Abstract
In this study, we analyzed the behavior of several neglected, ancestral, and domesticated wheat genotypes, including Ae. triuncialis, Ae. neglecta, Ae. caudata, Ae. umbellulata, Ae. tauschii, Ae. speltoides, T. boeoticum, T. urartu, T. durum, and T. aestivum under control and salinity stress to assess the mechanisms involved in salinity tolerance. Physiological and biochemical traits including root/shoot biomasses, root/shoot ion concentrations, activity of antioxidant enzymes APX, SOD, and GXP, and the relative expression of TaHKT1;5, TaSOS1, APX, GXP, and MnSOD genes were measured. Analysis of variance (ANOVA) revealed significant effects of the salinity treatments and genotypes for all evaluated traits. Salinity stress (350 mM NaCl) significantly decreased root/shoot biomasses, K+ concentration in root/shoot, and root/shoot K+/Na+ ratios. In contrast, salinity stress significantly increased Na+ concentration in root and shoot, activity of antioxidant enzymes (APX, SOD, and GPX) and relative expression of salt tolerance-related genes (TaHKT1;5, TaSOS1, APX, GPX, and MnSOD). Based on heat map and principal component analysis, the relationships among physiological traits and relative expression of salt-responsive genes were investigated. Remarkably, we observed a significant association between the relative expression of TaHKT1;5 with root K+ concentration and K+/Na+ ratio and with TaSOS1. Taken together, our study revealed that two neglected (Ae. triuncialis) and ancestral (Ae. tauschii) wheat genotypes responded better to salinity stress than other genotypes. Further molecular tasks are therefore essential to specify the pathways linked with salinity tolerance in these genotypes.
Keywords: Antioxidant enzymes, Na+ transporter genes, Salt tolerance, Wild relatives of wheat
Introduction
Salinity stress is a major environmental challenge that limits the productivity of crop production worldwide (Oyiga et al. 2016). More than 800 Mha of land are affected by salinity, which is equivalent to more than 6% of the world’s total land (Mickelbart et al. 2005). Hence, efforts to improve the salt tolerance of plants are of immense importance for sustainable agriculture and may also significantly improve crop yield (Goyal et al. 2016).
One of the main cellular events that occurs during salinity stress is an increase in the occurrence of reactive oxygen species (ROS) (Hossain et al. 2017). Members of the ROS family, include hydrogen peroxide (H2O2), singlet oxygen (1O2), superoxide (O2−), and hydroxyl radical (OH) as signal molecules in the response to salinity stress (Zhang et al. 2015). Excessive generation of ROS may lead to cellular damage, necrosis, metabolic disorders, and finally cell death (Hossain and Dietz 2016). Therefore, the overexpression of antioxidant genes could induce tolerance against oxidative stress. Regulation of gene expression controls several biochemical and physiological responses to stress (Yousfi et al. 2016). With advances in molecular biology and biotechnology tools, the expression and regulation of genes proposed as a powerful tool to improve crop resistance to various environmental stresses (Blum 2011).
Over recent decades, numerous transcription factors (TFs) and salt-responsive genes have been identified. These include late embryogenesis abundant (LEA) proteins, antioxidant proteins, osmoregulatory genes, transporters/antiporters, and signal-related protein kinases (Su et al. 2015). The interactions between these genes and TFs can form the basis for several pathways, such as the genes to salt overly sensitive (SOS) genes, the mitogen-activated protein kinase (MAPK), and the calcium-dependent protein kinase (CDPK) (Zhu 2003; Ludwig et al. 2004; Nakagami et al. 2005) pathways. As one of the large superfamily of ionic transporters, high-affinity potassium transporters (HKTs) play considerable physiological roles in salinity tolerance through the removal of Na+ from the xylem during stress (Su et al. 2015). HKT genes can be classified into two main classes based on their transport selectivity. Class 1 includes HKT genes of rice (OsHKT1;5) and Arabidopsis thaliana (AtHKT1;1), which reduce the transport of Na+ to aboveground tissues (Maser et al. 2002; Moller et al. 2009). Class 2 consists of several genes from rice (OsHKT2;1–4) and wheat (TaHKT2;1) that play a crucial role in Na+ and K+ transport (Ariyarathna et al. 2016). One of the main strategies to cope with salt stress is the maintenance of cellular ion homeostasis by limiting the accumulation of Na+ (Tester and Davenport 2003). The SOS signaling pathway controls cellular ion homeostasis through the activity of the genes SOS1, SOS2, and SOS3 (Zhu 2000). Of these, SOS1 is conserved in both mono- and dicot plants and its activity is regulated by a complex made up of SOS2 and SOS3 (Horie and Schroeder 2004). Ji et al. (2013) demonstrated that SOS1 activity significantly controls ion homeostasis. Oh et al. (2009) showed in Thellungiella salsuginea that reduction in SOS1 expression leads to accumulation of Na+ in cells and organs and finally loss of halophytism. Hence, the SOS signaling pathway is an important regulatory mechanism in ion homeostasis (Ji et al. 2013).
Among cereal crops, wheat (Triticum aestivum L.) is one of the main crops worldwide with an annual production of approximately 736 million metric tons (FAO 2015). Due to their longtime adaptation to different climates, it is well known that wild progenitors of wheat can overcome adverse growth conditions (Pagnotta et al. 2009). Several studies have indicated specific capabilities of wheat ancestral species to favorably respond to abiotic stresses like drought and salinity (Ramezani et al. 2013; Arabbeigi et al. 2014, 2018; Pour-Aboughadareh et al. 2017a; Ahmadi et al. 2018a, b, c). Thus, improving salt tolerance in cultivated wheat may be achieved by the allelic repertoire available in its germplasm (Rampino et al. 2006). Despite extensive research on salinity tolerance in the wild relatives of wheat, the physiological and molecular mechanisms in various species with alien genomes have not been studied in detail. Furthermore, it remains unclear how some wild wheats cope with salinity at the molecular level. Here, we compared the several biochemical traits of some wild wheats (Ae. tauschii Coss., Ae. speltoides Tausch, Ae. cylindrica Host, Ae. caudata L., Ae. triuncialis L., Ae. umbellulata Zhuk., T. urartu Thumanjan ex Gandilyan and T. boeoticum Boiss.) along with two bread (T. aestivum L.) and durum (T. durum Desf.) wheats when subjected to salinity stress. To dissect the differential responses to salinity stress in these materials, we estimated the relative change in expression levels of some salt tolerance-related genes, including guaiacol peroxidase (GPX), superoxide dismutase (MnSOD), ascorbate peroxidase (APX), TaSOS1, and TaHKT1;5.
Materials and methods
Plant material and experimental details
The materials used in this study included 11 Aegilops L. and Triticum L. genotypes along with two local varieties (T. aestivum cv. Arg as the tolerant and T. aestivum cv. Darya as the sensitive genotype). These genotypes were selected based on the results of previous research (Ahmadi et al. 2018c) that investigated 179 genotypes of wheat germplasm under control (0 mM NaCl) and severe salinity stress (300 mM NaCl) conditions for root and shoot biomasses and ion concentrations at the seedling stage. Based on the results obtained from this research, we selected one tolerant genotype from each species. Supplemental information on the selected genotypes for the current work is presented in Table 1. Ten seeds from each genotype were sowed in plastic pots with 40-cm height and 20-cm diameter filled with sand. The seedlings were grown in controlled conditions with an optimal growing photoperiod cycle (16 h day and 8 h night) and temperature (25 °C day and 20 °C night). The seedling plants were initially irrigated with tap water and then irrigated twice a week with half-strength Hoagland nutrient solution (Hoagland and Arnon 1950), which was supplemented with full-strength solution at the second leaf emergence. 14 days after germination, control seedlings were irrigated with tap water and a full-strength solution without NaCl, while in the plant-stressed-salinity seedlings, to prevent sudden shock to plants stress, NaCl was added in five gradual steps to reach 350 mM (EC ≈ 35 dS m−1, as severe salinity stress level). After 14 days of growing under salinity stress, leaf tissues were harvested and frozen in liquid nitrogen and stored at − 80 °C.
Table 1.
Passport of the Aegilops and Triticum genotypes assessed in this study
| No. | Genotype code† | Species | Genome | Province/region |
|---|---|---|---|---|
| 1 | IUGB-01695 | T. aestivum | ABD | Kermanshah/Qasr-e-Shirin |
| 2 | IUGB-00912 | T. durum | AB | Ilam/Mehran–Salehabad Road |
| 3 | IUGB-00407 | T. boeoticum | Ab | Kermanshah/Kerend-e-Gharb |
| 4 | IUGB-00423 | T. urartu | Au | Kermanshah/Kermanshah |
| 5 | IUGB-00369 | Ae. tauschii | D | Gilan/Kelachay |
| 6 | IUGB-00076 | Ae. umbellulata | U | Kermanshah/Kermanshah–Eslamabad-e Gharb |
| 7 | IUGB-00025 | Ae. speltoides | B | Kermanshah/Qasr-e-Shirin |
| 8 | IUGB-00304 | Ae. caudata | C | Lorestan/Shorab region |
| 9 | IUGB-00137 | Ae. neglecta | UM | Gilan/Hayran Ghaut |
| 10 | IUGB-00359 | Ae. cylindrica | DC | Kurdistan/Zarineh |
| 11 | IUGB-01146 | Ae. triuncialis | UC | Ilam/Ilam |
| 12 | Tolerant check | T. aestivum cv. Arg | ABD | Seed and Plant Improvement Institute (SPII), Karaj, Iran |
| 13 | Sensitive check | T. aestivum cv. Darya | ABD | Seed and Plant Improvement Institute (SPII), Karaj, Iran |
†IUGB Ilam University Genbank
Measurement of shoot and root biomasses and ion concentrations
At the end of the 14-day growth period, aboveground tissue was cut at the base and the root was washed carefully to remove sand. After determination of root and shoot fresh weights, the samples were dried for 72 h at 70 °C for determination of dry weights. To measure root and leaf K+ and Na+ concentrations, 10 mg of the ground root and shoot dried sample was mixed with 10 mL 0.5 N nitric acid. After incubation of samples in a bain-marie for 2 h at 85 °C, the extracted solutions were filtered and K+ and Na+ concentrations were analyzed using flame photometry (Corning 410; Sherwood Scientific, Cambridge, UK). The typical detection limits for Na+ and K+ was 0.043 and 0.25 mmol L−1, respectively.
Antioxidant enzyme activities
Antioxidant enzyme activities were measured according to Kang and Saltveit (2002). A total of 0.5 g of fresh leaf was homogenized in 1 mL extraction buffer containing 0.05 M Tris-HCl buffer (pH 7.5), 1.5% w/v polyvinylpyrrolidone (PVP), 3 mM MgCl2, and 1 mM EDTA. The extraction buffer used for the APX assay contained 0.2 mM ascorbate. The homogenate was filtered and centrifuged for 20 min at 12,000 RPM and the supernatant was used as crude extract for the GPX (EC 1.11.1.7), APX (EC 1.11.1.1), and SOD (EC 1.15.1.1) assay.
GPX activity was measured following the protocol suggested by Upadhyaya et al. (1985). The reaction mixture contained 20 μL enzyme extract, 2.27 mL 1.25 mM phosphate buffer (pH 6.1), 0.50 mL 1% guaiacol, and 0.50 mL 1% H2O2. The increased trend in absorbance at 420 nm was followed for 60 s. Activity of APX was recorded following the approach of Chen and Asada (1989). The reaction mixture contained 60 μL enzyme extract, 1.54 mM hydrogen peroxide, 1.7 mL 50 mM phosphate buffer (pH 7.0), 0.5 mM ascorbate, and 0.1 mM EDTA. The rate of APX activity was estimated at 290 nm. SOD activity was determined following the method of Dhindsa et al. (1981). The reaction mixture contained 40 μL enzyme extract, 50 mM phosphate buffer (pH 7.8), 25 μL riboflavin, 75 μM NBT and 0.1 mM EDTA. The reaction mixtures were then shaken before being placed 30 cm below 30-W fluorescent lamps for 15 min. SOD activity was recorded at 560 nm.
RT-qPCR assay
Total RNA was isolated from 100 mg of ground tissue using an RNX-Plus™ Kit (Sinaclon) according to the manufacturer’s instructions. To eliminate genomic DNA contamination, DNase treatment was performed using RNase-free DNase I (Fermentas, #N0521) according to the manufacturer’s instructions. Finally, the concentration of the extracted total RNA was determined with Nano Drop Spectrophotometers (Thermo Scientific-2000C, USA). Synthesis of cDNA from 1 μg of total RNA was performed using Hyper Script™ Reverse Transcriptase Kit (GeneAll, Korea) per the manufacturer’s instructions in a 20-μL final volume.
RT-qPCR was performed in a 15-μL volume containing 7.5 μL of 2 × RealQ Plus 2 × Master Mix Green (Ampliqon), 0.5 μL of (0.5 μM) forward and reverse primers, 1 μL of diluted cDNA (50 ng μL−1), and 5.5 μL of RNAse-free water. RT-qPCR was performed in a MiniOpticon™ Real-Time PCR System (Bio-Rad, USA). The PCR reactions were performed with the following thermal cycles: 95 °C for 3 min followed by 45 cycles of 95 °C for 15 s, 58 °C for 20 s, and 72 °C for 30 s. After 45 cycles, a melting curve analysis was performed (65–95 °C) to verify the specificity of the amplicons. The 18S rRNA gene was selected as a reference gene to normalize the expression data for the TaHKT1;5, TaSOS1, APX, MnSOD, and GPX genes. The sequences of genes primers are listed in Table 2. Based on the CT values for each sample, the relative expression of each gene was estimated based on the following formula (Pfaffl 2001):
Table 2.
Sequences of genes primers used for RT-qPCR reactions
| Gene | Primer sequence | References | |
|---|---|---|---|
| TaHKT1;5 | Forward | 5′-CTATCACGTGGTGGTGCACC-3′ | Zamani Bangohari et al. (2013) |
| Reverse | 5′-ACGGAGAAGGTGTGCAGGCTG-3′ | ||
| TaSOS1 | Forward | 5′-GTTGTCGGTGAGGTCGGAGGG-3′ | Ramezani et al. (2013) |
| Reverse | 5′-TCATCTTCTCCTACCGCCCTG C-3′ | ||
| MnSOD | Forward | 5′-CAGAGGGTGCTGCTTTACAA-3′ | Baek and Skinner (2003) |
| Reverse | 5′-GGTCACAAGAGGGTCCTGAT-3′ | ||
| GXP | Forward | 5′-CCCCCTGTACAAGTTCCTGA-3′ | Baek and Skinner (2003) |
| Reverse | 5′-GTCAACAACGTGACCCTCCT-3′ | ||
| APX | Forward | 5′-GCAGCTGCTGAAGGAGAAGT-3′ | Baek and Skinner (2003) |
| Reverse | 5′-CACTGGGGCCACTCACTAAT-3′ | ||
| 18S | Forward | 5′-GGCCGCTCCTAGCCCTAATTG-3′ | Yousfi et al. (2016) |
| Reverse | 5′-TGAGCACTCTAATTTCTTCAAAGTACG-3′ |
Data analysis
The experimental design was a factorial randomized complete block design with three replications. Two salt treatments and different genotypes were selected as the two main factors. Analysis of variance (ANOVA) was performed to examine the effects of salinity treatment on the measured physiological and biochemical traits, shoot and root biomass, and gene expression of leaf tissues in the different genetic materials using SAS software (SAS Institute 2011). To explore associations between measured traits, a heat map was rendered using the ‘gplot’ package of R software (Warnes et al. 2014). Principal component analysis (PCA) was performed with Kaiser’s criterion (i.e. eigenvalue more than 1) using XLSTAT software. The possible associations among the relative gene expression data and measured physiological and biochemical traits were determined through a PCA-based biplot derived using two first principal components.
Results
Effect of salinity on root and shoot biomass and ion concentrations
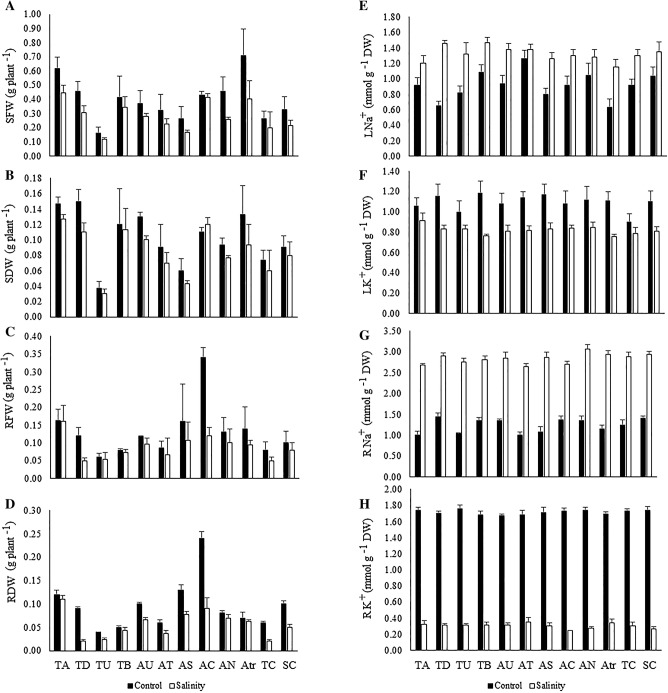
The studied genotypes showed differences in terms of root and shoot biomass and ion concentrations under different growth conditions. The results of ANOVA showed that shoot and root fresh weights exhibited ion concentrations significantly affected by salinity stress (Table 3). There were highly significant differences in the studied genotypes for all parameters except for leaf and root K+ concentrations. The interaction between genotypes and salinity stress were only significant for root dry weight and the K+/Na+ ratio of the roots. The percentage change of each parameter due to salinity stress is shown in Table 3. The overall mean of the 12 genotypes decreased by 92.31% (K+/Na+ ratio of root), 81.87% (root K+ concentration), 51.56% (K+/Na+ ratio of leaf), 47.23% (root fresh weight), 32.75% (root dry weight), 30.28% (shoot fresh weight), and 26.13% (leaf K+ concentration). On the other hand, salinity stress significantly increased root and leaf Na+ concentrations by 130.08% and 43.48%, respectively. In the salinity stress condition, the highest shoot and root fresh weights were recorded for the genotypes T. aestivum (0.45 and 0.16 g plant−1) and Ae. caudata (0.41 and 0.12 g plant−1) (Fig. 1a, c). Moreover, the genotypes T. aestivum (0.13 and 0.11 g plant−1) and Ae. caudata (0.12 and 0.09 g plant−1) had the highest root and shoot root dry weights of all the genotypes (Fig. 1b, d). The different studied genotypes varied significantly for leaf and root Na+ concentrations and the K+/Na+ ratios of their leaves and roots in both control and salinity stress conditions. In the salinity condition, the leaf Na+ concentrations across the studied genotypes ranged from 1.15 to 1.46 mmol g−1 DW (dry weight); the genotypes Ae. triuncialis (1.15 mmol g−1 DW) followed by T. aestivum (1.20 mmol g−1 DW) and Ae. speltoides (1.26 mmol g−1 DW) had the lowest Na+ concentrations in their leaves (Fig. 1e). In the same condition, root Na+ concentrations varied between 2.65 and 3.06 mmol g−1 DW; the genotypes Ae. tauschii (2.65 mmol g−1 DW), T. aestivum (2.68 mmol g−1 DW), and Ae. caudata (2.70 mmol g−1 DW) showed the lowest concentration of Na+ in their roots (Fig. 1g).
Table 3.
Analysis of variance, means (± SDE), and relative changes due to salinity stress for physiological traits, biochemical activities, and the relative expression of the studied genes in the different Aegilops and Triticum genotypes
| Variable | Source of variation | Mean | Relative change to control condition | |||||
|---|---|---|---|---|---|---|---|---|
| Replication | Stress (S) | Genotype (G) | S × G | Error | Control condition | Salinity condition | ||
| SFW | 0.02 | 0.26** | 0.93** | 9 × 10−3 | 0.21 | 0.40 ± 0.03 | 0.28 ± 0.02 | 30.28% |
| RFW | 1 × 10−3 | 0.04** | 0.02* | 7 × 10−3 | 4 × 10−3 | 0.07 ± 0.01 | 0.12 ± 0.01 | 47.23% |
| SDW | 1 × 10−3 | 2 × 10−3 | 6 × 10−3** | 1 × 10−4 | 1 × 10−3 | 0.10 ± 0.01 | 0.09 ± 0.01 | 10.68% |
| RDW | 1 × 10−4 | 4 × 10−4 | 1 × 10−3** | 4 × 10−4* | 1 × 10−4 | 0.05 ± 0.01 | 0.03 ± 0.01 | 32.75% |
| LN | 0.01 | 2.94** | 0.08* | 0.05 | 0.03 | 0.92 ± 0.04 | 1.32 ± 0.08 | − 43.48%† |
| LK | 0.05 | 1.52** | 4 × 10−3 | 0.01 | 0.02 | 1.11 ± 0.06 | 0.82 ± 0.09 | 26.13% |
| RN | 0.05 | 46.48** | 0.09** | 0.03 | 0.03 | 1.23 ± 0.05 | 2.83 ± 0.57 | − 130.08% |
| RK | 6 × 10−3 | 35.77** | 2 × 10−3 | 2 × 10−3 | 4 × 10−3 | 1.71 ± 1.07 | 0.31 ± 0.97 | 81.87% |
| LKN | 0.09 | 7.72** | 0.14* | 0.13 | 0.05 | 1.28 ± 0.03 | 0.62 ± 0.01 | 51.56% |
| RKN | 0.02 | 31.71** | 0.07** | 0.06** | 0.02 | 1.43 ± 0.01 | 0.11 ± 0.01 | 92.31% |
| APX activity | 2 × 10−3 | 0.02** | 0.01** | 0.02** | 1 × 10−3 | 0.10 ± 0.01 | 0.13 ± 0.01 | − 31.38% |
| GPX activity | 27.45 | 414.62* | 412.32** | 509.57** | 122.40 | 32.79 ± 2.61 | 37.59 ± 2.39 | − 14.64% |
| SOD activity | 2.88 | 120.92** | 100.02** | 132.21** | 3.56 | 8.48 ± 1.18 | 11.07 ± 0.87 | − 30.56% |
| APX gene | 15.41 | 5861.55** | 44.96** | 29.51 | 16.49 | 2.29 ± 0.31 | 20.33 ± 1.08 | 8.87-fold†† |
| GPX gene | 2.28 | 1098.24** | 32.42** | 28.15** | 3.68 | 1.72 ± 0.15 | 22.15 ± 1.57 | 12.87-fold |
| MnSOD gene | 35.37 | 7508.02** | 83.29** | 87.06** | 25.88 | 1.87 ± 0.22 | 9.68 ± 0.78 | 5.16-fold |
| TaSOS1 gene | 0.312 | 45.58** | 0.71** | 0.94** | 0.23 | 0.69 ± 0.07 | 2.28 ± 0.13 | 3.30-fold |
| TaHKT1;5 gene | 280.05 | 20,094.44** | 562.72** | 544.11** | 147.41 | 1.37 ± 016 | 34.78 ± 3.93 | 25.38-fold |
df value for replication, stress, genotypes, and interaction effect; error is 2, 1, 11, 11, and 46, respectively
SFW, shoot fresh weight; RFW, root fresh weight; SDW, shoot dry weight; RDW, root dry weight; LN, leaf Na+ concentration; LK, leaf K+ concentration; RN, root Na+ concentration; RK, root K+ concentration; LKN, K+/Na+ ratio of leaf; RKN, K+/Na+ ratio of root; APX activity, ascorbate peroxidase activity; GPX activity, guaiacol peroxidase activity; SOD activity, superoxide dismutase activity; APX gene, APX gene expression; GPX gene, GPX gene expression; SOD gene, MnSOD gene expression, SOS1 gene, TaSOS1 gene expression; TaHKT1;5 gene, TaHKT1;5 gene expression
*, **Significant at 0.05 and 0.01 probability levels, respectively
†Negative numbers indicate values higher than the control condition
††Relative change to the control condition
Fig. 1.
The means of shoot fresh weight (a), shoot dry weight (b), root fresh weight (c), root dry weight (d), leaf Na+ concentration (e), leaf K+ concentration (f), root Na+ concentration (g), and root K+ concentration in different neglected, ancestral, and domesticated wheat genotypes under control and salinity stress conditions. TA, T. aestivum; TD, T. durum; TU, T. urartu; TB, T. boeoticum; AU, Ae. umbellulata; AT, Ae. tauschii; AS, Ae. speltoides; AC, Ae. cylindrica; AN, Ae. neglecta; AC, Ae. caudata; Atr, Ae. triuncialis; TC, Tolerant control variety; SC, Sensitive control variety
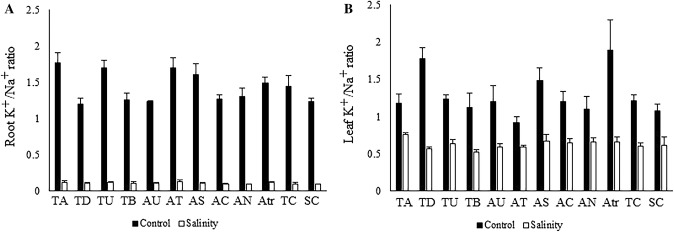
Under the salinity condition, T. aestivum (0.91 mmol g−1 DW), Ae. caudata (0.84 mmol g−1 DW), and Ae. caudata (0.83 mmol g−1 DW) had the highest K+ concentration in their leaves (Fig. 1f). The highest root K+ concentrations were observed for Ae. tauschii (0.35 mmol g−1 DW), Ae. triuncialis (0.34 mmol g−1 DW), and T. urartu (0.33 mmol g−1 DW) (Fig. 1h). The K+/Na+ ratios of leaves had a wide range of variability for different genotypes and varied between 0.52 and 0.75 under salinity condition. The highest ratio was observed in genotype T. aestivum (0.75) followed by Ae. neglecta (0.66) and Ae. caudata (0.64) (Fig. 2a). In saline conditions, there was variability in the K+/Na+ ratios of roots; these values ranged from 0.09 to 0.13. T. aestivum (0.13), Ae. tauschii (0.12), and Ae. triuncialis (0.11) showed higher values than the other genotypes (Fig. 2b).
Fig. 2.
The means of K+/Na+ ratio of root (a) and leaf (b) in different neglected, ancestral, and domesticated wheat genotypes under control and salinity stress conditions. TA, T. aestivum; TD, T. durum; TU, T. urartu; TB, T. boeoticum; AU, Ae. umbellulata; AT, Ae. tauschii; AS, Ae. speltoides; AC, Ae. cylindrica; AN, Ae. neglecta; AC, Ae. caudata; Atr, Ae. triuncialis; TC, tolerant control variety; SC, sensitive control variety
Effect of salinity on antioxidant metabolism
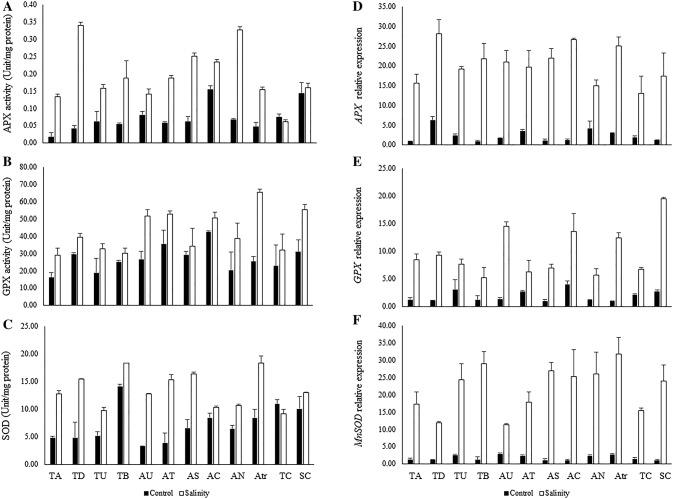
The interactions between salinity and genotype significantly affected the activity of guaiacol peroxidase (GPX), superoxide dismutase (SOD), and ascorbate peroxidase (APX) antioxidant enzymes (Table 3). Salinity considerably increased the activity of APX (31.38%), SOD (30.56%), and GPX (14.64%). Under salinity stress, the highest APX activity occurred in T. durum (0.34 Unit mg protein−1), Ae. neglecta (0.33 Unit mg protein−1), Ae. speltoides (0.25 Unit mg protein−1), and Ae. caudata (0.23 Unit mg protein−1), respectively (Fig. 3a). Salinity stress significantly changed the GXP activity. Ae. triuncialis (65.44 Unit mg protein−1), the tolerant control variety (55.41 Unit mg protein−1), Ae. tauschii (52.74 Unit mg protein−1), and Ae. umbellulata (51.46 Unit mg protein−1) showed higher GXP activities than the other genotypes (Fig. 3b). SOD activity showed a wide range of variability among the 12 genotypes. Under salinity, SOD activity varied between 9.10 and 18.35 Unit mg protein−1. The highest SOD activities were recorded for the genotypes Ae. umbellulata (18.35 Unit mg protein−1), T. boeoticum (18.33 Unit mg protein−1), Ae. tauschii (16.33 Unit mg protein−1), and T. durum (15.43 Unit mg protein−1) (Fig. 3c).
Fig. 3.
The means of ascorbate peroxidase activity (Unit mg protein−1) (a), guaiacol peroxidase activity (Unit mg protein−1) (b), superoxide dismutase activity (Unit mg protein−1) (c), the mean fold-change of APX gene expression (d), the mean fold-change of GPX gene expression (e), and the mean fold-change MnSOD gene expression (f) in different neglected, ancestral, and domesticated wheat genotypes under control and salinity stress conditions. TA, T. aestivum; TD, T. durum; TU, T. urartu; TB, T. boeoticum; AU, Ae. umbellulata; AT, Ae. tauschii; AS, Ae. speltoides; AC, Ae. cylindrica; AN, Ae. neglecta; AC, Ae. caudata; Atr, Ae. triuncialis; TC, tolerant control variety; SC, sensitive control variety
Change in expression levels of salt tolerance-related genes
The relative transcript abundance of APX, MnSOD, GPX, TaHKT1;5, and TaSOS1 in the leaf tissues of 12 genotypes under two growth conditions was determined by RT-qPCR. The results of ANOVA showed that salinity stress affected the expression profiles of five genes, with significant differences between the genotypes. The interaction between genotype and stress treatment for all genes was also significant (Table 3). Salinity stress significantly increased the mean transcript levels by 25.38 (TaHKT1;5), 12.87 (GPX), 8.87 (APX), 5.16 (MnSOD), and 3.30 (TaSOS1)-fold more than under the control condition (Table 3). In the control condition, no significant difference was observed for the expression patterns of all genes, while in the salinity condition expression of the TaHKT1;5 gene was much higher than the others (Table 3). Under salinity condition, a considerable increase in mRNA transcripts of the APX gene was observed in the leaves of T. durum (23.69-fold), Ae. caudata (23.65-fold), Ae. triuncialis (8.32-fold), and Ae. neglecta (3.70-fold) when compared to the control condition (Fig. 3d).
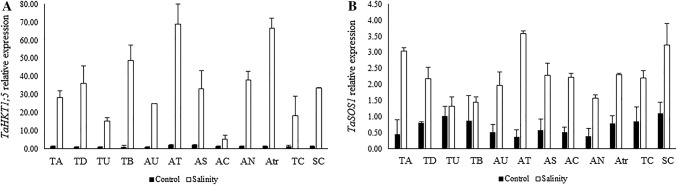
Under salinity conditions, the expression levels of the GPX gene showed that salinity stress increased the mRNA transcripts of this gene. Higher numbers of transcripts were found in the tolerant control varieties (7.21-fold), Ae. umbellulata (11.39-fold), Ae. caudata (3.43-fold), and Ae. triuncialis (13.42-fold) (Fig. 3e). The expression level of the MnSOD gene was significantly higher in Ae. speltoides (25.24-fold) followed by T. boeoticum (25.21-fold), Ae. triuncialis (12.09-fold) and Ae. neglecta (11.25-fold) than in unstressed plants (Fig. 3f). Ae. triuncialis (54.27-fold), Ae. tauschii (29.92-fold), Ae. neglecta (29.81-fold) and Ae. umbellulata (24.09-fold) showed the highest expression of TaHKT1;5 gene and compared to control conditions for these genotypes (Fig. 4a). Moreover, compared with the control condition, the maximum numbers of TaSOS1 gene transcripts were observed in Ae. tauschii (9.68-fold), T. aestivum (6.76-fold), the tolerant control variety (2.94-fold) and Ae. triuncialis (2.93-fold) (Fig. 4b).
Fig. 4.
Mean fold-change of TaHKT1;5 gene expression (a) and mean fold-change of TaSOS1 gene expression (b) in different neglected, ancestral, and domesticated wheat genotypes under control and salinity stress conditions. TA, T. aestivum; TD, T. durum; TU, T. urartu; TB, T. boeoticum; AU, Ae. umbellulata; AT, Ae. tauschii; AS, Ae. speltoides; AC, Ae. cylindrica; AN, Ae. neglecta; AC, Ae. caudata; Atr, Ae. triuncialis; TC, tolerant control variety; SC, sensitive control variety
Correlation between different traits and principal component analysis
A heatmap plot based on Pearson’s correlation was rendered to describe the associations between different physiological traits, biochemical activities, and transcriptional activities of salt tolerance-related genes (Fig. 5a). According to this plot, there was a strong association among RFW, RDW, SFW and SDW. Leaf K+ concentration and the leaf K+/Na+ ratio showed a positive correlation with both RFW and RDW. RFW, RDW, SFW and SDW positively correlated with the expression of TaSOS1 and GPX genes. Among ion concentrations, leaf K+ and Na+ concentrations showed a positive relation with the activity of the APX enzyme and expression of the TaSOS1 gene. Root K+ concentration and K+/Na+ ratio positively correlated with the expression of the TaHKT1;5 and TaSOS1 genes. The associations between three antioxidant enzyme activities were also strongly positive with each other. Other relations between different traits and expression profile of studied genes are shown in Fig. 5a.
Fig. 5.

Heatmap indicating association among different physiological traits, biochemical activities, and relative expression of studied genes under salinity stress (350 mM NaCl) condition (a). The biplot rendered is based on the first two components (PC1 and PC2) for physiological and biochemical-related traits and relative expression of the studied genes in neglected, ancestral, and domesticated wheat genotypes under salinity stress (350 mM NaCl) condition (b). SFW, shoot fresh weight; RFW, root fresh weight; SDW, shoot dry weight; RDW, root dry weight; LN, leaf Na+ concentration; LK, leaf K+ concentration; RN, root Na+ concentration; root K+ concentration; LKN, K+/Na+ ratio of leaf; RKN, K+/Na+ ratio of root; APX activity, ascorbate peroxidase activity; GPX activity, guaiacol peroxidase activity; SOD activity, superoxide dismutase activity; APX gene, APX gene expression; GPX gene, GPX gene expression; SOD gene, MnSOD gene expression, SOS1 gene, TaSOS1 gene expression; TaHKT1;5 gene, TaHKT1;5 TaHKT1;5 gene
In the principal component analysis (PCA), all the examined traits, APX, SOD, and GPX enzyme activities along with the transcription values of TaHKT1;5, TaSOS1, APX, GPX, and MnSOD were loaded into two major principal components (PC1 and PC2) that explained 43.35% of the variance (Fig. 5b). In the PCA loading plot, PC1 was positively affected by RFW, RDW, SFW and SDW, leaf K+ concentration, leaf K+/Na+ ratios, and expression of GPX and TaSOS1 genes. On the other hand, leaf and root Na+ concentrations, root K+ concentration, K+/Na+, expression of the APX and TaHKT1;5 genes, and activity of APX and SOD enzymes had a negative correlation, explaining 24.26% of the total physiological variation. Similarly, PC2 was significantly affected by SFW, SDW, root K+ concentration, root K+/Na+ ratio, expression of APX, TaHKT1;5, TaSOS1 genes, and activity of GPX and SOD enzymes. PC2 had a strongly negative relationship with leaf K+ concentration and root Na+ concentration, altogether accounting for 19.08% of the total variation. Figure 5b shows the angles between the trait vectors used to explore the relationships between different traits and the expression levels of the studied genes. In this way, small or large angles indicate a strong positive or weaker association, respectively. On the other hand, a 180° angle shows that there was a negative association between the traits, whereas a 90° angle indicates no association. For instance, the shoot and root biomasses showed a significant positive association with each other and with the expression of the TaSOS1, MnSOD, and GPX genes. The expression of TaHKT1;5 significantly correlated with SOD enzyme activity, root K+ concentration, and K+/Na+ ratio. The activity of the SOD enzyme had a significant positive relationship with the level of APX gene expression. The PCA plot differentiates between the different domesticated and wild wheat genotypes based on response to saline conditions. The biplot analysis indicated that shoot and root biomasses, expression of GPX gene, and leaf K+/Na+ ratio were positively associated with the T. aestivum genotype, whereas leaf K+ concentration was associated with Ae. caudata. Root K+ concentration and K+/Na+ ratio, SOD enzyme activity, and expression of the APX and TaHKT1;5 genes were also highly associated with the Ae. tauschii and Ae. triuncialis genotypes. Root Na+ concentration and APX enzyme activity were highly associated with T. urartu, Ae. speltoides, and the tolerant and sensitive control genotypes.
Discussion
Genetic erosion in our improved wheat genotypes and the small contribution to these genotypes from the genetic potential that could be gained by the development of new cultivars are reasons why further research should explore the source of ideal genes found in wild wheat (Pour-Aboughadareh et al. 2017b). The results of many comparative studies have revealed that some of the wild relatives of domestic wheat deserve better consideration and acknowledgement of the genetic potential of this gene pool due to the physiological mechanisms and molecular pathways involved in abiotic tolerance mechanisms (Byrt et al. 2007; Pour-Aboughadareh et al. 2017a; Ahmadi et al. 2018b, c; Arabbeigi et al. 2018). In this investigation, several ancestral and neglected wheat species with different genetic backgrounds were investigated for their shoot and root biomasses, leaf and root ion concentrations, antioxidant enzyme activities, and salinity-related gene expression under two control and severe salinity conditions to uncover genotypes and mechanisms that may be reliable for the selection of salt-tolerant genotypes. Our results showed that the impact of salinity stress on the response of plant seedlings was significantly dependent on genotype, as significant differences were found for shoot and root biomass production, ion concentrations, antioxidant activities, and gene expression (Table 3). Salinity stress significantly reduced the experimental plants’ root and shoot biomasses, K+ concentration, and K+/Na+ ratio in comparison with the control condition (Table 3). Similarly, salinity stress had a negative effect on the shoot and root seedling performances of common and wild species of wheat; the rate of reduction in these parameters was significantly associated with the increase in salt concentration (Arabbeigi et al. 2014; Ahmadi et al. 2018c). It is known that there is a strong association between salinity tolerance and the K+ concentration status in different plant’s organs. Increasing K+ supply in the root environment limits transportation of Na+ from vascular systems (Ahmadi et al. 2018c). Furthermore, the ability to maintain high tissue K+/Na+ ratios could be a crucial salinity tolerance discriminator (Shabala and Pottosin 2014). In this study, two genotypes T. aestivum (with ABD genome) and Ae. triuncialis (with UC genome) showed higher K+/Na+ ratios for leaf and root tissues than from other genotypes. This result is consistent with Arabbeigi et al. (2014) and Ahmadi et al. (2018c), who illustrated a considerable ability of species possessing the C genome to respond to salinity stress.
Upon exposure to salinity stress, plants undergo several changes from morpho-physiological adaptation to gene expression (Shinozaki and Yamaguchi-Shinozaki 2007). In such situations, the production of ROS is one of the unavoidable events that appears as a secondary stress. Scavenging excess ROS to protect plant cells and stabilizing an optimum level of ROS for signaling are important strategies in a tolerant genotype to cope with saline conditions (Verma et al. 2014). Hence, plants possess a specific defense mechanism for scavenging ROS that includes activation of both non-enzymatic antioxidants and antioxidant enzymes. Several antioxidant enzymes such as GPX, SOD, APX, and CAT are known to significantly decline the levels of ROS in plants (Johnson et al. 2003; Ali and Alqurainy 2006). In the present study, biochemical activities including APX, GPX, and SOD in the tested genotypes increased significantly under salinity stress conditions relative to the levels under control conditions, indicating serious damage to the plant cells (Table 3). A recent study by Ahmadi et al. (2018c) indicated that salinity stress significantly affected antioxidant activities in different Aegilops and Triticum species, and differences in these biochemical activities in response to salinity stress were detected among the tested accessions. Based on our results, salinity-stressed genotypes of T. durum followed by Ae. caudata, Ae. speltoides, and Ae. neglecta had higher APX activities than controls, indicating that genomes AB, C, B, and UM may be more successful in controlling intercellular ROS accumulation than others. These results were supported by RT-qPCR data (Fig. 3a, d). Similarly, several studies reported an increasing trend for the expression of the APX gene under salinity conditions in different crop plants such as tomatoes, tobacco, sweet potatoes, and Arabidopsis (Wang et al. 2005; Lee et al. 2007; Yan et al. 2016). Sharma et al. (2012) demonstrated that upregulation of this antioxidant enzyme increases the tolerance of plants. It is known that the SOD enzyme catalyzes the dismutation of O2− into H2O2 and this act is known as the main defense mechanism to cope with oxidative stress in plants (Bose et al. 2014). Suneja et al. (2017) indicated that the SOD antioxidant enzyme is one of the key biochemical parameters for assessing the tolerance mechanism of plants and can be used as an indirect selection criteria for identifying salt-tolerant plants. In the present study, the highest SOD activities were found in U (Ae. umbellulata), AB (T. durum), B (Ae. speltoides), and Ab (T. boeoticum) genomes; this might protect them from the negative effects of salinity stress (Fig. 3c). When the expression data was compared, the increasing trend for the MnSOD gene was Ae. triuncialis > T. boeoticum > Ae. speltoides > Ae. neglecta > other genotypes (Fig. 3f). SOD catalyze the dismutation of O.−2 into H2O2 and are considered as the vital of defense in confronting oxidative stress in plants (Bose et al. 2014). The MnSOD gene consists of three isoforms, which are activated in the mitochondria, peroxisome, chloroplast, and cytosol (Bowler et al. 1992). Hence, increased expression of the MnSOD gene in the aforementioned genotypes (especially B and Ab genomes) suggest a protective mechanism targeted to the mitochondria. Moreover, three wild genotypes with D (Ae. tauschii), UC (Ae. triuncialis), and U (Ae. umbellulata) genomes showed higher GPX enzyme activity and expression than the other genotypes (Fig. 3b, e), indicating that biosynthesis of lignin and scavenging H2O2 in chloroplasts are the main defense mechanisms against salinity in these genomes (Gill and Tuteja 2010). In general, the variation in biochemical activities among the Aegilops species showed the different capabilities of these genotypes to address oxidative damage by increasing antioxidant activities. In addition, the increased expression observed for the APX, GPX, and MnSOD genes in some wild relatives suggests that alien genomes possess a considerable ability to respond to saline conditions.
In plants, the sequestration and translocation of Na+ inside different organs is an important factor that supports plant growth in saline environments. The Na+ absorbed at the root environment will be moved to the xylem through several transporters and channels. In glycophytes—which include many crop plants—regulating the transportation of Na+ from the root to leaf is an important factor in adaptation to saline environments (Tester and Davenport 2003). Among the transporters, high-affinity K+ transporters (HKTs) are one of the important transporters that display specificity for K+ and Na+ (Assaha et al. 2017). In addition to HKT genes, the SOS pathway is commonly viewed as a key pathway involved in regulation of Na+ homeostasis in plants (Katschnig et al. 2015). This pathway system includes three components (SOS1, SOS2, and SOS3), of which SOS1 is a Na+/H+ antiporter and stimulates efflux Na+ in exchange for H+ (Gong et al. 2004). In the present investigation, salinity stress increased the relative expression of the TaHKT1;5 and TaSOS1 genes. Among different domesticated and wild wheat genotypes, Ae. tauschii, Ae. triuncialis, and T. aestivum showed high transcription for these genes (Fig. 4). This in turn indicates that Na+ exclusion may be one of the key salinity tolerance mechanisms in these genotypes. Hence, the existence of an efficient SOS pathway would make a crucial contribution to the superior tolerance in these genotypes (Gao et al. 2016). In a study conducted by Zhu et al. (2015), it was founded that the Nax2 locus coding for HKT1;5 is involved in the regulation of SOS1 activity in xylem loading of Na+. Furthermore, there is a strong association between SOS1 and HKTs pathways for salt tolerance (Assaha et al. 2017). In this regard, our results also indicated a significant association between the expression of TaHKT1;5 and TaSOS1 under salinity stress (Fig. 5a), suggesting an increase in the expression of SOS1 leads to the intake of Na+ into the xylem for translocation to the shoots for osmoregulation (Assaha et al. 2017). Considering the biplot rendered from the PCA analysis (Fig. 5b), we identified the superior genotypes in terms of physiological and biochemical responses to salinity stress. Based on this biplot, the wild genotypes Ae. tauschii and Ae. triuncialis exhibited a clearly better physiological potential than the control varieties and other genotypes. However, we could not show a clear grouping pattern for separation of the different genotypes through their responses to salinity stress. We thus surmise this unclear pattern is a consequence of the high rate of genetic variation and the differences between the genetic backgrounds in the tested materials.
Conclusion
In conclusion, our findings revealed a high level of physiological and biochemical variation among the wheat germplasm in response to salinity stress. These results may reveal new paths for comprehensive consideration of some of the neglected and ancestral species of wheat with salinity tolerance and for questioning the complex connections among alien genomes such as C, U, DC, MD and UC to cope with salinity stress. Remarkably, in the present study we identified two wild wheat genotypes (Ae. tauschii and Ae. triuncialis) which had a possible source of valuable physio-chemical traits for breeding as well as an interesting features for future investigations. Hence, investigation of genetic diversity within these species and further molecular studies are essential to determine the novel mechanisms and pathways associated with salinity tolerance in these genotypes.
Acknowledgements
The authors acknowledge with thanks the technical and lab facilities support from the Genetic and Genomic laboratory of Department of Genetics and Plant Breeding, Imam Khomeini International University, Qazvin, Iran.
Abbreviations
- SFW
Shoot fresh weight
- RFW
Root fresh weight
- SDW
Shoot dry weight
- RDW
Root dry weight
- LN
Leaf Na+ concentration
- LK
Leaf K+ concentration
- RN
Root Na+ concentration
- RK
Root K+ concentration
- LKN
K+/Na+ ratio of leaf
- RKN
K+/Na+ ratio of root
- APX activity
Ascorbate peroxidase activity
- GPX activity
Guaiacol peroxidase activity
- SOD activity
Superoxide dismutase activity
- APX gene
APX gene expression
- GPX gene
GPX gene expression
- SOD gene
MnSOD gene expression
- SOS1 gene
TaSOS1 gene expression
- TaHKT1;5 gene
TaHKT1;5 gene expression
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Ahmadi J, Pour-Aboughadareh A, Fabriki Ourang S, Mehrabi AA, Siddique KHM. Screening wheat germplasm for seedling root architectural traits under contrasting water regimes: potential sources of variability for drought adaptation. Arch Agron Soil Sci. 2018;64:1351–1365. doi: 10.1080/03650340.2018.1432855. [DOI] [Google Scholar]
- Ahmadi J, Pour-Aboughadareh A, Fabriki Ourang S, Mehrabi AA, Siddique KHM. Wild relatives of wheat: Aegilops–Triticum accessions disclose differential antioxidative and physiological responses to water stress. Acta Physiol Plant. 2018;40:90. doi: 10.1007/s11738-018-2673-0. [DOI] [Google Scholar]
- Ahmadi J, Pour-Aboughadareh A, Fabriki-Ourang S, Mehrabi AA, Siddique KHM. Screening wild progenitors of wheat for salinity stress at early stages of plant growth: insight into potential sources of variability for salinity adaptation in wheat. Crop Pasture Sci. 2018;69:649–658. doi: 10.1071/CP17418. [DOI] [Google Scholar]
- Ali AA, Alqurainy F. Activities of antioxidants in plants under environmental stress. In: Motohashi N, editor. The lutein-prevention and treatment for diseases. Trivandrum: Transworld Research Network; 2006. pp. 187–256. [Google Scholar]
- Arabbeigi M, Arzani A, Majidi MM, Kiani R, Tabatabaei BES, Habibi F. Salinity tolerance of Aegilops cylindrica genotypes collected from hyper-saline shores of Uremia Salt Lake using physiological traits and SSR markers. Acta Physiol Plant. 2014;36:2243–2251. doi: 10.1007/s11738-014-1602-0. [DOI] [Google Scholar]
- Arabbeigi M, Arzani A, Majidi MM, Seyed-Tabatabaei B, Saha P. Expression pattern of salt tolerance-related genes in Aegilops cylindrica. Physiol Mol Biol Plants. 2018;24:61–73. doi: 10.1007/s12298-017-0483-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariyarathna HACK, Oldach KH, Francki M. A comparative gene analysis with rice identified orthologous group II HKT genes and their association with Na+ concentration in bread wheat. BMC Plant Biol. 2016;16:1–21. doi: 10.1186/s12870-016-0714-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assaha DVM, Ueda A, Saneoka H, Al-Yahyai R, Yaish MW. The role of Na+ and K+ transporters in salt stress adaptation in glycophytes. Front Physiol. 2017;8:509. doi: 10.3389/fphys.2017.00509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek KH, Skinner DZ. Alteration of antioxidant enzyme gene expression during cold acclimation of near-isogenic wheat lines. Plant Sci. 2003;165:1221–1227. doi: 10.1016/S0168-9452(03)00329-7. [DOI] [Google Scholar]
- Blum A. Drought resistance—is it really a complex trait? Funct Plant Biol. 2011;38:753–757. doi: 10.1071/FP11101. [DOI] [PubMed] [Google Scholar]
- Bose J, Rodrigo-Moreno A, Shabala S. ROS homeostasis in halophytes in the context of salinity stress tolerance. J Exp Bot. 2014;65:1241–1257. doi: 10.1093/jxb/ert430. [DOI] [PubMed] [Google Scholar]
- Bowler C, Van Montagu M, Inze D. Superoxide dismutase and stress tolerance. Annu Rev Plant Physiol Plant Mol Biol. 1992;43:83–116. doi: 10.1146/annurev.pp.43.060192.000503. [DOI] [Google Scholar]
- Byrt CS, Platten JD, Spielmeyer W, James RA, Lagudah ES, Dennis ES, Tester M, Munns R. HKT1;5-like cation transporters linked to Na+ exclusion loci in wheat, Nax2 and Kna1. Plant Physiol. 2007;143:1918–1928. doi: 10.1104/pp.106.093476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen GX, Asada K. Ascorbate peroxidase in tea leaves: occurrence of two isozymes and the differences in their enzymatic and molecular properties. Plant Cell Physiol. 1989;30:987–998. doi: 10.1093/oxfordjournals.pcp.a077713. [DOI] [Google Scholar]
- Dhindsa RS, Plump-Dhindsa P, Thorpe TA. Leaf senescence: correlated with increased levels of membrane permeability and lipid peroxidation, and decreased levels of superoxide dismutase and catalase. J Exp Bot. 1981;32:93–101. doi: 10.1093/jxb/32.1.93. [DOI] [Google Scholar]
- Food Agriculture and Organization (FAO) (2015) Cereal supply and demand brief. http://www.fao.org/worldfoodsituation/csdb/en/. Accessed 20 Mar 2016
- Gao J, Sun J, Cao P, Ren L, Liu L, Chen S, Chen F, Jiang J. Variation in tissue Na+ concentration and the activity of SOS1 genes among two species and two related genera of Chrysanthemum. BMC Plant Biol. 2016;16:98–113. doi: 10.1186/s12870-016-0781-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill SS, Tuteja N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol Biochem. 2010;48:909–930. doi: 10.1016/j.plaphy.2010.08.016. [DOI] [PubMed] [Google Scholar]
- Gong D, Guo Y, Schumaker KS, Zhu JK. The SOS3 family of calcium sensors and SOS2 family of protein kinases in Arabidopsis. Plant Physiol. 2004;134:919–926. doi: 10.1104/pp.103.037440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyal E, Amit SK, Singh RS. Transcriptome profiling of the salt-stress response in Triticum aestivum cv. Kharchia Local. Sci Rep. 2016;6:27752. doi: 10.1038/srep27752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoagland DR, Arnon DI (1950) The water-culture method for growing plants without soil. California Agricultural Experiment Station, Circular No. 374. The College of Agriculture, University of California, Berkeley, CA, USA
- Horie T, Schroeder JI. Sodium transporters in plants diverse genes and physiological functions. Plant Physiol. 2004;136:2457–2462. doi: 10.1104/pp.104.046664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain MS, Dietz KJ. Tuning of redox regulatory mechanisms, reactive oxygen species and redox homeostasis under salinity stress. Front Plant Sci. 2016;7:548. doi: 10.3389/fpls.2016.00548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain MS, ElSayad AI, Moore M, Dietz KJ. Redox and reactive oxygen species network in acclimation for salinity tolerance in sugar beet. J Exp Bot. 2017;68:1283–1298. doi: 10.1093/jxb/erx019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji H, Pardo JM, Batelli G, Van Oseten MJ, Bressan RA, Li X. The salt overly sensitive (SOS) pathway: established and emerging roles. Mol Plant. 2013;6:275–286. doi: 10.1093/mp/sst017. [DOI] [PubMed] [Google Scholar]
- Johnson SM, Doherty SJ, Croy RRD. Biphasic superoxide generation in potato tubers. A self-amplifying response to stress. Plant Physiol. 2003;13:1440–1449. doi: 10.1104/pp.013300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H-M, Saltveit ME. Antioxidant enzymes and DPPH-radical scavenging activity in chilled and heat shocked rice (Oryza sativa L.) seedling radicles. J Agric Food Chem. 2002;50(3):513–518. doi: 10.1021/jf011124d. [DOI] [PubMed] [Google Scholar]
- Katschnig D, Bliek T, Rozema J, Schat H. Constitutive highlevel SOS1 expression and absence of HKT1; 1 expression in the salt accumulating halophyte Salicornia dolichostachya. Plant Sci. 2015;234:144–154. doi: 10.1016/j.plantsci.2015.02.011. [DOI] [PubMed] [Google Scholar]
- Lee YP, Kim SH, Bang JW, Lee HS, Kwon SY. Enhanced tolerance to oxidative stress in transgenic tobacco plants expressing three antioxidant enzymes in chloroplasts. Plant Cell Rep. 2007;26:591–598. doi: 10.1007/s00299-006-0253-z. [DOI] [PubMed] [Google Scholar]
- Ludwig AA, Romeis T, Jones JD. CDPK-mediated signaling pathways: specificity and cross talk. J Exp Bot. 2004;55:181–188. doi: 10.1093/jxb/erh008. [DOI] [PubMed] [Google Scholar]
- Maser P, Eckelmana BV, Vaidyanathan R, Horie T, Frbairn DJ, Kubo M, Yamagami M, Yamaguchi K, Nishimura M, Uozumi N, Robertson W, Mr Sussman, Schroeder JI. Altered shoot/root Na+ distribution and bifurcating salt sensitivity in Arabidopsis by genetic disruption of the Na+ transporter AtHKT1. FEBS Lett. 2002;531:157–161. doi: 10.1016/S0014-5793(02)03488-9. [DOI] [PubMed] [Google Scholar]
- Mickelbart MV, Hasegawa PM, Bailey-Serres J. Genetic mechanisms of abiotic stress tolerance that translate to crop yield stability. Nat Rev Genet. 2005;16:237–251. doi: 10.1038/nrg3901. [DOI] [PubMed] [Google Scholar]
- Moller IS, Gilliham M, Jha D, Mayo GM, Roy SJ, Coates JC, Haseloff J, Tester M. Shoot Na+ exclusion and increased salinity tolerance engineered by cell type-specific alteration of Na+ transport in Arabidopsis. Plant Cell. 2009;21:2163–2178. doi: 10.1105/tpc.108.064568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagami H, Pitzschke A, Hirt H. Emerging MAP kinase pathways in plant stress signaling. Trends Plant Sci. 2005;7:339–346. doi: 10.1016/j.tplants.2005.05.009. [DOI] [PubMed] [Google Scholar]
- Oh DH, Leidi E, Zhang Q, Hwang SM, Li Y, Quintero FH, Jiang X, Durzo MP, Lee SY, Zhao Y, Bahk JD, Bressan RA, Yun DJ, Pardo JM, Bohnert HJ. Loss of halophytism by interference with SOS1 expression. Plant Physiol. 2009;151:210–222. doi: 10.1104/pp.109.137802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyiga BC, Sharma RC, Shen J, Baum M. Identification and characterization of salt tolerance of wheat germplasm using multivariable screening. J Agron Crop Sci. 2016;202:472–485. doi: 10.1111/jac.12178. [DOI] [Google Scholar]
- Pagnotta MA, Mondini L, Porceddu E. Quantification and organization of WIS2-1A and BARE-1 retrotransposons in different genomes of Triticum and Aegilops species. Mol Genet Genomics. 2009;282:245–255. doi: 10.1007/s00438-009-0462-6. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucl Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pour-Aboughadareh A, Ahmadi J, Mehrabi AA, Moghaddam M, Etminan A. Physiological responses to drought stress in wild relatives of wheat: implications for wheat improvement. Acta Physiol Plant. 2017;39:106. doi: 10.1007/s11738-017-2403-z. [DOI] [Google Scholar]
- Pour-Aboughadareh A, Mohmoudi AM, Ahmadi J, Mehrabi AA, Alavikia SS. Agro-morphological and molecular variability in Triticum boeoticum accessions from Zagros Mountains, Iran. Genet Resour Crop Evol. 2017;64:545–556. doi: 10.1007/s10722-016-0381-4. [DOI] [Google Scholar]
- Ramezani A, Niazi A, Abolimoghadam AA, Zamani Bobgohari M, Deihimi T, Ebrahimi M, Akhtadanesh H, Ebrahimie E. Quantitative expression analysis of TaSOS1 and TaSOS4 genes in cultivated and wild wheat plants under salt stress. Mol Biotechnol. 2013;53:189–197. doi: 10.1007/s12033-012-9513-z. [DOI] [PubMed] [Google Scholar]
- Rampino P, Pataleo S, Gerardi C, Mita G, Perrotta C. Drought stress response in wheat: physiological and molecular analysis of resistant and sensitive genotypes. Plant, Cell Environ. 2006;29:2143–2152. doi: 10.1111/j.1365-3040.2006.01588.x. [DOI] [PubMed] [Google Scholar]
- SAS Institute . Base SAS 9.1 procedures guide. Cary: SAS Institute Inc; 2011. [Google Scholar]
- Shabala S, Pottosin I. Regulation of potassium transport in plants under hostile conditions: implications for abiotic and biotic stress tolerance. Physiol Plant. 2014;151:257–279. doi: 10.1111/ppl.12165. [DOI] [PubMed] [Google Scholar]
- Sharma P, Jha AB, Dubey RS, Pessarakli M. Reactive oxygen species, oxidative damage and antioxidative defense mechanism in plants under stressful conditions. J Bot. 2012;217037:1–26. [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K. Gene networks involved in drought stress response and tolerance. J Exp Bot. 2007;58:221–227. doi: 10.1093/jxb/erl164. [DOI] [PubMed] [Google Scholar]
- Su Y, Luo W, Lin W, Ma L, Kabir MH. Model of cation transportation mediated by high-affinity potassium transports (HKTs) in higher plants. Biol Proced Online. 2015;17:1–13. doi: 10.1186/s12575-014-0013-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suneja Y, Gupta AK, Bains NS. Bread wheat progenitors: Aegilops tauschii (DD genome) and Triticum dicoccoides (AABB genome) reveal differential antioxidative response under water stress. Physiol Mol Biol Plants. 2017;23:99–114. doi: 10.1007/s12298-016-0409-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tester M, Davenport R. Na+ tolerance and Na+ transport in higher plants. Ann Bot. 2003;91:503–527. doi: 10.1093/aob/mcg058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhyaya A, Sankhla D, Davis TD, Sankhla N, Smith BN. Effect of paclobutrazol on the activities of some enzymes of activated oxygen metabolism and lipid peroxidation in senescing soybean leaves. J Plant Physiol. 1985;121:453–461. doi: 10.1016/S0176-1617(85)80081-X. [DOI] [Google Scholar]
- Verma KK, Singh M, Gupta RK, Verma CL. Photosynthetic gas exchange, chlorophyll fluorescence, antioxidant enzymes, and growth responses of Jatropha curcas during soil flooding. Turk J Bot. 2014;38:130–140. doi: 10.3906/bot-1212-32. [DOI] [Google Scholar]
- Wang Y, Wisniewski M, Meilan R, Cui M, Webb R, Fuchigami L. Overexpression of cytosolic ascorbate peroxidase in tomato confers tolerance to chilling and salt stress. J Am Soc Hortic Sci. 2005;130:167–173. doi: 10.21273/JASHS.130.2.167. [DOI] [Google Scholar]
- Warnes GR, Bolker B, Bonebakker L, Gentlemean R, Huber W, Liaw A et al (2014) gplots: various R programming tools for plotting data. http://CRAN.R-project.org/package=gplots. Accessed 5 Feb 2019
- Yan H, Li Q, Park SC, Wang X, Liu YJ, Zhang YG, Tang W, Kou M, Ma DF. Overexpression of CuZnSOD and APX enhance salt stress tolerance in sweet potato. Plant Physiol Biochem. 2016;109:20–27. doi: 10.1016/j.plaphy.2016.09.003. [DOI] [PubMed] [Google Scholar]
- Yousfi S, Marquez AJ, Betti M. Gene expression and physiological responses to salinity and water stress of contrasting durum wheat genotypes. J Integr Plant Biol. 2016;58:48–66. doi: 10.1111/jipb.12359. [DOI] [PubMed] [Google Scholar]
- Zamani Bangohari M, Niazi A, Moghaddam AA, Deihimi T, Ebrahimie E. Genome-wide analysis of key salinitytolerance transporter (HKT;5) in wheat and wild wheat relatives (A and D genomes) Vitro Cell Dev Biol Plant. 2013;49:97–106. doi: 10.1007/s11627-012-9478-4. [DOI] [Google Scholar]
- Zhang Y, Li Y, Peng Y, Wang X, Peng D, Li Y, He X, Zhang X, Ma X, Huang L, Yan Y. Clones of FeSOD, MDHAR, DHAR genes from white clover and gene expression analysis of ROS-scavenging enzymes during abiotic stress and hormone treatments. Molecules. 2015;20:20939–20954. doi: 10.3390/molecules201119741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JK. Genetic analysis of plant salt tolerance using Arabidopsis. Plant Physiol. 2000;124:941–948. doi: 10.1104/pp.124.3.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JK. Regulation of ion homeostasis under salt stress. Curr Opin Plant Biol. 2003;6:441–445. doi: 10.1016/S1369-5266(03)00085-2. [DOI] [PubMed] [Google Scholar]
- Zhu M, Shabala L, Cuin TA, Huang X, Zhou M, Munns R, Shabala S. Nax loci affect SOS1-like Na+/H+ exchanger expression and activity in wheat. J Exp Bot. 2016;67(3):835–844. doi: 10.1093/jxb/erv493. [DOI] [PMC free article] [PubMed] [Google Scholar]