Abstract
The CCAAT sequence is a ubiquitous cis-element of eukaryotic promoters, and genes containing CCAAT sequences have been shown to be activated by the CCAAT-binding transcription factor complex in several eukaryotic model organisms. In general, CCAAT-binding transcription factors form heterodimers or heterotrimeric complexes that bind to CCAAT sequences within the promoters of target genes and regulate various cellular processes. To date, except Hap complex, CCAAT-binding complex has been rarely reported in fungi. In this study, we characterized two CCAAT-binding transcription factors (Fct1 and Fct2) in the plant pathogenic fungus Fusarium graminearum. Previously, FCT1 and FCT2 were shown to be related to DNA damage response among eight CCAAT-binding transcription factors in F. graminearum. We demonstrate that the nuclear CCAAT-binding complex of F. graminearum has important functions in various fungal developmental processes, not just DNA damage response but virulence and mycotoxin production. Moreover, the results of biochemical and genetic analyses revealed that Fct1 and Fct2 may form a complex and play distinct roles among the eight CCAAT-binding transcription factors encoded by F. graminearum. To the best of our knowledge, the results of this study represent a substantial advancement in our understanding of the molecular mechanisms underlying the functions of CCAAT-binding factors in eukaryotes.
Subject terms: Fungal genetics, Fungal pathogenesis
Introduction
Gene expression is primarily orchestrated by a set of transcription factors that bind to cis-elements in promoter regions1. In addition to the TATA-box, the CCAAT sequence is a ubiquitous cis-element of eukaryotic promoters that is present in the promoters of approximately 30% of eukaryotic genes2. Therefore, genes carrying a CCAAT-box are known to be primarily activated by conserved CCAAT-binding complexes in model eukaryotic organisms3.
The CCAAT-binding complex (CBC) typically consists of heterotrimeric core subunits and regulates primary/secondary metabolism, development, stress responses, and virulence in animals, plants, and fungi4–7. The heme activator protein (HAP) complex, which is also termed nuclear factor Y (NF-Y) or CCAAT-binding factor (CBF), was the first identified and is the most well studied CBC in various eukaryotic organisms. The Saccharomyces cerevisiae Hap complex consists of three essential CCAAT-binding factors (Hap2p, Hap3p, and Hap5p) that are indispensable for CCAAT-binding activity8,9, orthologues of which (NF-YA/CBF-B, NF-YB/CBF-A, and NF-YC/CBF-C, respectively) also comprise the mammalian CCAAT complex (NF-Y/CBF)10. Core elements of Hap3 and Hap5 display amino acid sequence similarities to the histone fold motifs of histones H2B and H2A, respectively, which are responsible for heterodimeric interactions. Another essential element of the complex, Hap2, contains a subunit association domain that allows for heterotrimer formation and nuclear localization signals (NLS)11,12. After the assembly of Hap2, Hap3, and Hap5, the heterotrimeric complex then recruits Hap4, an additional component that is only present in fungi, which allows for subsequent binding to the promoter of the target genes containing the CCAAT sequence3,12.
Similar to Hap5 and Hap3, two small components of DNA polymerase epsilon (Pol ε), DNA polymerase II subunit B3 (Dpb3) and Dpb4 are known that harbour H2A/H2B-like histone fold motifs in S. cerevisiae and Schizosaccharomyces pombe, as well as in plants and humans13–16. Pol ε plays crucial roles in chromosome replication, cell cycling, and the repair of damaged DNA17. Both Dpb3 and Dpb4 are non-essential proteins that form a heterodimeric complex and bind to double-stranded DNA18–20. The Dpb3-Dpb4 complex physically associates with the Pol ε catalytic subunit, Cdc20, and also interacts with proteins that are important for heterochromatin assembly. However, orthologues of Dpb3 and Dpb4 have rarely been identified in filamentous fungi to date.
The HAP complex is responsible for global transcriptional activation, and several genes directly regulated by the HAP complex have been characterized in filamentous fungi. The HAP complex of Aspergillus nidulans (AnCF) is involved in the utilization of carbon and nitrogen sources, and several enzyme-coding genes (e.g., amdS, taaG2, and ipnA) are positively regulated by AnCF3,21. The Aspergillus oryzae HAP complex, AoCP, is a direct activator of the Taka-amylase A gene (taa)22. In addition, activation of the gene encoding the cellobiohydrolase II cellulose degrading enzyme (cbh2) is mediated by the Hap3 orthologue (HapC) in the biomass-degrading fungus Trichoderma reesei23. Recently, a few orthologues of HAP complex components and the histone-like protein have been characterized in plant pathogenic fungi, including Fusarium species24–26. These proteins are involved in pathogenesis, fungal development, and various biological processes, yet interactions between CCAAT-binding factors have not been investigated in plant pathogenic fungi, including F. graminearum.
The ascomycete fungus F. graminearum is a prominent plant pathogen that causes Fusarium head blight (FHB) in cereal crops and ear and stalk rot on maize27, all of which result in severe yield losses and an accumulation of mycotoxins (e.g., trichothecenes and zearalenone) that are harmful to animals and humans28. Previously, we performed a genome-wide functional analysis of the complete repertoire of transcription factor-encoding genes in F. graminearum29, which resulted in the identification of eight transcription factors containing CCAAT-binding domains29. Interestingly, while the phenotypes of F. graminearum strains mutated for six out of eight genes were not significantly different from those of the wild-type strain, the deletion of two putative CCAAT-binding transcription factors resulted in defects in fungal development, including increased sensitivity to DNA damaging agents29,30. Furthermore, a recent study revealed that the expression of FgHLTF1, one of the two CCAAT-binding factors involved in the DNA damage response, was downregulated by the putative type 2A phosphatase FgPpg1 and was shown to be associated with the high osmolarity glycerol (HOG) pathway24.
In this study, we attempted to characterize the CCAAT complex structure and its biological functions in F. graminearum. Our results demonstrate that F. graminearum has two distinct CCAAT complex components containing histone-fold motifs, Fct1 and Fct2 (FgHltf1), which are required for DNA damage responses, sexual development, virulence, and trichothecene production. A protein-binding microarray analysis revealed that Fct2 binds to the consensus sequence CCAAT, and we also confirmed that Fct1 and Fct2 interact to form a complex. Moreover, this study provides the strong evidence supporting that the CCAAT-binding complex only has two distinct CCAAT-binding factors in F. graminearum.
Results
Identification and cellular localization of CCAAT-binding factors
In our previous study, we identified 16 transcription factors involved in DNA damage responses in F. graminearum30. Among them, strains carrying mutations in two CCAAT-binding factor-encoding genes, GzCCAAT002 (FGSG_01182) and GzCCAAT004 (FGSG_05304), were highly sensitive to DNA damaging agents compared to the wild-type strain. Recently, GzCCAAT004 was identified as a putative histone-like transcription factor, FgHLTF1, through a transcriptome analysis of a ΔFgppg1 strain24. In this study, we designated GzCCAAT002 and GzCCAAT004 as F. graminearum CCAAT-binding transcription factor 1 (FCT1) and FCT2, respectively. To generate complementation strains, the geneticin resistance gene cassette31 in each deletion mutant was replaced with FCT1 or FCT2 fused to the green fluorescent protein-encoding gene (GFP) and a hygromycin resistance gene cassette (HYG), yielding the strains FCT1c and FCT2c (Table 1 and Supplementary Fig. S1).
Table 1.
F. graminearum strains used in this study.
| Strain | Genotype | Source or reference |
|---|---|---|
| Z-3639 | Wild-type Fusarium graminearum | 53 |
| HK12 | GFP-HYG (GFP constitutive expresser in cytosol) | 63 |
| KM19 | Δmat1-1-1::GEN; GFP-HYG | 34 |
| mat1r | ∆mat1-1-1::GEN; hH1::hH1-RFP-GEN | 55 |
| fct1 | ∆fct1::GEN | ∆gzccaat0229 |
| fct2 | ∆fct2::GEN | ∆gzccaat0429 |
| fct1 fct2 | ∆fct1::GEN; ∆fct2::HYG | This study |
| FCT1c | ∆fct1::FCT1-GFP-HYG | This study |
| FCT2c | ∆fct2::FCT2-GFP-HYG | This study |
| FCT1c-r | ∆fct1::FCT1-GFP-HYG; hH1::hH1-RFP-GEN | mat1r × fct1 |
| FCT2c-r | ∆fct2::FCT2-GFP-HYG; hH1::hH1-RFP-GEN | mat1r × fct2 |
| fct1-g | ∆fct1::GEN; GFP-HYG | KM19 × fct1 |
| fct2-g | ∆fct2::GEN; GFP-HYG | KM19 × fct2 |
| fct1/2-g | ∆fct1::GEN; ∆fct2::GEN; GFP-HYG | KM19 × fct1 fct2 |
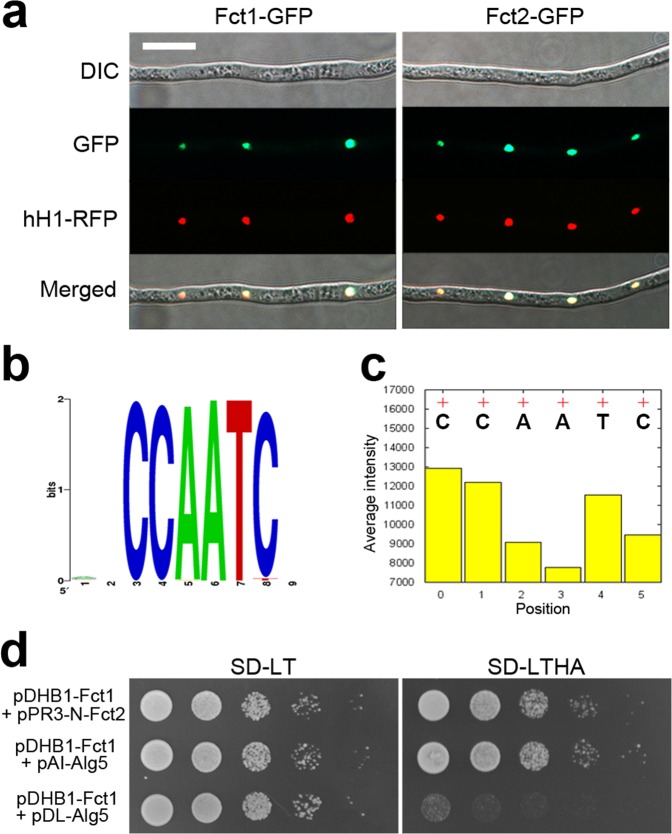
We examined the complementation strains FCT1c and FCT2c, which harbour single copies of FCT1-GFP or FCT2-GFP, respectively, and observed GFP signal in the nuclei of both strains (Fig. 1a). To confirm the nuclear localization of Fct1-GFP and Fct2-GFP, FCT1c-r (fct1::FCT1-GFP-HYG; hH1-RFP-GEN) or FCT2c-r (fct2::FCT2-GFP-HYG; hH1-RFP-GEN) strains were generated via outcrosses between mat1r and FCT1c or FCT2c (Table 1). Both Fct1-GFP and Fct2-GFP colocalized with hH1-RFP and were highly fluorescent in all of the tested developmental stages, including the mycelia, indicating that Fct1 and Fct2 are constitutively expressed nuclear proteins.
Figure 1.
Characterization of the nuclear CCAAT-binding complex of F. graminearum. (a) Nuclear localization of Fct1-GFP and Fct2-GFP. FCT1c-r/FCT2c-r strains carrying both Fct1-GFP or Fct2-GFP and hH1-RFP were used for the colocalization study. Scale bar = 20 µm. (b) Consensus binding sequence identified via the PBM assay. Consensus sequences that robustly bound to the Fct2-DsRed fusion protein. (c) The effects of positional mutations in CCAATC. To visualize the effects of the mutations on the binding intensities in the consensus binding motif, the average binding intensities (+) of the probes containing the core consensus 6-mer binding motif CCAATC relative to those of probes with mutations at each position (bar) are plotted. (d) Yeast two-hybrid analysis of the interaction between Fct1 and Fct2. The plasmid pairs pDHB1-Fct1/pAI-Alg5 and pDHB1-Fct1/pDL2-Alg5 served as positive and negative controls, respectively. The growth of the transformed yeast was assayed on synthetic dextrose medium lacking Leu and Trp (SD-LT) or Leu, Trp, His, and Ade (SD-LTHA). The columns in each panel represent serial decimal dilutions.
The CCAAT-binding protein Fct2 interacts with Fct1
Only two CCAAT-binding factor genes, FCT1 and FCT2, are important for the development of the fungus F. graminearum, whereas the HAP complexes of higher eukaryotic organisms comprise three CCAAT-binding factors. To characterize the CCAAT-binding complex of F. graminearum, we first attempted to identify the DNA-binding sequence of Fct2 via a protein binding microarray (PBM) analysis (Fig. 1b,c). This PBM uses the target probes that are synthesized as quadruples of all possible 9-mer combinations, resulting in robust identification of the DNA-binding sequences of transcription factors32. The quadruple 9-mer (Q9)-based PBM analysis using the Fct2-DsRed fusion protein identified 4,526 putative DNA-binding sequences, with CCAATC as the predominant sequence (Fig. 1b). Individual substitutions at each position of the CCAATC sequence markedly reduced its DNA-binding affinity, suggesting that Fct2 has CCAAT DNA-binding activity and that it is a subunit of the CCAAT-binding complex in F. graminearum (Fig. 1c).
To identify the other components of the CCAAT-binding complex in F. graminearum, proteins that copurified with Fct2-GFP were analysed via mass spectrometry (Table 2). We successfully identified 13 putative Fct2-interacting proteins, and among seven CCAAT-binding factors, only Fct1 was revealed as an Fct2 interaction partner. To validate the physical interaction between Fct1 and Fct2, we used the DUALhunter yeast two-hybrid (Y2H) assay, because conventional Y2H systems cannot be used to analyse integral membrane proteins or transcription factors33. A strong positive interaction between Fct1 and Fct2 was indicated by yeast colony growth on medium lacking leucine (Leu), tryptophan (Trp), histidine (His), and adenine (Ade) (SD-LTHA) (Fig. 1d).
Table 2.
Putative Fct2-interacting proteins identified via the affinity capture assay in Fusarium graminearum.
| Locus ID | PSMs | Predicted function or gene name |
|---|---|---|
| FGRAMPH1_01G00849 | 10 | Related to D-xylose reductase II,III protein |
| FGRAMPH1_01G03423 | 9 | Related to 3-isopropylmalate dehydrogenase |
| FGRAMPH1_01G02933 | 9 | Conserved hypothetical protein (GzCCAAT002/FCT1) |
| FGRAMPH1_01G19303 | 7 | Probable ribosomal protein S25 |
| FGRAMPH1_01G20773 | 6 | Probable nucleolar protein NOP58 |
| FGRAMPH1_01G12393 | 6 | Related to endo-polygalacturonase 6 |
| FGRAMPH1_01G11971 | 6 | Phytoene dehydrogenase |
| FGRAMPH1_01G26385 | 5 | Related to phosphomevalonate kinase |
| FGRAMPH1_01G02335 | 5 | Probable cytochrome-b5 reductase |
| FGRAMPH1_01G27519 | 5 | Probable hydroxymethylglutaryl-CoA synthase |
| FGRAMPH1_01G17119 | 5 | Probable dead-box protein precursor CYT-19 |
| FGRAMPH1_01G12125 | 5 | Related to sedoheptulose-1, 7-bisphosphatase |
| FGRAMPH1_01G27113 | 5 | Related to NIPSNAP protein |
PSMs, total number of identified peptide sequences (peptide spectrum matches) for the protein.
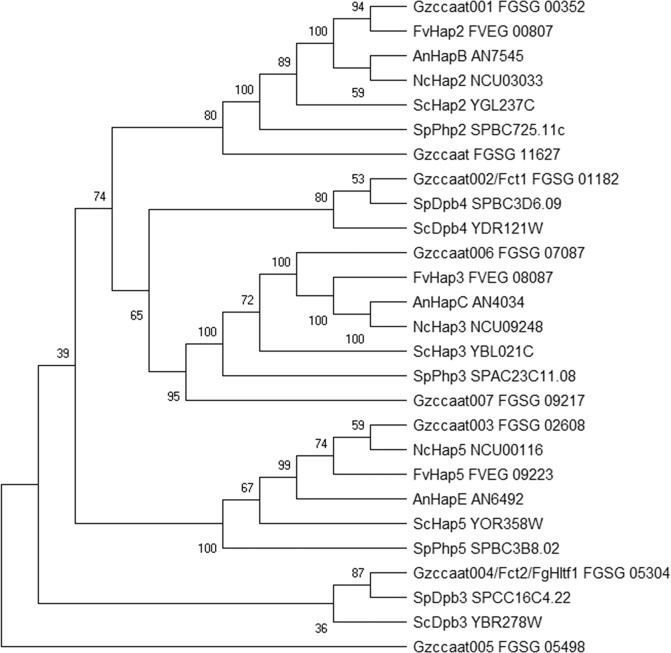
Subsequently, phylogenetic analyses of the CCAAT-binding factors of F. verticillioides, A. nidulans, Neurospora crassa, S. pombe, and S. cerevisiae were performed (Fig. 2). Gzccaat001, Gzccaat006, and Gzccaat003 were identified as putative orthologues of Hap2, Hap3, and Hap5, respectively. Gzccaat007 and Gzccaat008 also grouped with Hap3 and Hap2, although they shared lower sequence similarity with the S. cerevisiae Hap3 and Hap2 (30.1 and 35.7% identity) than Gzccaat6 and Gzccaat001, respectively. Fct1 and Fct2 were shown to be evolutionarily closer to Dpb4 and Dpb3, respectively, than the Hap complex components. Dpb3 and Dpb4, small components of yeast DNA polymerase epsilon (Pol ε) subunits, each have a H2A/H2B histone-fold motif and bind to DNA sequence as a complex20.
Figure 2.
Phylogenetic tree of fungal CCAAT-binding factors. The alignment was performed with ClustalW, and MEGA X was used to perform a 1,000-bootstrap phylogenetic analysis using the neighbour joining method62. Bootstrap support is shown for each node.
Fct1 and Fct2 are required for vegetative growth and perithecial development
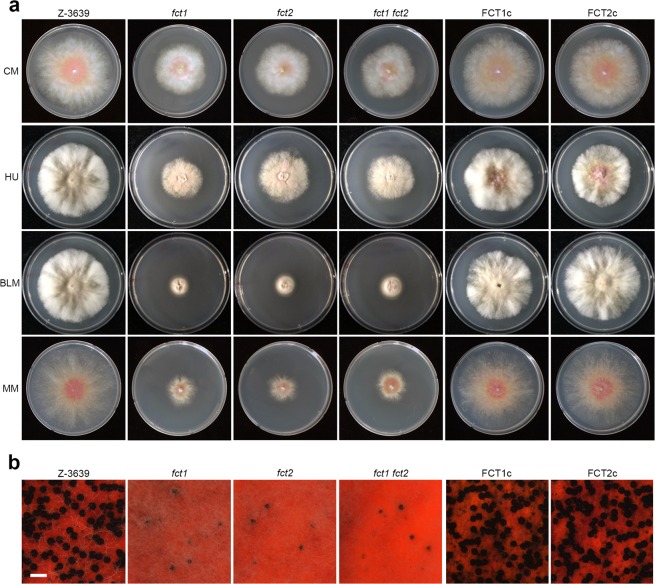
To confirm the genetic requirement of FCT1 and FCT2 for vegetative growth and sexual development, we compared the phenotypes of the deletion mutants with the wild-type and complementation mutant strains. The deletion mutants grew poorly on complete medium (CM) and minimal medium (MM), whereas complementation fully rescued the growth defects (Fig. 3a). The increased sensitivity of the deletion mutants to the DNA damaging agents, hydroxyurea (HU) and bleomycin (BLM), was also rescued in the complemented strains. During sexual development, the FCT1 and FCT2 deletion mutants lost self-fertility, while the wild-type and complemented strains produced normal perithecia (Fig. 3b).
Figure 3.
Vegetative growth and sexual development of F. graminearum strains. (a) The mycelial growth of F. graminearum strains on complete medium (CM), CM supplemented with 10 mM hydroxyurea (HU), 10 mU/ml bleomycin (BLM), and minimal medium (MM). The strains were imaged 5 days after inoculation. (b) Sexual development. A five-day-old culture on carrot agar medium was mock-fertilized to induce sexual reproduction, and the cultures were incubated for an additional 7 days. Scale bar = 500 µm.
FCT1 and FCT2 are important for virulence and total trichothecene production
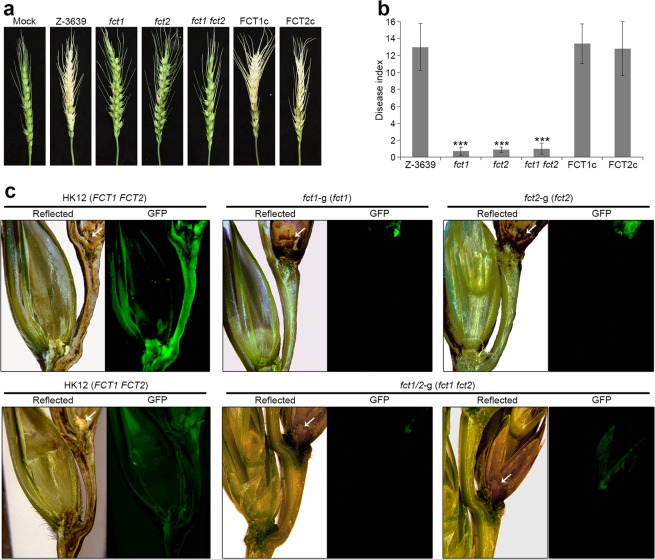
To evaluate the involvement of FCT1 and FCT2 in virulence on flowering wheat heads, conidial suspensions of strains were point-inoculated on a spikelet, and the plants were incubated in a greenhouse. The wild-type strain induced normal head blight symptoms, manifesting as discoloration at 21 days after inoculation, whereas the fct1 and fct2 strains were restricted to the initial infection sites and were unable to spread to adjacent spikelets on the head (Fig. 4a,b).
Figure 4.
Virulence of F. graminearum strains. (a) Virulence on wheat heads. The centre spikelet of each wheat head was injected with 10 μl of a conidial suspension. Images were captured 21 days after inoculation. (b) Disease index. The disease index was estimated as the number of diseased spikelets on each wheat head. Asterisks represent significant difference between the wild-type strain and each mutant (p < 0.001). (c) Micrographs of manually generated sections after the infection of wheat. Wheat spikelets were inoculated with conidial suspensions from strains expressing GFP in the cytoplasm (HK12, fct1-g, fct2-g, and fct1/2-g). Infected wheat heads were longitudinally dissected 6 days after inoculation and examined under a fluorescence microscope. GFP fluorescence indicates hyphal spreading from the inoculation points. Arrowheads mark the inoculated spikelets. Reflected, reflected light.
To further visualize the spread of the mycelia on wheat heads during infection, the strains fct1-g (∆fpo1::GEN; GFP-HYG) and fct2-g (∆fpo2::GEN; GFP-HYG) were generated via outcrossing of KM1934 with the fct1 or fct2 deletion mutants (Table 1). By 6 days after inoculation, hyphae of the HK12 strain (which carries wild-type FCTs and expresses cytosolic GFP) had spread to adjacent spikelets through rachis nodes (Fig. 4c). However, fluorescent hyphae of the fct1-g and fct2-g strains were only detected on the inoculated spikelets and failed to penetrate rachis nodes.
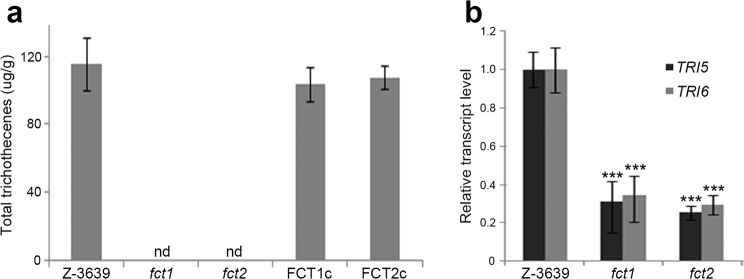
The levels of trichothecene synthesized by both the fct1 and fct2 deletion mutants were mostly undetectable, whereas the wild-type and complementation strains accumulated high amounts of trichothecenes (Fig. 5a). Furthermore, the transcriptional levels of the trichothecene biosynthetic genes TRI5 and TRI6 were also significantly reduced in the deletion mutants (Fig. 5b).
Figure 5.
Trichothecene production by the F. graminearum strains. (a) Total trichothecene production. Each strain was grown in minimal medium containing 5 mM agmatine (MMA) for 7 days. Trichothecenes were analysed via gas chromatography-mass spectrometry (GC-MS) and quantified based on the biomass of each strain. (b) Transcript levels of TRI5 and TRI6 in the F. graminearum strains. The transcript levels were analysed via quantitative real-time PCR (qRT-PCR) 4 days after inoculation in MMA. Asterisks represent significant differences in the relative transcript levels of TRI5 and TRI6 between the wild-type strain and each mutant (p < 0.001).
Phenotypes of double mutant revealed genetic interaction of FCT1 and FCT2
We hypothesized that if Fct1-Fct2 complex formation is crucial for the biological functions of these two proteins, then the phenotypic defects of the fct1 and fct2 single deletion mutants should be similar to those of an fct1 fct2 double deletion mutant. We generated fct1 fct2 double deletion mutants by deleting the FCT2 gene in an fct1 mutant (Supplementary Fig. S2), and the fct1 fct2 double deletion mutant showed indistinguishable phenotypes from those of the single deletion mutants (fct1 and fct2) under our tested assay conditions.
Discussion
In this study, we identified and characterized the CCAAT-binding transcription factors in the plant pathogenic fungus F. graminearum. Our biochemical, cytological, and genetic results demonstrated that the CCAAT-binding complex of F. graminearum comprises only two CCAAT-binding factors (Fct1 and Fct2) that are not highly conserved in other fungi or higher eukaryotes. Moreover, in-depth phenotypic analysis revealed that the Fct1-Fct2 complex is involved in various biological processes in F. graminearum, including vegetative growth, sexual reproduction, virulence, and mycotoxin production.
In most eukaryotic organisms, the heme activator protein (HAP), also known as nuclear factor Y (NF-Y) or CCAAT-binding factor (CBF), is composed of three subunits: Hap2 (also termed NF-YA, CBF-B, or HapB), Hap3 (NF-YB, CBF-A, or HapC), and Hap5 (NF-YC, CBF-C, or HapE), indicating that this complex structure is evolutionally conserved in eukaryotes35–37. Because each of these subunits is required for the DNA-binding activity of the complex, they are all essential for the function of the CCAAT-binding complex38–40. Biochemical studies of the interactions between these subunits has revealed that Hap3 and Hap5 form a tight dimer via a protein-protein interaction that is similar to the head to tail association of histones H2A/H2B and that this dimer offers a complex binding surface for Hap239,41. Interestingly, although the trimeric complex is sufficient to regulate gene expression in mammalian cells, orthologues of an additional component, Hap4 (HapX) or Hap4-like proteins, have been identified in fungi, including S. cerevisiae, S. pombe, Aspergillus, and Candida species3,42–44. Hap4 orthologues harbour a bZIP domain and a conserved 16-amino acid motif that is required for its interaction with the Hap2/Hap3/Hap5 complex. The Hap2, Hap3, and Hap5 complex assembles in the cytoplasm and is then transported to the nucleus via the nuclear localization signal (NLS) of Hap211,45,46. Subsequently, the heterotrimer complex binds to the promoters of target genes containing a CCAAT sequence and recruits Hap4 for subsequent gene activation46.
A histone fold is a structurally conserved motif identified near the C-terminus in core histones and is responsible for the ability of histones to bind and form heterodimers47. In addition to core histones, a similar secondary structure arrangement has been observed in several nuclear proteins involved in DNA metabolism, including TATA box-binding protein-associated factors, the DNA polymerase II subunits Dpb3 and Dpb4, and the CCAAT-binding complex proteins Hap3 (NF-YB) and Hap5 (NF-YC). The structure of the NF-YC-NF-YB complex resembles that of the H2A-H2B histone dimer48. Crystal structure analysis of the A. nidulans HAP complex showed that the complex specifically recognizes the CCAAT box of the promoter, and the S. cerevisiae Dpb3-Dpb4 heterodimer associates with double-stranded DNA without a preference for specific DNA sequences18,49. In this study, based on phylogenetic analysis, we determined that Fct1 and Fct2 are more closely related to Dpb4 and Dpb3, respectively, than to Hap complex components and that the two proteins interact with each other. In addition, protein binding microarray results showed that Fct2 specifically bound to the CCAAT sequence, unlike the Dpb3-Dpb4 complex, which binds to nonspecific double-stranded DNA. The deletion of FCT1 and FCT2, alone or in combination, resulted in an increased sensitivity of F. graminearum to DNA damaging agents. We speculate that Fct1 and Fct2 may be responsible for histone modification and DNA replication, although further investigation is needed to confirm this hypothesis.
In F. graminearum, among eight transcription factors identified as CCAAT-binding factors, Gzccaat001 (FGSG_00352), Gzccaat006 (FGSG_07087), and Gzccaat003 (FGSG_02608) are orthologous proteins of Hap2, Hap3, and Hap5, respectively (Fig. 2). However, deletion of these genes did not significantly affect F. graminearum phenotypes, such as mycelial growth, sexual development, toxin production, and virulence29. Thus, further in-depth genetic and biochemical analyses of these genes are needed to reveal the biological function of the HAP complex in F. graminearum.
Members of the CCAAT-binding factor gene family have diverse roles as transcriptional regulators for multiple cellular processes, including cell proliferation, apoptosis, differentiation, the control of metabolic pathways, the establishment of cell fate and identity, and stress responses in animals and plants35,50,51. In the yeast S. cerevisiae, CCAAT boxes are present in the promoters of cytochrome-encoding genes and in other genes involved in the use of nonfermentable carbon sources38 and in nitrogen metabolism52. CCAAT boxes are present in the promoters of genes involved in penicillin biosynthesis, and the HAP complex is also involved in the metabolism of carbon and nitrogen in the filamentous fungus A. nidulans40. Although F. graminearum has a simple CCAAT-binding complex structure compared to those of other eukaryotes, its Fct1-Fct2 complex has evolved to possess important functions in the development and virulence of F. graminearum.
In summary, in this study, we report that Fct1 and Fct2 are distinct components of the CCAAT-binding complex that have histone-fold motifs and are involved in various fungal developmental processes and virulence in F. graminearum. To the best of our knowledge, this is the first study on transcription factors containing CCAAT-binding domains in a plant pathogenic fungus, the results of which provide important information on the molecular mechanisms underlying the functions of CCAAT-binding factors in eukaryotes. Compared with what is known regarding the structure and function of the HAP complexes of model organisms, our knowledge on the CCAAT-binding complexes of plant pathogenic fungi, including F. graminearum, is lacking. Therefore, further studies will be needed to characterize the target genes and specialized functions of the CCAAT-binding complex in F. graminearum.
Methods
Fungal strains and media
All strains used in this study are listed in Table 1. The F. graminearum wild-type strain Z-363953 and mutants derived from this strain were maintained according to the Fusarium laboratory manual54. A transgenic strain, mat1r55, harbouring both a MAT1-1 deletion and red fluorescent protein (RFP)-tagged histone H1 was used in the colocalization study. Minimal liquid medium supplemented with 5 mM agmatine (MMA) was used for the trichothecenes analysis56.
Genetic manipulations, primers, and sequencing
Fungal genomic DNA was extracted according to the Fusarium laboratory manual54. Total RNA was isolated from mycelia ground in liquid nitrogen using an Easy-Spin Total RNA Extraction kit (Intron Biotech, Seongnam, Republic of Korea). Standard protocols were followed for restriction endonuclease digestion, agarose gel electrophoresis, and DNA gel blot hybridization with 32P labelled probes57. The PCR primers used in this study were synthesized at an oligonucleotide synthesis facility (Bionics, Seoul, Republic of Korea) (Supplementary Table S1).
Targeted deletion and complementation
The double-joint (DJ) PCR strategy was used to construct the fusion PCR products required to generate the targeted gene deletion and complementation58 via homologous recombination. The open reading frames (ORFs) of FCT1 or FCT2 in the F. graminearum wild-type strain Z-3639 were replaced with the geneticin resistance gene31 to create the deletion mutants. The fct1 and fct2 double mutant was generated by replacement of FCT2 gene with the hygromycin resistance gene (HYG) in the fct1 mutant. For the complementation and cellular localization assays, the 5′ flanking region, including the ORF with its own promoter, and the 3′ flanking region were amplified from genomic DNA of the wild-type strain using the primer pairs FCT1-5F/FCT1-5R GFP and FCT1-3F GFP/FCT1-3R, respectively. The GFP-HYG construct was amplified from the plasmid pIGPAPA using the primers pIGPAPA-sGFP/HYG-F1. The three amplicons were then fused via a second round of DJ PCR, after which the fusion constructs for transformation were amplified with nested primers using the second round PCR product as a template. Fungal transformation was performed as previously described55. The FCT2c strain, which was used to investigate cellular localization, was generated via the same strategy.
Microscopic observation
Microscopic observations were performed using a DE/Axio Imager A1 microscope (Carl Zeiss, Oberkochen, Germany) with the filter set 38HE (excitation 470/40; emission 525/50) for GFP and the filter set 15 (excitation 546/12; emission 590) for RFP.
Wheat heads inoculated with the GFP-expressing strains were observed as previously described34. Infected wheat heads were longitudinally dissected 6 days after inoculation and examined under a fluorescence microscope. Longitudinal sections cut through the centre of the spikelets were prepared freehand using a clean scalpel. The sectioned wheat heads were observed under reflected light and GFP fluorescence light (470 nm excitation and 525 nm emission wavelength filters) on a SteREO Lumar V12 microscope (Carl Zeiss).
Protein binding microarray (PBM) analysis
To determine the DNA-binding sequence of Fct2, a protein binding microarray assay was performed as previously described32,59. The full-length cDNA of FCT2 was inserted into the pET-DsRed expression vector to generate pET-Fct2Red, and the Fct2-DsRed fusion protein was expressed in the E. coli strain BL21-ColonPlus. Subsequently, the purified protein was incubated with a Q9 protein-binding microarray (Q9-PBM), which includes 232,145 quadruple probes, including 131,072 probes for all possible 9-mers, each of which was concatenated four times. Fluorescence images were captured using a GenePix 4000B microarray scanner (Molecular Devices, San Jose, CA, USA). The consensus binding sequence was determined based on the fluorescence signal intensities according to previously described methods32.
Affinity purification and mass spectrometry analysis
To capture Fct2-interacting proteins, cell lysates prepared from two independent FCT2c strains were incubated with magnetic beads conjugated to a mouse anti-GFP antibody (MBL International, Woburn, MA, USA) following the manufacturer’s instructions. After incubating at 4 °C for overnight, the magnetic beads were washed six times with phosphate-buffered saline (PBS; 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, and 1.8 mM KH2PO4, pH 7.4) prior to resolving the proteins via SDS-PAGE (12%). The proteins separated via SDS-PAGE were digested with trypsin in situ and then analysed using a Q Exactive™ nano high resolution LC/MS MS spectrometer (Thermo Scientific, Waltham, MA, USA). The resulting peptide amino acid sequences from the LC-MS/MS data were identified in sequences in the Fusarium graminearum database (https://fungidb.org/)60. The results obtained for the wild-type strain were used as a negative control.
Yeast two-hybrid assay (Y2H)
The Y2H assay was conducted using a DUALhunter kit (Dualsystems Biotech, Zurich, Switzerland) following the manufacturer’s instructions. To obtain cDNA, total RNA extracted from fungal cultures grown on carrot agar medium was reverse transcribed with SuperScriptIII reverse transcriptase (Invitrogen, Carlsbad, CA, USA). Each ORF was PCR amplified using primers with a SfiI restriction site (Supplementary Table S1). The full cDNAs of FCT1 and FCT2 were cloned into pDHB1, a Cub-based bait vector, and pRN3-N, a NubG-based prey vector, respectively (Dualsystems Biotech). After cotransformation of these vectors into S. cerevisiae NMY51 (MAT a his3Δ200 trp1-901 leu2-3, 112 ade2 LYS2::(lexApo)4- HIS3 ura3::(lexApo)8-lacZ ade2::(lexApo)8-ADE2 GAL4), a colony picked from the SD-Leu-Trp plates was grown in liquid SD-Leu-Trp medium, and the resulting cells were then spotted onto selective plates (SD-Leu-Trp-His-Ade). Strains carrying the empty vectors and pDL-Alg5 (−) were included as negative controls, while pAl-Alg5 (+) was included as a positive control.
Sexual crosses
Aerial mycelia were removed from cultures grown on carrot agar medium for 5 days with 0.5 ml of a 2.5% Tween 60 solution to induce sexual reproduction. The plates were incubated under a near-UV light (wavelength: 365 nm; Sankyo Denki Co., Ltd., Tokyo, Japan) at 25 °C for 7 to 10 days. For the outcrosses, mycelia from a female strain grown on carrot agar medium were fertilized with 1 ml of conidial suspension (106 conidia/ml) obtained from a male strain.
Virulence test and trichothecene analysis
For the virulence test, the point inoculation method was performed as previously described55. Conidial suspensions (105 conidia/ml) were prepared for each strain, and 10 μl of each suspension was injected into the centre spikelet of a wheat head (cultivar: Eunpamil). After inoculation, the wheat plants were incubated in a humidified chamber for 3 days and then transferred to a greenhouse. Spikelets exhibiting disease symptoms were counted 21 days after inoculation. The experiment was performed with five replicate inoculations per strain, and two independent mutant strains were used for the experiment.
Trichothecene analysis was performed as previously described29. Briefly, MMA cultures were extracted with ethyl acetate, and the extracts were concentrated to dryness. A portion of each extract was derivatized with Sylon BZT (BSA + TMCS + TMSI, 3:2:3 respectively, Supelco, Bellefonte, PA, USA) and analysed with a Shimadzu QP-5000 gas chromatograph mass spectrometer (GC-MS, Shimadzu, Kyoto, Japan) using the relevant ion-monitoring mode as previously described61. The trichothecenes were quantified based on the biomasses produced by each strain, and the experiment was repeated three times.
Quantitative real time (qRT)-PCR
Total RNA was prepared using an Easy-Spin Total RNA Extraction kit (Intron Biotech). The first strand cDNA was synthesized with SuperScriptIII reverse transcriptase (Invitrogen). Quantitative real-time PCR (qRT-PCR) was performed using SYBR Green Supermix (Bio-Rad, Hercules, CA, USA) and a 7500 real-time PCR system (Applied Biosystems, Foster City, CA, USA) with the corresponding primers (Supplementary Table S1). The endogenous housekeeping gene cyclophilin (CYP1) was used as an endogenous control for normalization. The qRT-PCR assay was repeated three times with three replicates per run, and the transcript levels relative to that of the housekeeping gene were expressed as 2−ΔΔCT31.
Supplementary information
Acknowledgements
This work was supported by the Korea Institute of Planning and Evaluation for Technology in Food, Agriculture and Forestry (IPET) via the Agricultural Microbiome R&D Program, funded by the Ministry of Agriculture, Food and Rural Affairs (MAFRA) (918012-4) and the National Research Foundation of Korea (2018R1C1B6002181 and 2018R1D1A1B07050702).
Author contributions
J.-E.K., H.N., Y.-W.L., and H.S. conceived and designed the experiments. J.-E.K., H.N., J.P., and G.J.C. performed experiments. G.J.C. performed the pathogenicity experiments. J.-E.K., H.N., Y.-W.L., and H.S. wrote the manuscript. All authors read, corrected and approved the final manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Jung-Eun Kim and Hyejin Nam.
Supplementary information
is available for this paper at 10.1038/s41598-020-61885-4.
References
- 1.Ludwig MZ, Bergman C, Patel NH, Kreitman M. Evidence for stabilizing selection in a eukaryotic enhancer element. Nature. 2000;403:564–567. doi: 10.1038/35000615. [DOI] [PubMed] [Google Scholar]
- 2.Périer RC, Praz V, Junier T, Bonnard C, Bucher P. The eukaryotic promoter database (EPD) Nucleic Acids Res. 2000;28:302–303. doi: 10.1093/nar/28.1.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kato M. An overview of the CCAAT-box binding factor in filamentous fungi: assembly, nuclear translocation, and transcriptional enhancement. Biosci. Biotechnol. Biochem. 2005;69:663–672. doi: 10.1271/bbb.69.663. [DOI] [PubMed] [Google Scholar]
- 4.Chakravarti A, Camp K, McNabb DS, Pinto I. The iron-dependent regulation of the Candida albicans oxidative stress response by the CCAAT-binding factor. Plos One. 2017;12:e0170649. doi: 10.1371/journal.pone.0170649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ly LL, Yoshida H, Yamaguchi M. Nuclear transcription factor Y and its roles in cellular processes related to human disease. Am. J. Cancer Res. 2013;3:339–346. [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao, H. et al. The Arabidopsis thaliana nuclear factor Y transcription factors. Front. Plant Sci. 7, 10.3389/fpls.2016.02045 (2017). [DOI] [PMC free article] [PubMed]
- 7.Thön M, et al. The CCAAT-binding complex coordinates the oxidative stress response in eukaryotes. Nucleic Acids Res. 2009;38:1098–1113. doi: 10.1093/nar/gkp1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McNabb DS, Xing Y, Guarente L. Cloning of yeast HAP5: a novel subunit of a heterotrimeric complex required for CCAAT binding. Genes Dev. 1995;9:47–58. doi: 10.1101/gad.9.1.47. [DOI] [PubMed] [Google Scholar]
- 9.Mantovani R. A survey of 178 NF-Y binding CCAAT boxes. Nucleic Acids Res. 1998;26:1135–1143. doi: 10.1093/nar/26.5.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maity SN, de Crombrugghe B. Role of the CCAAT-binding protein CBF/NF-Y in transcription. Trends Biochem. Sci. 1998;23:174–178. doi: 10.1016/S0968-0004(98)01201-8. [DOI] [PubMed] [Google Scholar]
- 11.Hortschansky P, Haas H, Huber EM, Groll M, Brakhage AA. The CCAAT-binding complex (CBC) in Aspergillus species. Biochim. Biophys. Acta Gene Regul. Mech. 2017;1860:560–570. doi: 10.1016/j.bbagrm.2016.11.008. [DOI] [PubMed] [Google Scholar]
- 12.Bolotin-Fukuhara M. Thirty years of the HAP2/3/4/5 complex. Biochim. Biophys. Acta Gene Regul. Mech. 2017;1860:543–559. doi: 10.1016/j.bbagrm.2016.10.011. [DOI] [PubMed] [Google Scholar]
- 13.Ohya T, Maki S, Kawasaki Y, Sugino A. Structure and function of the fourth subunit (Dpb4p) of DNA polymerase ε in Saccharomyces cerevisiae. Nucleic Acids Res. 2000;28:3846–3852. doi: 10.1093/nar/28.20.3846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chilkova O, Jonsson B-H, Johansson E. The quaternary structure of DNA polymerase ε from Saccharomyces cerevisiae. J. Biol. Chem. 2003;278:14082–14086. doi: 10.1074/jbc.M211818200. [DOI] [PubMed] [Google Scholar]
- 15.Li Y, Pursell ZF, Linn S. Identification and cloning of two histone fold motif-containing subunits of HeLa DNA polymerase ε. J. Biol. Chem. 2000;275:23247–23252. doi: 10.1074/jbc.M002548200. [DOI] [PubMed] [Google Scholar]
- 16.Sato H, et al. Arabidopsis DPB3-1, a DREB2A interactor, specifically enhances heat stress-induced gene expression by forming a heat stress-specific transcriptional complex with NF-Y subunits. Plant Cell. 2014;26:4954–4973. doi: 10.1105/tpc.114.132928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pursell, Z. F. & Kunkel, T. A. In Prog. Nucleic Acid Res. Mol. Biol. Vol. 82 (ed P. Michael Conn) 101–145 (Academic Press, 2008). [DOI] [PMC free article] [PubMed]
- 18.Tsubota T, et al. binding properties of Saccharomyces cerevisiae DNA polymerase ɛ and of the Dpb3p-Dpb4p subassembly. Genes Cells. 2003;8:873–888. doi: 10.1046/j.1365-2443.2003.00683.x. [DOI] [PubMed] [Google Scholar]
- 19.Tsubota T, et al. Double-stranded DNA binding, an unusual property of DNA polymerase ϵ, promotes epigenetic silencing in Saccharomyces cerevisiae. J. Biol. Chem. 2006;281:32898–32908. doi: 10.1074/jbc.M606637200. [DOI] [PubMed] [Google Scholar]
- 20.He H, et al. Coordinated regulation of heterochromatin inheritance by Dpb3–Dpb4 complex. Proc. Natl. Acad. Sci. USA. 2017;114:12524–12529. doi: 10.1073/pnas.1712961114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Heeswijck R, Hynes MJ. The amdR product and a CCAAT-binding factor bind to adjacent, possibly overlapping DNA sequences in the promoter region of the Aspergillus nidulans amdS gene. Nucleic Acids Res. 1991;19:2655–2660. doi: 10.1093/nar/19.10.2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tanaka A, et al. An Aspergillus oryzae CCAAT-binding protein, AoCP, is involved in the high-level expression of the Taka-amylase A gene. Curr. Genet. 2000;37:380–387. doi: 10.1007/s002940000125. [DOI] [PubMed] [Google Scholar]
- 23.Zeilinger S, Mach RL, Kubicek CP. Two adjacent protein binding motifs in the cbh2 (cellobiohydrolase II-encoding) promoter of the fungus Hypocrea jecorina (Trichoderma reesei) cooperate in the induction by cellulose. J. Biol. Chem. 1998;273:34463–34471. doi: 10.1074/jbc.273.51.34463. [DOI] [PubMed] [Google Scholar]
- 24.Lv W, et al. The putative histone-like transcription factor FgHltf1 is required for vegetative growth, sexual reproduction, and virulence in Fusarium graminearum. Curr. Genet. 2019;65:981–994. doi: 10.1007/s00294-019-00953-3. [DOI] [PubMed] [Google Scholar]
- 25.Ridenour JB, Bluhm BH. The HAP complex in Fusarium verticillioides is a key regulator of growth, morphogenesis, secondary metabolism, and pathogenesis. Fungal Genet. Biol. 2014;69:52–64. doi: 10.1016/j.fgb.2014.05.003. [DOI] [PubMed] [Google Scholar]
- 26.López-Berges MS, et al. HapX-mediated iron homeostasis is essential for rhizosphere competence and virulence of the soilborne pathogen Fusarium oxysporum. Plant Cell. 2012;24:3805–3822. doi: 10.1105/tpc.112.098624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goswami RS, Kistler HC. Heading for disaster: Fusarium graminearum on cereal crops. Mol. Plant Pathol. 2004;5:515–525. doi: 10.1111/j.1364-3703.2004.00252.x. [DOI] [PubMed] [Google Scholar]
- 28.Desjardins, A. E. Fusarium mycotoxins: chemistry, genetics, and biology. (APS Press, 2006).
- 29.Son H, et al. A phenome-based functional analysis of transcription factors in the cereal head blight fungus, Fusarium graminearum. Plos Pathog. 2011;7:e1002310. doi: 10.1371/journal.ppat.1002310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Son H, et al. A novel transcription factor gene FHS1 is involved in the DNA damage response in Fusarium graminearum. Sci. Rep. 2016;6:21572. doi: 10.1038/srep21572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 32.Kim M-J, et al. Quadruple 9-mer-based protein binding microarray with DsRed fusion protein. BMC Mol. Biol. 2009;10:91. doi: 10.1186/1471-2199-10-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Möckli N, Deplazes A, Auerbach D. Finding new protein interactions using the DUALhunter system. Nat. Methods. 2008;5:A1. doi: 10.1038/nmeth.f.204. [DOI] [Google Scholar]
- 34.Min K, et al. Peroxisome function is required for virulence and survival of Fusarium graminearum. Mol. Plant-Microbe Interact. 2012;25:1617–1627. doi: 10.1094/mpmi-06-12-0149-r. [DOI] [PubMed] [Google Scholar]
- 35.Laloum T, De Mita S, Gamas P, Baudin M, Niebel A. CCAAT-box binding transcription factors in plants: Y so many? Trends Plant Sci. 2013;18:157–166. doi: 10.1016/j.tplants.2012.07.004. [DOI] [PubMed] [Google Scholar]
- 36.Yang J, Xie Z, Glover BJ. Asymmetric Evolution of Duplicate Genes Encoding the CCAAT-Binding Factor NF-Y in Plant Genomes. New Phytol. 2005;165:623–631. doi: 10.1111/j.1469-8137.2004.01260.x. [DOI] [PubMed] [Google Scholar]
- 37.Li GL, et al. The animal nuclear factor Y: an enigmatic and important heterotrimeric transcription factor. Am. J. Cancer Res. 2018;8:1106–1125. [PMC free article] [PubMed] [Google Scholar]
- 38.McNabb DS, Tseng KA, Guarente L. The Saccharomyces cerevisiae Hap5p homolog from fission yeast reveals two conserved domains that are essential for assembly of heterotetrameric CCAAT-binding factor. Mol. Cell. Biol. 1997;17:7008. doi: 10.1128/MCB.17.12.7008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sinha S, Kim IS, Sohn KY, de Crombrugghe B, Maity SN. Three classes of mutations in the A subunit of the CCAAT-binding factor CBF delineate functional domains involved in the three-step assembly of the CBF-DNA complex. Mol. Cell. Biol. 1996;16:328. doi: 10.1128/MCB.16.1.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Steidl S, et al. AnCF, the CCAAT binding complex of Aspergillus nidulans, contains products of the hapB, hapC, and hapE genes and is required for activation by the pathway-specific regulatory gene amdR. Mol. Cell Biol. 1999;19:99. doi: 10.1128/MCB.19.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liberati C, di Silvio A, Ottolenghi S, Mantovani R. NF-Y binding to twin CCAAT boxes: role of Q-rich domains and histone fold helices. J. Mol. Biol. 1999;285:1441–1455. doi: 10.1006/jmbi.1998.2384. [DOI] [PubMed] [Google Scholar]
- 42.Mercier A, Watt S, Bähler J, Labbé S. Key function for the CCAAT-binding factor Php4 to regulate gene expression in response to iron deficiency in fission yeast. Eukaryot. Cell. 2008;7:493–508. doi: 10.1128/ec.00446-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jung WH, et al. HapX positively and negatively regulates the transcriptional response to iron deprivation in Cryptococcus neoformans. Plos Pathog. 2010;6:e1001209. doi: 10.1371/journal.ppat.1001209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gsaller F, et al. The Janus transcription factor HapX controls fungal adaptation to both iron starvation and iron excess. EMBO J. 2014;33:2261–2276. doi: 10.15252/embj.201489468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Steidl S, et al. A single subunit of a heterotrimeric CCAAT-binding complex carries a nuclear localization signal: piggy back transport of the pre-assembled complex to the nucleus. J. Mol. Biol. 2004;342:515–524. doi: 10.1016/j.jmb.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 46.McNabb DS, Pinto I. Assembly of the Hap2p/Hap3p/Hap4p/Hap5p-DNA complex in Saccharomyces cerevisiae. Eukaryot. Cell. 2005;4:1829–1839. doi: 10.1128/ec.4.11.1829-1839.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Arents G, Moudrianakis EN. The histone fold: a ubiquitous architectural motif utilized in DNA compaction and protein dimerization. Proc. Natl. Acad. Sci. USA. 1995;92:11170–11174. doi: 10.1073/pnas.92.24.11170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Romier C, Cocchiarella F, Mantovani R, Moras D. The NF-YB/NF-YC structure gives insight into DNA binding and transcription regulation by CCAAT factor NF-Y. J. Biol. Chem. 2003;278:1336–1345. doi: 10.1074/jbc.M209635200. [DOI] [PubMed] [Google Scholar]
- 49.Huber EM, Scharf, Daniel H, Hortschansky P, Groll M. & Brakhage, Axel A. DNA minor groove sensing and widening by the CCAAT-binding complex. Structure. 2012;20:1757–1768. doi: 10.1016/j.str.2012.07.012. [DOI] [PubMed] [Google Scholar]
- 50.Benatti P, et al. Specific inhibition of NF-Y subunits triggers different cell proliferation defects. Nucleic Acids Res. 2011;39:5356–5368. doi: 10.1093/nar/gkr128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bhattacharya A, et al. The B subunit of the CCAAT box binding transcription factor complex (CBF/NF-Y) is essential for early mouse development and cell proliferation. Cancer Res. 2003;63:8167. [PubMed] [Google Scholar]
- 52.Dang VD, Bohn C, Bolotin-Fukuhara M, Daignan-Fornier B. The CCAAT box-binding factor stimulates ammonium assimilation in Saccharomyces cerevisiae, defining a new cross-pathway regulation between nitrogen and carbon metabolisms. J. Bacteriol. 1996;178:1842. doi: 10.1128/jb.178.7.1842-1849.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bowden RL, Leslie JF. Sexual recombination in Gibberella zeae. Phytopathology. 1999;89:182–188. doi: 10.1094/PHYTO.1999.89.2.182. [DOI] [PubMed] [Google Scholar]
- 54.Leslie, J. F. & Summerell, B. A. The Fusarium laboratory manual. (Blackwell Pub., 2006).
- 55.Son H, Lee J, Park AR, Lee Y-W. ATP citrate lyase is required for normal sexual and asexual development in Gibberella zeae. Fungal Genet. Biol. 2011;48:408–417. doi: 10.1016/j.fgb.2011.01.002. [DOI] [PubMed] [Google Scholar]
- 56.Gardiner DM, Kazan K, Manners JM. Nutrient profiling reveals potent inducers of trichothecene biosynthesis in Fusarium graminearum. Fungal Genet. Biol. 2009;46:604–613. doi: 10.1016/j.fgb.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 57.Sambrook, J. & Russell, D. W. Molecular cloning: a laboratory manual, 4nd ed., (Cold Spring Harbor Laboratory Press, 2001).
- 58.Yu J-H, et al. Double-joint PCR: a PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal Genet. Biol. 2004;41:973–981. doi: 10.1016/j.fgb.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 59.Choonkyun J, et al. Quadruple 9-mer-based protein binding microarray analysis confirms AACnG as the consensus nucleotide sequence sufficient for the specific binding of AtMYB44. Mol. Cells. 2012;34:531–537. doi: 10.1007/s10059-012-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stajich JE, et al. FungiDB: an integrated functional genomics database for fungi. Nucleic Acids Res. 2011;40:D675–D681. doi: 10.1093/nar/gkr918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Seo J-A, Kim J-C, Lee D-H, Lee Y-W. Variation in 8-ketotrichothecenes and zearalenone production by Fusarium graminearum isolates from corn and barley in Korea. Mycopathologia. 1996;134:31–37. doi: 10.1007/bf00437050. [DOI] [PubMed] [Google Scholar]
- 62.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Son H, Min K, Lee J, Raju NB, Lee Y-W. Meiotic silencing in the homothallic fungus Gibberella zeae. Fungal Biol. 2011;115:1290–1302. doi: 10.1016/j.funbio.2011.09.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.