Abstract
Background/Aim: Prostate cancer (PCa) is the most frequent cancer found in males worldwide, and its mortality rate is increasing every year. However, there are no known molecular markers for advanced or aggressive PCa, and there is an urgent clinical need for biomarkers that can be used for prognosis and prediction of PCa. Materials and Methods: Mass spectrometry-based proteomics was used to identify new biomarkers in tissues obtained from patients with PCa who were diagnosed with T2, T3, or metastatic PCa in regional lymph nodes. Results: Among 1,904 proteins identified in the prostate tissues, 344 differentially expressed proteins were defined, of which 124 were up-regulated and 216 were down-regulated. Subsequently, based on the results of partial least squares discriminant analysis and Gene Ontology and Kyoto Encyclopedia of Genes and Genomes enrichment analyses, we proposed that spermidine synthase (SRM), nucleolar and coiled-body phosphoprotein 1 (NOLC1), and prostacyclin synthase (PTGIS) represent new protein biomarkers for diagnosis of advanced PCa. These proteomics results were verified by immunoblot assays in metastatic PCa cell lines and by indirect enzyme-linked immunosorbent assay in prostate specimens. Conclusion: SRM was significantly increased depending on the cancer stage, confirming the possibility of using SRM as a biomarker for prognosis and prediction of advanced PCa.
Keywords: Prostate cancer, quantitative proteomics, SRM, diagnostics, markers
According to a recent report from the American Cancer Society, prostate cancer (PCa) has the highest incidence of any cancer, and the mortality arising from PCa is second among all male cancers in the USA (1). Although most PCa is found at a local or regional stage, with a 5-year survival rate of close to 100%, the 5-year survival rate for those with late-stage PCa drops sharply to 30% (2,3). Advanced PCa, also called metastatic PCa, is an aggressive form that causes the prostatic adenocarcinoma to spread to other parts of the body, including pericapsular tissues, with lymph-node involvement, and distant metastases, which is histologically defined as stages T3 and T4 (4,5). Overall, PCa has one of the highest incidence and mortality rates among cancers, and most of the deaths due to PCa are a result of metastasis. Thus, finding a biomarker for advanced PCa associated with metastasis and aggression is a pressing need in clinical research aimed at treating PCa.
Tissue, blood, urine, and seminal fluids derived from patients are the typical sources used for the study of PCa biomarkers. Such bio-fluid samples are safe, owing to the non-invasive collection methods used, and can be collected quickly, lowering the cost of sample preparation. However, it is difficult to find low-abundance biomarkers in these samples, since most of the protein in the blood and urine is either albumin or uromodulin, respectively. In contrast, tissues collected after surgery can be used to directly observe the expression of tumor proteins, which might be used to indicate metastatic PCa. In particular, tissue-based biomarkers can be used as diagnostic, prognostic, and predictive markers (6). Advanced PCa has a poor prognosis, and most PCa deaths are caused by metastasis occurring after surgery (4). However, it is highly important to analyze tissues acquired directly from patients who have undergone surgery in order to predict metastasis. To date, there are no molecular markers available for accurate prediction and prognosis of advanced PCa.
During PCa diagnosis and treatment, decisions are based on a risk stratification strategy, which integrates three important prognostic and predictive factors: the pathological tumor, node, metastasis (TNM) score for determining the clinical stage; a biopsy Gleason Score (GS); and serum prostate-specific antigen (PSA) level (7). The PSA test is most often used for the early diagnosis of PCa and is the U.S. Food and Drug Administration-approved biomarker for PCa. However, the PSA level is not able to discriminate aggressive or advanced PCa, particularly at PSA levels below 20 ng/ml (8,9). Another difficulty with PSA is that it has a high false-positive rate, with only about 20% to 50% of men with positive PSA results (2.5-10 ng/ml serum PSA) being diagnosed with PCa after biopsy (10). A recent study also indicated that the early detection of PCa using the PSA test does not reduce the mortality rate of PCa. As such, the currently available methods cannot provide accurate parameters for prediction of metastatic potential.
Proteomics is an ideal and highly translatable technique for identifying novel biomarkers. Over the past 15 years, a number of potential tissue-based biomarkers for PCa have been ascertained based on proteomics approaches. Traditionally, two-dimensional difference gel electrophoresis (2D-DIGE) has been utilized to identify candidate protein biomarkers by comparing normal tissues with tissues representative of PCa or benign prostatic hyperplasia with PCa (11-14). Although 2D-DIGE is a familiar tool for proteomics analysis, recent mass spectrometry-based proteomics, also known as shotgun proteomics, employing nano liquid chromatography tandem mass spectrometry (LC-MS/MS) have been used extensively, and such analyses can identify up to 20 times more proteins relative to conventional 2D-DIGE analysis (15). In this study, we investigated the differences in protein expression patterns between normal, PCa, and advanced PCa tissues for the identification of differentially expressed proteins (DEPs) using a shotgun proteomics approach. The identified biomarkers were then verified for use in the prediction and prognosis of advanced PCa.
Materials and Methods
Clinical tissues of PCa from patients. The biospecimens and data used for this study were provided by the National Biobank of Korea-Kyungpook National University Hospital, a member of the Korea Biobank Network-KNUH, and were obtained (with informed consent) under Institutional Review Board-approved protocols (approval number: KNUMC 2016-05-021). All samples were obtained from 50 patients treated at the Kyungpook National University Chilgok Hospital. All tumor samples were acquired after acquiring patient consent for tissue sample donation and an examination was performed. The diagnosis of PCa was verified based on the outcomes of pathological analyses. For each patient, both normal and tumor tissues were collected and used in these studies. Both the normal tissues and PCa tissues (separated into T2 and T3 stages) were stored at −80˚C before use.
Sample preparation for quantitative proteomics. The prostate tissues were washed with 1 ml of PBS and then homogenized using a hand-held homogenizer in sodium dodecyl sulfate lysis buffer (50 mM Tris-Hcl, pH 6.8, 2% sodium dodecyl sulfate, 1% 2-mercaptoethanol, and 12.5 mM ethylenediaminetetra-acetic acid) including the Halt Protease Inhibitor Cocktail (Thermo Fisher Scientific, Rockford, IL, USA). For complete protein extraction, the homogenized tissues were sonicated for 2 min on ice and then incubated at 4˚C for 30 min on a rotating mixer. The samples were centrifuged at 12,000 ×g for 10 min at 4˚C, and the supernatant was transferred to low-protein binding E-tubes. The protein concentration was measured with a BCA Protein Assay kit (Thermo Fisher Scientific, Waltham, MA, USA). Proteins were reduced by the addition of 15 mM dithiothreitol and incubated at 56˚C for 30 min. The proteins were then alkylated with 60 mM iodoacetamide at room temperature for 30 min in the dark. To remove the detergent and chemical reagents, 10% trichloroacetic acid was added to the protein samples and incubated for 4 h at 4˚C. After centrifuging at 12,000 × g for 10 min at 4˚C, the protein pellets were washed twice with 500 μl of ice-cold acetone. The proteins were then centrifuged at 12,000 × g for 10 min at 4˚C, the pellets were then re-suspended in 100 mM triethylammonium bicarbonate, and the resulting protein concentrations were re-]measured using a BCA Protein Assay kit for tandem mass tag (TMT)-based quantitative proteomics. Proteins (100 μg) from the different prostate tissues were digested using trypsin at 37˚C overnight. For TMT labeling, 0.8 mg of the TMT sixplex reagent, dissolved in 41 μl of acetonitrile (ACN), was added to the peptides and incubated at room temperature for 1 h. Hydroxylamine (5%) in 50 mM triethylammonium bicarbonate was added to the labeled peptides and incubated for 15 min at room temperature to stop the TMT labeling reaction. Finally, all the labeled samples were combined into a low-protein binding E-tube.
To evaluate the number of proteins and peptides, the combined peptides were separated using both a High-PH RP fractionation kit and an OFFGEL fractionator (Agilent, Santa Clara, CA, USA). Both fractionations were performed using the manufacturer’s instructions. Briefly, eight different elution buffers were made in 0.1% triethylamine with 10%, 12.5%, 15%, 17.5%, 20%, 22.5%, 25%, and 50% ACN for the High-PH RP fractionation. After activation of the High-PH RP fractionation column with ACN and 0.1% trifluoroacetic acid (TFA), 100 μg of the peptides were dissolved in water containing 0.1% TFA and added to the column. For the washing step, 95% water in 5% ACN was added to the column, after which the peptides were eluted using eight different buffers. All of the samples were dried using a speed-vacuum system (Labconco, Kansas City, MO, USA). To perform the OFFGEL fractionation, we used high-resolution 24-well frame Immobiline DryStrip gels (IPG strips) at pH 3-10. Following the manufacturer’s procedure, 200 μg of the peptides were re-suspended in water including the OFFGEL buffer pH 3-10 and then loaded in each of the 24-well frames. Focusing was performed at a maximum current of 50 μA and stopped after the total voltage reached 50 kVh (approximately 18 h). The samples were recovered from each well and merged into groups of twelve consecutive fractions. All the fractionated samples were desalted using a C18 ziptip (Millipore, Milford, MA, USA) and then evaporated completely using a speed-vacuum system.
LC-MS/MS analysis. LC-MS/MS analysis was performed on both an LTQ-velos Orbitrap spectrometer with an Eksigent nanoLC chromatography system and a Q-Exactive spectrometer connected with an Easy nanoLC-1000 chromatograph (Thermo Fisher Scientific to reduce the technical variability. The samples were dissolved in solvent A (98% water in 0.1% formic acid). The 75 min gradient used for the Eksigent nanoLC was 5% solvent B (100% ACN in 0.1% formic acid) for 10 min, 5-30% solvent B for 50 min, 30-90% solvent B for 5 min, and 90% for 10 min at flow rate of 300 nl/min using a home-made C12 reverse-phase analytical column (75 μm × 100 mm, Jupiter C12 resin, 4 μm particle size, 90 Å pore size; Phenomenex Inc., Torrance, CA, USA). In contrast, the 60-min gradient for the Easy nanoLC-1000 was 2 min to 5% solvent B, 50 min to 23% solvent B, 3 min to 90% solvent B, and this was maintained for 5 min at a constant flow rate of 300 nl/min with a commercial C18 reverse-phase column (75 μm × 150 mm, 3 μm particle size, 100 Å pore size; Thermo Fisher Scientific).
The LTQ-velos Orbitrap was run using the following settings: source voltage 1.8 kV; MS range m/z 300-1,800; MS resolution 30,000; top 10 data-dependent mode; MS/MS resolution 7500; isolation width 3 m/z; normalized collision energy at 40% with higher energy collision dissociation (HCD) mode; 0.1 ms activation time; and first mass fixed at 100 m/z. For the Q-Exactive, full MS scans were acquired at m/z 300-1,800 with a resolution of 70,000; 1×106 of target value; and 120 ms of maximum IT. The top 15 data-dependent mode was fragmented with normalized collision energy at 27% in higher energy collision dissociation mode. MS/MS were obtained using the following settings: MS/MS resolution of 17,500; isolation width of 1.5 m/z; AGC target at 1e6; 60 ms of maximum IT; and first mass fixed at 100 m/z. The MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD013422 (16). Analysis was carried out by using (LTQ-velos Orbitrap) supported by Basic Science Research Capacity Enhancement Project through Korea Basic Science Institute (National Research Facilities and Equipment Center).
Data analysis and bioinformatics. MS/MS data were used to query the UniProtKB human database using MaxQuant 1.5.1.0 (downloaded from http://www.uniprot.org/proteomes/UP000005640; including 71,772 protein sequences) (17). To identify proteins and peptides, trypsin/P was used for the cleavage enzyme, and up to two missing cleavages were permitted. Additionally, carbamidomethylation on cysteine was set as a fixed modification, whereas oxidation of methionine and acetylation of the N-terminus were set as variable modifications. The mass tolerance was 5 ppm for full MS and 20 ppm for MS/MS. The reporter ions were calculated using the TMT sixplex tag on the peptide N-terminus and lysine residues; in addition, the minimum ratio count was 2. All of the other parameters were set to default values.
Search results were filtered at a MaxQuant score ≥40 and a false-discovery rate ≤0.01, as well as by removing incorrect proteins from the protein list that included reverse sequence, potential contaminants, or those only identified by a modification site. The protein ratio was calculated based on the reporter intensity of each PCa group per the reporter intensity of the control group. The ratios obtained were normalized to the median, and a log2-transformation of the protein ratios was obtained.
The lists of Gene Ontology (GO) classifications and the Kyoto Encyclopedia of Genes and Genomes (KEGG) for pathway analysis were generated using the Database for Annotation, Visualization, and Integrated Discovery (DAVID). The functional enrichment analysis of the GO and KEGG data was performed using DAVID with an EASE score and a modified Fisher’s exact test p-value of below 0.05 (18). The SIMCA P+ software (version 13.0; Metrics, Umea, Sweden) was used for partial least squares discriminant analysis (PLS-DA) to examine the extent of the differences among patient groups. The input variables were the PCa versus control ratios quantified with technical replication and featuring mean centering and Pareto scaling.
Unsupervised hierarchical clustering was performed within Perseus using Euclidian distance according to procedures previously described (19). Briefly, the log2-transformed ratios were normalized by their Z-score and clustered using a cluster number of 2 and a maximum of 100 iterations. In addition, a one-way t-test was used to distinguish the significantly changed proteins among the DEPs relative to the T-stage, and a volcano plot was constructed using Perseus according to the developer’s instructions (20). Gene expression profiling interactive analysis (GEPIA) and cBioPortal were used to compare the mRNA levels and protein ratios (21,22).
Results
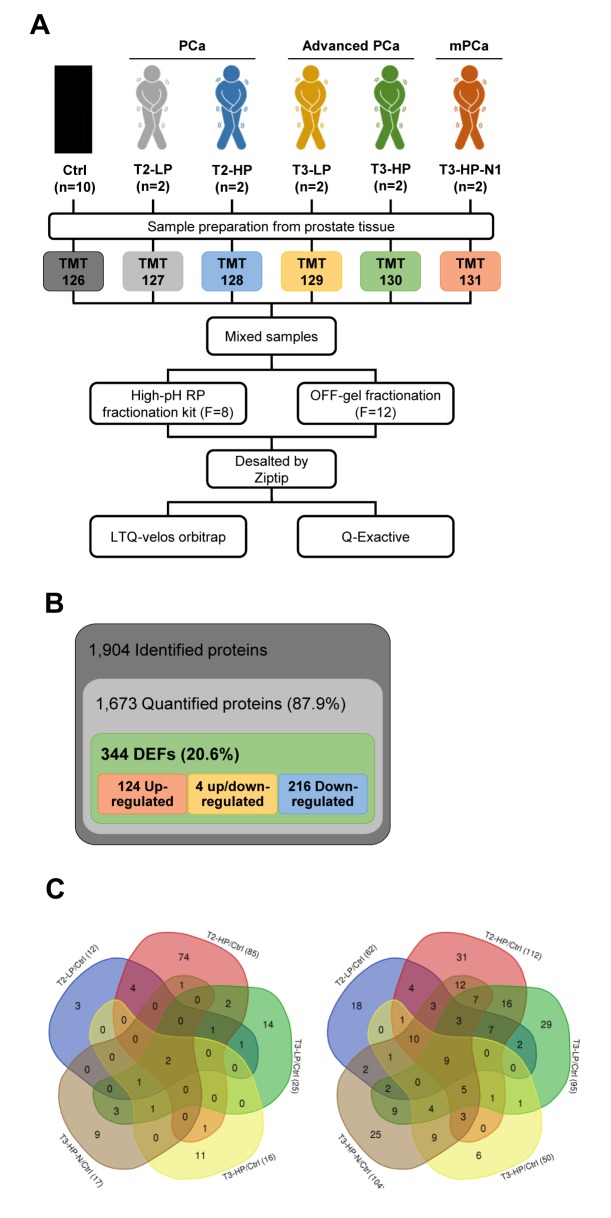
Quantitative tissue proteomics for profiling of prostate cancer. To profile proteome alterations occurring in the different stages of PCa, we used a MS-based quantitative proteomics approach based on TMT (Figure 1). All the tissues were obtained from the National Biobank of Korea-Kyungpook National University Hospital. For the proteomics analysis, we used 10 normal surrounding prostate tissues from the patients, and two PCa tissues from each group among 50 patients, as shown in Supplemental Table S1 which was selected for each stage with distinct characteristics. The PCa groups were divided based on the patient’s tumor T-stage (T2 as PCa or T3 as advanced PCa) and serum PSA level. In order to clearly distinguish PSA levels, a PSA value less than 5 ng/ml was defined as low (LP) and greater than 20 ng/ml PSA was defined as high (HP). Furthermore, a separate group demonstrating stage T3, HP, and metastasis in regional lymph nodes (N1) was defined as including patients with high-risk advanced PCa. Ultimately, the patients were classified into six groups, referred to as the control, T2-LP, T2-HP, T3-LP, T3-HP, and T3-HP-N1 (Figure 1A).
Figure 1. A: The experimental scheme for quantitative tissue-based proteomic profiling of prostate cancer (PCa) tissues. B: Overview of the frequency of differentially expressed proteins from prostate tissues identified using a proteomics approach. C: Venn diagrams depicting the up-regulated (left) and down-regulated proteins (right) among the patient’s with different T-stage (T2 as PCa, or T3 as advanced PCa) and serum prostate-specific antigen level (LP: low level; HP: high level). Ctrl: Control; DEPs: differentially expressed proteins.
Overall, we identified 1,904 proteins in the prostate tissues, among which we were able to quantify 1,673 (87.9%) proteins (Supplemental Table S2). To quantify the proteomics data, the ratio of each PCa reporter’s ion intensity to that of the control reporter’s was calculated and normalized using the median value (Supplemental Figure S1). We examined the Pearson correlation coefficients associated with the fractionation and LC-MS/MS data to ensure confidence in the quantification results. As expected, all the Pearson correlation coefficients between the technical replicates were very high, at 0.87 on average (Supplemental Figure S2). These data signified that our proteomics approach was technically sound. In total, there were 344 DEPs, including 124 up-regulated proteins, 216 down-regulated proteins that changed in their expression level by more than two-fold (log2 scale ≤−1 and ≥1) when comparing normal and PCa tissues (Figure 1B). Using a Venn diagram of the DEPs in each PCa group, cysteine-rich secretory protein 3 precursor (CRISP3) and alpha-N-acetylgalactosaminidase (NAGA) were found to be up-regulated in all the PCa groups relative to the normal tissues. In contrast, nine proteins, agamous-like MADS-box protein (AGL), CD9, antigen, glutamate receptor ionotropic NMDA 2D (GRIN2D), protein S100 A6 (S100A6), S100A8, S100A10, semenogelin-1 (SEMG1), SEMG2 and tropomyosin alpha-1 chain (TPM1), were reduced in all the PCa groups compared with normal prostate tissues (Figure 1C).
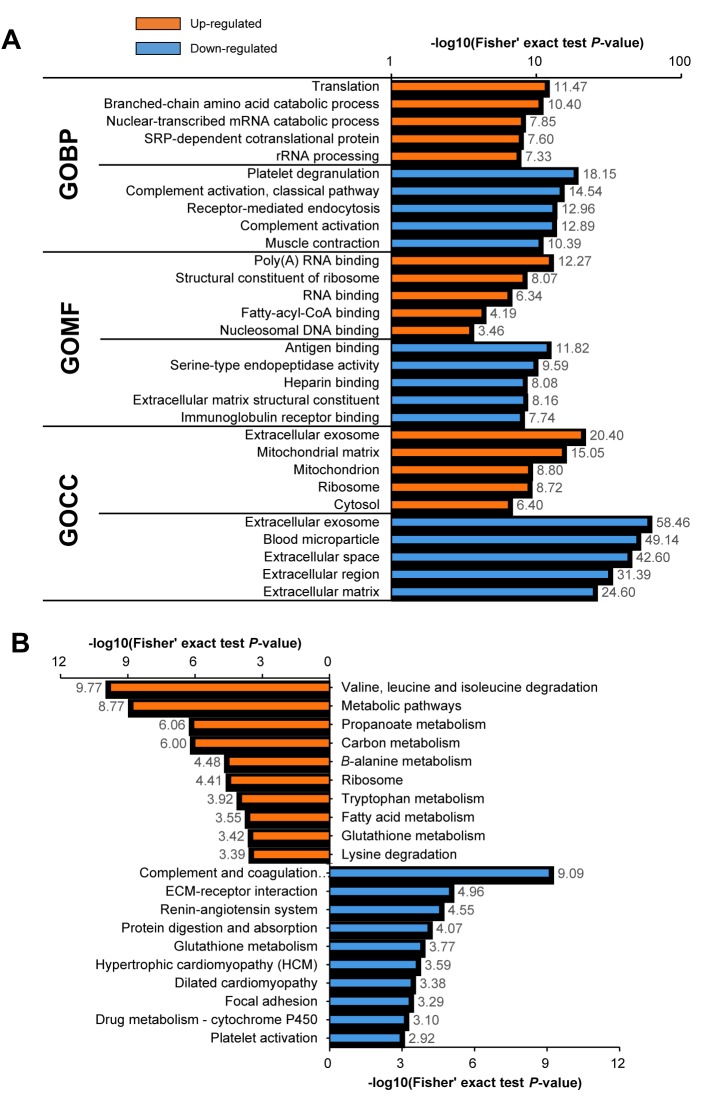
Bioinformatics analysis of the proteomics results. Next, we performed GO and KEGG enrichment analysis using DAVID to understand the potential functional implications of the DEPs identified in the PCa samples (Figure 2). We selected the top five GO terms with respect to biological process (GOBP), molecular function (GOMF), and cellular component (GOCC) (Figure 2A). The GOBP showed that the up-regulated proteins were involved in translation and rRNA processing, whereas platelet aggregation, complement activation, and muscle contraction were enriched in the down-regulated proteins. With respect to GOMF, the most highly enriched categories for the up-regulated proteins were binding to poly(A) RNA, RNA, and nucleosomal DNA. In contrast, antigen and heparin binding, as well as extracellular matrix (ECM) structural constituents, were associated with the down-regulated proteins. Lastly, in the GOCC categories among the proteins changed in PCa there was an equal representation of extracellular exosomes in both the up- and down-regulated proteins. The up-regulated proteins were also found to have GOCC categories that were partly mitochondrial and partly ribosomal. In contrast, the down-regulated proteins were exceptionally enriched for blood microparticles and extracellular related categories.
Figure 2. A: Gene Ontology (GO) functional enrichment analysis of the differentially expressed proteins found in prostate cancer. B: Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of the differentially expressed proteins found in prostate cancer. GOBP: Biological process; GOMF molecular function; GOCC: cellular component.
For the KEGG pathway enrichment analysis, the proteins increased in PCa were mainly associated with metabolic pathways, such as valine, leucine, and isoleucine degradation as well as propanoate, carbon, and fatty acid metabolism (Figure 2B). In contrast, the down-regulated proteins were associated with processing in the complement and coagulation pathways, the extracellular associated pathway, ECM–receptor interaction, and focal adhesion.
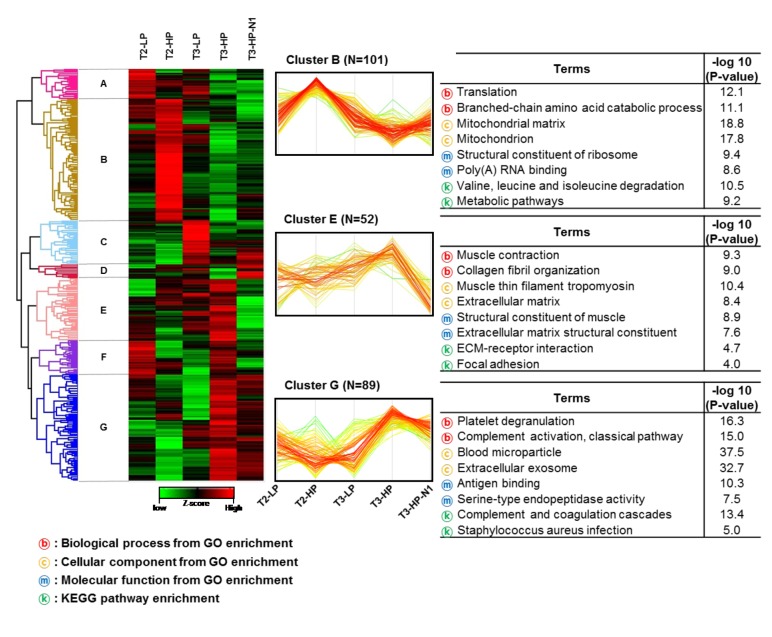
Next, we performed unsupervised hierarchical clustering of the quantitative protein data to produce clusters of proteins with similar patterns throughout the PCa development process (Figure 3). The quantitative ratio of DEPs was re-normalized using the Z-score. The horizontal axis was arranged in the order T2-LP, T2-HP, T3-LP, T3-HP, and T3-HP-N1, which was expected to reflect the order of increasingly advanced PCa. A red-green plot was used to improve the visualization. Based on this analysis, we generated seven clusters (A-G) that contained 25, 101, 37, 12, 52, 28, and 89 DEPs, respectively. Among these, three dominant clusters, clusters B (N=101), E (N=52), and G (N=89), are shown in Figure 3.
Figure 3. Unsupervised hierarchical clustering of proteins altered during the development of prostate cancer. T2/T3: T2/T3 stage; LP: low serum prostate-specific antigen level; HP: high serum prostate-specific antigen level. KEGG: Kyoto Encyclopedia of Genes and Genomes; GO: Gene Ontology.
Cluster B was present at relatively higher levels in the T2 stage relative to the T3 stage. Cluster E showed a gradually increasing pattern from T2 to T3, with the exception of T3-HP-N1. Cluster G showed relatively high expression levels in the groups T3-HP and T3-HP-N1, which would be expected to represent highly aggressive PCa. GO and KEGG enrichment analyses were performed using DAVID in order to understand the biological relevance of the proteins contained in these clusters. Interestingly, cluster B was highly associated with mitochondrial-related functions, including metabolic pathways, whereas cluster E was strongly related to muscle function, ECM, ECM–receptor interactions, and focal adhesion processes. Cluster G was enriched for proteins associated with platelet degranulation, blood microparticles, as well as the complement and coagulation cascades. The results of this unsupervised hierarchical clustering with functional enrichment suggested that mitochondrial function declines, whereas the performance of muscle, extracellular proteins, and blood microparticles increases relative to the progression of PCa.
Selection of candidate biomarkers in advanced PCa. In order to explore the separation of the PCa groups, a PLS-DA using the quantified ratio datasets was generated using SIMCA P+ software (Supplemental Figure S3). The PLS-DA gave R2X=0.71 and Q2=0.31 for the discrimination of the PCa groups. This result indicated that the technical replicates within the groups were largely clustered together and were clearly distinguishable from the other groups. In addition, the T-stage was clearly distinguished on the y-axis, with T2 on the upper side (red zone) and T3 on the lower side (blue zone). In contrast, the PSA groups did not separate well because the T2-LP group was found among the HP groups.
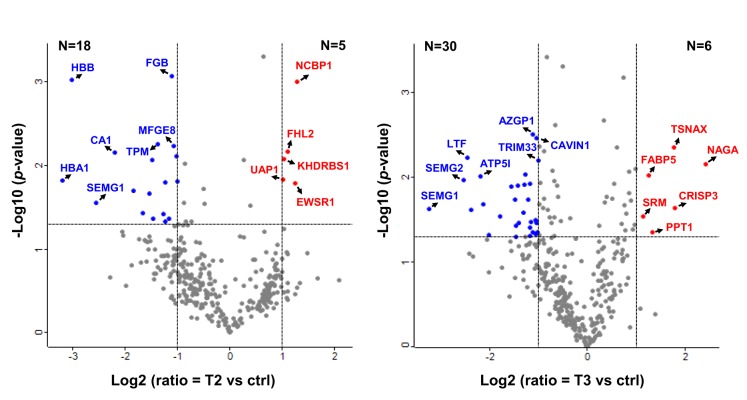
In order to find statistically significant DEPs in the T-stage groups, we carried out a one-way t-test on the 344 DEPs using Perseus, and the results are displayed as volcano plots (Figure 4). The DEPs in each of the T2 (T2-LP, T2-HP) and T3 (T3-LP, T3-HP, and T3-HP-N1) groups were individually used to apply a one-way t-test and were filtered using a p-value of less 0.05 and a greater than two-fold (log2 scale ≤−1 and ≥1) change in expression. Consequently, we found 23 and 36 statistically increased DEPs in the T2 and T3 groups, respectively.
Figure 4. Volcano plot depicting the T2-stage and T3-stage sets of differentially expressed proteins via the tandem mass tag analysis. ctrl: Control. ATP5I, ATP synthase subunit e, mitochondrial; AZGP1, zinc-alpha-2-glycoprotein; CA1, carbonic anhydrase 1; CAVIN1, caveolae-associated protein 1; CRISP3, cysteine-rich secretory protein 3; EWSR1, RNA-binding protein EWS; FABP5, fatty acid-binding protein, epidermal; FGB, fibrinogen beta chain; FHL2, four and a half LIM domains protein 2; HBA1, hemoglobin subunit alpha; HBB, hemoglobin subunit beta; KHDRBS1, KH domain-containing, RNA-binding, signal transduction-associated protein 1; LTF, lactotransferrin; MFGE8, lactadherin; NAG, alpha-Nacetylgalactosaminidase; NCBP1, nuclear cap-binding protein subunit 1; PPT1, palmitoyl-protein thioesterase 1; SEMG1, semenogelin-1; SEMG2, semenogelin-2; SRM, spermidine synthase; TPM, tropomyosin alpha-4 chain; TRIM33, E3 ubiquitin-protein ligase; TSNAX, translin-associated protein X; UAP1, UDP-N-acetylhexosamine pyrophosphorylase.
We had carefully selected the following criteria to identify reliable candidate biomarkers in advanced PCa tissues and to avoid false-positives due to chance correlations (Supplemental Figure S4A). Of the 344 DEPs identified in our proteomics analysis, 45 proteins that were changed in all the PCa groups, or only in the T3-HP-N1 group, were selected. Accordingly, we included 55 statistically significant DEPs found via a one-way t-test, displayed in a volcano plot, and a total of 89 DEPs were first selected (Supplemental Figure S4B). Among these 89 proteins, we manually sorted 31 proteins with the same expression pattern as the genomics database containing GEPIA and cBioPortal (Supplemental Table S3). With the exception of previously reported studies on 28 of these proteins in PCa, we singled out three proteins that consisted of the up-regulated proteins spermidine synthase (SRM) and nucleolar and coiled-body phosphoprotein 1 (NOLC1) as well as the down-regulated protein prostacyclin synthase (PTGIS) as being previously unknown candidate biomarkers for advanced PCa. Additionally, these three proteins also showed the greatest change in the T3-HP-N1 group and were significantly altered between the T2 and T3 stages. As such, these three proteins were chosen as potential protein candidate biomarkers for prognosis and prediction of advanced PCa. Furthermore, cysteine-rich secretory protein 3 (CRISP3), which had been previously shown to be altered in PCa, was selected as a positive control for subsequent assays, immunoblot analyses, and enzyme-linked immunosorbent assays (ELISAs) (23,24).
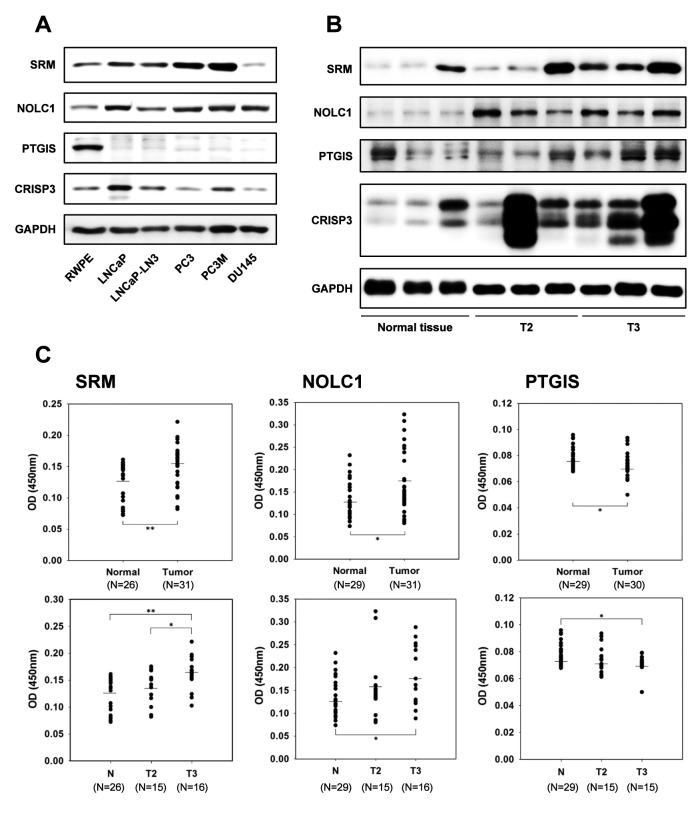
Verification of candidate biomarkers using immunoblot analysis and ELISA. To verify the proteomics results, we used an immunoblot assay to determine whether the three candidate proteins and the control protein could be detected in prostate specimens and in PCa cell lines, including LNCaP, LNCaP-LN3, PC-3, PC-3M, and DU-145, when compared to RWPE, a normal prostate cell line. As shown in Figure 5A, except for DU-145, the protein expression of SRM in all the PCa cells was observed to be significantly increased in the LNCaP, LNCaP-LN3, PC-3, and PC-3M cell lines. NOLC1 appeared to be slightly elevated in all the PCa cell lines. Another candidate biomarker, PTGIS, which was reduced in our proteomics analysis, was confirmed to be dramatically reduced in all the PCa cell lines compared with the normal prostate cell line. CRISP3, as the positive control, was elevated in the LNCaP, LNCaP-LN3, and PC-3M cell lines compared to the normal prostate cell line.
Figure 5. A: Immunoblot analysis verifying the expression of the protein candidate biomarkers for prostate cancer in prostate cancer cell lines. B: Randomly selected prostate tissues in T2 and T3 stages among 50 patient tissues. C: Confirmation of spermidine synthase (SRM), nucleolar and coiled-body phosphoprotein 1 (NOLC1) and prostacyclin synthase (PTGIS) as protein candidate biomarkers for prostate cancer using enzymelinked immunosorbent assay. CRISP3: Cysteine-rich secretory protein 3 precursor; GAPDH: glyceraldehyde 3-phosphate dehydrogenase. Significantly different at *p<0.05 and **p<0.01.
In addition, the candidate biomarkers were verified in normal and PCa tissues using immunoblot analyses (Figure 5B). As expected, SRM, NOLC1, and CRISP3 were significantly increased in the T2 and T3 PCa samples when compared with normal prostate tissues. However, for PTGIS there was no significant difference between the normal and PCa tissues. Interestingly, both SRM and CRISP3 were found to be increased in the T3 samples compared the T2 samples.
To confirm the immunoblot results, we carried out an indirect ELISA using the prostate tissues from patients with PCa and healthy controls (Figure 5C). As expected, the relative protein levels of SRM were significantly elevated in all PCa tissues (n=31, p<0.01) and from men who went on to be diagnosed with T2 stage (n=15) or T3 stage (n=16, p<0.01) PCa when compared with normal tissues. There was also an increase in SRM level between the T2 and T3 stages (p<0.05). NOLC1 was also elevated in tumor tissues (n=31, p<0.05); in addition, the expression of NOLC1 in the T3 (N=16, p<0.05) and T2 stages were also higher than in the control tissues. Unfortunately, the protein range for the T3 group appeared to be very wide. PTGIS, which was expected to be reduced in PCa, was in fact only slightly reduced in PCa (n=30, p<0.05) when compared to normal tissue, and the levels of PTGIS in the T3 group were slightly lower than those in the T2 group. Overall, we observed that the expression of SRM was more affected in advanced PCa than the expression of NOLC1 or PTGIS, indicating that SRM might be an important prognostic and predictive biomarker candidate for advanced PCa.
Discussion
We adopted an MS-based proteomics approach to identify potential prognostic and predictive protein biomarkers from PCa tissues. In addition, we attempted to reveal the relationship between serum PSA levels and protein expression levels in PCa, which had previously remained unexplored. Previous proteomics studies in PCa tissues did not consider serum PSA levels but instead concentrated on the TNM stage and biopsy GS only. Typically, most clinicians consider the serum PSA level to be normal at levels below 4 ng/nl and often recommend a prostate biopsy if the PSA level is above 4 ng/ml. The risk of PCa based on serum PSA level can be divided into three stages: low, intermediate, and high risk. For patients with low-risk PSA level (4 to 10 ng/ml), it is ambiguous as to whether they have PCa or not. In addition, if the PSA level is above 10, patients are classified as at intermediate risk, and above 20 as high risk (25,26). For this reason, we classified patients into low PSA (LP; ≤5 ng/ml) or high PSA (HP; ≥20 ng/ml) groups at each T-stage in order to be able to assess the protein changes in PCa based on the serum PSA level. Our original intention was to define a 4 ng/ml as being cut-off for the low PSA group. Unfortunately, this PSA level was extremely rare in the patients for whom we had PCa tissue samples. Accordingly, we selected 5 ng/ml as the cut-off value for the low PSA group.
Using a TMT-based proteomics approach, a total of 1,904 and 1,673 proteins were identified and the quantified from PCa tissues, respectively. Specifically, we found that there were 344 DEPs, of which 124 were up-regulated and 216 were down-regulated in the PCa samples. Among these, CRISP3 was found to be increased in all PCa groups, as has been presented in other previously published studies (23,24). CRISP3 is closely associated with cancer progression, including aggressive PCa, as based on ETS-related gene (ERG) status and phosphatase and tensin homolog (PTEN) deletion, as well as with castration-resistant PCa. In addition, S100A6, S100A8, and S100A10 were all down-regulated in all of the PCa groups. The S100 family of proteins consists of calcium-binding proteins that are known to play an important role in tumorigenesis by controlling cell differentiation, growth, migration, and invasion. Of the 21 members of the S100 family, S100A3, S100A4, S100A8, S100A9, and S100P have been reported to modulate tumor growth and invasion in PCa (27-30). However, the functions and mechanisms of S100A6 and S100A10 in PCa remain unknown.
We used a PLS-DA score plot with protein ratios for each PCa group to discriminate between the differences in the proteomics data from each of the different PCa groups. Interestingly, this PLS-DA analysis of the proteomics data showed that the T-stage to be clearly distinguished between T2 (red zone) and T3 (blue zone), while these PCa groups could not be distinguished based on the PSA level. Thus, although T-stage classification is possible using the proteomics results from the PCa tissues, serum PSA levels cannot be predicted using proteomics data. These results suggested that the relationship between serum PSA level and protein expression in prostate tissues is imperfect.
Next, we used functional enrichment analysis by DAVID to provide insight into the biological properties of the DEPs we identified in the PCa tissues. As shown in Figure 2, the proteins that were up-regulated or down-regulated in PCa clearly had functional differences. The up-regulated proteins in the tumors were predominantly mitochondrial proteins, leading to an increase in metabolic pathways, including amino acid and fatty acid metabolism. In contrast, numerous down-regulated proteins were extracellular-related proteins or associated with muscle contraction, ECM–receptor interactions, or focal adhesion. Another group of proteins decreased in the PCa tissues was related to blood microparticles, which influence the complement and coagulation cascades and platelet degranulation. These bioinformatic results are very similar to previous proteomics studies conducted in PCa (31-33). In fact, these three protein groups, including the mitochondrial proteins, extracellular-related proteins, and blood microparticle proteins, are known to be highly related to the occurrence of cancer.
The reprogramming of cellular metabolism in mitochondria is a well-known cause of tumorigenesis, and these reprogramming activities have been recently recognized as a hallmark of cancer (34,35). Previous metabolomics studies have revealed general amino acid metabolism changes in PCa, and that the metabolism of some amino acids, such as histidine, serine, threonine, alanine, and tryptophan, are activated in PCa cells following exposure to androgens (36,37). Moreover, cancer requires a substantial reorganization of the ECM during its development and progression. The ECM is often deregulated and becomes destroyed in various cancer types, leading to a remodeling of the tumor microenvironment (38,39). In particular, cell-cell and cell–ECM adhesions are known to be significantly associated with a number of biological functions involved in cell growth, morphogenesis, cell motility, and inflammation. Tumor cell adhesion to the ECM plays an important role in the invasion and metastasis associated with the progression of cancer (40,41). Additionally, blood microparticles can be spontaneously released from tumor cells, and their levels in plasma are related to cell stimulation, proliferation, and the activation of coagulation (42,43). Conversely, proteins associated with blood microparticles are expected to be less than normal in tissue. Complement derived from blood microparticles plays a central role in the immune system and is able to promote the growth of cancer cells in situations involving chronic inflammation. Consequently, complement proteins can help orchestrate invasion and metastasis (44). Therefore, changes in mitochondrial proteins, extracellular-related proteins, and blood microparticle proteins in PCa are essential factors and have been previously identified in other types of cancer.
We used several criteria to identify the significant biomarkers from our proteomics data, including statistical methods and comparing our results with previously generated genomic databases. First, a one-way t-test, visualized using a volcano plot, was performed using the Perseus software platform, where we divided the PCa tissues into the T2 and T3 groups to ascertain statistically significant DEPs among the 344 identified DEPs. As shown in Figure 4, for patients with T2 or T3 PCa, we were able to identify 23 and 36 significant DEPs, respectively, which satisfied the cut-off criterion of p<0.05. In addition, we were able to include 11 DEPs that were altered in all of the groups. These proteins were expected to be useful for the diagnosis and prognosis of PCa. Moreover, 34 DEPs that were specifically altered only in the T3-HP-N1 group were also included and were presumed to be biomarkers specific for advanced PCa. The 89 selected DEPs were compared with previous genomics results from GEPIA and cBioPortal, and this analysis confirmed our proteomics results. GEPIA compared the mRNA differences between the normal tissue and tumors using the TCGA and Get databases, and cBioPortal obtained comparative results between PCa and metastatic PCa based on two previous studies (45,46). We then excluded proteins that have previously been reported to have a relationship with PCa. With the exception of a few of the proteins we identified, most were already known, including CRISP3 and S100A8, and have been previously studied in relation to PCa. Eventually, we accepted three proteins, namely SRM, NOLC1, and PTGIS, as new candidate biomarkers for advanced PCa, none of which have been previously studied in relation to PCa as far as we are aware. In order to verify the proteomics data, we performed both immunoblot analysis and ELISA, and the data generated for these three candidate biomarkers were consistent with our proteomics results. As expected, SRM and NOLC1 were increased in PCa tissues in both the immunoblot analysis and ELISA, whereas PTGIS slightly decrease.
SRM is a polyamine biosynthetic enzyme. During polyamine biosynthesis, ornithine decarboxylase (ODC) produces putrescine, and SRM and spermine synthase sequentially generate spermidine from putrescine and spermine from spermidine, respectively (47). Polyamines modulate gene expression by altering DNA structure and regulating signal transduction pathways, and they are consequently associated with proliferation, tumor invasion, and metastasis (48,49). As such, increased levels of polyamines due to their enhanced biosynthesis have been found in many cancer types, including skin, breast, colon, lung, and prostate (50). Northern blotting of polyamine metabolism regulatory genes in human PCa specimens indicated that ODC and the other polyamine metabolism genes S-adenosylmethionine decarboxylase (AMD1) and spermidine/spermine N1-acetyltransferase (SAT1), with the exception of SRM, were relatively increased in highly aggressive PCa compared to normal tissues (51). In another meta-analysis study of microarray studies performed in PCa, the mRNA levels of the polyamine biosynthesis pathway containing SRM were found to be elevated (52). However, the role of SRM in PCa is not yet known. SRM has, however, been shown to be regulated by androgen in the rat prostate (53) and transcribed by the Myc gene (c-MYC) oncogene in B-cell lymphomas (54). Moreover, increased spermidine levels are involved in normal cellular growth and proliferation via a post-translational modification, termed hypusination, of lysine-50 in eukaryotic translation initiation factor 5A2 (eIF5A2) (55,56). Even though the role of SRM in PCa has not been clearly elucidated, our immunoblot analysis and ELISA results indicated that there was a significant difference in its expression among the T2, T3, and normal tissue groups. Thus, we suggest that SRM expression in prostate tissues might be used as a prognostic and predictive candidate biomarker for advanced PCa.
NOLC1 was initially identified as a nuclear localization signal binding protein that acts as a shuttling chaperone between the nucleolus and the cytoplasm (57,58). Previous studies have shown that NOLC1 is more highly expressed in nasopharyngeal carcinomas than in normal tissues and plays a role in tumorigenesis by working together with TP53 (59). In contrast, expression of NOLC1 is reduced in human hepatocellular carcinoma tissues (60). It is known that the overexpression of NOLC1 in hepatocellular carcinoma cells suppresses cell proliferation, whereas knockdown of NOLC1 can increase the growth of cancer cells (61). According to a recent report, nucleolar-localized NOLC1 interacts with telomeric repeat-binding factor 2 (TRF2), and the overexpression of NOLC1 inhibits cell proliferation by inducing apoptosis and cell-cycle arrest in HepG2 cells (62). Therefore, although the protein expression levels of NOLC1 differ depending on the type of cancer, NOLC1 is also expected to play a role in the development of PCa.
Lastly, PTGIS is known to functionally combine with inducible cyclo-oxygenase-2 and contribute to cancer progression. PTGIS is often down-regulated in many cancer types, but this has not been demonstrated for PCa (63,64). PTGIS is known to be down-regulated via epigenetic mechanisms in human non-small cell lung cancer (65), and genetic variations in PTGIS are also associated with breast cancer progression and survival (66). Furthermore, hypermethylation of the PTGIS gene promoter led to a decrease in its expression in colorectal cancer (67). In contrast, the protein expression levels of PTGIS were significantly higher in hepatic metastases compared with primary colon cancer tissues (68). Here, we demonstrated a reduction in the levels of the PTGIS protein in PCa.
Three PCa candidate protein markers were shown to be associated with tumors, and these are projected to play an important role in PCa. In particular, SRM was observed to be significantly increased at the T3 stage relative to the T2 stage via ELISA. Based on these results, SRM appears to be a promising new candidate biomarker for assessing advanced PCa.
Funding
This work was supported by Biomedical Research Institute grant, Kyungpook National University Hospital (2019).
Conflicts of Interest
The Authors declare that they have no competing interests.
Authors’ Contributions
OK, YS and AY carried out the experiment, and OK and AY wrote the article. TG and SY provided prostate specimens. JN and SK conceived the original idea and supervised the project. All Authors read and approved the final article.
Availability of Data and Materials
The mass spectrometric proteomics data have been deposited at the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD013422.
Supplementary Material
The supplementary material is available free of charge at https://drive.google.com/drive/folders/1Di0lT_1ZSgEBQW_KIrIJi4yb32Ocpftd?usp=sharing
References
- 1.American Cancer Society. Cancer Facts & Figures. 2018.
- 2.Ha YS, Bak DJ, Chung JW, Lee JN, Kwon SY, Choi SH, Kwon TG, Kim TH. Postoperative cystographic findings as an independent predictor of urinary incontinence three months after radical prostatectomy. Minerva Urol Nefrol. 2017;69(3):278–284. doi: 10.23736/S0393-2249.16.02763-6. PMID: 27681662. DOI: 10.23736/S0393-2249.16.02763-6. [DOI] [PubMed] [Google Scholar]
- 3.Kwon YS, Ha YS, Modi PK, Salmasi A, Parihar JS, Patel N, Faiena I, May M, Lee DI, Llukani E, Patrick T, Rha KH, Ahlering T, Skarecky D, Ahn H, Choi SK, Park S, Jeon SS, Ou YC, Eun D, Manucha V, Albala D, Badani K, Yuh B, Ruel N, Kim TH, Kwon TG, Marchalik D, Hwang J, Kim WJ, Kim IY. Oncologic outcomes in men with metastasis to the prostatic anterior fat pad lymph nodes: A multi-institution international study. BMC Urol. 2015;15:79. doi: 10.1186/s12894-015-0070-1. PMID: 26231860. DOI: 10.1186/s12894-015-0070-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moul JW. The evolving definition of advanced prostate cancer. Rev Urol. 2004;6 Suppl8:S10–17. PMID: 16985915. [PMC free article] [PubMed] [Google Scholar]
- 5.van Poppel H. Locally advanced and high risk prostate cancer: The best indication for initial radical prostatectomy. Asian J Urol. 2014;1(1):40–45. doi: 10.1016/j.ajur.2014.09.009. PMID: 29511636. DOI: 10.1016/j.ajur.2014.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pin E, Fredolini C, Petricoin EF 3rd. The role of proteomics in prostate cancer research: Biomarker discovery and validation. Clin Biochem. 2013;46(6):524–538. doi: 10.1016/j.clinbiochem.2012.12.012. PMID: 23266295. DOI: 10.1016/j.clinbiochem.2012.12.012. [DOI] [PubMed] [Google Scholar]
- 7.Gaudreau PO, Stagg J, Soulieres D, Saad F. The present and future of biomarkers in prostate cancer: Proteomics, genomics, and immunology advancements. Biomark Cancer. 2016;8(Suppl 2):15–33. doi: 10.4137/BIC.S31802. PMID: 27168728. DOI: 10.4137/BIC.S31802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Intasqui P, Bertolla RP, Sadi MV. Prostate cancer proteomics: Clinically useful protein biomarkers and future perspectives. Expert Rev Proteomics. 2018;15(1):65–79. doi: 10.1080/14789450.2018.1417846. PMID: 29251021. DOI: 10.1080/14789450.2018.1417846. [DOI] [PubMed] [Google Scholar]
- 9.Martin RM, Donovan JL, Turner EL, Metcalfe C, Young GJ, Walsh EI, Lane JA, Noble S, Oliver SE, Evans S, Sterne JAC, Holding P, Ben-Shlomo Y, Brindle P, Williams NJ, Hill EM, Ng SY, Toole J, Tazewell MK, Hughes LJ, Davies CF, Thorn JC, Down E, Davey Smith G, Neal DE, Hamdy FC. Effect of a low-intensity psa-based screening intervention on prostate cancer mortality: The CAP randomized clinical trial. JAMA. 2018;319(9):883–895. doi: 10.1001/jama.2018.0154. PMID: 29509864. DOI: 10.1001/jama.2018.0154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grossman DC, Curry SJ, Owens DK, Bibbins-Domingo K, Caughey AB, Davidson KW, Doubeni CA, Ebell M, Epling JW Jr., Kemper AR, Krist AH, Kubik M, Landefeld CS, Mangione CM, Silverstein M, Simon MA, Siu AL, Tseng CW. Screening for prostate cancer: US Preventive Services Task Force recommendation statement. JAMA. 2018;319(18):1901–1913. doi: 10.1001/jama.2018.3710. PMID: 29801017. DOI: 10.1001/jama.2018.3710. [DOI] [PubMed] [Google Scholar]
- 11.Ahram M, Best CJ, Flaig MJ, Gillespie JW, Leiva IM, Chuaqui RF, Zhou G, Shu H, Duray PH, Linehan WM, Raffeld M, Ornstein DK, Zhao Y, Petricoin EF 3rd, Emmert-Buck MR. Proteomic analysis of human prostate cancer. Mol Carcinog. 2002;33(1):9–15. doi: 10.1002/mc.10019. PMID: 11807953. DOI: 10.1002/mc.10019. [DOI] [PubMed] [Google Scholar]
- 12.Lexander H, Palmberg C, Auer G, Hellstrom M, Franzen B, Jornvall H, Egevad L. Proteomic analysis of protein expression in prostate cancer. Anal Quant Cytol Histol. 2005;27(5):263–272. PMID: 16447818. [PubMed] [Google Scholar]
- 13.Ummanni R, Barreto F, Venz S, Scharf C, Barett C, Mannsperger HA, Brase JC, Kuner R, Schlomm T, Sauter G, Sultmann H, Korf U, Bokemeyer C, Walther R, Brummendorf TH, Balabanov S. Peroxiredoxins 3 and 4 are overexpressed in prostate cancer tissue and affect the proliferation of prostate cancer cells in vitro. J Proteome Res. 2012;11(4):2452–2466. doi: 10.1021/pr201172n. PMID: 22424448. DOI: 10.1021/pr201172n. [DOI] [PubMed] [Google Scholar]
- 14.Davalieva K, Kostovska IM, Kiprijanovska S, Markoska K, Kubelka-Sabit K, Filipovski V, Stavridis S, Stankov O, Komina S, Petrusevska G, Polenakovic M. Proteomics analysis of malignant and benign prostate tissue by 2D DIGE/MS reveals new insights into proteins involved in prostate cancer. Prostate. 2015;75(14):1586–1600. doi: 10.1002/pros.23034. PMID: 26074449. DOI: 10.1002/pros.23034. [DOI] [PubMed] [Google Scholar]
- 15.de Godoy LM, Olsen JV, de Souza GA, Li G, Mortensen P, Mann M. Status of complete proteome analysis by mass spectrometry: Silac-labeled yeast as a model system. Genome Biol. 2006;7(6):R50. doi: 10.1186/gb-2006-7-6-r50. PMID: 16784548. DOI: 10.1186/gb-2006-7-6-r50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perez-Riverol Y, Csordas A, Bai J, Bernal-Llinares M, Hewapathirana S, Kundu DJ, Inuganti A, Griss J, Mayer G, Eisenacher M, Perez E, Uszkoreit J, Pfeuffer J, Sachsenberg T, Yilmaz S, Tiwary S, Cox J, Audain E, Walzer M, Jarnuczak AF, Ternent T, Brazma A, Vizcaino JA. The pride database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 2019;47(D1):D442–D450. doi: 10.1093/nar/gky1106. PMID: 30395289. DOI: 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cox J, Mann M. Maxquant enables high peptide identification rates, individualized p.P.B.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26(12):1367–1372. doi: 10.1038/nbt.1511. PMID: 19131956. DOI: 10.1038/ nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 18.uang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. PMID: 19131956. DOI: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 19.Tyanova S, Temu T, Sinitcyn P, Carlson A, Hein MY, Geiger T, Mann M, Cox J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat Methods. 2016;13(9):731–740. doi: 10.1038/nmeth.3901. PMID: 27348712. DOI: 10.1038/ nmeth.3901. [DOI] [PubMed] [Google Scholar]
- 20.Tyanova S, Cox J. Perseus: A bioinformatics platform for integrative analysis of proteomics data in cancer research. Methods Mol Biol. 2018;1711:133–148. doi: 10.1007/978-1-4939-7493-1_7. PMID: 29344888. DOI: 10.1007/978-1-4939-7493-1_7. [DOI] [PubMed] [Google Scholar]
- 21.Tang Z, Li C, Kang B, Gao G, Zhang Z. Gepia: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98–W102. doi: 10.1093/nar/gkx247. PMID: 28407145. DOI: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, Cerami E, Sander C, Schultz N. Integrative analysis of complex cancer genomics and clinical profiles using the cbioportal. Sci Signal. 2013;6(269):pl1. doi: 10.1126/scisignal.2004088. PMID: 23550210. DOI: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kosari F, Asmann YW, Cheville JC, Vasmatzis G. Cysteine-rich secretory protein-3: A potential biomarker for prostate cancer. Cancer Epidemiol Biomarkers Prev. 2002;11(11):1419–1426. PMID: 12433721. [PubMed] [Google Scholar]
- 24.Al Bashir S, Alshalalfa M, Hegazy SA, Dolph M, Donnelly B, Bismar TA. Cysteine- rich secretory protein 3 (CRISP3), ERG and PTEN define a molecular subtype of prostate cancer with implication to patients’ prognosis. J Hematol Oncol. 2014;7:21. doi: 10.1186/1756-8722-7-21. PMID: 24606912. DOI: 10.1186/1756-8722-7-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thompson IM, Pauler DK, Goodman PJ, Tangen CM, Lucia MS, Parnes HL, Minasian LM, Ford LG, Lippman SM, Crawford ED, Crowley JJ, Coltman CA Jr. Prevalence of prostate cancer among men with a prostate-specific antigen level < or =4.0 ng per milliliter. N Engl J Med. 2004;350(22):2239–2246. doi: 10.1056/NEJMoa031918. PMID: 15163773. DOI: 10.1056/NEJMoa031918. [DOI] [PubMed] [Google Scholar]
- 26.Roobol MJ, Carlsson SV. Risk stratification in prostate cancer screening. Nat Rev Urol. 2013;10(1):38–48. doi: 10.1038/nrurol.2012.225. PMID: 23247693. DOI: 10.1038/nrurol.2012.225. [DOI] [PubMed] [Google Scholar]
- 27.Hermani A, Hess J, De Servi B, Medunjanin S, Grobholz R, Trojan L, Angel P, Mayer D. Calcium-binding proteins S100A8 and S100A9 as novel diagnostic markers in human prostate cancer. Clin Cancer Res. 2005;11(14):5146–5152. doi: 10.1158/1078-0432.CCR-05-0352. PMID: 16033829. DOI: 10.1158/1078-0432.CCR-05-0352. [DOI] [PubMed] [Google Scholar]
- 28.Saleem M, Kweon MH, Johnson JJ, Adhami VM, Elcheva I, Khan N, Bin Hafeez B, Bhat KM, Sarfaraz S, Reagan-Shaw S, Spiegelman VS, Setaluri V, Mukhtar H. S100A4 accelerates tumorigenesis and invasion of human prostate cancer through the transcriptional regulation of matrix metalloproteinase 9. Proc Natl Acad Sci USA. 2006;103(40):14825–14830. doi: 10.1073/pnas.0606747103. PMID: 16990429. DOI: 10.1073/pnas.0606747103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kang M, Lee HS, Lee YJ, Choi WS, Park YH, Jeong CW, Ku JH, Kim HH, Kwak C. S100A3 suppression inhibits in vitro and in vivo tumor growth and invasion of human castration-resistant prostate cancer cells. Urology. 2015;85(1):273 e279–215. doi: 10.1016/j.urology.2014.09.018. PMID: 25440760. DOI: 10.1016/j.urology.2014.09.018. [DOI] [PubMed] [Google Scholar]
- 30.Bresnick AR, Weber DJ, Zimmer DB. S100 proteins in cancer. Nat Rev Cancer. 2015;15(2):96–109. doi: 10.1038/nrc3893. PMID: 25614008. DOI: 10.1038/nrc3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iglesias-Gato D, Wikstrom P, Tyanova S, Lavallee C, Thysell E, Carlsson J, Hagglof C, Cox J, Andren O, Stattin P, Egevad L, Widmark A, Bjartell A, Collins CC, Bergh A, Geiger T, Mann M, Flores-Morales A. The proteome of primary prostate cancer. Eur Urol. 2016;69(5):942–952. doi: 10.1016/j.eururo.2015.10.053. PMID: 26651926. DOI: 10.1016/j.eururo.2015.10.053. [DOI] [PubMed] [Google Scholar]
- 32.Muller AK, Foll M, Heckelmann B, Kiefer S, Werner M, Schilling O, Biniossek ML, Jilg CA, Drendel V. Proteomic characterization of prostate cancer to distinguish nonmetastasizing and metastasizing primary tumors and lymph node metastases. Neoplasia. 2018;20(2):140–151. doi: 10.1016/j.neo.2017.10.009. PMID: 29248718. DOI: 10.1016/j.neo.2017.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen C, Zhang LG, Liu J, Han H, Chen N, Yao AL, Kang SS, Gao WX, Shen H, Zhang LJ, Li YP, Cao FH, Li ZG. Bioinformatics analysis of differentially expressed proteins in prostate cancer based on proteomics data. Onco Targets Ther. 2016;9:1545–1557. doi: 10.2147/OTT.S98807. PMID: 27051295. DOI: 10.2147/OTT.S98807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pavlova NN, Thompson CB. The emerging hallmarks of cancer metabolism. Cell Metab. 2016;23(1):27–47. doi: 10.1016/j.cmet.2015.12.006. PMID: 26771115. DOI: 10.1016/j.cmet.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.DeBerardinis RJ, Chandel NS. Fundamentals of cancer metabolism. Sci Adv. 2016;2(5):e1600200. doi: 10.1126/sciadv.1600200. PMID: 27386546. DOI: 10.1126/sciadv.1600200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Putluri N, Shojaie A, Vasu VT, Nalluri S, Vareed SK, Putluri V, Vivekanandan-Giri A, Byun J, Pennathur S, Sana TR, Fischer SM, Palapattu GS, Creighton CJ, Michailidis G, Sreekumar A. Metabolomic profiling reveals a role for androgen in activating amino acid metabolism and methylation in prostate cancer cells. PLoS One. 2011;6(7):e21417. doi: 10.1371/journal.pone.0021417. PMID: 21789170. DOI: 10.1371/journal.pone.0021417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lucarelli G, Rutigliano M, Galleggiante V, Giglio A, Palazzo S, Ferro M, Simone C, Bettocchi C, Battaglia M, Ditonno P. Metabolomic profiling for the identification of novel diagnostic markers in prostate cancer. Expert Rev Mol Diagn. 2015;15(9):1211–1224. doi: 10.1586/14737159.2015.1069711. PMID: 26174441. DOI: 10.1586/14737159.2015.1069711. [DOI] [PubMed] [Google Scholar]
- 38.Lu P, Weaver VM, Werb Z. The extracellular matrix: A dynamic niche in cancer progression. J Cell Biol. 2012;196(4):395–406. doi: 10.1083/jcb.201102147. PMID: 22351925. DOI: 10.1083/jcb.201102147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pickup MW, Mouw JK, Weaver VM. The extracellular matrix modulates the hallmarks of cancer. EMBO Rep. 2014;15(12):1243–1253. doi: 10.15252/embr.201439246. PMID: 25381661. DOI: 10.15252/embr.201439246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stetler-Stevenson WG, Aznavoorian S, Liotta LA. Tumor cell interactions with the extracellular matrix during invasion and metastasis. Annu Rev Cell Biol. 1993;9:541–573. doi: 10.1146/annurev.cb.09.110193.002545. PMID: 8280471. DOI: 10.1146/annurev.cb.09.110193.002545. [DOI] [PubMed] [Google Scholar]
- 41.Okegawa T, Pong RC, Li Y, Hsieh JT. The role of cell adhesion molecule in cancer progression and its application in cancer therapy. Acta Biochim Pol. 2004;51(2):445–457. PMID: 15218541. DOI: 035001445. [PubMed] [Google Scholar]
- 42.Falanga A, Tartari CJ, Marchetti M. Microparticles in tumor progression. Thromb Res. 2012;129(Suppl 1):S132–136. doi: 10.1016/S0049-3848(12)70033-6. PMID: 22682124. DOI: 10.1016/S0049-3848(12)70033-6. [DOI] [PubMed] [Google Scholar]
- 43.Geddings JE, Mackman N. Tumor-derived tissue factor-positive microparticles and venous thrombosis in cancer patients. Blood. 2013;122(11):1873–1880. doi: 10.1182/blood-2013-04-460139. PMID: 23798713. DOI: 10.1182/blood-2013-04-460139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pio R, Corrales L, Lambris JD. The role of complement in tumor growth. Adv Exp Med Biol. 2014;772:229–262. doi: 10.1007/978-1-4614-5915-6_11. PMID: 24272362. DOI: 10.1007/978-1-4614-5915-6_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Robinson D, Van Allen EM, Wu YM, Schultz N, Lonigro RJ, Mosquera JM, Montgomery B, Taplin ME, Pritchard CC, Attard G, Beltran H, Abida W, Bradley RK, Vinson J, Cao X, Vats P, Kunju LP, Hussain M, Feng FY, Tomlins SA, Cooney KA, Smith DC, Brennan C, Siddiqui J, Mehra R, Chen Y, Rathkopf DE, Morris MJ, Solomon SB, Durack JC, Reuter VE, Gopalan A, Gao J, Loda M, Lis RT, Bowden M, Balk SP, Gaviola G, Sougnez C, Gupta M, Yu EY, Mostaghel EA, Cheng HH, Mulcahy H, True LD, Plymate SR, Dvinge H, Ferraldeschi R, Flohr P, Miranda S, Zafeiriou Z, Tunariu N, Mateo J, Perez-Lopez R, Demichelis F, Robinson BD, Schiffman M, Nanus DM, Tagawa ST, Sigaras A, Eng KW, Elemento O, Sboner A, Heath EI, Scher HI, Pienta KJ, Kantoff P, de Bono JS, Rubin MA, Nelson PS, Garraway LA, Sawyers CL, Chinnaiyan AM. Integrative clinical genomics of advanced prostate cancer. Cell. 2015;161(5):1215–1228. doi: 10.1016/j.cell.2015.05.001. PMID: 26000489. DOI: 10.1016/j.cell.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kumar A, Coleman I, Morrissey C, Zhang X, True LD, Gulati R, Etzioni R, Bolouri H, Montgomery B, White T, Lucas JM, Brown LG, Dumpit RF, DeSarkar N, Higano C, Yu EY, Coleman R, Schultz N, Fang M, Lange PH, Shendure J, Vessella RL, Nelson PS. Substantial interindividual and limited intraindividual genomic diversity among tumors from men with metastatic prostate cancer. Nat Med. 2016;22(4):369–378. doi: 10.1038/nm.4053. PMID: 26928463. DOI: 10.1038/nm.4053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Casero RA Jr., Marton LJ. Targeting polyamine metabolism and function in cancer and other hyperproliferative diseases. Nat Rev Drug Discov. 2007;6(5):373–390. doi: 10.1038/nrd2243. PMID: 17464296. DOI: 10.1038/nrd2243. [DOI] [PubMed] [Google Scholar]
- 48.Thomas T, Thomas TJ. Polyamine metabolism and cancer. J Cell Mol Med. 2003;7(2):113–126. doi: 10.1111/j.1582-4934.2003.tb00210.x. PMID: 12927050. DOI: 10.1111/j.1582-4934.2003.tb00210.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gerner EW, Meyskens FL Jr. Polyamines and cancer: Old molecules, new understanding. Nat Rev Cancer. 2004;4(10):781–792. doi: 10.1038/nrc1454. PMID: 15510159. DOI: 10.1038/nrc1454. [DOI] [PubMed] [Google Scholar]
- 50.Nowotarski SL, Woster PM, Casero RA Jr. Polyamines and cancer: Implications for chemotherapy and chemoprevention. Expert Rev Mol Med. 2013;15:e3. doi: 10.1017/erm.2013.3. PMID: 23432971. DOI: 10.1017/erm.2013.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bettuzzi S, Davalli P, Astancolle S, Carani C, Madeo B, Tampieri A, Corti A. Tumor progression is accompanied by significant changes in the levels of expression of polyamine metabolism regulatory genes and clusterin (sulfated glycoprotein 2) in human prostate cancer specimens. Cancer Res. 2000;60(1):28–34. PMID: 10646846. [PubMed] [Google Scholar]
- 52.Rhodes DR, Barrette TR, Rubin MA, Ghosh D, Chinnaiyan AM. Meta-analysis of microarrays: Interstudy validation of gene expression profiles reveals pathway dysregulation in prostate cancer. Cancer Res. 2002;62(15):4427–4433. PMID: 12154050. [PubMed] [Google Scholar]
- 53.Bataskov KL, Hariharan S, Horowitz MD, Neibart RM, Cox MM. Gonococcal endocarditis complicating pregnancy: A case report and literature review. Obstet Gynecol. 1991;78(3 Pt 2):494–496. PMID: 1870804. [PubMed] [Google Scholar]
- 54.Forshell TP, Rimpi S, Nilsson JA. Chemoprevention of B-cell lymphomas by inhibition of the MYC target spermidine synthase. Cancer Prev Res. 2010;3(2):140–147. doi: 10.1158/1940-6207.CAPR-09-0166. PMID: 20103729. DOI: 10.1158/1940-6207.CAPR-09-0166. [DOI] [PubMed] [Google Scholar]
- 55.Park MH. The post-translational synthesis of a polyamine-derived amino acid, hypusine, in the eukaryotic translation initiation factor 5a (EIF5A) J Biochem. 2006;139(2):161–169. doi: 10.1093/jb/mvj034. PMID: 16452303. DOI: 10.1093/jb/mvj034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Paz EA, Garcia-Huidobro J, Ignatenko NA. Polyamines in cancer. Adv Clin Chem. 2011;54:45–70. doi: 10.1016/b978-0-12-387025-4.00002-9. PMID: 21874756. DOI: 10.1016/b978-0-12-387025-4.00002-9. [DOI] [PubMed] [Google Scholar]
- 57.Meier UT, Blobel G. A nuclear localization signal binding protein in the nucleolus. J Cell Biol. 1990;111(6 Pt 1):2235–2245. doi: 10.1083/jcb.111.6.2235. PMID: 2177472. DOI: 10.1083/jcb.111.6.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Meier UT, Blobel G. NOPP140 shuttles on tracks between nucleolus and cytoplasm. Cell. 1992;70(1):127–138. doi: 10.1016/0092-8674(92)90539-o. PMID: 1623516. DOI: 10.1016/0092-8674(92)90539-o. [DOI] [PubMed] [Google Scholar]
- 59.Hwang YC, Lu TY, Huang DY, Kuo YS, Kao CF, Yeh NH, Wu HC, Lin CT. NOLC1, an enhancer of nasopharyngeal carcinoma progression, is essential for TP53 to regulate MDM2 expression. Am J Pathol. 2009;175(1):342–354. doi: 10.2353/ajpath.2009.080931. PMID: 19541936. DOI: 10.2353/ajpath.2009.080931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Duan X, Zhang J, Liu S, Zhang M, Wang Q, Cheng J. Methylation of nucleolar and coiled-body phosphoprotein 1 is associated with the mechanism of tumorigenesis in hepatocellular carcinoma. Oncol Rep. 2013;30(5):2220–2228. doi: 10.3892/or.2013.2676. PMID: 23970161. DOI: 10.3892/or.2013.2676. [DOI] [PubMed] [Google Scholar]
- 61.Yuan F, Zhang Y, Ma L, Cheng Q, Li G, Tong T. Enhanced NOLC1 promotes cell senescence and represses hepatocellular carcinoma cell proliferation by disturbing the organization of nucleolus. Aging Cell. 2017;16(4):726–737. doi: 10.1111/acel.12602. PMID: 28493459. DOI: 10.1111/acel.12602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yuan F, Li G, Tong T. Nucleolar and coiled-body phosphoprotein 1 (NOLC1) regulates the nucleolar retention of TRF2. Cell Death Discov. 2017;3:17043. doi: 10.1038/cddiscovery.2017.43. PMID: 28875039. DOI: 10.1038/cddiscovery.2017.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ueno N, Murakami M, Tanioka T, Fujimori K, Tanabe T, Urade Y, Kudo I. Coupling between cyclooxygenase, terminal prostanoid synthase, and phospholipase A2. J Biol Chem. 2001;276(37):34918–34927. doi: 10.1074/jbc.M100429200. PMID: 11418589. DOI: 10.1074/jbc.M100429200. [DOI] [PubMed] [Google Scholar]
- 64.Sasaki Y, Ochiai T, Takamura M, Kondo Y, Yokoyama C, Hara S. Role of prostacyclin synthase in carcinogenesis. Prostaglandins Other Lipid Mediat. 2017;133:49–52. doi: 10.1016/j.prostaglandins.2017.05.001. PMID: 28506876. DOI: 10.1016/j.prostaglandins.2017.05.001. [DOI] [PubMed] [Google Scholar]
- 65.Cathcart MC, Gray SG, Baird AM, Boyle E, Gately K, Kay E, Cummins R, Pidgeon GP, O’Byrne KJ. Prostacyclin synthase expression and epigenetic regulation in nonsmall cell lung cancer. Cancer. 2011;117(22):5121–5132. doi: 10.1002/cncr.26168. PMID: 21523772. DOI: 10.1002/cncr.26168. [DOI] [PubMed] [Google Scholar]
- 66.Abraham JE, Harrington P, Driver KE, Tyrer J, Easton DF, Dunning AM, Pharoah PD. Common polymorphisms in the prostaglandin pathway genes and their association with breast cancer susceptibility and survival. Clin Cancer Res. 2009;15(6):2181–2191. doi: 10.1158/1078-0432.CCR-08-0716. PMID: 19276290. DOI: 10.1158/1078-0432.CCR-08-0716. [DOI] [PubMed] [Google Scholar]
- 67.Frigola J, Munoz M, Clark SJ, Moreno V, Capella G, Peinado MA. Hypermethylation of the prostacyclin synthase (PTGIS) promoter is a frequent event in colorectal cancer and associated with aneuploidy. Oncogene. 2005;24(49):7320–7326. doi: 10.1038/sj.onc.1208883. PMID: 16007128. DOI: 10.1038/sj.onc.1208883. [DOI] [PubMed] [Google Scholar]
- 68.Lichao S, Liang P, Chunguang G, Fang L, Zhihua Y, Yuliang R. Overexpression of ptgis could predict liver metastasis and is correlated with poor prognosis in colon cancer patients. Pathol Oncol Res. 2012;18(3):563–569. doi: 10.1007/s12253-011-9478-4. PMID: 22109564. DOI: 10.1007/s12253-011-9478-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The mass spectrometric proteomics data have been deposited at the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD013422.