Abstract
Background/Aim: Mounting evidence has shown that miRNAs play a critical role in the regulation of hematopoiesis of cell proliferation and apoptosis as well as in tumorigenesis. The miR146a rs2910164 polymorphism, which is closely responsive for its expression, has been reported to associate with the risk of several solid cancers. The study aimed at examining the association of the it with susceptibility to childhood acute lymphoblastic leukemia (ALL) in Taiwan. Materials and Methods: We recruited 266 patients with childhood ALL and 266 healthy controls, and rs2910164 genotypes were determined by the polymerase chain reaction-restriction fragment length polymorphism methodology. Results: The allele G was associated with decreased childhood ALL risk (OR=0.66, 95%CI=0.52-0.85, p=0.0011). Consistently, the GG genotype was associated with a decreased susceptibility (OR=0.40, 95%CI=0.23-0.67, p=0.0004). Patients with CG and GG genotypes were of earlier onset than those with CC genotype (p=0.0255 and p=0.0001). Conclusion: MiR146a rs2910164 G allele serves as a protective marker for childhood ALL in Taiwan.
Keywords: Acute lymphoblastic leukemia, case–control study, genotype, miR146a, polymorphism, Taiwan
Acute lymphoblastic leukemia (ALL) is the most common type of malignancy among children worldwide. The molecular etiology of ALL is not completely understood. Genome-wide and candidate-gene association studies show that genetic polymorphisms of some susceptibility genes are associated with the personal susceptibility of childhood ALL (1-5). In recent years, it has been reported that microRNAs (miRNAs) may play an important role in the regulation of hematopoiesis (6). MiRNAs are those small non-coding RNAs of 17-24 nucleotides in length, which are capable of regulating gene expression by translational repression or mRNA degradation (7). Among them, miR146a in particular is identified as a modulator of cell differentiation, in addition to the innate and adaptive immunity. Given its importance in regulating key cellular functions, it is not surprising that up- or down-regulation of miRNA-146a is frequently observed in human disorders, such as inflammatory diseases and cancers (6,8,9).
The miR146a rs2910164 polymorphism involves a nucleotide substitution from G to C which causes an alteration from a G:U pair to a C:U mismatch in the stem structure of miR146a precursor that results in a reduced amount of mature miR146a (10,11). In the literature, it is also reported that miR146a rs2910164 can directly inhibit the expression of IRAK1 and IRAF6, impair nuclear factor (NF-ĸB) activity, and suppress several NF-ĸB targeted inflammatory genes, such as IL-6, IL-8, IL-Iβ, and TNF-α (6,8). The variant C allele at miR146a rs2910164 polymorphism was associated with an elevated risk of prostate cancer (12), breast cancer (13,14) and hepatocellular carcinoma (15). On the contrary, there is a meta-analysis report summarizing that the miR146a rs2910164 GG genotype, not CC or CG, was associated with increased gastric cancer risk (16) and breast cancer (17). There were also several negative reports suggesting that miR146a polymorphism was not associated with oral (18), gastric (19), colorectal (20,21) and lung cancer (22). The conflicting findings may be due to different ethnic populations investigated, and various case numbers with different analyzing power.
There exists limited literature examining the contribution of miR146a rs2910164 to childhood cancers. In 2014, Hasani and colleagues reported that CC genotypes at miR146a rs2910164 were associated with elevated risk of ALL among Thailand children (23). No study has investigated the association of those genotypes of miRNAs with Taiwan childhood leukemia. Therefore, we aimed at determining the genotyping of miR146a rs2910164 among Taiwanese children and comparing the genetic and allelic frequencies of miR146a rs2910164 between childhood ALL and healthy children groups.
Materials and Methods
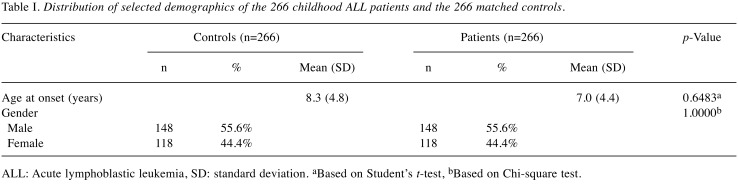
Specimens from childhood ALL patients and control subjects. The research design and detail procedures of the current study were approved by the Institutional Review Board of China Medical University Hospital with the number of DMR106-IRB-142. Written informed consent for each subject was obtained from one or both parents of all participants. Briefly, cases with childhood leukemia were identified and ascertained into the study by pediatric clinicians with pathologic confirmation, no matter at which ages or at what stages they were diagnosed. In brief, finally a total of 266 patients who had been diagnosed with childhood ALL were recruited from the general surgery outpatient clinics of the pediatric departments of China Medical University Hospital and the National Taiwan University Hospital, Taiwan, Republic of China during the period of 2005 to 2010. All basic and clinical characteristics of the recruited children, including their histological details, were identified by expert surgeons in the two hospitals. All investigated subjects voluntarily participating in this study, completed a questionnaire form with the help of their parents or guardians, and provided their peripheral blood samples less than 5 ml without any uncomfortable feelings. An equal number of age-matched healthy subjects without any cancer were recruited as the control group in accordance with the method for initial random sampling established by the Health Examination Cohort over the similar period of 2005 to 2010 as we previously published (3-5). Most healthy subjects underwent health examinations every 5 to 6 months. A total of 457 volunteers aged under 18 years were recruited in the study and were diagnosed as cancer-free in accordance with the criteria set by the International Classification of Disease (ninth revision, defined by World Health Organization). At last, 266 participants were included in the analysis to match the population structure (number, age, and gender) of our case population. All the participants are Taiwanese and the overall agreement rate in the study exceeded 85%. As shown in Table I, we have provided a concise summary and comparison of the selected recorded characteristics of the case and control groups.
Table I. Distribution of selected demographics of the 266 childhood ALL patients and the 266 matched controls.
ALL: Acute lymphoblastic leukemia, SD: standard deviation. aBased on Student’s t-test, bBased on Chi-square test.
DNA isolation. Genomic DNA from the peripheral blood samples of each participant was extracted, measured by the spectrophotometer, aliquoted, and stored at –20˚C prior to use or at –80˚C for a longer period as we performed routinely (24-26).
Genotyping of the miR146a (G>C) polymorphism. The miR146a (G>C) polymorphism was performed by polymerase chain reaction plus restriction fragment length polymorphism (PCR-RFLP) methodology. The PCR-RFLP genotyping of miR146a (G>C) polymorphism was conducted with forward and reverse primers of 5’-CATGGGTTGTGTCAGTGTCAGAGCT-3’ and 5’-TGCCTTCTGTCTCCAGTCTTCCAA-3’, respectively, as designed previously in 2011 (10). DNA was amplified in a 25 μl reaction mixture containing 100 ng of genomic DNA of specific subject, 20 μM of each primer, 5 μl of 10X buffer, 2.5 μl of 5X Q-solution, 10mM of each dNTP and 1 Ut of hotstart taq DNA polymerase (Qiagen, Hilden, Germany). PCR was performed in a PCR thermal cycle using the following conditions: initial denaturation 94˚C for 5 min, followed by denaturation at 94˚C for 30 s, annealing at 64˚C for 40 s, and extension at 72˚C for 45 s. After completion of 35 cycles, a final extension step was carried out at 72˚C for 10 min. The sizes of PCR amplicon were identified by 3% agarose gel electrophoresis for the successful rates of original PCR. Then the PCR amplicons were digested by Sac I at 37˚C for 16 h. The product sizes after restriction digested were verified on 3% agarose gel electrophoresis again, stained with ethidium bromide and visualized using the gel documentation. The product presented 3 different patterns: an intact single 147 bp fragment for the GG genotype; full-digested fragments of 122 and 25 bp for the CC genotype; and fragments of 147, 122 and 25 bp for the heterozygous GC genotype.
Statistical analysis. The Student’s t-test was used to test the difference of the age between the case and control groups. The Pearson’s χ-square test without Yates’ correction or Fisher’s exact test (when any cell number is less than 5) was used to compare the distribution of miR146a rs2910164 genotypes between case and control groups or any other two sub-groups. Crude odds ratios (OR) and 95% confidence intervals (CI) were also calculated. For some specific reasons, the ORs may be adjusted for possible confounding factors whenever it was needed. Any p-value less than 0.05 was considered statistically significant.
Results
This study consisted of 266 childhood ALL patients and the same amounts of healthy children. The ages of ALL patients and normal controls as well as genders of all the participants are shown in Table I. The genotypic frequencies for miR146a rs2910164 were determined in the childhood ALL and control groups, and compared with each other.
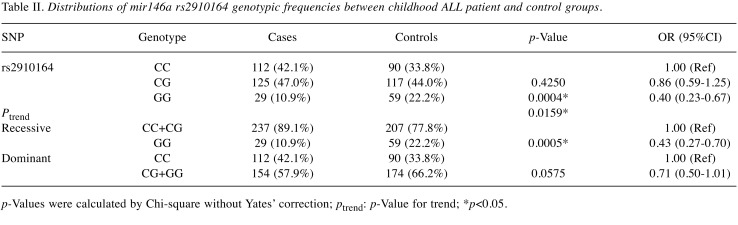
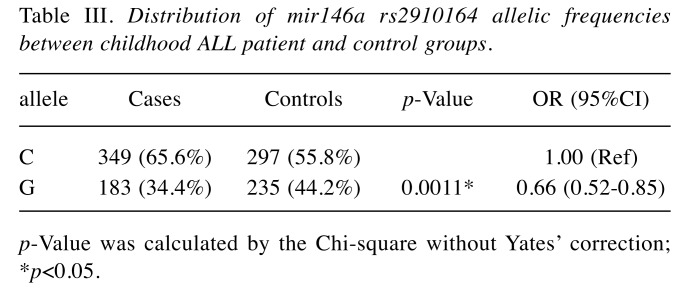
First, the genotypes of miR146a rs2910164 in the control groups matched well with the Hardy-Weinberg equation (p>0.05). Second, there was a significant association between miR146a rs2910164 polymorphism and susceptibility to childhood ALL (p-Value for trend=0.0159). In detail, the GG genotype carriers at miR146a rs2910164 were of 0.40-fold of risk of childhood ALL (95%CI=0.23-0.67, p=0.0004) (Table II). The findings are validated in the recessive model (OR=0.43, 95%CI=0.27-0.70, p=0.0005), and at a borderline in the dominant model (OR=0.71, 95%CI=0.50-1.01, p=0.0575) (Table II). The allele frequencies of miR146a G allele in controls was 44.2%, much higher than that of 34.4% in childhood ALL patients (Table III).
Table II. Distributions of mir146a rs2910164 genotypic frequencies between childhood ALL patient and control groups.
p-Values were calculated by Chi-square without Yates’ correction; ptrend: p-Value for trend; *p<0.05.
Table III. Distribution of mir146a rs2910164 allelic frequencies between childhood ALL patient and control groups.
p-Value was calculated by the Chi-square without Yates’ correction;*p<0.05.
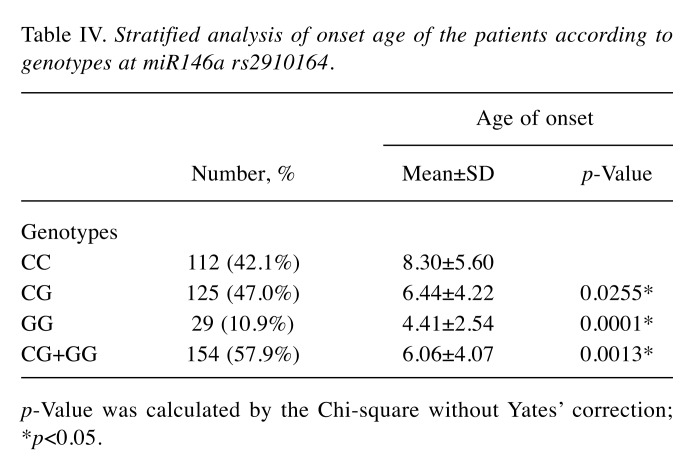
Moreover, we have also found that childhood ALL patients with CG and GG genotypes at miR146a rs2910164 tended to have earlier onset compared with those with wild-type CC genotype (average age: 6.44 and 4.41 vs. 8.30, p=0.0255 and 0.0001, respectively). After combination, CG+GG genotypes still tended to have earlier onset compared with those with wild-type CC genotype (average age: 6.06 vs. 8.30, p=0.0013) (Table IV).
Table IV. Stratified analysis of onset age of the patients according to genotypes at miR146a rs2910164.
p-Value was calculated by the Chi-square without Yates’ correction; *p<0.05.
Discussion
In recent years, mounting evidence showed that small non-coding miRNAs, play important roles in development and cellular processes such as cell proliferation, differentiation, apoptosis and tumorigenesis (8,27). Also, miRNAs can be further classified according to the criteria that for those with their expression levels increased in cancer patients are called oncogenic microRNAs, while those decreased in cancer patients are called tumor suppressor microRNAs (28). In 2008, Jazdaewski and his colleagues firstly reported that miR146a rs2910164 genotypes were associated with determination of personal susceptibility to thyroid papillary carcinoma. They found that C allele of mature miR146a would have lower capacity in inhibition target genes TRAF6 and IRAK1 than that of G allele (29). In addition, miR146a was reported by Tagnov and his colleagues to serve as a modulator of the well-known pro-apoptotic transcription factor nuclear-kappa B (NF-ĸB) (30).
In recent years, the miR146a rs2910164 polymorphism has been widely examined in various types of cancers. In several studies, the GG genotype was found to be associated with an increased risk of cancer including breast cancer (14), hepatocellular carcinoma (15). On the contrary, it was reported that the rs2910164 CG or GG genotype was associated with a significantly decreased risk for lung cancer compared to that of the CC genotype (31). Similarly, it was found that the miR146a rs2910164 GG genotype was significantly associated with gastric cancer susceptibility (16). These conflicting findings may depend on various sample sizes, ethnic groups and type of cancer. As for childhood leukemia, one report firstly examining the association between SNP (rs2910164), miR146a G>C and susceptibility to ALL in an Iranian population with 75 cases and 155 controls. They found that the rs2910164 C variants significantly increased the risk of ALL (23). In 2016, Chansing and his colleagues found that the frequencies of miR146a rs2910164 genotypes were not associated with demographic data or clinical outcomes for ALL in a Thailand population containing 100 childhood ALL cases and 200 healthy controls (32). In 2019, one group reported that no statistically significant association was found between miR146a rs2910164 genotypes and childhood ALL risk in an Indian population containing 71 cases and 74 control individuals (33). Compared to these studies, our sample size is much more convincing with 266 cases and 266 controls, and representative in an islandic country with conservative genetic background.
In the current study, the miR146a rs2910164 GG genotype was significantly associated with a decreased susceptibility to childhood ALL (Table II). In our population, the allele frequencies of miR146a C and G allele were 55.8% and 44.2%, respectively (Table III). The relative percentages of these allelic frequencies were similar to NCBI SNP database in Asian population, but quite different from European groups. The data showed that the frequency of C allele is significantly lower in control group than that in the childhood ALL group (Table III), thus the G allele at miR146a rs2910164 may serve as a protective marker for childhood ALL in Taiwan. Interestingly, we have found that the childhood ALL patients with CG and GG genotypes were of earlier onset ages than those with CC genotype (Table IV).
Although the story of how miR146a genotypes play a role in childhood ALL remain largely unrevealed, we have tried to depict the pathobiology behind miR146a rs2910164 GG genotype being significantly associated with a decreased susceptibility to childhood ALL via colleting the evidence from the literature. First, the genotype-phenotype correlation study showed that people of miR146a rs2910164 CG/GG genotypes have higher levels of miR146a than those of miR146a rs2910164 CC genotype (34-36). Second, overexpression of miR146a has been reported to inhibit the expression of TRAF-6 and IRAK-1 (29,35), together with their downstream molecule NF-ĸB (30), and involve in the suppression of carcinogenesis. Third, in 2018, Hosseini and colleagues have demonstrated that IRAK1 is overexpressed in primary acute myeloid leukemia (AML) cells (37), while inhibiting the activities of IRAK1 by pacritinib can inhibit the growth of the AML cells, reduce AML progenitors in vitro and the leukemia burden in AML xenograft model (37). The notion that IRAK1 signaling pathway regulates cancer stemness and drug resistance and may be a novel therapeutic target is keeping on being validated (38). Thus, it is very possible that miR146a and IRAK1 may also serve as therapeutic targets for childhood leukemia. It is very promising that translational scientists are adopting high-throughput methodologies such as proteomic analysis for childhood AML to reveal the signaling network underlying the etiology of childhood diseases (39).
In conclusion, miR146a rs2910164 genotypes were significantly associated with susceptibility in Taiwan childhood ALL and can serve as an early onset marker. Studies of larger sample sizes and investigating more populations are in need to validate the role of miR146a in cancer development.
Conflicts of Interest
The Authors declare no conflicts of interest.
Authors’ Contributions
Research design: Pei JS, Chang WS and Gong CL; Patient questionnaire Summarizing: Pei JS, Hsu PC and Chen CC; Experiment performance: Chin YT, Huang TL and Wang YC; Statistical analysis: Hsu YN and Kuo CC; Manuscript writing: Tsai CW and Bau DT; Reviewing and revising: Bau DT, Chang WS and Tsai CW.
Acknowledgements
The Authors are grateful to Hsin-Ting Li, Yu-Chen Hsiau, Yu-Hsin Lin and Yi-Ru Huang for their excellent technical assistance. All the participants including those who were not selected into the control group in the study are appreciated. This study was supported mainly by Taoyuan General Hospital, Ministry of Health and Welfare, Taiwan, ROC to Dr. Pei (Grant Number: PTH108012). The funders had no role in designing the study, data collection, statistical analysis, or decision to publish or preparation of the manuscript.
References
- 1.Trevino LR, Yang W, French D, Hunger SP, Carroll WL, Devidas M, Willman C, Neale G, Downing J, Raimondi SC, Pui CH, Evans WE, Relling MV. Germline genomic variants associated with childhood acute lymphoblastic leukemia. Nat Genet. 2009;41:1001–1005. doi: 10.1038/ng.432. PMID: 19684603. DOI: 10.1038/ng.432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hsu PC, Pei JS, Chen CC, Chang WS, Kuo CC, Cheng SP, Tsai CW, Bau DT, Gong CL. Association of matrix metallopeptidase-2 promoter polymorphisms with the risk of childhood leukemia. Anticancer Res. 2019;39:1185–1190. doi: 10.21873/anticanres.13228. PMID: 30842148. DOI: 10.21873/anticanres.13228. [DOI] [PubMed] [Google Scholar]
- 3.Hsu PC, Chen CC, Tzeng HE, Hsu YN, Kuo CC, Lin ML, Chang WS, Wang YC, Tsai CW, Pei JS, Bau DT. HOGG1 rs1052133 Genotypes and risk of childhood acute lymphoblastic leukemia in a Taiwanese population. In Vivo. 2019;33:1081–1086. doi: 10.21873/invivo.11576. PMID: 31280195. DOI: 10.21873/invivo.11576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pei JS, Chang WS, Hsu PC, Chen CC, Cheng SP, Wang YC, Tsai CW, Shen TC, Bau DT. The contribution of XRCC3 genotypes to childhood acute lymphoblastic leukemia. Cancer Manag Res. 2018;10:5677–5684. doi: 10.2147/CMAR.S178411. PMID: 30532590. DOI: 10.2147/CMAR.S178411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pei JS, Chou AK, Hsu PC, Tsai CW, Chang WS, Wu MF, Wu MH, Hsia TC, Cheng SP, Bau DT. Contribution of matrix metalloproteinase-7 genotypes to the risk of non-solid tumor, childhood leukemia. Anticancer Res. 2017;37:6679–6684. doi: 10.21873/anticanres.12126. PMID: 29187444. DOI: 10.21873/anticanres.12126. [DOI] [PubMed] [Google Scholar]
- 6.Iriyama N, Yoshino Y, Yuan B, Horikoshi A, Hirabayashi Y, Hatta Y, Toyoda H, Takeuchi J. Speciation of arsenic trioxide metabolites in peripheral blood and bone marrow from an acute promyelocytic leukemia patient. J Hematol Oncol. 2012;5:1. doi: 10.1186/1756-8722-5-1. PMID: 22272800. DOI: 10.1186/1756-8722-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. PMID: 14744438. DOI: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 8.Wang AX, Xu B, Tong N, Chen SQ, Yang Y, Zhang XW, Jiang H, Liu N, Liu J, Hu XN, Sha GZ, Chen M. Meta-analysis confirms that a common G/C variant in the pre-miR-146a gene contributes to cancer susceptibility and that ethnicity, gender and smoking status are risk factors. Genet Mol Res. 2012;11:3051–3062. doi: 10.4238/2012.August.31.2. PMID: 23007982. DOI: 10.4238/2012.August.31.2. [DOI] [PubMed] [Google Scholar]
- 9.He H, Jazdzewski K, Li W, Liyanarachchi S, Nagy R, Volinia S, Calin GA, Liu CG, Franssila K, Suster S, Kloos RT, Croce CM, de la Chapelle A. The role of microRNA genes in papillary thyroid carcinoma. Proc Natl Acad Sci USA. 2005;102:19075–19080. doi: 10.1073/pnas.0509603102. PMID: 16365291. DOI: 10.1073/pnas.0509603102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yue C, Wang M, Ding B, Wang W, Fu S, Zhou D, Zhang Z, Han S. Polymorphism of the pre-miR-146a is associated with risk of cervical cancer in a Chinese population. Gynecol Oncol. 2011;122:33–37. doi: 10.1016/j.ygyno.2011.03.032. PMID: 21529907. DOI: 10.1016/j.ygyno.2011.03.032. [DOI] [PubMed] [Google Scholar]
- 11.Palmieri A, Carinci F, Martinelli M, Pezzetti F, Girardi A, Cura F, Rubini C, Scapoli L. Role of the MIR146A polymorphism in the origin and progression of oral squamous cell carcinoma. Eur J Oral Sci. 2014;122:198–201. doi: 10.1111/eos.12121. PMID: 24612133. DOI: 10.1111/eos.12121. [DOI] [PubMed] [Google Scholar]
- 12.Mi Y, Ren K, Zou J, Bai Y, Zhang L, Zuo L, Okada A, Yasui T. The association between three genetic variants in microRNAs (rs11614913, rs2910164, rs3746444) and prostate cancer risk. Cell Physiol Biochem. 2018;48:149–157. doi: 10.1159/000491671. PMID: 30001553. DOI: 10.1159/000491671. [DOI] [PubMed] [Google Scholar]
- 13.Lian H, Wang L, Zhang J. Increased risk of breast cancer associated with CC genotype of Has-miR-146a Rs2910164 polymorphism in Europeans. PLoS One. 2012;7:e31615. doi: 10.1371/journal.pone.0031615. PMID: 22363684. DOI: 10.1371/journal.pone.0031615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qi P, Wang L, Zhou B, Yao WJ, Xu S, Zhou Y, Xie ZB. Associations of miRNA polymorphisms and expression levels with breast cancer risk in the Chinese population. Genet Mol Res. 2015;14:6289–6296. doi: 10.4238/2015.June.11.2. PMID: 26125831. DOI: 10.4238/2015.June.11.2. [DOI] [PubMed] [Google Scholar]
- 15.Wang Z, Zhang L, Shi X, Xu H, Wang T, Bian J. Association between two common polymorphisms and risk of hepatocellular carcinoma: evidence from an updated meta-analysis. Biomed Res Int. 2014;2014:468605. doi: 10.1155/2014/468605. PMID: 24860819. DOI: 10.1155/2014/468605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu Z, Zhang L, Cao H, Bai B. MiR-146a rs2910164 G/C polymorphism and gastric cancer susceptibility: a meta-analysis. BMC Med Genet. 2014;15:117. doi: 10.1186/s12881-014-0117-2. PMID: 25326754. DOI: 10.1186/s12881-014-0117-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nejati-Azar A, Alivand MR. miRNA 196a2(rs11614913) & 146a(rs2910164) polymorphisms & breast cancer risk for women in an Iranian population. Per Med. 2018;15:279–289. doi: 10.2217/pme-2017-0088. PMID: 29965793. DOI: 10.2217/pme-2017-0088. [DOI] [PubMed] [Google Scholar]
- 18.Chu YH, Tzeng SL, Lin CW, Chien MH, Chen MK, Yang SF. Impacts of microRNA gene polymorphisms on the susceptibility of environmental factors leading to carcinogenesis in oral cancer. PLoS One. 2012;7:e39777. doi: 10.1371/journal.pone.0039777. PMID: 22761899. DOI: 10.1371/journal.pone.0039777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang L, Gao J, Zhou D, Bao F. Lack of association of two common polymorphisms rs2910164 and rs11614913 with susceptibility to gastric cancer: A meta-analysis. Turk J Gastroenterol. 2015;26:378–385. doi: 10.5152/tjg.2015.6603. PMID: 26215064. DOI: 10.5152/tjg.2015.6603. [DOI] [PubMed] [Google Scholar]
- 20.Min KT, Kim JW, Jeon YJ, Jang MJ, Chong SY, Oh D, Kim NK. Association of the miR-146aC>G, 149C>T, 196a2C>T, and 499A>G polymorphisms with colorectal cancer in the Korean population. Mol Carcinog. 2012;51(Suppl 1):E65–73. doi: 10.1002/mc.21849. PMID: 22161766. DOI: 10.1002/mc.21849. [DOI] [PubMed] [Google Scholar]
- 21.Du W, Ma XL, Zhao C, Liu T, Du YL, Kong WQ, Wei BL, Yu JY, Li YY, Huang JW, Li ZK, Liu L. Associations of single nucleotide polymorphisms in miR-146a, miR-196a, miR-149 and miR-499 with colorectal cancer susceptibility. Asian Pac J Cancer Prev. 2014;15:1047–1055. doi: 10.7314/apjcp.2014.15.2.1047. PMID: 24568449. DOI: 10.7314/apjcp.2014.15.2.1047. [DOI] [PubMed] [Google Scholar]
- 22.Tian T, Shu Y, Chen J, Hu Z, Xu L, Jin G, Liang J, Liu P, Zhou X, Miao R, Ma H, Chen Y, Shen H. A functional genetic variant in microRNA-196a2 is associated with increased susceptibility of lung cancer in Chinese. Cancer Epidemiol Biomarkers Prev. 2009;18:1183–1187. doi: 10.1158/1055-9965.EPI-08-0814. PMID: 19293314. DOI: 10.1158/1055-9965.EPI-08-0814. [DOI] [PubMed] [Google Scholar]
- 23.Hasani SS, Hashemi M, Eskandari-Nasab E, Naderi M, Omrani M, Sheybani-Nasab M. A functional polymorphism in the miR-146a gene is associated with the risk of childhood acute lymphoblastic leukemia: a preliminary report. Tumour Biol. 2014;35:219–225. doi: 10.1007/s13277-013-1027-1. PMID: 23888320. DOI: 10.1007/s13277-013-1027-1. [DOI] [PubMed] [Google Scholar]
- 24.Wu MH, Tzeng HE, Wu CN, Yueh TC, Peng YC, Tsai CH, Wang YC, Ke TW, Pei JS, Chang WS, Tsai CW, Bau DT. Association of matrix metalloproteinase-9 rs3918242 promoter genotypes with colorectal cancer risk. Anticancer Res. 2019;39:6523–6529. doi: 10.21873/anticanres.13867. PMID: 31810917. DOI: 10.21873/anticanres.13867. [DOI] [PubMed] [Google Scholar]
- 25.Wu MH, Hung YW, Gong CL, Chao CC, Yueh TC, Wang SC, Lai YL, Hsu SW, Fu CK, Wang YC, Ke TW, Chang WS, Tsai CW, Bau DT. Contribution of caspase-8 genotypes to colorectal cancer risk in Taiwan. Anticancer Res. 2019;39:2791–2797. doi: 10.21873/anticanres.13406. PMID: 31177115. DOI: 10.21873/anticanres.13406. [DOI] [PubMed] [Google Scholar]
- 26.Lin KM, Yang MD, Tsai CW, Chang WS, Hsiao CL, Jeng LB, Yueh TC, Lee MC, Bau DT. The role of MTHFR genotype in colorectal cancer susceptibility in Taiwan. Anticancer Res. 2018;38:2001–2006. doi: 10.21873/anticanres.12438. PMID: 29599316. DOI: 10.21873/anticanres.12438. [DOI] [PubMed] [Google Scholar]
- 27.Hwang HW, Mendell JT. MicroRNAs in cell proliferation, cell death, and tumorigenesis. Br J Cancer. 2006;94:776–780. doi: 10.1038/sj.bjc.6603023. PMID: 16495913. DOI: 10.1038/sj.bjc.6603023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garzon R, Marcucci G, Croce CM. Targeting microRNAs in cancer: rationale, strategies and challenges. Nat Rev Drug Discov. 2010;9:775–789. doi: 10.1038/nrd3179. PMID: 20885409. DOI: 10.1038/nrd3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jazdzewski K, Murray EL, Franssila K, Jarzab B, Schoenberg DR, de la Chapelle A. Common SNP in pre-miR-146a decreases mature miR expression and predisposes to papillary thyroid carcinoma. Proc Natl Acad Sci USA. 2008;105:7269–7274. doi: 10.1073/pnas.0802682105. PMID: 18474871. DOI: 10.1073/pnas.0802682105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci USA. 2006;103:12481–12486. doi: 10.1073/pnas.0605298103. PMID: 16885212. DOI: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jeon HS, Lee YH, Lee SY, Jang JA, Choi YY, Yoo SS, Lee WK, Choi JE, Son JW, Kang YM, Park JY. A common polymorphism in pre-microRNA-146a is associated with lung cancer risk in a Korean population. Gene. 2014;534:66–71. doi: 10.1016/j.gene.2013.10.014. PMID: 24144839. DOI: 10.1016/j.gene.2013.10.014. [DOI] [PubMed] [Google Scholar]
- 32.Chansing K, Pakakasama S, Hongeng S, Thongmee A, Pongstaporn W. Lack of Association between the mir146a polymorphism and susceptibility to Thai childhood acute lymphoblastic leukemia. Asian Pac J Cancer Prev. 2016;17:2435–2438. PMID: 27268610. [PubMed] [Google Scholar]
- 33.Jemimah Devanandan H, Venkatesan V, Scott JX, Magatha LS, Durairaj Paul SF, Koshy T. MicroRNA 146a polymorphisms and expression in Indian children with acute lymphoblastic leukemia. Lab Med. 2019;50:249–253. doi: 10.1093/labmed/lmy074. PMID: 30576465. DOI: 10.1093/labmed/lmy074. [DOI] [PubMed] [Google Scholar]
- 34.Qi P, Wang L, Zhou B, Yao WJ, Xu S, Zhou Y, Xie ZB. Associations of miRNA polymorphisms and expression levels with breast cancer risk in the Chinese population. Genet Mol Res. 2015;14:6289–6296. doi: 10.4238/2015.June.11.2. PMID: 26125831. DOI: 10.4238/2015.June.11.2. [DOI] [PubMed] [Google Scholar]
- 35.Shao Y, Li J, Cai Y, Xie Y, Ma G, Li Y, Chen Y, Liu G, Zhao B, Cui L, Li K. The functional polymorphisms of miR-146a are associated with susceptibility to severe sepsis in the Chinese population. Mediators Inflamm. 2014;2014:916202. doi: 10.1155/2014/916202. PMID: 24701036. DOI: 10.1155/2014/916202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang B, Wang A, Xia C, Lin Q, Chen CA. Single nucleotide polymorphism in primary-microRNA-146a reduces the expression of mature microRNA-146a in patients with Alzheimer’s disease and is associated with the pathogenesis of Alzheimer’s disease. Mol Med Rep. 2015;12:4037–4042. doi: 10.3892/mmr.2015.3968. PMID: 26095531. DOI: 10.3892/mmr.2015.3968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hosseini MM, Kurtz SE, Abdelhamed S, Mahmood S, Davare MA, Kaempf A, Elferich J, McDermott JE, Liu T, Payne SH, Shinde U, Rodland KD, Mori M, Druker BJ, Singer JW, Agarwal A. Inhibition of interleukin-1 receptor-associated kinase-1 is a therapeutic strategy for acute myeloid leukemia subtypes. Leukemia. 2018;32:2374–2387. doi: 10.1038/s41375-018-0112-2. PMID: 29743719. DOI: 10.1038/s41375-018-0112-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cheng BY, Lau EY, Leung HW, Leung CO, Ho NP, Gurung S, Cheng LK, Lin CH, Lo RC, Ma S, Ng IO, Lee TK. IRAK1 augments cancer stemness and drug resistance via the AP-1/AKR1B10 signaling cascade in hepatocellular carcinoma. Cancer Res. 2018;78:2332–2342. doi: 10.1158/0008-5472.CAN-17-2445. PMID: 29483095. DOI: 10.1158/0008-5472.CAN-17-2445. [DOI] [PubMed] [Google Scholar]
- 39.Braoudaki M, Tzortzatou-Stathopoulou F, Anagnostopoulos AK, Papathanassiou C, Vougas K, Karamolegou K, Tsangaris GT. Proteomic analysis of childhood de novo acute myeloid leukemia and myelodysplastic syndrome/AML: correlation to molecular and cytogenetic analyses. Amino Acids. 2011;40:943–951. doi: 10.1007/s00726-010-0718-9. PMID: 20711619. DOI: 10.1007/s00726-010-0718-9. [DOI] [PubMed] [Google Scholar]