity by posttranslational modification at the Akt site. The Journal of clinical investigation. 2001;108(9):1341–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nishikawa T, Edelstein D, Du XL, et al. Normalizing mitochondrial superoxide production blocks three pathways of hyperglycaemic damage. Nature. 2000;404(6779):787–790. [DOI] [PubMed] [Google Scholar]
- 55.Du XL, Edelstein D, Rossetti L, et al. Hyperglycemia-induced mitochondrial superoxide overproduction activates the hexosamine pathway and induces plasminogen activator inhibitor-1 expression by increasing Sp1 glycosylation. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(22):12222–12226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Giacco F, Brownlee M. Oxidative stress and diabetic complications. Circ Res. 2010;107(9):1058–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fiorentino TV, Prioletta A, Zuo P, Folli F. Hyperglycemia-induced oxidative stress and its role in diabetes mellitus related cardiovascular diseases. Curr Pharm Des. 2013;19(32):5695–5703. [DOI] [PubMed] [Google Scholar]
- 58.Biarnes M, Montolio M, Nacher V, Raurell M, Soler J, Montanya E. Beta-cell death and mass in syngeneically transplanted islets exposed to short- and long-term hyperglycemia. Diabetes. 2002;51(1):66–72. [DOI] [PubMed] [Google Scholar]
- 59.Butler AE, Campbell-Thompson M, Gurlo T, Dawson DW, Atkinson M, Butler PC. Marked expansion of exocrine and endocrine pancreas with incretin therapy in humans with increased exocrine pancreas dysplasia and the potential for glucagon-producing neuroendocrine tumors. Diabetes. 2013;62(7):2595–2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Spijker HS, Song H, Ellenbroek JH, et al. Loss of β-Cell Identity Occurs in Type 2 Diabetes and Is Associated With Islet Amyloid Deposits. Diabetes. 2015;64(8):2928–2938. [DOI] [PubMed] [Google Scholar]
- 61.Lou H, Kaur K, Sharma AK, Singal PK. Adriamycin-induced oxidative stress, activation of MAP kinases and apoptosis in isolated cardiomyocytes. Pathophysiology. 2006;13(2):103–109. [DOI] [PubMed] [Google Scholar]
- 62.Giorgio M, Migliaccio E, Orsini F, et al. Electron transfer between cytochrome c and p66Shc generates reactive oxygen species that trigger mitochondrial apoptosis. Cell. 2005;122(2):221–233. [DOI] [PubMed] [Google Scholar]
- 63.Hekimi S, Wang Y, Noe A. Mitochondrial ROS and the Effectors of the Intrinsic Apoptotic Pathway in Aging Cells: The Discerning Killers! Front Genet. 2016;7:161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Friberg AS, Stahle M, Brandhorst H, Korsgren O, Brandhorst D. Human islet separation utilizing a closed automated purification system. Cell transplantation. 2008;17(12):1305–1313. [DOI] [PubMed] [Google Scholar]
- 65.Yang C, Loehn M, Jurczyk A, et al. Lixisenatide accelerates restoration of normoglycemia and improves human beta-cell function and survival in diabetic immunodeficient NOD-scid IL-2rg(null) RIP-DTR mice engrafted with human islets. Diabetes, metabolic syndrome and obesity: targets and therapy. 2015;8:387–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Paulo JA, Gygi SP. A comprehensive proteomic and phosphoproteomic analysis of yeast deletion mutants of 14-3-3 orthologs and associated effects of rapamycin. Proteomics. 2015;15(2–3):474–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Erickson BK, Mintseris J, Schweppe DK, et al. Active Instrument Engagement Combined with a Real-Time Database Search for Improved Performance of Sample Multiplexing Workflows. Journal of Proteome Research. 2019;18(3):1299–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kramer A, Green J, Pollard J Jr., Tugendreich S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics. 2014;30(4):523–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.