Abstract
Background
Ovarian cancer has a poor survival rate due to late diagnosis and improved methods are needed for its early detection. Our primary objective was to identify and incorporate additional biomarkers into longitudinal models to improve on the performance of CA125 as a first-line screening test for ovarian cancer.
Methods
This case–control study nested within UKCTOCS used 490 serial serum samples from 49 women later diagnosed with ovarian cancer and 31 control women who were cancer-free. Proteomics-based biomarker discovery was carried out using pooled samples and selected candidates, including those from the literature, assayed in all serial samples. Multimarker longitudinal models were derived and tested against CA125 for early detection of ovarian cancer.
Results
The best performing models, incorporating CA125, HE4, CHI3L1, PEBP4 and/or AGR2, provided 85.7% sensitivity at 95.4% specificity up to 1 year before diagnosis, significantly improving on CA125 alone. For Type II cases (mostly high-grade serous), models achieved 95.5% sensitivity at 95.4% specificity. Predictive values were elevated earlier than CA125, showing the potential of models to improve lead time.
Conclusions
We have identified candidate biomarkers and tested longitudinal multimarker models that significantly improve on CA125 for early detection of ovarian cancer. These models now warrant independent validation.
Subject terms: Diagnostic markers, Cancer screening, Ovarian cancer, Proteomic analysis
Background
Ovarian cancer is the sixth most common cancer in women, causing 152,000 deaths worldwide annually. The overall 5-year survival rate is ~40% due to late presentation, with the majority of cases diagnosed at stage III and IV, where the 5-year survival rate is only 3–19%. Stage I and II ovarian cancers have 5-year survival rates of 40–90%.1 Earlier detection of ovarian cancer, in particular aggressive tumours, is a possible way to improve outcomes.
Ovarian malignancies can be divided into two types differing by origin and molecular subtype and are associated with differing prognosis.2,3 Type I cancers can arise from tumours of low malignant potential, remain low grade, are slower growing and have a more favourable outcome. Type II cancers are typically high grade, aggressive, associated with poor survival, and are characterised by loss-of-function mutations of TP53 and BRCA1/2 and thus display genomic instability.4 It is well established that Type II cancers (mainly high-grade serous cancers) account for most ovarian cancer mortality and early detection of these tumours is likely to translate into mortality benefit.
Two tests used clinically to detect ovarian cancer are serum cancer antigen 125 (CA125) and transvaginal ultrasound. Both have limitations of specificity and sensitivity. CA125 is often elevated in benign conditions such as endometriosis and ovarian cysts and is not always detectable in early-stage disease. This has limited CA125’s potential as an accurate biomarker for early detection.5–8 Human epididymis secretory protein E4 (HE4/WFDC2), another Food and Drug Administration-approved biomarker for differential diagnosis of ovarian cancer, has similar limitations in detecting early and asymptomatic cancers.9–11
Multimarker tests have been shown to improve performance for ovarian cancer diagnosis compared to CA125 or HE4 alone.12,13 For example, the Risk of Ovarian Malignancy Algorithm, combining CA125 and HE4 for determining malignancy during pre-operative assessment, had a sensitivity of 88% compared to 63% and 78%, respectively, for CA125 and HE4 alone.14–16 Of more relevance, it has been shown that annual screening using serial serum measurements of CA125 in the Risk of Ovarian Cancer Algorithm (ROCA) within the UK Collaborative Trial of Ovarian Cancer Screening (UKCTOCS) provides an increased detection rate over CA125 cut-off models with a stage-shift in the detected cancers.17,18 Although the trial did not report a significant benefit in mortality through screening, there was an indication of a 15% mortality benefit, which awaits confirmation with further follow-up.19,20 Given the limitations of current ‘gold standards’ in ovarian cancer detection, there is an urgent need for new biomarkers. The incorporation of these into longitudinal algorithms with CA125 may be a promising strategy for improving early detection of ovarian cancer. We have previously described the Method of Mean Trends (MMT) algorithm, which provided high performance in predicting ovarian cancer based on serial CA125 measurements21 and have now adapted this to include multiple serial biomarker measurements.
Herein, we present the discovery of potential new serum biomarkers and longitudinal multimarker models capable of discriminating cancer-free controls and ovarian cancer cases prior to clinical diagnosis. We used 490 serial serum samples from 49 ovarian cancer cases taken at different times prior to diagnosis and 31 matched non-cancer controls nested within UKCTOCS, applying multidimensional liquid chromatography tandem mass spectrometry (LC-MS/MS) with tandem mass tagging for biomarker discovery. Promising candidates from this and the literature were assayed in all serial samples from these women and combined with CA125, and the data was used to generate longitudinal multimarker models. Using this novel approach, we show both increased sensitivity for ovarian cancer detection and earlier detection than using CA125 alone.
Methods
Sample set
The study set comprised serum from women recruited to UKCTOCS collected according to a standard operating procedure.17 Trial participants at enrolment were post-menopausal women aged 50–74 years who had no family history of ovarian cancer. Women subsequently diagnosed with ovarian cancer were identified by cross-referencing with the Health and Social Care Information Centre cancer registry and death codes, with diagnosis confirmed by review of histopathology reports. Forty-nine cases from the multimodal arm of UKCTOCS were selected comprising 10 borderline (BL) cases, 9 Type I cases and 30 Type II cases (Table 1). Thirty-one matched controls from the multimodal arm were selected who had no history of cancer and were matched to Type II cases based on age (±5 years), collection date (within 6 months), and collection centre. All serial samples from these women (n = 490) were retrieved from cryostorage, shipped on dry ice to the laboratory, and stored at −80 °C prior to analysis.
Table 1.
Sample set characteristics.
| (A) Characteristics of sample/patient cohort with cancer morphology, grade and Type | |||||||
|---|---|---|---|---|---|---|---|
| Cases | Controls | ||||||
| No. of individuals | 49 | 31 | |||||
| No. of samples | 315 | 175 | |||||
| Mean age at sample draw (years) (range) | 66.4 (50.6–79.7) | 65.0 (51.4–79.2) | |||||
| Median time to diagnosis (months) (range) | 26.27 (2.27–97.51) | na | |||||
| Morphology—grade (Type) | |||||||
| Serous—High grade (Type II) | 23 | ||||||
| Endometrioid—High grade (Type II) | 3 | ||||||
| Carcinosarcoma—High grade (Type II) | 1 | ||||||
| Not specified—High grade (Type II) | 3 | ||||||
| Endometrioid—Low grade (Type I) | 5 | ||||||
| Clear cell—High grade (Type I) | 3 | ||||||
| Not specified—Low grade (Type I) | 1 | ||||||
| Borderline (mixed morphologies) | 10 | ||||||
| (B) Characteristics of sample groups used for discovery profiling | |||||||
|---|---|---|---|---|---|---|---|
| Sample set | Sample group | No. of cases | No. of samples | Mean age (years) | Mean time to diagnosis (months) | Mean BMI | Modal FIGO stage |
| Discovery | Type I/BL ‘late’ | 9/10 | 9/10 | 68.1/70.1 | 8.8/4.7 | 28.4/26.04 | Ia/Ia |
| Type II ‘late’ | 30 | 30 | 67.1 | 5.5 | 26.8 | IIIc | |
| Control ‘late’ | 31 | 31 | 67.1 | na | 25.7 | na | |
| Type I/BL ‘early’ | 9/10 | 9/10 | 64.8/65.2 | 48.3/62.8 | 28.4/26.04 | Ia/Ia | |
| Type II ‘early’ | 30 | 30 | 62.5 | 60.4 | 26.8 | IIIc | |
| Control ‘early’ | 31 | 31 | 62.7 | NA | 25.5 | na | |
| All annual samples | Type I | 9 | 36 | 67.6 | 30.7 | 27.9 | Ia |
| Type II | 30 | 144 | 64.5 | 37.8 | 26.4 | IIIc | |
| Controls | 31 | 158 | 64.7 | na | 25.4 | na | |
Only annual screening samples were used for biomarker algorithm development
BL borderline ovarian cancers
LC-MS/MS discovery analysis
Samples were pooled into six groups for MS-based discovery, comprising ‘late’ (<14 months to diagnosis) and ‘early’ (>35 months to diagnosis) samples for each cancer case and control: Type I/BL early and late, Type II early and late, control early, and late (Table 1B). A pooling approach was taken, as the analysis of individual samples with extensive fractionation is not feasible in terms of time and cost-effectiveness. Details of the method can be found in Supplementary Materials and Methods. Briefly, pools were sequentially immunodepleted of the top 20 most abundant serum proteins, digested with trypsin, labelled in 6-plex using TMT reagents, and extensively fractionated (100 fractions) by strong anion exchange and high pH reversed-phase LC, prior to LC-MS/MS analysis on orbitrap instruments, essentially as described.22 Raw data files were combined and analysed using the Proteome Discoverer V1.4 software with database searching using the Mascot search engine V2.4. Data were filtered and reporter ion-based relative quantification of protein groups applied to compare expression across the six groups. A biomarker scoring system was applied to aid in candidate selection, ranking the proteins based on magnitude and consistency of expression differences, data quality, and biological function (see Supplementary Materials). The full data set and scoring system is available as Supplementary Data File S1.
Serum assays
Serum concentrations of biomarker candidates were quantified using commercial enzyme-linked immunosorbent assays (ELISA) or chemiluminescence immunoassays. Kits were first tested on pooled samples according to the manufacturers’ instructions to define optimal dilutions and assay reproducibility. The kits used, catalogue numbers, dilutions, and intra-assay coefficient of variations were: Human AGR2 ELISA Kit (ElabScience; E-EL-H0298; 1:20; 18%), CA125 ECLIA assay (Roche; Elecsys CA 125 II; 1:1; 4%), CHI3L1 Quantikine ELISA Kit (R&D Systems; DC3L10; 1:50; 14%), DNAH17 (human) ELISA Kit (EIAab; E5886h, 1:5; 17%), FSTL1 ELISA Kit (USCN; SEJ085Hu; 1:100; 11%), Glycodelin/PP14 Elisa Kit (Bioserv Diagnostics; BS-30-20; 1:2; 22%), HE4 ECLIA assay (Roche; Elecsys HE 4; 1:1; 8%), LRG1 ELISA Kit (IBL; 27769; 1:2000; 16%), Human PEBP4 ELISA Kit (ElabScience; E-EL-H5440; 1:200; 20%), and SLPI Quantikine ELISA Kit (R&D Systems; DP100, 1:50; 12%).
Model building, testing and statistical analysis
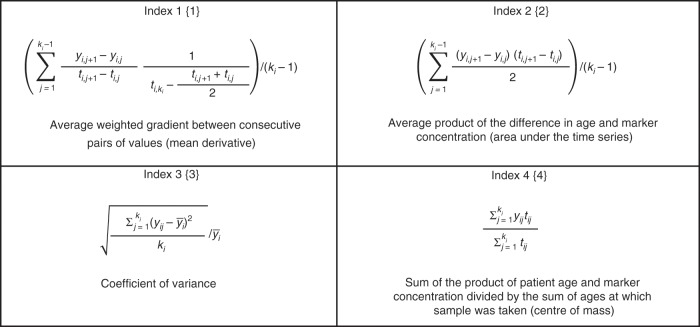
R software was used for model building and statistical analysis. T test or Mann–Whitney test was performed for parametric or non-parametric values, respectively, as determined by the D’Agostino and Pearson omnibus normality test. All marker values were log10-transformed for model generation. To generate longitudinal, multimarker models, for each individual, serial biomarker values from annual samples (taken <5 years to diagnosis) were first transformed into a single value that represented the degree of change over time of the candidate marker using one of the four indices (Fig. 1). Index 1{1} determines the average weighted gradient between consecutive pairs of values (mean derivative). Index 2{2} is the average product of the difference in age and marker concentration, representing the area under the time series. Index 3{3} is the coefficient of variance and does not use time as a factor. Index 4{4} is the sum of the product of patient age and marker concentration divided by the sum of ages at which the sample was taken (i.e., the centre of mass) and thus would reduce any effect of age on marker concentration, should such a relationship exist. We have previously described these indices and used them to build the MMT algorithm, based on serial CA125 measurements.21 Here indices were applied to all candidate measurements and, together with raw measurements (Index 5{5}), subjected to variable selection using a robust methodology that included Akaike Information Criterion (AIC), least absolute shrinkage and selection operator (lasso), bootstrapped lasso, mean accuracy decrease, and Gini impurity. AIC selection was performed using the standard implementation from package MASS in R. For lasso, the penalty parameter lambda was chosen as the first value which left only 3 variables in the model starting from 0 with 0.01 steps. In bootstrap lasso, 200 re-samplings were created from the original data and the traditional lasso approach applied to each of them. For each variable, the frequency of inclusion in the top three re-samplings was reported. Standard implementation of the mean accuracy decreases and Gini impurity was used from the randomForest package in R. This process identified the variables that could potentially serve as ‘good’ predictors in our models. All combinations of these predictors were then tested as logistic regression models, limiting the number of variables to three to avoid overfitting. The Caret package in R was used for leave-one-out cross-validation for selection of the best models. Models were then fitted on the whole data set and comparisons of sensitivity for the longitudinal models at fixed high specificity (>90% and >95%) were performed using McNemar’s test. Confidence intervals for sensitivity were generated by 2000 stratified bootstrap replicates23 for visual comparison. Model calibration was assessed with the Hosmer–Lemeshow goodness-of-fit test, with P values < 0.05 indicating a poorly calibrated model.
Fig. 1. Longitudinal trend indices used for transforming serial data.
For every ith patient, ki is the total number of serial measurements, yi,j is the jth serial measurement and ti,j and ti,ki represent, respectively, ages at which the current and the most recent measurements were taken. A simple cut-off for the final measurement of each candidate marker was used as Index 5{5}.
Results
MS-based profiling of pre-diagnosis serum samples
A set of 490 serum samples was sourced from the UKCTOCS biobank comprising serial samples from 49 volunteers who were later diagnosed with BL (n = 10), Type I (n = 9) or Type II (n = 30) ovarian malignancy (Table 1). Serial samples were also taken from matched cancer-free controls (n = 31). All women were aged >50 years and post-menopausal with no significant difference in age between the case and control groups. The majority of Type II cancers were high-grade serous (n = 23) with Type I cancers comprising low-grade endometrioid (n = 5), clear cell (n = 3) and low-grade unspecified (n = 1). Paired samples from these volunteers (n = 160) taken <14 months (‘late’) and >35 months (‘early’) prior to diagnosis and from matched controls (Table 1B) were pooled by cancer Type and time group and subjected to immunodepletion, tryptic digestion, 6-plex TMT-labelling and extensive peptide fractionation prior to LC-MS/MS-based proteomic profiling. TMT labelling efficiency was >99% with labelling in each TMT channel between 91.3% and 92.8%. This analysis yielded 748 protein groups quantified across all six sample groups. There were notable differences in protein expression and scores between the Type I/BL and Type II groups, possibly reflecting differences in tumour molecular profiles and the heterogeneity of the Type I/BL cases. Particularly, several acute-phase response proteins (e.g., A1AT, CRP, HP, ORM1, ORM2 and SAA1) were more highly upregulated in the Type I/BL group towards diagnosis, suggesting that Type I and/or BL disease is characterised by a more inflammatory phenotype.
Candidate selection and univariate testing
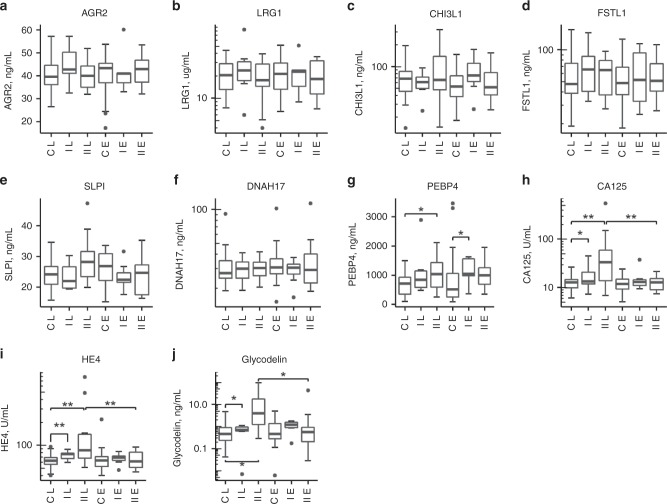
Five high-scoring candidates were selected for further testing based on functional assignment and availability of suitable commercial assays: chitinase-3-like protein 1 (CHI3L1/YKL40), dynein heavy chain 17 (DNAH17), follistatin-related protein 1 (FSTL1), leucine-rich alpha-2-glycoprotein (LRG1), and phosphatidylethanolamine-binding protein 4 (PEBP4). Our previous identification of CHI3L1 as a possible marker of early ovarian epithelial cell transformation24 also supported the choice of candidate. Four further proteins were selected based on previous studies: anterior gradient protein 2 (AGR2) homologue,25 human epididymis 4 (HE4/WFDC2),14,26 glycodelin (PAEP),27–29 and antileukoproteinase (SLPI).30,31 Assays were performed on all 490 serial samples from the 80 cases and controls to generate serial data (Supplementary Material; Fig. S1). Measurements for serum CA125 taken from UKCTOCS were also included in the analysis. We found no correlation between the concentration of any candidate and time from sample collection to spin (Pearson’s correlation coefficient, R < |0.2|), discounting any possible confounding effects of sample processing. In comparing late and early time groups used for LC-MS/MS discovery, CA125, HE4 and glycodelin distinguished cases from controls in the late time group and between the late and early time groups for Type II cases (Fig. 2h–j). PEBP4 distinguished Type II cases from controls in the late group (P = 0.02) and Type I cases from controls in the early group (P = 0.034) (Fig. 2g). No other candidate gave significant differences (P < 0.05), although trends were evident for some.
Fig. 2. Tukey box and whisker plots showing changes in pre-diagnostic serum candidate biomarker levels in cases and controls in two-time groups; <14 months to diagnosis (late) and >35 months to diagnosis (early).
a AGR2 serum levels; b LRG1 serum levels; c CHI3L1 serum levels; d FSTL1 serum levels; e SLPI serum levels; f DNAH17 serum levels; g PEBP4 serum levels; h CA125 serum levels; i HE4 serum levels; j glycodelin serum levels. *P < 0.05, **P < 0.001 from either t test or Mann–Whitney test. Data for borderline cases are omitted. Axis labels: C = controls; I = Type I; II = Type II; L = late; E = early.
Generation and testing of multimarker longitudinal models
Longitudinal analysis may increase the predictive capability of biomarkers.18 To address this, we first binned samples into pre-diagnosis time groups of ≤15, 15–30, 30–45 and >45 months and tested logistic regression models using log-transformed values, including up to 3 marker candidates or epidemiological variables (age, body mass index, oral contraceptive pill use, and hormone replacement therapy use) in the models. After leave-one-out cross-validation, these models failed to show any clinically significant improvement on CA125 alone (Supplementary Material; Table S1). We therefore undertook a novel longitudinal approach,21 whereby four trend indices ({1}–{4}) describing changes in concentration over time were used to transform the measurements for each candidate into a single value (Fig. 1). Only data from annual screening samples was used to eliminate potential bias from repeat CA125 testing triggered by ROCA in UKCTOCS and data for BL cases were also excluded. Summary statistics for the indexed biomarker values are presented in Tables S2 and S3.
Multimarker models may improve classification over single-marker tests. Therefore, following rigorous variable selection (Table S4), logistic regression was used to combine longitudinal index-transformed marker values, using a maximum of three main effects to avoid overfitting and applying leave-one-out cross-validation. Models were then evaluated using all cases whose final annual screening sample fell within 1 year to diagnosis. Samples taken >5 years before diagnosis were also excluded, leaving serial samples from 6 Type I and 22 Type II cases for comparison with serial samples from the 31 controls. Median sensitivities at fixed specificity (90.3% and 95.4%) were determined for each model with 95% confidence intervals (CIs) calculated by bootstrapping. Goodness-of-fit testing showed that the CA125{5} model was poorly calibrated (P = 0.04), while all multimarker models were well calibrated (P values ranging from 0.343 to 0.975). Model sensitivities were then compared to CA125 cut-off model CA125{5}, using McNemar’s exact test.
Considering all cases, three models combining CA125 with PEBP4, AGR2, CHI3L1 and/or HE4 gave significantly higher sensitivities (P < 0.05) at 90.3% specificity up to 1 year before diagnosis than CA125 alone (Table 2A, Fig. 3a, Table S5). At 95.4% specificity, while no model was significantly better than CA125 (best P value = 0.055), all had sensitivities >82%; the 2 best models (CA125{3}AGR2{3}CHI3L1{3} and CA125{3}CHI3L1{3}HE4{5}) had 87.5% sensitivity at 95.4% specificity, missing only 2 cases (Fig. 3d, e). Considering Type II cases only, 4 models gave 100% sensitivity at 90.3% specificity (Table 2 and Fig. 3a) and all models performed significantly better than CA125 alone (Table S5). The two best models (above) provided 95.5% sensitivity at 95.4% specificity (Table 2B and Fig. 3a), significantly outperforming CA125 (Table S5). Thus we demonstrate longitudinal multimarker models with significantly improved performance over CA125 alone for detecting ovarian cancer up to 1 year before diagnosis.
Table 2.
Performance of top models based on leave-one-out-cross-validation.
| Model | AUC (95% CI) | Sensitivity (95% CI) at 0.903 specificity | Sensitivity (95% CI) at 0.954 specificity |
|---|---|---|---|
| (A) All cases up to 1 year to diagnosis | |||
| CA125{5} | 0.869 (0.775–0.962) | 0.714 (0.536–0.857) | 0.643 (0.429–0.857) |
| CA125{3}CA125{4}PEBP4{5} | 0.96 (0.923–0.997) | 0.929 (0.75–1) | 0.821 (0.643–0.964) |
| CA125{3}CHI3L1{3}HE4{1} | 0.959 (0.916–1) | 0.929 (0.786–1) | 0.821 (0.643–0.964) |
| CA125{3}CHI3L1{3}HE4{5} | 0.96 (0.922–0.998) | 0.929 (0.786–1) | 0.857 (0.571–0.964) |
| CA125{3}AGR2{3}CHI3L1{3} | 0.966 (0.938–0.995) | 0.929 (0.786–1) | 0.857 (0.5–0.964) |
| CA125{3}HE4{4}HE4{5} | 0.974 (0.95–0.997) | 0.929 (0.786–1) | 0.821 (0.679–0.964) |
| (B) Type II cases up to 1 year to diagnosis | |||
| CA125{5} | 0.868 (0.756–0.979) | 0.727 (0.546–0.909) | 0.682 (0.455–0.864) |
| CA125{3}CA125{4}PEBP4{5} | 0.97 (0.934–1) | 0.955 (0.773–1) | 0.818 (0.636–1) |
| CA125{3}CHI3L1{3}HE4{1} | 0.986 (0.973–0.999) | 1.0 (0.909–1) | 0.909 (0.727-1) |
| CA125{3}CHI3L1{3}HE4{5} | 0.984 (0.97–0.998) | 1.0 (0.909–1) | 0.955 (0.636–1) |
| CA125{3}AGR2{3}CHI3L1{3} | 0.984 (0.971–0.998) | 1.0 (0.909–1) | 0.955 (0.636–1) |
| CA125{3}HE4{4}HE4{5} | 0.988 (0.976–1) | 1.0 (0.864–1) | 0.909 (0.727–1) |
| (C) All cases at 1–2 years to diagnosis | |||
| CA125{5} | 0.507 (0.382–0.631) | 0.094 (0–0.281) | 0 (0–0.094) |
| CA125{3}CA125{4}PEBP4{5} | 0.658 (0.543–0.772) | 0.313 (0.125–0.5) | 0.188 (0.063–0.375) |
| CA125{3}CHI3L1{3}HE4{1} | 0.648 (0.531–0.765) | 0.281 (0.125–0.438) | 0.156 (0.031–0.313) |
| CA125{3}CHI3L1{3}HE4{5} | 0.65 (0.54–0.789) | 0.25 (0.094–0.469) | 0.188 (0.031–0.344) |
| CA125{3}AGR2{3}CHI3L1{3} | 0.697 (0.595–0.8) | 0.375 (0.156–0.532) | 0.188 (0–0.406) |
| CA125{3}HE4{4}HE4{5} | 0.649 (0.532–0.766) | 0.313 (0.156–0.531) | 0.25 (0.063–0.438) |
| (D) Type II cases at 1–2 years to diagnosis | |||
| CA125{5} | 0.504 (0.361–0.648) | 0.08 (0–0.24) | 0 (0–0.12) |
| CA125{3}CA125{4}PEBP4{5} | 0.632 (0.499–0.765) | 0.24 (0.08–0.48) | 0.16 (0.04–0.32) |
| CA125{3}CHI3L1{3}HE4{1} | 0.65 (0.523–0.778) | 0.2 (0.04–0.44) | 0.08 (0–0.28) |
| CA125{3}CHI3L1{3}HE4{5} | 0.643 (0.519–0.766) | 0.24 (0.04–0.48) | 0.16 (0–0.32) |
| CA125{3}AGR2{3}CHI3L1{3} | 0.71 (0.6–0.82) | 0.36 (0.12–0.56) | 0.16 (0–0.4) |
| CA125{3}HE4{4}HE4{5} | 0.644 (0.517–0.772) | 0.24 (0.08–0.44) | 0.16 (0.04–0.36) |
Area under the ROC curve (AUC) is given with 95% confidence intervals (CI) calculated using the method of DeLong. Sensitivities are given at specificities of 0.903 and 0.954 with 95% CI calculated by bootstrapping with 2000 stratified replicates
Fig. 3. Top five longitudinal models.
a Sensitivity of models with 95% CI at 90.3% (left) and 95.4% (right) specificity at <1 year to diagnosis for all cases (top) or Type II cases only (bottom). Sensitivities were compared to CA125 cut-off model (CA125{5}) by one-tailed McNemar’s test; *P < 0.05. b Sensitivity of models 1–2 years before diagnosis. c–f Predictions are presented as logistic regression outcomes for four models plotted against model CA125{5}. Horizontal and vertical dashed lines represent cut-offs for classification at 90.3% sensitivity. Cases in the grey box were identified by the multimarker model and not by CA125{5}.
The capacity of models to predict ovarian cancer earlier than 1 year before diagnosis was next investigated. Performances were evaluated within 1–2 years of diagnosis after excluding samples taken within 1 year of diagnosis. All longitudinal models had sensitivities >25% at 90.3% specificity, although none were significantly better than CA125 alone (Table 2C, Fig. 3b, Table S5). The best model (CA125{3}AGR2{3}CHI3L1{3}) provided 37.5% sensitivity (all cases) and 36% sensitivity (Type II cases) at 90.3% specificity versus 9.4% and 8.0%, respectively, for CA125 alone. Plotting prediction values against time to diagnosis showed that the models detected cancer earlier than CA125 (Fig. 4). Models CA125{3}AGR2{3}CHI3L1{3} and CA125{3}HE4{4}HE4{5} were the best for Type II cases, providing test positivity 18 and 15 months earlier, respectively, than CA125. The data show the potential of the models to improve on the lead time of detection.
Fig. 4. Model predictions presented as logistic regression outcomes for multivariate and raw outcome for univariate analyses by time.
Time is years to diagnosis for cases and years since final sample for controls. Model cut-offs giving 90.3% specificity are indicated by horizontal dashed lines. Grey lines represent individuals and circles the time the sample was taken. The thick line is a Loess fit over all points with 95% CIs in grey shading. Vertical dashed lines indicate the timepoint at which the Loess curve intersects the threshold for model CA125{5}.
Discussion
While elevated serum CA125 is a reasonably robust marker for detecting ovarian cancer, it is not always raised in early-stage disease and can be elevated in benign conditions. This necessitates the discovery of complementary biomarkers and algorithms that together with CA125 will improve early detection of ovarian cancer. Herein we have identified new candidate biomarkers using a set of pre-diagnosis serum samples nested within UKCTOCS and generated longitudinal data for these markers. The data were then used to develop novel longitudinal models capable of identifying ovarian cancer cases that were undetectable using CA125 alone and gave a significant improvement in performance up to 1 year prior to diagnosis and with improved lead times of detection.
Initial proteomic profiling of pooled sera identified a number of potential biomarkers that were elevated in cancer cases compared to controls. One of these was PEBP4, a suppressor of proliferation and invasion in ovarian tumour cell lines,32 and a RAF1- and MEK1-binding protein that inhibits tumour necrosis factor-α-induced apoptosis.33 PEBP4 was significantly elevated in sera from cancer cases prior to diagnosis and featured in one of the top multivariable longitudinal models. CHI3L1 also featured in two of the best models, and while we showed it not to be discriminatory alone, it complemented CA125 when used longitudinally. This highlights the importance of not discounting candidates based on univariate analysis alone. CHI3L1 is a carbohydrate-binding lectin with a purported role in tissue remodelling, angiogenesis and survival that may function by modulating chemokine and inflammatory responses. It has been previously reported as a serum marker of ovarian cancer34 and, in one study, outperformed CA125 for detecting early-stage disease.35 Our previous work had identified elevated levels of secreted CHI3L1 in immortalised ovarian surface epithelial cells transformed with either MYC or MYC plus activated KRAS-G12V, suggesting that its overexpression may be an early event in epithelial cell transformation.24 AGR2 also complemented CA125 and CHI3L1 in one of the top models. AGR2 is a secreted and endoplasmic reticulum-resident chaperone protein required for folding, trafficking and assembly of cysteine-rich transmembrane receptors and mucins. Its overexpression in numerous cancer types has been linked with enhanced cell migration and proliferation, altered adhesion and differentiation and promotion of angiogenesis and metastasis.36 Previous studies have found elevated tissue expression of AGR2 to be an indicator of poor prognosis,37 and it has been reported as a putative blood-borne biomarker for the detection and/or prognosis of ovarian, pancreatic, prostate and lung cancer.25,38–40 Our findings support AGR2 as a biomarker for early ovarian cancer detection that complements CA125 (and CHI3L1) when used longitudinally.
A major challenge in early detection is how best to normalise a biomarker to natural variations in serum levels within individuals and across populations. By analysing longitudinal data using the described indices, we have defined changes in biomarker levels that deviate from individual baselines providing greatly improved performance compared to simple cut-off models. This type of approach was used successfully to develop the ROCA, where serial CA125 measurements were used to substantially improve on using single CA125 measurements.18 In agreement, the best single marker in our data set was longitudinal CA125 using Index 3 (CA125{3}), which provided higher sensitivity at fixed specificity compared to using the final CA125 measurement, although the increase was not significant. However, by combining longitudinally indexed markers (including CA125), we show significant improvement in sensitivity and lead time over using CA125 alone.
The sensitivity of the CA125 cut-off model used herein (71.4%, 95% CI 53.6–85.7) was similar to that reported previously using a validation set of 25,042 UKCTOCS women (73.1%, 95% CI 63.6–80.8) at 90% specificity.21 This demonstrates that the distribution of CA125 in our data set is representative of the whole of UKCTOCS. Comparison with performance of published serial CA125 algorithms applied to the whole UKCTOCS cohort as first-line screening tests shows our models gave higher sensitivity at similar specificity. Four of the models gave 92.9% sensitivity (95% CI 78.6–100) at 90.3% specificity, compared to MMT with 86.5% sensitivity (95% CI 78.4–91.9) at 89.5% specificity, PEB with 88.5% sensitivity (95% CI 80.6–93.4) at 89.5% specificity, and ROCA with 87.1% sensitivity at 87.6% specificity.18,21 The best model (CA125{3}AGR2{3}CHI3L1{3}) also substantially outperformed these serial CA125 algorithms at 1–2 years to diagnosis, with a sensitivity of 37.5% versus 23.3% for MMT and 26.7% for PEB,21 detecting cancers earlier. Moreover, for the more aggressive Type II cases, our models provided 100% sensitivity at 90.3% specificity at 1 year to diagnosis. Thus we have demonstrated that combining additional markers longitudinally improves detection rate and lead time compared to using CA125 alone.
The key strengths of our study are the use of a novel combination of longitudinal biomarker measurements using pre-diagnosis serial samples from a well-characterised cohort, with rigorous variable selection and model testing that compared poor prognosis cancers with all cancer cases. The main weakness of this discovery study was the small sample size used, with samples from 6 Type I and 22 Type II cases used for model testing; some cases were excluded as they lacked an annual UKCTOCS sample taken within 1 year to diagnosis, and we used controls only from women who had not been diagnosed with any cancer at sample donation and during follow-up; the latter may exaggerate specificity. The small sample size and heterogeneous nature of the cases also precluded any meaningful subset analysis. Despite rigorous variable selection and cross-validation, we cannot rule out the potential for overfitting and it is now essential to validate these models in a larger independent cohort of samples with more detailed investigation of lead time and model performance by stage and histological subtype. Nevertheless, the findings are encouraging and advocate the benefits of incorporating serial sampling into biomarker discovery and clinical testing studies.
In conclusion, we have generated multimarker longitudinal models for the early detection of ovarian cancer that significantly outperform CA125, detecting Type I and II cases that CA125 did not. Furthermore, we show the potential of these models to improve lead time. Blinded validation of these models in a larger, longitudinal sample set is now warranted to investigate the potential of these algorithms for early detection in ovarian cancer screening.
Supplementary information
Acknowledgements
We thank all trial participants and all staff involved in the UKCTOCS trial.
Author contributions
J.F.T., I.J., U.M. and A.Z. conceived the design of the study and secured funding. A.G.-M., J.K. and A.R. selected samples and provided associated data from UKCTOCS. H.J.W., J.W. and R.G. performed proteomic and biomarker analyses. H.J.W. and O.B. performed data analysis, model building and testing. J.T., H.J.W. and O.B. drafted the initial manuscript. All authors provided comments and edited the final version of the manuscript.
Ethics approval and consent to participate
This nested case–control study within UKCTOCS was approved by the Joint UCL/UCLH Research Ethics Committee A (Ref. 05/Q0505/57). Written informed consent was obtained from donors and no data allowing identification of patients was provided. The study was performed in accordance with the Declaration of Helsinki.
Consent to publish
Not applicable.
Data availability
Raw assay data, excepting CA125, are available upon request.
Competing interests
U.M. and I.J. declare financial interest through UCL Business and Abcodia Ltd in the third-party exploitation of clinical trial biobanks, which have been developed through research at UCL. The remaining authors declare no conflict of interest.
Funding information
This study, part of PROMISE, was funded through Cancer Research UK PRC Programme Grant A12677, Cancer Research UK EDC Project Award C12077/A26223 and by the Eve Appeal Gynaecological Cancer Research Trust. It was supported by the National Institute for Health Research (NIHR) University College London Hospitals (UCLH) Biomedical Research Centre. UKCTOCS was core funded by the Medical Research Council, Cancer Research UK and the Department of Health with additional support from the Eve Appeal, Special Trustees of Bart’s and the London and Special Trustees of UCLH. A.Z. acknowledges support by the grant of the Ministry of Education and Science of the Russian Federation Agreement No. 074-02-2018-330.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Harry J. Whitwell, Jenny Worthington, Oleg Blyuss
Supplementary information
Supplementary information is available for this paper at 10.1038/s41416-019-0718-9.
References
- 1.CRUK. Cancer statistics: ovarian cancer survival statistics. https://www.cancerresearchuk.org/health-professional/cancer-statistics/statistics-by-cancer-type/ovarian-cancer/survival (2019).
- 2.Kurman RJ, Shih IeM. The origin and pathogenesis of epithelial ovarian cancer: a proposed unifying theory. Am. J. Surg. Pathol. 2010;34:433–443. doi: 10.1097/PAS.0b013e3181cf3d79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koshiyama M, Matsumura N, Konishi I. Recent concepts of ovarian carcinogenesis: type I and type II. Biomed. Res. Int. 2014;2014:934261. doi: 10.1155/2014/934261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bowtell DD. The genesis and evolution of high-grade serous ovarian cancer. Nat. Rev. Cancer. 2010;10:803–808. doi: 10.1038/nrc2946. [DOI] [PubMed] [Google Scholar]
- 5.Jacobs I, Bast RC., Jr. The CA 125 tumour-associated antigen: a review of the literature. Hum. Reprod. 1989;4:1–12. doi: 10.1093/oxfordjournals.humrep.a136832. [DOI] [PubMed] [Google Scholar]
- 6.Daoud E, Bodor G. CA-125 concentrations in malignant and nonmalignant disease. Clin. Chem. 1991;37:1968–1974. doi: 10.1093/clinchem/37.11.1968. [DOI] [PubMed] [Google Scholar]
- 7.Collins WP, Bourne TH, Campbell S. Screening strategies for ovarian cancer. Curr. Opin. Obstet. Gynecol. 1998;10:33–39. doi: 10.1097/00001703-199802000-00007. [DOI] [PubMed] [Google Scholar]
- 8.Kitawaki J, Ishihara H, Koshiba H, Kiyomizu M, Teramoto M, Kitaoka Y, et al. Usefulness and limits of CA-125 in diagnosis of endometriosis without associated ovarian endometriomas. Hum. Reprod. 2005;20:1999–2003. doi: 10.1093/humrep/deh890. [DOI] [PubMed] [Google Scholar]
- 9.Van Gorp T, Cadron I, Despierre E, Daemen A, Leunen K, Amant F, et al. HE4 and CA125 as a diagnostic test in ovarian cancer: prospective validation of the Risk of Ovarian Malignancy Algorithm. Br. J. Cancer. 2011;104:863–870. doi: 10.1038/sj.bjc.6606092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sarojini S, Tamir A, Lim H, Li S, Zhang S, Goy A, et al. Early detection biomarkers for ovarian cancer. J. Oncol. 2012;2012:709049. doi: 10.1155/2012/709049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moore RG, Miller MC, Steinhoff MM, Skates SJ, Lu KH, Lambert-Messerlian G, et al. Serum HE4 levels are less frequently elevated than CA125 in women with benign gynecologic disorders. Am. J. Obstet. Gynecol. 2012;206:351 e1–351 e8. doi: 10.1016/j.ajog.2011.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nolen BM, Lokshin AE. Protein biomarkers of ovarian cancer: the forest and the trees. Future Oncol. 2012;8:55–71. doi: 10.2217/fon.11.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Whitwell HJ, Blyuss O, Menon U, Timms JF, Zaikin A. Parenclitic networks for predicting ovarian cancer. Oncotarget. 2018;9:22717–22726.. doi: 10.18632/oncotarget.25216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moore RG, McMeekin DS, Brown AK, DiSilvestro P, Miller MC, Allard WJ, et al. A novel multiple marker bioassay utilizing HE4 and CA125 for the prediction of ovarian cancer in patients with a pelvic mass. Gynecol. Oncol. 2009;112:40–46. doi: 10.1016/j.ygyno.2008.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Montagnana M, Danese E, Ruzzenente O, Bresciani V, Nuzzo T, Gelati M, et al. The ROMA (Risk of Ovarian Malignancy Algorithm) for estimating the risk of epithelial ovarian cancer in women presenting with pelvic mass: is it really useful? Clin. Chem. Lab. Med. 2011;49:521–525. doi: 10.1515/CCLM.2011.075. [DOI] [PubMed] [Google Scholar]
- 16.Dikmen ZG, Colak A, Dogan P, Tuncer S, Akbiyik F. Diagnostic performances of CA125, HE4, and ROMA index in ovarian cancer. Eur. J. Gynaecol. Oncol. 2015;36:457–462. [PubMed] [Google Scholar]
- 17.Menon U, Gentry-Maharaj A, Hallett R, Ryan A, Burnell M, Sharma A, et al. Sensitivity and specificity of multimodal and ultrasound screening for ovarian cancer, and stage distribution of detected cancers: results of the prevalence screen of the UK Collaborative Trial of Ovarian Cancer Screening (UKCTOCS) Lancet Oncol. 2009;10:327–40.. doi: 10.1016/S1470-2045(09)70026-9. [DOI] [PubMed] [Google Scholar]
- 18.Menon U, Ryan A, Kalsi J, Gentry-Maharaj A, Dawnay A, Habib M, et al. Risk algorithm using serial biomarker measurements doubles the number of screen-detected cancers compared with a single-threshold rule in the United Kingdom Collaborative Trial of Ovarian Cancer Screening. J. Clin. Oncol. 2015;33:2062–71.. doi: 10.1200/JCO.2014.59.4945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jacobs IJ, Menon U, Ryan A, Gentry-Maharaj A, Burnell M, Kalsi JK, et al. Ovarian cancer screening and mortality in the UK Collaborative Trial of Ovarian Cancer Screening (UKCTOCS): a randomised controlled trial. Lancet. 2016;387:945–56.. doi: 10.1016/S0140-6736(15)01224-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson N. Two large randomised trials show ovarian cancer screening has minimal impact on survival. BJOG. 2018;125:524–525. doi: 10.1111/1471-0528.14052. [DOI] [PubMed] [Google Scholar]
- 21.Blyuss O, Burnell M, Ryan A, Gentry-Maharaj A, Marino IP, Kalsi J, et al. Comparison of longitudinal CA125 algorithms as a first-line screen for ovarian cancer in the general population. Clin. Cancer Res. 2018;24:4726–4733. doi: 10.1158/1078-0432.CCR-18-0208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sinclair J, Timms JF. Quantitative profiling of serum samples using TMT protein labelling, fractionation and LC-MS/MS. Methods. 2011;54:361–369. doi: 10.1016/j.ymeth.2011.03.004. [DOI] [PubMed] [Google Scholar]
- 23.Carpenter J, Bithell J. Bootstrap confidence intervals: when, which, what? A practical guide for medical statisticians. Stat. Med. 2000;19:1141–64.. doi: 10.1002/(SICI)1097-0258(20000515)19:9<1141::AID-SIM479>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 24.Lawrenson K, Mhawech-Fauceglia P, Worthington J, Spindler TJ, O'Brien D, Lee JM, et al. Identification of novel candidate biomarkers of epithelial ovarian cancer by profiling the secretomes of three-dimensional genetic models of ovarian carcinogenesis. Int. J. Cancer. 2015;137:1806–17.. doi: 10.1002/ijc.29197. [DOI] [PubMed] [Google Scholar]
- 25.Edgell TA, Barraclough DL, Rajic A, Dhulia J, Lewis KJ, Armes JE, et al. Increased plasma concentrations of anterior gradient 2 protein are positively associated with ovarian cancer. Clin. Sci. (Lond.) 2010;118:717–25.. doi: 10.1042/CS20090537. [DOI] [PubMed] [Google Scholar]
- 26.Hellstrom I, Raycraft J, Hayden-Ledbetter M, Ledbetter JA, Schummer M, McIntosh M, et al. The HE4 (WFDC2) protein is a biomarker for ovarian carcinoma. Cancer Res. 2003;63:3695–3700. [PubMed] [Google Scholar]
- 27.Bischof A, Briese V, Richter DU, Bergemann C, Friese K, Jeschke U. Measurement of glycodelin A in fluids of benign ovarian cysts, borderline tumours and malignant ovarian cancer. Anticancer Res. 2005;25:1639–1644. [PubMed] [Google Scholar]
- 28.Havrilesky LJ, Whitehead CM, Rubatt JM, Cheek RL, Groelke J, He Q, et al. Evaluation of biomarker panels for early stage ovarian cancer detection and monitoring for disease recurrence. Gynecol. Oncol. 2008;110:374–82.. doi: 10.1016/j.ygyno.2008.04.041. [DOI] [PubMed] [Google Scholar]
- 29.Blyuss O, Gentry-Maharaj A, Fourkala EO, Ryan A, Zaikin A, Menon U, et al. Serial Patterns of ovarian cancer biomarkers in a prediagnosis longitudinal dataset. Biomed. Res. Int. 2015;2015:681416. doi: 10.1155/2015/681416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsukishiro S, Suzumori N, Nishikawa H, Arakawa A, Suzumori K. Use of serum secretory leukocyte protease inhibitor levels in patients to improve specificity of ovarian cancer diagnosis. Gynecol. Oncol. 2005;96:516–519. doi: 10.1016/j.ygyno.2004.10.036. [DOI] [PubMed] [Google Scholar]
- 31.Timms JF, Arslan-Low E, Kabir M, Worthington J, Camuzeaux S, Sinclair J, et al. Discovery of serum biomarkers of ovarian cancer using complementary proteomic profiling strategies. Proteomics Clin. Appl. 2014;8:982–993. doi: 10.1002/prca.201400063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li HZ, Wang Y, Gao Y, Shao J, Zhao XL, Deng WM, et al. Effects of raf kinase inhibitor protein expression on metastasis and progression of human epithelial ovarian cancer. Mol. Cancer Res. 2008;6:917–928. doi: 10.1158/1541-7786.MCR-08-0093. [DOI] [PubMed] [Google Scholar]
- 33.Wang X, Li N, Liu B, Sun H, Chen T, Li H, et al. A novel human phosphatidylethanolamine-binding protein resists tumor necrosis factor alpha-induced apoptosis by inhibiting mitogen-activated protein kinase pathway activation and phosphatidylethanolamine externalization. J. Biol. Chem. 2004;279:45855–45864. doi: 10.1074/jbc.M405147200. [DOI] [PubMed] [Google Scholar]
- 34.Chen, J., Liu, X., Pan, Y. Q., He, B. S., Ying, H. Q., Sun, H. L. et al. Analysis of diagnostic value of YKL-40 in ovarian cancer. Int. J. Gynecol. Cancer10.1097/IGC.0000000000000717 (2016). [DOI] [PubMed]
- 35.Dupont J, Tanwar MK, Thaler HT, Fleisher M, Kauff N, Hensley ML, et al. Early detection and prognosis of ovarian cancer using serum YKL-40. J. Clin. Oncol. 2004;22:3330–3339. doi: 10.1200/JCO.2004.09.112. [DOI] [PubMed] [Google Scholar]
- 36.Delom F, Nazaraliyev A, Fessart D. The role of protein disulphide isomerase AGR2 in the tumour niche. Biol. Cell. 2018;110:271–282. doi: 10.1111/boc.201800024. [DOI] [PubMed] [Google Scholar]
- 37.Tian SB, Tao KX, Hu J, Liu ZB, Ding XL, Chu YN, et al. The prognostic value of AGR2 expression in solid tumours: a systematic review and meta-analysis. Sci. Rep. 2017;7:15500. doi: 10.1038/s41598-017-15757-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Makawita S, Dimitromanolakis A, Soosaipillai A, Soleas I, Chan A, Gallinger S, et al. Validation of four candidate pancreatic cancer serological biomarkers that improve the performance of CA19.9. BMC Cancer. 2013;13:404. doi: 10.1186/1471-2407-13-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kani K, Malihi PD, Jiang Y, Wang H, Wang Y, Ruderman DL, et al. Anterior gradient 2 (AGR2): blood-based biomarker elevated in metastatic prostate cancer associated with the neuroendocrine phenotype. Prostate. 2013;73:306–15.. doi: 10.1002/pros.22569. [DOI] [PubMed] [Google Scholar]
- 40.Chung K, Nishiyama N, Yamano S, Komatsu H, Hanada S, Wei M, et al. Serum AGR2 as an early diagnostic and postoperative prognostic biomarker of human lung adenocarcinoma. Cancer Biomark. 2011;10:101–107. doi: 10.3233/CBM-2012-0234. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw assay data, excepting CA125, are available upon request.