Abstract
Breast cancer research has traditionally centred on genomic alterations, hormone receptor status and changes in cancer-related proteins to provide new avenues for targeted therapies. Due to advances in next generation sequencing technologies, there has been the emergence of long, non-coding RNAs (lncRNAs) as regulators of normal cellular events, with links to various disease states, including breast cancer. Here we describe our bioinformatic analyses of a previously published RNA sequencing (RNA-seq) dataset to identify lncRNAs with altered expression levels in a subset of breast cancer cell lines.
Using a previously published RNA-seq dataset of 675 cancer cell lines, a subset of 18 cell lines was selected for our analyses that included 16 breast cancer lines, one ductal carcinoma in situ line and one normal-like breast epithelial cell line. Principal component analysis demonstrated correlation with well-established categorisation methods of breast cancer (i.e. luminal A/B, HER2 enriched and basal-like A/B). Through detailed comparison of differentially expressed lncRNAs in each breast cancer sub-type with normal-like breast epithelial cells, we identified 15 lncRNAs with consistently altered expression, including three uncharacterised lncRNAs.
Utilising data from The Cancer Genome Atlas (TCGA) and The Genotype Tissue Expression (GETx) project via Gene Expression Profiling Interactive Analysis (GEPIA2), we assessed clinical relevance of several identified lncRNAs with invasive breast cancer. Lastly, we determined the relative expression level of six lncRNAs across a spectrum of breast cancer cell lines to experimentally confirm the findings of our bioinformatic analyses. Overall, we show that the use of existing RNA-seq datasets, if re-analysed with modern bioinformatic tools, can provide a valuable resource to identify lncRNAs that could have important biological roles in oncogenesis and tumour progression.
Keywords: Bioinformatics, Breast cancer, Ductal carcinoma in situ, Long non-coding RNAs (lncRNAs) RNA sequencing (RNA-seq), Quantitative reverse transcriptase polymerase chain reaction (qRT-PCR)
1. Introduction
Advances in next generation sequencing technologies over the past 15 years has led to an explosion of molecular information about the human transcriptome that previously was not possible to observe [1,2]. In particular, RNA sequencing (RNA-seq) has led to the discovery that the bulk of transcription in our cells is dedicated to producing RNAs that do not produce protein products [3,4]. The sheer abundance of non-coding RNAs and their identification by RNA-seq has largely outpaced their functional and biochemical characterisation. As the transcriptome is very dynamic and changes in normal versus disease states, non-coding RNAs have come into focus as potential disease modifiers and could be exploited as biomarkers and/or therapeutic targets [[5], [6], [7]]. There are many kinds of non-coding RNAs in human cells, including microRNAs (miRs) [8] PIWI-associated RNAs (piRNAs) [9] and circular RNAs (circRNAs) [10]. Another abundant group are the long, non-coding RNAs (lncRNAs) – defined as greater than 200 nucleotides and often resembling protein-coding messenger RNA (mRNA) [11]. With thousands of estimated lncRNAs in human cells [12], we are specifically interested in understanding how lncRNAs are altered and contribute to cancer, along with discovering their normal physiological roles.
Breast cancer remains the leading cause of cancer-related deaths among women worldwide, with incidence rates increasing globally (World Health Organization). Within the past ten years, numerous studies have implicated the mis-regulation of lncRNAs to breast cancer development and progression [[13], [14], [15], [16], [17]]. To begin to understand which lncRNAs are specifically be linked to breast cancer, it is important to examine their expression profiles in various cell lines, and ultimately, in patient samples. This information will become the basis for further investigations into the cellular context and processes that could be affected by altered lncRNA expression.
Many studies have focused on the classification of invasive breast lesions into molecular subtypes based on the presence or absence of receptors for hormones, oestrogen (ER) and progesterone (PR), along with human epidermal growth factor-2 (HER2/ERBB2). These distinctions have profound implications on staging and treatment management [18,19] and form the basis of the molecular classification of breast cancer into four major groups: luminal A, luminal B, HER2 enriched and basal-like [[20], [21], [22]]. Luminal A involves cancer cells that are ER and/or PR positive, HER2-negative and low levels of the cell cycle-regulated protein, Ki-67. These cancers tend to be lower grade, progress slowly and have the best prognosis [23]. Luminal B cancers exhibit lower ER/PR expression, with variable HER2 levels and high levels of protein Ki-67. Luminal B disease progression is slightly faster than luminal A, with a slightly worse prognosis [24]. HER2-enriched cancer cells are ER/PR negative but HER2 positive. These cancers progress faster than luminal cancers, although they are susceptible to targeted therapies against the HER-2 protein [25]. Basal-like breast cancers are negative for all three receptors and are also known as triple-negative. This type of breast cancer has the worst prognosis and presents a significant clinical challenge [26].
In addition to invasive carcinomas, there are also preinvasive forms of breast cancer - ductal carcinoma in situ (DCIS) [27] and lobular carcinoma in situ (LCIS) [28] – distinguished by their sites of origin within the ducts or the lobules of the breast. Interestingly all molecular subtypes of invasive breast cancer are also observed in DCIS [29]. Currently it is not clear which cases of in situ breast cancer will progress to invasive disease; therefore, a better molecular understanding of the events that occur during the transition to invasive carcinoma is warranted.
Similar to breast cancer tumours, breast cancer cell lines are also classified according to the same molecular subtypes as described above [[30], [31], [32]], with the basal-like lines being subdivided into basal A and basal B clusters that are not apparent in primary tumours [30]. While cell lines have limitations, the use of breast cancer cell lines to uncover the molecular details underlying the biological processes involved with cancer initiation and progression is undisputed.
Starting with an existing RNA-seq dataset of 675 cancer cell lines by Klijn et al. [33], here we re-analysed data from subset of breast cancer cell lines to specifically examine lncRNA expression. Importantly, the Klijn et al. dataset contains RNA-seq data from 148 cancer cell lines that were not present in two genomics studies from the Sanger Institute [34] and the Cancer Cell Line Encyclopedia [35]. The dataset also contained a DCIS cell line that is unavailable in CCLE and other RNA-seq datasets from breast cancer cell lines [31]. We reasoned that this dataset, in particular, would be a useful starting point for our study.
Based on molecular classification of breast cancer cell lines, we selected representative lines from luminal A, luminal B, HER2/ErbB2-enriched, basal-like (A and B) subtypes, along with one ductal carcinoma in situ line, to identify lncRNAs with altered expression in comparison to the normal-like, immortalized breast cell line, MCF10A. From this we identified several lncRNAs with altered expression, including lncRNAs previously associated with breast cancer, i.e. DSCAM-AS1 [15,36]. We also uncovered lncRNAs previously associated with other cancer types, but not breast cancer. Importantly, we also identified novel, uncharacterised lncRNAs, LOC101448202, LOC105372471 and LOC105372815. Using Gene Expression Profiling Interactive Analysis (GEPIA2) [37] and data from The Cancer Genome Atlas (TCGA) [38] and The Genotype-Tissue Expression (GTEx) project, we examined the distribution of expression of several identified lncRNAs in tumour versus normal samples and their correlation with patient outcomes. Lastly, quantitative, reverse transcriptase, polymerase chain reaction (qRT-PCR) was used to experimentally verified RNA expression of six lncRNAs from a panel of breast cancer cell lines. Overall, our study indicates that bioinformatic re-examination of an existing RNA-seq dataset can provide an avenue to discover potentially biologically relevant lncRNAs in breast cancer development and progression.
2. Materials and methods
2.1. RNA sequencing dataset
Prior to our study, permission to access the RNA-seq data in Klijn et al. (2015) was requested from the Genentech Data Access Committee (DAC). Consent was granted to make use of the data generated by Genentech/Genentech Research and Early Development to specifically examine lncRNAs. Data was retrieved from the EMBL-European Genome-Phenome Archive (EGA) servers under EGAD00001000725.
2.2. Selection of breast cancer cell lines
Using the Klijn et al. dataset as a starting point, breast cancer cell line RNA-seq data files were identified using the metadata file provided EGA [33]. This resulted in 68 breast cancer cell lines. Subsequently 18 lines were selected for our analyses based on their molecular classification namely, normal-like (MCF10A), ductal carcinoma in situ (MCF10DCIS.com) [39], luminal A (BT-483, CAMA-1, KPL-1, MCF-7), luminal B (MDA-MB-330, UACC-812, ZR-75-30), HER2 enriched (MDA-MB-453, SK-BR-3, UACC-893), basal-like type A (BT-20, MDA-MB-436, MFM-223), basal-like type B (CAL-120, MDA-MB-157, MDA-MB-231) [40,41].
2.3. Bioinformatics methodology to identify lncRNAs in RNA-seq datasets
Each cell line consisted of two RNA-seq data files encrypted in a zipped Fastq format, with a forward read and a reverse read. The forward reads were selected for the purpose of this project. Once downloaded, the RNA-seq data was then decrypted and unzipped into Fastq format. Download and decryption of the RNA-seq data was done via the Java shell provided by the EGA (EGA Download Client v2). The quality of the RNA-seq data was then rechecked with FastQC. The RNA-seq data was then aligned to the latest human genome reference sequence, GRCh38, as provided by the National Center for Biotechnology Information (NCBI), using Spliced Transcripts Alignment to a Reference (STAR) [42]. The reference genome annotation file was used for GRCh38 with the command “-t *lnc_RNA” to select for lncRNA. Next, HTSeq was used to perform read counts [43]. The counts for each cancer cell line were then compiled into a data frame using Excel and imported into R Studio for statistical analyses. The package DESeq2 [44] was then used to carry out statistical analysis of differential lncRNA expression between the breast cancer cell lines based on their molecular subtypes indicated above. Principal component analysis was used to review the distribution of differential lncRNA expression among the molecular sub-type groups, i.e. normal-like, DCIS, luminal A, luminal B, HER2-positive, basal A and basal B. Other packages utilised were pheatmap [45] and EnhancedVolcano [46]. We chose to trim our results by eliminating non-significant results by setting an adjusted p-value of 0.01. The resulting subsets of lncRNAs were then arranged from lowest to highest log2 fold change and represented the most downregulated and the most upregulated lncRNA respectively for each cell line.
2.4. Expression of lncRNAs in breast tumour samples and patient survival analyses
Expression in tumour samples and survival analysis in patients was examined with Gene Expression Profiling Interactive Analysis 2 (GEPIA2) [37], using data generated by The Cancer Genome Atlas Research Network https://www.cancer.gov/tcga and The Genotype-Tissue Expression Project.
2.5. Cell culture
For RNA analysis of selected lncRNAs, breast cancer cell lines were purchased or obtained as indicated: MCF10DCIS.com (purchased from Wayne State University, Michigan, USA); MCF10A, MCF7 and MDA-MB-231 cells (gift from Prof Rosemary O'Connor, University College Cork); SK-BR-3 (gift from Dr Kenneth Nally, University College Cork); ZR-75-30 (gift from Prof William Gallagher, University College Dublin). Cell lines were authenticated using short, tandem repeat (STR) profiling (Eurofins Genomics). Cells were cultured in dishes with the following media requirements. MCF-10A cells were maintained in DMEM/F12 supplemented with 5% horse serum, 10 μg/ml insulin, 20 ng/ml EGF, 100 ng/ml cholera toxin and 0.5 μg/ml hydrocortisone. MCF10DCIS.com were cultured in DMEM/F12 supplemented with 5% horse serum, 1.05 mM calcium chloride and 10 mM HEPES. MCF-7 and MDA-MB-231 cells were cultured in DMEM supplemented with 10% FBS and 1% penicillin/streptomycin. SK-BR3 cells were grown in RPMI +10% FBS +1% penicillin/streptomycin. ZR-75-30 cells were cultured in RPMI supplemented with 10% FBS and 1% penicillin/streptomycin. Cells were maintained at 37 °C with 5% CO2 and were mycoplasma-free.
2.6. RNA analysis by qRT-PCR
Total RNA was extracted from cells using TRIzol (Thermo Fisher Scientific). Briefly, 0.2 mL of chloroform was added per 1 ml of TRIzol reagent, samples were homogenized and then left at room temperature for 3 min. The aqueous phase was separated by centrifugation, and RNA was precipitated using isopropanol. After two washes using 75% ethanol, the RNA pellet was airdried, resuspended in water and incubated at 58 °C for 10 min.
1 μg of RNA was treated with DNase to eliminate contaminating DNA using TURBO DNase (Invitrogen) then used in a cDNA synthesis reaction using Superscript II (Thermo Fisher Scientific) as per manufacturer instructions. Reactions lacking reverse transcriptase enzyme were also run in the same condition as controls. The cDNA synthesized was diluted 1:5 and used for qRT-PCR. The diluted cDNA was used in qRT-PCR reactions. Briefly, 25 ng cDNA was combined with SYBR Green JumpStart Taq ReadyMix (Sigma-Aldrich) in 20 μl reactions and run using the following conditions:
-
-
95 °C 10 min
-
-
94 °C 30 s, 57 °C 45 s, 72 °C 1 min repeated 39 times
-
-
94 °C 30 s, 57 °C 45 s, 72 °C 15 min
-
-
Melting curve stage
qRT-PCR was performed using the StepOnePlus™ Real-Time PCR System and StepOnePlus software (Applied Biosystems). After analysis of the melting curve, results were normalized to the expression of glyceraldehyde 3-phosphate dehydrogenase, GAPDH, using the ΔΔCT method. Technical duplicates were done for each reaction, and three biologicals replicates were processed. qRT-PCR primers used for each lncRNA are listed below:
| lncRNA | Forward | Reverse |
|---|---|---|
| CCAT1 | GCAGGCAGAAAGCCGTATCT | TCCCAGGTCCTAGTCTGCTT |
| DSCAM-AS1 | ACCACAACAACAACAACAG | ATGATGAGACCAGAACTTCC |
| LINC00885 | CAGGGTTGGTGCTATGAATGAC | GAAGATTGTCCATGTTGGCAGTAT |
| LOC105372815 | TCTTCAACATGGCGGTCGAT | GTGGCAGAAGTGGAGTGGAG |
| MUC5B-AS1 | CTCTGTGAGGATCCAGTGGACG | TGTGCTTTGCTGTGACGACT |
| ZNF667-AS1 | TGTGACAAGTTCTTCAGGCG | GGATGAATGCCGATTGCAGAC |
| GAPDH | GAGTCAACGGATTTGGTCGT | TTCCCGTTCTCAGCCTTG |
2.7. Statistics and code availability
Most statistical analyses were performed in R (version 3.5.2). One-way ANOVA with multiple comparisons was done using GraphPad Prism v.8.3.0. Source codes and scripts are available upon request.
3. Results
3.1. Bioinformatic identification of lncRNA differentially expressed in malignant versus non-malignant breast cancer cell lines
The paper Klijn et al. (2015) described RNA-seq and single nucleotide polymorphism (SNP) array analysis of 675 human cancer cell lines. Using that dataset as a starting point, we focused on the 68 breast cancer cell lines using the metadata file provided by EGA. Next we narrowed this to 17 breast cancer cell lines based on their molecular subtypes, ensuring that we had at least three to four representative lines from each group, i.e. luminal A, luminal B, HER2 positive, basal A and basal B. Our analyses also included a single DCIS cell line (MCF10DCIS.com) and the immortalized, normal-line breast cell line, MCF10A. This resulted in our working RNA-seq dataset from 18 cell lines.
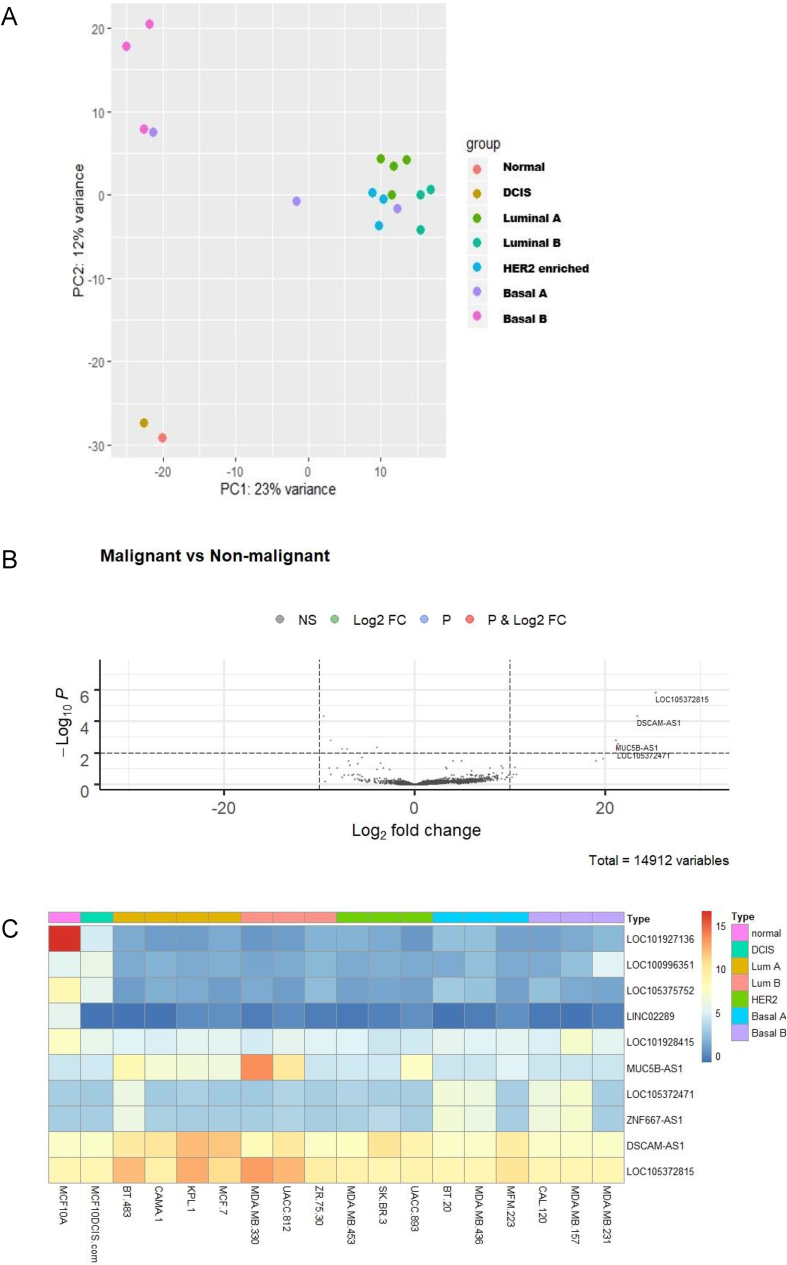
First, we examined the variation of the selected cell lines using the multivariate data analysis method, principal component analysis (PCA). The resulting plot (Fig. 1A) showed clustering among the luminal A, luminal B and HER2 enriched cell lines with respect to lncRNA expression. Basal B cell lines showed greater variance to other malignant subtypes; while basal A displayed degrees of variance to non-malignant and malignant cell lines. The normal-like line, MCF10A, and the DCIS line, MCF10DCIS.com, clustered closely and showed minimal variance to each other.
Fig. 1.
Breast cancer cell lines distinguished by malignant versus non-malignant show differential expression of lncRNAs (A) Principal component analysis of selected breast cancer cell lines grouped by molecular classification, normal-like, DCIS, luminal A, luminal B, HER2 enriched, basal A and basal B. PC1 (x-axis) is representative of the non-malignant cell line (MCF10A); PC2 (y-axis) is representative of the 17 malignant cell lines. (B) Volcano plot (log2 FC > 10, p ≤ 0.01) to filter differentially expressed lncRNAs in malignant cell lines versus normal-like, MCF10A. (C) Heatmap of differentially expressed lncRNAs in malignant versus non-malignant cell lines. DSCAM-AS1 and LOC105372815 were the most highly expressed lncRNAs in many of the cell lines examined.
We then categorised lncRNAs that were differentially expressed in malignant versus non-malignant cells lines. For this comparison, the DCIS cell line was included in the malignant group. A full list of read counts for lncRNAs from processed RNA-seq data from each cell line is available in Supplemental Table 1. We proceeded to visualise the distribution of differentially expressed lncRNAs between the malignant versus non-malignant lines using a volcano plot and heatmap (Fig. 1B and C). A total of ten lncRNAs were determined to be differentially expressed in the malignant cell lines when compared to the normal-like cell line, MCF10A, with five more highly expressed and five more lowly expressed. It was noted that in choosing a cutoff of 10 for the log2 fold change (Fig. 1B) there were no downregulated lncRNAs surpassing this limit. However, several highly expressed lncRNAs were identified, including DSCAM-AS1, LOC105372471, LOC105372815, MUC5B-AS1 and ZNF667-AS1.
3.2. Bioinformatic identification of lncRNAs differentially expressed in breast cancer cell lines divided by hormone/receptor status
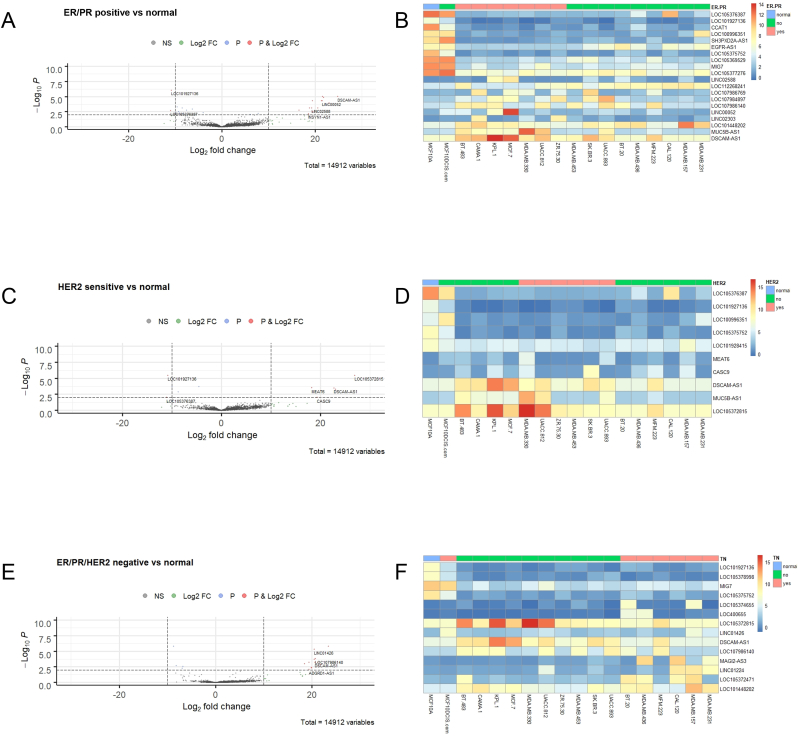
Next we divided the malignant cell lines into groups based on their hormone/receptor sensitivity, namely ER/PR positive, HER2 sensitive and ER/PR/HER2 negative. Our logic in dividing our data into these groups was to fit within the pre-existing paradigms of breast cancer risk stratification and treatment management in the clinical setting [47,48]. The visualisations of differentially expressed lncRNAs in ER/PR positive, HER2 sensitive and ER/PR/HER2 negative cell lines by volcano plots and heatmaps are shown in Fig. 2. ER/PR positive cell lines versus the normal-like cell line showed differential expression of 27 lncRNAs in total, with 16 lncRNAs at higher levels and 11 lncRNAs with lower expression. Notably, DSCAM1-AS1 was the most significant upregulated lncRNA; while LOC101927136 was the most significant downregulated lncRNA (Fig. 2A and B). Using the same procedure and visualisation methods, we found ten differentially expressed lncRNAs in the HER sensitive group (five over- and under-expressed; Fig. 2C and D); while the ER/PR/HER2 negative group contained 17 differentially expressed lncRNAs (13 over- and four under-expressed; Fig. 2E and F). Interestingly some specific lncRNAs that were differentially expressed emerged, including increased expression of LOC105372815 and reduced expression of LOC101927136 in the hormone receptor/HER2 positive groups, which was not observed in the ER/PR/HER2 negative group.
Fig. 2.
Differential expression of lncRNAs in ER/PR positive, HER2sensitiveand triple-negative breast cancer cell lines (A) Volcano plot (log2 FC > 10, p ≤ 0.01 indicated as dashed lines) and (B) Corresponding heatmap of differentially expressed lncRNAs in ER/PR positive breast cancer cell lines versus normal-like, MCF10A, analysed across all 18 cell lines examined. Similar analyses were done for HER2 sensitive lines (C) Volcano plot (log2 FC > 10, p ≤ 0.01), (D) Corresponding heatmap of lncRNA expression across all cell lines; and triple-negative breast cancer cell lines (those lacking ER/PR/HER2) (E) Volcano plot (log2 FC > 10, p ≤ 0.01), (F) Corresponding heatmap of lncRNA expression across all cell lines.
3.3. Evaluation of breast cancer cell lines by molecular subtypes for lncRNA expression
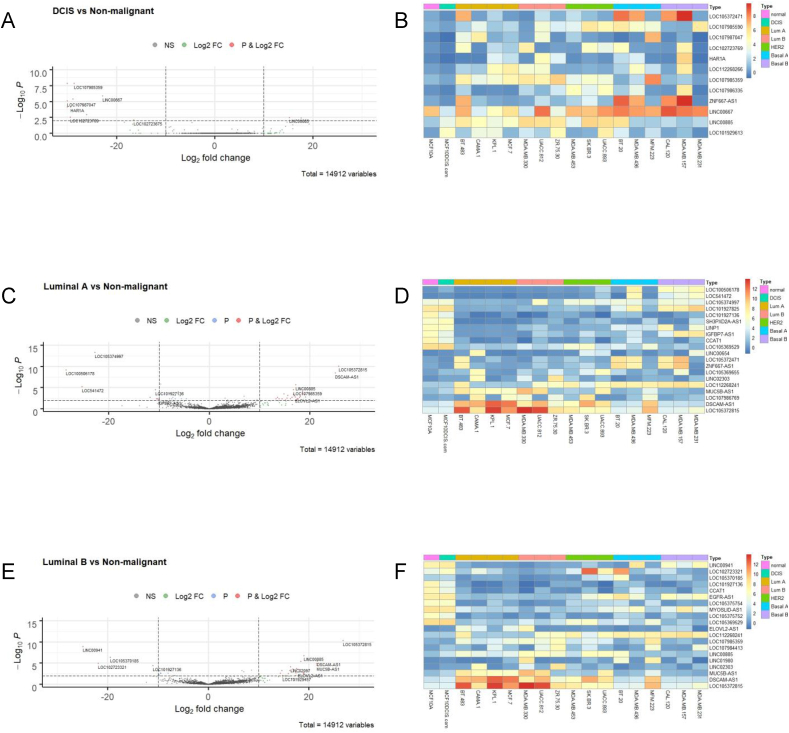
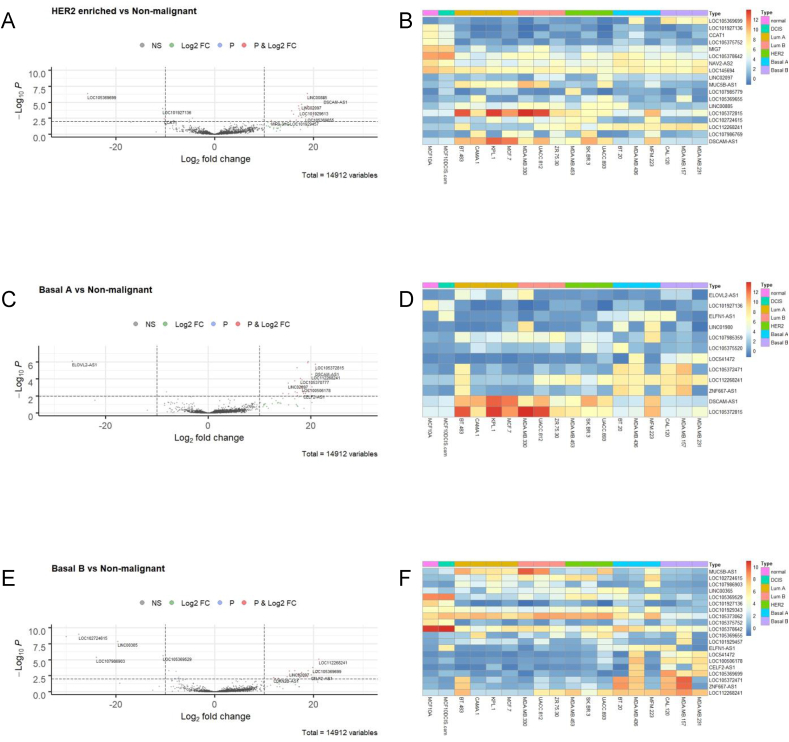
We then chose to explore the differences among cell lines based on their molecular classifications in more detail. For this purpose, we created a category in the design matrix where again the normal-like breast cell line, MCF10A, was chosen as the basis for comparison. Cell lines based on the molecular groups – DCIS, luminal A, luminal B, HER2 enriched, basal-like type A and basal-like type B – were compared in turn using our DESeq2 data (Supplemental Table 1) and visualised by a volcano plots and heatmaps as shown in Fig. 3 (DCIS, luminal A and luminal B) and Fig. 4 (HER2 positive, basal A and basal B). Using a fold change ≥2.0 and p value ≤ 0.01, we compiled lists of the top ten up- and downregulated lncRNAs for each molecular subtype. From those lists, we developed a curated list of lncRNAs that were differentially expressed in at least two molecular subtypes to identify lncRNAs with persistently higher or lower expression (Table 1). Following an extensive literature review, previous associations with breast and/or any other sites of cancer were also included in Table 1.
Fig. 3.
Differential expression of lncRNAs in DCIS, luminal A and luminal B breast cancer cell lines (A) Volcano plot (log2 FC > 10, p ≤ 0.01 indicated as dashed lines) and (B) Corresponding heatmap of differentially expressed lncRNAs in DCIS cell line, MCF10DCIS.com, versus normal-like, MCF10A, analysed across all 18 cell lines examined. Similar analyses were done for luminal A cell lines (BT-483, CAMA-1, KPL-1, MCF-7) (C) Volcano plot (log2 FC > 10, p ≤ 0.01), (D) Corresponding heatmap of lncRNA expression across all cell lines; and luminal B breast cancer cell lines (MDA-MB-330, UACC-812, ZR-75-30) (E) Volcano plot (log2 FC > 10, p ≤ 0.01), (F) Corresponding heatmap of lncRNA expression across all cell lines.
Fig. 4.
Differential expression of lncRNAs in HER2 enriched, basal-like type A (basal A) and basal-like type B (basal B) (A) Volcano plot (log2 FC > 10, p ≤ 0.01 indicated as dashed lines) and (B) Corresponding heatmap of differentially expressed lncRNAs in HER2 positive breast cancer cell lines (MDA-MB-453, SK-BR-3, UACC-893) versus normal-like, non-malignant line, MCF10A, analysed across all 18 cell lines examined. Similar analyses were performed for basal A breast cancer lines (BT-20, MDA-MB-436, MFM-223) (C) Volcano plot (log2 FC > 10, p ≤ 0.01), (D) Corresponding heatmap of lncRNA expression across all cell lines; and for basal B breast cancer cell lines (CAL-120, MDA-MB-157, MDA-MB-231) (E) Volcano plot (log2 FC > 10, p ≤ 0.01), (F) Corresponding heatmap of lncRNA expression across all cell lines.
Table 1.
Curated list of over- and under-expressed lncRNAs in selected breast cancer cell lines with their molecular subtypes. Previous associations with cancers are noted, along with publications.
| lncRNAs over-expressed in breast cancer cell lines examined | ||||
|---|---|---|---|---|
| lncRNA | RefSeq ID | Differentially expressed in: | Previous cancer association | References |
| CELF2-AS1 | NR_126062.1 | Basal A, Basal B | No cancer related publications | |
| DSCAM-AS1 | NR_038896.1 | LA, LB, HER2 enriched, Basal A, ER/PR +ve, HER2 sensitive, triple negative | Breast and lung cancer | [15,36,[49], [50], [51], [52]] |
| ELFN1-AS1 | NR_120508.1 | DCIS, Basal A, Basal B | Expressed in various tumour samples | [53] |
| LINC00885 | NR_034088.1 | DCIS, Luminal B | Bladder cancer | [54] |
| LOC101448202 | NR_103451.1 | ER/PR +ve, triple negative | Uncharacterised | |
| LOC105372471 | XR_001754022.1 | Basal A, Basal B, triple negative | Uncharacterised | |
| LOC105372815 | XR_937755.2 | LA, LB, HER2 enriched, Basal A, HER2 sensitive, triple negative | Uncharacterised | |
| MUC5B-AS1 | NR_157183.1 | LA, LB, HER2 enriched, ER/PR +ve, HER2 sensitive | Lung cancer | [55] |
| ZNF667-AS1 | NR_036521.1 | LA, Basal A, Basal B | Breast, cervical, oeosphageal, laryngeal cancer | [[56], [57], [58], [59], [60], [61]] |
| lncRNAs under-expressed in breast cancer cell lines examined | ||||
| CCAT1 | NR_108049.1 | LA, LB, HER2 enriched, ER/PR +ve |
Multiple cancers including acute myeloid leukaemia, breast, colon, gallbladder, liver and squamous cell carcinoma | [14,[62], [63], [64], [65], [66], [67], [68], [69], [70], [71]] |
| EGFR-AS1 | NR_047551.1 | Luminal B, ER/PR +ve, | Head & neck, lung, gastric and hepatocellular cancers | [[72], [73], [74], [75]] |
| LINC00885 | NR_034088.1 | Basal A | Bladder cancer | [54] |
| MIG7 | NR_148965.1 | HER2 enriched, ER/PR +ve, triple negative | Expressed in malignant cells; bone, hepatocellular and ovarian cancers | [[76], [77], [78], [79], [80]] |
| MUC5B-AS1 | NR_157183.1 | Basal B | Lung cancer | [55] |
| ZNF667-AS1 | NR_036521.1 | DCIS | Breast, cervical, oeosphageal, laryngeal cancer | [[56], [57], [58], [59], [60], [61]] |
From our curated list of lncRNAs differentially expressed in breast cancer cell lines, we confirmed increased expression of DSCAM-AS1 in multiple breast cancer cell lines. DSCAM-AS1 is regulated by ER and has been previously associated with breast cancer [15,36,50,51]. In a recent study, DSCAM-AS1 was shown to regulated the cell cycle at the G1/S transition, increasing cell proliferation [50]. We also identified several lncRNAs with previous associations to cancer types other than breast, namely LINC00885 and MUC5B-AS1; while CELF2-AS1 has no known cancer association. Most interestingly, we identified a few overexpressed lncRNAs that are uncharacterised, LOC101448202, LOC105372471 and LOC105372815.
3.4. Assessment of clinical relevance of lncRNAs in breast cancer using GEPIA2
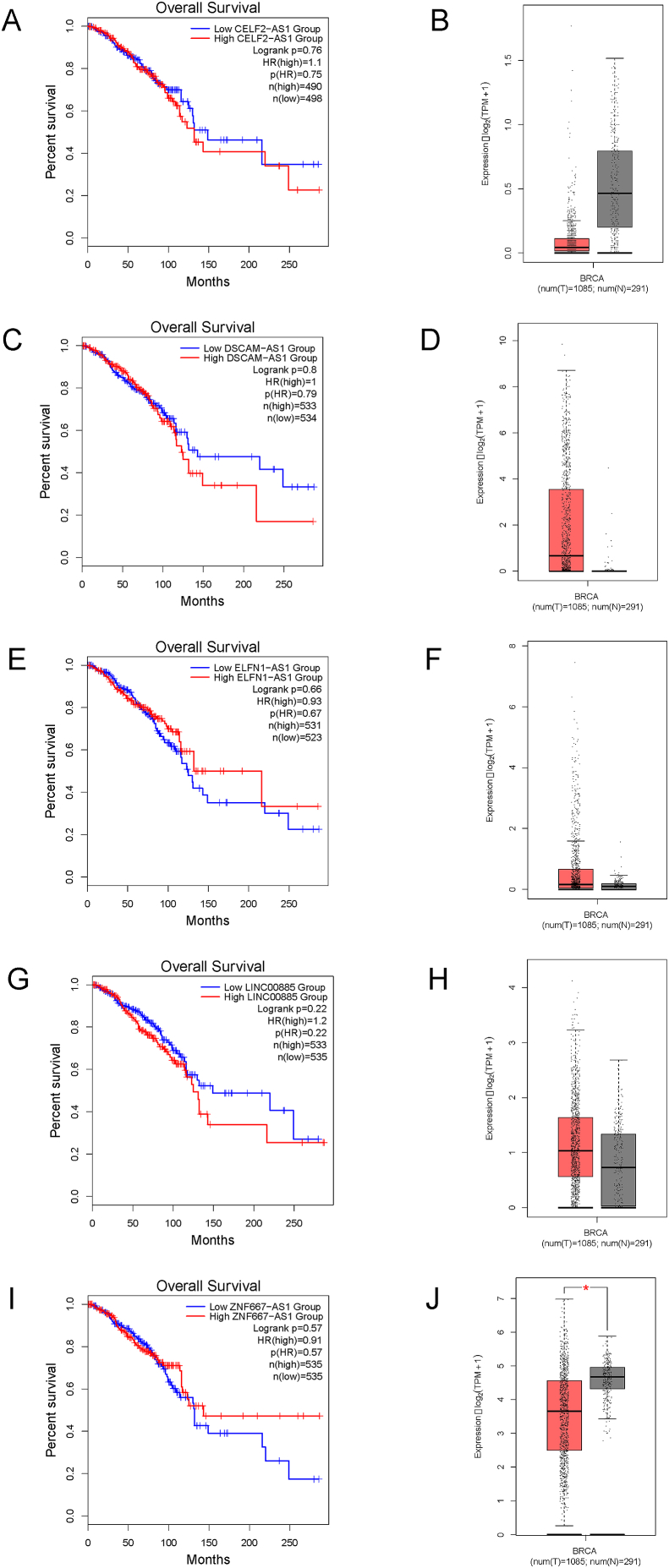
To examine the clinical significance of identified lncRNAs, we used GEPIA2 [37] to explore data from TCGA and GTEx databases. Using our curated list, five lncRNAs were found to have associations with breast cancer in GEPIA2, including CELF2-AS1, DSCAM-AS1, ELFN1-AS1, LINC00885 and ZNF667-AS1. Breast cancer survival and comparative expression (tumour vs. normal tissue) plots for each lncRNA are shown in Fig. 5. For CELF2-AS1, the Kaplan-Meier plot indicates higher expression is associated with poorer survival; however, its expression in tumour tissue appears lower (Fig. 5A and B). As expected, higher DSCAM-AS1 expression was correlated with poorer patient survival (Fig. 5C) and a corresponding increased expression in tumour versus normal tissue sample (Fig. 5D). Following a similar pattern, LINC00885 is associated with slightly poorer survival and higher tumour expression (Fig. 5G and H), perhaps indicating an oncogenic role. Lastly, both higher expression of ELFN1-AS1 and ZNF667-AS1 were associated with better patient survival (Fig. 5E and I). While the comparative expression of ZNF667-AS1 in tumour is lower (Fig. 5J), the expression of ELFN1-AS1 in tumour samples does not appear to be lower than normal tissue (Fig. 5F).
Fig. 5.
Clinical relevance of select lncRNAs identified bioinformatically using Gene Expression Profiling Interactive Analysis (GEPIA2). Breast cancer survival analysis plots (Kaplan-Meier) were generated for lncRNAs (A) CELF2-AS1; (C) DSCAM-AS1; (E) ELFN1-AS1; (G) LINC00885 and (I) ZNF667-AS1 using GEPIA2 [37]. Corresponding box plots of the comparative expression of the same lncRNAs (B) CELF2-AS1; (D) DSCAM-AS1; (F) ELFN1-AS1; (H) LINC00885 and (J) ZNF667-AS1 in breast cancer tumour samples (red) versus normal tissue samples (grey) generated using GEPIA2.
3.5. Experimental validation of lncRNA expression by qRT-PCR
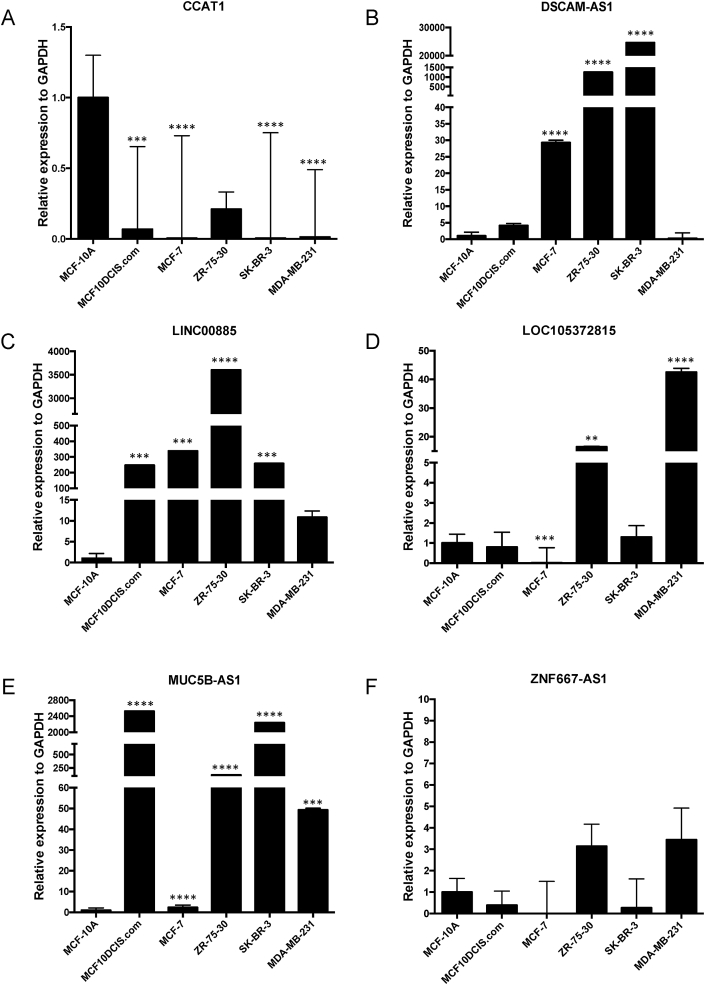
In effort to experimentally verify the expression patterns observed from our analysis of the RNA-seq data, we next examined lncRNA expression for six lncRNAs on our curated list (CCAT1, DSCAM-AS1, LINC00885, LOC105372815, MUC5B-AS1 and ZNF667-AS1) by qRT-PCR from breast cancer cell lines, representative of each molecular subtype, and the normal-like line, MCF10A. Breast cancer cell lines selected included: MCF10DCIS.com (DCIS); MCF7 (luminal A); ZR-75-30 (luminal B); SK-BR-3 (HER2 positive); and MDA-MB-231 (basal B). Total RNA isolated from cells was used for cDNA synthesis and qRT-PCR with lncRNA specific primers. Relative expression to GAPDH for each lncRNA is shown in Fig. 6.
Fig. 6.
Experimental confirmation of differential expression of selected lncRNAs in a breast cancer cell line panel, representing each molecular subtype. qRT-PCR was performed using cDNA synthesized from total RNA isolated from MCF10A (normal-like), MCF10DCIS.com (DCIS), MCF7 (luminal A), ZR-75-30 (luminal B), SK-BR-3 (HER2 positive), and MDA-MB-231 (basal B) cells. Relative expression of lncRNAs (A) CCAT1; (B) DSCAM-AS1; (C) LINC00885; (D) LOC105372815; (E) MUC5B-AS1 and (F) ZNF667-AS1, as compared to GAPDH, are shown, using one-way ANOVA (GraphPad Prism v.8.3.0). **** p-value < 0.0001; *** p-value < 0.001; ** p-value < 0.01.
Largely in agreement with our bioinformatic analyses, the qRT-PCR experiments validated lncRNA expression in the tested cell lines. Similar to the results from DESeq analysis, we observed that CCAT1 lncRNA was very lowly expressed in most breast cancer cell lines tested, with highest expression in the normal-like line, MCF10A (Fig. 6A). For DSCAM-AS1, the highest expression was in the luminal A (MCF7), luminal B (ZR-75-30) and HER2 positive (SK-BR-3) lines, with virtually no detection in the basal-like line (Fig. 6B). This seems contradictory, as DSCAM-AS1 was one of the most significant, highly expressed lncRNAs in the ER/PR/HER2 negative (Fig. 2E) and basal A (Fig. 4C) subtypes. Interestingly qRT-PCR analysis of LINC00885 shows lowest expression of this lncRNA in the basal-like line (MDA-MB-231) unlike the other breast cancer cell lines tested (Fig. 6C), agreeing with our bioinformatic analysis. For LOC105372815 and MUC5B-AS1, each lncRNA was expressed at an increased level in certain cell lines over MCF10A; however, there was consistent low expression for each of these lncRNAs in MCF7 cells (Fig. 6D and E). Given that MCF7 cells are a non-invasive breast cancer cell line, it is possible that low expression of these lncRNAs may be indicative of this phenotype, particularly since MUC5B-AS1 has been linked to metastasis in lung cancer [55]. Lastly, ZNF667-AS1 expression for any cell line failed to reach significance over MCF10A (Fig. 6F), indicating the over-expression observed with our bioinformatic analysis may reflective of cell line-specific effects, i.e. ZNF667-AS1 is more highly expressed in MDA-MB-157 versus MDA-MB-231 (Fig. 4F), despite both being classified as basal-like type B.
4. Discussion
Based on our bioinformatic analysis of a subset of breast cancer cell line RNA-seq data [33], certain lncRNAs were more persistently upregulated and downregulated (Table 1), with many of these experimentally verified using qRT-PCR (Fig. 6). These lncRNAs also correlated with the categorisation of breast cancer cell lines based on hormonal sensitivity (Fig. 2) and/or molecular classification (Fig. 3, Fig. 4). We chose to divide our study samples by hormonal/protein sensitivity and molecular classification, as most clinical treatment options are based on these parameters [47,48]. Our principal component analysis further supported this division, as clustering was evident based on the cell line subtypes (Fig. 1A). Interestingly, the basal A and B cell lines displayed the greatest variance in our assessment. This could reflect the observation that all molecular subtypes are observed across triple-negative disease, although the majority fall within the basal-like subtype [81].
Our analyses are based on the Klijn et al., 2015 dataset, where RNA-seq data was prepared via the poly-adenylate (poly-A) selection method. The two main approaches in the early stage of an RNA-seq protocol are either poly-A enrichment or selective degradation of ribosomal RNA (rRNA) [82]. The poly-A selection method almost exclusively selects for transcripts with 3’ poly-A tails; whereas, the rRNA depletion method is able to capture both poly-A+ and non-adenylated transcripts. It has been suggested that the quality of reads is higher using the poly-A selection method for protein-coding genes, as most mature messenger RNAs (mRNAs) are adenylated; while some lncRNAs, small RNAs and T-cell/B-cell receptor transcripts can only be detected via rRNA depletion [83]. For example, the lncRNA BC200 (brain cytoplasmic 200) has been shown to have strong association with invasive breast cancer [84,85], but as an RNA polymerase III transcript [86], it is not represented in this study. It is our opinion that re-running our pipeline on RNA-seq data prepared using the rRNA depletion method could improve the quality control of our analysis by incorporating non-adenylated transcripts.
Metadata of the Klijn et al., 2015 dataset as provided by the EGA, revealed 68 cancer cell lines which were of breast origin. Unfortunately, we were not able to fully analyse the whole dataset due to heavy computational requirements to carry out this task. Therefore, a selection of cell lines was chosen to represent our chosen subtypes. Unsurprisingly, we had very limited options when it came to cell lines to represent the normal-like and DCIS groups, with our only options MCF10A and MCF10DCIS.com cell lines, respectively. Other breast cancer cell line RNA-seq data exists [31,35]; however, unlike the Klijn et al., 2015 paper they do not have a cell line representative of DCIS disease. We also chose not to incorporate RNA-seq data from other sources and instead worked only from a single dataset for consistency. The other aim of keeping our study limited to the Klijn et al., 2015 dataset was to investigate the feasibility of re-analysing a previous dataset as a guide for further research. Ideally, we would have preferred more cell lines to represent the normal-like and in particular DCIS subtypes; however as it stands, there are only a limited number of DCIS cell lines [87], and they are not usually included in breast cancer cell line panels.
Since DSCAM-AS1 was highly expressed in most of our comparisons as shown in Table 1, we chose to examine the clinical relevance of the expression of this lncRNA using GEPIA2 [37]. Importantly, our results are in agreement with previous work describing the oncogenic role of this RNA [15]. The Kaplan-Meier survival plot generated via GEPIA2, using data from TCGA, supported our findings regarding DSCAM-AS1 with a lower 10-year survival in breast cancer cases with higher expression of DSCAM-AS1 (Fig. 5C).
In contrast to other lncRNAs identified, our analysis of lncRNA ZNF667-AS1 did not match with the survival plot generated with GEPIA2. Our bioinformatic analysis showed ZNF667-AS1 to be differentially upregulated in the luminal B and basal-like subtypes. However, the survival analysis indicated that high expression of ZNF667-AS1 was associated with increased survival rates at ten years (Fig. 5I). Even when we reviewed the survival of ZNF667-AS1 in the luminal B and basal-like subtypes, it showed better long-term survival with higher expression. However, for the initial eight to nine years in this breast cancer subtype, survival was slightly lower with higher expression (data not shown). Previous publications have explored low expression of ZNF667-AS1 in cervical cancer [58] where it was associated with poorer prognosis. Interestingly, another paper investigated ZNF667-AS1's downregulation in 16 cancer cell lines and proposed it played an important role as a tumour suppressor [57]. Unlike the survival curves, the differential expression of ZNF667-AS1 in tumour versus normal tissue presented here does show a lower expression in tumour samples (Fig. 5J), favouring support of a tumour suppressor role of this lncRNA in breast cancer.
Initially our analysis of the lncRNA ELFN1-AS1 also suggested that our analysis did not match the survival plot generated with GEPIA2. The survival plot showed that higher expression of ELFN1-AS1 was associated with improved survival rates (Fig. 5E). When we reviewed the survival plot in the basal-like subtype only, survival was slightly improved with lower expression of this lncRNA (data not shown). A previous publication on this lncRNA has shown higher expression in tumour tissue of various histological origin [53], but breast was not examined.
One of the lncRNAs downregulated in most of our breast cancer cell line subtypes was CCAT1 (Table 1 and Fig. 6A). CCAT1, colon cancer-associated transcript-1 (also CASC19, cancer susceptibility 19, CARLo-6 and LINC01245), was first reported to be highly expressed in colon cancer [62] and is present in a frequently amplified genomic region in colorectal cancer [88]. Other studies have linked elevated CCAT1 to other cancers including acute myeloid leukaemia [71], gallbladder [65], liver [89] and squamous cell carcinoma [68]. In 2015, Zhang et al. showed that higher CCAT1 expression was associated with aggressive disease progression and poor prognosis of breast cancer patients [14]. In a more recent study by Han et al. (2019), the authors reported increased expression of CCAT1 from triple-negative breast cancer tissues and cell lines, i.e. MDA-MB-231 cells [69]; however, this observation is not in agreement with our analysis of CCAT1 in breast cancer cell lines, in which lower expression was observed in the RNA-seq data and by qRT-PCR. It is unclear why our results are not in agreement, unless we have detected a different transcript variant. Since the specific CCAT1 qRT-PCR primer sequences used by Han et al. are not published, we were unable to compare this directly.
Among our most interesting findings, we uncovered several lncRNAs previously associated with other cancer types, and not breast cancer, as well as several uncharacterised lncRNAs. Of these, the lncRNA MUC5B-AS1 has been associated with promoting metastasis in lung cancer [55]; however, there are currently no studies linking MUC5B-AS1 to breast cancer. Given the very high expression of MUC5B-AS1 that we observed across multiple cell lines by qRT-PCR (Fig. 6E), this lncRNA will be of future interest. Similar to MUC5B-AS1, LINC00885 has been associated with bladder cancer, and not breast [54]. Given our consistent results across bioinformatic, GEPIA2 and qRT-PCR analyses (Figs. 3 and 5G and H and Fig. 6C), we propose that LINC00885 may have an oncogenic role; however, further research is necessary to assess LINC00885's biological role in the cell. Future work will also be required to elucidate the functions of currently uncharacterised lncRNAs identified in our study. This includes a very prominent lncRNA in our analysis, LOC105372815, along with LOC101448202 and LOC105372815, all of which are uncharacterised.
In conclusion, our study has successfully shown that an existing RNA-seq dataset can be re-analysed to provide further avenues of research. Although the scope of this work was focused on breast cancer, the methods used could easily be applied to other sites of primary tumours. There does indeed appear to be a strong argument for a correlation between the differential expression of lncRNAs and their hypothesised biological roles in oncogenesis and tumour progression, paving the way for lncRNAs to be used a disease biomarkers and/or therapeutic targets [17]. A recent publication by Ghandi et al. (2019), involving a re-examination of cancer cell line data provided by CCLE [90], further demonstrates that re-analysis of existing data is a powerful approach to gain new insights into cancer biology.
CRediT authorship contribution statement
Oza Zaheed: Software, Formal analysis, Data curation, Visualization, Writing - original draft. Julia Samson: Investigation, Formal analysis, Visualization, Writing - original draft. Kellie Dean: Conceptualization, Supervision, Writing - original draft, Writing - review & editing, Project administration, Funding acquisition.
Acknowledgments
We would like to thank Darren Fenton and Prof Pavel Baranov (School of Biochemistry and Cell Biology, University College Cork) for helpful discussions, technical assistance and server access during this project. We also thank Dr Orla Cox, Prof Rosemary O'Connor, Subhasree Rajaram, Dr Kenneth Nally (School of Biochemistry and Cell Biology, University College Cork); and Chowdhury Arif Jahangir, Dr Arman Rahman and Prof William Gallagher (Conway Institute for Biomolecular and Biomedical Research, University College Dublin) for the gifts of breast cancer cells lines. The results shown here are in part based upon data generated by the TCGA Research Network. The Genotype-Tissue Expression (GTEx) Project was supported by the Common Fund of the Office of the Director of the National Institutes of Health, and by NCI, NHGRI, NHLBI, NIDA, NIMH, and NINDS. The data from TCGA and GTEx used for the analyses described in this manuscript were obtained from the GEPIA2 portal.
This project was initially supported though the Translational Research Access Programme, School of Medicine, University College Cork (KD).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ncrna.2020.02.004.
Contributor Information
Oza Zaheed, Email: 118226079@umail.ucc.ie.
Julia Samson, Email: 116224719@umail.ucc.ie.
Kellie Dean, Email: k.dean@ucc.ie.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Wang Z., Gerstein M., Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goodwin S., McPherson J.D., McCombie W.R. Coming of age: ten years of next-generation sequencing technologies. Nat. Rev. Genet. 2016;17:333–351. doi: 10.1038/nrg.2016.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Birney E., Stamatoyannopoulos J.A., Dutta A., Guigó R., Gingeras T.R., Margulies E.H. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Djebali S., Davis C.A., Merkel A., Dobin A., Lassmann T., Mortazavi A. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Leeneer K., Claes K. Non coding RNA molecules as potential biomarkers in breast cancer. Adv. Exp. Med. Biol. 2015;867:263–275. doi: 10.1007/978-94-017-7215-0_16. [DOI] [PubMed] [Google Scholar]
- 6.Huarte M. The emerging role of lncRNAs in cancer. Nat. Med. 2015;21:1253–1261. doi: 10.1038/nm.3981. [DOI] [PubMed] [Google Scholar]
- 7.Waller P., Blann A. Non-coding RNAs – a primer for the laboratory scientist. Br. J. Biomed. Sci. 2019;76:157–165. doi: 10.1080/09674845.2019.1675847. [DOI] [PubMed] [Google Scholar]
- 8.Yates L.A., Norbury C.J., Gilbert R.J.C. The long and short of MicroRNA. Cell. 2013;153:516–519. doi: 10.1016/j.cell.2013.04.003. [DOI] [PubMed] [Google Scholar]
- 9.Iwasaki Y.W., Siomi M.C., Siomi H. PIWI-interacting RNA: its biogenesis and functions. Annu. Rev. Biochem. 2015;84:405–433. doi: 10.1146/annurev-biochem-060614-034258. [DOI] [PubMed] [Google Scholar]
- 10.Ebbesen K.K., Hansen T.B., Kjems J. Insights into circular RNA biology. RNA Biol. 2017;14:1035–1045. doi: 10.1080/15476286.2016.1271524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deveson I.W., Hardwick S.A., Mercer T.R., Mattick J.S. The dimensions, dynamics, and relevance of the mammalian noncoding transcriptome. Trends Genet. 2017;33:464–478. doi: 10.1016/j.tig.2017.04.004. [DOI] [PubMed] [Google Scholar]
- 12.Kopp F., Mendell J.T. Functional classification and experimental dissection of long noncoding RNAs. Cell. 2018;172:393–407. doi: 10.1016/j.cell.2018.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hansji H., Leung E.Y., Baguley B.C., Finlay G.J., Askarian-Amiri M.E. Keeping abreast with long non-coding RNAs in mammary gland development and breast cancer. Front. Genet. 2014;5:1–15. doi: 10.3389/fgene.2014.00379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang X.-F., Liu T., Li Y., Li S. vol. 8. 2015. (Overexpression of Long Non-coding RNA CCAT1 Is a Novel Biomarker of Poor Prognosis in Patients with Breast Cancer). [PMC free article] [PubMed] [Google Scholar]
- 15.Niknafs Y.S., Han S., Ma T., Speers C., Zhang C., Wilder-Romans K. The lncRNA landscape of breast cancer reveals a role for DSCAM-AS1 in breast cancer progression. Nat. Commun. 2016;7:12791. doi: 10.1038/ncomms12791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tracy K.M., Tye C.E., Ghule P.N., Malaby H.L.H., Stumpff J., Stein J.L. Mitotically-associated lncRNA (MANCR) affects genomic stability and cell division in aggressive breast cancer. Mol. Canc. Res. 2018;16:587–598. doi: 10.1158/1541-7786.MCR-17-0548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Slack F.J., Chinnaiyan A.M. The role of non-coding RNAs in oncology. Cell. 2019;179:1033–1055. doi: 10.1016/j.cell.2019.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giuliano A.E., Connolly J.L., Edge S.B., Mittendorf E.A., Rugo H.S., Solin L.J. Breast Cancer-Major changes in the American Joint Committee on Cancer eighth edition cancer staging manual. Ca - Cancer J. Clin. 2017;67:290–303. doi: 10.3322/caac.21393. [DOI] [PubMed] [Google Scholar]
- 19.Nicolini A., Ferrari P., Duffy M.J. Prognostic and predictive biomarkers in breast cancer: past, present and future. Semin. Canc. Biol. 2018;52:56–73. doi: 10.1016/j.semcancer.2017.08.010. [DOI] [PubMed] [Google Scholar]
- 20.Perou C.M., Sørlie T., Eisen M.B., van de Rijn M., Jeffrey S.S., Rees C.A. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 21.Sørlie T., Perou C.M., Tibshirani R., Aas T., Geisler S., Johnsen H. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl. Acad. Sci. U. S. A. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu Z., Fan C., Oh D.S., Marron J., He X., Qaqish B.F. The molecular portraits of breast tumors are conserved across microarray platforms. BMC Genom. 2006;7:96. doi: 10.1186/1471-2164-7-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao J.J., Swain S.M. Luminal A breast cancer and molecular assays: a review. Oncol. 2018;23:556–565. doi: 10.1634/theoncologist.2017-0535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ades F., Zardavas D., Bozovic-Spasojevic I., Pugliano L., Fumagalli D., de Azambuja E. Luminal B breast cancer: molecular characterization, clinical management, and future perspectives. J. Clin. Oncol. 2014;32:2794–2803. doi: 10.1200/JCO.2013.54.1870. [DOI] [PubMed] [Google Scholar]
- 25.Godoy-Ortiz A., Sanchez-Muñoz A., Chica Parrado M.R., Álvarez M., Ribelles N., Rueda Dominguez A. Deciphering HER2 breast cancer disease: biological and clinical implications. Front. Oncol. 2019;9:1124. doi: 10.3389/fonc.2019.01124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bianchini G., Balko J.M., Mayer I.A., Sanders M.E., Gianni L. Triple-negative breast cancer: challenges and opportunities of a heterogeneous disease. Nat. Rev. Clin. Oncol. 2016;13:674–690. doi: 10.1038/nrclinonc.2016.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hong Y.K., McMasters K.M., Egger M.E., Ajkay N. Ductal carcinoma in situ current trends, controversies, and review of literature. Am. J. Surg. 2018;216:998–1003. doi: 10.1016/j.amjsurg.2018.06.013. [DOI] [PubMed] [Google Scholar]
- 28.Wen H.Y., Brogi E. Lobular carcinoma in situ. Surg. Pathol. Clin. 2018;11:123–145. doi: 10.1016/j.path.2017.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mardekian S.K., Bombonati A., Palazzo J.P. Ductal carcinoma in situ of the breast: the importance of morphologic and molecular interactions. Hum. Pathol. 2016;49:114–123. doi: 10.1016/j.humpath.2015.11.003. [DOI] [PubMed] [Google Scholar]
- 30.Neve R.M., Chin K., Fridlyand J., Yeh J., Baehner F.L., Fevr T. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Canc. Cell. 2006;10:515–527. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marcotte R., Sayad A., Brown K.R., Sanchez-Garcia F., Reimand J., Haider M. Functional genomic landscape of human breast cancer drivers, vulnerabilities, and resistance. Cell. 2016;164:293–309. doi: 10.1016/j.cell.2015.11.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dai X., Cheng H., Bai Z., Li J. Breast cancer cell line classification and its relevance with breast tumor subtyping. J. Canc. 2017;8:3131–3141. doi: 10.7150/jca.18457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klijn C., Durinck S., Stawiski E.W., Haverty P.M., Jiang Z., Liu H. A comprehensive transcriptional portrait of human cancer cell lines. Nat. Biotechnol. 2015;33:306–312. doi: 10.1038/nbt.3080. [DOI] [PubMed] [Google Scholar]
- 34.Garnett M.J., Edelman E.J., Heidorn S.J., Greenman C.D., Dastur A., Lau K.W. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature. 2012;483:570–575. doi: 10.1038/nature11005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barretina J., Caponigro G., Stransky N., Venkatesan K., Margolin A.A., Kim S. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miano V., Ferrero G., Reineri S., Caizzi L., Annaratone L., Ricci L. Luminal long non-coding RNAs regulated by estrogen receptor alpha in a ligand-independent manner show functional roles in breast cancer. Oncotarget. 2016;7:3201–3216. doi: 10.18632/oncotarget.6420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tang Z., Kang B., Li C., Chen T., Zhang Z. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019;47:W556–W560. doi: 10.1093/nar/gkz430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koboldt D.C., Fulton R.S., McLellan M.D., Schmidt H., Kalicki-Veizer J., McMichael J.F. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miller F.R., Santner S.J., Tait L., Dawson P.J. MCF10DCIS.com xenograft model of human comedo ductal carcinoma in situ. JNCI J. Natl. Canc. Inst. 2000;92:1185a–1186. doi: 10.1093/jnci/92.14.1185a. [DOI] [PubMed] [Google Scholar]
- 40.Kao J., Salari K., Bocanegra M., Choi Y.-L., Girard L., Gandhi J. Molecular profiling of breast cancer cell lines defines relevant tumor models and provides a resource for cancer gene discovery. PloS One. 2009;4:e6146. doi: 10.1371/journal.pone.0006146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith S.E., Mellor P., Ward A.K., Kendall S., McDonald M., Vizeacoumar F.S. Molecular characterization of breast cancer cell lines through multiple omic approaches. Breast Cancer Res. 2017;19:1–12. doi: 10.1186/s13058-017-0855-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Anders S., Pyl P.T., Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kolde R. 2019. Pheatmap: Pretty Heatmaps; pp. 1–8. R package version 1.0.12. [Google Scholar]
- 46.Blighe, K; Rana, S; Lewis M. EnhancedVolcano: publication-ready volcano plots with enhanced colouring and labeling. R package version 1.4.0 2019.
- 47.Malhotra G.K., Zhao X. Band H, Band V. Histological, molecular and functional subtypes of breast cancers. Canc. Biol. Ther. 2010;10:955–960. doi: 10.4161/cbt.10.10.13879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Waks A.G., Winer E.P. Breast cancer treatment. J. Am. Med. Assoc. 2019;321:288. doi: 10.1001/jama.2018.19323. [DOI] [PubMed] [Google Scholar]
- 49.Zhao W., Luo J., Jiao S. Comprehensive characterization of cancer subtype associated long non-coding RNAs and their clinical implications. Sci. Rep. 2015;4:6591. doi: 10.1038/srep06591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sun W., Li A.-Q., Zhou P., Jiang Y.-Z., Jin X., Liu Y.-R. DSCAM-AS1 regulates the G 1/S cell cycle transition and is an independent prognostic factor of poor survival in luminal breast cancer patients treated with endocrine therapy. Canc. Med. 2018;7:6137–6146. doi: 10.1002/cam4.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Khorshidi H., Azari I., Oskooei V.K., Taheri M., Ghafouri-Fard S. DSCAM-AS1 up-regulation in invasive ductal carcinoma of breast and assessment of its potential as a diagnostic biomarker. Breast Dis. 2019;38:25–30. doi: 10.3233/BD-180351. [DOI] [PubMed] [Google Scholar]
- 52.Liang W.-H., Li N., Yuan Z.-Q., Qian X.-L., Wang Z.-H. DSCAM-AS1 promotes tumor growth of breast cancer by reducing miR-204-5p and up-regulating RRM2. Mol. Carcinog. 2019;58:461–473. doi: 10.1002/mc.22941. [DOI] [PubMed] [Google Scholar]
- 53.Polev D.E., Karnaukhova I.K., Krukovskaya L.L., Kozlov A.P. ELFN1-AS1: a novel primate gene with possible microRNA function expressed predominantly in human tumors. BioMed Res. Int. 2014;2014:398097. doi: 10.1155/2014/398097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li M., Liu Y., Zhang X., Liu J., Wang P. Transcriptomic analysis of high-throughput sequencing about circRNA, lncRNA and mRNA in bladder cancer. Gene. 2018 doi: 10.1016/j.gene.2018.07.041. [DOI] [PubMed] [Google Scholar]
- 55.Yuan S., Liu Q., Hu Z., Zhou Z., Wang G., Li C. Long non-coding RNA MUC5B-AS1 promotes metastasis through mutually regulating MUC5B expression in lung adenocarcinoma. Cell Death Dis. 2018;9:450. doi: 10.1038/s41419-018-0472-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vrba L., Garbe J.C., Stampfer M.R., Futscher B.W. A lincRNA connected to cell mortality and epigenetically-silenced in most common human cancers. Epigenetics. 2015;10:1074–1083. doi: 10.1080/15592294.2015.1106673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vrba L., Futscher B.W. Epigenetic silencing of MORT is an early event in cancer and is associated with luminal, receptor positive breast tumor subtypes. J. Breast Canc. 2017;20:198. doi: 10.4048/jbc.2017.20.2.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhao L.-P., Li R.-H., Han D.-M., Zhang X.-Q., Nian G.-X., Wu M.-X. Independent prognostic Factor of low-expressed LncRNA ZNF667-AS1 for cervical cancer and inhibitory function on the proliferation of cervical cancer. Eur. Rev. Med. Pharmacol. Sci. 2017;21:5353–5360. doi: 10.26355/eurrev_201712_13920. [DOI] [PubMed] [Google Scholar]
- 59.Meng W., Cui W., Zhao L., Chi W., Cao H., Wang B. Aberrant methylation and downregulation of ZNF667-AS1 and ZNF667 promote the malignant progression of laryngeal squamous cell carcinoma. J. Biomed. Sci. 2019;26:13. doi: 10.1186/s12929-019-0506-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li Y., Yang Z., Wang Y., Wang Y. Long noncoding RNA ZNF667‐AS1 reduces tumor invasion and metastasis in cervical cancer by counteracting microRNA‐93‐3p‐dependent PEG3 downregulation. Mol. Oncol. 2019;13:2375–2392. doi: 10.1002/1878-0261.12565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dong Z., Li S., Wu X., Niu Y., Liang X., Yang L. Aberrant hypermethylation-mediated downregulation of antisense lncRNA ZNF667-AS1 and its sense gene ZNF667 correlate with progression and prognosis of esophageal squamous cell carcinoma. Cell Death Dis. 2019;10:930. doi: 10.1038/s41419-019-2171-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nissan A., Stojadinovic A., Mitrani-Rosenbaum S., Halle D., Grinbaum R., Roistacher M. Colon cancer associated transcript-1: a novel RNA expressed in malignant and pre-malignant human tissues. Int. J. Canc. 2012;130:1598–1606. doi: 10.1002/ijc.26170. [DOI] [PubMed] [Google Scholar]
- 63.Alaiyan B., Ilyayev N., Stojadinovic A., Izadjoo M., Roistacher M., Pavlov V. Differential expression of colon cancer associated transcript1 (CCAT1) along the colonic adenoma-carcinoma sequence. BMC Canc. 2013;13:196. doi: 10.1186/1471-2407-13-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kam Y., Rubinstein A., Naik S., Djavsarov I., Halle D., Ariel I. Detection of a long non-coding RNA (CCAT1) in living cells and human adenocarcinoma of colon tissues using FIT-PNA molecular beacons. Canc. Lett. 2014;352:90–96. doi: 10.1016/j.canlet.2013.02.014. [DOI] [PubMed] [Google Scholar]
- 65.Ma M.-Z., Chu B.-F., Zhang Y., Weng M.-Z., Qin Y.-Y., Gong W. Long non-coding RNA CCAT1 promotes gallbladder cancer development via negative modulation of miRNA-218-5p. Cell Death Dis. 2015;6 doi: 10.1038/cddis.2014.541. e1583–e1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cabanski C.R., White N.M., Dang H.X., Silva-Fisher J.M., Rauck C.E., Cicka D. Pan-cancer transcriptome analysis reveals long noncoding RNAs with conserved function. RNA Biol. 2015;12:628–642. doi: 10.1080/15476286.2015.1038012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.McCleland M.L., Mesh K., Lorenzana E., Chopra V.S., Segal E., Watanabe C. CCAT1 is an enhancer-templated RNA that predicts BET sensitivity in colorectal cancer. J. Clin. Invest. 2016;126:639–652. doi: 10.1172/JCI83265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jiang Y., Jiang Y.Y., Xie J.J., Mayakonda A., Hazawa M., Chen L. Co-activation of super-enhancer-driven CCAT1 by TP63 and SOX2 promotes squamous cancer progression. Nat. Commun. 2018;9 doi: 10.1038/s41467-018-06081-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Han C., Li X., Fan Q., Liu G., Yin J. CCAT1 promotes triple-negative breast cancer progression by suppressing miR-218/ZFX signaling. Aging. 2019;11:4858–4875. doi: 10.18632/aging.102080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kalmár A., Nagy Z.B., Galamb O., Csabai I., Bodor A., Wichmann B. Genome-wide expression profiling in colorectal cancer focusing on lncRNAs in the adenoma-carcinoma transition. BMC Canc. 2019;19:1059. doi: 10.1186/s12885-019-6180-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.El-Khazragy N., Elayat W., Matbouly S., Seliman S., Sami A., Safwat G. The prognostic significance of the long non-coding RNAs CCAT1, PVT1 in t(8;21) associated Acute Myeloid Leukemia. Gene. 2019;707:172–177. doi: 10.1016/j.gene.2019.03.055. [DOI] [PubMed] [Google Scholar]
- 72.Qi H., Li C., Qian C., Xiao Y., Yuan Y., Liu Q. The long noncoding RNA, EGFR-AS1, a target of GHR, increases the expression of EGFR in hepatocellular carcinoma. Tumor Biol. 2016;37:1079–1089. doi: 10.1007/s13277-015-3887-z. [DOI] [PubMed] [Google Scholar]
- 73.Tan D.S.W., Chong F.T., Leong H.S., Toh S.Y., Lau D.P., Kwang X.L. Long noncoding RNA EGFR-AS1 mediates epidermal growth factor receptor addiction and modulates treatment response in squamous cell carcinoma. Nat. Med. 2017;23:1167–1175. doi: 10.1038/nm.4401. [DOI] [PubMed] [Google Scholar]
- 74.Hu J., Qian Y., Peng L., Ma L., Qiu T., Liu Y. Long noncoding RNA EGFR-AS1 promotes cell proliferation by increasing EGFR mRNA stability in gastric cancer. Cell. Physiol. Biochem. 2018;49:322–334. doi: 10.1159/000492883. [DOI] [PubMed] [Google Scholar]
- 75.Xu Y.-H., Tu J.-R., Zhao T.-T., Xie S.-G., Tang S.-B. Overexpression of lncRNA EGFR-AS1 is associated with a poor prognosis and promotes chemotherapy resistance in non-small cell lung cancer. Int. J. Oncol. 2018;54:295–305. doi: 10.3892/ijo.2018.4629. [DOI] [PubMed] [Google Scholar]
- 76.Crouch S., Spidel C.S., Lindsey J.S. HGF and ligation of αvβ5 integrin induce a novel, cancer cell-specific gene expression required for cell scattering. Exp. Cell Res. 2004;292:274–287. doi: 10.1016/j.yexcr.2003.09.016. [DOI] [PubMed] [Google Scholar]
- 77.Phillips T.M., Lindsey J.S. Carcinoma cell-specific Mig-7: a new potential marker for circulating and migrating cancer cells. Oncol. Rep. 2005;13:37–44. [PubMed] [Google Scholar]
- 78.Ren K., Yao N., Wang G., Tian L., Ma J., Shi X. Vasculogenic mimicry: a new prognostic sign of human osteosarcoma. Hum. Pathol. 2014;45:2120–2129. doi: 10.1016/j.humpath.2014.06.013. [DOI] [PubMed] [Google Scholar]
- 79.Huang B., Yin M., Li X., Cao G., Qi J., Lou G. Migration-inducing gene 7 promotes tumorigenesis and angiogenesis and independently predicts poor prognosis of epithelial ovarian cancer. Oncotarget. 2016;7:27552–27566. doi: 10.18632/oncotarget.8487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Qu B., Sheng G., Guo L., Yu F., Chen G., Lu Q. MIG7 is involved in vasculogenic mimicry formation rendering invasion and metastasis in hepatocellular carcinoma. Oncol. Rep. 2017;39:679–686. doi: 10.3892/or.2017.6138. [DOI] [PubMed] [Google Scholar]
- 81.Prat A., Pineda E., Adamo B., Galván P., Fernández A., Gaba L. Clinical implications of the intrinsic molecular subtypes of breast cancer. Breast. 2015;24:S26–S35. doi: 10.1016/j.breast.2015.07.008. [DOI] [PubMed] [Google Scholar]
- 82.Van Dijk E.L., Jaszczyszyn Y., Thermes C. Library preparation methods for next-generation sequencing: tone down the bias. Exp. Cell Res. 2014 doi: 10.1016/j.yexcr.2014.01.008. [DOI] [PubMed] [Google Scholar]
- 83.Zhao S., Zhang Y., Gamini R., Zhang B., von Schack D. Evaluation of two main RNA-seq approaches for gene quantification in clinical RNA sequencing: polyA+ selection versus rRNA depletion. Sci. Rep. 2018;8:4781. doi: 10.1038/s41598-018-23226-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Iacoangeli A., Lin Y., Morley E.J., Muslimov I.A., Bianchi R., Reilly J. BC200 RNA in invasive and preinvasive breast cancer. Carcinogenesis. 2004;25:2125–2133. doi: 10.1093/carcin/bgh228. [DOI] [PubMed] [Google Scholar]
- 85.Samson J., Cronin S., Dean K. BC200 (BCYRN1) – the shortest, long, non-coding RNA associated with cancer. Non-Coding RNA Res. 2018 doi: 10.1016/j.ncrna.2018.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Martignetti J.A., Brosius J. BC200 RNA : a neural RNA polymerase III product encoded by a monomeric alu element. Proc. Natl. Acad. Sci. Unit. States Am. 1993;90:11563–11567. doi: 10.1073/pnas.90.24.11563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Brock E.J., Ji K., Shah S., Mattingly R.R., Sloane B.F. In vitro models for studying invasive transitions of ductal carcinoma in situ. J. Mammary Gland Biol. Neoplasia. 2019;24:1–15. doi: 10.1007/s10911-018-9405-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ozawa T., Matsuyama T., Toiyama Y., Takahashi N., Ishikawa T., Uetake H. CCAT1 and CCAT2 long noncoding RNAs, located within the 8q.24.21 ‘gene desert’, serve as important prognostic biomarkers in colorectal cancer. Ann. Oncol. 2017;28:1882–1888. doi: 10.1093/annonc/mdx248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhu H., Zhou X., Chang H., Li H., Liu F., Ma C. CCAT1 promotes hepatocellular carcinoma cell proliferation and invasion. Int. J. Clin. Exp. Pathol. 2015;8:5427–5434. [PMC free article] [PubMed] [Google Scholar]
- 90.Ghandi M., Huang F.W., Jané-Valbuena J., Kryukov G.V., Lo C.C., McDonald E.R. Next-generation characterization of the cancer cell line Encyclopedia. Nature. 2019;569:503–508. doi: 10.1038/s41586-019-1186-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.