Abstract
Chimeric antigen receptor (CAR) T cells are considered genetically modified organisms (GMOs) and constitute gene therapy medicinal products. Thus, CAR T cell manufacturing for clinical application is strictly regulated. Appropriate methods to assess vector copy numbers (VCNs) in CAR T cell products and monitoring of CAR T cell frequencies in patients are required. Quantitative polymerase chain reaction (qPCR) is the preferred method for VCN assessment. However, no standardized procedure with high reproducibility has been described yet. Here, we report on a single copy gene (SCG)-based duplex (DP)-qPCR assay (SCG-DP-PCR) to determine VCN in CAR T cell products. SCG-DP-PCR was validated and compared to the absolute standard curve method (ACM) within the framework of a clinical trial treating patients with good manufacturing practice (GMP)-grade CAR T cells at the University Hospital Heidelberg. Methodologically, SCG-DP-PCR displayed technical advantages over ACM and minimized mathematical analysis. SCG-DP-PCR, as a highly reproducible approach, can be used for clinical follow-up of patients treated with CAR T cells or other GMOs and might replace established methods for VCN quantification. This work will enable clinicians to assess VCN, as well as CAR T cell frequencies, in patients as a basis for decisions on subsequent therapies, including repeated CAR T cell administration.
Keywords: vector copy number, single-copy gene, CAR T cells, qPCR, CAR T cell monitoring
Introduction
Chimeric antigen receptor (CAR) T cells constitute a highly promising adoptive immunotherapy for cancer. CAR T cells directed against CD19 have shown remarkable clinical results in heavily pretreated patients with relapsed or refractory lymphoid malignancies, including pediatric1,2 and adult3,4 acute lymphoblastic leukemia (ALL), chronic lymphocytic leukemia (CLL),5,6 and other non-Hodgkin’s lymphoma (NHL).7, 8, 9, 10 Development of CARs for treatment of other hematologic malignancies or solid tumors is ongoing.11 Anti-CD19 CAR T cells have been approved by both the US Food and Drug Administration (FDA) and the European Medicines Agency (EMA). The University Hospital Heidelberg initiated the first investigator-initiated trial (IIT) for CAR T cell therapy in Germany. The Heidelberg Chimeric Antigen Receptor T Cell Trial Number 1 (HD-CAR-1; European Union Drug Regulating Authorities Clinical Trials Database [EudraCT] no. 2016-004808-60; ClinicalTrials.gov: NCT03676504) is a monocentric, open-label, prospective phase I/II trial initiated in September 2018 with in-house leukapheresis as well as CAR T cell manufacturing for treating adult and pediatric ALL, adult CLL, and NHL patients with autologous T lymphocytes transduced with a third-generation CAR vector (RV-SFG.CD19.CD28.4-1BBzeta) targeting CD19.12 Due to the fact that CAR T cells are considered genetically modified organisms (GMOs), they constitute gene therapy medicinal products (GTMPs). Hence, CAR T cells are manufactured according to good manufacturing practice (GMP) standards. Regulatory authorities require extensive safety evaluation of advanced therapy medicinal products (ATMPs), i.e., cellular immune therapies, such as CAR T cells. To warrant safety, transgene copies within a CAR T cell product, i.e., vector copy numbers (VCNs), have to be assessed prior to patient administration. Additionally, response to CAR T cell treatment is associated with expansion,6 as well as persistence of CAR T cells in treated patients.10,13 Therefore, the assessment of CAR T cell levels in patients at different time points after CAR T cell administration is crucial for deciding on further patient therapies. CAR VCN assessment in CAR T cell products and monitoring CAR T cells in the peripheral blood (PB) of treated patients are usually performed via quantitative PCR (qPCR).14, 15, 16, 17. Here, we propose a duplex (DP) qPCR strategy based on a single-copy gene (SCG; SCG-DP-PCR), i.e. ribonuclease (RNase)P RNA component H1 gene (RPPH1; RNaseP in the following) (single copy per haploid human genome), for accurate and robust determination of VCN in CAR T cell products and in PB samples of treated patients. SCG-DP-PCR was compared to the absolute standard curve singleplex (SP) qPCR approach (absolute standard curve method [ACM]) and validated within the framework of HD-CAR-1.
Results
Method validation and evaluation of SCG-DP-PCR were performed as follows: efficiencies and correlation coefficients (R2) of PCR reactions within the duplex PCR (targeting the CAR transgene and SCG) were compared before similar efficiencies over the relevant transgene copy number range were confirmed. A final proof of similar efficiencies was achieved by the use of an efficiency control (EC) sample (description in Materials and Methods) as a direct testing method. Following this method validation, VCNs in healthy donor samples were assessed via ACM and SCG-DP-PCR, and both methods were directly compared. Subsequently, SCG-DP-PCR was applied on follow-up samples of patients treated with HD-CAR-1 CAR T cells. Differences and influencing factors of ACM and SCG-DP-PCR were determined.
Efficiencies and Linearity of PCR Reactions (ACM and SCG-DP-PCR)
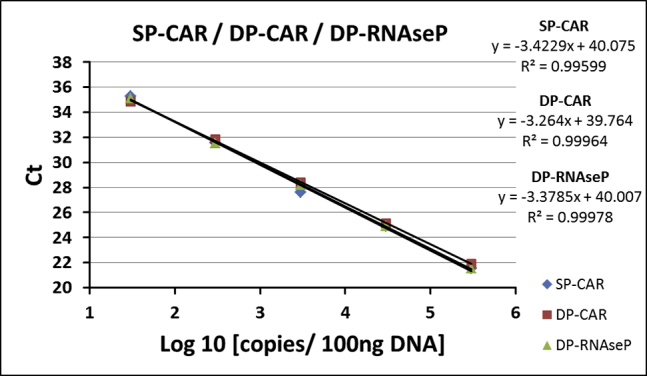
For method validation, efficiency and linearity (correlation coefficient) of PCR reactions of ACM (SP-CAR) and SCG-DP-PCR (DP-CAR, DP-RNaseP) were assessed by linear regression analysis of standards. Standard curves were only considered valid if a R2 of above 0.98 and mean efficiencies of 100% ± 10% were achieved. SP-CAR PCR reaction displayed an efficiency of 103.5 ± 7.1%; efficiencies of 104.2 ± 2.1% and 99.3 ± 1.6% were achieved for DP-CAR and DP-RNaseP PCR reactions, respectively (Table 1).
Table 1.
Comparison of Relevant Parameters of qPCR Reactions (Efficiency, Linearity) in the Singleplex Setup (SP-CAR) of ACM and the SCG-DP-PCR Duplex Setup (DP-CAR, DP-RNaseP)
| PCR Reaction | Efficiency | Correlation Coefficient (R2) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| ACM | SP-CAR | 103.5% ± 7.1% | 0.99 | ||||||
| SCG-DP-PCR | DP-CAR | 104.2% ± 2.1% | 0.99 | ||||||
| DP-RNaseP | 99.3% ± 1.6% | 0.99 | |||||||
| Copies/Std. | Ct (SP-CAR) | Ct (DP-CAR) | Ct (DP-RNaseP) | ||||||
| Val. 1 | Val. 2 | Val. 3 | Val. 1 | Val. 2 | Val. 3 | Val. 1 | Val. 2 | Val. 3 | |
| 3 × 105 | 20.9 ± 0.1 | 21.4 ± 0.1 | 21.5 ± 0.1 | 21.9 ± 0.1 | 21.9 ± 0.0 | 21.9 ± 0.1 | 21.5 ± 0.0 | 21.3 ± 0.0 | 21.5 ± 0.1 |
| 3 × 104 | 24.2 ± 0.1 | 24.2 ± 0.1 | 24.9 ± 0.2 | 25.1 ± 0.0 | 25.1 ± 0.0 | 25.1 ± 0.1 | 24.8 ± 0.0 | 24.6 ± 0.0 | 24.9 ± 0.0 |
| 3 × 103 | 27.5 ± 0.1 | 27.6 ± 0.1 | 27.6 ± 0.0 | 28.4 ± 0.0 | 28.4 ± 0.0 | 28.4 ± 0.0 | 28.1 ± 0.1 | 28.0 ± 0.0 | 28.2 ± 0.1 |
| 3 × 102 | 30.9 ± 0.0 | 31.1 ± 0.1 | 31.6 ± 0.3 | 31.8 ± 0.2 | 31.9 ± 0.2 | 31.8 ± 0.1 | 31.5 ± 0.1 | 31.3 ± 0.1 | 31.5 ± 0.1 |
| 30 | 33.1 ± 0.1 | 34.0 ± 0.3 | 35.3 ± 0.6 | 34.4 ± 0.6 | 34.7 ± 0.2 | 34.8 ± 0.2 | 34.9 ± 0.6 | 34.5 ± 0.0 | 35.1 ± 0.5 |
qPCR data from standards of three independent experiments were analyzed by linear regression (validation [Val.] 1, 2, and 3). Efficiencies of three experiments are represented as mean ± SD. Reactions were performed in triplicates. Ct values are represented as mean ± SD. Std., standard.
Standard curves were generated for SP-CAR, DP-CAR, and DP-RNaseP in three validation experiments. Figure 1 illustrates the results obtained in one of three validation experiments.
Figure 1.
Efficiencies and Linearity of PCR Reactions (ACM and SCG-DP-PCR)
Standard curves of qPCR reactions. Exemplary data from one validation experiment are shown. Mean Ct values from qPCR were used for linear regression. Reactions were performed in triplicates. Ct, threshold cycle.
Relative Efficiency Plot of SCG-DP-PCR
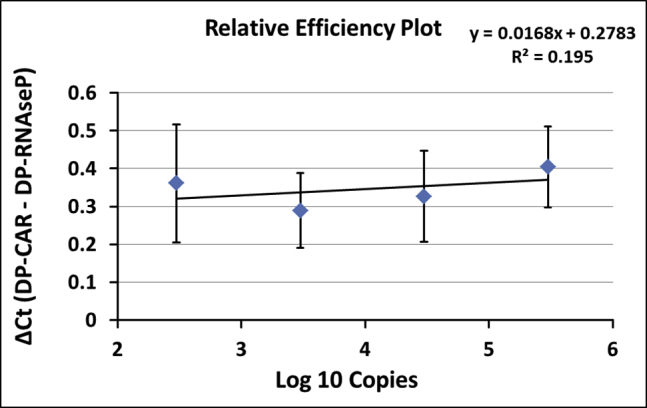
The relative efficiency plot compared simultaneous PCR reactions over the tested transgene copy number range by calculation of comparative threshold cycle (ΔCt; DP-CAR − DP-RNaseP) and graphical analysis (semi-logarithmic display; transgene copies [log10]: x axis; ΔCt: y axis). Figure 2 displays generated relative efficiency plots. ΔCt (DP-CAR − DP-RNaseP) in the DP-PCR standards was similar for the 4 higher copy standards (3 × 102 − 3 × 105 copies/reaction). The smallest standard (30 copies/reaction) was excluded from analysis due to high standard deviation in replicates. Consequently, diagnostic measuring range for this method setup was defined within 3 × 102 − 3 × 105 copies/reaction. In this range, the slope of the regression curve was 0.0168.
Figure 2.
Relative Efficiency Plot of SCG-DP-PCR
Graphical analysis of relative efficiencies from 4 higher duplex standards (3 × 102 – 3 × 105 copies) of three independent experiments (validation 1, 2, and 3). Results are represented as mean ± standard deviation (SD). Reactions were performed in triplicates. Smallest duplex standard, i.e., 30 copies, was excluded from analysis due to high SD.
Efficiency Control Sample
The EC sample was added to every duplex PCR experiment. The calculated ΔCt values (DP-CAR − DP-RNaseP) in this control sample ranged between Ct ≥ −0.31 and Ct ≤ 0.17. The application of 2−ΔCt (DP-CAR − DP-RNaseP), acceptable copy numbers of the CAR transgene relative to RNaseP gene of 0.89, 1.06, and 1.24 (1.06 ± 0.18) was achieved (Table 2). The amplification plot of the EC sample from one validation experiment (Figure S1; Supplemental Materials and Methods) illustrates similarity of the CAR transgene and RNaseP gene amplifications in the SCG-DP-PCR setup.
Table 2.
Analysis of EC Samples from Three Validation Experiments (Validation 1, 2, and 3)
| Validation 1 | Validation 2 | Validation 3 | |
|---|---|---|---|
| Ct (DP-CAR) | 24.74 ± 0.08 | 24.25 ± 0.01 | 25.19 ± 0.04 |
| Ct (DP-RNaseP) | 24.83 ± 0.04 | 24.56 ± 0.04 | 25.02 ± 0.09 |
| ΔCt | −0.09 | −0.31 | 0.17 |
| SCG-DP-PCR 2−ΔCt method | 1.06 | 1.24 | 0.89 |
Reactions were performed in triplicates. Ct values are represented as mean ± SD. Mean Ct values from qPCR were used for ΔCt calculation.
Vector Copy Numbers
VCNs were obtained by mathematical extrapolation of regression curves to sample signals via ACM and by relative SCG-DP-PCR applied. The application of SCG-DP-PCR on CAR T cell samples generated from healthy donors, an increase of 0.8 ± 0.2 VCN/cell, was observed when compared to ACM (Table 3).
Table 3.
Validation and Comparison of the VCN Determination by Different Strategies ACM and SCG-DP-PCR
| Validation 1 | Validation 2 | Validation 3 | |
|---|---|---|---|
| Ct (DP-CAR) | 24.77 ± 0.03 | 24.28 ± 0.03 | 25.31 ± 0.02 |
| Ct (DP-RNaseP) | 24.93 ± 0.05 | 24.80 ± 0.01 | 25.50 ± 0.02 |
| ΔCt | −0.16 | −0.52 | −0.19 |
| VCN/cell (SCG-DP-PCR) | 2.2 | 2.9 | 2.3 |
| VCN/cell (ACM) | 1.3 | 2.3 | 1.5 |
| Copies/Std. (ACM) | Ct (Validation 1) | Ct (Validation 2) | Ct (Validation 3) |
| 3 × 105 | 20.9 ± 0.1 | 21.4 ± 0.1 | 21.5 ± 0.1 |
| 3 × 104 | 24.2 ± 0.1 | 24.2 ± 0.1 | 24.9 ± 0.2 |
| 3 × 103 | 27.5 ± 0.1 | 27.6 ± 0.1 | 27.6 ± 0.0 |
| 3 × 102 | 30.9 ± 0.0 | 31.1 ± 0.1 | 31.6 ± 0.3 |
| 30 | 33.1 ± 0.1 | 34.0 ± 0.3 | 35.3 ± 0.6 |
Three independent experiments were performed (validation 1, 2, and 3). Mean Ct values from qPCR were used for linear regression (ACM) and ΔCt calculation (SCG-DP-PCR). Reactions were performed in triplicates. Ct values are represented as mean ± SD.
CAR T Cell Monitoring in Patients Using a Validated SCG-DP-PCR
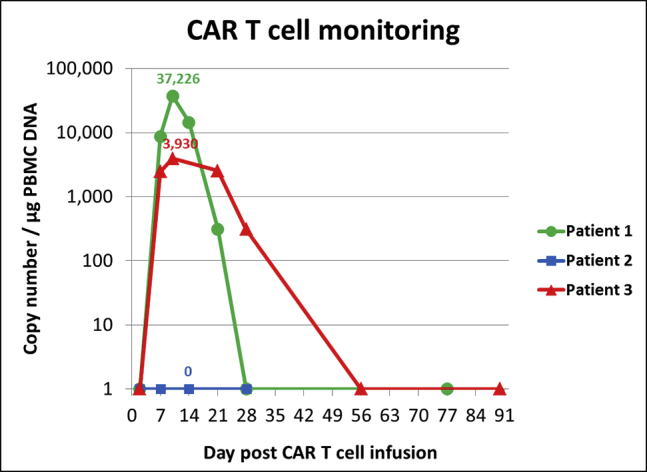
Following validation on healthy donor-derived CAR T cells, SCG-DP-PCR was also used to assess CAR T cell expansion in HD-CAR-1 patient samples. Figure 3 displays CAR T cell expansion within PB samples of 3 HD-CAR-1 patients at different time points after CAR T cell administration.
Figure 3.
CAR T Cell Monitoring in Patients Using a Validated SCG-DP-PCR
CAR T cell monitoring in peripheral blood (PB) samples of three different patients, assessed by validated SCG-DP-PCR. Patient 1 was assessed by absolute standard curve method (ACM) before SCG-DP-PCR was established in our GMP facility for all further quantification experiments. Patients were treated with a dose of 1 × 106 CD19+CAR+-transduced T cells per square meter body surface at day 0. Different kinetics of CAR T cells were observed. Determined peak copy numbers are included into the graph above peak data points. No CAR T cells were detected in samples of patient 2. The samples were not measured by other validated methods.
Discussion
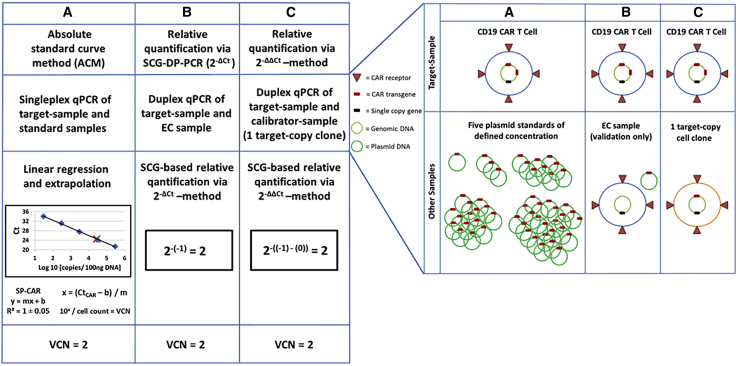
CAR-expressing T cells targeting CD19 are currently revolutionizing the treatment of patients with hematologic malignancies and are becoming an integral part of the clinical hematological practice.18 CAR-transduced T cells in clinical practice are mostly generated by the use of viral vectors. These vectors originate from the Orthoretrovirinae subfamily of Retroviridae, i.e., γ-retrovirus and lentivirus. They convert their RNA genome into cDNA and integrate this genetic information into the genome of the target cell, thus enabling long-term transgene expression. However, the use of viral vectors for CAR T cell manufacturing for therapeutic purposes requires strict biosafety testing. The exclusion of the presence of replication-competent retrovirus (RCR) within CAR T cell samples, as well as evaluation of the average number of integrated vector copies per transduced T cell, is mandatory. A variety of different strategies for VCN determination have been used, relying mainly on ACM,14 as well as relative quantification approaches.15, 16, 17. ACM is associated with potential inaccuracies, due to the need of standard curves. Relative quantification of VCN displays practical and technical issues. The 2−ΔΔCt method for relative quantification was described by Livak and Schmittgen19 and was originally used for qPCR analysis of fold changes in gene expression. It has been also applied to quantify transgene VCNs in CAR T cell samples.16,17 In addition to the general preconditions of approximately similar and optimal efficiencies of 100% for the target and the reference qPCR reactions, VCN quantification by the use of the 2−ΔΔCt method requires special conditions, i.e., a SCG as a reference gene and a special calibrator sample. Especially, the generation of a calibrator sample consisting of a CAR T cell clone with a defined and stable VCN of the CAR transgene constitutes a major issue in terms of practicability and feasibility. Hence, a quantitative approach that operates independently of impractical factors, such as standard curves or calibrator samples, additionally economizing material and time, is a highly desirable tool for clinical CAR T cell research. We developed and validated an adapted SCG-based quantification approach (SCG-DP-PCR) to address these issues. SCG-DP-PCR VCN assessment applies the 2−ΔCt method that is derived after implementation of the following assumption to the 2−ΔΔCt method:19 similar and optimal efficiencies of 100% for the target and the reference qPCR reactions result in similar Ct values of the EC sample (for mathematical deduction of the 2−ΔCt method see Supplemental Materials and Methods A2). Figure 4, left, summarizes ACM, SCG-DP-PCR (2−ΔCt), and the established 2−ΔΔCt19 methods. Corresponding required samples for VCN determination are depicted in Figure 4, right.
Figure 4.
Summary of ACM, SCG-DP-PCR (2−ΔCt), and the Established 2−ΔΔCt19 Methods
(Left) Schematic illustration of VCN determination with anti-CD19 CAR T cells harboring 2 CAR transgene copies under assumption of optimal PCR conditions. The figures and calculations refer to a haploid human genome to decrease complexity. Different qPCR strategies are illustrated. (A) Absolute standard curve method (ACM). (B) Relative quantification via the 2−ΔCt method (SCG-DP-PCR). (C) Relative quantification via the 2−ΔΔCt method. SCG, single copy gene; VCN, vector copy number; qPCR, quantitative polymerase chain reaction; Ct, threshold cycle; DP, duplex; EC, efficiency control. (Right) Corresponding required samples for VCN determination by different qPCR methods. Schematic illustration of samples required for different qPCR-strategies. (A) Absolute quantification via the standard curve method. (B) Relative quantification via the 2−ΔCt method. (C) Relative quantification via the 2−ΔΔCt method.
The validation of SCG-DP-PCR was performed by testing the following: (1) the efficiency and linearity of PCR-reactions, (2) the constancy of PCR efficiencies within the relevant transgene copy range, and (3) the similarity of PCR efficiencies, as well as RNaseP SCG status, by using an EC sample and applying the 2−ΔCt method.
SCG-DP-PCR reactions displayed similar efficiencies close to 100% and almost optimal linearities (R2 > 0.98) (Table 1; Figure 1). Constant PCR efficiencies over the relevant concentration range, i.e., similar ΔCt (DP-CAR − DP-RNaseP) in every duplex standard sample, were confirmed by a relative efficiency plot. Here, log10 copies (x axis) were plotted against ΔCt (DP-CAR − DP-RNaseP) (y axis). Via linear regression, a slope of <0.1 (optimal 0) was obtained, proving constancy (Figure 2). Efficiency validation was completed, verifying similar amplification efficiencies of DP-CAR and DP-RNaseP PCR reactions and confirming the SCG status of RNaseP using the EC sample. CAR transgene copies relative to the RNaseP gene of 0.89, 1.06, and 1.24 (1.06 ± 0.18) for the EC sample were achieved, lying within our accepted range (Table 2; Figure S1). Consequently, we established a relative qPCR approach that is independent from a calibrator sample. RNaseP could be detected in all samples we assessed. In the unlikely case of a missing RNaseP signal, a repetition of the PCR reaction is highly advised.
After validation, SCG-DP-PCR was compared to ACM by measuring VCN of CAR T cells generated from healthy donors (Table 3). For SCG-DP-PCR, a higher VCN 0.8 ± 0.2/cell was assessed when compared to ACM. We assume that via ACM VCN might be under-represented due to the influence of differing reaction conditions within an experimental setup with standard samples (standard curve; no genomic DNA) and GMO samples (target sample; genomic DNA). Additional factors, such as well-to-well variations or errors in DNA concentration measurement, influence ACM results. Moreover, sequential dilution of standards, as well as mathematical extrapolation, affects ACM and might contribute to observed VCN variations. However, underlying reasons for VCN discrepancy of ACM and SCG-DP-PCR were not analyzed any further. In SCG-DP-PCR, reactions are performed within one well. Hence, a main technical requirement for SCG-DP-PCR is the use of highly efficient and compatible primer and probe sets targeting the CAR transgene and the SCG. Methodical differences and influencing factors on ACM and SCG-DP-PCR are summarized in Table 4. SCG-DP-PCR was subsequently applied on HD-CAR-1 patient samples achieving clinical relevant data on CAR T cell expansion (Figure 3). No CAR T cells could be detected in samples of peripheral blood in patient 2. Given that the patient displayed a partial response to treatment, CAR T cells might have accumulated at the cancer cell site without circulating into the peripheral blood.
Table 4.
Summary of Differences/Influencing Factors of Experimentally Compared Two Strategies ACM and SCG-DP-PCR
| Factors | Absolute Standard Curve Method (ACM) | SCG-DP-PCR |
|---|---|---|
| Experiment | multiple wells (single PCR reaction/well) | single well (duplex PCR reactions) |
| standard curve | no standard curve required | |
| no genomic DNA within PCR reaction | genomic DNA within PCR reactions | |
| no internal control | RNaseP signal as internal control | |
| extensive experimental setup | fast experimental setup | |
| VCN analysis | extensive: linear regression | fast: relative calculation |
| mathematical extrapolation | 2−ΔCt (DP-Car − DP-RNaseP) | |
| Method validation | standard curve validation | extensive efficiency validation |
Overall, validated SCG-DP-PCR represents a less error-prone method to address the regulatory safety release criterion VCN in CAR T cell products compared to ACM. It is highly suitable to follow up CAR T cells in the peripheral blood of patients. Additionally, RNaseP represents an internal control for every PCR reaction of patient samples. Our main aim, to develop a suitable approach for standardization of VCN assessment in the field of clinical CAR therapy, was achieved. Importantly, the lack of standardized detection methods for the monitoring of CAR T cells or other GMOs in patients post-treatment could be overcome by SCG-DP-PCR. Moreover, given the lack of a calibrator sample, this relative quantification approach can be easily transferred and established in other laboratories. Subsequently, the important ability to monitor the expansion of CAR T cells or other GMOs in patients could be extended to many hospitals. This might improve the assessment of the course of diseases of patients in the field of gene therapy, particularly CAR T cell therapy.
Materials and Methods
Manufacturing of CAR T Cells
Clinical-grade CAR T cells were produced in the GMP Core Facility of our institution from healthy donors, as well as from patients enrolled in the HD-CAR-1 trial. Standardized CAR T cell manufacturing was established and validated before initiating HD-CAR-1.12 Informed consent was obtained from all healthy donors and HD-CAR-1 patients, according to the Declaration of Helsinki. Approval from the Ethics Committee of the University Heidelberg (AFmu-405/2017), the Paul-Ehrlich-Institut (PEI) competent authority (EudraCT no. 2016-004808-60), and the responsible regional authority (federal authority no. 3148/02) was granted in October 2017, September 2018, and August 2018, respectively. The first HD-CAR-1 patient was included in September 2018 and dosed in October 2018. Currently, 14 patients have been screened for treatment, and 12 patients have been enclosed.
In brief, healthy donors and patients following enrollment underwent unstimulated leukapheresis for collection of peripheral blood mononuclear cells (PBMCs). PBMCs were transduced with the RV-SFG.CD19.CD28.4-1BBzeta retroviral vector (kindly provided by Professor Malcolm Brenner, Baylor College of Medicine, Houston, Texas) at our GMP Core Facility after activation with anti-CD3 and anti-CD28 antibodies (MACS GMP Pure; Miltenyi Biotec, Bergisch Gladbach, Germany) and culturing with interleukin (IL)-7 (10 ng/mL) (R&D System, Minneapolis, USA) and IL-15 (5 ng/mL) (Cellgenix, Freiburg, Germany). RV-SFG.CD19.CD28.4-1BBzeta carries an anti-CD19 single-chain variable fragment (scFv) derived from the FMC63 antibody inserted within the SFG retroviral backbone. The transmembrane domain is derived from CD28, the hinge domain from the human immunoglobulin G1 (IgG1)-CH2CH3 domain. 4-1BB is inserted between the CD28 and the CD3ζ domain. CAR T cells derived from healthy donors were collected and directly analyzed for VCN validation experiments. HD-CAR-1 patients were treated with escalating CAR T cell doses (1–20 × 106-transduced cells/m2 body surface area [BSA]) after lymphodepletion with fludarabine (30 mg/ m2 BSA) and cyclophosphamide (500 mg/m2 BSA) for 3 days. PB samples from HD-CAR-1 patients were collected for analysis on days 1, 2, 3, 7, 14, 21, 28, 56, and 90 after CAR T cell administration.
Isolation of PBMCs
PBMCs containing CAR T cells from healthy donors and HD-CAR-1 patient PB samples were isolated by Ficoll density gradient (Linaris, Dossenheim, Germany) and genomic DNA extracted (cat. #51104, QIAamp DNA Blood Mini; QIAGEN). DNA concentration was measured by UV spectroscopy (NanoDrop OneC; Thermo Fisher Scientific, Applied Biosystems). Samples were diluted to a final concentration of 20 ng/μL in nuclease-free H2O.
qPCR Methods
SCG-DP-PCR and ACM qPCR were performed on genomic DNA derived from PBMCs. To determine the number of integrated CD19.CD28.4-1BBzeta copies in CAR T cells, i.e., VCN, 100 ng genomic DNA isolated from patients and healthy-donor PBMCs was amplified using the StepOnePlus real-time PCR system (Applied Biosystems). Whereas ACM was performed as the SP PCR targeting only the CAR transgene (SP-CAR), SCG-DP-PCR amplifies the CAR transgene (DP-CAR) and the SCG RNaseP (DP-RNaseP) simultaneously.
Thermal cycling for all PCR experiments was performed using the following amplification conditions: 2 min for 50°C, 10 min for 95°C, followed by 40 cycles of 15 s 95°C and 1 min 60°C. Primers, probes, and TaqMan Gene Expression Master Mix were purchased from Applied Biosystems. Detailed parameters of the qPCR experiments are comprised within Supplemental Materials and Methods B). Nontemplate control (NTC), biological negative control (nontransduced donor cells), and the RV-SFG.CD19.CD28.4-1BBzeta plasmid, as positive control, were included within all experiments.
Absolute Standard Curve Method
ACM was performed using conventional plasmid standard curves generated via serial dilution of RV-SFG.CD19.CD28.4-1BBzeta plasmid DNA in nuclease-free H2O (30, 3 × 102, 3 × 103, 3 × 104, 3 × 105 plasmid copies per PCR reaction). The following primers and probe were used for the SP-CAR qPCR: CAR transgene forward primer (FP): 5′-AGCTGCCGATTTCCAGAAGA-3′, reverse primer (RP): 5′-GCGCTCCTGCTGAACTTCA-3′, and probe: FAM-5′-AAGGAGGATGTGAACTGAGA-3′-MGB/NFQ. FP binds within the 4-1BB sequence, RP within the CD3ζ sequence, and the probe in the transition between 4-1BB and CD3ζ.
SCG-DP-PCR
SCG-DP-PCR was established and validated on CAR T cells generated from 3 healthy donors within the framework of the approval of the HD-CAR-1 trial for the regulatory authorities.
For SCG-DP-PCR validation, genomic DNA was added to RV-SFG.CD19.CD28.4-1BBzeta plasmid DNA in a 1:1 ratio of copies. Via serial dilution, a duplex standard curve (30, 3 × 102, 3 × 103, 3 × 104, 3 × 105 copies per PCR reaction) was generated to target the CAR transgene, as well as the RNaseP gene.
For SCG-DP-PCR VCN calculation, the 2−ΔCt method, based on the previously described 2−ΔΔCt method,19 was used. The mathematical evaluation of experimentally generated qPCR data applying SCG-DP-PCR using the 2−ΔCt method or via ACM is described in detail within the Supplemental Materials and Methods A.
The following primer sets were used for SCG-DP-PCR reactions:
-
(1)
Sequences of forward, reverse primer, and probe targeting the CAR transgene were used, as described before for the SP-CAR qPCR (ACM).
-
(2)
RNaseP: Copy Number Reference Assay, RNaseP (cat. #4403326, TaqMan; Applied Biosystems), containing RNaseP gene-specific forward primer, reverse primer, and probe (VIC/TAMRA) within a reaction mix.
Besides NTC, a biological negative control, as well as the RV-SFG.CD19.CD28.4-1BBzeta plasmid as a positive control, an EC sample was included within each experiment for SCG-DP-PCR validation to verify similar amplification efficiencies of CAR transgene and RNaseP. The EC sample consisted of genomic DNA from a nontransduced cell sample (comprising RNaseP) combined with the RV-SFG.CD19.CD28.4-1BBzeta plasmid in a 1:1 ratio. Calculations for EC sample generation and preparation are described within the Supplemental Materials and Methods. Besides testing similar PCR efficiencies, the EC sample verifies the SCG status of RNaseP in genomic DNA when a VCN of 1 is achieved.
The accepted range for ΔCt (DP-CAR − DP-RNaseP) in the EC sample was defined per calculation between ΔCt ≥ −0.4 and ΔCt ≤ 0.56. Application of 2−ΔCt (DP-CAR − DP-RNaseP) results in an accepted variance for the copy number of 1 ± 0.32 for Ct values of DP-CAR relative to DP-RNaseP.
Author Contributions
A.K., M.L.S., and M.S. conceived the qPCR validation study and wrote the manuscript. U.G., B.N., L.W., and A.H.K. supported study conception. A.K. designed, performed and analyzed qPCR experiments. U.G. and B.M. processed patient samples. M.S., A.S., S.H., C.M.T., and P.D. treated patients and provided clinical data. All authors reviewed and edited the manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
We thank Prof. Malcolm Brenner and Dr. Olga Dakhova from the Center for Cell and Gene Therapy (CAGT), Baylor College of Medicine, Houston, TX, USA, for providing material and technical and methodological support for the ACM.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtm.2020.02.003.
Supplemental Information
References
- 1.Maude S.L., Laetsch T.W., Buechner J., Rives S., Boyer M., Bittencourt H., Bader P., Verneris M.R., Stefanski H.E., Myers G.D. Tisagenlecleucel in Children and Young Adults with B-Cell Lymphoblastic Leukemia. N. Engl. J. Med. 2018;378:439–448. doi: 10.1056/NEJMoa1709866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gardner R.A., Finney O., Annesley C., Brakke H., Summers C., Leger K., Bleakley M., Brown C., Mgebroff S., Kelly-Spratt K.S. Intent-to-treat leukemia remission by CD19 CAR T cells of defined formulation and dose in children and young adults. Blood. 2017;129:3322–3331. doi: 10.1182/blood-2017-02-769208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Park J.H., Rivière I., Gonen M., Wang X., Sénéchal B., Curran K.J., Sauter C., Wang Y., Santomasso B., Mead E. Long-Term Follow-up of CD19 CAR Therapy in Acute Lymphoblastic Leukemia. N. Engl. J. Med. 2018;378:449–459. doi: 10.1056/NEJMoa1709919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davila M.L., Riviere I., Wang X., Bartido S., Park J., Curran K., Chung S.S., Stefanski J., Borquez-Ojeda O., Olszewska M. Efficacy and toxicity management of 19-28z CAR T cell therapy in B cell acute lymphoblastic leukemia. Sci. Transl. Med. 2014;6:224ra25. doi: 10.1126/scitranslmed.3008226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Porter D.L., Hwang W.T., Frey N.V., Lacey S.F., Shaw P.A., Loren A.W., Bagg A., Marcucci K.T., Shen A., Gonzalez V. Chimeric antigen receptor T cells persist and induce sustained remissions in relapsed refractory chronic lymphocytic leukemia. Sci. Transl. Med. 2015;7:303ra139. doi: 10.1126/scitranslmed.aac5415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Turtle C.J., Hay K.A., Hanafi L.A., Li D., Cherian S., Chen X., Wood B., Lozanski A., Byrd J.C., Heimfeld S. Durable Molecular Remissions in Chronic Lymphocytic Leukemia Treated With CD19-Specific Chimeric Antigen Receptor-Modified T Cells After Failure of Ibrutinib. J. Clin. Oncol. 2017;35:3010–3020. doi: 10.1200/JCO.2017.72.8519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kochenderfer J.N., Somerville R.P.T., Lu T., Yang J.C., Sherry R.M., Feldman S.A., McIntyre L., Bot A., Rossi J., Lam N., Rosenberg S.A. Long-Duration Complete Remissions of Diffuse Large B Cell Lymphoma after Anti-CD19 Chimeric Antigen Receptor T Cell Therapy. Mol. Ther. 2017;25:2245–2253. doi: 10.1016/j.ymthe.2017.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Turtle C.J., Hanafi L.A., Berger C., Hudecek M., Pender B., Robinson E., Hawkins R., Chaney C., Cherian S., Chen X. Immunotherapy of non-Hodgkin’s lymphoma with a defined ratio of CD8+ and CD4+ CD19-specific chimeric antigen receptor-modified T cells. Sci. Transl. Med. 2016;8:355ra116. doi: 10.1126/scitranslmed.aaf8621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schuster S.J., Svoboda J., Chong E.A., Nasta S.D., Mato A.R., Anak Ö., Brogdon J.L., Pruteanu-Malinici I., Bhoj V., Landsburg D. Chimeric Antigen Receptor T Cells in Refractory B-Cell Lymphomas. N. Engl. J. Med. 2017;377:2545–2554. doi: 10.1056/NEJMoa1708566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Neelapu S.S., Locke F.L., Bartlett N.L., Lekakis L.J., Miklos D.B., Jacobson C.A., Braunschweig I., Oluwole O.O., Siddiqi T., Lin Y. Axicabtagene Ciloleucel CAR T-Cell Therapy in Refractory Large B-Cell Lymphoma. N. Engl. J. Med. 2017;377:2531–2544. doi: 10.1056/NEJMoa1707447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krebs S., Rodríguez-Cruz T.G., Derenzo C., Gottschalk S. Genetically modified T cells to target glioblastoma. Front. Oncol. 2013;3:322. doi: 10.3389/fonc.2013.00322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schubert M.L., Schmitt A., Sellner L., Neuber B., Kunz J., Wuchter P., Kunz A., Gern U., Michels B., Hofmann S. Treatment of patients with relapsed or refractory CD19+ lymphoid disease with T lymphocytes transduced by RV-SFG.CD19.CD28.4-1BBzeta retroviral vector: a unicentre phase I/II clinical trial protocol. BMJ Open. 2019;9:e026644. doi: 10.1136/bmjopen-2018-026644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maude S.L., Frey N., Shaw P.A., Aplenc R., Barrett D.M., Bunin N.J., Chew A., Gonzalez V.E., Zheng Z., Lacey S.F. Chimeric antigen receptor T cells for sustained remissions in leukemia. N. Engl. J. Med. 2014;371:1507–1517. doi: 10.1056/NEJMoa1407222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Till B.G., Jensen M.C., Wang J., Qian X., Gopal A.K., Maloney D.G., Lindgren C.G., Lin Y., Pagel J.M., Budde L.E. CD20-specific adoptive immunotherapy for lymphoma using a chimeric antigen receptor with both CD28 and 4-1BB domains: pilot clinical trial results. Blood. 2012;119:3940–3950. doi: 10.1182/blood-2011-10-387969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hollyman D., Stefanski J., Przybylowski M., Bartido S., Borquez-Ojeda O., Taylor C., Yeh R., Capacio V., Olszewska M., Hosey J. Manufacturing validation of biologically functional T cells targeted to CD19 antigen for autologous adoptive cell therapy. J. Immunother. 2009;32:169–180. doi: 10.1097/CJI.0b013e318194a6e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maiti S.N., Huls H., Singh H., Dawson M., Figliola M., Olivares S., Rao P., Zhao Y.J., Multani A., Yang G. Sleeping beauty system to redirect T-cell specificity for human applications. J. Immunother. 2013;36:112–123. doi: 10.1097/CJI.0b013e3182811ce9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Singh H., Figliola M.J., Dawson M.J., Olivares S., Zhang L., Yang G., Maiti S., Manuri P., Senyukov V., Jena B. Manufacture of clinical-grade CD19-specific T cells stably expressing chimeric antigen receptor using Sleeping Beauty system and artificial antigen presenting cells. PLoS ONE. 2013;8:e64138. doi: 10.1371/journal.pone.0064138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.June C.H., Sadelain M. Chimeric Antigen Receptor Therapy. N. Engl. J. Med. 2018;379:64–73. doi: 10.1056/NEJMra1706169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.