Group B Streptococcus is an important human pathogen that causes serious infections during pregnancy which can lead to chorioamnionitis, funisitis, premature rupture of gestational membranes, preterm birth, neonatal sepsis, and death. GBS is evolving antimicrobial resistance mechanisms, and the work presented in this paper provides evidence that prebiotics such as human milk oligosaccharides can act as adjuvants to restore the utility of antibiotics.
KEYWORDS: group B Streptococcus, human milk oligosaccharides, resistance, adjuvants, antifolate drugs
ABSTRACT
Adjuvants can be used to potentiate the function of antibiotics whose efficacy has been reduced by acquired or intrinsic resistance. In the present study, we discovered that human milk oligosaccharides (HMOs) sensitize strains of group B Streptococcus (GBS) to trimethoprim (TMP), an antibiotic to which GBS is intrinsically resistant. Reductions in the MIC of TMP reached as high as 512-fold across a diverse panel of isolates. To better understand HMOs’ mechanism of action, we characterized the metabolic response of GBS to HMO treatment using ultrahigh-performance liquid chromatography–high-resolution tandem mass spectrometry (UPLC-HRMS/MS) analysis. These data showed that when challenged by HMOs, GBS undergoes significant perturbations in metabolic pathways related to the biosynthesis and incorporation of macromolecules involved in membrane construction. This study represents reports the metabolic characterization of a cell that is perturbed by HMOs.
INTRODUCTION
The development of antibiotics is arguably one of the most important advances in modern medicine. Antibiotics can be organized according to the cellular component or system they engage and whether they inhibit cell growth (bacteriostatic) or induce cell death (bactericidal). Although antibiotics that target cellular viability are effective, these agents impose selective pressures that foster the evolution of resistant phenotypes (1). Combination therapy has emerged as a powerful solution to resistance issues that plague monotherapy (2). This approach, which involves codosing an antibiotic with an adjuvant that potentiates its function or a second antibiotic with a different target, can improve efficacy and suppress resistance evolution (2–7).
One bacterial pathogen group that showcases multidrug resistance is group B Streptococcus (GBS) (8). GBS is a leading cause of neonatal sepsis, pneumonia, and meningitis (9–14). Recent data also suggest that GBS is a frequent cause of chorioamnionitis, endometritis, pneumonia, and urosepsis in adults with underlying medical conditions (i.e., diabetes mellitus or immunosuppression) (15–19). As these patterns of pathogenesis suggest, GBS is considered a saprophytic organism, i.e., invasive GBS disease is most commonly observed in weakened hosts.
Treatment of GBS disease relies primarily on penicillin and ampicillin, followed by first-generation cephalosporins and vancomycin (20). Alternative antibiotics, such as lincosamides, are used for patients with β-lactam allergies. Due to resistance evolution, macrolides, aminoglycosides, and tetracyclines are no longer clinically efficacious (21–24). While our group and others have observed that GBS is resistant to a wide range of antibiotics, GBS resistance remains poorly characterized and is a frontier of concern in the clinic.
In the early stages of this program, we hypothesized that human milk oligosaccharides (HMOs) possess antimicrobial and antivirulence properties (25). Indeed, we discovered that heterogeneous HMOs modulate growth and biofilm production for a number of bacterial pathogens (26, 27). We also determined the identities of several single-entity HMOs with potent antimicrobial activity against GBS (28–31). In addition to structure-activity relationship (SAR) studies, we found that HMOs potentiate the activity of select intracellular targeting antibiotics (32). This included three antibiotics to which GBS has evolved resistance, aminoglycosides, macrolides, and tetracyclines (33–42). At their 50% inhibitory concentration (IC50), HMO extracts reduced the MICs of certain intracellular targeting antibiotics up to 32-fold (Table 1). Interestingly, HMO treatment did not affect β-lactam or glycopeptide activity; antibiotics that interfere with cell wall synthesis. Based on this activity pattern, we hypothesized that HMOs function by increasing membrane permeability, which would be an unprecedented mode of action in GBS. This hypothesis was validated when HMOs were found to increase membrane permeability by ca. 30% using a LIVE/DEAD BacLight assay (32).
TABLE 1.
Established patterns of HMO potentiation of antibiotic activity
| Antibiotic in THB medium (strain) | MIC (μg/ml) |
Fold reduction | |
|---|---|---|---|
| Overall | With 5.0 mg/ml HMOs | ||
| Penicillin (CNCTC 10/84) | 0.03 | 0.015 | 2 |
| Vancomycin (CNCTC 10/84) | 2 | 1 | 2 |
| Clindamycin (GB2) | 0.0312 | 0.0078 | 4 |
| Gentamicin (GB590) | 16 | 1 | 16 |
| Erythromycin (GB590) | 0.0312 | 0.001 | 32 |
| Minocycline (CNCTC 10/84) | 0.0625 | 0.0019 | 32 |
Based on their ability to increase cellular permeability, a second-generation combination study was initiated to further characterize HMO enhancement of intracellular targeting antibiotics in the GBS model. We took particular interest in trimethoprim (TMP), an antifolate used in the treatment of enteric, respiratory, skin, and urinary tract infections (43). Mechanistically, TMP is a bacteriostatic agent that inhibits dihydrofolate reductase (DHFR), an enzyme within the folate biosynthesis pathway (44). Importantly, interference with this pathway inhibits pyrimidine and purine biosynthesis, with downstream effects on bacterial DNA synthesis. Furthermore, a wide range of streptococcal strains, including GBS, are intrinsically resistant to TMP (45–51). Resistance is typically mediated by one of the following five mechanisms: (i) poor membrane permeability, (ii) an impervious DHFR, (iii) mutations in the inherent DHFR, (iv) upregulation of gene expression or gene duplication to increase DHFR production, and (v) horizontal transfer of dfr genes that encode resistant DHFRs (45). We hypothesized that if TMP has difficulty gaining penetrance into the GBS cell, HMOs could be used to sensitize GBS to TMP. Described herein are the results of testing this hypothesis using heterogeneous HMO extracts. To further evaluate the mechanism of HMO sensitization, ultrahigh-performance liquid chromatography–high-resolution tandem mass spectrometry analysis (UPLC-HRMS/MS) was used to characterize the immediate metabolic response of GBS to HMO-induced perturbations.
RESULTS AND DISCUSSION
HMOs demonstrate synergy with TMP against group B Streptococcus.
HMOs were isolated from donor breast milk and pooled to create two HMO cocktails; the first (HMO-1) used milk from 10 donors, while the second (HMO-2) used milk from 7 donors. Prior to potentiation studies, the MIC of the HMOs and TMP were determined separately in each strain of GBS grown in Todd-Hewitt broth (THB) using a broth microdilution assay (Table 2). HMOs were assayed against five strains of GBS of various serotypes to determine the strain specificity of antibiotic potentiation. The strains selected are all clinical isolates. CNCTC 10/84 is commercially available (52). Isolates GB00590, GB00002, GB00651, and GB00083 were recovered from colonized pregnant women (53, 54). GBS strains are divided into 10 serotypes (1a, 1b, and II to IX) based on a serological reaction against their capsular polysaccharides (55). GB2, GB590, and CNCTC 10/84 are serotypes Ia, III, and V, respectively. These three serotypes are the most common isolates associated with early-onset disease in the United States and together account for over 80% of all isolates (56). GB651 and GB83 are serotypes Ib and IV, respectively. Globally, the five strains represent 85% of all isolate serotypes (57).
TABLE 2.
HMO potentiation of TMP
| Strain in THB medium | MIC (μg/ml) for: |
Fold reduction | ||
|---|---|---|---|---|
| HMOs | TMP | TMP with 1.42 mg/ml HMO | ||
| CNCTC 10/84 | 5.12a | >1,024 | 8a | ≥256 |
| GB2 | 2.56a | 1,024 | 2a | 512 |
| GB590 | 5.12a | >1,024 | 32a | ≥64 |
| GB651 | 5.12b | 512 | 32b | 16 |
| GB83 | 5.12b | >1,024 | 128b | ≥16 |
HMO-1.
HMO-2.
HMOs were dosed at their 25% inhibitory concentrations (IC25s) in CNCTC 10/84 and GB2. The growth of the remaining GBS strains were so rapidly affected by HMO treatment that subsequent IC50 curve fitting yielded immeasurable confidence limits. For these strains, the IC25 from a similar strain having a superior fit dose-response curve was used (see Fig. S1A to E in the supplemental material). In each strain, the MIC of TMP was 512 μg/ml or higher (Table 2). In GB2, a 512-fold reduction in MIC was observed. A 256-fold reduction in MIC was observed in CNCTC 10/84. For the three remaining strains (GB651, GB83, and GB590), the fold reductions in MIC were 16, 16, and 64, respectively. The potentiation patterns described above are remarkable for several reasons. First, they represent the greatest magnitude of antibiotic enhancement that we have observed. Second, GBS is not susceptible to antifolate antibiotics, so the chemotherapeutic regime is effective at sensitizing GBS to TMP.
(A) IC50 curve of HMO cocktail for GB2 in THB. Bacterial growth (OD600) was recorded after 24 h of HMO treatment at increasing HMO concentrations. MIC, 2.56 mg/ml; IC50, 1.897 mg/ml; IC25, 1.43 mg/ml; Hill slope, −3.894. (B) IC50 curve of HMO cocktail for GB590 in THB. Bacterial growth (OD600) was recorded after 24 h of HMO treatment at increasing HMO concentrations. MIC = 5.125 mg/ml, IC50 ∼ 2.417 mg/ml, IC25 ∼2.30 mg/ml, hill slope = −22.37. (C) IC50 curve of HMO cocktail for GB CNCTC 10/84 in THB. Bacterial growth (OD600) was recorded after 24 h of HMO treatment at increasing HMO concentrations. MIC = 5.125 mg/ml, IC50 = 2.05 mg/ml, IC25 = 1.42 mg/ml, hill slope = −2.98. (D) IC50 curve of HMO cocktail for GB83 in THB. Bacterial growth (OD600) was recorded after 24 h of HMO treatment at increasing HMO concentrations. MIC = 5.125 mg/ml, IC50 = 3.616 mg/ml, IC25 = 3.51 mg/ml, hill slope = −39.06. (E) IC50 curve of HMO cocktail for GB651 in THB. Bacterial growth (OD600) was recorded after 24 h of HMO treatment at increasing HMO concentrations. MIC = 5.125 mg/ml, IC50 = 2.616 mg/ml, IC25 = 2.33 mg/ml, hill slope = −9.639. Download FIG S1, DOCX file, 0.1 MB (59KB, docx) .
Copyright © 2020 Chambers et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Next, checkerboard assays were conducted with GB2 and GB590, stains for which strong and weak potentiation of TMP were observed, respectively, to determine if the HMO-TMP combination was synergistic or additive in nature (Fig. S2A and B). Synergy is measured using the fractional inhibitory concentration (FIC) index value and is defined when the FIC is ≤0.5 for each combination of compounds. It was demonstrated that in GB590, synergy was achieved when dosing HMOs from 1.28 to 2.56 mg/ml in combination with TMP dosed at 8 to 128 μg/ml (ΣFIC values, 0.281 to 0.508). In GB2, the combination was synergistic with treatment of HMOs between 0.64 and 1.28 mg/ml in conjunction with TMP at 4 to 32 μg/ml (ΣFIC values, 0.281 to 0.508). These assays firmly demonstrate the HMO-TMP combination to be synergistic, and they characterize the dosing windows required to achieve this effect.
(A) Checkerboard assay in GB2 showing the synergistic combinations between TMP and the HMO cocktail (bolded). (B) Checkerboard assay in GB590 showing the synergistic combinations between TMP and the HMO cocktail (bold). The combination is considered synergistic when the ΣFIC is ≤0.5, additive or indifferent when the ΣFIC is >0.5 to <4, and antagonistic when the ΣFIC is ≥4. Download FIG S2, DOCX file, 0.1 MB (24.2KB, docx) .
Copyright © 2020 Chambers et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
After evaluating the level of synergy in the cocktail, we conducted an experiment to validate whether growth inhibition was due to HMO enablement of cognate engagement of TMP with the folate pathway. By inhibiting folate biosynthesis, antifolates inhibit the de novo biosynthesis of thymidylate and purine nucleotides. However, in addition to de novo synthesis, cells can produce these nucleotides via salvage pathways that use free thymidine or purine bases as precursors for the corresponding nucleotides. Thus, we hypothesized that if HMOs facilitate TMP inhibition of the de novo synthesis pathway, the addition of the preformed nucleotide precursors thymidine or hypoxanthine would dampen the growth inhibitory capabilities of the HMO-TMP combination, as thymidine and hypoxanthine serve as precursors in the pyrimidine and purine salvage pathways, respectively (58).
In the experiment, we evaluated the MIC of the HMO-TMP cocktail in the presence of thymidine (Table 3). The experiment was conducted in strains GB2 and GB590. Against GB2, HMO supplementation decreased the MIC of TMP from 1,024 μg/ml to 2 μg/ml (512-fold reduction). Against GB590, the MIC of TMP was reduced from >1,024 μg/ml to 32 μg/ml (at least a 64-fold reduction) (Table 2). In the presence of added thymidine, the MICs of TMP in the HMO-TMP combination increased 8-fold to 16 μg/ml in GB2 and 4-fold to 128 μg/ml in GB590. These results support the hypothesis that supplemental thymidine mitigates the effects of the HMO-TMP combination and is able to partially salvage the folate biosynthetic pathway. Importantly, the MIC of the HMO cocktail individually did not change in the presence of added thymidine. This indicates that the folate pathway is not a target for HMOs. We therefore conclude that, in the presence of HMOs, TMP gains penetrance into the group B streptococcal cell and exhibits on-target inhibition of the folate cycle.
TABLE 3.
HMO potentiation of TMP in the presence of thymidine
| Strain in THB medium plus 20 μg/ml thymidine | MIC (μg/ml) for: |
Fold reduction | |
|---|---|---|---|
| TMP | TMP with HMO-1 (dose [mg/ml]) | ||
| GB2 | 1,024 | 16 (1.43) | 64 |
| GB590 | >1,024 | 128 (1.42) | 8 |
The final assay in this study was a comparison of the HMO-TMP combination with the clinically useful TMP-sulfadiazine (SDZ) combination (Table S2). In both GB590 and GB2, the TMP-sulfadiazine combination was largely ineffective, with an MIC of ≥512 μg/ml, while the HMO-TMP combination sees the potentiation profile described above (Table 2). This result demonstrates that while antifolate-based antibiotic combination treatments remain largely ineffective against GBS, the HMO-TMP combination is operative. This insight offers new consideration for the use of existing combination therapies in patient care.
Characterizing the HMO mode of action using untargeted metabolomics.
The mode of action of an antimicrobial agent cannot accurately be described in terms of a single static target; rather, the complete induced response must be evaluated. In theory, a single chemotherapeutic could have a wide range of direct and indirect targets, simultaneously interfering with multiple enzymes or pathways. Accordingly, the final stage of the study focused on utilizing global, untargeted metabolomic analysis to characterize the early response of GBS to HMO-mediated perturbations. The analysis described provides HMO-mediated perturbations. For this study, strain GB2 was used, as it was most susceptible to treatment with HMOs (MIC, 2.56 μg/ml). Two groups were analyzed and compared, with the first being an untreated GB2 control and the second being GB2 treated with HMOs dosed at 1 mg/ml. This concentration promoted cellular death (ca. 20 to 40%) compared to the untreated controls but also provided enough remaining cellular mass for analysis (minimum, 200 μg).
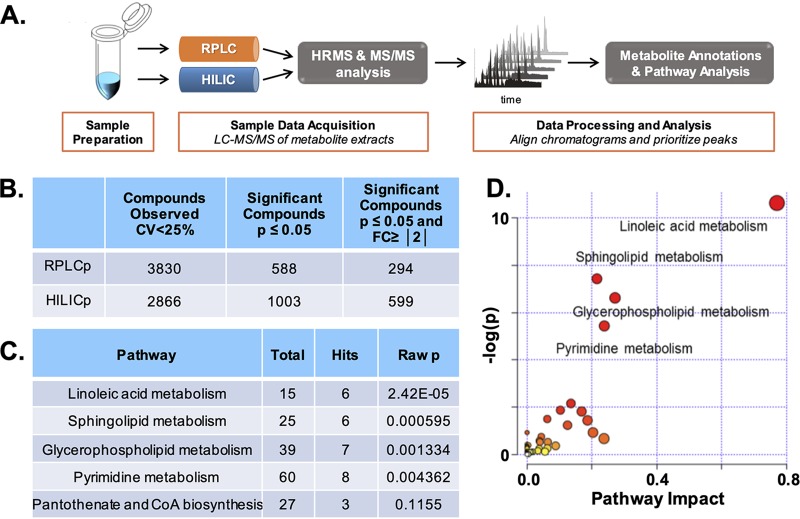
Our experimental design from sample collection through data analysis is depicted in Fig. 1A. The annotated and statistically significant metabolites observed in the experiment (Fig. 1B) were subjected to traditional pathway analysis (Fig. S3). The results showed the most statistically perturbed metabolic pathways to be linoleic acid metabolism, sphingolipid metabolism, glycerophospholipid metabolism, and pyrimidine metabolism (Fig. 1C and D). Characterized below are perturbations to both linoleic acid and glycerophospholipid metabolism (Fig. 2 and 3). We focus on these pathways, as each is critical to membrane formation and structural integrity, i.e., each pathway contributes to the synthesis of membrane-bound macromolecules and their corresponding precursors (59).
FIG 1.
Workflow and pathway analysis using global, untargeted metabolomics data analysis. (A) Overview of global, untargeted metabolomic workflow. (B) Global output of identified metabolites from RPLC and HILIC methods and subsequent filtering for significance according to a P value of ≤0.05 and fold change of ≥|2|. (C) Table output of metabolic pathway enrichment analysis. The number of total metabolites in the pathway, the number of hits, and the P value were calculated using MetaboAnalyst 4.0. CoA, coenzyme A. (D) Metabolomic pathway analysis visualization. Shown is a graphical representation analysis using the statistically significant metabolite compounds (P ≤ 0.05; fold change, ≥|2|) annotated from RPLC and HILIC analyses. Matched pathways were arranged by P values (from pathway enrichment analysis) on the y axis, and pathway impact values (from pathway topology analysis) are shown on the x axis; node color is based on pathway P value, and node radius is determined based on pathway impact values; individual nodes represent individual pathways.
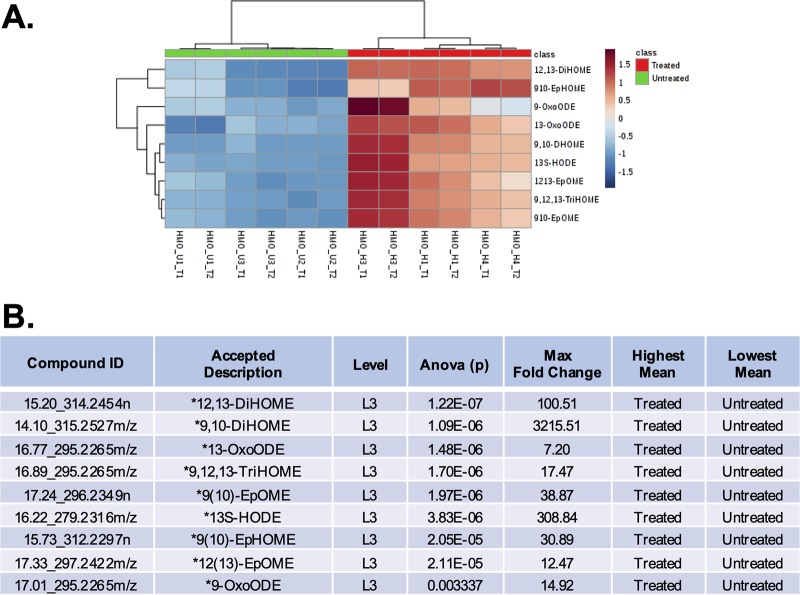
FIG 2.
Linoleic acid-associated metabolite identification and statistical representation. (A) Heat map visualization of the significantly differently regulated linoleic acid metabolic pathway upon HMO treatment. Linoleic acid metabolism members shown here were detected by RPLC-positive LC-MS/MS analysis. Samples (columns) and metabolite compounds (rows) were processed using Euclidean average clustering via MetaboAnalyst 4.0. The heat map was generated for Pareto-scaled, log-transformed data, and colors are displayed by relative abundance, ranging from low (blue) to high (red), as shown in the legend. (B) Corresponding data table of linoleic acid metabolites, where the asterisk (*) denotes significance with a P value of ≤0.05 and fold change of ≥|2|. ODE, octadecadienoic acid; 13-HOTE, 13-hydroxyoctadeca-9,11,15-trienoic acid; 9,10-DiHOME, 9,10-dihydroxyoctadec-12-enoic acid; 9,12,13-TriHOME, 9,12,13-trihydroxyoctadecanoic acid; 13S-HODE, 13S-octadecadienoic acid; 9(10)-EpHOME, 9(10)-epoxyhydroxyoctadecanoic acid; ID, identifier.
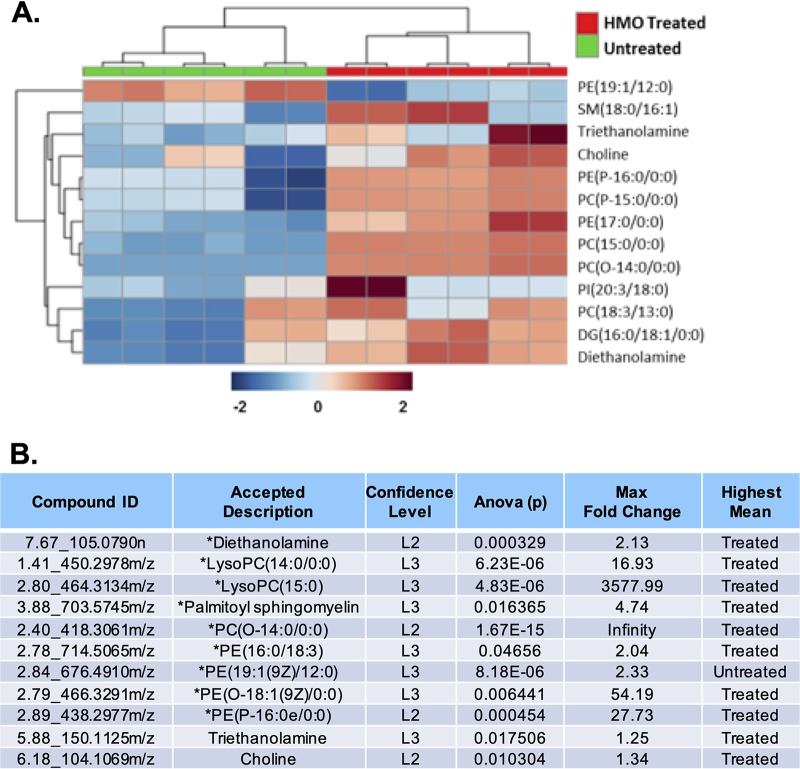
FIG 3.
Glycerophospholipid-associated metabolite identification and statistical representation. (A) Heat map visualization of the significantly differently regulated glycerophospholipid metabolism pathway upon HMO treatment. Glycerophospholipid members shown here were detected by HILIC-positive LC-MS/MS analysis. Samples (columns) and metabolite compounds (rows) were processed using Euclidean average clustering via MetaboAnalyst 4.0. The heat map was generated for Pareto-scaled, log-transformed data, and colors are displayed by relative abundance, ranging from low (blue) to high (red), as shown in the legend. SM, sphingomyelin; PC, phosphocholine; PI, phosphoinositol; DG, diglyceride; LysoPC, lysophosphatidylcholine. (B) Corresponding data table of glycerophospholipid metabolites, where the asterisk (*) denotes significance with a P value of ≤0.05 and fold change of ≥|2|.
Global principal-component analysis (PCA) of cellular extracted metabolites for the two biological conditions is illustrating a distinct shift in metabolic profiles between groups. Presenting the abundance data in PC space allows us to separate the run samples based on overall variability and shows a clear separation of untreated samples versus HMO-treated samples across the first PC dimension (x axis). Download FIG S3, DOCX file, 0.1 MB (88KB, docx) .
Copyright © 2020 Chambers et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Based on statistical significance, linoleic acid metabolism is the metabolic pathway most impacted when GBS is exposed to HMOs (Fig. 2A and S4). Linoleic acid metabolites play a critical role in both cellular signaling and the stress response. Each is also critical to proper membrane construction (60, 61). All identified linoleic acid metabolites were accumulated in the HMO-treated population, with several metabolites having a >100-fold increase from the untreated controls (Fig. 2B). Two epoxyoctadecanoic acid metabolites were of particular interest, epoxyoctadecanoic acids (EpOMEs) and dihydroxyoctadecanoic acids (DiHOMEs). Accumulation of these metabolites is linked to changes in Na+ and K+ ion channels and, subsequently, cell membrane fluidity (62). In addition to the roles of EpOMEs and DiHOMEs in cell membrane construction, linoleic acid metabolites have been shown to have a critical role in cellular signaling and the stress response.
Linoleic acid metabolism supplementary images. (A) Heat map visualization of the differently regulated members within the linoleic acid metabolic pathway upon HMO treatment. Linoleic acid metabolism members shown here were detected by HILIC-positive LC-MS/MS analysis. (B) Red nodes reveal individual significant metabolites matched to the linoleic acid metabolic pathway. The KEGG C number is shown within a node. The individual significant metabolites (P < 0.05) identified at confidence level L3 are highlighted in red for the linoleic acid metabolism, generated by the MetaboAnalyst 4.0 software. Download FIG S4, DOCX file, 0.2 MB (206.4KB, docx) .
Copyright © 2020 Chambers et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Glycerophospholipid metabolism was also significantly impacted, and in general, we observed an accumulation of these metabolites compared to the control (Fig. 3A and S5). Glycerophospholipid metabolites were observed with significant fold changes compared to the control. For example, PE(17:0/0:0), PE(P-16:0/0:0), and PE(19:1/12:0), known degradation products of phosphatidylethanolamine (PE), one of three major components of the cellular membrane, were observed to have up to a 50-fold change increase compared to the untreated control (63). This observed accumulation of lipid metabolites indicates an increased rate of breakdown of critical cell membrane components when bacteria are dosed with HMOs. In fact, this type of metabolite accumulation has been previously observed in other model organisms when exposed to antibiotics (64–67).
Glycerophospholipid metabolism supplementary images. (A) Heat map visualization of the differently regulated members within the glycerophospholipid metabolic pathway upon HMO treatment. Glycerophospholipid metabolism members shown here were detected by RPLC-positive LC-MS/MS analysis. (B) Red nodes reveal individual significant metabolites matched to the glycerophospholipid metabolic pathway. The KEGG C number is shown within a node. The individual significant metabolites (P < 0.05) identified at confidence level L3 are highlighted in red for the glycerophospholipid metabolism, generated by the MetaboAnalyst 4.0 software. Download FIG S5, DOCX file, 0.4 MB (370.4KB, docx) .
Copyright © 2020 Chambers et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
In addition to dysregulating pathways directly related to the production of membrane components, HMOs perturbed additional pathways related to essential cellular function. These include, for example, increased accumulation of purine and pyrimidine nucleotide precursors. An inability to synthesize nucleotides would lead to perturbations in DNA and RNA synthesis. HMO treatment also led to an accumulation of metabolites in the cysteine and methionine metabolic pathway. Cysteine biosynthesis is the primary pathway for incorporating sulfur into cellular components. In addition to serving as a precursor of methionine, cysteine is also the direct precursor to biotin, thiamine, and lipoic acid. Methionine is an essential amino acid in all organisms, as it is both proteinogenic and a component of the cofactor S-adenosyl methionine. Interestingly, sphingolipid metabolism was observed to be significantly perturbed upon HMO treatment. While GBS does not synthesize sphingolipids directly, it does rely on the host for access to these compounds for the biosynthesis of cell membrane components. This metabolic change could suggest that HMO treatment has an impact on host-microbe interactions (68, 69). Finally, several cell wall synthesis-associated metabolites were also identified as being accumulated in the treated sample, but a higher experimental mass range would be needed to identify a more significant amount of these precursors and better identify a global trend. The variety of metabolic pathways perturbed not only illuminates the synergistic nature of the TMP and HMO combination treatment but demonstrates that antimicrobial agents have broad effects on cellular biology (Fig. S6).
Heat map visualization of the differently regulated members within the cell wall synthesis-associated metabolites upon HMO treatment. Metabolites shown here were detected by HILIC-positive LC-MS/MS analysis. Download FIG S6, DOCX file, 0.1 MB (40.8KB, docx) .
Copyright © 2020 Chambers et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
In summary, this study demonstrates that HMOs potentiate trimethoprim in group B Streptococcus bacteria, with a synergistic profile that spans the most prevalent serotypes worldwide. This potentiation profile makes antifolate antibiotics of potential use in an organism where they have been long considered resistant. Moreover, this combination could represent an alternative treatment for GBS-positive mothers with penicillin allergies given the high rates of resistance observed with alternative agents.
To characterize the mechanism of HMO-mediated antimicrobial activity, we have presented the first global, untargeted metabolomic analysis of HMO-mediated perturbations within any cell type and have shown significant impacts on cell membrane-affiliated macromolecules. While a number of high-throughput methods have been performed to elucidate an antibiotic’s mode of action (e.g., cytological profiling, genetic screens, or gene expression and proteomic profiling), direct experimental evidence that rapid metabolic changes are causal in facilitating the microbial response to antibiotics is lacking. In fact, little is described about the downstream phenotypic changes induced by antimicrobial agents. In the future, metabolomic experiments will be employed to better describe the phenotypic response of GBS to HMO-induced effects. We hypothesize that metabolomics will enable the characterization of the indirect connections critical to HMOs’ mechanism of action. Since metabolites present the final phenotypic manifestation of an organism and the final endpoint of biochemical reactions reflects the interplay between gene expression, protein function, and the environment, we argue that further metabolomics analyses are necessary to understand the HMO mode of action (70, 71).
MATERIALS AND METHODS
Antibiotics and additional chemicals.
Trimethoprim lactate 98% was purchased from Alfa Aesar. β-Galactosidase from Kluyveromyces lactis, at 2,600 units/g, was purchased from Sigma-Aldrich. Acetonitrile (ACN; catalog no. A955-1), methanol (MeOH; catalog no. A456-1), and water (catalog no. W6-1, liquid chromatography-mass spectrometry [LC-MS] grade; Optima) for the mass spectrometry analysis were obtained from Thermo Fisher Scientific.
HMO isolation.
Human milk was obtained from 17 healthy, lactating women between 3 days and 3 months postpartum and stored between −80 and –20°C. Deidentified milk was provided by Jörn-Hendrik Weitkamp from the Vanderbilt Department of Pediatrics, under a collection protocol approved by the Vanderbilt University institutional review board (IRB no. 100897), or from Medolac. Milk samples were thawed and then centrifuged for 45 min. Following centrifugation, the resultant top lipid layer was removed. The proteins were then removed by diluting the remaining sample with roughly 1:1 (vol/vol) 180 or 200 proof ethanol, chilling the sample briefly, and centrifuging for 45 min, followed by removal of the resulting HMO-containing supernatant. Following concentration of the supernatant in vacuo, the HMO-containing extract was dissolved in 0.2 M phosphate buffer (pH 6.5) and heated to 37°C. β-Galactosidase from Kluyveromyces lactis was added, and the reaction mixture was stirred until lactose hydrolysis was complete. The reaction mixture was diluted with roughly 1:0.5 (vol/vol) 180 or 200 proof ethanol, chilled briefly, and then centrifuged for 30 min. The supernatant was removed and concentrated in vacuo, and the remaining salts, glucose, and galactose were separated from the oligosaccharides using size exclusion chromatography with P-2 gel (H2O eluent). The oligosaccharides were then dried by lyophilization. Correspondingly, HMO isolates from donors were combined and solubilized in water to reach a final concentration of 102.6 mg/ml.
Bacterial strains and culture conditions.
The bacterial strains are shown in Table S1. All strains were grown on tryptic soy agar plates supplemented with 5% sheep blood (blood agar plates) at 37°C in ambient air overnight. All strains were subcultured from blood agar plates into 5 ml of Todd-Hewitt broth (THB) and incubated under shaking conditions at 180 rpm at 37°C overnight. Following overnight incubation, bacterial density was quantified through absorbance readings at 600 nm (OD600) using a Promega GloMax-Multi detection system plate reader. Bacterial numbers were determined using the predetermined coefficient of an OD600 of 1, equal to 109 CFU/ml.
Group B Streptococcus strains evaluated in the study. *ST, sequence type as defined by multilocus sequence typing (MLST). Download Table S1, DOCX file, 0.1 MB (13.3KB, docx) .
Copyright © 2020 Chambers et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
TMP-sulfadiazine antibiotic combination assay. aTHB medium. bShown in μg/ml. c1:4.85 ratio of TMP-sulfadiazine. Download Table S2, DOCX file, 0.1 MB (13KB, docx) .
Copyright © 2020 Chambers et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Broth microdilution method for determination of MICs.
All strains were grown overnight as described above and used to inoculate fresh THB or THB plus 20 μg/ml thymidine to achieve 5 × 105 CFU/ml. To 96-well tissue culture-treated, sterile polystyrene plates was added the inoculated medium in the presence of increasing concentrations of antibiotic or HMO cocktail to achieve a final volume of 100 μl per well. Bacteria grown in medium in the absence of any compounds served as the controls. The plates were incubated under static conditions at 37°C in ambient air for 24 h. Bacterial growth was quantified through absorbance readings (OD600). The MICs were assigned at the lowest concentration of compound at which no bacterial growth was observed.
Broth microdilution method for antibiotic combination.
All strains were grown overnight as described above and the subcultures used to inoculate fresh THB or THB plus 20 μg/ml thymidine to achieve 5 × 105 CFU/ml. Freshly inoculated medium was then supplemented with HMOs at their IC25. To 96-well tissue culture-treated, sterile polystyrene plates was added the inoculated medium supplemented with HMOs in the presence of increasing concentrations of antibiotic. Bacteria grown in medium in the absence of any compounds served as one control. Bacteria grown in medium supplemented with HMOs in the absence of any antibiotic served as a second control. MICs were determined as previously described.
Synergy assay.
Group B Streptococcus strains (GB2 and GB590) were grown overnight as described above and used to inoculate fresh THB to achieve 5 × 105 CFU/ml. One hundred microliters per well of inoculated medium was added to 96-well tissue culture-treated, sterile polystyrene plates. Trimethoprim was 2-fold serially diluted descending down the plate to achieve a final volume of 100 μl per well. The final row was left without any trimethoprim. The HMO cocktail was 2-fold serially diluted going from right to left across the plate. The far-left column was left without any HMO cocktail. Bacteria grown in medium in the absence of either compound served as the controls. The plates were incubated under static conditions at 37°C in ambient air for 24 h. Bacterial growth was quantified through absorbance readings (OD600). The MICs were assigned at the lowest concentration of compound at which no bacterial growth was observed. The fractional inhibitory concentration (FIC) index was used to evaluate synergy. The calculation of the FIC index is as follows: ΣFIC = FIC A + FIC B = (MIC of drug A in the combination/MIC of drug A alone) + (MIC of drug B in the combination/MIC of drug B alone), where A is trimethoprim and B is the HMO cocktail. The combination is considered synergistic when the ΣFIC is ≤0.5, additive or indifferent when the ΣFIC is >0.5 to <4, and antagonistic when the ΣFIC is ≥4.
Statistical analysis.
The data for the HMO antimicrobial and combination assays represent 3 biological replicates, each with 3 technical replicates. The data for the synergy assays represent 3 biological replicates. Data are expressed as the mean biomass ± standard error of the mean (SEM). Statistical analyses were performed in the GraphPad Prism software v. 7.0c. Statistical significance was determined using one-way analysis of variance (ANOVA) with post hoc Dunnett’s multiple-comparison test comparing growth in the presence of ca. 5 mg/ml HMOs to growth in medium alone. HMO IC50 curves were generated in the GraphPad Prism software v. 7.0c using an inhibition dose-response nonlinear regression curve fit for log(inhibitor) versus normalized response with a variable slope.
Sample preparation for metabolomic analysis.
Group B Streptococcus strain GB2 was grown overnight as described above and used to inoculate 10 ml of fresh THB medium to achieve 5 × 105 CFU/ml. Untreated GB2 in 10 ml of medium served as a control, while other GB2 cultures were treated with HMOs at 1.00 mg/ml. After 24 h, the samples were centrifuged at 1,500 rpm for 20 min to generate a bacterial pellet. The medium was removed and the pellet washed with 200 μl of 50 mM ammonium formate buffer. The pellet was then resuspended in 200 μl of 50 mM ammonium formate buffer and transferred to a sterile Eppendorf tube. This was then centrifuged at 1,500 rpm for 10 min to generate a bacterial pellet. The buffer was removed and the pellet flash frozen in liquid N2 and stored until use.
The bacterial cell pellets were lysed using 400 μl ice-cold lysis buffer (1:1:2, AcCN:MeOH:ammonium bicarbonate 0.1 M [pH 8.0], LC-MS grade) and vortexed. Individual samples were sonicated using a probe tip sonicator, with 10 pulses at 30% power and cooling down in ice between samples. A bicinchoninic acid (BCA) protein assay was used to determine the protein concentration for each individual sample and adjusted to a total amount of protein of 200 μg in 200 μl of lysis buffer. Isotopically labeled standard molecules phenylalanine-D8 (CDN Isotopes, Quebec, CA) and biotin-D2 (CIL, MA, USA) were added to each sample to assess sample preparation reproducibility. Metabolites were extracted from untreated control and HMO-treated cultures using protein precipitation by the addition of 800 μl of ice-cold methanol (4× by volume) and incubated overnight at –80°C. Following incubation, samples were centrifuged at 10,000 rpm for 10 min to eliminate precipitated proteins, and the metabolite-containing supernatant was dried in vacuo and stored at –80°C until further UPLC-HRMS/MS analysis.
Global untargeted metabolomic analyses.
Metabolite extracts were analyzed using reverse-phase liquid chromatography (RPLC) and hydrophilic interaction liquid chromatography (HILIC), followed by subsequent mass spectrometry analysis using a high-resolution Q-Exactive high-fidelity (HF) hybrid quadrupole-Orbitrap mass spectrometer (Thermo Fisher Scientific, Bremen, Germany) equipped with a Vanquish ultrahigh-performance liquid chromatography (UHPLC) binary system and autosampler (Thermo Fisher Scientific, Bremen, Germany). A quality control sample was prepared by pooling equal volumes of each sample. Isotopically labeled standards tryptophan-D3, carnitine-D9 (CDN Isotopes, Quebec, CA), valine-D8, and inosine-4N15 (CIL, MA, USA) were added to each sample to assess MS instrument reproducibility.
Metabolite extracts (10-μl injection volume) were separated on a SeQuant ZIC-HILIC 3.5-μm, 2.1-mm by 100-mm column (Millipore Corporation, Darmstadt, Germany) held at 40°C for the HILIC analysis. Liquid chromatography was performed at 200 μl/min using solvent A (5 mM ammonium formate in 90% water, 10% acetonitrile) and solvent B (5 mM ammonium formate in 90% acetonitrile, 10% water) with the following gradient: 95% B for 2 min, 95 to 40% B over 16 min, 40% B held for 2 min, and 40 to 95% B over 15 min, and 95% B held for 10 min (gradient length, 45 min). For the RPLC analysis, metabolite extracts (10 μl injection volume) were separated on a Hypersil Gold, 1.9 μm, 2.1-mm by 100-mm column (Thermo Fisher) held at 40°C. Liquid chromatography was performed at 250 μl/min using solvent A (0.1% formic acid [FA] in water) and solvent B (0.1% FA in acetonitrile [ACN]) with the following gradient: 5% B for 1 min, 5 to 50% B over 9 min, 50 to 70% B over 5 min, 70 to 95% B over 5 min, 95% B held for 2 min, 95 to 5% B over 3 min, and 5% B held for 5 min (gradient length, 30 min).
MS analyses were acquired over a mass range of m/z 70 to 1,050 using electrospray ionization positive mode. MS scans were analyzed at a resolution of 120,000, with a scan rate of 3.5 Hz. The automatic gain control (AGC) target was set to 1 × 106 ions, and the maximum injection time (IT) was at 100 ms. Source ionization parameters were optimized, and these include spray voltage, 3.0 kV; transfer temperature, 280°C; S-lens level, 40; heater temperature, 325°C; sheath gas, 40; aux gas, 10; and sweep gas flow, 1. Tandem spectra were acquired using a data-dependent acquisition (DDA) in which one MS scan is followed by 2, 4, or 6 tandem MS (MS/MS) scans. MS/MS scans are acquired using an isolation width of m/z 1.3, stepped normalized collision energy (NCE) of 20 and 40, and a dynamic exclusion for 6 s. MS/MS spectra were collected at a resolution of 15,000, with an AGC target set at 2 × 105 ions and maximum IT of 100 ms. Instrument performance and reproducibility in the run sequence were assessed by monitoring the retention times and peak areas for the heavy labeled standards added to the individual samples prior to and after metabolite extraction to assess sample processing steps and instrument variability (Table S3).
Quality metrics obtained for the heavy labeled standard molecules used for this study to assess the metabolite extraction, instrument performance, and injection volume reproducibility. Download Table S3, DOCX file, 1.1 MB (1.1MB, docx) .
Copyright © 2020 Chambers et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Metabolomics data processing.
UPLC-HRMS/MS raw data were imported, processed, normalized, and reviewed using Progenesis QI v.2.1 (Nonlinear Dynamics, Newcastle, UK). All MS and MS/MS sample runs were aligned against a quality control (pooled) reference run, and peak picking was performed on individual aligned runs to create an aggregate data set. Following peak picking, unique spectral features (retention time and m/z pairs) were grouped based on adducts and isotopes, and individual features or metabolites were normalized to all features. Compounds with <25% coefficient of variance (CV) were retained for further analysis. P values were calculated by Progenesis QI using variance-stabilized measurements achieved through log normalization, and metabolites with a P value of <0.05 calculated by one-way analysis of variance (ANOVA) and with a fold change (FC) of >|2| were considered significant.
Tentative and putative identifications were performed within Progenesis QI using accurate mass measurements (<5 ppm error), isotope distribution similarity, and fragmentation spectrum matching based on database searches against the Human Metabolome Database (HMDB), METLIN and National Institute of Standards and Technology (NIST) databases, and an in-house database (72–76). Annotations from both RPLC and HILIC analyses were performed for all significant compounds (P < 0.05, FC > |2|). Annotations were further analyzed using pathway overrepresentation analysis using MetaboAnalyst 4.0 (77, 78). The level system for metabolite identification confidence was used. The level 3 (L3) of confidence for the metabolite identifications was assigned for those molecules that showed minimal experimental evidence compared to level 2 (L2) but do prioritize a top candidate. These are accepted by the metabolomics community and represent families of molecules that cannot be distinguished by the data acquired, predominantly because there are too many isomers as possible candidate metabolites, but the family trends can be informative as well.
ACKNOWLEDGMENTS
This work was supported by the National Science Foundation (CAREER award to S.D.T., CHE-1847804). S.D.T. is supported by a Dean’s Faculty Fellowship from the College of Arts & Science at Vanderbilt University. J.A.G., D.M.A., and S.D.M. acknowledge support from the NIH under grants R01-HD090061, R01-AI134036, and U01-TR02398, and from the March of Dimes Foundation. H.C.T. was supported by an undergraduate research fellowship from the Fleischer family.
The donor mothers are acknowledged for their generous contributions.
Footnotes
This article is a direct contribution from David M. Aronoff, a Fellow of the American Academy of Microbiology, who arranged for and secured reviews by Christian Melander, University of Notre Dame; Amit Basu, Brown University; and Xin Zhang, The Pennsylvania State University.
Citation Chambers SA, Moore RE, Craft KM, Thomas HC, Das R, Manning SD, Codreanu SG, Sherrod SD, Aronoff DM, McLean JA, Gaddy JA, Townsend SD. 2020. A solution to antifolate resistance in group B Streptococcus: untargeted metabolomics identifies human milk oligosaccharide-induced perturbations that result in potentiation of trimethoprim. mBio 11:e00076-20. https://doi.org/10.1128/mBio.00076-20.
REFERENCES
- 1.Laxminarayan R, Duse A, Wattal C, Zaidi AK, Wertheim HF, Sumpradit N, Vlieghe E, Hara GL, Gould IM, Goossens H, Greko C, So AD, Bigdeli M, Tomson G, Woodhouse W, Ombaka E, Peralta AQ, Qamar FN, Mir F, Kariuki S, Bhutta ZA, Coates A, Bergstrom R, Wright GD, Brown ED, Cars O. 2013. Antibiotic resistance-the need for global solutions. Lancet Infect Dis 13:1057–1098. doi: 10.1016/S1473-3099(13)70318-9. [DOI] [PubMed] [Google Scholar]
- 2.Melander RJ, Melander C. 2017. The challenge of overcoming antibiotic resistance: an adjuvant approach? ACS Infect Dis 3:559–563. doi: 10.1021/acsinfecdis.7b00071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Worthington RJ, Melander C. 2013. Combination approaches to combat multidrug-resistant bacteria. Trends Biotechnol 31:177–184. doi: 10.1016/j.tibtech.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wright GD. 2016. Antibiotic adjuvants: rescuing antibiotics from resistance. Trends Microbiol 24:862–871. doi: 10.1016/j.tim.2016.06.009. [DOI] [PubMed] [Google Scholar]
- 5.Calza L, Manfredi R, Marinacci G, Fortunato L, Chiodo F. 2003. Ampicillin, gentamicin and teicoplanin as antimicrobial therapy for recurrent Streptococcus agalactiae and Enterococcus faecalis endocarditis in an intravenous drug abuser with HIV infection. Chemotherapy 49:206–208. doi: 10.1159/000071146. [DOI] [PubMed] [Google Scholar]
- 6.Barnes AI, Herrero IL, Albesa I. 2005. New aspect of the synergistic antibacterial action of ampicillin and gentamicin. Int J Antimicrob Agents 26:146–151. doi: 10.1016/j.ijantimicag.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 7.Fouhy F, Guinane CM, Hussey S, Wall R, Ryan CA, Dempsey EM, Murphy B, Ross RP, Fitzgerald GF, Stanton C, Cotter PD. 2012. High-throughput sequencing reveals the incomplete, short-term recovery of infant gut microbiota following parenteral antibiotic treatment with ampicillin and gentamicin. Antimicrob Agents Chemother 56:5811–5820. doi: 10.1128/AAC.00789-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tsai MH, Hsu JF, Lai MY, Lin LC, Chu SM, Huang HR, Chiang MC, Fu RH, Lu JJ. 2019. Molecular characteristics and antimicrobial resistance of group B Streptococcus strains causing invasive disease in neonates and adults. Front Microbiol 10:264. doi: 10.3389/fmicb.2019.00264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fox BC. 1994. Delayed-onset postpartum meningitis due to group B streptococcus. Clin Infect Dis 19:350. doi: 10.1093/clinids/19.2.350. [DOI] [PubMed] [Google Scholar]
- 10.Krause JC, Ghandil P, Chrabieh M, Casanova JL, Picard C, Puel A, Creech CB. 2009. Very late-onset group B Streptococcus meningitis, sepsis, and systemic shigellosis due to interleukin-1 receptor-associated kinase-4 deficiency. Clin Infect Dis 49:1393–1396. doi: 10.1086/630206. [DOI] [PubMed] [Google Scholar]
- 11.Lazarus JM, Sellers DP, Marine WM. 1965. Meningitis due to the group B beta-hemolytic Streptococcus. N Engl J Med 272:146–147. doi: 10.1056/NEJM196501212720308. [DOI] [PubMed] [Google Scholar]
- 12.Nakstad B, Sonerud T, Solevag AL. 2016. Early detection of neonatal group B streptococcus sepsis and the possible diagnostic utility of IL-6, IL-8, and CD11b in a human umbilical cord blood in vitro model. Infect Drug Resist 9:171–179. doi: 10.2147/IDR.S106181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Faro S. 1980. Group B streptococcus and puerperal sepsis. Am J Obstet Gynecol 138:1219–1220. doi: 10.1016/S0002-9378(16)32795-8. [DOI] [PubMed] [Google Scholar]
- 14.Jawa G, Hussain Z, da Silva O. 2013. Recurrent late-onset group B Streptococcus sepsis in a preterm infant acquired by expressed breastmilk transmission: a case report. Breastfeed Med 8:134–136. doi: 10.1089/bfm.2012.0016. [DOI] [PubMed] [Google Scholar]
- 15.Ying Q, Wang S, Lou X, Ding J, Ding J. 2019. Burden and risk factors of invasive group B Streptococcus disease among neonates in a Chinese maternity hospital. BMC Infect Dis 19:123. doi: 10.1186/s12879-018-3660-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seale AC, Baker CJ, Berkley JA, Madhi SA, Ordi J, Saha SK, Schrag SJ, Sobanjo-Ter Meulen A, Vekemans J. 2019. Vaccines for maternal immunization against group B Streptococcus disease: WHO perspectives on case ascertainment and case definitions. Vaccine 37:4877–4885. doi: 10.1016/j.vaccine.2019.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Edwards JM, Watson N, Focht C, Wynn C, Todd CA, Walter EB, Heine RP, Swamy GK. 2019. Group B streptococcus (GBS) colonization and disease among pregnant women: a historical cohort study. Infect Dis Obstet Gynecol 2019:5430493. doi: 10.1155/2019/5430493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Gier B, van Kassel MN, Sanders EAM, van de Beek D, Hahné SJM, van der Ende A, Bijlsma MW. 2019. Disease burden of neonatal invasive group B Streptococcus infection in the Netherlands. PLoS One 14:e0216749. doi: 10.1371/journal.pone.0216749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Berardi A, Spada C, Reggiani MLB, Creti R, Baroni L, Capretti MG, Ciccia M, Fiorini V, Gambini L, Gargano G, Papa I, Piccinini G, Rizzo V, Sandri F, Lucaccioni L, GBS Prevention Working Group of Emilia-Romagna. 2019. Group B Streptococcus early-onset disease and observation of well-appearing newborns. PLoS One 14:e0212784. doi: 10.1371/journal.pone.0212784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mahieu LM, De Dooy JJ, Leys E. 2000. Obstetricians’ compliance with CDC guidelines on maternal screening and intrapartum prophylaxis for group B streptococcus. J Obstet Gynaecol 20:460–464. doi: 10.1080/014436100434596. [DOI] [PubMed] [Google Scholar]
- 21.Moraleda C, Benmessaoud R, Esteban J, Lopez Y, Alami H, Barkat A, Houssain T, Kabiri M, Bezad R, Chaacho S, Madrid L, Vila J, Munoz-Almagro C, Bosch J, Soto SM, Bassat Q. 2018. Prevalence, antimicrobial resistance and serotype distribution of group B streptococcus isolated among pregnant women and newborns in Rabat, Morocco. J Med Microbiol 67:652–661. doi: 10.1099/jmm.0.000720. [DOI] [PubMed] [Google Scholar]
- 22.Li YP, Kuok CM, Lin SY, Hsieh WS, Shyu MK. 2016. Group B streptococcus antimicrobial resistance in neonates born to group B streptococcus-colonized mothers: single-center survey. J Obstet Gynaecol Res 42:1471–1475. doi: 10.1111/jog.13082. [DOI] [PubMed] [Google Scholar]
- 23.Phares CR, Lynfield R, Farley MM, Mohle-Boetani J, Harrison LH, Petit S, Craig AS, Schaffner W, Zansky SM, Gershman K, Stefonek KR, Albanese BA, Zell ER, Schuchat A, Schrag SJ, Active Bacterial Core Surveillance/Emerging Infections Program Network. 2008. Epidemiology of invasive group B streptococcal disease in the United States, 1999–2005. JAMA 299:2056–2065. doi: 10.1001/jama.299.17.2056. [DOI] [PubMed] [Google Scholar]
- 24.Johri AK, Paoletti LC, Glaser P, Dua M, Sharma PK, Grandi G, Rappuoli R. 2006. Group B Streptococcus: global incidence and vaccine development. Nat Rev Microbiol 4:932–942. doi: 10.1038/nrmicro1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Craft KM, Townsend SD. 2019. Mother knows best: deciphering the antibacterial properties of human milk oligosaccharides. Acc Chem Res 52:760–768. doi: 10.1021/acs.accounts.8b00630. [DOI] [PubMed] [Google Scholar]
- 26.Ackerman DL, Craft KM, Doster RS, Weitkamp JH, Aronoff DM, Gaddy JA, Townsend SD. 2018. Antimicrobial and antibiofilm activity of human milk oligosaccharides against Streptococcus agalactiae, Staphylococcus aureus, and Acinetobacter baumannii. ACS Infect Dis 4:315–324. doi: 10.1021/acsinfecdis.7b00183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ackerman DL, Doster RS, Weitkamp JH, Aronoff DM, Gaddy JA, Townsend SD. 2017. Human milk oligosaccharides exhibit antimicrobial and antibiofilm properties against group B Streptococcus. ACS Infect Dis 3:595–605. doi: 10.1021/acsinfecdis.7b00064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Craft KM, Thomas HC, Townsend SD. 2018. Interrogation of human milk oligosaccharide fucosylation patterns for antimicrobial and antibiofilm trends in group B Streptococcus. ACS Infect Dis 4:1755–1765. doi: 10.1021/acsinfecdis.8b00234. [DOI] [PubMed] [Google Scholar]
- 29.Craft KM, Thomas HC, Townsend SD. 2018. Sialylated variants of lacto-N-tetraose exhibit antimicrobial activity against group B Streptococcus. Org Biomol Chem 17:1893–1900. doi: 10.1039/c8ob02080a. [DOI] [PubMed] [Google Scholar]
- 30.Craft KM, Townsend SD. 2017. Synthesis of lacto-N-tetraose. Carbohydr Res 440–441:43–50. doi: 10.1016/j.carres.2017.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Craft KM, Townsend SD. 2019. 1-Amino-2′-fucosyllactose inhibits biofilm formation by Streptococcus agalactiae. J Antibiot (Tokyo) 72:507–512. doi: 10.1038/s41429-019-0151-6. [DOI] [PubMed] [Google Scholar]
- 32.Craft KM, Gaddy JA, Townsend SD. 2018. Human milk oligosaccharides (HMOs) sensitize group B Streptococcus to clindamycin, erythromycin, gentamicin, and minocycline on a strain specific basis. ACS Chem Biol 13:2020–2026. doi: 10.1021/acschembio.8b00661. [DOI] [PubMed] [Google Scholar]
- 33.Morales WJ, Dickey SS, Bornick P, Lim DV. 1999. Change in antibiotic resistance of group B streptococcus: impact on intrapartum management. Am J Obstet Gynecol 181:310–314. doi: 10.1016/S0002-9378(99)70553-3. [DOI] [PubMed] [Google Scholar]
- 34.Back EE, O'Grady EJ, Back JD. 2012. High rates of perinatal group B Streptococcus clindamycin and erythromycin resistance in an upstate New York hospital. Antimicrob Agents Chemother 56:739–742. doi: 10.1128/AAC.05794-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Buu-Hoï A, Le Bouguenec C, Horaud T. 1990. High-level chromosomal gentamicin resistance in Streptococcus agalactiae (group B). Antimicrob Agents Chemother 34:985–988. doi: 10.1128/aac.34.6.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mariani-Kurkdjian P, Benayoun E, Bingen E, Aujard Y. 2003. Group B streptococcus and high level resistance to aminoglycosides: therapeutic implications for neonates. Arch Pediatr 10:736–737. (In French.) doi: 10.1016/S0929-693X(03)00344-0. [DOI] [PubMed] [Google Scholar]
- 37.Clifford V, Heffernan HM, Grimwood K, Garland S, Australasian GBS Resistance Study Group. 2011. Variation in erythromycin and clindamycin resistance patterns between New Zealand and Australian group B streptococcus isolates. Aust N Z J Obstet Gynaecol 51:328–332. doi: 10.1111/j.1479-828X.2011.01302.x. [DOI] [PubMed] [Google Scholar]
- 38.Ji W, Liu H, Jin Z, Wang A, Mu X, Qin X, Wang W, Gao C, Zhu Y, Feng X, Lei J, She S, Jiang L, Liu J, Yang S, Liu Z, Li G, Li Q, Guo D, Aziz MM, Gillani AH, Fang Y. 2017. Disease burden and antimicrobial resistance of invasive group B streptococcus among infants in China: a protocol for a national prospective observational study. BMC Infect Dis 17:377. doi: 10.1186/s12879-017-2475-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Manning SD, Pearlman MD, Tallman P, Pierson CL, Foxman B. 2001. Frequency of antibiotic resistance among group B Streptococcus isolated from healthy college students. Clin Infect Dis 33:E137–E139. doi: 10.1086/324588. [DOI] [PubMed] [Google Scholar]
- 40.Manning SD, Schaeffer KE, Springman AC, Lehotzky E, Lewis MA, Ouellette LM, Wu G, Moorer GM, Whittam TS, Davies HD. 2008. Genetic diversity and antimicrobial resistance in group B streptococcus colonizing young, nonpregnant women. Clin Infect Dis 47:388–390. doi: 10.1086/589864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Campisi E, Rosini R, Ji W, Guidotti S, Rojas-Lopez M, Geng G, Deng Q, Zhong H, Wang W, Liu H, Nan C, Margarit I, Rinaudo CD. 2016. Genomic analysis reveals multi-drug resistance clusters in group B Streptococcus CC17 hypervirulent isolates causing neonatal invasive disease in southern mainland China. Front Microbiol 7:1265. doi: 10.3389/fmicb.2016.01265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lambiase A, Agangi A, Del Pezzo M, Quaglia F, Testa A, Rossano F, Martinelli P, Catania MR. 2012. In vitro resistance to macrolides and clindamycin by group B Streptococcus isolated from pregnant and nonpregnant women. Infect Dis Obstet Gynecol 2012:913603. doi: 10.1155/2012/913603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wormser GP, Keusch GT, Heel RC. 1982. Co-trimoxazole (trimethoprim-sulfamethoxazole): an updated review of its antibacterial activity and clinical efficacy. Drugs 24:459–518. doi: 10.2165/00003495-198224060-00002. [DOI] [PubMed] [Google Scholar]
- 44.Quinlivan EP, McPartlin J, Weir DG, Scott J. 2000. Mechanism of the antimicrobial drug trimethoprim revisited. FASEB J 14:2519–2524. doi: 10.1096/fj.99-1037com. [DOI] [PubMed] [Google Scholar]
- 45.Bergmann R, van der Linden M, Chhatwal GS, Nitsche-Schmitz DP. 2014. Factors that cause trimethoprim resistance in Streptococcus pyogenes. Antimicrob Agents Chemother 58:2281–2288. doi: 10.1128/AAC.02282-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bergmann R, Sagar V, Nitsche-Schmitz DP, Chhatwal GS. 2012. First detection of trimethoprim resistance determinant dfrG in Streptococcus pyogenes clinical isolates in India. Antimicrob Agents Chemother 56:5424–5425. doi: 10.1128/AAC.01284-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kärpänoja P, Nyberg ST, Bergman M, Voipio T, Paakkari P, Huovinen P, Sarkkinen H, Finnish Study Group for Antimicrobial Resistance (FiRe Network). 2008. Connection between trimethoprim-sulfamethoxazole use and resistance in Streptococcus pneumoniae, Haemophilus influenzae, and Moraxella catarrhalis. Antimicrob Agents Chemother 52:2480–2485. doi: 10.1128/AAC.01118-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dueger EL, Asturias EJ, Matheu J, Gordillo R, Torres O, Halsey N. 2008. Increasing penicillin and trimethoprim-sulfamethoxazole resistance in nasopharyngeal Streptococcus pneumoniae isolates from Guatemalan children, 2001–2006. Int J Infect Dis 12:289–297. doi: 10.1016/j.ijid.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schmitz FJ, Perdikouli M, Beeck A, Verhoef J, Fluit AC, European SENTRY participants. 2001. Resistance to trimethoprim-sulfamethoxazole and modifications in genes coding for dihydrofolate reductase and dihydropteroate synthase in European Streptococcus pneumoniae isolates. J Antimicrob Chemother 48:935–936. doi: 10.1093/jac/48.6.935. [DOI] [PubMed] [Google Scholar]
- 50.Rudolph KM, Parkinson AJ, Roberts MC. 2001. Mechanisms of erythromycin and trimethoprim resistance in the Alaskan Streptococcus pneumoniae serotype 6B clone. J Antimicrob Chemother 48:317–319. doi: 10.1093/jac/48.2.317. [DOI] [PubMed] [Google Scholar]
- 51.Lovgren M, Dell'Acqua L, Palacio R, Echániz-Aviles G, Soto-Noguerón A, Castañeda E, Agudelo CI, Heitmann I, Brandileone MC, Zanella RC, Rossi A, Pace J, Talbot JA. 1999. Determination of trimethoprim-sulfamethoxazole resistance in Streptococcus pneumoniae by using the E test with Mueller-Hinton agar supplemented with sheep or horse blood may be unreliable. The Pneumococcal Study Group. J Clin Microbiol 37:215–217. doi: 10.1128/JCM.37.1.215-217.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wilkinson HW. 1977. Nontypable group B streptococci isolated from human sources. J Clin Microbiol 6:183–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Davies HD, Adair C, McGeer A, Ma D, Robertson S, Mucenski M, Kowalsky L, Tyrell G, Baker CJ. 2001. Antibodies to capsular polysaccharides of group B Streptococcus in pregnant Canadian women: relationship to colonization status and infection in the neonate. J Infect Dis 184:285–291. doi: 10.1086/322029. [DOI] [PubMed] [Google Scholar]
- 54.Manning SD, Lewis MA, Springman AC, Lehotzky E, Whittam TS, Davies HD. 2008. Genotypic diversity and serotype distribution of group B streptococcus isolated from women before and after delivery. Clin Infect Dis 46:1829–1837. doi: 10.1086/588296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cieslewicz MJ, Chaffin D, Glusman G, Kasper D, Madan A, Rodrigues S, Fahey J, Wessels MR, Rubens CE. 2005. Structural and genetic diversity of group B streptococcus capsular polysaccharides. Infect Immun 73:3096–3103. doi: 10.1128/IAI.73.5.3096-3103.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Johri AK, Lata H, Yadav P, Dua M, Yang Y, Xu X, Homma A, Barocchi MA, Bottomley MJ, Saul A, Klugman KP, Black S. 2013. Epidemiology of group B Streptococcus in developing countries. Vaccine 31(Suppl 4):D43–D45. doi: 10.1016/j.vaccine.2013.05.094. [DOI] [PubMed] [Google Scholar]
- 57.Melin P, Efstratiou A. 2013. Group B streptococcal epidemiology and vaccine needs in developed countries. Vaccine 31(Suppl 4):D31–D42. doi: 10.1016/j.vaccine.2013.05.012. [DOI] [PubMed] [Google Scholar]
- 58.Corominas-Faja B, Quirantes-Piné R, Oliveras-Ferraros C, Vazquez-Martin A, Cufí S, Martin-Castillo B, Micol V, Joven J, Segura-Carretero A, Menendez JA. 2012. Metabolomic fingerprint reveals that metformin impairs one-carbon metabolism in a manner similar to the antifolate class of chemotherapy drugs. Aging (Albany NY) 4:480–498. doi: 10.18632/aging.100472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Strahl H, Errington J. 2017. Bacterial membranes: structure, domains, and function. Annu Rev Microbiol 71:519–538. doi: 10.1146/annurev-micro-102215-095630. [DOI] [PubMed] [Google Scholar]
- 60.Salsinha AS, Pimentel LL, Fontes AL, Gomes AM, Rodríguez-Alcalá LM. 2018. Microbial production of conjugated linoleic acid and conjugated linolenic acid relies on a multienzymatic system. Microbiol Mol Biol Rev 82:e00019-18. doi: 10.1128/MMBR.00019-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Devillard E, McIntosh FM, Duncan SH, Wallace RJ. 2007. Metabolism of linoleic acid by human gut bacteria: different routes for biosynthesis of conjugated linoleic acid. J Bacteriol 189:2566–2570. doi: 10.1128/JB.01359-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ha J, Dobretsov M, Kurten RC, Grant DF, Stimers JR. 2002. Effect of linoleic acid metabolites on Na+/K+ pump current in N20.1 oligodendrocytes: role of membrane fluidity. Toxicol Appl Pharmacol 182:76–83. doi: 10.1006/taap.2002.9435. [DOI] [PubMed] [Google Scholar]
- 63.Dalebroux ZD. 2017. Cues from the membrane: bacterial glycerophospholipids. J Bacteriol 199:e00136-17. doi: 10.1128/JB.00136-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Epand RM, Walker C, Epand RF, Magarvey NA. 2016. Molecular mechanisms of membrane targeting antibiotics. Biochim Biophys Acta 1858:980–987. doi: 10.1016/j.bbamem.2015.10.018. [DOI] [PubMed] [Google Scholar]
- 65.Zampieri M, Szappanos B, Buchieri MV, Trauner A, Piazza I, Picotti P, Gagneux S, Borrell S, Gicquel B, Lelievre J, Papp B, Sauer U. 2018. High-throughput metabolomic analysis predicts mode of action of uncharacterized antimicrobial compounds. Sci Transl Med 10:eaal3973. doi: 10.1126/scitranslmed.aal3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vincent IM, Ehmann DE, Mills SD, Perros M, Barrett MP. 2016. Untargeted metabolomics to ascertain antibiotic modes of action. Antimicrob Agents Chemother 60:2281–2291. doi: 10.1128/AAC.02109-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Baptista R, Fazakerley DM, Beckmann M, Baillie L, Mur LAJ. 2018. Untargeted metabolomics reveals a new mode of action of pretomanid (PA-824). Sci Rep 8:5084. doi: 10.1038/s41598-018-23110-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Heaver SL, Johnson EL, Ley RE. 2018. Sphingolipids in host-microbial interactions. Curr Opin Microbiol 43:92–99. doi: 10.1016/j.mib.2017.12.011. [DOI] [PubMed] [Google Scholar]
- 69.Heung LJ, Luberto C, Del Poeta M. 2006. Role of sphingolipids in microbial pathogenesis. Infect Immun 74:28–39. doi: 10.1128/IAI.74.1.28-39.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dunn WB, Broadhurst DI, Atherton HJ, Goodacre R, Griffin JL. 2011. Systems level studies of mammalian metabolomes: the roles of mass spectrometry and nuclear magnetic resonance spectroscopy. Chem Soc Rev 40:387–426. doi: 10.1039/b906712b. [DOI] [PubMed] [Google Scholar]
- 71.Lin VY, Fain MD, Jackson PL, Berryhill TF, Wilson LS, Mazur M, Barnes SJ, Blalock JE, Raju SV, Rowe SM. 2019. Vaporized E-cigarette liquids induce ion transport dysfunction in airway epithelia. Am J Respir Cell Mol Biol 61:162–173. doi: 10.1165/rcmb.2017-0432OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wishart DS, Tzur D, Knox C, Eisner R, Guo AC, Young N, Cheng D, Jewell K, Arndt D, Sawhney S, Fung C, Nikolai L, Lewis M, Coutouly MA, Forsythe I, Tang P, Shrivastava S, Jeroncic K, Stothard P, Amegbey G, Block D, Hau DD, Wagner J, Miniaci J, Clements M, Gebremedhin M, Guo N, Zhang Y, Duggan GE, Macinnis GD, Weljie AM, Dowlatabadi R, Bamforth F, Clive D, Greiner R, Li L, Marrie T, Sykes BD, Vogel HJ, Querengesser L. 2007. HMDB: the Human Metabolome Database. Nucleic Acids Res 35:D521–D526. doi: 10.1093/nar/gkl923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wishart DS, Knox C, Guo AC, Eisner R, Young N, Gautam B, Hau DD, Psychogios N, Dong E, Bouatra S, Mandal R, Sinelnikov I, Xia J, Jia L, Cruz JA, Lim E, Sobsey CA, Shrivastava S, Huang P, Liu P, Fang L, Peng J, Fradette R, Cheng D, Tzur D, Clements M, Lewis A, De Souza A, Zuniga A, Dawe M, Xiong Y, Clive D, Greiner R, Nazyrova A, Shaykhutdinov R, Li L, Vogel HJ, Forsythe I. 2009. HMDB: a knowledgebase for the human metabolome. Nucleic Acids Res 37:D603–D610. doi: 10.1093/nar/gkn810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wishart DS, Jewison T, Guo AC, Wilson M, Knox C, Liu Y, Djoumbou Y, Mandal R, Aziat F, Dong E, Bouatra S, Sinelnikov I, Arndt D, Xia J, Liu P, Yallou F, Bjorndahl T, Perez-Pineiro R, Eisner R, Allen F, Neveu V, Greiner R, Scalbert A. 2013. HMDB 3.0–the Human Metabolome Database in 2013. Nucleic Acids Res 41:D801–D807. doi: 10.1093/nar/gks1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wishart DS, Feunang YD, Marcu A, Guo AC, Liang K, Vázquez-Fresno R, Sajed T, Johnson D, Li C, Karu N, Sayeeda Z, Lo E, Assempour N, Berjanskii M, Singhal S, Arndt D, Liang Y, Badran H, Grant J, Serra-Cayuela A, Liu Y, Mandal R, Neveu V, Pon A, Knox C, Wilson M, Manach C, Scalbert A. 2018. HMDB 4.0: the human metabolome database for 2018. Nucleic Acids Res 46:D608–D617. doi: 10.1093/nar/gkx1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Smith CA, O'Maille G, Want EJ, Qin C, Trauger SA, Brandon TR, Custodio DE, Abagyan R, Siuzdak G. 2005. METLIN: a metabolite mass spectral database. Ther Drug Monit 27:747–751. doi: 10.1097/01.ftd.0000179845.53213.39. [DOI] [PubMed] [Google Scholar]
- 77.Xia J, Wishart DS. 2016. Using MetaboAnalyst 3.0 for comprehensive metabolomics data analysis. Curr Protoc Bioinformatics 55:14.10.1–14.10.91. doi: 10.1002/cpbi.11. [DOI] [PubMed] [Google Scholar]
- 78.Schrimpe-Rutledge AC, Codreanu SG, Sherrod SD, McLean JA. 2016. Untargeted metabolomics strategies-challenges and emerging directions. J Am Soc Mass Spectrom 27:1897–1905. doi: 10.1007/s13361-016-1469-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) IC50 curve of HMO cocktail for GB2 in THB. Bacterial growth (OD600) was recorded after 24 h of HMO treatment at increasing HMO concentrations. MIC, 2.56 mg/ml; IC50, 1.897 mg/ml; IC25, 1.43 mg/ml; Hill slope, −3.894. (B) IC50 curve of HMO cocktail for GB590 in THB. Bacterial growth (OD600) was recorded after 24 h of HMO treatment at increasing HMO concentrations. MIC = 5.125 mg/ml, IC50 ∼ 2.417 mg/ml, IC25 ∼2.30 mg/ml, hill slope = −22.37. (C) IC50 curve of HMO cocktail for GB CNCTC 10/84 in THB. Bacterial growth (OD600) was recorded after 24 h of HMO treatment at increasing HMO concentrations. MIC = 5.125 mg/ml, IC50 = 2.05 mg/ml, IC25 = 1.42 mg/ml, hill slope = −2.98. (D) IC50 curve of HMO cocktail for GB83 in THB. Bacterial growth (OD600) was recorded after 24 h of HMO treatment at increasing HMO concentrations. MIC = 5.125 mg/ml, IC50 = 3.616 mg/ml, IC25 = 3.51 mg/ml, hill slope = −39.06. (E) IC50 curve of HMO cocktail for GB651 in THB. Bacterial growth (OD600) was recorded after 24 h of HMO treatment at increasing HMO concentrations. MIC = 5.125 mg/ml, IC50 = 2.616 mg/ml, IC25 = 2.33 mg/ml, hill slope = −9.639. Download FIG S1, DOCX file, 0.1 MB (59KB, docx) .
Copyright © 2020 Chambers et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
(A) Checkerboard assay in GB2 showing the synergistic combinations between TMP and the HMO cocktail (bolded). (B) Checkerboard assay in GB590 showing the synergistic combinations between TMP and the HMO cocktail (bold). The combination is considered synergistic when the ΣFIC is ≤0.5, additive or indifferent when the ΣFIC is >0.5 to <4, and antagonistic when the ΣFIC is ≥4. Download FIG S2, DOCX file, 0.1 MB (24.2KB, docx) .
Copyright © 2020 Chambers et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Global principal-component analysis (PCA) of cellular extracted metabolites for the two biological conditions is illustrating a distinct shift in metabolic profiles between groups. Presenting the abundance data in PC space allows us to separate the run samples based on overall variability and shows a clear separation of untreated samples versus HMO-treated samples across the first PC dimension (x axis). Download FIG S3, DOCX file, 0.1 MB (88KB, docx) .
Copyright © 2020 Chambers et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Linoleic acid metabolism supplementary images. (A) Heat map visualization of the differently regulated members within the linoleic acid metabolic pathway upon HMO treatment. Linoleic acid metabolism members shown here were detected by HILIC-positive LC-MS/MS analysis. (B) Red nodes reveal individual significant metabolites matched to the linoleic acid metabolic pathway. The KEGG C number is shown within a node. The individual significant metabolites (P < 0.05) identified at confidence level L3 are highlighted in red for the linoleic acid metabolism, generated by the MetaboAnalyst 4.0 software. Download FIG S4, DOCX file, 0.2 MB (206.4KB, docx) .
Copyright © 2020 Chambers et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Glycerophospholipid metabolism supplementary images. (A) Heat map visualization of the differently regulated members within the glycerophospholipid metabolic pathway upon HMO treatment. Glycerophospholipid metabolism members shown here were detected by RPLC-positive LC-MS/MS analysis. (B) Red nodes reveal individual significant metabolites matched to the glycerophospholipid metabolic pathway. The KEGG C number is shown within a node. The individual significant metabolites (P < 0.05) identified at confidence level L3 are highlighted in red for the glycerophospholipid metabolism, generated by the MetaboAnalyst 4.0 software. Download FIG S5, DOCX file, 0.4 MB (370.4KB, docx) .
Copyright © 2020 Chambers et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Heat map visualization of the differently regulated members within the cell wall synthesis-associated metabolites upon HMO treatment. Metabolites shown here were detected by HILIC-positive LC-MS/MS analysis. Download FIG S6, DOCX file, 0.1 MB (40.8KB, docx) .
Copyright © 2020 Chambers et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Group B Streptococcus strains evaluated in the study. *ST, sequence type as defined by multilocus sequence typing (MLST). Download Table S1, DOCX file, 0.1 MB (13.3KB, docx) .
Copyright © 2020 Chambers et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
TMP-sulfadiazine antibiotic combination assay. aTHB medium. bShown in μg/ml. c1:4.85 ratio of TMP-sulfadiazine. Download Table S2, DOCX file, 0.1 MB (13KB, docx) .
Copyright © 2020 Chambers et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Quality metrics obtained for the heavy labeled standard molecules used for this study to assess the metabolite extraction, instrument performance, and injection volume reproducibility. Download Table S3, DOCX file, 1.1 MB (1.1MB, docx) .
Copyright © 2020 Chambers et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.