Abstract
Background & Aims
The protective intestinal mucosal barrier consists of multiple elements including mucus and epithelial layers and immune defense; nonetheless, barrier dysfunction is common in various disorders. The imprinted and developmentally regulated long noncoding RNA H19 is involved in many cell processes and diseases. Here, we investigated the role of H19 in regulating Paneth and goblet cells and autophagy, and its impact on intestinal barrier dysfunction induced by septic stress.
Methods
Studies were conducted in H19-deficient (H19-/-) mice, mucosal tissues from patients with sepsis, primary enterocytes, and Caco-2 cells. Septic stress was induced by cecal ligation and puncture (CLP), and gut permeability was detected by tracer fluorescein isothiocyanate–dextran assays. The function of Paneth and goblet cells was examined by immunostaining for lysozyme and mucin 2, respectively, and autophagy was examined by microtubule-associated proteins 1A/1B light chain 3 II immunostaining and Western blot analysis. Intestinal organoids were isolated from H19-/- and control littermate mice and treated with lipopolysaccharide (LPS).
Results
Intestinal mucosal tissues in mice 24 hours after exposure to CLP and in patients with sepsis showed high H19 levels, associated with intestinal barrier dysfunction. Targeted deletion of the H19 gene in mice enhanced the function of Paneth and goblet cells and promoted autophagy in the small intestinal mucosa. Knockout of H19 protected Paneth and goblet cells against septic stress, preserved autophagy activation, and promoted gut barrier function after exposure to CLP. Compared with organoids from control littermate mice, intestinal organoids isolated from H19-/- mice had increased numbers of lysozyme- and mucin 2–positive cells and showed increased tolerance to LPS. Conversely, ectopic overexpression of H19 in cultured intestinal epithelial cells prevented rapamycin-induced autophagy and abolished the rapamycin-induced protection of the epithelial barrier against LPS.
Conclusions
In investigations of mice, human tissues, primary organoids, and intestinal epithelial cells, we found that increased H19 inhibited the function of Paneth and goblet cells and suppressed autophagy, thus potentially contributing to barrier dysfunction in intestinal pathologies.
Keywords: Long Noncoding RNAs, Gut permeability, Mucosal Defense, Paneth Cells, Goblet cells, Autophagy
Abbreviations used in this paper: AJ, adherens junction; CLP, cecal ligation and puncture; FITC, fluorescein isothiocyanate; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; IEC, intestinal epithelial cell; lncRNA, long noncoding RNA; LPS, lipopolysaccharide; miR, microRNA (miR)-675; miRNA, microRNA; TEER, transepithelial electrical resistance; TJ, tight junction; ZO-1, zonula occludens 1
Graphical abstract
Summary.
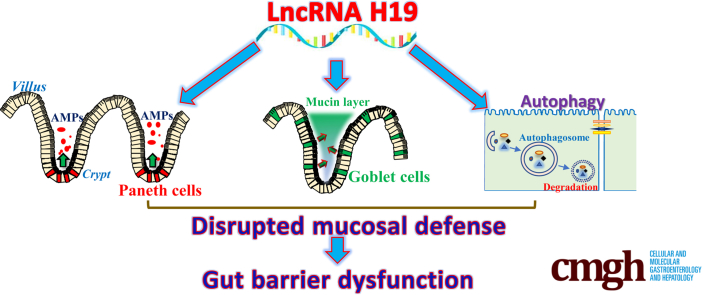
The intestinal barrier protects the subepithelial tissue against pathogens and noxious substances in the lumen. Here, we show that increased levels of long noncoding RNA H19 disrupt the intestinal barrier by inhibiting autophagy and repressing the function of Paneth and goblet cells, whereas targeted deletion of the H19 gene promotes the barrier function in response to septic stress.
The mammalian intestinal barrier is a specialized domain responding to and interacting with different luminal stimuli and the microbiome. The intestinal barrier consists of multiple elements, including a mucus layer, an epithelial layer, and a complex immune defense network that depends on the functions of innate and acquired immunity cells in the lamina propria.1,2 Surface mucus, predominantly composed of mucin 2 in the small and large intestine, is secreted by goblet cells and serves as the first physical defense in the barrier that prevents toxins, antigens, and bacteria from direct contact with the epithelium.3 Intestinal epithelial cells (IECs), connected by apical intercellular junctional complexes named tight junctions (TJs) and adherens junctions (AJs), establish a selectively permeable barrier that protects the subepithelial tissue against luminal noxious substances, but they also react to noxious stimuli by secreting different antimicrobial peptides and proteins.4,5 Paneth cells that reside at the bottom of the crypts produce high quantities of defensins and other antibiotic proteins such as lysozyme, Reg3 lectins, and phospholipase A2 when exposed to pathogenic bacteria and bacterial products such as lipopolysaccharide (LPS).6 In response to bacterial infection of the intestines, Paneth cells secrete lysozyme through secretory autophagy7 and their function is tightly regulated at the posttranscriptional level by the RNA binding protein HuR.8 Autophagy is a conserved intracellular pathway that sequesters cytoplasmic structures and pathogens targeted for degradation.9,10 Intestinal barrier dysfunction occurs commonly in various pathologies, leading to leaky gut and structural abnormalities of the epithelium.2
Many regions of the mammalian genome are transcribed into vast numbers of noncoding RNAs with active roles in gene regulation.11 Long noncoding RNAs (lncRNAs) are defined as transcripts spanning more than 200 nucleotides in length that share structural features with messenger RNAs such as the presence of a 5’-cap and a 3’-poly(A) tail.12-14 Although some lncRNAs are ubiquitous, lncRNAs often are expressed in specific tissues, differentiation stages, and cell types, and the levels of cellular lncRNAs can be altered rapidly in response to stressful environments.13 LncRNAs modulate a variety of biological functions and are involved in diverse human diseases by controlling gene expression at different levels, including chromatin remodeling, transcriptional and posttranscriptional processes, and protein metabolism.12,13 LncRNAs can modulate gene transcription, messenger RNA stability, or translation, and can work jointly with microRNAs (miRNAs), RNA binding proteins, and occasionally other molecules.15,16 Recent evidence has indicated that lncRNAs are an emerging class of master regulators of intestinal epithelium homeostasis and participate in the control of gut permeability, mucosal growth, and adaptation.5,17, 18, 19
Transcribed from the conserved imprinted H19/igf2 gene cluster, lncRNA H19 is implicated in different cellular processes.20,21 During embryogenesis, H19 expression levels increase in extraembryonic tissues, in the embryo itself, and in most fetal tissues, but its levels decrease after birth.22 During fetal development, H19 promotes expression of imprinted genes and inhibits embryonic placental growth.23 In adult tissues, H19 increases in disease conditions such as cancer,24,25 after exposure to hypoxia or estrogens,26,27 and in situations of inflammation.28 Targeted deletion of H19 in mice causes an overgrowth phenotype with increased body weight.20 The role of H19 in cancer development is complex because it can be tumor-suppressive or pro-oncogenic, depending on the cellular context of H19 and tumor types.24,27,29 The levels of H19 increase markedly in the inflamed human and murine intestinal mucosa, predominantly resulting from an increase in the inflammatory cytokine interleukin 22.28 We have reported that H19 overexpression inhibits expression of the TJ protein zonula occludens 1 (ZO-1) and the AJ protein E-cadherin at the post-transcriptional level via release of microRNA (miR)-675 embedded in H19 exon 1, and disrupts epithelial barrier function in an in vitro model using cultured IECs.18 Here, we investigated the in vivo function of H19 in the intestinal epithelium and show that ablating H19 in mice protects the intestinal barrier against septic stress by enhancing the function of Paneth and goblet cells and by promoting autophagy.
Results
H19 Levels Increase During Gut Barrier Dysfunction After Septic Stress
To determine the impact of H19 on gut barrier dysfunction in critical surgical conditions, we first examined changes in the levels of H19 in the small intestine of wild-type mice exposed to septic stress induced by cecal ligation and puncture (CLP). Twenty-four hours after CLP, mucosal H19 abundance increased dramatically in the small intestine, reaching 20-fold higher levels than in sham mice (Figure 1A). CLP did not alter the levels of U6 RNA in the intestinal mucosa but it decreased lncRNA uc.173 levels, which served as negative and positive controls, respectively. Sham surgery did not significantly affect mucosal H19 levels in the small intestine compared with mice without any surgery (data not shown). CLP stress also led to acute gut barrier dysfunction, as indicated by an increased mucosal permeability to fluorescein isothiocyanate (FITC) dextran (Figure 1B). In addition, focal villous necrosis in the small intestinal mucosa was observed 24 hours after CLP, but the damage was repaired within the next 48 hours, as reported previously.30 Second, we examined changes in the levels of H19 in the small intestinal mucosa from patients with sepsis. Tissue samples from patients without increased gut permeability, mucosal inflammation, or injury/erosions served as controls. As shown in Figure 1C (top), the basal levels of H19 in normal small intestinal mucosa obtained from control individuals were relatively low and almost undetectable as measured by RNA fluorescence in situ hybridization analysis. However, the intestinal mucosa from patients with sepsis showed a significant increase in the levels of H19, particularly in the villous area of the mucosa. The increase in mucosal H19 abundance in the intestine of patients with sepsis was associated with the epithelial barrier dysfunction, as indicated by a marked loss of the AJ protein E-cadherin (Figure 1C, middle) and the TJ protein ZO-1 (Figure 1C, bottom). These results suggest that increased levels of mucosal H19 in the small intestine are involved in the pathogenesis of gut barrier dysfunction in patients with critical surgical illnesses.
Figure 1.
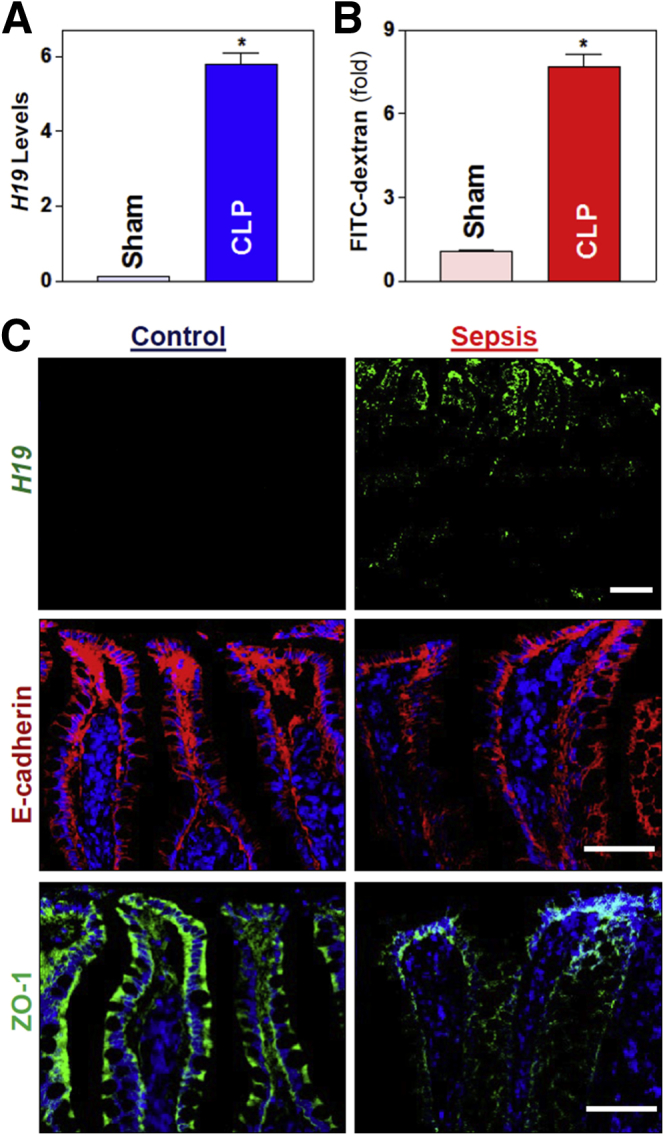
Septic stress increases mucosal H19 levels and disrupts epithelial barrier function. (A) Levels of H19 in the small intestinal mucosa of mice exposed to CLP for 24 hours as measured by reverse-transcription quantitative PCR analysis. Values are the means ± SEM (n = 5). *P < .05 compared with sham-treated mice. (B) Gut permeability in mice described in panel A. FITC dextran was given orally, and blood samples were collected 4 hours thereafter for measurement. (C) Association of increased H19 with reduction in the levels of intercellular junction proteins E-cadherin and ZO-1 in patients with sepsis. Top: in situ hybridization of H19 with fluorescent Locked nucleic acids-RNA detection probe in the small intestinal mucosa, as shown in green; middle: immunostaining of E-cadherin (red); and bottom: immunostaining of ZO-1 (green). Experiments were repeated in samples obtained from 3 patients with sepsis or controls and showed similar results. Scale bars: 50 μm.
H19 Deletion Enhances Paneth and Goblet Cell Numbers in the Intestinal Epithelium
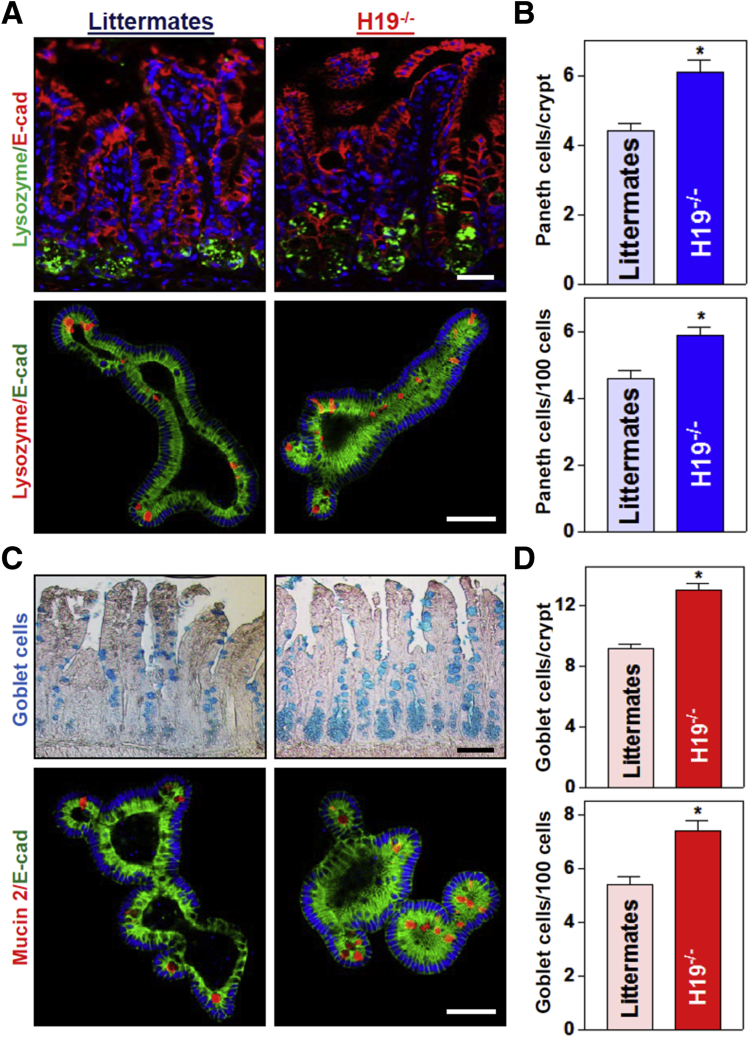
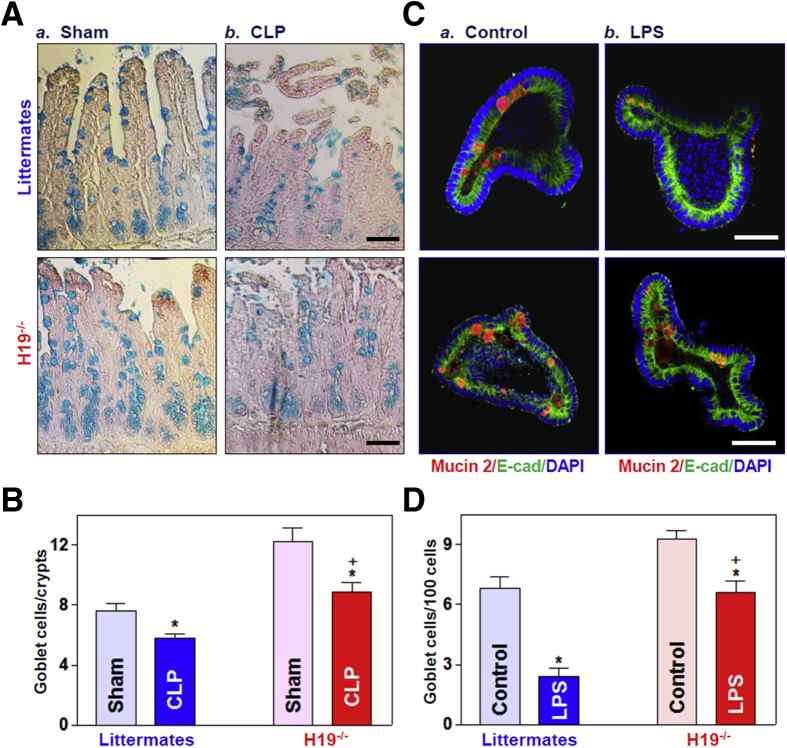
To define the in vivo function of H19 in the intestinal epithelium, we used H19ΔEx1/+ (H19-/-) mice bearing a maternal deletion of exon 1 in the H19 gene (obtained from Dr Karl Pfeifer, National Institutes of Health).31,32 Consistent with previous observations,31 targeted deletion of H19 did not alter gut development or mucosal growth under physiological conditions (data not shown). H19-/- mice looked normal overall and did not show significant differences in gastrointestinal gross morphology, mucosal histology, body weight, or general appearances as compared with control littermates. Interestingly, H19 knockout enhanced the functions of Paneth cells (lysozyme-positive cells) and goblet cells (determined by Alcian blue staining in vivo and mucin 2 immunostaining ex vivo) in the small intestinal mucosa. As reported,7,8 Paneth cells normally were located at the base of the crypt in littermate mice, but the numbers of these lysozyme-positive cells increased significantly in H19-/- mice relative to control littermate mice (Figure 2A and B, top panel). The number of lysozyme-positive cells in the intestinal epithelium from H19-deficient mice increased by approximately 20% compared with those from control littermate mice. In an ex vivo model, we also found increased numbers of Paneth cells in intestinal organoids isolated from H19-/- mice (Figure 2A, bottom panel). Intestinal organoids were grown from a single proliferating cell, but by 5 days after culture, the structures of organoids consisted of multiple cells and buds in the organoids from both H19-/- mice and control littermate mice. H19 deletion did not alter the growth of the intestinal organoids because the sizes and numbers of buds of intestinal organoids isolated from H19-/- mice were similar to those observed in organoids generated from control littermate mice. However, consistent with the observations in vivo, there were many more lysozyme-positive cells in organoids from H19-/- mice than in organoids from control littermate mice.
Figure 2.
Targeted deletion of H19 in mice increases the numbers of Paneth and goblet cells in the intestinal epithelium. (A) Immunostaining of Paneth cells (lysozyme-positive cells). Top: Paneth cells in the small intestinal mucosa of H19-/- and control littermate mice (green); bottom: Paneth cells in small intestinal organoids isolated from H19-/- and littermate mice (red). Scale bars: 50 μm. (B) Quantitative data of lysozyme-positive cells in the small intestinal epithelium described in panel A. Values are the means ± SEM (n = 5). *P < .05 compared with control littermate mice. (C) Immunostaining of Goblet cells (Alcian blue staining in vivo; mucin 2–positive cells ex vivo). Top: Goblet cells in the small intestinal mucosa of H19-/- and control littermate mice (blue); bottom: Goblet cells in small intestinal organoids isolated from H19-/- and littermate mice (red). Scale bars: 50 μm. (D) Quantitative data of mucin 2–positive cells in the small intestinal epithelium described in panel C. Values are the means ± SEM (n = 5). *P < .05 compared with control littermate mice. E-cad, E-cadherin.
Moreover, H19 deletion in mice also promoted goblet cell function in the small intestinal epithelium in vivo as well as ex vivo (Figure 2C and D). There were significant increases in the numbers of Alcian blue–positive cells in the small intestinal mucosa of H19-/- mice and mucin 2–positive cells in the H19-deficient intestinal organoids isolated from H19-/- mice when compared with those in control littermate mice. We observed that targeted deletion of H19 particularly increased the number of goblet cells in the crypt region rather than the villous area (Figure 2C, top right). On the other hand, H19 knockout did not affect function of mouse tuft cells in the small intestinal mucosa, as measured by Double cortin-like kinase 1 (DCLK1) immunostaining analysis (data not shown). Together, these results indicate that knockout of H19 enhances functions of Paneth and goblet cells in the small intestinal epithelium.
H19 Overexpression Blocks Increased Paneth and Goblet Cells in the H19-Deficient Intestinal Organoids
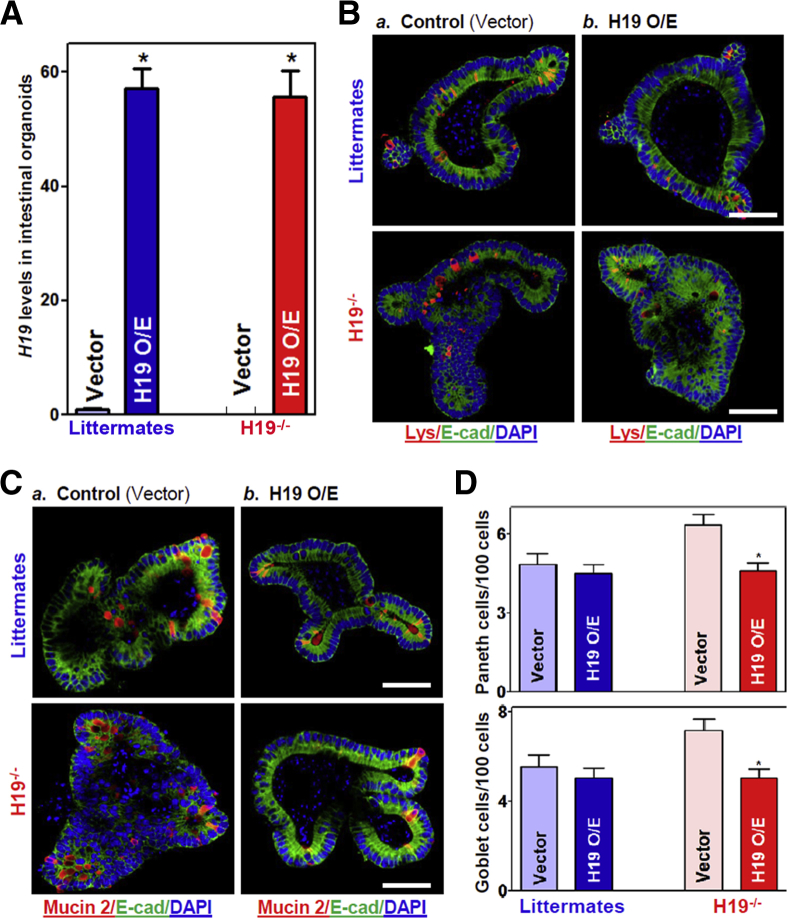
To examine the effect of increasing the levels of H19 on Paneth and goblet cell function, intestinal organoids isolated from both control littermate and H19-/- mice were transfected with a plasmid vector expressing H19 under control of the cytomegalovirus immediate early promoter/enhancer (pCMV) promoter as reported previously.18 The levels of H19 increased dramatically by transfection of the H19 expression vector; 48 hours later, organoid H19 levels increased strongly and specifically (Figure 3A), but lncRNA uc.230 and U6 RNA did not (data not shown). In the intestinal organoids generated from control littermate mice, increasing H19 levels slightly decreased the numbers of lysozyme- and mucin 2–positive cells, but this minor inhibition by H19 overexpression was not statistically significant (Figure 3B and C, top panels; D, left). In the H19-deficient organoids isolated from H19-/- mice, however, ectopically expressed H19 prevented the increase in the numbers of Paneth and goblet cells, as evidenced by a significant decrease in the numbers of lysozyme- and mucin 2–positive cells after transfection with the H19 expression vector compared with the empty control vector (Figure 3B and C, bottom panels; D, right). These data indicate that transient H19 overexpression prevented the increase in the numbers of Paneth and goblet cells in the H19-deficient intestinal organoids, but did not alter the levels of Paneth and goblet cells in intestinal organoids from control littermate mice.
Figure 3.
Ectopic overexpression of H19 prevents an increase in the numbers of Paneth and goblet cells in intestinal organoids isolated from H19-/-mice. (A) Levels of H19 in intestinal organoids 48 hours after transfection with an H19 expression vector or a control empty vector. Values are the means ± SEM (n = 3). *P < .05 compared with control vector. (B) Immunostaining of Paneth cells (lysozyme-positive cells, red) in intestinal organoids described in panel A: a) control; and b) H19 O/E. Scale bars: 50 μm. (C) Immunostaining of Goblet cells (mucin 2–positive cells, red) in intestinal organoids described in panel A. Scale bars: 50 μm. (D) Quantitative data of Paneth (top) and goblet cells (bottom) in the intestinal organoids described in panels B and C: a) control; and b) H19 O/E. Values are the means ± SEM (n = 12). *P < .05 compared with control vector. DAPI, 4′,6-diamidino-2-phenylindole; E-cad, E-cadherin; Lys, lysozyme; O/E, overexpression.
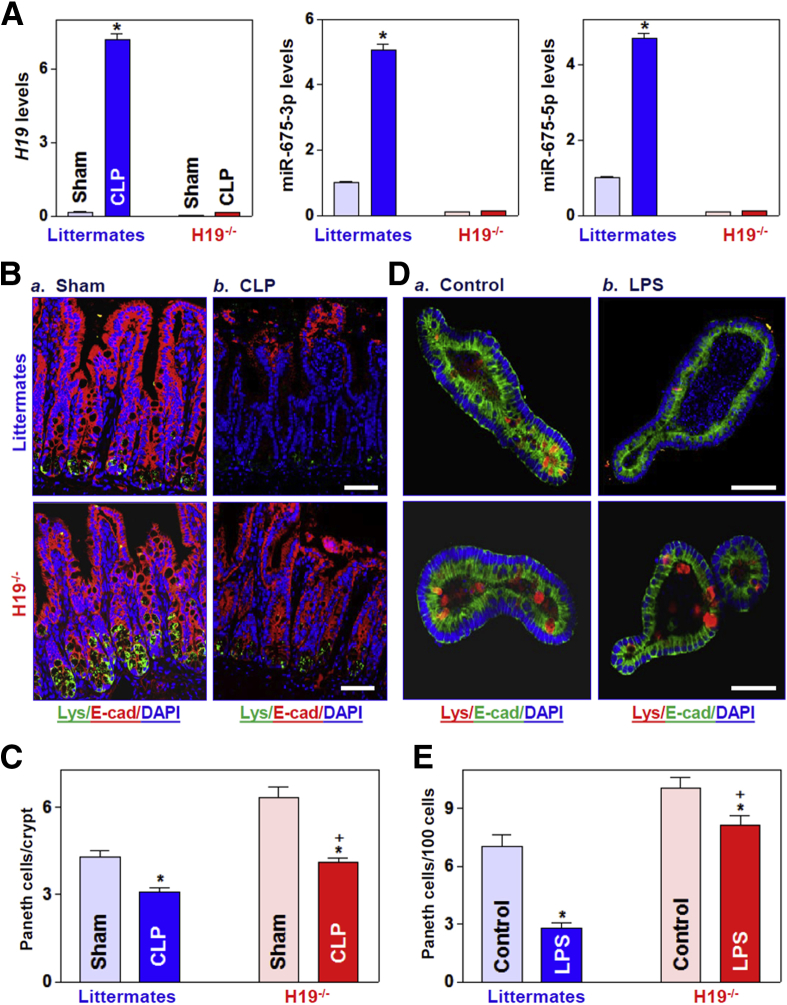
Preventing the Increase in H19 Levels Protects Paneth and Goblet Cells From Septic Stress Consistent with observations in wild-type mice, the levels of mucosal H19 in the small intestine of littermate mice increased by 24 hours after CLP, accompanied by a significant increase in the abundance of miR-675-3p and miR-675-5p (Figure 4A). In contrast, the levels of mucosal H19 and miR-675 were almost undetectable in the intestinal mucosa of H19-/- mice with or without CLP stress. As shown (Figure 4B and C), exposure to CLP inhibited Paneth cell function in both H19-/- and littermate mice, as indicated by decreases in the number of lysozyme-positive cells and the number of lysozyme granules per Paneth cell. However, H19 deletion partially but significantly prevented the CLP-elicited loss of Paneth cells because many more lysozyme-positive cells were found in the mucosa of H19-/- mice compared with littermates by 24 hours after exposure to CLP. In the ex vivo model, treatment with LPS (20 ng/mL for 5 days) also suppressed the formation of Paneth cells, as shown by a dramatic decrease in the number of lysozyme-positive cells in intestinal organoids from control littermate mice, but this inhibition was alleviated in the organoids from H19-/- mice (Figure 4D and E). In fact, exposure of organoids from H19-/- mice to the same dose of LPS for 5 days only marginally decreased the number of Paneth cells.
Figure 4.
H19 deletion promotes Paneth cell function in vivo and ex vivo. (A) Levels of mucosal H19 (left), miR-675-3p (middle), and miR-675-5p (right) in the small intestine of mice exposed to CLP for 24 hours. Values are the means ± SEM (n = 5). *P < .05 compared with sham-treated mice. (B) Immunostaining of lysozyme-positive cells in the intestinal mucosa of mice treated as described in panel A: a) Sham; and b) CLP. Signals evidence the presence of lysozyme (green), E-cadherin (red), and nuclei (stained with 4′,6-diamidino-2-phenylindole [DAPI]) (blue). Scale bars: 50 μm. (C) Quantitative data of lysozyme-positive cells in the small intestinal epithelium described in panel B. Values are the means ± SEM (n = 5). *,+P < .05 compared with sham-treated mice and littermate mice exposed to CLP, respectively. (D) Immunostaining of lysozyme-positive cells in intestinal organoids isolated from H19-/- and control littermate mice (a) Control; and b) LPS) after treatment with LPS (20 ng/mL) for 5 days (red). E-cadherin, green; nuclei, blue. Scale bar: 50 μm. (E) Quantitative results of lysozyme-positive cells in intestinal organoids treated as described in panel D. Values are the means ± SEM (n = 5). *,+P < .05 compared with controls (nontreatment) and organoids (from littermate mice) exposed to LPS, respectively. E-cad, E-cadherin.
Similarly, H19 knockout also protected goblet cells in the intestinal epithelium responding to septic stress. Both exposure of mice to CLP and treatment of intestinal organoids with LPS decreased the number of goblet cells in the presence or absence of cellular H19 as measured by Alcian blue staining in vivo (Figure 5A and B) and mucin 2 immunostaining ex vivo (Figure 5C and D). However, the reduction of goblet cells by CLP or LPS was rescued by knockout of H19. These results strongly suggest that preventing the induction in H19 in response to septic stress preserves Paneth and goblet cells in the intestinal epithelium.
Figure 5.
H19 deletion protects Goblet cells against CLP- and LPS-induced stress in vivo and ex vivo. (A) Goblet cells were examined by Alcian blue staining in the small intestinal mucosa of H19-/- and control littermate mice (a) Sham; and b) CLP) exposed to CLP for 24 hours. Scale bars: 50 μm. (B) Quantitative data of goblet cells in the small intestinal mucosa of mice treated as described in panel A. Values are the means ± SEM (n = 5). *,+P < .05 compared with sham-treated mice and littermate mice exposed to CLP, respectively. (C) Immunostaining of goblet cells (mucin 2–positive cells) in intestinal organoids exposed to LPS (20 ng/mL) for 5 days (red). Scale bar: 50 μm. (D) Quantitative results of goblet cells in intestinal organoids treated as described in panel C: a) Control; and b) LPS. Values are the means ± SEM (n = 5). *,+P < .05 compared with controls (without LPS treatment) and organoids (from littermate mice) exposed to LPS, respectively. DAPI, 4′,6-diamidino-2-phenylindole; E-cad, E-cadherin.
H19 Knockout Increases Autophagy in the Intestinal Epithelium
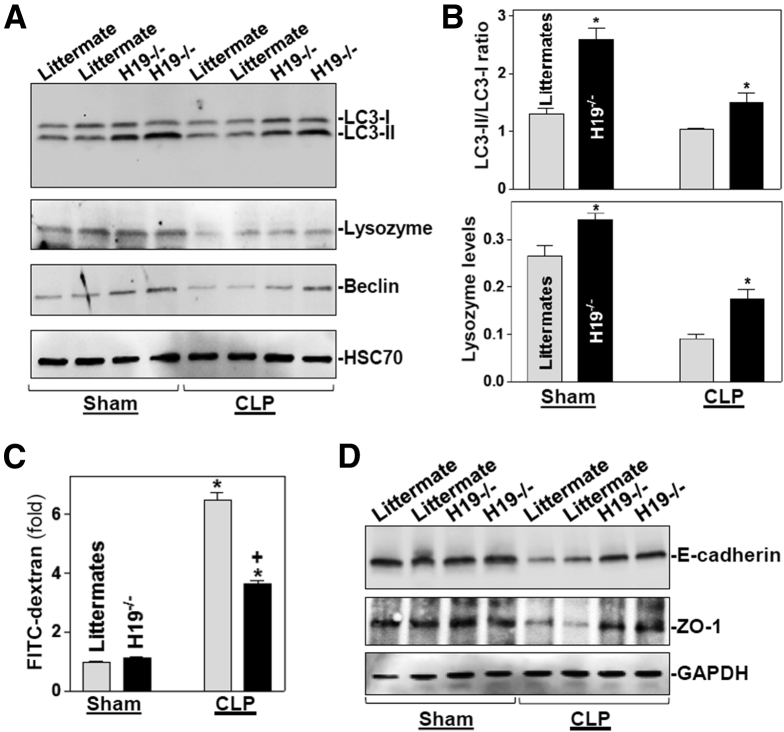
Because Paneth cells secrete lysozyme via secretory autophagy in the intestinal epithelium7 and activation of autophagy is involved in the regulation of goblet cell function and mucosal defense,33 we examined changes in autophagy in H19-/- mice exposed to CLP. Basal levels of autophagy proteins LC3-II, lysozyme, and beclin in the small intestinal mucosa were increased in H19-ablated mice compared with control littermate mice (Figure 6A, left). Autophagy was suppressed in the mucosa of littermate mice by 24 hours after CLP, as indicated by decreased levels of these autophagy proteins, but the CLP-inhibited autophagy was abolished in H19-/- mice (Figure 6A and B, right). Although H19 deletion did not alter the basal level of gut permeability to FITC dextran, both H19-/- and control littermate mice showed gut barrier dysfunction by 24 hours after CLP (Figure 6C). Notably, however, the levels of gut permeability resulting from CLP in H19-/- mice were much lower (∼3.5-fold) than those in control littermate mice (>6-fold), as compared with sham mice. As expected, the levels of E-cadherin and ZO-1 proteins also decreased in the small intestinal mucosa of littermate mice by 24 hours after CLP, but this reduction was partially prevented by H19 knockout (Figure 6D). Taken together, these findings indicate that H19 knockout promotes Paneth and goblet cell function, and induces autophagy, thus protecting the intestinal barrier against septic stress.
Figure 6.
H19 deletion in mice activates autophagy and protects the gut barrier function. (A) Immunoblots of autophagy proteins LC3, lysozyme, and beclin in the small intestinal mucosa of mice exposed to CLP for 24 hours. Equal loading was monitored by assessing heat shock cognate 71 kDa protein 70 levels. (B) Quantitative analysis derived from densitometric scans of immunoblots. Top: changes in LC3 activity, as quantified by examining the ratio of LC3-II and LC3-I; bottom: changes in the levels of lysozyme as quantified by the ratio of lysozyme signals to heat shock cognate 71 kDa protein 70 signals. Values are means ± SEM (n = 3). *P < .05 compared with control littermates. (C) Gut permeability in H19-/- and control littermate mice treated as described in panel A. Values are the means ± SEM (n = 5). *,+P < .05 compared with sham-treated mice and littermate mice exposed to CLP, respectively. (D) Immunoblots of E-cadherin and ZO-1 in the small intestinal mucosa of mice treated as described in panel A. Equal loading was monitored by measuring GAPDH levels. Three separate experiments were performed that showed similar results.
H19 Overexpression Suppresses Autophagy and Disrupts Epithelial Barrier Function In Vitro
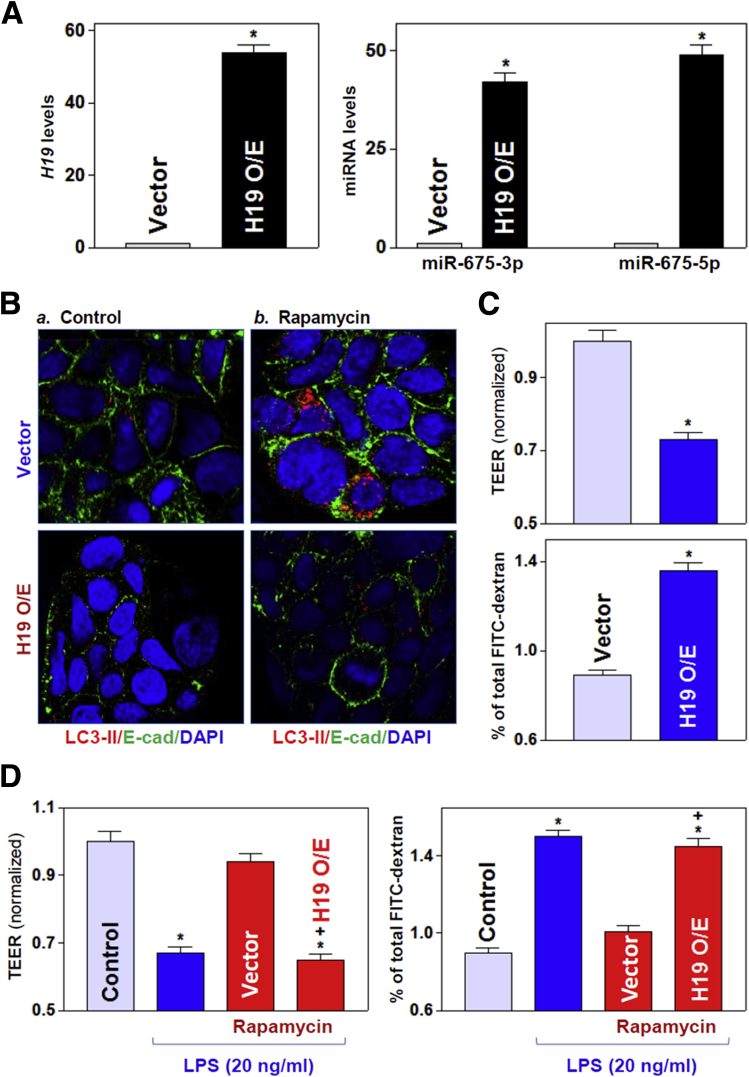
To define further the role of H19-mediated inhibition of autophagy in the regulation of intestinal epithelial barrier function, we used Caco-2 cells as an in vitro model for gut epithelial barrier study as reported previously.34,35 As shown (Figure 7A), the levels of cellular H19 increased dramatically in cells that were transfected with the H19 expression vector as compared with cells transfected with an empty vector (control). Ectopically expressed H19 specifically induced the expression levels of miR-675-3p and miR-675-5p, but did not alter the expression of other miRNAs such as miR-29b or miR-222 (data not shown). Increasing the levels of H19 by transfection with the H19 expression vector prevented the activation of autophagy by the pharmacologic inducer rapamycin. In control IECs, treatment with rapamycin activated autophagy, as indicated by increased levels of LC3-II on Western blot (Figure 7B, top), but this activation was abolished in cells overexpressing H19 (Figure 7B, bottom).
Figure 7.
Ectopically expressed H19 prevents induced autophagy and disrupts the epithelial barrier function in vitro. (A) Levels of H19 (left) and miR-675 (right) in cultured IECs transfected with the H19 expression vector and measured 48 hours later. Values are relative to those for the control (vector) and are the means ± SEM from triplicate experiments. *P < .05 compared with the vector. (B) Immunostaining of LC3-II in cells exposed to rapamycin (50 ng/mL) for 24 hours (red). Twenty-four hours after transfection with a control vector or a vector to overexpress H19, cells were exposed to rapamycin: a) Coontrol; and b) Rapamycin. Three experiments were performed that showed similar results. (C) Changes in epithelial barrier function were assessed by monitoring changes in TEER (top) and FITC-dextran paracellular permeability (bottom) in cells treated as described for panel A. TEER assays were performed on 12-mm Transwell filters; paracellular permeability was assayed by using the membrane-impermeable trace molecule FITC dextran, which was added to the insert medium. Values are the means ± SEM from triplicate experiments. *P < .05 compared with the vector. (D) Changes in barrier function in cells treated for 24 hours with LPS (20 ng/mL) alone or with LPS plus rapamycin in control cells as in cells overexpressing H19. *,+P < .05 compared with control and cells treated with LPS plus rapamycin, respectively. O/E, overexpression. DAPI, 4′,6-diamidino-2-phenylindole; E-cad, E-cadherin.
The increase of H19 abundance also resulted in epithelial barrier dysfunction in vitro as shown by a decrease in transepithelial electrical resistance (TEER) and an increase in the levels of paracellular flux of FITC dextran (Figure 7C). As expected, exposure to LPS also disrupted the epithelial barrier in cultured cells; both the decreased TEER and increased paracellular permeability induced by LPS were prevented by treatment with rapamycin (Figure 7D). Moreover, the rapamycin-induced protective effects on the epithelial barrier in LPS-treated cells was abolished by overexpressing H19, as shown by the finding that TEER values and paracellular permeability in H19-transfected cells treated with LPS plus rapamycin were indistinguishable from those observed in cells treated with LPS alone (Figure 7D). On the other hand, neither H19 overexpression nor treatment with rapamycin affected cell viability, as measured by trypan blue staining (data not shown). These data indicate that the increase in H19 levels after stress disrupts epithelial barrier function at least partially by inhibiting autophagy in the intestinal epithelium.
Discussion
Our previous study showed that ectopically expressed H19 in cultured IECs disrupted epithelial barrier function in an in vitro model by inhibiting expression of ZO-1 and E-cadherin post-transcriptionally,18 but the exact in vivo function of H19 in the intestinal epithelium remains to be fully elucidated. By using mice bearing a conditional ablation of H19 exon 1, we found strong genetic evidence that H19 plays an important suppressive role in the intestinal barrier by reducing autophagy and the function of Paneth and goblet cells. Targeted deletion of H19 in mice not only promoted the functions of Paneth and goblet cells in the intestinal mucosa but also activated autophagy. Preventing the induction of H19 levels in mice exposed to CLP protected Paneth and goblet cell function and preserved autophagy, thus promoting intestinal barrier function. These findings advance our understanding of the biological function of H19 in the intestinal epithelium and highlight a crucial role of increased H19 in the pathogenesis of intestinal barrier dysfunction. Because H19 is highly conserved in mammalian tissues and its levels increase markedly in human intestinal mucosa from patients with sepsis, our present results provide a strong rationale for considering therapeutic strategies directed at the H19/miR-675 axis to protect intestinal barrier function in clinical settings.
Our results reported here indicate that an increase of tissue H19 levels in the small intestinal mucosa impairs the effectiveness and integrity of barrier function in response to stressful environments. The mucosal H19 content increased dramatically in mice exposed to CLP and in patients with sepsis, associated with intestinal barrier dysfunction. In contrast, H19 knockout in mice protected intestinal barrier function against septic stress induced by CLP, as shown by much lower gut permeability after CLP in H19-/- mice than in littermate mice. H19 elicits multiple biological functions by targeting different signals through distinct molecular mechanisms, so its specific roles in physiology and disease states depend on a range of factors.36,37 For example, H19 represses embryonic placental growth during fetal development through processing of miR-675,23 but it promotes mucosal regeneration in inflamed intestinal tissues by inhibiting p53 expression and interacting with miR-34a and let-7.28 The H19/miR-675 axis inhibits prostate cancer metastasis by targeting transforming growth factor-β–induced protein,38 and it promotes breast cancer cell plasticity by differentially sponging miR-200b/c and let-7b.39 Deletion of the H19 gene also affects the expression levels of the Igf2 gene and 8 other genes in the imprinted gene network.20,40 Our study provides additional evidence showing an in vivo function for H19 in the regulation of intestinal barrier function.
We present evidence that H19 regulates intestinal barrier function partially by modulating the levels of Paneth and goblet cells that are responsible for mucosal defense and host protection from enteric pathogens.41,42 Paneth and goblet cells increased in the absence of H19 in vivo as well as ex vivo, and H19 knockout also prevented CLP-induced inhibition of Paneth and goblet cells and improved intestinal barrier function. In an ex vivo model, H19-deficient enteroids from H19-/- mice showed a protective effect on Paneth and goblet cells against LPS. Paneth and goblet cells are specialized secretory cell types of the intestinal epithelium and share similar characteristics, but they have distinct biological roles. During intestinal development, the secretory lineages in mammals differentiate from a shared progenitor located above the stem cell niche. During differentiation, Paneth cells move back to the base of the crypts and become interspersed between the stem cells, whereas goblet cells migrate into both crypts and villi.43 Paneth cells produce abundant antibacterial peptides/proteins that confer mucosal protection and provide signals for the maintenance of stem cells for normal mucosal renewal.44 Goblet cells synthesize and release various mucin proteins that are major components of the unstirred mucus layer covering the epithelium.45 Defects in Paneth and goblet cells disrupt the intestinal barrier function and compromise epithelium homeostasis.6,8 The exact mechanisms by which H19 regulates Paneth and goblet cell levels remain unknown and are under intensive investigation in our laboratory.
Another significant finding of this study was the increase in autophagy in the H19-deficient intestinal epithelium, which further contributed to the protective effect on barrier function in H19-/- mice exposed to CLP. Although the role of H19 in regulating autophagy is not clear at this time,46,47 the basal levels of LC3-II, lysozyme, and beclin increased in the small intestinal mucosa of H19-/- mice relative to control littermates. Septic stress–induced suppression of autophagy was prevented in H19-ablated mice after CLP. Conversely, overexpression of H19 in cultured IECs prevented an increase in autophagy mediated by rapamycin and also disrupted epithelial barrier function in an in vitro permeability model. Moreover, ectopically expressed H19 in IECs abolished rapamycin-induced protection of epithelial barrier function. Autophagy is a conserved intracellular pathway that is characterized by the formation of double-membrane vesicles called autophagosomes, which contain cytoplasmic structures and pathogens targeted for degradation.9,48 Autophagy is believed have a generally favorable impact on cell, tissue, and organ homeostasis, and participates in various physiological processes including the maintenance of intestinal barrier function.49,50 Induced expression of the autophagy protein ATG16L1 prevents necroptosis in the intestinal epithelium.26 Paneth cells secrete lysosome through secretory autophagy,7 and autophagy proteins also control function of goblet cells.33 Deficiency of autophagy proteins and/or reduction in autophagy also reduced Paneth and goblet cell levels and function.6,8,50,51 Defective autophagy is observed commonly in mice with destructive mucosal inflammatory erosions,51 and disrupted expression of autophagy genes such as Atg16l1, Atg5, and Atg7 also occurs in patients with inflammatory bowel diseases and other mucosal disorders.6,50 Although H19 deletion activates autophagy in the intestinal epithelium, the exact mechanisms and consequences of the altered autophagic response in Paneth and goblet cell function remain be investigated using appropriate mouse models.
Finally, the results from this study have clinical significance because human intestinal mucosa from patients with sepsis has shown both increased levels of H19 and intestinal barrier dysfunction. As reported,28 tissue levels of H19 increased significantly in the intestinal mucosa of mice exposed to LPS or dextran sulfate sodium and in human mucosal tissues from patients with inflammatory bowel disease, associated with massive mucosal inflammation, injury/erosions, and increased gut permeability. Deregulated expression of H19 also commonly has been observed in patients with other diseases including various cancers and liver fibrosis.52,53 In this study, we identified a novel role for H19 in regulating Paneth and goblet cell function and in increasing autophagy in the intestinal epithelium in vivo, ex vivo, as well as in vitro. We also established the importance of increased Paneth and goblet cell levels and autophagy in preserving intestinal barrier function in H19-/- mice exposed to septic stress. These findings shed light on how H19 regulates intestinal barrier function in response to pathologic stress, and highlight the possible value of targeting H19 therapeutically and its interacting miRNAs in patients with compromised intestinal barrier function.
Materials and Methods
Culture Cells and Mice
Caco-2 cells were purchased from the American Type Culture Collection (Manassas, VA) and were maintained under standard culture conditions.54,55 The culture medium and fetal bovine serum were purchased from Invitrogen (Carlsbad, CA) and biochemicals were from Sigma (St. Louis, MO). Antibodies recognizing E-cadherin, ZO-1, lysozyme, mucin 2, LC3-II, beclin, heat shock cognate 71 kDa protein (HSC)70, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were obtained from Santa Cruz Biotechnology (Santa Cruz, CA) and BD Biosciences (Sparks, MD), and the secondary antibody conjugated to horseradish peroxidase was from Sigma. All antibodies used in this study were thoroughly validated for species specificity. Antibody dilutions used for Western blot of E-cadherin, ZO-1, lysozyme, mucin 2, LC3-II, beclin, and GAPDH were 1:1000 (first Ab) and 1:2000 (second Ab), respectively, whereas antibody dilutions for immunostaining were 1:200 (first) and 1:2000 (second). Relative protein levels were analyzed using Bio-Rad (Hercules, CA) Chemidoc and the XRS system equipped with Image lab software (version 4.1). We also used the Quantity (Hercules, CA) tool to determine the band intensity volume; the values were normalized with internal loading control of either GAPDH or HSC70.
Wild-type C57BL/6J mice (males and females; age, 6–9 wk) were purchased from The Jackson Laboratory (Bar Harbor, ME). H19 knockout (H19Δexon1/+) mice were kindly provided by Dr Karl Pfeifer (National Institutes of Health),31 and they carried a 1-kb deletion of H19 exon 1 that removes almost the entire exon 1, including the region encoding miR-675, and reduces levels of even the partial H19 transcript by more than 100-fold globally.31,32 All animal experiments were performed in accordance with National Institutes of Health guidelines and were approved by the Institutional Animal Care and Use Committee of the University Maryland School of Medicine and the Baltimore Veterans Affairs Hospital. Wild-type C57BL/6J and H19 knockout and their control littermate mice were housed and handled in a specific pathogen-free breeding barrier and cared for by trained technicians and veterinarians. Animals were deprived of food but allowed free access to tap water for 24 hours before experiments. After different treatments, two 4-cm segments taken from the middle of the small intestine were removed in each animal as described previously.54 One segment was for chemical analysis, and the other segment was for histologic examination.
Intestinal Organoid Culture
Isolation and culture of primary enterocytes were conducted following the method described previously.8,56 Briefly, primary crypts were released from the small intestinal mucosa in mice, then the isolated crypts were mixed with Matrigel (Corning Inc, Corning, NY) and cultured in Advanced Dulbecco’s modified Eagle medium/F12 medium. The growth of organoids was examined under phase-contrast microscopy.
Plasmid Construction
An expression vector containing a 1.5-kb fragment flanking the human H19 locus (including the entire exon 1) under the control of pCMV promoter was purchased from Origene (Rockville, MD) and used to increase cellular H19 as described previously.18 Transient transfections were performed using the Lipofectamine reagent following the manufacturer’s recommendations (Invitrogen). Forty-eight hours after transfection using Lipofectamine, cells were harvested for analysis.
Reverse Transcription and Real-Time Quantitative PCR Analysis
Total RNA was isolated using the RNeasy mini kit (Qiagen, Valencia, CA), and reverse transcription and PCR amplification reactions were performed as described.17 The levels of Gapdh PCR product were examined to monitor the evenness in RNA input in reverse transcription PCR samples. Real-time quantitative PCR analysis was conducted using 7500-Fast Real-Time PCR Systems with specific primers, probes, and software (Applied Biosystems, Foster City, CA). For miRNA studies, the levels of miR-675 and miR-29b also were quantified by quantitative PCR by using the TaqMan (Aplied Biosystems, Foster City, CA) MicroRNA assay; small nuclear RNA U6 was used as endogenous control.
Western Blot Analysis
Whole-cell lysates were prepared using 2% sodium dodecyl sulfate, sonicated, and centrifuged (12,000 rpm) at 4°C for 15 minutes. The supernatants were boiled for 5 minutes and size-fractionated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (7.5% acrylamide). After transferring proteins onto nitrocellulose filters, the blots were incubated with primary antibodies recognizing different proteins; after incubations with secondary antibodies, immunocomplexes were developed by using chemiluminescence.
Immunofluorescence Staining
The immunofluorescence staining procedure was performed according to the method described in our previous publications.8,30 For experiments using mucosal tissue samples from human beings and mice, dissected and opened intestines were mounted onto a solid surface and fixed in formalin and paraffin. In each tissue sample, more than 5 slides (5-μm thickness per section) were prepared for immunofluorescence staining. For studies in cultured intestinal organoids and Caco-2 cells, the slides were fixed in 3.7% formaldehyde in phosphate-buffered saline and rehydrated. All slides were incubated with the primary antibody against lysozyme or mucin 2 in the block buffer at a concentration of 1:200 or 1:300 dilution at 4ºC overnight and then incubated with secondary antibody conjugated with Alexa Fluor-594 (Molecular Probes, Eugene, OR) for 2 hours at room temperature. After rinsing 3 times, the slides were incubated with 4′,6-diamidino-2-phenylindole (Molecular Probes) at a concentration of 1 μmol/L for 10 minutes to stain cell nuclei. Finally, the slides were washed, mounted, and viewed through a Zeiss (Oberkochen, Germany) confocal microscope (model LSM710). Slides were examined in a blinded fashion by coding them, and only after the examination was complete were they decoded. Images were processed using Adobe Photoshop software (San Jose, CA).
Measurement of Epithelial Barrier Function In Vitro
The epithelial barrier function in vitro was examined by paracellular tracer flux assays using the 12-well Transwell plate (surface area, 1.12 cm2) as described.30 FITC dextran (70 kilodaltons; Sigma), a membrane-impermeable molecule, served as the paracellular tracer and was added to the apical bathing wells. The basal bathing well had no added tracers and contained the same flux assay medium as in the apical compartment. All flux assays were performed at 37°C, and the basal medium was collected at different times after addition of the FITC dextran. The concentration of the FITC dextran in the basal medium was determined using a fluorescence plate reader with an excitation wavelength of 490 nm and an emission wavelength of 530 nm. TEER was measured with an epithelial voltmeter under open-circuit conditions (WPI, Sarasota, FL) as described,5 and the TEER of all monolayers was normalized to that of control monolayers in the same experiment.
Surgical Procedures and Measurement of Gut Permeability In Vivo
Mice were anesthetized by Pentobarbitol (5.5 mg/100-g wt, intraperitoneally), and CLP was performed as described.57 The distal portion of the cecum (1 cm) was ligated with 5-0 silk suture. The ligated cecum then was punctured with a 25-gauge needle and slightly compressed with an applicator until a small amount of stool appeared. In sham-operated animals, the cecum was manipulated but without ligation and puncture, and was placed back in the peritoneum. The incision was closed using a 2-layer procedure: 5-0 silk suture on the muscle layer and the skin. Mice received 1 mL of saline intraperitoneally for fluid resuscitation at the time of closure and 0.1 mg/100 g weight buprenorphine subcutaneously 4 times at 12-hour intervals to minimize distress.
Gut permeability in vivo was determined by examining the appearance in blood of FITC dextran administered by gavage as described.35 Briefly, mice were gavaged with FITC dextran at a dose of 60 mg/100 g wt 4 hours before harvest. Blood samples were collected by cardiac puncture. The serum concentration of the FITC dextran was determined using a fluorescence plate reader as described earlier.
Statistical Analysis
All values were expressed as the means ± SEM from 5 animals or 3 separate experiments. An unpaired, 2-tailed Student t test was used when indicated with a P value less than .05 considered statistically significant. When assessing multiple groups, 1-way analysis of variance was used with the Tukey post hoc test.58 The statistical software used was GraphPad Instat Prism 5 (San Diego, CA). For nonparametric analysis rank comparison, the Kruskal–Wallis test was conducted.
Footnotes
Author contributions Ting-Xi Yu, Hee K. Chung, and Lan Xiao performed most experiments and summarized the data; Jun-Jie Piao and Shaoyang Lan participated in performing experiments in vivo and immunoprecipitation assays; Suraj K. Jaladanki and Douglas J. Tuner were involved in the experiments conducted on intestinal organoids and cultured intestinal epithelial cells; Jean-Pierre Raufman and Myriam Gorospe participated in experiments using human tissues and performed data analysis; and Jian-Ying Wang designed the experiments, analyzed data, prepared figures, and drafted the manuscript.
Conflicts of interest This author discloses the following: Jian-Ying Wang is a Senior Research Career Scientist at the Biomedical Laboratory Research and Development Service (US Department of Veterans Affairs). The remaining authors disclose no conflicts.
Funding This work was supported by Merit Review Awards (J.-Y.W., D.J.T., J.-P.R.) from the US Department of Veterans Affairs; grants DK57819, DK61972, and DK68491 from the National Institutes of Health (J.-Y.W.); and funding from the National Institute on Aging Intramural Research Program, National Institutes of Health (M.G).
References
- 1.Ahmad R., Sorrell M.F., Batra S.K., Dhawan P., Singh A.B. Gut permeability and mucosal inflammation: bad, good or context dependent. Mucosal Immunol. 2017;10:307–317. doi: 10.1038/mi.2016.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Camilleri M. Leaky gut: mechanisms, measurement and clinical implications in humans. Gut. 2019;68:1516–1526. doi: 10.1136/gutjnl-2019-318427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pelaseyed T., Bergström J.H., Gustafsson J.K., Ermund A., Birchenough G.M., Schütte A., van der Post S., Svensson F., Rodríguez-Piñeiro A.M., Nyström E.E., Wising C., Johansson M.E., Hansson G.C. The mucus and mucins of the goblet cells and enterocytes provide the first defense line of the gastrointestinal tract and interact with the immune system. Immunol Rev. 2014;260:8–20. doi: 10.1111/imr.12182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peterson L.W., Artis D. Intestinal epithelial cells: regulators of barrier function and immune homeostasis. Nat Rev Immunol. 2014;14:141–153. doi: 10.1038/nri3608. [DOI] [PubMed] [Google Scholar]
- 5.Wang J.Y., Cui Y.H., Xiao L., Chung H.K., Zhang Y., Rao J.N., Gorospe M., Wang J.Y. Regulation of intestinal epithelial barrier function by long noncoding RNA uc.173 through interaction with microRNA 29b. Mol Cell Biol. 2018;38:e00010–e00018. doi: 10.1128/MCB.00010-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cadwell K., Liu J.Y., Brown S.L., Miyoshi H., Loh J., Lennerz J.K., Kishi C., Kc W., Carrero J.A., Hunt S., Stone C.D., Brunt E.M., Xavier R.J., Sleckman B.P., Li E., Mizushima N., Stappenbeck T.S., Virgin H.W., 4th A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature. 2008;456:259–263. doi: 10.1038/nature07416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bel S., Pendse M., Wang Y., Li Y., Ruhn K.A., Hassell B., Leal T., Winter S.E., Xavier R.J., Hooper L.V. Paneth cells secrete lysozyme via secretory autophagy during bacterial infection of the intestine. Science. 2017;357:1047–1052. doi: 10.1126/science.aal4677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xiao L., Li X., Chung H.K., Kalakonda S., Cai J.Z., Cao S., Chen N., Liu Y., Rao J.N., Wang H.Y., Gorospe M., Wang J.Y. RNA-binding protein HuR regulates Paneth cell function by altering membrane localization of TLR2 via post-transcriptional control of CNPY3. Gastroenterology. 2019;157:731–743. doi: 10.1053/j.gastro.2019.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boya P., Reggiori F., Codogno P. Emerging regulation and functions of autophagy. Nat Cell Biol. 2013;15:713–720. doi: 10.1038/ncb2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gukovskaya A.S., Gukovsky I., Algul H., Habtezion A. Autophagy, inflammation, and immune dysfunction in the pathogenesis of pancreatitis. Gastroenterology. 2017;153:1212–1226. doi: 10.1053/j.gastro.2017.08.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mercer T.R., Dinger M.E., Mattick J.S. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 12.Batista P.J., Chang H.Y. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152:1298–1307. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morris K.V., Mattick J.S. The rise of regulatory RNA. Nat Rev Genet. 2014;15:423–437. doi: 10.1038/nrg3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Quinn J.J., Chang H.Y. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. 2016;17:47–62. doi: 10.1038/nrg.2015.10. [DOI] [PubMed] [Google Scholar]
- 15.Wang K.C., Chang H.Y. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ulitsky I. Evolution to the rescue: using comparative genomics to understand long non-coding RNAs. Nat Rev Genet. 2016;17:601–614. doi: 10.1038/nrg.2016.85. [DOI] [PubMed] [Google Scholar]
- 17.Xiao L., Rao J.N., Cao S., Liu L., Chung H.K., Zhang Y., Zhang J., Liu Y., Gorospe M., Wang J.Y. Long noncoding RNA SPRY4-IT1 regulates intestinal epithelial barrier function by modulating the expression levels of tight junction proteins. Mol Biol Cell. 2016;27:617–626. doi: 10.1091/mbc.E15-10-0703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zou T., Jaladanki S.K., Liu L., Xiao L., Chung H.K., Wang J.Y., Xu Y., Gorospe M., Wang J.Y. H19 long noncoding RNA regulates intestinal epithelial barrier function via microRNA 675 by interacting with RNAbinding protein HuR. Mol Cell Biol. 2016;36:1332–1341. doi: 10.1128/MCB.01030-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xiao L., Gorospe M., Wang J.Y. Long noncoding RNAs in intestinal epithelium homeostasis. Am J Physiol Cell Physiol. 2019;17:C93–C100. doi: 10.1152/ajpcell.00092.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leighton P.A., Ingram R.S., Eggenschwiler J., Efstratiadis A., Tilghman S.M. Disruption of imprinting caused by deletion of the H19 gene region in mice. Nature. 1995;375:34–39. doi: 10.1038/375034a0. [DOI] [PubMed] [Google Scholar]
- 21.Juan V., Crain C., Wilson C. Evidence for evolutionarily conserved secondary structure in the H19 tumor suppressor RNA. Nucleic Acids Res. 2000;28:1221–1227. doi: 10.1093/nar/28.5.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gabory A., Ripoche M.A., Le Digarcher A., Watrin F., Ziyyat A., Forné T., Jammes H., Ainscough J.F., Surani M.A., Journot L., Dandolo L. H19 acts as a trans regulator of the imprinted gene network controlling growth in mice. Development. 2009;136:3413–3421. doi: 10.1242/dev.036061. [DOI] [PubMed] [Google Scholar]
- 23.Keniry A., Oxley D., Monnier P., Kyba M., Dandolo L., Smits G., Reik W. The H19 lincRNA is a developmental reservoir of miR-675 that suppresses growth and Igf1r. Nat Cell Biol. 2012;14:659–665. doi: 10.1038/ncb2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yan L., Zhou J., Gao Y., Ghazal S., Lu L., Bellone S., Yang Y., Liu N., Zhao X., Santin A.D., Taylor H., Huang Y. Regulation of tumor cell migration and invasion by the H19/let-7 axis is antagonized by metformin induced DNA methylation. Oncogene. 2015;34:3076–3084. doi: 10.1038/onc.2014.236. [DOI] [PubMed] [Google Scholar]
- 25.Luo M., Li Z., Wang W., Zeng Y., Liu Z., Qiu J. Long non-coding RNA H19 increases bladder cancer metastasis by associating with EZH2 and inhibiting E-cadherin expression. Cancer Lett. 2013;333:213–221. doi: 10.1016/j.canlet.2013.01.033. [DOI] [PubMed] [Google Scholar]
- 26.Ghazal S., McKinnon B., Zhou J., Mueller M., Men Y., Yang L., Mueller M., Flannery C., Huang Y., Taylor H.S. H19 lncRNA alters stromal cell growth via IGF signaling in the endometrium of women with endometriosis. EMBO Mol Med. 2015;7:996–1003. doi: 10.15252/emmm.201505245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matouk I.J., DeGroot N., Mezan S., Ayesh S., Abu-lail R., Hochberg A., Galun E. The H19 non-coding RNA is essential for human tumor growth. PLoS One. 2007;2:e845. doi: 10.1371/journal.pone.0000845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Geng H., Bu H.F., Liu F., Wu L., Pfeifer K., Chou P.M., Wang X., Sun J., Lu L., Pandey A., Bartolomei M.S., De Plaen I.G., Wang P., Yu J., Qian J., Tan X.D. In inflamed intestinal tissues and epithelial cells, interleukin 22 signaling increases expression of H19 long noncoding RNA, which promotes mucosal regeneration. Gastroenterology. 2018;155:144–155. doi: 10.1053/j.gastro.2018.03.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yoshimizu T., Miroglio A., Ripoche M.A., Gabory A., Vernucci M., Riccio A., Colnot S., Godard C., Terris B., Jammes H., Dandolo L. The H19 locus acts in vivo as a tumor suppressor. Proc Natl Acad Sci U S A. 2008;105:12417–12422. doi: 10.1073/pnas.0801540105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chung H.K., Chen Y., Rao J.N., Liu L., Xiao L., Turner D.J., Yang P., Gorospe M., Wang J.Y. Transgenic expression of miR-222 disrupts intestinal epithelial regeneration by targeting multiple genes including Frizzled-7. Mol Med. 2015;21:676–687. doi: 10.2119/molmed.2015.00147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park K.Y., Sellars E.A., Grinberg A., Huang S.P., Pfeifer K. The H19 differentially methylated region marks the parental origin of a heterologous locus without gametic DNA methylation. Mol Cell Biol. 2004;24:3588–3595. doi: 10.1128/MCB.24.9.3588-3595.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Katz D.J., Beer M.A., Levorse J.M., Tilghman S.M. Functional characterization of a novel Ku70/80 pause site at the H19/Igf2 imprinting control region. Mol Cell Biol. 2005;25:3855–3863. doi: 10.1128/MCB.25.10.3855-3863.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Patel K.K., Miyoshi H., Beatty W.L., Head R.D., Malvin N.P., Cadwell K., Guan J.L., Saitoh T., Akira S., Seglen P.O., Dinauer M.C., Virgin H.W., Stappenbeck T.S. Autophagy proteins control goblet cell function by potentiating reactive oxygen species production. EMBO J. 2013;32:3130–3144. doi: 10.1038/emboj.2013.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chung H.K., Wang S.R., Xiao L., Rathor N., Turner D.J., Yang P., Gorospe M., Rao J.N., Wang J.Y. α4 coordinates small intestinal epithelium homeostasis by regulating stability of HuR. Mol Cell Biol. 2018;38 doi: 10.1128/MCB.00631-17. e00631–e00617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu T.X., Wang P.Y., Rao J.N., Zou T., Liu L., Xiao L., Gorospe M., Wang J.Y. Chk2-dependent HuR phosphorylation regulates occludin mRNA translation and epithelial barrier function. Nucleic Acids Res. 2011;39:8472–8487. doi: 10.1093/nar/gkr567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kallen A.N., Zhou X.B., Xu J., Qiao C., Ma J., Yan L., Lu L., Liu C., Yi J.S., Zhang H., Min W., Bennett A.M., Gregory R.I., Ding Y., Huang Y. The imprinted H19 lncRNA antagonizes let-7 microRNAs. Mol Cell. 2013;52:101–112. doi: 10.1016/j.molcel.2013.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Monnier P., Martinet C., Pontis J., Stancheva I., Ait-Si-Ali S., Dandolo L. H19 lncRNA controls gene expression of the Imprinted Gene Network by recruiting MBD1. Proc Natl Acad Sci U S A. 2013;110:20693–20698. doi: 10.1073/pnas.1310201110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu M., Chen Q., Liu X., Sun Q., Zhao X., Deng R., Wang Y., Huang J., Xu M., Yan J., Yu J. lncRNA H19/miR-675 axis represses prostate cancer metastasis by targeting TGFBI. FEBS J. 2014;281:3766–3775. doi: 10.1111/febs.12902. [DOI] [PubMed] [Google Scholar]
- 39.Zhou W., Ye X.L., Xu J., Cao M.G., Fang Z.Y., Li L.Y., Guan G.H., Liu Q., Qian Y.H., Xie D. The lncRNA H19 mediates breast cancer cell plasticity during EMT and MET plasticity by differentially sponging miR-200b/c and let-7b. Sci Signal. 2017;10 doi: 10.1126/scisignal.aak9557. [DOI] [PubMed] [Google Scholar]
- 40.Gabory A., Jammes H., Dandolo L. The H19 locus: role of an imprinted non-coding RNA in growth and development. Bioessays. 2010;32:473–480. doi: 10.1002/bies.200900170. [DOI] [PubMed] [Google Scholar]
- 41.Van der Sluis M., De Koning B.A., De Bruijn A.C., Velcich A., Meijerink J.P., Van Goudoever J.B., Büller H.A., Dekker J., Van Seuningen I., Renes I.B., Einerhand A.W. Muc2-deficient mice spontaneously develop colitis, indicating that MUC2 is critical for colonic protection. Gastroenterology. 2006;131:117–129. doi: 10.1053/j.gastro.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 42.Riba A., Olier M., Lacroix-Lamande S., Lencina C., Bacquié V., Harkat C., Gillet M., Baron M., Sommer C., Mallet V., Salvador-Cartier C., Laurent F., Théodorou V., Ménard S. Paneth cell defects induce microbiota dysbiosis in mice and promote visceral hypersensitivity. Gastroenterology. 2017;153:1594–1606. doi: 10.1053/j.gastro.2017.08.044. [DOI] [PubMed] [Google Scholar]
- 43.van Es J.H., Sato T., van de Wetering M., Lyubimova A., Yee Nee A.N., Gregorieff A., Sasaki N., Zeinstra L., van den Born M., Korving J., Martens A.C.M., Barker N., van Oudenaarden A., Clevers H. Dll1+ secretory progenitor cells revert to stem cells upon crypt damage. Nat Cell Biol. 2012;14:1099–1104. doi: 10.1038/ncb2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schuijers J., Clevers H. Adult mammalian stem cells: the role of Wnt, Lgr5 and R-spondins. EMBO J. 2012;31:2685–2696. doi: 10.1038/emboj.2012.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Heuberger J., Kosel F., Qi J., Grossmann K.S., Rajewsky K., Birchmeier W. Shp2/MAPK signaling controls goblet/Paneth cell fate decisions in the intestine. Proc Natl Acad Sci U S A. 2014;111:3472–3477. doi: 10.1073/pnas.1309342111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhuo C., Jiang R., Lin X., Shao M. LncRNA H19 inhibits autophagy by epigenetically silencing of DIRAS3 in diabetic cardiomyopathy. Oncotarget. 2017;8:1429–1437. doi: 10.18632/oncotarget.13637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang M., Han D., Yuan Z., Hu H., Zhao Z., Yang R., Jin Y., Zou C., Chen Y., Wang G., Gao X., Wang X. Long non-coding RNA H19 confers 5-Fu resistance in colorectal cancer by promoting SIRT1-mediated autophagy. Cell Death Dis. 2018;9:1149. doi: 10.1038/s41419-018-1187-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bento C.F., Renna M., Ghislat G., Puri C., Ashkenazi A., Vicinanza M., Menzies F.M., Rubinsztein D.C. Mammalian autophagy: how does it work? Annu Rev Biochem. 2016;85:685–713. doi: 10.1146/annurev-biochem-060815-014556. [DOI] [PubMed] [Google Scholar]
- 49.Wu Y., Tang L., Wang B., Sun Q., Zhao P., Li W. The role of autophagy in maintaining intestinal mucosal barrier. J Cell Physiol. 2019;234:19406–19419. doi: 10.1002/jcp.28722. [DOI] [PubMed] [Google Scholar]
- 50.Khor B., Gardet A., Xavier R.J. Genetics and pathogenesis of inflammatory bowel disease. Nature. 2011;474:307–317. doi: 10.1038/nature10209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Marchiando A.M., Ramanan D., Ding Y., Gomez L.E., Hubbard-Lucey V.M., Maurer K., Wang C., Ziel J.W., van Rooijen N., Nuñez G., Finlay B.B., Mysorekar I.U., Cadwell K. A deficiency in the autophagy gene Atg16L1 enhances resistance to enteric bacterial infection. Cell Host Microbe. 2013;14:216–224. doi: 10.1016/j.chom.2013.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu R., Li X., Zhu W., Wang Y., Zhao D., Wang X., Gurley E.C., Liang G., Chen W., Lai G., Pandak W.M., Robert Lippman H., Bajaj J.S., Hylemon P.B., Zhou H. Cholangiocyte-derived exosomal long noncoding RNA H19 promotes hepatic stellate cell activation and cholestatic liver fibrosis. Hepatology. 2019;70:1317–1335. doi: 10.1002/hep.30662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nakagawa H., Chadwick R.B., Peltomaki P., Plass C., Nakamura Y., de La Chapelle A. Loss of imprinting of the insulin-like growth factor II gene occurs by biallelic methylation in a core region of H19-associated CTCF-binding sites in colorectal cancer. Proc Natl Acad Sci U S A. 2001;98:591–596. doi: 10.1073/pnas.011528698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xiao L., Wu J., Wang J.Y., Chung H.K., Kalakonda S., Rao J.N., Gorospe M., Wang J.Y. Long noncoding RNA uc.173 promotes renewal of the intestinal mucosa by inducing degradation of microRNA 195. Gastroenterology. 2018;154:599–611. doi: 10.1053/j.gastro.2017.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu L., Xiao L., Chung H.K., Kwon M.S., Li X.X., Wu N., Rao J.N., Wang J.Y. RNA-binding protein HuR regulates Rac1 nucleocytoplasmic shuttling through nucleophosmin in the intestinal epithelium. Cell Mol Gastroenterol Hepatol. 2019;8:475–486. doi: 10.1016/j.jcmgh.2019.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lindemans C.A., Calafiore M., Mertelsmmann A.M., O’Connor M.H., Dudakov J.A., Jeng R.R., Velardi E., Young L.F., Smith O.M., Lawrence G., Ivanov J.A., Fu Y.Y., Takashima S., Hua G., Martin M.L., O’Rourke K.P., Lo Y.H., Mokry M., Romera-Hernandez M., Cupedo T., Dow L., Nieuwenhuis E.E., Shroyer N.F., Liu C., Kolesnick R., van den Brink M.R.M., Hanash A.M. Interleukin-22 promotes intestinal stem cell-mediated epithelial regeneration. Nature. 2015;528:560–564. doi: 10.1038/nature16460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hubbard W.J., Choudhry M., Schwacha M.G., Kerby J.D., Rue L.W., 3rd, Bland K.I., Chaudry I.H. Cecal ligation and puncture. Shock. 2005;24(Suppl 1):52–57. doi: 10.1097/01.shk.0000191414.94461.7e. [DOI] [PubMed] [Google Scholar]
- 58.Harter H.L. Critical values for Duncan’s new multiple range test. Biometrics. 1960;16:671–685. [Google Scholar]