Abstract
In this study, we compared genome-wide transcriptome profile of two rice hybrids, one with (test hybrid IR79156A/IL50-13) and the other without (control hybrid IR79156A/KMR3) O. rufipogon introgressions to identify candidate genes related to grain yield in the test hybrid. IL50-13 (Chinsurah Nona2 IET21943) the male parent (restorer) used in the test hybrid, is an elite BC4F8 introgression line of KMR3 with O. rufipogon introgressions. We identified 2798 differentially expressed genes (DEGs) in flag leaf and 3706 DEGs in panicle. Overall, 78 DEGs were within the major yield QTL qyld2.1 and 25 within minor QTL qyld8.2. The DEGs were significantly (p < 0.05) enriched in starch synthesis, phenyl propanoid pathway, ubiquitin degradation and phytohormone related pathways in test hybrid compared to control hybrid. Sequence analysis of 136 DEGs from KMR3 and IL50-13 revealed 19 DEGs with SNP/InDel variations. Of the 19 DEGs only 6 showed both SNP and InDel variations in exon regions. Of these, two DEGs within qyld2.1, Phenylalanine ammonia- lyase (PAL) (Os02t0626400-01, OsPAL2) showed 184 SNPs and 11 InDel variations and Similar to phenylalanine ammonia- lyase (Os02t0627100-01, OsPAL4) showed 205 SNPs and 13 InDel variations. Both PAL genes within qyld2.1 and derived from O. rufipogon are high priority candidate genes for increasing grain yield in rice.
Subject terms: Gene expression profiling, Plant breeding
Introduction
Rice (Oryza sativa) is an important food crop providing 20% of daily calories to more than 50 percent of global population. Nearly 90% of rice is produced and consumed in Asia. The continuous increase in human population, especially in Asia, poses a major challenge to food security. Therefore, enhancing grain yield is the primary thrust area of plant breeders. Hybrid rice can help increase productivity by 10–20% more than conventional varieties1. Currently, the highest-yielding rice hybrids are developed from inter-subspecific crosses between indica and japonica2–5. Wild species have been used to breed parental lines for yield improvement in derived rice hybrids6–10.
Flag leaf is the most essential functional organ to produce a large proportion of photo-assimilates that are stored in grains11,12. It is estimated that it contributes around 32.3% of total carbohydrates during grain filling13. The panicle morphology also directly affects the number and size of seeds and also determines grain yield in rice14–16. Flag leaf and panicles have been used previously at different developmental stages to unravel gene expression in pollen development and genetic networks that control panicle branching and architecture17,18. The spatial and temporal expression profiles of genes during 19 vegetative and reproductive stages of organ development were analysed to identify stage-preferential/stage-specific genes in IR64 variety and anther-specific genes in Pusa Basmati1 variety19,20.
A whole-genome oligonucleotide microarray of super hybrid LYP9 (Liangyoupeijiu) and its parents 9311 and PA64s in 7 different tissues showed that differentially expressed genes for energy metabolism in first 3 stages of flag leaf and genes for transportation in next 3 stages of flag leaf were enriched in between the hybrid and parents rather than in between the parents21. Likewise, serial analysis of gene expression (SAGE) in roots, leaves and panicles of LYP9 showed that genes related to enhancing carbon-nitrogen assimilation pathways in leaves, nitrogen uptake in roots, protein biosynthesis and peptide transport were up-regulated in panicle of hybrids compared to respective tissues in parents22. SAGE analysis in leaves at grain filling stage of another super hybrid rice Liangyou-2186 and its parental lines SE21s and MH86 (Minghui86) showed differentially up-regulated genes related to photosynthesis and carbon fixation pathways in hybrid23. The whole-genome oligonucleotide microarray of flag leaves in three super-hybrid rice combinations LY2163 (SE21s x MH63), LY2186 (SE21s x MH86) and LYP9 (PA64s x 93-11) and their respective parental lines at flowering and grain filling stages showed that DEGs in all three super-hybrid combinations were significantly enriched in carbon fixation pathway, starch and sucrose metabolic pathway and flavonoid biosynthesis pathway24. The analysis showed heterotic gene RH8/DTH8/Ghd8/LHD1 is one of the loci contributing to yield heterosis in hybrid rice LYP925. Thus, gene expression in hybrids points to some relationship between DEGs in carbon fixation pathway and heterosis. However, these studies were carried out only in inter-subspecific hybrids and comparison was made between parents and hybrids. The higher yielding hybrids derived from improved restorer lines having introgressions from wild species have not been used for whole-genome expression profiling previously. In this study we compare transcriptome of two hybrids one with O. rufipogon introgressions in its restorer and the other without O. rufipogon introgressions in the restorer to know what effect the O. rufipogon introgressions in the restorer have on the derived hybrid.
In our previous work, we mapped two yield-enhancing QTLs (quantitative trait loci) qyld2.1 and qyld8.2 from BC2 testcross progeny from IR58025A/O. rufipogon//IR580325B///IR58025B////KMR326. IR58025A is a popular CMS line and KMR3 is restorer for the well adapted popular hybrid KRH2. The major yield enhancing QTL qyld2.1 from O. rufipogon was dissected into 8 subQTL regions27,28. A total of 67 hybrids were developed using 27 of KMR3/O. rufipogon elite introgression lines (ILs) as restorers and 6 CMS lines as recipients. The highest yielding top 11 of 67 hybrids which had introgressions from qyld2.1 showed 40.0–48.06 g yield per plant compared with respective control hybrids with 30.2–35.5.9 g9. An elite hybrid IR79156A/IL50-13 showed significantly high combining ability and standard heterosis for yield over popular hybrid DRRH29,10. The test hybrid IR79156A/IL50-13 (with O. rufipogon introgression) showed yield advantage of 32% over the control hybrid IR79156A/KMR3 (without O. rufipogon introgression) during kharif season 2012 at IIRR (Indian Institute of Rice Research) farm, Rajendranagar, Hyderabad.
In addition the test hybrid IR79156A × IL50-13 [IET 24441 (DRRH 102)] was tested in multilocation trials of All India Coordinated Rice Improvement Program (AICRIP) in 2014. It gave mean grain yield of 2.2 t/ha and showed 14% yield advantage over the best salinity check variety CSR10 and 24% over the yield check variety Jaya and was therefore promoted to advanced variety trial 1 - Coastal Saline Tolerant Variety Trial (AVT 1 -CSTVT)29. In moderate salinity, the hybrid ranked first (5796 kg/ha) in overall mean yield and showed 57.9% yield advantage over CSR10. However, in severe coastal saline stress it gave mean grain yield of only 2.1t/ha but still showed 59.4% yield advantage over CSR1030. So the test hybrid is demonstrated to be high yielding and moderately tolerant to coastal salinity stress in field conditions. The control hybrid was not tested in AICRIP trials.
We selected this elite hybrid as test hybrid for further functional validation of qyld2.1 and other heterotic loci derived from O. rufipogon associated with high yield and compared it with the respective control hybrid IR79156A/KMR3. This study was aimed to uncover the genes, molecular mechanisms and metabolic pathways which are involved in higher yield in test hybrid IR79156A/IL50-13 (with O. rufipogon introgressions) compared to control hybrid IR79156A/KMR3 (without O. rufipogon introgressions). Thus the difference is assumed to be due to heterotic loci from O. rufipogon introgressions.
Results
Transcriptome analysis
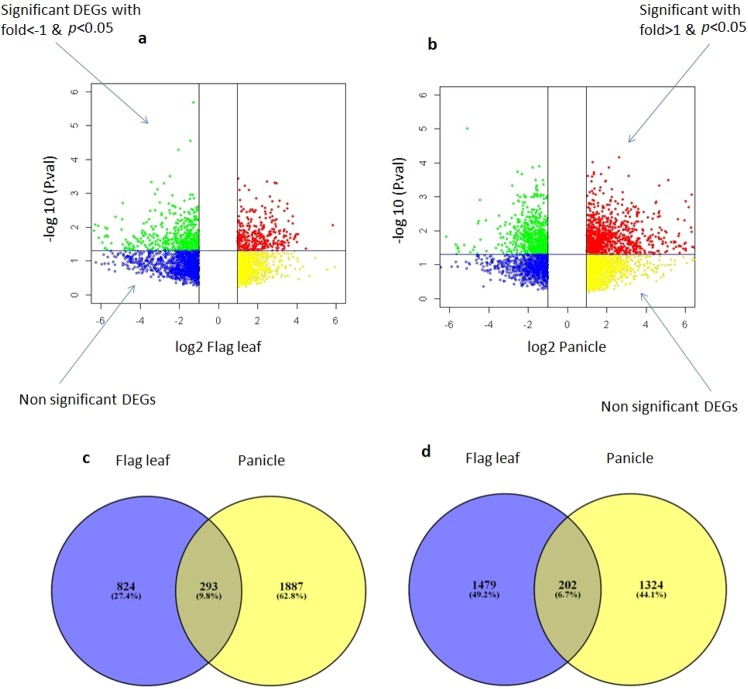
The transcriptome of flag leaf of test hybrid was compared with that of control hybrid to know the effect of O. rufipogon introgressions in the hybrid. Likewise the panicle of test hybrid was compared with that of control hybrid. In all, 1117 up-regulated and 1681 down-regulated differentially expressed genes (DEGs) were identified in flag leaf of test hybrid (IR79156A/IL50-13) compared to control hybrid (IR79156A/KMR3). Of these, 363 genes were significantly up-regulated and 458 were significantly down-regulated (p < 0.05). Similarly, 2180 up-regulated and 1526 down-regulated DEGs were identified in panicle of test hybrid compared to control hybrid. In panicle, 931 genes were significantly up-regulated and 647 were significantly down-regulated (Supplementary Table S1). The number of DEGs showing an expression change of >2 fold with a significance of p < 0.05 were lower in flag leaf compared to panicle (Fig. 1a). However, more genes showed up-regulation in panicle than in flag leaf of test hybrid (Fig. 1b). The maximum up-regulation was 5.96 fold (Os02t0616100 - similar to protein binding protein) in flag leaf and 8.21 fold (Os08t0474000 - similar to AP2 domain containing protein RAP2.6) in panicle. The maximum down- regulation was −8.7 fold (Os01t0538000 - conserved hypothetical protein) in flag leaf and −10.02 fold (Os01t0579000 - conserved hypothetical protein) in panicle of test hybrid. Since our focus was on the yield enhancing QTLs mapped previously, we first analyzed DEGs within qyld2.1 on chromosome 2 and qyld8.2 on chromosome 8 in more detail.
Figure 1.
Volcano plot showing gene expression differences in (a) flag leaf and (b) panicles of test hybrid IR79156A/IL50-13 with P-values and intensity ratios as log-scaled axes. Significant differences at p < 0.05 with >1-fold and <1 fold intensity ratios are shown. Blue: Fold <= −1 & p-value >0.05, Yellow: Fold >= 1 & p-value > 0.05, Green: Fold <= −1 & p-value <0.05, Red: Fold >= 1 & p-value <0.05. (c) Venn diagrams of up-regulated genes (d) down-regulated genes in flag leaf and panicle of test hybrid in comparison with control hybrid.
A Venn diagram was constructed to demonstrate common and exclusively expressed DEGs in flag leaf and panicle using total number of DEGs. Panicle had highest number (1887 = 86.5%) of exclusively up-regulated DEGs (Fig. 1c). On the other hand, flag leaf had highest number (1479 = 87.9%) of exclusively down-regulated DEGs (Fig. 1d). There were 293 up-regulated and 202 down-regulated DEGs common in flag leaf and panicle of test hybrid when compared to control hybrid. The details of DEGs with fold change are given in Supplementary Table S2.
DEGs within yield QTLs qyld2.1 and qyld8.2
Comparing the test hybrid with control hybrid, we found 78 DEGs within the major yield QTL qyld2.1 (5 Mb) and 25 DEGs within the minor yield QTL qyld8.2 (2 Mb). Of the 78 DEGs within qyld2.1, 12 DEGs were up-regulated and 21 were down-regulated in flag leaf while 29 DEGs were up-regulated and 16 were down-regulated in panicle (Supplementary Table S3). It is important to note that 3 DEGs within qyld2.1 - Os02t0596300-01 (Cytochrome P450 family protein), Os02t0618700 (Transmembrane receptor, eukaryota domain containing protein) and Os02t0571800-00 (Terpene synthase-like domain containing protein) were down-regulated in both flag leaf and panicle of test hybrid, whereas four DEGs Os02t0559800-01 (E3 ubiquitin ligase EL5), Os02t0624300-01 (Similar to Y19 protein), Os02t0629000-01 (Protein of unknown function DUF584 family protein) and Os02t0627100-01 (Similar to phenylalanine ammonia- lyase) were down-regulated in flag leaf but up-regulated in panicle. Only two DEGs Os02t0552700-01 (Zinc finger, CCHC-type domain containing protein) and Os02t0596200-01 (Glycoside hydrolase family 5 protein) showed up- regulation in both flag leaf and panicle of test hybrid. Similarly, analysis of 25 DEGs within qyld8.2 showed 5 DEGs were up-regulated and 4 DEGs were down-regulated in leaf, whereas 9 DEGs were up-regulated and 7 DEGs were down-regulated in panicle of test hybrid compared to control hybrid respectively. It is significant that 3 DEGs Os08t0468100-01, Os08t0468100-02 and Os08t0468100-03, all ‘Similar to nitrate reductase’ were up-regulated in both flag leaf and panicle of test hybrid.
Expression of DEGs associated with grain yield and abiotic stress
There were 52 transcription factors in flag leaf and 59 in panicle which showed more than 2- fold differential expression in test hybrid (Supplementary Table S4). The maximum up-regulation in flag leaf (3.8 fold) was shown by squamosa promoter binding protein-like transcription factor (Os08t0509600-01) and maximum up-regulation in panicle (8.2 fold) was shown by similar to AP2 domain containing protein RAP2.6 (Os08t0474000-01) within the minor yield QTL qyld8.2. It is interesting to note that genes known to be associated with abiotic stress tolerance eg. encoding salt-induced protein (Os01g0348900), similar to OsNAC6 protein (Os03g0815100), WRKY transcription factor 74 (Os09g0334500), late embryogenesis abundant protein 3 family protein (Os01g0314800) and similar to KUP related potassium transporter (Os08g0466200) were significantly up-regulated in panicle of test hybrid even though the plants were not exposed to any abiotic stress (Supplementary Table S5). It is likely that the higher expression of these genes in panicle contributes to its salinity tolerance and high yield in multilocation trials in coastal saline areas.
In addition, two DEGs Os06t0248300-03 (LOC_Os06g13850.1, Conserved hypothetical protein, F-box protein), Os01t0842500-01 (LOC_Os01g62490.1, Similar to laccase) were down-regulated in panicle and Os03t0150800-01 (LOC_Os03g05640.1, inorganic phosphate (Pi) transporter, Pi homeostasis, Selenite uptake) was up-regulated in panicle and down-regulated in flag leaf. These 3 genes known to be associated with plant architecture and grain yield were identified as microRNA target genes in test hybrid.
Functional annotation and classification of DEGs based on gene ontology (GO)
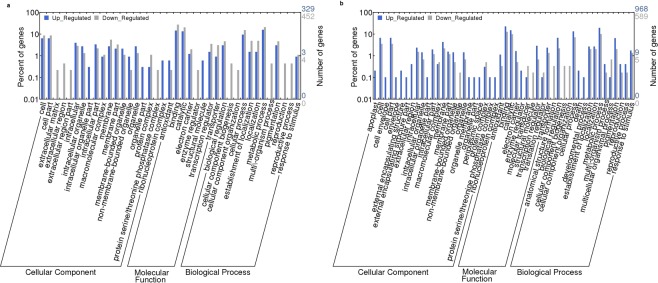
Gene ontology analysis was performed to classify the functional categories of DEGs in both tissues of test hybrid. There were 109 terms for up-regulated DEGs and 91 terms for down-regulated DEGs enriched in flag leaf (Supplementary Table S6). Similarly 143 terms for up-regulated DEGs and 111 for down-regulated DEGs were enriched in panicle of test hybrid (Supplementary Table S7). Out of these, 53 terms were expressed only in flag leaf and 93 terms were expressed only in panicle. There were 104 terms that were common to both flag leaf and panicle of test hybrid (Supplementary Table S8).
Further, the top GO terms that were significantly (p < 0.05) down-regulated in flag leaf were membrane-bounded vesicle (GO: 0031988, P = 0.0486) and cytoplasmic membrane-bounded vesicle (GO: 0016023, P = 0.0486) in cellular component category, while for molecular function category, catalytic activity (GO: 0003824, P = 0.0221) was most predominant term (Fig. 2a). Likewise, in panicle hemi cellulose metabolic process (GO: 0010410, P = 0.0202), xylan catabolic process (GO: 0045493, P = 0.0202), cell wall polysaccharide metabolic process (GO: 0010383, P = 0.0202), xylan metabolic process (GO: 0045491, P = 0.0202) and response to stress (GO: 0006950, P = 0.0584) were most significantly up-regulated terms in biological process category (Fig. 2b). This indicates that significantly over represented genes in this category play an extensive role in xylan and cell wall metabolic processes that may be involved in grain yield in test hybrid.
Figure 2.
Gene Ontology enrichment analysis of differentially expressed genes by WEGO (Web Gene Ontology Annotation Plot -http://wego.genomics.org.cn/cgi-bin/wego/index.pl). The P-value is below the significance level of p < 0.05. Percentage of enrichment is also shown (a) flag leaf (b) panicle.
KEGG (kyoto encyclopedia of genes and genomes) enrichment analysis
Pathway analysis was performed using KEGG database to understand the biological functions and enriched metabolic pathways of DEGs in test hybrid. The annotations revealed that 100 metabolic pathways (182 DEGs) were up-regulated and 196 pathways (473 DEGs) down-regulated in flag leaf (Supplementary Table S9). Likewise, 166 pathways (342 DEGs) were up-regulated and 121 pathways (229 DEGs) were down-regulated in panicle of test hybrid (Supplementary Table S10). There were 232 pathways common between leaf and panicle. Four major pathways - starch synthesis, ionic stress related calmodulin pathway, transcription factor related and proteasome related (E3 Ubiquitin ligase) showed high fold up-regulation in panicle but down-regulation in flag leaf of test hybrid compared to control hybrid.
Mapman analysis allowed the exploration of metabolic pathways which were activated specifically in test hybrid. The analysis revealed that the DEGs involved in major metabolic pathways such as carbohydrate metabolism, phenyl propanoid pathway, ubiquitin - dependent degradation and jasmonic acid pathways play a key role in contributing to grain yield in test hybrid. Since our interest was on grain yield in test hybrid as compared to control hybrid, we performed the Mapman analysis for DEGs within qyld2.1 (Supplementary Fig. S1a,b) and qyld8.2 (Supplementary Fig. S2a,b).
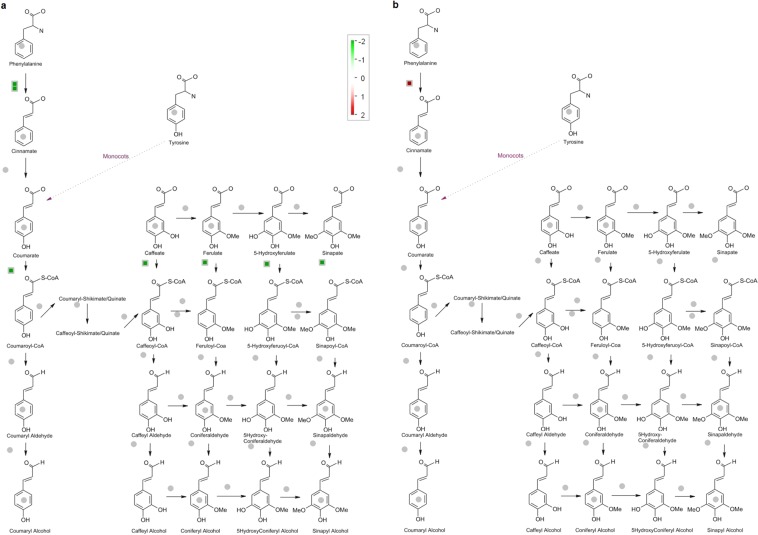
Carbohydrate metabolism overview revealed involvement of the down-regulated gene Os03t0401300-01 (Sucrose synthase 2) in sucrose-starch metabolism (Supplementary Fig. 3a), plant glycolysis, cell wall precursors and raffinose metabolism in flag leaf while in panicle, the up-regulated gene Os10t0465700-01 (Similar to beta amylase PCT-BMYI) which acts on α-1, 4 glycosidic bonds showed involvement in plant glycolysis and sucrose-starch metabolism (Supplementary Fig. 3b). Phenyl propanoid metabolism overview revealed that down-regulated DEGs Os02t0626400-01 (Phenyl alanine ammonia-lyase), Os02t0626100-01 (Similar to phenyl alanine ammonia-lyase), Os08t0498100-01 (Similar to caffeoyl coA methyl transferase 2) in flag leaf and up-regulated DEG Os08t0448000-01 (Similar to coumarate coA ligase 1) in panicle were involved in secondary metabolism such as biosynthesis of lignin, suberin, salicylate, piperidine and pyridine alkaloid, ubiquinone and other terpenoid-quinone and threonine (Fig. 3a,b). The ubiquitin metabolism overview clearly revealed the involvement of Os02t0559800-01 (E3 ubiquitin ligase) in protein ubiquitination. This is one of the important pathways for improving grain yield in rice and also regulates cell cycle. Notably this gene was down-regulated in flag leaf (Fig. 4a) and up-regulated in panicle (Fig. 4b). Jasmonic acid pathway analysis showed that Os12t0559200-01 (lipoxygenase-LOX) is down-regulated in flag leaf. It is involved in the conversion of linolenic acid to 13(S) hydroperoxylinolenic acid a precursor for jasmonic acid biosynthesis. The expression of LOX gene also depends on the functional activity of ubiquitin ligase which was down-regulated in flag leaf of test hybrid (Supplementary Fig. S4).
Figure 3.
Phenyl propanoid biosynthesis pathway in rice. The green squares represent down-regulation and red squares represent up-regulation in test hybrid (a) flag leaf (b) panicle.
Figure 4.
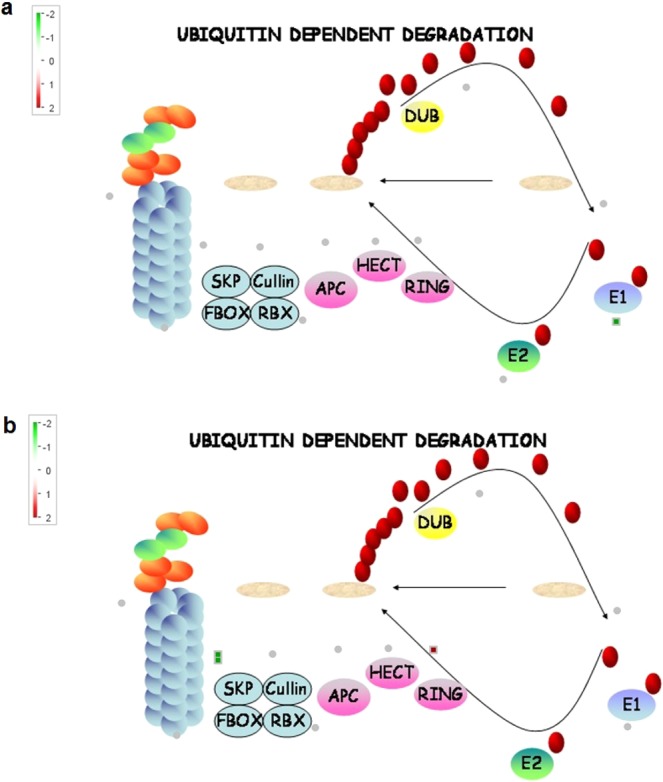
Schematic representation of the ubiquitination reaction involving a HECT-type E3. The green squares represent down-regulation and red squares represent up-regulation in test hybrid (a) flag leaf (b) panicle.
qRT-PCR validation of differentially expressed genes
A set of 22 DEGs were selected to validate the microarray results with quantitative reverse transcription PCR (qRT-PCR) including 5 genes within qyld2.1, five genes within qyld8.2, and 12 DEGs associated with yield but located on other chromosomes (Table 1). All 22 DEGs showed reproducibility and were significantly correlated with the microarray data. In these 22 genes, 8 were differentially down-regulated and one gene was up-regulated only in flag leaf (Supplementary Fig. S5a), 9 genes were differentially up-regulated in panicle (Supplementary Fig. 5b) and 4 genes showed differential expression in both flag leaf and panicle (Supplementary Fig. 5c). The degree of expression was higher in panicle than in flag leaf of test hybrid. The expression patterns of these 22 yield related genes were in good agreement with the quantification of transcripts done through microarray, though the degree of expression varied.
Table 1.
Primer sequences of 22 selected genes used for qRT-PCR.
| S.No | Locus Id | Gene Name | Primer sequence (5′-3′) |
|---|---|---|---|
| 1 |
Os02t0559800-01 chr02:21250083..21251343 |
E3 ubiquitin ligase EL5 |
F: GTGATCGAGATCCCCGAAT R: CCAGAGCCTTCTCAGTGACC |
| 2 |
Os02t0624300-01 chr02:24878777..24879932 |
Similar to Y19 protein |
F: TCCGGTGGATCAACTACCTC R: GTTGCCGAGAAGGTCGTG |
| 3 |
Os08t0482600-01 chr08:23853339..23854314 |
Cupredoxin domain containing protein. |
F: TCTGATCGCAATGCTCCTC R: CGTTAGCCTCGAAGGTTTTG |
| 4 |
Os08t0473900-03 chr08:23341357..23343240 |
Similar to Alpha-amylase isozyme 3D |
F: AGCTTGTGTTTGCTGCTCCT R: CTGGAAGAGGACCTGTGCTT |
| 5 |
Os08t0473900-01 chr08:23341289..23343273 |
Alpha-amylase isozyme 3D precursor (EC 3.2.1.1) (1,4-alpha-D-glucan glucanohydrolase) |
F: GTGAAGATCGGGACGAGGTA R: CCTTCTCCCAGACGCTGTAG |
| 6 |
Os08t0466200-01 chr08:22904756..22908287 |
Similar to KUP-related potassium transporter |
F: CTTCAGCAAGAGGTCCATCC R: GTTGATCAGGTACGCCGTCT |
| 7 |
Os02t0569900-01 chr02:21740304..21741960 |
Cytochrome P450 family protein-1500bp |
F: ACACGTTCGAGTGGAAGCTC R: CAAGCACATTGGCAGACTTG |
| 8 |
Os02t0626400-01 chr02:24985294..24989388 |
Phenylalanine ammonia-lyase (EC 4.3.1.5) |
F: ACCACCTGACACACAAGCTG R: TACGAGCTGCCTTCCAAGAT |
| 9 | Os03t0753100-01 chr03:31048351..31055017 | MADS-box transcription factor, Inflorescence and spikelet development |
F: GATCGACGTAGAGGCAGCTC R: GGAGGCTCACTGGAAAACAC |
| 10 | Os10t0531400-01 chr10:20645931..20646880 | Glutathione S-transferase GST 30 (EC 2.5.1.18) |
F: GAAGCTACTGGGGATGTGGA R: CTCGTACGGCAGTGACTTGA |
| 11 | Os03t0401300-01 chr03:16301279..16306089 | Sucrose synthase 2 (EC 2.4.1.13) (Sucrose-UDP glucosyltransferase 2) |
F: GCTGAAGGACAGGAACAAGC R: CAGCTCAACCAGACCAGTCA |
| 12 | Os01t0348900-01 chr01:13903285..13904626 | SalT gene product (Salt-induced protein) |
F: CTGGAGTCCCAAATGGAAAG R: CGTTCCAGACCTTCCAAAGA |
| 13 | Os01t0314800-01 chr01:11863682..11864412 | Late embryogenesis abundant protein 3 family protein. |
F: GGCGGACGAGAAGAAGGT R: GCCGGTAGTACCCGGTCA |
| 14 | Os03t0815100-01 chr03:34166100..34167521 | Similar to OsNAC6 protein |
F: GATGATGGTGCCCAAGAAAG R: AACAGGCTGCTGTTGTTCCT |
| 15 | Os04t0180400-01 chr04:5484865..5486703 | Similar to Cytochrome P450 99A2 |
F: GCTCCTACCCAAAGCTGATG R: CATTATCCGGGGACAAACAT |
| 16 | Os09t0334500-01 chr09:10128837..10131086 | WRKY transcription factor 74 |
F: ACGGTGTTCGACGTGATCTA R: CGTGTCCGTCTCCGTCTC |
| 17 | Os12t0559200-01 chr12:22854749..22860198 | Lipoxygenase (EC 1.13.11.12) |
F: CTCCATCAAGGAGTGGGTGT R: CTGGAGCTCCTTGTCCATGT |
| 18 | Os08t0498100-01 chr08:24586613..24587908 | Similar to Caffeoyl-CoA O-methyltransferase 2 (EC 2.1.1.104) |
F: ATCGAGGTGGGTGTCTTCAC R: TCTCGTAGCTCTCCCTGTCC |
| 19 | Os02t0626100-01 chr02:24973450..24977287 | Similar to Phenylalanine ammonia-lyase |
F: AGCGAGTGGATCCTCAACTG R: CTGAGGAGCTCGACTTGGA |
| 20 | Os10t0465700-01 chr10:17180759..17183189 | Similar to Beta-amylase PCT-BMYI (EC 3.2.1.2) |
F: GTGGTGGAGGAGATGGAGAA R: CCGAGGGAGATGTACTCGAA |
| 21 | Os08t0448000-01 chr08:21873076..21875497 | Similar to 4 coumarate coA ligase1 |
F: TCCACGTCTACTCCCTCCAC R: GCAGCATCTTGACGGTGTC |
| 22 | Os02t0738200-01 chr02:30805988..30807610 | Zinc finger, RING/FYVE/PHD-type domain containing protein |
F: AGCTGCTCAGGCTTCTCAAC R: GTCTCTTCAGCCCCTTGAAA |
| 23 | OsACT1 Lee et al. (2011) | actin |
F: TGGAAGCTGCGGGTATCCAT R: TACTCAGCCTTGGCAATCsCACA |
Genes underlying yield QTLs qyld2.1 and qyld8.2 are shown in bold.
Annotation of 22 validated DEGs
The KEGG annotation of 22 DEGs used for qRT-PCR validation showed 11 DEGs were involved in various metabolic pathways in test hybrid (Table 2). These included, 4 DEGs (Os08t0473900-03, Os08t0473900-01, Os03t0401300-01, Os10t0465700-01) related to sucrose and starch metabolism, 4 DEGs (Os02t0626400-01, Os02t0626100-01, Os08t0498100-01, Os08t0498100-01) related to phenyl propanoid biosynthesis, JA biosynthesis pathway gene (Os12t0559200-01), momilactone biosynthesis gene (Os04t0180400-01), and glutathione mediated detoxification pathway gene (Os10t0531400-01). In these, Os02t0626400-01 (Phenyl alanine ammonia- lyase) within qyld2.1 is involved in phenyl propanoid biosynthesis, suberin synthesis and salicylate biosynthesis pathways. Two DEGs Os08t0473900-03 and Os08t0473900-01 (Similar to alpha-amylase isozyme 3D and alpha-amylase isozyme 3D precursor (1,4-alpha-D-glucan glucanohydrolase) within qyld8.2 are involved in starch degradation.
Table 2.
The expression pattern of 22 DEGs validated through qRT-PCR and KEGG pathway analysis results in test hybrid, ↓ denotes down-regulation and ↑ denotes up-regulation.
| S. No | Gene name | Description | Microarray | qRT-PCR | KEGG pathway analysis | ||
|---|---|---|---|---|---|---|---|
| Leaf | Panicle | Leaf | Panicle | ||||
| 1 | Os02t0559800-01 | E3 ubiquitin ligase EL5 | ↓ | ↑ | ↓ | ↑ | — |
| 2 | Os02t0624300-01 | Similar to Y19 protein | ↓ | ↑ | ↓ | ↑ | — |
| 3 | Os08t0482600-01 | Cupredoxin domain containing protein. | ↑ | ↑ | |||
| 4 | Os08t0473900-03 | Similar to Alpha-amylase isozyme 3D | ↑ | ↑ | Starch degradation | ||
| 5 | Os08t0473900-01 | Alpha-amylase isozyme 3D precursor | ↑ | ↑ | Starch degradation | ||
| 6 | Os08t0466200-01 | Similar to KUP-related potassium transporter. | ↑ | ↑ | |||
| 7 | Os02t0569900-01 | Cytochrome P450 family protein-1500bp | ↓ | ↓ | |||
| 8 | Os02t0626400-01 | Phenylalanine ammonia-lyase | ↓ | ↓ | Suberin biosynthesis, phenyl propanoid bio synthesis and initial reactions, salycilate biosynthesis | ||
| 9 | Os03t0753100-01 | MADS-box transcription factor, Inflorescence and spikelet development | ↑ | ↑ | |||
| 10 | Os10t0531400-01 | Glutathione S-transferase GST 30 | ↓ | ↓ | Glutathion mediated detoxification | ||
| 11 | Os03t0401300-01 | Sucrose synthase 2 | ↓ | ↓ | Sucrose degradation to ethanol and lactate (anaerobic), galactose degradation II, starch and sucrose metabolism, sucrose degradation III, sucrose biosynthesis | ||
| 12 | Os01t0348900-01 | SalT gene product (Salt-induced protein). | ↑ | ↑ | |||
| 13 | Os01t0314800-01 | Late embryogenesis abundant protein 3 family protein | ↑ | ↑ | |||
| 14 | Os03t0815100-01 | Similar to OsNAC6 protein | ↑ | ↑ | |||
| 15 | Os04t0180400-01 | Similar to Cytochrome P450 99A2 | ↓ | ↑ | ↓ | ↑ | Momilactone biosynthesis, nicotine degradation II, nicotine degradation III, bupropion degradation, acetone degradation to methyl glyoxal |
| 16 | Os09t0334500-01 | WRKY transcription factor 74. | ↑ | ↑ | |||
| 17 | Os12t0559200-01 | Lipoxygenase | ↓ | ↓ | Biosynthesis of plant harmones, JA biosynthesis, alpha-lenolenic acid metabolism, 13-LOX and 13 HPL pathway, divinyl ether biosynthesis II, lenoleic acid metabolism | ||
| 18 | Os08t0498100-01 | Similar to Caffeoyl-CoA O-methyltransferase 2 | ↓ | ↓ | Phenyl propanoid biosynthesis, suberin biosynthesis | ||
| 19 | Os02t0626100-01 | Similar to Phenylalanine ammonia-lyase. | ↓ | ↓ | Phenyl propanoid biosynthesis, suberin biosynthesis, phenyl alanine metabolism, phenyl propanoid biosynthesis initial reactions, salicylate biosynthesis, Tropane, piperidine and pyridine alkaloid biosynthesis, nitrogen metabolism | ||
| 20 | Os08t0448000-01 | Similar to 4-coumarate-CoA ligase 1 | ↑ | ↑ | Biosynthesis of phenyl propanoids, Biosynthesis of alkaloids derived from shikimate pathway, phenyl propanoid biosynthesis, metheonine biosynthesis II, Ubiquinone and other terpenoid-quinone biosynthesis, threonine biosynthesis from homoserine | ||
| 21 | Os10t0465700-01 | Similar to Beta-amylase PCT-BMYI | ↓ | ↑ | ↓ | ↑ | starch degradation |
| 22 | Os02t0738200-01 | Zinc finger, RING/FYVE/PHD-type domain containing protein. | ↓ | ↓ | |||
In panicle, two DEGs within qyld8.2 were up-regulated. Os08t0473900-03 (Similar to alpha-amylase isozyme 3D) was classified in metabolic process, carbohydrate metabolic process and primary metabolic process in biological process terms, and hydrolase activity, hydrolyizing O-glycosyl compounds, acting on glycosyl bonds, binding, ion binding, metal ion binding, cation binding, calcium ion binding, catalytic activity, amylase activity, alpha-amylase activity were enriched terms in molecular function category. Another DEG Os08t0482600-01 (Cupredoxin domain containing protein) was classified in cation binding activity, transition metal ion binding, electron carrier activity and copper ion binding activity which were enriched terms in molecular function category.
Sequence polymorphism of DEGs and other reported yield-related genes (not DEGs) between the restorer lines KMR3 and IL50-13 used for producing hybrids
Since the genome sequences of restorer lines KMR3 and IL50-13 were available (unpublished), they were compared only for DEGs within qyld2.1 (78 DEGs), qyld8.2 (25 DEGs) and 12 other DEGs based on significance (p < 0.05) and fold change value above 2. In addition, we also investigated the sequence differences of 21 reported yield-related genes between KMR3 and IL50-13 on different chromosomes. Out of 136 genes analysed, 117 sequences were identical between the two genomes (KMR3 and IL50-13) and only 19 genes (16 DEGs and 3 yield-related genes) showed differences between KMR3 and IL50-13 in terms of SNPs (single nucleotide polymorphism) and InDels (insertion and deletion polymorphism) (Supplementary Table 11). Of the 19 genes, one DEG Os08t0468100-03 (Similar to nitrate reductase) had unique InDels and 9 DEGs had unique SNPs and the other 9 genes showed both SNPs and InDel variations (Table 3). We obtained a total of 1073 variants in the 19 genes, of which 959 were SNPs and 114 were InDels (Table 4). The maximum number of variants were observed for the gene Os02t0592000-00, which is a similar to OSIGBa0106G07.8 protein and there were seven genes that had only one SNP/InDel variation. We compared the SNPs and InDels present in different regions (coding DNA sequence CDS (exons), untranslated regions UTRs (5′ and 3′UTRs) and introns) of these 19 genes. Of the 959 SNPs in 19 genes, CDS had the maximum number of SNPs (541) followed by introns (413) and the least number was found in UTRs (5). But considering 114 InDels found in these 19 genes, introns had the maximum number of SNPs (77) followed by CDS (36) and UTRs had the least number (1) of SNPs. Six DEGs Os02t0626400-03 (84), Os02t0569800-00 (100), Os02t0616600-01 (108), Os02t0592000-00 (195), Os02t0626400-01 (184) and Os02t0627100-01 (205) within qyld2.1 showed high number of SNP variations in IL50-13 compared to KMR3 (Table 4). The study of SNPs in coding region and 2 kb upstream/downstream of 19 genes (16 DEGs and 3 yield reported genes) identified only one gene Os02t0831500-01 (Similar to sucrose synthase) which showed differences in terms of SNPs. We obtained three unique SNPs in the upstream region of Os02t0831500-01 in IL50-13 that showed a modifying effect on the gene (Supplementary Table S12).
Table 3.
DEGs and reported yield related genes showing difference between the sequences of KMR3 and IL50-13.
| S. No | Category | Gene Name | Description | Gene length | KMR3 scaffold | Start position of scaffold | End position of scaffold | 50-13 scaffold | Start position of scaffold | End position of scaffold | Differences between KMR3 and 50-13 scaffolds |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | A28 | Os02t0589000-01 | Lecithin: cholesterol acyltransferase family protein | 10391 | scaffold9446_size10065 | 984 | 3580 | scaffold14856_size6220 | 1 | 2790 | SNPs + InDels |
| 2 | A61 | Os02t0626400-03 | Similar to Phenylalanine ammonia-lyase | 1589 | scaffold13385_size7025 | 3655 | 4920 | scaffold12144_size7524 | 3315 | 2050 | SNPs + InDels |
| 3 | A11 | Os02t0569800-00 | Hypothetical genes | 4024 | scaffold2303_size25555 | 19748 | 21711 | scaffold17631_size5156 | 2282 | 3988 | SNPs + InDels |
| 4 | A51 | Os02t0616600-01 | Conserved hypothetical protein | 3728 | scaffold15492_size5752 | 3500 | 5752 | scaffold26405_size2842 | 474 | 2842 | SNPs + InDels |
| 5 | A32 | Os02t0592000-00 | Similar to OSIGBa0106G07.8 protein | 9817 | scaffold8341_size11245 | 7965 | 10401 | scaffold17585_size5169 | 1236 | 2636 | SNPs + InDels |
| 6 | A16 | Os02t0574800-01 | Ethylene insensitive 3 domain containing protein | 739 | scaffold14052_size6604 | 4 | 276 | scaffold21804_size3915 | 3915 | 3667 | SNPs |
| 7 | A1 | Os02t0552500-00 | 1185 | scaffold23485_size2774 | 2774 | 1848 | scaffold30709_size2103 | 415 | 1 | SNPs | |
| 8 | A12 | Os02t0569900-01 | Cytochrome P450 family protein | 1657 | scaffold7129_size12786 | 9197 | 10493 | scaffold24827_size3169 | 3169 | 2828 | SNPs |
| 9 | A62 | Os02t0626400-01 | Phenylalanine ammonia-lyase (EC 4.3.1.5) | 2593 | scaffold13385_size7025 | 2826 | 4920 | scaffold12144_size7524 | 4141 | 2050 | SNPs + InDels |
| 10 | A63 | Os02t0627100-01 | Similar to Phenylalanine ammonia-lyase (EC 4.3.1.5) | 2405 | scaffold7515_size12279 | 1251 | 3210 | scaffold12144_size7524 | 4200 | 2051 | SNPs + InDels |
| 11 | B8 | Os08t0468100-03 | Similar to Nitrate reductase. | 2053 | scaffold32698_size2633 | 1589 | 1 | scaffold5531_size12871 | 3714 | 1775 | InDels |
| 12 | B21 | Os08t0487301-00 | - | 485 | scaffold243_size48649 | 4120 | 3822 | scaffold6702_size11543 | 11094 | 11491 | SNPs + InDels |
| 13 | B23 | Os08t0490100-01 | Similar to PBF protein | 1261 | scaffold8993_size10535 | 8750 | 9323 | scaffold3687_size21453 | 15734 | 15885 | SNPs + InDels |
| 14 | C1 | Os01t0314800-01 | Late embryogenesis abundant protein 3 family protein. | 731 | scaffold2182_size25019 | 20065 | 19335 | scaffold17822_size5094 | 3714 | 4149 | SNPs |
| 15 | C4 | Os02t0831500-01 | Similar to Sucrose synthase | 6271 | scaffold8577_size10987 | 1 | 2889 | scaffold5992_size12325 | 12325 | 9137 | SNPs |
| 16 | C10 | Os06t0229800-01 | Similar to Starch synthase IIA. | 4905 | scaffold1456_size29059 | 29059 | 24569 | scaffold8945_size9534 | 1 | 3623 | SNPs |
| 17 | D6 | Os03t0407400-00 | - | 945 | scaffold14776_size6169 | 2241 | 1683 | scaffold6119_size12184 | 6218 | 6724 | SNPs |
| 18 | D8 | Os03t0117900-01 | - | 955 | scaffold9265_size10248 | 5473 | 6424 | scaffold21042_size4123 | 1 | 446 | SNPs |
| 19 | D19 | Os08t0509600-01 | Rice squamosa promoter binding protein-like 14. | 4156 | scaffold3452_size19961 | 487 | 4642 | scaffold2693_size19548 | 108 | 3677 | SNPs |
Category A refers to genes within qyld2.1, B refers to genes within qyld8.2, C refers to genes showing high fold expression and D refers to reported yield related genes.
Table 4.
Polymorphism observed in terms of SNPs and InDels between sequences of KMR3 and IL50-13 for the DEGs and reported yield related genes.
| S. No. | Category | Gene Name | No. of variants | No. of SNPs | No. of SNPs (CDS) | No. of SNPs (UTRs) | No. of SNPs (Introns) | No.of InDels | No.of InDels (CDS) | No.of InDels (UTRs) | No.of InDels (Introns) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | A28 | Os02t0589000-01 | 13 | 10 | 0 | 0 | 10 | 3 | 0 | 0 | 3 |
| 2 | A61 | Os02t0626400-03 | 86 | 84 | 84 | 0 | 0 | 2 | 2 | 0 | 0 |
| 3 | A11 | Os02t0569800-00 | 118 | 100 | 4 | 0 | 96 | 18 | 0 | 0 | 18 |
| 4 | A51 | Os02t0616600-01 | 123 | 108 | 0 | 5 | 103 | 15 | 0 | 1 | 14 |
| 5 | A32 | Os02t0592000-00 | 230 | 195 | 0 | 0 | 195 | 35 | 0 | 0 | 35 |
| 6 | A16 | Os02t0574800-01 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| 7 | A1 | Os02t0552500-00 | 3 | 3 | 3 | 0 | 0 | 0 | 0 | 0 | 0 |
| 8 | A12 | Os02t0569900-01 | 36 | 34 | 34 | 0 | 0 | 2 | 2 | 0 | 0 |
| 9 | A62 | Os02t0626400-01 | 195 | 184 | 184 | 0 | 0 | 11 | 11 | 0 | 0 |
| 10 | A63 | Os02t0627100-01 | 218 | 205 | 205 | 0 | 0 | 13 | 13 | 0 | 0 |
| 11 | B8 | Os08t0468100-03 | 7 | 1 | 0 | 0 | 1 | 6 | 0 | 0 | 6 |
| 12 | B21 | Os08t0487301-00 | 11 | 9 | 5 | 0 | 4 | 2 | 2 | 0 | 0 |
| 13 | B23 | Os08t0490100-01 | 23 | 17 | 17 | 0 | 0 | 6 | 6 | 0 | 0 |
| 14 | C1 | Os01t0314800-01 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| 15 | C4 | Os02t0831500-01 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 |
| 16 | C10 | Os06t0229800-01 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| 17 | D6 | Os03t0407400-00 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| 18 | D8 | Os03t0117900-01 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| 19 | D19 | Os08t0509600-01 | 4 | 4 | 0 | 0 | 4 | 0 | 0 | 0 | 0 |
| Total | 1073 | 959 | 541 | 5 | 413 | 114 | 36 | 1 | 77 |
Category A refers to genes within qyld2.1, B refers to genes within qyld8.2, C refers to genes showing high fold expression and D refers to reported yield related genes. The candidate genes (PAL) showing maximum SNPs in CDS and within qyld2.1 are shown in bold.
Discussion
To explore the genes and mechanisms involved in heterosis for yield in test hybrid, we profiled the genome-wide transcriptomes of flag leaves and young panicles in test hybrid IR79156A/IL50-13 and control hybrid IR79156A/KMR3. Both hybrids were developed using same cytoplasmic male sterile (CMS) line IR79156A, but two cognate restorer lines one with O. rufipogon introgression (IL50-13) and other without O. rufipogon introgression (KMR3). Such a comparative analysis of hybrids has not been made before. Our previous work showed that the hybrid IR79156A/IL50-13 gave significantly high yield 43 g/plant and showed highest specific combining ability (SCA) for yield (6.04) among 36 hybrids and high standard heterosis over KRH2 (61.79%) and DRRH2 (50.23%) indicating predominance of non-additive gene action for grain yield heterosis in test hybrid9,10. Several studies showed that non-additive gene action is important for yield and yield related traits in rice hybrids31–33. Similar results were obtained for yield heterosis in maize hybrids34–36. Based on the above analysis, we hypothesised that this yield heterosis might be due to the new genetic variation introduced from O. rufipogon via the restorer line and which enhances/regulates the yield potential of test hybrid.
Since, the restorer line IL50-13 is derived from an interspecific cross of KMR3/O. rufipogon, we focussed primarily on the DEGs within major yield QTL qyld2.1 and minor effect QTL qyld8.2 reported previously26. We indeed found 78 DEGs within qyld2.1 and 25 DEGs within qyld8.2 (Supplementary Table S3). The prominent DEGs within qyld2.1 were E3 ubiquitin ligases, phenylalanine ammonia-lyases and cytochrome P450 family proteins and DEGs within qyld8.2 were alpha-amylases, nitrate reductases and similar to KUP related potassium ion transporters. Interestingly, the highest (5.96) fold up-regulated gene Os02t0616100-01 (Similar to protein binding protein) in flag leaf lies within qyld2.1, and the highest (8.21) fold up-regulated DEG in panicle Os08t0474000-01 (Similar to AP2 domain containing protein RAP2.6) lies within qyld8.2. These two DEGs are worth further study. However, the GO enrichment analysis revealed cytoplasmic membrane-bound vesicle and catalytic activity are most significant terms in flag leaf whereas, hemi cellulose metabolic process, cell wall polysaccharide metabolic process, xylan metabolic process, antioxidant and response to stress are most significant terms in panicle of test hybrid (Supplementary Tables S6 and S7). Thus these processes are associated with grain yield and tolerance to salt stress (since the hybrid gave high yield in coastal salinity). Starch synthesis, phenyl propanoid pathway, ubiquitin dependent degradation and phytohormone related pathways were highly enriched pathways in test hybrid according to KEGG analysis (Supplementary Tables S9 and S10). The transcriptomic analysis of super hybrid rice LYP9 and its parents 93-11 and PA64s showed that the genes in the categories of energy metabolism and transport are enriched in between the hybrid and its parents rather than in between the parents21. Similarly the comparative transcriptional profile of three super hybrids LY2163, LY2186, LYP9 showed the DEGs were significntly (p < 0.01) enriched in carbon fixation pathway in all 3 super-hybrid combinations compared to its parents24.
We found some genes related to salinity tolerance and other abiotic stresses were also differentially expressed in the test hybrid even though the plants were not exposed to any abiotic stress. It is pertinent to note that the restorer IL50-13 (IET21943, RPBio4919-50-13-CN2079, IC616879) was released as Chinsurah Nona2 (Gosaba 6) in 2016 for coastal saline areas of West Bengal state of India and notified by central sub-committee on crop standards notification release of varieties in 2019. It gave mean yield 2.8 t/h during 2010-2013 in 4 years multi-location testing of AICRIP (All India Coordinated Rice Improvement Project)37. Its yield was similar to that of KMR3 under normal non stress conditions, but in salinity stress IL50-13 showed both seedling stage and reproductive stage tolerance and grain yield was not affected even at 150 mM NaCl38,39. Likewise, it also showed drought tolerance under direct seeded conditions40. The test hybrid using this restorer line IR79156A/IL50-13 (with O. rufipogon introgressions) also showed higher yield than salinity check CSR10 and gave mean grain yield of 5t/h in advanced varietal trial of AICRIP29. The genome of IL50-13 and parent line KMR3 were re-sequenced at CCMB (i-Life Discoveries Ltd, data information available, seq deposited at DDBJ at DDBJ/ENA/GenBank under accessions LVCG00000000 for KMR3 and LVCH00000000 for IL50-13).
In this study we explored the link between the DEGs within the yield QTLs, their sequence difference and grain yield. Five DEGs Os02t0559800-01 (E3 ubiquitin ligase EL5), Os02t0624300-01 (Similar to Y19 protein), Os02t0569900-01 (Cytochrome P450 family protein) and Os02t0626400-01 (Phenylalanine ammonia-lyase), Os02t0626100-01 (Similar to phenylalanine ammonia-lyase) within qyld2.1 and 5 DEGs Os08t0482600-01 (Cupredoxin domain containing protein), Os08t0473900-03 (Similar to alpha-amylase isozyme 3D), Os08t0473900-01 (Alpha-amylase isozyme 3D precursor) and Os08t0466200-01 (Similar to KUP-related potassium transporter), Os08t0448000-01 (Similar to 4 coumarate CoA ligase1) within qyld8.2 were validated using qRT-PCR (Supplementary Fig. S5). In addition, 12 other DEGs were also validated and results were consistent with microarray results. The DEGs within qyld2.1 were mostly down-regulated in flag leaf whereas the DEGs within qyld8.2 were up-regulated in panicle of test hybrid.
Further, we found that only 19 DEGs out of 136 showed sequence differences between the restorer lines KMR3 and IL50-13. Of the 19 DEGs only 6 showed SNP/InDel variations in CDS region and these 6 DEGs also showed high fold change in test hybrid compared to control hybrid. In accordance with these results the candidate genes within qyld2.1 were mainly Os02t0626400-03 (Phenylalanine ammonia-lyase), Os02t0626400-01 (Phenyl alanine ammonia-lyase) and Os02t0627100-01 (Similar to PAL). Os02t0626400-03 showing 84 SNPs and 2 InDels, Os02t0626400-01 showing 184 SNPs and 11 InDels and Os02t0627100-01 showing 205 SNPs and 13 InDel variations in CDS regions (Supplementary Table S11). Another DEG within qyld2.1 Os02t0569900-01 (Cytochrome P450 family protein) showed 34 SNPs and 2 InDel variations in CDS region. These 4 DEGs are involved in suberin biosynthesis, phenyl propanoid biosynthesis and initial reactions in salicylate biosynthesis pathways. On the other hand 2 DEGs within qyld8.2 were Os08t0487301-00 (Conserved hypothetical protein) showing 5 SNPs and 2 InDels and Os08t0490100-01 (Similar to PBF protein) showing 17 SNPs and 6 InDel variations in CDS regions. In addition, Os08t0509600-01 (OsSPL14-Squamosa promoter-binding-like transcription factor) near qyld8.2 also showed 4 SNP variations in intron regions between KMR3 and IL50-13 but not in exons (Table 4). However, these SNP/InDel variations in introns, UTR regions and CDS regions may not change the function of the final protein, that were involved in metabolic processes, but as modifiers may significantly affect the expression of genes and translation. Alternatively, it is also possible that variations in a promoter sequence acting in trans and away from CDS region confers differential gene expression. But we only considered the genome sequence in the 2 kb upstream and 2 kb downstream of the 136 target genes/DEGs. In general, OsSPL14 regulates primary panicle branching and vegetative shoot branching. A single point mutation in the micro RNA OsmiR156-targeted site in the third exon of OsSPL14 leads to generation of ideal plant architecture with low tiller number at vegetative stage, lodging resistance and increased grain yield41,42.
Interestingly Os06t0248300-03 (conserved hypothetical protein, OsFbox protein 305) on chromosome 6, Os01t0842500-01 (Similar to laccase) on chromosome 1 and Os03t0150800-01 (Pi transporter, Pi homeostasis, Selenite uptake, OsPT2) on chromosome 3 were identified as miRNA target genes in test hybrid. Of these, Os06t0248300-03 was down-regulated in panicle and controls anther/pollen development in P3 and P4 stages of rice panicle43,44. In addition, 2 pollen-specific cis regulatory elements GTGANTG10 (GTGA) and POLLEN1LeLAT52 (AGAAA) were reported in the regulatory region of OsFbox gene that showed maximum activity in meiotic anther stage and controls early anther development in rice20. Many OsFbox protein-encoding genes control plant growth and various stages of panicle and seed development in rice. Os03t0150800-01 (OsPht1;2) the miRNA target gene on chromosome 3 was down-regulated (2.36 fold) in leaf and up-regulated (1.60 fold) in panicle in our study and also reported up-regulated in response to both nitrogen and phosphate starvation in rice roots and shoots45. OsPht1;2 mediates inorganic phosphate (Pi) uptake and transport in root and shoots under Pi deprivation in rice46. The third gene OsLAC5 (Os01t0842500-01, similar to laccase) was up-regulated in panicle and down-regulated in flag leaf. High accumulation of miR397 target laccases that regulate lignification process in wild species O. nivara and domestication associated phenotypes such as yield related traits in cultivated rice47. The over expression of OsmiR397 in young panicles and grains enlarges the grain size and promotes panicle branching, leading to an increase in grain yield of up to 25% by down-regulating its target gene OsLAC (laccase like protein) on chromosome 548. However, the target DEG laccase in our studies is on chromosome 1.
We considered the possible role of the DEGs within yield QTLs in improving yield and abiotic stress tolerance. The DEG Alpha-amylase within qyld8.2 was up-regulated in panicle and plays a major role in starch degradation. These are starvation induced genes which provide energy for elongation of shoot by hydrolysing starch into sugars49,50. This was also induced during anoxia as well as stress conditions especially in cold stress in rice shoots51. Salinity is another major abiotic stress which affects around 7% of land area of world52. In higher plants similar to KUP related potassium transporters are up-regulated during K deficiency or salt stress. In our results it was up-regulated in panicle of test hybrid.
Three DEGs (E3 Ubiquitin ligase, Similar to Y19 protein and Cytochrome P450) in qyld2.1 showed down-regulation in leaf and up-regulation in panicle. Among these sequence difference was found only in Cytochrome P450 and it was in CDS region. E3 ubiquitin ligases are candidate genes for yield as they have been reported to enhance yield53 and are up-regulated during different abiotic stresses54–60. Reduced expression of RING-type E3 ubiquitin ligase on short arm of chromosome 2 in rice increased grain size and weight53 and seed size in Arabidopsis61. Though E3 ubiquitin ligase showed high (4.68) fold change in both flag leaf and panicle of test hybrid but there was no sequence difference between KMR3 and IL50-13. However, it is also likely that trans-acting elements influence its expression. Os11Gsk from O. rufipogon was previously shown to act in trans and increase yield in IL50-7, a sister line of IL50-1362. Similar to Y19 protein on chromosome 10 was reported to be induced during drought stress63,64. Cytochrome P450 are involved in brassinosteroid pathway and phenyl propanoid pathway and one gene Os04g0469800 of the CytP450 family on chromosome 4 is reported to control panicle structure and seed size65,66.
Among the high priority candidate genes within qyld2.1 which were differentially expressed only in flag leaf was Phenylalanine ammonia-lyase (PAL) which also showed sequence difference between KMR3 and IL50-13. PAL is an important enzyme in phenyl propanoid biosynthesis pathway and catalyzes the conversion of L-phenylalanine to cinnamic acid a precursor of salicylic acid which protects plants against various pests and diseases67. The phenolic compounds derived during phenyl propanoid biosynthesis pathway scavenge the reactive oxygen species to protect plants against abiotic stresses also68. The cytosolic enzymes such as PAL, 4 coumarate CoA ligase (4CL) and caffeoyl-CoA 3-O-methyl-transferase (CCoAOMT) of phenyl propanoid pathway influence cell wall lignin content, and are positively correlated with lodging resistance in culm of buckwheat69. Our results showed that 4 coumarate CoA ligases Os4CL2 and Os4CL3 on chromosome 2 and Os4CL4 on chromosome 6 were down-regulated in leaf but Os4CL5 on chromosome 8 was significantly up-regulated in panicle, whereas caffeoyl-CoA 3-O-methyl-transferase (CCoAOMT-2) was significantly down-regulated in leaf. Over expression of 4CL (OsAAE3) on chromosome 4 reduces floret development, fertility rate of anther, lignin biosynthesis and rice blast resistance in rice70. High lignin content decreases cell wall expansion and cell extensibility thereby it can limit water loss and prevent cell collapse during abiotic stress71. There is a link between lignin biosynthesis and grain yield as laccase which catalyzes the oxidative polymerization of monolignols into lignin was a DEG in our study. Lignin in plants is synthesised from mono lignols derived from phenylalanine in phenylpropanoid pathway66. Laccase is also the target of miRNA397 and a key regulator of domestication phenotype in rice48. In our results laccase OsLAC5 was down-regulated in flag leaf and OsLAC4, a microRNA target gene on chromosome 1 was up-regulated in panicle. The PAL genes OsPAL6 and OsPAL8 mediate brown planthopper resistance by controlling lignin biosynthesis and its accumulation in rice72. Phenylalanine ammonia-lyase (OsPAL4) on chromosome 2 is associated with broad spectrum disease resistance in rice. The 750 base pair deletion in second exon of OsPAL4 in the mutant line leads to down-regulation of OsPAL4 which up-regulates the expression of OsPAL2 on chromosome 2 and down-regulates the expression of un-linked OsPAL6 on chromosome 473. It is significant that in our study the same two genes OsPAL2 (Phenylalanine ammonia-lyase- Os02t0626400-01) and OsPAL4 (Similar to phenylalanine ammonia- lyase - Os02t0627100-01) were down-regulated (2.36 to 2.59) in flag leaf and are thus candidate genes derived from O. rufipogon that increase grain yield in test hybrid. Both the genes are located within subQTL-7 region (RM6318-RM1920) of qyld2.19. Both expression and sequence analysis indicates that OsPAL2 and OsPAL4 influence grain yield in test hybrid. Thus phenyl propanoid pathway and PAL genes in particular are at centre stage of not only broad spectrum disease74, pest resistance72 and tolerance to abiotic stress75 but also for increasing grain yield.
In conclusion, carbohydrate metabolism, phenyl propanoid pathway, ubiquitin-dependent degradation and phytohormone related pathways were most enriched in test hybrid with O. rufipogon introgressions compared to control hybrid without O. rufipogon introgressions and play a major role in high yield. The yield enhancing role of introgressions from wild species is thus established and key differentially expressed genes underlying major yield enhancing QTL qyld2.1 identified. Two DEGs OsPAL2, (Os02t0626400-01 Phenyl alanine ammonia-lyase) and OsPAL4 (Os02t0627100-01 Similar to PAL) within qyld2.1 showed high SNP and InDel variations in CDS regions and were also differentially expressed in test hybrid. We report for the first time that PAL genes within qyld2.1 from Oryza rufipogon are high priority candidate genes or heterotic loci for higher grain yield in test hybrid.
Materials and Methods
Plant material
The seedlings of control hybrid [CMS line IR79156A x restorer line KMR3] and its test hybrid [CMS line IR79156A x restorer line IL50-13] were planted in clay pots (one plant/pot). Please note that in control hybrid the restorer male parent used is without O. rufipogon introgression and in test hybrid the restorer male parent used is IL50-13 which is a stable elite backcross introgression line (IL) derived from KMR3 x O. rufipogon and thus is with O. rufipogon introgression. The same cms line was used as the female parent in both control and test hybrid. Plants were grown in green house at Indian Institute of Rice Research (IIRR). Flag leaves and young panicles at P2 stage (<5 cm long) were collected at panicle initiation stage (Supplementary Fig. S6). The samples were harvested from two biological replicates.
RNA extraction
Total RNA was extracted from flag leaves and panicles of both control and experimental hybrids (4 samples × 2 biological replications) using Trizol-RNA lysis method (Cat. No. 15596018, Invitrogen, Thermo Fisher Scientific Inc., USA). The extracted RNA was treated with RNase-free DNase I (Qiagen) to prevent genomic DNA contamination. The purity and quantity of the RNA was determined using Nanodrop Spectrophotometer (ND-1000, Thermo Scientific, USA) and integrity of RNA was measured using Agilent 2100 Bioanalyzer (Agilent technologies, Santa Clara, CA95051, USA).
cDNA labeling
All 8 samples were labeled using Agilent Quick-Amp (single color) labeling kit (p/n5190-0442). 500 ng of each sample RNA was reverse transcribed at 40 °C using oligo dT primer tagged to a T7 polymerase promoter and converted to cDNA. The cRNA was synthesized using cDNA as template by in vitro transcription and Agilent dye Cy3 CTP was incorporated during this step. Labeled cRNA was cleaned up using Qiagen RNeasy mini kit columns (Qiagen, Cat No: 74106), quality and quantity was measured using Nanodrop ND-1000.
Hybridization and scanning
The labeled cRNA samples were fragmented at 60 °C and hybridized on to a genotypic designed rice gene expression microarrays, 8 × 60 k format which contains 60,045 Oryza sativa probes (Agilent Technologies, In situ hybridization kit, part number 5190-0404, AMADID No: G4102A_048014). Hybridization was carried out in Agilent’s Surehyb Chambers at 65 °C for 16 hours. The hybridized slides were washed using Agilent gene expression wash buffers (Agilent Technologies, Part Number 5188-5327) and scanned using the Agilent Microarray Scanner (Agilent Technologies, Part Number G2600D).
Statistical analysis
The array data was extracted from images and quantified using feature extraction software version 11.5 and 12.6 Agilent). The raw data was analyzed and normalized with the help of GeneSpring GX software (Agilent technologies, Inc., Santa Clara, CA, 95051, USA) using 75th percentile shift [Percentile shift normalization is a global normalization, where the locations of all the spot intensities in an array are adjusted. This normalization takes each column in an experiment independently, and computes the nth percentile of the expression values for this array, across all spots (where n has a range from 0–100 and n = 75 is the median). It subtracts this value from the expression value of each entity, and fold expression values were obtained with respect to specific control Samples] (http://genespring-support.com/category/faqcategories/microarray-data-analysis/normalization). Differential expression patterns were identified among the samples. Significant genes up-regulated fold >1 (logbase2) and down-regulated < −1 (logbase2) in the test samples with respect to control sample were identified. Statistical student t-test is used to calculate the p-values among the replicates. Volcano plot was constructed based on p-values from a t-test and fold-change values76. Venn diagram was generated to look at the differential expression of common and specific genes in test hybrid compared to control hybrid.
Functional enrichment analysis
Gene ontology (GO) and KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway analysis was performed to annotate and classify the functional categories and pathways of differentially expressed genes in test hybrid compared to control hybrid. Pathway analysis was performed using Genotypic Biointerpreter-Biological Analysis Software. (Genotypic Technology Private Limited, Bangalore). Genes were also classified based on their functional category, and pathways using Biological Analysis tool DAVID (Database for Annotation, Visualization, and Integrated Discovery) (http://david.abcc.ncifcrf.gov/) (LIB, Frederick, MD, 21702, USA). GO annotations for DEGs were plotted using WEGO 2.0 (Web Gene Ontology Annotation Plot) online tool77. Pathway analysis was also performed for some of the important pathways within the major yield QTL qyld2.1 and minor QTL qyld8.2 regions using Mapman tool78. Wilcoxon Rank Sum Test was used to calculate the P-values for significant pathways in test hybrid.
Primer designing
Few differentially expressed genes underlying the major yield QTL qyld2.1 and minor QTL qyld8.2 were selected for real time PCR validation. In addition, several other yield related genes were selected based on their role in different metabolic pathways. Gene sequences were retrieved from NCBI (http://www.ncbi.nlm.nih.gov). Primers for quantitative polymerase chain reaction (qPCR) (Table 1) were designed using Primer 3 software (http://frodo.wi.mit.edu/).
qRT-PCR validation of DEGs
First-strand cDNA was synthesized from 2 μg of total RNA in a 25 μl reaction mixture with M-MLV reverse transcriptase (Promega, Madison, WI, USA) and oligo dT primers. cDNA was treated with RNase and normalized to obtain similar concentration. qRT-PCR was performed on a ABI 7500 real-time analyzer (Applied Biosystems), using a SYBR premix ExTaq kit (Takara Bio). The levels of OsActin1 served to normalize the expression ratio for each gene. Each reaction was run in duplicate (with three biological replicates) and the melting curves were constructed using Dissociation Curves Software (Applied Biosystems), to ensure that only a single specific product is amplified.
Changes in expression were calculated via the ‘comparative Ct method’ (Applied Biosystems). The mean threshold cycle (Ct) value obtained after each reaction was normalized to the Ct value of reference gene whose expression was consistent across the conditions. Further ΔΔCT values were calculated using the formulae ΔΔCT = ΔCT of test sample − ΔCT control sample, and then fold difference was calculated from 2−ΔΔCt. Similarly, ΔCT standard deviation was calculated as given at www3.appliedbiosystems.com/…/general documents/cms_042380.pdf.
Sequence comparison of DEGs and other reported yield related genes for polymorphism
In order to identify polymorphism between KMR3 and IL50-13, we performed BLASTN analysis of 136 DEGs including 78 DEGs of qyld2.1, 25 DEGs of qyld8.2, 12 DEGs showing high fold expression and 21 reported yield related gene sequences against the scaffolds obtained by whole-genome sequencing of the restorers KMR3 (without O. rufipogon) and IL50-13 [IET21943 = RPBio4919-50-13] (with O. rufipogon). The comparison was done by considering the gene sequence corresponding to Nipponbare and its gene sequence was used to obtain the corresponding gene sequences from the scaffolds of the two restorers KMR3 and IL50-13. The orthologous genomic regions were available for all genes analysed. Further we also analyzed the effect of identified SNPs/InDels in the coding region that could affect the expression of the genes showing polymorphism and also the effect of the variants in the 2 kb upstream and downstream regions of the given genes using the SnpEff software (Wayne State University, Detroit MI USA)79. The DEGs along with their gene and chromosome names and their corresponding coordinates are given in Supplementary Table S3.
Ethics statement
The authors declare that the experiments comply with the current laws of the country in which they were performed and in compliance with ethical standards.
Supplementary information
Acknowledgements
Financial support of Department of Biotechnology, Government of India (DBT No. BT/PR13357/AGR/02/695/2009), (BT/AB/FG-2(PHII)(IA/2009) and National Professor Project, Indian Council of Agricultural Research (ICAR-F.NoEdn/27/4/NP/2012-HRD) to NS is gratefully acknowledged. We thank Genotypic Technology Private Limited, Bangalore, India for the microarray processing of hybrids using Agilent platform.
Author contributions
N.S. and G.H. designed the study. G.H. analyzed microarray data and wrote the manuscript with contributions from S.K.M. S.A. and G.H. designed primers and did qRT-PCR. T.S. did bioinformatic analysis. S.K.M., R.R. and N.S. guided the work and revised the manuscript. All authors reviewed and approved the final manuscript.
Data availability
All data generated or analysed during this study are included in this published article (and its Supplementary Information files). The sequence data is available on request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Satendra K Mangrauthia, Email: skmdrr@gmail.com.
Sarla Neelamraju, Email: sarla_neelamraju@yahoo.com.
Supplementary information
is available for this paper at 10.1038/s41598-020-60922-6.
References
- 1.Cheng SH, et al. Super hybrid rice breeding in China: achievements and prospects. J. Integr. Plant Biol. 2007;49:805–810. doi: 10.1111/j.1744-7909.2007.00514.x. [DOI] [Google Scholar]
- 2.Dwivedi DK, et al. Heterosis in inter and intra sub specific crosses over three environments in rice. Euphytica. 1998;99:155–165. doi: 10.1023/A:1018357300923. [DOI] [Google Scholar]
- 3.Virmani, S. S. et al. Two-line hybrid rice breeding manual. Los Baños, Phillipines: International Rice Research Institute, 88p (2003).
- 4.Yuan L. Development of hybrid rice to ensure food security. Rice Sci. 2014;21:1–2. doi: 10.1016/S1672-6308(13)60167-5. [DOI] [Google Scholar]
- 5.Cheng SH, Liao XY, Min SK. Super rice research in China: Background, target and some considerations. China Rice. 1998;1:3–5. [Google Scholar]
- 6.Liang F, et al. Molecular marker-assisted selection for yield-enhancing genes in the progeny of 9311 x O. rufipogon using SSR. Euphytica. 2004;139:159–165. doi: 10.1007/s10681-004-2560-1. [DOI] [Google Scholar]
- 7.Fu Q, et al. Analysis of QTLs for yield-related traits in Yuanjiang common wild rice (Oryza rufipogon Griff.) J Genet Genomics. 2010;37:147–157. doi: 10.1016/S1673-8527(09)60033-3. [DOI] [PubMed] [Google Scholar]
- 8.Gaikwad KB, et al. Yield-enhancing heterotic QTL transferred from wild species to cultivated rice Oryza sativa L. PloS one. 2014;9:e96939. doi: 10.1371/journal.pone.0096939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thalapati, S. et al. Introgressions from Oryza rufipogon into restorer line KMR3 increase hybrid rice yield. In: Xie, F. & Hardy, B. (eds) Public-private partnership for hybrid rice: Proceedings of 6th International Hybrid Rice Symposium, 10–12 September 2012, Hyderabad, India, Los Baños (Philippines): International Rice Research Institute, pp 227–234 (2014).
- 10.Thalapati S, et al. Heterosis and combining ability in rice as influenced by introgressions from wild species Oryza rufipogon including qyld2.1 sub-QTL into the restorer line KMR3. Euphytica. 2015;202:81–95. doi: 10.1007/s10681-014-1222-1. [DOI] [Google Scholar]
- 11.Sanchez-Bragado R, et al. Photosynthetic contribution of the ear to grain filling in wheat: a comparison of different methodologies for evaluation. J Exp Bot. 2016;67:2787–2798. doi: 10.1093/jxb/erw116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yin C, et al. Genetic dissection of top three leaf traits in rice using progenies from a japonica x indica cross. J Integr Plant Biol. 2017;59:866–880. doi: 10.1111/jipb.12597. [DOI] [PubMed] [Google Scholar]
- 13.Zheng H, et al. Contributions of photosynthetic organs to the seed yield of hybrid rice: the effects of gibberellin application examined by carbon isotope technology. Seed Science and Technology. 2018;46:533–546. doi: 10.15258/sst.2018.46.3.10. [DOI] [Google Scholar]
- 14.Kellogg EA, et al. Early inflorescence development in the grasses (Poaceae) Front Plant Sci. 2013;4:250. doi: 10.3389/fpls.2013.00250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu Z, et al. Genetic control of inflorescence architecture during rice domestication. Nat Commun. 2013;4:2200. doi: 10.1038/ncomms3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao XQ, et al. Architecture of Wheat Inflorescence: Insights from Rice. Trends Plant Sci. 2019;24:802–809. doi: 10.1016/j.tplants.2019.06.002. [DOI] [PubMed] [Google Scholar]
- 17.Furutani I, et al. Genome-wide analysis of spatial and temporal gene expression in rice panicle development. Plant J. 2006;46:503–511. doi: 10.1111/j.1365-313X.2006.02703.x. [DOI] [PubMed] [Google Scholar]
- 18.Yamburenko MV, et al. Dynamic patterns of expression for genes regulating cytokinin metabolism and signaling during rice inflorescence development. PloS one. 2017;12:e0176060. doi: 10.1371/journal.pone.0176060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sharma R, et al. Expression dynamics of metabolic and regulatory components across stages of panicle and seed development in indica rice. Funct Integr Genomics. 2012;12:229–48. doi: 10.1007/s10142-012-0274-3. [DOI] [PubMed] [Google Scholar]
- 20.Khurana R, et al. Spatial and temporal activity of upstream regulatory regions of rice anther-specific genes in transgenic rice and Arabidopsis. Transgenic res. 2013;22:31–46. doi: 10.1007/s11248-012-9621-3. [DOI] [PubMed] [Google Scholar]
- 21.Wei G, et al. A transcriptomic analysis of super hybrid rice LYP9 and its parents. Proc. Nat. Acad. Sci. USA. 2009;106:7695–701. doi: 10.1073/pnas.0902340106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bao J, et al. Serial analysis of gene expression study of a hybrid rice strain (LYP9) and its parental cultivars. Plant Physiol. 2005;138:1216–1231. doi: 10.1104/pp.105.060988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Song GS, et al. Comparative transcriptional profiling and preliminary study on heterosis mechanism of super-hybrid rice. Mol Plant. 2010;3:1012–1025. doi: 10.1093/mp/ssq046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peng Y, et al. Comparative transcriptional profiling of three super-hybrid rice combinations. Int. J. Mol. Sci. 2014;15:3799–3815. doi: 10.3390/ijms15033799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li D, et al. Integrated analysis of phenome, genome, and transcriptome of hybrid rice uncovered multiple heterosis-related loci for yield increase. Proc. Nat. Acad. Sci. USA. 2016;113:E6026–E6035. doi: 10.1073/pnas.1610115113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marri PR, et al. Identification and mapping of yield and yield related QTLs from an Indian accession of Oryza rufipogon. BMC Genet. 2005;6:33. doi: 10.1186/1471-2156-6-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Babu, A. P. Genomics of yield enhancing QTLs from wild species of rice. PhD Thesis, Jawaharlal Nehru Technological University, Hyderabad (2009).
- 28.Babu AP, et al. Marker aided selection of yield-enhancing QTL yld2.1 into restorer KMR3 and fine mapping a genomic region on chromosome 2. J. Exp. Agric. Int. 2017;17:1–14. doi: 10.9734/JEAI/2017/34039. [DOI] [Google Scholar]
- 29.AICRIP (All India Coordinated Rice Improvement Project) Varietal improvement. Indian Institute of Rice Research Annual Progress Report. 2014;1(1):469–1.485. [Google Scholar]
- 30.AICRIP (All India Coordinated Rice Improvement Project) Varietal improvement. Indian Institute of Rice Research Annual Progress Report. 2015;1(1):315–1.341. [Google Scholar]
- 31.Zhang HY, et al. A genome-wide transcription analysis reveals a close correlation of promoter INDEL polymorphism and heterotic gene expression in rice hybrids. Mol Plant. 2008;1:720–731. doi: 10.1093/mp/ssn022. [DOI] [PubMed] [Google Scholar]
- 32.Zhou G, et al. Genetic composition of yield heterosis in an elite rice hybrid. Proc. Nat. Acad. Sci. U S A. 2012;109:15847–15852. doi: 10.1073/pnas.1214141109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhen G, et al. Genome-wide dissection of heterosis for yield traits in two-line hybrid rice populations. Sci. Rep. 2017;7:7635. doi: 10.1038/s41598-017-06742-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gissa DW, et al. Heterosis and combining ability for grain yield and its components in selected maize inbred lines. S. Afr. J. Plant Soil. 2007;24:133–137. doi: 10.1080/02571862.2007.10634795. [DOI] [Google Scholar]
- 35.Hoecker N, et al. Comparison of maize (Zea mays L.) F1-hybrid and parental inbred line primary root transcriptomes suggests organ-specific patterns of nonadditive gene expression and conserved expression trends. Genetics. 2008;179:1275–1283. doi: 10.1534/genetics.108.088278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Talukder MZA, et al. Combining ability and heterosis on yield and its component traits in maize (Zea mays L.) Bangladesh J. Agril. Res. 2016;41:565–577. doi: 10.3329/bjar.v41i3.29727. [DOI] [Google Scholar]
- 37.AICRIP (All India Coordinated Rice Improvement Project) Varietal improvement. Indian Institute of Rice Research Annual Progress Report. 2013;1(1):359–1.371. [Google Scholar]
- 38.Ganeshan P, et al. Identification of salt tolerant rice lines among interspecific BILs developed by crossing Oryza sativa x O. rufipogon and O. sativa x O. nivara. AJCS. 2016;10:220–228. [Google Scholar]
- 39.Chowdhury AD, et al. Haplotyping of rice genotypes using simple sequence repeat markers associated with salt tolerance. Rice Sci. 2016;23:317–325. doi: 10.1016/j.rsci.2016.05.003. [DOI] [Google Scholar]
- 40.Rai V, et al. Swarna/Oryza nivara and KMR3/O. rufipogon introgression lines tolerant to drought and salinity. DRR Newsletter. 2010;8:4. [Google Scholar]
- 41.Jiao Y, et al. Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat Genet. 2010;42:541. doi: 10.1038/ng.591. [DOI] [PubMed] [Google Scholar]
- 42.Miura K. OsSPL14 promotes panicle branching and higher grain productivity in rice. Nat Genet. 2010;42:545. doi: 10.1038/ng.592. [DOI] [PubMed] [Google Scholar]
- 43.Lai Z, et al. An F-box gene linked to the self-incompatibility (S) locus of Antirrhinum is expressed specifically in pollen and tapetum. Plant Mol Biol. 2002;50:29–42. doi: 10.1023/A:1016050018779. [DOI] [PubMed] [Google Scholar]
- 44.Kim OK, et al. An Arabidopsis F-box protein regulates tapetum degeneration and pollen maturation during anther development. Planta. 2010;232:353–366. doi: 10.1007/s00425-010-1178-x. [DOI] [PubMed] [Google Scholar]
- 45.Liu F, et al. OsSPX1 suppresses the function of OsPHR2 in the regulation of expression of OsPT2 and phosphate homeostasis in shoots of rice. Plant J. 2010;62:508–517. doi: 10.1111/j.1365-313X.2010.04170.x. [DOI] [PubMed] [Google Scholar]
- 46.Ai P, et al. Two rice phosphate transporters, OsPht1; 2 and OsPht1; 6, have different functions and kinetic properties in uptake and translocation. Plant J. 2009;57:798–809. doi: 10.1111/j.1365-313X.2008.03726.x. [DOI] [PubMed] [Google Scholar]
- 47.Swetha C, et al. Major domestication-related phenotypes in indica rice are due to loss of miRNA-mediated laccase silencing. The Plant Cell. 2018;30:2649–2662. doi: 10.1105/tpc.18.00472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang YC, et al. Overexpression of microRNA OsmiR397 improves rice yield by increasing grain size and promoting panicle branching. Nat. Biotechnol. 2013;31:848. doi: 10.1038/nbt.2646. [DOI] [PubMed] [Google Scholar]
- 49.Beck E, Ziegler P. Biosynthesis and degradation of starch in higher plants. Annu. Rev. Plant Biol. 1989;40:95–117. doi: 10.1146/annurev.pp.40.060189.000523. [DOI] [Google Scholar]
- 50.Perata P, et al. Effect of anoxia on starch breakdown in rice and wheat seeds. Planta. 1992;188:611–618. doi: 10.1007/BF00197056. [DOI] [PubMed] [Google Scholar]
- 51.Lasanthi-Kudahettige R, et al. Transcript profiling of the anoxic rice coleoptile. Plant Physiol. 2007;144:218–231. doi: 10.1104/pp.106.093997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shabala S, Cuin TA. Potassium transport and plant salt tolerance. Physiol. Plant. 2008;133:651–669. doi: 10.1111/j.1399-3054.2007.01008.x. [DOI] [PubMed] [Google Scholar]
- 53.Song XJ, et al. QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nature Genet. 2007;39:623. doi: 10.1038/ng2014. [DOI] [PubMed] [Google Scholar]
- 54.Yee D, Goring DR. The diversity of plant U-box E3 ubiquitin ligases: from upstream activators to downstream target substrates. J. Exp. Bot. 2009;60:1109–1121. doi: 10.1093/jxb/ern369. [DOI] [PubMed] [Google Scholar]
- 55.Lyzenga WJ, Stone SL. Abiotic stress tolerance mediated by protein ubiquitination. J. Exp. Bot. 2012;63:599–616. doi: 10.1093/jxb/err310. [DOI] [PubMed] [Google Scholar]
- 56.Stone SL. The role of ubiquitin and the 26 S proteasome in plant abiotic stress signaling. Front Plant Sci. 2014;5:135. doi: 10.3389/fpls.2014.00135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu ZB, et al. A novel membrane-bound E3 ubiquitin ligase enhances the thermal resistance in plants. Plant Biotechnol. J. 2014;12:93–104. doi: 10.1111/pbi.12120. [DOI] [PubMed] [Google Scholar]
- 58.Byun MY, et al. Homologous U-box E3 ubiquitin ligases OsPUB2 and OsPUB3 are involved in the positive regulation of low temperature stress response in rice (Oryza sativa L.) Front Plant Sci. 2017;8:16. doi: 10.3389/fpls.2017.00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Takiguchi H, et al. Discovery of E3 ubiquitin ligases that alter responses to Nitrogen deficiency using rice full-length cDNA overexpressor (FOX)-hunting system. Plant Mol Biol Report. 2017;35:343–54. doi: 10.1007/s11105-017-1027-1. [DOI] [Google Scholar]
- 60.Chapagain S, Park YC, Jang CS. Functional diversity of RING E3 ligases of major cereal crops in response to abiotic stresses. JCSB. 2017;20:351–357. [Google Scholar]
- 61.Xia T, et al. The ubiquitin receptor DA1 interacts with the E3 ubiquitin ligase DA2 to regulate seed and organ size in Arabidopsis. Plant Cell. 2013;25:3347–3359. doi: 10.1105/tpc.113.115063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thalapati S, et al. Os11Gsk gene from a wild rice, Oryza rufipogon improves yield in rice. Funct Integr Genomics. 2012;12:277–289. doi: 10.1007/s10142-012-0265-4. [DOI] [PubMed] [Google Scholar]
- 63.Lee TH. Rice Array Net: a database for correlating gene expression from transcriptome profiling, and its application to the analysis of coexpressed genes in rice. Plant Physiol. 2009;151:16–33. doi: 10.1104/pp.109.139030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shin, S. J. et al. Novel drought-responsive regulatory coding and non-coding transcripts from Oryza sativa L. Genes Genom38, 949–960 (2016).
- 65.Wu Y, et al. CLUSTERED PRIMARY BRANCH 1, a new allele of DWARF11, controls panicle architecture and seed size in rice. Plant Biotechnol J. 2016;14:377–386. doi: 10.1111/pbi.12391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dixon RA, Barros J. Lignin biosynthesis: old roads revisited and new roads explored. Open Biol. 2019;9:190215. doi: 10.1098/rsob.190215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Herrmann KM, Weaver LM. The shikimate pathway. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1999;50:473–503. doi: 10.1146/annurev.arplant.50.1.473. [DOI] [PubMed] [Google Scholar]
- 68.Sharma A, et al. Response of phenylpropanoid pathway and the role of polyphenols in plants under abiotic stress. Molecules. 2019;24:2452. doi: 10.3390/molecules24132452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang C, et al. Relationship between lignin metabolism and lodging resistance of culm in buckwheat. J Agric Sci. 2014;6:29. [Google Scholar]
- 70.Liu H, et al. 4-Coumarate-CoA ligase-like gene OsAAE3 negatively mediates the rice blast resistance, floret development and lignin biosynthesis. Front. Plant Sci. 2017;7:2041. doi: 10.3389/fpls.2016.02041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Le Gall H. Cell wall metabolism in response to abiotic stress. Plants. 2015;4:112–166. doi: 10.3390/plants4010112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.He, J. et al. An R2R3 MYB transcription factor confers brown planthopper resistance by regulating the phenylalanine ammonia-lyase pathway in rice. Proc Natl Acad Sci USA.117, 271–277. [DOI] [PMC free article] [PubMed]
- 73.Tonnessen BW, et al. Rice phenylalanine ammonia-lyase gene OsPAL4 is associated with broad spectrum disease resistance. Plant Mol. Biol. 2015;87:273–286. doi: 10.1007/s11103-014-0275-9. [DOI] [PubMed] [Google Scholar]
- 74.Ning Y, Wang GL. Breeding plant broad-spectrum resistance without yield penalties. Proc Natl Acad Sci USA. 2018;115:2859–2861. doi: 10.1073/pnas.1801235115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gho YS, et al. Phenylalanine ammonia-lyase family is closely associated with response to phosphate deficiency in rice. Genes Genom. 2020;42:67–76. doi: 10.1007/s13258-019-00879-7. [DOI] [PubMed] [Google Scholar]
- 76.Patterson TA, et al. Performance comparison of one-color and two-color platforms within the microarray quality control (MAQC) project. Nat Biotechnol. 2006;24:1140–1150. doi: 10.1038/nbt1242. [DOI] [PubMed] [Google Scholar]
- 77.Ye J, et al. WEGO 2.0: a web tool for analyzing and plotting GO annotations, 2018 update. Nucleic Acids Res. 2018;46:W71–W75. doi: 10.1093/nar/gky400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Thimm O, et al. MAPMAN: a user - driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant Journal. 2004;37:914–939. doi: 10.1111/j.1365-313X.2004.02016.x. [DOI] [PubMed] [Google Scholar]
- 79.Cingolani P, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin). 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analysed during this study are included in this published article (and its Supplementary Information files). The sequence data is available on request.