Abstract
Host organism offers an environment for a parasite, and this environment is heterogenous within the host, variable among individual as well as between the hosts, and changing during the host's lifetime. This heterogeneity may act as a prerequisite for parasite species divergence. Intraspecific variability related to a certain type of heterogeneity may indicate an initial stage of speciation, and thus poses an evolutionary importance. Here we analyzed genetic and morphologic variation of trematode metacercariae of Microphallus piriformes (Trematoda, Microphallidae). Genetic variability of trematodes was assessed from sequences of cytochrome c oxidase subunit 1 (COI) and internal transcribed spacer region (ITS-1). Morphological variation of metacercarial body shape was for the first time analyzed using geometric morphometrics. Parasites from the White Sea and the Barents Sea coasts demonstrated partial genetic divergence (according to COI sequence analysis) and had significantly different body shape. Neither genetic nor morphological variation of metacercariae was related to intermediate host species. We discuss possible causes of the observed genetic divergence of parasite populations in different geographic regions.
Keywords: Microphallus piriformes, Trematoda, Paraxenia, Molecular markers, Geometric morphometrics, Developmental stability
Graphical abstract
Highlights
-
•
Nuclear marker proves species integrity of a trematode Microphallus piriformes.
-
•
Mitochondrial marker reveals gene flow restriction between distant populations.
-
•
The White Sea M. piriformes metacercariae have distinct body shape and size.
-
•
Morphological variation of M. piriformes metacercariae is high in the White Sea region.
-
•
Suboptimal conditions for hosts can lead to developmental instability in a parasite.
1. Introduction
Intraspecies variability (due to both genetic polymorphism and phenotypic plasticity) is a fundamental attribute of living organisms, making them able to survive under variable conditions within species ranges (Lewontin, 1957; Selander and Kaufman, 1973; Meyers and Bull, 2002). In turn, local adaptation is an evolutionary important process, acting as a prerequisite for ecological speciation (both allo- and sympatric) (Mayr, 1970; Thibert-Plante and Hendry, 2011; Lenormand, 2012; Nosil, 2012). Although the terminology of ecological speciation and related concepts were developed mainly for free-living organisms, all this reasoning is quite fair for parasites as well (Combes, 2001). Host organism acts as a first-order environment for parasite (Pavlovsky, 1934; Dogiel, 1964). This environment is heterogeneous within a host, variable among individual hosts, and changing during the host's lifetime.
Some parasites, being at the same ontogenetic stage, are able to infect several host species. Such taxonomically different hosts functionally similar for the life cycle of a parasite are called paraxenic, and the phenomenon is called paraxenia (Granovitch, 1996, 1999; 2009, 2016). Evidently, hosts belonging to different species provide different conditions for a parasite (due to their ecological and physiological dissimilarities) contributing to the general variability of parasites' environment. Adaptation to a certain host species represents the parasitic version of local adaptation. This might become a prerequisite for alloxenic speciation (as adaptation to hosts of different species), a parasitic version of ecological speciation (Combes, 2001). It is fair not exclusively for definitive hosts (where sexual reproduction occurs) but for intermediate hosts as well. For example, association with certain intermediate host species may facilitate transmission to certain definitive host species, prompt partially independent parasite's life cycle functioning of these subsets and finally drive their divergence. Paraxenia at the level of intermediate host may cause genetic subdivision of parasite population also due to differential survival of parasites in different host species, which is crucial for supporting of parasites genetic diversity and essential for the whole parasitic system stability (Granovitch, 1999; Granovitch and Mikhailova, 2004). Importantly, first-order environment for a parasite is not continuous but “patchy”, being comprised of many individual hosts. Owing to this, a population of a parasite (all the individuals at all ontogenetic stages within some geographical region, Combes, 2001) is hence subdivided by (a) host individuals (infrapopulations, Margolis et al., 1982),(b) host species harboring the same parasitic stages (“paraxenic hemipopulations” sensu Beklemishev, 1960; Granovitch, 1996, or “xenopopulations” sensu Combes, 2001); (c) host species harboring different ontogenetic stages of a parasite due to complexity of parasite life-cycle (“metaxenic hemipopulations”, Granovitch, 1996).
On the other hand, geographically distant host populations can strongly differentiate due to either local adaptation or neutral genetic shift or both; they will not be evenly matched as an environment for a parasite. Such a host “non-equivalence” within a geographic range is expected to affect polymorphism in a parasite population and might contribute to divergence processes in case of restricted gene flow between parasite populations. Thus, polymorphism and the discrete genetic structure in a parasite population may be related to heterogeneity of both the first-order (set of paraxenic hosts, their sex, age and immune competence etc.; e.g. Granovitch et al., 1987; Granovitch and Gorbushin, 1995; Lively et al., 2004; Koehler and Poulin, 2012; Levakin et al., 2013) and second-order environment (distant geographic regions, contrasting biotopes etc.; e.g. Blair et al., 1997; Semyenova et al., 2006; Sithithaworn et al., 2007; Webster et al., 2007; Krapivin et al., 2018). Study of polymorphism and population genetic structure due to influence of the first and second-order environment can bring new insight in parasitic systems functioning in nature and evolutionary processes occurring there.
The trematode Microphallus piriformes (Odhner, 1905) (Plagiorchiida, Microphallidae) from the North Atlantic intertidal has a dixenic life cycle. Five periwinkle species of the Littorina Ferussac, 1822 genus (L. saxatilis (Olivi, 1792), L. arcana Hannaford Ellis, 1978, L. compressa Jeffreys, 1865 – “saxatilis” group; L. obtusata (Linnaeus, 1758), L. fabalis Turton, 1825 – “obtusata” group) serve as paraxenic intermediate hosts; several seabird species (gulls, e.g. Larus argentatus Pontoppidan, 1763, eiders, most commonly Somateria mollissima (Linnaeus, 1758), oystercatchers, e.g. Haematopus ostralegus Linnaeus, 1758) are definitive hosts (Galaktionov et al., 2012). M. piriformes, like other species from “pygmeus” group, does not have a free-living stage: metacercariae are formed within daughter sporocysts. Metacercariae within mature daughter sporocysts look like adult worms which have not started egg production yet. They have almost completely developed somatic organs, but do not possess a mature reproductive system. This accelerates maturation and potentiates reproduction efficacy in case of definitive host infection (Galaktionov, 1993; Galaktionov and Dobrovolskij, 2003). Infective metacercariae are remarkably variable in their body size and shape among hosts individuals (our own observations). In addition, this trematode species has a high level of genetic diversity (Galaktionov et al., 2004, 2005, 2008). We consider M. piriformes as a convenient model to study genetic and morphological variation related to paraxenia and geographical distribution, since this parasite is widely distributed along the coasts of the Northern Europe and infects several intermediate hosts.
2. Material and methods
2.1. Sampling
Sporocysts with metacercariae of Microphallus piriformes were sampled during summer expeditions of 2017 and 2018 from wild populations at 4 geographic locations (Fig. 1; Table 1). In total 72 samples were collected.
Fig. 1.
The map of the study region (image: TerraMetrics, map data: Google). Sample collection sites (Tromsø city, Kiberg settlement, Dalnie Zelentsy settlement, Sredny Island) are shown.
Table 1.
Information on sampling sites. Only M. pygmeaus which was used for genetic analysis as an outgroup was collected in the Yakovleva inlet.
| Collection sites | Water area | Year | Coordinates |
|---|---|---|---|
| Korga-Islet in the Levina inlet, vicinities of Sredniy Island | Kandakaksha Bay, White Sea |
2018 | 66°18′04.09″N 33°27′27.02″E |
| Yakovleva inlet, vicinities of Sredniy Island | Kandakaksha Bay, White Sea |
2018 | 66°18′50.01″N 33°50′20.06″E |
| Vicinities of Dalnie Zeletsy | Barents Sea | 2017 | 69°06′56″N 36°04′10″E |
| Vicinities of Kiberg | Varanger Fjord, Barents Sea | 2017 | 70°17′07″N 30°59′53″E |
| Vicinities of Tromsø | Norwegian Sea | 2018 | 69°40′58″N 18°56′34″E |
2.2. Identification and photography
Adult individuals of littorinid snails L. arcana, L. compressa, L. fabalis, L. obtusata and L. saxatilis were collected from intertidal area and transported to the laboratory. For sample preparation snails were dissected under MBI-10 binocular microscope to identify species, sex and possible trematode infection. Infected hepatic tissues were separated and soaked in distilled water, where parasites were carefully washed out from host tissues using glass pipettes. Washed parasites were used for (1) temporary preparations for identification and photographing; (2) fixation in 96% ethanol, final DNA isolation and molecular analysis.
To complete relax and flattening of metacercariae on temporary preparation, glass slides were heated in a drop of distilled water for 2 min at 70 °C using heating table (Galaktionov, 1980, 1993) (Supplementary Fig. 1). Photographic pictures were taken with MBI-10 binocular microscope coupled with MFU photo adapter and Canon EOS 1200D camera; camera settings remained unaltered throughout the study.
2.3. Molecular markers analysis
DNA isolation was performed using minispin collumns for genomic DNA extraction (ExtractDNA Blood, Evrogen) following recommendations of the manufacturer (http://evrogen.ru/kit-user-anuals/ExtractDNA_Blood.pdf). DNA samples were stored at −20 °C until use.
Two molecular markers: fragments of the mitochondrial cytochrome oxidase subunit I (COI) and internal transcribed spacer (ITS-1) were used for the analysis. The primer sequences used for amplification are in Table 2.
Table 2.
Sequences of primers used for amplification of molecular markers.
| Direction | Marker | Sequence 5’ -> 3′ | Fragment length | Reference |
|---|---|---|---|---|
| Fw | COI | TTTTTTGGGCATCCTGAGGTTTAT | 368 bp | Bowles et al. (1992) |
| Rev | COI | TAAAGAAAGAACATAATGAAAATG | Bowles et al. (1992) | |
| Fw | ITS-1 | ACACCGCCCGTCGCTACTA | 347 bp | Galaktionov et al. (2012) |
| Rev | ITS-1 | TGGACGAAACTGCGCGCTTC | Authors |
Ready PCR mixture (iQТМ SYBR® Green Supermix, Bio-Rad) was used for amplification following manufacturer's recommendations. PCR was performed using Veriti 96-Well Thermal Cycler (ThermoFisher Scientific). Amplification method was initial denaturation at 95 °C for 3 min, in cycle denaturation at 95 °C for 30 s, primers annealing at 56 °C for 30 seс, in cycle elongation at 72 °C for 30 seс, final elongation at 72 °C for 4 min; 30 cycles.
Presence of PCR products of the expected length was assessed by 1%-agarose gel electrophoresis in Tris-acetate-EDTA buffer, pH 8.3 (TAE, Evrogen); SYBR green (50x SYBR® Green I for PCR, Evrogen) was added to samples while in-gel loading for visualization; electropherogram was visualized with BioRad ChemiDoc MP at wavelength 497 nm.
Amplicons were sequenced by Sanger in both directions using 3500xL Genetic Analyzer (ThermoFisher Scientific) and BigDyeТМ Terminator v3.1 Cycle Sequencing Kit (ThermoFisher Scientific) with technical support of Alexey Masharsky. Sequencing results were analyzed using ChromasPro v.1.7.4 software (Technelysium Pty Ltd); sequence alignment was performed with MEGA X v.10.0.4 software (Kumar et al., 2018).
2.4. Phylogenetic analysis
We used Bayesian inference for phylogenetic reconstruction in MrBayes v.3.2 software (Ronquist et al., 2012). Nucleotide substitution model was GTR + G + I (Lanave et al., 1984). Optimal nucleotide substitution model was assorted using PAUP4 (Swofford, 2002) and MrModeltest v.2.3 (Nylander, 2004) software packages. Analysis was performed as two independent runs, five chains in each (four heated and one cold; the first 25% samples from the cold chain were discarded) for 15 000 000 generations with a sample frequency of 1000, print frequency of 1000 and diagnostics calculated every 1000 generations. The convergence between two runs was tested by comparison of statistical parameters in the Tracer Software (Rambaut et al., 2018). 65 COI sequences of 72 were used for analysis, as we failed to get sequences of acceptable quality for other 7 samples. We also used sequences of Microphallus pygmaeus (Levinsen, 1881), Microphallus triangulatus Galaktionov, 1984 and Microphallus similis (Jägerskiöld, 1900) as outgroups in our analysis. Results of phylogenetic analysis were visualized using FigTree v.1.4.3 software (Rambaut, 2009).
M. piriformes haplotypes network was constructed according to TCS algorithm (Clement et al., 2000) using PopART software (Leigh and Bryant, 2015).
2.5. Geometric morphometrics
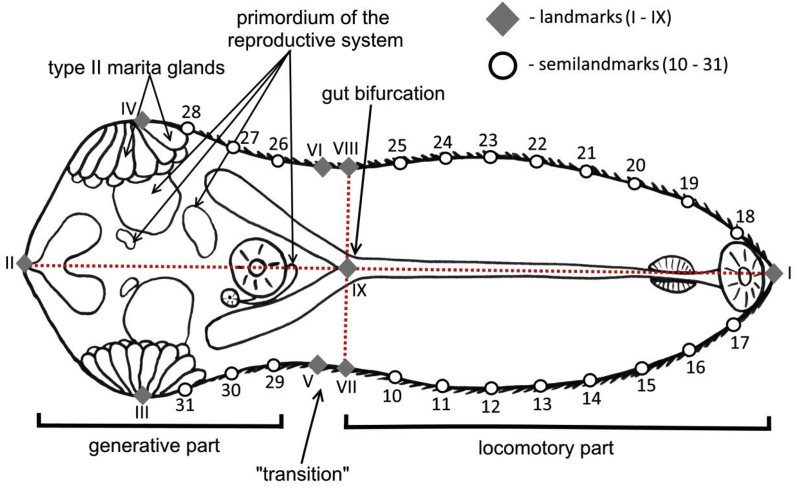
Tps-software package (Rohlf, 2004; The Stony Brook Morphometrics) was used for geometric morphometrics analysis. Two-dimensional images were compiled to tps format using tpsUtil v.1.74, the coordinate locations of landmarks and semilandmarks were digitised in tpsDig2 v.2.3. We used nine landmarks to model metacercarial body shape: two along middle body line at anterior and posterior poles, two at edges of most wide part of posterior body compartment, two at edges of most narrow body part (at the transition between anterior locomotory and posterior generative body parts), one at the gut bifurcation, and two on the body edges at the level of the gut bifurcation. Additionally, 22 semilandmarks forming four curves were used to describe the contour of the body (Fig. 2). Images of 46 clones (clone = metacercariae from different daughter sporocysts within 1 host individual) were analyzed (10 metacercariae from each clone).
Fig. 2.
Scheme of landmarks and semilandmarks on the metacercarial body.
To minimize the effect of allometric variability only fully formed metacercariae with all anatomical details visible were used in the analysis. These metacercariae are characterized by completely formed type II marita glands (the final stage of maturation, no morphogenic changes are known after this stage up to maritogony process starting in the final host; Galaktionov, 1991, 1993).
2.6. Statistical analysis
A geometric morphometric analysis was carried out in R (R v.3.5.3; R Core Team, 2019) using geomorph (Adams et al., 2019) and abind (Plate, Heiberger, 2016) packages. Shape variables were derived from a Generalized Procrustes Analysis (GPA; Gower, 1975), which superimposes and aligns landmark configurations (through translation, rotation and isometric scaling) removing the information not related to shape. Aligned procrustes coordinates were subjected to Principal Component Analysis (PCA) to visualize shape variation of metacercariae. Proportion of variance explained by each principal component was computed from eigenvalues.
Non-parametric Multivariate Analysis of Variance (perMANOVA; Anderson, 2001) on the matrix of procrustes coordinates was used (1) to compare the shape of metacercariae from L. saxatilis and L. obtusata from the White and Barents Seas; and (2) to test for possible allometric relationships of shape and size of metacercariae. The design included the effect of the host individual nested within a particular species to account for non-independence of the observations belonging to the same clone of metacercariae. We tested significance of shape variations using randomized residual permutation procedure (RRPP; Collyer and Adams, 2018, Collyer and Adams, 2019; RRPP package v.0.4.2). Post-hoc pairwise comparisons were held using perMANOVA; p-values were corrected using Holm-Bonferroni procedure (Holm, 1979). Morphological disparity was assessed using morphol.disparity () function from the geomorph package for groups of meracercariae from different host species and sampling locations. Relative morphological disparity was computed as a percentage of the sum of within-group disparities.
To compare centroid size of metacercariae from different hosts and sites, we applied a General Linear Mixed Model with Gaussian error distribution (GLMM; Pinheiro, Bates, 2000) using lme4 package (Bates et al., 2015). The model included one fixed predictor (group of metacercariae defined by host species and site) and one random effect (host individual). The model assumptions were checked visually on the plots of residuals. Effect significance was tested with Likelihood Ratio Test (LRT). Intraclass Correlation Coefficient (ICC) was used to assess the size variability of metacercariae from an individual snail. Tukey's test was used for post hoc comparisons (Quinn, Keough, 2002).
The results of analysis were visualized using ggplot2 (Wickham, 2016) and cowplot (Wilke, 2019) packages. Tables were produced using knitr (Xie, 2019) and huxtable (Hugh-Jones, 2019) packages.
3. Results
3.1. Genetic polymorphism in populations of M. piriformes. ITS-1
All analyzed ITS-1 sequences were identical. This supports, on one hand, the unity of M. piriformes as a species and, on the other, it justifies the affiliation of all the samples to the same species. P-distance between sequences of M. piriformes and closely related species M. pygmaeus was 3.75% (12 SNP, 1 indel).
3.2. Genetic polymorphism in populations of M. piriformes. COI
COI gene proved to be rather polymorphic in M. piriformes: among 64 samples sequenced we have found 29 different haplotypes of COI fragment; three were observed most often. The Bayesian inference based on COI-sequences (Fig. 3) implies a limited gene flow between populations of the White and Barents Seas, as there are several location-specific clades on the tree, e.g., clades including samples exclusively from ether Korga-islet (White Sea) or Kiberg (Barents Sea).
Fig. 3.
Bayesian inference based on COI sequence (369 bp); 15000000 generation; GTR + I + G substitution model; A posteriori probabilities are indicated by node shapes; sample name includes parasite species (pir – M. piriformes, pyg – M. pygmaeus, tri – M. triangulatus, sim – M. similis), sample number, geographic region and location (WSk – White Sea, Korga-Islet; WSy – White Sea, Yakovleva; DZe – Barents Sea, Dalnie Zelentsy; Kib - Barents Sea, Kiberg; Tro - Norwegian Sea, Tromsø), host species (sax – L. saxatilis; arc – L. arcana; comp – L. compressa; obt – L. obtusata; fab – L. fabalis). Identical haplotypes and the FST-value of differentiation between populations (Weir, and Cockerham, 1984) are shown in Supplementary Table 1. Branch color reflects geographic region.
Strong effect of geographic location on haplotypes distribution is obvious also in the haplotypes network (Fig. 4A): although most often haplotypes were detected in all three examined regions, there were also many location-specific haplotypes, including several singletons, clustering according geographic region. As evidenced by both the Bayesian inference (Fig. 3) and haplotypes network (Fig. 4B) there was no detectable population subdivision based on host species: haplotypes of M. piriformes parasitizing hosts of different species were interspersed after both analyses.
Fig. 4.
Haplotype networks, COI sequence (369 bp); TCS algorithm; dashes correspond to mutations. A: color reflects sampling location. B: color reflects host species.
3.3. Intrapopulational polymorphism of metacercarial body shape
Parasite samples from L. saxatilis and L. obtusata hosts only, collected at the White and Barents (Kiberg and Dalnie Zelentsy) Sea shores, were used for analysis: there were five groups in comparison (L. saxatilis was the only host in Dalnie Zelentsy populations). Metacercarial body shape variation related to sampling site and host species is visible on the PCA ordination plot (Fig. 5); PerMANOVA accounting for intraclonal variability confirmed the effects of the factors “location” and “host species” (p < 0.01) on metacercarial body shape. Significant differences were found between all combinations of locations (except samples from Dalnie Zelentsy vs Kiberg, L. saxatilis as a host), but there were no differences in body shape between metacercariae from different hosts within the same location; this result is also visible on the general PCA-ordination plot (Fig. 6, Supplementary Table 2).
Fig. 5.
PCA-ordination of individual M. piriformes metacercariae body shapes grouped by host species. PC1 can be interpreted as a deepness of a “waist” between locomotory and generative body parts; PC2 can be interpreted as a width of locomotory body part. B: Pairwise post-hoc comparison; significant value are shown as bold (considering Holmes correction for multiple comparison); host species: sax – L. saxatilis; obt – L. obtusata; sampling site: Kib - Barents Sea, Kiberg; Kor – White Sea, Korga-Islet; Zel – Barents Sea, Dalnie Zelentsy.
Fig. 6.
A: PCA-ordination of individual M. piriformes metacercariae body shapes. Ellipses show 95%-confidence intervals for the five groups in comparison. B: Mean body shapes of M. piriformes according to sampling site and host species. Transformation grids reflect deviation from the overall mean body shape.
Morphological disparity of metacercariae within the same hemipopulation (hemipopulation is the totality of parasite individuals at the same stage of the life cycle within all host individuals within a given area (Beklemishev, 1960)) and within the same infrapopulation at different sampling sites (separately for different host species) was also assessed. This analysis revealed that metacercarial body shape varied most strongly within the White Sea hemipopulation; this was fair for the both host species. The same tendency was observed in case of infrapopulations. The metacercarial body shape variability was comparatively low within the Barents Sea hemi- and infrapopulations (both Kiberg and Dalnie Zelentsy) (Fig. 7, Supplementary Tables 3 and 4).
Fig. 7.
Variability of metacercarial body shape within hemipopulations and infrapopulations of M. piriformes. A: Absolute and relative morphological disparity (MD) of metacercariae within hosts of the same species. B: Distribution of morphological disparity (MD) within individual snails grouped by host species and sampling location.
3.4. Interactions of metacercarial body shape and size
Theoretically, differences in body shape might originate from allometry – i.e. age-related changes in proportions of the whole body and particular organs during ontogenesis (Klingenberg, 1996, 2016). To avoid allometry effects, only metacercariae with completely formed anatomical structures (see methods) were included into analysis. Nevertheless, the question of changes in body shape proportions after maturation (when metacercariae are ready to infect a final host and when transition to adult worm occurs) is still vague.
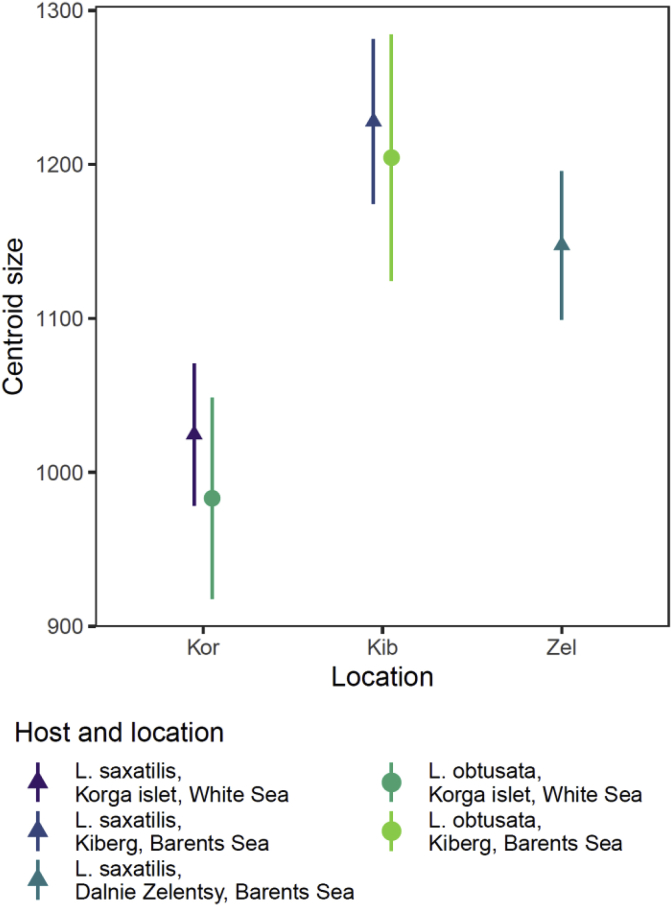
With respect to this, we evaluated interactions between body shape and size (using centroid size as a proxy) in M. piriformes metacercariae and found no significant allometric effect (perMANOVA). However, metacercarial body size varied significantly between the hosts species and locations (LRT = 39.5, df = 4, p < 0.01). Notably, the size of metacercariae from different hosts within the same location did not significantly differ (Fig. 8, Supplementary Table 5). High intraclass correlation coefficient (ICC = 0.71) implies that metacercariae of the same infrapopulation are of very similar size; this fits well with data on synchrony of microphallid development within daughter sporocysts (Galaktionov, 1993).
Fig. 8.
Body size of M. piriformes metacercariae from different host species and sampling locations. Mean centroid size and 95% confidence interval obtained via bootstrap.
4. Discussion
In this study we evaluated genetic and morphological intraspecific variability in metacercariae of Microphallus piriformes at several levels of variability: between populations (effects of different geographic locations), between paraxenic hemipopulations (effects of intermediate host species), between clones (effect of a host individual), and between individuals (intraclonal variability).
4.1. Geography-related genetic polymorphism within M. piriformes populations
Our results indicate some gene flow limitations between populations of different geographic regions (strong between the White and Barents Sea populations; moderate between the two populations from the Barents Sea). Both intermediate (periwinkles) and final (birds) hosts of M. piriformes are widely distributed along the Northern Atlantic sea coasts. Hypothetically, gene flow between spatially disconnected populations might be maintained by bird migrations. Consistently, isolates of M. piriformes from the Vaygach Island (Barents Sea) and Iceland coast (straight distance exceeds 3000 km) demonstrated almost identical UP-PCR fingerprinting patterns (Galaktionov et al., 2008), while UP-PCR fingerprinting did not reveal any geography-related subdivision in any other species of the “pygmaeus” group (Galaktionov et al., 2005). This might be a consequence of a bird migration-maintained gene flow. Shown in our study prevalence of limited number of haplotypes shared by all studied populations agrees well with such a scenario. On the other hand, some heterogeneity of M. piriformes populations related to sampling site was also recorded in previous studies. In particular, genetic differences between several populations of M. piriformes from the White Sea were detected by RAPD approach (Khalturin et al., 2000). Similarly, genetic divergence between populations from the White and Barents seas (but not between the White and Norwegian seas) was demonstrated with UP-PCR approach (Galaktionov et al., 2004). Thus, our results agree well with the earlier data. Some irregularity in the patterns of population genetic differentiation, in our opinion, might be explained by migratory activity of final host birds funneled to specific routes. Gulls, eiders and waders regularly migrate along the North Atlantics shore (Bakken et al., 2003; Bianki, 1967, Bianki, 1989; Coulson et al., 1984; Dementiev and Vuchetich, 1947; Haftorn, 1971; Kokhanov and Skokova, 1967; Krasnov et al., 2015; Mineyev, 2003; Strom, 2006; Tatarinkova, 1970; Anker-Nilssen et al., 2000; Noskov et al., 2016). The schemes of bird migratory routes, reconstructed from published data, are presented in the Supplementary Fig. 2. Obviously, not all the populations of M. piriformes can be connected via bird migration, but only those lying along migratory routes. Locations of bird settlement sites are another factor, exacerbating genetic breach between parasite populations: wintering sites at the coasts of the White and Barents seas were recorded for gulls and eiders (Koryakin and Kondratiev, 1983; Kohanov and Shkliarevitch, 1985; Shkliarevitch, 1979; Сrаmр and Simmons, 1983; Scott and Rose, 1996). Final host settlement provides an opportunity for a year-round autonomous sustainability of local parasite populations, and as a consequence, their divergence from each other. Short adult longevity (no longer than 10 days) also strongly limits transmission capacity via bird migrations (Belopol'skaya, 1983; Galaktionov, 1993; Galaktionov and Dobrovolskij, 2003). Thus, spatially limited and transient connection of trematode populations by hosts migrations cause partial differentiation of the former along the geographical range. It is worth noting that the formation of such partial subdivision in the genetic structure of a parasite species can act as a prerequisite for allopatric speciation (Mayr, 1970; Huyse et al., 2005).
Evidence of partial isolation between different geographic regions was detected by the mitochondrial marker (COI) only, not the nuclear one (ITS-1). Mitochondrial genomes are expected to be more strongly affected by the factors discussed below. Although adaptive neutrality and clonality of mitochondrial genomes were questioned during the last decade, these are still expected to evolve faster than nuclear ones due to haploidy and maternal germline bottleneck, and thus are better indicators of shallow evolutionary events (rev. in Zink and Barrowclough, 2008; Galtier et al., 2009). Mitochondrial marker COI is widely used (often in combination with other molecular markers for more reliable results) in studies of genetic variation and genetic structure of trematode groups (Blasco-Costa et al., 2016). Often, these studies reveal the presence of several genetic lineages and a high haplotype diversity in fluke populations. Sometimes, genetic structure of a parasite species is consistent with the geographical distance between its populations (e.g. Blair et al., 1997; Semyenova et al., 2006; Stothard et al., 2009). Nevertheless, in other cases, such clear dependence can't be reliably identified (e.g. Lopatkin et al., 2010; Tatonova et al., 2013) for different reasons (influence of autogenetic processes, hosts migrations, insufficient sampling, etc.). In many studies, including ours, the authors conclude that genetic structure of trematode species and genetic lineages they form are significantly affected by features of parasites biology and life cycle and movements of their hosts (especially definitive) (Criscione and Blouin, 2004; Huyse et al., 2005; Keeney et al., 2009; Blasco-Costa et al., 2011).
Host species-related genetic polymorphism within M. piriformes populations was not observed in this study. Importantly, 3 species (Littorina arcana, L. compressa, L. fabalis) were relatively rarely detected as hosts of M. piriformes due to their low densities. Therefore, no informative conclusions on genetic specialization of the parasites to these hosts may be done. Most of data were gathered for parasites obtained from L. saxatilis and L. obtusata hosts, and will be discussed in detail below.
All the five species of paraxenic hosts of M. piriformes belong to the subgenus Littorina (Neritrema Recluz, 1869) (Littorinidae, Caenogastropoda). They are phylogenetically closely related and comparatively recently diverged from each other (estimated divergence time does not exceed 5 Ma) (Reid, 1996, 2012). These species differ in several sets of morphological, physiological and ecological characters. Thus, geographic ranges of Littorina arcana and L. compressa are narrower and more patchy compared to other species; they were not recorded from several regions where L. saxatilis, L. fabalis and L. obtusata are present, e.g. the White and North Seas, the Iberian Peninsula coast, etc. (Reid, 1996). There are also differences in the microbiotopic distribution, e.g. L. compressa, L. fabalis and L. obtusata inhabit the fucoid area only, while L. saxatilis and L. arcana can live in the upper part of the intertidal where fucoids are absent; L. fabalis occupies lower intertidal and upper subtidal area and more often is associated with Fucus serratus, in contrast to L. obtusata usually preferring F. vesiculosus and Ascophyllum nodosum (Reid, 1996; Sergievsky et al., 1997; Iastchenko and Granovitch, 2002; Granovitch et al., 2004, 2013). Littorina spp. also exhibit functional differences in tolerance to low salinity, desiccation and other stressors, as well as physiological plasticity (e.g. Todd, 1964; Clarke et al., 2000; Sokolova et al., 2000; Maltseva et al., 2016; Muraeva et al., 2017; Maltseva et al., 2020). Altogether, these differences might form preconditions for host-related speciation among paraxenic hemipopulations of parasites. It is known that subdivision of trematode populations at the genetic level may emerge in connection with paraxenia after expansion of host range via either new host species invasion (usually phylogenetically closely related) or cospeciation (e.g. LoVerde et al., 1985; Theron and Combes, 1988; Semyenova et al., 1995; Blair et al., 1997; Galaktionov et al., 2008, Koehler et al., 2011).
Nevertheless, the most probable divergence scenario of the microphallids from the “pygmaeus” species group assumes final host species and geographic isolation as drivers, and not the intermediate host species (Galaktionov et al., 2012). Despite a number of differences between the species of intermediate hosts, the possibility of host species-driven evolution of M. piriformes seems unlikely for several reasons:
-
(1)
The common ancestor of M. piriformes and other “pygmaeus” species expanded to the North Atlantic from the North Pacific between 3.5 Myr and 2.4 Myr BP (together with the Littorina (Neritrema) species and gull hosts) and infected several available Neritrema species (ancestors of the “obtusata” and “saxatilis” groups had already been diverged by then, Reid, 1996; Reid et al., 2012). The northern communication between Arctic-Atlantic and Pacific via Bering Strait closed again, and this promoted speciation of M. piriformes in the North Atlantic and M. calidris in the North Pacific; the branch leading to other “pygmaeus” species specified due to switch to seaducks species as final hosts (Galaktionov et al., 2012). Speciation of L. compressa, L. fabalis and L. arcana occurred in the North Atlantic after the Bering Landbridge emergence (Reid, 1996), and this did not cause partitioning of M. piriformes, which is able to successfully infect all those species. Accordingly, no evidence of the intermediate host-related subdivision in modern populations of M. piriformes was revealed in this study, while there was an evidence for geography-related subdivision.
-
(2)
It is also worth mentioning, that some Neritrema species are very similar at the both genetic and functional levels, e.g. L. saxatilis and L. arcana with indirect evidence of regular hybridization (Mikhailova et al., 2008, 2009; Granovitch et al., 2013; Maltseva et al., 2016, 2020), therefore hardly providing any background for parasite differentiation.
-
(3)
Additionally, some facts suggest that this trematode species is rather sensitive to host's physiological properties, which in turn is a prerequisite for host-driven speciation. In comparison to other species from “pygmaeus” group, M. piriformes is characterized by relatively narrow range of intermediate hosts: it can infect only snails from Littorina (Neritrema) subgenera. In contrast, e.g. M. pygmaeus can also heavily infect L. littorea (Linnaeus, 1758) (Werding, 1969; Pohley, 1976; Lauckner, 1984), while M. pseudopygmaeus Galaktionov, 2009 is known for its ability to infect a set of hosts of several families and orders (Galaktionov, 2009; Galaktionov et al., 2012). Such narrow specialization of M. piriformes reduces the likelihood of the formation of subdivision in connection with the species of intermediate host.
Affirming this, previous studies detected paraxenia-related genetic variability of M. piriformes. Trematodes from either L. saxatilis or L. obtusata (the hosts significantly different at both ecological and functional levels) collected from the same geographic location demonstrated distinct RAPD patterns depending on host species (Khalturin et al., 2000). Our findings are not necessarily inconsistent with this data due to the lesser sensitivity of our approach (only two markers were used in this study, so our conclusion that there is no intermediate host species related subdivision may not be approved by further whole-genome scans). Instead, together these results suggest that geography-related differences are more profound than those related to the intermediate host species.
4.2. Metacercarial body shape variability
Data on metacercariae (and also other ontogenetic stages) body shape variability in M. piriformes are scarce. Significant variability of the metacercarial body shape was shown for M. pseudopygmaeus which is closely phylogenetically related to M. piriformes, yet, there was no clear relations to different host species (represented by a rather diverse set of L. saxatilis, Onoba aculeus, Margarites helicinus, Solariella varicosa, Epheria vincta, Lacuna neritoides, Cryptonatica clausa) with exception of metacercariae from M. groenlandicus, which were relatively distinct in their body shape. Noteworthy, the same study revealed that the body shape of metacercariae from M. groenlandicus tended to correlate with geographic location (Dalnezelenetskaya bay vs Kolguev Island; Galaktionov, 1993). Still, the author stressed that the variability revealed was not constrained to discrete variants but rather continuous with all kinds of intermediate states. This is congruent with our results: there was a metacercarial body shape continuum, and those from either different host species or geographic locations tended to form certain variants more often with no break between these variants (Fig. 5, Fig. 6). The physiological and genetic background of this body shape variability is completely unclear.
Quite interestingly, not only the variability in body shape per se was revealed in our study, but also the differences between geographic locations in the degree of morphological disparity: it was maximal within the White Sea hemi- and infrapopulations. Our genetic data do not allow to suspect a higher genetic diversity of the White sea population (compared to the other two from the Barents Sea) as a possible explanation for an increased level of inter-hemipopulational variability. The higher degree of intraclonal shape variability (based on identical or very similar genetic background) might be interpreted similar to fluctuating asymmetry. Generally, there are two fundamental properties of any organism development restricting its phenotypic variation: canalization (among individuals) and developmental stability (within individuals) (Waddington, 1942; Schmalhausen, 1949; Debat and David, 2001; Willmore et al., 2007). It is well known that under pressure of environmental stressors the accuracy of the molecular and physiological background for both canalization and developmental stability may be compromised, which in turn, would cause an increase in phenotypic variation within and among individuals (e.g. Scharloo, 1991; Parsons, 1997; Hoffmann and Hercus, 2000; Lazic et al., 2015). The increase in both types of phenotypic variation was detected in the White Sea population which might indicate some kind of disturbance. Importantly, high level of phenotypic variation was accompanied with decreasing of the average body size, which might be due to the same reason. The organism size is a function of the growth rate and duration, which depend on temperature, nutrition and oxygen availability in ectotherms (rev. in Shingleton, 2011). The White Sea has a relatively low salinity (24–28 ppm), high seasonal salinity variation in the surface layer, long ice cover period (up to 8 month) (Naumov and Fediakov, 1993). These conditions are likely not optimal for periwinkles, the M. piriformes intermediate hosts Littorina saxatilis and L. obtusata (L. compressa and L. arcana are absent in this area), as evidenced by the sizes of these snails from the Barents and White Sea populations (Granovitch et al., 2004). Temperature together with oxygen availability (which solubility in water depends on temperature) form a latitudinal gradient shaping the size increase at high latitudes demonstrated in different groups of marine invertebrates, e.g. benthic amphipod crustaceans (Chapelle and Peck, 1999), benthic gastropods of the Turridae family (McClain and Rex, 2001) and oceanic nematodes (Soetaert et al., 2002). Body size reducing effects of hypoxia on growth were also described in not aquatic insects, e.g. Drosophila (rev. in Shingleton, 2011). The size differences in the Littorina snails between the Barents and White Sea populations are fully in line with such reasoning, as the White Sea population is the most southern among the studied. Suboptimal functioning of hosts at the White Sea is expected to affect parasite, since it raises basal metabolic costs (rev. in Sokolova, 2013). M. piriformes metacercarial body size forms a trend collinear with latitude increase (Fig. 8). This trend also corresponds well to the hosts’ size distribution on one hand, and to degree of metacercarial shape disparity within hemi- and infrapopulations on the other. Altogether, our results imply that fundamental environment-development interaction patterns known for free-living organisms are also legitimate for parasites, and that phenotypic variation in a parasite may act as an indicator of host functioning.
5. Conclusion
In our study we applied both genetic and morphometric approach to evaluate the degree of populations subdivision of the dixenic trematode M. piriformes. Both methods showed consistent results: there were signs of gene flow restriction between parasite populations from remote locations where genetic exchange is limited due to both absence of regular bird migration and short parasite lifetime within the bird. Additionally, a strongly increased level of intra- and interclonal metacerarian body shape disparity in the White Sea region together with a reduced mean body size were revealed using geometric morphometrics. We suggest that this may be result of a restricted nutrient supply, development destabilization and decanalization emerging from suboptimal environmental conditions for the intermediate hosts in that region. We failed to find any differences related to the intermediate host species, which contrasts with some earlier data (Khalturin et al., 2000). This implies, that inter-region differences are more pronounced than those between paraxenic hemipopulations; more sensitive methods are needed to detect the latter.
Acknowledgements
This research was funded by the Russian Foundation for Basic Research grants number 19-04-00392; the purchase of basic laboratory equipment was financed by the St. Petersburg State University grant number 0.40.491.2017. The opportunities for DNA purification, amplification and sequencing were provided by the Resource Center “Molecular and Cell Technologies”; data analysis was performed in the Computing Centre of the St. Petersburg State University; experimental animals were maintained in the Resource Center “Observatory of environmental safety” of St. Petersburg State University. Sampling in the White Sea coast was logistically based on the Educational and research station “Belomorskaia” of the St. Petersburg State University. We thank Eugeniy L. Yakovis for thorough proofreading and language editing.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ijppaw.2020.02.004.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Adams D.C., Collyer M.L., Kaliontzopoulou A. Geomorph: software for geometric morphometric analyses. 2019. https://cran.rproject.org/package=geomorph R package version 3.1.0.
- Anderson M.J. A new method for non-parametric multivariate analysis of variance. Austral. Ecol. 2001;26(1):32–46. [Google Scholar]
- Anker-Nilssen T., Bakken V., Strøm H., Golovkin A.N., Bianki V.V., Tatarinkova I.P., editors. The Status of Marine Birds Breeding in the Barents Sea Region. Norsk Polarinstitutt; Polarmiljøsenteret, NO-9296 Tromso: 2000. [Google Scholar]
- Bakken V., Runde O., Tjorve E. vol. 1. Stavanger Museum; Stavanger: 2003. (Norwegian Bird Ringing Atlas). [Google Scholar]
- Bates D., Maechler M., Bolker B., Walker S. Fitting linear mixed-effects models using lme4. J. Stat. Software. 2015;67(1):1–48. [Google Scholar]
- Beklemishev V.N. Spatial and functional structure of populations. Byull. Mosk. Obshch. Ispyt. Prir. 1960;65:41–45. (In Russian) [Google Scholar]
- Belopol’skaya M.M. Biology and geography of helminths of migrating birds. In: Poliansky YuI., editor. vol. 34. Leningrad University Press; Leningrad: 1983. pp. 174–189. (Free Living and Parasitic Invertebrates (Morphology, Biology and Evolution). Tr. Biol. Nauchno.-Issled. Inst. Leningr. Univ.). (In Russian) [Google Scholar]
- Bianki V.V. Trans. Kandalaksha State Reserve. Vol. 6. 1967. Waders, gulls and alcids of Kandalaksha bay; p. 364. (In Russian) [Google Scholar]
- Bianki V.V. Migration of Birds of the Eastern and Northern Asia: Anseriformes. Nauka; Moscow: 1989. Migration of common eiders of the Barents sea; pp. 205–208. (In Russian) [Google Scholar]
- Blair D., Agatsuma T., Watanobe T., Okamoto M., Ito A. Geographical genetic structure within the human lung fluke, Paragonimus westermani, detected from DNA sequences. Parasitology. 1997;115(4):411–417. doi: 10.1017/s0031182097001534. [DOI] [PubMed] [Google Scholar]
- Blasco-Costa I., Waters J.M., Poulin R. Swimming against the current: genetic structure, host mobility and the drift paradox in trematode parasites. Mol. Ecol. 2011;21(1):207–217. doi: 10.1111/j.1365-294X.2011.05374.x. [DOI] [PubMed] [Google Scholar]
- Blasco-Costa I., Cutmore S.C., Miller T.L., Nolan M.J. Molecular approaches to trematode systematics: «best practice» and implications for future study. Syst. Parasitol. 2016;93(3):295–306. doi: 10.1007/s11230-016-9631-2. [DOI] [PubMed] [Google Scholar]
- Bowles J., Blair D., McManus D.P. Genetic variants within the genus Echinococcus identified by mitochondrial DNA sequencing. Mol. Biochem. Parasitol. 1992;54(2):165–173. doi: 10.1016/0166-6851(92)90109-w. [DOI] [PubMed] [Google Scholar]
- Chapelle G., Peck L.S. Polar gigantism dictated by oxygen availability. Nature. 1999;399:114–115. [Google Scholar]
- Clarke A.P., Mill P.J., Grahame J. Biodiversity in Littorina species (Mollusca: Gastropoda): a physiological approach using heat-coma. Mar. Biol. 2000;137(3):559–565. [Google Scholar]
- Clement M., Posada D., Crandall K.A. TCS: a computer program to estimate gene genealogies. Mol. Ecol. 2000;9(10):1657–1659. doi: 10.1046/j.1365-294x.2000.01020.x. [DOI] [PubMed] [Google Scholar]
- Collyer M.L., Adams D.C. RRPP: an R package for fitting linear models to high dimensional data using residual randomization. Methods Ecol. Evol. 2018;9(7):1772–1779. [Google Scholar]
- Collyer M.L., Adams D.C. 2019. RRPP: Linear Model Evaluation with Randomized Residuals in a Permutation Procedure.https://CRAN.R-project.org/package=RRPP [Google Scholar]
- Combes C. University of Chicago Press; Chicago and London: 2001. Parasitism. The Ecology and Evolution of Intimate Interactions. [Google Scholar]
- Coulson J.C., Monaghan P., Butterfield J.E.L., Duncan N., Ensor K., Shedden C., Thomas C. Scandinavian herring gulls wintering in Britain. Ornis Scand. 1984;15(2):79–88. [Google Scholar]
- Criscione C.D., Blouin M.S. Life cycles shape parasite evolution: comparative population genetics of salmon trematodes. Evolution. 2004;58(1):198–202. doi: 10.1111/j.0014-3820.2004.tb01587.x. [DOI] [PubMed] [Google Scholar]
- Debat V., David P. Mapping phenotypes: canalization, plasticity and developmental stability. Trends Ecol. Evol. 2001;16:555–561. [Google Scholar]
- Dementiev G.P., Vuchetich V.N. The seasonal distribution and migration of gulls in the USSR on the basis of ringing data. Trudy Central'nogo byoro kol'tsevaniya. 1947;5:1–31. (In Russian) [Google Scholar]
- Dogiel V.A. Oliver and Boyd; London: 1964. General Parasitology; p. 516. (English translation) [Google Scholar]
- Galaktionov K.V. Four types of metacercariae species in molluscs, Littorina saxatilis and L. obtusata from the Barents and White Seas. Vestn. Leningr. Univ. Ser. Biol. 1980;3(1):21–28. (In Russian) [Google Scholar]
- Galaktionov K.V. The development of the metacercariae of Microphallus pirum (Syn. Levinseniella somateria) (Trematoda: Microphallidae) Parazitologiya. 1991;25:116–124. (In Russian) [Google Scholar]
- Galaktionov K.V. Kola Scientific Centre; Apatity: 1993. The Life Cycles of Trematodes as the Components of Ecosystems. (In Russian) [Google Scholar]
- Galaktionov K.V. Description of the maritae and determination of the species status of Microphallus pseudopygmaeus sp. nov. (Trematoda: Microphallidae) Parazitologiya. 2009;43:289–299. (In Russian) [PubMed] [Google Scholar]
- Galaktionov K.V., Dobrovolskij A.A. Kluwer Academic Publisher; The Netherlands: 2003. The Biology and Evolution of Trematodes. An Essay on the Biology, Morphology, Life Cycles, Transmissions, and Evolution of Digenetic Trematodes. [Google Scholar]
- Galaktionov K.V., Bulat S.A., Alekhina I.A., Saville D.H., Fitzpatrick S.M., Irwin S.W.B. Evolutionary relationships within “pygmaeus” group microphallids using genetic analysis and scanning electron microscopy. J. Helminthol. 2004;78:231–236. doi: 10.1079/joh2004245. [DOI] [PubMed] [Google Scholar]
- Galaktionov K.V., Irwin S.W.B., Bulat S.A., Alekhina I.A., Mokrousov I.V., Skirnisson K., Bustnes J.O., Saville D.H., Fitzpatrick S.M. Population divergence and speciation in digenetic trematodes with two-host life cycles the “pygmaeus” microphallids. Bull. Scand. Baltic Soc. Parasitol. 2005;14:57–58. [Google Scholar]
- Galaktionov K.V., Bulat S.A., Alekhina I.A., Mokrousov I.V. Proceedings of the IV Congress of the Russian Society of Parasitologists – Russian Academy of Sciences “Parasitology in XXI century – problems, methods, solutions”. Vol. 1. 2008. Intraspecific genetic variability in microphallids of the “pygmaeus” group (Trematoda, Microphallidae) and the possible reasons its determining; pp. 154–159. (In Russian) [Google Scholar]
- Galaktionov K.V., Blasco-Costa I., Olson P.D. Life cycles, molecular phylogeny and historical biogeography of the “pygmaeus” microphallids (Digenea: Microphallidae): widespread parasites of marine and coastal birds in the Holarctic. Parasitology. 2012;139(10):1346–1360. doi: 10.1017/S0031182012000583. [DOI] [PubMed] [Google Scholar]
- Galtier N., Nabholz B., Glemin S., Hurst G.D.D. Mitochondrial DNA as a marker of molecular diversity: a reappraisal. Mol. Ecol. 2009;18(22):4541–4550. doi: 10.1111/j.1365-294X.2009.04380.x. [DOI] [PubMed] [Google Scholar]
- Gower J.C. Generalized procrustes analysis. Psychometrika. 1975;40(1):33–51. [Google Scholar]
- Granovitch A.I. Parasitare systems and the population structure of parasites. Parazitologiya. 1996;30(4):343–356. (In Russian) [PubMed] [Google Scholar]
- Granovitch A.I. Parasitic systems and the structure of parasite populations. Helgol. Mar. Res. 1999;53(1):9–18. [Google Scholar]
- Granovitch A.I. Parasitic system reflects population structure of a parasite: conception and terms. Tr. Zool. Inst. RAS. 2009;313(3):329–337. (In Russian) [Google Scholar]
- Granovitch A.I. From host–parasite systems to parasitic systems. Interactions of littoral mollusks of the genus Littorina with their trematode parasites. Zool. Zh. 2016;95(3):252–266. (In Russian) [Google Scholar]
- Granovitch A.I., Gorbushin A.M. Differences in the Trematode parthenite infection rate in males and females of the littoral snail genus Littorina and Hydrobia in the Kandalaksha Bay of White Sea. Parazitologiya. 1995;29(3):167–178. (In Russian) [Google Scholar]
- Granovitch A.I., Mikhailova N.A. Rocky shore trematodes of the west coast of Sweden: distribution and life cycle strategies. Acta Parasitol. 2004;49(3):228–236. [Google Scholar]
- Granovitch A.I., Mikhailova N.A., Sergivsky S.O. Age-specific peculiarities of prevalence of Littorina obtusata and L. saxatilis populations by the Trematode parasites. Parazitologiya. 1987;21:721–729. (In Russian) [PubMed] [Google Scholar]
- Granovitch A.I., Mikhailova N.A., Znamenskaya O., Petrova YuA. Species complex of mollusks of the genus Littorina (Gastropoda, Prosobranchia) from the eastern Murman coast. Zool. Zh. 2004;83(11):1305–1316. (In Russian) [Google Scholar]
- Granovitch A.I., Maximovich A.N., Avanesyan A.V., Starunova Z.I., Mikhailova N.A. Micro-spatial distribution of two sibling periwinkle species across the intertidal indicates hybridization. Genetica. 2013;141(7–9):293–301. doi: 10.1007/s10709-013-9728-3. [DOI] [PubMed] [Google Scholar]
- Haftorn S. 1971. Norges Fugler. Universitetsforlaget, Oslo, Вегgen and Tromso. [Google Scholar]
- Hoffmann A.A., Hercus M.J. Environmental stress as an evolutionary force. Bioscience. 2000;50:217–226. [Google Scholar]
- Holm S. A simple sequentially rejective multiple test procedure. Scand. Stat. Theory Appl. 1979;6:65–70. [Google Scholar]
- Hugh-Jones D. 2019. Huxtable: Easily Create and Style Tables for LaTeX, HTML and Other Formats. R Package Version 4.5.0.https://CRAN.R-project.org/package=huxtable [Google Scholar]
- Huyse T., Poulin R., Theron A. Speciation in parasites: a population genetics approach. Trends Parasitol. 2005;21(10):469–475. doi: 10.1016/j.pt.2005.08.009. [DOI] [PubMed] [Google Scholar]
- Iastchenko, V.V., Granovitch A.I. Littorina fabalis as another littoral gastropode species of the White Sea. Vestnik of St. Petersburg State University. Ser. Biol. 4(27), 34-45. (In Russian).
- Keeney D.B., King T.M., Rowe D.L., Poulin R. Contrasting mtDNA diversity and population structure in a direct-developing marine gastropod and its trematode parasites. Mol. Ecol. 2009;18(22):4591–4603. doi: 10.1111/j.1365-294X.2009.04388.x. [DOI] [PubMed] [Google Scholar]
- Khalturin K.V., Mikhailova N.A., Granovitch A.I. Genetic heterogeny in the natural populations of Microphallus piriformes and M. pygmaeus parthenites (Trematoda: Microphallidae) Parazitologiya. 2000;34(6):486–501. (In Russian) [PubMed] [Google Scholar]
- Klingenberg C.P. Multivariate allometry. In: Marcus L.F., Corti M., Loy A., Naylor G.J.P., Slice D.E., editors. vol. 284. Springer; Boston, MA: 1996. (Advances in Morphometrics. NATO ASI Series (Series A: Life Sciences)). [Google Scholar]
- Klingenberg C.P. Size, shape, and form: concepts of allometry in geometric morphometrics. Dev. Gene. Evol. 2016;226(3):113–137. doi: 10.1007/s00427-016-0539-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehler A.V., Poulin R. Clone-specific immune reactions in a trematode-crustacean system. Parasitology. 2012;139(1):128–136. doi: 10.1017/S0031182011001739. [DOI] [PubMed] [Google Scholar]
- Koehler A.V., Gonchar A.G., Poulin R. Genetic and environmental determinants of host use in the trematode Maritrema novaezealandensis (Microphallidae) Parasitology. 2011;138:100–106. doi: 10.1017/S0031182010001022. 01. [DOI] [PubMed] [Google Scholar]
- Kokhanov V.D., Shklyarevich F.N. Wintering of waterfowl and gulls on the Murman coast and on the White sea. In: Kumari E., editor. vol. 17. 1985. pp. 28–44. (Communications of the Baltic Comission for the Study of Bird Migration). Tartu. (In Russian with English summary) [Google Scholar]
- Kokhanov V.D., Skokova N.N. Trans. Kandalaksha State Reserve. Vol. 5. 1967. The avifauna in the Ainov islands; pp. 185–268. (In Russian) [Google Scholar]
- Koryakin A.S., Kondratiev A.V. Common Eider in the Bird Community of the Islets. Abstracts of 3rd All-Union Meeting on Eiders (Matsalu State Nature Reserve, August 24-26 1983), Tallinn. 1983. Wintering of the common eider in the area of the Veliky Island (Kandalaksha bay of the White sea) pp. 59–62. (In Russian and English) [Google Scholar]
- Krapivin V.A., Bagrov S.V., Varfolomeeva M.A. Effect of tidal level on abundance of symbionts in the White Sea blue mussel. Dis. Aquat. Org. 2018;130(2):131–144. doi: 10.3354/dao03259. [DOI] [PubMed] [Google Scholar]
- Krasnov YuV., Gavrilo M.V., Shavykin A.A. Status, number and monitoring of the common eider (Somateria mollissima) population in the Barents and White Seas. Zool. Zh. 2015;94:62–67. (In Russian) [Google Scholar]
- Kumar S., Stecher G., Li M., Knyaz C., Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018;35(6):1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanave C., Preparata G., Sacone C., Serio G. A new method for calculating evolutionary substitution rates. J. Mol. Evol. 1984;20(1):86–93. doi: 10.1007/BF02101990. [DOI] [PubMed] [Google Scholar]
- Lauckner G. Brackish-water submergence of the common periwinkle, Littorina littorea, and its digenean parasites in the Baltic Sea and in the Kattegat. Helgol. Mar. Res. 1984;37:177–184. [Google Scholar]
- Lazic M.M., Carretero M.A., Crnobrnja-Isailovic J., Kaliontzopoulou A. Effects of environmental disturbance on phenotypic variation: an integrated assessment of canalization, developmental stability, modularity, and allometry in lizard head shape. Am. Nat. 2015;185(1):44–58. doi: 10.1086/679011. [DOI] [PubMed] [Google Scholar]
- Leigh J.W., Bryant D. PopART: full-feature software for haplotype network construction. Methods Ecol. Evol. 2015;6(9):1110–1116. [Google Scholar]
- Lenormand T. From local adaptation to speciation: specialization and reinforcement. Int. J. Ecol. 2012;2012:1–11. [Google Scholar]
- Levakin I.A., Losev E.A., Nikolaev K.E., Galaktionov K.V. In vitro encystment of Himasthla elongata cercariae (Digenea, Echinostomatidae) in the haemolymph of blue mussels Mytilus edulis as a tool for assessing cercarial infectivity and molluscan susceptibility. J. Helminthol. 2013;87(2):180–188. doi: 10.1017/S0022149X1200017X. [DOI] [PubMed] [Google Scholar]
- Lewontin R.C. The adaptations of populations to varying environments. Cold Spring Harbor Symp. Quant. Biol. 1957;22:395–408. doi: 10.1101/sqb.1957.022.01.037. [DOI] [PubMed] [Google Scholar]
- Lively C.M., Dybdahl M.F., Jokela J., Osnas E.E., Delph L.F. Host sex and local adaptation by parasites in a snail‐trematode interaction. Am. Nat. 2004;164(S5):S6–S18. doi: 10.1086/424605. [DOI] [PubMed] [Google Scholar]
- Lopatkin A.A., Chrisanfova G.G., Voronin M.V., Zazornova O.P., Beer S.A., Semyenova S.K. Polymorphism of the cox1 gene in cercariae isolates of bird schistosomes (Trematoda: Schistosomatidae) from ponds of Moscow and Moscow region. Russ. J. Genet. 2010;46(7):873–880. [PubMed] [Google Scholar]
- LoVerde P.T., DeWald J., Minchella D.J., Bosshardt S.C., Damian R.T. Evidence for host-induced selection in Schistosoma mansoni. J. Parasitol. 1985;71(3):297–301. [PubMed] [Google Scholar]
- Maltseva A.L., Varfolomeeva M.A., Lobov A.A., Mikhailova N.A., Renaud P.E., Volovik K., Grishankov A.V., Granovitch A.I. Measuring physiological similarity of closely related littorinid species: a proteomic insight. Mar. Ecol. Prog. Ser. 2016;552:177–193. [Google Scholar]
- Maltseva A.L., Varfolomeeva M.A., Lobov A.A., Tikanova P.O., Panova M., Mikhailova N.A., Granovitch A.I. Proteomic similarity of the Littorinid snails in the evolutionary context. PeerJ. 2020;8 doi: 10.7717/peerj.8546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolis L., Esch G., Holmes J., Kuris A., Schad G. The use of ecological terms in parasitology (Report of an Ad hoc Committee of the American Society of Parasitologists) J. Parasitol. 1982;68(1):131–133. [Google Scholar]
- Mayr E. Belknap Press of Harvard University Press; Cambridge, Massachusetts and London: 1970. Populations, Species and Evolution. An Abridgment of Animal Species and Evolution. [Google Scholar]
- McClain C.R., Rex M.A. The relationship between dissolved oxygen concentration and maximum size in deep-sea turrid gastropods: an application of quantile regression. Mar. Biol. 2001;139:681–685. [Google Scholar]
- Meyers L.A., Bull J.J. Fighting change with change: adaptive variation in an uncertain world. Trends Ecol. Evol. 2002;17(12):551–557. [Google Scholar]
- Mikhailova N.A., Gracheva YuA., Granovitch A.I. Analysis of the interspecific copulations frequency in the mating pairs of marine gastropod molluscs genus Littorina of “saxatilis” complex. Vestnik SPbU. Ser. Biol. 2008;4:5–9. (In Russian) [Google Scholar]
- Mikhailova N.A., Gracheva YuA., Backeljau T., Granovitch A.I. A potential species-specific molecular marker suggests interspecific hybridization between sibling species Littorina arcana and L. saxatilis (Mollusca, Caenogastropoda) in natural populations. Genetica. 2009;137(3):333–340. doi: 10.1007/s10709-009-9397-4. [DOI] [PubMed] [Google Scholar]
- Mineyev Y.N. Ural Branch of the Russian Academy of Science; Ekaterinburg: 2003. Anseriformes of the Eastern European Tundra. (In Russian) [Google Scholar]
- Muraeva O.A., Maltseva A.L., Varfolomeeva M.A., Mikhailova N.A., Granovitch A.I. Mild osmotic stress in intertidal gastropods Littorina saxatilis and Littorina obtusata (Mollusca: Caenogastropoda): a proteomic analysis. Biol. Commun. 2017;62(3):202–213. [Google Scholar]
- Naumov A.D., Fedyakov V.V. 1993. Ever-living the White Sea. St. Petersburg. (In Russian) [Google Scholar]
- Nosil P. Oxford University Press; Oxford: 2012. Ecological Speciation. [Google Scholar]
- Noskov G.A., Rymkevich T.A., Gaginskaya A.R., editors. Migrations of Birds of Northwest of Russia. Non-passerines. ANO LA “Professional”; St. Petersburg: 2016. (In Russian) [Google Scholar]
- Nylander J.A. Uppsala University; 2004. MrModeltest, Version 2. Program Distributed by the Author. Evolutionary Biology Centre. [Google Scholar]
- Parsons P.A. Cambridge University Press; Cambridge, New York: 1997. Extreme Environmental Change and Evolution. [Google Scholar]
- Pavlovsky E.N. The organism as the environment. Priroda. 1934;1:80–91. (In Russian) [Google Scholar]
- Pinheiro J.C., Bates D.M. Springer-Verlag New York; New York: 2000. Mixed-effects Models in Sand S-PLUS. Statistics and Computing. [Google Scholar]
- Plate T., Heiberger R. 2016. Abind: Combine Multidimensional Arrays. R Package Version 1.4-5.https://CRAN.R-project.org/package=abind [Google Scholar]
- Pohley W.J. Relationships among three species of Littorina and their larval digenea. Mar. Biol. 1976;37:179–186. [Google Scholar]
- Quinn G.P., Keough M.J. Cambridge University Press; New York: 2002. Experimental Design and Analysis for Biologists. [Google Scholar]
- R Core Team . R Foundation for Statistical Computing; Vienna, Austria: 2019. R: A Language and Environment for Statistical Computing.http://www.r-project.org/index.html 3.5.3. [Google Scholar]
- Rambaut A. Institute of Evolutionary Biology, University of Edinburgh; Edinburgh: 2009. FigTree, Version 1.4.3.http://tree.bio.ed.ac.uk [Google Scholar]
- Rambaut A., Drummond A.J., Xie D., Baele G., Suchard M.A. Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 2018;67(5):901–904. doi: 10.1093/sysbio/syy032. http://tree.bio.ed.ac.uk/software/tracer [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid D.G. The Ray Society; London: 1996. Systematics and Evolution of Littorina. [Google Scholar]
- Reid D.G., Dyal P., Williams S.T. A global molecular phylogeny of 147 periwinkle species (Gastropoda, Littorininae) Zool. Scripta. 2012;41(2):125–136. [Google Scholar]
- Rohlf F.J. Department of Ecology and Evolution, State University of New York at Stony Brook; New York: 2004. tpsDig, Digitize Landmarks and Outlines, Version 2.0.http://life.bio.sunysb.edu/morph [Google Scholar]
- Ronquist F., Teslenko M., Van Der Mark P., Ayres D.L., Darling A., Höhna S., Larget B., Liu L., Suchard M.A., Huelsenbeck J.P. Mrbayes 3.2: efficient bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012;61(3):539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharloo W. Canalization: genetic and developmental aspects. Annu. Rev. Ecol. Systemat. 1991;22:65–93. [Google Scholar]
- Schmalhausen I.I. University of Chicago Press; Chicago: 1949. Factors of Evolution. [Google Scholar]
- Scott D.A., Rose P.M. Wageningen Wetlands International; Wageningen: 1996. Atlas of Anatidae Populations in Africa and Western Eurasia. [Google Scholar]
- Selander R.K., Kaufman D.W. Genic variability and strategies of adaptation in animals. Proc. Natl. Acad. Sci. U.S.A. 1973;70(6):1875–1877. doi: 10.1073/pnas.70.6.1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semyenova S.K., Romanova E.A., Benediktov I.I., Ryskov A.P. Analysis of genetic variability of Fasciola hepatica using polymerase chain reaction with random primers. Genetica. 1995;31(2):273–275. (In Russian) [Google Scholar]
- Semyenova S.K., Morozova E.V., Chrisanfova G.G., Gorokhov V.V., Arkhipov I.A., Moskvin A.S., Movsessyan S.O., Ryskov A.P. Genetic differentiation in Eastern European and Western Asian populations of the liver fluke, Fasciola hepatica, as revealed by mitochondrial nad1 and cox1 genes. J. Parasitol. 2006;92(3):525–530. doi: 10.1645/GE-673R.1. [DOI] [PubMed] [Google Scholar]
- Sergievsky S.O., Granovitch A.I., Sokolova I.M. Long-term studies of Littorina obtusata and Littorina saxatilis populations in the White sea. Oceanol. Acta. 1997;20(1):259–265. [Google Scholar]
- Shingleton A.W. Evolution and the regulation of growth and body size. In: Flatt T., Heyland A., editors. Mechanisms of Life History Evolution. Oxford University Press; New York: 2011. pp. 43–55. [Google Scholar]
- Shklyarevich F.N. Common eider wintering in the White sea. In: Kistchinskii A.A., editor. Ecology and Morphology of Eiders in the USSR. Nauka; Moscow: 1979. pp. 61–67. (In Russian) [Google Scholar]
- Sithithaworn P., Nuchjungreed C., Srisawangwong T., Ando K., Petney T.N., Chilton N.B., Andrews R.H. Genetic variation in Opisthorchis viverrini (Trematoda: Opisthorchiidae) from northeast Thailand and Laos PDR based on random amplified polymorphic DNA analyses. Parasitol. Res. 2007;100(3):613–617. doi: 10.1007/s00436-006-0304-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soetaert K., Muthumbi A., Heip C. Size and shape of ocean margin nematodes: morphological diversity and depth-related patterns. Mar. Ecol. Prog. Ser. 2002;242:179–193. [Google Scholar]
- Sokolova I.M. Energy-limited tolerance to stress as a conceptual framework to integrate the effects of multiple stressors. Integr. Comp. Biol. 2013;53(4):597–608. doi: 10.1093/icb/ict028. [DOI] [PubMed] [Google Scholar]
- Sokolova I.M., Bock C., Pörtner H.O. Resistance to freshwater exposure in White Sea Littorina spp. I: anaerobic metabolism and energetics. J. Comp. Physiol. B. 2000;170(2):91–103. doi: 10.1007/s003600050264. [DOI] [PubMed] [Google Scholar]
- Stothard J.R., Webster B.L., Weber T., Nyakaana S., Webster J.P., Kazibwe F., Kabatereine N.B., Rollinson D. Molecular epidemiology of Schistosoma mansoni in Uganda: DNA barcoding reveals substantial genetic diversity within Lake Albert and Lake Victoria populations. Parasitology. 2009;136:1813–1824. doi: 10.1017/S003118200999031X. [DOI] [PubMed] [Google Scholar]
- Strom H. Birds of Svalbard. In: Kovacs K.M., Lydersen C., editors. Birds and Mammals of Svalbard. Norwegian Polar Institutt. Tromso; 2006. pp. 86–191. [Google Scholar]
- Swofford D.L. Sinauer Associates; Sunderland, Massachusetts: 2002. PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods). Version 4.https://paup.phylosolutions.com [Google Scholar]
- Tatarinkova I.P. Trans. Kandalaksha State Reserve. Vol. 8. 1970. Results of ringing of great black-backed and herring gulls on the Murman coast; pp. 149–181. (In Russian) [Google Scholar]
- Tatonova Y.V., Chelomina G.N., Besprozvannykh V.V. Genetic diversity of Clonorchis sinensis (Trematoda: Opisthorchiidae) in the Russian southern Far East based on mtDNA cox1 sequence variation. Folia Parasitol. 2013;60(2):155–162. doi: 10.14411/fp.2013.017. [DOI] [PubMed] [Google Scholar]
- Theron A., Combes C. Genetic analysis of cercarial emergence rhythms of Schistosoma mansoni. Behav. Genet. 1988;18(2):201–209. doi: 10.1007/BF01067842. [DOI] [PubMed] [Google Scholar]
- Thibert-Plante X., Hendry A.P. The consequences of phenotypic plasticity for ecological speciation. J. Evol. Biol. 2011;24(2):326–342. doi: 10.1111/j.1420-9101.2010.02169.x. [DOI] [PubMed] [Google Scholar]
- Todd M.E. Osmotic balance in Littorina littorea, L. littoralis, and L. saxatilis (Littorinidae) Physiol. Zool. 1964;37(1):33–44. [Google Scholar]
- Waddington C.H. Canalization of development and the inheritance of acquired characters. Nature. 1942;150:563–565. [Google Scholar]
- Webster J.P., Shrivastava J., Johnson P.J., Blair L. Is host-schistosome coevolution going anywhere? BMC Evol. Biol. 2007;7:1–11. doi: 10.1186/1471-2148-7-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir B., Cockerham C. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38(6):1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- Werding B. Morphologie, entwicklung und ökologie digener trematoden-larven der strandschnecke Littorina littorea. Mar. Biol. 1969;3:306–333. [Google Scholar]
- Wickham H. Springer International Publishing; New York: 2016. ggplot2: Elegant Graphics for Data Analysis. [Google Scholar]
- Wilke C.O. 2019. Cowplot: Streamlined Plot Theme and Plot Annotations for “Ggplot2”. R Package Version 0.9.99.https://github.com/wilkelab/cowplot [Google Scholar]
- Willmore K.E., Young N.M., Richtsmeier J.T. Phenotypic variability: its components, measurement and underlying developmental processes. Evol. Biol. 2007;34:99–120. [Google Scholar]
- Xie Y. 2019. Knitr: a General-Purpose Package for Dynamic Report Generation in R. R Package Version 1.22.https://CRAN.R-project.org/package=knitr [Google Scholar]
- Zink R.M., Barrowclough G.F. Mitochondrial DNA under siege in avian phylogeography. Mol. Ecol. 2008;17(9):2107–2121. doi: 10.1111/j.1365-294X.2008.03737.x. [DOI] [PubMed] [Google Scholar]
- Сrаmр S., Simmons К.E.L. Oxford University Press; Oxford: 1983. The Birds of the Wester Palearctic. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.