Abstract
Saccharomyces cerevisiae expressing heterologous pathways for xylose, arabinose, and galacturonic acid metabolism has been constructed by a Cas9-based genome editing technology [1]. The fermentation performance of the final strain (YE9) was tested under various substrate conditions, and the fermentation parameters were calculated. The dataset can be used for designing bioprocesses for pectin-rich biomass.
Keywords: Citrus peel waste, Sugar beet pulp, Pectin, Metabolic engineering, CRISPR/Cas9, Bioethanol
Specifications Table
| Subject | Applied Microbiology and Biotechnology |
| Specific subject area | Yeast metabolic engineering |
| Type of data | Tables and Figures |
| How data were acquired | The fermentation data were obtained by HPLC (Agilent Technologies 1260 series). |
| Data format | Raw and Analysed |
| Parameters for data collection | Fermentation conditions at 30oC and 130 rpm. |
| Description of data collection | Time series analysis of fermentation samples. |
| Data source location | Institution: Kyungpook National University City/Town/Region: Daegu Country: Korea |
| Data accessibility | With the article |
| Related research article | Author’s name: Deokyeol Jeong, Suji Ye, Heeyoung Park, and Soo Rin Kim Title: Simultaneous fermentation of galacturonic acid and five-carbon sugars by engineered Saccharomyces cerevisiae Journal: Bioresource Technology https://doi.org/10.1016/j.biortech.2019.122259 |
Value of the Data
|
1. Data
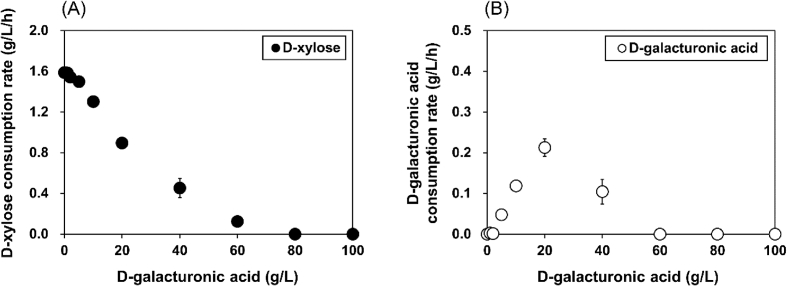
This dataset contains 1) the construction of engineered Saccharomyces cerevisiae strain (YE9) capable of fermenting galacturonic acid, arabionse, and xylose, and 2) its fermentation data with different carbon sources (galacturonic acid, arabinose, xylose, galactose, glucose, and fructose) and their mixtures, all of which present in pectin-rich biomass. In Fig. 1, the fermentation patterns of the YE9 strain with natively fermentable sugars (glucose, fructose, and galactose) as a sole carbon source are presented. In Table 1, the fermentation profiles of the YE9 strain with xylose, arabinose, and galacturonic acid in comparison to its wild type strain (D452-2). In Fig. 2, the YE9 strain was tested for xylose and galacturonic acid consumption rates in a mixture of 40 g/L xylose and various galactornic acid concentrations. In Table 2, the fermentation parameters of the YE9 strain with a mixture of galacturonic acid and co-substrates.
Fig. 1.
Fermentation profiles of the YE9 strain in a complex medium containing (A) 40 g/L d-glucose, (B) 40 g/L d-fructose, and (C) 40 g/L d-galactose as the sole carbon sources. Fermentations were performed under oxygen-limited conditions (130 rpm) with a starting cell density of 25 g/L. All experiments were performed in biological triplicate, and the error bars indicate the standard deviations.
Table 1.
Fermentation profiles of the native S. cerevisiae strain (D452-2) and engineered strain (YE9) expressing heterologous pathways for metabolizing d-xylose, l-arabinose, and d-galacturonic acid (galUA).
| Strain | Substrate | Substrate consumed (g/L) | Substrate consumption rate (g/L/h) | Products (g/L) |
Parametersb) |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| Glycerol | Ethanol | YGlycerol | YEthanol | PEthanol∗ | ||||||
| D452-2 | D-xylosea) | 5.9 ± 0.2 | 0.19 ± 0.01 | 0.3 ± 0.0 | n. d. | 0.07 ± 0.02 | n. d. | n. d. | ||
| l-arabinose | 1.3 ± 0.6 | 0.08 ± 0.03 | n. d. | n. d. | n. d. | n. d. | n. d. | |||
| galUA | < 0.0 | < 0.00 | n. d. | n. d. | n. d. | n. d. | n. d. | |||
| YE9 | d-xylose | 33.7 ± 0.5 | 1.41 ± 0.02 | 0.6 ± 0.1 | 11.3 ± 0.1 | 0.02 ± 0.00 | 0.34 ± 0.01 | 0.05 ± 0.00 | ||
| l-arabinose | 30.2 ± 0.1 | 0.63 ± 0.07 | n. d. | 1.9 ± 0.1 | n. d. | 0.07 ± 0.00 | <0.00 | |||
| galUA | 6.7 ± 0.7 | 0.27 ± 0.01 | 0.3 ± 0.1 | 0.3 ± 0.0 | 0.04 ± 0.01 | 0.08 ± 0.02 | < 0.00 | |||
Fermentations were performed in a complex medium containing 40 g/L d-xylose, 40 g/L l-arabinose, or 20 g/L d-galacturonic acid under oxygen-limited conditions (130 rpm) with a starting cell density of 25 g/L. Substrate consumption rate was calculated for 24 h and the others were calculated for 72 h.
YGlycerol, glycerol yield (g glycerol/g substrate); YEthanol, ethanol yield (g ethanol/g substrate); PEthanol∗, specific ethanol productivity (g ethanol/g cell/h); n. d., not detected.
Fig. 2.
Effect of d-galacturonic acid on the rate of d-xylose consumption in the YE9 strain. Consumption rate of d-xylose (A) and d-galacturonic acid (B) was evaluated under 40 g/L D-xylose and different d-galacturonic acid concentrations (0–100 g/L). All experiments were performed in biological triplicate, and error bars indicate standard deviations and were not visible when smaller than the symbol size.
Table 2.
Fermentation profiles of mixed culture by engineered S. cerevisiae YE9 strain expressing heterologous pathways metabolizing d-xylose, l-arabinose, and d-galacturonic acid (galUA).
| Mediuma) | Substrate consumed (g/L) |
galUA consumption rate (g/L/h) | Products (g/L) |
Parametersb) |
|||||
|---|---|---|---|---|---|---|---|---|---|
| galUA | Sugars | Glycerol | Ethanol | YGlycerol | YEthanol | PGlycerol∗ | PEthanol∗ | ||
| galUA | 6.7 ± 0.7 | – | 0.27 ± 0.01 | 0.3 ± 0.1 | 0.3 ± 0.0 | 0.04 ± 0.01 | 0.08 ± 0.02 | < 0.00 | < 0.00 |
| galUA + Glucose | 3.3 ± 0.2 | 36.7 ± 0.1 | 0.14 ± 0.01 | 2.4 ± 0.3 | 16.9 ± 0.2 | 0.06 ± 0.01 | 0.40 ± 0.01 | 0.06 ± 0.00 | 0.66 ± 0.01 |
| galUA + Fructose | 4.5 ± 0.3 | 36.1 ± 0.8 | 0.18 ± 0.02 | 2.9 ± 0.1 | 16.9 ± 0.5 | 0.07 ± 0.00 | 0.36 ± 0.01 | < 0.00 | 0.65 ± 0.03 |
| galUA + Galactose | 4.6 ± 1.2 | 25.4 ± 7.3 | 0.17 ± 0.03 | 1.6 ± 0.7 | 2.4 ± 1.1 | 0.04 ± 0.01 | 0.05 ± 0.02 | < 0.00 | < 0.00 |
| galUA+ Xylose | 13.1 ± 0.4 | 33.3 ± 0.5 | 0.49 ± 0.02 | 4.5 ± 0.1 | 12.8 ± 0.3 | 0.08 ± 0.00 | 0.23 ± 0.01 | 0.01 ± 0.00 | 0.04 ± 0.00 |
| galUA+ Arabinose | 11.9 ± 0.7 | 28.4 ± 0.1 | 0.32 ± 0.03 | 4.2 ± 0.2 | 4.1 ± 0.5 | 0.11 ± 0.01 | 0.11 ± 0.02 | < 0.00 | < 0.00 |
| galUA +Xylose (X) +Arabinose (A) |
15.3 ± 0.6 | 33.7 ± 0.1 (X) 25.9 ± 4.4 (A) |
0.49 ± 0.04 | 5.3 ± 0.6 | 16.5 ± 1.2 | 0.07 ± 0.00 | 0.22 ± 0.01 | < 0.00 | 0.02 ± 0.00 |
Fermentations were performed in a complex medium containing 20 g/L d-galacturonic acid (galUA) and 40 g/L sugar (d-glucose, d-fructose, d-galactose, d-xylose, l-arabinose, and mixture of d-xylose and l-arabinose) under oxygen-limited conditions (130 rpm) with a starting cell density of 25 g/L. d-galacturonic acid consumption rate was calculated for 24 h and the others were calculated for 72 h.
YGlycerol, glycerol yield (g glycerol/g substrates); YEthanol, ethanol yield (g ethanol/g substrates); PGlycerol∗, specific glycerol productivity (g glycerol/g cell/h); PEthanol∗, specific ethanol productivity (g ethanol/g cell/h).
2. Experimental design, materials, and methods
2.1. Strain construction by Cas9-based genome editing
To construct the YE9 strain, four consecutive transformations were performed as summarized in Fig. 3 using strains listed in Table 3. Briefly, the strain construction includes three parts: 1) guide RNA (gRNA) plasmid construction, 2) donor DNA preparation, and 3) yeast transformation.
-
1)
Guide RNA (gRNA) plasmid construction
Fig.3.
Construction of engineered S. cerevisiae YE9 strains expressing heterologous d-xylose, d-galacturonic acid, and l-arabinose pathways. (A) Strain construction using Cas9-based in vivo assembly and genome integration strategy. (B) Confirmation primers for correct assembly and integration by yeast colony PCR. The primer sequences are listed in Table S5.
Table 3.
Saccharomyces cerevisiae strains used for the construction of YE9.
| Strains | Description/relevant genotypea | Ref. |
|---|---|---|
| D452-2 | Wild type; Matα leu2 his3 ura3 | [7] |
| DY02 | Expressing the heterologous d-xylose pathway; D452-2 ald6::TDH3P-XYL1-TDH3T-PGK1P-XYL2-PGK1Tpho13::TEF1P-XYL3-TEF1T |
|
| YE3 | DY02 int#4::CCW12P-gaaA-CCW12T | |
| YE4 | DY02 int#4::PGK1P-lgd1-PGK1T | |
| YE5 | DY02 int#4::TDH3P-gaaC-TDH3T | |
| YE6 | Expressing the heterologous D-xylose and d-galacturonic acid pathway; DY02 int#4::CCW12P-gaaA-CCW12T-PGK1P-lgd1-PGK1T-TDH3P-gaaC-TDH3T |
|
| YE6 YPR1 | YE6 CCW12P-YPR1 | |
| YE6 gaaD | YE6 int#6::CCW12P-gaaD-CCW12T | |
| YE01 | Expressing the heterologous d-xylose, and l-arabinose pathway; D452-2 ald6::TDH3P-XYL1-TDH3T-PGK1P-XYL2-PGK1Tint#1::TEF1P-XYL3-TEF1Tsor1::FBA1P-LAD1-FBA1T-PGK1P-ALX1-CYC1T |
[8] |
| YE9 | Expressing the heterologous d-xylose, l-arabinose, and d-galacturonic acid pathway;YE6 int#7::FBA1P-lad1-FBA1T-PGK1P-alx1-CYC1T |
XYL1, XYL2, and XYL3 are derived from Pichia stipitis; gaaA, gaaC, and gaaD are derived from Aspergillus niger; lgd1 and lad1 are derived from Trichoderma reesei; alx1 is derived from Ambrosiozyma monospora.
gRNA sequences are designed to be target cut site-specific and 20-bp long, as listed in Table 4. The plasmids expressing each gRNA sequence were constructed by the fast cloning method [2], which is a PCR-based protocol for plasmid mutagenesis. To construct the pRS42H-ALD6.1 plasmid, for example, the pRS42H-GND1.1 plasmid (a template plasmid) [3] was amplified with the primers Kim044/Kim045 (Table 5). The PCR products were treated with DpnI and used to transform E. coli TOP10 (Invitrogen, Carlsbad, CA, USA). The transformants were selected on an LBA (5 g/L yeast extract, 10 g/L tryptone, 10 g/L NaCl, and 100 μg/mL ampicillin) agar plate. The gRNA sequence of the resulting plasmid was confirmed by Sanger sequencing using a universal primer for the T3 promoter. All other gRNA plasmids were constructed using the same procedure but different primers, as listed in Table 5.
-
2)
Donor DNA preparation
Table 4.
Guide RNA (gRNA) plasmids.
| gRNA | Target cut site | gRNA and PAM sequences (5’-) | Plasmid name |
|---|---|---|---|
| ALD6.1 | ALD6 | GTCAAGATCACACTTCCAAA TGG | pRS42H-ALD6.1 |
| PHO13.1 | PHO13 | TCCCTTATCTATTAACTTTC CGG | pRS42H-PHO13.1 |
| YPR1.1 | YPR1 | CATGGTAGATTATTATCTGT GGG | pRS42H-YPR1.1 |
| INT#4 | Intergenic region upstream ASF1 | CTCTCGAAGTGGTCACGTGC GGG | pRS42H-INT#4 |
| INT#6 | Intergenic region upstream ATG33 | TTGTCACAGTGTCACATCAG CGG | pRS42H-INT#6 |
| INT#7 | Intergenic region downstream YGR190C | GATACTTATCATTAAGAAAA TGG | pRS42H-INT#7 |
Table 5.
Primers used for construction of guide RNA plasmids.
| Plasmid name | Primers | Sequences (5’-) |
|---|---|---|
| pRS42H-ALD6.1 | Kim044 | AAGATCACACTTCCAAAGTTTTAGAGCTAGAAATAGCAAG |
| Kim045 | TTGGAAGTGTGATCTTGACGATCATTTATCTTTCACTGCG | |
| pRS42H-PHO13.1 | Kim624 | CTTATCTATTAACTTTCGTTTTAGAGCTAGAAATAGCAAG |
| Kim625 | AAAGTTAATAGATAAGGGAGATCATTTATCTTTCACTGCG | |
| pRS42H-YPR1.1 | Kim535 | GGTAGATTATTATCTGTGTTTTAGAGCTAGAAATAGCAAG |
| Kim536 | CAGATAATAATCTACCATGGATCATTTATCTTTCACTGCG | |
| pRS42H-INT#4 | Kim310 | TCGAAGTGGTCACGTGCGTTTTAGAGCTAGAAATAGCAAG |
| Kim311 | CACGTGACCACTTCGAGAGGATCATTTATCTTTCACTGCG | |
| pRS42H-INT#6 | Kim314 | TCACAGTGTCACATCAGGTTTTAGAGCTAGAAATAGCAAG |
| Kim315 | TGATGTGACACTGTGACAAGATCATTTATCTTTCACTGCG | |
| pRS42H-INT#7 | Kim486 | AGGAATTATGTTCGCCCGTTTTAGAGCTAGAAATAGCAAG |
| Kim487 | GGCGAACATAATTCCTTACGATCATTTATCTTTCACTGCG |
Donor DNA fragments were prepared by PCR using the primers listed in Table 6. Each of the fragments was flanked by 40–50 bp to allow in vivo assembly and genome integration through homologous recombination. Each assembly was an expression cassette of a heterologous gene as described in Fig. 3A. The donor DNAs for the xylose expression cassettes were designed to achieve complete removal of a target gene when genome integrated. On the other hand, the expression cassettes of the arabinose pathway and galacturonic acid pathway were integrated into an intergenic region without interfering neighboring genes.
-
3)
Yeast transformation
Table 6.
Primers used for construction of donor DNA fragments.
| Template genomic DNAa | Donor DNA fragments | Primers | Sequences (5’-) |
|---|---|---|---|
| XYL1 and XYL2 expression cassettes for deleting ALD6 (ald6::TDH3P-XYL1-TDH3T-PGK1P-XYL2-PGK1T) | |||
| S. cerevisiae | TDH3P | Kim626 | TAACATACACAAACACATACTATCAGAATACACTATTTTCGAGGACCTTGTC |
| SOO384 | TCAACTTAATAGAAGGCATTTTTAGATCTCCTAGGTTTGTTTGTTTATGTGTGTTTAT TC | ||
| P. stipitis | XYL1 | SOO385 | ATAAACACACATAAACAAACAAACCTAGGAGATCTAAAAATGCCTTCTATTAAGTTGA AC |
| SOO386 | AAT GCAAGATTTAAAGTAAATTCACTGTTAACGCATGCTTAGACGAAGATAGGAATCTTG | ||
| S. cerevisiae | TDH3T | SOO387 | GGA CAAGATTCCTATCTTCGTCTAAGCATGCGTTAACAGTGAATTTACTTTAAATCTTGC |
| SOO388 | ATTCTTTGAAGGTACTT CTTCGAAAAATTCGCGTCTGCTAGCTCCTGGCGGAAAAAATTC | ||
| S. cerevisiae | PGK1P | SOO389 | TTTTAAAGTTTACAAAT GAATTTTTTCCGCCAGGAGCTAGCAGACGCGAATTTTTCGAAG |
| SOO390 | CACCAA GGAAGGGTTAGCAGTCATTTTTTCTAGATGTTTTATATTTGTTGTAAAAAGTAG | ||
| P. stipitis | XYL2 | SOO391 | AATTAT CTACTTTTTACAACAAATATAAAACATCTAGAAAAAATGACTGCTAACCCTTCC |
| SOO392 | AAAAAATTGAT CTATCGATTTCAATTCAATTCAATACTAGTTTACTCAGGGCCGTCAATG | ||
| S. cerevisiae | PGK1T | SOO393 | GTCAAGTGTCT CATTGACGGCCCTGAGTAAACTAGTATTGAATTGAATTGAAATCGATAG |
| Kim627 | GTATATGACGGAAAGAAATGCAGGTTGGTACA AAATAATATCCTTCTCGAAAG | ||
| XYL3 expression cassette for deleting PHO13 (pho13::TDH3P-XYL1-TDH3T-PGK1P-XYL2-PGK1T) | |||
| S. cerevisiae | TDH3P | Kim628 | ATGTGACATCTTTACTATTCTCCAGCACGTTT CTTCATCGGTATCTTCGC |
| SOO374 | AA TGGGGTAGTGGTCATTTTTAAGCTTGAATTCTTTGTAATTAAAACTTAGATTAGATTG | ||
| P. stipitis | XYL3 | SOO375 | AT CTAATCTAAGTTTTAATTACAAAGAATTCAAGCTTAAAAATGACCACTACCCCATTTG |
| SOO376 | GCAACTA GAAAAGTCTTATCAATCTCCGTCGACATCGATTTAGTGTTTCAATTCACTTTC | ||
| S. cerevisiae | TDH3T | SOO377 | CAAGATG GAAAGTGAATTGAAACACTAAATCGATGTCGACGGAGATTGATAAGACTTTTC |
| Kim629 | CTATAACTCATTATTGGTTAAGGTGTAGATG AAGTTGGGTAACGCCAGG | ||
| gaaA expression cassette (int#4::CCW12P-gaaA-CCW12T) | |||
| S. cerevisiae | CCW12P | Kim379 | TTCCTCGGGCAGAGAAACTCGCAGGCAACTTG CACGCAAAAGAAAACCTT |
| Kim380 | TCAACA CAGCTGGGGGAGCCATTTTTTATTGATATAGTGTTTAAGCGAAT | ||
| A. niger | gaaA | Kim381 | TCTGTC ATTCGCTTAAACACTATATCAATAAAAAATGGCTCCCCCAGCTG |
| Kim382 | TAGA ATGTATAAATAATAATAAACTAAGTCTACTTCAGCTCCCACTTTCC | ||
| S. cerevisiae | CCW12T | Kim383 | GGAT GGAAAGTGGGAGCTGAAGTAGACTTAGTTTATTATTATTTATACAT |
| Kim384 | TGTGAGGGCCGATTATGCAGGCCTAGA TGTTCTAGTGTGTTTATATTATC | ||
| lgd1 expression cassette (int#4::PGK1P-lgd1-PGK1T) | |||
| S. cerevisiae | PGK1P | Kim385 | CCTCGGGCAGAGAAACTCGCAGGCAACTTG GTGAGTAAGGAAAGAGTGAG |
| Kim386 | GTGATGGTGACTTCAGACATTTTTTGTTTTATATTTGTTGTAAAAAGTAG | ||
| T. reesei | lgd1 | Kim387 | CTACTTTTTACAACAAATATAAAACAAAAAATGTCTGAAGTCACCATCAC |
| Kim388 | ATTGATCTAT CGATTTCAATTCAATTCAATTCAGATCTTCTCTCCGTTCA | ||
| S. cerevisiae | PGK1T | Kim389 | CTGCCCATCT TGAACGGAGAGAAGATCTGAATTGAATTGAATTGAAATCG |
| Kim390 | CTCTGTGAGGGCCGATTATGCAGGCCTAGA AAATAATATCCTTCTCGAAA | ||
| gaaC expression cassette (int#4::TDH3P-gaaC-TDH3T) | |||
| S. cerevisiae | TDH3P | Kim391 | CTCGGGCAGAGAAACTCGCAGGCAACTTG GAATAAAAAACACGCTTTTTC |
| Kim392 | GACTCCGGGGCG GAGCGGGGTAAAAGGCATTTTTTTTGTTTGTTTATGTGTGTT | ||
| A. niger | gaaC | Kim393 | TTCGAATA AACACACATAAACAAACAAAAAAAATGCCTTTTACCCCGCTC |
| Kim394 | ATTTAAAT GCAAGATTTAAAGTAAATTCACCTAAGCAATATCCGGCAACG | ||
| S. cerevisiae | TDH3T | Kim395 | TGAGAAGT CGTTGCCGGATATTGCTTAGGTGAATTTACTTTAAATCTTGC |
| Kim396 | CCTCTGTGAGGGCCGATTATGCAGGCCTAGA ATCCTGGCGGAAAAAATTC | ||
| gaaA, lgd1, and gaaC expression cassettes (int#4::CCW12P-gaaA-CCW12T-PGK1P-lgd1-PGK1T-TDH3P-gaaC-TDH3T) | |||
| S. cerevisiae YE3 | CCW12P-gaaA-CCW12T | Kim410 | TCTTTAGGTTAATTGTCGCTGTTATTGTCTA GATTTTTTCTCGGAGATGG |
| Kim411 | TAGTTC CTCACTCTTTCCTTACTCACTGTTCTAGTGTGTTTATATTATCC | ||
| S. cerevisiae YE4 | PGK1P-lgd1-PGK1T | Kim412 | AGCCAA GGATAATATAAACACACTAGAACA GTGAGTAAGGAAAGAGTGAG |
| Kim413 | AAACTCGAA CTGAAAAAGCGTGTTTTTTATTCCCGATTATGCAGGCCTAG | ||
| S. cerevisiae YE5 | TDH3P-gaaC-TDH3T | Kim414 | TATTATTTT CTAGGCCTGCATAATCGGGAATAAAAAACACGCTTTTTCAG |
| Kim415 | CTACTCTCTTCCTAGTCGCCCGGTTGTT GAAAGTTTAATTGTGGGTTTTC | ||
| lad1 and alx1 expression cassettes (int#7::FBA1P-lad1-FBA1T-PGK1P-alx1-CYC1T) | |||
| S. cerevisiae YE01 | FBA1P-lad1-FBA1T-PGK1P-alx1-CYC1T | Kim553 | CTTACACTTGTGTAATGACAAATGTTTTT TGAACAACAATACCAGCCTTC |
| Kim554 | TGTTTCACGTTATCAAGATTATGTCATCTATT GGCCGCAAATTAAAGCCT | ||
| Overexpression of YPR1 (CCW12P-YPR1) | |||
| S. cerevisiae | CCW12P | Kim537 | GTAACTTTGCAATATAATCAGGTCGCAAATAT CACGCAAAAGAAAACCTT |
| Kim538 | GAAGAATTCTTTAACGTAGCAGGCAT TATTGATATAGTGTTTAAGCGAAT | ||
| gaaD expression cassette (int#6::CCW12P-gaaD-CCW12T) | |||
| S. cerevisiae | CCW12P | Kim541 | CGGAGGAGACCGCTATAACCGGTTTGAATTTA CACGCAAAAGAAAACCTT |
| Kim542 | TA ACCTTCTTTCCGAGAGACATTTTTTATTGATATAGTGTTTAAGCGAAT | ||
| A. niger | gaaD | Kim543 | TC ATTCGCTTAAACACTATATCAATAAAAAATGTCTCTCGGAAAGAAGGT |
| Kim544 | GT ATAAATAATAATAAACTAAGTTTATTAAACAATCACCTTATGACCAGC | ||
| S. cerevisiae | CCW12T | Kim545 | TG GTCATAAGGTGATTGTTTAATAAACTTAGTTTATTATTATTTATACAT |
| Kim546 | CTTGCTTGCTGTCAAACTTCTGAGTTG TGTTCTAGTGTGTTTATATTATC | ||
The flanking region is underlined.
Saccharomyces cerevisiae D452-2; Pichia stipitis CBS 6054; Aspergillus niger CBS 120.49; Trichoderma reesei ATCC 5676.
For yeast transformation, a gRNA plasmid (4 μg) and donor DNA fragments (4 μg each) were used to transform a designated strain harboring pRS41N-Cas9 [3]. The resulting transformants were selected on a YPD agar plate supplemented with 100 μg/mL nourseothricin sulfate (Gold Biotechnology, St. Louis, MO, USA) and 300 μg/mL hygromycin B (Invitrogen, Carlsbad, CA, USA). Selected transformants were serially sub-cultured in YPD medium supplemented with 100 μg/mL nourseothricin sulfate to only remove the existing gRNA plasmids. Correct assembly and integration was then confirmed by yeast colony PCR with the primers listed in Table 7. Through four consecutive transformations, as described in Fig. 3, the YE9 strain was finally constructed.
Table 7.
Primers used for confirmation of correct assembly and integration.
| Primers | Sequences (5’-) | Primers | Sequences (5’-) |
|---|---|---|---|
| Introduction of d-xylose pathway | Introduction of d-galacturonic acid pathway | ||
| Kim049 | GGAACGGTGAGTGCAACG | Kim322 | GCGCATCTATTTGCCGTC |
| Kim427 | AAACTGTTCACCCAGACACC | Kim397 | GCTGGGGGAGCCATTTTTTATTG |
| Kim194 | AGCGCAACTACAGAGAACAGG | Kim398 | GTGGGAGCTGAAGTAGACTTAG |
| Kim100 | CGGCACCGTCGAACAATCTG | Kim323 | TCACGACACACCTCACTG |
| Kim101 | CCGCTTACTCTTCGTTCGGTCC | Kim399 | CCTGTGATGGTGACTTCAGAC |
| Kim193 | CTCAGCATCCACAATGTATCAG | Kim401 | GAACGGAGAGAAGATCTGAATTG |
| Kim426 | GCGCTATTGCATTGTTCTTGTC | Kim400 | ACAGCCTGTTCTCACACAC |
| Kim547 | AGGTATGCGATAGTTCCTCAC | Kim402 | GCGGGGTAAAAGGCATTTTTTTTG |
| Kim125 | TGCAGCTTCCAATTTCGTCAC | Kim408 | GCCGGATATTGCTTAGGTG |
| Kim630 | GAGGTGACACCCTTACCAAC | ||
| Kim631 | CTGCTACTCACACCTTCAACTC | Introduction of l-arabinose pathway | |
| Kim632 | CGCTGAACCCGAACATAGAAATATC | Kim490 | GGCACTAGGAGCATTTGTCG |
| Kim633 | TCGATATTTCTATGTTCGGGTTCAG | Kim304 | GCTTCGCTAATCCAGAGGTC |
| Kim078 | GATTGGAATTGGTTCGCAGTG | Kim400 | ACAGCCTGTTCTCACACAC |
| Kim048 | GAGGAAGACGTTGAAGGTGG | Kim491 | GTCCCTTAGGGTGCGTATAATG |
| Kim149 | TTTGAAGTGGTACGGCGATG | ||
| Kim577 | CACCCAAGCACAGCATAC | Overexpression of YPR1 | |
| Kim634 | TGGCTCGATAACGAAGATTCAG | Kim539 | CAATTCCGTGAAACCCTTTTCTT |
| Kim635 | GTCTTGTAGATTGAGAACTGGTCC | Kim540 | CTGCCAACTTCTTCTTCATTCAA |
| Kim636 | TCTATGAGGCAAGTAAGAGGCAC | ||
| Kim492 | AACAGGCGACAGTCCAAATG | Introduction of gaaD gene cassette | |
| Kim077 | TTGGAGTTCAAACTGGCGAG | Kim326 | GGTTCTGACTCCTACTGAGC |
| Kim093 | GCAAAGATAGCGGCGTAGGTG | ||
| Kim549 | GCATCCTTTGCCTCCGTTC | ||
| Kim327 | AGCATCGAGTACGGCAGTTC | ||
2.2. Fermentation
For fermentation of the YE9 strain, one colony was pre-cultured in YP medium (10 g/L yeast extract and 20 g/L peptone) supplemented with 20 g/L of glucose for 36 h at 30oC and 250 rpm. Cells were centrifuged, washed twice, and re-suspended in YP medium supplemented with desired carbon sources. The initial cell density of fermentation was 25 g/L dry weight, which corresponds to approximately 125 g/L wet weight, and this conversion factor was obtained from a prior study [4]. In the industrial bioethanol processes, >90% cells are recycled in repeated batch-type fermentation; therefore, very high cell density of up to 170 g/L wet weight [5] is often achieved. The concentrations of the carbon sources were selected to reflect the typical chemical composition of pectin-rich biomass (Table 8).
Table 8.
Chemical composition of pectin-rich biomass.
2.3. HPLC analysis
Quantitation of glucose, fructose, galactose, xylose, arabinose, galacturonic acid, glycerol, and ethanol was performed by high-performance liquid chromatography (HPLC; Agilent Technologies, 1260 series, USA) device equipped with a RI detector and a Rezex-ROA Organic Acid H+ (8%) (150 mm × 4.6 mm) column (Phenomenex Inc., Torrance, CA, USA). The column was eluted with 0.005 N H2SO4 at 0.6 mL/min and 50oC [1,6].
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.dib.2020.105359.
Conflict of Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Jeong D., Ye S., Park H., Kim S.R. Simultaneous fermentation of galacturonic acid and five-carbon sugars by engineered Saccharomyces cerevisiae. Bioresour. Technol. 2020;295:122259. doi: 10.1016/j.biortech.2019.122259. [DOI] [PubMed] [Google Scholar]
- 2.Li C., Wen A., Shen B., Lu J., Huang Y., Chang Y. FastCloning: a highly simplified, purification-free, sequence-and ligation-independent PCR cloning method. BMC Biotechnol. 2011;11(1):92. doi: 10.1186/1472-6750-11-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim S.R., Xu H., Lesmana A., Kuzmanovic U., Au M., Florencia C., Oh E.J., Zhang G., Kim K.H., Jin Y.-S. Deletion of PHO13, encoding haloacid dehalogenase type IIA phosphatase, results in upregulation of the pentose phosphate pathway in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2015;81(5):1601–1609. doi: 10.1128/AEM.03474-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aon J.C., Sun J., Leighton J.M., Appelbaum E.R. Hypoxia-elicited impairment of cell wall integrity, glycosylation precursor synthesis, and growth in scaled-up high-cell density fed-batch cultures of Saccharomyces cerevisiae. Microb. Cell Factories. 2016;15(1):142. doi: 10.1186/s12934-016-0542-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Basso L.C., De Amorim H.V., De Oliveira A.J., Lopes M.L. Yeast selection for fuel ethanol production in Brazil. FEMS Yeast Res. 2008;8(7):1155–1163. doi: 10.1111/j.1567-1364.2008.00428.x. [DOI] [PubMed] [Google Scholar]
- 6.Park J.-B., Kim J.-S., Jang S.-W., Kweon D.-H., Hong E.K., Shin W.C., Ha S.-J. Sequence analysis of KmXYL1 genes and verification of thermotolerant enzymatic activities of xylose reductase from four Kluyveromyces marxianus strains. Biotechnol. Bioproc. Eng. 2016;21(5):581–586. [Google Scholar]
- 7.Hosaka K., Nikawa J.-i., Kodaki T., Yamashita S. A dominant mutation that alters the regulation of INO1 expression in Saccharomyces cerevisiae. J. Biochem. 1992;111(3):352–358. doi: 10.1093/oxfordjournals.jbchem.a123761. [DOI] [PubMed] [Google Scholar]
- 8.Ye S., Jeong D., Shon J.C., Liu K.-H., Kim K.H., Shin M., Kim S.R. Deletion of PHO13 improves aerobic l-arabinose fermentation in engineered Saccharomyces cerevisiae. J. Ind. Microbiol. Biotechnol. 2019:1–7. doi: 10.1007/s10295-019-02233-y. [DOI] [PubMed] [Google Scholar]
- 9.Li P.P.-j., Xia J.-l., Nie Z.-y., Shan Y. Pectic oligosaccharides hydrolyzed from orange peel by fungal multi-enzyme complexes and their prebiotic and antibacterial potentials. LWT - Food Sci. Technol. (Lebensmittel-Wissenschaft -Technol.) 2016;69:203–210. [Google Scholar]
- 10.Zykwinska A., Boiffard M.-H.l.n., Kontkanen H., Buchert J., Thibault J.-F.o., Bonnin E. Extraction of green labeled pectins and pectic oligosaccharides from plant byproducts. J. Agric. Food Chem. 2008;56(19):8926–8935. doi: 10.1021/jf801705a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.